Abstract
Background
The role of nasopharyngeal bacteria in respiratory syncytial virus (RSV) disease has been underestimated. We measured the frequency and burden of respiratory bacteria in the upper respiratory tract of infants with RSV infection over 7 respiratory seasons, and their impact on clinical outcomes.
Methods
Children <2 years old with mild (outpatients, n=115) or severe (inpatients, n=566) RSV infection, and matched healthy controls (n=161) were enrolled. Nasopharyngeal samples were obtained for RSV, Streptococcus pneumoniae, Staphylococcus aureus, Moraxella catarrhalis, and Haemophilus influenzae detection and quantitation by PCR. Multivariable models were constructed to identify variables predictive of severe disease.
Results
S. pneumoniae, H. influenzae, and M. catarrhalis, but not S. aureus, were detected more frequently in RSV-infected children (84%) than healthy controls (46%; P<.001). Detection of S. pneumoniae and/or H. influenzae was associated with fever, more frequent antibiotic treatment, worse radiologic findings, and higher neutrophil counts (P<.01). In adjusted analyses, S. pneumoniae/H. influenzae codetection was independentlyassociated with greater odds of hospitalization, higher disease severity scores, need for supplemental oxygen, and longer hospitalization.
Conclusions
Nasopharyngeal codetection of S. pneumoniae and H. influenzae in infants with RSV infection is associated with increased disease severity.
Keywords: RSV, nasopharyngeal bacterial colonization, bacterial PCR, disease severity, infants
In a large cohort of previously healthy children <2 years of age with RSV infection spanning the disease spectrum, codetection of H. influenzaeand S. pneumoniaein the upper respiratory tract was independently associated with worse clinical outcomes.
Respiratory syncytial virus (RSV) is the leading cause of bronchiolitis in infants, of pneumonia in children <5 years of age, and the second cause of death in the first year of life in resource-limited countries [1, 2]. There are groups of children at high risk for severe RSV disease; however, the majority of infants hospitalized with RSV infection are previously healthy [3, 4]. Studies have shown that viral factors, a dysregulated host immune response, and genetic predisposition play a role in RSV disease severity [5, 6]. Nevertheless, none of these factors can entirely explain the wide differences in clinical presentations and outcomes observed in infants with RSV infection.
Our previous studies in small cohorts of children with RSV infection, also confirmed by others, suggested that detection of Streptococcus pneumoniae or Haemophilus influenzae in the upper respiratory tract was associated with differences in host immune responses and clinical outcomes [7–10]. Nevertheless, there are still significant knowledge gaps regarding the frequency, potential additive effect, and clinical implications of bacterial detection in the respiratory tract of children with RSV infection. In particular, the role of these bacteria alone or in combination, and whether bacterial loads influence RSV disease pathogenesis and severity, have not been fully defined.
The aims of this study were (1) to define the frequency of nasopharyngeal bacterial detection in young children with mild (outpatients) and severe (inpatients) RSV infection, compared to healthy age- and season-matched controls across several respiratory seasons; and (2) to define whether specific bacteria, bacterial loads, and/or combination of specific bacteria were associated with enhanceddisease severity.
METHODS
Study Design
From December 2010 to December 2018, we conducted a prospective observational study and enrolled a convenience sample of previously healthy children <2 years of age hospitalized or evaluated in the outpatient setting with RSV infection. RSV was diagnosed per standard of care by rapid antigen or polymerase chain reaction (PCR) test. We excluded premature children (≤36 weeks’ gestation), those with previous history of wheezing or chronic medical conditions (ie, congenital heart disease, chronic lung disease, immunodeficiency). Inpatients were enrolled within medians 25%–75% interquartile range (IQR)24 (18–34) hours of hospitalization to Nationwide Children’s Hospital (NCH) inpatient units, and outpatients at the time of presentation at the emergency department or primary care clinics.
Each respiratory season, we also enrolled healthy control children of similar age, sex, and race/ethnicity compared to RSV cases, with no respiratory symptoms or treated with antibiotics within 2 weeks of enrollment. Controls were enrolled in the operating room while undergoing minor scheduled surgical procedures, or at primary care offices during well-child visits. The study was approved by the institutional review board at NCH, and informed consent obtained from legal guardians before study participation.
Data and Sample Collection
In all study subjects, we collected demographic and clinical information using a standardized questionnaire designed for the study. We also collected 2 upper respiratory swabs for bacterial detection and RSV quantitation respectively, and a blood sample for white blood cell (WBC) count with differential analyses. Disease severity was assessed by the need for hospitalization and a standardized clinical disease severity score (CDSS) that included 5 parameters (transcutaneous oxygen saturation, respiratory rate, chest wall retractions, auscultation, and level of activity) [11]. The CDSS ranged from 0 (normal) to 15 (most severe disease) and has been validated internally and externally in other studies [12–15]. In hospitalized infants, we analyzed additional criteria of severity including duration of hospitalization, oxygen administration, and need for pediatric intensive care unit (PICU) care.
Nasopharyngeal bacterial swabs were obtained in RSV patients and healthy controls as described [9] and aliquots stored at −80°C until further quantitation by real time PCR (qRT-PCR) for S. pneumoniae, Staphylococcus aureus, Moraxella catarrhalis, and H. influenzae using published primers (Supplementary Table 1) [8]. Bacterial loads were measured in copies/mL and log10 transformed for analyses purposes. These bacteria were selected based on previous studies that showed their relevant role in acute and long-term respiratory morbidity in children [9, 16–18]. Midturbinate swabs were also obtained in RSV patients and controls for RSV detection, quantitation, and typing using qRT-PCR targeting the N gene as described [5, 9, 13, 14].
Statistical Analysis
Baseline characteristics are reported using means with standard deviation, medians with 25%–75% interquartile ranges, or frequencies with percentages. For continuous variables, group comparisons were assessed using 2-tailed t test with a Satterthwaite correction for unequal group variance, Mann-Whitney test, 1-way analysis of variance (ANOVA), or Kruskal-Wallis tests with Dunn correction for multiplicity when appropriate. Categorical data were analyzed using χ2 or Fisher exact tests.
To assess the role of nasopharyngeal bacteria on clinical outcomes we conducted multivariate logistic regression for binary outcomes (need for hospitalization and oxygen administration) and ordinal logistic regression for continuous outcomes (duration of hospitalization and the CDSS). For analyses purposes, children were categorized based on the CDSS as mild (≤3), which represents the mildest disease phenotype and did not include administration of supplemental oxygen, moderate (4–7), or severe (≥8–15). Covariates were included in the models if they were clinically meaningful or had univariate P values of <.15, and were retained if they had an adjusted P value of <.1 or if their inclusion had a substantial impact on the models’ goodness of fit, based on Akaike information criterion (AIC) [19]. These covariates included age, sex, race, antibiotic use at enrollment, blood neutrophil counts, RSV loads, and bacterial detection. Statistical analyses were conducted using GraphPad Prism version 8.0 and SAS version 9.4, with a 2-sided P value<.05 considered statistically significant.
