Abstract
Cloud-based bioinformatic platforms address the fundamental demands of creating a flexible scientific environment, facilitating data processing and general accessibility independent of a countries’ affluence. These platforms have a multitude of advantages as demonstrated by omics technologies, helping to support both government and scientific mandates of a more open environment.
Keywords: Systems biology, multi-omics, metabolomics, XCMS Online/METLIN, open science, big data
Moving to the Cloud
We are increasingly surrounded by cloud computing, whether it is private or scientific in nature; thousands of computers and servers are used to process and handle our information [1]. The benefits of cloud-based computing over downloadable desktop-based software include straightforward data sharing, transfer, management and archiving, standardized data formats, distributed data processing, and global access that is independent of local high-end hardware [2, 3]. Moreover, the cloud provides a universal platform for data analysis across communities and locations, enabling outside researchers to perform modified analyses and ask new questions utilizing the same datasets. These cloud-based platforms also make computational power available to academic institutions with limited resources, which otherwise may not be able to afford the infrastructure required to analyze complex datasets. This was recently demonstrated by the establishment of a low-cost and scalable experimentation platform to address professional training needs in basic biology [4]. Even high-end labs are starting to purchase time from cloud providers. Furthermore, these advantages coincide with recent government mandates seeking to make science more accessible and transparent [5].
The advantages of cloud computing are numerous, though it is not without its challenges; it requires reliable and fast internet connections, creates a time lag between uploading and processing data, lacks flexibility and control over resources, and requires high-level security to ensure that proprietary information is safe [6, 7]. Particularly, in environmental, microbiological, medical, or toxicological applications, data protection is an ongoing and controversial issue. The user does not know who might gain access to data and, once uploaded, cannot control its protection nor can track its usage. Moreover, missing availability of the source code can make cloud services a black box. Yet, even with these challenges, cloud computing has recently emerged as a cost-effective and more ‘open’ way to perform research in the life sciences. In fact, biology and cloud computing have become irrevocably intertwined, particularly for the omic disciplines, dealing with data sets reaching the terabyte range [2, 7] and driven by the increasing performance of high throughput methods, such as next generation sequencing or high resolution mass spectrometry-based metabolomics and proteomics [8]. Responding to the massive amounts of data generated, NIH initiatives, including ‘Big Data to Knowledge,’ and bioinformatics hubs are now exploring cloud-computing options. Here, we describe cloud-based computing in metabolomics, which is currently facing the key challenge of integrating within the broader context of systems biology (Figure 1).
Figure 1. Cloud-based computing provides an interconnectedness with infrastructure, data sharing and the ability to integrate multiple resources.
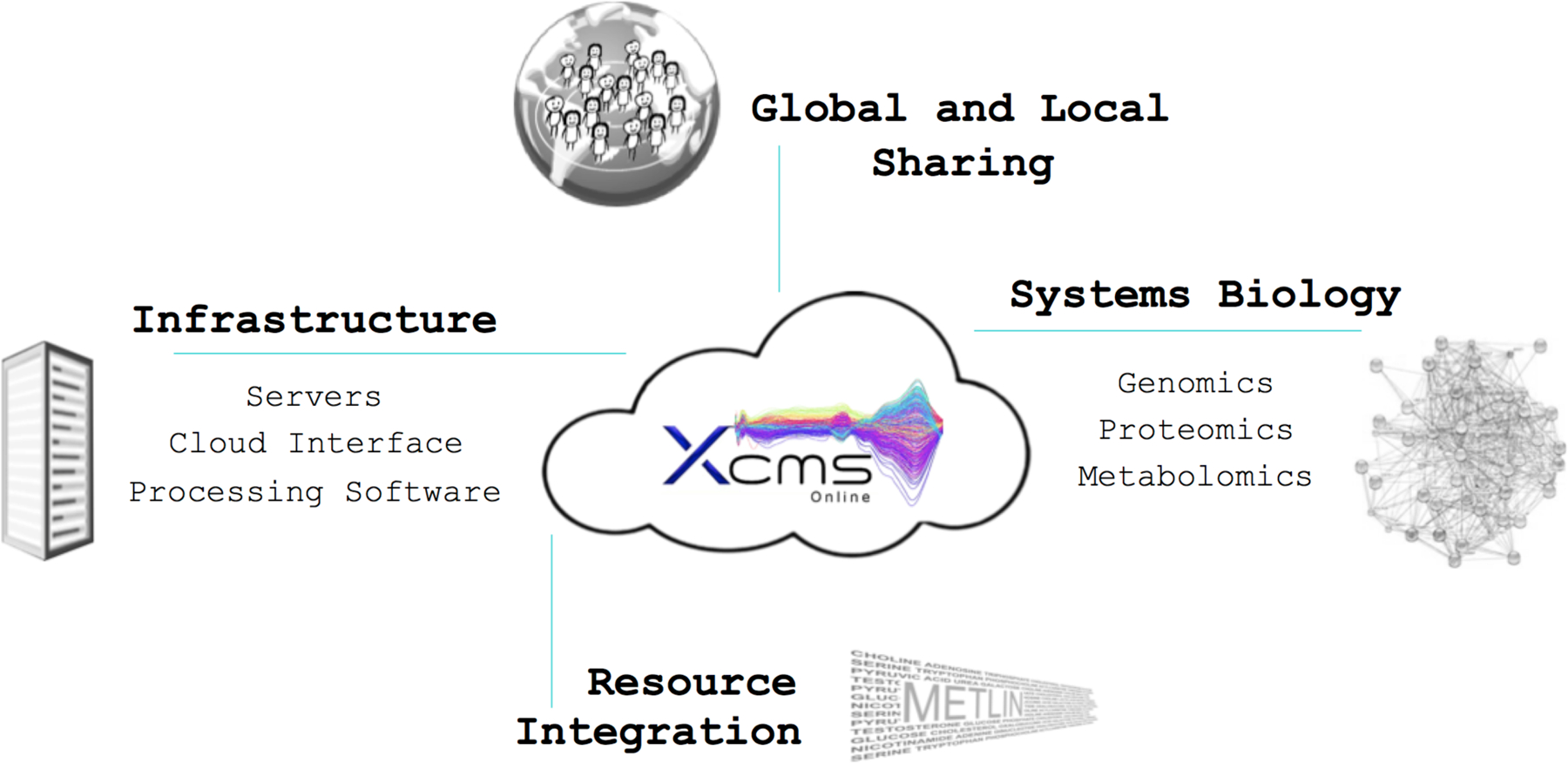
The multi-omics XCMS Online platform with over 12,500 registered users represents a freely available, cloud-based resource offering data processing, archiving and sharing, easy-to-use statistical tools, intuitive visualization, pathway analysis and, in combination with the METLIN database, metabolite identification.
