Abstract
This review focuses on critical scientific barriers that the field of point-of-care (POC) testing of SARS-CoV-2 is facing and possible solutions to overcome these barriers using functional nucleic acid (FNA)-based technology. Beyond the summary of recent advances in FNA-based sensors for COVID-19 diagnostics, our goal is to outline how FNA might serve to overcome the scientific barriers that currently available diagnostic approaches are suffering. The first introductory section on the operationalization of the COVID-19 pandemic in historical view and its clinical features contextualizes essential SARS-CoV-2-specific biomarkers. The second part highlights three major scientific barriers for POC COVID-19 diagnosis, that is, the lack of a general method for (1) designing receptors of SARS-CoV-2 variants; (2) improving sensitivity to overcome false negatives; and (3) signal readout in resource-limited settings. The subsequent part provides fundamental insights into FNA and technical tricks to successfully achieve effective COVID-19 diagnosis by using in vitro selection of FNA to overcome receptor design barriers, combining FNA with multiple DNA signal amplification strategies to improve sensitivity, and interfacing FNA with portable analyzers to overcome signal readout barriers. This review concludes with an overview of further opportunities and emerging applications for FNA-based sensors against COVID-19.
Graphical abstract
Keywords: Aptamer, Nucleic acid enzyme, COVID-19, Nanomaterial, Portable analyzer
Introduction
Since the identification of the first case of coronavirus disease 2019 (COVID-19) stemming from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus infection in December 2019, the virus has been proliferating globally at an extremely high rate of contagion [1], with a cumulative total of over 280 million confirmed cases and over 5.4 million deaths in 200 countries and territories worldwide (https://covid19.who.int/, up to 29 December 2021). The reported global mortality rate is approximately 2%, which is significantly lower than the 10% for SARS-CoV and the 35% for MERS-CoV [2]. Despite rapid vaccination efforts worldwide, the number of confirmed cases per week has rebounded repeatedly around the world, and the continued emergence of new mutated strains contributes to the current high global outbreak. Given the global perspective, controlling the outbreak remains a daunting long-term challenge owing to the rapid mutability and high rate of transmission of the SARS-CoV-2 virus, which poses a huge threat to public health.
Belonging to the beta genus coronavirus, SARS-CoV-2 has a single-stranded positive-sense RNA genome with an envelope structure, which is generally round or oval in shape, with a size of 60 ~ 140 nm [3]. The genome is ~ 30 kb in length, encoding four conserved structural proteins, named as the spike protein (S), the envelope protein (E), the nucleocapsid protein (N), and the membrane protein (M) (Fig. 1A) [3]. These structural proteins are not only individually responsible for essential physiological roles, but also serve as components of the viral replication machinery or interact with many host cytokines. On the other hand, thanks to advances in structural biology, it is now possible to visualize the entire virus and related structural proteins quickly using transmission electron microscope (TEM) and cryo-electron microscopy (cryo-EM), often within a matter of weeks (Fig. 1B) [4, 5]. Specifically, the viral S protein mediates the receptor binding to angiotensin-converting enzyme 2 (ACE2), assisting the viral entry into host cells via TMPRSS2 protease handling [6]. Two functional subunits, S1 and S2, constitute the S protein; the S1 subunit binds to ACE2, whereas the S2 subunit incorporates membranes (viral and host cells). N protein is responsible for mediating the transcription and replication of viral RNA, which contributes to encapsulated genome packaging into viral particles and the host cell cycle. This structural information results in a rapid development of strategies for COVID-19 diagnostics, therapeutics, and vaccination.
Fig. 1.
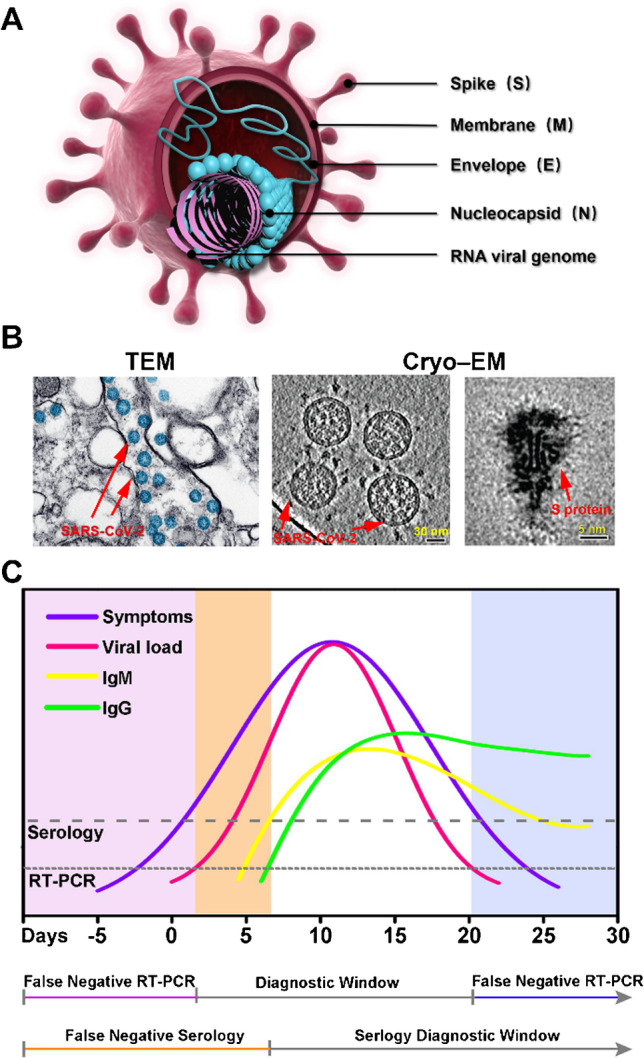
A Viral structure of SARS-CoV-2. B Transmission electron microscopy (TEM) and cryo-electron microscopy (cryo-EM) images of SARS-CoV-2. TEM image: reproduced with permission from [4], Copyright 2020, American Chemical Society. Cryo-EM images: reproduced from [5], open access. C Schematic representation of the relationship between viral load and clinical symptoms during SARS-CoV-2 infection, showing the diagnostic window for reverse transcription-polymerase chain reaction (RT–PCR) and serological testing
During each period of SARS-CoV-2 infection, a range of biomarkers, including viral nucleic acids, protein antigens, and antibodies, have been identified in several biological fluids, including blood and lower and upper respiratory fluids of infected people (Table 1), which are extremely critical for disease diagnosis and therapeutic purposes. Despite their importance, the detection of these biomarkers poses a considerable challenge because of their distinct diagnostic window in SARS-CoV-2 infection (Fig. 1C). Specifically, in the incubation period, viral RNA and antigen, gathered in nasal and pharyngeal swabs, were the primary biomarkers. After 7 days of onset, IgG and IgM antibodies are detectable in blood samples from patients. Within recovering patients, neutralizing immune antibodies, as in CA1 and CB6, are present, of which CB6 is a potential COVID-19 therapeutic agent [12]. The metabolic products, molecular lipids, and additional detective elements comprising the count of leukocyte eosinophils also showed utmost clinical importance. Nevertheless, the concentrations of all the above biomarkers are low, and increasingly, the development of biosensors is dedicated to the rapid and accurate diagnosis of COVID-19. Over the past 3 years, many routine clinical and laboratory assays, such as reverse transcription-polymerase chain reaction (RT–PCR), enzyme-linked immunosorbent assay (ELISA), and computed tomography (CT) imaging, have been developed. Apart from the accuracy problems of false positives/negatives, these methods, however, often rely on expensive and complex laboratory instruments, complicated operating procedures, and trained technicians, becoming a problem that requires consideration and solution for mass screening in public health centers and remote rural areas [13]. At the same time, the decline in viral load as the disease progressed led to unreliable molecular diagnostic assays. Therefore, there is an acute necessity for developing a high-performing, cost-effective, point-of-care (POC) diagnostic system for large-scale screening.
Table 1.
Concentrations of general biomarkers of SARS-CoV-2 in patients with varying degrees of infection
| Types of biomarkers | Asymptomatic infection | Common (mild/moderate) infection | Severe infection |
|---|---|---|---|
| Viral loads | 102.0 ~ 315.0 copies mL−1 (N gene) [7] | 104.0 ~ 107.0 copies mL−1 [8] | 106.0 ~ 108.0 copies mL−1 [9] |
| Antibodies |
IgG: 44.4 ~ 58.8 U mL−1 IgM < 1.0 COI [7] |
IgG: 285.8 U mL−1 (peak) IgM: ~ 4.7 COI [7] |
IgA: 2.8 ~ 3.2 g L−1 IgG: 7.1 ~ 8.2 g L−1 [10] |
| Antigens | - | 103.9 ~ 106.0 fg mL−1 [11] | 106.1 ~ 107.2 fg mL−1 [11] |
Functional nucleic acids (FNAs) are structurally specific nucleic acid molecules that fulfill specific biological functions of nucleic acids and nucleic acid analogs. In this case, FNAs are programmable, cleavable, biocompatible, and molecular recognition-specific stimulus-responsive assembly size control [14]. The structural diversity of nucleic acids allows compatibility with a variety of targeting, encompassing inorganic molecules (e.g., metal ions and pesticide molecules), inorganic nanomaterials (e.g., graphene biomolecules), biological systems (e.g., microbes and cultured cells), and biomaterials (e.g., magnetic liposomes and exosomes) [15]. These features have made FNAs a promising candidate for the combination of biological signatures and probe-finding procedures, and can be readily integrated with multiple signaling amplification and transduction regimes to accelerate biosensing engineering. As a result, FNA has been applied as a key component in POC biosensors for efficient COVID-19 diagnostics, and is emerging as a major active area of research with many exciting results [15].
Recently, an astonishing explosion of researches on COVID-19 has emerged with several review articles from different perspectives. Asghari et al. [3] focused on the state-of-the-art biosensing optical platform for POC COVID-19 diagnosis. Gowri et al. [16] summarized research advances in POC-based optical and electrochemical biosensors, and provided an outlook on the application of wearable devices and smart nano-biosensors for real-time monitoring of COVID-19. Loeffelholz et al. [17] provided a comparative review of POC COVID-19 tests, including their performance, settings for which they might be used, their impact, and future directions. Despite the progress, none of the abovementioned review papers focused on the engineering and application of FNAs for the diagnosis and therapeutics of COVID-19, which is an important and a burgeoning field.
In this review, we focus on state-of-the-art POC strategies targeting COVID-19 and discuss the merits and possible limitations of each method. Additional sections describe the scientific barriers that current POC sensing technologies are facing and summarize the fundamental insights into FNAs and technical tricks toward the successful realization of effective POC detection of SARS-CoV-2. Finally, we conclude with an overview of further opportunities and emerging applications for FNA-based sensors against COVID-19.
