Summary
Comprehensive understanding of a protein’s function depends on having reliable, sophisticated tools to study protein structural dynamics in physiologically-relevant conditions. Here, we present an effective, robust step-by-step protocol to monitor the structural dynamics (including hydration dynamics) of a protein utilizing various site-directed fluorescence (SDFL) approaches. This protocol should be widely applicable for studying soluble proteins, intrinsically-disordered proteins, and membrane proteins.
For complete details on the use and execution of this protocol, please refer to Das et al. (2020), Das and Raghuraman (2021), and Chatterjee et al. (2021).
Subject areas: Biophysics, Molecular/Chemical Probes, Protein Biochemistry, Structural Biology
Graphical abstract

Highlights
-
•
A step-by-step protocol to monitor the structural dynamics of proteins using SDFL
-
•
Applicable to proteins with intrinsic Trp and labeled with extrinsic fluorophores
-
•
This protocol should be widely applicable for soluble and membrane proteins
Comprehensive understanding of a protein’s function depends on having reliable, sophisticated tools to study protein structural dynamics in physiologically-relevant conditions. Here, we present an effective, robust step-by-step protocol to monitor the structural dynamics (including hydration dynamics) of a protein utilizing various site-directed fluorescence (SDFL) approaches. This protocol should be widely applicable for studying soluble proteins, intrinsically-disordered proteins, and membrane proteins.
Before you begin
Intrinsic tryptophan (Trp) fluorescence is a widely used tool to monitor conformational changes in proteins and to gain information regarding local structure and dynamics (Biswas et al., 2020; Chatterjee et al., 2021). SDFL has been proved powerful in determining protein and other macromolecular structural information in complex biological systems (Raghuraman et al., 2019). Labeling of a protein with external fluorophores like N,N′-dimethyl-N-(iodoacetyl)-N’-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine (IANBD amide) and bimane has been extensively used, given its spatial resolution and high sensitivity, in understanding complex conformational variations in protein structure by site-selective probing of residues in the protein where intrinsic fluorescence is not adequate (Mansoor and Farrens, 2004; Raghuraman et al., 2019). This requires insertion of a cysteine (Cys) by site-directed mutagenesis (SDM). The reason for the preference of single-Cys mutants is that, compared to other amino acids, Cys is underrepresented in all organisms. The occurrence frequency of Cys, on average, is ∼1.4% of all amino acids in proteins from different domains of life (Wiedemann et al., 2020). Even this value is only ∼2.5% in mammalian proteins (Hansen et al., 2009; Wiedemann et al., 2020). Further, some proteins do not even contain Cys residue so it becomes easy to generate single-Cys mutants as in the case of KcsA potassium channel, CorA and MgtE magnesium channels etc. In proteins with multiple Cys residues, the multiplicity is often small enough that it is feasible to obtain single-Cys mutants by site-directed mutagenesis without significant disruption of the structure or function of the protein.
Here, we present a detailed working protocol starting with thiol-reactive fluorophore labeling for single-Cys mutants and utilizing various easy-to-perform fluorescence approaches, which are applicable for proteins with intrinsic Trp and/or labeled with extrinsic fluorophores.
Preparation of growth media, LB agar plate, and competent cells
Timing: 1–2 days
-
1.
Details for the preparation of different types of growth media such as Luria-Bertani (LB), 2YT, Terrific broth, minimal media etc. and LB agar plate are available online.
-
2.
Prepare competent cells
The widely popular ‘one-step method of competent cell preparation’ (Chung and Miller, 1993) is followed using the sterile Transformation and Storage Solution (TSS).
TSS is LB broth with 10% PEG (molecular weight 3350 or 8000), 5% DMSO, and 40 mM Mg2+ (MgSO4 or MgCl2) at a final pH of 6.5. Perform the following steps aseptically.-
a.Inoculate 2–5 mL of LB media with E. coli strain of choice in a polypropylene (PP) tube with cap and incubate ∼12–16 h at 37°C in shaking condition (250 rpm).
 CRITICAL: Make sure that the PP tubes containing 2–5 ml LB media are previously autoclaved, which can be stored for weeks. However, check for any growth in the tubes before starting the procedure.
CRITICAL: Make sure that the PP tubes containing 2–5 ml LB media are previously autoclaved, which can be stored for weeks. However, check for any growth in the tubes before starting the procedure. -
b.Take 25 mL of LB media in a sterile 100 mL Erlenmeyer flask, and inoculate with 250 μL of the saturated overnight culture (1:100 dilution) and incubate at 37°C with uniform rotation at 250 rpm.
 CRITICAL: TSS needs to be prepared beforehand and stored at −20°C in aliquots. Working aliquots can be stored at 4°C.
CRITICAL: TSS needs to be prepared beforehand and stored at −20°C in aliquots. Working aliquots can be stored at 4°C. -
c.Grow the cells until the optical density of the culture at 600 nm (OD600) is 0.4 (0.4 ≤ OD600 ≤ 0.5). This will take ∼ 2–3 h depending on the E. coli strain.
 CRITICAL: Make sure that OD600 does not exceed above 0.5. TSS should be ice-cold by this time.
CRITICAL: Make sure that OD600 does not exceed above 0.5. TSS should be ice-cold by this time. -
d.Once proper OD600 has been achieved, transfer 25 mL of culture into a pre-chilled 50 mL Falcon tube and keep it in ice for 20 min.
-
e.Centrifuge at 1200–1500×g for 10 min at 4°C.
-
f.Resuspend the cells gently in 2.5 mL (1:10 dilution) of ice-cold TSS.
-
g.Prepare/label 25 × 1.5 mL Eppendorf tubes during this time and pre-chill them in ice.
-
h.Distribute 100 μL of TSS suspended competent cells to each pre-chilled Eppendorf tube while ensuring the cells remain well mixed.
-
i.Cells can be used immediately or stored at −80°C freezer.
 CRITICAL: To prevent any further growth of the cells, and preparation of good quality competent cells, make sure that the 50 ml Falcon tube, and the Eppendorf tubes are pre-chilled in ice for at least 20 min.
CRITICAL: To prevent any further growth of the cells, and preparation of good quality competent cells, make sure that the 50 ml Falcon tube, and the Eppendorf tubes are pre-chilled in ice for at least 20 min.
-
a.
Generation of single-Trp or single-Cys mutants
Timing: 1–2 weeks
-
3.Primer design
-
a.Basic rules
-
i.Generally, PCR primers (oligonucleotide between 18–35 bases) work very well if the annealing temperature of the PCR reaction is set within a few degrees of the primer melting temperature (Tm). In our lab, we use primer length of 33 bases (Braman et al., 1996).
-
ii.PCR primers should maintain a reasonable GC content between 40 and 60% with the 3′ of a primer ending in G or C to promote binding. However, care should be taken not to have too many repeating G or C bases.
-
iii.Try to keep the Tm of the forward (Fw) and reverse (Rv) primers between 50°C to 60°C, and within 5°C of each other.
-
iv.Try to avoid runs of 4 or more of same base or dinucleotide repeats (for example, ACCCC or ATATATAT).
-
v.Avoid intra-primer homology (i.e., more than 3 bases that complement within the primer) or inter-primer homology (i.e., Fw and Rv primers having complementary sequences). These circumstances can lead to self-dimers or primer-dimers instead of annealing to the desired DNA sequences.
-
i.
-
b.Designing the Fw and Rv primers for generating single-Cys mutants
-
i.Copy the 5′ DNA sequence of 20–40 base pairs with the target mutation site in the middle.
 CRITICAL: Make sure that the target mutation site is in the middle of the primer with approx. 10–15 bases of correct sequence on either side.
CRITICAL: Make sure that the target mutation site is in the middle of the primer with approx. 10–15 bases of correct sequence on either side. -
ii.Change the mutation site to TGC or TGT (codons for Cys) for generating single-Cys mutant at the desired site.
-
iii.If basic rules are satisfied, reverse complement the Fw primer sequence to obtain the Rv primer. Otherwise, adjust the sequence length slightly (make sure to keep the mutation site in the sequence).
-
i.
-
c.Designing the Fw and Rv primers for generating single-Trp mutants
-
i.Copy the 5′ DNA sequence of 20–40 base pairs with the target mutation site in the middle.
-
ii.Change the mutation site to TGG (codons for Trp) for generating single-Trp mutant at the desired site from the Trp-less mutant plasmid.Note: To obtain single-Trp mutants from multi-Trp containing protein, it will be better to make Trp-less mutant by replacing all the existing native Trp residues to phenylalanine (Phe). Then, one Trp at a time can be introduced at desired locations in a protein.
-
iii.If basic rules are satisfied, reverse complement the Fw primer sequence to obtain the Rv primer. Otherwise, adjust the sequence length slightly (make sure to keep the mutation site in the sequence).
-
i.
-
d.Order primers for each mutation (depending on the desired site in the protein) from commercial resources. In our case, we order from IDT DNA technologies (https://www.idtdna.com/pages).
-
e.Dissolve the primers in PCR grade water to a final concentration of 40 mM and store at −20°C.
-
a.
-
4.
Perform SDM as per the DNA polymerase manufacturer’s protocol. Generally, PfuUltra High-Fidelity or KOD DNA polymerases are used. The following link provides the Instruction Manual for using the QuikChange II Site-Directed Mutagenesis Kit:
-
5.
Transform the mutated plasmid after SDM into XL1 Blue competent cells and isolate the plasmid DNA via mini-prep using the standard procedure and the commercial kit: QIAprep Spin Miniprep Kit (qiagen.com).
-
6.
Confirm the sequence for the desired mutation via any reliable commercial DNA sequencing service.
-
7.
The plasmid with the confirmed mutation can be stored at −20°C for future use.
Preparation of buffer
Timing: 1 h
-
8.
Requirement of buffers depends on the type and nature of the proteins under investigation. Several types of buffers, with different composition at different pH, are typically used for purifying various proteins. Further, a guide for salt selection for buffer preparation is available online.
However, a few things need to be kept in mind while preparing buffers as it is one of the critical steps in protein purification.-
a.Prepare buffers at the working temperature to ensure the correct pH.
-
b.Filter (use 0.22 μm filter unit) buffers after all salts and additives, if any, have been properly dissolved.
-
c.Degasing of buffers is extremely important to remove air bubbles which might cause problems in clogging the columns while using in size-exclusion chromatography.
-
d.Use a buffer concentration, typically 20–50 mM, that is sufficient to maintain buffering capacity at the desired pH.
-
e.Examples of a few commonly used buffers for protein purification are: 50 mM HEPES, 150 mM NaCl, pH 7–8; 20 mM Tris, 150 mM KCl, pH 7–8.
-
a.
Large-scale expression and purification of proteins
Timing: 1 week
Note: The growth media, competent cells and expression conditions (such as inducer concentration, temperature, expression time etc.) are protein-specific (Kaur et al., 2018; Rosano et al., 2019). The following are the general steps for IPTG-induced expression and purification of histidine (His)-tagged proteins.
-
9.Expression
-
a.Grow pre-culture by inoculating a single transformed colony (from LB agar plate) in 25 mL of LB media and incubate ∼12–15 h at 37°C with 250 rpm rotation in a shaker incubator. Do not forget to add a respective selection antibiotic.
-
b.Inoculate 1 L of LB broth with 10 mL of pre-culture (1:100 dilution).
-
c.Add respective selection antibiotic at appropriate concentration and incubate at 37°C at 250 rpm big volume shaker (like stackable shaking incubator) until the desired induction OD600 has reached.
-
d.Harvest cells by centrifugation at 3800 rpm for 40 min at 4°C, and resuspend the pellet in 30 mL of suitable resuspension buffer.
-
a.
Note: Alternatively, Beckman Coulter Avanti J-20 floor super speed centrifuge (rotor: J-LITE JLA-1.8000) can be used at 6500 rpm for 10 min to harvest cells in lesser time.
Pause Point: The resuspended cells can be stored at −80°C for several weeks.
-
10.Purification
-
a.Add protease inhibitors cocktail based on manufacturer’s instructions and make up the final volume of the resuspended cells to 45 mL.
