Abstract
Survival data with competing or semi-competing risks are common in observational studies. As an alternative to cause-specific and subdistribution hazard ratios, the between-group difference in cause-specific restricted mean times lost (RMTL) gives the mean difference in life expectancy lost to a specific cause of death or in disease-free time lost, in the case of a nonfatal outcome, over a pre-specified period. To adjust for covariates, we introduce an inverse probability weighted estimator and its variance for the marginal difference in RMTL. We also introduce an inverse probability of censoring weighted regression model for the RMTL. In simulation studies, we examined the finite sample performance of the proposed methods under proportional and nonproportional subdistribution hazards scenarios. We illustrated both methods with competing risks data from the Framingham Heart Study. We estimated sex differences in atrial fibrillation (AF)-free times lost over 40 years. We also estimated sex differences in mean lifetime lost to cardiovascular disease (CVD) and non-CVD death over 10 years among individuals with AF.
Keywords: competing risks, cumulative incidence, restricted mean survival, restricted mean time lost, survival analysis
1 |. BACKGROUND
The difference in restricted mean survival times (RMST) has recently become of great interest as an alternative measure of association to the hazard ratio in time-to-event analyses.1,2 Marginal between-group differences in RMST up to a given timepoint τ can be estimated nonparametrically by integration of the survival function or, in the presence of covariates, of the inverse probability weighted (IPW) survival function.3 Moreover, regression models have been introduced to estimate the difference in RMST conditional on covariates.4–6
Competing risks time-to-event data arise commonly in biomedical research, when a nonterminal event may be of interest but its observation is subject to a terminal event. Or, time to death from a specific disease may be of interest, but individuals can die from other causes. In this context, between-group comparisons for a specific event have been studied using the cumulative incidence, cause-specific hazard, or subdistribution hazard.7–9
In the presence of competing risks, the difference in RMST is inadequate. Andersen10 and Zhao et al11 proposed instead to measure the difference in restricted mean times lost (RMTL). With a single cause of failure, RMTL is equal to τ minus RMST. With competing risks, the RMTL is the area under the cause-specific cumulative incidence function. As a consequence, the difference in RMTL can be interpreted as the increase or decrease in lost life expectancy due to a specific cause of death or, in the case of a nonfatal outcome, the mean difference in disease-free time, up to time τ.
Methods for inference on the difference in RMTL with competing risks data and adjustment for covariates are scarce. Andersen described a generalized linear model for the cause-specific RMTL using the pseudo-observation technique.10 In this article, we propose an IPW-adjusted estimator of the difference in cause-specific RMTL. We also present a regression model for the RMTL conditional on covariates estimated with inverse probability of censoring weighting (IPCW). We provide R code to apply our methods with working examples and to replicate our simulation study on GitHub at https://github.com/s-conner/rmtl.
In Section 2, we discuss estimation of the difference in cause-specific RMTL, without adjustment for covariates. In Section 3, we present estimation of the marginal RMTL adjusted for covariates with IPW and its variance. In Section 4, we describe an IPCW regression model for RMTL conditional on covariates. In Section 5, we present a simulation study to assess the IPW estimator’s finite sample performance. In Section 6, we apply the proposed methods to incident atrial fibrillation (AF) and cause-specific mortality after AF with data from the Framingham Heart Study.
2 |. DIFFERENCE IN CAUSE-SPECIFIC RMTL
Without loss of generality, we focus on a terminal or nonterminal event of interest ϵ = 1, whose occurrence is subject to a competing event ϵ = 2. Our notations extend to scenarios with k competing types of event. Let T be the time to event, C be the censoring time, X = T ∧ C be the observed time to event, and δ = I(T ≤ C) be an indicator of noncensoring. We assume that C is independent of T. We observe (Xi, δiϵi) for individuals i =1, …, n.
The cumulative incidence function is defined as where is the overall survival function, αϵ(u) is the cause-specific hazard function for event ϵ, and is the cumulative all-cause hazard function. Let R = T ∧ τ be the restricted time to event. We define the cause-specific RMTL as the area under the cause-specific cumulative incidence function up to timepoint τ. In Appendix 1, we prove that . We estimate the RMTL by integrating the Aalen-Johansen estimator of the cumulative incidence function
| (1) |
We approximate Equation (1) with Riemann sums, where tj : t1, …, tD are the ordered, unique event times prior to τ and tD+1 = τ. In Appendix 2, we show the variance of the RMTL can be estimated by
| (2) |
With two groups defined by A =1 and A =0, the difference in cause-specific RMTL is given by , with variance .
With a single group, the timepoint τ should be less than or equal to the greatest observed time, τ ≤ max(Xi), and may be chosen clinically or empirically, for example, the 95th percentile of observed times or the maximum observed time, max(Xi), which approximates infτ{P(C ≥ τ) = 0}.12 When taking the difference in RMTL between groups, τ should be less than or equal to the minimum of the largest event time across groups, τ ≤ min{max(Xi|Ai = 1), max(Xi|Ai = 0)}. Tian et al12 demonstrated max(Xi) may be an inappropriate choice for τ if nearby observed times are sparse, as evidenced by a flat tail in the cumulative incidence curve, and should be chosen with caution.
3 |. ADJUSTED DIFFERENCE IN CAUSE-SPECIFIC RMTL
In observational studies, we must adjust for covariates. Here, we demonstrate how to estimate the between-group difference in cause-specific RMTL adjusted for covariates. We extend Equation (1) by considering the area under the cumulative incidence function adjusted for covariates with IPW.
We again consider two groups defined by A = 1 and A = 0. Let Z denote a n × p matrix of covariates and pia = P(Ai = a|Zi). So pi1 is the propensity score for exposure. The inverse probability weight is defined as . Then, the IPW Aalen-Johansen estimator of the cumulative incidence function in exposure group A = a is given by
where tj : t1, …, tD are the ordered, unique event times prior to τ in group a, , is the weighted number of events of cause k in group a, is the increment in weighted events in group a, is the weighted number at risk in group a, and is the weighted overall survival function for being alive and event-free in group a.13–15 We define the estimated variance and covariance of the IPW Aalen-Johansen estimator as
and
where
and, for tj < tk,
where is the effective sample size, and assuming .
We estimate the RMTL in exposure group a by integrating the IPW Aalen-Johansen estimator of the cumulative incidence function
| (3) |
We approximate Equation (3) as . We estimate the variance of the IPW estimator of the RMTL in exposure group a by
| (4) |
Then, the adjusted difference in RMTL between exposure groups A =1 and A =0 is
with variance
assuming the weights are known. In practice, we estimate pia when deriving the weights wi used in the quantities introduced above.
4 |. INVERSE PROBABILITY OF CENSORING WEIGHTED GENERALIZED LINEAR MODEL FOR THE RMTL
We describe an IPCW regression model to assess associations between covariates and the cause-specific RMTL.6 Let and Z* = (1, A, Z) denote the n × (p + 2) matrix of predictors allowing an intercept term. We can model the RMTL for event ϵ = 1 with a generalized linear model,
with link function g(·). Model coefficients β represent differences in cause-specific RMTL when using the identity link, and log ratios of cause-specific RMTL when using the log link. Tian et al6 also suggested a logit-like link function, , which restricts the predicted RMTL to (0, τ).
