Abstract
Despite extensive research, there is still no treatment that would lead to remission in all patients with rheumatoid arthritis as our understanding of the affected site, the synovium, is still incomplete. Recently, single-cell technologies helped to decipher the cellular heterogeneity of the synovium; however, certain synovial cell populations, such as endothelial cells or peripheral neurons, remain to be profiled on a single-cell level. Furthermore, associations between certain cellular states and inflammation were found; whether these cells cause the inflammation remains to be answered. Similarly, cellular zonation and interactions between individual effectors in the synovium are yet to be fully determined. A deeper understanding of cell signalling and interactions in the synovium is crucial for a better design of therapeutics with the goal of complete remission in all patients.
Keywords: RA, synovium, single-cell transcriptomics, cellular localization, experimental arthritis
Rheumatology key messages.
Single-cell technologies have identified the diverse immune and structural cell composition of the synovium.
Many human synovial cell populations appear to have their counterparts in the murine synovium.
The cellular localisation and cell–cell interactions in the synovium are yet to be fully deciphered.
Introduction
RA is a chronic inflammatory disease that mainly affects the synovium that nourishes and supports the joint during homeostasis (disease pathology was reviewed, for example, in [1, 2]). A better understanding of synovial biology could help in identifying novel drugable targets for the pursuit of achieving sustained remission or, better yet, finding a cure for RA [3]. RA environment and its biomarkers were originally studied indirectly in peripheral blood or synovial fluid; however, better synovial sampling through arthroscopic or ultrasonography-guided biopsies allowed for immunohistochemical and biochemical profiling of the synovial composition leading to better understanding of the disease site (reviewed in [4]). Still, profiling cellular heterogeneity, localisation, and interactions in the synovium and integrating those data is the way forward in disentangling the underlying mechanisms of inflammation as well as those underlying resolution.
Single-cell technologies allowed for deep characterisation of the synovial tissue heterogeneity; however, recent reviews have been almost exclusively focussed on fibroblasts, macrophages/monocytes, T cells and B cells and omitted other structural and supporting cells such as endothelial cells or peripheral neurons. In this review, we aim to comprehensively describe synovial cell populations, their activation states and association with synovial pathology, as well as identify knowledge gaps and populations remaining to be profiled. Subsequently, we will discuss the positioning (zonation) of individual subpopulations of cells in the synovium and how the single-cell profiling can assist in discovering the localisation of those cells. Finally, we will provide a summary of the known intercellular interactions as well as identify which tools can be used to predict and confirm further signalling pathways.
Cellular heterogeneity in the synovium
Methods to study single-cell heterogeneity
The field of single-cell sequencing is rapidly growing, and several new methods have been developed to facilitate the in-depth characterisation of different cell types. These novel technologies, their applications, and their integration into existing research frameworks have been recently reviewed (for example in [5–7]). Single-cell transcriptomics has been extensively used to profile synovial tissue and infer the cellular function and contribution to inflammation (Fig. 1). Several methods such as Smart-seq2 [8] or 10X Genomics [9] are in routine use and others, such as the low-cost microfluidic instrument, were also reported for profiling the synovium in RA [10]. However, some challenges remain in the use of these technologies (reviewed in [11]). Following the best practices during the analysis is crucial to ensure that results are valid and reproducible [12].
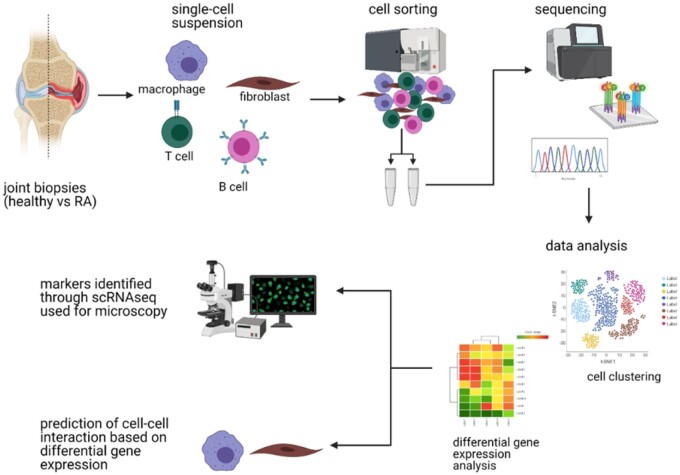
Fig. 1.
Simplified workflow of a sequencing experiment
The sample acquisition (e.g. human or murine joint biopsies) is followed by the processing of the sample into a single-cell suspension. Population of interest is then sorted by FACS and prepared for sequencing (e.g. barcoding the mRNA to identify cell source, reverse transcription, preparation of cDNA library). Raw sequencing data need to be pre-processed and analysed. The findings from single-cell RNA sequencing experiments should be experimentally validated. Figure created with BioRender.com.
Immune cells
Mononuclear phagocytic cells
Mononuclear phagocytes are important contributors to both homeostasis and inflammation. The role of synovial macrophages in the context of RA has been extensively studied in patients as well as in animals (recent reviews: [13–15]). Previously, tissue-macrophages were found to restrict excessive neutrophil recruitment [16]; however, recent single-cell experiments revealed substantial heterogeneity among synovial myeloid cells in both mice and humans [17–20].
Single-cell profiling of synovial macrophages in healthy controls, treatment-naïve/-resistant patients with RA, and patients in sustained remission revealed nine distinct subpopulations (Table 1, Fig. 2) [17]. The association of these subsets to inflammation vs remission was largely determined by the expression of MER tyrosine kinase (MERTK); MerTKpos macrophages were more abundant in stable remission and healthy controls, while MerTKneg subset was relatively scarce in healthy synovium and over-abundant in active RA [17]. Similarly, MERTK-expressing macrophages were upregulated with anti-inflammatory medication in RA patients [20] and expression of Mertk itself was elevated during resolution of inflammation in a murine model of arthritis [21]. Furthermore, the ratio of MerTKpos to MerTKneg macrophages was also found to be predictive of flares’ occurrences [17] showing MerTK as a potential predictive tool for clinicians. Conversely, MerTKneg subpopulations show similarity to previously identified pro-inflammatory macrophages [17, 19]. Another pro-inflammatory population was identified in RA patients, marked by high expression of heparin binding EGF-like growth factor (HBEGF), that interacted with synovial fibroblasts and was predicted to increase fibroblast invasiveness and neutrophil recruitment [20].
Table 1.
