Abstract
A hypervirulent pathotype of A. hydrophila (vAh) is responsible for Motile Aeromonas Septicemia (MAS) and causes mass mortalities among farmed carp and catfish species in the USA and China. One unique phenotype for vAh among other A. hydrophila strains is the ability to utilize myo-inositol as a sole carbon source. While screening for Aeromonas isolates from diseased fish that can grow using myo-inositol as a sole carbon source, A. dhakensis 1P11S3 was isolated from the spleen of striped catfish (Pangasianodon hypopthalmus) displaying clinical MAS symptoms from a freshwater farm in Malaysia. Aeromonas dhakensis is also an important pathogen in aquaculture, and in this study, we report the draft genome sequence for A. dhakensis 1P11S3, that utilize myo-inositol as a sole carbon source.
Keywords: Draft genome, myo-inositol, A. dhakensis, Malaysia
Specifications Table
| Subject | Biology |
| Specific subject area | Microbiology, Genomics, Biotechnology |
| Type of data | Table, figures |
| How data were acquired | The draft genome sequence was processed using Illumina HiSeq instrument |
| Data format | Raw, analyzed and deposited |
| Parameters for data collection | Aeromonas dhakensis 1P11S3 was isolated from the spleen of cultured striped catfish (Pangasianodon hypopthalmus) in Malaysia. Genomic DNA extraction and sequencing were performed. |
| Description of data collection | Aeromonas dhakensis 1P11S3 was able to grow in M9 agar with myo-inositol as a sole carbon source. Genomic DNA was isolated from a pure culture of A.dhakensis 1P11S3 |
| Data source location | Aeromonas dhakensis 1P11S3 was isolated from the spleen of cultured striped catfish (Pangasianodon hypopthalmus) in Tumpat, Kelantan, Malaysia. Latitude and longitude: 6.196106 N 102.148057 E |
| Data accessibility | Data are publicly available at NCBI GenBank https://www.ncbi.nlm.nih.gov/assembly/GCA_015666195.1 https://www.ncbi.nlm.nih.gov/biosample/SAMN16824286 https://www.ncbi.nlm.nih.gov/bioproject/PRJNA679132 https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA679132 |
| Related research article | M. Azzam-Sayuti, M.Y. Ina-Salwany, M. Zamri-Saad, M.T. Yusof, S. Annas, M.Y. Najihah, et al., The prevalence, putative virulence genes, and antibiotic resistance profiles of Aeromonas spp. isolated from cultivated freshwater fish in Peninsular Malaysia, Aquaculture 540 (2021) 736719. DOI: https://doi.org/10.1016/j.aquaculture.2021.736719 |
Value of the Data
-
•
The draft genome of A. dhakensis 1P11S3, isolated from cultured freshwater fish will be useful for further research on the influence of myo-inositol catabolism to the virulence of A. dhakensis.
-
•
Data from A. dhakensis 1P11S3 will facilitate understanding of lateral gene transfer of myo-inositol catabolism genes
-
•
Data on the genome sequence of A. dhakensis 1P11S3 can be used for comparative genomic studies with other Aeromonas spp. disease isolates, including vAh strains.
-
•
Data on the genome sequence of A. dhakensis 1P11S3 could be used to identify and characterize important virulence factors that contribute to pathogenesis.
1. Data Description
A hypervirulent pathotype of A. hydrophila, vAh strains have been responsible for huge losses suffered by the catfish and cyprinid fish industry in USA and China [1,2]. The ability to utilize myo-inositol as a growth substrate has been reported to be present in all vAh strains and has not previously been reported among non-vAh strains [3]. This ability has also been linked to contributing to its virulence since myo-inositol utilization could allow persistence of A. hydrophila strains, and the transcriptional regulator IolR, an important negative regulator of myo-inositol pathway has been found to regulate autoaggregation and biofilm formation [3,4]. Therefore, this trait has been used as a key determinant to identify vAh that is also considered to be a primary pathogen [1], unlike typical A. hydrophila globally that are considered opportunistic and secondary pathogens [1,2]. Aeromonas dhakensis has also been recognized as a significant pathogen in the field of aquaculture. The bacterium is as one of the Aeromonas species that is responsible for motile Aeromonas septicemia (MAS) [1], and have been reported to be more virulent than some A. hydrophila strains [5]. To the author's knowledge, there are no studies that have reported A. dhakensis that could utilize myo-inositol.
