Keywords: leech, proboscis, de novo transcriptome assembly, differentially expressed genes
Abstract
Cephalization refers to the evolutionary trend towards the concentration of neural tissues, sensory organs, mouth and associated structures at the front end of bilaterian animals. Comprehensive studies on gene expression related to the anterior formation in invertebrate models are currently lacking. In this study, we performed de novo transcriptional profiling on a proboscis-bearing leech (Helobdella austinensis) to identify differentially expressed genes (DEGs) in the anterior versus other parts of the body, in particular to find clues as to the development of the proboscis. Between the head and the body, 132 head-specific DEGs were identified, of which we chose 11 to investigate their developmental function during embryogenesis. Analysis of the spatial expression of these genes using in situ hybridization showed that they were characteristically expressed in the anterior region of the developing embryo, including the proboscis. Our results provide information on the genes related to head formation and insights into the function of proboscis-related genes during organogenesis with the potential roles of genes not yet characterized.
1. Introduction
In the development of bilaterian animals, a broadly conserved genetic toolkit underlies the specification of positional identity along the body axis, usually including the establishment of a centralized nerve system, concentration of sensory organs and development of specialized mouth parts at the most anterior region [1–3]. The mouth parts of animals have undergone tremendous specialization and diversification as they have evolved to capture and ingest food according to their diverse life styles and ecosystems [4–7], in large part by modifying the regulation and deployment of the broadly conserved toolkit. Invertebrates especially show a wide variety of mouthpart structures such as tooth, mandible/maxilla and proboscis, depending on diverse food sources, at the end of the head part [7–12]. Among various mouthpart structures in invertebrates, the organ called the proboscis has been known as a specialized apparatus for piercing and holding the host's gut wall [13], capturing prey through injecting toxin [14,15] and fluid sucking [16,17], and the proboscis as a feeding apparatus has a tube for sucking up food such as nectar, body fluid and cellular constituents [18,19]. Fluid sucking of insects is generated in the canalized proboscis by suction pressure produced from the proboscis–cranial sucking pump complex with capillary activity which directs the flow of fluid into the oesophagus [19–22]. On the other hand, the feeding tube of the leech, another representative proboscis-bearing animal, consists only of a muscle complex. The force generated by this complex induces peristalsis for moving food in an aboral direction [11,16].
Cell lineage experiments performed with a well-studied leech model, Helobdella (family Glossiphoniidae), have provided information on the development of the leech proboscis [23,24]. The formation of germ layer-specific tissues in the proboscis is achieved through orchestrated proliferation and interdigitation of clones arising from cells in the early lineage of the mesodermal proteoblast DM' (‘4d cell' in spiralian nomenclature), which mainly contribute to the formation of non-segmental mesoderm that can develop into the digestive tract during organogenesis [23]. At stage 10 (220 h to 245 h after zygotic development), the proboscis develops in an everted. During stage 11, the proboscis gradually inverts to its resting adult position within the foregut region, by which time it consists of a complex array of longitudinal, radial and circular muscles [23,25]. These radical and dynamic developmental changes raise the question of whether essential factors are actively expressed at stage 10 for the formation of the head or specific structures such as the proboscis.
Over the past few years, many studies have been performed to understand the develpomental involvement of various transcription factors [26–28] and signalling molecules [29–31], including physiological substances in the anterior region of the leech [32]. Previous studies have suggested that anterior development involves a combination of numerous signalling pathways and transcription factors and that the anterior head is a vast hub of gene expression. However, large-scale studies on expressed genes have not been reported. Towards this end, we used stage 10 leech embryos and conducted RNA sequencing (RNA-seq), a successful method for understanding differential gene expression in specific tissues or under different conditions in a wide variety of animals and plants [33–35]. In this study, we provide detailed information on the stage 10 leech embryo transcriptome and present proboscis-specific genes identified via differentially expressed gene (DEG) analysis between the head and body. Our results will be used as comparative data for the evolutionary relevance of the proboscis and also fill the gap in transcriptional information about the missing stage during leech embryo development.
2. Results and discussion
2.1. Internal structure development of the proboscis during organogenesis
The harmonious proliferation and differentiation of the progeny of lineage-specific blast cells and micromeres give rise to the development of epidermis, ganglia and musculature with the formation of primary mouthparts that can develop into a future digestive tract [23–25,36]. After epiboly, the anterior germinal plate develops into the precursor of the mouthpart (stomodeum), surrounded by primary nerves and muscle fibres (figure 1a–d) [23]. At stage 10, the proboscis gradually protrudes, covered with epithelial tissues which develop into the proboscis sheath. This process involves the active development of a complex of innervated muscle layers in the proboscis structure and oesophagus (figure 1a,e–h″). By stage 11, the proboscis invaginates into the mouth pore located in the foregut region with placement of a presumptive circumferential muscle layer and spanning of the radial muscle along with development of longitudinal and oesophageal muscle (figure 1a,i–l′). The timing of the morphological differentiation of the proboscis thus suggests that stage 10 might be a crucial step in the development of the internal structure and that proboscis-related genes might be highly expressed during this dynamic event.
Figure 1.
Proboscis formation and internal structure differentiation. The formation and internal structure differentiation of the leech proboscis. All embryos are oriented anterior to the left and dorsal to the top of the panel. White dotted rectangles indicate the magnified region in (e) and (i). White dotted lines indicate the sectional regions. (a) Schematic of organogenesis stages. Between stages 10 and 11, the proboscis invaginates (red arrows) into the mouth pore and locates in the foregut region. (b–d) At stage 9, the oral precursor stomodeum (white dotted circles) develops in the anterior region. (e,f) Lateral view of the stage 11 embryo showing the everted proboscis. (g–h) Sectioned views showing the primordium of innervated muscle fibres (white arrowheads). (i–j) Lateral view of the stage 11 embryo showing ganglia and the invaginated proboscis located in the foregut region. (k–l) Sectioned views showing differentiated muscles and nerve fibres (blue arrowheads). Scale bar, 200 µm in panels b, e, i; 20 µm in panels g and k. as, anterior sucker; AZD, after zygotic development; cc, coelomic cavity; cm, circular muscle; ep, everted proboscis; gp, germinal plate; l, lumen; lm, longitudinal muscle; o, oesophagus; om, oesophageal muscle; p, proboscis; pc, proboscis chamber; ps, proboscis sheath; rm, radial muscle; s, stomodeum; vg, ventral ganglion; vlm, ventral longitudinal muscle; y, yolk.
2.2. De novo transcriptome assembly of a stage 10 leech embryo
We used RNA-seq to find specific genes in the proboscis compared with the body region at stage 10 (figure 2a and electronic supplementary material, figure S1). More than 85 million raw reads of the body and 79 million raw reads of the proboscis were obtained, yielding 84 million and 78 million filtered reads, respectively, with high-quality values (filtered-out ratio greater than 98%; Q20 > 98%; for details, see electronic supplementary material, table S1). We combined these reads and subjected them to de novo transcriptome assembly using Trinity v. 2.1.1. The transcriptome assembly yielded 127 484 transcripts with an N50 length of 1002 bp and an average length of 655.1 bp (for details, see electronic supplementary material, table S1). Subsequently, we annotated the assembled transcripts using BLASTP (e-value ≤1 × 10−10) based on Uniprot/SwissProt and UniRef50 databases to obtain complete sequence coverage. We obtained 13 735 annotated transcripts, including 8003 genes annotated based on the Uniprot/SwissProt database and 5732 genes annotated based on the UniRef50 database (electronic supplementary material, tables S1 and S2). To examine the completeness of this Helobdella austinensis embryo transcriptome, we assessed annotated protein-coding sequences. A total of 82.5% (250/303 genes) and 78.4% (767/978 genes) of the eukaryote and metazoan single-copy orthologues were identified, respectively (figure 2b).