Synovial myeloid cell heterogeneity (based on [17])
| Markers defining this population | Associated with inflammation/ remission | Possible function, associated expression profile | Mouse equivalent (Culemann et al. 2019) | ||
|---|---|---|---|---|---|
| MerTKpos | TREM2pos | TREM2low | Increased in early undifferentiated arthrits | May represent early activated TREM2high | |
| TREM2high | Remission | Restraining inflammation; expression associated with complement and defensin pathways, Treg differentiation, restriction of effector T-cell function, scavenger receptor etc. | lining Trem2pos Cx3cr1pos | ||
| FOLR2pos | ID2pos | Similar abundance in all conditions | May represent in situ precursors of resident macrophages | ||
| LYVE1pos | Remission | Immuno-regulation; gene expression associated with collagen turnover, antiprotease enzymes, coagulation factors and regulators of vascular endothelial growth factor | RELMα+ sublining mph | ||
| ICAM1pos | Similar abundance in all conditions | May represent first-line defence against pathogens; high expression of proinflammatory cytokine genes | |||
| MerTKneg | HLApos |
|
Interferon signalling | ||
|
Similar abundance in all conditions | May represent synovial tissue-resident antigen-presenting cells | |||
| CD48pos | S100A12pos | Inflammation | Proinflammatory phenotype; expression of alarmin S100A9 correlated with disease severity | ||
|
Inflammation | Proinflammatory phenotype; osteopontin expression correlated with disease severity | |||
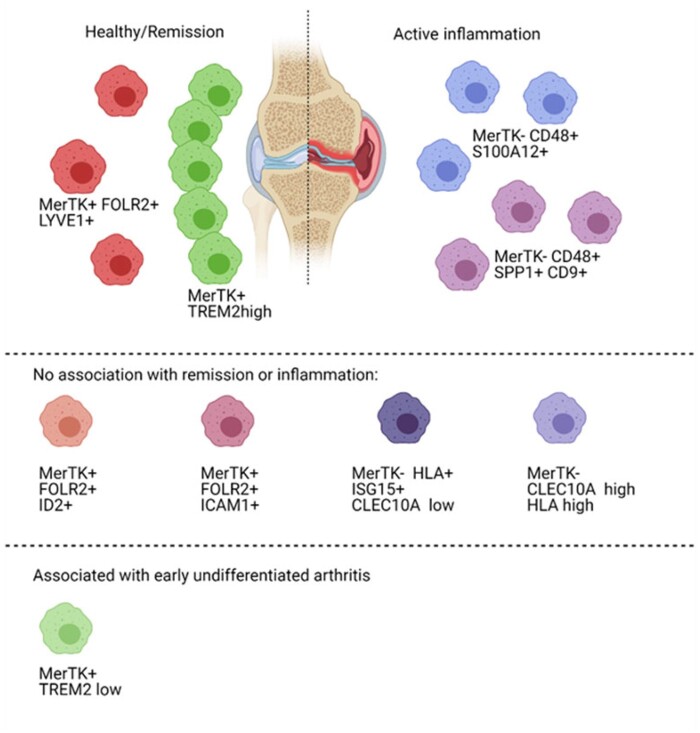
Fig. 2.
Macrophage populations identified in human synovium (based on [17])
Macrophage populations characterised as MerTKposFOLR2pos+LYVE1pos or MerTKposTREM2high were associated with remission, while MerTKnegCD48posS100A12pos or MerTKnegCD48posSPP1posCD9pos macrophages were preferentially found in actively inflamed synovium. Other populations had no clear association with either remission or inflammation. Interestingly, an increased number of MerTKposTREM2low macrophages was found in early undifferentiated arthritis. Figure created with BioRender.com.
Similarly, macrophages in a murine RA model, serum transfer-induced arthritis (STIA), were identified as a heterogeneous population displaying dynamic changes in their respective abundances over the first 5 days of the disease [18]. This was largely caused by the macrophage infiltration during disease development. At the initial stage, seven subpopulations of macrophages were identified: C-X3-C motif chemokine receptor 1 (CX3CR1)+ lining, resistin-like alpha (RELM-α)+ interstitial, aquaporin 1 (AQP1)+ interstitial, C-C chemokine receptor type 2 (CCR2)+ infiltrating, major histocompatibility complex class II (MHCII)+ interstitial, stathmin 1 (STMN1)+ proliferating macrophages and lymphocyte antigen 6 complex, locus C2 (LY6C2)+ monocytes [18]. CX3CR1+ lining macrophages expressed genes associated with immune-regulation, clearance of apoptotic cells, and barrier formation [18], suggesting important homeostatic function within the synovium.
Monocytes are important for the development of STIA; their absence is associated with lower leukocyte recruitment and restriction of inflammation [21]. Furthermore, compared with OA samples that are not defined by immune cell abundance, RA biopsies have a strong myeloid expression signature [22]. Thus, profiling of myeloid cells in inflammatory arthritis contributes to a better understanding of this disease. Synovial macrophage subpopulations, their associations with pathology, predicted functions and comparison between humans and mice are summarized in Table 1.
Granulocytes
Due to the high levels of intracellular RNases, granulocytes are difficult to profile by transcriptomic approaches. Despite those difficulties, several studies were able to profile heterogeneity of granulocytes, mainly neutrophils, during both homeostasis and inflammation [23, 24]. Such transcriptomic advances should be applied to investigate the synovial granulocytes considering the importance of neutrophils at the onset of arthritis in the murine models [25–27]. Diversification of neutrophil phenotypes could be of particular interest as low-density neutrophils have been recently identified to correlate with pathology in multiple inflammatory diseases (for example [28–30]). Interestingly, type 2 responses, including an increased number of eosinophils within the joint, were found protective in murine models of arthritis [31, 32] and were mediated by IL-4/IL-13-induced signal transducer and activator of transcription 6 (STAT6) activation [31]. Granulocyte-associated gene patterns, such as markers of phagocytosis, pattern recognition receptor and complement activation, were apparent in synovial samples of RA patients [22, 33]. Therefore, closer profiling of granulocytes within the synovium would be beneficial to determine their contribution to inflammatory arthritis.
B lymphocytes
Autoantibodies against IgG antibodies (RF) and citrullinated proteins (ACPA) are characteristic of RA in certain patients but their presence is not universal, a phenomenon indicative of heterogeneous B-cell responses. Gene patterns associated with classical B-cell function have been detected only in synovial tissue of patients with RA, not those with OA [22]. Moreover, B-cell activation and differentiation-associated gene signature was predictive of increased bone erosion [33].
Single-cell characterisation of B cells in the synovium of patients with OA or RA revealed four distinct subpopulations of B cells: naïve B cells (called SC-B1), IGHG+CD27+ memory B cells (SC-B2), autoimmune-associated B cells (SC-B3) and plasmablasts (SC-B4) [19]. The SC-B3 cluster had an elevated expression of markers associated with B-cell activation, interferon stimulation and autoimmunity, and was overabundant in inflamed synovium compared with OA controls [19]. A subset of receptor activator of nuclear factor kappa-Β ligand (RANKL)-producing B cells was also identified in the synovial fluid and patient tissue [34]. Further validation of the functionality of these cells will be necessary to confirm their proinflammatory and autoimmune role in RA. In-depth B-cell profiling may be valuable to a subset of patients; however, as patients manifest with discrete levels of lymphocyte infiltration [33, 35, 36], the effectivity of drugs targeting B-cell activation may be limited.
