Abstract
Prostate-specific antigen (PSA) is the gold-standard marker to screen prostate cancer (PCa) nowadays. Unfortunately, its lack of specificity and sensitivity makes the identification of novel tools to diagnose PCa an urgent medical need. In this context, microRNAs (miRNAs) have emerged as potential sources of non-invasive diagnostic biomarkers in several pathologies. Therefore, this study was aimed at assessing for the first time the dysregulation of the whole plasma miRNome in PCa patients and its putative implication in PCa from a personalized perspective (i.e., obesity condition). Plasma miRNome from a discovery cohort (18 controls and 19 PCa patients) was determined using an Affymetrix-miRNA array, showing that the expression of 104 miRNAs was significantly altered, wherein six exhibited a significant receiver operating characteristic (ROC) curve to distinguish between control and PCa patients (area under the curve [AUC] = 1). Then, a systematic validation using an independent cohort (135 controls and 160 PCa patients) demonstrated that miR-107 was the most profoundly altered miRNA in PCa (AUC = 0.75). Moreover, miR-107 levels significantly outperformed the ability of PSA to distinguish between control and PCa patients and correlated with relevant clinical parameters (i.e., PSA). These differences were more pronounced when considering only obese patients (BMI > 30). Interestingly, miR-107 levels were reduced in PCa tissues versus non-tumor tissues (n = 84) and in PCa cell lines versus non-tumor cells. In vitro miR-107 overexpression altered key aggressiveness features in PCa cells (i.e., proliferation, migration, and tumorospheres formation) and modulated the expression of important genes involved in PCa pathophysiology (i.e., lipid metabolism [i.e., FASN] and splicing process). Altogether, miR-107 might represent a novel and useful personalized diagnostic and prognostic biomarker and a potential therapeutic tool in PCa, especially in obese patients.
Keywords: miRNome, miR-107, prostate cancer, obesity, non-invasive biomarker, PSA, FASN
Graphical abstract

This study demonstrates that plasma levels of miR-107 represents a novel and useful personalized diagnostic and prognostic biomarker in the pathological association between prostate cancer and obesity. Moreover, this study provides new, compelling evidence demonstrating that a transcriptomic intervention of miR-107 might represent a potential therapeutic tool in prostate cancer patients.
Introduction
Prostate cancer (PCa) has emerged as the most frequent tumor type among men and represents a severe health problem worldwide.1 A key limitation in PCa management is that the gold-standard screen test is based on the plasma levels of prostate-specific antigen (PSA), a biomarker that exhibits profound drawbacks, especially in the so-called “gray zone” (defined as a PSA range of 3–10 ng/mL).2 In fact, PSA test displays low specificity in that multiple factors can increase PSA levels without necessarily indicating the presence of a tumor, such as benign prostatic hyperplasia or inflammatory conditions. In addition, PSA test is not able to accurately distinguish clinically relevant tumors from indolent cases.3 For these reasons, the anatomo-pathological analysis of prostate biopsies, which represent a highly invasive technique, is still necessary to appropriately diagnose PCa nowadays. Therefore, there is an important unmet clinical need for the identification and validation of new, reliable, and specific non-invasive diagnostic biomarkers, ideally showing prognostic and/or therapeutic potential.
In this context, microRNAs (miRNAs) have emerged as promising clinical tools, especially due to their potential as diagnostic and therapeutic targets.4 miRNAs are RNA transcripts that lack protein-coding capacity. Specifically, miRNAs are short non-coding RNAs (20–22 nt) that act at post-transcriptional level, negatively regulating translation of target mRNAs by altering their stability, resulting either in the degradation of the target or in the translational repression through different mechanisms.5 In fact, miRNAs are attractive biomarker candidates as they can be reproducibly extracted from a wide range of biologic samples and are generally stable and resistant to various storage conditions.6,7 Indeed, recent studies have suggested a relationship between circulating miRNAs and PCa presence and outcome.8, 9, 10 Specifically, recent studies have identified some miRNAs differentially present in plasma samples from PCa patients; however, only some of them seem to be specifically derived from PCa tissues.11, 12, 13 Unfortunately, although various studies have identified some putative PCa-specific miRNAs (e.g., miR-141,11,14 miR-375,8,15 and miR-2116), there is not a consensus in the utility of plasma miRNAs as circulating non-invasive biomarkers for PCa. In fact, to the best of our knowledge, there are no studies describing the dysregulation of the whole miRNome in PCa versus healthy patients. For these reasons, this field requires further investigation in order to ascertain whether a specific miRNA or a plasma or serum miRNA signature could be associated with PCa risk and provide diagnostic and prognostic value through a fast, easy, and non-invasive test.
Likewise, it should be noted that, during the last years, many studies have demonstrated the implication of the metabolic status, especially the obese condition, in the development, progression, or aggressiveness of several pathologies, including PCa.17, 18, 19 However, the metabolic status of the patients has not been taking into account in the previous studies implemented to define a diagnostic and prognostic model based on miRNAs expression profile, suggesting the necessity of considering the metabolic status of the patients, especially the obesity condition, when developing and validating new biomarkers to improve the diagnostic and/or prognostic models in PCa.
Therefore, this study was aimed to explore for the first time the dysregulation of the human miRNome in PCa patients, considering the obesity condition of the patients (normoweight, overweight, or obesity) in order to discover and develop new and more personalized diagnostic, prognostic, and therapeutic tools for the management of PCa. In addition, we explored the potential pathophysiological role and the molecular mechanisms underlying the role of miR-107 in PCa, the most consistently altered miRNA in PCa patients found in our study.
Results
Plasma miRNAs landscape is dysregulated in PCa subjects
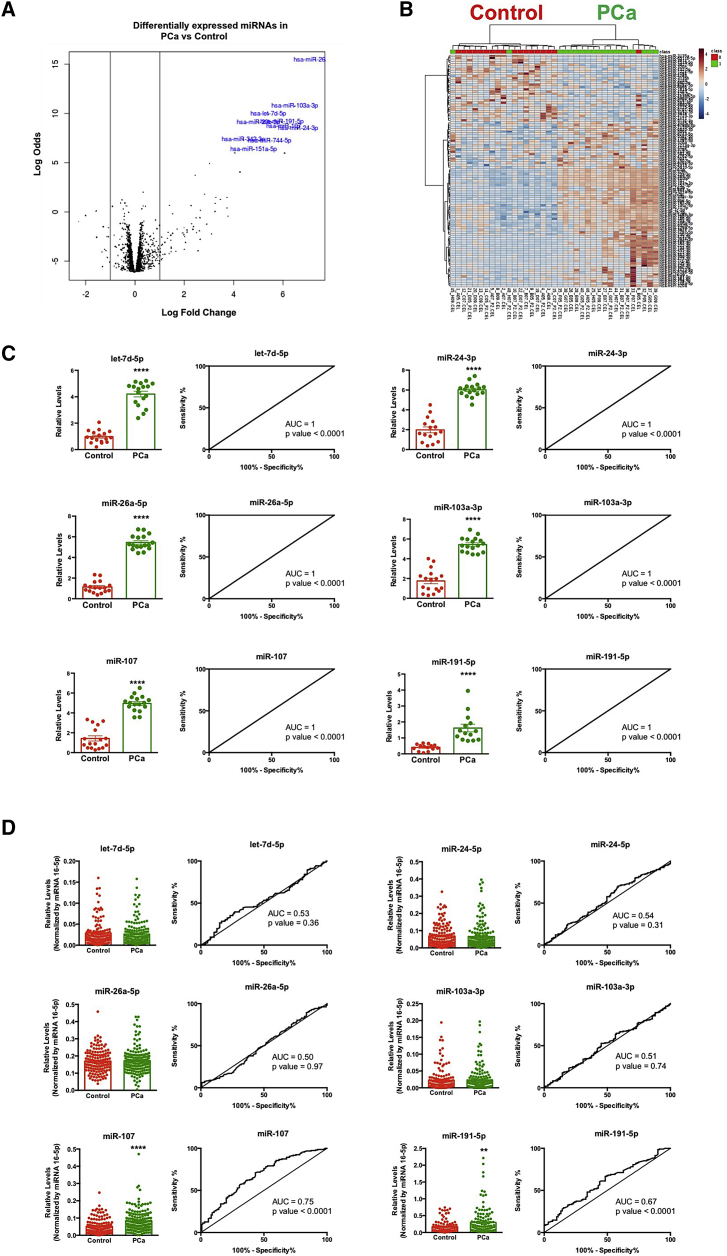
Results obtained from the Affymetrix miRNA array 4.1 (which analyzes the complete list of mature human miRNAs from the miRNome; n = 2,578) using plasma samples from PCa and healthy volunteers revealed that all miRNAs, with the exception of 18 transcripts, were detected in all the plasma samples analyzed. Specifically, this analysis revealed a significant dysregulation (p < 0.01) in the circulating pattern of 104 miRNAs, wherein 74 miRNAs showed higher levels and 30 miRNAs exhibited lower levels in PCa compared with control patients (Figure 1A). An unsupervised clustering of healthy volunteers and PCa patients revealed that the plasma levels of these 104 differentially expressed miRNAs effectively discriminated between both groups (Figure 1B). Of note, six of these significantly altered miRNAs (i.e., let-7d-5p, miR-24-5p, miR-26a-5p, miR-103a-3p, miR-107, and miR-191-5p) accurately distinguished between healthy volunteers and PCa patients, showing an area under the curve (AUC) = 1 (Figure 1C).