Current trends in COVID-19 diagnostics: from centralized laboratories to POC settings
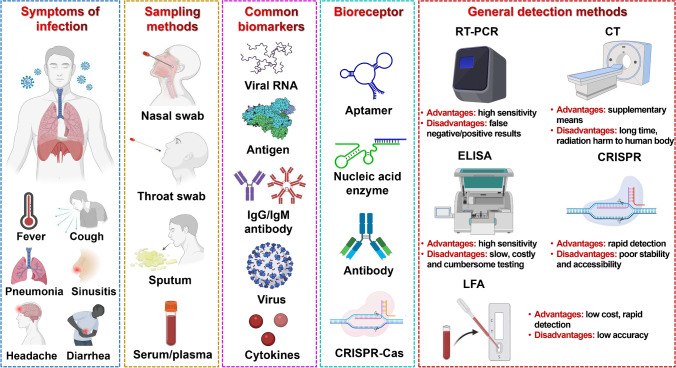
POC diagnostics, defined as diagnostics performed near-patient care, represent an intelligent strategy used for real-time, rapid, accurate, and on-site testing at the point of patient need. Biosensors are regarded as remarkably versatile POC diagnostic platforms, with various biosensors currently being developed on the basis of antibodies, proteins, enzymes, aptamers, cells, and nucleic acids as receptor probes, incorporating optical, electrochemical, mechanical, magnetic, and thermal modes of transmission [16] (Fig. 2). Rapid advances in emerging technologies are propelling the trend of in vitro diagnostics from centralized laboratories to POC testing, with the emergence of lab-on-a-chip (LOC), lab-on-a-disk (LOAD), microfluidic paper-based assays (μPADs), lateral flow assays (LFAs), miniaturized PCR, and isothermal nucleic acid amplification (INAA).
Fig. 2.
Representative SARS-CoV-2 infections, symptoms, clinical sampling methods, biomarkers and bioreceptors for diagnosis, and conceptualized overview on general detection methods, their advantages and disadvantages
Concentrated laboratory assays for COVID-19 consist of nucleic acid amplification, antigen testing, and antibody or serological assays. In comparison to time-consuming and labor-intensive traditional diagnostic platforms, POC testing technologies feature rapid (typically results within minutes), high responsiveness and flexibility, low-cost advantages, and traceable testing (repeatable, real-time testing). In terms of SARS-CoV-2, the POC diagnostic setting is defined as physician offices, emergency departments, urgent care facilities, school health clinics, and pharmacies [18]. By highlighting the significance of exploiting POC diagnostic technologies for preventing and controlling pandemics in a timely manner, the global pandemic of COVID-19 has exposed clear discrepancies in POCT techniques, particularly the requirement for affordable portable diagnostic devices capable of offering fast and precise outcomes. Possible solutions are proposed to fulfill this need in Table 2.
Table 2.
POC diagnostic strategies of COVID-19
| Type | Feature | Target | Sample | Method | LOD | Time (min) | Sensitivity/specificity | Ref |
|---|---|---|---|---|---|---|---|---|
| Nucleic acid testing |
Advantages: Sensitivity varies depending on the method, but is generally high Disadvantages: Some methods are time-consuming, costly and technically demanding, and not user-friendly |
M or N genes | Clinical swab | RT-LAMP, hCG-P, and PTSs | 0.5 copies μL−1 | 120 |
100%/100% (1 positive/16 negative) |
[19] |
| As1e, N, E genes | Nasopharyngeal swab | IoT, RT-eLAMP, and LoC | 20 copies μL−1 | 13 ~ 51 |
100%/100% (4 clinical samples) |
[20] | ||
| N gene | Saliva and nasopharyngeal samples | miSHERLOCK | 1.24 copies μL−1 | 60 |
96%/95% (27 positive/21 negative) |
[21] | ||
| E and N genes | Nasopharyngeal swab and sputum | NESBA | 0.5 copies μL−1 | 30 |
100%/100% (98 clinical samples) |
[27] | ||
| Antigen testing |
Advantages: rapid, low cost, simple operation, and high-throughput Disadvantages: false negatives based on viral load |
N protein | Viral samples | Antibody-coated GNPs and colorimetric readout | 150 ng mL−1 | 5 | - | [28] |
| N protein | Plasma samples | Self-powered microfluidic chip | 0.0031 ng mL−1 | 15 | - | [22] | ||
| S protein | Serum | LSG | 2.9 ng mL−1 | 60 |
100%/- (23 clinical samples) |
[29] | ||
| Antibody/serology testing |
Advantages: simple process and high accuracy Disadvantages: Detection time and sensitivity cannot be taken into account at the same time |
Total SARS-CoV-2 antibody | Serum | SQS-LFIA |
IgG: 0.10 ng mL−1 IgM: 0.04 ng mL−1 IgA: 0.06 ng mL−1 |
15 |
100%/100% (97 positive/300 negative) |
[23] |
| Anti-N antibody | Whole blood | ELISA | 5 ng mL−1 | ~ 20 | - | [24] | ||
| Antibody | Serum | MID | - | 21 |
97%/92% (170 clinical samples) |
[25] | ||
| IgM and IgG | Serum | MCIA |
IgM: 0.06 ng mL−1 IgG: 0.10 ng mL−1 |
30 | - | [26] |
RT-LAMP reverse transcription loop-mediated thermal amplification; hCG-P human chorionic gonadotropin (hCG)-conjugated toehold-mediated strand exchange (TMSE) probes; PTSs pregnancy test strips; IoT Internet of Things; RT-eLAMP real-time loop-mediated isothermal amplification; LoC lab-on-chip device; miSHERLOCK minimally instrumented SHERLOCK; NESBA nicking and extension chain reaction; LSG laser-scribed graphene; SQS-LFIA dSiO2/QD/SiO2 lateral flow immunoassay; ELISA enzyme-linked immunosorbent assay; MID magnetic immune detection; MCIA microflow cytometry-based agglutination immunoassay
Currently, the FDA’s authorization of emergency usage of the POC diagnostic platform for nucleic acid amplification comprises two quantitative approaches, Abbott ID NOW COVID-19 and Xpert Xpress SARS-CoV-2, and a qualitative approach, the AcculaSARS-CoV-2 Test (Mesa Biotech Inc.). While all the above devices offer rapid sample-to-result diagnostics, their lack of portability may make them unaffordable by the majority of users within resource-limited environments. Motivated by the cost of diagnostics, a semiautomated field assay was developed via a human chorionic gonadotropin-conjugated toehold-mediated strand exchange probe, which was conducted onto a portable commercial pregnancy test strip [19]. The Internet of Things-based reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) POC device analyzes three target genes (As1e, N, and E genes) of SARS-CoV-2 to prevent false-positive results [20]. Moreover, another platform enables multiplex detection of coronavirus and mutations associated with B.1.1.7, B.1.351, and P.1 variants [21].
For POC diagnosis of antigens, integrated electrochemical biochips are a foreseeable development direction. The self-powered microfluidic platform provides a good equipment base for automated virus diagnosis. The microfluidic automated electrochemical immune-sensing platform was designed for the SARS-CoV-2 N protein assay within 15 min without any liquid handling [22]. The gold-based lateral flow immunoassay (GLFIA) commonly applied in antibody/serological assays and risks exposing false-negative and false-positive interpretations is not suitable for large-scale population screening. Jia et al. [23] constructed a novel fluorescent lateral flow immunoassay (LFIA), which, to overcome the problem of false negatives, using hydrophobic quantum dots to ensure single-labeled high-intensity fluorescence and to overcome the problem of false positives, a double antigen sandwich construction utilizing labeled/immobilized SARS-CoV-2 RBD. Analysis of nearly 400 clinical samples yielded 100% sensitivity and 100% specificity. Electrochemical capillary osmosis devices [24], hematological magnetic immunoassay [25], and microflow cytometry-based agglutination immunoassay [26] were also developed for COVID-19 diagnosis, combined with smartphones to obtain faster test results.
The current development of POC diagnostic platforms is attempting to advance in the direction of (1) multiplexing multiple targets to improve detection accuracy and sensitivity, (2) nesting existing portable microanalytical devices to reduce costs, and (3) collaborating with smartphone terminals to enhance user-friendliness. However, it is still difficult in the process of moving from a central laboratory to a commercial POC.
Critical scientific barriers to developing successful sensors for POC detection of SARS-CoV-2
Over the past decade, the field of sensors has been revolutionized by rapid advancements in synthetic molecular recognition units, which have paved new ways to remarkably improve the analytical performance of sensors and realize commercial success for a variety of practical applications. From the perspective of POC sensors, current sensing technology poses three major scientific barriers.
Scientific barrier I: lack of a general method for receptor design to meet the need to recognize a broad range of SARS-CoV-2 variants
An essential component of POC sensors is the receptor molecule for selective target recognition, and such a feature is especially important to meet the demand of practical applications because new targets can emerge constantly. For example, multiple SARS-CoV-2 variants of concern have emerged, and mutations in S or N proteins of these variants have further led to a significant challenge for accurate and POC identification of new positives [30]. One recent study reported that the lambda variant in South America is not only more infectious but can also evade neutralizing antibodies and reduce vaccine protection. Among them, the RSYLTPGD246-253 N mutation is the only 7-amino acid deletion variant in the S protein NTD, which is in charge of evading neutralizing antibodies. Additional variants, such as delta, continue to emerge, and its L452R mutation may not only increase binding ability with ACE2 protein (enhance infectivity) but also lead to some degree of immune escape. Both D614G and P681R mutations may enhance the binding ability of viral S protein and human ACE2, resulting in increased infectivity of the virus. To date, no sufficient proof exists that the presently established mutations in concern have escaped the vitally significant function of vaccines—to prevent serious illness. In particular, there are increasing concerns that SARS-CoV-2 genetic variants can escape commercially available serological antigen detection tests [31]. As the genome of SARS-CoV-2 antibodies is extremely similar to that of antibodies produced from other coronaviruses, there is still an underlying cross-reactivity in the majority of serological assays, resulting in a significant risk of false positives [32]. Given that SARS-CoV-2 variants will emerge for the duration of the pandemic of COVID-19, a general method to obtain molecules to recognize any variants of concern specifically is highly desirable yet remains a major barrier for sensing applications.