RESULTS
Nasopharyngeal Detection of Potentially Pathogenic Bacteria in Infants with RSV Infection
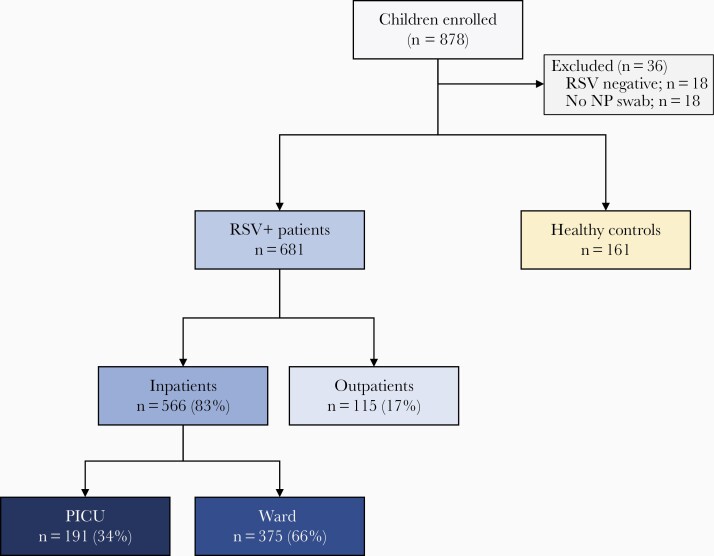
From December 2010 to December 2018, we enrolled 878 children <2 years of age, and excluded 36 children for the reasons outlined in Figure 1, leaving a total of 681 children with RSV infection and 161 healthy controls. Within the RSV cohort, 566 children (83%) were hospitalized (ward n=375, 55%; PICU n=191, 28%) and 115 (17%) were managed as outpatients (Table 1). The majority of RSV patients (n=620, 91%) were <12 months of age. Median age for healthy controls and RSV outpatients was comparable (6–7 months), but higher than for inpatients (2.5 months). Inpatients also showed significantly lower RSV loads than outpatients (P<.001).
Figure 1.
Flowchart of study participants. The upper boxes in gray indicate the number of patients and controls enrolled and the reasons for exclusion. Children with RSV infection included in the study (blue boxes) were classified according to disease severity in RSV outpatients and RSV inpatients (ward and PICU). Yellow box indicates the number of healthy controls enrolled. Abbreviations: NP, nasopharyngeal; PICU, pediatric intensive care unit; RSV, respiratory syncytial virus.
Table 1.
Demographic Characteristics of RSV Patients and Healthy Controls
| Healthy Controls (n=161) | All RSV Patients (n=681) | RSV Inpatients (n=566) | RSV Outpatients (n=115) | P Valuea | P Valueb | |
|---|---|---|---|---|---|---|
| Age, mo, median (25%–75% IQR) | 7.0 (4–10.2) | 2.9 (1.5–6.5) | 2.5 (1.4–5.4) | 6.0 (3.4–10) | <.0001 | <.0001 |
| Sex, male | 117 (73) | 372 (55) | 315 (56) | 57 (50) | <.0001 | .258 |
| Race | ||||||
| White | 103 (64) | 454 (67) | 401 (71) | 53 (46) | .251 | <.0001 |
| Black | 38 (24) | 124 (18) | 81 (14) | 43 (37) | ||
| Other | 20 (12) | 103 (15) | 84 (15) | 19 (16) | ||
| Delivery, vaginal | 118 (73) | 509 (75) | 419 (74) | 90 (78) | .689 | .409 |
| Daycare attendance | 31 (19) | 202 (30) | 159 (28) | 43 (37) | .008 | .056 |
| Smoke exposure | 31 (19) | 233 (34) | 199 (35) | 34 (30) | .0002 | .281 |
| Breastfeeding | 60 (37) | 237 (35) | 188 (33) | 49 (43) | .582 | .067 |
| Immunizations | 146 (91) | 578 (85) | 481 (85) | 97 (84) | .058 | .886 |
| Parental asthma | 82 (51) | 347 (51) | 287 (51) | 60 (52) | 1.000 | .838 |
| RSV A | … | 393 (59) | 343 (63) | 50 (43) | .0002 | |
| RSV B | … | 270 (41) | 205 (37) | 65 (57) | ||
| RSV loads, log10 copies/mL, median (25%–75% IQR)c | Negative | 7.5 (5–7.6) | 7.0 (6.2–7.8) | 7.9 (6.9–8.4) | <.0001 |
Data are No. (%) except where indicated. We used the χ² or Fisher exact tests for categorical variables, and Mann-Whitney U or Kruskal-Wallis tests for continuous variables.
Abbreviations: IQR, interquartile range; RSV, respiratory syncytial virus.
P value for comparisons between healthy controls and RSV patients.
P value for comparisons between RSV inpatients and RSV outpatients.
Data not available for 18 RSV inpatients.
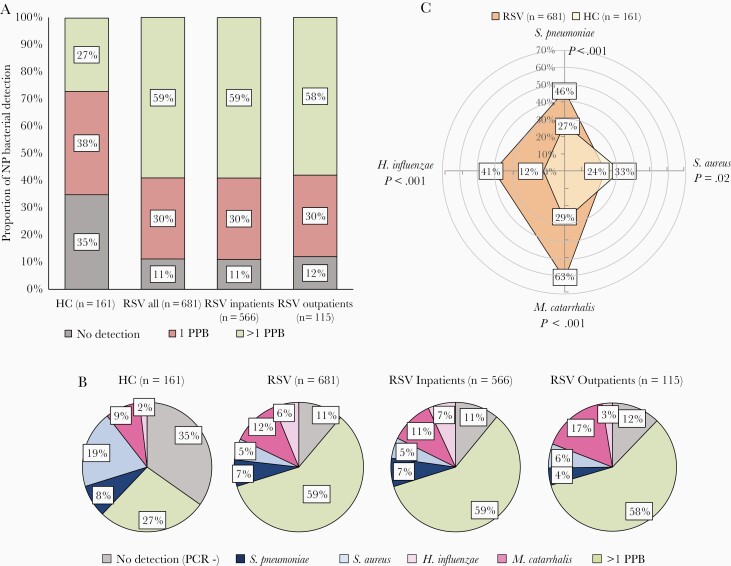
First, we compared the frequency of S. aureus, S. pneumoniae, H. influenzae, and M. catarrhalis detection by qRT-PCR in children with RSV infection versus healthy controls. Detection of at least 1 of the 4 potentially pathogenic bacteria was more common in RSV patients, irrespective of the need for hospitalization, than in healthy controls (89% vs 65%; P<.0001; Figure 2A). Detection of >1 of these 4 potentially pathogenic bacteria was alsomore common in children with RSV infection (58% inpatients, 59% outpatients) than in controls (27%; P<.0001; Figure 2A and 2B).
