Abstract
Cytomegalovirus (CMV) is an important pathogen in transplant recipients and human immunodeficiency virus (HIV)-infected individuals. Major progress has been made in developing quantitative detection methods for CMV in recent years. Due to their high sensitivity, these assays can detect CMV early, and quantitation may be useful in predicting the patient’s risk for disease and in monitoring the effect of antiviral therapy. This review discusses methodological aspects of currently used quantitative assays for CMV (i.e., viral culture techniques, antigen detection assays, DNA detection assays including PCR, branched-DNA assay, and the DNA hybrid capture assay) and addresses the correlation of systemic and site-specific CMV load and CMV disease in different populations of immunosuppressed patients as well as the response to antiviral treatment. To date, direct antigen detection and molecular techniques have largely replaced traditional culture-based techniques for CMV quantitation. In general, a high systemic CMV load is correlated with CMV disease. This correlation is strong in the HIV-infected population and in solid-organ transplant recipients but less clear in allogeneic marrow transplant recipients. Measuring the viral load at specific anatomic sites may be an alternative way to assess disease activity in situations where the systemic viral load correlates poorly with disease activity. A reduction of the systemic CMV load also correlates with a response to antiviral treatment, but more research is needed to evaluate the role of viral load as a surrogate marker for drug resistance. Due to the widespread use of quantitative CMV detection techniques to direct and monitor antiviral treatment, there is a great need for an assessment of the reproducibility of test results and better standardization of the assays.
Cytomegalovirus (CMV) causes significant morbidity and mortality in severely immunosuppressed individuals. Although antiviral prophylaxis has led to a reduction of both morbidity and mortality of CMV disease in recent years, the toxicity associated with currently available antiviral agents (i.e., ganciclovir, foscarnet, and cidofovir) remains a significant problem. Thus, efforts have been aimed at developing highly sensitive and quantitative detection methods to identify patients at risk for disease prior to the onset of disease, thereby focusing antiviral treatment to patients at risk for disease only. This strategy has been termed “preemptive” or “early” therapy (76, 141, 150). Several aspects of the biology of CMV support such an approach. CMV dissemination in the blood occurs during active infection (143), and viremia has been recognized as the major virologic risk factor for the progression to clinical disease (117). Thus, quantitation of the systemic CMV load may provide a highly sensitive and specific method to predict the development of CMV disease.
Efforts to quantify CMV date back several decades. Early assays for CMV quantitation included the traditional plaque assay (182) and the determination of the 50% tissue culture infective dose (TCID50) (151). These assays and modified tissue culture-based methods are hampered by time-consuming procedures, poor reproducibility, and/or a relatively low sensitivity. DNA hybridization techniques provided the first major advance in quantifying CMV with a high degree of reproducibility but were also relatively time-consuming and insensitive (37, 38, 144). There are several requirements for an optimal assay for CMV monitoring. The most important characteristics are (i) a high sensitivity that allows early detection in individuals at high risk for disease, (ii) the potential to quantify the results to increase the positive predictive value and to measure viral load during antiviral treatment, (iii) rapidity to allow early initiation or change of treatment, and (iv) a high degree of reproducibility. Until recently, rapid and reproducible quantitative results have been difficult to obtain. However, quantitative antigen detection methods and molecular amplification methods which fulfill these requirements to some extent are now available. In particular, PCR has revolutionized diagnostic virology by providing a powerful tool to detect and quantify viral DNA and RNA in various clinical specimens.
This review will discuss methodological aspects of quantitative CMV assays with an emphasis on recently developed antigen detection assays and molecular methods. In addition, the role of the CMV load in the pathogenesis of CMV disease in different patient settings, the influence of antiviral treatment on viral load, and the significance of viral load measurement at specific anatomic sites are reviewed.
PATHOPHYSIOLOGY OF CMV DISSEMINATION
Dissemination of CMV in the blood is an important factor in the pathogenesis of disease in both primary infection and reactivation. During active infection, CMV dissemination in the blood occurs in most immunocompromised patients. Evidence that CMV dissemination in blood is a significant risk factor for the progression from infection to disease comes from studies with allogeneic marrow transplant recipients which showed that viremia was highly predictive for the development of CMV disease (50, 117, 150). In solid-organ transplant recipients and human immunodeficiency virus (HIV)-infected patients, this association also exists but viremia is somewhat less predictive of disease (53, 61, 133, 188). Studies involving more sensitive techniques for the detection of CMV in blood such as the antigenemia assay or PCR for CMV DNA show that CMV can be detected in almost all patients with CMV disease. Due to the high sensitivity of these assays, CMV is also detectable in a substantial number of patients with asymptomatic infection who never progress to disease (16, 179). However, patients with disease often have a higher viral load than those who remain asymptomatic (31, 64, 169). CMV DNA can be detected in different fractions of leukocytes during active infection, including mononuclear leukocytes (MN) and polymorphonuclear leukocytes (PMN) (27, 70, 144). While the major amount of DNA in PMN seems to originate from phagocytosis, there is also evidence of active replication (70, 78, 138). Although CMV is a cell-associated virus, CMV DNA can also be detected in plasma or serum (2, 33, 123, 161). Longitudinal studies indicate that DNA is detected in plasma coincident with or after being detected in PMN (16). It has been hypothesized that cell-free viral DNA in plasma could be derived from lysis of CMV-infected cells such as PMN and endothelial cells (16, 64). The correspondence of the glycoprotein B (gB) and glycoprotein H (gH) variants in cells and extracellular fluid further indicates that extracellular DNA is derived from the cells supporting viral replication (186). Additional support for endothelial cells as an important source of cell-free CMV DNA comes from the observation that CMV DNA can be detected in plasma from patients with absolute neutropenia early after marrow transplantation (16). CMV-infected endothelial cells have also been detected in the peripheral blood during active infection in both transplant recipients and HIV-infected patients (79, 128, 147).
METHODS FOR CMV QUANTIFICATION
CMV quantitation can be performed in different fractions of the blood (i.e., cellular fractions and plasma) and organ fluids (e.g., cerobrospinal fluid [CSF], urine, throat wash, and semen). The optimal specimen depends on the test method and patient setting. Although CMV is recovered more frequently by culture of samples enriched for PMN than by culture of samples enriched for MN (22, 142), the amount of CMV DNA recovered is virtually identical in PMN and MN from subjects with CMV disease (27, 144). More recently, CMV DNA has been found in cell-free plasma or serum by PCR (2, 16, 33, 60, 64, 86, 123, 126, 161). In contrast, isolation of CMV from plasma in cell culture is extremely infrequent. Although plasma has been used more extensively, serum was found to be an equivalent substrate for the detection of CMV DNA by PCR in liver transplant recipients (126). Most studies show that the quantity of viral DNA in leukocytes is generally greater than in plasma for both transplant recipients (64) and subjects with AIDS (24, 64, 133, 186). The positive predictive value of PCR of plasma for the development of CMV disease has been evaluated in different settings with somewhat contradictory results. Although CMV infection has been shown to precede the development of clinical disease (86, 161), PCR of plasma reflected the kinetics of CMV infection poorly, especially during therapy (compared to PCR of leukocytes) in subjects with AIDS (64). In liver transplant recipients, PCR of serum was the best method of predicting the development of symptomatic CMV infection (125). PCR of plasma appeared to be an earlier and more sensitive marker of serious CMV infection in marrow transplant recipients than did PCR of leukocytes in one study (123), while others found either similar (87) or lower (16) sensitivity than PCR of leukocytes.
Thus, the use of leukocytes may be more appropriate in situations where the viral DNA load is low (i.e., during antiviral therapy) (64) or when preemptive therapy is envisaged in a setting where there is rapid progression from first detection to CMV disease, such as after allogeneic marrow transplantation (14, 16). In contrast, quantitation of CMV in plasma seems to be useful in HIV-infected individuals, in whom the progression from first detection of CMV DNA to disease often is several weeks or months (31, 156, 162). The detection of CMV DNA in plasma may also be useful when leukocytes counts are low, especially in marrow transplant recipients (16).
Quantitative Viral Cultures
Principle and assay characteristics.
There are three major methods of quantitating CMV in cultures: the plaque assay, determination of TCID50, and shell vial centrifugation cultures. The traditional method of CMV quantitation in culture is the plaque assay (182). In a plaque assay, serial dilutions of the specimen are inoculated onto fibroblast monolayers and after infection the cells are overlaid with a semisolid medium. The virus spreads from cell to cell, resulting in a localized plaque. After an incubation period, the infectious plaques are enumerated under microscopic examination. The plaque assay and modifications of it have been used frequently for CMV quantitation in a wide variety of clinical specimens and in animal models (37, 38, 103, 152, 182) (Table 1). Results are obtained by calculating a logarithmic viral titer from the plaque counts in the dilution giving about 200 plaques. Absorption can be enhanced by centrifugation during the inoculation period (37).
TABLE 1.
Summary of nonmolecular quantitative CMV assays
| Assay | Turnaround time | Sample processing (blood) | Reporting of results | Batch testing | Reproducibility (reference) | Equipment and facilities needed | Advantages | Disadvantages |
|---|---|---|---|---|---|---|---|---|
| Conventional cell culture | 2–4 wk | Recovery of PMN within a few hours | No. of PFU or TCID50 | NAa | Low (38) | Cell culture facility, light microscopy | Virus isolate available for susceptibility testing | Low sensitivity; very slow CPEc; risk of bacterial or fungal contamination; large set of dilutions needed; rapid loss of viability in clinical specimens |
| Shell vial assay | 16–48 h | Recovery of PMN within a few hours | No. of infectious foci (p72 antigen) | NA | Unknown | Cell culture facility, IFb or light microscopy | Can be used with nonblood samples; detection of infectious virus; rapidity of procedure | Low sensitivity; risk of cell toxicity with blood samples; not well suited for large numbers of samples; rapid loss of viability in clinical specimens |
| pp65 antigenemia assay | 5 h | Recovery of PMN within 4–6 h | No. of positive cells (pp65 antigen per 1.5 × 105 or 2 × 105 cells) | Variable (e.g., 20 to 30 samples) | Few data, relatively low (177) | Cytospin (facultative); IF or light microscopy | Rapidity of procedure | Requires rapid sample processing for reliable quantitation; not well suited for large numbers of samples |
NA, not applicable.
IF, immunofluorescent.
CPE, cytopathic effect.
A somewhat less precise method of quantifying CMV in tissue culture is determination of the TCID50. With this method, the virus titer can be determined by determining the highest dilution of the specimen which produces a cytopathic effect in 50% of the cell cultures or wells inoculated (151). The TCID50 has been used to quantify CMV in lung tissue and urine (134, 153, 181). Serial log or half-log dilutions of the specimen are inoculated onto human foreskin fibroblasts and observed for 6 weeks.
Recently, traditional tissue culture methods have been replaced by the more rapid shell vial centrifugation culture assay system. CMV is quantified in shell vial-based assays either by keeping the viral inoculum constant and counting the number of infectious foci per shell vial (67) or by inoculating serial dilutions of the sample and determining the titer, which is defined as the reciprocal of the final dilution in which CMV can be detected (158). Shell vial centrifugation culture-based assays have been used for the quantitation of viremia and of CMV in bronchoalveolar lavage (BAL) fluid (6, 37, 49, 67, 158, 164).
Overall, all culture-based assays do not require a highly specialized laboratory. Problems that may occur include the poor ability of some CMV strains to form plaques, rapid loss of viability, lack of staining by monoclonal antibodies (MAbs), and nonspecific monolayer toxicity (Table 1). The principal advantage of the shell centrifugation culture system is the early availability of the results.
Assay performance.
The plaque assay has been compared with DNA hybridization, shell vial-based methods, and the branched-DNA (bDNA) assay. The virus titers determined by plaque assay showed a moderate correlation with the quantity of CMV DNA measured by DNA hybridization (correlation coefficient, r = 0.588) (38). A high degree of correlation between the plaque assay and a shell vial-based rapid assay was reported when a MAb against the immediate-early antigen was used (37). The results of DNA hybridization experiments correlated reasonably well with the results of TCID50 experiments (correlation coefficient, r = 0.77) on urine specimens having a viral titer of 2.5 TCID50/0.2 ml or higher (38).
The shell vial-based quantitative viremia assay has been correlated with quantitative pp65 antigenemia, quantitation of CMV-DNA in plasma and PMN, and the development of CMV disease in both transplant recipients and AIDS patients. Although the results of the quantitative viremia assay correlate well with those of quantitative assays of pp65 antigenemia and CMV DNA in peripheral blood leukocytes (PBL) and plasma, the sensitivity of the assay is significantly lower than that of the antigenemia assay and PCR-based assays (63, 64, 67). To optimize sensitivity, at least 4 × 106 leukocytes should be used (34). Due to this lower sensitivity, CMV is generally detected in blood cultures later during the course of infection than by the antigenemia assay or by PCR-based methods (17, 169). Similarly, blood cultures are of limited value for monitoring the response to antiviral therapy, because they often become negative regardless of the outcome (12). Also, a low systemic viral load cannot be quantified by this method.
Reproducibility and test accuracy.
The reproducibility of the plaque assay was compared with that of DNA hybridization (Table 1). The coefficient of variation of the plaque assay was significantly greater than that of the DNA hybridization method (15.05 and 5.27, respectively) (38). This result suggests that quantitation of CMV by culture may be less accurate than quantitation of CMV DNA. Confidence limits for CMV quantitation by the shell vial method have been reported (34).
Summary.
Available data suggest that culture-based assays can provide an estimate of CMV quantity. The practical significance of these assays has been limited by the relatively low sensitivity, the time-consuming nature of the methods, and the rapid loss of viability in stored specimens. Therefore, these methods have been largely replaced by direct antigen or DNA detection methods. While these assays may be useful for specific research questions, their relatively low sensitivity limits their value in early treatment strategies and in monitoring antiviral treatment. The characteristics of these quantitative culture methods are summarized in Table 1.
Quantitative pp65 Antigenemia Assay
Principle.
The CMV antigenemia assay consists of direct staining of PMN with monoclonal antibodies directed against the lower matrix protein pp65 (UL83) (70, 81, 138). The results are expressed as the number of antigen-positive cells relative to the number of cells used to prepare the slide (Table 1).
Assay characteristics.
The antigenemia assay consists of four steps: (i) isolation of the PMN by dextran separation and preparation of slides, (ii) fixation, (iii) immunostaining, and (iv) reading and quantification. Modifications concerning all four procedural steps have been proposed to optimize and simplify the original procedure. The technical aspects of the CMV antigenemia assay are summarized in Table 2.
TABLE 2.
Technical aspects of the CMV antigenemia assay
| Aspect of the assay | Original method (177) | Published modifications | Modification evaluated in comparative trial (reference) | Comments on modifications |
|---|---|---|---|---|
| Preparation of slides | ||||
| Type of blood sample | EDTA | Heparin | Equivalent to EDTA (166) | |
| Citrate | Equivalent to EDTA (104) | Small sample size, but a trend to a smaller number of positive cells with citrate | ||
| Isolation method | Dextran sedimentation | Whole blood | Inferior (106), equivalent (91) | Time-saving |
| Ficoll sedimentation | Inferior (177) | pp65 found predominantly in granulocytes | ||
| Slide preparation | Centrifugation | Methanol spreading | NDa | |
| Adherence | ND | |||
| Flow cytometry | ND | No comparison with cytospin method (93) | ||
| No. of cells used per slide | 150,000 | 50,000 | ND | |
| 100,000 | ND | |||
| 200,000 | ND | |||
| 250,000 | ND | |||
| 300,000 | ND | |||
| 1,000,000 | ND | |||
| No. of slides stained per sample | Three | Two | ND | Two slides most commonly used |
| One | ND | |||
| Fixation | Acetone | Methanol-acetone | Inferior to formaldehyde–NP-40 with immunofluorescence (66) | |
| Formaldehyde–NP-40 | Superior to acetone with immunofluorescence (20, 51, 66, 105, 129) | |||
| Formaldehyde | Equivalent to formaldehyde–NP-40 (129) | |||
| Methanol-acetic acid | ND | |||
| Formol acetone | ND | |||
| Immunostaining | C10-C11-C12 | C10-C11 | Equivalent to C10-C11-C12 (177) | |
| Monoclonal antibodies | C12 | Superior to C10-C11 (28) | ||
| 2A6, 1C3, 4C1 | ||||
| Individually | Equivalent to C10-C11 (66); 1C3 superior to C10-C11 by flow cytometry (93) | More positive foci | ||
| Pooled | Superior to C10-C11 (66) | More positive foci | ||
| AAC10 | ND | |||
| HRP-C7 | ND | |||
| Monofluo kit CMV | Superior to C10-C11 and 1C3 (163) | |||
| Detection system | Indirect immunoperoxidase | Indirect immunofluorescence | Superior to immunoperoxidase (20, 66, 105) | More positive foci |
| APAAP | Inferior to immunofluorescence (66) | Fewer positive foci | ||
| Immunogold-silver | ND | |||
| ABCb | Inferior to immunofluorescence (66) | Fewer positive foci per slide | ||
| Quantification | Per 50,000 PBL (counted on each slide) | Per slide, with reporting of the number of cells used to prepare the slides | ND | Most commonly used |
ND, not determined.
ABC, avidin-biotin peroxidase complex.
(i) Isolation and preparation of slides.
Heparin, EDTA, and citrate appear to be equivalent as anticoagulants in blood samples submitted to the laboratory for antigenemia testing; however, most data were obtained in experiments with heparin or EDTA. During the various steps of the procedure, a substantial loss of cells may occur (as high as 50 to 60% of the original concentration) (10, 171). Although a decrease in quantitative antigenemia after storage of blood specimens has been reported, overnight storage only rarely results in the designation of a truly positive specimen as false-negative (20, 149). Nevertheless, for optimal results, slides should be prepared on the same day, preferably within 6 h of blood collection (149). The slides can be stained the following day without resulting in a decline of quantitative antigenemia (10). Dextran is most commonly used to separate the leukocytes in the antigenemia assay. Two recent comparative studies suggest that direct leukocyte lysis may be equivalent to dextran separation and also reduces the specimen-processing time to less than 3 h (52, 91). Because pp65 antigenemia is found mainly in granulocytes, methods that select for the MN fraction such as Ficoll gradient separation should not be used. Since there is high variability between slides in samples with low viral load (177) and since the use of only one slide is likely to compromise the sensitivity of the assay, most investigators use two or three slides per sample. The optimal number of cells per slide has not been established, but most investigators use either 150,000 or 200,000 cells (14, 64, 168). The point at which alteration in the number of cells per slide results in a clinically significant difference in virus quantification is unknown.