Coefficients are estimated with the IPCW technique by solving the weighted estimating equation,
| (5) |
where is the Kaplan-Meier estimator of the noncensoring distribution G(t). The outcome for each uncensored individual is either τ − Ri or 0 due to the event type indicator, I(ϵi = 1); essentially, individuals with a competing event do not lose any time to event ϵ = 1. The noncensoring indicator equals 0 for censored observations and 1 for uncensored observations; thus, censored observations have a weight of 0 and do not contribute to the estimating equation. However, the censoring distribution is estimated from the sample of all individuals. In Appendix 3, we show Equation (5) is based on an unbiased estimating equation. In Appendix 4, we show the estimated coefficients are asymptotically normally distributed using counting process theory, similar to Tian et al6 and Bang and Tsiatis.16,17 Then, the estimated covariance matrix that accounts for the variability in estimating can be obtained from the sandwich estimator where
and
In Appendix 7, we demonstrate that the model coefficients are collapsible when g(·) is the identity or log link. In other words, adjusting for a covariate that is independent of exposure does not affect the model coefficient of the exposure.
5 |. SIMULATION STUDY
We conducted Monte Carlo simulation studies to assess the performance of the IPW method in estimating the marginal difference in RMTL between exposure groups in the presence of covariates, and the IPCW regression method in estimating the difference in RMTL conditional on covariates. We generated data with proportional and nonproportional subdistribution hazards (crossing cumulative incidence functions).8,18,19 We estimated the mean bias, mean relative bias, root mean squared error, relative standard error (SE), and empirical coverage rate of both methods. We define the relative SE as the ratio of the mean estimated SE and the Monte Carlo empirical SE.
5.1 |. IPW adjusted difference in RMTL
5.1.1 |. Data generation
We first generated five independent covariates Z = (Z1, Z2, Z3, Z4, Z5), each . Then we generated the binary exposure, A, from a Bernoulli distribution with p = expit(Z′α), where α = {α0, ln(0.7), ln(0.6), ln(0.5), ln(0.9), ln(0.8)}T and Z′ = (1, Z). We set the intercept α0 to allow 25% and 50% exposure.
To generate event times, we let the baseline subdistribution hazard function λ10(t) = γ exp(ρt) follow a Gompertz distribution. We then defined the subdistribution hazard function as , where , ψ1 = {ψ11, ln(0.9), ln(0.8), ln(0.7), ln(0.6), ln(0.5)}T, and ψ2 = (ψ21, 0, 0, 0, 0, 0)T, in which ψ11 is the main effect of A and ψ21 is the time-varying effect of A. We then defined the cumulative incidence function for event ϵ = 1 as , and the cumulative incidence function for event ϵ = 2 is , where ψ3 = {ln(0.67), ln(0.5), ln(0.6), ln(0.7), ln(0.8), ln(0.9)}T is the vector of effects for on the subdistribution hazard of event 2.
We assigned the event type by generating a Bernoulli variable with probability where , which is the limit of with respect to t. We generate the event times by simulating a random variable U ∼Uniform(0, 1) and determining the inverse of the conditional cumulative incidence function with respect to t. We generate event times from the respective conditional cumulative incidence functions: for event ϵ = 1, ; and for event .
We applied independent right-censoring by generating censoring times C ~Uniform(0, c), where the parameter c was chosen to yield mild and heavy right censoring (10% and 25% proportion of individuals). Then, we determined if the event time was censored, δ = I(T ≤ C), and the final observed event type, δ · ϵ.
5.1.2 |. Scenarios and true values
We considered both proportional, (γ, ρ, ψ11, ψ21) = (1.2,−1.5,−0.3,0), and nonproportional, (γ, ρ, ψ11, ψ21) = (1.5,−1.5,0.5,−2) subdistribution hazards settings. In total, there were 16 unique scenarios, with varying censoring (10%, 25%), proportion of exposure (25%, 50%), pattern of cumulative incidence functions (proportional vs nonproportional subdistribution hazards), and sample size (500, 1000). We generated 6000 datasets for each scenario.
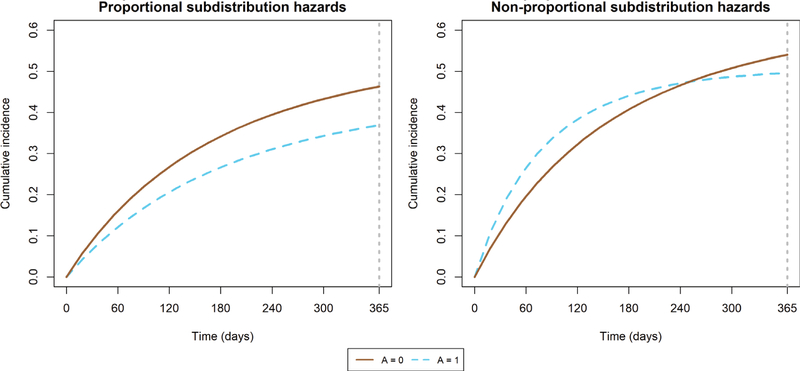
We determined the true marginal difference in RMTL at τ = 365 days by generating Z for a sample of size n = 1,000,000.20 We first set everyone to be exposed (A = 1), and generated the event type and corresponding event time as described in 5.1.1. Then, we repeated the process where everyone was unexposed (A = 0), yielding two event sets of outcomes per person (event type and time). The true marginal difference in RMTL for event ϵ = 1 at τ = 1 year is the unadjusted difference in RMTL for event ϵ = 1 when A = 1 and A = 0; it was equal to −22.63 days for the proportional subdistribution hazards setting and 8.03 days for the nonproportional subdistribution hazards setting. The true cumulative incidence functions are depicted in Figure 1.
FIGURE 1.
True cumulative incidence functions in simulation study of inverse probability weighted estimator
To apply the IPW method, we first derived the weights by estimating the probability of exposure with a logistic regression model, logit{P(A = 1 Z′)} = Z′α. We then estimated the IPW-adjusted RMTL in each exposure group, A = 1 and A = 0, and the adjusted difference between groups.
5.1.3 |. Results
Overall, the IPW-estimator demonstrated little bias in all settings (Table 1). The bias was slightly greater in the presence of nonproportional subdistribution hazards in scenarios with smaller sample size (n = 500). The mean squared error was smaller in scenarios with larger sample size (n = 1000) and equal exposure groups (50% exposed). The coverage proportion of 95% confidence intervals (CIs) was greater than the nominal value, indicating that the CIs tend to be conservative. The coverage proportion was sometimes slightly better in scenarios with balanced exposure groups (50% exposed) compared with settings with unbalanced exposure groups (25% exposed). The relative SE was greater than 1 in all scenarios, which indicates that the estimated SE was greater than the Monte Carlo empirical SE and likely explains the observed overcoverage.
