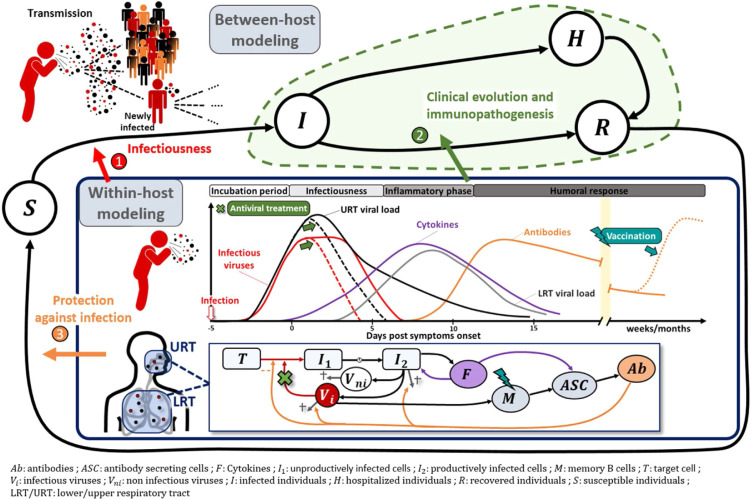
Viral dynamics is a field of research that develops mathematical models based on biological knowledge to characterise the evolution of virological and immunological markers during infection. Analogous to epidemiological models, these models view the human body as a series of compartments composed of different cell or virus types that evolve over time (Fig. 1 , panel “within-host modelling”).
Fig. 1.
Within-host models during the dynamics of an acute viral infection.
Models are used to understand the evolution of the viro-immunological response during an infection. As such, they can be used to disentangle the factors associated with (1) enhanced infectiousness during acute infection (2) progression towards a severe disease, and how to prevent it via pharmacological interventions (3) protection against infection via previous infection or vaccination.
In its most basic formulation, the model assumes three compartments: free virus particles infect target cells, which become productively infected cells, that are then gradually lost as a result of viral cytopathicity and/or immune response. Analogous to epidemiological models, one can define a “within-host” basic reproductive ratio (R0) that quantifies how many infected cells arise from a single infected cell at the beginning of infection and depends on the balance between the processes of virus production and elimination. When R0 > 1, the virus grows exponentially until most target cells are depleted. Thereafter, the virus declines rapidly at a rate that reflects the loss of infected cells that can either be eliminated or cured of their viral content. Over the past 30 years, many models have been developed that go beyond this simple “target-cell limited model” and incorporate more biological processes: the intracellular life cycle, the role of the innate and adaptive immune response, virus mutation, or pharmacological interventions [1] for a comprehensive set of reviews for various infectious diseases. In the context of SARS-CoV-2 infection, viral dynamic models are used for three main purposes: (i) characterising the association of viral load with transmission and with clinical evolution, (ii) optimising treatment and vaccine strategies, and (iii) understanding the viral-host interactions.
From a public health perspective, these models can quantify the parameters that determine the kinetics of viral load and its distribution in the population. An important observation that has been made is that the peak of viral load coincides with the symptom onset, at least in the pre-vaccination and Omicron era [2], [3], [4]. This means that patients may shed large amounts of virus in the few days that precede their symptom onset, and underscores the problem of infections occurring during the pre-symptomatic phase of the disease. When longitudinal studies are nested within epidemiological studies, longitudinal profiles of individuals can be used to assess the heterogeneity of viral dynamics, and evaluate the impact of variants of concern or vaccination [5], [6], or suggest strategies to improve testing strategies [3], [5], [7]. If both the index and its high risk contacts are followed prospectively [8], modelling can also be used to reconstruct viral load at the time of contact, and explore the accrued risk of transmission caused by increased levels of viral shedding and temporal changes in infectiousness [9] (Fig. 1 arrow 1).