Figure 2.
Detection of potentially pathogenic bacteria (PPB) in the upper respiratory tract of young children with respiratory syncytial virus (RSV) infection and healthy age controls. A, Frequency of nasopharyngeal (NP) bacterial detection in study patients. The horizontal axis represents the study groups and the vertical axis represents the percentage of NP bacterial detection (Streptococcus pneumoniae, Staphylococcus aureus, Moraxella catarrhalis, Haemophilus influenzae): negative detection for any of the 4 bacteria (gray), detection of 1 of these 4 PPB (red), and detection of >1 PPB (green). B, Frequency of NP detection of specific bacteria. Pie charts for healthy controls and children with RSV infection (all, inpatients, and outpatients) showing the percentage detection of none of the 4 bacteria by PCR (gray), S. pneumoniae only (dark blue), S. aureus only (light blue), M. catarrhalis only (dark pink), H. influenzae only (light pink), and >1 PPB (green). C, Radar plot depicting the frequency of any detection of S. pneumoniae, S. aureus, M. catarrhalis, and H. influenzae in children with RSV infection (orange) compared to healthy controls (HC; light yellow). P values indicate the comparisons between RSV and HC for any detection (alone and in combination with other bacteria) of each of the 4 bacteria.
Analyses of the contribution of these bacteria as a single pathogen or combined with other bacteria (any detection) is shown in Figure 2B and 2C. S. aureus detection alone was more common in healthy controls than RSV patients (19% vs 5%, respectively; P<.001), while H. influenzae alone was more frequently detected in RSV patients than controls (6% vs 2%, respectively; P=.03; Figure 2B). Any detection of S. aureus was also more common in healthy controls than RSV patients (33% vs 24%, respectively; P=.02; Figure 2C). On the other hand, any detection of S. pneumoniae (46% vs 27%; P<.001) and M. catarrhalis (63% vs 29%; P<.001) was more common in RSV patients that in healthy controls. We also assessed the types of bacterial associations. The likelihood of S. pneumoniae, M. catarrhalis, and H. influenzae codetections was high, while S. aureus detection was inversely associated with H. influenzae and not associated with M. catarrhalis or S. pneumoniae detection (Supplementary Figure 1).
Nasopharyngeal Bacterial Detection According to Age
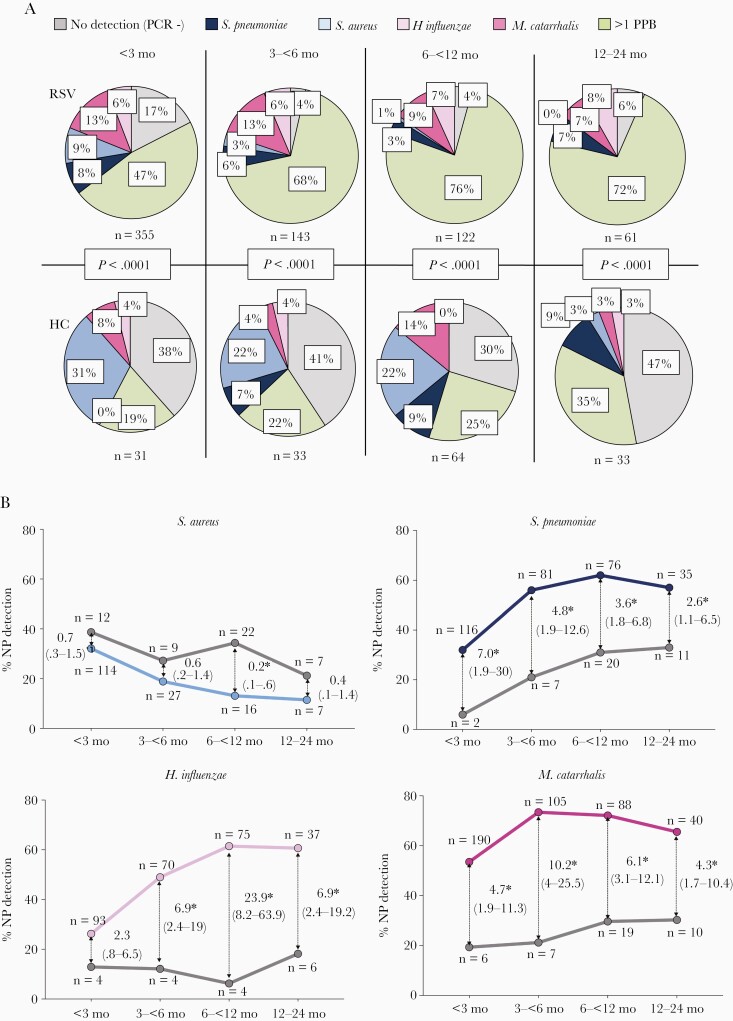
To evaluate differences in bacterial detection according to age, RSV patients and healthy controls were stratified in 4 age groups: <3 months, 3 to <6 months, 6 to <12 months, and 12–24 months (Figure 3). Single detection of S. pneumoniae, M. catarrhalis, and H. influenzae remained stable in RSV patients and healthy controls across age groups. S. aureus detection decreased significantly with increasing age in RSV patients (9% in <3 months vs 0% in >12 months; P=.005) and controls (31% in <3 months vs 3% in >12 months; P<.0001). Detection of more than 1 of the 4 potentially pathogenic bacteria remained higher in RSV patients than in controls across all age groups, and increased with age with 47% detection of >1 bacterium in RSV infants <3 months and 72% in the >12–24 months age group (P<.0001).
Figure 3.
Nasopharyngeal (NP) bacterial detection stratified by age in children with respiratory syncytial virus (RSV) infection in relation to healthy controls. A, Pie charts depicting the proportion of nasopharyngeal bacterial detected in RSV patients (upper panels) and healthy controls (HC; lower panels) stratified by age: no detection of the 4 bacteria (PCR−; gray), single detection of Streptococcus pneumoniae (dark blue), Staphylococcus aureus (light blue), Moraxella catarrhalis (dark pink), Haemophilus influenzae (light pink), and >1 potentially pathogenic bacteria (PPB; green). Comparisons by χ2 for each age group. B, Odds of S. aureus, S. pneumoniae, H. influenzae, and M. catarrhalis detection in RSV patients (colored lines) in relation to healthy controls (gray lines) according to age. Thin arrows indicate the odds ratio (95% confidence interval) for each age group comparison. Asterisks indicate the time points that are significantly different. The numbers of RSV patients and controls per bacteria and age group are depicted in each plot. Analyses by χ2 test for trends.