-
b.For cell lysis, either homogenization or sonication can be used on ice. In the former, homogenize the resuspended cells for at least 2 times at 10,000 psi. For sonication using a 7 mm probe size, use 20 s on:20 s off cycle; ∼ 4–6 times at 40 % amplitude.
-
c.Centrifuge the homogenized cell suspension
-
i.In case of soluble proteins, separate the supernatant from the cell debris by centrifugation at 14000×g at 4°C for 45 min, and collect it. The supernatant contains the expressed soluble protein. After this, follow Procedure step 10e below.
-
ii.For membrane proteins, ultracentrifuge the homogenized cell suspension at 100,000×g at 4°C for 45 min, and collect the pellet, which contains the membrane with the expressed membrane protein.
-
i.
-
d.The following extra step is needed only for membrane proteins
-
i.Membrane solubilization by the suitable detergent is required to extract the expressed membrane protein by resuspending the pellet (this will take ∼1–2 h). Typically, at least 10× CMC (Critical Micelle Concentration) of detergent is used for membrane solubilization (Newby et al., 2009; Arachea et al., 2012). The experimental conditions for extracting the membrane proteins are protein-specific.
-
ii.Ultracentrifuge the membrane-solubilized suspension at 1,00,000 g at 4°C for 45 min, and collect the supernatant, which contains detergent-solubilized membrane protein. After this, follow Procedure step 10e below.
-
i.
-
e.Add the respective supernatant to immobilized metal affinity chromatography (IMAC) column (∼2 mL of packed Ni2+-NTA or Co2+ resin can be used) to specifically purify the desired protein that contains the 6×-His tag. The His-tag has a high affinity for these metal ions and binds strongly to the IMAC column.
-
f.Wash 20× column volume (i.e., ∼40 mL) with an appropriate washing buffer that contains low concentration of imidazole (varies between 5 to 50 mM depending on the protein of interest). This will help in removing the non-specifically bound proteins to IMAC column, leaving only the His-tagged protein attached to the column.
-
g.Elute the His-tagged protein using the appropriate elution buffer containing high concentration of imidazole (300–500 mM).Note: For membrane proteins, both the wash and elution buffers should contain the appropriate detergent (3–5× CMC).
-
h.Concentrate the eluted protein using a refrigerated table-top centrifuge and quantify using Bradford method or Microvolume spectrophotometer.
-
i.Check the purity of protein by SDS-PAGE and its homogeneity using size exclusion chromatography (SEC).
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides,and recombinant proteins | ||
| DMSO | Sigma-Aldrich | Cat#D8418 |
| PEG 3350 | Merck | Cat#1546547 |
| KI | Merck | Cat#221945 |
| LB media | VWR Life Science | Cat#J104 |
| LB Agar | Merck | Cat#L3027 |
| XL1-Blue cells | Agilent | Cat#200150 |
| IPTG | VWR Life Science | Cat#0487-100G |
| PMSF | Gold Biotechnology | Cat#P-470-25 |
| Protease inhibitor cocktail | Roche | Cat#11873580001 |
| IANBD amide | Thermo Fisher Scientific | Cat#D2004 |
| DTT | Gold Biotechnology | Cat#DTT25 |
| TCEP | Sigma-Aldrich | Cat#C4706 |
| Bradford Assay Reagent | Bio-Rad | Cat#500-0006 |
| Acrylamide | Merck | Cat#110784 |
| Na2S2O3.5H2O | Merck | Cat#1615107 |
| Co2+ affinity resin | Takara Bio | Cat#635504 |
| Ni-NTA affinity resin | QIAGEN | Cat#30230 |
| Ludox | Sigma-Aldrich | Cat#420832 |
| NATA | Merck | Cat#A6501 |
| Imidazole | VWR Lifescience | Cat#97064-626 |
| Critical commercial assays | ||
| PCR Kit | Merck | Cat#71086-3 |
| Miniprep Kit | QIAGEN | Cat#27106 |
| Software and algorithms | ||
| GC content and Tm calculation | Integrated DNA Technologies | https://www.idtdna.com/pages |
| DNA secondary structure predictor | Integrated DNA Technologies | https://eu.idtdna.com/calc/analyzer |
| Confirmation of DNA sequence | Eurofins | https://www.eurofins.com |
| OriginPro 8 | Origin Lab | https://www.originlab.com/demodownload.aspx |
| Decay Analysis version 2018 | Smith et al. (2017) | https://doi.org/10.7488/ds/2093 |
| Matlab 2017a | MathWorks | https://www.mathworks.com/products/matlab.html |
| Other | ||
| Quick guide to different kinds of buffer solution at different pH. | Sigma-Aldrich | https://www.sigmaaldrich.com/IN/en/technical-documents/protocol/protein-biology/protein-concentration-and-buffer-exchange/buffer-reference-center |
| Quick guide to salt selection for buffer preparation at different pH. | Sigma-Aldrich | https://www.sigmaaldrich.com/IN/en/technical-documents/technical-article/protein-biology/protein-concentration-and-buffer-exchange/salt-selection-and-buffer-preparation |
| PD10 columns | Cytiva | Cat#GE 17-0851-01 |
| PCR grade water | Sigma-Aldrich | Cat#W4502 |
| Syringes | Merck | Cat#Z683531 |
| Syringe filter (0.22 μM) | Merck | Cat#SLGV033RS |
| Filter unit (0.22 μM) | Merck | Cat#S2GPU02RE |
| Fernbach culture flask | Corning | Cat#4423-2XL |
| Filtration (Amicon) concentrator | Merck | Cat#UFC903024 Cat#UFC901024 |
| Autoclave tape | Sigma-Aldrich | Cat#A2549 |
| Amber glass vials | Merck | Cat#854172 |
| Amber eppendorf tubes | Tarsons | Cat#500013 |
| Primer resource | Integrated DNA Technologies | https://www.idtdna.com/pages |
| SDS-PAGE Protein Standard | Bio-Rad | Cat#1610374 |
| Ultracentrifuge tube | Thermo Scientific | Cat#314460 |
| Quartz Cuvette | Hellma Analytics | Cat#104-10-40-QS |
| Shaker incubator | Incubation and cell growth | Scigenics Biotech (Orbitek LT) |
| Big volume shaker | Incubation, cell growth, and protein expression | Scigenics Biotech (LE4676-AH) |
| Rotator | Mixing samples | Labnet H5500 |
| Sonicator | Cell lysis | Hielscher (UP100H) |
| Sonicator probe | Cell lysis | Sonotrode MS7 |
| Homogenizer/ Microfluidizer | Cell lysis or homogenization | Microfluidics LM-20 |
| Ultracentrifuge | Separation | Thermofisher Scientific (Sorvall wx+) |
| Ultracentrifuge rotor | Separation | Thermo Fisher Scientific (T647.5) |
| Refrigerated table-top centrifuge | Spinning down cells and for concentrating proteins | Thermo Scientific Sorvall ST4 Plus (75009911) |
| Refrigerated table-top centrifuge rotor | Spinning down cells | TX-1000 Swinging Bucket Rotor (75003017) |
| Refrigerated table-top centrifuge rotor | Concentrating proteins | Fiberlite F15-6X 100Y Fixed angle rotor (75003698) |
| Microvolume spectrophotometer | Protein quantification | DeNovix (DS-11+) |
| SEC | Protein purification | Cytiva (AktaPure 25M) |
| Spectrofluorometer∗ | Fluorescence measurements | Hitachi F7000 |
| NanoLED# | Light source for lifetime measurements | HORIBA (NanoLED-280; NanoLED-295; NanoLED-460) |
Alternatives:∗Other spectrofluorometers such as HORIBA PTI QuantaMaster (QM-8075-11-C) can also be used; #laser light source can also be used.
Optional: The resources listed above were only based on our experience. In principle, the chemicals and resources can be obtained from any reliable commercial sources and do not need to be limited to those listed in our table.
Step-by-step method details
Site-specific labeling of single-Cys mutants of proteins with thiol-reactive fluorophores
Timing: 1–2 days
Fluorophores with thiol-reactive groups such as iodoacetamides, maleimides etc. react by S-alkylation of protein thiols to generate stable thio-ether products. Although there is a wide array of such fluorophores (Haugland, 2005; Molecular Probes Handbook), small, non-perturbing fluorophores such as NBD and bimane serve as excellent tools for spectroscopic and structural mapping of proteins (Raghuraman et al., 2019). A schematic representation of labeling a single-Cys mutant of protein with thiol-reactive fluorophores is given in Figure 1.
-
1.
Add 5 mM dithiothreitol (DTT) to 1–2 mg of the purified single-Cys mutant of the protein (∼0.5–1 mL) and incubate at 4°C for 1 h.
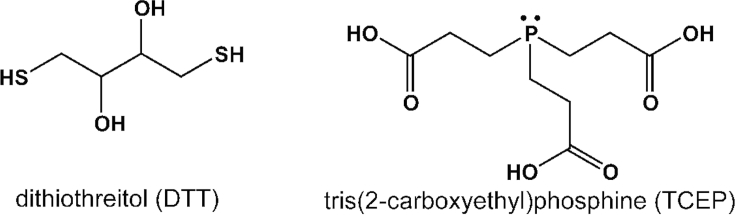
CRITICAL: DTT is a thiol-containing reagent (see Figure 2), which might compete with the Cys in the protein for conjugation with the thiol-reactive probe. Therefore, DTT should be completely removed before proceeding with fluorophore labeling.
Figure 1.
Site-specific labeling of single-Cys residue by a thiol-reactive fluorophore
Schematic representation of fluorophore labeling (top) along with the chemical structures of widely used thiol-reactive fluorophores (NBD and Bimane) for protein structural dynamics studies. The fluorophore that is attached to single-Cys is depicted as a green circle.
Figure 2.
Chemical structures of commonly used reducing agents during fluorophore labeling
Optional: Tris-(2-carboxyethyl) phosphine (TCEP) can also be used to reduce the disulfide bonds between Cys residues.
Note: For the thiol-reactive fluorophore to efficiently react with the free thiol group of the purified single-Cys mutant protein, the disulfide bonds formed (if any) must be chemically reduced before adding the fluorophore.
Unlike DTT, TCEP does not have any thiol and therefore downstream thiol labeling reactions do not generally require prior removal of TCEP (Getz et al., 1999). However, TCEP is highly reactive towards haloalkyl derivatives such as Bimane, IANBD, etc., and therefore must be removed before conjugation with these fluorescent dyes.
-
2.Remove excess DTT before protein labeling with a fluorophore.
-
a.Equilibrate PD10 desalting column with 4 column volumes (∼20 mL) of respective protein buffer without DTT.
-
b.Add 500–1,000 μL of DTT-treated protein through a pre-equilibrated PD10 desalting column and make sure the entire protein solution has gone into the column. Add 2 mL of desired protein buffer and let it pass through the column completely.
-
c.Add 3 mL of desired protein buffer and collect the flow through, and this eluent should contain the protein without DTT.
-
a.
Optional: Dialysis or SEC can also be used to remove the excess reducing agent if a desalting column is not available.
Note: PD10 desalting column is generally preferred since it is less time-consuming than doing SEC or dialysis. Further, dialysis might cause the protein to aggregate leading to a significant loss.
-
3.
Wash the PD10 column with plenty of Milli-Q water to completely remove the free DTT trapped in column.
Note: This column is now ready for reuse, if needed (follow steps 2 and 3).
-
4.
Concentrate the protein using Amicon centrifugal concentrators (with a desired MW cut-off) in a table-top centrifuge at 2000×g at 4°C. Check the concentration using microvolume spectrophotometer.
-
5.
Prepare a stock concentration of the fluorophore in DMSO (∼30–40 mM).
CRITICAL: Since the fluorophores are light-sensitive, the stock solutions should be protected from light as much as possible. Use amber glass vial covered with aluminum foil for storing the fluorophore stock solution.
Optional: Instead of amber glass vial, normal glass vials wrapped in an aluminum foil can also be used.
Pause Point: The stock solutions of the fluorophores can be aliquoted and stored at −20°C for months.
-
6.