(ii) Fixation.
Although acetone fixation gives a strong staining signal, stained cells have poor morphology and artifacts occur with immunoperoxidase staining due to endogenous peroxidase. Endogenous peroxidase can be blocked (96, 169), but this adds an additional step to the procedure. A number of alternative fixation methods have been proposed. There is good evidence that formaldehyde–Nonidet P-40 (NP-40) fixation provides a clearer signal, fewer artifacts, and a higher sensitivity with immunofluorescence staining (Table 2). One study reported that the permeabilization step with NP-40 may not be required when formaldehyde is used (129).
(iii) Immunostaining.
MAbs directed against the lower matrix protein pp65 are used in the assay (80). Most studies have been performed with the C10-C11 mixture but newly developed MAbs show promising results (Table 2). The original protocol used acetone fixation in conjunction with immunoperoxidase staining for visualization (177), but the relatively high rate of artifacts led to the evaluation of alternative staining methods such as immunofluorescence and alkaline phosphatase anti-alkaline phosphate (APAAP). A lower rate of artifacts, a shorter processing time, and a higher sensitivity with immunofluorescence staining than with immunoperoxidase staining has been reported (Table 2). Although the APAAP method requires a somewhat longer processing time, interpretation of slides may be easier and less time-consuming because the high signal-to-noise ratio of this technique allows reading of the slides at a low magnification (8). There has been no systematic comparative study evaluating the APAAP technique for the detection of quantitative antigenemia.
(iv) Quantification.
Two methods are currently used to quantify antigenemia results. The more commonly used method is to report the number of positive cells per slide or per number of cells used to prepare the slide. The other method is to quantify the result per 50,000 cells by estimating the number of cells on each slide with a grid (173, 177). An advantage of the latter method is that more accurate results are obtained because potential cell loss during the procedure (e.g., due to inadequate adherence to the slide) is taken into account; a disadvantage is that it is very time-consuming. Whether counting the actual number of cells on the slide is relevant for reporting quantitative antigenemia under clinical laboratory conditions is unknown.
In an effort to automate the procedure, recent modifications of the antigenemia assay have included the use of flow cytometry (93) and image cytometry (28, 95). With flow cytometry, the 1C3 MAb was superior to the C10-C11 pool; however, the overall sensitivity appeared to be somewhat lower than that of the cytospin method (93). While these initial results are promising, systematic comparative evaluations of quantitative aspects of antigenemia testing with these techniques are needed.
Assay performance.
Unfortunately, the impact of assay modifications on the detection of quantitative antigenemia have not always been studied systematically. Some modifications (e.g., formaldehyde fixation, immunofluorescence staining, new MAbs) were reported to increase the sensitivity of the assay (Table 2), thereby possibly altering the predictive values for disease. Studies show consistently that the antigenemia assay is more sensitive than culture methods (64, 105, 176). The assay is either as sensitive as (87) or less sensitive than (14, 64, 169) PCR, depending on the method used for the antigenemia or PCR assay. The quantitative aspect of antigenemia testing has been compared with quantitative PCR, the DNA hybrid capture assay, and the bDNA assay. Quantitative antigenemia correlates well with DNA content in both plasma and leukocytes (23, 63, 64). Only a few studies have compared the antigenemia assay with the bDNA and hybrid capture assays (see below). Longitudinal studies of transplant recipients indicate that CMV infection is detected earlier by the antigenemia assay than by the shell vial centrifugation culture technique, i.e., an average 7 to 14 days before the onset of disease (12, 169). There are currently no longitudinal studies of HIV-infected individuals evaluating the time pattern of quantitative antigenemia.
Quantitation of antigenemia can be used to predict CMV disease: a higher level of antigenemia has a higher positive predictive value for disease. However, the significant threshold for predicting disease seems to differ among patient settings (Tables 4 and 5). Since the level of antigenemia decreases when antiviral treatment is initiated, although an intermittent rise may occur (14, 82), the measurement of antigenemia may be useful to monitor antiviral treatment.
TABLE 4.
Results of quantitative antigenemia in different settings
| Setting | Reference | No. of patients | No. of PMN per slide/staining methoda | Level of antigenemia per slide in patients with CMV disease or syn- drome (mean or median) | Threshold considered high risk for disease requiring antiviral treatment (per slide or per 50,000 PMN) |
|---|---|---|---|---|---|
| Heart transplant | Koskinen et al. (99) | 68 | 150,000/IP | 500 | 100 |
| Grossi et al. (83) | 294 | 200,000/IF | |||
| All patients | 385 | 200 | |||
| Recipient positive | 366 | ||||
| Recipient negative | 450 | ||||
| Ghisetti et al. (71) | 30 | 200,000/IF | 310 | NRc | |
| Liver transplant | van den Berg et al. (172) | 45 | 150,000/IP | 257b | >100 |
| Kidney transplant | van den Berg et al. (174) | 130 | 150,000/IP | 54b | >10 |
| Halwachs et al. (85) | 59 | 300,000/IP | >10 | >10 | |
| Mixed solid-organ transplant | Niubo et al. (122) | 127 | 200,000/IP | 28–250 | >20 |
| Autologous marrow transplant | Boeckh et al. (19) | 67 | 150,000/IF | >50 | >5 |
| Allogeneic marrow transplant | Boeckh et al. (12) | 59 | 150,000/IF | NR | ≥2 |
| Locatelli et al. (112) | 48 | 200,000/IF | 51 | ≥2 | |
| Gondo et al. (74) | 15 | 150,000/IP | >10 | NR | |
| HIV and AIDS | Gerna et al. (64) | 52 | 200,000/IF | >100 | NR |
| Francisci et al. (59) | 49 | 200,000/IF | 59 | NR | |
| Salzberger et al. (146) | 144 | 50,000/APAAP | |||
| Retinitis | 28 | NR | |||
| Colitis | 44 | NR | |||
| Esophagitis | 171 | NR | |||
| Bek et al. (9) | 144 | 200,000/IF | NR | >5 | |
| Wetherill et al. (183) | 22 | 100,000/IF | 693 | >48 | |
| Reynes et al. (140) | 138 | 200,000/IF | 695 | >100 | |
| Dodt et al. (45) | 200 | 50,000/APAAP | 15d | NR | |
| Podzamczer (130) | 241 | 200,000/IF | >20d | >20d |
IP, immunoperoxidase staining; IF, immunofluorescence staining.
Numbers are per 50,000 PMN (counted).
NR, not reported.
Numbers adjusted to number of cells per slide.
TABLE 5.
Positive and negative predictive value of pp65 antigenemia for CMV disease in different patient settings
| Setting | Reference/antigenemia level | No. of patients | Prevalence of CMV disease (%) | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|---|
| HIV-infected patients | Bek et al. (9) | 144 | 30 | ||
| Any positive test | 47 | 95 | |||
| More than 5 positive cells/slidea | 63 | 84 | |||
| Reynes et al. (140) | 138 | 27 | |||
| Any positive test | 45 | 94 | |||
| More than 10 positive cells/slide | 49 | 86 | |||
| More than 50 positive cells/slide | 78 | 80 | |||
| More than 100 positive cells/slide | 93 | 80 | |||
| More than 500 positive cells/slide | 100 | 77 | |||
| Francisci et al. (58) | 49 | 27 | |||
| Any positive test | 46 | 92 | |||
| More than 20 positive cells/slide | 91 | 92 | |||
| Podzamczer et al. (130) | 241 | 12 | |||
| Any positive test | 26 | 97 | |||
| More than 10 positive cells per 100,000 cells | 65 | 95 | |||
| Allogeneic marrow transplant recipients | Boeckh et al. (12, 17) | 33 | 27 | ||
| Any positive test | 53 | 91 | |||
| More than 1 positive cell/slide | 75 | 72 | |||
| Kidney transplant recipients | van den Berg et al. (174) | 130 | 17 | ||
| Any positive test | 41 | 100 | |||
| More than 1 positive cell per 50,000 cells | 53 | 100 | |||
| More than 10 positive cells per 50,000 cells | 69 | 96 | |||
| More than 100 positive cells per 50,000 cells | 75 | 88 |
Numbers of cells used to prepare slides are listed in Table 4.
Because the antigenemia assay is cell based and antigenemia is a low-frequency event, a sufficient number of granulocytes is required. Therefore, the test may be difficult to perform early after marrow or stem cell transplantation or during a period of severe neutropenia (12, 109). In a study of allogeneic marrow transplant recipients, 2.8% of 570 samples were deemed noninterpretable due to low granulocyte counts (12). Absolute neutrophil counts (ANC) correlated with the ability to perform the assay: 96, 70, and 11% of samples yielded interpretable results when the ANC was between 1.0 × 109 and 0.5 × 109/liter, between 0.5 × 109 and 0.2 × 109/liter, and less than 0.2 × 109/liter, respectively. Thus, an ANC of at least 0.2 × 109/liter is required to obtain reliable results (10). Strategies to increase the PMN yield in patients with a low ANC include increasing the blood volume and reducing the number of slides to get at least one good slide. Alternatively, PCR for CMV DNA in plasma could be performed in this situation (discussed below).
Reproducibility and test accuracy.
Because of the relatively low frequency of pp65 antigen-positive cells in most patients, a substantial degree of intra-assay variability is to be expected. When acetone fixation and immunoperoxidase staining were used, the coefficient of variation between duplicate slides with more than 50, 5 to 50, and less than 5 antigen-positive cells per 50,000 PMN was 9, 22, and 101%, respectively (177). No data exist on interassay variability and variability with respect to other antigenemia protocols. The interobserver variability was 6% in one study (177) and 1.8% in another (9). A comparison of results from several different laboratories, including some with no or limited experience with the antigenemia assay, showed a concordance rate for qualitative results of 97.5% (51).
Summary.
The pp65 antigenemia assay is a sensitive method of estimating the systemic CMV load. Advantages include a short processing time (less than 6 h) and the lack of requirement of a highly specialized laboratory, although some experience in slide preparation and reading is required. The assay is a good choice for laboratories with low- to medium-volume testing. Disadvantages include the need for immediate processing, the relatively time-consuming nature of the different assay steps with large specimen numbers, and the subjective component of slide interpretation. Nevertheless, numerous studies indicate that the antigenemia assay provides a good estimate of the systemic CMV burden, which correlates well with results obtained by quantitative PCR of leukocytes and plasma. Because there are various modifications of the assay, some of which influence sensitivity, standardization of the assay would greatly improve the comparability of study results (168). Available data suggest that a method with 150,000 to 200,000 PMN per slide on two slides combined with immunofluorescence staining and formaldehyde fixation provides optimal results.
Quantitative PCR Assays
General principles.
In the last 10 years, PCR technology has become an invaluable diagnostic tool in virology because of its ability to detect minute amounts of viral nucleic acids in clinical specimens. A more recent application of this technique, called quantitative PCR, has been used to determine relative or absolute amounts of target DNA or cDNA subjected to the amplification reaction. The following relationship is used to relate the initial concentration of target DNA to the concentration of the amplified target DNA:
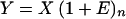 |
where Y is the amount of PCR-amplified DNA, X is the amount of target DNA prior to PCR, E is the average efficiency for each PCR cycle, and n is the number of amplification cycles.
The concentration of the unknown quantity of target DNA in a clinical sample can be estimated from a standard curve, generated by plotting the (logarithmic) values of the PCR products (expressed in any units of measurement) against the (logarithmic) values of known quantities of target DNA (standards). The original number of viral copies present in clinical samples can then be determined by interpolating the (logarithmic) values of amplified products into the standard curve. This strategy is valid if PCR is performed in the exponential phase of the reaction, i.e., for a given number of cycles, E is close to unity and the slope of the curve is also close to unity. However, in most PCR procedures, the overall efficiency is less than 100% and the increase in the amount of amplicons stays exponential for a limited number of cycles, after which the amplification rate reaches a plateau (39, 54). Many factors can affect the efficiency of amplification and hence the yield of PCR products, including primer-annealing kinetics; concentrations of DNA, deoxynucleoside triphosphates, MgCl2, and primers; number of cycles, incomplete DNA strand separation; and PCR product strand reannealing.
Semiquantitative PCR protocols have been designed by performing titer determinations of the target template or of an external control before amplification (113) or by coamplification of an endogenous sequence to control for tube-to-tube variability (119, 124). For these methods to be quantitatively reliable, many conditions must be fulfilled, including minimal variation in the efficiency of amplification, analysis of the data before the plateau phase of the reaction, and a good knowledge of the expression of the reference standard gene in the case of an endogenous control (39, 54, 73).
The competitive PCR method is based on coamplification of an exogenous template as an internal standard that competes with the target sequence for the same primers so that any variable affecting amplification has the same effect on both. In competitive PCR, the equation of PCR amplification is as follows:
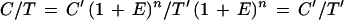 |
where C and T are the product yields of the competitor and the target templates, respectively, and C′ and T′ are the initial amounts of both templates. Since E and n are theoretically identical for both templates in competitive PCR, the relative product ratio (C/T) depends directly on the ratio of the initial amount of the two templates (C′/T′) (39, 40).
Two basic types of competitors can be used in the quantitative-competitive PCR (QC-PCR) approach. Homologous competitors are identical to the target sequence except for a small difference in sequence length (internal deletions or insertions) or for the presence of small introns (26, 56, 73). Differences between the two templates are resolved by gel electrophoresis or after restriction enzyme digestion of the PCR products (a mismatch is created in the competitor for generating or obliterating a restriction site). While homologous competitors theoretically have the same amplification efficiency as their corresponding target, the formation of heteroduplexes between the target and the competitor when the reaction has reached its plateau is a possible pitfall. Generally, enzyme digestion of amplified sequences should be avoided because an additional step, whose efficiency may vary, is introduced into the assay (39). In contrast, heterologous internal standards have the same primer target sequences but contain a different intervening sequence (62, 132, 185). This type of competitor cannot form heteroduplexes, but its amplification efficiency must be shown to be equivalent to that of the target so that reliable quantitative results are obtained. In particular, the size and the G+C content of the competitor must be as similar to those of the target template as possible. Different capture probes are then used to discriminate between the two PCR products.
In general, amplified products are detected and quantified by densitometric evaluation of ethidium bromide-stained gels or after isotopic or nonisotopic hybridization procedures. The amount of PCR products expressed as optical density values, counts per minute, or densitometric values are used for the construction of the standard curve for the assay.
A relatively new concept in quantitation of DNA or RNA, which has not been evaluated for CMV quantitation, is the detection of PCR products as they accumulate, thereby avoiding disadvantages of traditional quantitative PCR methods including post-PCR processing and a limited linear dynamic range. This technique is based on a fluorogenic 5′ nuclease assay (88, 89) (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). A fluorogenic probe (TaqMan; Roche Molecular Systems) that is complementary to the target sequence is added to the PCR mixture. This probe consists of an oligonucleotide with a reporter and quencher dye attached which anneals specifically between the upstream and downstream primer sites. The oligomer is blocked at the 3′ end and therefore cannot be extended. When the probe is cleaved by the 5′ nuclease activity of the Taq DNA polymerase, the reporter is physically separated from the quencher dye and a sequence-specific signal is generated. With each cycle, additional reporter dye molecules are cleaved from their respective probes, and the fluorescence intensity is continuously monitored during the PCR cycling. The light output is directly proportional to the amount of DNA in the sample.
Protocols and assay performance.
Many in-house quantitative CMV PCR assays have been described in the last 5 years. Protocols differ in many aspects including the specimen types and preparation, primers and targets, quantitation standards and controls, reaction and amplification protocols, prevention of contamination, signal generation systems, and methods for calculating copy numbers. For example, Fox et al. developed a QC-PCR assay based on a homologous gB competitor with the exception of a unique restriction site (56, 57, 59). Zipeto et al. have designed a PCR protocol in which a heterologous internal competitor, containing at its ends a sequence identical to the primer-binding regions of exon 4 of the CMV IE-1 gene, was used (185). More recently, Boivin et al. have developed a QC-PCR assay with a gH homologous competitor (containing a small deletion) and a rapid fluorescence-based detection system involving an automated DNA sequencer coupled to a software program (26). Additional protocols have been reported (131, 132, 148, 149, 156, 180). Although only a few comparative studies exist, there is evidence that differences in the assay protocol may affect assay performance. For example, the primer choice has been reported to influence sensitivity (116). However, delayed specimen processing has no major effect on CMV quantitation by PCR (149).
In general, most QC-PCR protocols described so far have not been of immediate use in diagnostic virology laboratories because of their expensive and cumbersome detection procedures and because of the limited potential for batch testing. At present, two detection kits are commercially available (although not Food and Drug Administration approved) for the qualitative detection of CMV DNA in clinical samples. The Sharp Signal System (Digene Corp., Silver Spring, Md.) uses PMN or plasma as source of viral DNA, whereas the AMPLICOR CMV test (Roche Diagnostic Systems, Branchburg, N.J.) uses plasma (90, 100). Both systems use nonisotopic hybridization for viral target detection in a convenient microplate assay. The AMPLICOR CMV test also includes an internal control that is coamplified with each specimen to check for the presence of PCR inhibitors (90). A quantitative PCR assay, the AMPLICOR MONITOR CMV test, is currently under evaluation for use with both plasma and PMN (167). The Sharp Signal System can also be adapted for reliable quantitative results (23).
Reproducibility and test accuracy.
Boivin et al. reported reliable results over a large dynamic range (25 to 25,000 copies of CMV DNA in a background of 105 leukocyte extracts) with intra- and interassay variabilities of 15 and 24%, respectively, by using a noncompetitive quantitative assay (23) (Table 3). Similarly, Rasmussen et al. found good reproducibility (r = 0.996) when using noncompetitive and colorimetric detection in a microplate assay (132).
TABLE 3.