TABLE 1.
Performance of IPW estimator of the difference in RMTL in simulation studies, τ = 365 days
|
Proportional subdistribution hazards, ν = −22.63 days at τ = 365 days
| |||||||
| Sample size | Exposed | Censoring | Bias, days | Rel bias | RMSE, days | Rel SE | Coverage |
| 500 | 25% | 10% | −0.026 | 0.001 | 14.269 | 1.262 | 0.984 |
| 25% | 0.231 | −0.010 | 15.046 | 1.224 | 0.982 | ||
| 50% | 10% | 0.118 | −0.005 | 12.568 | 1.173 | 0.976 | |
| 25% | 0.197 | −0.009 | 13.179 | 1.146 | 0.974 | ||
| 1000 | 25% | 10% | 0.134 | −0.006 | 10.153 | 1.248 | 0.984 |
| 25% | 0.424 | −0.019 | 10.455 | 1.237 | 0.981 | ||
| 50% | 10% | −0.319 | 0.014 | 8.949 | 1.158 | 0.977 | |
| 25% | −0.182 | 0.008 | 9.267 | 1.146 | 0.974 | ||
|
Nonproportional subdistribution hazards, ν = 8.03 days at τ = 365 days
| |||||||
| Sample size | Exposed | Censoring | Bias, days | Rel bias | RMSE, days | Rel SE | Coverage |
| 500 | 25% | 10% | 0.721 | 0.090 | 16.702 | 1.215 | 0.980 |
| 25% | 0.759 | 0.095 | 17.433 | 1.196 | 0.978 | ||
| 50% | 10% | 0.716 | 0.089 | 13.291 | 1.206 | 0.981 | |
| 25% | 0.616 | 0.077 | 14.163 | 1.163 | 0.980 | ||
| 1000 | 25% | 10% | 0.344 | 0.043 | 11.679 | 1.219 | 0.982 |
| 25% | 0.264 | 0.033 | 12.070 | 1.213 | 0.980 | ||
| 50% | 10% | 0.359 | 0.045 | 9.495 | 1.186 | 0.979 | |
| 25% | 0.379 | 0.047 | 9.872 | 1.174 | 0.977 | ||
Note: To assess the IPW method, we generated competing risks data dependent on a binary covariate A and covariates Z. We obtained ν, the true adjusted difference in RMTL between A = 1 and A = 0 at τ = 365 days with a counterfactual approach in a sample of n = 1,000,000.
Abbreviations: IPW, inverse probability weighted; Rel bias, mean bias relative to true parameter; Rel SE, mean estimated standard error/Monte Carlo empirical error; RMSE, root of mean squared error; RMTL, restricted mean time lost.
5.2 |. Inverse probability of censoring weighted model for the RMTL
5.2.1 |. Data generation
To assess the IPCW model, we generated data according two binary covariates, Z = (Z1, Z2). We generated each binary covariate from independent Bernoulli distributions. We let the subdistribution hazard function follow a Gompertz distribution, λ1(t|Z) = γz exp(ρzt), where (γz,ρz) were set according to the 4 strata of (Z1, Z2).
We defined the cumulative incidence function for event 1 as , and the cumulative incidence function for event 2 as . We assigned the event type by generating a Bernoulli variable with probability where ρz < 0. We again generated times using the inversion method, with conditional cumulative incidence functions and P(T ≤ t|ϵ = 2,Z) = 1 − exp(−t). We generated right censoring and determined the final observed event type as described in Section 5.1.1.
5.2.2 |. Scenarios and true values
Similar to 5.1.2, we included 16 unique scenarios with varying censoring (10%, 25%), proportion of exposure (both P(Z1 =1) and P(Z2 =1) equal to 25% or 50%), pattern of cumulative incidence functions (proportional vs nonproportional subdistribution hazards), and sample size (500, 1000). We generated 6000 datasets for each scenario.
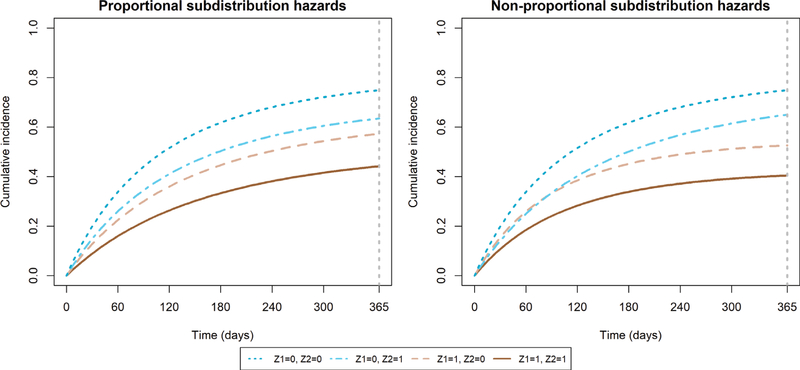
We define (β1, β2) as the true adjusted differences in RMTL at τ = 1 year for covariates Z = (Z1, Z2). First we let . We set Gompertz parameters (ρ, γ) that yielded RMTLs for cause 1 of {μ1(τ|Z1 = 1, Z2 = 1), μ1(τ|Z1 = 1, Z2 =|0), μ1(τ|Z1 = 0, Z2 = 1), μ1(τ|Z1 = 0, Z2 = 0)} = (0.3,0.4,0.45,0.55) years. Then, the effect of| Z1 and Z2 on the RMTL for cause 1 is additive such that (β1,β2) = (−0.15,−0.1) years, or (−54.75,−36.5) days. The Gompertz parameters are provided in Table A1, and the true cumulative incidence functions are depicted in Figure 2.
FIGURE 2.
True cumulative incidence functions in simulation study of inverse probability of censoring weighting regression model
We estimated IPCW regression models g{μ1(τ|Z)} = β0 + β1Z1 + β2Z2 with an identity link. We then evaluated the performance of to estimate (β1,β2).
5.2.3 |. Results
The IPCW model demonstrated very little bias in all settings (Table 2). The mean squared error decreased as sample size increased, censoring decreased, and when the proportion exposed was balanced compared with imbalanced. The coverage proportion of 95% CIs ranged from 93% to 95%, which indicates slight undercoverage. The relative SE was less than 1 in all scenarios, which indicates that the estimated SE was smaller than the Monte Carlo empirical SE and likely explains the observed undercoverage.
TABLE 2.