Any detection of S. pneumoniae, H. influenzae, or M. catarrhalis was consistently higher in RSV patients versus controls (P<.0001). While S. pneumoniae and H. influenzae detection increased with age, detection of M. catarrhalis remained stable (Figure 3B). Rates of bacterial detection across the 7 respiratory seasons was not significantly different (Supplementary Figure 2).
Nasopharyngeal Bacterial Loads and Clinical Outcomes in Children with RSV Infection
Next, we evaluated whether the pattern and loads of nasopharyngeal bacteria were associated with clinical parameters (Table 2). Children with RSV infection with any of the bacteria identified had fever at presentation (>38°C) more frequently (67% vs 30% for negative PCR; P<.0001), higher CDSS (P=.03), higher WBC counts and neutrophil percentages (P<.0001), and atelectasis and/or consolidation by chest X-ray (P=.02). Similar results were identified in children with RSV infection with S. pneumoniae or H. influenzae detection, but not in those with S. aureus detection or negative PCR results for the 4 bacteria. In fact, children with S. aureus detection or negative bacterial PCR had the lowest CDSS compared to children with other bacteria (P=.02; Supplementary Figure 3).
Table 2.
Demographic and Clinical Characteristics of RSV Patients Stratified by Potentially Pathogenic Bacteria
| Characteristic | PCR− (n=76) | Any Bacteria (n=605) | S. pneumoniae (n=45) | S. aureus (n=36) | M. catarrhalis (n=81) | H. influenzae (n=42) | >1 Bacteria (n=401) | P Valuea | P Valueb |
|---|---|---|---|---|---|---|---|---|---|
| Demographic characteristics | |||||||||
| Age, mo, median (25%–75% IQR) | 1.4 (0.9–2.3) | 3.2 (1.6–6.8) | 2.2 (1–4) | 1.6 (0.9–2.5) | 2.4 (1.6–4.8) | 3.2 (1.6–7.1) | 3.8 (2–7.8) | <.0001 | <.0001 |
| Sex (male) | 44 (58) | 328 (54) | 23 (51) | 14 (39) | 45 (56) | 27 (64) | 219 (55) | .625 | .330 |
| Race, white | 57 (75) | 397 (66) | 36 (80) | 24 (67) | 49 (61) | 35 (83) | 253 (63) | .047 | .018 |
| Vaginal delivery | 53 (70) | 456 (75) | 30 (67) | 26 (72) | 59 (74) | 34 (81) | 307 (76) | .326 | .528 |
| Daycare | 11 (14) | 191 (32) | 9 (20) | 2 (6) | 24 (30) | 11 (26) | 145 (36) | <.0001 | .002 |
| Smoke exposure | 26 (34) | 207 (34) | 12 (27) | 11 (31) | 27 (33) | 20 (48) | 137 (34) | 1.000 | .451 |
| Breastfeeding | 37 (49) | 200 (33) | 21 (47) | 17 (47) | 27 (33) | 14 (33) | 121 (30) | .010 | .008 |
| Parental asthma | 28 (37) | 319 (53) | 23 (51) | 18 (50) | 38 (47) | 23 (55) | 217 (54) | .010 | .133 |
| Immunizations | 66 (87) | 513 (85) | 38 (84) | 32 (89) | 71 (88) | 39 (93) | 333 (83) | .734 | .502 |
| Clinical parameters | |||||||||
| Duration of symptoms, d,median (25%–75% IQR)c | 5 (4–6) | 5 (3–6) | 5 (3.5–5.5) | 4 (3–5.7) | 5 (4–6) | 5.5 (4–6) | 4 (3–6) | .095 | .088 |
| Presence of feverc | 23 (30) | 403 (67) | 34 (76) | 10 (28) | 39 (48) | 33 (79) | 287 (72) | <.0001 | <.0001 |
| Hospitalized | 62 (81) | 504 (83) | 40 (89) | 29 (81) | 62 (76) | 39 (93) | 334 (83) | .745 | .243 |
| Supp. O2 need | 41 (54) | 369 (61) | 33 (73) | 19 (53) | 43 (53) | 31 (74) | 243(60) | .263 | .060 |
| Ventilatory supportd | 17 (42) | 134 (36) | 10 (30) | 4 (21) | 19 (45) | 9 (29) | 92 (38) | .609 | .423 |
| Supp. O2 duration, h, median (25%–75% IQR) | 48 (22–76) | 48 (24–95) | 36 (23–114) | 24 (12–72) | 48(35–73) | 45 (24–72) | 48 (24–96) | .622 | .584 |
| CDSS, median (25%–75% IQR) | 6 (4–9) | 7 (5–10) | 8 (5–10.5) | 6 (3–8) | 7 (4–9) | 7 (5–10) | 7 (5–10) | .037 | .027 |
| Antibiotic treatmentc | 30 (39) | 271 (45) | 28 (62) | 9 (25) | 30 (37) | 28 (67) | 176 (44) | .393 | .001 |
| PICU need | 23 (30) | 168 (28) | 14 (31) | 6 (17) | 20 (25) | 14 (33) | 114 (28) | .684 | .583 |
| PICU stay, h, median (25%–75% IQR) | 54 (32–101) | 88 (54–146) | 98(53–154) | 63 (46–156) | 89 (39–110) | 76 (41–175) | 92 (57–159) | .002 | .049 |
| Hospitalization, d, median (25%–75% IQR) | 2.7 (1.7–4) | 2.8 (1.8–4.5) | 2.6 (1.8–4.7) | 2 (1.5–3.5) | 2.7 (1.6–4.1) | 2.7 (1.–5.8) | 2.9 (1.8–4.5) | .625 | .560 |
| Laboratory and radiologic studies | |||||||||
| WBC 103/mL, median (25%–75% IQR) | 8.5 (7–10) | 10.6 (8–13) | 11.5 (7–13) | 8.2 (7–11) | 10 (8–12) | 11 (9–15) | 11.2 (9–14) | <.0001 | <.0001 |
| Neutrophils %, median (25%–75% IQR) | 19 (14–28) | 35 (25–47) | 36.5 (29–49) | 21 (13–31) | 29 (21–45) | 42 (29–56) | 36 (26–48) | <.0001 | <.0001 |
| Lymphocytes %, median (25%–75% IQR) | 65 (56–73) | 51 (39–61) | 49 (39–55) | 65 (55–75) | 54 (38–64) | 51 (33–58) | 50 (39–60) | <.0001 | <.0001 |
| Radiology | |||||||||
| Normal | 7 (14) | 26 (6 | 1 (3) | 1 (6) | 5 (11) | 0 (0) | 19 (7) | .026 | .043 |
| BWT/hyperinflation | 29 (57) | 187 (45 | 21 (55) | 12 (71) | 22 (47) | 13 (41) | 119(42) | ||
| Atelectasis | 7 (13) | 118 (28) | 6 (16) | 4 (23) | 11 (24) | 11 (34) | 86 (30) | ||
| Lobar consolidation | 8 (16) | 86 (21) | 10 (26) | 0 (0) | 8 (18) | 8 (25) | 60 (21) | ||
| RSV load, log10 copies/mL, median (25%–75% IQR) | 7.5 (6.5–8.2) | 7.1 (6.3–7.9) | 6.8 (5.8–7.9) | 7.5 (6.4–8.4) | 7.1 (6.4–7.9) | 6.8 (6.1–7.8) | 7.1 (6.3–7.9) | .185 | .178 |
Data are No. (%) except where indicated. We used the χ² test for categorical variables, and the Kruskal-Wallis test for continuous variables to calculate P values.