Figure 1.
Landscape of circulating miRNAs in prostate cancer (PCa)
(A) Volcano plot representing the alteration in the circulating levels of the whole human miRNome. (B) Heatmap of the dysregulated miRNAs (n = 104) in PCa patients compared with healthy volunteers is shown. (C) Plasma level and ROC curve analysis of the selected miRNAs (let-7d-5p, 24-3p, 26a-5p, 103a-3p, 107, and 191-5p) comparing plasma samples from PCa patients and healthy volunteers (cohort A) are shown. All these data are derived from the array analysis. (D) Plasma level and ROC curve analysis of the selected miRNAs (let-7d-5p, 24-3p, 26a-5p, 103a-3p, 107, and 191-5p) comparing plasma samples from PCa and control individuals (cohort B) are shown. These data derive from quantitative PCR analysis. AUC, area under the curve. Asterisks indicate significant differences between compared groups (∗∗p < 0.01; ∗∗∗∗p < 0.0001).
The expression profile of these six miRNAs was analyzed using quantitative real-time PCR (PCR) in an ampler cohort of subjects (validation cohort B). This analysis confirmed that the levels of miR-107 and miR-191-5p were higher in plasma from PCa patients compared with control patients (Figure 1D). In addition, both miR-107 and miR-191-5p significantly distinguished between PCa and control patients, exhibiting AUCs of 0.75 and 0.67, respectively (Figure 1D). Based on these data, miR-107 was selected for further analyses in order to more profoundly characterize its potential value as biomarker.
Potential value of miR-107 as a non-invasive biomarker in PCa
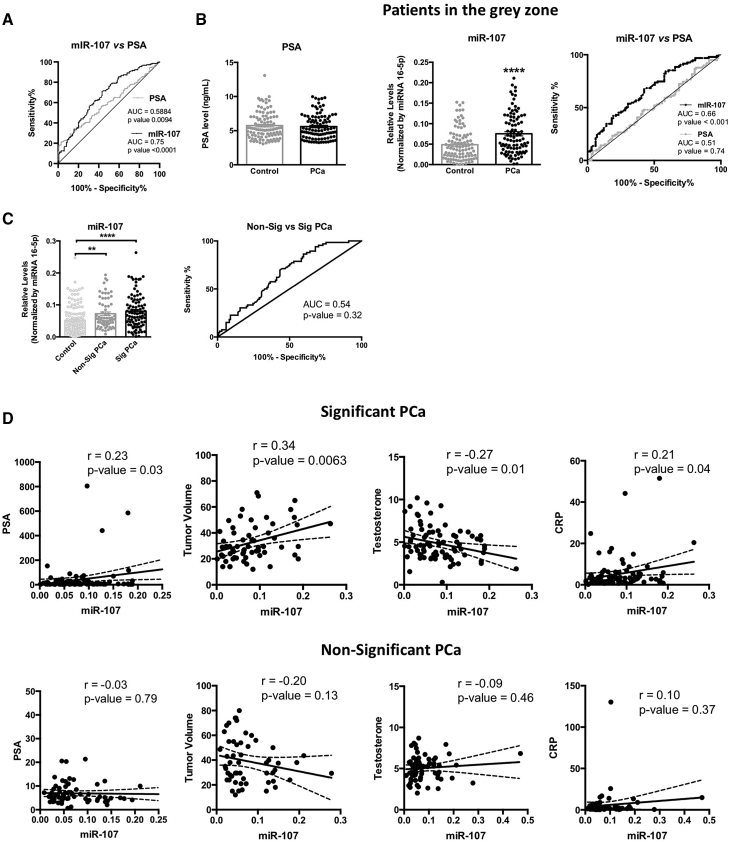
Further analyses of miR-107 plasma levels in the validation cohort B revealed a positive association with plasma PSA levels in PCa patients (Figure S1), but not with glucose level or other available clinical parameters (such as insulin level or plasma lipids [cholesterol and triglycerides (TGs)]; data not shown). Interestingly, miR-107 outperformed the capacity of PSA to distinguish between control and PCa patients (miR-107 [AUC = 0.75; p < 0.0001] versus PSA [AUC = 0.5884; p = 0.0094]; Figure 2A). Remarkably, when the same analysis was performed considering only patients with PSA levels in the gray zone (3–10 ng/mL; n = 238), we found that plasma levels of miR-107 were significantly higher in PCa patients compared with control patients (Figure 2B, middle panel), while no changes were observed when comparing PSA levels (Figure 2B, left panel). Consequently, miR-107 (but not PSA) levels are able to significantly distinguish between control and PCa patients (AUC = 0.66; p < 0.001; Figure 2B, right panel).
Figure 2.
Circulating levels of miR-107 are altered in PCa and associated to oncogenic parameters
(A) ROC curve analysis of miR-107 and PSA comparing plasma samples from PCa and control patients (cohort B). (B) Plasma level and ROC curve analysis of miR-107 and PSA comparing plasma sample from PCa and control patients included in the gray zone of PSA are shown. (C) Plasma level and ROC curve analysis of miR-107 comparing control, non-significant, and significant PCa patients are shown. (D) Correlation between plasma level of miR-107 and PSA, tumor volume, testosterone, and CRP in non-significant and significant PCa patients is shown. Data represent mean ± SEM. Asterisks indicate significant differences between compared groups (∗∗p < 0.01; ∗∗∗∗p < 0.0001).
Remarkably, plasma miR-107 levels were higher in patients with significant PCa (Sig PCa; defined as Gleason > 6) as compared with NonSig PCa patients (defined as Gleason = 6; Figure 2C). Interestingly, although the levels of miR-107 could not significantly discriminate between NonSigPCa and SigPCa patients (Figure 2C), circulating levels of miR-107 were clearly associated with several key clinical parameters, such as PSA levels, tumor volume, testosterone, and C-reactive protein (CRP) levels in SigPCa patients, but not in NonSigPCa subjects (Figure 2D).
miR-107 exerts a dual suppressive/oncogenic role in PCa
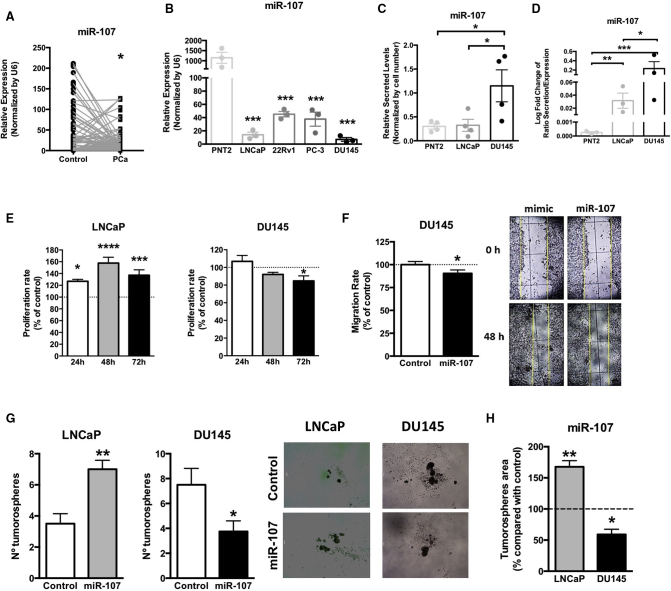
We then compared the expression levels of miR-107 between tumor regions and the adjacent non-tumor regions from PCa patients included in cohort C and found that miR-107 was downregulated in PCa tissues (Figure 3A). Similarly, all PCa cell lines analyzed herein exhibited lower expression levels of miR-107 compared with the normal-like, prostate-derived cell line PNT2 (Figure 3B). Specifically, LNCaP, an androgen-sensitive PCa cell, and DU145, a more aggressive PCa cell line, showed the lowest expression level of miR-107 (Figure 3B). In order to further explore this apparent contradiction between plasma and tissue levels of miR-107, we analyzed miR-107 levels in the conditioned and secreted media from PNT2, LNCaP, and DU145 cells. Interestingly, miR-107 secretion in DU145 was significantly higher compared with PNT2 and LNCaP cell lines (PNT2 = LNCaP < DU145; Figure 3C). Moreover, the ratio between the secretion and the expression of miR-107 was significantly higher in PCa DU145 and LNCaP compared with normal-like PNT2 cells (PNT2 < LNCaP < DU145; Figure 3D), suggesting that the low levels observed in PCa tissue and the high levels observed in plasma from PCa patients could be due to the prominent secretion of miR-107 from PCa cells, especially in the most aggressive models (SigPCa patients [observed in Figure 2C] and DU145 cells [Figure 3C]).
Figure 3.