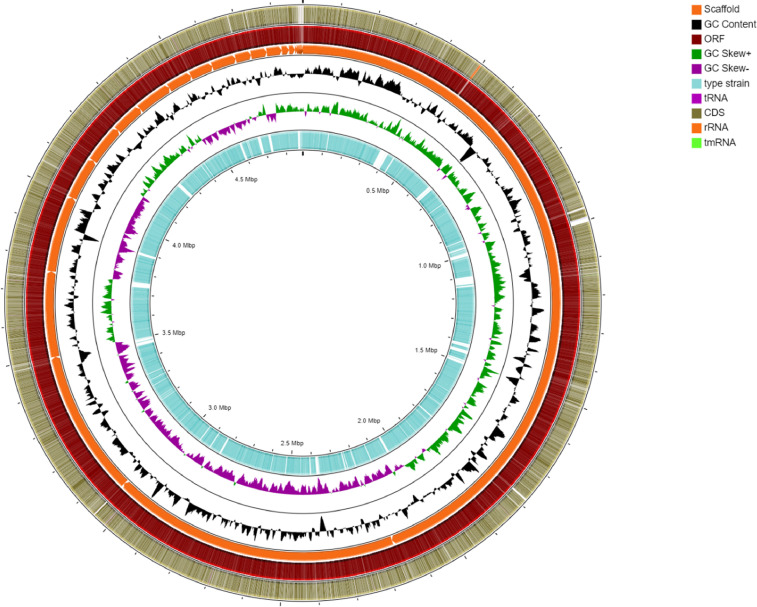
The draft genome sequence for A. hydrophila 1P11S3 was deposited in GenBank under accession number JADPIC000000000. In total, 4,884,279 bp of total bases were assembled with 43 total scaffolds with a N50 value of 885,649 with 61.55% G+C content (Table 1, Fig. 1). The genome was predicted to encode 4,391 coding genes, whereas the total non-coding RNA was 182. The major class of carbohydrate-active enzyme was found to be glycosyl transferase (GT; 47.5%), followed by glycoside hydrolases (GH; 25.7%), carbohydrate-binding molecules (CBM; 18.1%), carbohydrate esterases (CE; 7.8%), auxiliary activities (AA; 0.6%) and polysaccharide lyases (PL; 0.2%). The percentage of gene functions present in A. dhakensis 1P11S3 in each of six major categories is as follows: general function (11.7%), amino acid transport and metabolism (9.6%), signal transduction mechanisms (7.5%), transcription (7.4%), energy production and conversion (6.2%), and carbohydrate transport and conversion (5.6%). The least categories (0.1%) were extracellular structures, chromatin structure and dynamics, and RNA processing and modification.
Table 1.
Genome features of A. dhakensis 1P11S3.
| Attribute | A. dhakensis 1P11S3 value |
|---|---|
| Genome size (bp) | 4,884,279 |
| Number of scaffolds | 43 |
| N50 (bp) | 885649 |
| GC content (%) | 61.55 |
| Predicted coding genes | 4391 |
| Predicted non-coding RNA | 182 |
| GenBank accession | JADPIC000000000 |
| BioSample accession | SAMN16824286 |
| BioProject accession | PRJNA679132 |
Fig. 1.
Circular representation of the A. dhakensis 1P11S3 genome. From the outside to the center of the diagram, the circles show the following: circle 1, coding region (CDS) (tRNA shown in purple, rRNA shown in orange and tmRNA shown in light green); circle 2, open reading frame (ORF); circle 3, scaffolds; circle 4, GC contents; circle 5, GC skew (G+C shown in green, G-C shown in purple); circle 6, blast comparison with A. dhakensis CIP 107500 (T).
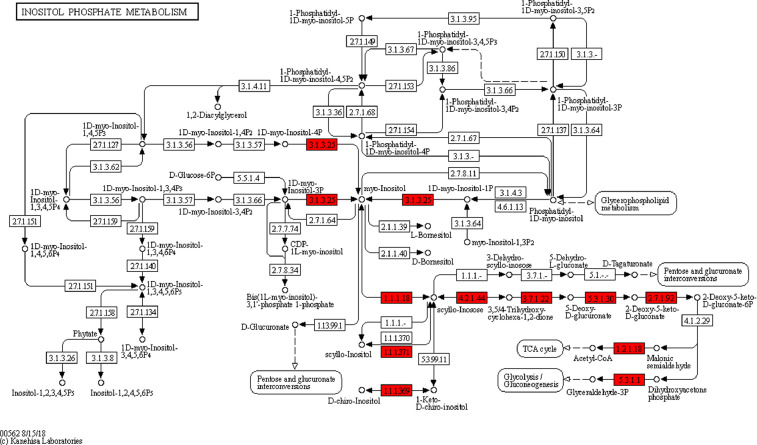
KEGG pathway analysis revealed that A. dhakensis 1P11S3 has all the proteins necessary for a complete myo-inositol metabolism (Fig. 2), in concordance with A. hydrophila ML09-119 (vAh) (accession no: CP005966.1, https://www.kegg.jp/kegg-bin/show_pathway?ahy00562), with the extra predicted ability to utilize scyllo-inositol, which was apparently not present in A. hydrophila ML09-119.
Fig. 2.
The proposed KEGG pathway for inositol catabolism. Highlighted red boxes indicate the proteins expressed by the A. dhakensis 1P11S3 that are involved in the utilization of myo-inositol.