Abbreviations: BWT, bronchial wall thickening; CDSS, clinical disease severity score; IQR, interquartile range; PCR, polymerase chain reaction; PICU, pediatric intensive care unit; PPB, potentially pathogenic bacteria; RSV, respiratory syncytial virus; WBC, white blood cell.
P value for comparisons between PCR− and any PPB.
P value for comparisons between PCR−, individual bacteria, and >1 PPB (6 groups).
Presence of fever, duration of symptoms, and antibiotic treatment at enrollment.
Indicates invasive and noninvasive ventilatory support.
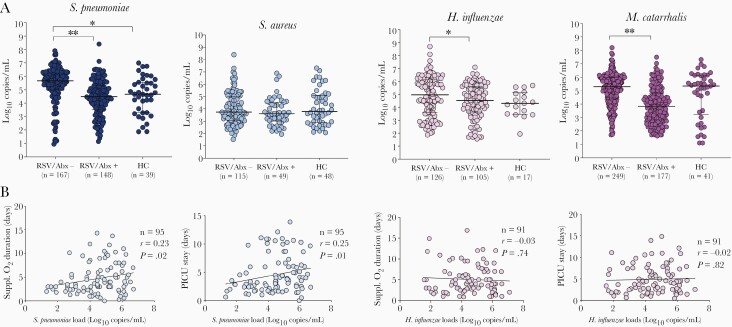
We then analyzed the role of bacterial loads on clinical outcomes. Although bacterial detection was conducted by PCR and patients were enrolled early in the disease course, we assessed whether prior antibiotic treatment (any dose) had an impact on bacterial detection and quantitation. Forty-four percent of RSV patients, the majority in the PICU, had received antibiotics for a median of 25%–75% interquartile rangeof24 (24–48) hours at the time of study enrollment (Supplementary Table 2). Rates of bacterial detection in children with RSV infection were similar, irrespective of antibiotic treatment (antibiotic treated, 87% and nontreated, 91%; P=.14). None of the healthy controls had received antibiotics as this was an exclusion criterion. S. pneumoniae, H. influenzae, and M. catarrhalis bacterial loads, measured in log10 copies/mL, were lower in children that received antibiotics with activity against these bacteria (Figure 4A and Supplementary Table 3). Excluding children who had received antibiotics, we found that S. pneumoniae loads were significantly higher (P=.0003) and H. influenzae loads modestly higher (P=.05) in children with RSV infection than in healthy controls (Figure 4A), with no differences in M. catarrhalis and S. aureus loads. In addition, S. pneumoniae, but not H. influenzae, loads in children with severe RSV infection that required PICU care were significantly correlated with duration of oxygen and PICU length of stay (Figure 4B).
Figure 4.
Bacterial loads in children with respiratory syncytial virus (RSV) infection and healthy controls. A, Bacterial loads in healthy controls and children with RSV infection according to antibiotic use at study enrollment. The horizontal axis represents RSV-infected children not treated with antibiotics (RSV/Abx−), those treated (RSV/Abx+), and healthy controls (HC). The Y-axis represent bacterial loads in log10 copies/mL for each bacterium. Comparisons by Mann-Whitney U test between RSV/Abx− and RSV/Abx+, and between RSV/Abx− and HC. ∗P<.05; ∗∗P<.01. B, Spearman correlations between Streptococcus pneumoniae (blue) and Haemophilus influenzae (pink) loads and duration of supplemental oxygen and pediatric intensive care unit (PICU) stay among patients admitted to the PICU. The number of patients (n), correlation coefficient (r), and P value are included in each plot.
Influence of S. pneumoniae and H. influenzae Detection on RSV Clinical Outcomes
As we found that S. pneumoniae and/or H. influenzae were associated with differences in clinical parameters, we analyzed the possible synergistic effect of these bacteria on disease severity. To this end, RSV patients were stratified according to the presence/absence of these bacteria: no detection (S. pneumoniae negative/H. influenzae negative; n=232), detection of one but not the other (S. pneumoniae positive/H. influenzae negative; n=172) or (S. pneumoniae negative/H. influenzae positive; n=134), and codetection of both (S. pneumoniae positive/H. influenzae positive; n=143; Supplementary Table 4).
RSV patients with S. pneumoniae/H. influenzae codetection, followed by the H. influenzae or S. pneumoniae groups had fever more frequently (88% vs 75% vs 73% respectively)compared with the S. pneumoniae negative/H. influenzae negative group (35%; P<.0001). Oxygen administration was also more frequent in the codetection (66%) and the S. pneumoniae negative/H. influenzae positive (66%) groups versus the S. pneumoniae negative/H. influenzae negative group (53%; P=.03). In addition, children with RSV infection with S. pneumoniae/H. influenzae codetection versus no detection had higher CDSS (8 vs 6; P=.001), increased rates of PICU admission (37% vs 25%; P=.03), higher WBC and neutrophil counts (P<.0001), and atelectasis/consolidation documented by chest X-ray more frequently (56% vs 31%; P<.001).
To confirm these findings, we conducted multivariable analyses and calculated the odds ratio (OR) and 95% confidence interval (CI) of hospitalization and a high CDSS in all RSV patients (Supplementary Table 5), and the need for oxygen and duration of hospitalization in RSV inpatients (Figure 5 and Supplementary Table 6). We included in all models demographic data, clinical variables, RSV loads, and detection of S. pneumoniae and H. influenzae according to the scheme described above.