Dual suppressive and oncogenic role of miR-107 in PCa
(A) miR-107 expression level in PCa tissues compared with adjacent non-tumor regions from PCa patients included in cohort C. (B) miR-107 expression level in PCa cells (LNCaP, 22Rv1, PC-3, and DU145) compared with the normal-like prostate cell line (PNT2) is shown. (C) miR-107 secretion levels in media obtained from PNT2, LNCaP, and DU145 cell lines are shown. (D) Ratio between secretion and expression of miR-107 in PNT2, LNCaP, and DU145 cell lines is shown. (E) Proliferation rate analysis of LNCaP and DU145 cell lines overexpressing miR-107 is shown. (F) Migration rate and representative images of DU145 cells overexpressing miR-107 are shown. Data are expressed as percentage of control (set at 100%). (G) Analysis of tumorosphere formation assay and representative images of LNCaP and DU145 cells overexpressing miR-107 is shown. Data represent number of tumorospheres. (H) Comparison of tumorospheres area in LNCaP and DU145 cells overexpressing miR-107 compared with control cells is shown. Data are expressed as percentage of control (set at 100%). Asterisks indicate significant differences between compared groups (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
Next, and in order to study the potential role of miR-107 in functional parameters of aggressiveness, overexpression of miR-107 in DU145 and LNCaP cells were performed by transfection. The intracellular overexpression of miR-107 in DU145 cells (validation results are shown in Figure S2) significantly reduced proliferation (at 72 h; Figure 3E, right panel), migration (Figure 3F), and the number and area of tumorospheres (Figures 3G, right graphic, and 3H, respectively). However, miR-107 overexpression increased proliferation and the number and area of tumorospheres in LNCaP cells (Figures 3E, left panel; 3G, left panel; and 3H, respectively).
In silico and in vitro analyses of targets and pathways regulated by miR-107
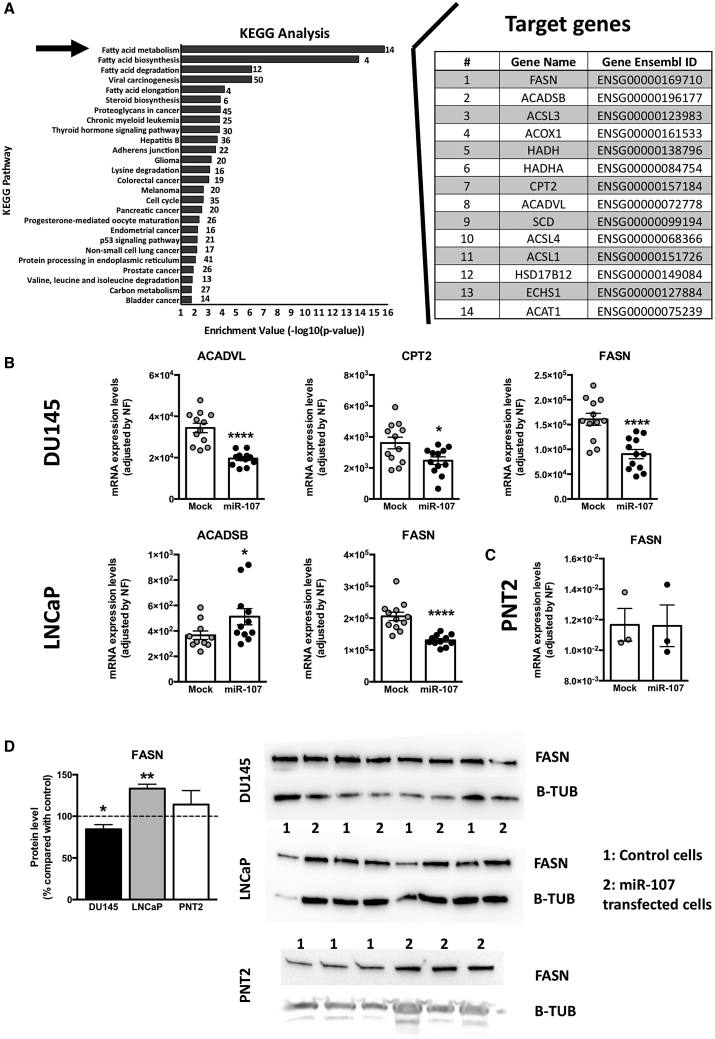
The predicted target genes of miR-107 were obtained by implementing different bioinformatics enrichment analyses to gain novel insights on the putative role of miR-107. Firstly, a Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis showed that miR-107 might be implicated in 26 pathways (Figure 4A, left panel). In fact, most of these pathways could be grouped in the main pathway observed, the fatty acid metabolism, which is represented by the putative targeting of 14 genes (Figure 4A, right panel). Complementary Gene Ontology (GO) analysis also confirmed that miR-107 might be involved, among others processes, in cellular lipid metabolic process (Figure 5A, black arrow). In order to further analyze the regulatory role of miR-107 in this complex metabolic pathway, the expression of these 14 genes was analyzed in LNCaP and DU145 cell lines overexpressing miR-107, being represented in Figure 4B the genes significantly altered (results of the genes that were not altered are shown in Figure S3). Remarkably, a dysregulation in acyl-coenzyme A (CoA) dehydrogenase (ACAD) chains was observed in these cells, resulting in an upregulation of the short chain (ACADSB) in LNCaP and a downregulation of the long chain (ACADVL) in DU145 (Figure 4B). A downregulation of carnitine palmitoyltransferase 2 (CPT2) was also observed in DU145 (Figure 4B). Remarkably, the main gene involved in this cellular pathway, fatty acid synthase (FASN), was significantly downregulated in both prostate cancer cell lines (Figure 4B), but not in the normal-like prostate cell model (PNT2; Figure 4C), in response to miR-107 overexpression. However, at protein level, a distinct regulation was observed in both PCa cell lines since FASN protein levels were increased in LNCaP and decreased in DU145 (Figure 4D). FASN protein levels were not altered in the normal-like prostate cell model (Figure 4D).
Figure 4.
Fatty acid metabolism pathway alteration by miR-107
(A) (Left) Data represent −log of the p value of each category and pathway. The arrow indicates fatty acid metabolism, the main pathway associated to miR-107. (Right) Representation of target genes of this pathway and gene ensemble ID are shown. (B) Effect of miR-107 overexpression in the modulation of the expression level of genes related to the fatty acid metabolism pathway in LNCaP and DU145 cell lines is shown. (C) Effect of miR-107 overexpression in the modulation of the expression level of FASN in the normal-like prostate cell model (PNT2) is shown. mRNA levels of (B) and (C) were determined by quantitative PCR and normalized by a normalization factor calculated from ACTB, GAPDH, and HPRT expression levels. (D) Representative western blots and quantification of FASN and B-TUB in LNCaP, DU145, and PNT2 cell lines after overexpression of miR-107 are shown. Data are expressed as percentage of control cells (set at 100%). In (B)–(D), values represent the mean ± SEM of at least n = 3 independent experiments. Asterisks indicate significant differences versus controls (∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001).
Figure 5.
GO terms associated with miR-107 target genes
(A) Data represent −log of the p value of each category and pathway. The black arrow indicates the association of miR-107 with the category of the cellular lipid metabolic process, while gray arrows indicate the association of miR-107 with the category of the splicing process. (B) Effect of miR-107 overexpression in the modulation of the expression level of key genes related with the category of splicing process in LNCaP and DU145 cell lines is shown. mRNA levels were determined by quantitative PCR and normalized by a normalization factor calculated from the expression levels of three housekeeping genes (ACTB, GAPDH, and HPRT). Values represent the mean ± SEM. Asterisks indicate significant differences versus controls (∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001).
Interestingly, GO analysis also showed that miR-107 might be also involved in the RNA splicing process (Figure 5A), a molecular event that we have recently demonstrated to be critically altered in PCa and associated with PCa aggressiveness.20 For this reason, we analyzed the expression of key components of the splicing machinery and associated splicing factors in LNCaP and DU145 cell lines overexpressing miR-107. Intriguingly, a profound dysregulation of several genes was observed in both cell lines (11 altered genes; Figure 5B).
miR-107 as a personalized non-invasive biomarker in the pathophysiological crosstalk between PCa and obesity
Based on the results obtained from the in silico and in vitro analyses linking the dysregulation of miR-107 with the metabolic milieu of PCa cells, we further analyzed the plasma levels of miR-107 in the patients included in cohort B, considering the metabolic status of the patients (i.e., obesity state). To that end, control and PCa patients from cohort B were subdivided according to their body mass index (BMI) in three different subgroups (normoweight [18.5 < BMI < 25], overweight [25 ≤ BMI < 30], and obese [BMI ≥ 30]), as shown in Table 3. This analysis showed a clear influence of the obese status in miR-107 plasma levels, especially in PCa patients (Figure 6A). Specifically, miR-107 levels were not affected by obesity in control patients, while in PCa patients, miR-107 levels were significantly higher in overweight and obese patients (Figure 6A, left panel). This was confirmed by the receiver operating characteristic (ROC) curve analysis, wherein the maximal difference, represented by an AUC = 0.73, was found in the comparison between obese PCa and obese control patients (Figure 6A, right panel).
Table 3.