Scientific barrier II: lack of a general method for improved sensitivity to overcome false negatives
Following the progress of molecular biology over the past few years, novel POC assays are available on the basis of conserved genome sequences of SARS-CoV-2. As the gold standard for assaying SARS-CoV-2, RT-PCR still carries the challenge of inducing false-negative outcomes, owing to the combination of low amounts of primary material, extraction and process instability, and the presence of suppressive interference in the samples [33]. A number of serological assays for COVID-19 diagnosis, involving LFIA, chemiluminescent immunoassays, and ELISA, have been developed without being useful to diagnose infection with the virus within a single subject [34]. Kits for immunoassays and molecular diagnostics, such as Abbott’s ID NOWTM, Lumex Instruments’ Microchip RT–PCR, and Roche’s Cobas, were available for low-cost diagnostic screening for COVID-19 [35]. Notwithstanding the advances achieved, false-negative outcomes from POC testing are now being consistently reported among suspected patients who have typical COVID-19 clinical features and specific computed tomography scans of the same. Furthermore, a number of mild or asymptomatic infections that were initially negatively diagnosed have taken 2nd, 3rd, or several repeat assays to confirm the diagnosis of COVID-19 infection [36]. A primary reason for these misdiagnoses is the dynamic viral loads in different sites during the progression of the disease, while the limit of detection or sensitivity of various POC diagnostic approaches cannot be met. Therefore, a general POC method that provides the required sensitivity without compromising the selectivity while still efficiently converting the target recognition event into a physically detectable signal is highly desirable but remains another barrier for the successful development of consumer-level POC sensors.
Scientific barrier III: lack of a user-friendly signal readout method in resource-limited settings
In addition to the barriers of sensitivity and specificity discussed above, the emphasis in relation to the development of novel COVID-19 assays has moved toward cost-effectiveness and user-friendliness for unskilled practitioners in resource-constrained settings. In order to accomplish this goal, a number of portable sensors were designed and adapted to POC for fast and flexible analysis of COVID-19 [37]. Despite the promise of these sensors, their lack of user-friendliness and cost to meet the vital demands for users to test themselves in resource-limited environments limits their market demand. Hence, the development and commercialization of a de novo signaling readout method possessing the desired accuracy in terms of sensitivity, specificity, generality and adaptation to different objectives, cost-effectiveness, accessibility, and, more importantly, adaptability to self-test still represent a significant barrier that cannot be overcome by the presently existing diagnostic methodologies available.
Advances in functional nucleic acid-based technology to overcome critical scientific barriers
FNAs, including aptamers and nucleic acid enzymes (NAEs), generate significant interest within the healthcare field owing to distinctive capabilities of identifying targets and catalysis [15]. The attractive superiority and versatility of FNAs have catalyzed the appearance of FNA-based functional probes to combat COVID-19. FNAs may be utilized to overcome the scientific barriers that currently available diagnostic approaches are suffering.
Using in vitro selection of functional nucleic acids to overcome barriers of receptor design
FNAs are typically isolated via a combinatorial technique known as in vitro selection, that is, systematic evolution of ligands by exponential enrichment (SELEX). In contrast, the identification for novel FNAs to target SARS-CoV-2 not only represents significant progress in the domain of virus biology but also revolutionizes the design of functional probes to be used in COVID-19 diagnostic applications. In vitro SELEX employs a chemically synthesized initial random library of short single-stranded DNA/RNA sequences to generate FNAs that are highly specific to their target over a range of concentrations depending on the selection conditions [38]. In addition, in vitro selection using highly diverse DNA/RNA libraries can deliver artificial FNAs for almost any desired target of interest. Therefore, the major advantage of the in vitro selection system is that the generality of the in vitro selection process is then carried over to the synthesis of FNAs or in other words that provides a general method for bioreceptor synthesis and thereby overcomes the first barrier to successful receptor design.
In the last 2 years, aptamers with high affinity and specificity were screened against the RBD [39, 40] or N-terminal domain (NTD) of the wild-type S protein [41], B.1.1.7 SARS-CoV-2 S1 protein [42], N protein [43], and Mpro [44]. Exceptionally, xeno-nucleic acid (XNA) has been adopted as a nucleotide analog to displace DNA or RNA with an altered sugar, base or phosphate backbone, low immunogenicity, and high stability [45]. Aptamers were generated via a revised SELEX approach utilizing a mutase capable of switching between DNA and FANA. Following eight rounds of screening, FANA-R8-9 was selected to bind to the S1 protein of RBD, even the delta mutant RBD protein [40].
A multicomponent XNAzyme X10 − 23 targeting SARS-CoV-2-specific RNA was reported in recent times, providing a novel strategy for improving the velocity, sensibility, and specifically the assay of COVID-19 [46]. XNAzymes, based on cross-chemistry selective enrichment by exponential amplification (X-SELEX) for in vitro screening, were retrotranscribed into cDNA by XNA-dependent DNA polymerase and subsequently PCR amplified for the generation of templates to follow rounds or deep sequencing [47]. X10-23 is an XNA-modified version of DNAzyme 10–23. We will summarize reported SARS-CoV-2-specific FNAs and their chemical and biophysical characteristics to help further design POC sensors against COVID-19 in Table 3.
Table 3.
A brief overview of FNA probes against COVID-19
| Aptamer | |||||
| Type | Name | Molecular target | Kd (nM) | LOD/IC50 | Ref |
| ssDNA | S14 | NTD of S protein | 21.8 | 2 nM | [39] |
| SNAP1 | NTD of S protein | 39.32 ± 0.12 |
250,000 nM (with LFA) 10,000 nM (with ELISA) |
[41] | |
| MSA1 | S1 protein | 1.8 ± 0.4 | 500 ~ 20,000 nM | [42] | |
| Np-A48 | N protein | 0.49 | 1 ng mL−1 | [43] | |
| cb-CoV2-6C3 | RBD of S protein | 0.13 | 0.42 nM (authentic virus) | [48] | |
| Aptamer-1 | RBD of S protein | 6.05 ± 2.06 | 5.2 nM (inhibition of binding) | [49] | |
| Aptamer-2 | 6.95 ± 1.10 | 4.4 nM (inhibition of binding) | |||
| SP6 | S protein (not RBD) | 21 ± 46 | 0.0002 ~ 0.001 nM (pseudovirus) | [50] | |
| FANA | FANA-R8-9 | RBD of S protein | 1.4 ± 0.4 (delta variant) | 1300 ng mL−1 | [40] |
| NAE | |||||
| Name/type | Molecular target | Detection strategy | LOD | Ref | |
| XNAzyme X10 − 23 | S gene | Fluorescence lateral flow readout | 20 aM | [46] | |
| HRPZyme | RNA |
RT–PCR Colorimetric readout |
100 copies | [51] | |
| Exo III and DNAzyme | SARS-CoV-2 genome |
Logic DNA circuit Fluorescent |
3300 aM | [52] | |
| RT–PCR-based DNAzyme | N gene | Colorimetric readout | 1000 copies | [53] | |
Kd dissociation constant values; IC50 half-maximal inhibitory concentration value.
Interfacing functional nucleic acids with diverse DNA-based signal amplification strategies to overcome barriers of improved sensitivity
While the abovementioned FNAs can specifically recognize SARS-CoV-2 and its variants, transducing the binding events into detectable signals is still challenging, and thus, a number of signal amplification strategies have been developed by integrating FNA-based functional probes with diverse DNA-based reactions. Depending on the functionality of FNAs, the features of FNA-based probes can be categorized into three types, that is, biometric recognition, signaling amplification, and neutralizer. Recently, two main branches of FNAs have been used for the diagnosis of COVID-19, that is, aptamers and NAEs, which play an exciting role in early diagnostics, in-population screening, and on-site testing toward outbreak control.
Representative examples of aptamer-based diagnostic strategies
Aptamers represent a category of potent functional recognition tools. Their high specificity and affinity, rapid and stable synthesis, and easily modifiable and excellent viability for binding to other DNA reactions to perform signal amplification make aptamers a promising bioreceptor with broad utility in COVID-19 diagnostics.
The development of highly sensitive amplification approaches is essential toward the assay of COVID-19. Several amplification techniques have been advanced as replacements for PCR. Isothermal amplification techniques with a single temperature request simplify the POC molecule detection requirements. Among them, recombinant polymerase amplification (RPA), performed at 37 ~ 42 °C, has the shortest sample preparation time, amplifying 1 ~ 10 target DNA copies within 20 min. This method has high sensitivity, good selectivity, and rapid response and has been used to amplify a variety of targets, comprising RNA, miRNA, ssDNA, and dsDNA, from a variety of organisms and samples [54]. A COVID-19 diagnostic approach that integrates RT-RPA and universal lateral flow test strips into an individual microfluidic chip has been reported with fast, highly sensitive, 30-min acquisition results [55]. Kang et al. [56] reported an ultrasensitive protein biomarker detection platform based on immunoassays combined with RPA. Only the DNA bound to the target in the form of a heterosandwich was selectively amplified with high specificity and sensitivity. The platform already offers successful applications for the detection of protein biomarkers of various infectious diseases, including SARS-CoV-2.
Likewise, another isothermal amplification technique, the ligation-dependent isothermal reaction, serves as a sequencing-specific assay relying upon two independent probes ligated at adjacent positions to the target sequence. Woo et al. [57] used a one-pot, ligation-dependent isothermal reaction cascade for rapid and sensitive detection of RNA, involving two brief enzymatic reactions, namely, splint R ligase and subsequent transcription by T7 RNA polymerase. The final transcript is formed into an RNA adaptor that is bound to a fluorescent dye that fluoresces solely upon the presence of the target RNA within the specimen.
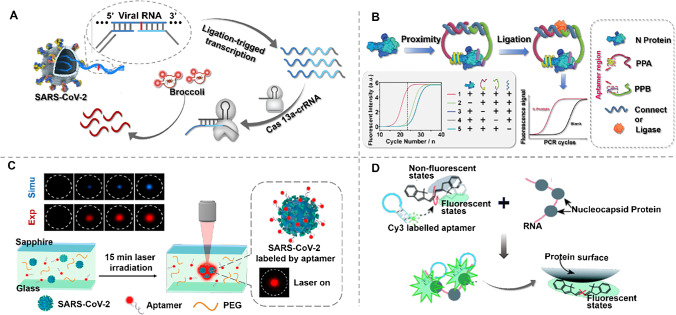
The quantitative properties of isothermal amplification approaches can be affected by nonspecific amplification of nontarget sequences or primer dimers, whereas the high specificity of clustered regularly interspaced short palindromic repeats (CRISPR) technology can improve the performance of diagnostic strategies. A variety of CRISPR-Cas-based detection platforms have been exploited to sense different analytes ranging from nucleic to non-nucleic-acids. Various assay platforms are currently used as diagnostic platforms for COVID-19. Among them, the Cas13 effector protein is able to target RNA and is a promising vehicle toward the diagnosis and treatment of SARS-CoV-2, particularly against mutations of the virus. The nucleic acid detection techniques currently in use have not yet adequately resolved mutations in SARS-CoV-2. Possible avenues to address this issue include the use of an aptamer with specific affinity as a molecular recognition factor or overriding the binding of this aptamer to the virus through an extremely sensitive signaling strategy. Wang et al. [58] reported a combination of luminescent RNA aptamer and CRISPR-Cas13 amplification for the analysis of SARS-CoV-2 and its mutants (Fig. 3A). The luminescent RNA aptamer is a special NA affinity that binds specifically to dyes and fixes its structure to form luminescent aptamer dye complexes. A linkage strategy is introduced by initiating transcriptional amplification. The SARS-CoV-2 was successfully tested in throat swabs and serum samples and in food packaging and seafood.