Figure 2.
Schematic of the analysis and DEGs between the proboscis and body. (a) At stage 10, the head and body of embryos were cut into sections. Each section was used to obtain raw reads after RNA extraction followed by DEG analysis (for detailed flow information, see electronic supplementary material, figure S1). (b) Results of BUSCO analysis. (c) Scatterplot showing global gene expression profiles (log2-transformed FPKM+1) between the proboscis and body. Red, DEGs; yellow, highly expressed genes; green, specifically expressed genes. ρ is Spearman's correlation coefficient. FPKM, fragments per kilobase of transcript per million mapped reads.
2.3. Identification of differentially expressed genes and gene ontology analysis
Of the 13 735 annotated transcripts, 263 were DEGs in the head and the body (false discovery rate [FDR] < 0.05) (figure 2c). Of these 263 DEGs, 132 and 131 were specifically upregulated in the head and the body, respectively (electronic supplementary material, tables S1 and S3). Characteristically, more than half of these DEGs were uncharacterized genes covered by Uniprot reference clusters [37] (70 of 132), suggesting that anterior development involved a larger proportion of putatively leech-specific genes during anterior formation.
Gene ontology (GO) terms associated with DEGs were used to identify differences within representative classification (molecular function, biological process, protein class) (figure 3). Genes related to binding (GO:0005488), cellular process (GO:0009987) and gene-specific transcription factor (PC00264) accounted for the highest at 43.5% (27/62), 51.6% (32/62) and 22.6% (14/62), respectively. Interestingly, among these genes, homeodomain transcription factors (OTX1, SIX3, SIX6, Nkx1–2, Hmx) were the most common, followed by basic helix–loop–helix transcription factors (SIM1, SIM2, PTF1A, FER3). These results indicate that the expression of common anterior developmental factors might be conserved in the anterior of leech embryos [3,38]. They also suggest that various transcription factors can activate crucial signalling pathways and give rise to organization of the nerve system in the anterior region [3,39–43].
Figure 3.
GO annotation in head-specific DEGs. PANTHER analysis was performed using 62 optimized DEGs annotated from the Uniprot database for functional annotation. The bar chart shows the distribution of genes within each representative group (molecular function, biological process, protein class).
For further investigation, we focused on genes that were both highly and specifically expressed in the head, assuming that highly expressed genes and those most differentially expressed would mainly contribute to head formation, including proboscis development. We selected carefully based on criteria (highly expressed genes annotated, head fragments per kilobase of transcript per million mapped reads (FPKM) >100; highly expressed genes uncharacterized, head FPKM >100; differentially expressed genes annotated, head FPKM >10 and body FPKM <1), and 11 transcripts including three uncharacterized genes were identified (table 1). Among these head-related DEGs, genes encoding secreted frizzled-related protein (sFRP) homologues were highly and specifically expressed based on both criteria. These findings suggest that a variety of genes are related to the development of the anterior region. Among them, the high and characteristic expression of sFRPs might be correlated with anterior formation [3,38,44].
Table 1.
Genes of interest selected based on the three criteria. Asterisks indicate previously studied genes (for detailed phylogeny, see electronic supplementary material, figure S2).
| gene | expression level (FPKM) |
q-value | source | |
|---|---|---|---|---|
| body | head | |||
| highly expressed genes annotated (head FPKM >100) | ||||
| SFRP2 (sFRP1/2/5a*) | 6.36 | 124.08 | 6.79 × 10−6 | Uniprot-Swissprot |
| CML23 | 5.26 | 101.45 | 6.83 × 10−6 | |
| HS71L | 29.7 | 401.82 | 7.88 × 10−5 | |
| SCOT1 | 15.97 | 116.86 | 4.61 × 10−3 | |
| highly expressed genes uncharacterized (head FPKM > 100) | ||||
| UniRef50_T1FEH8 | 1.05 | 119.36 | 8.26 × 10−11 | UniRef50 |
| UniRef50_A7TSG4 | 16.24 | 182.23 | 3.04 × 10−5 | |
| UniRef50_T1F354 | 51.5 | 246.87 | 4.90 × 10−2 | |
| differentially expressed genes annotated (head FPKM >10 and body < 1) | ||||
| ZCH24 | 0.03 | 28.61 | 8.44 × 10−14 | Uniport-Swissprot |
| DFP3 | 0.21 | 19.42 | 3.92 × 10−9 | |
| DMTA2 (DMRT93B*) | 0.41 | 22.09 | 1.17 × 10−8 | |
| SFRP1 (sFRP1/2/5b*) | 0.68 | 21.69 | 5.22 × 10−8 | |
In addition to transcriptional expression analysis, we also investigated spatial expression patterns of these 11 transcripts to examine their embryological contribution during organogenesis.
2.4. Widespread expression of the hs71l and scot1 homologue
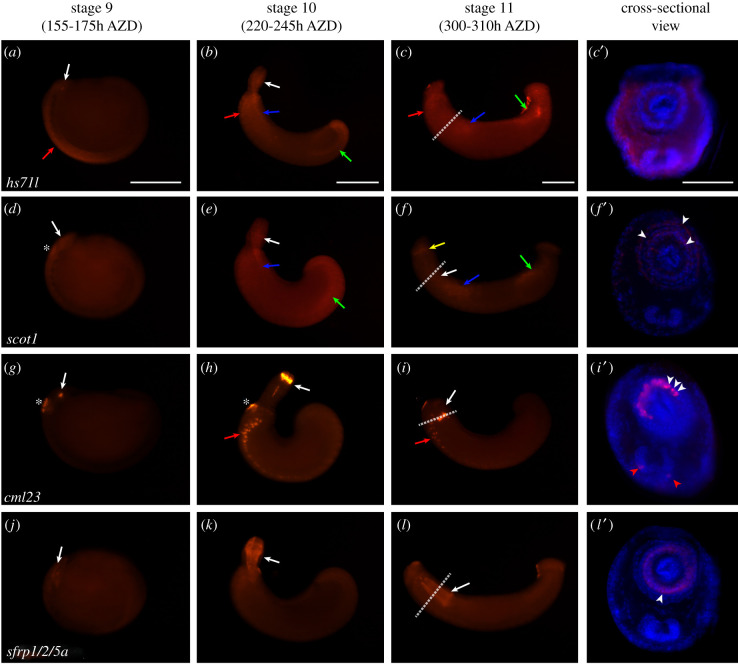
In stage 9, a homologue of the heat shock protein (HSP) hs71l was expressed in the stomodeum and ventral germinal plate (figure 4a). Its expression in the mouthpart persisted. Its expression was also detected in the rostral ganglia, salivary gland and visceral muscle, including the precursor of the posterior sucker (figure 4b). After proboscis invagination, it was expressed ubiquitously in the whole body with discriminate expression in the rostral ganglia, salivary gland and hindgut (figure 4c-c′). HSPs are multifunctional proteins related to protein maturation, protein refolding, protein import and translocation. HSPs facilitate proteolytic degradation of unstable proteins by targeting proteins to lysosomes or proteasomes under normal and stress conditions [45–47]. Although some HSPs have been shown to have embryological functions such as neurodevelopment in previous studies [48,49], comparable expression patterns in lophotrochozoans have not to our knowledge been previously described. Our results for the first time suggest that hs71l plays multifunctional roles in tissues or organs undergoing dynamic structural changes during leech embryogenesis.