General distribution of patients from validation cohort (cohort B) based on body mass index (BMI) (normoweight [BMI < 25], overweight [BMI ≥ 25 and <30], and obese [BMI ≥ 30])
| Control patients (n = 135) |
Prostate cancer patients (n = 160) |
p value | |||||
|---|---|---|---|---|---|---|---|
| Normoweighta | Overweightb | Obesityc | Normoweighta | Overweightb | Obesityc | ||
| n | 52 | 50 | 33 | 40 | 74 | 46 | |
| Age | 65.24 ± 6.31 | 67.3 ± 5.58 | 63.26 ± 8.12 | 65.65 ± 7.5 | 62.8 ± 5.12 | 62.43 ± 8.31 | 0.99a; 0.9b; 0.83c |
| BMI | 23.44 ± 1.46 | 27.8 ± 1.47 | 33.17 ± 2.73 | 23.83 ± 1.1 | 27.65 ± 1.13 | 32.76 ± 2.84 | 0.91a; 0.99b; 0.93c |
| PSA levels (ng/mL) | 7.66 ± 6.09 | 6.89 ± 3.98 | 6.64 ± 4.04 | 37.75 ± 127.5 | 32.35 ± 91.66 | 12.37 ± 23.98 | 0.26a; 0.29b; 0.99c |
| Sig PCa (n [%]) | – | – | – | 26 (65%) | 35 (47.29%) | 29 (63.04) | – |
Control patients represent subjects with suspect of prostate cancer but with a negative biopsy result. Data are represented as no. total, mean ± SD. or no. total (% [no./total]).
Refers to the comparison between normoweight control patients and normoweight prostate cancer patients.
Refers to the comparison between overweight control patients and overweight prostate cancer patients.
Refers to the comparison between obese control patients and obese prostate cancer patients.
Figure 6.
Circulating levels of miR-107 represent a personalized biomarker in the pathological association between PCa and obesity
(A) Plasma level and ROC curve analysis of miR-107 comparing plasma samples from PCa and control patients categorized in normoweight, overweight, and obese groups (cohort B). a, b, and c indicate significant differences compared with normoweight (NW), overweight (OW), and obese OB) control groups, respectively (a, b, c, p < 0.05; aa, bb, cc, p < 0.01). (B) Plasma level and ROC curve analysis of PSA and miR-107 comparing plasma sample from PCa and control patients included in the gray zone of PSA subdividing in NW, OW, and OB groups (cohort B) is shown. Asterisks indicate significant differences between compared groups (∗p < 0.05; ∗∗p < 0.01). (C) Plasma level and ROC curve analysis of miR-107 comparing between control, non-significant, and significant PCa patients and subdividing in NW, OW, and OB groups (cohort B) are shown. a and b indicate significant differences compared with control and non-significant PCa groups, respectively (a, b, p < 0.05; aaa, p < 0.001; aaaa, p < 0.0001). GZ means gray zone. Data represent mean ± SEM.
Moreover, when considering those patients included in the gray zone of the PSA (3–10 ng/mL), plasma miR-107 levels exhibited significantly improved ROC curve analysis compared with PSA, when these comparative analyses were carried out in overweight (n = 100; Figure 6B, middle right panel) and obese (n = 69; Figure 6B, bottom right panel) conditions, but not in the normoweight condition (n = 69; Figure 6B, top right panel). Notably, the highest statistical difference with the ROC curve analyses was observed when comparing PSA and miR107 in obese PCa patients (Figure 6B, right panels). Specifically, we observed an increasing AUC of the miR-107 ROC curve analysis dependent on the obesity state (AUC normoweight [NW] = 0.53, overweight [OW] = 0.65, and obese [OB] = 0.74), a result that was not observed on the PSA ROC curve analyses (AUC NW = 0.68, OW = 0.51, and OB = 0.52; Figure 6B, right panels).
Remarkably, when comparing PCa patients stratified by Gleason score and obese status, we found that the levels of miR-107 were significantly higher in obese-SigPCa patients compared with obese-NonSigPCa patients (Figure 6C, bottom panel). In fact, obese patients with SigPCa exhibited the highest AUC in the ROC analysis performed herein (AUC = 0.8025; p < 0.0001; Figure 6C). In this sense, the plasma levels of miR-107 were able to discriminate between SigPCa and NonSigPCa in obese patients, exhibiting a potential prognostic value (Figure 6C).
Discussion
Plasma PSA levels remain the current gold-standard biomarker to diagnose PCa, which represents one of the tumor types with the highest incidence worldwide.21 Unfortunately, PSA continues to show important limitations (especially in the range of 3–10 ng/mL, also named the “gray zone”), including compromised specificity, inasmuch as non-tumor conditions (e.g., infections and inflammation) can also increase PSA levels. Therefore, considerable research efforts have been focused on the identification of novel biomarkers that could complement or even replace plasma PSA in order to improve the diagnosis of PCa. In this line, we explored herein for the first time the dysregulation of the whole known miRNome in human plasma samples from PCa patients compared with those from healthy volunteers in order to identify novel and useful personalized diagnostic and prognostic biomarkers and potential therapeutic tools in PCa.
In particular, results from the analysis of the whole miRNome demonstrated a significant dysregulation of the plasmatic levels of 104 miRNAs, wherein six of them exhibited a great capacity to discriminate between PCa patients and healthy volunteers. The dysregulation of these miRNAs in PCa patients further supports previous data reporting the alteration of some of these miRNAs, including miR-107,9,10,22, 23, 24, 25 in certain studies. However, any of these studies have explored its putative role as a diagnostic biomarker as well as prognostic or therapeutic tool. Interestingly, the dysregulation of miR-107 and miR-191-5p was corroborated by qPCR in a second, ampler, and independent cohort of subjects (n = 295). The fact that the dysregulation of other miRNAs was not corroborated may be explained by the fact that control patients from this ampler cohort are patients with suspect of PCa but negative results in the biopsy instead of healthy volunteers, as used in cohort A. Nonetheless, we demonstrated that diagnostic capacity of plasma miR-107 levels to discriminate between tumor and control patients significantly outperformed that of PSA levels. Most importantly, this improvement in the diagnostic ability of miR-107 persists when comparing only patients in the gray zone (wherein the capacity of PSA is extremely low). Remarkably, although miR-107 levels were higher in SigPCa patients compared with controls, these levels did not allow to discriminate between SigPCa and NonSigPCa patients; however, miR-107 levels correlated with relevant oncogenic parameters, such as PSA levels,26 tumor volume,27 testosterone levels,28 and CRP29 in SigPCa patients, but not in NonSigPCa patients, reinforcing also a putative prognostic capacity of plasma miR-107 levels.
Despite miR-107 plasma levels being increased in PCa patients, this miRNA exhibits an apparently controversial reduced expression in PCa tissue samples compared with their non-tumoral adjacent tissues as well as in all PCa cell lines analyzed compared with a non-tumor prostate cell line. These results are consistent with a previous study, which also suggests a tumor suppressor role of miR-107 in PCa cells.25 However, it has not been demonstrated the reason why the levels of this miRNA increase in plasma samples from PCa patients while their levels are reduced in PCa cells. Here, we found that this discrepancy could be possibly explained by the higher capacity of PCa cells to secrete miR-107 to the extracellular medium compared with control prostate cells (possibly through a direct extracellular vesicle release and/or by a PCa-derived exosomal miRNA release), resulting in a lower level of miR-107 in PCa tissues compared with the plasma levels. These results are in line with several studies demonstrating the same cellular phenomenon in the case of other miRNAs.30,31 Moreover, this is also consistent with the previously suggested tumor suppressor role of miR-107 in PCa,25 which was corroborated in the present study. Indeed, when miR-107 is overexpressed in androgen-independent PCa cells (i.e., DU145), several oncogenic features, including cell proliferation, migration, and tumorosphere formation, were significantly reduced. However, it should be noted that the overexpression of miR-107 in an androgen-dependent PCa cell line (i.e., LNCaP) triggered an increase in cell proliferation and tumorosphere formation. These results obtained from androgen-dependent versus androgen-independent PCa cells have not been reported before and suggest the idea that miR-107 could play a different endogenous role in PCa cells, depending on the degree of tumor aggressiveness, maybe due to the sophisticated molecular mechanism of regulation of miRNAs in different cell types.5 Therefore, although differential responses in androgen-dependent versus androgen-independent PCa cells have been previously reported,32, 33, 34 this particular phenomenon observed in our study warrants further investigation in order to unveil its clinical implication.