From the stock solution, add 10-fold molar excess of the fluorophore with respect to the single-Cys mutant of the protein in a 1.5 mL amber Eppendorf tube. Wrap the vial in aluminum foil to protect from light, and keep it on a rotator for ∼12 h at 4°C.
Optional: Labeling can also be performed at ∼25°C. For that, the reaction mixture (light protected) should be kept for only 2 h on a rotator. Further, if amber Eppendorf tubes are not available, normal 1.5 mL Eppendorf tubes wrapped in an aluminum foil can also be used.
-
7.Remove the excess unbound/free fluorescent dye.
-
a.Equilibrate PD10 desalting column with 4 column volumes (∼20 mL) of respective protein buffer.
-
b.Add the fluorophore-containing protein solution through a pre-equilibrated PD10 desalting column and make sure the entire protein solution has gone into the column. Add 2 mL of desired protein buffer and let it pass through the column completely.
-
c.Add 3 mL of desired protein buffer and collect the flow through, and this eluent should be devoid of unbound/free fluorophore and contain the protein labeled with the fluorophore at the desired single-Cys residue.
-
a.
Optional: Dialysis or SEC can also be used to remove the excess unbound/free fluorescent dye if a PD10 desalting column is not available. See ‘Note’ in step 2.
-
8.
Concentrate the protein using Amicon centrifugal concentrators (with a desired MW cut-off) in a table-top centrifuge at 2000×g at 4°C. Check the concentration using microvolume spectrophotometer.
-
9.Check the labeling efficiency.
-
a.Use the following formula to calculate the labeling efficiency:
-
a.
| Equation 1 |
where Ax is the absorbance value of the dye at the absorption maximum wavelength. ε is molar extinction coefficient of the dye at the absorption maximum wavelength. Use ε478 = 25,000 M-1cm-1 for NBD. Ensure that the labeling efficiency of the protein with the fluorophore is at least 50% before performing fluorescence measurements.
Note: A few points that should be taken care of during site-directed fluorescence labeling with extrinsic probes:
The size and structure of the fluorophore should be non-perturbing, and small enough for efficient insertion even in relatively hydrophobic regions. NBD and Bimane probes are small and good for this purpose, whereas Alexa, BODIPY and cyanine dyes (Cy3 and Cy5) can only be used for labeling at the exposed sites.
The fluorophore should be sensitive to changes in environmental polarity. This is an important criterion for monitoring the protein folding, membrane binding events and topology of the protein (whether solvent or lipid exposed) etc.
Excitation and corresponding emission wavelengths should ideally be in the visible range so as to avoid fluorescence contributions from intrinsic Trp and Tyr residues.
Since a single fluorophore per molecule is expected, the fluorophore should have a high quantum yield to get a good signal-to-noise (S/N) ratio.
-
10.
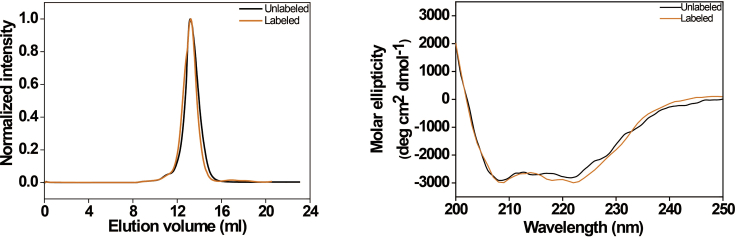
After labeling the protein, ensure the labeled protein retains the proper structural integrity by checking the homogeneity of the protein with FPLC and the secondary structure of the labeled protein with circular dichroism (CD) spectroscopy (see Figure 3).
Figure 3.
Checking the structural integrity of a purified protein after fluorophore labeling
Shown are the SEC profiles (left) and Far-UV circular dichroism (CD) spectra (right) of the purified KvAP voltage sensor before (black) and after (orange) NBD labeling. Figure modified from Das et al. (2020).
Fluorescence approaches to study labeled and Trp-containing proteins
Timing: 1–2 days/sample
-
11.General tips for measuring fluorescence (intrinsic and extrinsic) in proteins.
-
a.Quartz cuvette should be thoroughly cleaned. See the following link for cleaning suggestions: https://www.starnacells.com/d_tech/tech01.html
-
b.The slits of the excitation and emission monochromators can be adjusted to let more light through. The signal should be high to obtain a good S/N ratio, but it should be below the detector saturation limit, which otherwise can alter the detector behavior and distort the data.
-
c.Slit-width of the fluorometer should be set depending upon the protein concentration as well as labeling efficiency of protein with an external probe. Typically, excitation and emission slits with a nominal bandpass of 5 nm would suffice.
-
d.Another common problem in fluorescence spectroscopy affecting spectral measurements is the inner filter effect, which can be avoided by reducing the concentration of the sample. Typically, micromolar concentration of the native (for intrinsic fluorescence) or labeled protein can be used.
-
e.Buffers and protein samples should be filtered using a 0.22 μm syringe filter.
-
f.Once the conditions have been optimized, the measurement parameters can be set in the spectrometer software as per the need.
-
g.We use Hitachi F-7000 spectrofluorometer which allows setting the wavelength range, step size, integration time etc. All fluorescence parameters mentioned below are done at ∼25°C using a sample volume of 1 mL.
-
a.
Note: Any other Fluorescence spectrometer can be used for taking measurements.
-
12.Taking the fluorescence emission scans.
-
a.Take the emission scan of the Blank.
-
i.For soluble proteins, take the emission scan of only the buffer (same buffer in which the protein is present). This will be the Blank.
-
ii.In case of membrane proteins, the Blank contains buffer components along with micelles or liposomes. So, basically Blank should have everything except the protein.
-
i.
-
b.Now, take the emission scan of the protein sample (∼3–6 μM).Note: To measure the intrinsic fluorescence of a protein, which has only Trp and no Tyr residues, use excitation wavelength of 280 nm, whereas for proteins that contain both Trp and Tyr residues, use excitation wavelength of 295 nm to predominantly excite Trp residues and remove the contributions from Tyr. Then take the emission scan from 305 to 400 nm. Use ‘corrected spectra’ option.If the single-Cys mutant is labeled with an extrinsic fluorophore like NBD, use excitation wavelength of 465 nm and take the emission scan from 475 to 600 nm.
 CRITICAL: Rayleigh scattering is an elastic scattering and appears at the same wavelength as that of the excitation wavelength. Rayleigh scattering is very intense, and hence start the emission scan range at least 10 nm red shifted from the respective excitation wavelength.
CRITICAL: Rayleigh scattering is an elastic scattering and appears at the same wavelength as that of the excitation wavelength. Rayleigh scattering is very intense, and hence start the emission scan range at least 10 nm red shifted from the respective excitation wavelength. -
c.Subtract the buffer Blank spectrum from the emission spectrum of the protein sample using the in-built software of the respective fluorometer.
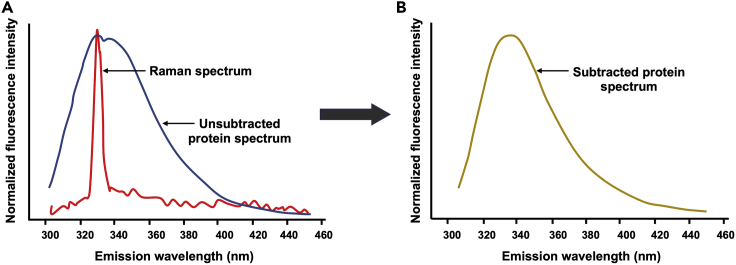
 CRITICAL: Blank subtraction from the protein emission spectrum is extremely important to avoid potential artifacts caused by the Raman scattering, which is an inelastic scattering and appears after the excitation wavelength. Raman scattering is a big problem in measuring Trp fluorescence especially at excitation wavelengths of 295–310 nm (see REES experiments below) since the Raman peak overlaps with the spectral contribution from Trp residues (see Figure 4).Note: For measuring the emission spectrum of NBD-labeled protein, Raman scattering is generally not an issue. However, it is always advisable to do the Blank subtraction.
CRITICAL: Blank subtraction from the protein emission spectrum is extremely important to avoid potential artifacts caused by the Raman scattering, which is an inelastic scattering and appears after the excitation wavelength. Raman scattering is a big problem in measuring Trp fluorescence especially at excitation wavelengths of 295–310 nm (see REES experiments below) since the Raman peak overlaps with the spectral contribution from Trp residues (see Figure 4).Note: For measuring the emission spectrum of NBD-labeled protein, Raman scattering is generally not an issue. However, it is always advisable to do the Blank subtraction. -
d.Take the emission scans of the other labeled single-Cys mutants as described above.Note: Use the same Blank spectrum for obtaining the Blank-subtracted emission spectra for all the mutants.
 CRITICAL: It is possible that the labeled protein might have weak or reduced fluorescence. Possible reasons could either be aggregation due to prolonged storage or high working concentration.
CRITICAL: It is possible that the labeled protein might have weak or reduced fluorescence. Possible reasons could either be aggregation due to prolonged storage or high working concentration. -
e.From the Blank-subtracted spectra, note down the fluorescence emission maximum (λem) as well as the intensity values at λem from the respective sample.
-
a.
-
13.Steady-state fluorescence anisotropy/polarization measurements
-
a.Place the excitation and emission polarizers in the respective slot. Rotate both polarizers to 90°(Horizontal, H) orientation in the spectrofluorometer. Excite at 280 nm or 295 nm for Trp residues depending on the protein composition for intrinsic fluorescence (see ‘Note 12b’). For NBD-labeled protein, use 465 nm for excitation. Measure the fluorescence intensity at respective emission maximum. This is the value for IHH.
-
b.Now, rotate only the emission polarizer to 0° (Vertical, V) orientation and measure the fluorescence intensity to get the value for IHV.
-
c.Then, rotate only the excitation polarizer to 0° (V orientation) and measure the fluorescence intensity to get the value for IVV.
-
d.Finally, rotate only the emission polarizer back to 90° (H orientation) and measure the fluorescence intensity to get the value for IVH.Note: Perform the above steps (13a-d) for the Blank before triggering measurements for samples.
-
e.Calculate the fluorescence anisotropy (r) values using the equation (Lakowicz, 2006):
where IVV and IVH are the measured fluorescence intensities (after appropriate Blank subtraction) with the excitation polarizer vertically oriented and emission polarizer vertically and horizontally oriented, respectively. G is the grating correction factor and is the ratio of the efficiencies of the detection system for vertically and horizontally polarized light.Equation 2 Equation 3 Note: Fluorescence anisotropy (r) values can be interchanged to fluorescence polarization (P) values using the equation P = 3r/(2+r). -
f.Calculate the apparent rotational correlation times (τc) from the Perrin’s equation (Lakowicz, 2006):
where ro is the limiting anisotropy of the fluorophore, r is the steady-state anisotropy, and <τ> is the mean fluorescence lifetime of the fluorophore. Please see Step 16 for how to measure lifetimes.Equation 4 Note: The ro for Trp is 0.16 (Eftink et al., 1990), and for NBD is 0.354 (Mukherjee et al., 2004).
-
a.
-
14.Red Edge Excitation Shift (REES).
-
a.Excite the sample at its respective excitation maximum and take the emission scan.
-
i.For intrinsic Trp fluorescence, excite the protein sample at 280 nm or 295 nm depending on the protein composition (see ‘Note 12b’).
-
ii.For NBD-labeled protein, use 465 nm for excitation.
-
i.
-
b.Now, excite the sample going towards the red edge of the absorption band, and take the respective emission scans.
-
i.For Trp, use 295, 298, 301, 303, 305, 307 and 310 nm excitation wavelengths.