Summary of molecular quantitative CMV assays
| Assay | Turnaround time (h) | Sample processing (blood) | Reporting of results | Batch testing | Lower limit of detection | Dynamic range for quantitation | Reproducibility (coefficient of variation) | Equipment and facilities needed | Advantages | Disadvantages |
|---|---|---|---|---|---|---|---|---|---|---|
| bDNA assay | 24 | Recovery of ≥2 × 106 PMN | No. of CMV copies per ml | ≤41 specimens (+3 controls) (plate format) | 4 × 103 copies/106 leukocytes (version 1); 9 × 102 copies/106 leukocytes (version 2) | 3 log10 (36) | Intra- and interassay variabilities of 5.9 and 9.7% (high copy number) and 24.9 and 26.2% (low copy number) (121) | Quantiplex bDNA System | High reproducibility | Requires large number of PMNs; long initial incubation period (16–18 h) |
| Hybrid capture DNA assay | 6 | Whole blood (3.5 ml); delayed processing possible | pg of CMV DNA or no. of CMV copies per ml | ≤48 specimens (+12 controls) per quantitative run (tube format) | 5 × 103 copies/ml of whole blood (version 1); 7 × 102 copies/ml of whole blood (version 2) | 2 log10 (16.6–1,660 pg of DNA [version 1]; 2.1–830 pg of DNA [version 2]) | Intra- and interassay variabilities of 17.8 and 16.3%, respectively (115) | Luminometer | Rapidity of procedure (6 h); simple sample processing | Many controls required for quantitative testing |
| PCR assays | 24–48a | Variable (PMN or plasma); delayed processing possible | No. of CMV copies per μl of plasma, per 105 PMN, or per μg of DNA | Variable (e.g., ≤21 samples and 3 controls per run in plate format for AMPLICOR MONITOR CMV test) | Variable (e.g., 5 × 102 [qualitative], 2.5 × 103 [quantitative] per ml of plasma [162]; 1 × 103 [qualitative], 4 × 102 [quantitative] per ml of plasma [AMPLICOR CMV test] [167]) | 2–3 log10 (23) | No data; higher if competitive assay used | Thermal cycler; detection devices (spectrophotometer, phosphorimager, etc.) | High sensitivity | Amplicon contamination; not well standardized; time-consuming |
Amplification and detection.
Comparison of quantitative PCR assay results has been elusive due to marked differences in PCR methods and reporting of results. Standardization of various parts of the assay and the development of a universal standard of CMV DNA that is amplifiable by any CMV primer set have been proposed (84, 133). Recommendations for standardizing the PCR detection of CMV in leukocytes include the processing of 2 × 106 leukocytes for the assay and the addition of DNA equivalent of 2 × 105 leukocytes per reaction (47). A method described by Schäfer et al. that measures the amount of cellular DNA might be useful in standardizing the assay results (149).
Summary.
Major progress has been made in developing quantitative assays for CMV DNA. Quantitative PCR can accurately and reproducibly determine the systemic and site-specific CMV load. Most methods are still time-consuming and require a fair amount of expertise, but newer, faster methods which allow batch testing of large numbers of specimens are being developed. Efforts should be made to standardize key elements of the assay as well as reporting of the results.
Branched-DNA Signal Amplification Assay
Principle.
The bDNA signal amplification assay (Chiron Corp., Emeryville, Calif.) quantifies CMV directly from clinical specimens. The assay relies on signal amplification with bDNA multimers. The bDNA molecule provides multiple binding sites for an enzyme-labeled probe. The complex is detected with a chemiluminescent substrate in which the light output is directly proportional to the sample input and the amount of DNA in the original sample.
Assay characteristics.
The bDNA signal amplification assay has been used with blood, CSF, and semen samples (51, 55, 98) but is not commercially available to date. At least 2 × 106 PBL or 1 to 1.5 ml of ultracentrifuged CSF are used. PMN should be separated within 8 h of collection, at which time they can be frozen for subsequent testing. The PMN pellet or CSF sample is incubated with proteinase K in a lysis buffer, and the target probes are added. After incubation for 16 to 18 h in a microtiter plate, the bDNA is added; then the enzyme-labeled probe and the chemiluminescent substrate are added. Finally, the light emission is measured (Table 3). The amount of DNA per sample is quantified with a standard curve containing known quantities of CMV DNA. Recently, an improved version of the assay with a higher sensitivity was developed (121). This was accomplished by incorporating nonnatural nucleotides into the generic sequences of the assay components, which results in a lower background signal, and by the addition of preamplifiers, which increases the amount of alkaline phosphatase-labeled probes bound to the target. No cross-reactivity was reported with herpes simplex virus type 1 (HSV-1), HSV-2, human herpesvirus 6 (HHV-6), Epstein-Barr virus, HHV-8, and adenovirus or with a variety of commonly used drugs (36). Samples from CMV-seropositive and -seronegative normal donors did not give positive results.
Assay performance.
To date, there is only limited published information on the bDNA assay. Results of the original version of the assay indicate that the assay is more sensitive than viral cultures but less sensitive than the antigenemia assay. A second generation of the assay seems to be more sensitive than viral cultures and as sensitive as the antigenemia assay (121). With this improved assay, the lower limit of detection has been increased by almost 1 log10 (Table 3) (97). The original version of the bDNA assay proved to equivalent to the antigenemia assay in detecting CMV in CSF from patients with polyneuropathy (55). In a small number of patients with polyneuropathy, the DNA content declined with antiviral treatment. The assay was also used in parallel with a traditional plaque assay in semen specimens and showed a parallel reduction of viral load in five of six patients (103). Comparative longitudinal studies with quantitative antigenemia or PCR assays have not been reported.
Reproducibility and test accuracy.
The coefficients of variation of the second-generation assay measured by three different operators over a 4-day period with two different lots was 5.9% for a high viral load and 24.9% for a viral load close to the detection limit of the assay (Table 3) (121). The bDNA assay is sufficient to discern 1.1- to 1.5-fold changes of CMV DNA levels (36).
Summary.
The bDNA signal amplification assay is a new standardized method for CMV quantification that is in an early stage of evaluation. It can be used in PMN, CSF, and semen. The lack of amplification makes the assay less susceptible to contamination. As with PCR assays and the hybrid capture assay, quantitation can be performed with stored or frozen specimens. Disadvantages of the present version of the assay are the requirement for large numbers of PMN, a possible limitation in patients with low leukocyte counts, and the relatively long processing time (Table 3). Initial data suggest that the assay may be a reliable tool for monitoring antiviral treatment, but more comparative data with antigenemia testing and quantitative PCR are required, especially with the improved version of the assay.
Hybrid Capture CMV DNA Assay
Principle.
The hybrid capture system (Digene Corp.) is a solution hybridization assay that involves amplified chemiluminescent detection. Specimens containing the target DNA hybridize with a specific CMV RNA probe complementary to approximately 40,000 bp or 17% of the CMV genome. The resultant RNA-DNA hybrids are captured onto the surface of a tube coated with antibodies specific for RNA-DNA hybrids. The immobilized hybrids are then reacted with alkaline phosphatase-conjugated antibodies specific for the hybrids and detected with a chemiluminescent substrate. Because each 38-kb RNA-DNA hybrid binds approximately 1,000 antibody conjugate molecules, each of which is bound to about 3 alkaline phosphatase molecules, the resulting signal is amplified at least 3,000-fold. The amount of light emitted, which is measured as relative light units on a luminometer, is proportional to the amount of target DNA in the specimen.
Assay characteristics.
A 1- to 3.5-ml volume of whole blood may be processed and tested by the hybrid capture CMV DNA assay, although the sensitivity may be reduced with smaller sample sizes as specified by the manufacturer. Collected blood can be held at 4°C for 6 days from the time of collection. Cell pellets may be tested immediately or stored at −20°C for future testing. For qualitative detection, samples are tested in separate tubes and up to 54 specimens (plus the controls) per batch can be processed within 6 h. With the calibration curve based on the three external standards of the kit, up to 48 specimens per run (plus controls) can be quantified over a magnitude of 2 log10 (i.e., from 16.6 to 1,660 pg of DNA per ml).
Assay performance.
The lower detection limit of the assay is approximately 5,000 copies per ml of whole blood, according to the manufacturer. A second-generation assay is now available with a claimed lower detection limit of approximately 700 copies per ml of whole blood. No cross-reactivity of the CMV probe to related viruses (HSV-1, HSV-2, Epstein-Barr virus, varicella-zoster virus, and HHV-6) has been observed. Six recent studies compared the hybrid capture DNA assay to conventional culture, the shell vial assay, and the antigenemia assay (30, 92, 106, 114, 115, 178). Veal et al. (178) found concordant results in 78.3 and 82.7% of the samples in comparisons of the hybrid capture assay to culture methods and antigenemia, respectively. Overall, there was a very good correlation between pp65 antigenemia levels and the amounts of CMV DNA as measured by the hybrid capture CMV DNA assay. In contrast, Mazulli et al. found the antigenemia assay to be more sensitive than the original hybrid capture system, especially with samples having small numbers of pp65-positive cells, i.e., less than 20 per slide (115). Lazzarotto et al. compared the hybrid capture assay with the quantitative antigenemia assay and qualitative PCR of CMV DNA in PMN (106); the hybrid capture assay was less sensitive than both the antigenemia and PCR assays. A positive though moderate correlation was found between the hybrid capture and antigenemia assays (r = 0.734) when the number of CMV DNA copies detected per milliliter in PBL was compared with the number of antigen-positive PMN per ml of blood in 32 samples (106). A decline in the number of DNA copies after start of antiviral treatment was reported in several studies (92, 106, 114, 127, 178).
Reproducibility and assay accuracy.
The intra- and interassay coefficients of variation of the hybrid capture CMV DNA system were estimated to be 17.8 and 16.3%, respectively (115) (Table 3).
Summary.
Overall, the hybrid capture technique is simple, rapid (less than 6 h), and not prone to contamination risks associated with target amplification methods such as PCR. Specimens can be held for 6 days before testing. The hybrid capture CMV DNA assay is well suited for large-volume testing. Available data indicate that the assay is specific for CMV disease and may be useful for monitoring solid-organ transplant recipients. More comparative data with antigenemia and PCR assays are needed for the second-generation hybrid capture CMV DNA system, especially for HIV-infected patients and hematopoietic stem cell recipients, who often have low systemic viral loads.
CORRELATION OF SYSTEMIC VIRAL LOAD AND CMV DISEASE
Numerous studies have investigated the correlation of CMV load and symptomatic CMV disease in immunocompromised patients by using the antigenemia and quantitative PCR assays. The question whether systemic viral load can be used to predict the subsequent development of CMV disease or relapse of CMV disease before the onset of clinical symptoms is best addressed in longitudinal studies that evaluate the time pattern of occurrence of systemic viral load relative to the onset of symptoms. Well-defined disease criteria are a critical component of these studies (110).
Solid-Organ Transplantation
Relationship between systemic viral load and CMV disease.
Studies consistently demonstrate that patients with symptomatic CMV infection have a higher systemic CMV load than those without symptoms (Tables 4 to 6). However, the numerical values reported for antigenemia and DNA copy numbers in different studies sometimes differ substantially, even when the same patient population is studied (Tables 4 to 6). These differences in quantitation make the interpretation of results difficult. Differences may be due to differences in the test method, the method of quantification (especially for antigenemia), definition of CMV disease, and the immunosuppression method. Nevertheless, antigenemia as well as DNA levels associated with disease seem to differ among patient populations (Tables 4 to 6), with heart and liver transplant recipients demonstrating a higher viral load than kidney and especially marrow transplant recipients.
TABLE 6.
Quantitative PCR: thresholds and outcomes in different patient settings
| Setting | Reference | No. of patients | Breakpoints or associationsa | Outcome |
|---|---|---|---|---|
| Renal transplant | Fox et al. (57) | 103 | >106.5 DNA copies per ml of urine | Higher association with CMV disease |
| Kühn et al. (102) | 58 | >1,000 DNA copies per 106 copies of cellular DNA | Highly predictive for CMV disease | |
| Cope et al. (43) | 196 | Each 0.25 log10 increase in baseline CMV DNA load in urine | 2.8-fold increase in CMV disease risk | |
| Toyoda et al. (170) | 25 | >500 DNA copies per 1 μg of total DNA | Increased risk of CMV disease | |
| Liver transplant | Cope et al. (42) | 162 | Each 0.25 log10 increase in baseline CMV DNA load in whole blood | 2.7-fold increase in CMV disease risk |
| 104.75–105.25 DNA copies per ml | Increased disease probability | |||
| Cardiac transplant | Toyoda et al. (170) | 95 | >500 DNA copies per 1 μg of total DNA | Increased risk of CMV disease |
| Allogeneic marrow transplant | Zaia et al. (184) | 117 | >104 DNA copies per ml of plasma | Increased risk of CMV disease after day 100 posttransplant |
| Gor et al. (77) | 110 | >104 DNA copies per ml of whole blood | Odds ratio for disease, 6.46 (95% CIb, 1.5–27.4) | |
| >105 DNA copies per ml of whole blood | Odds ratio for disease 10.66 (95% CI, 1.8–60.5) | |||
| HIV | Shinkai et al. (156) | 94 | >100 DNA copies per μl of plasma | High predictive values for CMV disease |
| Rasmussen et al. (133) | 75 | >320 copies per μg of DNA | Sustained level associated with CMV retinitis | |
| >32 copies per 25 μl of plasma | ||||
| Bowen et al. (31) | 97 | Each 0.25 log10 increase in baseline CMV DNA load in whole blood | 1.37-fold increase in risk in CMV disease | |
| Spector et al. (162) | 201 | Each log10 increase in baseline CMV DNA load in plasma | 3.1-fold increase in risk in CMV disease; 2.2-fold increase in mortality |
Shown are specific breakpoints or relative increases in quantitative CMV load that are associated with an increased risk of disease.
CI, confidence interval.
A longitudinal study performed with kidney transplant patients found that high initial levels of antigenemia correlated with higher positive predictive values for disease (Table 6). This study suggests that patients with an antigenemia of greater than 10 positive cells per slide are at high risk for the development of CMV-related symptoms (174). Therefore, these patients might benefit from antiviral treatment. Another pattern seen in many studies is that patients who have rising levels of antigenemia are likely to progress to disease (69, 83, 172, 173). To date, a trial that uses quantitative antigenemia to initiate antiviral treatment at a certain level has not been reported.
Studies that evaluated quantitative PCR of CMV DNA in solid-organ transplant patients consistently showed that patients with symptomatic CMV infection have a higher viral load than do those with asymptomatic infection (42, 43, 57, 102, 120, 170). However, occasionally symptomatic infection also occurs in patients with a low CMV DNA load (170). Several longitudinal studies using quantitative PCR indicated that patients with higher CMV viral load have a higher risk of CMV disease (Table 6). Two studies used the hybrid capture assay for CMV quantification; in both, symptomatic CMV infection was associated with higher CMV DNA levels (92, 114). Levels of more than 50 and 60 pg of CMV DNA per ml of whole blood, respectively, were predictive of more severe CMV disease.
Summary.
There is substantial evidence that weekly measurement of the systemic CMV load during the first 3 months after transplant is useful to predict CMV disease in solid-organ transplant recipients. Although good data have been obtained for renal, liver, and heart transplant recipients using the CMV antigenemia assay, more data are needed to define breakpoints for CMV DNA in different patient populations by using molecular methods. Randomized trials are also required to evaluate whether early treatment strategies based on CMV load are effective in the prevention of CMV disease. Breakpoints of 10 positive cells per slide in kidney transplant recipients and 100 positive cells per slide in liver and heart transplant recipients in the antigenemia assay may be used to institute early antiviral treatment. Available data on DNA copy numbers from different laboratories are difficult to compare due to differences in PCR methods and in reporting of results.
Marrow and Stem Cell Transplantation
Role of viremia.
Without prophylaxis, CMV disease occurs in up to 35% of allogeneic marrow transplant recipients and carries a high mortality rate despite treatment. CMV viremia is highly predictive of CMV disease in marrow transplant recipients (positive predictive value, 68%), but the predictive value is hampered by the low sensitivity of the CMV blood culture methods (117). Up to 30% of CMV-seropositive allogeneic recipients develop CMV disease without positive blood cultures before the onset of disease (48, 75), thereby limiting the use of viremia in early treatment strategies. Although highly effective in preventing CMV disease (14, 75), ganciclovir prophylaxis given at engraftment is associated with severe marrow toxicity, increased invasive fungal infections (145), and delays CMV-specific T-cell reconstitution (108), thereby increasing the risk for late CMV disease (18). Therefore, there has been considerable interest in evaluating new methods to detect and quantify CMV in blood for early treatment strategies.
Relationship of systemic viral load and CMV disease.
Studies correlating CMV load with disease have shown conflicting results. For example, a significant correlation of high levels of antigenemia and DNA in plasma and PMN with CMV disease has been found in marrow transplant patients with documented disease (Tables 4 and 6). Also, a high CMV load in plasma during the first 3 months after transplant is associated with a higher risk of late-onset CMV disease (184) (Table 6). These results are in contrast with results of other studies of marrow transplant patients, which did not report a quantitative correlation with visceral disease (27, 143). One study also found a correlation between transplant-related mortality and high systemic viral load measured by antigenemia (4). These results have not been confirmed by other groups to date.
The CMV load may be helpful in predicting CMV disease, thereby reducing the number of patients who require antiviral treatment (Table 6). In a recent study by Gor et al., the CMV load in whole blood was associated with a higher risk of CMV disease (Table 6), which was independent of acute graft-versus-host disease (GVHD) (77). The positive and negative predictive values for CMV disease in a study cohort with a prevalence of disease of 31% were 43 and 100%, respectively, for 3.5 log10 genome copies per ml, 46% and 86%, respectively, for 4.0 log10 genome copies per ml, and 86 and 79%, respectively for 5.0 log10 genome copies per ml (77).
However, even a low CMV load may be associated with CMV disease (12, 77), since it may be the first indication of CMV infection in patients with rapidly progressing CMV disease. In an earlier study, 4 of 10 patients who developed CMV disease had initial low-grade antigenemia (i.e., fewer than two positive cells per slide) (12). Subsequently, several studies with both solid organ and marrow transplant patients reported that some patients with initial low-grade antigenemia who eventually developed disease often had rapidly rising antigenemia level before the onset of disease. Thus, repeated testing shortly after the detection of a low viral load might identify patients who have rising levels of antigenemia.
Early antiviral treatment based on viral load.