2. Experimental Design, Materials and Methods
Aeromonas dhakensis 1P11S3 was previously isolated from the spleen of cultured striped catfish (Pangasianodon hypopthalmus) in a fish farm in Kelantan, Malaysia, which displayed clinical signs of MAS such as fin and tail rot, hemorrhagic scale and eye, and loss of scale [6]. For the genome analysis, the purified strain of A. dhakensis 1P11S3 was inoculated in 1 mL of TSB and incubated overnight at 30°C. Later, DNA extraction was done using a DNA purification kit (Promega, Inc., Madison, WI, USA) according to the manufacturer's protocol. The DNA concentration and quality were determined using Qubit 3.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). Libraries with different indices were multiplexed and loaded onto an Illumina HiSeq instrument according to the manufacturer's instructions (Illumina, San Diego, CA, USA) and the genome was sequenced using a 2 × 150 paired-end sequencing kit. A total of 19,649,882 clean reads (out of 19,688,664 raw reads) were generated and were trimmed of low-quality bases and reads using cutadapt v1.9.1. The final scaffold was then assembled using velvet, and the gaps were filled with SSPACE v3.0 and GapFiller v1-10 [7], [8], [9], [10]. A reference genome, A. dhakensis CIP 107500 (type strain) (accession no: CDBH00000000.1), was used to assemble the A. dhakensis 1P11S3 genome into scaffolds. Using the default setting of the program tRNAscan-SE v2.0, transfer RNAs (tRNAs) were detected [11]. The coding genes of the bacteria were predicted using the Prodigal v2.6.3 and were annotated with National Center for Biotechnology (NCBI) (https://github.com/hyattpd/Prodigal). Their functions were annotated by Gene Ontology (GO) database, pathways were annotated by Kyoto Encyclopedia of Genes and Genomes (KEGG) and phylogenetic classification of the proteins encoded by genes was done by the Cluster of Orthologous Groups [12], [13], [14].
CRediT Author Statement
Mohamad Azzam-Sayuti: Methodology, Investigation, Formal analysis, Writing – original draft; Md Yasin Ina-Salwany: Supervision, Data Curation, Writing – review & editing; Mohd Zamri-Saad: Supervision, Data Curation, Writing – review & editing; Salleh Annas: Supervision, Writing – review & editing; Mark R. Liles: Data Curation, Writing – Review & editing; Tingbi Xu: Methodologies, Data Curation; Mohammad Noor Azmai Amal: Supervision, Writing – review & editing; Mohd Termizi Yusof: Supervision, Data Curation, Writing – review & editing.
Ethical Statement
Not applicable.
Declaration of Competing Interest
The authors declared that they have no known competing interests or personal relationships that could have affected the work reported in this paper.
Acknowledgments
This research was supported by using the grants of Long Term Research Grant Scheme (LRGS), code: LRGS/1/2019/UPM/01/1/2; Higher Institution Centre of Excellence (HiCoE), vot no.: 6369100 provided by the Ministry of Higher Education Malaysia.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2022.107974.
Appendix. Supplementary materials
References
- 1.Bebak J., Wagner B., Burnes B., Hanson T. Farm size, seining practices, and salt use: risk factors for Aeromonas hydrophila outbreaks in farm-raised catfish, Alabama, USA. Preventive Veterinary Medicine. 2015;118:161–168. doi: 10.1016/j.prevetmed.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Pang M., Jiang J., Xie X., Wu Y., Dong Y., Kwok A.H.Y., et al. Novel insights into the pathogenicity of epidemic Aeromonas hydrophila ST251 clones from comparative genomics. Sci. Rep. 2015;5:983. doi: 10.1038/srep09833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasmussen-Ivey C.R., Hossain M.J., Odom S.E., Terhune J.S., Hemstreet W.G., Shoemaker C.A., et al. Classification of a hypervirulent Aeromonas hydrophila pathotype responsible for epidemic outbreaks in warm-water fishes. Frontiers in Microbiology. 2016;7:1615. doi: 10.3389/fmicb.2016.01615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong Y., Li S., Zhao D., Liu J., Ma S., Geng J., et al. IolR, a negative regulator of the myo-inositol metabolic pathway, inhibits cell autoaggregation and biofilm formation by downregulating RpmA in Aeromonas hydrophila. Npj Biofilms and Microbiomes. 2020;6:1–12. doi: 10.1038/s41522-020-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P.L., Lamy B., Ko W.C. Aeromonas dhakensis, an increasingly recognized human pathogen. Frontiers in Microbiology. 2016;7:1–8. doi: 10.3389/fmicb.2016.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azzam-Sayuti M., Ina-Salwany M.Y., Zamri-Saad M., Yusof M.T., Annas S., Najihah M.Y., et al. The prevalence, putative virulence genes, and antibiotic resistance profiles of Aeromonas spp. isolated from cultivated freshwater fish in Peninsular Malaysia. Aquaculture. 2021;540 [Google Scholar]
- 7.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boetzer M., Henke C.V., Jansen H.J., Butler D., Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 9.Boetzer M., Pirovano W. Toward almost closed genomes with GapFiller. Genome Biology. 2012;13:R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt M., Newbold C., Berriman M., Otto T.D. A comprehensive evaluation of assembly scaffolding tools. Genome Biology. 2014;15:R42. doi: 10.1186/gb-2014-15-3-r42. [J] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowe T.M., Chan P.P. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2013;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2014;43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.