-
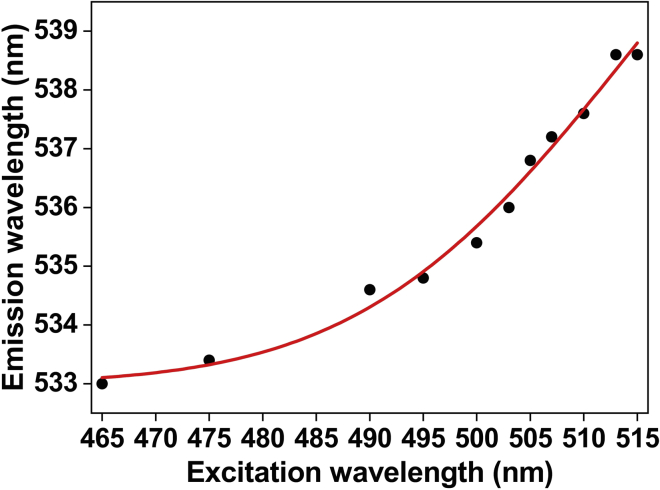
ii.In case of NBD (see Figure 5), emission scans as a function of increasing excitation wavelengths 465, 475, 490, 495, 500, 503, 505, 507, 510, 513 and 515 nm can be measured.
 CRITICAL: REES experiments require a polar fluorophore. Therefore, bimane is not a suitable probe for REES due to its nonpolar nature.Note: Perform the above steps (14a, b) for the Blank before triggering measurements for samples.
CRITICAL: REES experiments require a polar fluorophore. Therefore, bimane is not a suitable probe for REES due to its nonpolar nature.Note: Perform the above steps (14a, b) for the Blank before triggering measurements for samples.
-
i.
-
c.Make sure to subtract the corresponding Blank spectrum associated with a particular excitation wavelength from the respective protein emission spectra.
-
d.From the Blank-subtracted spectra, note down the respective fluorescence emission maximum (λem) at different excitation wavelengths.Note: The S/N ratio will dramatically decrease as a function of increasing excitation wavelengths due to reduction in both molar absorption coefficient and the quantum yield. Due to this, sometimes it will be difficult to get the emission maximum from the Blank-subtracted spectrum for 310 nm excitation (for Trp) and 515 nm excitation (for NBD) especially for membrane proteins.
-
e.The magnitude of REES is calculated by taking the difference between the λem obtained upon exciting at the most red-edged excitation (i.e., low energy excitation) and at the excitation maximum (i.e., high-energy excitation). See Figure 5 for a representative example, in which the magnitude of REES is 5.5 nm (538.5–533).
-
a.
-
15.Fluorescence quenching measurements.
-
a.Take 1 mL of native protein or labeled mutant protein (1–2 μM) in a clean cuvette and measure the fluorescence intensity at the desired emission wavelength upon exciting at the wavelength of excitation maximum. This is fluorescence intensity in the absence of the quencher (i.e., Fo).
-
b.Add a small aliquot of a quencher (Acrylamide for Trp fluorescence; Iodide (KI) for NBD) from a freshly prepared stock solution (8 M and 2.5 M for Acrylamide and KI, respectively) to the protein sample while stirring and incubate for 2 min in the dark sample compartment (shutters closed) in the fluorometer.
 CRITICAL: Acrylamide is known to cause neurotoxic effects in humans. Care must be taken while handling acrylamide.
CRITICAL: Acrylamide is known to cause neurotoxic effects in humans. Care must be taken while handling acrylamide. CRITICAL: Always prepare fresh stock solution of quencher for the quenching measurements. When iodide (I-) is used as a fluorescence quencher, the I- gets oxidized to I2. This is prevented by the addition of 1 mM sodium thiosulfate (Na2S2O3). Also, keeping the solution on ice helps to inhibit I2 formation for some time.
CRITICAL: Always prepare fresh stock solution of quencher for the quenching measurements. When iodide (I-) is used as a fluorescence quencher, the I- gets oxidized to I2. This is prevented by the addition of 1 mM sodium thiosulfate (Na2S2O3). Also, keeping the solution on ice helps to inhibit I2 formation for some time. -
c.Measure the fluorescence emission intensity.
-
d.Using a UV-Vis spectrophotometer, measure the absorbance of the protein-quencher mixture at the respective excitation and emission wavelengths for inner filter effect correction.Note: As a rule of thumb, absorbance should be < 0.1. In case the absorbance is more, reduce the sample concentration.
-
e.Repeat b-d for subsequent addition of quencher until the Fo reduces by ∼50%.
-
f.Subtract the corresponding fluorescence intensities obtained from Blank in the absence and presence of quencher from the sample’s fluorescence intensities.
-
g.Get the dilution-corrected fluorescence intensities.
-
h.Correct for inner filter effect, if any, by using the following equation (Lakowicz, 2006):
where F is the corrected fluorescence intensity and Fobs is the background subtracted fluorescence intensity of the sample. Aex and Aem are the measured absorbance at the excitation and emission wavelengths, respectively.Equation 5 -
i.Get the Fo/F ratio for each quencher concentration. This value is 1.0 for samples that do not contain the quencher (see Figure 6).
-
j.Analyze the quenching data by fitting to the following Stern-Volmer equation (Lakowicz, 2006) as schematically shown above:
where Fo and F are the fluorescence intensities in the absence and presence of the quencher, respectively, KSV is the Stern-Volmer quenching constant (unit is M-1), and [Q] is the molar quencher concentration. The Stern-Volmer quenching constant (KSV) is equal to kqτo, where kq is the bimolecular quenching constant (unit is M-1s-1) and τo is the lifetime of the fluorophore in the absence of quencher (see step 16 for the lifetime measurements).Equation 6 -
k.Calculate the kq values using KSV and τo
Equation 7
-
a.
-
16.Measuring fluorescence lifetimes.
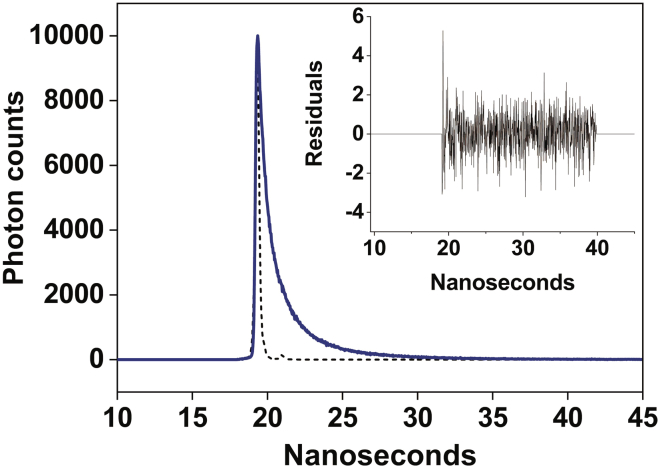
-
a.Turn on the NanoLED. Measure the instrument response function (IRF, also called Prompt) at the excitation wavelength of the desired NanoLED attached. The emission wavelength should also be the same. This is generally done by using 1 mL of very diluted solution of colloidal silica (1% Ludox) as the scatterer in a quartz cuvette. This will take ∼5 min.Note: There is no Blank measurement in lifetime. Instead, the IRF is required for deconvolution analysis that considers the distortion caused by the instrumental response on the decay. Basically, IRF reflects the distribution of photons from the excitation pulse in a non-fluorescent scattering medium.
 CRITICAL: Do not use ‘non-dairy café creamer’ or glycogen as the scatterer as mentioned in older literature since they might contain fluorescent impurities.
CRITICAL: Do not use ‘non-dairy café creamer’ or glycogen as the scatterer as mentioned in older literature since they might contain fluorescent impurities. CRITICAL: We use Time-Correlated Single Photon Counting (TCSPC) decay measurements using HORIBA Fluoromax steady-state and time-resolved table-top spectrofluorometer. Make sure to use the nominal emission bandpass (typically 3 nm) so that the α value (which is the stop-to-start ratio) is not more than 2%. This is to reduce the likelihood that more than one photon will be detected in response to any one excitation event.Note: The scattering solution must be really weak, so if the signal from Ludox is too strong, dilute it further.
CRITICAL: We use Time-Correlated Single Photon Counting (TCSPC) decay measurements using HORIBA Fluoromax steady-state and time-resolved table-top spectrofluorometer. Make sure to use the nominal emission bandpass (typically 3 nm) so that the α value (which is the stop-to-start ratio) is not more than 2%. This is to reduce the likelihood that more than one photon will be detected in response to any one excitation event.Note: The scattering solution must be really weak, so if the signal from Ludox is too strong, dilute it further. -
b.To optimize the S/N ratio, at least 10,000 photon counts are collected in the peak channel.
-
c.Set up a new decay, and put the protein (native or labeled sample) inside.
-
d.Depending on the concentration of the protein (native or labeled), the emission bandpass is generally kept between 5-8 nm. Change the emission wavelength to the respective wavelength required for the sample. Start acquiring the fluorescence decay.
-
e.After the acquisition is complete, save the decay file along with the IRF.
-
f.The analysis is generally done in a Decay Analysis software provided by the manufacturer.
-
g.The fitted data can be exported as text files, which can be read by graphing software for plotting the decay profile, and calculation of mean lifetime.
-
h.Discrete analysis of lifetime decay:
-
i.It is a model-dependent analysis of fluorescence lifetime decay.
-
ii.Fluorescence intensity decay curves so obtained were deconvoluted with the instrument response function and analyzed as a sum of exponential terms:
where F(t) is the fluorescence intensity at time t and αi is a pre-exponential factor representing the fractional contribution to the time-resolved decay of the component.Equation 8 -
iii.The decay can be fitted by a mono, bi, or tri-exponential fit, depending on the type of fluorescent probe and its local environment (see Figure 7).
-
iv.Check the goodness of the fit: The fit is considered good if the reduced χR2 is close to 1. Also, the goodness of fit is visually inspected using weighted residuals (which should be randomly distributed about zero), and autocorrelation function (which should show ‘high frequency low amplitude oscillations’ about zero).
-
v.The mean (average) lifetimes <τ> of the fluorophore is calculated from the decay times and pre-exponential factors in two ways (Lakowicz, 2006) as is shown below for a tri-exponential decay of fluorescence:Intensity-weighted mean lifetime
Equation 9 Amplitude-weighted mean lifetimeEquation 10
-
i.
-
i.Histogram analysis of lifetime decay:
-
i.The fluorescence lifetime can be directly calculated using a model-independent approach from the histogram of photons obtained during fluorescence lifetime measurements for a protein sample using the following equation (Fiserova and Kubala, 2012; Chatterjee et al., 2019):
where Ni and ti denote the number of detected photons in the ith channel and the corresponding value on the time axis, respectively, n is the total number of channels in the histogram, p is the channel with the highest number of detected photons (peak of the decay) and tp is the corresponding time (see Figure 8).Equation 11 -
ii.Find the maximal value in the histogram and identify tp (Figure 8).
-
iii.Calculate the average value of noise from the noise-domain (for data with t<tp, while ignoring a few channels from the rising edge) and subtract it from all data.
-
iv.For data with t>tp calculate the average time as (∑(Ni- noise)ti)/∑(Ni-noise).
-
v.Subtracting of tp value from the average time yields the mean fluorescence lifetime.Note: The lifetime obtained from histogram analysis is similar to intensity-weighted mean fluorescence lifetime obtained from discrete analysis.
-
i.
-
j.Maximum Entropy Method (MEM) analysis of lifetime decay.
-
i.MEM is a model-independent analysis of fluorescence lifetime decay.
-
ii.In MEM, the fluorescence intensity decay [I(t)] is analyzed using the model of continuous distribution of lifetimes:
where α(τ) represents the amplitude corresponding the lifetime τ in the intensity decay.Equation 12 -
iii.The open access AnalyseDistribution MATLAB code (Smith et al., 2017) is used for analyzing the distribution by MEM.
-
iv.To fit the data, rearrange the lifetime decay text file such that there are only 3 columns consisting of the Time, Decay and Prompt, in the exact same order.
-
v.In MATLAB, run the AnalyseDistribution code. Load the lifetime decay text file.
-
vi.Set the analysis as Rate and the lower and upper limits are set to 0.1 ns-1 and 10 ns-1, respectively (Figure 9).