Fig. 3.
A Schematic of SARS-CoV-2 and its variation detected by the CRISPR-Cas13 transcriptional amplification technique. Single-stranded RNA from SARS-CoV-2 serves as a template for the ligation of presubstrate probes, yielding a template for transcription amplification. Unlabeled light-up RNA aptamer allows sensitive output of amplification signals via the target-activated ribonuclease activity of CRISPR-Cas13a. Reproduced with permission from [58], Copyright 2021, American Chemical Society. B Schematic of the aptamer-assisted proximity ligation assay and workflow for nucleocapsid protein. The design of the ssDNA connector for the two proximity probes (PPA and PPB) ensures that subsequent ligation-dependent qPCR amplification can only be initiated in the presence of PPA, PPB, connector, and N protein. Reproduced with permission from [59], Copyright from author. C Schematic of the one-step aptamer-based thermophoretic assay for rapid detection of SARS-CoV-2 viral particles. Binding of the Cy5-conjugated aptamer to the S protein and PEG-enhanced thermal migration accumulation of the viral particles occurred simultaneously under a temperature gradient of local infrared laser heating. The fluorescence intensity of the enriched aptamer-virus complex after 15 min of laser irradiation indicates the level of S protein on the viral particles. Reproduced with permission from [60], Copyright 2021, American Chemical Society. D Schematic of the PIFE-based aptasensor for a mix-and-read, 1-min SARS-CoV-2 diagnostic assay. In close proximity to a protein, Cy3 is preferentially locked into its trans conformational state, resulting in fluorescence enhancement. Reproduced with permission from [62] , Copyright 2022, Elsevier
In addition to techniques for nucleic acid amplification that can improve the performance of the assay, proximity ligation immunoassay (PLA) has recently been designed for the highly sensitive diagnosis of COVID-19. PLA is performed by converting antibody protein-binding events into amplifiable DNA sequences for subsequent real-time PCR quantitative detection of protein targets. Our group [59] reported a biosensor for detecting COVID-19 serum on the basis of an aptamer-assisted proximity ligation test. As shown in Fig. 3B, the biosensor binds two aptamer probes to the same protein target, bringing the connected DNA regions close to each other and thus initiating ligation-dependent qPCR amplification. Utilizing this strategy, the protein recognition of serum nucleocapsid protein can be transformed into a detectable qPCR signal in 2 h through a simple, uniform, and rapid assay process with high sensitivity and specificity. Such characteristics enable clinical laboratories to perform nucleic acid and serological antigen tests at the same time, thereby improving diagnostic accuracy, especially in asymptomatic populations.
As an identification element, it is well suited to thermocapacitive sensors based on the small size of the aptamer, its low spatial potential resistance, and its good thermal stability. Thermal electrophoresis is an effective strategy for handling and detecting objects based on the directional migration of objects along a temperature gradient. An aptamer-based thermophoretic accumulation sensor combines aptamer recognition and thermoswimming signal amplification to quantitatively analyze biomolecules under a temperature gradient. Deng et al. [60] described a one-step thermal stroking assay in Fig. 3C for the quantitative assay of pseudo SARS-CoV-2 virus particles in use of PEG-enhanced thermal stroking accumulation based on a SARS-CoV-2 S protein-binding aptamer. The assay has a 100-fold lower LOD than the lateral flow assay, 100% accuracy for clinical samples, and, more importantly, a cost of $0.40 per test, making it an attractive strategy for large-scale testing in resource-limited settings. Furthermore, for the direct detection of intact virions, Peinetti et al. [61] selected DNA aptamers that bind intact infectious viruses. The aptamer is integrated into a solid nanopore, creating a strong constraint on the virus and strongly enhancing the sensitivity to SARS-CoV-2 intact virions.
For the sake of simplifying steps, protein-induced fluorescence enhancement (PIFE), a distance-dependent liquid platform fluorescence enhancement technique, offers higher spatial resolution and specificity compared to fluorescence resonance energy transfer (FRET) for the detection of direct protein-DNA binding. Lee et al. [62] integrated the PIFE method with an aptamer as a 1-min, one-step accurate assay system for SARS-CoV-2 N protein, which is more sensitive and specific than conventional antibody testing (Fig. 3D).
Aptamers possess the merits of fast synthesis, low cost, small size, and high stability. As shown with the analytical performance in Table 4, utilizing the existing in vitro screening of various high affinity aptamers for SARS-CoV-2 (including ssDNA, ssRNA, and peptide aptamers), in conjunction with signal transduction strategies (PLA, CRISPR-based transcription amplification, RPA, isothermal ligation and transcription, thermophoretic accumulation, and PIFE), holds great promise for satisfying the demand for a sensitive, rapid, cost-effective, portable, and user-friendly diagnostic platform that can translate COVID-19 assays from centralized laboratories to POC locations.
Table 4.
Aptamer-based diagnostic strategies for COVID-19
| Amplification strategies | Target | LOD | Time (min) | Portability | Ref |
|---|---|---|---|---|---|
| RPA | N gene | 30 copies reaction−1 | 30 | Portable | [55] |
| Ligation-dependent isothermal reaction | RNA-dependent RNA polymerase (RdRp) gene | 1 aM | 30 ~ 50 | Nonportable | [57] |
| CRISPR-Cas13 amplification | N gene | 80 copies reaction−1 | 20 | Nonportable | [58] |
| PLA | N protein | 0.0309 ng mL−1 | 120 | Nonportable | [59] |
| Thermoswimming signal amplification | S protein | 26,000 aM | 15 | Nonportable | [60] |
| PIFE | N protein | 50 ng mL−1 | 2 | Portable | [62] |
Representative examples of nucleic acid enzyme-based diagnostic strategies
NAEs have been identified to catalyze many chemical reactions, including cleavage, ligation, and phosphorylation of DNA or RNA, and applications in analytical chemistry, gene therapy, and nanotechnology have been most widely studied. These features have been successfully applied in various platforms for COVID-19 diagnosis. In general, according to their functions, NAEs can be categorized into three types: RNA-cleaving DNAzymes [52, 53], horseradish peroxidase-mimicking DNAzymes [63], and a multicomponent split XNAzyme [46]. NAE-based sensors for SARS-CoV-2 detection have rapidly progressed and can serve as either receptors of RNA or signal amplifiers.
RNA cleavage DNA enzymes are the first discovered catalytic DNA, and they can cleave RNA in a sequence-specific manner. Of current significance in drug therapy, virus control, and gene silencing, its relevance in the diagnostic context is being explored. Pan et al. [52] proposed a novel coronavirus detection logic DNA circuit based on exonuclease III and DNAzyme dual identification element control. Synergistic signal amplification combined with exonuclease III and DNAzyme effectively improves the detection sensitivity. Anantharaj et al. [53] used peroxidase DNAzyme as a catalyst to induce the generation of a DNAzyme sensor by PCR and reported a visible colorimetric approach for the assay of SARS-CoV-2 RNA toward a traditional thermal cyclometer, which can detect up to 1000 viral RNAs.
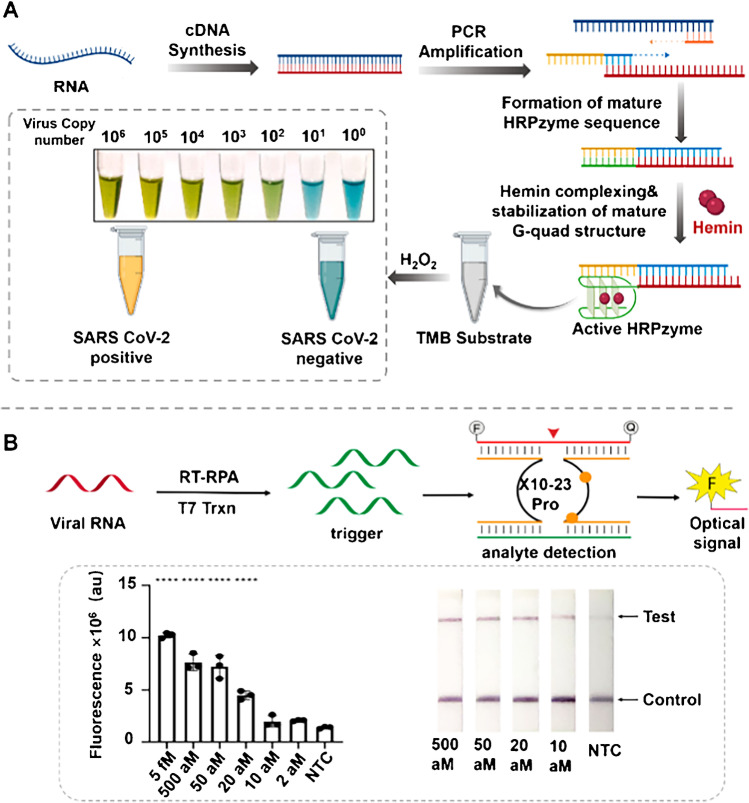
Horseradish peroxidase-mimicking DNAzyme (HRPzyme) is a short DNA sequence composed of guanine (G)-rich sequences. It can fold into a stable G-quadruple complex structure with hemin [64]. In addition, HRPzyme can catalyze the peroxidation of various HRP substrates, which also makes HRPzyme have good application prospects in the biomedical field. Ahmad et al. [51] proposed a novel optical measurement HRPZyme-assisted assay (HARIOM) of COVID-19 (Fig. 4A), which combines the specificity of NATs with the sensitivity of enzyme determination to greatly improve the signal strength and sensitivity of colorimetry. The method detects up to 10 RNA components of the synthetic virus.
Fig. 4.