To date, only one randomized study has evaluated the initiation of antiviral therapy based on systemic CMV load (14). In this double-blind study, 226 marrow transplant recipients were randomized at engraftment to receive placebo (antigenemia-ganciclovir group) or ganciclovir (ganciclovir group) until day 100 after transplant. Quantitative antigenemia assay and PCR in PMN, as well as shell vial blood cultures, were performed weekly; patients in whom a low-grade antigenemia was detected were retested within 2 to 4 days. In patients with antigenemia of two or more positive cells per slide and/or viremia by shell vial culture from blood, ganciclovir therapy was started and continued for at least 3 weeks or until 6 days after a negative CMV antigenemia test (whichever occurred later) and was resumed only if antigenemia recurred. More patients in the antigenemia-ganciclovir group developed CMV disease before day 100 posttransplantation than in the ganciclovir group (14 and 2.7%, respectively; P = 0.002). An analysis of the effect of delaying treatment until the antigenemia threshold used in this study showed a correlation with the degree of immunosuppression. Untreated low-grade antigenemia (fewer than two positive cells per slide) progressed to CMV disease in 19% of patients with grade 3 to 4 acute GVHD compared to 0% of patients with grade 0 to 2 acute GVHD (P = 0.04). Overall, there was no significant difference between the two groups in CMV disease, CMV-related death, and transplant survival after day 180 posttransplantation. Although intermittent early ganciclovir treatment based on systemic viral load as measured by antigenemia was less effective in preventing CMV disease during the first 100 days posttransplantation than was ganciclovir prophylaxis, important conclusions could be drawn from this study. Regarding the use of systemic viral load to direct antiviral treatment, these data suggest that measuring viral load is of limited value in patients with severe GVHD because of the rapid progression (i.e., sometimes less than 1 week) to CMV disease. Thus, using the viral load to limit the amount of antiviral drug treatment does not appear to be safe in a clinical setting of rapid progression from first detection of CMV to disease, and even low levels of antigenemia should be treated. In contrast, in patients without or with mild GVHD, measurement of viral load appears to be a good strategy to target antiviral drug treatment.
There are additional uncontrolled studies which have reported using antigenemia-guided early ganciclovir treatment based on quantitative antigenemia (two positive cells per slide) (63, 112, 179). These studies were performed in patients with a relatively low incidence of GVHD and showed antigenemia-guided therapy to be effective in preventing CMV disease. No studies have been reported on the amount of DNA or RNA needed to direct the antiviral treatment in hematopoietic stem transplant recipients.
Systemic viral load in autologous transplantation.
CMV disease in seropositive autologous transplant recipients is rare, but the fatality rate is similar to that in allogeneic marrow transplant recipients (15, 50). In a study of seropositive marrow autograft recipients, low-grade antigenemia was common (39%) and self-limited in most of the cases (19). Two patients with fatal CMV pneumonia had antigenemia levels of greater than 50 at the time of diagnosis, which were preceded by rising levels prior to the onset of disease. A conclusion of this study was that early antiviral treatment of antigenemia of greater than five positive cells may prevent CMV disease; there is no controlled study investigating such an approach. Quantitative molecular methods have not been evaluated in autograft recipients. Whether it is necessary to monitor seropositive autograft recipients for CMV load is currently controversial due to the low incidence of disease.
Summary.
There is only a moderate association between high systemic CMV load with CMV disease in allogeneic hematopoietic stem cell transplant patients. A significant portion of patients develop CMV disease with low systemic viral load or progress rapidly from low levels to overt disease. The latter occurs in the setting of severe acute GVHD requiring treatment with high-dose steroids. In this situation, delaying early treatment until a certain viral load threshold is reached may not be safe. Viral load should be measured at least once a week; in patients with a low viral load, testing should be repeated after 2 days. In patients with no or mild GVHD or in autograft recipients, early antiviral treatment based on systemic viral load (i.e., at least two positive cells per slide in allograft recipients and at least five positive cells per slide in autograft recipients) can prevent disease. More data are required defining specific breakpoints for quantitative PCR in this setting.
HIV and AIDS
Relationship of systemic viral load and CMV disease.
Gerna and coworkers first showed that CMV-related symptoms in AIDS patients were generally found when the number of infected PMN was greater than 50 per 2 × 105 cells as determined by the shell vial or the antigenemia assay, although such symptoms occurred sporadically with levels of 10 to 50 per 2 × 105 cells (65, 187). These studies have been subsequently confirmed by other groups (Tables 4 and 5).
By using quantitative PCR, two studies showed that the CMV DNA load in leukocytes of AIDS patients with CMV retinitis was significantly higher than the viral burden in cells of subjects with asymptomatic CMV infection and low CD4 cell counts (less than 100/μl) (23, 132). In these two studies, there was a 24- to 34-fold difference in the mean number of CMV copies between the two groups of subjects. Patients with CMV retinitis had mean DNA copy numbers of 26,433 per μg of DNA (132) and 20,453 per 105 PMN (23), compared to 1,119 and 603, respectively, in asymptomatic individuals. Boivin et al. have determined the clinical utility of a PMN PCR for the diagnosis of CMV disease in that population (23, 24). The sensitivity, specificity, and positive and negative predictive values of a qualitative PCR assay with PMN for the detection of CMV disease were 100, 62.4, 15.1, and 100%, respectively. By using a cutoff of 1,000 copies per 105 PMN, these values increased to 87.5, 96.2, 60.9, and 99.1%, respectively. In that context, almost 50% of false-positive results were from asymptomatic subjects who subsequently developed CMV disease within 1 year of testing.
Shinkai et al. have used a more convenient type of sample (plasma) as a source of viral DNA in their PCR assay (156). By comparison with urine and blood cultures, the detection of CMV DNA in plasma by PCR was associated with a higher relative risk (i.e., 23) for the development of CMV disease (versus 2.3 for urine cultures and 9.2 for blood cultures). In agreement with previous data obtained with PMN, the number of CMV copies per microliter of plasma from patients who developed CMV disease was significantly greater than that per microliter of plasma from those who remained asymptomatic. The sensitivity, specificity, and positive and negative predictive values of qualitative and quantitative (greater than 100 copies per μl) PCR of plasma for development of CMV disease were 89 and 73%, 75 and 90%, 58 and 73%, and 94 and 90%, respectively. In that study, the median time between the first positive PCR assay of plasma and CMV disease was 6 months, versus 5.7 and 5.3 months for urine and blood cultures, respectively (156).
Four additional recent studies evaluated the use of CMV load for the prediction of CMV disease (Table 6). Bowen et al. reported a high risk of CMV disease in HIV-infected patients with CD4 counts of less than 50/μl who had a positive qualitative PCR result for the CMV DNA in whole blood (relative hazard of 20.15) (31). In PCR-positive patients, each 0.25 log10 increase in viral load over time increased the risk of CMV disease by 37%. Both effects were independent of CD4 count and age (31). In a longitudinal study of patients with CD4 counts of less than 100/μl by Rasmussen et al. (133), a sustained high viral load in both plasma and leukocytes was associated with a higher risk of CMV retinitis. Progression to retinitis was not consistently accompanied by increases in the CMV burden, but isolation of CMV from blood was significantly associated with the development of retinitis; CMV load did not predict survival. In a study by Podzamczer et al. (130), both any level and a high level of pp65 CMV antigenemia were significant risk factors for CMV disease. Antigenemia positivity was also an independent risk factor for poor survival (130). Spector et al. examined the baseline CMV load in plasma in patients who participated in prospective a randomized trial of oral ganciclovir versus placebo (160, 162). PCR positivity at baseline was a strong predictor for CMV disease in both groups. Among PCR-positive patients, a high CMV load at baseline was associated with a higher risk of CMV disease, i.e., with a 3.1-fold risk for each log10 in baseline viral load. CMV positivity by plasma PCR was also associated with poor survival (adjusted relative risk, 2.5). Among patients with detectable baseline CMV DNA in plasma, each log10 increase in CMV load was associated with a 2.2-fold increase in the risk of death (162). Neither study controlled for HIV load, which might explain the inconsistent association of CMV load with poor survival. Thus, the use of quantitative PCR assays with PMN, whole blood, or plasma as well as the antigenemia assay appears to be helpful in confirming the presence of CMV disease or in identifying high-risk persons for preemptive therapy. Additional studies are needed to define the optimal frequency for viral load testing.
Lastly, it has been shown that the measure of the systemic viral DNA load does not correlate perfectly with the presence of CMV disease; i.e., some AIDS patients develop CMV retinitis at a time when the viral load in their blood is low or not detectable (9, 11, 31, 32, 130, 132, 133, 143, 156, 162). Thus, other factors may be important for the development of CMV disease, including viral virulence factors such as the possible ocular tropism or limited ability of dissemination in blood (13) of certain gB genotypes (154), as well as currently poorly defined host factors, such as CMV-specific immunity. In addition, differences in the sensitivity of the test method as well as an incorrect diagnosis of CMV retinitis may be responsible for this phenomenon (11, 62).
Summary.
There is an association between a high systemic CMV load and CMV disease in HIV-infected individuals. The CMV load is also a predictor for the development of CMV disease and response to treatment, and a high CMV load is an independent predictor of poor survival in most studies. Whether these effects of CMV load are independent of HIV load requires further study. Although this correlation was observed by using different methods of CMV load measurement (i.e., whole-blood PCR, plasma PCR, and the CMV antigenemia assay), it is unknown whether any one of these methods is superior. Thus, a prospective, longitudinal, comparative study of different methods of CMV load measurement is needed. The CMV load in HIV-infected individuals seems to be generally higher than in transplant recipients. However, some patients with a low or undetectable systemic viral load develop CMV retinitis. The optimal monitoring schedule for the CMV load has not been defined. However, monitoring of the CMV load every 2 months in patients at high risk for disease (e.g., those with CD4 counts of less than 50 per μl) seems reasonable and should identify patients who might benefit from early antiviral treatment or prophylaxis (162). Available data suggest that patients with CMV DNA at any level in plasma or whole blood might benefit from antiviral treatment. For more sensitive PMN-based assays, a higher threshold may apply (e.g., 10 positive cells per slide for antigenemia). It is also possible that more aggressive suppression of CMV load to nondetectable levels (i.e., by combination antiviral therapy) will improve survival. Randomized trails are needed to test these hypotheses.
RESPONSE TO TREATMENT, SURROGATE MARKER FOR ANTIVIRAL RESISTANCE
Solid-Organ Transplantation
A decline in the systemic viral load occurs after initiation of antiviral treatment in solid-organ transplant recipients (64, 69). In heart and heart-lung transplant recipients, a reduction of the DNA copy number in PMN by more than 2 log10 within 2 weeks was observed in two patients treated with foscarnet and three patients treated with ganciclovir (64, 69). In the same study, DNA levels in plasma were generally lower after treatment and the observed reductions were more than 1 log10. A study of renal transplant recipients by van den Berg et al. showed a sharp decline of antigenemia during treatment in 10 of 14 patients while 4 patients showed an intermittent rise of antigenemia for 1 week which was followed by a decrease during the second and third weeks of treatment (175). Antigenemia became undetectable in 50% of treatment courses after a median of 12 days (range, 7 to 21 days), while in the remaining treatment courses antigenemia remained detectable at low levels. An antigenemia level of more than 10 positive cells per 50,000 PMN was also highly predictive of a subsequent relapse of disease in patients after discontinuation of ganciclovir treatment (175). An intermittent increase of antigenemia was observed in liver transplant recipients who received early treatment with antigenemia-guided ganciclovir (82).
Marrow and Stem Cell Transplantation
Levels of DNA and antigenemia in marrow and stem cell recipients consistently decline after the start of antiviral treatment; however, there are differences in the time to viral clearance among patients. Gerna reported a permanent clearance of DNA in 9 of 14 patients treated with ganciclovir or foscarnet for antigenemia while the other 5 patients had recurrence of both antigenemia and DNAemia requiring additional treatment courses (63). The effect of foscarnet compared to ganciclovir on CMV antigenemia was evaluated in allogeneic marrow transplant recipients in a nonrandomized trial (5). This study suggested that foscarnet was as effective as ganciclovir in clearing antigenemia. In a study by Boeckh et al., 75% of patients had a negative antigenemia test result after 4 weeks of treatment with ganciclovir for asymptomatic antigenemia, and 18, 14, 9, and 5% of patients were still testing positive after 5, 6, 7, and 8 weeks, respectively, of continued ganciclovir treatment (14). As in organ transplant recipients, approximately 30% of marrow transplant recipients may experience an intermittent rise of antigenemia for up to 3 weeks after the start of ganciclovir therapy (14). This initial rise does not appear to indicate antiviral resistance in an asymptomatic patient and does usually not require a change of antiviral treatment. Although the mechanism of this phenomenon has not been evaluated, it may be due to ongoing phagocytosis by PMN of degrading CMV matrix protein pp65 from lysed, infected leukocytes (70) or from circulating CMV-infected endothelial cells (79, 128, 147). Concurrent studies measuring antigenemia and CMV DNA by PCR in these patients have not been published.
Whether antiviral therapy should be stopped once the viral load has declined to nondetectable levels is controversial. Einsele et al. reported that a short-term course of PCR-guided ganciclovir is safe in patients who test negative by PCR by the end of treatment (48). Similar results were reported by other investigators (111, 112). In contrast, in a large randomized trial in which ganciclovir was given for antigenemia at a level of 2 positive cells per slide and discontinued 6 days after a negative test result was obtained, early relapsing CMV pneumonia occurred in 7% of the patients (14). Importantly, all patients with a negative antigenemia at the end of treatment were also negative by PMN PCR in that study and only 25% of those with early relapsing pneumonia had detectable DNA before the onset of disease (14); these results are consistent with the results of an earlier study by Vlieger et al. (179). It is conceivable that in some cases CMV is cleared from the blood faster than from the lungs, which may lead to overt lung disease in patients with persistently poor CMV-specific T-cell immunity.
HIV and AIDS
Results in HIV-infected individuals and AIDS patients are in general agreement with those in the transplant setting. Antiviral treatment results in a decline of systemic viral burden both in CMV disease and in asymptomatic CMV infection (9, 27, 32). The CMV load was also assessed during foscarnet therapy in AIDS patients with asymptomatic viremia by using end-point cell dilution microcultures, pp65 antigenemia, and QC-PCR (7). During 10 days of dosing, the levels of CMV viremia, antigenemia, and DNAemia decreased from 117.5 to 12.7 TCID50 (P = 0.001), from 14.9 to 1.6 positive cells per 5 × 104 leukocytes (P = 0.008), and from 20,328 to 622 copies per 1.5 × 105 leukocytes (P = 0.02), respectively. Thus, a 10-day course of foscarnet resulted in an almost 1 log10 reduction in the CMV load in antigenemia and quantitative viremia and more than a 1 log10 reduction in CMV DNA. Moreover, a significant pharmacodynamic relationship was found between the peak foscarnet concentration and a decrease in the level of CMV antigenemia. Interestingly, foscarnet was also shown to reduce the HIV-1 burden of these subjects as determined by p24 antigen and HIV RNA assays. Bowen et al. studied the CMV load in blood and urine from 45 patients with CMV retinitis (32) and found that 85% of the patients with detectable DNA at study entry became negative for CMV DNA after 21 days of ganciclovir induction therapy. Patients who failed to clear CMV DNA by day 21 had a higher viral load at presentation than those who cleared the DNA (6.18 log10 and to 4.79 log10 DNA copies/ml, respectively). There was also a shorter time to progression of retinitis and death in patients with a higher DNA level at presentation. Quantitation of CMV DNA in blood proved superior to quantitation of CMV DNA in urine (32). The effect of foscarnet on the quantity of CMV DNA in PMN and aqueous humors of AIDS patients with CMV retinitis was first described by Gerna et al. (62). After a 3-week course of foscarnet induction therapy, 41 of 56 patients (73%) whose blood was initially CMV DNA positive became negative by PCR. Most of the subjects who had persistent CMV DNA in their blood at the end of induction had high viral loads (more than 5,000 copies/2 × 105 PMNL) before therapy. In addition, a marked reduction in viral DNA load or a negative PCR result was found in most aqueous humor samples (10 of 12) analyzed after induction therapy. This brief period of treatment was not associated with emergence of foscarnet-resistant strains or with clinical progression of retinitis. In patients with CMV gastrointestinal disease, a significant reduction of the viral load in the blood as well as in gastrointestinal biopsy specimens could be documented after induction treatment with either ganciclovir or foscarnet (44, 68).
Systemic Viral Load and Resistance
Relationship between systemic viral load and resistance.
The relationships between the viral DNA load in the blood, the emergence of drug-resistant CMV strains, and progression of CMV disease have been recently studied. The emergence of drug-resistant CMV isolates is of particular relevance for subjects with AIDS requiring lifelong maintenance antiviral therapy. Indeed, the prevalence of ganciclovir-resistant CMV isolates recovered from the urine of AIDS patients receiving this drug for more than 3 months was estimated to be about 8% (46). Mutations in the UL97 gene were detected in 30% of patients who received ganciclovir for more than 3 months (72). The systemic CMV load relative to the emergence of specific CMV UL97 mutations conferring ganciclovir resistance was evaluated in four subjects who died of progressive CMV disease (21). In this study, the viral DNA burden correlated strongly with drug resistance and disease progression in the two subjects with AIDS but not in the two persons with chronic lymphocytic leukemia. In other words, the recovery of ganciclovir-resistant CMV isolates from the blood and the detection of specific resistance-associated UL97 mutations (codons 460, 594, and 595) directly in PMN of AIDS patients was found to correlate with a high viral DNA load in the same samples. Thus, the viral DNA load in PMN may be useful to monitor AIDS patients while on therapy. Whether the use of plasma is equivalent to PMN as a source of viral DNA in this context has not been extensively studied. In a single longitudinal molecular analysis of the CMV load and viral mutations in blood compartments of an AIDS patient with progressive CMV disease (22), PCR of PMN was found to be a better (earlier) predictor of clinical and virological resistance than was PCR of plasma. An association of high viral load with both genotypic and phenotypic resistance to ganciclovir was found in a patients who was monitored by the bDNA assay (36). An increasing viral DNA load in plasma was also found in association with excretion of ganciclovir-resistant isolates from the blood and CSF before clinical manifestations of CMV polyradiculopathy (159).
Summary.
CMV load measurement appears to be a promising method for comparing the antiviral effects of different drugs, determining the length of induction therapy, and possibly assessing the emergence of drug-resistant CMV strains in subjects with AIDS. To date, only anecdotal reports exist for the last application. There is an intermittent increase of quantitative antigenemia during the first 3 weeks of antiviral treatment in some patients, which does not appear to indicate treatment failure; whether the DNA load is also intermittently increased in these patients has not been reported. Nevertheless, in antiviral agent-naive patients only increases in CMV viral load occurring after 3 weeks of treatment may be due to antiviral resistance. Prospective longitudinal studies are needed to further determine the utility of the systemic CMV DNA load for predicting clinical outcome, as well as the frequency of viral load determination and the viral load threshold associated with clinical progression.
VIRAL LOAD AT SPECIFIC ORGAN SITES
There has been considerable interest in assessing the CMV load at specific sites. The evaluation of site-specific CMV load may be particularly helpful in understanding the pathogenesis of CMV disease manifestation and in diagnosing disease and monitoring treatment effects in individuals with a low initial systemic viral load, insufficient penetration of the antiviral drug to the specific site, or competing causes of organ damage.