T lymphocytes
T cells form an important part of the adaptive immune system and transcripts associated with T cell function were detected in synovial tissue isolated from patients with RA but not in OA, suggesting their role in inflammation [22]. Conversely, Th2 responses were found to be protective in murine models of RA [31, 32], a discrepancy potentially explained by synovial T-cell heterogeneity [19, 33, 37].
ScRNAseq of the synovial tissue isolated from patients with either OA or RA revealed three subpopulations of CD4+ [CCR7+ (called SC-T1), forkhead box P3 (FOXP3)+ regulatory (SC-T2), PDCD1+ peripheral helper (TPH) and follicular helper T cells (TFH) (SC-T3)] and three subpopulations of CD8+ T cells [GZMK+ (SC-T4), GNLY+GZMB+ cytotoxic (SC-T5), GZMK+GZMB+ T cells (SC-T6)] [19]. SC-T3 TPH and TFH cells were found to produce high levels of a chemokine C-X-C motif chemokine ligand 13 (CXCL13), inhibitory receptors [T cell immunoreceptor with Ig and ITIM domains (TIGIT), cytotoxic T-lymphocyte-associated protein 4 (CTLA4)] and were associated with inflammation in the synovium [19]. The population of TPH cells, characterised as PD1hiCXCR5-CD4+, was found to be expanded in the joints of seropositive RA patients and supported B-cell differentiation and antibody production [37]. Similarly, PD-1high TPH cells producing PDCD1, TIGIT and CXCR6 were associated with strong inflammation in treatment-naïve patients with RA [33]. Overall, TPH cells may represent a pathological player in RA.
Studies in psoriatic arthritis showed that expanded T cells are likely retained in the synovium by an increased expression of CXCR3 or CXCR6 whose ligands CXCL10 and CXCL9 or CXCL16 respectively are present in an increased level in the synovial fluid during the disease [38, 39]. Although untested, similar retention could apply to PD-1high TPH in RA as they also express an increased level of CXCR6 [33]. Inhibiting the retention of these proinflammatory T cells may present a potential therapeutic target; however, further research is required to validate T-cell relationships to pathology identified by single-cell experiments.
Structural cells
Tissue maintenance: fibroblasts
Stromal cells including fibroblasts are important contributors to synovial homeostasis and have a substantial role in the pathology of RA (recently reviewed in [40–42]). Recent single-cell studies revealed vast heterogeneity among this population and the differential role of distinct fibroblast clusters in RA pathogenesis [19, 43–45].
ScRNAseq profiling of synovial fibroblasts isolated from patients with RA or OA revealed three major subsets: CD34-THY1-, CD34-THY1+ and CD34+ [43] (Fig. 3). CD34-THY1+ were more abundant in RA than in OA, positively correlated with leukocyte infiltration, histological synovitis and hypertrophy [43]. Furthermore, CD34-THY1+ fibroblasts expressed factors that drive osteoclastogenesis and triggered the generation of triiodothyronine receptor auxiliary protein (TRAP)+ osteoclasts in vitro [43]. CD34+ fibroblasts were characterized by high expression of interleukin 6 (IL6), CXCL12, C-C motif chemokine ligand 2 (CCL2). In vitro experiments confirmed their ability to recruit a high number of peripheral blood monocytes [43].
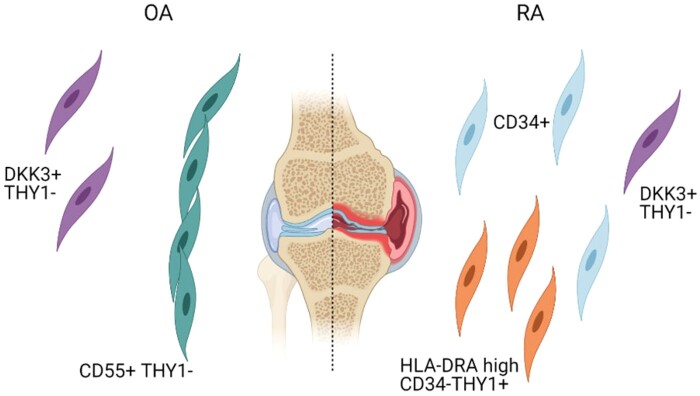
Fig. 3.
Fibroblast populations identified in human synovium (based on [19])
CD55+THY1- lining fibroblast were found to be associated with OA while CD34+ and HLA-DRAhiCD34-THY1+ sublining fibroblasts were preferentially found in RA. Population of sublining fibroblasts defined as DKK3+THY1- was not clearly associated with either. Figure created with BioRender.com.
A similar analysis of synovial tissue from RA and OA patients by single-cell technologies identified four fibroblast subpopulations: CD34+ sublining fibroblasts (termed SC-F1), human leukocyte antigen (HLA)-DRAhi sublining fibroblasts (SC-F2), Dickkopf WNT signalling pathway inhibitor 3 (DKK3+) sublining fibroblasts (SC-F3) and CD55+ lining fibroblasts (SC-F4) [19]. The HLA-DRAhi population of fibroblasts (SC-F2) [19] seems to correspond to the previously defined inflammatory CD34-THY1+ fibroblasts [43]. They expressed genes associated with antigen presentation, IFNγ signalling, high levels of IL6 and CXCL12, and were overabundant in leukocyte-rich RA samples compared with leukocyte-poor RA and OA samples [19].
Furthermore, RA patients have a higher number of fibroblast activation protein alpha (FAPα)+ fibroblasts compared with patients without active inflammation [44]. Also, FAPα+ fibroblasts were significantly associated with inflammation, leukocyte infiltration, joint damage and pannus formation in STIA [44]. In this murine model, scRNA-sequencing identified five distinct fibroblast populations [44]. FAPα+THY1+ were found to represent sublining fibroblasts which increased the severity and persistence of inflammation, while FAPα+THY1- fibroblasts were located to the synovial lining and had a role in bone and cartilage destruction [44].
Overall, sublining THY1+ fibroblasts are expanded in both RA and animal models of this disease and associate with increased leukocyte infiltration [19, 43–45] representing a potential therapeutic target. The comparison of murine and human synovial fibroblasts, their identifying markers and predicted functions are summarized in Table 2.
Table 2.