Performance of IPCW model of the difference in RMTL in simulation studies, τ = 365 days
|
β1 = −54.75 days |
β2 = −36.5 days |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PSH | Sample size | Exposed | Censoring | Bias, days | Rel bias | RMSE, days | Rel SE | Cov | Bias, days | Rel bias | RMSE, days | Rel SE | Cov |
| Yes | 500 | 50% | 10% | 0.076 | −0.001 | 13.241 | 0.956 | 0.938 | 0.049 | −0.001 | 13.251 | 0.956 | 0.938 |
| 25% | 0.135 | −0.002 | 14.340 | 0.955 | 0.939 | 0.088 | −0.002 | 14.282 | 0.955 | 0.942 | |||
| 25% | 10% | 0.307 | −0.006 | 15.096 | 0.977 | 0.942 | 0.116 | −0.003 | 15.332 | 0.977 | 0.936 | ||
| 25% | 0.035 | −0.001 | 16.661 | 0.957 | 0.935 | 0.481 | −0.013 | 17.037 | 0.957 | 0.930 | |||
| 1000 | 50% | 10% | −0.039 | 0.001 | 9.251 | 0.968 | 0.943 | −0.055 | 0.002 | 9.106 | 0.968 | 0.945 | |
| 25% | −0.149 | 0.003 | 10.132 | 0.957 | 0.937 | 0.097 | −0.003 | 10.095 | 0.957 | 0.940 | |||
| 25% | 10% | −0.254 | 0.005 | 10.772 | 0.969 | 0.942 | 0.068 | −0.002 | 10.852 | 0.969 | 0.933 | ||
| 25% | 0.006 | 0.000 | 11.811 | 0.954 | 0.937 | −0.086 | 0.002 | 11.781 | 0.954 | 0.938 | |||
| No | 500 | 50% | 10% | −0.204 | 0.004 | 13.411 | 0.963 | 0.942 | −0.133 | 0.004 | 13.327 | 0.963 | 0.943 |
| 25% | 0.097 | −0.002 | 14.458 | 0.967 | 0.943 | −0.341 | 0.009 | 14.375 | 0.967 | 0.943 | |||
| 25% | 10% | 0.077 | −0.001 | 15.833 | 0.963 | 0.941 | −0.276 | 0.008 | 15.311 | 0.963 | 0.941 | ||
| 25% | −0.152 | 0.003 | 17.372 | 0.958 | 0.938 | 0.175 | −0.005 | 17.405 | 0.958 | 0.938 | |||
| 1000 | 50% | 10% | 0.189 | −0.003 | 9.304 | 0.982 | 0.942 | −0.087 | 0.002 | 9.386 | 0.982 | 0.944 | |
| 25% | −0.163 | 0.003 | 10.366 | 0.954 | 0.938 | −0.134 | 0.004 | 10.363 | 0.954 | 0.937 | |||
| 25% | 10% | −0.056 | 0.001 | 11.058 | 0.974 | 0.944 | 0.016 | 0.000 | 10.818 | 0.974 | 0.941 | ||
| 25% | 0.034 | −0.001 | 12.289 | 0.957 | 0.937 | −0.190 | 0.005 | 12.066 | 0.957 | 0.937 | |||
Note: To assess the IPCW model, we generated competing risks data dependent on two binary covariates (Z1, Z2). We set the true unadjusted difference in RMTL at τ = 365 days, .
Abbreviations: IPCW, inverse probability of censoring weighted; PSH, proportional subdistribution hazards; Rel SE, mean estimated standard error/Monte Carlo empirical error; RMSE, root of mean squared error; RMTL, restricted mean time lost.
6 |. ILLUSTRATIVE EXAMPLES
We present illustrative examples on AF in the Framingham Heart Study.21–23 We first evaluated sex differences in incident AF. We also evaluated sex differences in cause-specific mortality after AF. In both examples, we apply the IPW and IPCW methods. We compared results with and without accounting for competing risks. The IPW method estimates the marginal RMTL in each sex and the difference in RMTL between sexes, adjusted for covariates, while the IPCW model estimates differences in RMTLs conditional on covariates. We also estimated subdistribution hazard ratios with Fine-Gray models. To protect the confidentiality of the Framingham Heart Study participants, the data from our illustrative examples are not publicly available. Participant level data from the Framingham Heart Study are available at the database of Genotypes and Phenotypes (https://www.ncbi.nlm.nih.gov/gap/) and BioLINCC (https://biolincc.nhlbi.nih.gov/home/).
6.1 |. Sex differences in incident AF
When assessing incident AF, individuals may die before developing AF; thus, death is considered a competing risk. Up to timepoint τ, the RMTL for AF is the mean AF-free time lost, or the mean time diagnosed with AF.
We included 6016 participants who were free of AF at index age 55 years and attended an examination between ages 50 and 60 years. Risk factors were measured at the examination closest to age 55 years. Participants were followed until AF or atrial flutter, death, loss to follow-up, the last visit or medical contact. We set τ = 40 years. We adjusted for age, height, weight, current smoking status, systolic and diastolic blood pressure, hypertensive medication, current diabetes status, prior heart failure, and prior myocardial infarction.22 To address highly influential observations, leading to the instability of the IPW estimator, we replaced weights above the 99th percentile of the distribution by the 99th percentile.24
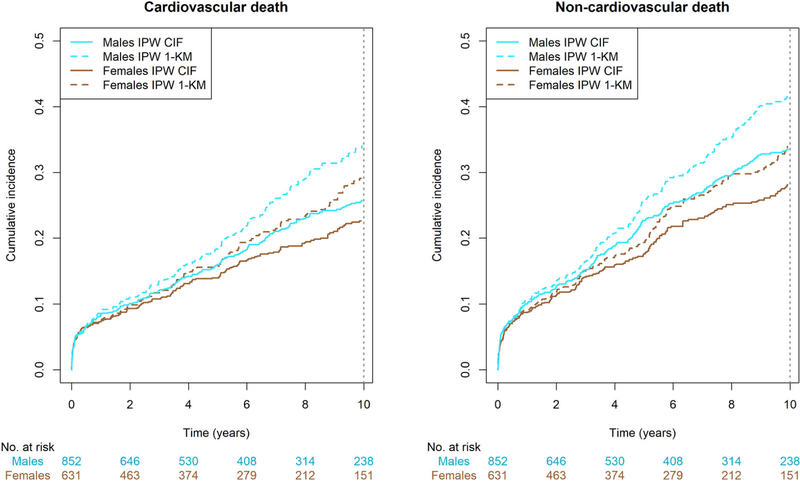
In Figure 3, we show the IPW cumulative incidence curves and the complement of the IPW Kaplan-Meier curves (1-KM). In Table A2 (Appendix 5), we show the IPW-adjusted RMTL in males and females, separately.
FIGURE 3.
Inverse probability weighted cumulative incidence curves for incident atrial fibrillation in the Framingham Heart Study, with and without accounting for competing risks. CIF, cumulative incidence function; IPW, inverse probability weighted; KM, Kaplan-Meier. The Kaplan-Meier method does not account for competing risks, while the cumulative incidence function does
With the IPW method, we found evidence of a marginal difference between sexes in RMTL for AF over 40 years (Table 3). Men lost an additional 1.4 (95% CI: 0.5, 2.4) years of AF-free time as compared with women. With the IPCW model, there was no evidence of difference in the conditional difference in RMTL. On average females lost 0.4 (−0.92, 1.78) additional years of AF-free time as compared with males. When interpreting the results, it is important to note the two methods estimate different quantities. The IPW method estimates the population difference in RMTL between sexes marginalized over covariates, or the difference of the population being male or female.25 By contrast, the IPCW regression model estimates the individual difference in RMTL between sexes conditional on covariates, or the difference in an individual being male or female.25 The adjusted subdistribution hazard ratio for AF was 1.2 (1.0, 1.5), also indicating males have an increased incidence of AF compared with females. As for the competing event, death without AF, males lost more AF-free life expectancy than females(IPW: 2.3 [1.2, 3.3], IPCW: 1.2 [0, 2.5]). The adjusted subdistribution hazard ratio for death without AF was 1.5 (1.3, 1.8).
