Abstract
Understanding the evolution of metallo-β-lactamases (MBLs) is fundamental to deciphering the mechanistic basis of resistance to carbapenems in pathogenic and opportunistic bacteria. Presently, these MBL-producing pathogens are linked to high rates of morbidity and mortality worldwide. However, the study of the biochemical and biophysical features of MBLs in vitro provides an incomplete picture of their evolutionary potential, since this limited and artificial environment disregards the physiological context where evolution and selection take place. Herein, we describe recent efforts aimed to address the evolutionary traits acquired by different clinical variants of MBLs in conditions mimicking their native environment (the bacterial periplasm) and considering whether they are soluble or membrane-bound proteins. This includes addressing the metal content of MBLs within the cell under zinc starvation conditions and the context provided by different bacterial hosts that result in particular resistance phenotypes. Our analysis highlights recent progress bridging the gap between in vitro and in-cell studies.
Keywords: antibiotic resistance, metallo-β-lactamase, protein evolution, periplasmic space, outer membrane vesicles, Zn(II) limitation
Abbreviations: BcII, Bacillus cereus type II; CP, calprotectin; IMP-1, imipenemase 1; kcat, β-lactamase's turnover rate; Kd, dissociation constant; KM, Michaelis-Menten constant; MBL, metallo-β-lactamase; MIC, minimum inhibitory concentration; NDM, New Delhi metallo-β-lactamase; OMV, outer membrane vesicle; SBL, serine-β-lactamase; SP, signal peptide; SPM-1, São Paulo metallo-β-lactamase 1; VIM-2, Verona imipenemase 2
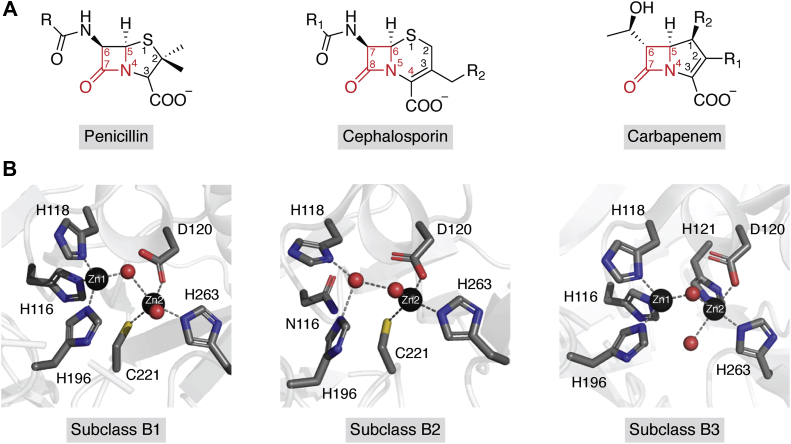
The scourge of antimicrobial resistance represents a growing threat for public health. The indiscriminate use and abuse of antibiotics has fostered the selection of bacteria resistant even to our most potent antibiotics such as carbapenems (Fig. 1A) (1, 2, 3, 4, 5, 6, 7). β-Lactamases represent the most important mechanism of resistance against these “life-saving” drugs, because of the dissemination of genes coding for enzymes with carbapenemase activity (8, 9, 10, 11, 12). Metallo-β-lactamases (MBLs) are Zn(II)-dependent enzymes, that represent one of the largest groups of carbapenemases for which clinical inhibitors are not yet commercially available (13, 14, 15, 16). Since MBLs are highly divergent in sequence, metal ligands at the active site, and Zn(II) stoichiometry, they have been divided into three subclasses: B1, B2, and B3 (Fig. 1B) (14, 17, 18, 19). Most acquired, clinically relevant MBLs present in Gram-negative pathogens belong to subclass B1 (16, 20, 21, 22, 23). B1 enzymes are active with two metal ions: Zn1 coordinated to three His ligands: His116, His118, and His196 (3H site) and Zn2 bound to Asp120, Cys221, and His263 (also called DCH site) (Fig. 1B) (15, 19, 24, 25). The coordination sphere is completed by strategically positioned water molecules (Fig. 1B). B3 enzymes are also binuclear, with a 3H ligand set in the Zn1 site, but with different ligands at the Zn2 site: Asp120, His121, and His263 (DHH site) (Fig. 1B). Finally, B2 MBLs are active in their mono-Zn(II) form, in which the metal ion is bound to the DCH site (Fig. 1B). In these enzymes, uptake of a second Zn(II) ion inhibits their activity (15). For a more comprehensive description of the structural and biochemical features of MBLs, the reader is referred to a recent review covering these aspects (15).
Figure 1.
A, chemical scaffold of the main classes of β-lactam antibiotics hydrolyzed by MBLs. The β-lactam ring is highlighted in red. B, representative active site for subclasses B1, B2, and B3 MBLs. The Zn(II) ions are represented by black spheres, and water molecules are represented by red spheres. In each panel, metal ligand are highlighted in sticks and the gray dashed lines show coordination geometry spheres for the Zn1 and Zn2 sites. The Protein Data Bank files used are: 3SPU (B1—NDM-1), 3SD9 (B2—SfhI), and 3LVZ (B3–BJP-1). BJP-1, MBL from Bradyrhizobium japonicum; MBL, metallo-β-lactamase; NDM-1, New Delhi metallo-β-lactamase 1; SfhI, MBL from Serratia fonticola.
β-Lactamases are a unique model to study protein evolution, since the survival of the bacterial cell challenged with antibiotics can be correlated with the biochemical and biophysical traits of a single protein (26). The serine-β-lactamase (SBL) TEM (named for patient Temoneira) has been an excellent model for the study of protein evolution (27, 28, 29, 30, 31, 32, 33). The essential Zn(II) ions in MBLs represent an additional constraint for protein fitness that does not allow to extrapolate evolutionary studies on SBLs to MBLs (15, 34, 35, 36, 37).
The study of the biochemical and biophysical features of purified proteins has provided clues for understanding the impact of substitutions in the evolution of β-lactamases (27, 28, 29, 30, 31, 32, 33). However, it provides an incomplete and sometimes biased picture, since the impact of the cellular environment is disregarded. In the case of Gram-negative organisms, survival of the bacterial cell in the presence of β-lactam antibiotics depends on the successful expression, translocation, processing, and the appropriate localization of these enzymes. Moreover, MBLs can be present in different bacterial hosts with different resistance phenotypes. The in vitro study of the purified mature protein also overlooks the impact of mutations in the signal peptide (SP) that may affect the translocation and levels of functional protein in the periplasm (38, 39), their cellular localization, and even the correct folding or stability. The description of protein evolution based solely on in vitro studies in purified proteins is an example of the “streetlight effect,” in which a person who is intoxicated is looking under a lamppost for the keys lost somewhere else because the light is there. The understanding of the differences between the features displayed by a protein in vitro and within the cell is crucial for disentangling the driving forces in evolution.
Evolution results from the accumulation of mutations and the selection of particular traits under defined conditions. However, bacteria are continuously exposed to changes in their environment. Two significant factors are usually disregarded when describing MBL evolution: the exposure of bacteria to antibiotics is not permanent, and bacteria cycle between restrictive and permissive conditions (with and without antibiotics, respectively) (2, 39, 40, 41) at the sites of infection, the immune system response elicits a massive metal starvation, limiting the amount of available Zn(II), which impacts on MBL-mediated resistance (42, 43, 44, 45). These conditions define the driving forces in the evolutionary landscape of MBLs.
Here, we present and discuss recent advances and new challenges in the study of the evolution of MBLs, encompassing methodological approaches that enable a quantitative measurement of biochemical and biophysical parameters in cell-like environments. We also review the “state of the art” regarding the physiology (and life cycle) of MBLs in bacterial cells, which is intimately related to their evolutionary landscape. In the last section, we also discuss the biochemical and biophysical traits that have been optimized in the clinical evolution of different MBLs.
MBLs: A journey from the periplasm to the test tube
The first challenge in extrapolating in vitro to in cell data is the attempt to correlate the catalytic activities of the β-lactamases with the resistance phenotype in different bacteria. The resistance phenotype is quantitated based on the minimum inhibitory concentration (MIC) of an antibiotic that inhibits bacterial growth. Most B1 MBLs exhibit catalytic efficiencies (kcat/KM) within a range of 105–106 M−1s−1 (15, 17), differing at the most by one order of magnitude. In contrast, MIC values are quite variable, particularly among different hosts (38, 39). These different resistance phenotypes are due in most cases to changes in the cell permeability and the presence of other, complementary, and resistance mechanisms (15). In this review, we ask the reader to also appreciate the different environments present in each host that impact on the expression levels, processing, and activity of the MBLs. This notion regards the so-called “quinary structure of proteins,” a term coined by McConkey (46), who considers macromolecular interactions within the cell, and it has been addressed by Pielak, Gruebele, Shekhtman, and others (47, 48, 49) by measuring the biochemical and biophysical properties of proteins under conditions mimicking the physiological environment.
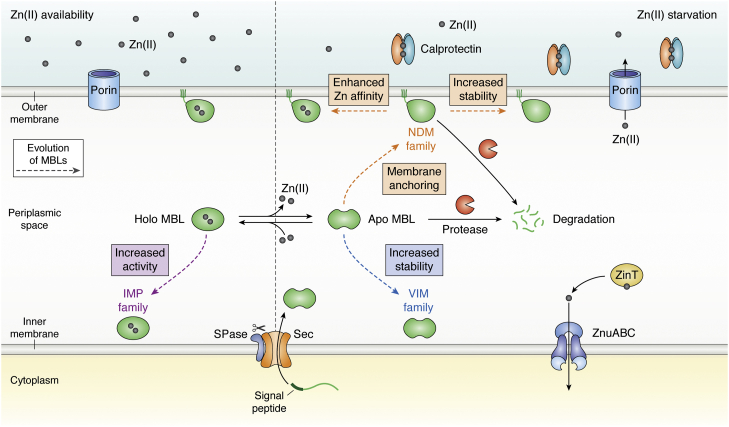
In Gram-negative bacteria, MBLs are synthesized as cytoplasmic precursors with an N-terminal signal sequence (the SP), which directs them to the secretion machinery for their translocation into the cell envelope, where they perform their hydrolytic activity (Fig. 2). So far, all known MBLs are exported by the SecA–SecYEG system as unfolded polypeptides, recognized and processed by type I signal peptidase (SPase I) or by type II lipoprotein signal peptidase (SPase II), depending on whether their final localization is the periplasm or the outer membrane, respectively (Figs. 2 and 3) (50, 51, 52, 53, 54). Finally, the enzymes fold and bind Zn(II) in the periplasmic space (55). Most MBLs are soluble periplasmic enzymes, except for New Delhi metallo-β-lactamase (NDM) enzymes, which are lipoproteins anchored to the inner leaflet of the outer membrane (Fig. 2) (44, 56). The functional levels of MBLs in the periplasmic space vary considerably among different bacterial hosts, finally impacting on the resistance phenotype (38, 44).
Figure 2.