Figure 4.
Spatial expression of the highly expressed gene (annotated) during organogenesis. Whole-mount in situ hybridization results of highly expressed genes (annotated). All embryos are oriented anterior to the left and dorsal to the top of the panel. The rightmost panel shows a cross-sectional view of a stage 11 embryo. White arrows indicate expression in the developing mouth part. Red arrows point to nervous expression of the candidate genes. Blue arrows indicate expression in the salivary gland. Green arrows point to the visceral muscle precursor. Yellow arrow indicates the expression in the intrinsic muscle. White asterisks indicate the expression in the adhesion site for attachment to the maternal venter. White dotted lines indicate the sectioned region seen in the rightmost panel. (a) hs71l transcripts are expressed in the stomodeum and the germinal plate. (b) hs71l expression is seen in the proboscis, rostral ganglia, salivary gland and developing posterior region involving the visceral muscle at stage 10. (c,c′) At stage 11, hs71l transcripts are expressed in the rostral ganglia, salivary gland and the developing hindgut, including whole mesodermal tissues in the body. (d) scot1 is expressed in the adhesion site for attachment to the maternal venter and around the stomodeum. (e) At stage 10, scot1 is expressed in the proboscis, salivary gland and posterior region involving the visceral muscle. (f,f′) At stage 11, scot1 is expressed in the anterior intrinsic muscle, salivary gland and hindgut. In a sectioned view, its expression is found in the proboscis sheath, longitudinal muscle and radial muscle (white arrowheads). (g) At stage 9, cml23 is expressed in the adhesion site for attachment to the maternal venter and in the stomodeum. (h) At stage 10, cml23 is expressed in the apical proboscis, adhesion region and ventral ganglia. (i) cml23 expression remains in the same region. (i′) In a sectioned view, cml23 is expressed in cells of radial muscles (white arrowheads) and ventral ganglia (red arrowheads). (j) At stage 9, sfrp1/2/5a is expressed in the stomodeum. (k) At stage 10, sfrp1/2/5a is expressed in the radial muscle precursor in the everted proboscis. (l) sfrp1/2/5a expression is maintained at stage 11. (l′) sfrp1/2/5a is detected in the developing radial muscle (white arrowheads). Scale bar, 200 µm in panels a–c; 100 µm in panel c'. AZD, after zygotic development; scot1, succinyl-coenzymeA (CoA):3-ketoacid-CoA transferase 1; cml23, calmodulin-like 23; sfrp1/2/5a, secreted frizzled protein 1/2/5a.
Succinyl-CoA-3-oxaloacid CoA transferase 1 (SCOT1) is related to ketone metabolism, which catalyses acetoacetate to acetoacyl–coenzyme A (acetoacyl-CoA) in the mitochondria [50,51]. At stage 9, the scot1 homologue was expressed around the stomodeum and in the adhesive zone, by which the embryo attaches to the maternal venter during the period of development after hatching from the fertilization membrane and cocoon and before the rear sucker becomes functional (figure 4d). At stage 10, scot1 was expressed in the proboscis, salivary gland and visceral muscle (figure 4e). Even after proboscis invagination, its overall expression pattern in the proboscis persisted along with the developing head, salivary glands and hindgut (figure 4f,f′ and electronic supplementary material, figure S3). We believe that, during embryogenesis, scot1 is expressed in structures and developing regions in which ketone metabolism might be required [52–54]. These results suggest that the stage of organogenesis might be a period in which tissues and organs undergo dynamic changes with organ-specific metabolic process of ketone.
2.5. Genes expressed in specific tissue layers in the proboscis
Previous lineage analyses [23,24,36] of leech proboscis development and our morphostructural analysis revealed that the development of the proboscis is marked by orderly expansion of radial muscles and differentiation of longitudinal muscles. We hypothesized that characteristic core signalling genes might contribute to this process.
Calmodulin (CaM) and calmodulin-like (CML) proteins known as major regulators of Ca2+-dependent signalling are key regulators of various mechanisms such as cell proliferation, motility and cell cycle progression [55–57]. In addition, CaM is an upstream regulator of calmodulin kinase (CaMK), which activates CaMK signalling to induce the development of the central nervous system [58,59]. At stage 9, transcripts of cml23 homologue were expressed in the furrow of the stomodeum and the adhesion site for attachment to the maternal venter (figure 4g). The adhesion site develops into the future epithelium of the anterior sucker. Transcripts of the cm123 homologue were continuously expressed in the region, including nuclei of radial muscle and ventral ganglia (figure 4h–i′). Our results indicate that the cml23 homologue is related to the cellular process of the developing proboscis and central nerve system during organogenesis.
sFRPs are key modulators of the Wnt signalling system, which determines the anteroposterior axis according to the distribution of Wnt ligands and their frizzled receptors; Wnt signalling is essential for bilaterian head formation [3,44]. In this study, sfrp orthologues showed an anterior-specific expression pattern in embryos during organogenesis (figures 4j–l′ and 5a–c′). At stage 9, these homologues were expressed around the stomodeum. At stages 10 and 11, these two paralogues showed different expression patterns. Transcripts of sfrp1/2/5a were expressed in the radial muscle precursor in the proboscis but weakly expressed in the posterior body (figure 4k–l′). However, sfrp1/2/5b was expressed in the posteriormost proboscis sheath and the anterior intrinsic muscle [60] (figure 5b-c′). Based on these spatial evidences, sfrp orthologues might play a role as conserved anterior morphogens with proboscis differentiation factors [3,44,61].
Figure 5.
Spatial expression of DEGs (annotated) during organogenesis. Whole-mount in situ hybridization results of differentially expressed genes (annotated). All embryos are oriented anterior to the left and dorsal to the top of the panel. The rightmost column shows a cross-sectional view of a stage 11 embryo. White arrows indicate expression in the developing mouth part. Red arrows point to nervous expression of candidate genes. Yellow arrow indicates expression in the intrinsic muscle. White dotted lines indicate the sectioned region seen in the rightmost column. (a) By stage 10, sfrp1/2/5b is expressed in and around the stomodeum. (b) At stage 10, sfrp1/2/5b is expressed in the posterior epithelial tissues of the everted proboscis. (c,c′) After proboscis invagination, sfrp1/2/5b remains expressed in the proboscis sheath and the intrinsic muscle. (d) At stage 9, zch24 is expressed inside the stomodeum. (e) After proboscis eversion, zch24 is expressed in the radial muscle precursor. (f,f′) After proboscis invagination, zch24 is expressed in the radial muscle layer (white arrowheads). (g) Dmrt93B is expressed in the ectodermal lineage in the germinal plate and the stomodeum. (h) Dmrt93B is expressed in the proboscis, ganglia and the visceral muscle region. (i) At stage 11, Dmrt93B expression remains in the rostral ganglia, proboscis and posterior sucker. (i′) In a cross-sectional view, Dnrt93B is detected in the epithelial tissue of the body, ventral ganglia (red arrowhead) and proboscis innervation (white arrowheads). (j) No dfp3 expression is visible at stage 9. (k) At stage 10, dfp3 transcripts start to be detected in the everted proboscis. (l,l′) By stage 11, dfp3 is expressed in the longitudinal muscle layer (white arrowhead). Scale bar, 200 µm in panels a–c; 100 µm in panel c. AZD, after zygotic development; sfrp1/2/5b, secreted frizzled protein 1/2/5b; zch24, zinc finger CCHC domain-containing protein 24; Dmrt93B, doublesex- and male-abnormal-3-related transcription factor 93B; dfp3, defence protein 3.