TABLE 3.
Adjusted differences in RMTL between males and females in 40-year incident atrial fibrillation
|
RMTL difference, years
|
||
| Estimators accounting for competing risks | AF | Death without AF |
| IPW | 1.41 (0.45, 2.38) | 2.26 (1.21, 3.30) |
| IPCW regression model | −0.43 (−1.78, 0.92) | 1.24 (0.00, 2.48) |
|
RMTL difference, years
|
||
| Estimators not accounting for competing risks | AF | Death without AF |
| IPW | 2.83 (1.64, 4.01) | 2.93 (1.81, 4.04) |
| IPCW regression model | −1.23 (−3.10, 0.64) | 1.97 (0.57, 3.37) |
Note: Data are differences in cause-specific RMTL (95% confidence intervals) for men vs women. The IPW method directly yields marginal differences in RMTL adjusting for covariates, while the IPCW regression models yield differences in RMTL conditional on covariates.
Abbreviations: AF, atrial fibrillation; IPW, inverse probability weighted; IPCW, inverse probability of censoring weighted; RMTL, restricted mean time lost.
When ignoring competing risks, both the IPW and IPCW methods provided estimates larger in magnitude for both events (Table 3). In addition, both methods provided larger estimates of the RMTL in each sex, particularly the IPCW method (Appendix 6, Table A2).
6.2 |. Sex differences in cause-specific mortality after AF
We included 1483 individuals with incident AF who attended an exam within 10 years of diagnosis with nonmissing covariates. We categorized death causes into cardiovascular disease (CVD), non-CVD, and unknown causes of death. We assessed sex differences in CVD and non-CVD mortality, while unknown cause of death was considered an additional competing risk. Individuals entered the analysis at AF diagnosis, and were followed until death or loss to follow-up. We estimated differences in cause-specific RMTL between men and women, adjusted for age at AF and risk factors measured at the closest examination prior to AF: body mass index, smoking status, systolic and diastolic blood pressure, hypertensive medication, and diabetes status. We also adjusted for prior heart failure and prior myocardial infarction, assessed at time of AF onset. We set τ = 10 years.
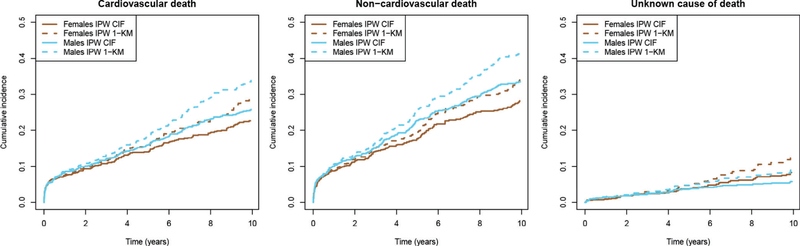
In Figure 4, we show the IPW cumulative incidence curves and the complement of the IPW Kaplan-Meier curves (1-KM). In Table A3 (Appendix 6), we show the IPW-adjusted RMTLs for CVD death and CVD death in males and females, separately.
FIGURE 4.
Inverse probability weighted cumulative incidence curves for cause-specific mortality among individuals with AF in the Framingham Heart Study, with and without accounting for competing risks. CIF, cumulative incidence function; IPW, inverse probability weighted; KM, Kaplan-Meier. The Kaplan-Meier method does not account for competing risks, while the cumulative incidence function does
We did not find evidence of differences between sexes in 10-year cause-specific mortality (Table 4). When accounting for competing risks, both methods gave similar estimates for the difference in RMTL. The IPCW method provided slightly larger CIs. On average, males had greater lost life expectancy to CVD (IPW: 2.2 [−1.9, 6.4] months) and non-CVD causes (IPW: 4.0 [−0.5, 8.5] months) as compared with females. The adjusted subdistribution hazard ratios were consistent in direction with the differences in RMTLs (CVD-death: 1.2 [1.0, 1.5], non-CVD death: 1.3 [1.1, 1.5]).
TABLE 4.
Adjusted differences in RMTL between males and females in 10-year cause-specific mortality after atrial fibrillation
|
RMTL difference, years
|
||
| Estimators accounting for competing risks | CVD death | Non-CVD death |
| IPW | 0.18 (−0.16, 0.53) | 0.33 (−0.04, 0.71) |
| IPCW regression model | 0.19 (−0.28, 0.66) | 0.37 (−0.08, 0.81) |
|
RMTL difference, years
|
||
| Estimators not accounting for competing risks | CVD death | Non-CVD death |
| IPW | 0.28 (−0.10, 0.67) | 0.43 (0.02, 0.84) |
| IPCW regression model | 0.34 (−0.02, 0.70) | 0.52 (0.12, 0.92) |
Note: Data are differences in cause-specific RMTL (95% confidence intervals) for men vs women. The IPW methods yield marginal differences in RMTL adjusting for covariates, while the IPCW regression models yield differences in RMTL conditional on covariates.
Abbreviations: CVD, cardiovascular disease; IPW, inverse probability weighted; IPCW, inverse probability of censoring weighted; RMTL, restricted mean time lost.
When ignoring competing risks, both methods provided slightly larger estimates of the difference in RMTL for both CVD and non-CVD causes. However, the change in estimates when accounting or not accounting for competing risks was much smaller than the previous example. Again, both methods also provided larger estimates of the RMTLs in each sex (Appendix 6, Table A3).
7 |. DISCUSSION
In summary, we introduced methods to estimate and model the difference in RMTL in the presence of competing risks. We derived an IPW adjusted estimator of the difference in RMTL between exposure groups and its variance, and a generalized linear model for the RMTL conditional on covariates estimated with IPCW. In our illustrative examples, we demonstrate how ignoring competing risks leads to exaggerated estimates.
Ignoring competing events will lead to biased inference. The complement of the Kaplan-Meier curve overestimates the cumulative incidence of an event and similarly, estimators of the RMTL must also account for the competing risk to avoid bias. In our illustrative examples, the estimated RMTLs and their differences were considerably larger when ignoring competing risks. The IPCW method was particularly prone to giving larger estimates when the competing risk was ignored. This may be due to the twofold bias of the IPCW model: if competing events are considered censored, the outcome is estimated only based on the event of interest rather than taking a value of 0 for competing events, and the censoring distribution G(t) used to weight individuals is actually the distribution of a composite endpoint of censoring and the competing event.