A Schematic illustration of the working principle of the HARIOM assay. Reproduced with permission from [51], Copyright 2021, Elsevier. B XNAzyme-mediated sensor detection of SARS-CoV-2. Reproduced with permission from [46], Copyright 2021, American Chemical Society
XNAs have the ability to substitute backbone chemicals that do not exist in nature and can fold into well-defined structures and bind ligands, raising the possibility that they may also form catalysts (XNAzymes). As shown in Fig. 4B, Yang et al. [46] reported a multicomponent nucleic acid testing system based on XNAzyme, which combines preamplification of analytes and X10-23-mediated catalysis to detect SARS-CoV-2. The platform uses traditional fluorescence and paper-based lateral flow readout modalities within an LOD of 20 aM within 1 h, which provides a method toward the fast diagnosis of COVID-19.
Few DNAzyme-based diagnostic applications for COVID-19 exist. By rationally engineering responsive DNAzymes that can exploit the complementary nature of DNA for controlled capture and release while introducing a signal amplification strategy, it is promising to build highly sensitive, highly specific systems for POC applications. Not to be overlooked is the issue of stability of DNAzymes in complex samples (clinical samples), for which there are currently little data, but this is a critical step in the translation of DNAzyme-based assays into commercial products.
Representative examples of diagnostic strategies incorporating nanomaterials
Nanomaterial-based biosensors are constructed with fast, highly sensitive, selective, user-friendly, scalable, realistic, and portable properties, which fits our needs for POC diagnostics for COVID-19. Various types of nanomaterials, for instance metal nanoparticles (NPs), graphene oxide (GO), graphene, quantum dots (QDs), and nanocomposites, have been exploited within the diagnostics of virally infected illnesses. These nano-sensing platforms offer great sensitivity and minimal false-positive/negative responses [65]. Nanomaterials can be functionalized by antibodies/nucleic acids and are ideally suited for highly sensitive COVID-19 assays due to their excellent surface-to-volume ratio, which allows for high interaction strength.
Commonly used nanomaterials for COVID-19 diagnosis include gold nanoparticles, quantum dots, carbon-based nanoparticles, and magnetic nanoparticles. Gold nanoparticles are one of the most commonly used nanomaterials in disease diagnosis owing to their excellent optical properties, catalytic properties, and easy combination of multiple substances. Pan et al. [66] reported a colorimetric bioassay on basis of AuNPs that utilized an all-inclusive targeting approach mediated via four of the thiol-modified antisense oligonucleotide sequences covering two regions of the SARS-CoV-2 N gene and then diagnosed positive cases into 10 min from the isolated RNA biosamples. Upon inclusion with RNase H, splits in the RNA strand away from the complex heterodimer of RNA and Au-ASOmix result from visible precipitation in solution being mediated through added clustering between the AuNPs. The method has high selectivity and can be viewed with the naked eye and does not require any complex instrument technology, which is worth advocating and further studying.
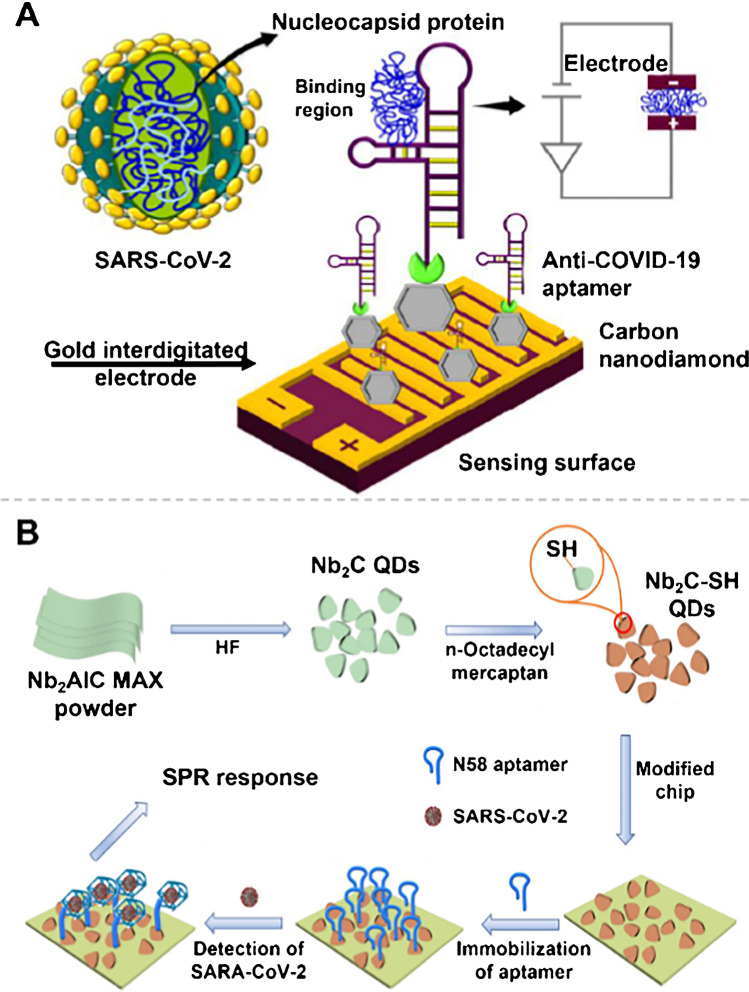
Nanomaterials serve as labels that allow for significant signal improvements. Ramanathan et al. [67] deposited ~ 20 nm diamond on the surface of a 10-μm notched gold interdigitated electrode (AuIDE), which enhanced the sensing of SARS-CoV-2 N protein within an LOD of 0.389 fM (Fig. 5A). As a result, the accomplished modification of nanomaterials on microsized AuIDE provides reliable assay support for immobilizing anti-SARS-CoV-2 aptamers at high density and offers opportunities for future research efforts to develop ultrasensitive sensing surfaces using multidimensional nanomaterials and aptamers.
Fig. 5.
A Schematic of N protein detection using an anti-N protein aptamer on nanodiamond-enhanced AuIDE. Reproduced with permission from [67], Copyright 2022, Elsevier. B Schematic of the Nb2C-SH QD-based SPR aptasensor for detecting the N gene of SARS-CoV-2. Reproduced with permission from [68], Copyright 2021, Spring Nature
The incorporation of nanomaterials can broaden the detector and signal readout; for example, Chen et al. [68] constructed a novel aptamer-based unlabeled surface plasmon resonance (SPR) approach to detect the SARS-CoV-2 N gene (Fig. 5B). Among others, SPR is frequently applied to a variety of immunoassay strategies for the determination of interactions between various biomolecules (e.g., antigen/antibody, DNA/RNA, and protein). Thiol-modified MXene quantum dots (Nb2C-SH QDs) as the bioplatform were utilized with an LOD of 4.9 pg mL−1.
Interfacing functional nucleic acids with portable analyzers to overcome barriers of signal readout
While it has been widely recognized that affordable and pocket-size signal readout devices play a vital role in COVID-19 diagnostics in resource-limited settings, it often takes a tremendous amount of resources to develop such devices. A major challenge is the requirement of enormous financial and personnel expenses as well as time during the research and development (R&D) process, making it difficult to be affordable to the general public. In addition, devices that have been developed are often dedicated devices that can detect only one or a few targets. In this context, an alternative method is to integrate well-developed bioassays with current commercially available portable analyzers via new signal transduction strategies, which has emerged as a leading and active field for research with numerous encouraging outcomes.
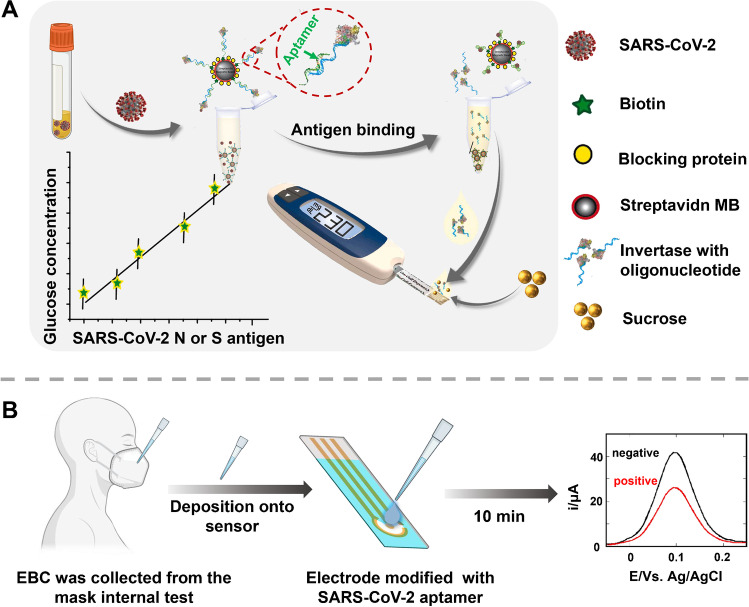
A glucose meter is a kind of electronic medical equipment used to measure glucose concentration in blood and has been widely used in clinical diagnosis. Due to the advantages of fast detection speed, ease of carry, low cost, and simple operation, the glucose meter has been successfully applied in-home testing. Combining a simple and convenient glucose meter with a COVID-19 test has been a challenging and interesting study. Singh et al. [69] successfully detected SARS-CoV-2 viral antigen in saliva samples using an off-the-shelf portable glucose meter. As shown in Fig. 6A, a single-strand aptamer targeting a viral S or N protein first binds to a complementary strand of an aptamer that binds high-efficiency invertase. In the presence of an antigen, the aptamer binds to the antigen, and the double-stranded structure opens, freeing the invertase. Invertase hydrolyzes sucrose into glucose and then generates a detection signal, and the test developed detects SARS-CoV-2 infection within a variety of viral loads in a patient’s saliva within 1 h. Our research group also tried this work, and we describe concept validation for a target response CRISPR-portable glucose meter (PGM) platform that converts SARS-CoV-2 signals into glucose signals for assay. Employing this device, the N gene, N protein, and pseudovirus of SARS-CoV-2 were quantified [70].
Fig. 6.