Mechanisms of MBL evolution. Periplasmic Zn(II) levels depend on the passive diffusion through porins in Escherichia coli. In the presence of Zn(II), represented by gray spheres (left panel), the external concentration is in equilibrium with periplasmic Zn(II) levels (62, 63), and MBLs bind the cofactor and hydrolyze β-lactams. The dashed arrows highlight the different mechanism of MBL evolution (79, 130, 139). IMP families have evolved increasing their activity in response of antibiotic evolutionary pressure. When the innate immune response is active (right panel), neutrophils secrete calprotectin (CP) that chelates Zn(II) ions (43), reducing the periplasmic Zn(II) levels and activating expression of the Zn(II) uptake mechanisms to the E. coli cytosol (ZnuABC system and ZinT chaperone). MBLs lose the Zn(II) cofactor leading to formation of apo-MBLs. The nonmetallated form is susceptible to degradation in the periplasm by proteases (shown as Pac-Man-like orange objects) (44). Different mechanisms of evolution are indicated with dashed arrows of different colors, such as outer membrane anchoring (44), stabilization toward degradation, or enhancement of the Zn(II) affinity (15, 73). IMP, imipenemase; MBL, metallo-β-lactamase; SPase, signal peptidase.
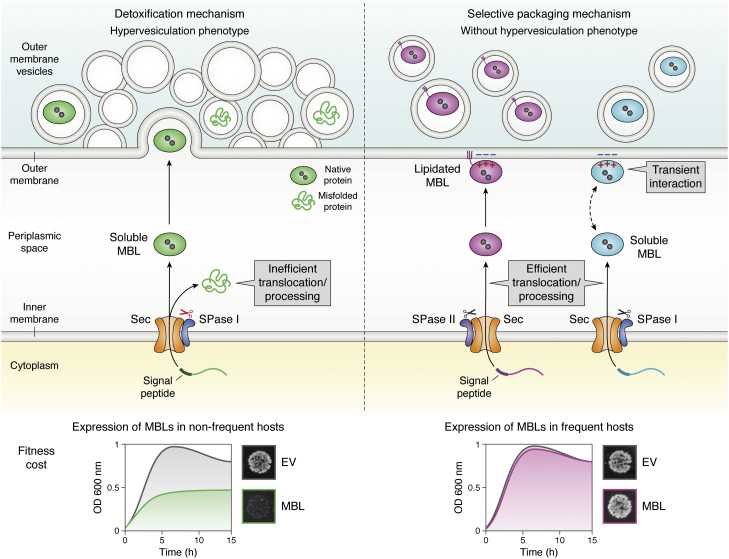
Figure 3.
The adaptability of MBLs to different bacterial hosts depends on the impact of protein expression on biological fitness. This model describes the adaptability of MBLs to different bacterial hosts based on the processing of their SPs (39). An MBL is confined to a narrow range of bacteria (left panel) when its expression in nonfrequent hosts causes the accumulation of toxic precursor forms (shown as green misfolded protein) in the periplasmic face of the inner membrane because of an inefficient translocation and processing that compromises the bacterial growth (bottom left panel). This toxicity correlates with an increase in the production of outer membrane vesicles (OMVs) that incorporate both the mature (folded protein) and the precursor protein (misfolded) to alleviate the envelope stress; thus acting as a vesicle-mediated detoxification mechanism. By contrast, a broad host MBL (right panel) is produced by different bacteria without fitness cost (bottom right panel). These MBLs are efficiently translocated and processed. In these cases, the level of selective packaging into OMVs depends on the interaction of the MBL with the outer membrane, either by membrane anchoring (violet folded protein) or by specific electrostatic interactions (cyan folded protein) through a positively charged patch. The double-headed dashed arrow represents the binding equilibrium of soluble periplasmic MBL with the outer membrane, which favors protein packaging into OMVs. EV, cells carrying the empty vector; MBL, metallo-β-lactamase; SP, signal peptide.
Based on this knowledge, one simple strategy to fill this gap is to measure the catalytic efficiency of the enzymes in periplasmic extracts, using immunoblotting quantitation to retrieve apparent kcat values. kcat/KM values measured in the periplasm show a significant correlation with MIC values (57, 58, 59). This approach has been useful to describe the stepwise evolution of an enzyme evolved in the laboratory (57), to account for differences in the activities in vitro and MICs in MBL mutants (57, 58, 59), and to study the resistance phenotype of a series of MBLs (either chromosomal or acquired) in different bacterial hosts (38). In all these cases, the activity measured in these extracts showed a tight correlation with MIC data. In the case of membrane-bound lactamases, such as NDM-1, similar experiments have been optimized for the measurement of kinetic parameters in spheroplasts (60).
This approach can also be applied to quantitate the thermodynamic stability by measuring the enzymatic activity after incubating the periplasmic extracts at different temperatures (57, 59), providing an apparent Tm value that reflects the thermal stability of the MBLs in conditions mimicking the physiological ones (57, 59). This parameter describes how the equilibrium between the folded and unfolded forms of the protein is affected by the temperature. However, this information does not provide clues regarding the kinetic stability of the protein. The kinetic stability, instead, is related to the energetic barrier separating the native functional state from nonfunctional forms that can accumulate within the cell because of irreversible cellular processes (proteolysis, aggregation, etc.). In this regard, the kinetic stability is more relevant to describe the in-cell behavior, since it may anticipate possible events leading to the loss of the native functional state (61).
Impact of Zn(II) availability on the kinetic stability of MBLs in the periplasm
MBLs bind Zn(II) in the periplasm, since they are secreted as unfolded polypeptides (55). To date, specific metallochaperones for MBLs have not been identified (15), and different studies suggest that the metalation state of MBLs depends mostly on the Zn(II) availability in the external milieu (15, 57, 58, 59, 62, 63, 64).
During an infection, the immune system in mammals elicits a nutritional immunity response, in which metal-binding proteins are targeted to the infection site, withholding essential metal ions at the host–pathogen interface (65). Calprotectin (CP) is the most relevant protein in this process, by sequestering Zn(II) with a subnanomolar affinity (Fig. 2) (42, 43, 45, 66, 67). In Escherichia coli, this process triggers derepression of the Zur regulon and expression of high-affinity transporters in the inner membrane (ZnuABC) and periplasmic metallochaperones that deliver Zn(II) ions to ZnuA (such as ZinT) (Fig. 2), among other proteins (15, 68). However, active zinc importers at the outer membrane in E. coli are not reported. Thus, Zn(II) influx to the periplasm is assumed to occur mainly by passive diffusion through nonselective porins: OmpF and OmpC in E. coli (Fig. 2) (61, 62). The mechanisms for Zn(II) accumulation and transport in the bacterial periplasm have not been fully dissected yet. An in-depth discussion on these mechanisms is provided in these reviews (68, 69).
In the presence of a strong metal chelator, (pKd >8), such as EDTA, dipicolinic acid, or CP (70, 71, 72), MBLs are inhibited by removal of the metal ions, giving rise to stable apo (nonmetalated) variants in vitro. The Zn(II)-binding affinities of B1 and B3 MBLs are in the nanomolar range, whereas B2 MBLs display picomolar binding affinities, based on in vitro measurements (15). The impact of a chelator in the resistance phenotype of MBL producers generally shows a trend that follows the in vitro metal-binding affinities, but there are clearly many other factors that determine the outcome of the treatment with chelators. In the bacterial cell, external Zn(II) depletion results in reduced resistance by eliciting metal dissociation from the MBLs in the periplasm (Fig. 2) (44, 73, 74). This phenomenon raised many questions regarding the level of antibiotic resistance conferred by MBLs expressed in clinical strains depending on the Zn(II) availability in infection models (75) (this discussion is beyond the scope of our review). In any case, infection models and clinical studies should consider that the Zn(II) availability impacts differently depending on the infection site, the bacterial host, and the MBL being expressed (76, 77). The optimization of the conditions to test MIC values in physiological and relevant conditions is currently debated (77). Assessing the metal content of MBLs in the periplasm is essential to better understand these processes. The recent development of a selective fluorescent sensor has been a major advance in this direction since it enables a dynamic monitoring of the metalation state of NDM-1 in the periplasm of E. coli cells (78).
Apo-MBLs have been historically regarded as stable proteins, based on their high thermal stability (57, 74, 79, 80) that has enabled different biophysical studies (81), including resolution of several crystal structures (82). In stark contrast to this in vitro stability, apo-MBLs are not as stable in the periplasm, being subjected to degradation or (possibly) aggregation (Fig. 2) (44, 73). The kinetic stability of apo-MBLs can be quantitated by measuring the half-life time of decay in the periplasm upon zinc deprivation, which follows a single-exponential behavior (73). Different MBLs also show distinct kinetic stabilities in the periplasm: São Paulo metallo-β-lactamase 1 (SPM-1) and imipenemase 1 (IMP-1) are much more stable than Verona imipenemase 2 (VIM-2) (44). The situation of NDM-1 is unique since membrane anchoring stabilizes the apoenzyme (Fig. 2), as evidenced by the rapid degradation of an engineered soluble variant of NDM-1 (44). Overall, this supports the importance of studying the kinetic stability of MBLs under physiological conditions. The understanding of the factors governing the distinct kinetic stabilities of apo-MBLs is crucial to identify the evolutionary traits operative in clinical variants (61).
Evolution of MBLs in different bacterial hosts: Genetic and biochemical determinants of specificity
Table 1 summarizes the different families of acquired MBLs. Among them, IMP, VIM, and NDM types (from B1 subclass) are the most clinically relevant enzymes produced by human pathogens. Although MBLs associated with mobile genetic elements can spread rapidly among different bacteria, they have different host specificities. For example, NDM and IMP enzymes have spread rapidly in Enterobacterales and nonfermenters, whereas SPM-1, Adelaide imipenemase 1 (AIM-1), and VIM-2 are mainly found in Pseudomonas aeruginosa (Table 1) (13, 83). The elucidation of the molecular determinants of host specificity has been subject of recent studies.
Table 1.
Differential distribution of plasmid-associated MBLs among bacterial hosts
| MBL host range | Acquired MBL | Ambler subclass | Bacterial hostsa |
|---|---|---|---|
| MBLs found in a narrow range of bacteria | VIM-2 | B1 | Pseudomonas spp. (P. aeruginosa, P. putida, P. stutzeri, P. juntendi, P. asiatica), Serratia marcescens, Acinetobacter baumanii |
| SPM-1 | B1 | Pseudomonas aeruginosa | |
| AIM-1 | B3 | Pseudomonas aeruginosa | |
| SMB-1 | B3 | Serratia marcescens | |
| DIM-1 | B1 | Pseudomonas aeruginosa, Pseudomonas stutzeri | |
| FIM-1 | B1 | Pseudomonas aeruginosa | |
| KHM-1 | B1 | Citrobacter freundii, Enterobacter hormaechei subsp. hoffmannii | |
| SIM-1 | B1 | Acinetobacter spp. (A. baumannii, A. bereziniae and A. baylyi) | |
| MBLs found in a broad range of bacteria | VIM-1 | B1 | Enterobacterales (Enterobacter hormaechei, Klebsiella pneumoniae, Klebsiella michiganensis, Escherichia coli, Kluyvera cryocrescens, Enterobacter cloacae, Salmonella enterica), Pseudomonas aeruginosa |
| IMP-1 | B1 | Pseudomonas aeruginosa, Enterobacterales (Serratia marcescens, Enterobacter cloacae, Enterobacter roggenkampii, Klebsiella pneumoniae, Escherichia coli), Acinetobacter baumannii | |
| NDM-1 | B1 | Enterobacterales (Klebsiella pneumoniae, Salmonella enterica, Escherichia coli, Enterobacter hormaechei, Enterobacter cloacae, Proteus mirabilis, Proteus vulgaris, Citrobacter freundii, Providencia rettgeri), Acinetobacter spp., Pseudomonas aeruginosa, Vibrio cholera | |
| GIM-1 | B1 | Pseudomonas spp. and Enterobacterales (Enterobacter cloacae, Klebsiella oxytoca, Serratia marcescens, Escherichia coli, and Citrobacter freundii) | |
| TMB-1 | B1 | Achromobacter spp., Acinetobacter spp., Enterobacterales (Enterobacter hormaechei and Citrobacter freundii) |
MBLs are shown according to their bacterial host range: in the first block, those confined to one or a few bacterial hosts are indicated, and in the second block, the enzymes widely distributed among different bacteria are indicated.