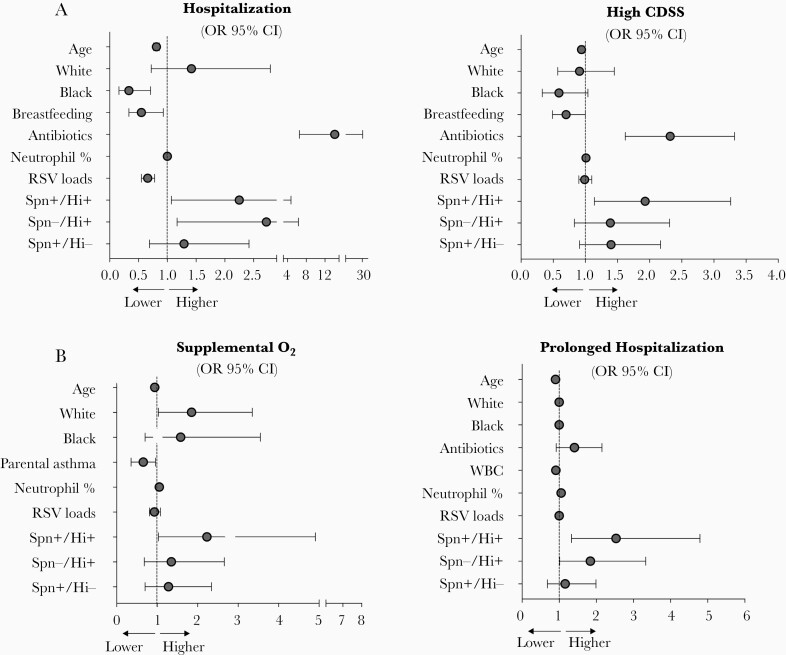
Figure 5.
Predictors of disease severity in children with RSV infection. A, Adjusted need for hospitalization and high CDSS evaluated in the whole cohort of RSV patients. High CDSS was defined as a CDSS >3. B, Adjusted odds of oxygen use and prolonged hospital stay defined by a duration of hospitalization of ≥3 days. Reference group for race is other race. Reference group for bacterial classification is no detection of Streptococcus pneumoniae and Haemophilus influenzae (Spn−/Hi−). Data are adjusted ORs with point estimates and 95% CIs. Abbreviations: CDSS, clinical disease severity score; CI, confidence interval; Hi, H. influenzae; OR, odds ratio; RSV, respiratory syncytial virus; Spn, S. pneumoniae.
After controlling for other factors, children with RSV infection in the S. pneumoniae negative/H. influenzae positive group (OR, 2.72; 95% CI, 1.17–6.33), and in the codetection S. pneumoniae positive/H. influenzae positive group (OR, 2.25; 95% CI, 1.07–4.74) had greater odds of hospitalization. Codetection of S. pneumoniae/H. influenzae was also associated with high CDSS (OR, 1.93; 95% CI, 1.14–3.26). In hospitalized children, S. pneumoniae/H. influenzae codetection was associated with higher odds of oxygen administration (OR, 2.23; 95% CI, 1.01–4.91). In addition, S. pneumoniae negative/H. influenzae positive (OR, 1.84; 95% CI, 1.01–3.33) and S. pneumoniae positive/H. influenzae positive (OR, 2.53; 95% CI, 1.33–4.79) detection were associated with increase odds of prolonged hospitalization. Together, these data indicate that detection of H. influenzae, and especially the simultaneous detection of S. pneumoniae and H. influenzae, in children with RSV infection is associated with increased disease severity, even after adjusting for other covariables such as age and viral load.
DISCUSSION
In this study we analyzed a large cohort of young children with mild or severe RSV infection over several respiratory seasons to explore whether the detection and loads of specific bacteria in the upper respiratory tract were associated with clinical outcomes. We found that compared with healthy controls of similar demographic characteristics, M. catarrhalis, S. pneumoniae, and H. influenzae, but not S. aureus, were frequently identified in RSV-infected children, both outpatients and inpatients. We also found that the detection of S. pneumoniae or H. influenzae was associated with worse laboratory, radiologic, and clinical parameters. However, it was the codetection of S. pneumoniae and H. influenzae that consistently increased the risk for severe disease defined by greater odds of hospitalization, clinical scores, oxygen administration, and prolonged hospital stay.
There is an increasing interest in understanding the interplay between the host, RSV, and the respiratory microbiota, which has been recognized as an important contributor to respiratory morbidity in children [20–22]. Studies conducted in selected and unselected populations of children showed the presence of a stable respiratory microbiome during health that was altered with incursions of Streptococcus spp., Moraxella spp., or Haemophilus spp. during acute respiratory infections. These changes in microbiome composition were also influenced by age and seasonality [21, 23–25]. Our study design differs from the aforementioned studies, as we studied exclusively children with acute RSV infection in a cross-sectional mode and compared them with contemporary heathy controls of similar ages. Despite these differences, we also found that detection of M. catarrhalis, and especially of S. pneumoniae and H. influenzae, was more frequent in RSV-infected children and increased with age. On the other hand, S. aureus detection was not common in children with RSV infection and it was not associated with worse clinical outcomes. This is different from other respiratory viral infections, like influenza, where S. aureus is a risk factor for severe pneumonia [26].
The interactions between RSV and S. pneumoniae or H. influenzae have been studied in in vitro systems, animal models, and in children [27–31]. In human bronchial epithelial cells, S. pneumoniae colonization enhanced RSV replication [32, 33]. In animal models, a preceding RSV infection predisposed to invasive pneumococcal disease [33]. In children, epidemiologic studies showed an association between the peak activity of RSV and the incidence of invasive pneumococcal disease [34, 35]. In addition, a reduction in pneumococcal carriage through vaccination led to decreased rates of hospitalization for RSV pneumonia [36, 37]. Also, S. pneumoniae- or H. influenzae-dominated microbiome profiles in children have been associated with enhanced mucosal proinflammatory responses [22, 29–31, 36, 38].
Initial studies conducted in small numbers of low- and high-risk children hospitalized with RSV infection in the PICU showed high rates of bacterial detection in the lower respiratory tract that was associated with worse clinical outcomes [10, 39–41]. Subsequent studies in children hospitalized with bronchiolitis showed that nasopharyngeal detection of H. influenzae was associated with longer duration of hospitalization and increased rates of PICU admission [42–44]. Nevertheless, invasive bacterial infections, such as bacteremia or meningitis, are rare in children with RSV infection, thus antibiotics are not routinely recommended [3, 45]. A recent large retrospective study conducted in children <2 years of age with RSV lower respiratory tract infection in the PICU showed that early antibiotic treatment was associated with shorter duration of mechanical ventilation and hospital stay [46]. It is possible that PICU patients represent a unique subset, in whom respiratory bacteria play a more significant role in disease severity, and that the benefit associated with antibiotic therapy could be related to a reduction in bacterial burden. Nevertheless, this observation deserves further prospective randomized studies.