-
vii.A regularization parameter, λ, is set to a value of 0.001 or 0.01 depending on the fluorophore system, such that the χ2 is minimized while maximizing the entropy (S). The expression used for S is the Shannon-Jaynes entropy function, which is:
where the set of values b represent a default model for the system. However, in the absence of a default model for our system, bi are generally set to a constant value. Using constant bi value favors equal contribution from all lifetimes which means that the introduction of structure into the distribution is discouraged.Equation 13 -
viii.Click ‘Fit Data”.
-
ix.MEM initially starts with a flat distribution of amplitudes α (τ), i.e., each lifetime has equal contribution in the beginning and arrives at the amplitude distribution which best describes the observed experimental fluorescence intensity decay.
-
x.The analysis is terminated when χ2 reaches the specified lower limit or when χ2 and α(τ) show no change in successive iterations. Click ‘Analyse data’ as highlighted in Figure 10A.
-
xi.Select the peaks to be analyzed (see Figure 10B) and press enter.
-
xii.Fitted peaks and relative peak areas (shown as red dots) will be displayed. Click ‘Press to save fit’ (see Figure 10C).
-
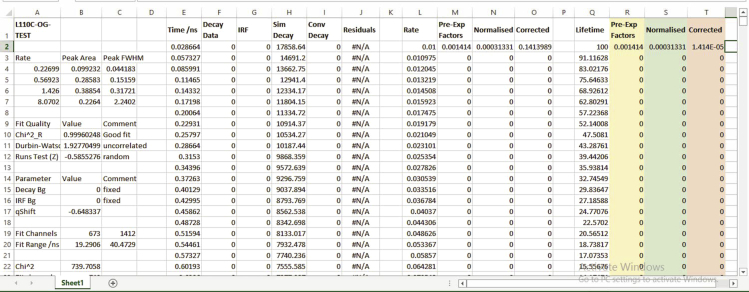
xiii.The analysis results get saved as excel file (see Figure 11), which is used for plotting the lifetime distributions.Note: The normalized values are the pre-exponential values divided by the sum of all the pre-exponential values. The normalized plots are generally used as a representation of a decay fit of a sample because the peak sizes visually match with the respective areas.
-
i.
-
a.
Figure 4.
Blank-subtracted fluorescence emission spectrum
(A) Shown are the Raman peak (red trace) of Blank, and the emission spectrum of the unsubtracted protein (blue trace) sample.
(B) shows the Blank-subtracted protein spectrum (Trp fluorescence). Excitation wavelength used was 295 nm.
Figure 5.
REES measurement
The effect of changing excitation wavelength on the wavelength of maximum emission for a mutant (134-NBD) of a potassium channel KvAP in membrane (unpublished data). Red line is merely a visual guide.
Figure 6.
Schematic representation of a quenching experiment
Left: The fluorescence emission spectrum in the absence (broken line) and the presence (solid lines) of quencher (+Q), and increasing concentrations of quencher is denoted by an arrow. Right: The trend of Fo/F as a function of increasing quencher concentration is shown. Fitting this with a line equation yields the slope, that is the KSV value.
Figure 7.
Time-resolved fluorescence intensity decay of NBD-labeled KvAP voltage sensor at residue 116 in micelles
The sharp peak (dotted line) is the IRF. The decay profile (solid blue line), is fitted to a tri-exponential function. The plot in the inset shows the weighted residuals. Excitation wavelength is 460 nm using NanoLED and emission is monitored at 535 nm. Figure modified from Das et al. (2020).
Figure 8.
The temporal-histogram of detected photons from a sample
The vertical lines divide the histogram into noise-domain (N), rising-edge-domain (R) and decay-domain (D), tp denotes the channel with the highest number of detected photons.
Figure 9.
Setting the parameters in AnalyseDistribution for MEM analysis using MATLAB
See the highlighted box (in red) and text for details.
Figure 10.
Analysis windows in AnalyseDistribution for MEM lifetime distribution analysis
(A) shows the decay fit along with its statistics;
(B) shows the window for selecting peaks to be analysed, and (C) shows the fitted peaks along with respective peak areas (red dots). See text for more details.
Figure 11.
The output of the AnalyseDistribution fit is generated as an excel file
The highlighted columns indicate the Pre-exp factors (yellow), Normalised values (green), and Corrected values (pink) of lifetime distributions.
The number of peaks indicate the number of discrete conformations, and the peak areas denote the relative populations. The full-width at half maximum (FWHM) of each peak when compared can give an idea of the heterogeneity.
The corrected values give an estimate of the true contribution of a component/peak at a particular lifetime of decay of a sample. Since the MEM data is obtained in log scale, it biases the contribution of longer rates. This is corrected by dividing the calculated pre-exponential factors by the rates (or lifetime). The area under each peak and full width at half maximum (FWHM) are calculated from the corrected plots.
Expected outcomes
The examples given below are to highlight the possible use of various fluorescence parameters to monitor various biologically-relevant processes. Several of the described parameters can be used to probe a particular phenomenon, e.g., protein folding, however we give representative examples for potential applicability of each fluorescence parameter.
Fluorescence emission maximum and intensity changes
Several factors affect the fluorescence emission spectrum and quantum yield of a fluorophore, which include solvent polarity and viscosity, rate of solvent relaxation, probe conformational changes, rigidity of the local environment. This means that emission maximum is sensitive to the probe’s surrounding microenvironment. In this regard, intrinsic Trp fluorescence is the most widely used tool to monitor the changes in local structure and dynamics in proteins (Vivian and Callis, 2001; Raghuraman and Chattopadhyay, 2003; Raghuraman et al., 2005; Callis and Tusell, 2014; Chatterjee et al., 2021). Further, the environment sensitivity of Trp fluorescence has been extensively used to monitor protein folding/unfolding transitions, membrane partitioning, lipid-protein interactions etc. (Raghuraman et al., 2019; Biswas et al., 2020; Raghuraman and Chattopadhyay, 2004a,b,c; Raghuraman et al., 2006; Haldar et al., 2010).
Example 1: Monitoring protein unfolding
The following example (Figure 12) shows how one can use the changes in fluorescence emission maximum and fluorescence intensity of Trp fluorescence to monitor the protein unfolding mediated by chemical denaturants such as urea (Biswas et al., 2020). In general, Trp exhibits an emission maximum of ∼350 nm when fully exposed to aqueous media, whereas the emission maximum is significantly blue shifted (∼325–335 nm) when the Trp is localized in a hydrophobic environment, like when buried inside the protein or partitions into the membrane (Vivian and Callis, 2001; Raghuraman and Chattopadhyay, 2004a,b,c). As can be seen from the figure, the average fluorescence emission maximum of Trp residues of native LmPIN1 is 336 nm, suggesting only a partial exposure of Trp to surrounding solvent. Urea-mediated denaturation/unfolding of this protein results in significant red-shifted emission maximum (from 336 to 349 nm). The overall change in the emission maximum of LmPIN1 corresponds to a redshift of 13 nm that is accompanied by ∼60% reduction in fluorescence intensity. This indicates that the Trp residues of LmPIN1 are predominantly exposed to surrounding aqueous medium upon denaturation.
Figure 12.
Monitoring protein folding/unfolding transition
Left: Homology model of LmPIN1 showing the location of native Trp residues and are indicated by arrows. Middle: Fluorescence emission spectra of LmPIN1 at various urea concentrations. Right: Plot of emission maxima (red line) and relative fluorescence intensity (blue line) as a function of urea concentration. Adapted and modified from Biswas et al. (2020).
Like Trp, the extrinsic fluorophores (NBD and bimane) are small fluorophores (comparable to the size of Trp) and are environment-sensitive (Raghuraman et al., 2019) with well-characterized spectral properties, and are widely used to map the structure, topology and monitor the structural dynamics of proteins (Shatursky et al., 1999; Johnson, 2005; Raghuraman and Chattopadhyay, 2007a,b; Raghuraman et al., 2014; Mansoor et al., 2010; Ho et al., 2013).
Example 2: Monitoring membrane binding of amphipathic peptides
Melittin is an amphipathic peptide with diverse cellular functions (Raghuraman and Chattopadhyay, 2007a,b). In solution, low concentration of melittin exists in a random coil conformation, and it spontaneously binds to phospholipid membranes and adopts an α-helical conformation. Raghuraman and Chattopadhyay (2004c) has showed that the binding of melittin to membrane causes a blue shift in the sole Trp of melittin (356 nm in buffer to 336 nm in membranes) along with changes in fluorescence intensity due to the change in the polarity of the surrounding medium. In this study, the increase in fluorescence intensity ratio (F336/F352) has effectively been utilized to monitor the cholesterol effect on the complete membrane-binding of melittin (Figure 13).
Figure 13.
Partitioning of melittin into membranes probed by intensity ratio measurement
Effect of cholesterol on membrane-binding of melittin monitored using the ratio of fluorescence intensity of melittin Trp at 336 nm (membrane-bound) and 352 nm (in buffer). Adapted from Raghuraman and Chattopadhyay (2004c). See this reference for more details.
Example 3: Predicting the α-helical structure
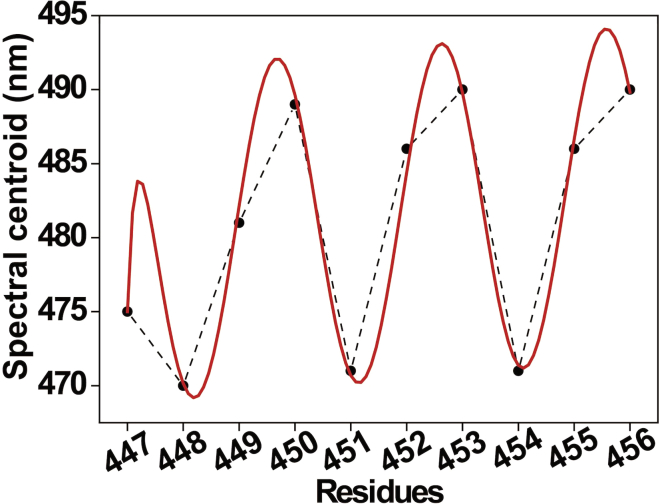
The following is the example to predict the membrane-bound topology of an amphipathic H6 helix in a Colicin E1 ion channel protein from changes in emission maximum of site-directed bimane labeling (see Figure 14). Results using helical periodicity analysis method shows that H6 is α-helical in soluble form, whereas it might undergo conformational change from α-helical to 310 helix upon membrane binding. It should be noted that the spectral centroid (also called center of spectral mass, CSM) has been used instead of fluorescence emission maximum for this analysis. However, in most cases, there is no difference between CSM and emission maximum values.
Figure 14.
Changes in emission maximum of bimane-labeled Colicin E1 at the indicated residues
The dashed line is merely a visual guide. The fit according to the helical periodicity analysis to predict the α-helical nature is shown in red. Data adapted from Ho et al. (2013). See this reference for more details.
Fluorescence polarization/anisotropy
The polarization (P) and anisotropy (r) values are related by r = 2P/(3-P). Fluorophores that are freely mobile (i.e., either in bulk solution or on the surface of a protein) will display low polarization values. For example, Trp and NBD display a low polarization value of ∼0.004 in solution. However, fluorophores that are membrane-bound or at buried sites exhibit a higher anisotropy/polarization value due to hindered rotational mobility of the probe. (Raghuraman and Chattopadhyay, 2004c; Das et al., 2020; Chatterjee et al., 2021). The limiting anisotropy value for Trp is 0.16 (Eftink et al., 1990) and the corresponding value for NBD is 0.354 (Mukherjee et al., 2004). Since fluorescence polarization/anisotropy provides information on molecular flexibility and rotational motion, it is a powerful tool to monitor the structural dynamics of soluble and membrane proteins (Jameson and Ross, 2010; James and Jameson, 2014). The dynamics of the fluorophore is influenced by the motion of its attached region in a protein thus providing valuable information regarding protein structure, dynamics and its conformational changes.
Example 1: Monitoring the folding and aggregation of proteins in solution
Folding and aggregation (self-association) of proteins in solution is a complex process (Othon et al., 2009). The following example (Figure 15) shows how fluorescence polarization values can be used to probe the ionic strength mediated folding and self-association of melittin to tetramer in solution. The Trp polarization of melittin increases with increasing ionic strength, which implies restricted mobility of Trp during folding and self-association.