Bacterial strains frequently producing each MBL are shown and ordered by decreasing frequency in cases where the MBL is not confined to a single bacterium host.
NDM-1 is more prevalent than IMP-1 in carbapenem-resistant Enterobacterales despite exhibiting similar activities in vitro (15, 17). Avison et al. (84) have shown that this is because of higher expression levels of NDM-1, since the blaNDM-1 gene is found downstream an ISAba125 insertion sequence, which provides a conserved strong promoter. In contrast, blaIMP-1 genes are located downstream of variable promoter sequences with different strengths, resulting in lower protein levels and, consequently, reduced resistance (84). An E. coli isolate carrying the blaIMP-6 gene was shown to be susceptible to imipenem and meropenem, as the result of the transcriptional repression of blaIMP-6. This repression was due to the binding of a protein-encoded upstream of blaIMP-6 (85). However, the mechanisms regulating the production of MBLs warrant further studies, since there is little known about this aspect on acquired MBLs (15).
The finding of a strong promoter can account for the higher frequency of NDM-1 in Enterobacterales based on genetic determinants (84). However, this cannot explain why some blaMBL genes are confined to one host, whereas others can adapt to a wide variety of bacteria (Table 1). A high-throughput analysis of the expression of 200 antibiotic resistance genes in E. coli revealed that the protein–host compatibility depends mostly on the biochemical interaction between the resistance mechanism and the physiology of the host, rather than on genetic factors (86). This insight has been addressed recently for MBLs in two studies, from Tokuriki's (38) and Vila's (39) groups. With complementary approaches, these studies have disclosed a key role of the SP processing in host specificity.
The efficiency of each step in the journey from the gene to the periplasm (transcription, translation, and translocation of an MBL) impacts on the resistance phenotype in a given bacterial host, mostly by defining the functional periplasmic levels of MBLs within the cell (38). In contrast, the guanine–cytosine content, codon usage, or the mRNA stability (87) are crucial in determining neither the resistance phenotype nor the expression levels in different hosts.
Expression of different MBLs (the chromosomal enzymes Bacillus cereus type II [BcII], cefoxitin and carbapenem resistant [CcrA], and IND-1 [Chryseobacterium (Flavobacterium) indologenes], and the acquired MBLs, NDM-1, VIM-1, VIM-2, IMP-1, and SPM-1) with a common SP (PelB from Erwinia carotovora) in different hosts provided a better correlation between MICs and the catalytic efficiencies in vitro, suggesting that the SP and its processing could give rise to the host-dependent phenotypes (38). A deep mutational study on full-length blaVIM-2 by Tokuriki et al. (88) revealed that its SP is highly tolerant to substitutions, except for residue Lys3, which is conserved in all but three VIM allelic variants (88). Synonymous mutation on the SP affected the resistance in vivo. These experiments also revealed that the SP of VIM-2 is not optimized for expression in E. coli, an observation in agreement with the fact that this bacterium is a nonfrequent host for VIM-2 (see later).
The fitness cost elicited in a particular bacterial host upon expression of a resistance determinant is a main driver of enzyme–host adaptability. López et al. (39) showed that the expression of MBLs in less frequent hosts under permissive conditions (in the absence of antibiotics) compromised the biological fitness by impairing bacterial growth (Fig. 3). The expression of SPM-1 and VIM-2 is well tolerated in P. aeruginosa (a frequent host of these enzymes). Instead, both MBLs induced a significant fitness cost when expressed in E. coli or Acinetobacter baumannii (nonfrequent hosts of these proteins). The expression of NDM-1 (an enzyme ubiquitously found in Enterobacterales and nonfermenters) does not cause a fitness cost in any of these three strains (39). The observed growth defects in nonfrequent or less common hosts were accompanied by the accumulation of the precursor proteins, triggering a strong stress envelope response. Thus, MBL expression in nonfrequent hosts saturates the Sec translocon, leading to the accumulation of toxic precursor species that eventually impact negatively on biological fitness (Fig. 3). These toxic species are eliminated from the cell as partially inactive species in outer membrane vesicles (39) (OMVs, see later) (Fig. 3).
These studies reveal that the adaptation of MBLs is not exclusively governed by the genetic determinants, since there are many biochemical factors playing key roles. In this regard, the SP sequence dictates this adaptability based on the processing efficiency (see model in Fig. 3). SP swapping between different MBLs led to a change in the fitness cost and adaptability in different hosts, being able to revert the deleterious impact of the inefficient processing (39). More studies are required to identify sequence patterns in the SPs that can assist in predicting the enzyme adaptability to different hosts.
MBL resistance is strongly dependent on the efficient processing by the corresponding signal peptidases. Therefore, signal peptidases can be also considered as targets to combat carbapenem resistance. Inhibitors of the translocation systems, such as arylomycins (89) or globomycins (90), as well as optimized derivatives (91), can be successfully combined with carbapenems to tackle MBL-mediated resistance.
MBLs beyond the bacterial cell: Protein sorting into vesicles
We have discussed the impact of Zn(II) binding and processing linked to the translocation of MBLs to their native environment, the periplasm. Interestingly, the periplasm is not necessarily the final localization of MBLs, since some of them can be packed into OMVs. All Gram-negative bacteria produce OMVs in response to a wide variety of environments (92, 93, 94, 95, 96). Bacterial OMVs, possessing a diameter of 20 to 250 nm, are spherical portions of the outer membrane that protrude and detach from growing cells. The content of the vesicles resembles that of the producing bacteria, but there are cargo selection mechanisms that results in the enrichment or exclusion of certain molecules in the vesicles (97, 98) (proteins, metabolites, and even nucleic acids) (99, 100, 101). Bacteria employ OMVs to improve their chances of survival and induce changes in their immediate environment (92, 102, 103, 104), such as the detoxification mechanism discussed previously. Cell envelope stress generated by the accumulation of toxic species (such as MBLs processed inefficiently in nonfrequent hosts) leads to a hypervesiculating phenotype, and these enzymes are eliminated by OMVs, mostly in misfolded and inactive forms (Fig. 3) (39).
OMVs can also transport β-lactamases in their folded and active form. The encapsulation of these enzymes in vesicles protects them from degradation in the extracellular environment maintaining a favorable milieu in which the enzymes can exert their catalytic activity beyond the cell boundaries. OMVs transport SBLs and MBLs in their active form as well as plasmids coding for these enzymes (39, 44, 99, 100, 105, 106, 107, 108). OMVs incorporating β-lactamases can protect populations of susceptible bacteria against antibiotics (44, 106, 109), as recently shown in vivo (110, 111), and have been postulated as a novel mechanism of plasmid transfer (112, 113). While the secretion of toxic species is nonspecific, the packaging of folded and active MBLs is selective. In this regard, there is an emerging picture of the cargo selection of MBLs into vesicles.
The membrane-anchored enzyme NDM-1 is packaged in vesicles produced by different bacteria in an active form because of its cellular localization (39, 44). In addition to the lipid anchor, a positively charged patch on the surface of the soluble domain of NDM-1 makes attractive electrostatic interactions with the bacterial membrane (114) and favors their inclusion into vesicles (115). The same mechanism is valid for soluble MBLs: a positively charged patch in the surface of IMP-1 favors its inclusion into OMVs (115). These residues are conserved among NDM and IMP enzymes, suggesting that the phenomenon is common to most allelic variants from these families. Instead, the lack of electrostatic interactions of VIM-2 with the membrane accounts for the small levels of this protein into OMVs under nonstressful conditions (115). This reveals that some MBLs are tuned or poised to interact with the bacterial membrane and therefore are selectively exported into OMVs in a folded, metalated, and active form (Fig. 3). The conserved nature of residues that favor the interaction with the membrane suggests that export of MBLs is common to many enzymes and could be a trait selected in evolution.
Biochemical and biophysical traits selected in the evolution of acquired MBLs
The three largest families of acquired MBLs (IMP, VIM, and NDM) comprise almost 200 allelic variants (83, 74, and 38, respectively) now (15, 116). This large diversity reflects the impact of antibiotic prescription in selecting different variants according to the evolutionary pressures in different environments (such as host, infection sites, and type of antibiotic). In the case of β-lactamase-mediated resistance, selection takes place at the level of bacterial cell survival, depending on the ability of these enzymes to hydrolyze the β-lactam antibiotics (26). This depends not only on the catalytic efficiency but also on the amount of folded and active lactamase in the periplasm (39, 44). Thus, issues discussed before, such as the optimization of expression levels and host adaptability, play a role in evolution, together with the efficacy of the secretion and processing that leads to the ultimate steps of folding and Zn(II) binding in the periplasm. Substitutions are pleiotropic in nature, able to act on the enzyme activity, stability, cofactor binding, among other factors (117). Moreover, when mutations accumulate, epistatic effects are usually observed and determine the possibility of incorporating more substitutions (31, 57, 117, 118). Thus, identification of the biochemical and biophysical traits influenced by the substitutions within the periplasm is crucial to understand the driving forces of evolution in each type of enzyme.
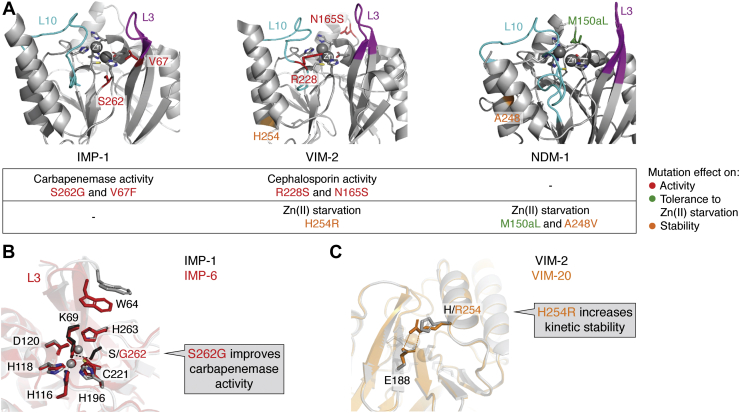
As previously discussed, the thermodynamic stability and kinetic parameters in vitro under nonphysiological conditions can be misleading. Here, we will discuss the current knowledge of the driving forces of evolution in the IMP, VIM, and NDM families. Active-site residues acting as metal ligands are fully conserved within these enzymes, and substitutions are found in second sphere residues (those interacting with the zinc ligands), in the active-site loops L3 and L10 (that flank the active site and define the substrate profile); and in more distant locations in the protein structure (Fig. 4A) (15, 116, 119). The latter substitutions are usually more difficult to rationalize, but (as discussed later), many of them are highly relevant in defining the resistance phenotype.