Zinc finger CCHC-type containing protein 24 (ZCH24) is one of the zinc finger domain proteins [62] known to be related to the development of somitogenic mesoderm in vertebrates [63], although it is largely unknown in invertebrates. In developing leech embryos, the zch24 homologue was only expressed inside the stomodeum at stage 9 (figure 5d) and in radial muscle precursors from the anterior to the posterior of the proboscis by stage 11 (figure 5e–f′). These restricted expressions of zch24 transcripts in proboscis indicate that zch24 is a possible proboscis-specific gene in H. austinensis as well as related to the radial muscle differentiation of the developing proboscis.
2.6. Nervous system-specific expression of the Dmrt93B homologue
At stage 9, Dmrt93B was expressed around and inside the stomodeum and in the ectodermal region of the germinal plate (figure 5g). At stage 10, its transcripts were mainly expressed in the rostral segment of ganglia and the proboscis including ventral ganglia (figure 5h). After proboscis invagination, Dmrt93B transcripts were expressed in the rostral segments, epidermis, ventral ganglion, nerve canal in the proboscis cavity [16] and the posterior sucker (figure 5i,i′). Dmrt genes are well known to be mainly sex dimorphic factors [64,65]. They have been actively studied in various fields such as neuronal differentiation and brain development, in addition to differentiation of animal sex determinants and reproductive organs [66,67]. Furthermore, in Panarthropoda, Dmrt93B is expressed in the nervous system including mouth development [68]. Taken together, these results indicate that this highly conserved gene also plays a role as a transcription factor for the development of the mouth and nervous system during leech embryogenesis.
2.7. Developmental relevance of the defence-related gene
Defence protein is characterized by a reeler domain. It is an innate immune substance that protects against pathogens [69,70]. In this study, dfp3 homologue was not expressed at stage 9 (figure 5j). However, it began to be expressed in the epithelium region of the proboscis at stage 10 (figure 5k). Its expression in the epithelium surrounding the proboscis was maintained even after proboscis invagination (figure 5l,l′). We suggest that this expression pattern is related to the development process of leech embryos. Helobdella embryos are surrounded by a vitelline membrane until stage 9. After hatching, they attach to the maternal venter and take off from it [25]. Based on the developmental evidence of spatial expression, the innate immune system in the leech might begin its protective function as the leech breaks through the vitelline membrane and becomes exposed to the environment.
2.8. Developmental contribution of uncharacterized genes during organogenesis
At stage 9, UniRef50_T1FEH8 transcripts were expressed in the periphery of the stomodeum along with overall expression in the germinal band Then, transcripts were expressed in the epithelium of the everted proboscis and rostral ganglia at stage 10. After proboscis invagination, UniRef50_T1FEH8 transcripts were expressed in the proboscis chamber, proboscis sheath and ventral ganglion (figure 6a–c′). From these results, we found that UniRef50_T1FEH8 is related to development of the epithelium of the proboscis and ganglia formation.
Figure 6.
Spatial expression of the highly expressed gene (uncharacterized) during organogenesis. Whole-mount in situ hybridization results of highly expressed genes (uncharacterized). All embryos are oriented anterior to the left and dorsal to the top of the panel. The rightmost panel shows a cross-sectional view of a stage 11 embryo. Turquoise arrows indicate the somatic muscle fibres. Green arrows point to the visceral muscle precursor. Magenta arrows indicate the developing proboscis chamber. Red arrows point to nervous expression of candidate genes. White arrows indicate expression in the developing mouth part. White dotted lines indicate the sectioned region seen in the rightmost column. (a) At stage 9, transcripts of UniRef50_T1FEH8 are expressed broadly in the stomodeum and the anterior germinal plate. (b) At stage 10, UniRef50_T1FEH8 transcripts are expressed in the everted proboscis epithelium and rostral ganglia. (c,c′) At stage 11, the expression of UniRef50_T1FEH8 is detected in the longitudinal muscle and proboscis sheath region. (d) UniRef50_A7TSG4 is expressed in the stomodeum. (e) At stage 10, transcripts of UniRef50_A7TSG4 are expressed in the proboscis epithelium, oesophagus and gut boundary. (f,f′) At stage 11, transcripts of UniRef50_A7TSG4 are expressed in the developing hindgut, proboscis sheath and oesophageal funnel. (g) UniRef50_T1F354 is expressed in the whole body and strongly expressed in the coelomic cavity, visceral muscle and stomodeum. (h) At stage 10, UniRef50_T1F354 is expressed in the proboscis, muscle fibres of the body and visceral muscle. (i,i′) At stage 11, transcripts of UniRef50_T1F354 are expressed in the whole mesodermal tissue, including the proboscis sheath, radial muscle precursor and somatic muscle. Scale bar, 200 µm in panels a–c; 100 µm in panel c′. AZD, after zygotic development.
UniRef50_A7TSG4 transcripts were expressed in the foregut precursor region and around the stomodeum at stage 9 and in the proboscis epithelium, oesophageal funnel and overall gut boundaries at stage 10. At stage 11, the expression at stage 10 was maintained, in addition to the expression in the proboscis chamber, proboscis sheath, oesophagus and the developing hindgut (figure 6d–f′). Therefore, we found that UniRef50_A7TSG4 is related to the epithelium of the proboscis along with overall gut development.
UniRef50_T1F354 transcripts were expressed in the stomodeum, coelomic cavity and visceral muscle precursors at stage 9 (figure 6g). At stage 10, these transcripts were expressed in the radial muscle precursor in the anterior proboscis, fibres of the proboscis sheath, body and visceral muscles (figure 6h). The expression was maintained in the same region at stage 11 (figure 6i,i′). Therefore, UniRef50_T1F354 is related to the overall mesodermal tissue development during organogenesis. Their intense expression in the proboscis with major organs of these uncharacterized or maybe leech-specific genes indicates that these unique genes are not only important for embryogenesis but also are specifically related to the proboscis development.