In the past, these models have also been widely used to optimise antiviral therapies (Fig. 1, arrow 2). By fitting frequent nasopharyngeal viral load data, it was found that R0 was close to 10, albeit with variability across studies [10], [11]. As the within-host equivalent of the herd immunity threshold, antiviral treatment must be above 1-1/R0 = 90% to dramatically reduce viral replication. Given the in vitro estimated drug EC50 (half maximal effective concentration) and their pharmacological properties, we have shown that repurposed antiviral agents (e.g., hydroxychloroquine, lopinavir, remdesivir, favipiravir) are unlikely to achieve this pharmacodynamic target, anticipating the negative outcome of all trials evaluating these molecules [10]. In the new era of monoclonal antibodies (mAbs), we still lack exhaustive analyses of viral kinetics during treatment. Analyses in the macaque model of COVA1-18 suggest that the efficacy of mAbs in blocking de novo cell infection in the upper and lower respiratory tract may exceed 95% [12], which is consistent with the impressive results of several mAbs in phase 3 clinical trials. Modelling analyses are now urgently needed to explore the causal pathways between antiviral effect, viral dynamics and clinical benefit. In particular, it is still unclear whether monoclonal antibodies might also be beneficial when administered at a later stage of infection. In an analysis of hospitalised patients from the French COVID-19 cohort, we showed that viral dynamics after admission was an independent factor associated with mortality [4]. With a treatment reducing viral production by 90%, the time to viral clearance can be shortened by more than 2 days on average, which could lead to a reduction in mortality of approximately 25% in high-risk patients (age ≥ 65 years with risk factors). Interestingly, the results of the Recovery trial supported these predictions; REGN-COV reduced mortality by 30% in patients who were seronegative at inclusion, a marker strongly associated with high viral load. Studies such as the randomised clinical trial Discovery (NCT 04315948) are now ongoing to examine the extent to which antiviral treatment may be relevant in hospitalised patients who had a high viral load at inclusion (Fig. 1, arrow 2).
Depending on the data collected, models may also provide insights into the role of the immune response on viral clearance. Several models have challenged the hypothesis of peak viral load being primarily caused by cell depletion, and have proposed models in which the innate [3] or adaptive immune response [4], [11] plays a key role in viral clearance. These models can also be used to reproduce patterns observed in some patients, such as bimodal peak in viral load. However, these models are often limited by the difficulty of collecting frequent viral load and immunological data, which poses problems in parameter identifiability and leads to large uncertainty in parameter estimates.
For this reason, characterisation of the adaptive immune response within the host is often considered a separate problem. First, integrative analyses of multiple immunological markers can help understand the hyper inflammation and cytokine storm associated with clinical complications [13]. For instance, markers such as CD177 have been shown to be associated with disease progression and predict the course of patient hospitalisation trajectory [14]. Then, modelling the cellular and molecular determinants of the duration of antibody response is key to realistic prediction of epidemic dynamics, taking into account waning immunity (Fig. 1 arrow 3). Hence, the within-host modelling of Ebola vaccine response based on early phase 1 trial data has revealed the long-term duration of the response [15]. In what follows, we will focus on vaccination, but similar considerations can be applied to natural infection. Of note, natural immunity confers at least similar [16] or longer-lasting and stronger protection against infection [17]. Binding and neutralising antibodies are clearly associated with protection against infection [18], [19]. This means that the effect of vaccines on disease transmission and/or severity could be captured by these immunological markers, called correlates of protection (CoP). Defining such markers is critical to accelerate the development of new vaccines and vaccination strategies [20]. The use of within-host mathematical models can help in this investigation to define mechanistic CoP, which is directly related to the protective mechanism triggered by the vaccine, and thus causally responsible [21]. Therefore, within-host models help quantifying the effect of vaccines on the infection of new cells by SARS-CoV-2 through neutralisation [22]. Recent studies have also shown that neutralisation measured in vitro or in vivo is a reasonable mechanistic correlate of protection [19], [23]. Finding good correlates of protection is particularly challenging in the context of an ever-evolving virus, where variants of concern (VoC) frequently emerge. Indeed, for each VoC, vaccine protection against transmission and hospitalisation needs to be rapidly assessed [24]. In addition, multiple mechanisms, and thus multiple biomarkers, could contribute to the protective efficacy of a vaccine. In the case of SARS-CoV-2, the role of T-cell responses has also been highlighted [25]. Therefore, expanding the framework for within-host modelling to integrate high-dimensional data generated in current immunological studies is promising research.
Conflicts of interest
The authors declare that they have no conflict of interest.
Acknowledgement
This work has received funding from the French Agency for Research on AIDS and Emerging Infectious Diseases via the EMERGEN project (ANRS0151).