Figure 4.
A, key mutations and evolutionary traits in the main families of acquired MBLs: IMP-1, VIM-2, and NDM-1. Figures were rendered with PyMol based on the PDB IDs 4C1G, 4NQ2, and 3SPU, respectively. The active-site loops L10 and L3 are shown in cyan and magenta, respectively; the metal ligands as sticks, and the Zn(II) ions as spheres. The mutational “hot spots” are indicated in different colors according to their impact on catalytic activity (red) (79, 130, 139) or in the tolerance against Zn(II) starvation by improving metal binding (green) or the kinetic stability (orange) (73, 74, 79). B, a conformational change in loop L3 introduced by substitution S262G in IMP improves carbapenemase activity. The interaction between S262 and K69 marked with black dashes in IMP-1 (gray structure) is disrupted by the substitution in IMP-6 (red; PDB: 6LVJ). C, the substitution H254R increases the kinetic stability of VIM-20 (PDB: 6OP6) by the formation of a salt bridge between R254 and E188. IMP-1, imipenemase 1; MBL, metallo-β-lactamase; NDM-1, New Delhi metallo-β-lactamase 1; PDB, Protein Data Bank; VIM-2, Verona imipenemase 2.
IMPs
IMP-1 was the first acquired MBLs to be identified in Japan (120). As such, it has been used as a model to assess the role of active-site loops and second sphere residues on the activity of MBLs (Fig. 4A) (121, 122, 123). At the moment, 83 natural IMP variants are divided into six subgroups (124), as the result of the accumulation of more than 54 substitutions distributed in different regions of the protein. The large diversity within this family suggests a polyphyletic origin of IMP enzymes (125), thus limiting a global comparison among variants.
Randomization of selected positions on IMP-1 revealed that second sphere residues (such as Ser262) and residues present in the mobile loop L3 (like Val67) (Fig. 4) shape the substrate profile (122). This pioneering randomization experiment by the Palzkill laboratory also identified positions 67 and 262 as “hot spots” that were found replaced in the clinical variants (126, 127, 128, 129). This study did not explore newer antibiotics, such as ertapenem, or more recent combination treatments that may result in distinct requirements.
A comprehensive study of 20 variants from the IMP-1 subgroup (the largest one) revealed that substitutions Ser262Gly (214 in IMP-1) and Val67Phe (49 in IMP) increased the resistance against certain carbapenems at the expense of ampicillin resistance in E. coli (130). This improvement is the result of an increment in the kcat values for carbapenems and a decrease for ampicillin in agreement with previous reports on isolated variants (127, 128, 131, 132, 133). Ser262Gly is present in other subgroups of the IMP family, eliciting the same effect, that is, without showing epistatic effects with other mutations (cf. IMP-7 and IMP-51 or IMP-11 and IMP-68 that differ only by the Ser262Gly substitution) (134, 135). The Ser262Gly substitution disrupts a hydrogen-bonding interaction with the base of L3 loop, altering its conformation (Fig. 4B) (136). Intriguingly, this substitution is not present in any variant from the second largest subgroup, that of IMP-2. The Val67Phe substitution, instead, is more distributed among the IMP family, and it is present within the subgroup IMP-2. This suggests that different IMP subgroups could be evolving against carbapenems.
The resistance profile of IMP variants under zinc starvation conditions revealed that occurring substitutions do not improve the enzyme ability to resist metal limitation (130). This can be attributed to the observation that IMP-1 is more resistant to zinc limitation than VIM-2 and NDM-1 (44, 76, 80, 130), despite the Zn(II) ions in IMP-1 are kinetically labile (137). It is not unlikely, however, that some mutations that improve the carbapenemase activity may reduce the zinc-binding affinity and require a compensatory mutation. This phenomenon has been reported in a dissection of the evolutionary pathways of the in vitro evolution of BcII (57).
More studies on IMP variants from different subgroups are required to better understand the impact of mutations, their adaptation to different conditions, and eventually relate this information with reported outbreaks (23).
VIMs
The VIM family presents a different evolutionary landscape compared with IMP enzymes. First, the 74 reported alleles form a monophyletic group (125) divided into five clusters (15). The main clusters are the ones related to VIM-1/4 and to VIM-2. Second, VIM alleles accumulate mutations in different regions of the structure, many of them in the active-site loop L10, which determine the substrate profile of MBLs (Fig. 4A) (15). Third, analysis of the resistance profile of different variants suggests that mutations such as R228S and N165S have been selected to optimize the activity against some cephalosporins, as shown by the Galán, Bonomo, and Crowder groups (79, 138, 139). In particular, the substitution R228S was identified as the responsible of the diversification event that distinguishes VIM-1 from VIM-4. Both the VIM-2 and VIM-1/4 subgroups have evolved resistance against cephalosporins with bulky C-3 substituents such as ceftazidime. In contrast to the IMP family, these mutations do not seem to enhance the carbapenemase activity (79, 139, 140, 141). A deep mutational scanning study of VIM-2 (88) identified the essential positions and those tolerant to mutagenesis and identified beneficial mutations occurring in the natural VIM variants (79). Some single mutations giving rise to deleterious effects present in natural mutants may be compensated by other mutations in clinical alleles. In vitro studies comparing phylogenetically distant VIM variants (VIM-2, VIM-1, and VIM-5) suggested that the thermal stability could have been improved upon evolution of this family (142, 143). However, it is not known how this observation could impact on the kinetic stability of the VIM MBL in the periplasm.
A recent study on VIM variants revealed that many of them (10 of 45 studied) exhibit an increased tolerance to zinc starvation conditions (79). Preservation of enzymatic activity under these conditions was observed for VIM-20 (VIM-2 His254Arg) (Fig. 4), VIM-28 (VIM-4 His224Leu), VIM-37 (VIM-4 Ala57Ser), and VIM-59 (VIM-1 His254Arg). Notably, VIM-20 is much less susceptible to proteolysis in vivo than VIM-2 under zinc deprivation. This has been accounted for by the formation of the salt bridge between Arg254 (229 in VIM sequence) and Glu188 (171 in VIM) (Fig. 4C) (79). VIM-20 exhibits a higher thermal stability than VIM-2, but the most relevant trait optimized upon this mutation is the stabilization of the apo form in the periplasm (79). This study by the Crowder and Bonomo laboratories suggests that a group of VIM variants is evolving to better endure zinc starvation (79).
More studies on VIM variants are required, including aspects previously discussed such as the analysis of the expression levels and the impact of the SP in the processing in different hosts, to obtain a complete description of the effect of the occurring mutations in the resistance phenotype and the adaptation to the environmental challenges. Of note, VIM-2 is mostly present in P. aeruginosa, but most evolutionary studies in the laboratory use E. coli as host. As previously highlighted, it is important to interrogate the behavior of the proteins in a context as close as possible to the native one, such as in their most frequent host.
NDMs
The 38 known NDM alleles reveal amino acid substitutions outside the active site, distributed over all the protein structures. The number of substitutions is smaller than those present in the IMP and VIM families, since in most of the cases, they differ by three amino acid changes (the exceptions are NDM-10, NDM-18, and NDM-36) (116). In general, substitutions are located outside the active site and the second sphere. All NDM variants contain the lipobox sequence in the SP that targets them to the outer membrane (44). Residues Arg39 (45 in NDM) and Arg46 (52 in NDM) form a positively charged patch that favors the electrostatic interaction with the membrane and are conserved in all NDM variants (114, 115).
Notably, the activity and resistance profile of NDM alleles does not suggest an adaptation to a particular antibiotic, in contrast with the IMP and VIM families (73, 74, 144). So far, the kinetic characterization and antimicrobial resistance profile of the first 17 variants is very similar in Zn(II)-replete conditions (73, 74, 144). A deep mutational study on NDM-1 by the Palzkill laboratory identified essential residues as well as other residues contributing to substrate specificity. This in-depth analysis revealed that carbapenem hydrolysis by NDM involves more stringent conditions than penicillin or cephalosporin hydrolysis. Based on these findings, the Palzkill group designed the Lys224Arg/Gly232Ala/Asn233Gln (211/219/220 in NDM numbering) triple mutant that lost the imipenemase activity, while retaining the ability to hydrolyze penicillins (145). The authors conclude that, if an inhibitor of NDM-1 is developed, its use in combination with a carbapenem (instead of penicillins or cephalosporins) would be more efficient in restricting the possible escape mutations.
Most of NDM variants are able to confer more resistance than NDM-1 under Zn(II) limitation conditions, suggesting that this enzyme, instead of optimizing the catalytic efficiency, is evolving to endure the metal restriction imposed by the immune system during the infection (73, 74). The variants that proved to be more refractory to Zn(II) starvation were double mutants, all of them containing the substitution Met150aLeu (154 in NDM numbering), which is the most frequent mutation, present in almost one-third of the variants. This reveals that overcoming Zn(II) deprivation is a trait selected in the evolution of NDMs (73, 74).
This evolution has taken place by two independent mechanisms: an increase of the Zn(II)-binding affinity to maintain high levels of active metalated enzyme elicited by the substitution Met150aLeu (Fig. 4A) as in NDM-4 (73, 74); and enhancement of the kinetic stability of the apo form in the periplasm, by substitutions Ala248Val (233 in NDM numbering) in NDM-6 (Fig. 4A) or Glu149Lys (152 in NDM numbering) in NDM-9 (73). Both mechanisms are independent, since the effects are additive, with no epistatic interactions. This is the case of NDM-15 (M150aL-A248V), exhibiting enhanced tolerance to Zn(II) starvation compared with most alleles, including single mutants NDM-4 and NDM-6 (73). Interestingly, NDM-15 retains high catalytic efficiency at subnanomolar Zn(II) levels against ampicillin, whereas NDM-1 did not show activity under these conditions (74). In vitro studies reported an improved thermal stability for most NDM variants (74, 144, 146). However, this stabilization does not correlate with the kinetic stability of the apo variants in the periplasm (73), which seems to be a trait selected in evolution.
Direct competition of E. coli cells expressing NDM-15 or NDM-4 versus NDM-1 at sub-MIC concentrations in Zn(II)-rich medium led to a prevalence of NDM-1. However, in the presence of the chelator dipicolinic acid or CP, at concentrations that do not impair bacterial growth, NDM-15 prevailed over NDM-1 or the single mutants (73). This result suggests that mutations enhancing the zinc-binding affinity or the kinetic stability of apo-NDM variants are shaping the evolution of this MBL family providing more fitness upon metal starvation. The available crystal structures have revealed no clue accounting neither for the increase in zinc-binding affinity because of the Met150aLeu substitution in NDM-4 nor for the kinetic stability provided by the Ala248Val substitution in NDM-6.