Central Nervous System
CMV has been implicated in distinct neurological syndromes in patients with AIDS, i.e., retinitis, myelitis/polyradiculopathy, encephalitis with dementia, ventriculoencephalitis, and mononeuritis multiplex. Autopsy studies indicate that CMV encephalitis occurs in as many as 19 to 28% of patients with AIDS and often remains undiagnosed during the life of the patient. PCR has greatly improved the diagnosis of CMV infection of the central nervous system (CNS). However, both the bDNA assay and the antigenemia assay seem to be useful as well.
CMV quantitation has been used to study the viral load in CSF in different CNS manifestations of CMV infection and to monitor treatment. Revello et al. examined the CSF of AIDS patients with neurologic symptoms by the antigenemia assay and PCR (139). Three patients with polyradiculopathy had pp65 antigen-positive PMN in the CSF at a frequency of 1.2 to 20.8%. All patients had more than 50,000 DNA copies per 5 μl of CSF. After the start of antiviral treatment, the viral load in the CSF remained elevated longer than that in blood PMN. Quantitation of CMV by the antigenemia assay and PCR showed a high degree of concordance (94%) (139). Shinkai and Spector examined 12 AIDS patients with CMV polyneuropathy and 9 patients with CMV encephalitis and found higher viral load in the patients with polyradiculopathy than in those with encephalitis (median 4,632 [range, 1 to 48,240] versus 612 [range, 1 to 8,048] DNA copies/μl of CSF) (157). In the same study, three patients showed a decline in the amount of CMV DNA in the CSF after 1 to 3 weeks of treatment with ganciclovir, ranging from 4-fold after 7 days to 70-fold after 3 weeks. Cinque et al. studied CSF from 7 AIDS patients with CMV-related CNS disease before and after 3 weeks of ganciclovir treatment. After therapy, PCR was negative in three patients who had low baseline CMV DNA levels, while four patients with higher baseline levels remained positive but with decreased levels of DNA (3.3- to 47-fold) (157). Kühn et al. evaluated the CMV load in brain autopsy specimens from patients with AIDS (101). Patients with CMV encephalitis had a 10- to 1,000-fold-higher CMV load than did patients without histologic evidence of encephalitis. CMV DNA copy numbers of greater than 6,000 per 106 copies of cellular DNA were associated with CMV encephalitis. The bDNA assay was compared with the pp65 antigen assay in the detection of CMV DNA in AIDS patients with CMV polyradiculopathy (55). Both assays detected CMV in 15 of 16 patients. Of 10 patients with follow-up specimens, 9 showed a decline in CMV DNA levels after treatment but only 2 improved clinically. Using a semiquantitative assay, Arribas et al. found CMV DNA in 12 of 13 patients with autopsy-proven CMV CNS disease (1). Patients with more than 103 DNA copies per 8 μl of CSF had severe CNS disease.
Thus, quantitation of CMV DNA in CSF by PCR and the bDNA assay in both CSF and brain tissue are sensitive tools to diagnose and monitor antiviral treatment, and they may provide more relevant results than measurement of the systemic viral load. The pp65 antigenemia assay appears to be useful as well, especially for patients with polyradiculopathy. More data are required on the correlation between changes in viral load, development of resistance, and clinical outcome.
Lungs
Because CMV can be detected in the lungs of immunocompromised patients without clinical symptoms, several studies have attempted to quantify CMV in the lung tissue or in BAL fluid to determine whether viral load is a determining factor for viral pathogenicity. Earlier studies involving quantitative culture methods or direct fluorescent-antibody assays found inconsistent results regarding the correlation between the CMV load in lung tissue or BAL fluid and symptomatic disease. Some studies showed a correlation between viral load and disease severity or outcome (38, 49, 165), and others found no such correlation (152, 158). These differences may be due in part to method used and the patient setting. Recent studies by quantitative PCR strongly support the hypothesis that the CMV load in the lungs is an important factor in the pathogenesis of CMV pneumonia (25, 35, 155). Boivin et al. measured the CMV load by quantitative PCR and CMV gene expression in 18 patients with asymptomatic pulmonary shedding and 19 patients with definite or probable CMV pneumonitis (25). There was greater than a 2 log10 unit difference in the amount of DNA between patients with asymptomatic shedding and those with definite or probable CMV pneumonia (Table 7). In addition, all patients with definite or probable CMV pneumonitis had detectable glycoprotein H mRNA compared to none of the asymptomatic shedders. These data suggest that CMV load in the lungs plays an important role in the pathogenesis of CMV pneumonia.
TABLE 7.
Quantitation of CMV glycoprotein H gene and detection of CMV transcripts in cells from BAL from immunocompromised patientsa
| Category | No. of BAL samples | Presence of IE-I mRNAb | Presence of gH mRNAb | gH copy no./μg of cell DNA
|
||
|---|---|---|---|---|---|---|
| Mean | Median | Range | ||||
| CMV seronegativeb | 8 | 0/8 | 0/8 | 0 | 0 | |
| CMV seropositive onlyc | 6 | 0/6 | 0/6 | 0 | 0 | |
| Asymptomatic CMV shedders | 18 | 7/18 | 0/18 | 72 | 20 | 0–352d |
| Definite CMV pneumonitis | 15 | 7/7 | 7/7 | 267,580 | 57,000 | 1,332–1,596,208 |
| Probable CMV pneumonitis | 4 | 4/4 | 4/4 | 16,128 | 11,992 | 1,002–40,095 |
Reprinted from reference 25 with permission of the publisher.
IE-I, immediate-early gene 1. Results are number of positive samples/total number.
BAL culture negative for CMV.
No viral DNA was detected in 6 of 18 samples.
Eyes
Testing for CMV DNA in the eyes may provide an objective virologic measurement of the response to treatment, especially in patients with a low or undetectable systemic viral load, and may provide a way to study the pathogenesis of disease (29, 118). In addition, because other viral causes of retinitis have been reported, it is useful to confirm CMV as the cause of the pathologic findings in the eyes, especially when the retinal lesions are atypical. The aqueous humor provides an accessible sample for DNA quantitation and may reflect the viral load in the eyes. The CMV load in the aqueous humor was measured in 15 AIDS patients with retinitis at the time of diagnosis; follow-up samples were obtained from 12 patients after 3 weeks of foscarnet induction therapy (62). All the patients had detectable DNA at diagnosis in both the aqueous humor and the blood, including four patients who had significantly higher levels in the aqueous humor than in the peripheral blood. A decline in the viral load both in the blood and aqueous humor was observed in four patients, while the viral load in four patients remained positive at both sites and that in four patients remained positive in the eyes but became negative in the blood. Quantitative comparison between the viral load in aqueous humor and PMN at the time of diagnosis and after induction therapy indicated a correlation between local and systemic viral load in some but not all patients. A comparison of viral load and clinical response has not been reported.
Thus, measuring the CMV load is feasible and may be useful for monitoring the efficacy of antiviral treatment in selected clinical situations. More data are required with regard to the correlation of local viral load in different compartments of the eye with progression of retinitis and development of drug resistance.
CONCLUDING REMARKS
We have reviewed the currently available methods for CMV quantitation (Tables 1 and 3). Quantification of CMV will undoubtedly play an important role in the future not only in identifying patients at high risk for disease but also in assessing the response to antiviral treatment. Transplant recipients are now monitored routinely for the CMV viral load at many centers, and HIV-infected individuals are likely to benefit from such an approach as well. Because increasing levels of systemic CMV load precede overt CMV disease in the majority of patients, preventive strategies increasingly use the CMV load as a surrogate marker for disease and initiate antiviral treatment based on systemic viral load. Indeed, ongoing prophylaxis studies in allogeneic blood and marrow transplant recipients use the CMV load rather than disease as the primary end point because CMV disease is associated with significant morbidity and a high fatality rate. It can be expected that this trend will be extended to other patient populations. HIV-infected individuals who are treated with highly effective antiretroviral therapy consisting of protease inhibitors may experience an increase in the CD4 counts (3). Although the effect of such treatment on the risk of CMV disease is currently unknown, recent data suggest that the T-cell repertoire is not immediately restored after effective therapy (41), which may lead to continued susceptibility to CMV in some patients, even those with relatively high CD4 counts (94). In this situation, where patients live longer and the traditional CD4 count may be a poor guide for risk stratification, measurement of the CMV load could provide a predictive means of identifying patients at high risk for CMV disease.
Several longitudinal natural history studies have already established breakpoints for a high risk of CMV disease in different settings. However, there is currently an almost complete lack of randomized, controlled studies validating the concept of using the CMV load in early-treatment strategies (14). Such studies are needed in different patient settings to assess the cost-effectiveness and toxicity of such an approach. Also, because a high CMV load in HIV patients is a predictor of poor survival, suppression of the systemic CMV load may have a positive effect on survival. Finally, measurement of reduction in the systemic viral load will probably be used increasingly to assess antiviral treatment strategies or to determine treatment failures. However, more data are needed for the different methods of CMV quantitation to define quantitatively the association of decline in viral load and outcome.
There is good evidence that a high CMV load is associated with a higher risk of progression to CMV disease, especially in HIV-infected hosts and in solid-organ transplant recipients. The association is less clear for allogeneic marrow transplant recipients because a substantial proportion of patients develop CMV disease with a low or undetectable CMV load. Although less common, this phenomenon has also been observed in HIV-infected patients with CMV retinitis (9, 31, 32, 132, 156). The pathogenic mechanism for these differences between settings is currently unknown, but studies performed with marrow transplant recipients suggest that it is probably due to the degree of the underlying T-cell immunodeficiency (18, 108, 136, 137) and possibly viral factors such as strain differences (13). These studies indicate that there is an inverse correlation between functional CMV-specific T-cell immunity and the CMV load (18, 136). More research is needed to confirm these data in HIV-infected individuals.
Methodologically, major improvements in the technology have occurred and simplified, faster, and optimized techniques of DNA quantitation will become available in the near future. Therefore, more data are needed on reproducibility, test accuracy, and comparability of newly developed or modified test protocols, and multi-institutional quality control programs are necessary, similar to those for HIV load testing. Also, evaluation of test modifications that alter the sensitivity or performance of tests must be evaluated comparatively with a well-characterized method involving well-defined end points (110) and appropriate sample sizes (135).
The studies summarized in this review indicate that there has been major progress over the last decade in developing and evaluating assays for CMV quantitation. What is needed now is the systematic comparative evaluation of treatment strategies involving these methods in different patient settings.
ACKNOWLEDGMENTS
Michael Boeckh was supported in part by NIH grants CA 18029 and AI 41754-01. Guy Boivin was supported in part by a grant (MA-13924) from the Medical Research Council of Canada.
We thank Alex Ryncarz for manuscript review, Terry Stevens-Ayers for editorial comments, and Jennifer Shotwell for manuscript preparation.
REFERENCES
- 1.Arribas J R, Clifford D B, Fichtenbaum C J, Commins D L, Powderly W G, Storch G A. Level of cytomegalovirus (CMV) DNA in cerebrospinal fluid of subjects with AIDS and CMV infection of the central nervous system. J Infect Dis. 1995;172:527–531. doi: 10.1093/infdis/172.2.527. [DOI] [PubMed] [Google Scholar]
- 2.Aspin M M, Gallez-Hawkins G M, Giugni T D, Tegtmeier B, Lang D J, Schmidt G M, Forman S J, Zaia J A. Comparison of plasma PCR and bronchoalveolar lavage fluid culture for detection of cytomegalovirus infection in adult bone marrow transplant recipients. J Clin Microbiol. 1994;32:2266–2269. doi: 10.1128/jcm.32.9.2266-2269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Autran B, Carcelain G, Li T S, Blanc C, Mathez D, Tubiana R, Katlama C, Debre P, Leibowitch J. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science. 1997;277:112–116. doi: 10.1126/science.277.5322.112. [DOI] [PubMed] [Google Scholar]
- 4.Bacigalupo A, Tedone E, Sanna M A, Moro F, Van Lint M T, Grazi G, Balestreri M, Frassoni F, Occhini D, Gualandi F, et al. CMV infections following allogeneic BMT: risk factors, early treatment and correlation with transplant related mortality. Haematologica. 1992;77:507–513. [PubMed] [Google Scholar]
- 5.Bacigalupo A, van Lint M T, Tedone E, Moro F, Sanna M A, Longren M, Trespi G, Frassoni F, Occhini D, Gualandi F, et al. Early treatment of CMV infections in allogeneic bone marrow transplant recipients with foscarnet or ganciclovir. Bone Marrow Transplant. 1994;13:753–758. [PubMed] [Google Scholar]
- 6.Bailey T C, Buller R S, Ettinger N A, Trulock E P, Gaudreault-Keener M, Langlois T M, Fornoff J E, Cooper J D, Storch G A. Quantitative analysis of cytomegalovirus viremia in lung transplant recipients. J Infect Dis. 1995;171:1006–1010. doi: 10.1093/infdis/171.4.1006. [DOI] [PubMed] [Google Scholar]
- 7.Balfour H H, Fletcher C V, Erice A, Henry W K, Acosta E P, Smith S A, Holm M A, Boivin G, Shepp D H, Crumpacker C S, Eaton C A, Martinmunley S S. Effect of foscarnet on quantities of cytomegalovirus and human immunodeficiency virus in blood of persons with AIDS. Antimicrob Agents Chemother. 1996;40:2721–2726. doi: 10.1128/aac.40.12.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bein G, Bitsch A, Hoyer J, Kirchner H. The detection of human cytomegalovirus immediate early antigen in peripheral blood leucocytes. J Immunol Methods. 1991;137:175–180. doi: 10.1016/0022-1759(91)90022-8. [DOI] [PubMed] [Google Scholar]
- 9.Bek B, Boeckh M, Lepenies J, Bieniek B, Arasteh K, Heise W, Deppermann K M, Bornhoft G, Stöffler-Meilicke M, Schuller I, Höffken G. High-level sensitivity of quantitative pp65 cytomegalovirus (CMV) antigenemia assay for diagnosis of CMV disease in AIDS patients and follow-up. J Clin Microbiol. 1996;34:457–459. doi: 10.1128/jcm.34.2.457-459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeckh, M. Unpublished results.
- 11.Boeckh M, Bek B, Lepenies J, Höffken G. Interest of quantitative pp65 cytomegalovirus antigenemia assay for human immunodeficiency virus-positive patients. J Clin Microbiol. 1996;34:3255–3256. doi: 10.1128/jcm.34.12.3255-3256.1996. . (Reply.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boeckh M, Bowden R A, Goodrich J M, Pettinger M, Meyers J D. Cytomegalovirus antigen detection in peripheral blood leukocytes after allogeneic marrow transplantation. Blood. 1992;80:1358–1364. [PubMed] [Google Scholar]
- 13.Boeckh M, Gooley T, Myerson D, Torok-Storb B. 8th European Congress of Clinical Microbiology and Infectious Diseases, Lausanne, Switzerland. 1997. Specific CMV gB genotypes are associated with viremia in patients with CMV disease after allogeneic marrow transplant, abstr. 1508. [Google Scholar]
- 14.Boeckh M, Gooley T A, Myerson D, Cunningham T, Schoch G, Bowden R A. Cytomegalovirus pp65 antigenemia-guided early treatment with ganciclovir versus ganciclovir at engraftment after allogeneic marrow transplantation: a randomized double-blind study. Blood. 1996;88:4063–4071. [PubMed] [Google Scholar]
- 15.Boeckh M, Gooley T A, Reusser P, Buckner C D, Bowden R A. Failure of high-dose acyclovir to prevent cytomegalovirus disease after autologous marrow transplantation. J Infect Dis. 1995;172:939–943. doi: 10.1093/infdis/172.4.939. [DOI] [PubMed] [Google Scholar]
- 16.Boeckh M, Hawkins G, Myerson D, Zaia J, Bowden R A. Plasma PCR for cytomegalovirus DNA after allogeneic marrow transplantation: comparison with PCR using peripheral blood leukocytes, pp65 antigenemia, and viral culture. Transplantation. 1997;64:108–113. doi: 10.1097/00007890-199707150-00020. [DOI] [PubMed] [Google Scholar]
- 17.Boeckh M, Myerson D, Bowden R A. Early detection and treatment of cytomegalovirus infections in marrow transplant patients: methodological aspects and implications for therapeutic interventions. Bone Marrow Transplant. 1994;14:S66–S70. [PubMed] [Google Scholar]
- 18.Boeckh, M., S. R. Riddell, T. Cunningham, D. Myerson, M. Flowers, and R. A. Bowden. 1996. Increased incidence of late CMV disease in allogeneic marrow transplant recipients after ganciclovir prophylaxis is due to a lack of CMV-specific T cell responses. Blood 88(Suppl. 1):302a.