References
- 1.Perelson A.S., Ribeiro R.M. Introduction to modeling viral infections and immunity. Immunol Rev. 2018;285:5–8. doi: 10.1111/imr.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He X., Lau E.H., Wu P., Deng X., Wang J., Hao X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 3.Ke R., Zitzmann C., Ho D.D., Ribeiro R.M., Perelson A.S. In vivo kinetics of SARS-CoV-2 infection and its relationship with a person’s infectiousness. Proc Natl Acad Sci. 2021;118(49) doi: 10.1073/pnas.2111477118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Néant N., Lingas G., Le Hingrat Q., Ghosn J., Engelmann I., Lepiller Q., et al. Modeling SARS-CoV-2 viral kinetics and association with mortality in hospitalized patients from the French COVID cohort. Proc Natl Acad Sci. 2021;118(8) doi: 10.1073/pnas.2017962118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kissler S.M., Fauver J.R., Mack C., Olesen S.W., Tai C., Shiue K.Y., et al. Viral dynamics of acute SARS-CoV-2 infection and applications to diagnostic and public health strategies. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kissler S.M., Fauver J.R., Mack C., Tai C.G., Breban M.I., Watkins A.E., et al. Viral dynamics of SARS-CoV-2 variants in vaccinated and unvaccinated persons. N Engl J Med. 2021;385:2489–2491. doi: 10.1056/NEJMc2102507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ejima K., Kim K.S., Iwanami S., Fujita Y., Li M., Zoh R.S., et al. Time variation in the probability of failing to detect a case of polymerase chain reaction testing for SARS-CoV-2 as estimated from a viral dynamics model. J R Soc Interface. 2021;18 doi: 10.1098/rsif.2020.0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marks M., Millat-Martinez P., Ouchi D., h Roberts C., Alemany A., Corbacho-Monné M., et al. Transmission of COVID-19 in 282 clusters in Catalonia, Spain: a cohort study. Lancet Infect Dis. 2021;21(5):629–636. doi: 10.1016/S1473-3099(20)30985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marc A., Kerioui M., Blanquart F., Bertrand J., Mitjà O., Corbacho-Monné M., et al. Quantifying the relationship between SARS-CoV-2 viral load and infectiousness. eLife. 2021;10 doi: 10.7554/eLife.69302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonçalves A., Bertrand J., Ke R., Comets E., de Lamballerie X., Malvy D., et al. Timing of antiviral treatment initiation is critical to reduce SARS-CoV-2 viral load. CPT Pharmacometrics Syst Pharmacol. 2020;9:509–514. doi: 10.1002/psp4.12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goyal A., Cardozo-Ojeda E.F., Schiffer J.T. Potency and timing of antiviral therapy as determinants of duration of SARS-CoV-2 shedding and intensity of inflammatory response. Sci Adv. 2020;6 doi: 10.1126/sciadv.abc7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maisonnasse P., Aldon Y., Marc A., Marlin R., Dereuddre-Bosquet N., Kuzmina N.A., et al. COVA1-18 neutralizing antibody protects against SARS-CoV-2 in three preclinical models. Nat Commun. 2021;12 doi: 10.1038/s41467-021-26354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lévy Y., Wiedemann A., Hejblum B.P., Durand M., Lefebvre C., Surénaud M., et al. CD177, a specific marker of neutrophil activation, is associated with coronavirus disease 2019 severity and death. iScience. 2021;24 doi: 10.1016/j.isci.2021.102711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasin C., Balelli I., Van Effelterre T., Bockstal V., Solforosi L., Prague M., et al. Dynamics of the humoral immune response to a prime-boost ebola vaccine: quantification and sources of variation. J Virol. 2019;93 doi: 10.1128/JVI.00579-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kojima N., Klausner J.D. Protective immunity after recovery from SARS-CoV-2 infection. Lancet Infect Dis. 2022;22:12–14. doi: 10.1016/S1473-3099(21)00676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gazit S., Shlezinger R., Perez G., Lotan R., Peretz A., Ben-Tov A., et al. Comparing SARS-CoV-2 natural immunity to vaccine-induced immunity: reinfections versus breakthrough infections (preprint) MedRXiv. 2021 doi: 10.1101/2021.08.24.21262415. [DOI] [Google Scholar]
- 18.Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 20.Jin P., Li J., Pan H., Wu Y., Zhu F. Immunological surrogate endpoints of COVID-2019 vaccines: the evidence we have versus the evidence we need. Sig Transduct Target Ther. 2021;6:48. doi: 10.1038/s41392-021-00481-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plotkin S.A., Gilbert P.B. Nomenclature for immune correlates of protection after vaccination. Clin Infect Dis. 2012;54:1615–1617. doi: 10.1093/cid/cis238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandre M., Marlin R., Prague M., Coleon S., Kahlaoui N., Cardinaud S., et al. SARS-CoV-2 mechanistic correlates of protection: insight from modelling response to vaccines (preprint) BioRXiv. 2021 doi: 10.1101/2021.10.29.466418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higdon M.M., Wahl B., Jones C.B., Rosen J.G., Truelove S.A., Baidya A., et al. A systematic review of COVID-19 vaccine efficacy and effectiveness against SARS-CoV-2 infection and disease (preprint) MedRXiv. 2021 doi: 10.1101/2021.09.17.21263549. [DOI] [Google Scholar]
- 25.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184(4):861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]