Gly63Ser (69 in NDM numbering) and Ala68Thr (74 in NDM numbering) are the only substitutions in the active-site loop 3, present in variant NDM-10 (both) and NDM-21 (G63S V82L M150aL). However, deep scanning mutagenesis revealed that a Gly residue was not essential for ampicillin, imipenem, and cefotaxime hydrolysis (145). The impact of other substitutions in NDM variants as well as the possible epistatic effects among them remains to be studied.
In conclusion, Zn(II) deprivation is shaping the adaptive landscape of the NDM family of proteins, through the accumulation of mutations that either increase the Zn(II) affinity or the kinetic stability of the apoenzyme in the periplasm. At the moment, zinc-binding affinities in the low nanomolar range seem to be required for MBLs to provide resistance (57, 58).
Other MBLs
SPM-1 is a B1 MBL restricted to P. aeruginosa (39), intriguingly with a single variant after almost 2 decades of its first report (147). However, SPM-1 is endowed with a unique set of second sphere residues (differing from most B1 enzymes) that optimize its resistance under Zn(II) deprivation and were selected to confer resistance against antipseudomonal β-lactam antibiotics in detriment of the catalytic efficiency against other β-lactams (59). This case reveals an evolutionary tradeoff in the catalytic profile in which residues surrounding the active site were selected to match the requirements of a particular bacterial host.
B2 enzymes represent the smallest group of MBLs, with a reduced number of allelic variants (116, 148). This may be because B2 enzymes are not present in primary human pathogens (23), being less exposed to prescribed antibiotics. The B2 enzyme CphA (carbapenem-hydrolyzing Aeromonas hydrophila) has the strongest affinity toward Zn(II) among MBLs (149) (Kd = 6 pM), suggesting that their possible dissemination to pathogenic bacteria could be a concern based on their ability to resist metal starvation.
A large allelic diversity (118 variants) was recently reported for the B3 enzyme L1 (150), from the opportunistic pathogen Stenotrophomonas maltophilia. L1 is unique among MBLs in being a tetrameric enzyme (15, 151). Residues involved in tetramerization are conserved in all alleles, confirming that the quaternary arrangement is essential for the enzyme activity (150). Second sphere, loop residues, and positions surrounding the tetramerization patch vary among the different alleles. Zn(II) is essential for the tetramerization of L1 in the periplasm of E. coli (152). This result prompts for the study of the impact on Zn(II) starvation on L1 in S. maltophilia as well as for the evolutionary pathways leading to this large allelic diversity.
Concluding remarks
The early characterization of BcII, the MBLs from Bacillus cereus as a zinc-dependent lactamase in 1966 (153), spurred the curiosity of talented biochemists such as Professor Stephen Waley (154) to describe the features of this enzyme as an outlier within the realm of β-lactamases. Three decades later, genes coding for MBLs were identified in mobile genetic elements, leading to a new era in which efforts were devoted to understanding the genetics of dissemination, their diversity, and mechanism of action with the final aim to design an MBL inhibitor. In recent years, the finding of membrane-bound MBLs, their secretion into vesicles, and understanding that their metal load in the periplasm is variable has required a different approach that involves the description of their physiology, their bacterial host specificity, and the identification of different driving forces of evolution operative in different MBL families. A new era has started in the study of MBLs that requires a more comprehensive approach, integrating clinical and basic microbiology, biochemistry, biophysics, and cell biology, which we expect will provide a better understanding in the evolution of these fascinating enzymes in their native context or closely approaching their natural environment. Each family of acquired MBLs accumulates mutations in different positions of their structure and experiences different evolutionary trajectories with distinct traits being selected. These traits cannot always be described based on studies on purified proteins. The study of MBL evolution in a context as close as possible as the native one requires: (1) studying the resistance phenotype in model host systems related to the more frequent bacterial hosts in the clinics, instead of resorting only to E. coli as a model organism; (2) express each MBL with their native SPs; (3) quantitate the expression levels of active protein in the periplasm, either in the soluble form or in the membrane-bound form; (4) explore the impact of Zn(II) deprivation in the resistance phenotype and in the kinetic stability of the apovariant; (5) correlate these data with in vitro studies of catalytic efficiency and zinc-binding affinity; and (6) determine apparent kinetic parameters in periplasmic extracts or spheroplast to attempt filling the gap between in vitro and in cell experiments. In addition, insights into MBL folding linked to metal binding in the periplasm will require the use of new experimental (78, 155, 156, 157) and computational tools, particularly those that encompass different timescales (158, 159). We hope that these efforts can provide to a faster “bench-to-bedside” translation of novel therapies.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to our colleagues of the Metalloprotein laboratory and collaborators for scientific contributions and discussions. We thank the support of our home institutions, CONICET and the University of Rosario.
Author contributions
C. L., J. D., and A. J. V. conceptualization; C. L., J. D., R. A. B., and A. J. V. writing–original draft; R. A. B. and A. J. V. funding acquisition.
Funding and additional information
This work was supported by a grant from ANPCyT (grant no.: PICT-2016-1657; to A. J. V.) and a National Institute of Allergy and Infectious Diseases, National Institutes of Health, grant (grant no.: 2R01AI100560-06A1; to A. J. V. and R. A. B). C. L. and J. D. are recipients of a postdoctoral and doctoral fellowships from CONICET, respectively. A. J. V. is a staff member from CONICET. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (to R. A. B.) under Award number R01AI100560. This study was also supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs (Award number: 1I01BX001974; to R. A. B.) from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center (Veterans Integrated Service Networks 10). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
Edited by Wolfgang Peti
References
- 1.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 2.Fisher J.F., Meroueh S.O., Mobashery S. Bacterial resistance to β-lactam antibiotics: Compelling opportunism, compelling opportunity. Chem. Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 3.Sugden R., Kelly R., Davies S. Combatting antimicrobial resistance globally. Nat. Microbiol. 2016;1:16187. doi: 10.1038/nmicrobiol.2016.187. [DOI] [PubMed] [Google Scholar]
- 4.Bush K., Bradford P.A. β-lactams and β-lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med. 2016;6 doi: 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes A.H., Moore L.S.P., Sundsfjord A., Steinbakk M., Regmi S., Karkey A., Guerin P.J., Piddock L.J.V. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387:176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 6.Meletis G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2016;3:15–21. doi: 10.1177/2049936115621709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papp-Wallace K.M., Endimiani A., Taracila M.A., Bonomo R.A. Carbapenems: Past, present, and future. Antimicrob. Agents Chemother. 2011;55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Philippon A., Dusart J., Joris B., Frère J.M. The diversity, structure and regulation of β-lactamases. Cell. Mol. Life Sci. 1998;54:341–346. doi: 10.1007/s000180050161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frère J.M. Beta-lactamases and bacterial resistance to antibiotics. Mol. Microbiol. 1995;16:385–395. doi: 10.1111/j.1365-2958.1995.tb02404.x. [DOI] [PubMed] [Google Scholar]
- 10.Patel G., Bonomo R.A. “Stormy waters ahead”: Global emergence of carbapenemases. Front. Microbiol. 2013;4:1–17. doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush K. Proliferation and significance of clinically relevant β-lactamases. Ann. N. Y. Acad. Sci. 2013;1277:84–90. doi: 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- 12.Bonomo R.A. β-Lactamases: A focus on current challenges. Cold Spring Harb. Perspect. Med. 2017;7 doi: 10.1101/cshperspect.a025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh T.R., Toleman M.A., Poirel L., Nordmann P. Metallo-β-lactamases: The quiet before the storm? Clin. Microbiol. Rev. 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordmann P., Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 2014;20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 15.Bahr G., González L.J., Vila A.J. Metallo-β-lactamases in the age of multidrug resistance: From structure and mechanism to evolution, dissemination, and inhibitor design. Chem. Rev. 2021;121:7957–8094. doi: 10.1021/acs.chemrev.1c00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mojica M.F., Rossi M.-A., Vila A.J., Bonomo R.A. The urgent need for metallo-β-lactamase inhibitors: An unattended global threat. Lancet Infect. Dis. 2022;22:e28–e34. doi: 10.1016/S1473-3099(20)30868-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bebrone C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007;74:1686–1701. doi: 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Garau G., García-Sáez I., Bebrone C., Anne C., Mercuri P., Galleni M., Frère J.M., Dideberg O. Update of the standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 2004;48:2347–2349. doi: 10.1128/AAC.48.7.2347-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berglund F., Johnning A., Larsson D.G.J., Kristiansson E. An updated phylogeny of the metallo-β-lactamases. J. Antimicrob. Chemother. 2021;76:117–123. doi: 10.1093/jac/dkaa392. [DOI] [PubMed] [Google Scholar]
- 20.Mojica M.F., Bonomo R.A., Fast W. B1-metallo-β-lactamases: Where do we stand? Curr. Drug Targets. 2015;17:1029–1050. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palzkill T. Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013;1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall S., Hujer A.M., Rojas L.J., Papp-Wallace K.M., Humphries R.M., Spellberg B., Hujer K.M., Marshall E.K., Rudin S.D., Perez F., Wilson B.M., Wasserman R.B., Chikowski L., Paterson D.L., Vila A.J., et al. Can ceftazidime-avibactam and aztreonam overcome β-lactam resistance conferred by metallo-β-lactamases in Enterobacteriaceae? Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush K., Bradford P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen B.A., Bush K. Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 1997;41:223–232. doi: 10.1128/aac.41.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tooke C.L., Hinchliffe P., Bragginton E.C., Colenso C.K., Hirvonen V.H.A., Takebayashi Y., Spencer J. β-Lactamases and β-lactamase inhibitors in the 21st century. J. Mol. Biol. 2019;431:3472–3500. doi: 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson D.I., Balaban N.Q., Baquero F., Courvalin P., Glaser P., Gophna U., Kishony R., Molin S., Tønjum T. Antibiotic resistance: Turning evolutionary principles into clinical reality. FEMS Microbiol. Rev. 2021;44:171–188. doi: 10.1093/femsre/fuaa001. [DOI] [PubMed] [Google Scholar]