Synovial fibroblast heterogeneity [19]
| Markers defining this population | Name in Zhang et al. 2019 | Associated with RA/OA | Possible function, associated expression profile | Position in the synovium | Mouse equivalent (Croft et al. 2019) |
|---|---|---|---|---|---|
| CD34+ | SC-F1 | Associated with inflammation in RA | Expression of IL6, CXCL12, CCL2; potential role in monocyte recruitment (Mizoguchi et al. 2018) | Sublining: observed in both superficial and deeper sublining areas (Mizoguchi et al. 2018) | |
| HLA-DRAhi | SC-F2 | Associated with inflammation in RA | MHC II antigen presentation and IFNγ signalling; production of proinflammatory cytokines; potential role in leukocyte infiltration (Mizoguchi et al. 2018) | Sublining | FAPα+THY1+CD34- (STIA Col11a1+ F1 population) |
| DKK3 | SC-F3 | No association found | Identified as a novel subpopulation with high expression of DKK3, CAMD1, COL8A2 | Sublining | |
| CD55 | SC-F4 | OA associated | murine FAPα+THY- lining fibroblast may be potentially associated with bone and cartilage erosion (Croft et al. 2019) | Lining | Similarity to FAPα+THY- lining fibroblasts, (STIA F5 lining population) |
Vascularisation: endothelial cells
Increased vascular permeability is a feature of inflammatory models of arthritis and endothelial dysfunction is a common complication of RA (reviewed, for example, in [27, 46, 47]). Synovial endothelial cells, therefore, seem to have an impact on the propagation of the inflammation.
Endothelial cells are a heterogeneous population in other tissues with organ-specific characteristics supporting the needs of the local microenvironment [48–52]. These differences are apparent in ECs’ appearance, morphology and transcriptional profile [48]. Both human and murine endothelial cells even show sex-specific transcriptome differences across organs such as the aorta, heart and lung [49, 53].
Human synovial arterial (PODXL+) and venous [Duffy blood group, atypical chemokine receptor (DARC+)] endothelial cells were found to contribute differential cues to the synovial microenvironment [45]. Arterial endothelial cells, not venous ones, support the differentiation of synovial sublining fibroblasts [45]. Endothelial cells also assist local cell functionality in other organs such as promoting hepatocyte survival and albumin production in the liver [48]. Furthermore, capillaries traversing cortical bone from the bone marrow have been recently identified [54]; however, their contribution to the synovial environment remains to be established.
Regrettably, endothelial heterogeneity in the joint has not been sufficiently studied. Considering the importance of vascular permeability in the immunopathology of RA, it would be interesting to investigate endothelial cell functions of the synovium in steady-state and inflammation. Understanding synovial endothelium could lead to the development of novel therapeutics that target vascular permeability and oedema.
Innervation: peripheral nervous system
Joint-associated pain is common to both RA and OA [1, 2, 55, 56]. In a murine model of OA, osteoclasts induced axonal growth in the subchondral bone by secreting netrin-1, which was predicted to mediate increased sensitivity of the peripheral nervous system and pain perception [57]. As aberrant innervation of the cartilage is seen in an animal model of inflammatory arthritis [58], similar effector pathways could mediate joint pain in RA as well. Changes in the dorsal horn of the central nervous system in inflammatory arthritis were also detected [58]. Furthermore, the loss of sensory innervation in the joint was found to be protective in a patient with psoriatic arthritis [59]. This effect of protective sensory denervation was confirmed in an animal model of inflammatory arthritis [59]. Of note, partial denervation of the wrist resulted in self-reported improvement of pain perception in patients with inflammatory arthritis [60].
The peripheral nervous system was found to be formed by a highly heterogeneous population of cells (among many, for example, [61–64]). In addition to heterogeneous populations of myelinating and non-myelinating Schwann cells, neural-associated endothelial cells, fibroblasts and leukocytes were also identified [64]. Leukocytes of the peripheral nervous system consist of CX3CR1-expressing myeloid cells with CXCL4+ macrophages, also identified in the mouse brain [64, 65]. Changes in the populations of cells associated with peripheral nerves were observed in an animal model of spontaneous chronic peripheral neuritis [64]. Similarly, sensory and sympathetic neurons innervating the draining lymph nodes were found to be highly heterogeneous, able to communicate with surrounding stromal and immune cells and expand after toll-like receptor 4 stimulation [63].
In conclusion, the heterogeneity of synovial innervation and its contribution to the pathology of RA remain incompletely understood. However, both peripheral and central nervous system seem to be altered in inflammatory arthritis [58], providing the rationale for the development of pain-modifying drugs that interfere with neural effector pathways to improve the quality of life of patients with both RA and OA [66].
Cellular zonation in the synovium
Methods to study cellular zonation
Fluorescence imaging and its implication for deciphering organ function have been recently reviewed [67]. Advances in confocal microscopy, for example by the addition of Airyscan detectors, and in 3D imaging were beneficial for the field of rheumatology as well. However, spatiotemporal localisation of different subsets of immune and structural cells within the synovium has been difficult due to the lack of markers characterizing different subpopulations. As described in the previous section, for most synovial cells, single-cell transcriptomics helped to overcome this issue by providing a characterisation of differentially expressed genes across populations. Furthermore, the recent development of spatial transcriptomics such as sequential fluorescence in situ hybridization [68], Slide-seq [69] or commercially available 10X Genomics’ Visium and nanoString is becoming instrumental to further understand the spatiotemporal changes in the synovium. Spatial transcriptomics has been used already to study cellular niches and networks in Alzheimer’s disease [70] or carcinomas [71] and very recently also to uncover signalling and localisation of infiltrating leukocytes in RA [35, 36].
Immune cells
Two major interstitial macrophage niches were recently identified in mice by combining transcriptional profiling and fluorescence imaging, namely vasculature and nerve-associated macrophages [72]. Lyve1loMHCIIhi tissue-resident macrophages expressed higher levels of genes associated with antigen presentation and were more closely associated with peripheral nerves [72]. Conversely, Lyve1hiMHCIIlo macrophages were found to contribute to tissue repair and resided close to the vasculature [72]. These macrophage populations were identified in murine lung, fat, heart and dermis, but also in several human tissues [72]. We compared the expression of markers defining these two interstitial populations to the murine synovial macrophages from Culemann et al. [18] and found that synovial RELMα+ sublining macrophages resembled Lyve1hiMHCIIlo, while synovial MHCII+ interstitial macrophages were more like Lyve1loMHCIIhi. This suggests that RELMα+ sublining macrophages are localized closer to the blood vessels and MHCII+ interstitial macrophages near the synovial innervation. This hypothesis is further supported by the resemblance of synovial RELMα+ macrophages to human MerTKposFOLR2pos LYVE1pos macrophages, which produce high levels of perivascular marker lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1) and express markers associated with regulation of vascular endothelial growth factor [17]. However, the exact positioning of synovial interstitial macrophages requires further investigation by imaging technologies.
CX3CR1+ lining macrophages were identified as a dense protective barrier which is Csf1R- and gets replenished by Csf1R+ sublining macrophages [18]. Based on published confocal microscopy images by Culemann et al. [18] and our independent observations, lining macrophages form the uppermost lining layer; however, it is not a dense layer similar to epithelial cells, but it can be rather fragmented.