3. Conclusion
The feeding organs of invertebrates have undergone extensive diversification and specialization over the course of evolution. We performed a comprehensive study to expand our knowledge about proboscis evolution and to discover genes related to the development of this specialized organ. Through transcriptome profiling, DEG analysis and spatial analysis, we identified specific genes related to proboscis development including highly conserved anterior formation factors. This not only provides novel information that has not been previously elucidated but also suggests that leeches also undergo anterior formation by conserved morphogens along with diverse transcription factors and that the developing proboscis is a vast hub of signal molecules. Furthermore, the presence of numerous uncharacterized genes in the anterior formation suggests a possible involvement of specific genes in proboscis development, including head formation in leeches, raising questions about the evolution of mouth formation in other proboscis-bearing animals. Taken together, our comprehensive survey will be used for comparative analysis of feeding organ diversity and proboscis formation in invertebrates.
4. Material and methods
4.1. Animals
Adult H. austinensis specimens were bred in the Laboratory of Cellular and Development Biology (Department of Biology, Chungbuk National University, Republic of Korea). They were bred in bowls containing artificial freshwater placed in a biological oxygen demand (BOD) incubator at 22°C. Embryos were placed in Petri dishes with lids containing Helobdella triserialis (HTR) saline medium (4.8 mM NaCl, 1.2 mM KCl, 2 mM MgCl2, 8 mM CaCl2 and 1 mM maleic acid) according to the protocol for handling leech embryos.
4.2. RNA isolation and RNA-seq library preparation
Embryos were cultured in clean HTR medium until stage 10 (220–245 h after zygotic development). To obtain a sufficient amount of RNA, about 400 embryos were used in the experiment. The head region was dissected using insect pins (Shiga, no. 2) under a Leica ZOOM 2000 stereomicroscope (Leica, Wetzlar, Germany). Next, the proboscis region was transferred to a 1.7 ml tube containing 200 µl of TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was isolated from each sample using TRIzol reagent according to the manufacturer's instructions. The purity and integrity of the total RNA isolated from dissected embryo samples were examined using a Nanodrop 2000C spectrophotometer (Thermo Scientific, Waltham, MA, USA) and a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA). Then, total RNA concentration was calculated using Quant-IT RiboGreen (Invitrogen, R11490). To assess the integrity of the total RNA, samples were run on a TapeStation RNA screentape (Agilent, 5067–5576). Only high-quality RNA (RNA integrity number greater than 7.0) was used to construct an RNA library. A library was independently prepared with 1 µg of total RNA for each sample using Illumina TruSeq Stranded Total RNA Sample Prep Kit v. 2 (Illumina, Inc., San Diego, CA, USA). Ribosomal RNA in total RNA was depleted using a Ribo-Zero kit (Epicentre Biotechnologies, Madison, WI, USA). After the rRNA depletion, the remaining RNA was purified, fragmented and primed for cDNA synthesis. Cleaved RNA fragments were copied into first-strand cDNAs using reverse transcriptase and random hexamers followed by second-strand cDNA synthesis using DNA polymerase I, RNase H and dUTP. Next, cDNA fragments underwent end repair, addition of a single A base and adapter ligation. Products were then purified and enriched with polymerase chain reaction (PCR) to create the final cDNA library. All libraries were quantified using quantitative PCR according to the qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms) and qualified using a TapeStation D1000 ScreenTape (Agilent Technologies, Waldbronn, Germany). Finally, library samples were sequenced using an Illumina HiSeq 2000 system (Illumina, Inc., San Diego, CA, USA) to generate paired-end reads.
4.3. De novo assembly, differential expression and gene ontology analyses
Transcriptome assembly was performed using Trinity v. 2.1.1 [71]. After assembly, protein-coding sequences (CDSs) within assembled transcripts were predicted using the Uniprot/Swiss-Prot database (http://www.uniprot.org), BLASTP v. 2.2.31+ and TransDecoder v. 3.0.0 (http://transdecoder.sourceforge.net) included in the Trinity software. Redundant CDSs with identity greater than 99% were clustered and the longest CDS was left using CD-HIT v. 4.6.5 [72]. Finally, a set of non-redundant coding sequences (NRCDS) was generated. To quantify each transcript's expression level, RNA-seq reads for each sample were aligned against the transcriptome sequences using Bowtie v. 2.2.6 [73] and abundances of reads according to transcripts were estimated with RSEM v. 1.2.26 [74]. For annotated NRCDS, homology search was performed against the Uniprot/SwissProt database using BLASTP with an E-value cut-off of 10−10 and the best blast hit. For one-to-one protein and gene mapping, the one with the highest expression level was selected if multiple NRCDS were annotated in a single gene. To minimize the loss of unannotated NRCDS, it was reannotated with the UniRef50 database, an automatically annotated database and clustered sets of sequences from UniProt Knowledgebase and selected UniParc records [37]. Differentially expressed genes between the head and body were identified using the edgeR program with an FDR cut-off of less than 0.05. GO terms of differentially expressed genes were identified using PANTHER (Protein Analysis Through Evolutionary Relationships, http://pantherdb.org/) [75].
4.4. Gene cloning and probe synthesis
Specific primers (electronic supplementary material, table S4) for genes of interest were designed based on transcriptome sequences. Genes were amplified from cDNA of H. austinensis at stage 10 using a TaKaRa E×Taq kit (Takara Bio Inc., Kusatsu, Japan) according to the manufacturer's instructions with the following cycling conditions: pre-denaturation at 94°C for 5 min, denaturation at 94°C for 30 s, variable annealing temperature for 30 s, variable extension time at 72°C and post-extension at 72°C for 5 min. These amplified fragments were cloned into pGEM T vector (Promega, Madison, WI, USA). RNAprobes labelled with digoxigenin were constructed using a MEGAscript kit (Ambion, Austin, TX, USA) and a DIG RNA Labeling Mix (Roche, Basel, Switzerland) according to each manufacturer's instruction.
4.5. Whole mount in situ hybridization
Fluorescence in situ hybridization (FISH) was performed as previously described [29,32] as follows: embryos were treated with protease from Streptomyces griseus (Sigma-Aldrich, St. Louis, MO, USA) in 1× phosphate-buffered saline (PBS), rinsed three times with glycine dissolved in PBS at room temperature (RT) for 5 min, fixed with 4% paraformaldehyde at RT for 40 min and rinsed with PBT (1× PBS+0.1% Tween-20) three times. Next, prehybridization was performed at 64.7°C for one day in hybridization buffer (50% formamide, 5× saline-sodium citrate (SSC), 1× Denhardt's solution, 0.1% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 100 mg ml−1 of heparin, 0.1% Tween 20 and 100 mg ml−1 of transfer RNA (tRNA)). After incubation, the prehybridized buffer was replaced with fresh hybridization buffer containing 2 ng ml−1 of corresponding probe. Embryos were then hybridized at 64.7°C for 2 days. After washing embryos with PBT, they were preincubated with maleic acid buffer (100 mM maleic acid, 150 mM NaCl, pH 7.5) at RT for 15 min, blocked with 1% blocking reagent for nucleic acid hybridization and detection (Roche, Basel, Switzerland) at RT for 2 h and incubated with anti-DIG/ POD (1/1000 dilution) in 1% blocking reagent at 4°C for 16 h. After incubation, embryos were rinsed twice with TNT buffer (0.1 M Tris–HCl (pH 7.5), 0.15 M NaCl, 0.1% Tween 20) at RT and washed with amplification solution provided by the NEN Tyramide Signal Amplification (TSA) Plus Kit (PerkinElmer, Wellesley, MA, USA). A colour reaction was initiated by adding a 1 : 50 dilution of reconstituted cyanine-3 tyramide in amplification solution. After checking the signal, the embryos were labelled with 4′,6-diamidino-2-phenylindole (DAPI) in PBT (1 : 100) at RT for 20 min in the dark. These embryos were imaged using a Leica DM6 B with a Leica DFC450 C camera (Leica, Wetzlar, Germany). Stained embryos were then dehydrated in ethanol, mounted in plastic embedding solution (PolyBed, Polysciences, Inc.) and used for sectioned images.