- 27.Orencia M.C., Yoon J.S., Ness J.E., Stemmer W.P.C., Stevens R.C. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat. Struct. Biol. 2001;8:238–242. doi: 10.1038/84981. [DOI] [PubMed] [Google Scholar]
- 28.Sideraki V., Huang W., Palzkill T., Gilbert H.F. A secondary drug resistance mutation of TEM-1 β-lactamase that suppresses misfolding and aggregation. Proc. Natl. Acad. Sci. U. S. A. 2001;98:283. doi: 10.1073/pnas.011454198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Minasov G., Shoichet B.K. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 30.Bershtein S., Segal M., Bekerman R., Tokuriki N., Tawfik D.S. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 31.Weinreich D.M., Delaney N.F., DePristo M.A., Hartl D.L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 32.Salverda M.L.M., Dellus E., Gorter F.A., Debets A.J.M., van der Oost J., Hoekstra R.F., Tawfik D.S., de Visser J.A.G.M. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong L.I., Bloom J.D. Epistatically interacting substitutions are enriched during adaptive protein evolution. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambler R.P., Daniel M., Fleming J., Hermoso J.M., Pang C., Waley S.G. The amino acid sequence of the zinc-requiring β-lactamase II from the bacterium Bacillus cereus 569. FEBS Lett. 1985;189:207–211. doi: 10.1016/0014-5793(85)81024-3. [DOI] [PubMed] [Google Scholar]
- 35.Hussain M., Carlino A., Madonna M.J., Lampen J.O. Cloning and sequencing of the metallothioprotein β-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J. Bacteriol. 1985;164:223–229. doi: 10.1128/jb.164.1.223-229.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baier F., Tokuriki N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 2014;426:2442–2456. doi: 10.1016/j.jmb.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Meini M.R., Llarrull L.I., Vila A.J. Overcoming differences: The catalytic mechanism of metallo-β-lactamases. FEBS Lett. 2015;589:3419–3432. doi: 10.1016/j.febslet.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Socha R.D., Chen J., Tokuriki N. The molecular mechanisms underlying hidden phenotypic variation among metallo-β-lactamases. J. Mol. Biol. 2019;431:1172–1185. doi: 10.1016/j.jmb.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 39.López C., Ayala J.A., Bonomo R.A., González L.J., Vila A.J. Protein determinants of dissemination and host specificity of metallo-β-lactamases. Nat. Commun. 2019;10:3617. doi: 10.1038/s41467-019-11615-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes D., Andersson D.I. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol. Rev. 2017;41:374–391. doi: 10.1093/femsre/fux004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pereira C., Larsson J., Hjort K., Elf J., Andersson D.I. The highly dynamic nature of bacterial heteroresistance impairs its clinical detection. Commun. Biol. 2021;4:1–12. doi: 10.1038/s42003-021-02052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corbin B.D., Seeley E.H., Raab A., Feldmann J., Miller M.R., Torres V.J., Anderson K.L., Dattilo B.M., Dunman P.M., Gerads R., Caprioli R.M., Nacken W., Chazin W.J., Skaar E.P. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 43.Kehl-Fie T.E., Skaar E.P. Nutritional immunity beyond iron: A role for manganese and zinc. Curr. Opin. Chem. Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.González L.J., Bahr G., Nakashige T.G., Nolan E.M., Bonomo R.A., Vila A.J. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-β-lactamase. Nat. Chem. Biol. 2016;12:516–522. doi: 10.1038/nchembio.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antelo G.T., Vila A.J., Giedroc D.P., Capdevila D.A. Molecular evolution of transition metal bioavailability at the host–pathogen interface. Trends Microbiol. 2021;29:441–457. doi: 10.1016/j.tim.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McConkey E.H. Molecular evolution, intracellular organization, and the quinary structure of proteins. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3236–3240. doi: 10.1073/pnas.79.10.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monteith W.B., Cohen R.D., Smith A.E., Guzman-Cisneros E., Pielak G.J. Quinary structure modulates protein stability in cells. Proc. Natl. Acad. Sci. U. S. A. 2015;112:1739–1742. doi: 10.1073/pnas.1417415112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen R.D., Pielak G.J. A cell is more than the sum of its (dilute) parts: A brief history of quinary structure. Protein Sci. 2017;26:403–413. doi: 10.1002/pro.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guin D., Gruebele M. Weak chemical interactions that drive protein evolution: Crowding, sticking, and quinary structure in folding and function. Chem. Rev. 2019;119:10691–10717. doi: 10.1021/acs.chemrev.8b00753. [DOI] [PubMed] [Google Scholar]
- 50.Pradel N., Delmas J., Wu L.F., Santini C.L., Bonnet R. Sec- and Tat-dependent translocation of β-lactamases across the Escherichia coli inner membrane. Antimicrob. Agents Chemother. 2009;53:242–248. doi: 10.1128/AAC.00642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du Plessis D.J.F., Nouwen N., Driessen A.J.M. The Sec translocase. Biochim. Biophys. Acta Biomembr. 2011;1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 52.Denks K., Vogt A., Sachelaru I., Petriman N.A., Kudva R., Koch H.G. The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 2014;31:58–84. doi: 10.3109/09687688.2014.907455. [DOI] [PubMed] [Google Scholar]
- 53.Paetzel M., Karla A., Strynadka N.C.J., Dalbey R.E. Signal peptidases. Chem. Rev. 2002;102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 54.Natale P., Brüser T., Driessen A.J.M. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane-distinct translocases and mechanisms. Biochim. Biophys. Acta Biomembr. 2008;1778:1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 55.Morán-Barrio J., Limansky A.S., Viale A.M. Secretion of GOB metallo-β-lactamase in Escherichia coli depends strictly on the cooperation between the cytoplasmic DnaK chaperone system and the Sec machinery: Completion of folding and Zn(II) ion acquisition occur in the bacterial periplasm. Antimicrob. Agents Chemother. 2009;53:2908–2917. doi: 10.1128/AAC.01637-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.King D., Strynadka N. Crystal structure of New Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011;20:1484–1491. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meini M.-R., Tomatis P.E., Weinreich D.M., Vila A.J. Quantitative description of a protein fitness landscape based on molecular features. Mol. Biol. Evol. 2015;32:1774–1787. doi: 10.1093/molbev/msv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.González J.M., Meini M.-R., Tomatis P.E., Medrano Martín F.J., Cricco J.A., Vila A.J. Metallo-β-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions. Nat. Chem. Biol. 2012;8:698–700. doi: 10.1038/nchembio.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.González L.J., Moreno D.M., Bonomo R.A., Vila A.J. Host-specific enzyme-substrate interactions in SPM-1 metallo-β-lactamase are modulated by second sphere residues. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giannini E., González L.J., Vila A.J. A simple protocol to characterize bacterial cell-envelope lipoproteins in a native-like environment. Protein Sci. 2019;28:2004–2010. doi: 10.1002/pro.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchez-Ruiz J.M. Protein kinetic stability. Biophys. Chem. 2010;148:1–15. doi: 10.1016/j.bpc.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 62.Sigdel T.K., Cilliers R., Gursahaney P.R., Thompson P., Easton J.A., Crowder M.W. Probing the adaptive response of Escherichia coli to extracellular Zn(II) Biometals. 2006;19:461–471. doi: 10.1007/s10534-005-4962-5. [DOI] [PubMed] [Google Scholar]
- 63.Easton J.A., Thompson P., Crowder M.W. Time-dependent translational response of E. coli to excess Zn(II) J. Biomol. Tech. 2006;17:303. [PMC free article] [PubMed] [Google Scholar]
- 64.Meini M.R., González L.J., Vila A.J. Antibiotic resistance in Zn(II)-deficient environments: Metallo-β-lactamase activation in the periplasm. Future Microbiol. 2013;8:947–979. doi: 10.2217/fmb.13.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hood M.I., Skaar E.P. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kehl-Fie T.E., Chitayat S., Hood M.I., Damo S., Restrepo N., Garcia C., Munro K.A., Chazin W.J., Skaar E.P. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang S., Song R., Wang Z., Jing Z., Wang S., Ma J. S100A8/A9 in inflammation. Front. Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lonergan Z.R., Skaar E.P. Nutrient zinc at the host-pathogen interface. Trends Biochem. Sci. 2019;44:1041. doi: 10.1016/j.tibs.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bahr G., González L.J., Vila A.J. Metallo-β-lactamases and a tug-of-war for the available zinc at the host–pathogen interface. Curr. Opin. Chem. Biol. 2022;66:102103. doi: 10.1016/j.cbpa.2021.102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao Z., Wedd A.G. The challenges of determining metal-protein affinities. Nat. Prod. Rep. 2010;27:768–789. doi: 10.1039/b906690j. [DOI] [PubMed] [Google Scholar]
- 71.Brophy M.B., Hayden J.A., Nolan E.M. Calcium ion gradients modulate the zinc affinity and antibacterial activity of human calprotectin. J. Am. Chem. Soc. 2012;134:18089–18100. doi: 10.1021/ja307974e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chung L., Rajan K.S., Merdinger E., Grecz N. Coordinative binding of divalent cations with ligands related to bacterial spores: Equilibrium studies. Biophys. J. 1971;11:469–482. doi: 10.1016/S0006-3495(71)86229-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bahr G., Vitor-Horen L., Bethel C.R., Bonomo R.A., Gonzalez L.J., Vila A.J. Clinical evolution of New Delhi metallo-β-lactamase (NDM) optimizes resistance under Zn(II) deprivation. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.01849-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheng Z., Thomas P.W., Ju L., Bergstrom A., Mason K., Clayton D., Miller C., Bethel C.R., VanPelt J., Tierney D.L., Page R.C., Bonomo R.A., Fast W., Crowder M.W. Evolution of New Delhi metallo-β-lactamase (NDM) in the clinic: Effects of NDM mutations on stability, zinc affinity, and mono-zinc activity. J. Biol. Chem. 2018;293:12606–12618. doi: 10.1074/jbc.RA118.003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Asempa T.E., Abdelraouf K., Nicolau D.P. Activity of β-lactam antibiotics against metallo-β-lactamaseproducing enterobacterales in animal infection models: A current state of affairs. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.02271-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lasko M.J., Gill C.M., Asempa T.E., Nicolau D.P. EDTA-modified carbapenem inactivation method (eCIM) for detecting IMP metallo-β-lactamase–producing Pseudomonas aeruginosa: An assessment of increasing EDTA concentrations. BMC Microbiol. 2020;20:1–5. doi: 10.1186/s12866-020-01902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belanger C.R., Hancock R.E.W. Testing physiologically relevant conditions in minimal inhibitory concentration assays. Nat. Protoc. 2021;16:3761–3774. doi: 10.1038/s41596-021-00572-8. [DOI] [PubMed] [Google Scholar]