In our previous studies using bacterial 16S-rRNA sequencing in a small cohort of children with RSV infection, we found that compared to S. aureus or M. catarrhalis, nasopharyngeal detection of Haemophilus spp. or Streptococcus spp. enriched profiles were associated with distinct systemic immune transcriptional profiles and more frequent hospitalization [8]. The present study confirms and expands those findings as we targeted using qRT-PCRthe four most common respiratory bacteria that have been identified in children with RSV infection. We found that detection of S. pneumoniae, H. influenzae, M. catarrhalis, but not S. aureus, was frequent and consistent in children with RSV infection, and that the vast majority had more than one bacterium identified. The number of RSV patients and healthy controls enrolled allowed us to define the differences in rates of bacterial detection (alone or in combination) based on age. In addition, univariate followed by multivariable analysis showed that although S. pneumoniae or H. influenzae, were associated with significant clinical and laboratory changes, it was the concomitant detection of both bacteria that was associated with worse clinical outcomes. Last, we also found that antibiotic treatment was associated with greater odds of hospitalization and a higher initial CDSS. At the same time, antibiotic treatment was more frequently administered to children with RSV infection and S. pneumoniae and/or H. influenzae detection, by physicians who were unaware of the results of bacterial detection. Further controlled studies are needed to understand the impact of antibiotics on clinical outcomes in young children with RSV infection.
Our study has limitations. The use of antibiotics before sample collection may have altered the bacterial composition or nasopharyngeal bacteria loads. However, we used targeted qRT-PCR that increased the yield of bacterial detection and quantitation, and the multivariable models were adjusted for antibiotic therapy. In addition, we conducted a sensitivity analysis that showed that antibiotic administration decreased bacterial loads but did not influence the rates of bacterial detection. We did not collect sequential samples during the acute disease to assess whether delay of bacterial clearance could have influenced our results. Nevertheless, we prospectively enrolled a large cohort of young children with RSV infection over multiple respiratory seasons, including a representative number of outpatients, that allowed us to compute fair comparisons. While the current study did not allow us to assess whether RSV infection favored the acquisition of the bacteria analyzed, or whether the preexisting microbiota composition predisposed to a more severe RSV infection, we were able to identify a significant association between S. pneumoniae and H. influenzae nasopharyngeal detection and RSV disease pathogenesis and severity, especially when detected together. Last, although biomarkers indicative of a proinflammatory state in the blood or respiratory mucosa were not studied, the proportion of blood neutrophils was consistently increased in infants with RSV infection and codetection of S. pneumoniae and/or H. influenzae, suggesting that nasopharyngeal detection of specific bacteria, in the context of RSV infection, is more than a passive phenomenon.
In summary, our findings contribute to the growing evidence of the complex interactions between RSV and respiratory bacterial communities in children with acute RSV infection. Further studies are needed to help elucidate the mechanisms involved in these interactions and which ones contribute to greater disease severity, which may be amenable to targeted interventions.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Presented in part: Infectious Diseases Society of America 56th Annual Meeting, 3–7 October 2018, San Francisco, CA; and the Pediatric Academic Societies Annual Meeting, 24 April–1 May 2019, Baltimore, MD.
Acknowledgments. We thank all research nurses as part of Nationwide Children’s Hospital Clinical Research Services for their extraordinary efforts enrolling patients and controls, Cynthia Burch at NCH Department of Laboratory Medicine for her help with bacterial PCR processing, the Rush and Ireland family for their generosity and continuous support for our studies, and specially our patients and their families for agreeing to participate in the study.
Disclaimer . The sponsors had no role in study design, data collection, data analysis, data interpretation, decision to publish, or preparation of the manuscript.
Financial support . This work was supported by the National Institutes of Health (grant number AI112524 to A. M. and O. R.); and the Infectious Diseases Intramural Grant Consortium at Nationwide Children’s Hospital (to A. M. and J. N.). Financial support was received from the Rush and Ireland Family.
Potential conflicts of interest. A. M. has received research grants from Janssen; and fees for participation in advisory boards from Janssen, Sanofi-Pasteur, Merck, and Roche. O. R. has received research grants from the Bill and Melinda Gates Foundation, Janssen, and Merk; fees for participation in advisory boards from Merck, Sanofi-Pasteur, Lilly, Pfizer, and Adagio; and fees for lectures from Pfizer, unrelated to the current work. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Shi T, McAllister DA, O’Brien KL, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 2017; 390:946–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pneumonia Etiology Research for Child Health (PERCH) Study Group. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet 2019; 394:757–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. García CG, Bhore R, Soriano-Fallas A, et al. Risk factors in children hospitalized with RSV bronchiolitis versus non-RSV bronchiolitis. Pediatrics 2010; 126:e1453–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hall CB, Weinberg GA, Iwane MK, et al. The burden of respiratory syncytial virus infection in young children. N Engl J Med 2009; 360:588–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mella C, Suarez-Arrabal MC, Lopez S, et al. Innate immune dysfunction is associated with enhanced disease severity in infants with severe respiratory syncytial virus bronchiolitis. J Infect Dis 2013; 207:564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heinonen S, Velazquez VM, Ye F, et al. Immune profiles provide insights into respiratory syncytial virus disease severity in young children. Sci Transl Med 2020; 12:eaaw0268. [DOI] [PubMed] [Google Scholar]
- 7. Hasegawa K, Mansbach JM, Ajami NJ, et al. Serum cathelicidin, nasopharyngeal microbiota, and disease severity among infants hospitalized with bronchiolitis. J Allergy Clin Immunol 2017; 139:1383–6.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Steenhuijsen Piters WA, Heinonen S, Hasrat R, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 2016; 194:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suárez-Arrabal MC, Mella C, Lopez SM, et al. Nasopharyngeal bacterial burden and antibiotics: influence on inflammatory markers and disease severity in infants with respiratory syncytial virus bronchiolitis. J Infect 2015; 71:458–69. [DOI] [PubMed] [Google Scholar]