Finally, as the peripheral helper T cells were found to have a pathological role in B-cell activation in inflammatory arthritis [19, 33, 37], these cells need to be localized close to each other. By immunohistochemistry, TPH cells in synovial lymphoid aggregates were found in proximity to B cells [37]. In ∼40% of patients with RA, leukocyte infiltrates manifest as tertiary lymphoid organ-like structure (reviewed, for example, in [73, 74]). B cells and T cells seem to have an organized positioning within these structures with CCL21+ cells on the inside and marginal zone B and B1 cell specific protein (MZB1)+ plasma cells on the outside [35]. Vickovic et al. [35] hypothesized that tertiary lymphoid organ-like structures retain newly recruited leukocytes; however, the positioning of these structures in respect to the vasculature or other synovial immune and structural cells is unknown.
Structural cells
Similar to immune cells, different subsets of fibroblasts localize to different regions of the synovium [19, 43–45]. CD34-THY1+ fibroblasts were found to reside in the sublining where they surrounded the capillaries [43]. This layer was a few cells thin in OA patients and expanded in the synovium isolated from patients with active inflammation [43]. CD34+ fibroblasts could also be observed in the sublining layer, scattered across both deep and superficial interstitium [43]. Finally, CD34-THY1- produced lubricin and formed the synovial lining layer [19, 43, 44]. Overall, THY1 expression determines sublining populations of fibroblasts [19, 43–45]. Furthermore, the distance from endothelial cells seems to affect the phenotype of fibroblasts; cells closest to the vasculature express high levels of THY1 and cells furthest express high levels of lubricin [45]. In conclusion, local microenvironment cues are crucial for cell positioning, identity and function.
Cell–cell interactions in the synovium
Methods to study cellular interactions
Possible methods of how cellular interactions can be studied based on gene expression were nicely summarized in a recent review [75]. In brief, a variety of methods have been developed such as NicheNet [76], SoptSC [77, 78], CellPhoneDB [79], CellChat [80], and many others. Each method is using a different algorithm for assessing cell–cell interactions, such as using differential gene expression between and within cell clusters to search for ligand-receptor pairs. These approaches also use different data sources to access ligand-receptor pairs; for example, NicheNet is using KEGG as its main data source [76]. The assessment of individual strengths and weaknesses of the available methods is not the subject of this review. Nevertheless, as emphasized in the recent review [75], these approaches should be considered mainly for hypothesis generation and require subsequent experimental validation. Additionally, spatial transcriptomics can help with understanding cellular interactions by providing site-specific expression profile of the synovium [35, 36].
Identified cellular interactions in the synovium
The ligand-receptor analysis was used to identify the Notch signalling axis between fibroblasts and endothelial cells, which shapes fibroblast identity in the synovium [45]. Based on this analysis, Wei et al. [45] identified that synovial arterial endothelial cells express Notch ligands jagged canonical Notch ligand 1 (JAG1) and delta-like canonical Notch ligand 4 (DLL4), which interact with NOTCH3 on surrounding mural cells and fibroblasts (Fig. 4A). Furthermore, the endothelial cells seemed to direct the differentiation of THY1+ sublining fibroblast, which was confirmed in mixed synovial organoids [45].
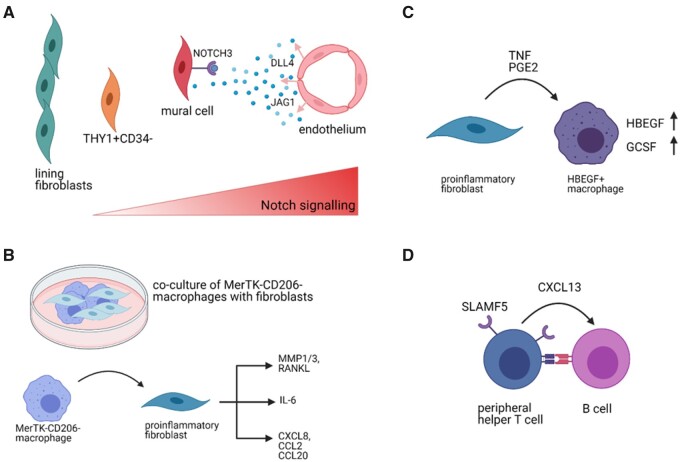
Fig. 4.
Examples of interactions identified in human or murine synovium
(A) Arterial endothelial cells were found to release NOTCH3 ligands DLL4 and JAG1 that bind NOTCH3 receptor on mural cells and sublining fibroblasts. The Notch signalling gradient shapes the identity of fibroblasts where fibroblasts closer to the endothelium will develop into THY1+CD34- fibroblasts and cells further from it will become THY1- lining fibroblasts. (Based on [45]) (B) Co-culture of proinflammatory MerTKnegCD206neg macrophages with fibroblasts caused the fibroblasts to adopt a proinflammatory phenotype and secrete bone and cartilage destructing mediators (MMP1/3, RANKL) and inflammatory cytokines (IL6) or chemokines (CXCL8, CCL2, CCL20) [17]. (C) Proinflammatory fibroblasts were found to secrete TNF or PGE2, which induce expression of HBEGF and GCSF in macrophages, inducing pro-inflammatory phenotype [20]. (D) Peripheral helper T cells and B cells communicate in the synovium through multiple pathways, for example via secretion of CXCL13 by T cells [19, 37, 81] or via upregulation of SLAMF5 [37]. Figure created with BioRender.com.
Fibroblasts were also found to be affected by synovial macrophages in an in vitro co-culture [17]. MerTKneg synovial macrophages induced an increased expression of mediators of bone and cartilage destruction, inflammation and leukocyte recruitment in fibroblasts [17] (Fig. 4B). Vice versa, in the resolution phase of RA, THY1posCXCL14pos sublining fibroblasts were found to abundantly express an activator of MerTK, growth arrest specific 6 (GAS6) [17]. MerTK was previously found to be significantly associated with the resolution of inflammation [17, 20, 21].
Synovial fibroblasts further impact the transcriptome of macrophages by secreting pro-inflammatory factors such as TNF and prostaglandin E2 (PGE2) and support their development into inflammatory HBEGF+ macrophages [20] (Fig. 4C). Medications, such as leflunomide, dexamethasone, naproxen and triple therapy could inhibit the fibroblast induced HBEGF+ macrophage polarization [20]. For example, naproxen functionally blocked the prostaglandin generation and in turn abrogated HBEGF mRNA and granulocyte colony-stimulating factor (GCSF) protein production [20], showing the importance of both cellular heterogeneity and interactions in understanding the molecular action of anti-inflammatory drugs.