- 78.Mehta R., Rivera D.D., Reilley D.J., Tan D., Thomas P.W., Hinojosa A., Stewart A.C., Cheng Z., Thomas C.A., Crowder M.W., Alexandrova A.N., Fast W., Que E.L. Visualizing the dynamic metalation state of New Delhi metallo-β-lactamase-1 in bacteria using a reversible fluorescent probe. J. Am. Chem. Soc. 2021;143:8314–8323. doi: 10.1021/jacs.1c00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng Z., Shurina B.A., Bethel C.R., Thomas P.W., Marshall S.H., Thomas C.A., Yang K., Kimble R.L., Montgomery J.S., Orischak M.G., Miller C.M., Tennenbaum J.L., Nix J.C., Tierney D.L., Fast W., et al. A single salt bridge in VIM-20 increases protein stability and antibiotic resistance under low-zinc conditions. mBio. 2019;10 doi: 10.1128/mBio.02412-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Horton L.B., Shanker S., Mikulski R., Brown N.G., Phillips K.J., Lykissa E., Venkataram Prasad B.V., Palzkill T. Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-β-lactamase active site. Antimicrob. Agents Chemother. 2012;56:5667–5677. doi: 10.1128/AAC.01276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacquin O., Balbeur D., Damblon C., Marchot P., De Pauw E., Roberts G.C.K., Frère J.M., Matagne A. Positively cooperative binding of zinc ions to Bacillus cereus 569/H/9 β-lactamase II suggests that the binuclear enzyme is the only relevant form for catalysis. J. Mol. Biol. 2009;392:1278–1291. doi: 10.1016/j.jmb.2009.07.092. [DOI] [PubMed] [Google Scholar]
- 82.Kim Y., Tesar C., Mire J., Jedrzejczak R., Binkowski A., Babnigg G., Sacchettini J., Joachimiak A. Structure of apo- and monometalated forms of NDM-1—a highly potent carbapenem-hydrolyzing metallo-β-lactamase. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nordmann P., Poirel L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin. Infect. Dis. 2019;69:S521–S528. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheung C.H.P., Alorabi M., Hamilton F., Takebayashi Y., Mounsey O., Heesom K.J., Williams P.B., Williams O.M., Albur M., MacGowan A.P., Avison M.B. Trade-offs between antibacterial resistance and fitness cost in the production of metallo-β-lactamases by enteric bacteria manifest as sporadic emergence of carbapenem resistance in a clinical setting. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.02412-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Segawa T., Sekizuka T., Suzuki S., Shibayama K., Matsui M., Kuroda M. The plasmid-encoded transcription factor ArdK contributes to the repression of the IMP-6 metallo-β-lactamase gene blaIMP-6, leading to a carbapenem-susceptible phenotype in the blaIMP-6-positive Escherichia coli strain A56-1S. PLoS One. 2018;13 doi: 10.1371/journal.pone.0208976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Porse A., Schou T.S., Munck C., Ellabaan M.M.H., Sommer M.O.A. Biochemical mechanisms determine the functional compatibility of heterologous genes. Nat. Commun. 2018;9:1–11. doi: 10.1038/s41467-018-02944-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mehlhoff J.D., Stearns F.W., Rohm D., Wang B., Tsou E.-Y., Dutta N., Hsiao M.-H., Gonzalez C.E., Rubin A.F., Ostermeier M. Collateral fitness effects of mutations. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11597–11607. doi: 10.1073/pnas.1918680117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen J.Z., Fowler D.M., Tokuriki N. Comprehensive exploration of the translocation, stability and substrate recognition requirements in vim-2 lactamase. Elife. 2020;9 doi: 10.7554/eLife.56707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith P.A., Romesberg F.E. Mechanism of action of the arylomycin antibiotics and effects of signal peptidase I inhibition. Antimicrob. Agents Chemother. 2012;56:5054–5060. doi: 10.1128/AAC.00785-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dev I.K., Harvey R.J., Ray P.H. Inhibition of prolipoprotein signal peptidase by globomycin. J. Biol. Chem. 1985;260:5891–5894. [PubMed] [Google Scholar]
- 91.Kulanthaivel P., Kreuzman A.J., Strege M.A., Belvo M.D., Smitka T.A., Clemens M., Swartling J.B., Minton K.L., Zheng F., Angleton E.L., Mullen D., Jungheim L.N., Klimkowski V.J., Nicas T.I., Thompson R.C., et al. Novel lipoglycopeptides as inhibitors of bacterial signal peptidase I ∗. J. Biol. Chem. 2004;279:36250–36258. doi: 10.1074/jbc.M405884200. [DOI] [PubMed] [Google Scholar]
- 92.Schwechheimer C., Kuehn M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pathirana R.D., Kaparakis-Liaskos M. Bacterial membrane vesicles: Biogenesis, immune regulation and pathogenesis. Cell. Microbiol. 2016;18:1518–1524. doi: 10.1111/cmi.12658. [DOI] [PubMed] [Google Scholar]
- 94.Zlatkov N., Nadeem A., Uhlin B.E., Wai S.N. Eco-evolutionary feedbacks mediated by bacterial membrane vesicles. FEMS Microbiol. Rev. 2021;45:1–26. doi: 10.1093/femsre/fuaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Toyofuku M., Nomura N., Eberl L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2018;17:13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 96.Weber B.S., Kinsella R.L., Harding C.M., Feldman M.F. The secrets of acinetobacter secretion. Trends Microbiol. 2017;25:532–545. doi: 10.1016/j.tim.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bonnington K.E., Kuehn M.J. Protein selection and export via outer membrane vesicles. Biochim. Biophys. Acta Mol. Cell Res. 2014;1843:1612–1619. doi: 10.1016/j.bbamcr.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guerrero-Mandujano A., Hernández-Cortez C., Ibarra J.A., Castro-Escarpulli G. The outer membrane vesicles: Secretion system type zero. Traffic. 2017;18:425–432. doi: 10.1111/tra.12488. [DOI] [PubMed] [Google Scholar]
- 99.Chatterjee S., Mondal A., Mitra S., Basu S. Acinetobacter baumannii transfers the blaNDM-1 gene via outer membrane vesicles. J. Antimicrob. Chemother. 2017;72:2201–2207. doi: 10.1093/jac/dkx131. [DOI] [PubMed] [Google Scholar]
- 100.Rumbo C., Fernández-Moreira E., Merino M., Poza M., Mendez J.A., Soares N.C., Mosquera A., Chaves F., Bou G. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: A new mechanism of dissemination of carbapenem resistance genes in acinetobacter baumannii. Antimicrob. Agents Chemother. 2011;55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sjöström A.E., Sandblad L., Uhlin B.E., Wai S.N. Membrane vesicle-mediated release of bacterial RNA. Sci. Rep. 2015;5:15329. doi: 10.1038/srep15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kulp A., Kuehn M.J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jan A.T. Outer membrane vesicles (OMVs) of gram-negative bacteria: A perspective update. Front. Microbiol. 2017;8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McBroom A.J., Kuehn M.J. Release of outer membrane vesicles by gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ciofu O., Beveridge T.J., Kadurugamuwa J., Walther-Rasmussen J., Høiby N. Chromosomal β-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000;45:9–13. doi: 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- 106.Stentz R., Horn N., Cross K., Salt L., Brearley C., Livermore D.M., Carding S.R. Cephalosporinases associated with outer membrane vesicles released by Bacteroides spp. protect gut pathogens and commensals against β-lactam antibiotics. J. Antimicrob. Chemother. 2015;70:701–709. doi: 10.1093/jac/dku466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liao Y.T., Kuo S.C., Chiang M.H., Lee Y.T., Sung W.C., Chen Y.H., Chen T.L., Fung C.P. Acinetobacter baumannii extracellular OXA-58 is primarily and selectively released via outer membrane vesicles after sec-dependent periplasmic translocation. Antimicrob. Agents Chemother. 2015;59:7346–7354. doi: 10.1128/AAC.01343-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schaar V., Nordström T., Mörgelin M., Riesbeck K. Moraxella catarrhalis outer membrane vesicles carry β-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob. Agents Chemother. 2011;55:3845–3853. doi: 10.1128/AAC.01772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Devos S., Stremersch S., Raemdonck K., Braeckmans K., Devreese B. Intra-and interspecies effects of outer membrane vesicles from Stenotrophomonas maltophilia on beta-lactam resistance. Antimicrob. Agents Chemother. 2016;60:2516–2518. doi: 10.1128/AAC.02171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Martínez M.M.B., Bonomo R.A., Vila A.J., Maffía P.C., González L.J. On the offensive: The role of outer membrane vesicles in the successful dissemination of New Delhi metallo-β-lactamase (NDM-1) mBio. 2021;12 doi: 10.1128/mBio.01836-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bielaszewska M., Daniel O., Nyč O., Mellmann A. In vivo secretion of β-lactamase-carrying outer membrane vesicles as a mechanism of β-lactam therapy failure. Membranes (Basel) 2021;11:806. doi: 10.3390/membranes11110806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tran F., Boedicker J.Q. Genetic cargo and bacterial species set the rate of vesicle-mediated horizontal gene transfer. Sci. Rep. 2017;7:8813. doi: 10.1038/s41598-017-07447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Domingues S., Nielsen K.M. Membrane vesicles and horizontal gene transfer in prokaryotes. Curr. Opin. Microbiol. 2017;38:16–21. doi: 10.1016/j.mib.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 114.Prunotto A., Bahr G., González L.J., Vila A.J., Dal Peraro M. Molecular bases of the membrane association mechanism potentiating antibiotic resistance by New Delhi metallo-β-lactamase 1. ACS Infect. Dis. 2020;6:2719–2731. doi: 10.1021/acsinfecdis.0c00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.López C., Prunotto A., Bahr G., Bonomo R.A., González L.J., Dal Peraro M., Vila A.J. Specific protein-membrane interactions promote packaging of metallo-β-lactamases into outer membrane vesicles. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.00507-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Naas T., Oueslati S., Bonnin R.A., Dabos M.L., Zavala A., Dortet L., Retailleau P., Iorga B.I. Beta-lactamase database (BLDB)–structure and function. J. Enzyme Inhib. Med. Chem. 2017;32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.DePristo M.A., Weinreich D.M., Hartl D.L. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat. Rev. Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 118.Starr T.N., Thornton J.W. Epistasis in protein evolution. Protein Sci. 2016;25:1204–1218. doi: 10.1002/pro.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oelschlaeger P. Outsmarting metallo-β-lactamases by mimicking their natural evolution. J. Inorg. Biochem. 2008;102:2043–2051. doi: 10.1016/j.jinorgbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 120.Laraki N., Franceschini N., Rossolini G.M., Santucci P., Meunier C., De Pauw E., Amicosante G., Frère J.M., Galleni M. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 1999;43:902–906. doi: 10.1128/aac.43.4.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Materon I.C., Palzkill T. Identification of residues critical for metallo-β-lactamase function by codon randomization and selection. Protein Sci. 2001;10:2556–2565. doi: 10.1110/ps.40884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Materon I.C., Beharry Z., Huang W., Perez C., Palzkill T. Analysis of the context dependent sequence requirements of active site residues in the metallo-β-lactamase IMP-1. J. Mol. Biol. 2004;344:653–663. doi: 10.1016/j.jmb.2004.09.074. [DOI] [PubMed] [Google Scholar]