In analyses of competing risks data, subdistribution and cause-specific hazard ratios are frequently used to measure associations.26 However, these measures can be difficult to interpret. First, they rely on proportionality over time, which often does not hold. Second, the subdistribution hazard for a given cause is a function of all cause-specific hazards, which allows the possibility that an exposure can indirectly influence the subdistribution hazard ratio through a competing event.26 Moreover, the cause-specific hazard ratio does not have a one-to-one relationship with the cumulative incidence function. This point also applies to RMTL-based measures of association, as a covariate can affect the cumulative incidence curve through the competing event. This can present difficulty in interpretation if the competing events are not also analyzed. To address this, we recommended reporting RMTL-based measures for both the event of interest and the competing event. Third, the subdistribution and cause-specific hazard ratios are not collapsible.27,28 However, the IPCW regression model for the cause-specific RMTL yields collapsible coefficients with the identity or log link.
In a randomized experiment, Weir et al2 demonstrated how clinicians may reach different conclusions when hazard ratios or differences in RMST are reported. Similarly, reporting the difference in RMTL as a complement to subdistribution and cause-specific hazard ratios may give additional insight to the data and offer a clinically meaningful interpretation. Unlike either hazard ratio, the unadjusted, and IPW adjusted differences in RMTL directly give absolute measures in each group, which provides crucial background information.29 Furthermore, the difference in RMTL is not subject to any proportionality assumptions. The assumption of proportional hazards is complex in the competing risk setting, as subdistribution hazards and cause-specific hazards cannot be simultaneously proportional and both must be explicitly assessed.30
Other methods for the RMTL exist. In previous work, Calkins et al31 estimated differences in RMST adjusted with IPW, and the bootstrap for CIs. Andersen10 developed pseudo-observation model for cause-specific life years lost. Kipourou et al32 also used the pseudo-observation technique to model life-years lost to various causes when cause of death is unreliable or missing. Sabathé et al33 used the pseudo-observation approach to account for interval censoring in an illness-death model framework. Pan and Gastwirth34 modeled the restricted mean job duration, or “lifetime,” using Cox models for cause-specific event processes.
There are several limitations to our methods. First, if the IPW estimator weights are in fact estimated by estimating pia, the proposed variance of the RMTL does not account for variability in the estimated weights. In our simulation study, the coverage proportion of the IPW method was greater than 95%. Estimation of the weights can be accounted for using the bootstrap method.31 Second, when predicting the RMTL for a covariate profile using the IPCW model, one must be careful not to extrapolate outside the window of (0, τ), which may occur when using the identity or log link functions.10 By contrast, the logit-like link function, suggested by Tian et al, restricts the predicted RMTL to (0, τ).6 Third, both of our proposed methods assume the event time and censoring are independent, which may not always hold. The IPCW framework has been extended to accommodate dependent censoring in other settings.35 The IPCW model also exhibits a trade-off between estimation of the censoring distribution G(t) and the number of uncensored observations. Ideally, one would have a large sample with sufficient censoring. One could improve estimation of the censoring distribution and overall model by including an augmentation term in the estimating equation.36 Finally, we only considered covariates at baseline but it is possible that covariates are measured repeatedly over time. One could use g-computation and g-estimation to estimate the difference in RMTL while accounting for time-varying confounding.37,38
ACKNOWLEDGEMENTS
SCC received funding from the National Heart, Lung, and Blood Institute (NHLBI) (F31 HL145904-01). LT received funding from the American Heart Association (18SFRN34150007). The Framingham Heart Study is supported by NHLBI (N01-HC25195, HHSN268201500001I; 75N92019D00031) and Boston University School of Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Katia Bulekova for her support with Boston University’s Shared Computing Cluster, the Framingham Heart Study participants for their commitment, Dr. Sangbum Choi for communication about the IPW Aalen-Johansen estimator, and two anonymous reviewers for their insightful comments.
APPENDIX A
A.1. Derivation of RMTL
Here, we show that E [(τ − R) · I(ϵ = 1)] is the area under the cause-specific cumulative incidence function, or cause-specific RMTL. While we cannot take the expectation of the subdistribution failure time, T · I(ϵ = 1), we can find the expectation of the random variable R · I(ϵ = 1),
which is the time alive and free of cause 1, but still includes the expected time lost to cause 2. To isolate the time lost due to cause 1, we take the expectation of (τ − R) · I(ϵ = 1),
Since P(τ − T ≤ t,ϵ = 1) = 1 − F1(τ − t) and ,
By integration by parts, it follows that
This relationship can be extended to the cause-specific RMTL conditional on covariates Z∗, which we leverage in Appendix 3.
A.2. Variance of difference in RMTL
Let be the number of events of cause k by time t, N(t)= N1(t)+ N2(t) is the number of total events by time t, ΔNk(t) be the increment Nk(t)− Nk(t−), and be the number at risk just before t. Let be the Kaplan Meier estimator of being alive and event-free at time t, , and . The cumulative incidence function is then estimated by the Aalen-Johansen estimator , where tj are the unique event times.39 The variance and covariance are estimated using Greenwood’s formula,40,41
and
where
and
for tj < tk. Then, the variance of the cause-specific RMTL is
which is Equation (2).
A.3. Unbiased IPCW estimating equation
Here, we show that the IPCW estimating equation (3) is unbiased. In the generalized linear model, we assume . Following the conditional expectation arguments of Bang and Tsiatis16,17 and Scheike et al,42 we have
It follows that
Since G(t) is unknown, we propose estimating G(t) by the Kaplan-Meier estimator and solving the following equation to estimate β,
A.4. Asymptotic variance of IPCW estimating equation
Following Tian et al6 and Bang and Tsiatis,16,17 we leverage martingale identities
and
The estimating equation evaluated at the truth β0 can be written as asymptotically linear,
where
and
The first two terms are independent due to martingale properties, so
| (A1) |
Then, where B is the variance of the estimating equation (A1) and
This covariance matrix can be estimated by in which
and
where , , and .
A.4. Parameters for data generation in simulation study of IPCW regression model
TABLE A1.
Gompertz parameters for data generation in simulation study of IPCW regression model
|
Proportional subdistribution hazards, τ = 365 days
| |||||
| ρ | γ | Z 1 | Z 2 | RMTL, years | RMTL, days |
| −2.8 | 1.546588 | 1 | 1 | 0.30 | 109.5 |
| −2.9 | 2.291237 | 1 | 0 | 0.40 | 146.0 |
| −1.4 | 1.952647 | 0 | 1 | 0.45 | 164.25 |
| −1.7 | 2.879494 | 0 | 0 | 0.55 | 200.75 |
|
Nonproportional subdistribution hazards, τ = 365 days
| |||||
| ρ | γ | Z 1 | Z 2 | RMTL, years | RMTL, days |
| −1.7 | 1.213444 | 1 | 1 | 0.30 | 109.5 |
| −1.7 | 1.770851 | 1 | 0 | 0.40 | 146.0 |
| −1.7 | 2.096025 | 0 | 1 | 0.45 | 164.25 |
| −1.7 | 2.879494 | 0 | 0 | 0.55 | 200.75 |
Note: To assess the IPCW model, we generated competing risks data dependent on two binary covariates (Z1, Z2). We set the true conditional difference in RMTL at τ = 365 days, (β1, β2) = {μ1,1z2(τ) − μ1,0z2(τ), (μ1,z11(τ) − μ1,z10(τ)}.