Interestingly, more profound in silico (KEGG analysis) and in vitro studies demonstrated for the first time that miR-107 is implicated in the modulation of fatty acid metabolism, one of the main energy pathways used by tumor cells and whose dysregulation represents a hallmark of PCa.35 In this sense, we observed several transcriptional dysregulations in genes implicated in this pathway in response to the overexpression of miR-107 in LNCaP and DU145, such as ACADSB, ACADVL, and CPT2. Remarkably, only the expression level of FASN, the main driver of fatty acid metabolism,36 was decreased at mRNA level in both cancer cell lines. However, it should be noted that the protein levels of FASN were decreased in the androgen-independent PCa cell line DU145, while its levels were increased in LNCaP. Notably, these alterations could explain, at least in part, the differential functional in vitro results previously discussed in DU145 versus LNCaP cells, as FASN is a well-known oncogenic driver.37,38 Interestingly, the alteration in the expression of FASN (at mRNA and protein levels) observed in both prostate cancer cell models in response to the overexpression of miR-107 was not found in the normal-like prostate cell model (PNT2). These results might suggest that the association of miR-107 with the cellular lipid metabolic process could be specific of prostate cancer cells that reinforce the idea that this metabolic cellular process could be one of the main energy pathways used by prostate tumor cells to enhance their aggressiveness features. Likewise, the in silico GO analysis revealed that miR-107 could be also associated with another relevant cellular process, the RNA splicing, which has been recently reported by our laboratory to be tightly implicated in PCa aggressiveness.39 In support of this idea, we found that several relevant splicing-related genes (such as PTBP1, SRRM1, and SRSF6) were significantly altered in response to miR-107 overexpression in prostate cancer cells. Although more studies are necessary to better understand the cellular and molecular implication of miR-107 in PCa, our results clearly suggest that the dysregulation of the expression of this miRNA triggers defects in the splicing process but especially in the fatty acid metabolism (possible due to the dysregulation of FASN), which are two relevant cellular and molecular processes that represent hallmarks of cancer.40,41 Therefore, these observations unveiled new conceptual and functional avenues, with potential therapeutic implications, which are worth to be explored in future studies.
Finally, our study also revealed that miR-107 is tightly associated with obese condition in patients, which suggests its putative role as a personalized diagnostic biomarker based on the obese status of the patients. In this sense, it should be emphasized that PCa is strongly influenced by metabolic dysregulations, including obesity.17,42 Particularly, we observed that the diagnostic capacity of miR-107 significantly increased together with the increase of BMI, reaching its higher AUC value when comparing obese PCa patients versus obese control patients. Furthermore, miR-107 significantly outperformed the diagnostic capacity of the gold-standard PSA, especially when considering patients in the gray zone of PSA. Indeed, in the case of patients with obesity, miR-107 did not only discriminate between tumor and non-tumor patients but also between NonSigPCa and SigPCa, which further reinforces its potential value as prognostic biomarker for this pathology.
In conclusion, this study represents the first demonstration that the plasma levels of miR-107 might represent a useful personalized diagnostic biomarker of PCa since its levels are increased in plasma from PCa patients compared with control subjects using two independent cohorts. In addition, we found that plasma miR-107 levels are associated with key oncogenic parameters, such as PSA levels, tumor volume, testosterone, or CRP levels, suggesting that high plasma miR-107 levels could also be related to PCa aggressiveness and progression. Moreover, in vitro and in silico data revealed that miR-107 is implicated in the regulation of the splicing process and the fatty acid metabolism, altering the expression of the main driver FASN, resulting in a reduction of aggressiveness features in androgen-independent cells. Interestingly, this study also shows novel evidence demonstrating that miR-107 could represent a promising personalized biomarker for PCa, in that miR-107 levels were higher in plasma from obese patients with PCa compared with PCa patients with normoweight. Indeed, miR-107 also allowed a strong discriminatory capacity between SigPCa and NonSigPCa in obese PCa patients, thus representing not only a diagnostic but also a potential prognostic biomarker in that an obesity condition has been reported to represent a risk factor for PCa development, aggressiveness, and mortality.43,44 Altogether, our results provide new, compelling evidence supporting the contention that miR-107 represents a promising diagnostic, prognostic, and/or therapeutic tool, worth to be further explored, in the pathological association between PCa and obesity.
Materials and methods
Patients and samples
The study protocol was approved by the Reina Sofia University Hospital Ethics Committee, according to institutional and Good Clinical Practice guidelines (protocol number 20052020) and in compliment with the Declaration of Helsinki. Informed consent was obtained from all patients or their relatives. Plasma samples from two cohorts of male patients were collected: (1) cohort A or discovery cohort (n = 37; Table 1) divided in controls (healthy volunteers, n = 18) and PCa patients (n = 19) and (2) cohort B or validation cohort (n = 295; Table 2) divided in controls (patients with suspect of PCa but with a negative result in the biopsy; n = 135) and PCa patients (n = 160; who also were divided in patients with non-significant PCa [NonSigPCa; defined as Gleason score of 6 in the biopsy; n = 70] or with significant PCa [SigPCa; defined as Gleason score ≥7 on the biopsy; n = 90]). In addition, patients included in cohort B were also classified according to their BMI in normoweight (BMI < 25), overweight (BMI ≥ 25 and <30), and obese (BMI ≥ 30) for additional analyses as shown in Table 3. All samples were obtained through the Andalusian Biobank (Nodo Cordoba, Servicio Andaluz de Salud, Spain). Inclusion and exclusion criteria have been reported previously.45 This represents a retrospective analysis wherein patients were enrolled between 2013 and 2015 by consecutive recruitment of individuals with suspicion of PCa that underwent a transrectal ultrasound (TRUS)-guided prostate biopsy according to clinical practice in the Urology Service of Reina Sofia Hospital (Córdoba, Spain). Specifically, blood and plasma samples were collected early in the morning after an overnight fast and just before the prostate biopsy. Recommendations for biopsy indication were suspicious findings on digital rectal examination (DRE), PSA > 10 ng/mL, or PSA 3–10 ng/mL if free PSA ratio was low (usually <25%–30%) and in patients with previous biopsies, a persistent suspicion of PCa (i.e., persistently elevated PSA, suspicious DRE, etc.). For transrectal prostate biopsy, 12 biopsy cores were obtained from patients undergoing the first biopsy procedure and a minimum of 16 biopsy cores for those who had a previous biopsy. All biopsy specimens were analyzed by experienced urologic pathologists according to the International Society of Urological Pathology 2005 modified criteria.46
Table 1.
General characteristics of the samples included in the discovery cohort (cohort A)
| Control patients | Prostate cancer patients | p value | |
|---|---|---|---|
| n | 18 | 19 | |
| Age | 59.61 ± 7.29 | 67 ± 8.01 | 0.90 |
| Body mass index (BMI) | 27.73 ± 3.72 | 28.76 ± 3.85 | 0.41 |
| PSA levels (ng/mL) | 0.7 ± 0.41 | 5.51 ± 2.11 | <0.0001 |
Control patients represent healthy volunteers who donated blood samples. Data are represented as mean ± SD.
Table 2.
General characteristics of the samples included in the validation cohort (cohort B)
| Control patients | Prostate cancer patients | p value | |
|---|---|---|---|
| n | 135 | 160 | |
| Age | 66 ± 5.67 | 64 ± 7.21 | 0.90 |
| BMI | 27.43 ± 0.36 | 28.16 ± 0.3 | 0.11 |
| PSA levels (ng/mL) | 7.13 ± 0.42 | 27.87 ± 7.14 | 0.008 |
| Sig PCa (n [%]) | – | 90 [56.25%] | – |
| Tumor volume (cm3) | – | 35.64 ± 1.14 | – |
| Testosterone (ng/mL) | 5.30 ± 0.17 | 4.95 ± 0.13 | 0.11 |
| CRP (mg/L) | 4.05 ± 0.59 | 5.37 ± 0.95 | 0.26 |
Control patients represent subjects with suspect of prostate cancer but with a negative biopsy result. Data are represented as mean ± SD or no. total (% [no./total]). BMI, body mass index; CRP, C-reactive protein; PSA, prostate-specific antigen; Sig PCa, significant prostate cancer.
In addition, a set of available formalin-fixed paraffin-embedded (FFPE) tissue samples were used from a cohort of 84 patients with clinically localized PCa subjected to radical prostatectomies (PCa tumor regions [n = 84] and their adjacent non-tumor region [used as control tissues; n = 84] isolated by expert urologic pathologists; cohort C; Table 4) to isolate RNA and perform gene expression analyses, as previously reported.47,48
Table 4.
Demographic, biochemical, and clinical parameters of the patients with low aggressive PCa (cohort C)
| General characteristic | |
|---|---|
| n | 84 |
| Age, years (median [interquartile range (IQR)]) | 61 (57–66) |
| PSA levels, ng/mL (median [IQR]) | 5.2 (4.2–8.0) |
| Sig PCa (n [%]) | 76 (90.5%) |
| pT ≥ 3a (n [%]) | 59 (70.2%) |
| PI (n [%]) | 72 (85.7%) |
| VI (n [%]) | 8 (9.52%) |
| Recurrence (n [%]) | 35 (41.7%) |
| Metastasis (n [%]) | 0 (0%) |
PI, perineural invasion; pT, pathological primary tumor staging; VI, vascular invasion.
Determination of plasma PSA, testosterone, and C-reactive protein (CRP) levels
As previously reported,29,39 measurement of PSA, testosterone, and CRP levels were performed in the laboratory service of the Reina Sofia University Hospital of Cordoba using technology of Chemiluminescent Microparticle Immunoassays (References 7k70, 7k73, and 6k26-30/41, respectively; Abbott, Madrid, Spain), following the manufacturer's instructions.
miRNA extraction and retrotranscription
All plasma and FFPE samples were processed using the Maxwell 16 miRNA Tissue Kit and Maxwell 16 miRNA FFPE kit (Promega, Wisconsin, USA) respectively, and extracted using the Maxwell 16 MDx Instrument (Promega) in order to isolate and purify miRNAs as previously described.49 The quality and concentration of RNA extracted from these samples were evaluated using Nanodrop One Spectrophotometer (Thermo Fisher Scientific). Retrotranscription of these samples was performed using the miRCURY LNA RT Kit (Qiagen, Hilden, Germany), following manufacturer instructions.