Figure 15.
Effect of increasing ionic strength on the fluorescence polarization of melittin in aqueous solution
Inset shows the self-association of melittin from random coil to tetramer. Figure adapted and modified from Raghuraman and Chattopadhyay (2006). See this reference for more details.
Example 2: Monitoring the structural dynamics of proteins in different functional states
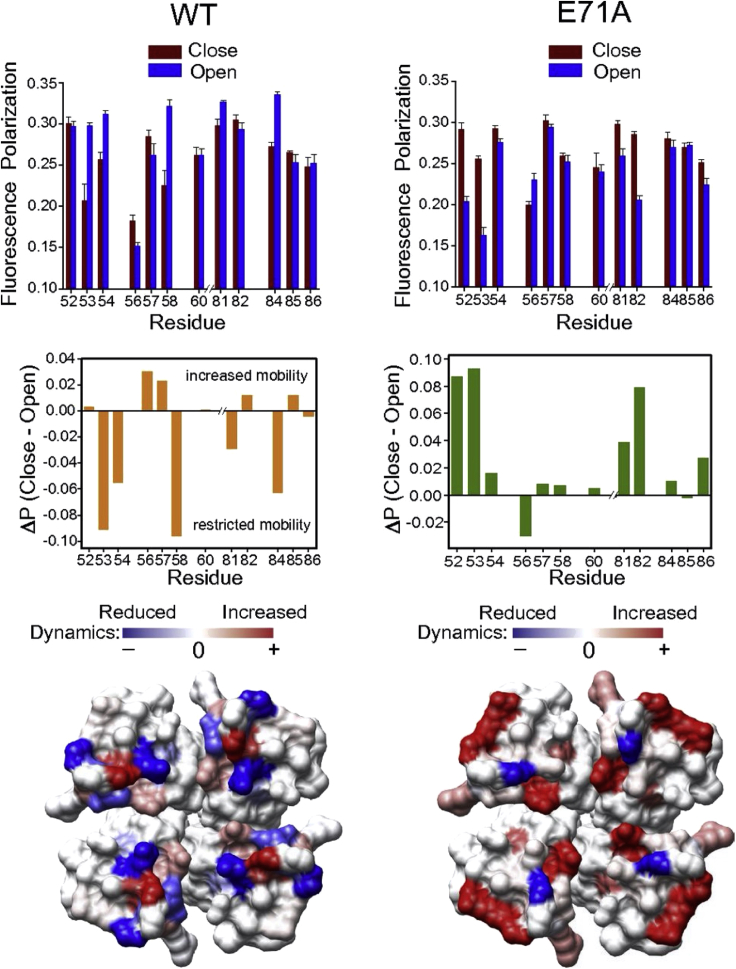
In pH-gated potassium channel, KcsA (Raghuraman et al., 2012, 2014; Kratochvil et al., 2016), site-directed NBD labeling has been done for outer vestibule residues of KcsA. The steady-state polarization measurements have been carried out in inactivating (WT background) and non-inactivating/conductive (E71A mutant background) functional states of KcsA in both closed and open states. Mapping the difference in NBD polarization values (ΔP) between the closed and open states in the crystal structure of KcsA (Figure 16) has demonstrated that the outer vestibule of the open/conductive conformation is highly dynamic compared to the outer vestibule during inactivation gating (Raghuraman et al., 2014).
Figure 16.
The outer vestibule of open/non-inactivating conformation of a potassium channel is highly dynamic
Steady-state polarization of NBD fluorescence monitored for the outer vestibule residues in different functional states of KcsA. Data adapted from Raghuraman et al. (2014). See this reference for more details.
Red edge excitation shift (REES)
REES is a wavelength-selective fluorescence approach, which is a convenient tool to monitor solvent relaxation dynamics (i.e., hydration dynamics) and environment-induced restriction and dynamics around organized molecular assemblies like membranes, proteins etc. The REES phenomenon is observed when the dipolar relaxation time for the solvent (water in biological systems) molecules around the excited-state polar fluorophore is comparable to or longer than the fluorescence lifetime (Demchenko, 2002; Demchenko, 2008; Raghuraman et al., 2003, 2005). This generally happens in motionally-restricted polar environments such as viscous solutions, condensed phases, membrane-mimetic systems etc. The presence or absence of REES can therefore distinguish between the restricted/bound and bulk/free water molecules around the fluorophore’s microenvironment. Since hydration dynamics plays a key role in membrane dynamics and in fine tuning the functionality of proteins in a variety of ways (Raghuraman et al., 2014; Bellisent-Funel et al., 2016; Ball, 2017; Pal and Chattopadhyay, 2021), REES has been utilized to monitor the folding and aggregation of proteins, lipid-protein interactions, membrane dynamics and heterogeneity, function-dependent hydration dynamics, discrete protein conformational states etc. Please refer to recent review (Brahma and Raghuraman, 2021) for REES and its applications.
Fluorescence quenching
When an excited fluorophore collides with a quencher (like acrylamide and iodide ions), this facilitates the deactivation rates of the excited state of the fluorophore. This collisional or dynamic quenching (in which the distance between the fluorophore and quencher changes rapidly) results in decreased fluorescence intensity (see Eftink, 1991 for detailed theory of fluorescence quenching). Since the magnitude of fluorescence quenching is dependent on the accessibility of the fluorophore to the quencher, the Stern-Volmer constant (KSV) can give an idea of the degree of accessibility of the fluorophore. High KSV means increased accessibility by the quencher, which, in case of aqueous quenchers like acrylamide and iodide ions, directly implies increased water accessibility and vice versa. For example, the KSV is ∼18 M-1 for a fully exposed Trp to aqueous medium (Raghuraman and Chattopadhyay, 2004a), and this value dramatically reduces to as low as ∼2–7 M-1 when Trp residue(s) is partitioned in micelles or lipid bilayer (Chatterjee et al., 2019, 2021). Comparing results between samples only on KSV values needs caution, because these values are only relevant if the fluorescence lifetime does not change significantly between the samples. For this reason, the bimolecular quenching constant (kq) should be used over KSV as the former corrects for differences in fluorescence lifetimes in the absence of quencher (Raghuraman et al., 2019; Lakowicz, 2006).
Example 1: Gating-induced changes in water accessibility
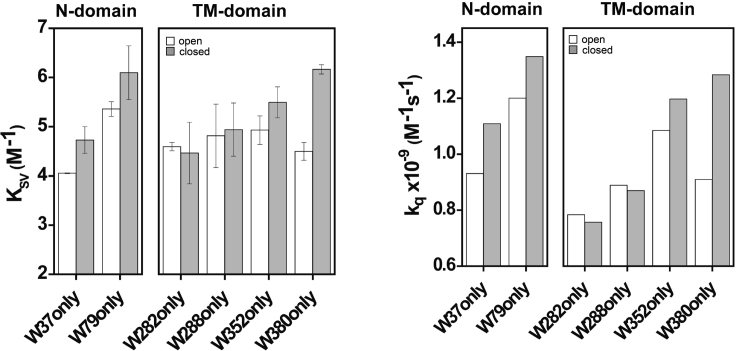
The following example (Figure 17) highlights the use of Trp quenching by acrylamide to probe the accessibility and changes in localization of native Trp residues in MgtE magnesium channel when the channel is shuttled between open (in the absence of Mg2+) and closed (in presence of Mg2+) states. The quenching results from single-Trp mutants of MgtE show that water accessibility is increased for most of the Trp residues in the closed state compared to open state, and supports the gating-induced ‘conformational wave’.
Figure 17.
Water accessibility of single-Trp mutants of MgtE magnesium channel during gating in membranes
Left: Stern-Volmer constants (KSV) and Right: bimolecular quenching constants (kq) for acrylamide quenching of Trp fluorescence for the single-Trp mutants of MgtE in the open and closed states in membranes. Data adapted and modified from Chatterjee et al. (2021). See this reference for more details.
In case of NBD quenching by iodide ions, kq value of ∼8 M-1ns-1 is expected if NBD is fully exposed to aqueous environment (Crowley et al., 1993) and this reduces significantly to <<1.5 M-1ns-1 when it is relatively inaccessible to the quencher. Please note that for NBD, apart from iodide ions, cobaltous ions also act as efficient collisional quenchers (Morris et al., 1985; Raghuraman et al., 2004; Raghuraman and Chattopadhyay, 2007b).
Example 2: Organization of voltage sensor loop in membrane-mimetic systems
The following is the example of iodide quenching of NBD-labeled residues of the voltage sensor loop of KvAP in micelles and membranes. As can be seen from Figure 18, the accessibility of iodide ions is significantly decreased for voltage sensor loop residues in the membrane environment compared to micelles, suggesting the reduced water accessibility due to partitioning of this loop into membrane interface.
Figure 18.
Differential organization of the voltage sensor loop in membrane-mimetic systems using iodide quenching of NBD fluorescence
Left: Stern-Volmer constants (KSV) and Right: bimolecular quenching constants (kq) for iodide quenching of NBD-labeled residues of the KvAP voltage sensor loop in micelles (black circles) and membranes (red circles). The lines joining the data points are provided merely as viewing guides. Data adapted and modified from Das et al. (2020). See this reference for more details.
Fluorescence lifetime
After excitation, the fluorophore stays in the excited state briefly (called fluorescence lifetime, τ, and is typically nanoseconds for most fluorophores) before relaxing back to the ground state with an emission. Compared to steady-state fluorescence intensity, τ is independent of sample concentration, light scattering from turbid samples etc. (Krishnamoorthy, 2018a). Importantly, τ is an intrinsic property of the fluorophore and therefore is an excellent indicator of fluorophore’s microenvironment (Berezin and Achilefu, 2010), and can be used to monitor increased hydration and conformational changes in proteins (Raghuraman et al., 2019). Generally, the lifetimes of most fluorophores are higher in nonpolar environment compared to aqueous environment. The fluorescence decay data analysis by MEM represents a convenient, robust, model-free and realistic approach of data analysis (Smith et al., 2017; Brochon, 1994; Krishnamoorthy, 2018b). MEM has been widely used to monitor the unfolding transitions in small soluble proteins (Lakshmikanth et al., 2001; Jha et al., 2009) and conformational heterogeneity in membrane proteins (Das and Raghuraman, 2021; Chatterjee et al., 2021), and to monitor membrane heterogeneity (Mukherjee et al., 2007).
Example 1: Effect of increasing amounts of water on mean fluorescence lifetimes of Trp
Unlike biological and model membranes (like micelles, liposomes and Nanodiscs), reverse micelles are excellent tool to control water content to study hydration mediated dynamic changes (Chattopadhyay et al., 2002; Raghuraman and Chattopadhyay, 2003). When melittin is placed in AOT (sodium bis(2-ethylhexyl)sulfosuccinate) reverse micelles and upon increasing the water content in the reverse micelles, there is a continuous reduction in the mean fluorescence lifetimes of melittin Trp (Figure 19). This example highlights the sensitivity of Trp lifetimes to increased polarity due to increased water content in its immediate vicinity.
Figure 19.
Trp lifetime is sensitive to environmental polarity
Sensitivity of mean fluorescence lifetimes obtained from discrete analysis of lifetime decay of melittin Trp in reverse micelles upon increasing water content. Data adapted and modified from Raghuraman and Chattopadhyay (2003).