Metabolomics - Leading from Behind
Metabolomics, a multi-disciplinary technology and the newest and fastest growing omic domain, aims to decipher the complexity in a given biological system. Despite overcoming its early stages of development and establishing community standards in the last decade [9], the field of metabolomics may still be regarded as an emerging scientific discipline. Given the scope of these efforts, significant time and funding resources are required to establish a comprehensive metabolomics lab, making the technology inaccessible to a large share of biological scientists without the required instrumentation or substantial bioinformatic support. To address the informatic challenge, XCMS Online [10] and the associated METLIN metabolite database [11] represent a model of how community oriented cloud-computing can be implemented on a global scale. This free, cloud-based platform for untargeted metabolomics data provides users with data processing tools, data streaming, statistical analysis, metabolite identification via high-resolution MS/MS library searching, pathway-based multi-omic analysis, and downloadable results for any alternative analyses. Originally developed as a command-line driven software (XCMS)[12], the online version neither requires programming skills nor expensive hardware. The challenges of cloud computing can be overcome by streamlining data uploads, using redundant grid-based processing and redundant secure data storage, and providing a user-friendly platform with enterprise-grade security that is compliant with established regulations [6]. Since data transfer still represents a major bottleneck in the metabolomics arena, we recently launched XCMS Stream [13]. This data streaming platform holds the potential to reduce data analysis time from days to minutes.
The most significant value of the universal access options provided by cloud computing is the fast and simple sharing of resources (Figure 2a). In the case of XCMS Online, uploaded liquid chromatography-mass spectrometry (LC-MS) raw data files, as well as the results of any specific data processing job including the experimental parameters and settings, can be shared either privately with any collaborator or publicly with the entire community. Both options require only a few mouse clicks. Thus, this type of cloud-based platform also increases the field’s ability to compare results with those of publicly shared jobs. Alongside XCMS Online, similar initiatives underpin the expanding role of cloud-based workflows in metabolomics and beyond. For example, MetaboAnalyst is a valuable collection of online tools for data analysis and interpretation, [14] and Galaxy, a platform with roots in genomics and transcriptomics, is currently diversifying into metabolomics [15]. The recently funded $8M Horizon2020 project PhenoMeNal aims to build a community-supported e-infrastructure based on cloud resources for medical metabolomics applications (Edmunds S. https://blogs.biomedcentral.com/gigablog/2016/07/19/guest-posting-building-phenomenal-metabolomics-e-infrastructure). It will support data processing and analysis for molecular phenotype data and leverage existing cloud infrastructures and data repositories. Although this platform is so far dedicated to the European biomedical community, these efforts again confirm the potential of cloud computing in the omic area in general and the primary role of metabolomics in particular.
Figure 2. XCMS Online exemplifies the value of cloud-based computing.