Total RNA from the human normal-like, prostate-derived PNT2 and the PCa-derived LNCaP, 22Rv1, PC-3, and DU145 cell lines was isolated using TRI Reagent (Sigma-Aldrich, Madrid, Spain), followed by DNAse treatment using the RNase-Free Dnase Kit (Qiagen), as previously reported.50 The amount and purity of RNA recovered was determined using the Nanodrop One Spectrophotometer (Thermo Scientific). One microgram of RNA was retrotranscribed to cDNA, using random hexamer primers and the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). To analyze the expression level of miR-107, retrotranscription of the samples was performed using the miRCURY LNA RT Kit (Qiagen), following the manufacturer instructions.
Moreover, miRNAs secreted by PNT2, LNCaP, and DU145 cells were evaluated by isolating miRNAs from the cell culture media (conditioned media) using the miRNeasy Serum/Plasma kit (Qiagen) following the manufacturer's protocol. Retrotranscription of these samples was performed using the miRCURY LNA RT Kit (Qiagen) as described below.
miRNome analysis
Circulating levels of the whole miRNome were determined and compared between control (healthy volunteers; n = 18) and PCa patients (n = 19) from cohort A using 50 ng of the extracted plasma miRNAs and the Affymetrix miRNA 4.1 array (Affymetrix, Santa Clara, USA), as previously described.49 This array is able to detect all mature human miRNAs identified to date (total of 2,578 miRNAs). Biotin-labeled RNA was synthesized using the FlashTag Biotin HSR RNA Labeling Kit (Thermo Fisher Scientific). Each sample was hybridized onto a GeneChip miRNA 4.1 Array Plate (Thermo Fisher Scientific) following the manufacturer's protocol.
In order to determine the most stably expressed miRNA and therefore the best housekeeping gene for our study, several standard criteria were established: (1) sufficiently high levels in order to allow its quantification by qPCR; (2) lower difference in the levels between different experimental groups, attending to higher (closer to 1) p value; and (3) lower standard error of the mean. According to these parameters, the results revealed that miR-16-5p was the most stable miRNA and the most appropriate housekeeping for the normalization of miRNA levels in these samples. Subsequently, the most altered miRNAs in plasma from PCa patients compared with healthy volunteers were selected according to lower fold-change p value (<0.0001) and higher AUC of the ROC curve analysis.
Analysis of miRNA levels by quantitative real-time PCR
Quantitative real-time PCR was used to determine the levels of selected miRNAs in the validation cohort of human patients (cohort B), in the different human cell lines, and in the conditioned media of the specific cell lines mentioned above. Particularly, to validate the previous results obtained from the Affymetrix miRNA 4.1 array, the levels of six miRNAs (let-7d-5p, miR-24-3p, 26a-5p, 103a-3p, 107, and 191-5p; based on a p < 0.0001 and ROC curve with AUC = 1) were measured. Specific primers for these miRNAs (miRCURY miRNA assay, Qiagen) were used in combination with GoTaq qPCR Master Mix (Promega) using the Stratagene Mx3000P (Agilent Technologies, Madrid, Spain), following manufacturer's instructions. mRNA levels were normalized according to the levels of miR-16-5p, whose levels did not significantly vary among the different experimental groups in both human cohorts A and B (data not shown). Similar approaches were used to determine the expression of miR-107 in the different cell lines (PNT2, LNCaP, 22Rv1, PC-3, and DU145) as well as in the conditioned media collected from PNT2, LNCaP, and DU145 cells. In these cases, data were normalized using RNU6 expression, the most accepted housekeeping for tissues and cells.
Cell cultures, reagents, and transfection with miR-107
PNT2 was a kindly gift from Prof. De Bono lab (London), and LNCaP, 22Rv1, PC-3, and DU145 cell lines were obtained from ATCC, cultured, and maintained under manufacturer's recommendations, as previously reported.18,20 These cell lines were validated by analysis of short tandem repeats (GenePrint 10 System; Promega, Barcelona, Spain) and systematically checked for mycoplasma contamination.49,51
Moreover, LNCaP and DU145 cells were seeded onto 12-well tissue culture plates and serum starved for 24 h previous to the transfection with miR-107 Mimic (1 nM; Qiagen, Hilden, Germany) using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, USA) and following the guidelines provided by the manufacturer. Proliferation, migration, and tumorospheres assays were performed in transfected (mock versus miR-107 overexpression) cells. Transfected cells were collected for RNA and protein isolation. Conditioned media (incubated during 24 h) from transfected cells were also collected for miRNA isolation.
Cell proliferation, migration, and tumorospheres formation assays in response to miR-107 overexpression
Cell proliferation in LNCaP and DU145 cells was evaluated using Alamar-Blue assay (Bio-Source International, Camarillo, CA, USA), as previously reported.20,52 Briefly, cells were seeded in 96-well culture plates at a density of 3,000–5,000 cells/well and serum starved for 24 h. Then, fluorescence (560 nm) was evaluated using the FlexStation III system (Molecular Devices, Sunnyvale, CA, USA) after 3 h of incubation with Alamar-Blue compound at 10%. Cell proliferation was measured at 24, 48, and 72 h after miR-107 overexpression. Cell culture media was replaced by Alamar-Blue free fresh media after each measurement.
Cell migration was evaluated by wound-healing assay in DU145 cells, as previously reported.20 Briefly, cells were serum starved for 24 h to achieve cell synchronization, and then the “wound” was made using a 200-μL sterile pipette tip. Wells were rinsed with sterile PBS, and cells were then incubated for 48 h with supplemented medium without FBS. Wound healing was compared with the area just after the wound was performed. Images were acquired along the “wound” to calculate the area by ImageJ software. Results were expressed as percentage referred to control.
Tumorospheres formation assay was carried out in LNCaP and DU145 cells cultured in a Corning Costar ultra-low attachment plate (no. CLS3473) with DMEM F-12 (Gibco, no. 11320033) with epidermal growth factor (EGF) (20 ng/μL; no. SRP3027) for 10 days (refreshed every 48 h), as previously reported.47 After 10 days of incubation, an inverted microscope coupled to a digital camera was used to take photographs to visualize and measure tumorospheres morphology and area in order to calculate the number of generated tumorospheres.
qPCR dynamic array based on microfluidic technology
A qPCR dynamic array based on microfluidic technology (Fluidigm, San Francisco, CA, USA) was used to simultaneously determine the gene expression of 14 components of the fatty acid metabolism, as well as of different components of the major spliceosome (n = 13), minor spliceosome (n = 4), and associated splicing factors (n = 14) in LNCaP and DU145 cell lines overexpressing miR-107 (Table S1). The expression of FASN was also determined in the prostate normal-like cell model (PNT2) in response to miR-107 overexpression. Moreover, the expression of three housekeeping genes (ACTB, GAPDH, and HPRT) was analyzed in the same samples. Specific primers for these transcripts were specifically designed with the Primer3 software and StepOne Real-Time PCR System software v.2.3 (Applied Biosystems, Foster City, CA, USA). Preamplification, exonuclease treatment, and qPCR dynamic array based on microfluidic technology were implemented as previously reported,49,52 following manufacturer's instructions, using the Biomark System and the Real-Time PCR Analysis Software (Fluidigm).
Western blotting
Prostate cancer cells (LNCaP and DU145) and the prostate normal-like cell model (PNT2) were processed to analyze protein levels by western blot after transfection with miR-107 using methods previously reported.20,53 Specifically, 200,000 cells were seeded in 12-well plates, and 48 h after transfection, proteins were collected using prewarmed (65°C) SDS-dithiothreitol (DTT) buffer (62.5 mM Tris-HCl, 2% SDS, 20% glycerol, 100 mM DTT, and 0.005% bromophenol blue). Then, proteins were sonicated for 10 s and boiled for 5 min at 95°C. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (Millipore, Billerica, MA). Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline/0.05% Tween 20 and incubated overnight with the specific antibodies for FASN (sc-55580 mouse monoclonal immunoglobulin G [IgG], Santa Cruz Biotechnology) and β-tubulin (rabbit monoclonal antibody [mAb] 2128S, Cell Signaling Technology) and secondary horseradish peroxidase (HRP)-conjugated antibodies (anti-mouse IgG no. 7076S and anti-rabbit IgG antibody no. 7074S, Cell Signal). Proteins were detected using an enhanced chemiluminescence detection system (GE Healthcare, Madrid, Spain). Densitometric analysis of the bands obtained was carried out with ImageJ software.
In silico analyses
The GO enrichment analysis for biological process and KEGG pathway were generated by DIANA-miRPath v.3.0. Predicted miR-107 target genes were obtained from top KEGG pathway and evaluated using the different in vitro approaches described above.