- 10. Brealey JC, Chappell KJ, Galbraith S, et al. Streptococcus pneumoniae colonization of the nasopharynx is associated with increased severity during respiratory syncytial virus infection in young children. Respirology 2018; 23:220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mejias A, Dimo B, Suarez NM, et al. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med 2013; 10:e1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garcia-Mauriño C, Moore-Clingenpeel M, Thomas J, et al. Viral load dynamics and clinical disease severity in infants with respiratory syncytial virus infection. J Infect Dis 2019; 219:1207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brenes-Chacon H, Garcia-Mauriño C, Moore-Clingenpeel M, et al. Age-dependent interactions among clinical characteristics, viral loads and disease severity in young children with respiratory syncytial virus infection. Pediatr Infect Dis J 2021; 40:116–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mejias A, Brenes-Chacon H, Garcia-Maurino C, Moore-Clingenpeel M, Ramilo O.. Clinical disease severity scores and viral loads in children with RSV infection. Clin Infect Dis 2021; 72:e1160–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haddadin Z, Beveridge S, Fernandez K, et al. Respiratory syncytial virus disease severity in young children [published online ahead of print 23 October 2020]. Clin Infect Dis doi: 10.1093/cid/ciaa1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bisgaard H, Hermansen MN, Buchvald F, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med 2007; 357:1487–95. [DOI] [PubMed] [Google Scholar]
- 17. Tsai MH, Huang SH, Chen CL, et al. Pathogenic bacterial nasopharyngeal colonization and its impact on respiratory diseases in the first year of life: the PATCH Birth Cohort Study. Pediatr Infect Dis J 2015; 34:652–8. [DOI] [PubMed] [Google Scholar]
- 18. Jartti T, Kuneinen S, Lehtinen P, et al. Nasopharyngeal bacterial colonization during the first wheezing episode is associated with longer duration of hospitalization and higher risk of relapse in young children. Eur J Clin Microbiol Infect Dis 2011; 30:233–41. [DOI] [PubMed] [Google Scholar]
- 19. Pan W. Akaike’s information criterion in generalized estimating equations. Biometrics 2001; 57:120–5. [DOI] [PubMed] [Google Scholar]
- 20. Man WH, van Houten MA, Mérelle ME, et al. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir Med 2019; 7:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015; 17:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grier A, Gill AL, Kessler HA, et al. Temporal dysbiosis of infant nasal microbiota relative to respiratory syncytial virus infection. J Infect Dis 2021; 223:1650–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brealey JC, Young PR, Sloots TP, et al. Bacterial colonization dynamics associated with respiratory syncytial virus during early childhood. Pediatr Pulmonol 2020; 55:1237–45. [DOI] [PubMed] [Google Scholar]
- 24. Salter SJ, Turner C, Watthanaworawit W, et al. A longitudinal study of the infant nasopharyngeal microbiota: the effects of age, illness and antibiotic use in a cohort of South East Asian children. PLoS Negl Trop Dis 2017; 11:e0005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van den Bergh MR, Biesbroek G, Rossen JW, et al. Associations between pathogens in the upper respiratory tract of young children: interplay between viruses and bacteria. PLoS One 2012; 7:e47711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Randolph AG, Vaughn F, Sullivan R, et al. Critically ill children during the 2009–2010 influenza pandemic in the United States. Pediatrics 2011; 128:e1450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hament JM, Aerts PC, Fleer A, et al. Enhanced adherence of Streptococcus pneumoniae to human epithelial cells infected with respiratory syncytial virus. Pediatr Res 2004; 55:972–8. [DOI] [PubMed] [Google Scholar]
- 28. McGillivary G, Mason KM, Jurcisek JA, Peeples ME, Bakaletz LO.. Respiratory syncytial virus-induced dysregulation of expression of a mucosal beta-defensin augments colonization of the upper airway by non-typeable Haemophilus influenzae. Cell Microbiol 2009; 11:1399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shilts MH, Rosas-Salazar C, Turi KN, et al. Nasopharyngeal Haemophilus and local immune response during infant respiratory syncytial virus infection. J Allergy Clin Immunol 2021; 147:1097–101.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sande CJ, Njunge JM, Mwongeli Ngoi J, et al. Airway response to respiratory syncytial virus has incidental antibacterial effects. Nat Commun 2019; 10:2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hasegawa K, Mansbach JM, Ajami NJ, et al. The relationship between nasopharyngeal CCL5 and microbiota on disease severity among infants with bronchiolitis. Allergy 2017; 72:1796–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hament JM, Aerts PC, Fleer A, et al. Direct binding of respiratory syncytial virus to pneumococci: a phenomenon that enhances both pneumococcal adherence to human epithelial cells and pneumococcal invasiveness in a murine model. Pediatr Res 2005; 58:1198–203. [DOI] [PubMed] [Google Scholar]
- 33. Stark JM, Stark MA, Colasurdo GN, LeVine AM.. Decreased bacterial clearance from the lungs of mice following primary respiratory syncytial virus infection. J Med Virol 2006; 78:829–38. [DOI] [PubMed] [Google Scholar]
- 34. Ampofo K, Bender J, Sheng X, et al. Seasonal invasive pneumococcal disease in children: role of preceding respiratory viral infection. Pediatrics 2008; 122:229–37. [DOI] [PubMed] [Google Scholar]
- 35. Techasaensiri B, Techasaensiri C, Mejías A, McCracken GH Jr, Ramilo O.. Viral coinfections in children with invasive pneumococcal disease. Pediatr Infect Dis J 2010; 29:519–23. [DOI] [PubMed] [Google Scholar]
- 36. Weinberger DM, Klugman KP, Steiner CA, Simonsen L, Viboud C.. Association between respiratory syncytial virus activity and pneumococcal disease in infants: a time series analysis of US hospitalization data. PLoS Med 2015; 12:e1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Greenberg D, Givon-Lavi N, Faingelernt Y, et al. Nasopharyngeal pneumococcal carriage during childhood community-acquired alveolar pneumonia: relationship between specific serotypes and coinfecting viruses. J Infect Dis 2017; 215:1111–6. [DOI] [PubMed] [Google Scholar]
- 38. Ederveen THA, Ferwerda G, Ahout IM, et al. Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome 2018; 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kneyber MC, Blussé van Oud-Alblas H, van Vliet M, Uiterwaal CS, Kimpen JL, van Vught AJ.. Concurrent bacterial infection and prolonged mechanical ventilation in infants with respiratory syncytial virus lower respiratory tract disease. Intensive Care Med 2005; 31:680–5. [DOI] [PubMed] [Google Scholar]
- 40. Thorburn K, Harigopal S, Reddy V, Taylor N, van Saene HK.. High incidence of pulmonary bacterial co-infection in children with severe respiratory syncytial virus (RSV) bronchiolitis. Thorax 2006; 61:611–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Levin D, Tribuzio M, Green-Wrzesinki T, et al. Empiric antibiotics are justified for infants with respiratory syncytial virus lower respiratory tract infection presenting with respiratory failure: a prospective study and evidence review. Pediatr Crit Care Med 2010; 11:390–5. [DOI] [PubMed] [Google Scholar]
- 42. Hasegawa K, Mansbach JM, Ajami NJ, et al. ; the MARC-35 Investigators. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J 2016; 48:1329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mansbach JM, Hasegawa K, Piedra PA, et al. Haemophilus-dominant nasopharyngeal microbiota is associated with delayed clearance of respiratory syncytial virus in infants hospitalized for bronchiolitis. J Infect Dis 2019; 219:1804–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang W, Wang T, Li L, Ji W, Wang Y, Yan Y.. Impact of bacteria in nasal aspirates on disease severity of bronchiolitis. Infect Dis (Lond) 2016; 48:82–6. [DOI] [PubMed] [Google Scholar]
- 45. Ralston SL, Lieberthal AS, Meissner HC, et al. ; American Academy of Pediatrics. Clinical practice guideline: the diagnosis, management, and prevention of bronchiolitis. Pediatrics 2014; 134:e1474–502. [DOI] [PubMed] [Google Scholar]
- 46. Shein SL, Kong M, McKee B, O’Riordan M, Toltzis P, Randolph AG.. Antibiotic prescription in young children with respiratory syncytial virus-associated respiratory failure and associated outcomes. Pediatr Crit Care Med 2019; 20:101–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.