Usage of spatial transcriptomics [81] revealed an upregulated expression of genes such as lymphotoxin beta (LTB), CCL19, CXCL13, CD52, membrane spanning 4-domains A1 (MS4A1) and CD79A, which are associated with T-cell and B-cell function and crosstalk [35, 36]. Especially, CXCL13 expression by TPH to activate B cells is of interest as it is confirmed in other sequencing studies [19, 37]. Furthermore, signalling lymphocytic activation molecule 5 (SLAMF5), required for T-cell and B-cell interaction, was upregulated on peripheral and follicular helper T cells and a blockade of SLAMF5 abrogated PD1hiCXCR5-CD4+ T-cell effect on B cells in an in vitro co-culture [37] suggesting an additional therapeutic target (Fig. 4D). However, this approach would most likely be restricted to seropositive patients. Additionally, patients with inflammatory arthritis have a varying degree of B-cell and T-cell infiltration [33], further complicating this approach.
Conclusion
Recent advances in single-cell technologies allowed for deeper profiling of synovial cells, identification of their respective markers, activation states and provided predictions of their functionality (Fig. 5). Studying cellular organization is virtually impossible without an appreciation of the characteristic cellular markers; therefore, we envisage that the field of rheumatology will soon be enriched with many studies profiling cellular zonation in the synovium. The spatial characterisation will further uncover which cells are likely to interact with each other, leading to a better understanding of signalling pathways in the synovium both in the steady-state and during inflammation. Single-cell transcriptomics already led to the development of computational methods allowing for the prediction of ligand-receptor interactomes but, as mentioned before, these tools should be used for hypothesis generation and subsequently experimentally verified. Apart from cellular heterogeneity, the extracellular matrix composition, function and localisation creating unique niches was recently reviewed as well [82]. Thus, the interactions between synovial cells and the extracellular matrix should also be considered to fully comprehend the maintenance and function of this complex tissue. Furthermore, bulk RNAseq was already shown to be able to stratify patients better than histological assessment [83]. However, scRNAseq, once the analysis becomes cheaper and more abundant, could provide even more precise patient classification into treatment groups as it can identify presence/absence of specific pathological cell subsets.
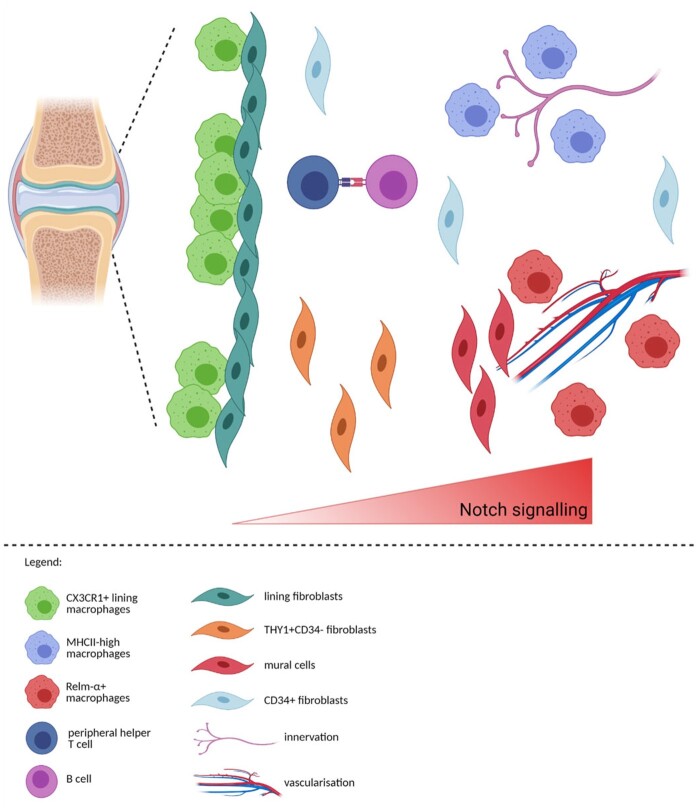
Fig. 5.
Prediction of the zonation of immune and structural cells in the synovium
CX3CR1+ lining macrophages and lining fibroblast create the ‘barrier’ layer of the synovium [18, 19]. Based on our analysis, MHCIIhigh macrophages could be positioned near the innervation, while RELM-α+ macrophages could be located near the vasculature, similarly to the lung interstitium [72]. Notch signalling axis is instructing the positioning of fibroblasts with mural cells closest to the vasculature, THY1+CD34- fibroblasts near the vasculature and mural cells with THY1-CD34- lining fibroblast at the opposing end [43, 45]. CD34+ fibroblasts are positioned both in the immediate sublining and deep interstitium [43]. Figure created with BioRender.com.
We identified several limitations in the current literature. Firstly, single-cell transcriptomics is uncovering a correlative relationship between cellular states and sustained inflammation or remission. The role and contributions of each subpopulation should be investigated in greater detail to decipher causal effects. Nevertheless, single-cell profiling can provide an excellent overview of the cell distribution in tissues, describe markers instrumental for imaging, and aid in hypothesis generation. Secondly, the selection of markers used for sorting prior to single-cell RNA sequencing can affect the subsequent analysis and some cellular populations can be unknowingly disregarded. Furthermore, different markers for identification of cells, different clustering strategies and analyses make the comparison of different datasets rather challenging. Potentially, an integrated meta-analysis comparing various datasets could verify current findings. Lastly, certain murine models of inflammatory arthritis are dependent only on an innate immune system; thus, profiling of adaptive immune cells in such cases is missing.
Single-cell transcriptomics is revolutionizing the rheumatology field. We believe that the extensive amount of literature identifying single-cell heterogeneity of the synovium will lead to better spatial characterisation of the synovium and identification of pathological interactions, which can be blocked by medical interventions to the therapeutic benefit of patients.
Acknowledgements
B.S. planned, researched data, and wrote the manuscript. K.Z. and I.A.U discussed the content, contributed to the planning, reviewed and edited the manuscript.
Funding: No specific funding was received from any bodies in the public, commercial or not-for-profit sectors to carry out the work described in this article.
Disclosure statement: The authors have declared no conflicts of interest.
Data availability statement
Data are available upon reasonable request by any qualified researchers who engage in rigorous, independent scientific research, and will be provided following review and approval of a research proposal and Statistical Analysis Plan (SAP) and execution of a Data Sharing Agreement (DSA). All data relevant to the study are included in the article.