Statistical analysis
Kolmogorov-Smirnov test was used to analyze the normality of the datasets. Parametric data were compared by two-tailed t test, while nonparametric data were compared by Mann-Whitney test. AUC from ROC curves were compared by DeLong test.54 Correlations were studied using Spearman's correlation test. Representation of volcano plot and clustering analysis by heatmaps were created using MetaboAnalyst.55 All data were obtained from at least three independent experiments from different cellular passages and expressed as mean ± SEM. p values lower than 0.05 were considered statistically significant. All statistical analyses were performed using the GraphPad Prism 6 (La Jolla, CA, USA) and SPSS v.17.0 (SPSS, Chicago, IL, USA).
Acknowledgments
This work was funded by MINECO/MECD (PID2019-105564RB-I00, FPU16/06190, FPU17/00263, FPU18/02485, FPU18/06009, and PRE2020-094225), University of Córdoba (Programa Operativo FEDER Andalucía: 27416), Junta de Andalucía (BIO-0139, PI-0094-2020, and P20_00442), European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 847468, and CIBERobn. CIBER is an initiative of Instituto de Salud Carlos III, Ministerio de Sanidad, Servicios Sociales e Igualdad, Spain.
Author contributions
V.H.-A., P.S.-M., J.M.J.V., A.D.H.-M., A.S.-C., M.D.G., and R.M.L. designed the project. V.H.-A., P.S.-M., J.M.J.V., M.T.M.-M., A.J.M.-H., J.M.P.-G., J.L.L.-C., F.P.-P., and A.I.-C. carried out the experiments and analyzed data. J.C.-V., F.J.A., E.G.-G., and E.M.Y.-S. organized, collected, and analyzed human samples. A.D.H.-M. and A.S.-C. performed formal analysis. M.D.G. and R.M.L. supervised and acquired funding resources. V.H.-A., P.S.-M., J.M.J.V., M.D.G., and R.M.L. wrote the original draft. All authors revised and edited the submitted manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.02.010.
Contributor Information
Manuel D. Gahete, Email: bc2gaorm@uco.es.
Raúl M. Luque, Email: raul.luque@uco.es.
Supplemental information
References
- 1.Culp M.B., Soerjomataram I., Efstathiou J.A., Bray F., Jemal A. Recent global patterns in prostate cancer incidence and mortality rates. Eur. Urol. 2020;77:38–52. doi: 10.1016/j.eururo.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Ross T., Ahmed K., Raison N., Challacombe B., Dasgupta P. Clarifying the PSA grey zone: the management of patients with a borderline PSA. Int. J. Clin. Pract. 2016;70:950–959. doi: 10.1111/ijcp.12883. [DOI] [PubMed] [Google Scholar]
- 3.Loeb S., Bjurlin M.A., Nicholson J., Tammela T.L., Penson D.F., Carter H.B., Carroll P., Etzioni R. Overdiagnosis and overtreatment of prostate cancer. Eur. Urol. 2014;65:1046–1055. doi: 10.1016/j.eururo.2013.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catela Ivkovic T., Voss G., Cornella H., Ceder Y. microRNAs as cancer therapeutics: a step closer to clinical application. Cancer Lett. 2017;407:113–122. doi: 10.1016/j.canlet.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 6.Balzano F., Deiana M., Dei Giudici S., Oggiano A., Baralla A., Pasella S., Mannu A., Pescatori M., Porcu B., Fanciulli G., et al. miRNA stability in frozen plasma samples. Molecules. 2015;20:19030–19040. doi: 10.3390/molecules201019030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji C., Guo X. The clinical potential of circulating microRNAs in obesity. Nat. Rev. Endocrinol. 2019;15:731–743. doi: 10.1038/s41574-019-0260-0. [DOI] [PubMed] [Google Scholar]
- 8.Huang X., Yuan T., Liang M., Du M., Xia S., Dittmar R., Wang D., See W., Costello B., Quevedo F., et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015;67:33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzo M., Berti G., Russo F., Fazio S., Evangelista M., D’Aurizio R., Pellegrini M., Rainaldi G. Discovering the miR-26a-5p targetome in prostate cancer cells. J. Cancer. 2017;8:2729–2739. doi: 10.7150/jca.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge J., Mao L., Xu W., Fang W., Wang N., Ye D., Dong Z., Guan H., Guan C. miR-103a-3p suppresses cell proliferation and invasion by targeting tumor protein D52 in prostate cancer. J. Invest. Surg. 2020;34:1–9. doi: 10.1080/08941939.2020.1738602. [DOI] [PubMed] [Google Scholar]
- 11.Brase J.C., Johannes M., Schlomm T., Fälth M., Haese A., Steuber T., Beissbarth T., Kuner R., Sültmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer. 2011;128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 12.Corcoran C., Rani S., O’Driscoll L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate. 2014;74:1320–1334. doi: 10.1002/pros.22848. [DOI] [PubMed] [Google Scholar]
- 13.Fabris L., Ceder Y., Chinnaiyan A.M., Jenster G.W., Sorensen K.D., Tomlins S., Visakorpi T., Calin G. The potential of MicroRNAs as prostate cancer biomarkers. Eur. Urol. 2016;70:312–322. doi: 10.1016/j.eururo.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haldrup C., Kosaka N., Ochiya T., Borre M., Høyer S., Orntoft T.F., Sorensen K.D. Profiling of circulating microRNAs for prostate cancer biomarker discovery. Drug Deliv. Transl. Res. 2014;4:19–30. doi: 10.1007/s13346-013-0169-4. [DOI] [PubMed] [Google Scholar]
- 15.Kachakova D., Mitkova A., Popov E., Popov I., Vlahova A., Dikov T., Christova S., Mitev V., Slavov C., Kaneva R. Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015;34:189–200. doi: 10.1089/dna.2014.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yaman Agaoglu F., Kovancilar M., Dizdar Y., Darendeliler E., Holdenrieder S., Dalay N., Gezer U. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol. 2011;32:583–588. doi: 10.1007/s13277-011-0154-9. [DOI] [PubMed] [Google Scholar]
- 17.Jiménez-Vacas J.M., Montero-Hidalgo A.J., Gómez-Gómez E., Fuentes-Fayos A.C., Ruiz-Pino F., Guler I., Camargo A., Anglada F.J., Carrasco-Valiente J., Tena-Sempere M., et al. In1-ghrelin splicing variant as a key element in the pathophysiological association between obesity and prostate cancer. J. Clin. Endocrinol. Metab. 2021;106:e4956–e4968. doi: 10.1210/clinem/dgab516. [DOI] [PubMed] [Google Scholar]
- 18.Hormaechea-Agulla D., Gahete M.D., Jimenez-Vacas J.M., Gomez-Gomez E., Ibanez-Costa A., L-Lopez F., Rivero-Cortés E., Sarmento-Cabral A., Valero-Rosa J., Carrasco-Valiente J., et al. The oncogenic role of the In1-ghrelin splicing variant in prostate cancer aggressiveness. Mol. Cancer. 2017;16:146. doi: 10.1186/s12943-017-0713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L-Lopez F., Sarmento-Cabral A., Herrero-Aguayo V., Gahete M.D., Castano J.P., Luque R.M. Obesity and metabolic dysfunction severely influence prostate cell function: role of insulin and IGF1. J. Cell. Mol. Med. 2017;21:1893–1904. doi: 10.1111/jcmm.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiménez-Vacas J.M., Herrero-Aguayo V., Montero-hidalgo A.J., Gómez-Gómez E., Fuentes-Fayos A.C., León-González A.J., Sáez-Martínez P., Alors-Perez E., Pedraza-Arévalo S., González T., et al. Dysregulation of the splicing machinery is directly associated to aggressiveness of prostate cancer. EBioMedicine. 2020;51:102547–102551. doi: 10.1016/j.ebiom.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 22.Li X., Han X., Wei P., Yang J., Sun J. Knockdown of lncRNA CCAT1 enhances sensitivity of paclitaxel in prostate cancer via regulating miR-24-3p and FSCN1. Cancer Biol. Ther. 2020;21:452–462. doi: 10.1080/15384047.2020.1727700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramberg H., Alshbib A., Berge V., Svindland A., Taskén K.A. Regulation of PBX3 expression by androgen and Let-7d in prostate cancer. Mol. Cancer. 2011;10:50. doi: 10.1186/1476-4598-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X., Shi Z., Liu X., Su Y., Li W., Dong H., Zhao L., Li M., Wang Y., Jin X., et al. Upregulation of miR-191 promotes cell growth and invasion via targeting TIMP3 in prostate cancer. J. BUON. 2018;23:444–452. [PubMed] [Google Scholar]
- 25.Zhang X., Jin K., Luo J.D., Liu B., Xie L.P. MicroRNA-107 inhibits proliferation of prostate cancer cells by targeting cyclin E1. Neoplasma. 2019;66:704–716. doi: 10.4149/neo_2018_181105N825. [DOI] [PubMed] [Google Scholar]