Example 2: Potential advantage of combining discrete and MEM analysis
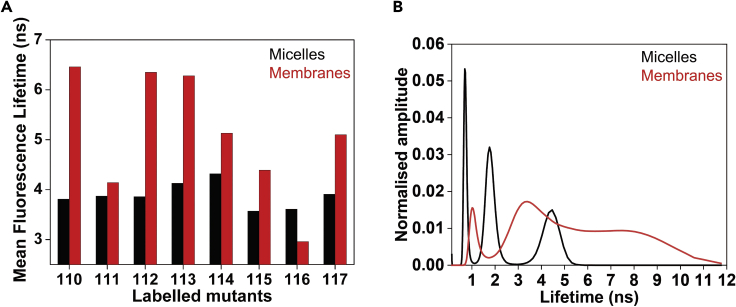
The fluorescence lifetime of NBD is highly sensitive to its local environment (Lin and Struve, 1991; Chattopadhyay et al., 2002; Raghuraman et al., 2019; Das et al., 2020; Das and Raghuraman, 2021). The fluorophore IANBD amide in buffer undergoes a monoexponential decay and gives a sub-nanosecond lifetime of 0.58 ns, and this corresponds well with the predominantly single peak in model-independent MEM analysis (Das and Raghuraman, 2021). Basically, NBD has a very short lifetime when fully exposed to aqueous medium (Fery-Forgues et al., 1993), whereas it has a much longer lifetimes (∼5–10 ns) in a nonpolar environment (Chattopadhyay et al., 2002; Johnson, 2005; Raghuraman et al., 2019). The mean fluorescence lifetimes therefore can give information on environmental heterogeneity. Interestingly, MEM analysis of decay curves of NBD-labeled residues of proteins can give crucial information on changes in conformational heterogeneity. The following example shown for NBD-labeled loop residues of KvAP voltage sensor illustrates that the voltage sensor experiences different structural organization in micelles and membranes (Figure 20). Further, combining the discrete and MEM analyses of NBD decay curves reveal that environmental heterogeneity is strongly correlated with conformational heterogeneity (see Das and Raghuraman, 2021 for more details).
Figure 20.
Environmental heterogeneity correlates with conformational heterogeneity of the voltage sensor loop
(A) Mean fluorescence lifetimes obtained from discrete analysis of lifetime decay of NBD-labeled residues of the KvAP voltage sensor loop in micelles (black) and membranes (red). Data adapted and modified from Das et al. (2020).
(B) MEM lifetime distribution for NBD-labeled loop residue at position 110 in micelles (black) and membranes (red). Figure adapted and modified from Das and Raghuraman (2021).
Limitations
Any fluorescence parameter gives only local dynamic information at the site of fluorophore attachment. Therefore, information derived only from one labeled site is not sufficient to get structural insights. In order to get comprehensive details on structural changes in proteins, many residues throughout the protein need to be labeled in a site-specific manner and all the fluorescence changes from all the sites should be taken into consideration. Further, in the labeled protein, the fluorophore dipole is generally few angstroms away from the protein backbone and hence experimental data coupled with molecular dynamics simulations will reveal the structural changes at the backbone level.
The fluorescence intensity ratio may not a reliable parameter when it comes to evaluating protein unfolding transitions because it might lead to incorrect determination of thermodynamic parameters (Zoldak et al., 2017).
Steady-state polarization (anisotropy, r) values are strongly influenced by the excited-state fluorescence lifetime. Care must therefore be taken to make sure that the polarization values are not due to changes in mean fluorescence lifetimes (<τ>). This can be easily done by calculating the apparent rotational correlation times (τc) as shown earlier. If there are no lifetime-induced artifacts, then the changes in both the anisotropy and rotational correlation times data will follow the same trend.
While steady-state polarization (anisotropy) provides information about the overall dynamics of the protein, it cannot provide individual information regarding the local, segmental and global motions. Since fluorescent dyes in complex heterogenous systems generally have many lifetime components due to various dynamic modes like global motion (tumbling of whole protein with attached probe), segmental motion (probe mobility mediated by protein’s local dynamics), and local motion (rotational motion of tethered probe with respect to attached protein), time-resolved anisotropy measurements are performed to resolve the various dynamic modes in their timescale (Krishnamoorthy, 2018a).
The magnitude of REES is qualitative and can be comparable under different conditions for a particular protein. However, it varies from system to system.
In case of discrete analysis, decay profiles are commonly fitted with bi- and tri-exponentials to obtain fit with good statistical parameters. Unless there is a physical meaning, bi- and tri-exponential fitting do not necessarily mean the number of components/conformations of the molecule.
Trp fluorescence decay is a complex process. This is because even the free L-Trp in aqueous solution does not undergo mono-exponential decay process and the mean fluorescence lifetime is ∼3 ns (Szabo and Rayner, 1980). Although Trp lifetimes decrease with increasing environmental polarity (see Figure 19), one cannot assign a specific lifetime value for a fully exposed Trp compared to Trp in nonpolar environments unlike NBD, whose lifetime is too short when fully exposed and dramatically increase in nonpolar/hydrophobic environments.
Troubleshooting
Problem 1
Low or poor fluorescence labeling efficiency (step 9).
Potential solution
Check the construct to make sure that it contains the desired single-Cys mutant.
If the construct is fine, then check the amount of fluorophore added to the protein. It should be 10-fold molar excess to that of the protein.
If the fluorophore addition is fine, check the amount and quality of DTT added. It is possible that an old stock of DTT will fail to reduce the disulphide bonds, if any, and further prevents the conjugation of the thiol-reactive probe. Make sure DTT is freshly prepared.
It is possible that the desired labeling site is deeply buried and cannot be accessible by the fluorophore. This is generally a problem when dealing with oligomeric proteins. In this situation, the protein can be labeled in denaturing conditions and refolded back. This method of refolding may be applicable for most of the soluble proteins, but it is difficult in case of membrane proteins.
Problem 2
No or weak fluorescence signal after fluorophore labeling (step 12).
Potential solution
A trivial mistake could be that the shutter of the fluorometer is closed while acquiring the fluorescence data. Always ensure that the shutter of the fluorometer is open while collecting data.
On a serious note, we recommend to check the construct to make sure that it contains the desired single-Cys mutant.
If the construct is fine, then check the fluorophore labeling efficiency carefully.
It is possible that the labeled protein has aggregated due to prolonged storage. Make sure to do the fluorescence experiments as soon as the labeled protein is ready.
Reduce the concentration of the labeled protein. The reduced or poor fluorescent signal could also be due to self-quenching and inner filter effects.
Problem 3
Making single-Cys mutants in case of Cys-rich proteins might seriously disturb structure and function.
Potential solution
One might try ‘cysteine metal protection and labeling’ (CyMPL) method, which has been shown to be useful to specifically label the Cys of interest in proteins containing several Cys residues (Puljung and Zagotta, 2011).
Problem 4
Polarization/anisotropy values are unusually high (step 13).
Potential solution
Always remember that the polarization/anisotropy value can theoretically be as high as 0.5/0.4 (Lakowicz, 2006). Also it is important to have the knowledge of limiting anisotropy for a particular fluorophore. If the obtained values are very high, this could be due to scattering problems since scattered light is highly polarized. And it is a problem when dealing with membrane proteins or using high concentrations of proteins.
We recommend using lower concentrations of proteins, yet keeping good S/N ratio. For membrane proteins, use unilamellar vesicles and lipid-to-protein ratios in such a way that the sample is optically transparent.
Problem 5
Lifetime values do not match with the published literature (step 16 h).
Potential solution
Check whether the lifetime instrument is working properly by measuring the lifetime of known standards such as NATA (N-Acetyl-L-tryptophanamide) in aqueous solution, which gives a mono-exponential decay with a lifetime of 3 ns (Excitation wavelength: 280 or 295 nm; Emission wavelength: 350 nm).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, H. Raghuraman (h.raghuraman@saha.ac.in).
Materials availability
The protocol did not generate new unique reagents.
Acknowledgments
This work was supported by the Department of Atomic Energy, Government of India, and India Alliance DBT-Wellcome Trust Intermediate Fellowship (IA/I/17/2/503321) awarded to H.R. We thank Dr. Darren Smith for helpful discussions regarding the analysis of MEM data. R.B. and A.D. thank the Department of Atomic Energy, Government of India, and Department of Biotechnology, Government of India, for the award of a Senior Research Fellowship, respectively. We thank Arpan Bysack for critically reading the manuscript.
Author contributions
R.B. and A.D. contributed to preparing figures. H.R., R.B., and A.D. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Data and code availability
Original/source data for all figures given in the expected outcome of the paper are available in the literature.
References
- Arachea B.T., Sun Z., Potente N., Malik R., Isailovic D., Viola R.E. Detergents selection for enhanced extraction of membrane proteins. Protein Expr. Purif. 2012;86:12–20. doi: 10.1016/j.pep.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Ball P. Water is an active matrix of life for cell and molecular biology. Proc. Natl. Acad. Sci. USA. 2017;114:13327–13335. doi: 10.1073/pnas.1703781114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellisent-Funel M.C., Hassanali A., Havenith M., Henchman R., Pohl P., Sterpone F., Van Der Spoel D., Xu Y., Garcia A.E. Water determines the structure and dynamics of proteins. Chem. Rev. 2016;116:7673–7697. doi: 10.1021/acs.chemrev.5b00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezin M.Y., Achilefu S. Fluorescence lifetime measurements and biological imaging. Chem. Rev. 2010;110:2641–2684. doi: 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G., Ghosh S., Raghuraman H., Banerjee R. Probing conformational transitions of PIN1 from L. major during chemical and thermal denaturation. Int. J. Biol. Macromol. 2020;154:904–915. doi: 10.1016/j.ijbiomac.2020.03.166. [DOI] [PubMed] [Google Scholar]
- Brahma R., Raghuraman H. Novel insights in linking solvent relaxation dynamics and protein conformations utilizing red edge excitation shift approach. Emerg. Top. Life Sci. 2021;5:89–101. doi: 10.1042/ETLS20200256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braman J., Papworth C., Greener A. Site-directed mutagenesis using double-stranded plasmid DNA templates. Methods Mol. Biol. 1996;57:31–44. doi: 10.1385/0-89603-332-5:31. [DOI] [PubMed] [Google Scholar]
- Brochon J.C. Maximum entropy method of data analysis in time-resolved spectroscopy. Methods Enzymol. 1994;240:262–311. doi: 10.1016/s0076-6879(94)40052-0. [DOI] [PubMed] [Google Scholar]
- Callis P.R., Tusell J.R. In: Fluorescence Spectroscopy and Microscopy: Methods and Protocols, Methods in Molecular Biology. Engelborghs Y., Visser A.J.W.G., editors. Springer Science+Business Media, LLC; 2014. MD + QM correlations with tryptophan fluorescence spectral shifts and lifetimes; pp. 171–214. [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Brahma R., Raghuraman H. Gating-related structural dynamics of the MgtE magnesium channel in membrane-mimetics utilizing site-directed tryptophan fluorescence. J. Mol. Biol. 2021;433:66691. doi: 10.1016/j.jmb.2020.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Das A., Raghuraman H. Biochemical and biophysical characterization of a prokaryotic Mg2+ ion channel: implications for cost-effective purification of membrane proteins. Protein Expr. Purif. 2019;161:8–16. doi: 10.1016/j.pep.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay A., Mukherjee S., Raghuraman H. Reverse micellar organization and dynamics: a wavelength-selective fluorescence approach. J. Phys. Chem. B. 2002;106:13002–13009. [Google Scholar]
- Chung C.T., Miller R.H. Preparation and storage of competent Escherichia coli cells. Methods Enzymol. 1993;218:621–627. doi: 10.1016/0076-6879(93)18045-e. [DOI] [PubMed] [Google Scholar]