Abbreviations: IPCW, inverse probability of censoring weighting; RMTL, restricted mean time lost.
A.5. Adjusted RMTL by sex from illustrative examples
For the IPCW models, we predicted the RMTL for each sex with a least squares means approach by summing the estimated model coefficients at the mean value for continuous covariates and the sample proportion for binary covariates.
A.6. Collapsibility of IPCW regression model
Non collapsibility is a disadvantage of the subdistribution hazard ratio.28 In other words, adjusting for a covariate may affect the subdistribution hazard ratio even if the covariate is independent of exposure. By contrast, an advantage of the IPCW regression model for the cause-specific RMTL is that it is a generalized linear model, and therefore yields collapsible coefficients when g(·) is the identity or log link. We demonstrate that the identity and log links yield collapsible differences and ratios, respectively, in cause-specific RMTL. Assume A is a binary exposure, Z is a covariate, A and Z are independent, and the true model is
| (A2) |
TABLE A2.
Adjusted RMTLs for 40-year incident atrial fibrillation by sex
|
RMTL, years |
|||
| Estimators accounting for competing risks | AF | Death without AF | |
| Males | IPW | 6.74 (5.98, 7.49) | 9.74 (8.90, 10.57) |
| IPCW regression model | 6.64 (5.78,7.51) | 8.95 (8.17, 9.74) | |
| Females | IPW | 5.33 (4.73, 5.93) | 7.48 (6.86, 8.10) |
| IPCW regression model | 7.07 (6.37, 7.78) | 7.71 (7.06, 8.37) | |
|
RMTL, years |
|||
| Estimators not accounting for competing risks | AF | Death without AF | |
| Males | IPW | 9.50 (8.55, 10.45) | 12.07 (11.18, 12.97) |
| IPCW regression model | 10.85 (9.87,11.82) | 12.80 (12.00, 13.60) | |
| Females | IPW | 6.67 (5.96, 7.39) | 9.15 (8.48, 9.81) |
| IPCW regression model | 12.08 (10.86, 13.29) | 10.83 (10.03, 11.63) | |
Abbreviations: AF, atrial fibrillation; IPW, inverse probability weighted; IPCW, inverse probability of censoring weighted; RMTL, restricted mean time lost.
TABLE A3.
Adjusted RMTLs for 10-year cause-specific death after atrial fibrillation by sex
|
RMTL, years |
|||
| Estimators accounting for competing risks | CVD death | Non-CVD death | |
| Males | IPW | 1.62 (1.40,1.84) | 2.13 (1.88, 2.37) |
| IPCW regression model | 1.58 (1.28, 1.89) | 2.13 (1.85, 2.41) | |
| Females | IPW | 1.44 (1.17, 1.70) | 1.79 (1.51, 2.08) |
| IPCW regression model | 1.39 (1.05, 1.73) | 1.76 (1.44, 2.09) | |
| RMTL, years | |||
| Estimators not accounting for competing risks | CVD death | Non-CVD death | |
| Males | IPW | 1.95 (1.70, 2.19) | 2.47 (2.20, 2.73) |
| IPCW regression model | 2.50 (2.28, 2.72) | 3.01 (2.76, 3.25) | |
| Females | IPW | 1.66 (1.37, 1.95) | 2.04 (1.73,2.35) |
| IPCW regression model | 2.16 (1.89, 2.43) | 2.49 (2.20, 2.78) | |
Abbreviations: CVD, cardiovascular disease; IPW, inverse probability weighted; IPCW, inverse probability of censoring; RMTL, restricted mean time lost.
When g(·) is the identity link, βA is the conditional difference in RMTL between exposure groups A = 1 and A = 0. Then, the marginal difference in RMTL is
which is the conditional difference in RMTL from (A2). Thus, the RMTL regression model with an identity link yields collapsible differences in RMTL. When g(·) is the log link, is the conditional ratio of RMTL of exposure groups A =1 vs A =0. Similarly, the marginal ratio in RMTL is
which is the conditional ratio. Thus, the RMTL regression model with a log link is also collapsible. These arguments apply to the RMST regression model as well. However, similar to other generalized linear models, other link functions such as the logit link are prone to noncollapsible coefficients.27,43
DATA AVAILABILITY STATEMENT
We provide R code to apply our methods with working examples and to replicate our simulation studies on GitHub at https://github.com/s-conner/rmtl. To protect the confidentiality of the Framingham Heart Study participants, the data from our illustrative examples are not on our GitHub page. Participant level data from the Framingham Heart Study are available at the database of Genotypes and Phenotypes (https://www.ncbi.nlm.nih.gov/gap/) and BioLINCC (https://biolincc.nhlbi.nih.gov/home/).