A Overview of the proposed point-of-care, aptamer-based COVID-19 assay. SARS-CoV-2 N or S protein-specific biotinylated aptamer is conjugated to a streptavidin-coated magnetic bead (MB) and pre-hybridized with a complementary antisense oligonucleotide strand that is covalently attached to an invertase enzyme. Saliva samples are added and invertase-antisense oligo released upon binding to viral antigens or SARS-CoV-2 viral particles. The solution containing the released antisense invertase is then collected and incubated with sucrose. Invertase converts sucrose to glucose that is directly readout using a glucometer. Reproduced with permission from [69], Copyright 2021, Elsevier. B Exhaled breath condensate (EBC)-based diagnostic strategy for SARS-CoV-2 infectivity. Laboratory-engineered mask allows collection of EBC by first cooling the mask for 30 min in the freezer, putting on the cooled mask, and breathing into it for 5 min. EBS formed in the Teflon lining of the inside of the mask is collected and directly deposited onto an electrochemical sensing modified with SARS-CoV-2-specific aptamer targeting the RBD region of the S1 spike protein as surface receptor. Using ferrocenemethanol as a redox meditator before and after viral interaction allows discrimination between positive and negative EBC samples. Reproduced with permission from [71], Copyright 2021, Elsevier
The continuous development of electrochemical biosensors and smartphones has promoted the development of smartphone-based electrochemical meters. First, various electrochemical biosensors are very suitable for POC diagnosis owing to their merits of simple operation, fast detection speed, and high sensitivity. The combination of smartphones and POC devices makes portable and inexpensive biosensors possible. Currently, smartphone-based electrochemical meters have been reported to detect various major disease markers, and people are gradually considering combining the COVID-19 test with smartphone-based electrochemical meters. Daniels et al. [71] prepared an electrochemical biosensor based on an adaptor and successfully combined the sensor with a smartphone (Fig. 6B). Targeting the S protein embedded in the lipid membrane of the outer wall of the SARS-CoV-2 virus, the sensor can detect SARS-CoV-2 virus particles in cultured SARS-CoV-2 suspension below 10 pfu mL−1.
To further advance FNA-based diagnostic devices from laboratory demonstration to large-scale industrial production, the use of commercially available readers is currently the most accessible option. Personal glucose meters, smartphone-based electrochemical meters, and handheld Raman analyzers have accelerated the development of POC devices for early diagnosis and mass screening for COVID-19. At the same time, the development of more commercially available POC devices has facilitated increased portability and accuracy in rapid disease diagnosis. Highly scalable portable analyzer-based testing, we believe that smartphones are excellent terminals for data collection, processing, and storage. Based on the Internet of Things, many devices can be connected to smartphones, allowing people to more easily access their personal test results and track their health status in real time.
Conclusions and outlook
The speedy and precise diagnostics for COVID-19 in POC settings performed a vital function within the current outbreak management. Given the progress made in the past 3 years, we have summarized the major scientific barriers that current POC technologies are facing from the perspective of POC assay of SARS-CoV-2, and we subsequently provided fundamental insights into FNAs and technical tricks for molecular engineering of FNAs, comprising aptamers and NAEs, in order to overcome these barriers. FNAs possess fascinating features and are competitive nominees in designing of diagnostic stratagems. By way of illustration, diverse signaling tactics that include electrochemistry, fluorescence, SERS, thermal conductivity, and PGM have been incorporated into aptamers and NAEs to detect various COVID-19 biomarkers. The emergence of these burgeoning FNA-based diagnosis platforms represents a valuable kit for the POC assay of COVID-19. As illustrated in Fig. 7, we conclude by outlining further opportunities and emerging applications for FNA-based sensors against COVID-19.
Fig. 7.
Further opportunities and emerging applications for FNA-based POC sensors against COVID-19
Better understanding of structure and mechanism
Despite the successful POC applications of FNAs to detect SARS-CoV-2, we still know very little about the structural features and reaction mechanisms of these FNAs. Theoretically, high affinity aptamers were obtained by in vitro SELEX; however, the outcome of SELEX was still highly unpredictable, and the ability to design FNAs from first principles was still available. Moreover, several in vivo requirements may have been overlooked owing to the in vitro SELEX procedure. Such an unknown character affects the structural arrangement of the aptamer to some extent and may lead to a lack of affinity and specificity of the aptamer in real samples. In addition, the low chemical diversity of aptamers and instability may contribute to a decrease in sensitivity. From the perspective of improving the stability of FNAs, it is urgent to explore more ways. With a deeper understanding of the structural features responsible for FNA-virus (biomarker) interactions, we identified the key contributing factors and mechanisms of FNA-related unique target recognition and catalytic properties [72], thereby developing next-generation diagnostic techniques with improved rapidity, simplicity, sensitivity, and specificity.
Home testing for daily COVID-19 screening: a potential game changer
Home testing represents a compelling tactic for combating widespread COVID-19 epidemics and preventing medical resource overload, which is an essential tool for isolating at home, screening each day, and surveillance for critical symptomatology. Moreover, while large-scale vaccinations have been initiated, routinely monitored against SARS-CoV-2 remains essential, yet vital problems remain attributable to mutations of the virus that make the vaccine less effective. In this case, the transfer of SARS-CoV-2 testing from centralized laboratories to self-monitoring by individuals at home is essential to meet the need for high-frequency COVID-19 monitoring and increase prevention and treatment effectiveness. To this end, a number of portable equipment have been designed with applications to quickly and sensitively diagnose COVID-19 in a POC format. Despite their considerable promise, the inability of these devices to satisfy the key demands upon home self-testing in terms of user-friendliness and expense has reduced their market expectations and value. Accordingly, the development and commercialization of a singular instrument that satisfies the requirements of sensitivity, specificity, generality, and adaptability to varying objectives, low cost, portability, and, crucially, self-test adaptability remain a necessity that cannot be fulfilled within the presently existing diagnostic platforms.
Wearable COVID-19 sensors for continuous vital sign monitoring
Wearable sensors that conform to the skin surface and allow continuous monitoring of user physiological signals can play an important role in the early warning of COVID-19 infections as well as the process of treatment and recovery [73]. While wearable sensors have ignited tremendous interest over the past decade, their potential for tackling COVID-19 is still in its infancy. Existing wearable sensors commonly monitor vital physiological signs, including temperature, respiratory rate, peripheral oxygen saturation, and metabolic biomarkers, which are regarded as critical signatures necessary for the diagnosis and monitoring of COVID-19. Multiple groups of researchers, in the period since the outbreak of the disease, have explored the strategic applications of wearable sensors that have recently been designed to detect and monitor the status of patients with SARS-CoV-2 infection [74]. Although these tools can offer an earlier intervention in serious cases and efficient allocation of medical resources to improve patient outcomes, unfortunately, at present, these technologies have not met the future demand of telemedicine to better control the COVID-19 pandemic. In designing wearable devices for preventive and therapeutic monitoring of COVID-19, there is first a need to overcome the problem of material selection, i.e., high-performance detection. Second, the integration of multiple targets is an issue to be considered, and data analysis is challenging. Finally, noninvasive and comfortable technology is a high requirement for users.
An explorative journey from POC diagnostics to therapeutics
Despite the considerable advantages of FNAs as diagnostic candidates for COVID-19, FNA-based therapy has emerged as a compelling and promising strategy in the fight against the COVID-19 epidemic. Currently, aptamers for COVID-19 therapy can be performed through strategies such as inhibition of the RBD of S protein and ACE2 interaction to inhibit viral infection [75], RBD-independent mechanisms to inhibit pseudoviral infection, and binding to the SARS-CoV-2 main protease to interfere with viral replication. Although not in direct usage of NAEs as therapeutic agents against COVID-19, alternative strategies for the treatment of the SARS-CoV-2 RNA genome have been devised, including ribonuclease targeting chimera degraders (RIBOTAC), RNA G-quadruplex, targeting bulge structures in the 5′ UTR of the RNA genome of SARS-CoV-2, and siRNA-nanoparticle-based therapeutic against COVID-19.
Despite the remarkable progress in engineering FNAs against SARS-CoV-2, there are still several challenges that restrict their commercial application, and we need to consider the further execution of FNA-based research against COVID-19 for therapeutic purposes:
-
i.
The genomic targeted domains of SARS-CoV-2 RNA are supposed to be extremely well conserved and essential to viral replication and, crucially, for NAE binding and subsequent efficient shearing with affinity.
-
ii.
Therapeutic aptamers suffer on entry into treatment regimens with high kidney clearances, security, and pharmaceutical delivery concerns, and underlying issues of FNA circulation, cleavage, and in vivo biological distribution should be taken into account.
-
iii.
FNAs that act as neutralizing antibodies can be designed to combine with nanomaterials, multivalent multisite, and spatial site resistance, particularly aptamer-based nanostructures, synergistically resulting in increased blocking efficiency.
-
iv.
Delivery of FNA formulations in a precisely controlled manner also demands additional innovative methodologies, with effective delivery methods being a key ingredient. Nanomaterial-based carriers, rapidly engineered for efficient delivery of FNAs, hold a promising direction for next-level FNA-based anti-COVID-19 drug therapy.
Concluding remarks
In view of the current pace of developments and the tremendous market opportunities for POC assays of SARS-CoV-2, the continued development of FNA-based diagnostic or therapeutic tools is expected to revolutionize management to better control the COVID-19 pandemic. Overcoming the critical scientific barriers through an extensive collaboration of experts in chemistry, engineering, and computer science would further broaden the scope of FNA-based diagnostic strategies and potentially facilitate their translation into POC clinical applications to combat COVID-19.