4.6. Immunostaining
Whole-mount immunostaining was performed according to previously published protocols [16]. Briefly, fixed embryos were washed with PBS three times. After washing with PBS, the embryos were rinsed with 1% Triton in 0.1 M Tris–HCl (pH 7.5) several times for 1 h, incubated with diluted block solution (1 : 9 = 10× Roche Western Blocking Reagent:PBT) for 2 h and then incubated with primary antibodies (anti-acetylated-α-tubulin produced in mouse; Sigma Aldrich; T-7451) in diluted blocking solution (1 : 500) at 4°C for 48 h. After five consecutive washes with PBT, embryos were incubated with a secondary antibody (goat anti-mouse IgG H&L Alexa Fluor 488, Abcam, ab150113) in diluted blocking solution (1 : 1000) at 4°C for 24 h. After checking the signal, embryos were washed five times with PBT and then stained with Texas Red-X Phalloidin (Thermo Scientific, Waltham, MA, USA) for 24 h to visualize F-actin. After checking the signal, embryos were washed with PBT five times and labelled with DAPI in PBT (1 : 100) at room temperature in the dark for 20 min. Fluorescence-stained embryos were imaged using a LSM 710 confocal microscope (Carl Zeiss, Oberkochen, BW, Germany). The obtained images were edited using ZEN software (Carl Zeiss). Fluorescence-labelled embryos were then dehydrated in ethanol series (70%, 90% and 100% diluted in 1 × PBS) and propylene oxide, followed by infiltration with plastic embedding solution (PolyBed). Embryos were cut with a microtome blade (Leica 818; Leica, Wetzlar, Germany) under an Olympus SZ-STS microscope (Olympus, Tokyo, Japan). These sections were then mounted with Fluoromount-G (SouthernBiotech, Birmingham, AL, USA). Stained embryos and slide samples were imaged using a LEICA DM6 B with a LEICA DFC450 C camera (Leica, Wetzlar, Germany). Obtained images were edited using a Las X software (Leica), and prepared as figure plates using Adobe Illustrator CS6 (Adobe, San Jose, CA, USA).
Contributor Information
Chungoo Park, Email: chungoo@jnu.ac.kr.
Sung-Jin Cho, Email: sjchobio@chungbuk.ac.kr.
Data accessibility
Sequences generated in this study are deposited in GenBank (electronic supplementary material, table S4). Raw sequencing data are deposited in the NCBI Sequence Read Archive (SRA) database with accession numbers of SRR15013283 and SRR15013284 under BioProject PRJNA742851.
The data are provided in electronic supplementary material [76].
Authors' contributions
H.-J.K.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, validation, visualization, writing—original draft, writing—review and editing; S.-G.L.: data curation, formal analysis, methodology, resources, software, validation, visualization, writing—original draft, writing—review and editing; S.C.P.: conceptualization, funding acquisition, project administration, supervision, writing—original draft, writing—review and editing; J.-H..: formal analysis, methodology, writing—original draft, writing—review and editing; D.A.W.: conceptualization, investigation, writing—original draft, writing—review and editing; C.P.: data curation, methodology, software, supervision, validation, visualization, writing—original draft, writing—review and editing; S.-J.C.: conceptualization, data curation, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing—original draft, writing—review and editing
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A1A01041898). This research was supported by the Chung-Ang University Research Grants in 2019. This work was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR202130202).
References
- 1.Erwin DH. 2009. Early origin of the bilaterian developmental toolkit. Phil. Trans. R. Soc. B 364, 2253-2261. ( 10.1098/rstb.2009.0038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DuBuc TQ, Stephenson TB, Rock AQ, Martindale MQ. 2018. Hox and Wnt pattern the primary body axis of an anthozoan cnidarian before gastrulation. Nat. Commun. 9, 1-12. ( 10.1038/s41467-018-04184-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo Y-J, Kanda M, Koyanagi R, Hisata K, Akiyama T, Sakamoto H, Sakamoto T, Satoh N. 2018. Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nat. Ecol. Evol. 2, 141-151. ( 10.1038/s41559-017-0389-y) [DOI] [PubMed] [Google Scholar]
- 4.Martinez Q, Lebrun R, Achmadi AS, Esselstyn JA, Evans AR, Heaney LR, Miguez RP, Rowe KC, Fabre P-H. 2018. Convergent evolution of an extreme dietary specialisation, the olfactory system of worm-eating rodents. Sci. Rep. 8, 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeMiguel D. 2016. Disentangling adaptive evolutionary radiations and the role of diet in promoting diversification on islands. Sci. Rep. 6, 1-11. ( 10.1038/srep29803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamichhaney S, et al. 2015. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature 518, 371-375. ( 10.1038/nature14181) [DOI] [PubMed] [Google Scholar]
- 7.Tzetlin A, Purschke G. 2005. Pharynx and intestine. In Morphology, molecules, evolution and phylogeny in polychaeta and related taxa, pp. 199-225. Berlin, Germany: Springer. [Google Scholar]
- 8.Resh VH, Cardé RT. 2009. Encyclopedia of insects. San Diego, CA: Academic Press. [Google Scholar]
- 9.May RM. 1988. How many species are there on earth? Science 241, 1441-1449. ( 10.1126/science.241.4872.1441) [DOI] [PubMed] [Google Scholar]
- 10.Barnes RD. 1987. Invertebrate zoology. Philadelphia, PA: WB Saunders Co. [Google Scholar]
- 11.Sawyer RT. 1986. Leech biology and behaviour. Oxford, UK: Clarendon Press. [Google Scholar]
- 12.Blanke A, Rühr PT, Mokso R, Villanueva P, Wilde F, Stampanoni M, Uesugi K, Machida R, Misof B. 2015. Structural mouthpart interaction evolved already in the earliest lineages of insects. Proc. R. Soc. B 282, 20151033. ( 10.1098/rspb.2015.1033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Divers SJ, Mader DR. 2005. Reptile medicine and surgery (e-book). London, UK: Elsevier Health Sciences. [Google Scholar]
- 14.Kapil S, Hendriksen S, Cooper JS. 2017. Cone snail toxicity. Treasure Island, FL: StatPearls Publishing. [PubMed] [Google Scholar]
- 15.Göransson U, Jacobsson E, Strand M, Andersson HS. 2019. The toxins of nemertean worms. Toxins 11, 120. ( 10.3390/toxins11020120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwak H-J, et al. 2021. Behavioral variation according to feeding organ diversification in glossiphoniid leeches (Phylum: Annelida). Sci. Rep. 11, 1-13. ( 10.1038/s41598-020-79139-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krenn HW. 2010. Feeding mechanisms of adult Lepidoptera: structure, function, and evolution of the mouthparts. Annu. Rev. Entomol. 55, 307-327. ( 10.1146/annurev-ento-112408-085338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingsolver J, Daniel T. 1995. Mechanics of food handling by fluid-feeding insects. In Regulatory mechanisms in insect feeding, pp. 32-73. Berlin, Germany: Springer. [Google Scholar]