- Crowley K., Reinhart G.D., Johnson A.E. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- Das A., Raghuraman H. Conformational heterogeneity of the voltage sensor of KvAP in micelles and membranes: a fluorescence approach. Biochim. Biophys. Acta. 2021;1863:183568. doi: 10.1016/j.bbamem.2021.183568. https://doi: 10.1016/j.bbamem.2021.183568 [DOI] [PubMed] [Google Scholar]
- Das A., Chatterjee S., Raghuraman H. Structural dynamics of the paddle motif loop in the activated conformation of KvAP voltage sensor. Biophys. J. 2020;118:873–884. doi: 10.1016/j.bpj.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko A.P. The red-edge effects: 30 years of exploration. Luminescence. 2002;17:19–42. doi: 10.1002/bio.671. [DOI] [PubMed] [Google Scholar]
- Demchenko A.P. Site-selective red-edge effects. Methods Enzymol. 2008;450:59–78. doi: 10.1016/S0076-6879(08)03404-6. [DOI] [PubMed] [Google Scholar]
- Eftink M.R. In: Lakowicz J.R., editor. Vol. 2. Plenum Press; 1991. Fluorescence quenching: theory and applications; pp. 53–126. (Top. Fluorescence Spectrosc.). [Google Scholar]
- Eftink M.R., Selvidge L.A., Callis P.R., Rehms A.A. Photophysics of indole derivatives: experimental resolution of La and Lb transitions and comparison with theory. J. Phys. Chem. 1990;94:3469–3479. [Google Scholar]
- Fery-Forgues S., Fayet J.-P., Lopez A.,J. Drastic changes in the fluorescence properties of NBD probes with the polarity of the medium: involvement of a TICT state? J. Photochem. Photobiol. A. 1993;70:229–243. [Google Scholar]
- Fiserova E., Kubala M. Mean fluorescence lifetime and its error. J. Lumin. 2012;132:2059–2064. [Google Scholar]
- Getz E.B., Xiao M., Chakrabarty T., Cooke R., Selvin P.R. A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal. Biochem. 1999;273:73–80. doi: 10.1006/abio.1999.4203. [DOI] [PubMed] [Google Scholar]
- Haldar S., Raghuraman H., Namani T., Rajarathnam K., Chattopadhyay A. Membrane interaction of the N-terminal domain of chemokine receptor CXCR1. Biochim. Biophys. Acta. 2010;1798:1056–1061. doi: 10.1016/j.bbamem.2010.02.029. [DOI] [PubMed] [Google Scholar]
- Hansen R.E., Roth D., Winter J.R. Quantifying the global cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA. 2009;106:422–427. doi: 10.1073/pnas.0812149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland R.P. Molecular Probes; 2005. The Handbook – A Guide to Fluorescent Probes and Labeling Technologies, 10th ed. [Google Scholar]
- Ho D., Lugo M.R., Merrill A.R. Harmonic analysis of the fluorescence response of bimane adducts of colicin E1 at helices 6, 7, and 10. J. Biol. Chem. 2013;288:5136–5148. doi: 10.1074/jbc.M112.436303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James N.G., Jameson D.M. In: Fluorescence Spectroscopy and Microscopy: Methods and Protocols, Methods in Molecular Biology. Engelborghs Y., Visser A.J.W.G., editors. Spinger Science+Business Media, LLC; 2014. Steady-state fluorescence polarization/anisotropy for the study of protein interactions; pp. 29–42. [Google Scholar]
- Jameson D.M., Ross J.A. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 2010;110:2685–2708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S.K., Dhar D., Krishnamoorthy G., Udgaonkar J.B. Continuous dissolution of structure during the unfolding of a small protein. Proc. Natl. Acad. Sci. USA. 2009;106:11113–11118. doi: 10.1073/pnas.0812564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.E. Fluorescence approaches for determining protein conformations, interactions and mechanisms at membranes. Traffic. 2005;6:1078–1092. doi: 10.1111/j.1600-0854.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- Kaur J., Kumar A., Kaur J. Strategies for optimization of heterologous protein expression in E.Coli: roadblocks and reinforcements. Int. J. Biol. Macromol. 2018;106:803–822. doi: 10.1016/j.ijbiomac.2017.08.080. [DOI] [PubMed] [Google Scholar]
- Kratochvil H.T., Carr J.K., Matulef K., Anne A.W., Li H., Maj M., Ostmeyer J., Serrano A.L., Raghuraman H., Moran S.D., et al. Instantaneous ion configurations in the K+ ion channel selectivity filter revealed by 2D IR spectroscopy. Science. 2016;353:1040–1044. doi: 10.1126/science.aag1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy G. In: Reviews in Fluorescence 2017. Geddes C.D., editor. Springer; 2018. Fluorescence lifetime distribution brings out mechanisms involving biomolecules while quantifying population heterogeneity; pp. 75–98. [Google Scholar]
- Krishnamoorthy G. Fluorescence spectroscopy for revealing mechanisms in biology: strengths and pitfalls. J. Biosci. 2018;43:555–567. [PubMed] [Google Scholar]
- Lakowicz J.R. Springer; 2006. Principles of Fluorescence Spectroscopy, 3rd ed. [Google Scholar]
- Lakshmikanth G.S., Sridevi K., Krishnamoorthy G., Udgaonkar J.B. Structure is lost incrementally during the unfolding of barstar. Nat. Struct. Biol. 2001;8:799–804. doi: 10.1038/nsb0901-799. [DOI] [PubMed] [Google Scholar]
- Lin S., Struve W.S. Time-resolved fluorescence of nitrobenzoxadiazol-aminohexanoic acid: effect of intermolecular hydrogen bonding on non-radiative decay. Photochem. Photobiol. 1991;54:361–365. doi: 10.1111/j.1751-1097.1991.tb02028.x. [DOI] [PubMed] [Google Scholar]
- Mansoor S.E., Farrens D.L. High-throuoghput protein structural analysis using site-directed fluorescence labeling and the bimane derivative (2-pyridyl)dithiobimane. Biochemistry. 2004;43:9426–9438. doi: 10.1021/bi036259m. [DOI] [PubMed] [Google Scholar]
- Mansoor S.E., DeWitt M.A., Farrens D.L. Distance mapping in proteins using fluorescence spectroscopy: the tryptophan-induced quenching (TrIQ) method. Biochemistry. 2010;49:9722–9731. doi: 10.1021/bi100907m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S.J., Bradley D., Blumenthal R. The use of cobalt ions as collisional quencher to probe surface charge and stability of fluorescently labelled bilayer vesicles. Biochim. Biophys. Acta. 1985;818:365–372. doi: 10.1016/0005-2736(85)90011-2. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Kombrabail M., Krishnamoorthy G., Chattopadhyay A. Dynamics and heterogeneity of bovine hippocampal membranes: role of cholesterol and proteins. Biochim. Biophys. Acta. 2007;1768:2130–2144. doi: 10.1016/j.bbamem.2007.05.025. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Raghuraman H., Dasgupta S., Chattopadhyay A. Organization and dynamics of N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-labeled lipids: a fluorescence approach. Chem. Phys. Lipids. 2004;127:91–101. doi: 10.1016/j.chemphyslip.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Newby Z.E.R., O’Connell J.D., Gruswitz F., Hays F.A., Harries W.E.C., Harwood I.M., Ho J.D., Lee J.K., Savage D.F., Miercke L.J.W., Stroud R.M. A general protocol for the crystallization of membrane proteins for X-ray structural investigation. Nat. Protoc. 2009;4:619–637. doi: 10.1038/nprot.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othon C.M., Kwon O.H., Lin M.M., Zewail A.H. Solvation in protein (un)folding of melittin tetramer-monomer transition. Proc. Natl. Acad. Sci. USA. 2009;106:12593–12598. doi: 10.1073/pnas.0905967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Chattopadhyay A. Hydration dynamics in biological membranes: emerging applications of terahertz spectroscopy. J. Phys. Chem. Lett. 2021;12:9697–9709. doi: 10.1021/acs.jpclett.1c02576. [DOI] [PubMed] [Google Scholar]
- Puljung M.C., Zagotta W.N. Labeling of specific cysteines in proteins using reversible metal protection. Biophys. J. 2011;100:2513–2521. doi: 10.1016/j.bpj.2011.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman H., Chattopadhyay A. Organization and dynamics of melittin in environments of graded hydration: a fluorescence approach. Langmuir. 2003;19:10332–10341. [Google Scholar]
- Raghuraman H., Chattopadhyay A. Effect of micellar charge on the conformation and dynamics of melittin. Eur. Biophys. J. 2004;33:611–622. doi: 10.1007/s00249-004-0402-7. [DOI] [PubMed] [Google Scholar]
- Raghuraman H., Chattopadhyay A. Influence of lipid chain unsaturation on membrane-bound melittin: a fluorescence approach. Biochim. Biophys. Acta. 2004;1665:29–39. doi: 10.1016/j.bbamem.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Raghuraman H., Chattopadhyay A. Interaction of melittin with membrane cholesterol: a fluorescence approach. Biophys. J. 2004;87:2419–2432. doi: 10.1529/biophysj.104.043596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman H., Chattopadhyay A. Effect of ionic strength on folding and aggregation of the hemolytic peptide melittin in solution. Biopolymers. 2006;83:111–121. doi: 10.1002/bip.20536. [DOI] [PubMed] [Google Scholar]
- Raghuraman H., Chattopadhyay A. Melittin: a membrane-active peptide with diverse functions. Biosci. Rep. 2007;27:189–223. doi: 10.1007/s10540-006-9030-z. [DOI] [PubMed] [Google Scholar]
- Raghuraman H., Chattopadhyay A. Orientation and dynamics of melittin in membranes of varying composition utilizing NBD fluorescence. Biophys. J. 2007;92:1271–1283. doi: 10.1529/biophysj.106.088690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman H., Chatterjee S., Das A. Site-directed fluorescence approaches for dynamic structural biology of membrane peptides and proteins. Front. Mol. Biosci. 2019;6:96. doi: 10.3389/fmolb.2019.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman H., Cordero-Morales J.F., Jogini V., Pan A.C., Kollewe A., Roux B., Perozo E. Mechanism of Cd2+ coordination during slow inactivation in potassium channels. Structure. 2012;20:1332–1342. doi: 10.1016/j.str.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman H., Ganguly S., Chattopadhyay A. Effect of ionic strength on the organization and dynamics of membrane-bound melittin. Biophys. Chem. 2006;124:115–124. doi: 10.1016/j.bpc.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Raghuraman H., Islam S.M., Mukherjee S., Roux B., Perozo E. Dynamics transitions at the outer vestibule of the KcsA potassium channel during gating. Proc. Natl. Acad. Sci. USA. 2014;111:1831–1836. doi: 10.1073/pnas.1314875111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman H., Kelkar D.A., Chattopadhyay A. Novel insights into membrane protein structure and dynamics utilizing the wavelength-selective fluorescence approach. Proc. Indian Natl. Sci. Acad. A. 2003;69:25–35. [Google Scholar]
- Raghuraman H., Kelkar D.A., Chattopadhyay A. In: Reviews in Fluorescence. Geddes C.D., Lakowicz J.R., editors. Springer; 2005. Novel insights into protein structure and dynamics utilizing the red edge excitation shift approach; pp. 199–214. [Google Scholar]
- Raghuraman H., Pradhan S.K., Chattopadhyay A. Effect of urea on the organization and dynamics of Triton X-100 micelles: a fluorescence approach. J. Phys. Chem. B. 2004;108:2489–2496. [Google Scholar]
- Rosano G.L., Morales E.S., Ceccarelli E.A. New tools for recombinant protein production in Escherichia coli: a 5-year update. Protein Sci. 2019;28:1412–1422. doi: 10.1002/pro.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatursky O., Heuck A.P., Shepard L.A., Rossjohn J., Parker M.W., Johnson A.E., Tweten R.K. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: a novel paradigm for pore-forming toxins. Cell. 1999;99:293–299. doi: 10.1016/s0092-8674(00)81660-8. [DOI] [PubMed] [Google Scholar]
- Smith D.A., McKenzie G., Jones A.C., Smith T.A. Analysis of time-correlated single photon counting data: a comparative evaluation of deterministic and probabilistic approaches. Methods Appl. Fluoresci. 2017;5:042001. doi: 10.1088/2050-6120/aa8055. [DOI] [PubMed] [Google Scholar]
- Szabo A.G., Rayner D.M. Fluorescence decay of tryptophan conformers in aqueous solution. J. Am. Chem. Soc. 1980;102:554–563. [Google Scholar]
- Vivian J.T., Callis P.R. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann C., Kumar A., Lang A., Ohlenschlager O. Cysteines and disulfide bonds as structure-forming units: insights from different domains of life and the potential for characterization by NMR. Front. Chem. 2020;8:280. doi: 10.3389/fchem.2020.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoldak G., Jancura D., Sedlak E. The fluorescence intensities ratio is not a reliable parameter for evaluation of protein unfolding transitions. Protein Sci. 2017;26:1236–1239. doi: 10.1002/pro.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original/source data for all figures given in the expected outcome of the paper are available in the literature.