Funding
We received financial support from the National Natural Science Foundation of China (no. 22004063) and Natural Science Foundation of Jiangsu Province (no. 20200303).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Almaghaslah D, Kandasamy G, Almanasef M, Vasudevan R, Chandramohan S. Review on the coronavirus disease (COVID-19) pandemic: its outbreak and current status. Int J Clin Pract. 2020;74:e13637. doi: 10.1111/ijcp.13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ. Coronavirus disease 2019 (COVID-19) a perspective from China. Radiology. 2020;296:E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asghari A, Wang C, Yoo KM, Rostamian A, Xu X, Shin J-D, Dalir H, Chen RT. Fast, accurate, point-of-care COVID-19 pandemic diagnosis enabled through advanced lab-on-chip optical biosensors: opportunities and challenges. Appl Phys Rev. 2021;8:031313. doi: 10.1063/5.0022211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, Chen H, Mubareka S, Gubbay JB, Chan WCW. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 5.Turonova B, Sikora M, Schuermann C, Hagen WJH, Welsch S, Blanc FEC, von Buelow S, Gecht M, Bagola K, Hoerner C, van Zandbergen G, Landry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Muehlebach MD, Hummer G, Locker JK, Beck M. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mousavizadeh L, Ghasemi S. Review article genotype and phenotype of COVID-19: their roles in pathogenesis. J Microbiol Immunol Infect. 2021;54:159–163. doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sui Z, Dai X, Lu Q, Zhang Y, Huang M, Li S, Peng T, Xie J, Zhang Y, Wu C, Xia J, Dong L, Yang J, Huang W, Liu S, Wang Z, Li K, Yang Q, Zhou X, Wu Y, Liu W, Fang X, Peng K. Viral dynamics and antibody responses in people with asymptomatic SARS-CoV-2 infection. Signal Transduct Target Ther. 2021;6:181. doi: 10.1038/s41392-021-00596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/s1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Ren L, Zhang L, Wang Y, Guo L, Wang C, Xiao Y, Wang Y, Rao J, Wang X, Liu Y, Huang C, Gu X, Fan G, Li H, Lu B, Cao B, Wang J. High anal swab viral load predisposes adverse clinical outcomes in severe COVID-19 patients. Emer Microbes Infect. 2020;9:2706–2713. doi: 10.1080/22221751.2020.1858700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasan Ali O, Bomze D, Risch L, Brugger SD, Paprotny M, Weber M, Thiel S, Kern L, Albrich WC, Kohler P, Kahlert CR, Vernazza P, Bühler PK, Schüpbach RA, Gómez-Mejia A, Popa AM, Bergthaler A, Penninger JM, Flatz L. Severe coronavirus disease 2019 (COVID-19) is associated with elevated serum immunoglobulin (Ig) A and antiphospholipid IgA antibodies. Clin Infect Dis. 2020;73:e2869–e2874. doi: 10.1093/cid/ciaa1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Hogan CA, Verghese M, Solis D, Sibai M, Huang C, Röltgen K, Stevens BA, Yamamoto F, Sahoo MK, Zehnder J, Boyd SD, Pinsky BA. SARS-CoV-2 nucleocapsid plasma antigen for diagnosis and monitoring of COVID-19. Clin Chem. 2021;68:204–213. doi: 10.1093/clinchem/hvab216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, Gao G, Hu X, Zhang Y, Tong Z, Huang W, Liu WJ, Wu G, Zhang B, Wang L, Qi J, Feng H, Wang F-S, Wang Q, Gao GF, Yuan Z, Yan J. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 13.Huff HV, Singh A. Asymptomatic transmission during the coronavirus disease 2019 pandemic and implications for public health strategies. Clin Infect Dis. 2020;71:2752–2756. doi: 10.1093/cid/ciaa654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alsaafin A, McKeague M. Functional nucleic acids as in vivo metabolite and ion biosensors. Biosens Bioelectron. 2017;94:94–106. doi: 10.1016/j.bios.2017.02.030. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Lan T, Lu Y. Molecular engineering of functional nucleic acid nanomaterials toward in vivo applications. Adv Healthcare Mater. 2019;8:1801158. doi: 10.1002/adhm.201801158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gowri A, Kumar NA, Anand BSS. Recent advances in nanomaterials based biosensors for point of care (PoC) diagnosis of Covid-19-a minireview. Trac-Trends Anal Chem. 2021;137:116205. doi: 10.1016/j.trac.2021.116205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen G, Marino J, Wang Z-X, Beavis KG, Rodrigo J, Labog K, Westblade LF, Jin R, Love N, Ding K, Garg S, Huang A, Sickler J, Tran NK. Clinical performance of the point-of-care cobas Liat for detection of SARS-CoV-2 in 20 minutes: a multicenter study. J Clin Microbiol. 2021;59:e02811. doi: 10.1128/jcm.02811-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeffelholz MJ, Tang Y-W. Detection of SARS-CoV-2 at the point of care. Bioanalysis. 2021;13:1213–1223. doi: 10.4155/bio-2021-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M, Tang Y, Qi L, Zhang S, Liu Y, Lu B, Yu J, Zhu K, Li B, Du Y. SARS-CoV-2 point-of-care (POC) diagnosis based on commercial pregnancy test strips and a palm-size microfluidic device. Anal Chem. 2021;93:11956–11964. doi: 10.1021/acs.analchem.1c01829. [DOI] [PubMed] [Google Scholar]
- 20.Huynh Quoc N, Bui HK, Vu Minh P, Seo TS. An internet of things-based point-of-care device for direct reverse-transcription-loop mediated isothermal amplification to identify SARS-CoV-2. Biosens Bioelectron. 2022;195:113655. doi: 10.1016/j.bios.2021.113655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Puig H, Lee RA, Najjar D, Tan X, Soekensen LR, Angenent-Mari NM, Donghia NM, Weckman NE, Ory A, Ng CF, Nguyen PQ, Mao AS, Ferrante TC, Lansberry G, Sallum H, Niemi J, Collins JJ. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Science Advances. 2021;7:eabh294. doi: 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haghayegh F, Salahandish R, Zare A, Khalghollah M, Sanati-Nezhad A. Immuno-biosensor on a chip: a self-powered microfluidic-based electrochemical biosensing platform for point-of-care quantification of proteins. Lab Chip. 2021;22:108–120. doi: 10.1039/d1lc00879j. [DOI] [PubMed] [Google Scholar]
- 23.Jia J, Ao L, Luo Y, Liao T, Huang L, Zhuo D, Jiang C, Wang J, Hu J. Quantum dots assembly enhanced and dual-antigen sandwich structured lateral flow immunoassay of SARS-CoV-2 antibody with simultaneously high sensitivity and specificity. Biosens Bioelectr. 1800;198:113810. doi: 10.1016/j.bios.2021.113810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samper IC, Sanchez-Cano A, Khamcharoen W, Jang I, Siangproh W, Baldrich E, Geiss BJ, Dandy DS, Henry CS. Electrochemical capillary-flow immunoassay for detecting anti-SARS-CoV-2 nucleocapsid protein antibodies at the point of care. ACS Sens. 2021;6:4067–4075. doi: 10.1021/acssensors.1c01527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pietschmann J, Voepel N, Voss L, Rasche S, Schubert M, Kleines M, Krause H-J, Shaw TM, Spiegel H, Schroeper F. Development of fast and portable frequency magnetic mixing-based serological SARS-CoV-2-specific antibody detection assay. Front Microbiol. 2021;12:643275. doi: 10.3389/fmicb.2021.643275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu J, Chenier M, Zhang Y, Xu C-q. A microflow cytometry-based agglutination immunoassay for point-of-care quantitative detection of SARS-CoV-2 IgM and IgG. Micromachines. 2021;12:443. doi: 10.3390/mi12040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ju Y, Kim J, Park Y, Lee CY, Kim K, Hong KH, Lee H, Yong D, Park HG. Rapid and accurate clinical testing for COVID-19 by nicking and extension chain reaction system-based amplification (NESBA) Biosens Bioelectron. 2022;196:113689. doi: 10.1016/j.bios.2021.113689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behrouzi K, Lin L. Gold nanoparticle based plasmonic sensing for the detection of SARS-CoV-2 nucleocapsid proteins. Biosens Bioelectron. 2022;195:113669. doi: 10.1016/j.bios.2021.113669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beduk T, Beduk D, de Oliveira Filho JI, Zihnioglu F, Cicek C, Sertoz R, Arda B, Goksel T, Turhan K, Salama KN, Timur S. Rapid point-of-care COVID-19 diagnosis with a gold-nanoarchitecture-assisted laser-scribed graphene biosensor. Anal Chem. 2021;93:8585–8594. doi: 10.1021/acs.analchem.1c01444. [DOI] [PubMed] [Google Scholar]
- 30.Zahradnik J, Marciano S, Shemesh M, Zoler E, Harari D, Chiaravalli J, Meyer B, Rudich Y, Li C, Marton I, Dym O, Elad N, Lewis MG, Andersen H, Gagne M, Seder RA, Douek DC, Schreiber G. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol. 2021;6:1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- 31.Del Vecchio C, Brancaccio G, Brazzale AR, Lavezzo E, Onelia F, Franchin E, Manuto L, Bianca F, Cianci V, Cattelan A, Toppo S, Crisanti A. Emergence of N antigen SARS-CoV-2 genetic variants escaping detection of antigenic tests. medRxiv. 2021;413:49. doi: 10.1101/2021.03.25.21253802. [DOI] [Google Scholar]