- 19.Kornev KG, Salamatin AA, Adler PH, Beard CE. 2017. Structural and physical determinants of the proboscis-sucking pump complex in the evolution of fluid-feeding insects. Sci. Rep. 7, 1-18. ( 10.1038/s41598-017-06391-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monaenkova D, Lehnert MS, Andrukh T, Beard CE, Rubin B, Tokarev A, Lee W-K, Adler PH, Kornev KG. 2012. Butterfly proboscis: combining a drinking straw with a nanosponge facilitated diversification of feeding habits. J. R. Soc. Interface 9, 720-726. ( 10.1098/rsif.2011.0392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauder JA-S, Handschuh S, Metscher BD, Krenn HW. 2013. Functional morphology of the feeding apparatus and evolution of proboscis length in metalmark butterflies (Lepidoptera: Riodinidae). Biol. J. Linn. Soc. 110, 291-304. ( 10.1111/bij.12134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson W, Peacock L, Hutchinson R. 2017. Microarchitecture of the tsetse fly proboscis. Parasites Vectors 10, 1-9. ( 10.1186/s13071-017-2367-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gline SE, Nakamoto A, Cho S-J, Chi C, Weisblat DA. 2011. Lineage analysis of micromere 4d, a super-phylotypic cell for Lophotrochozoa, in the leech Helobdella and the sludgeworm Tubifex. Dev. Biol. 353, 120-133. ( 10.1016/j.ydbio.2011.01.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang FZ, Kang D, Ramirez-Weber F-A, Bissen ST, Weisblat DA. 2002. Micromere lineages in the glossiphoniid leech Helobdella. Development 129, 719-732. ( 10.1242/dev.129.3.719) [DOI] [PubMed] [Google Scholar]
- 25.Weisblat DA, Kuo D-H. 2014. Developmental biology of the leech Helobdella. Int. J. Dev. Biol. 58, 429. ( 10.1387/ijdb.140132dw) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J-S, Kwak H-J, Jiménez BIM, Park SC, Xiao P, Weisblat DA, Cho S-J. 2017. Expression patterns of duplicated snail genes in the leech Helobdella. Dev. Genes Evol. 227, 415-421. ( 10.1007/s00427-017-0598-z) [DOI] [PubMed] [Google Scholar]
- 27.Kim J-S, Jiménez BIM, Kwak H-J, Park SC, Xiao P, Weisblat DA, Cho S-J. 2017. Spatiotemporal expression of a twist homolog in the leech Helobdella austinensis. Dev. Genes Evol. 227, 245-252. ( 10.1007/s00427-017-0585-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwak HJ, Ryu KB, Medina Jiménez BI, Park SC, Cho SJ. 2018. Temporal and spatial expression of the Fox gene family in the leech Helobdella austinensis. J. Exp. Zool. B: Mol. Dev. Evol. 330, 341-350. ( 10.1002/jez.b.22828) [DOI] [PubMed] [Google Scholar]
- 29.Cho S-J, Vallès Y, Giani VC Jr, Seaver EC, Weisblat DA. 2010. Evolutionary dynamics of the wnt gene family: a lophotrochozoan perspective. Mol. Biol. Evol. 27, 1645-1658. ( 10.1093/molbev/msq052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang D, Huang F, Li D, Shankland M, Gaffield W, Weisblat DA. 2003. A hedgehog homolog regulates gut formation in leech (Helobdella). Development 130, 1645-1657. ( 10.1242/dev.00395) [DOI] [PubMed] [Google Scholar]
- 31.Kuo D-H, Weisblat DA. 2011. A new molecular logic for BMP-mediated dorsoventral patterning in the leech Helobdella. Curr. Biol. 21, 1282-1288. ( 10.1016/j.cub.2011.06.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwak H-J, Park J-S, Medina Jiménez BI, Park SC, Cho S-J. 2019. Spatiotemporal expression of anticoagulation factor Antistasin in freshwater leeches. Int. J. Mol. Sci. 20, 3994. ( 10.3390/ijms20163994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graveley BR, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471, 473-479. ( 10.1038/nature09715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson PD, et al. 2018. Genome-wide transcriptome profiling and spatial expression analyses identify signals and switches of development in tapeworms. EvoDevo. 9, 1-29. ( 10.1186/s13227-018-0110-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruan J, Guo F, Wang Y, Li X, Wan S, Shan L, Peng Z. 2018. Transcriptome analysis of alternative splicing in peanut (Arachis hypogaea L). BMC Plant Biol. 18, 1-11. ( 10.1186/s12870-017-1213-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CM, Weisblat DA. 1994. Micromere fate maps in leech embryos: lineage-specific differences in rates of cell proliferation. Development 120, 3427-3438. ( 10.1242/dev.120.12.3427) [DOI] [PubMed] [Google Scholar]
- 37.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. 2007. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282-1288. ( 10.1093/bioinformatics/btm098) [DOI] [PubMed] [Google Scholar]
- 38.Martín-Durán JM, Passamaneck YJ, Martindale MQ, Hejnol A. 2016. The developmental basis for the recurrent evolution of deuterostomy and protostomy. Nat. Ecol. Evol. 1, 1-10. [DOI] [PubMed] [Google Scholar]
- 39.Kiecker C, Graham A, Logan M. 2016. Differential cellular responses to hedgehog signalling in vertebrates—what is the role of competence? J. Dev. Biol. 4, 36. ( 10.3390/jdb4040036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Lo P, Frasch M, Lufkin T. 2000. Hmx: an evolutionary conserved homeobox gene family expressed in the developing nervous system in mice and Drosophila. Mech. Dev. 99, 123-137. ( 10.1016/S0925-4773(00)00488-3) [DOI] [PubMed] [Google Scholar]
- 41.Segev E, Halachmi N, Salzberg A, Ben-Arie N. 2001. Nato3 is an evolutionarily conserved bHLH transcription factor expressed in the CNS of Drosophila and mouse. Mech. Dev. 106, 197-202. ( 10.1016/S0925-4773(01)00437-3) [DOI] [PubMed] [Google Scholar]
- 42.Nambu JR, Franks RG, Hu S, Crews ST. 1990. The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell 63, 63-75. ( 10.1016/0092-8674(90)90288-P) [DOI] [PubMed] [Google Scholar]
- 43.Millen KJ, Steshina EY, Iskusnykh IY, Chizhikov VV. 2014. Transformation of the cerebellum into more ventral brainstem fates causes cerebellar agenesis in the absence of Ptf1a function. Proc. Natl Acad. Sci. USA 111, E1777-E1786. ( 10.1073/pnas.1315024111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darras S, et al. 2018. Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol. 16, e2003698. ( 10.1371/journal.pbio.2003698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JY, Han Y, Lee JE, Yenari MA. 2018. The 70-kDa heat shock protein (Hsp70) as a therapeutic target for stroke. Expert Opin. Ther. Targets 22, 191-199. ( 10.1080/14728222.2018.1439477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon YM, Kim HJ, Lee JH, Lee SH. 2019. Melatonin enhances mitophagy by upregulating expression of heat shock 70 kDa protein 1 L in human mesenchymal stem cells under oxidative stress. Int. J. Mol. Sci. 20, 4545. ( 10.3390/ijms20184545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murthy VS, Ravishankar KV. 2016. Molecular mechanisms of heat shock proteins and thermotolerance in plants. In Abiotic stress physiology of horticultural crops, pp. 71-83. Berlin, Germany: Springer. [Google Scholar]