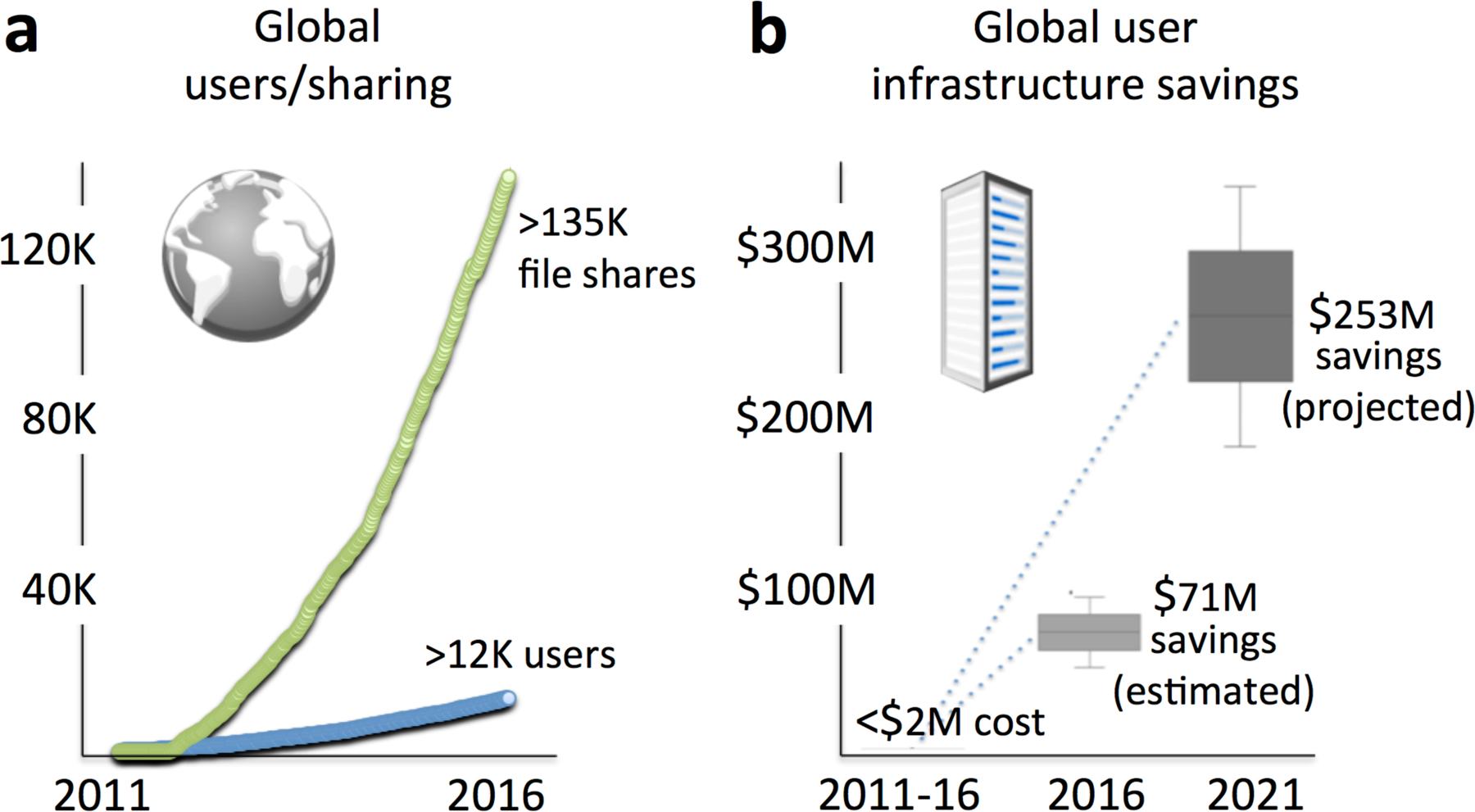
This resource has demonstrated an ability to analyze biological systems using multi-omic technologies, with its growing usage a result of (a) ease of data sharing and (b) significant cost savings. A conservative estimated cost savings (b) to the community is about $70 million, as calculated based on the average cost of comparable downloadable software from different vendors over the last five years and the number of XCMS Online users (assuming three users per purchased program). The increasing importance of data sharing (a) is demonstrated by the ratio of registered users to shared data sets.
Common alternatives to cloud computing are downloadable, vendor-specific software tools to process and evaluate metabolomic data sets. While these software solutions offer advantages, they are typically very costly and sometimes not compatible with data formats acquired on competitor instruments. With an XCMS Online user population of over 12,500 scientists in 120+ countries, we estimate the cost savings of the scientific community to be over $70 million USD since its launch in 2011 (Figure 2b), while the cost for development and maintenance of XCMS Online has been below $2 million USD, resulting in a community return on investment of approximately 97%. These savings are expected to increase to over $250 million USD by 2021 by extrapolating across the current user base. These calculations are based on the average cost from four different vendors over the course of the last five years and the assumption that there would be one license for every three XCMS Online users. Importantly, these estimates are conservative, as additional costs for software updates or the need for the acquisition of more than one program were not taken into account.