- 32.Telenti A, Arvin A, Corey L, Corti D, Diamond MS, Garcia-Sastre A, Garry RF, Holmes EC, Pang PS, Virgin HW. After the pandemic: perspectives on the future trajectory of COVID-19. Nature. 2021;596:495–504. doi: 10.1038/s41586-021-03792-w. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Li X, Li T, Zhang S, Wang L, Wu X, Liu J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur J Clin Microbiol Infect Dis. 2020;39:1629–1635. doi: 10.1007/s10096-020-03899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krajewski R, Golebiowska J, Makuch S, Mazur G, Agrawal S. Update on serologic testing in COVID-19. Clin Chim Acta. 2020;510:746–750. doi: 10.1016/j.cca.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu H, Zhang H, Ni S, Korabecna M, Yobas L, Neuzil P. The vision of point-of-care PCR tests for the COVID-19 pandemic and beyond. Trac-Trends Anal Chem. 2020;130:115984. doi: 10.1016/j.trac.2020.115984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green DA, Zucker J, Westblade LF, Whittier S, Rennert H, Velu P, Craney A, Cushing M, Liu D, Sobieszczyk ME, Boehme AK, Sepulveda JL. Clinical performance of SARS-CoV-2 molecular tests. J Clin Microbiol. 2020;58:e00995–e1020. doi: 10.1128/jcm.00995-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eissa S, Alhadrami HA, Al-Mozaini M, Hassan AM, Zourob M. Voltammetric-based immunosensor for the detection of SARS-CoV-2 nucleocapsid antigen. Mikrochim Acta. 2021;188:199–199. doi: 10.1007/s00604-021-04867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kruger A, de Jesus Santos AP, de Sa V, Ulrich H, Wrenger C. Aptamer applications in emerging viral diseases. Pharmaceuticals. 2021;14:622. doi: 10.3390/ph14070622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta A, Anand A, Jain N, Goswami S, Ananthraj A, Patil S, Singh R, Kumar A, Shrivastava T, Bhatnagar S, Medigeshi GR, Sharma TK, Research DBTICfC A novel G-quadruplex aptamer-based spike trimeric antigen test for the detection of SARS-CoV-2. Molec Ther Nucl Acid. 2021;26:321–332. doi: 10.1016/j.omtn.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferreira-Bravo IA, DeStefano JJ. Xeno-nucleic acid (XNA) 2’-fluoro-arabino nucleic acid (FANA) aptamers to the receptor-binding domain of SARS-CoV-2 S protein block ACE2 binding. Viruses-Basel. 2021;13:1983. doi: 10.3390/v13101983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kacherovsky N, Yang LF, Dang HV, Cheng EL, Cardle II, Walls AC, McCallum M, Sellers DL, DiMaio F, Salipante SJ, Corti D, Veesler D, Pun S. Discovery and characterization of spike N-terminal domain-binding aptamers for rapid SARS-CoV-2 detection. Angew Chem Int Ed. 2021;60:21211–21215. doi: 10.1002/anie.202107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Zhang Z, Gu J, Stacey HD, Ang JC, Capretta A, Filipe CDM, Mossman KL, Balion C, Salena BJ, Yamamura D, Soleymani L, Miller MS, Brennan JD, Li Y. Diverse high-affinity DNA aptamers for wild-type and B.1.1.7 SARS-CoV-2 spike proteins from a pre-structured DNA library. Nucleic Acids Res. 2021;49:7267–7279. doi: 10.1093/nar/gkab574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Fang X, Liu X, Ou H, Zhang H, Wang J, Li Q, Cheng H, Zhang W, Luo Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem Commun. 2020;56:10235–10238. doi: 10.1039/d0cc03993d. [DOI] [PubMed] [Google Scholar]
- 44.Morena F, Argentati C, Tortorella I, Emiliani C, Martino S. De novo ssRNA aptamers against the SARS-CoV-2 main protease: in silico design and molecular dynamics simulation. Int J Mol Sci. 2021;22:6874. doi: 10.3390/ijms22136874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinheiro VB, Holliger P. The XNA world: progress towards replication and evolution of synthetic genetic polymers. Curr Opin Chem Biol. 2012;16:245–252. doi: 10.1016/j.cbpa.2012.05.198. [DOI] [PubMed] [Google Scholar]
- 46.Yang K, Chaput JC. REVEALR: a multicomponent XNAzyme-based nucleic acid detection system for SARS-CoV-2. J Am Chem Soc. 2021;143:8957–8961. doi: 10.1021/jacs.1c02664. [DOI] [PubMed] [Google Scholar]
- 47.Taylor AI, Holliger P. Directed evolution of artificial enzymes (XNAzymes) from diverse repertoires of synthetic genetic polymers. Nat Protoc. 2015;10:1625–1642. doi: 10.1038/nprot.2015.104. [DOI] [PubMed] [Google Scholar]
- 48.Sun M, Liu S, Wei X, Wan S, Huang M, Song T, Lu Y, Weng X, Lin Z, Chen H, Song Y, Yang C. Aptamer blocking strategy inhibits SARS-CoV-2 virus infection. Angew Chem Int Ed. 2021;60:10266–10272. doi: 10.1002/anie.202100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Wang Y-l, Wu J, Qi J, Zeng Z, Wan Q, Chen Z, Manandhar P, Cavener VS, Boyle NR, Fu X, Salazar E, Kuchipudi SV, Kapur V, Zhang X, Umetani M, Sen M, Willson RC, Chen S-h, Zu Y. Neutralizing aptamers block S/RBD-ACE2 interactions and prevent host cell infection. Angew Chem Int Ed. 2021;60:10273–10278. doi: 10.1002/anie.202100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitz A, Weber A, Bayin M, Breuers S, Fieberg V, Famulok M, Mayer G. A SARS-CoV-2 spike binding DNA aptamer that inhibits pseudovirus infection by an RBD-independent mechanism. Angew Chem Weinheim Bergstr Ger. 2021;133:10367–10373. doi: 10.1002/ange.202100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmad M, Sharma P, Kamai A, Agrawal A, Faruq M, Kulshreshtha A. HRPZyme assisted recognition of SARS-CoV-2 infection by optical measurement (HARIOM) Biosens Bioelectron. 2021;187:113280. doi: 10.1016/j.bios.2021.113280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan J, He Y, Liu Z, Chen J. Dual recognition element-controlled logic DNA circuit for COVID-19 detection based on exonuclease III and DNAzyme. Chem Commun. 2021;57:1125–1128. doi: 10.1039/d0cc06799g. [DOI] [PubMed] [Google Scholar]
- 53.Anantharaj A, Das SJ, Sharanabasava P, Lodha R, Kabra SK, Sharma TK, Medigeshi GR. Visual detection of SARS-CoV-2 RNA by conventional PCR-induced generation of DNAzyme sensor. Front Mol Biosci. 2020;7:586254. doi: 10.3389/fmolb.2020.586254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Magrina Lobato I, O’Sullivan CK. Recombinase polymerase amplification: basics, applications and recent advances. Trac-Trends Anal Chem. 2018;98:19–35. doi: 10.1016/j.trac.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu D, Shen H, Zhang Y, Shen D, Zhu M, Song Y, Zhu Z, Yang C. A microfluidic-integrated lateral flow recombinase polymerase amplification (MI-IF-RPA) assay for rapid COVID-19 detection. Lab Chip. 2021;21:2019–2026. doi: 10.1039/d0lc01222j. [DOI] [PubMed] [Google Scholar]
- 56.Kang J, Jang H, Yeom G, Kim M-G. Ultrasensitive detection platform of disease biomarkers based on recombinase polymerase amplification with H-sandwich aptamers. Anal Chem. 2021;93:992–1000. doi: 10.1021/acs.analchem.0c03822. [DOI] [PubMed] [Google Scholar]
- 57.Woo CH, Jang S, Shin G, Jung GY, Lee JW. Sensitive fluorescence detection of SARS-CoV-2 RNA in clinical samples via one-pot isothermal ligation and transcription. Nat Biomed Eng. 2020;4:1168–1179. doi: 10.1038/s41551-020-00617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Zhang Y, Chen J, Wang M, Zhang T, Luo W, Li Y, Wu Y, Zeng B, Zhang K, Deng R, Li W. Detection of SARS-CoV-2 and its mutated variants via CRISPR-Cas13-based transcription amplification. Anal Chem. 2021;93:3393–3402. doi: 10.1021/acs.analchem.0c04303. [DOI] [PubMed] [Google Scholar]
- 59.Liu R, He L, Hu Y, Luo Z, Zhang J. A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chem Sci. 2020;11:12157–12164. doi: 10.1039/D0SC03920A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng J, Tian F, Liu C, Liu Y, Zhao S, Fu T, Sun J, Tan W. Rapid one-step detection of viral particles using an aptamer-based thermophoretic assay. J Am Chem Soc. 2021;143:7261–7266. doi: 10.1021/jacs.1c02929. [DOI] [PubMed] [Google Scholar]
- 61.Peinetti AS, Lake RJ, Cong W, Cooper L, Wu Y, Ma Y, Pawel GT, Toimil-Molares ME, Trautmann C, Rong L, Mariñas B, Azzaroni O, Lu Y. Direct detection of human adenovirus or SARS-CoV-2 with ability to inform infectivity using DNA aptamer-nanopore sensors. Sci Adv. 2021;7:eabh2848. doi: 10.1126/sciadv.abh2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JM, Kim CR, Kim S, Min J, Lee M-H, Lee S. Mix-and-read, one-minute SARS-CoV-2 diagnostic assay: development of PIFE-based aptasensor. Chem Commun. 2021;57:10222–10225. doi: 10.1039/d1cc04066a. [DOI] [PubMed] [Google Scholar]
- 63.Zhang R, Wu J, Ao H, Fu J, Qiao B, Wu Q, Ju H. A rolling circle-amplified G-quadruplex/hemin DNAzyme for chemiluminescence immunoassay of the SARS-CoV-2 protein. Anal Chem. 2021;93:9933–9938. doi: 10.1021/acs.analchem.1c02229. [DOI] [PubMed] [Google Scholar]
- 64.Travascio P, Li YF, Sen D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem Biol. 1998;5:505–517. doi: 10.1016/s1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- 65.Iravani S. Nano- and biosensors for the detection of SARS-CoV-2: challenges and opportunities. Mater Adv. 2020;1:3092–3103. doi: 10.1039/d0ma00702a. [DOI] [Google Scholar]
- 66.Moitra P, Alafeef M, Dighe K, Frieman MB, Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramanathan S, Gopinath SCB, Ismail ZH, Arshad MKM, Poopalan P. Aptasensing nucleocapsid protein on nanodiamond assembled gold interdigitated electrodes for impedimetric SARS-CoV-2 infectious disease assessment. Biosens Bioelectron. 2022;197:113735. doi: 10.1016/j.bios.2021.113735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen R, Kan L, Duan F, He L, Wang M, Cui J, Zhang Z, Zhang Z. Surface plasmon resonance aptasensor based on niobium carbide MXene quantum dots for nucleocapsid of SARS-CoV-2 detection. Microchim Acta. 2021;188:316. doi: 10.1007/s00604-021-04974-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh NK, Ray P, Carlin AF, Magallanes C, Morgan SC, Laurent LC, Aronoff-Spencer ES, Hall DA. Hitting the diagnostic sweet spot: point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens Bioelectron. 2021;180:113111. doi: 10.1016/j.bios.2021.113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu R, Hu Y, He Y, Lan T, Zhang J. Translating daily COVID-19 screening into a simple glucose test: a proof of concept study. Chem Sci. 2021;12:9022–9030. doi: 10.1039/d1sc00512j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daniels J, Wadekar S, DeCubellis K, Jackson GW, Chiu AS, Pagneux Q, Saada H, Engelmann I, Ogiez J, Loze-Warot D, Boukherroub R, Szunerits S. A mask-based diagnostic platform for point-of-care screening of Covid-19. Biosens Bioelectron. 2021;192:113486–113486. doi: 10.1016/j.bios.2021.113486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lake RJ, Yang Z, Zhang J, Lu Y. DNAzymes as activity-based sensors for metal ions: recent applications, demonstrated advantages, current challenges, and future directions. Acc Chem Res. 2019;52:3275–3286. doi: 10.1021/acs.accounts.9b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ray TR, Choi J, Bandodkar AJ, Krishnan S, Gutruf P, Tian L, Ghaffari R, Rogers JA. Bio-integrated wearable systems: a comprehensive review. Chem Rev. 2019;119:5461–5533. doi: 10.1021/acs.chemrev.8b00573. [DOI] [PubMed] [Google Scholar]
- 74.Ding X, Clifton D, Ji N, Lovell NH, Bonato P, Chen W, Yu X, Xue Z, Xiang T, Long X, Xu K, Jiang X, Wang Q, Yin B, Feng G, Zhang Y-T. Wearable sensing and telehealth technology with potential applications in the coronavirus pandemic. IEEE Rev Biomed Eng. 2021;14:48–70. doi: 10.1109/rbme.2020.2992838. [DOI] [PubMed] [Google Scholar]
- 75.Pramanik A, Gao Y, Patibandla S, Mitra D, McCandless MG, Fassero LA, Gates K, Tandon R, Ray PC. Aptamer conjugated gold nanostar-based distance-dependent nanoparticle surface energy transfer spectroscopy for ultrasensitive detection and inactivation of corona virus. J Phys Chem Lett. 2021;12:2166–2171. doi: 10.1021/acs.jpclett.0c03570. [DOI] [PMC free article] [PubMed] [Google Scholar]