- 48.Jagla T, Dubińska-Magiera M, Poovathumkadavil P, Daczewska M, Jagla K. 2018. Developmental expression and functions of the small heat shock proteins in Drosophila. Int. J. Mol. Sci. 19, 3441. ( 10.3390/ijms19113441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller DJ, Fort PE. 2018. Heat shock proteins regulatory role in neurodevelopment. Front. Neurosci. 12, 821. ( 10.3389/fnins.2018.00821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fukao T, et al. 2011. Clinical and molecular characterization of five patients with succinyl-CoA: 3-ketoacid CoA transferase (SCOT) deficiency. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 1812, 619-624. ( 10.1016/j.bbadis.2011.01.015) [DOI] [PubMed] [Google Scholar]
- 51.Newman JC, Verdin E. 2014. Ketone bodies as signaling metabolites. Trends Endocrinol. Metab. 25, 42-52. ( 10.1016/j.tem.2013.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui W, et al. 2019. Dysregulation of ketone body metabolism is associated with poor prognosis for clear cell renal cell carcinoma patients. Front. Oncol. 9, 1422. ( 10.3389/fonc.2019.01422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García-Caballero M, et al. 2019. Role and therapeutic potential of dietary ketone bodies in lymph vessel growth. Nat. Metab. 1, 666-675. ( 10.1038/s42255-019-0087-y) [DOI] [PubMed] [Google Scholar]
- 54.Linares A, Caamaño G, Diaz R, Gonzalez F, Garcia-Peregrin E. 1993. Utilization of ketone bodies by chick brain and spinal cord during embryonic and postnatal development. Neurochem. Res. 18, 1107-1112. ( 10.1007/BF00966692) [DOI] [PubMed] [Google Scholar]
- 55.Chin D, Means AR. 2000. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 10, 322-328. ( 10.1016/S0962-8924(00)01800-6) [DOI] [PubMed] [Google Scholar]
- 56.Toutenhoofd SL, Foletti D, Wicki R, Rhyner JA, Garcia F, Tolon R, Strehler EE. 1998. Characterization of the human CALM2 calmodulin gene and comparison of the transcriptional activity of CALM1, CALM2 and CALMS. Cell Calcium 23, 323-338. ( 10.1016/S0143-4160(98)90028-8) [DOI] [PubMed] [Google Scholar]
- 57.Berchtold MW, Villalobo A. 2014. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 1843, 398-435. ( 10.1016/j.bbamcr.2013.10.021) [DOI] [PubMed] [Google Scholar]
- 58.Wayman GA, Lee Y-S, Tokumitsu H, Silva A, Soderling TR. 2008. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron 59, 914-931. ( 10.1016/j.neuron.2008.08.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burkert P, Duch C. 2006. Developmental changes of CaMKII localization, activity and function during postembryonic CNS remodelling in Manduca sexta. Eur. J. Neurosci. 23, 335-349. ( 10.1111/j.1460-9568.2005.04562.x) [DOI] [PubMed] [Google Scholar]
- 60.Brunet T, Fischer AH, Steinmetz PR, Lauri A, Bertucci P, Arendt D. 2016. The evolutionary origin of bilaterian smooth and striated myocytes. Elife 5, e19607. ( 10.7554/eLife.19607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamaguchi TP. 2001. Heads or tails: Wnts and anterior–posterior patterning. Curr. Biol. 11, R713-R724. ( 10.1016/S0960-9822(01)00417-1) [DOI] [PubMed] [Google Scholar]
- 62.Aceituno-Valenzuela U, Micol-Ponce R, Ponce MR. 2020. Genome-wide analysis of CCHC-type zinc finger (ZCCHC) proteins in yeast, Arabidopsis, and humans. Cell. Mol. Life Sci. 77, 3991-4014. ( 10.1007/s00018-020-03518-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vitorino M, Correia E, Serralheiro A-R, De-Jesus A-C, Inácio JM, Belo JA. 2014. Expression pattern of zcchc24 during early Xenopus development. Int. J. Dev. Biol. 58, 45-50. ( 10.1387/ijdb.130261jb) [DOI] [PubMed] [Google Scholar]
- 64.Kopp A. 2012. Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28, 175-184. ( 10.1016/j.tig.2012.02.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matson CK, Zarkower D. 2012. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163-174. ( 10.1038/nrg3161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young FI, Keruzore M, Nan X, Gennet N, Bellefroid EJ, Li M. 2017. The doublesex-related Dmrta2 safeguards neural progenitor maintenance involving transcriptional regulation of Hes1. Proc. Natl Acad. Sci. USA 114, E5599-E5607. ( 10.1073/pnas.1705186114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu S, Xia W, Zohar Y, Gui J-F. 2013. Zebrafish dmrta2 regulates the expression of cdkn2c in spermatogenesis in the adult testis. Biol. Reprod. 88, 11-12. [DOI] [PubMed] [Google Scholar]
- 68.Panara V, Budd GE, Janssen R. 2019. Phylogenetic analysis and embryonic expression of panarthropod Dmrt genes. Front. Zool. 16, 1-18. ( 10.1186/s12983-019-0322-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gandhe AS, Arunkumar K, John SH, Nagaraju J. 2006. Analysis of bacteria-challenged wild silkmoth, Antheraea mylitta (lepidoptera) transcriptome reveals potential immune genes. BMC Genom. 7, 1-10. ( 10.1186/1471-2164-7-184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qian C, Wang F, Zhu B-J, Wei G-Q, Sun Y, Li S, Wang L, Liu C-L. 2015. Identification and expression analysis of a novel defense gene from Actias selene. Orient. Insects 49, 264-274. ( 10.1080/00305316.2015.1081648) [DOI] [Google Scholar]
- 71.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644-652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150-3152. ( 10.1093/bioinformatics/bts565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, 1-10. ( 10.1186/gb-2009-10-1-r1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 1-16. ( 10.1186/1471-2105-12-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mi H, Thomas P. 2009. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. In Protein networks and pathway analysis, pp. 123-140. Berlin, Germany: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kwak H-J, Lee S-G, Park S-C, Kim J-H, Weisblat DA, Park C, Cho S-J. 2022. Head transcriptome profiling of glossiphoniid leech (Helobdella austinensis) reveals clues of proboscis development. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kwak H-J, Lee S-G, Park S-C, Kim J-H, Weisblat DA, Park C, Cho S-J. 2022. Head transcriptome profiling of glossiphoniid leech (Helobdella austinensis) reveals clues of proboscis development. Figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
Sequences generated in this study are deposited in GenBank (electronic supplementary material, table S4). Raw sequencing data are deposited in the NCBI Sequence Read Archive (SRA) database with accession numbers of SRR15013283 and SRR15013284 under BioProject PRJNA742851.
The data are provided in electronic supplementary material [76].