- 19.Boeckh M, Stevens-Ayers T, Bowden R A. Cytomegalovirus pp65 antigenemia after autologous marrow and peripheral blood stem cell transplantation. J Infect Dis. 1996;174:907–912. doi: 10.1093/infdis/174.5.907. [DOI] [PubMed] [Google Scholar]
- 20.Boeckh M, Woogerd P M, Stevens-Ayers T, Ray C G, Bowden R A. Factors influencing detection of quantitative cytomegalovirus antigenemia. J Clin Microbiol. 1994;32:832–834. doi: 10.1128/jcm.32.3.832-834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boivin G, Chou S, Quirk M R, Erice A, Jordan M C. Detection of ganciclovir resistance mutations quantitation of cytomegalovirus (CMV) DNA in leukocytes of patients with fatal disseminated CMV disease. J Infect Dis. 1996;173:523–528. doi: 10.1093/infdis/173.3.523. [DOI] [PubMed] [Google Scholar]
- 22.Boivin G, Gilbert C, Morissette M, Handfield J, Goyette N, Bergeron M G. A case of ganciclovir-resistant cytomegalovirus (CMV) retinitis in a patient with AIDS—longitudinal molecular analysis of the CMV viral load and viral mutations in blood compartments. AIDS. 1997;11:867–873. doi: 10.1097/00002030-199707000-00005. [DOI] [PubMed] [Google Scholar]
- 23.Boivin G, Handfield J, Murray G, Toma E, Lalonde R, Lazar J G, Bergeron T G. Quantitation of cytomegalovirus (CMV) DNA in leukocytes of human immunodeficiency virus-infected subjects with and without CMV disease by using PCR and the sharp signal detection system. J Clin Microbiol. 1997;35:525–526. doi: 10.1128/jcm.35.2.525-526.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boivin G, Handfield J, Toma E, Murray G, Lalonde R, Bergeron M G. Comparative evaluation of the cytomegalovirus DNA load in polymorphonuclear leukocytes and plasma of human immunodeficiency virus-infected subjects. J Infect Dis. 1998;177:355–360. doi: 10.1086/514190. [DOI] [PubMed] [Google Scholar]
- 25.Boivin G, Olson C A, Quirk M R, Kringstad B, Hertz M I, Jordan M C. Quantitation of cytomegalovirus DNA and characterization of viral gene expression in bronchoalveolar cells of infected patients with and without pneumonitis. J Infect Dis. 1996;173:1304–1312. doi: 10.1093/infdis/173.6.1304. [DOI] [PubMed] [Google Scholar]
- 26.Boivin G, Olson C A, Quirk M R, St. Cyr S M, Jordan M C. Quantitation of human cytomegalovirus glycoprotein H gene in cells using competitive PCR and a rapid fluorescence-based detection system. J Virol Methods. 1995;51:329–342. doi: 10.1016/0166-0934(94)00128-4. . (Erratum, 53:165, 1995.) [DOI] [PubMed] [Google Scholar]
- 27.Boivin G, Quirk M R, Kringstad B A, Germain M, Jordan M C. Early effects of ganciclovir therapy on the quantity of cytomegalovirus DNA in leukocytes of immunocompromised patients. Antimicrob Agents Chemother. 1997;41:860–862. doi: 10.1128/aac.41.4.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boland G J, Mesker W E, Doorn R, Ploem-Zaaijer J J, Tank H J, de Gast G C. Detection improvement of cytomegalovirus antigen in human peripheral blood using monoclonal antibodies and automated reading of cell preparations. Eur J Clin Invest. 1995;25:639–646. doi: 10.1111/j.1365-2362.1995.tb01979.x. [DOI] [PubMed] [Google Scholar]
- 29.Bongarts A, Vonlaer D, Vogelberg C, Ebert K, Vanlunzen J, Garweg J, Vaith P, Hufert F T, Haller O, Meyerkönig U. Glycoprotein B genotype of human cytomegalovirus—distribution in HIV-infected patients. Scand J Infect Dis. 1996;28:447–449. doi: 10.3109/00365549609037937. [DOI] [PubMed] [Google Scholar]
- 30.Bossart W, Bienz K, Wunderli W. Surveillance of cytomegalovirus after solid-organ transplantation—comparison of pp65 antigenemia assay with a quantitative DNA hybridization assay. J Clin Microbiol. 1997;35:3303–3304. doi: 10.1128/jcm.35.12.3303-3304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowen E F, Sabin C A, Wilson P, Griffiths P D, Davey C C, Johnson M A, Emery V C. Cytomegalovirus (CMV) viraemia detected by polymerase chain reaction identifies a group of HIV-positive patients at high risk of CMV disease. Aids. 1997;11:889–893. doi: 10.1097/00002030-199707000-00008. [DOI] [PubMed] [Google Scholar]
- 32.Bowen E F, Wilson P, Cope A, Sabin C, Griffiths P, Davey C, Johnson M, Emery V. Cytomegalovirus retinitis in AIDS patients: influence of cytomegaloviral load on response to ganciclovir, time to recurrence and survival. AIDS. 1996;10:1515–1520. doi: 10.1097/00002030-199611000-00009. [DOI] [PubMed] [Google Scholar]
- 33.Brytting M, Xu W, Wahren B, Sundqvist V A. Cytomegalovirus DNA detection in sera from patients with active cytomegalovirus infections. J Clin Microbiol. 1992;30:1937–1941. doi: 10.1128/jcm.30.8.1937-1941.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buller R S, Bailey T C, Ettinger N A, Keener M, Langlois T, Miller J P, Storch G A. Use of a modified shell vial technique to quantitate cytomegalovirus viremia in a population of solid-organ transplant recipients. J Clin Microbiol. 1992;30:2620–2624. doi: 10.1128/jcm.30.10.2620-2624.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cagle P T, Buffone G, Holland V A, Samo T, Demmler G J, Noon G P, Lawrence E C. Semiquantitative measurement of cytomegalovirus DNA in lung and heart-lung transplant patients by in vitro DNA amplification. Chest. 1992;101:93–96. doi: 10.1378/chest.101.1.93. [DOI] [PubMed] [Google Scholar]
- 36.Chernoff D N, Miner R C, Hoo B S, Shen L P, Kelso R J, Jekicmcmullen D, Lalezari J P, Chou S W, Drew W L, Kolberg J A. Quantification of cytomegalovirus DNA in peripheral blood leukocytes by a branched-DNA signal amplification assay. J Clin Microbiol. 1997;35:2740–2744. doi: 10.1128/jcm.35.11.2740-2744.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chou S W, Scott K M. Rapid quantitation of cytomegalovirus and assay of neutralizing antibody by using monoclonal antibody to the major immediate-early viral protein. J Clin Microbiol. 1988;26:504–507. doi: 10.1128/jcm.26.3.504-507.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Churchill M A, Zaia J A, Forman S J, Sheibani K, Azumi N, Blume K G. Quantitation of human cytomegalovirus DNA in lungs from bone marrow transplant recipients with interstitial pneumonia. J Infect Dis. 1987;155:501–509. doi: 10.1093/infdis/155.3.501. [DOI] [PubMed] [Google Scholar]
- 39.Clementi M, Menzo S, Bagnarelli P, Manzin A, Valenza A, Varaldo P E. Quantitative PCR and RT-PCR in virology. PCR Methods Appl. 1993;2:191–196. doi: 10.1101/gr.2.3.191. [DOI] [PubMed] [Google Scholar]
- 40.Clementi M, Menzo S, Bagnarelli P, Valenza A, Paolucci S, Sampaolesi R, Manzin A, Varaldo P E. Clinical use of quantitative molecular methods in studying human immunodeficiency virus type 1 infection. Clin Microbiol Rev. 1996;9:135–147. doi: 10.1128/cmr.9.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connors M, Kovacs J S, Krevat S, Gea-Banacloche J C, Sneller M C, Flanigan M, Metcalf J A, Walker R E, Falloon J, Baseler M, Stevens R, Feuerstein I, Masur H, Lane H C. HIV infection induces changes in CD4+ T-cell phenotype and depletions within the CD4+ T-cell repertoire that are not immediately restored by antiretroviral or immune-base therapy. Nat Med. 1997;3:533–540. doi: 10.1038/nm0597-533. [DOI] [PubMed] [Google Scholar]
- 42.Cope A V, Sabin C, Burroughs A, Rolles K, Griffiths P D, Emery V C. Interrelationships among quantity of human cytomegalovirus (HCMV) DNA in blood, donor-recipient serostatus, and administration of methylprednisolone as risk factors for HCMV disease following liver transplantation. J Infect Dis. 1997;176:1484–1490. doi: 10.1086/514145. [DOI] [PubMed] [Google Scholar]
- 43.Cope A V, Sweny P, Sabin C, Rees L, Griffiths P D, Emery V C. Quantity of cytomegalovirus viruria is a major risk factor for cytomegalovirus disease after renal transplantation. J Med Virol. 1997;52:200–205. [PubMed] [Google Scholar]
- 44.Cotte L, Drouet E, Bailly F, Vitozzi S, Denoyel G A, Trepo C. Cytomegalovirus DNA level on biopsy specimens during treatment of cytomegalovirus gastrointestinal disease. Gastroenterology. 1996;111:439–444. doi: 10.1053/gast.1996.v111.pm8690210. [DOI] [PubMed] [Google Scholar]
- 45.Dodt K K, Jacobsen P H, Hofmann B, Meyer C, Kolmos H J, Skinhoj P, Norrild B, Mathiesen L. Development of cytomegalovirus (CMV) disease may be predicted in HIV-infected patients by CMV polymerase chain reaction and the antigenemia test. AIDS. 1997;11:F21–F28. doi: 10.1097/00002030-199703110-00001. [DOI] [PubMed] [Google Scholar]
- 46.Drew W L, Miner R C, Busch D F, Follansbee S E, Gullett J, Mehalko S G, Gordon S M, Owen W F, Jr, Matthews T R, Buhles W C, et al. Prevalence of resistance in patients receiving ganciclovir for serious cytomegalovirus infection. J Infect Dis. 1991;163:716–719. doi: 10.1093/infdis/163.4.716. [DOI] [PubMed] [Google Scholar]
- 47.Ehrnst A, Einsele H. Towards standardizations of CMV DNA PCR assays. Scand J Infect Dis Suppl. 1995;99:16–18. [PubMed] [Google Scholar]
- 48.Einsele H, Ehninger G, Hebart H, Wittkowski K M, Schuler U, Jahn G, Mackes P, Herter M, Klingebiel T, Loffler J, et al. Polymerase chain reaction monitoring reduces the incidence of cytomegalovirus disease and the duration and side effects of antiviral therapy after bone marrow transplantation. Blood. 1995;86:2815–2820. [PubMed] [Google Scholar]
- 49.Emanuel D, Peppard J, Stover D, Gold J, Armstrong D, Hammerling U. Rapid immunodiagnosis of cytomegalovirus pneumonia by bronchoalveolar lavage using human and murine monoclonal antibodies. Ann Intern Med. 1986;104:476–81. doi: 10.7326/0003-4819-104-4-476. [DOI] [PubMed] [Google Scholar]
- 50.Enright H, Haake R, Weisdorf D, Ramsay N, McGlave P, Kersey J, Thomas W, McKenzie D, Miller W. Cytomegalovirus pneumonia after bone marrow transplantation. Risk factors and response to therapy. Transplantation. 1993;55:1339–1346. doi: 10.1097/00007890-199306000-00024. [DOI] [PubMed] [Google Scholar]
- 51.Erice A, Crumpacker C, Britt W, Drew W L, Hillam R, Kao S, Kolberg J, Landry M, Lurain N, Manischewitz J, Nokta M, Spector S, Weinberg A, Yen-Lieberman B, Brambilla D, Reichelderfer P. Quantitation of cytomegalovirus load in blood fractions of patients with AIDS: a comparative study of different laboratory methods. 1996. p. 83. , abstr. 166. In 3rd Conference on Retroviruses and Opportunistic Infections. [Google Scholar]
- 52.Erice A, Holm M A, Sanjuan M V, Dunn D L, Gill P C, Balfour H H., Jr Evaluation of CMV-vue antigenemia assay for rapid detection of cytomegalovirus in mixed-leukocyte blood fractions. J Clin Microbiol. 1995;33:1014–1015. doi: 10.1128/jcm.33.4.1014-1015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falagas M, Snydman D R, Ruthazer R, Werner B G, Griffith J The Boston Center for Liver Transplantation CMVIG Study Group. Surveillance of blood, urine, and throat specimens are not valuable for predicting cytomegalovirus disease in liver transplant recipients. Clin Infect Dis. 1997;24:824–829. doi: 10.1093/clinids/24.5.824. [DOI] [PubMed] [Google Scholar]
- 54.Ferre F. Quantitative or semi-quantitative PCR: reality versus myth. PCR Methods Appl. 1992;2:1–9. doi: 10.1101/gr.2.1.1. [DOI] [PubMed] [Google Scholar]
- 55.Flood J, Drew W L, Miner R, Jekic-McMullen D, Shen L P, Kolberg J, Garvey J, Follansbee S, Posher M. Diagnosis of cytomegalovirus (CMV) polyradiculopathy and documentation of in vivo anti-CMV activity in cerobrospinal fluid using branched DNA amplification and antigen assays. J Infect Dis. 1997;176:348–352. doi: 10.1086/514051. [DOI] [PubMed] [Google Scholar]
- 56.Fox J C, Griffiths P D, Emery V C. Quantification of human cytomegalovirus DNA using the polymerase chain reaction. J Gen Virol. 1992;73:2405–2408. doi: 10.1099/0022-1317-73-9-2405. [DOI] [PubMed] [Google Scholar]
- 57.Fox J C, Kidd I M, Griffiths P D, Sweny P, Emery V C. Longitudinal analysis of cytomegalovirus load in renal transplant recipients using a quantitative polymerase chain reaction: correlation with disease. J Gen Virol. 1995;76:309–319. doi: 10.1099/0022-1317-76-2-309. [DOI] [PubMed] [Google Scholar]
- 58.Francisci D, Tosti A, Baldelli F, Stagni G, Pauluzzi S. The Pp65 antigenaemia test as a predictor of cytomegalovirus-induced end-organ disease in patients with AIDS. AIDS. 1997;11:1341–1345. doi: 10.1097/00002030-199711000-00007. [DOI] [PubMed] [Google Scholar]
- 59.Francisci D, Tosti A, Preziosi R, Baldelli F, Stagni G, Pauluzzi S. Role of antigenemia assay in the early diagnosis and prediction of human cytomegalovirus organ involvement in AIDS patients. Eur J Clin Microbiol Infect Dis. 1995;14:498–503. doi: 10.1007/BF02113427. [DOI] [PubMed] [Google Scholar]
- 60.Freymuth F, Gennetay E, Petitjean J, Eugene G, Hurault de Ligny B, Ryckelynck J P, Legoff C, Hazera P, Bazin C. Comparison of nested PCR for detection of DNA in plasma with pp65 leukocytic antigenemia procedure for diagnosis of human cytomegalovirus infection. J Clin Microbiol. 1994;32:1614–1618. doi: 10.1128/jcm.32.6.1614-1618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerard J, Leport C, Flandre P, Houhou N, Salmon-Ceron D, Pepin J M, Mandet C, Brun-Vezinet F, Vilde J L. Cytomegalovirus (CMV) viremia and the CD4+ lymphocyte count as predictors of CMV disease in patients infected with human immunodeficiency virus. Clin Infect Dis. 1997;24:836–840. doi: 10.1093/clinids/24.5.836. [DOI] [PubMed] [Google Scholar]
- 62.Gerna G, Baldanti F, Sarasini A, Furione M, Percivalle E, Revello M G, Zipeto D, Zella D. Effect of foscarnet induction treatment on quantitation of human cytomegalovirus (HCMV) DNA in peripheral blood polymorphonuclear leukocytes and aqueous humor of AIDS patients with HCMV retinitis. The Italian Foscarnet Study Group. Antimicrob Agents Chemother. 1994;38:38–44. doi: 10.1128/aac.38.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerna G, Furione M, Baldanti F, Percivalle E, Comoli P, Locatelli F. Quantitation of human cytomegalovirus DNA in bone marrow transplant recipients. Br J Haematol. 1995;91:674–683. doi: 10.1111/j.1365-2141.1995.tb05368.x. [DOI] [PubMed] [Google Scholar]
- 64.Gerna G, Furione M, Baldanti F, Sarasini A. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J Clin Microbiol. 1994;32:2709–2717. doi: 10.1128/jcm.32.11.2709-2717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerna G, Parea M, Percivalle E, Zipeto D, Silini E, Barbarini G, Milanesi G. Human cytomegalovirus viraemia in HIV-1-seropositive patients at various clinical stages of infection. AIDS. 1990;4:1027–1031. doi: 10.1097/00002030-199010000-00014. [DOI] [PubMed] [Google Scholar]
- 66.Gerna G, Revello M G, Percivalle E, Morini F. Comparison of different immunostaining techniques and monoclonal antibodies to the lower matrix phosphoprotein (pp65) for optimal quantitation of human cytomegalovirus antigenemia. J Clin Microbiol. 1992;30:1232–1237. doi: 10.1128/jcm.30.5.1232-1237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerna G, Revello M G, Percivalle E, Zavattoni M, Parea M, Battaglia M. Quantification of human cytomegalovirus viremia by using monoclonal antibodies to different viral proteins. J Clin Microbiol. 1990;28:2681–2688. doi: 10.1128/jcm.28.12.2681-2688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gerna G, Sarasini A, Baldanti F, Percivalle E, Zella D, Revello M G, Sacco O L, Moroni M, Darminio A, Antinori S, Milazzo F, Coen M, Cargnel A, Fasan A, Carosi G P, Castelli F, Tommasoni L, Alberici F, Viale P, Cadrobbi P, Crivellaro P, Carnevale G, Pan A, Suter F, Marchetti G, et al. Quantitative systemic and local evaluation of the antiviral effect of ganciclovir and foscarnet induction treatment on human cytomegalovirus gastrointestinal disease of patients with AIDS. Antiviral Res. 1997;34:39–50. doi: 10.1016/s0166-3542(96)01020-0. [DOI] [PubMed] [Google Scholar]
- 69.Gerna G, Zipeto D, Parea M, Revello M G, Silini E, Percivalle E, Zavattoni M, Grossi P, Milanesi G. Monitoring of human cytomegalovirus infections and ganciclovir treatment in heart transplant recipients by determination of viremia, antigenemia, and DNAemia. J Infect Dis. 1991;164:488–498. doi: 10.1093/infdis/164.3.488. [DOI] [PubMed] [Google Scholar]
- 70.Gerna G, Zipeto D, Percivalle E, Parea M, Revello M G, Maccario R, Peri G, Milanesi G. Human cytomegalovirus infection of the major leukocyte subpopulations and evidence for initial viral replication in polymorphonuclear leukocytes from viremic patients. J Infect Dis. 1992;166:1236–1244. doi: 10.1093/infdis/166.6.1236. [DOI] [PubMed] [Google Scholar]
- 71.Ghisetti V, Barbui A, Rocci M P, Donegani E, Bobbio M, Pucci A, Papandrea C, Pansini S, Zattera G, Mollo F, Di Summa M, Marchiaro G. Detection of human cytomegalovirus myocardial involvement by polymerase chain reaction during systemic infection and correlation with pp65 antigenemia and DNAemia in infected heart recipients. Transplantation. 1996;61:1072–1075. doi: 10.1097/00007890-199604150-00015. [DOI] [PubMed] [Google Scholar]