REFERENCES
- 1.Trinquart L, Jacot J, Conner SC, Porcher R. Comparison of treatment effects measured by the hazard ratio and by the ratio of restricted mean survival times in oncology randomized controlled trials. J Clin Oncol. 2016;34(15):1813–1819. [DOI] [PubMed] [Google Scholar]
- 2.Weir I, Marshall G, Schneider J, et al. Interpretation of time-to-event outcomes in randomized trials: an online randomized experiment. Ann Oncol. 2019;30(1):96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conner SC, Sullivan LM, Benjamin EJ, La Valley MP, Galea S, Trinquart L. Adjusted restricted mean survival times in observational studies. Stat Med. 2019;38(20):3832–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Royston P, Parmar MK. Flexible parametric proportional-hazards and proportional-odds models for censored survival data, with application to prognostic modelling and estimation of treatment effects. Stat Med. 2002;21(15):2175–2197. [DOI] [PubMed] [Google Scholar]
- 5.Andersen PK, Hansen MG, Klein JP. Regression analysis of restricted mean survival time based on pseudo-observations. Lifetime Data Anal. 2004;10(4):335–350. [DOI] [PubMed] [Google Scholar]
- 6.Tian L, Zhao L, Wei L. Predicting the restricted mean event time with the subject’s baseline covariates in survival analysis. Biostatistics. 2014;15(2):222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16(3):1141–1154. [Google Scholar]
- 8.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94(446):496–509. [Google Scholar]
- 9.Kulathinal S, Gasbarra D. Testing equality of cause-specific hazard rates corresponding to m competing risks among K groups. Lifetime Data Anal. 2002;8(2):147–161. [DOI] [PubMed] [Google Scholar]
- 10.Andersen PK. Decomposition of number of life years lost according to causes of death. Stat Med. 2013;32(30):5278–5285. [DOI] [PubMed] [Google Scholar]
- 11.Zhao L, Tian L, Claggett B, et al. Estimating treatment effect with clinical interpretation from a comparative clinical trial with an endpoint subject to competing risks. JAMA Cardiol. 2018;3(4):357–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian L, Jin H, Uno H, et al. On the empirical choice of the time window for restricted mean survival time. Biometrics. 2020;76(4):1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole SR, Lau B, Eron JJ, et al. Estimation of the standardized risk difference and ratio in a competing risks framework: application to injection drug use and progression to AIDS after initiation of antiretroviral therapy. Am J Epidemiol. 2015;181(4):238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann A, Billionnet C. Covariate adjustment of cumulative incidence functions for competing risks data using inverse probability of treatment weighting. Comput Methods Prog Biomed. 2016;129:63–70. [DOI] [PubMed] [Google Scholar]
- 15.Choi S, Kim C, Zhong H, Ryu ES, Han SW. Adjusted-crude-incidence analysis of multiple treatments and unbalanced samples on competing risks. Stat Interface. 2019;12(3):423–437. [Google Scholar]
- 16.Bang H, Tsiatis AA. Estimating medical costs with censored data. Biometrika. 2000;87(2):329–343. [Google Scholar]
- 17.Bang H, Tsiatis AA. Median regression with censored cost data. Biometrics. 2002;58(3):643–649. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Scheike TH, Zhang MJ. Checking fine and gray subdistribution hazards model with cumulative sums of residuals. Lifetime Data Anal. 2015;21(2):197–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou B, Fine J, Laird G. Goodness-of-fit test for proportional subdistribution hazards model. Stat Med. 2013;32(22):3804–3811. [DOI] [PubMed] [Google Scholar]
- 20.Austin PC, Fine JP. Propensity-score matching with competing risks in survival analysis. Stat Med. 2019;38(5):751–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staerk L, Wang B, Preis SR, et al. Lifetime risk of atrial fibrillation according to optimal, borderline, or elevated levels of risk factors: cohort study based on longitudinal data from the framingham heart study. British Med J. 2018;k1453:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staerk L, Preis SR, Lin H, et al. Novel risk modeling approach of atrial fibrillation with restricted mean survival times: application in the framingham heart study community-based cohort. Circ Cardiovasc Qual Outcomes. 2020;13(4):e005918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinter N, Huang Q, Fenger-Gron M, Frost L, Benjamin E, Trinquart L. 45-year trend in excess mortality associated with atrial fibrillation in the community-based Framingham heart study cohorts. BMJ. 2020;370:m2724. 10.1136/bmj.m2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole SR, Hernán MA. Constructing inverse probability weights for marginal structural models. Am J Epidemiol. 2008;168(6):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Austin PC. The performance of different propensity score methods for estimating marginal hazard ratios. Stat Med. 2013;32(16): 2837–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Latouche A, Allignol A, Beyersmann J, Labopin M, Fine JP. A competing risks analysis should report results on all cause-specific hazards and cumulative incidence functions. J Clin Epidemiol. 2013;66(6):648–653. [DOI] [PubMed] [Google Scholar]
- 27.Greenland S, Robins JM, Pearl J. Confounding and collapsibility in causal inference. Stat Sci. 1999;14(1):29–46. [Google Scholar]
- 28.Martinussen T, Vansteelandt S. On collapsibility and confounding bias in Cox and Aalen regression models. Lifetime Data Anal. 2013;19(3):279–296. [DOI] [PubMed] [Google Scholar]
- 29.Trinquart L, Jacot J, Conner SC, Porcher R. Reply to KH Eng et al. J Clin Oncol. 2017;35(4):466. [DOI] [PubMed] [Google Scholar]
- 30.Beyersmann J, Schumacher M. Misspecified regression model for the subdistribution hazard of a competing risk. Stat Med. 2007;26(7):1649. [DOI] [PubMed] [Google Scholar]
- 31.Calkins KL, Canan CE, Moore RD, Lesko CR, Lau B. An application of restricted mean survival time in a competing risks setting: comparing time to ART initiation by injection drug use. BMC Med Res Methodol. 2018;18(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kipourou DK, Perme MP, Rachet B, Belot A. Direct modeling of the crude probability of cancer death and the number of life years lost due to cancer without the need of cause of death: a pseudo-observation approach in the relative survival setting. Biostatistics. 2020;kxaa017. 10.1093/biostatistics/kxaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabathé C, Andersen PK, Helmer C, Gerds TA, Jacqmin-Gadda H, Joly P. Regression analysis in an illness-death model with interval-censored data: a pseudo-value approach. Stat Methods Med Res. 2020;29(3):752–764. [DOI] [PubMed] [Google Scholar]
- 34.Pan Q, Gastwirth JL. Estimating restricted mean job tenures in semi-competing risk data compensating victims of discrimination. Ann Appl Stat. 2013;7(3):1474–1496. [Google Scholar]
- 35.Wang X, Schaubel DE. Modeling restricted mean survival time under general censoring mechanisms. Lifetime Data Anal. 2018;24(1):176–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steingrimsson JA, Diao L, Molinaro AM, Strawderman RL. Doubly robust survival trees. Stat Med. 2016;35(20):3595–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conner SC, Lodi S, Lunetta KL, et al. Refining the association between body mass index and atrial fibrillation: G-formula and restricted mean survival times. J Am Heart Assoc. 2019;8(16):e013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagiwara Y, Shinozaki T, Matsuyama Y. G-estimation of structural nested restricted mean time lost models to estimate effects of time-varying treatments on a failure time outcome. Biometrics. 2020;76(3):799–810. [DOI] [PubMed] [Google Scholar]
- 39.Aalen OO, Johansen S. An empirical transition matrix for non-homogeneous Markov chains based on censored observations. Scand J Stat. 1978;5(3):141–150. [Google Scholar]
- 40.Dinse GE, Larson MG. A note on semi-Markov models for partially censored data. Biometrika. 1986;73(2):379–386. [Google Scholar]
- 41.Gaynor JJ, Feuer EJ, Tan CC, et al. On the use of cause-specific failure and conditional failure probabilities: examples from clinical oncology data. J Am Stat Assoc. 1993;88(422):400–409. [Google Scholar]
- 42.Scheike TH, Zhang MJ, Gerds TA. Predicting cumulative incidence probability by direct binomial regression. Biometrika. 2008;95(1):205–220. [Google Scholar]
- 43.Neuhaus JM, Jewell NP. A geometric approach to assess bias due to omitted covariates in generalized linear models. Biometrika. 1993;80(4):807–815. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We provide R code to apply our methods with working examples and to replicate our simulation studies on GitHub at https://github.com/s-conner/rmtl. To protect the confidentiality of the Framingham Heart Study participants, the data from our illustrative examples are not on our GitHub page. Participant level data from the Framingham Heart Study are available at the database of Genotypes and Phenotypes (https://www.ncbi.nlm.nih.gov/gap/) and BioLINCC (https://biolincc.nhlbi.nih.gov/home/).