It is worth noting that in an alternative scenario, in which a lab invests in trained informatics personnel applying open-source software or creating in-house scripts, the anticipated budget far exceeds that of purchased software solutions. Additionally, in-house software further increases the cost to the field because many academic developers release software with little to no documentation. Besides the financial burden, exporting and sharing data and results with colleagues and the public requires additional efforts and is less convenient or even impossible when using vendor programs or in-house scripts. Often, side-step analyses and crucial parameters can be hidden or forgotten, causing issues for replicating published findings. Hence, sharing standardized datasets including underlying raw data utilizing cloud-based solutions clearly supports the increasing demand for reproducibility of scientific data and the request of publishers and grant bodies to make data and software publically accessible.
The Path Upward
Overall, the value of cloud-based bioinformatic platforms goes beyond convenience and addresses the fundamental demands of putting analytical raw data into a biological context and creating a more open scientific environment for rapidly translating science. As stated by Schadt, Linderman, Sorenson, Lee and Nolan [2], the future success of cloud computing in the biomedical and life sciences will depend on the ability of investigators to properly interpret large-scale, multi-dimensional data sets that are generated by high-throughput technologies. From this perspective, freely accessible, cloud-based platforms are invaluable to facilitate appropriate data processing and handling. Making more metabolic data accessible on the cloud may trigger the integration with other high content datasets such as those existing in genomics and proteomics. One could image the creation of software to promote the integration and analysis of these datasets to generate more in depth insights. Having a central “hub” for these large data-sets and a way to integrate the information contained would be an invaluable asset to the scientific community.
The success of metabolomic platforms will likely evolve and expand into other fields, such as environmental and exposure sciences, microbial research, clinical diagnostics, pharmacokinetics, molecular modeling, education, and the social sciences, to name only a few. However, challenges in data protection will certainly remain to some extent. Because cloud-based tools are inherently easier to access, newly launched platforms should also stimulate synergies through smartly connecting resources. One particularly interesting possibility is the adoption of these technologies in less-affluent countries, where the necessary resources are unavailable. Given these unique advantages, it is inevitable that cloud-based computing will become an integral part of scientific communities.
Acknowledgements
The authors would like to thank their colleagues from the Siuzdak lab for valuable discussions and acknowledge the following for funding assistance: The Austrian Science Fund (FWF): J-3808 (Erwin Schrödinger Fellowship awarded to B.W.), Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory for the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under contract number DE-AC02–05CH11231, and the National Institutes of Health (NIH) grants R01 GMH4368 and PO1 A1043376–02S1.
References
- 1.Marx V. (2013) Biology: The big challenges of big data. Nature 498 (7453), 255–260. [DOI] [PubMed] [Google Scholar]
- 2.Schadt EE. et al. (2010) Computational solutions to large-scale data management and analysis. Nat Rev Genet 11 (9), 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buyya R. et al. (2009) Cloud computing and emerging IT platforms: Vision, hype, and reality for delivering computing as the 5th utility. Future Generation Computer Systems 25 (6), 599–616. [Google Scholar]
- 4.Hossain Z. et al. (2016) Interactive and scalable biology cloud experimentation for scientific inquiry and education. Nat Biotech 34 (12), 1293–1298. [DOI] [PubMed] [Google Scholar]
- 5.Levin N. and Leonelli S. (2016) How Does One “Open” Science? Questions of Value in Biological Research. Science, Technology and Human Values in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datta S. et al. (2016) Secure cloud computing for genomic data. Nat Biotech 34 (6), 588–591. [DOI] [PubMed] [Google Scholar]
- 7.Rinehart D. et al. (2014) Metabolomic data streaming for biology-dependent data acquisition. Nat Biotech 32 (6), 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson CH. et al. (2016) Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 17 (7), 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiehn O. et al. (2007) The metabolomics standards initiative (MSI). Metabolomics 3 (3), 175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tautenhahn R. et al. (2012) XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Analytical Chemistry 84 (11), 5035–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tautenhahn R. et al. (2012) An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotech 30 (9), 826–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith CA. et al. (2006) XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Analytical Chemistry 78 (3), 779–787. [DOI] [PubMed] [Google Scholar]
- 13.Montenegro-Burke JM. et al. (in revision) Data Streaming for Metabolomics: Accelerating Data Processing and Analysis from Days to Minutes. Analytical Chemistry [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia J. et al. (2015) MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Research 43 (W1), W251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boekel J. et al. (2015) Multi-omic data analysis using Galaxy. Nat Biotech 33 (2), 137–139. [DOI] [PubMed] [Google Scholar]