- 72.Gilbert C, Handfield J, Toma E, Lalonde R, Bergeron M G, Boivin G. Emergence and prevalence of cytomegalovirus UI97 mutations associated with ganciclovir resistance in AIDS patients. AIDS. 1998;12:125–129. doi: 10.1097/00002030-199802000-00001. [DOI] [PubMed] [Google Scholar]
- 73.Gilliland G, Perrin S, Blanchard K, Bunn H F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gondo H, Minematsu T, Harada M, Akashi K, Hayashi S, Taniguchi S, Yamasaki K, Shibuya T, Takamatsu Y, Teshima T, et al. Cytomegalovirus (CMV) antigenaemia for rapid diagnosis and monitoring of CMV-associated disease after bone marrow transplantation. Br J Haematol. 1994;86:130–137. doi: 10.1111/j.1365-2141.1994.tb03263.x. [DOI] [PubMed] [Google Scholar]
- 75.Goodrich J M, Bowden R A, Fisher L, Keller C, Schoch G, Meyers J D. Ganciclovir prophylaxis to prevent cytomegalovirus disease after allogeneic marrow transplant. Ann Intern Med. 1993;118:173–178. doi: 10.7326/0003-4819-118-3-199302010-00003. [DOI] [PubMed] [Google Scholar]
- 76.Goodrich J M, Mori M, Gleaves C A, Du Mond C, Cays M, Ebeling D F, Buhles W C, DeArmond B, Meyers J D. Early treatment with ganciclovir to prevent cytomegalovirus disease after allogeneic bone marrow transplant. N Engl J Med. 1991;325:1601–1607. doi: 10.1056/NEJM199112053252303. [DOI] [PubMed] [Google Scholar]
- 77.Gor D, Sabin C, Prentice H G, Yyas N, Man S, Griffith P D, Emery V C. Longitudinal fluctuation in cytomegalovirus load in bone marrow transplant patients: relationship between peak virus load, donor/recipient serostatus, acute GVHD and CMV disease. Bone Marrow Transplant. 1998;21:597–605. doi: 10.1038/sj.bmt.1701139. [DOI] [PubMed] [Google Scholar]
- 78.Grefte A, Harmsen M C, van der Giessen M, Knollema S, van Son W J, The T H. Presence of human cytomegalovirus (HCMV) immediate early mRNA but not ppUL83 (lower matrix protein pp65) mRNA in polymorphonuclear and mononuclear leukocytes during active HCMV infection. J Gen Virol. 1994;75:1989–1998. doi: 10.1099/0022-1317-75-8-1989. [DOI] [PubMed] [Google Scholar]
- 79.Grefte A, van der Giessen M, van Son W, The T H. Circulating cytomegalovirus (CMV)-infected endothelial cells in patients with an active CMV infection. J Infect Dis. 1993;167:270–277. doi: 10.1093/infdis/167.2.270. [DOI] [PubMed] [Google Scholar]
- 80.Grefte J M, van der Gun B T, Schmolke S, van der Giessen M, van Son W J, Plachter B, Jahn G, The T H. Cytomegalovirus antigenemia assay: identification of the viral antigen as the lower matrix protein pp65. J Infect Dis. 1992;166:683–684. doi: 10.1093/infdis/166.3.683. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 81.Grefte J M, van der Gun B T, Schmolke S, van der Giessen M, van Son W J, Plachter B, Jahn G, The T H. The lower matrix protein pp65 is the principal viral antigen present in peripheral blood leukocytes during an active cytomegalovirus infection. J Gen Virol. 1992;73:2923–2932. doi: 10.1099/0022-1317-73-11-2923. [DOI] [PubMed] [Google Scholar]
- 82.Grossi P, Kusne S, Rinaldo C, St George K, Magnone M, Rakela J, Fung J, Starzl T E. Guidance of ganciclovir therapy with pp65 antigenemia in cytomegalovirus-free recipients of livers from seropositive donors. Transplantation. 1996;61:1659–1660. doi: 10.1097/00007890-199606150-00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grossi P, Minoli L, Percivalle E, Irish W, Vigano M, Gerna G. Clinical and virological monitoring of human cytomegalovirus infection in 294 heart transplant recipients. Transplantation. 1995;59:847–851. [PubMed] [Google Scholar]
- 84.Grundy J E, Ehrnst A, Einsele H, Emery V C, Hebart H, Prentice H G, Ljungman P. A three-center European external quality control study of PCR for detection of cytomegalovirus DNA in blood. J Clin Microbiol. 1996;34:1166–1170. doi: 10.1128/jcm.34.5.1166-1170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Halwachs G, Zach R, Pogglitsch H, Holzer H, Tiran A, Iberer F, Wasler A, Tscheliessnigg H P, Lanzer G, Folsch B, et al. A rapid immunocytochemical assay for CMV detection in peripheral blood of organ-transplanted patients in clinical practice. Transplantation. 1993;56:338–342. doi: 10.1097/00007890-199308000-00016. [DOI] [PubMed] [Google Scholar]
- 86.Hansen K K, Ricksten A, Hofmann B, Norrild B, Olofsson S, Mathiesen L. Detection of cytomegalovirus DNA in serum correlates with clinical cytomegalovirus retinitis in AIDS. J Infect Dis. 1994;170:1271–1274. doi: 10.1093/infdis/170.5.1271. [DOI] [PubMed] [Google Scholar]
- 87.Hebart H, Müller C, Löffler J, Jahn G, Einsele H. Monitoring of CMV infection: a comparison of PCR from whole blood, plasma-PCR, pp65-antigenemia and virus culture in patients after bone marrow transplantation. Bone Marrow Transplant. 1996;17:861–868. [PubMed] [Google Scholar]
- 88.Higuchi R, Dollinger G, Walsh P S, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Bio/Technology. 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 89.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 90.Hiyoshi M, Tagawa S, Takubo T, Tanaka K, Nakao T, Higeno Y, Tamura K, Shimaoka M, Fujii A, Higashihata M, Yasui Y, Kim T, Hiraoka A, Tatsumi N. Evaluation of the Amplicor CMV test for direct detection of cytomegalovirus in plasma specimens. J Clin Microbiol. 1997;35:2692–2694. doi: 10.1128/jcm.35.10.2692-2694.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ho S K N, Lo C-Y, Cheng I K P, Chan T-M. Rapid cytomegalovirus pp65 antigenemia assay by direct erythrocyte lysis and immunofluorescence staining. J Clin Microbiol. 1998;36:638–640. doi: 10.1128/jcm.36.3.638-640.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Imbertmarcille B M, Cantarovich D, Ferreaubineau V, Richet B, Soulillou J P, Billaudel S. Usefulness of DNA viral load quantification for cytomegalovirus disease monitoring in renal and pancreas/renal transplant recipients. Transplantation. 1997;63:1476–1481. doi: 10.1097/00007890-199705270-00018. [DOI] [PubMed] [Google Scholar]
- 93.Imbertmarcille B M, Robillard N, Poirier A S, Costeburel M, Cantarovich D, Milpied N, Billaudel S. Development of a method for direct quantification of cytomegalovirus antigenemia by flow cytometry. J Clin Microbiol. 1997;35:2665–2669. doi: 10.1128/jcm.35.10.2665-2669.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jacobson M A, Zegans M, Pavan P R, JJ O D, Sattler F, Rao N, Owens S, Pollard R. Cytomegalovirus retinitis after initiation of highly active antiretroviral therapy. Lancet. 1997;349:1443–1445. doi: 10.1016/S0140-6736(96)11431-8. [DOI] [PubMed] [Google Scholar]
- 95.Jiwa N M, Mesker W E, Ploem-Zaaijer J J, van Dorp W, The T H, Raap A K. Quantification of low frequency white blood cells expressing human cytomegalovirus antigen by image cytometry. Cytometry. 1994;16:69–73. doi: 10.1002/cyto.990160110. [DOI] [PubMed] [Google Scholar]
- 96.Jiwa N M, van de Rijke F M, Mulder A, van der Bij W, The T H, Rothbarth P H, Velzing J, van der Ploeg M, Raap A K. An improved immunocytochemical method for the detection of human cytomegalovirus antigens in peripheral blood leucocytes. Histochemistry. 1989;91:345–349. doi: 10.1007/BF00493011. [DOI] [PubMed] [Google Scholar]
- 97.Kolberg, J. Personal communication.
- 98.Kolberg J, Miner R, Hoo B, Kelso R, Wiegand J, Jekicmcmullen D, Drew W L. Development of a branched DNA signal amplification assay to monitor patients on anti-CMV therapies. Biologicals. 1996;24:216. [Google Scholar]
- 99.Koskinen P K, Krogerus L A, Nieminen M S, Mattila S P, Hayry P J, Lautenschlager I T. Cytomegalovirus infection-associated generalized immune activation in heart allograft recipients: a study of cellular events in peripheral blood and endomyocardial biopsy specimens. Transplant Int. 1994;7:163–71. doi: 10.1007/BF00327082. [DOI] [PubMed] [Google Scholar]
- 100.Krajden M, Shankaran P, Bourke C, Lau W. Detection of cytomegalovirus in blood donors by PCR using the digene SHARP signal system assay: effects of sample preparation and detection methodology. J Clin Microbiol. 1996;34:29–33. doi: 10.1128/jcm.34.1.29-33.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kühn J E, Wendland T, Eggers H J, Lorentzen E, Wieland U, Eing B, Kiessling M, Gass P. Quantitation of human cytomegalovirus genomes in the brain of AIDS patients. J Med Virol. 1995;47:70–82. doi: 10.1002/jmv.1890470114. [DOI] [PubMed] [Google Scholar]
- 102.Kühn J E, Wendland T, Schäfer P, Möhring K, Wieland U, Elgas M, Eggers H J. Monitoring of renal allograft recipients by quantitation of human cytomegalovirus genomes in peripheral blood leukocytes. J Med Virol. 1994;44:398–405. doi: 10.1002/jmv.1890440416. [DOI] [PubMed] [Google Scholar]
- 103.Lalezari J P, Drew W L, Glutzer E, James C, Miner D, Flaherty J, Fisher P E, Cundy K, Hannigan J, Martin J C, et al. (S)-1-[3-hydroxy-2-(phosphonylmethoxy)propyl]cytosine (cidofovir): results of a phase I/II study of a novel antiviral nucleotide analogue. J Infect Dis. 1995;171:788–796. doi: 10.1093/infdis/171.4.788. [DOI] [PubMed] [Google Scholar]
- 104.Landry M L, Cohen S, Huber K. Comparison of EDTA and acid-citrate-dextrose collection tubes for detection of cytomegalovirus antigenemia and infectivity in leukocytes before and after storage. J Clin Microbiol. 1997;35:305–306. doi: 10.1128/jcm.35.1.305-306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Landry M L, Ferguson D. Comparison of quantitative cytomegalovirus antigenemia assay with culture methods and correlation with clinical disease. J Clin Microbiol. 1993;31:2851–2856. doi: 10.1128/jcm.31.11.2851-2856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lazzarotto T, Campisi B, Dalmonte P, Galli S, Spezzacatena P, Guglielmi P, Landini M P. A quantitative test (HCMV-Hybrid-Capture(TM)) to detect human cytomegalovirus DNA in the blood of immunocompromised patients compared with antigenemia and polymerase chain reaction. Microbiologica. 1996;19:193–201. [PubMed] [Google Scholar]
- 107.Reference deleted.
- 108.Li C R, Greenberg P D, Gilbert M J, Goodrich J M, Riddell S R. Recovery of HLA-restricted cytomegalovirus (CMV)-specific T-cell responses after allogeneic bone marrow transplant: correlation with CMV disease and effect of ganciclovir prophylaxis. Blood. 1994;83:1971–1979. [PubMed] [Google Scholar]
- 109.Limaye A, Bowden R A, Myerson D, Boeckh M. Cytomegalovirus disease before engraftment in marrow transplant patients. Clin Infect Dis. 1997;24:830–835. doi: 10.1093/clinids/24.5.830. [DOI] [PubMed] [Google Scholar]
- 110.Ljungman P, Griffith P. Definitions of cytomegalovirus infection and disease. In: Michelson S, Plotkin S A, editors. Multidisciplinary approach to understanding cytomegalovirus disease. Amsterdam, The Netherlands: Excerpta Medica; 1993. pp. 233–237. [Google Scholar]
- 111.Ljungman P, Lore K, Aschan J, Klaesson S, Lewensohn-Fuchs I, Lonnqvist B, Ringden O, Winiarski J, Ehrnst A. Use of a semi-quantitative PCR for cytomegalovirus DNA as a basis for pre-emptive antiviral therapy in allogeneic bone marrow transplant patients. Bone Marrow Transplant. 1996;17:583–587. [PubMed] [Google Scholar]
- 112.Locatelli F, Percivalle E, Comoli P, Maccario R, Zecca M, Giorgiani G, De Stefano P, Gerna G. Human cytomegalovirus (HCMV) infection in paediatric patients given allogeneic bone marrow transplantation: role of early antiviral treatment for HCMV antigenaemia on patients’ outcome. Br J Haematol. 1994;88:64–71. doi: 10.1111/j.1365-2141.1994.tb04978.x. [DOI] [PubMed] [Google Scholar]
- 113.Lu W, Andrieu J M. Early identification of human immunodeficiency virus-infected asymptomatic subjects susceptible to zidovudine by quantitative viral coculture and reverse transcription-linked polymerase chain reaction. J Infect Dis. 1993;167:1014–1020. doi: 10.1093/infdis/167.5.1014. [DOI] [PubMed] [Google Scholar]
- 114.Macartney M, Gane E J, Portmann B, Williams R. Comparison of a new quantitative cytomegalovirus DNA assay with other detection methods. Transplantation. 1997;63:1803–1807. doi: 10.1097/00007890-199706270-00017. [DOI] [PubMed] [Google Scholar]
- 115.Mazzulli T, Wood S, Chua R, Walmsley S. Evaluation of the Digene Hybrid Capture System for detection and quantitation of human cytomegalovirus viremia in human immunodeficiency virus-infected patients. J Clin Microbiol. 1996;34:2959–2962. doi: 10.1128/jcm.34.12.2959-2962.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mendez J C, Espy M J, Smith T F, Wilson J A, Paya C V. Evaluation of PCR primers for early diagnosis of cytomegalovirus infection following liver-transplantation. J Clin Microbiol. 1998;36:526–530. doi: 10.1128/jcm.36.2.526-530.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meyers J D, Ljungman P, Fisher L D. Cytomegalovirus excretion as a predictor of cytomegalovirus disease after marrow transplantation: importance of cytomegalovirus viremia. J Infect Dis. 1990;162:373–380. doi: 10.1093/infdis/162.2.373. [DOI] [PubMed] [Google Scholar]
- 118.Mitchell S. Diagnostic assays in cytomegalovirus retinitis. Br J Ophthalmol. 1996;80:195–196. doi: 10.1136/bjo.80.3.195. . (Editorial.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Murphy L D, Herzog C E, Rudick J B, Fojo A T, Bates S E. Use of the polymerase chain reaction in the quantitation of mdr-1 gene expression. Biochemistry. 1990;29:10351–10356. doi: 10.1021/bi00497a009. [DOI] [PubMed] [Google Scholar]
- 120.Mutimer D, Matyi-Toth A, Shaw J, Elias E, O’Donnell K, Stalhandske P. Patterns of viremia in liver transplant recipients with symptomatic cytomegalovirus infection. Transplantation. 1997;63:68–73. doi: 10.1097/00007890-199701150-00013. [DOI] [PubMed] [Google Scholar]
- 121.Myerow S, Lee M, Arruda J, Shen L P, Madej R, Collins M, Mlner R, Drew W L, Kolberg J. Presented at the 13th Annual Clinical Virology Symposium, Clearwater, Fla. 1997. Development of a branched DNA assay with improved sensitivity for monitoring CMV viral load in immunocompromised patients. [Google Scholar]
- 122.Niubo J, Perez J L, Martinez-Lacasa J T, Garcia A, Roca J, Fabregat J, Gil-Vernet S, Martin R. Association of quantitative cytomegalovirus antigenemia with symptomatic infection in solid organ transplant patients. Diagn Microbiol Infect Dis. 1996;24:19–24. doi: 10.1016/0732-8893(95)00248-0. [DOI] [PubMed] [Google Scholar]
- 123.Nolte F S, Emmens R K, Thurmond C, Mitchell P S, Pascuzzi C, Devine S M, Saral R, Wingard J R. Early detection of human cytomegalovirus viremia in bone marrow transplant recipients by DNA amplification. J Clin Microbiol. 1995;33:1263–1266. doi: 10.1128/jcm.33.5.1263-1266.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Noonan K E, Beck C, Holzmayer T A, Chin J E, Wunder J S, Andrulis I L, Gazdar A F, Willman C L, Griffith B, Von Hoff D D, et al. Quantitative analysis of MDR1 (multidrug resistance) gene expression in human tumors by polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:7160–7164. doi: 10.1073/pnas.87.18.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Patel R, Smith T F, Espy M, Portela D, Wiesner R H, Krom R A, Paya C V. A prospective comparison of molecular diagnostic techniques for the early detection of cytomegalovirus in liver transplant recipients. J Infect Dis. 1995;171:1010–1014. doi: 10.1093/infdis/171.4.1010. [DOI] [PubMed] [Google Scholar]
- 126.Patel R, Smith T F, Espy M, Wiesner R H, Krom R A, Portela D, Paya C V. Detection of cytomegalovirus DNA in sera of liver transplant recipients. J Clin Microbiol. 1994;32:1431–1434. doi: 10.1128/jcm.32.6.1431-1434.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Payan C, Veal N, Sarol L, Villarme M, Ngohou C, Riberi P, Francois S, Ifrah N, Loison J, Chennebault J M, Pichard E, Kouyoumdjian S, Lunel F. Human cytomegalovirus DNA kinetics using a novel HCMV DNA quantitative assay in white blood cells of immunocompromised patients under ganciclovir therapy. J Virol Methods. 1997;65:131–138. doi: 10.1016/s0166-0934(97)02180-0. [DOI] [PubMed] [Google Scholar]
- 128.Percivalle E, Revello M G, Vago L, Morini F, Gerna G. Circulating endothelial giant cells permissive for human cytomegalovirus (HCMV) are detected in disseminated HCMV infections with organ involvement. J Clin Invest. 1993;92:663–670. doi: 10.1172/JCI116635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Perez J L, De Ona M, Niubo J, Villar H, Melon S, Garcia A, Martin R. Comparison of several fixation methods for cytomegalovirus antigenemia assay. J Clin Microbiol. 1995;33:1646–1649. doi: 10.1128/jcm.33.6.1646-1649.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Podzamczer D, Ferrer E, Garcia A, Ramon J M, Niubo J, Santin M, Rufi G, Perez J L, Martin R, Gudiol F. pp65 antigenemia as a marker of future CMV disease and mortality in HIV-infected patients. Scand J Infect Dis. 1997;29:223–227. doi: 10.3109/00365549709019032. [DOI] [PubMed] [Google Scholar]