References
- 1. Weyand CM, Goronzy JJ.. The immunology of rheumatoid arthritis. Nat Immunol 2021;22:10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smolen JS, Aletaha D, Barton A. et al. Rheumatoid arthritis. Nat Rev Dis Primers 2018;4:18001. [DOI] [PubMed] [Google Scholar]
- 3. Schett G, Tanaka Y, Isaacs JD.. Why remission is not enough: underlying disease mechanisms in RA that prevent cure. Nat Rev Rheumatol 2021;17:135–44. [DOI] [PubMed] [Google Scholar]
- 4. Orr C, Vieira-Sousa E, Boyle DL. et al. Synovial tissue research: a state-of-the-art review. Nat Rev Rheumatol 2017;13:463–75. [DOI] [PubMed] [Google Scholar]
- 5. Tang X, Huang Y, Lei J, Luo H, Zhu X.. The single-cell sequencing: new developments and medical applications. Cell Biosci 2019;9:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stuart T, Satija R.. Integrative single-cell analysis. Nat Rev Genet 2019;20:257–72. [DOI] [PubMed] [Google Scholar]
- 7. Kashima Y, Sakamoto Y, Kaneko K. et al. Single-cell sequencing techniques from individual to multiomics analyses. Exp Mol Med 2020;52:1419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Picelli S, Björklund ÅK, Faridani OR. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 2013;10:1096–8. [DOI] [PubMed] [Google Scholar]
- 9. Zheng GXY, Terry JM, Belgrader P. et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stephenson W, Donlin LT, Butler A. et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat Commun 2018;9:791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lähnemann D, Köster J, Szczurek E. et al. Eleven grand challenges in single-cell data science. Genome Biol 2020;21:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luecken MD, Theis FJ.. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol 2019;15:e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kurowska-Stolarska M, Alivernini S.. Synovial tissue macrophages: friend or foe? RMD Open 2017;3:e000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tu J, Wang X, Gong X. et al. Synovial macrophages in rheumatoid arthritis: the past, present, and future. Mediators Inflamm 2020;2020:1583647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Udalova IA, Mantovani A, Feldmann M.. Macrophage heterogeneity in the context of rheumatoid arthritis. Nat Rev Rheumatol 2016;12:472–85. [DOI] [PubMed] [Google Scholar]
- 16. Uderhardt S, Martins AJ, Tsang JS, Lämmermann T, Germain RN.. Resident macrophages cloak tissue microlesions to prevent neutrophil-driven inflammatory damage. Cell 2019;177:541–55.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alivernini S, MacDonald L, Elmesmari A. et al. Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis. Nat Med 2020;26:1295–306. [DOI] [PubMed] [Google Scholar]
- 18. Culemann S, Grüneboom A, Nicolás-Ávila JÁ. et al. Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature 2019;572:670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang F, Wei K, Slowikowski K. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol 2019;20:928–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuo D, Ding J, Cohn IS. et al. HBEGF+ macrophages in rheumatoid arthritis induce fibroblast invasiveness. Sci Transl Med 2019;11:eaau8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Misharin AV, Cuda CM, Saber R. et al. Nonclassical Ly6C(-) monocytes drive the development of inflammatory arthritis in mice. Cell Rep 2014;9:591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smiljanovic B, Grützkau A, Sörensen T. et al. Synovial tissue transcriptomes of long-standing rheumatoid arthritis are dominated by activated macrophages that reflect microbial stimulation. Sci Rep 2020;10:7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liao M, Liu Y, Yuan J. et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 2020;26:842–4. [DOI] [PubMed] [Google Scholar]
- 24. Xie X, Shi Q, Wu P. et al. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat Immunol 2020;21:1119–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wright HL, Moots RJ, Edwards SW.. The multifactorial role of neutrophils in rheumatoid arthritis. Nat Rev Rheumatol 2014;10:593–601. [DOI] [PubMed] [Google Scholar]
- 26. Wright HL, Lyon M, Chapman EA, Moots RJ, Edwards SW.. Rheumatoid arthritis synovial fluid neutrophils drive inflammation through production of chemokines, reactive oxygen species, and neutrophil extracellular traps. Front Immunol 2021;11:3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyabe Y, Lian J, Miyabe C, Luster AD.. Chemokines in rheumatic diseases:pathogenic role and therapeutic implications. Nat Rev Rheumatol 2019;15:731–46. [DOI] [PubMed] [Google Scholar]
- 28. Wang L, Ai Z, Khoyratty T. et al. ROS-producing immature neutrophils in giant cell arteritis are linked to vascular pathologies. JCI Insight 2020;5:e139163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rahman S, Sagar D, Hanna RN. et al. Low-density granulocytes activate T cells and demonstrate a non-suppressive role in systemic lupus erythematosus. Ann Rheum Dis 2019;78:957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ostendorf L, Mothes R, van Koppen S. et al. Low-density granulocytes are a novel immunopathological feature in both multiple sclerosis and neuromyelitis optica spectrum disorder. Front Immunol 2019;10:2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Z, Andreev D, Oeser K. et al. Th2 and eosinophil responses suppress inflammatory arthritis. Nat Commun 2016;7:11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Biton J, Khaleghparast Athari S, Thiolat A. et al. In vivo expansion of activated Foxp3+ regulatory T cells and establishment of a type 2 immune response upon IL-33 treatment protect against experimental arthritis. J Immunol 2016;197:1708–19. [DOI] [PubMed] [Google Scholar]
- 33. Lewis MJ, Barnes MR, Blighe K. et al. Molecular portraits of early rheumatoid arthritis identify clinical and treatment response phenotypes. Cell Rep 2019;28:2455–70.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yeo L, Lom H, Juarez M. et al. Expression of FcRL4 defines a pro-inflammatory, RANKL-producing B cell subset in rheumatoid arthritis. Ann Rheum Dis 2015;74:928–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vickovic S, Schapiro D, Carlberg K. et al. Three-dimensional spatial transcriptomics uncovers cell type dynamics in the rheumatoid arthritis synovium. bioRxiv 2020:2020.12.10.420463. Preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- 36. Carlberg K, Korotkova M, Larsson L. et al. Exploring inflammatory signatures in arthritic joint biopsies with Spatial Transcriptomics. Sci Rep 2019;9:18975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rao DA, Gurish MF, Marshall JL. et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 2017;542:110–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Steel KJA, Srenathan U, Ridley M. et al. Polyfunctional, proinflammatory, tissue-resident memory phenotype and function of synovial interleukin-17A+CD8+ T cells in psoriatic arthritis. Arthritis Rheumatol 2020;72:435–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Penkava F, Velasco-Herrera MDC, Young MD. et al. Single-cell sequencing reveals clonal expansions of pro-inflammatory synovial CD8 T cells expressing tissue-homing receptors in psoriatic arthritis. Nat Commun 2020;11:4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nygaard G, Firestein GS.. Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes. Nat Rev Rheumatol 2020;16:316–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu Z, Ma D, Yang H. et al. Fibroblast-like synoviocytes in rheumatoid arthritis: surface markers and phenotypes. Int Immunopharmacol 2021;93:107392. [DOI] [PubMed] [Google Scholar]