- 123.Yamaguchi Y., Kuroki T., Yasuzawa H., Higashi T., Jin W., Kawanami A., Yamagata Y., Arakawa Y., Goto M., Kurosaki H. Probing the role of Asp-120(81) of metallo-β-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and x-ray crystallography. J. Biol. Chem. 2005;280:20824–20832. doi: 10.1074/jbc.M414314200. [DOI] [PubMed] [Google Scholar]
- 124.Softley C.A., Zak K.M., Bostock M.J., Fino R., Zhou R.X., Kolonko M., Mejdi-Nitiu R., Meyer H., Sattler M., Popowicz G.M. Structure and molecular recognition mechanism of IMP-13 metallo-β-lactamase. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00123-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pal A., Tripathi A. An in silico approach for understanding the molecular evolution of clinically important metallo-beta-lactamases. Infect. Genet. Evol. 2013;20:39–47. doi: 10.1016/j.meegid.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 126.Iyobe S., Kusadokoro H., Ozaki J., Matsumura N., Minami S., Haruta S., Sawai T., O'Hara K. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 2000;44:2023–2027. doi: 10.1128/aac.44.8.2023-2027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iyobe S., Kusadokoro H., Takahashi A., Yomoda S., Okubo T., Nakamura A., O'Hara K. Detection of a variant metallo-β-lactamase, IMP-10, from two unrelated strains of Pseudomonas aeruginosa and an Alcaligenes xylosoxidans strain. Antimicrob. Agents Chemother. 2002;46:2014–2016. doi: 10.1128/AAC.46.6.2014-2016.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yano H., Kuga A., Okamoto R., Kitasato H., Kobayashi T., Inoue M. Plasmid-encoded metallo-beta-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob. Agents Chemother. 2001;45:1343–1348. doi: 10.1128/AAC.45.5.1343-1348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu E.M., Pegg K.M., Oelschlaeger P. The sequence-activity relationship between metallo-β-lactamases IMP-1, IMP-6, and IMP-25 suggests an evolutionary adaptation to meropenem exposure. Antimicrob. Agents Chemother. 2012;56:6403–6406. doi: 10.1128/AAC.01440-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cheng Z., Bethel C.R., Thomas P.W., Shurina B.A., Alao J.P., Thomas C.A., Yang K., Marshall S.H., Zhang H., Sturgill A.M., Kravats A.N., Page R.C., Fast W., Bonomo R.A., Crowder M.W. Carbapenem use is driving the evolution of imipenemase 1 variants. Antimicrob. Agents Chemother. 2021;65:1–19. doi: 10.1128/AAC.01714-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oelschlaeger P., Mayo S.L. Hydroxyl groups in the ββ sandwich of metallo-β-lactamases favor enzyme activity: A computational protein design study. J. Mol. Biol. 2005;350:395–401. doi: 10.1016/j.jmb.2005.04.044. [DOI] [PubMed] [Google Scholar]
- 132.Pegg K.M., Liu E.M., George A.C., LaCuran A.E., Bethel C.R., Bonomo R.A., Oelschlaeger P. Understanding the determinants of substrate specificity in IMP family metallo-β-lactamases: The importance of residue 262. Protein Sci. 2014;23:1451. doi: 10.1002/pro.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.LaCuran A.E., Pegg K.M., Liu E.M., Bethel C.R., Ai N., Welsh W.J., Bonomo R.A., Oelschlaeger P. Elucidating the role of residue 67 in IMP-type metallo-β-lactamase evolution. Antimicrob. Agents Chemother. 2015;59:7299–7307. doi: 10.1128/AAC.01651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tada T., Nhung H., Miyoshi-akiyama T., Shimada K., Phuong M., Anh Q. IMP-51, a novel IMP-type metallo- beta-lactamase with increased doripenem- and meropenem-hydrolyzing activities, in a carbapenem-resistant Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 2015;59:7090–7093. doi: 10.1128/AAC.01611-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kubota H., Suzuki Y., Okuno R., Uchitani Y., Ariyoshi T., Takemura N., Mihara F., Mezaki K., Ohmagari N., Matsui M., Suzuki S., Sekizuka T., Kuroda M., Yokoyama K., Sadamasu K. IMP-68, a novel IMP-type metallo-β-lactamase in imipenem-susceptible Klebsiella pneumoniae. mSphere. 2019;4 doi: 10.1128/mSphere.00736-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Oelschlaeger P., Pleiss J. Hydroxyl groups in the betabeta sandwich of metallo-beta-lactamases favor enzyme activity: Tyr218 and Ser262 pull down the lid. J. Mol. Biol. 2007;366:316–329. doi: 10.1016/j.jmb.2006.11.027. [DOI] [PubMed] [Google Scholar]
- 137.Siemann S., Badiei H.R., Karanassios V., Viswanatha T., Dmitrienko G.I. 68Zn isotope exchange experiments reveal an unusual kinetic lability of the metal ions in the di-zinc form of IMP-1 metallo-β-lactamase. Chem. Commun. (Camb.) 2006;5:532–534. doi: 10.1039/b514227j. [DOI] [PubMed] [Google Scholar]
- 138.Mojica M.F., Mahler S.G., Bethel C.R., Taracila M.A., Kosmopoulou M., Papp-Wallace K.M., Llarrull L.I., Wilson B.M., Marshall S.H., Wallace C.J., Villegas M.V., Harris M.E., Vila A.J., Spencer J., Bonomo R.A. Exploring the role of residue 228 in substrate and inhibitor recognition by VIM metallo-β-lactamases. Biochemistry. 2015;54:3183–3196. doi: 10.1021/acs.biochem.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Martínez-García L., González-Alba J.M., Baquero F., Cantón R., Galán J.C. Ceftazidime is the key diversification and selection driver of VIM-type carbapenemases. mBio. 2018;9 doi: 10.1128/mBio.02109-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lassaux P., Traoré D.A.K., Loisel E., Favier A., Docquier J.D., Sohier J.S., Laurent C., Bebrone C., Frère J.M., Ferrer J.L., Galleni M. Biochemical and structural characterization of the subclass B1 metallo-β-lactamase VIM-4. Antimicrob. Agents Chemother. 2011;55:1248–1255. doi: 10.1128/AAC.01486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Docquier J.D., Lamotte-Brasseur J., Galleni M., Amicosante G., Frère J.M., Rossolini G.M. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 2003;51:257–266. doi: 10.1093/jac/dkg067. [DOI] [PubMed] [Google Scholar]
- 142.Makena A., Düzgün A., Brem J., McDonough M.A., Rydzik A.M., Abboud M.I., Saral A., Çiçek A., Sandalli C., Schofield C.J. Comparison of verona integron-borne metallo-β-lactamase (VIM) variants reveals differences in stability and inhibition profiles. Antimicrob. Agents Chemother. 2016;60:1377–1384. doi: 10.1128/AAC.01768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Borgianni L., Vandenameele J., Matagne A., Bini L., Bonomo R.A., Frère J.M., Rossolini G.M., Docquier J.D. Mutational analysis of VIM-2 reveals an essential determinant for metallo-β-lactamase stability and folding. Antimicrob. Agents Chemother. 2010;54:3197–3204. doi: 10.1128/AAC.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Makena A., Brem J., Pfeffer I., Geffen R.E.J., Wilkins S.E., Tarhonskaya H., Flashman E., Phee L.M., Wareham D.W., Schofield C.J. Biochemical characterization of New Delhi metallo-β-lactamase variants reveals differences in protein stability. J. Antimicrob. Chemother. 2015;70:463–469. doi: 10.1093/jac/dku403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sun Z., Hu L., Sankaran B., Prasad B.V.V., Palzkill T. Differential active site requirements for NDM-1 β-lactamase hydrolysis of carbapenem versus penicillin and cephalosporin antibiotics. Nat. Commun. 2018;9:1–14. doi: 10.1038/s41467-018-06839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stewart A.C., Bethel C.R., Vanpelt J., Bergstrom A., Cheng Z., Miller C.G., Williams C., Poth R., Morris M., Lahey O., Nix J.C., Tierney D.L., Page R.C., Crowder M.W., Bonomo R.A., et al. Clinical variants of New Delhi metallo-β-lactamase are evolving to overcome zinc scarcity. ACS Infect. Dis. 2017;3:927–940. doi: 10.1021/acsinfecdis.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Toleman M.A., Simm A.M., Murphy T.A., Gales A.C., Biedenbach D.J., Jones R.N., Walsh T.R. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: Report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 2002;50:673–679. doi: 10.1093/jac/dkf210. [DOI] [PubMed] [Google Scholar]
- 148.Valladares M.H., Felici A., Weber G., Adolph H.W., Zeppezauer M., Rossolini G.M., Amicosante G., Frère J.M., Galleni M. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-β-lactamase activity and stability. Biochemistry. 1997;36:11534–11541. doi: 10.1021/bi971056h. [DOI] [PubMed] [Google Scholar]
- 149.Hernandez Valladares M., Kiefer M., Heinz U., Soto R.P., Meyer-Klaucke W., Nolting H.F., Zeppezauer M., Galleni M., Frère J.M., Rossolini G.M., Amicosante G., Adolph H.W. Kinetic and spectroscopic characterization of native and metal-substituted β-lactamase from Aeromonas hydrophila AE036. FEBS Lett. 2000;467:221–225. doi: 10.1016/s0014-5793(00)01102-9. [DOI] [PubMed] [Google Scholar]
- 150.Mojica M.F., Rutter J.D., Taracila M., Abriata L.A., Fouts D.E., Papp-Wallace K.M., Walsh T.J., Lipuma J.J., Vila A.J., Bonomo R.A. Population structure, molecular epidemiology, and β-lactamase diversity among stenotrophomonas maltophilia isolates in the United States. mBio. 2019;10 doi: 10.1128/mBio.00405-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saino Y., Kobayashi F., Inoue M., Mitsuhashi S. Purification and properties of inducible penicillin β-lactamase isolated from Pseudomonas maltophilia. Antimicrob. Agents Chemother. 1982;22:564–570. doi: 10.1128/aac.22.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Periyannan G., Shaw P.J., Sigdel T., Crowder M.W. In vivo folding of recombinant metallo-β-lactamase L1 requires the presence of Zn(II) Protein Sci. 2004;13:2236–2243. doi: 10.1110/ps.04742704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sabath L.D., Abraham E.P. Zinc as a cofactor for cephalosporinase from Bacillus cereus 569. Biochem. J. 1966;98:11C–13C. doi: 10.1042/bj0980011c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bicknell R., Schaffer A., Waley S.G., Auld D.S. Changes in the coordination geometry of the active-site metal during catalysis of benzylpenicillin hydrolysis by Bacillus cereus .beta.-lactamase II. Biochemistry. 2002;25:7208–7215. doi: 10.1021/bi00370a066. [DOI] [PubMed] [Google Scholar]
- 155.Aron A.T., Petras D., Schmid R., Gauglitz J.M., Büttel I., Antelo L., Zhi H., Nuccio S.P., Saak C.C., Malarney K.P., Thines E., Dutton R.J., Aluwihare L.I., Raffatellu M., Dorrestein P.C. Native mass spectrometry-based metabolomics identifies metal-binding compounds. Nat. Chem. 2022;14:100–109. doi: 10.1038/s41557-021-00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brawley H.N., Lindahl P.A. Direct detection of the labile nickel pool in Escherichia coli: New perspectives on labile metal pools. J. Am. Chem. Soc. 2021;143:18571–18580. doi: 10.1021/jacs.1c08213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Foster A.W., Young T.R., Chivers P.T., Robinson N.J. Protein metalation in biology. Curr. Opin. Chem. Biol. 2021;66:102095. doi: 10.1016/j.cbpa.2021.102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kuzmanic A., Bowman G.R., Juarez-Jimenez J., Michel J., Gervasio F.L. Investigating cryptic binding sites by molecular dynamics simulations. Acc. Chem. Res. 2020;53:654–661. doi: 10.1021/acs.accounts.9b00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hart K.M., Ho C.M.W., Dutta S., Gross M.L., Bowman G.R. Modelling proteins’ hidden conformations to predict antibiotic resistance. Nat. Commun. 2016;7:1–10. doi: 10.1038/ncomms12965. [DOI] [PMC free article] [PubMed] [Google Scholar]