- 26.Bañez L.L., Hamilton R.J., Partin A.W., Vollmer R.T., Sun L., Rodriguez C., Wang Y., Terris M., Aronson W., Presti J., Jr., et al. Obesity-related plasma hemodilution and PSA concentration among men with prostate cancer. JAMA. 2007;298:2275–2280. doi: 10.1001/jama.298.19.2275. [DOI] [PubMed] [Google Scholar]
- 27.Wenzel M., Würnschimmel C., Nocera L., Collà Ruvolo C., Tian Z., Shariat S.F., Saad F., Briganti A., Tilki D., Graefen M., et al. Overall survival after systemic treatment in high-volume versus low-volume metastatic hormone-sensitive prostate cancer: systematic review and network meta-analysis. Eur. Urol. Focus. 2021 doi: 10.1016/j.euf.2021.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Claps M., Petrelli F., Caffo O., Amoroso V., Roca E., Mosca A., Maines F., Barni S., Berruti A. Testosterone levels and prostate cancer prognosis: systematic review and meta-analysis. Clin. Genitourin. Cancer. 2018;16:165–175.e2. doi: 10.1016/j.clgc.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Gómez-gómez E., Carrasco-valiente J., Campos-hernández J.P., Maria A., Manuel B.J., Ruiz-garcía J., Valero-Rosa J., Luque R.M., Requena-Tapia M.J. Clinical association of metabolic syndrome , C - reactive protein and testosterone levels with clinically significant prostate cancer. J. Cell. Mol. Med. 2019;23:934–942. doi: 10.1111/jcmm.13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sohel M.M.H. Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci. 2020;248:117473. doi: 10.1016/j.lfs.2020.117473. [DOI] [PubMed] [Google Scholar]
- 31.Makarova J., Turchinovich A., Shkurnikov M., Tonevitsky A. Extracellular miRNAs and cell-cell communication: problems and prospects. Trends Biochem. Sci. 2021;46:640–651. doi: 10.1016/j.tibs.2021.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Wang G., Wang Y., Feng W., Wang X., Yang J.Y., Zhao Y., Wang Y., Liu Y. Transcription factor and microRNA regulation in androgen-dependent and -independent prostate cancer cells. BMC Genomics. 2008;9:S22. doi: 10.1186/1471-2164-9-S2-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muñoz-Fonseca M.B., Vidal-Limon A., Fernández-Pomares C., Rojas-Durán F., Hernández-Aguilar M.E., Espinoza C., Trigos A., Suárez-Medellín J. Ergosterol exerts a differential effect on AR-dependent LNCaP and AR-independent DU-145 cancer cells. Nat. Prod. Res. 2020;35:1–4. doi: 10.1080/14786419.2020.1737054. [DOI] [PubMed] [Google Scholar]
- 34.Cheng H., Tang S., Lian X., Meng H., Gu X., Jiang J., Li X. The differential antitumor activity of 5-Aza-2’-deoxycytidine in prostate cancer DU145, 22RV1, and LNCaP cells. J. Cancer. 2021;12:5593–5604. doi: 10.7150/jca.56709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X., Daniels G., Lee P., Monaco M.E. Lipid metabolism in prostate cancer. Am. J. Clin. Exp. Urol. 2014;2:111–120. [PMC free article] [PubMed] [Google Scholar]
- 36.Menendez J.A., Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 37.Fhu C.W., Ali A. Fatty acid synthase: an emerging target in cancer. Molecules. 2020;25:3935. doi: 10.3390/molecules25173935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang B., Zhang J., Zhang X., Yuan J., Shi Y., Qiao L. Inhibition of lipogenesis and induction of apoptosis by valproic acid in prostate cancer cells via the C/EBPα/SREBP-1 pathway. Acta Biochim. Biophys. Sin. (Shanghai). 2021;53:354–364. doi: 10.1093/abbs/gmab002. [DOI] [PubMed] [Google Scholar]
- 39.Gómez-Gómez E., Jiménez-Vacas J.M., Carrasco-Valiente J., Herrero-Aguayo V., Blanca-Pedregosa A.M., León-González A.J., Valero-Rosa J., Fernández-Rueda J.L., González-Serrano T., López-Miranda J., et al. Plasma ghrelin O-acyltransferase (GOAT) enzyme levels: a novel non-invasive diagnosis tool for patients with significant prostate cancer. J. Cell. Mol. Med. 2018;22:5688–5697. doi: 10.1111/jcmm.13845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 41.Oltean S., Bates D.O. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 42.Hsing A.W., Sakoda L.C., Chua S.J. Obesity, metabolic syndrome, and prostate cancer. Am. J. Clin. Nutr. 2007;86:s843–s857. doi: 10.1093/ajcn/86.3.843S. [DOI] [PubMed] [Google Scholar]
- 43.Rivera-Izquierdo M., Pérez de Rojas J., Martínez-Ruiz V., Pérez-Gómez B., Sánchez M.-J., Khan K.S., Jiménez-Moleón J.J. Obesity as a risk factor for prostate cancer mortality: a systematic review and dose-response meta-analysis of 280,199 patients. Cancers (Basel) 2021;13:4169. doi: 10.3390/cancers13164169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adesunloye B.A. Mechanistic insights into the link between obesity and prostate cancer. Int. J. Mol. Sci. 2021;22:3935. doi: 10.3390/ijms22083935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiménez-Vacas J.M., Gómez-Gómez E., Montero-hidalgo A.J., Herrero-Aguayo V., L-López F., Sánchez- Sánchez R., Guler I., Blanca A., Méndez-Vidal M.J., Carrasco J., et al. Clinical utility of ghrelin-O-acyltransferase ( GOAT ) enzyme as a diagnostic tool and potential therapeutic target in prostate cancer. J. Clin. Med. 2019;8:2056. doi: 10.3390/jcm8122056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Epstein J.I., Allsbrook W.C.J., Amin M.B., Egevad L.L. The 2005 international society of urological pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. Am. J. Surg. Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 47.Jiménez-Vacas J.M., Herrero-Aguayo V., Gómez-Gómez E., León-González A.J., Sáez-Martínez P., Alors-Pérez E., Fuentes-Fayos A.C., Martínez-López A., Sánchez- Sánchez R., González-Serrano T., et al. Spliceosome component SF3B1 as novel prognostic biomarker and therapeutic target for prostate cancer. Transl. Res. 2019;212:89–103. doi: 10.1016/j.trsl.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Hormaechea-Agulla D., Jimenez-Vacas J.M., Gomez-Gomez E., L-Lopez F., Carrasco-Valiente J., Valero-Rosa J., Moreno M.M., Sánchez- Sánchez R., Ortega-Salas R., Gracia-Navarro F., et al. The oncogenic role of the spliced somatostatin receptor sst5TMD4 variant in prostate cancer. FASEB J. 2017;31:4682–4696. doi: 10.1096/fj.201601264RRR. [DOI] [PubMed] [Google Scholar]
- 49.Herrero-Aguayo V., Jiménez-Vacas J.M., Sáez-Martínez P., Gómez-Gómez E., López-Canovas J.L., Garrido-Sánchez L., Herrera-Martínez A.D., García-Bermejo L., Macías-González M., López-Miranda J., et al. Influence of obesity in the miRNome : miR-4454 , a key regulator of insulin response via splicing modulation in prostate. J. Clin. Endocrinol. Metab. 2021;106:e469–e484. doi: 10.1210/clinem/dgaa580. [DOI] [PubMed] [Google Scholar]
- 50.Del Rio-Moreno M., Alors-Perez E., Gonzalez-Rubio S., Ferrin G., Reyes O., Rodriguez-Peralvarez M., Sánchez-Frías M.E., Sánchez- Sánchez R., Ventura S., López-Miranda J., et al. Dysregulation of the splicing machinery is associated to the development of nonalcoholic fatty liver disease. J. Clin. Endocrinol. Metab. 2019;104:3389–3402. doi: 10.1210/jc.2019-00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saéz-Martínez P., Jiménez-Vacas J.M., León-González A.J., Herrero-Aguayo V., Montero-hidalgo A.J., Gómez-Gómez E., Sánchez- Sánchez R., Requena-Tapia M.J., Castaño J.P., Gahete M.D., et al. Unleashing the diagnostic , prognostic and therapeutic potential of the neuronostatin/GPR107 system in prostate cancer. J. Clin. Med. 2020;9:1703. doi: 10.3390/jcm9061703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuentes-Fayos A.C., Vazquez M.C., Jiménez-Vacas J.M., Bejarano L., Pedraza-Arevalo S., López-L F., Blanco-Acevedo C., Sánchez-Sánchez R., Reyes O., Ventura S., et al. Splicing machinery dysregulation drives glioblastoma development/aggressiveness : oncogenic role of SRSF3. Brain. 2020;143:3273–3293. doi: 10.1093/brain/awaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiménez-Vacas J.M., Herrero-Aguayo V., Montero-Hidalgo A.J., Sáez-Martínez P., Gómez-Gómez E., León-González A.J., Fuentes-Fayos A.C., Yubero-Serrano E.M., Requena-Tapia M.J., López M., et al. Clinical , cellular , and molecular evidence of the additive antitumor effects of biguanides and statins in prostate cancer. J. Clin. Endocrinol. Metab. 2021;106:696–710. doi: 10.1210/clinem/dgaa877. [DOI] [PubMed] [Google Scholar]
- 54.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 55.Xia J., Wishart D.S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics. 2016;55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.