- 42. Mousavi MJ, Karami J, Aslani S. et al. Transformation of fibroblast‐like synoviocytes in rheumatoid arthritis; from a friend to foe. Auto Immun Highlights 2021;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mizoguchi F, Slowikowski K, Wei K. et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun 2018;9:789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Croft AP, Campos J, Jansen K. et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature 2019;570:246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei K, Korsunsky I, Marshall JL. et al. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature 2020;582:259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Szekanecz Z, Koch AE.. Vascular involvement in rheumatic diseases: ‘vascular rheumatology’. Arthritis Res Ther 2008;10:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang X, Chang Y, Wei W.. Endothelial dysfunction and inflammation: immunity in rheumatoid arthritis. Mediators Inflamm 2016;2016:6813016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marcu R, Choi YJ, Xue J. et al. Human organ-specific endothelial cell heterogeneity. iScience 2018;4:20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Paik David T, Tian L, Williams Ian M. et al. Single-cell RNA sequencing unveils unique transcriptomic signatures of organ-specific endothelial cells. Circulation 2020;142:1848–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cleuren ACA, van der Ent MA, Jiang H. et al. The in vivo endothelial cell translatome is highly heterogeneous across vascular beds. Proc Natl Acad Sci 2019;116:23618–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rohlenova K, Goveia J, García-Caballero M. et al. Single-cell RNA sequencing maps endothelial metabolic plasticity in pathological angiogenesis. Cell Metabol 2020;31:862–77.e14. [DOI] [PubMed] [Google Scholar]
- 52. Feng W, Chen L, Nguyen PK, Wu SM, Li G.. Single cell analysis of endothelial cells identified organ-specific molecular signatures and heart-specific cell populations and molecular features. Front Cardiovasc Med 2019;6:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cattaneo MG, Banfi C, Brioschi M, Lattuada D, Vicentini LM.. Sex-dependent differences in the secretome of human endothelial cells. Biol Sex Differ 2021;12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grüneboom A, Hawwari I, Weidner D. et al. A network of trans-cortical capillaries as mainstay for blood circulation in long bones. Nat Metabol 2019;1:236–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pap T, Korb-Pap A.. Cartilage damage in osteoarthritis and rheumatoid arthritis—two unequal siblings. Nat Rev Rheumatol 2015;11:606–15. [DOI] [PubMed] [Google Scholar]
- 56. Martel-Pelletier J, Barr AJ, Cicuttini FM. et al. Osteoarthritis. Nat Rev Dis Primers 2016;2:16072. [DOI] [PubMed] [Google Scholar]
- 57. Zhu S, Zhu J, Zhen G. et al. Subchondral bone osteoclasts induce sensory innervation and osteoarthritis pain. J Clin Invest 2019;129:1076–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Locke S, Yousefpour N, Mannarino M. et al. Peripheral and central nervous system alterations in a rat model of inflammatory arthritis. Pain 2020;161:1483–96. [DOI] [PubMed] [Google Scholar]
- 59. Kane D, Lockhart JC, Balint PV. et al. Protective effect of sensory denervation in inflammatory arthritis (evidence of regulatory neuroimmune pathways in the arthritic joint). Ann Rheum Dis 2005;64:325–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peltz TS, Yapp LZ, Elherik FK, Breusch SJ.. Patient satisfaction and outcomes of partial wrist denervation in inflammatory arthritis. Clin Rheumatol 2019;38:2995–3003. [DOI] [PubMed] [Google Scholar]
- 61. Usoskin D, Furlan A, Islam S. et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015;18:145–53. [DOI] [PubMed] [Google Scholar]
- 62. Sharma N, Flaherty K, Lezgiyeva K. et al. The emergence of transcriptional identity in somatosensory neurons. Nature 2020;577:392–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huang S, Ziegler CGK, Austin J. et al. Lymph nodes are innervated by a unique population of sensory neurons with immunomodulatory potential. Cell 2021;184:441–59.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wolbert J, Li X, Heming M. et al. Redefining the heterogeneity of peripheral nerve cells in health and autoimmunity. Proc Natl Acad Sci 2020;117:9466–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jordão MJC, Sankowski R, Brendecke SM. et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 2019;363:eaat7554. [DOI] [PubMed] [Google Scholar]
- 66. Ghouri A, Conaghan PG.. Update on novel pharmacological therapies for osteoarthritis. Ther Adv Musculoskelet Dis 2019;11:1759720X19864492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ranjit S, Lanzanò L, Libby AE, Gratton E, Levi M.. Advances in fluorescence microscopy techniques to study kidney function. Nat Rev Nephrol 2021;17:128–44. [DOI] [PubMed] [Google Scholar]
- 68. Eng C-HL, Lawson M, Zhu Q. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 2019;568:235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rodriques SG, Stickels RR, Goeva A. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019;363:1463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen W-T, Lu A, Craessaerts K. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer's disease. Cell 2020;182:976–91.e19. [DOI] [PubMed] [Google Scholar]
- 71. Ji AL, Rubin AJ, Thrane K. et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 2020;182:497–514.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chakarov S, Lim HY, Tan L. et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 2019;363:eaau0964. [DOI] [PubMed] [Google Scholar]
- 73. Rivellese F, Pontarini E, Pitzalis C.. Tertiary lymphoid organs in rheumatoid arthritis. In: Kabashima K, Egawa G, eds. Inducible lymphoid organs. Cham: Springer International Publishing, 2020:119–41. [DOI] [PubMed] [Google Scholar]
- 74. Pipi E, Nayar S, Gardner DH. et al. Tertiary lymphoid structures: autoimmunity goes local. Front Immunol 2018;9:1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Armingol E, Officer A, Harismendy O, Lewis NE.. Deciphering cell–cell interactions and communication from gene expression. Nat Rev Genet 2021;22:71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Browaeys R, Saelens W, Saeys Y.. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods 2020;17:159–62. [DOI] [PubMed] [Google Scholar]
- 77. Wang S, Drummond ML, Guerrero-Juarez CF. et al. Single cell transcriptomics of human epidermis identifies basal stem cell transition states. Nat Commun 2020;11:4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang S, Karikomi M, MacLean AL, Nie Q.. Cell lineage and communication network inference via optimization for single-cell transcriptomics. Nucleic Acids Res 2019;47:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R.. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 80. Jin S, Guerrero-Juarez CF, Zhang L. et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021;12:1088. [DOI] [PMC free article] [PubMed]
- 81. Ståhl PL, Salmén F, Vickovic S. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016;353: 78–82. [DOI] [PubMed] [Google Scholar]
- 82. Buckley CD, Ospelt C, Gay S, Midwood KS.. Location, location, location: how the tissue microenvironment affects inflammation in RA. Nat Rev Rheumatol 2021;17:246. [DOI] [PubMed] [Google Scholar]
- 83. Humby F, Durez P, Buch MH. et al. Rituximab versus tocilizumab in anti-TNF inadequate responder patients with rheumatoid arthritis (R4RA): 16-week outcomes of a stratified, biopsy-driven, multicentre, open-label, phase 4 randomised controlled trial. Lancet 2021;397:305–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available upon reasonable request by any qualified researchers who engage in rigorous, independent scientific research, and will be provided following review and approval of a research proposal and Statistical Analysis Plan (SAP) and execution of a Data Sharing Agreement (DSA). All data relevant to the study are included in the article.