- 131.Poiriertoulemonde A S, Imbertmarcille B M, Ferreaubineau V, Besse B, Leroux M G, Cantarovich D, Billaudel S. Successful quantification of cytomegalovirus DNA by competitive PCR and detection with capillary electrophoresis. Mol Cell Probes. 1997;11:11–23. doi: 10.1006/mcpr.1996.0071. [DOI] [PubMed] [Google Scholar]
- 132.Rasmussen L, Morris S, Zipeto D, Fessel J, Wolitz R, Dowling A, Merigan T C. Quantitation of human cytomegalovirus DNA from peripheral blood cells of human immunodeficiency virus-infected patients could predict cytomegalovirus retinitis. J Infect Dis. 1995;171:177–182. doi: 10.1093/infdis/171.1.177. [DOI] [PubMed] [Google Scholar]
- 133.Rasmussen L, Zipeto D, Wolitz R A, Dowling A, Efron B, Merigan T C. Risk for retinitis in patients with AIDS can be assessed by quantitation of threshold levels of cytomegalovirus DNA burden in blood. J Infect Dis. 1997;176:1146–1155. doi: 10.1086/514106. [DOI] [PubMed] [Google Scholar]
- 134.Reed E C, Bowden R A, Dandliker P S, Lilleby K E, Meyers J D. Treatment of cytomegalovirus pneumonia with ganciclovir and intravenous cytomegalovirus immunoglobulin in patients with bone marrow transplants. Ann Intern Med. 1988;109:783–788. doi: 10.7326/0003-4819-109-10-783. [DOI] [PubMed] [Google Scholar]
- 135.Reid M C, Lachs M S, Feinstein A R. Use of methodological standards in diagnostic test research. Getting better but still not good. JAMA. 1995;274:645–651. [PubMed] [Google Scholar]
- 136.Reusser P, Attenhofer R, Hebart H, Helg C, Chapuis B, Einsele H. Cytomegalovirus-specific T-cell immunity in recipients of autologous peripheral blood stem cell or bone marrow transplant. Blood. 1997;89:3873–3879. [PubMed] [Google Scholar]
- 137.Reusser P, Riddell S R, Meyers J D, Greenberg P D. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood. 1991;78:1373–1380. [PubMed] [Google Scholar]
- 138.Revello M G, Percivalle E, Di Matteo A, Morini F, Gerna G. Nuclear expression of the lower matrix protein of human cytomegalovirus in peripheral blood leukocytes of immunocompromised viraemic patients. J Gen Virol. 1992;73:437–442. doi: 10.1099/0022-1317-73-2-437. [DOI] [PubMed] [Google Scholar]
- 139.Revello M G, Percivalle E, Sarasini A, Baldanti F, Furione M, Gerna G. Diagnosis of human cytomegalovirus infection of the nervous system by pp65 detection in polymorphonuclear leukocytes of cerebrospinal fluid from AIDS patients. J Infect Dis. 1994;170:1275–1279. doi: 10.1093/infdis/170.5.1275. [DOI] [PubMed] [Google Scholar]
- 140.Reynes J, Montes B, Atoui N, Segondy M. Significance of cytomegalovirus (CMV)-pp65 antigenemia in the diagnosis of CMV disease in human immunodeficiency virus-infected patients. J Med Virol. 1996;49:195–198. doi: 10.1002/(SICI)1096-9071(199607)49:3<195::AID-JMV6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 141.Rubin R H. Preemptive therapy in immunocompromised hosts. N Engl J Med. 1991;324:1057–1059. doi: 10.1056/NEJM199104113241509. [DOI] [PubMed] [Google Scholar]
- 142.Saltzman R L, Quirk M R, Jordan M C. Disseminated cytomegalovirus infection. Molecular analysis of virus and leukocyte interactions in viremia. J Clin Invest. 1988;81:75–81. doi: 10.1172/JCI113313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Saltzman R L, Quirk M R, Jordan M C. High levels of circulating cytomegalovirus DNA reflect visceral organ disease in viremic immunosuppressed patients other than marrow recipients. J Clin Invest. 1992;90:1832–1838. doi: 10.1172/JCI116059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Saltzman R L, Quirk M R, Mariash C N, Jordan M C. Quantitation of cytomegalovirus DNA by blot hybridization in blood leukocytes of viremic patients. J Virol Methods. 1990;30:67–77. doi: 10.1016/0166-0934(90)90044-g. [DOI] [PubMed] [Google Scholar]
- 145.Salzberger B, Bowden R A, Hackman R, Davis C, Boeckh M. Neutropenia in allogeneic marrow transplant recipients receiving ganciclovir for prevention of CMV disease: risk factors and outcome. Blood. 1997;90:2502–2508. [PubMed] [Google Scholar]
- 146.Salzberger B, Franzen C, Fatkenheuer G, Cornely O, Schwenk A, Rasokat H, Diehl V, Schrappe M. CMV-antigenemia in peripheral blood for the diagnosis of CMV disease in HIV-infected patients. J Acquired Immun Defic Syndr Hum Retrovirol. 1996;11:365–369. doi: 10.1097/00042560-199604010-00006. [DOI] [PubMed] [Google Scholar]
- 147.Salzberger B, Myerson D, Boeckh M. Circulating CMV-infected endothelial cells in marrow transplant patients with CMV disease and CMV infection. J Infect Dis. 1997;176:778–781. doi: 10.1086/517300. [DOI] [PubMed] [Google Scholar]
- 148.Schäfer P, Braun R W, Möhring K, Henco K, Kang J, Wendland T, Kühn J E. Quantitative determination of human cytomegalovirus target sequences in peripheral blood leukocytes by nested polymerase chain reaction and temperature gradient gel electrophoresis. J Gen Virol. 1993;74:2699–2707. doi: 10.1099/0022-1317-74-12-2699. [DOI] [PubMed] [Google Scholar]
- 149.Schäfer P, Tenschert W, Gutensohn K, Laufs R. Minimal effect of delayed sample processing on results of quantitative PCR for cytomegalovirus DNA in leukocytes compared to results of an antigenemia assay. J Clin Microbiol. 1997;35:741–744. doi: 10.1128/jcm.35.3.741-744.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schmidt G M, Horak D A, Niland J C, Duncan S R, Forman S J, Zaia J A. A randomized, controlled trial of prophylactic ganciclovir for cytomegalovirus pulmonary infection in recipients of allogeneic bone marrow transplants. The City of Hope-Stanford-Syntex CMV Study Group. N Engl J Med. 1991;324:1005–1111. doi: 10.1056/NEJM199104113241501. [DOI] [PubMed] [Google Scholar]
- 151.Schmidt N J. Cell culture procedures for diagnostic virology. In: Schmidt N J, Emmons R W, editors. Diagnostic procedures for viral, rickettsial and chlamydial infections. 5th ed. Washington, D.C: American Public Health Association, Inc.; 1989. pp. 78–79. [Google Scholar]
- 152.Shanley J D, Pesanti E L, Nugent K M. The pathogenesis of pneumonitis due to murine cytomegalovirus. J Infect Dis. 1982;146:388–396. doi: 10.1093/infdis/146.3.388. [DOI] [PubMed] [Google Scholar]
- 153.Shepp D H, Dandliker P S, de Miranda P, Burnette T C, Cederberg D M, Kirk L E, Meyers J D. Activity of 9-[2-hydroxy-1-(hydroxymethyl)ethoxymethyl]guanine in the treatment of cytomegalovirus pneumonia. Ann Intern Med. 1985;103:368–373. doi: 10.7326/0003-4819-103-3-368. [DOI] [PubMed] [Google Scholar]
- 154.Shepp D H, Match M E, Ashraf A B, Lipson S M, Millan C, Pergolizzi R. Cytomegalovirus glycoprotein B groups associated with retinitis in AIDS. J Infect Dis. 1996;174:184–187. doi: 10.1093/infdis/174.1.184. [DOI] [PubMed] [Google Scholar]
- 155.Shibata M, Terashima M, Kimura H, Kuzushima K, Yoshida J, Horibe K, Morishima T. Quantitation of cytomegalovirus DNA in lung tissue of bone marrow transplant recipients. Human Pathology. 1992;23:911–915. doi: 10.1016/0046-8177(92)90404-q. [DOI] [PubMed] [Google Scholar]
- 156.Shinkai M, Bozzette S A, Powderly W, Frame P, Spector S A. Utility of urine and leukocyte cultures and plasma DNA polymerase chain reaction for identification of AIDS patients at risk for developing human cytomegalovirus disease. J Infect Dis. 1997;175:302–308. doi: 10.1093/infdis/175.2.302. [DOI] [PubMed] [Google Scholar]
- 157.Shinkai M, Spector S A. Quantitation of human cytomegalovirus (HCMV) DNA in cerebrospinal fluid by competitive PCR in AIDS patients with different HCMV central nervous system diseases. Scand J Infect Dis. 1995;27:559–561. doi: 10.3109/00365549509047067. [DOI] [PubMed] [Google Scholar]
- 158.Slavin M A, Gleaves C A, Schoch H G, Bowden R A. Quantification of cytomegalovirus in bronchoalveolar lavage fluid after allogeneic marrow transplantation by centrifugation culture. J Clin Microbiol. 1992;30:2776–2779. doi: 10.1128/jcm.30.11.2776-2779.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Smith I L, Shinkai M, Freeman W R, Spector S A. Polyradiculopathy associated with ganciclovir-resistant cytomegalovirus in an AIDS patient: phenotypic and genotypic characterization of sequential virus isolates. J Infect Dis. 1996;173:1481–1484. doi: 10.1093/infdis/173.6.1481. [DOI] [PubMed] [Google Scholar]
- 160.Spector S A, McKinley G F, Lalezari J P, Samo T, Andruczk R, Follansbee S, Sparti P D, Havlir D V, Simpson G, Buhles W, Wong R, Stempien M. Oral ganciclovir for the prevention of cytomegalovirus disease in persons with AIDS. Roche Cooperative Oral Ganciclovir Study Group. N Engl J Med. 1996;334:1491–1497. doi: 10.1056/NEJM199606063342302. [DOI] [PubMed] [Google Scholar]
- 161.Spector S A, Merrill R, Wolf D, Dankner W M. Detection of human cytomegalovirus in plasma of AIDS patients during acute visceral disease by DNA amplification. J Clin Microbiol. 1992;30:2359–2365. doi: 10.1128/jcm.30.9.2359-2365.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Spector S A, Wong R, Hsia K, Pilcher M, Stempien M J. Plasma cytomegalovirus (CMV) DNA load predicts CMV disease and survival in AIDS patients. J Clin Invest. 1998;101:497–502. doi: 10.1172/JCI1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.St. George K, Rinaldo C R. Comparison of commercially available antibody reagents for the cytomegalovirus pp65 antigenemia assay. Clin Diagn Virol. 1997;7:147–152. doi: 10.1016/s0928-0197(96)00264-4. [DOI] [PubMed] [Google Scholar]
- 164.Storch G A, Buller R S, Bailey T C, Ettinger N A, Langlois T, Gaudreault-Keener M, Welby P L. Comparison of PCR and pp65 antigenemia assay with quantitative shell vial culture for detection of cytomegalovirus in blood leukocytes from solid-organ transplant recipients. J Clin Microbiol. 1994;32:997–1003. doi: 10.1128/jcm.32.4.997-1003.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Storch G A, Ettinger N A, Ockner D, Wick M R, Gaudreault-Keener M, Rossiter J, Trulock E P, Cooper J D. Quantitative cultures of the cell fraction and supernatant of bronchoalveolar lavage fluid for the diagnosis of cytomegalovirus pneumonitis in lung transplant recipients. J Infect Dis. 1993;168:1502–6. doi: 10.1093/infdis/168.6.1502. [DOI] [PubMed] [Google Scholar]
- 166.Storch G A, Gaudreault-Keener M, Welby P C. Comparison of heparin and EDTA transport tubes for detection of cytomegalovirus in leukocytes by shell vial assay, pp65 antigenemia assay, and PCR. J Clin Microbiol. 1994;32:2581–2583. doi: 10.1128/jcm.32.10.2581-2583.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tevere, V. Personal communication.
- 168.The T H, van den Berg A P, Harmsen M C, van der Bij W, van Son W J. The cytomegalovirus antigenemia assay: a plea for standardization. Scand J Infect Dis Suppl. 1995;99:25–29. [PubMed] [Google Scholar]
- 169.The T H, van der Ploeg M, van den Berg A P, Vlieger A M, van der Giessen M, van Son W J. Direct detection of cytomegalovirus in peripheral blood leukocytes—a review of the antigenemia assay and polymerase chain reaction. Transplantation. 1992;54:193–198. doi: 10.1097/00007890-199208000-00001. [DOI] [PubMed] [Google Scholar]
- 170.Toyoda M, Carlos J B, Galera O A, Galfayan K, Zhang X M, Sun Z L, Czer L S C, Jordan S C. Correlation of cytomegalovirus DNA levels with response to antiviral therapy in cardiac and renal allograft recipients. Transplantation. 1997;63:957–963. doi: 10.1097/00007890-199704150-00009. [DOI] [PubMed] [Google Scholar]
- 171.Van den Berg, A. Personal communication.
- 172.van den Berg A P, Klompmaker I J, Haagsma E B, Scholten-Sampson A, Bijleveld C M, Schirm J, van der Giessen M, Slooff M J, The T H. Antigenemia in the diagnosis and monitoring of active cytomegalovirus infection after liver transplantation. J Infect Dis. 1991;164:265–270. doi: 10.1093/infdis/164.2.265. [DOI] [PubMed] [Google Scholar]
- 173.van den Berg A P, Tegzess A M, Scholten-Sampson A, Schirm J, van der Giessen M, The T H, van Son W J. Monitoring antigenemia is useful in guiding treatment of severe cytomegalovirus disease after organ transplantation. Transplant Int. 1992;5:101–106. doi: 10.1007/BF00339224. [DOI] [PubMed] [Google Scholar]
- 174.van den Berg A P, van der Bij W, van Son W J, Anema J, van der Giessen M, Schirm J, Tegzess A M, The T H. Cytomegalovirus antigenemia as a useful marker of symptomatic cytomegalovirus infection after renal transplantation—a report of 130 consecutive patients. Transplantation. 1989;48:991–995. doi: 10.1097/00007890-198912000-00019. [DOI] [PubMed] [Google Scholar]
- 175.van den Berg A P, van Son W J, Haagsma E B, Klompmaker I J, Tegzess A M, Schirm J, Dijkstra G, van der Giessen M, Slooff M J, The T H. Prediction of recurrent cytomegalovirus disease after treatment with ganciclovir in solid-organ transplant recipients. Transplantation. 1993;55:847–851. doi: 10.1097/00007890-199304000-00031. [DOI] [PubMed] [Google Scholar]
- 176.van der Bij W, Schirm J, Torensma R, van Son W J, Tegzess A M, The T H. Comparison between viremia and antigenemia for detection of cytomegalovirus in blood. J Clin Microbiol. 1988;26:2531–2535. doi: 10.1128/jcm.26.12.2531-2535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.van der Bij W, Torensma R, van Son W J, Anema J, Schirm J, Tegzess A M, The T H. Rapid immunodiagnosis of active cytomegalovirus infection by monoclonal antibody staining of blood leucocytes. J Med Virol. 1988;25:179–188. doi: 10.1002/jmv.1890250208. [DOI] [PubMed] [Google Scholar]
- 178.Veal N, Payan C, Fray D, Sarol L, Blanchet O, Kouyoumdjian S, Lunel F. Novel DNA assay for cytomegalovirus detection: comparison with conventional culture and pp65 antigenemia assay. J Clin Microbiol. 1996;34:3097–3100. doi: 10.1128/jcm.34.12.3097-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Vlieger A M, Boland G J, Jiwa N M, de Weger R A, Willemze R, de Gast G C, Falkenburg J H. Cytomegalovirus antigenemia assay or PCR can be used to monitor ganciclovir treatment in bone marrow transplant recipients. Bone Marrow Transplant. 1992;9:247–253. [PubMed] [Google Scholar]
- 180.Vogel J U, Cinatl J, Lux A, Weber B, Driesel A J, Doerr H W. New PCR assay for rapid and quantitative detection of human cytomegalovirus in cerebrospinal fluid. J Clin Microbiol. 1996;34:482–483. doi: 10.1128/jcm.34.2.482-483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wade J C, McGuffin R W, Springmeyer S C, Newton B, Singer J W, Meyers J D. Treatment of cytomegaloviral pneumonia with high-dose acyclovir and human leukocyte interferon. J Infect Dis. 1983;148:557–562. doi: 10.1093/infdis/148.3.557. [DOI] [PubMed] [Google Scholar]
- 182.Wentworth B B, French L. Plaque assay of cytomegalovirus strains of human origin. Proc Soc Exp Biol Med. 1970;135:253–258. doi: 10.3181/00379727-135-35031. [DOI] [PubMed] [Google Scholar]
- 183.Wetherill P E, Landry M L, Alcabes P, Friedland G. Use of a quantitative cytomegalovirus (CMV) antigenemia test in evaluating HIV+ patients with and without CMV disease. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;12:33–37. doi: 10.1097/00042560-199605010-00005. [DOI] [PubMed] [Google Scholar]
- 184.Zaia J A, Gallez-Hawkins G M, Tegtmeier B R, ter Veer A, Li X, Niland J C, Forman S J. Late cytomegalovirus disease in marrow transplantation is predicted by virus load in plasma. J Infect Dis. 1997;176:782–785. doi: 10.1086/517301. [DOI] [PubMed] [Google Scholar]
- 185.Zipeto D, Baldanti F, Zella D, Furione M, Cavicchini A, Milanesi G, Gerna G. Quantification of human cytomegalovirus DNA in peripheral blood polymorphonuclear leukocytes of immunocompromised patients by the polymerase chain reaction. J Virol Methods. 1993;44:45–55. doi: 10.1016/0166-0934(93)90006-d. [DOI] [PubMed] [Google Scholar]
- 186.Zipeto D, Morris S, Hong C, Dowling A, Wolitz R, Merigan T C, Rasmussen L. Human cytomegalovirus (CMV) DNA in plasma reflects the quantity of CMV DNA present in leukocytes. J Clin Microbiol. 1995;33:2607–2611. doi: 10.1128/jcm.33.10.2607-2611.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zipeto D, Revello M G, Silini E, Parea M, Percivalle E, Zavattoni M, Milanesi G, Gerna G. Development and clinical significance of a diagnostic assay based on the polymerase chain reaction for detection of human cytomegalovirus DNA in blood samples from immunocompromised patients. J Clin Microbiol. 1992;30:527–530. doi: 10.1128/jcm.30.2.527-530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zurlo J J, O’Neill D, Polis M A, Manischewitz J, Yarchoan R, Baseler M, Lane H C, Masur H. Lack of clinical utility of cytomegalovirus blood and urine cultures in patients with HIV infection. Ann Intern Med. 1993;118:12–17. doi: 10.7326/0003-4819-118-1-199301010-00003. [DOI] [PubMed] [Google Scholar]


