Abstract
The rapid spread of SARS-CoV-2 infection throughout the world led to a global public health and economic crisis triggering an urgent need for the development of low-cost vaccines, therapies and high-throughput detection assays. In this work, we used a combination of Ideal-Filter Capillary Electrophoresis SELEX (IFCE-SELEX), Next Generation Sequencing (NGS) and binding assays to isolate and validate single-stranded DNA aptamers that can specifically recognize the SARS-CoV-2 Spike glycoprotein. Two selected non-competing DNA aptamers, C7 and C9 were successfully used as sensitive and specific biological recognition elements for the development of electrochemical and fluorescent aptasensors for the SARS-CoV-2 Spike glycoprotein with detection limits of 0.07 fM and 41.87 nM, respectively.
Keywords: Aptamers, SELEX, SARS-CoV-2, COVID-19, Aptasensor, Capillary electrophoresis
Graphical abstract
1. Introduction
Over two years have passed since the initial outbreak of a new Coronavirus disease (COVID-19). The original report to the World Health Organization (WHO), anticipated that COVID-19 would present a challenge for the public health systems across the world due to the rapid spread of the disease [1]. To date, over 323 million confirmed COVID-19 cases have been reported worldwide leading to the death of more than 5.5 million people by the severe acute respiratory syndrome associated with COVID-19 [2,3]. Although vaccines are already available in several countries, COVID-19 remains a major public health concern due to the uprising of SARS-CoV-2 variants and the lack of epidemiological surveillance and vaccination programs in impoverished countries [4].
The etiological agent for COVID-19 is the new coronavirus SARS-CoV-2, a single-stranded positive sense RNA (+ssRNA) enveloped virus with a genome of approximately 30 Kb encoding four structural proteins: Spike (S), membrane (M), envelope (E) and nucleocapsid (N) [5]. The trimeric S protein is recognized as the main virulence factor [6,7]. This is a type I transmembrane protein consisting of a large ectodomain, a single-pass transmembrane anchor and a short C-terminal intracellular tail [8]. The role of the S protein is crucial for viral adherence and entry to the host cell, where the receptor binding domain (RBD) within S protein mediates the interaction with the angiotensin-converting enzyme 2 (ACE2) attached to the cell membrane [9]. In addition, the S protein appears highly immunogenic, making it a suitable candidate for vaccine development and theranostic applications [[10], [11], [12], [13]].
Quantitative RT-PCR (RT-qPCR) or serological SARS-CoV-2 tests have become the standard COVID-19 diagnostic methods, however, they are costly, time-consuming and require specialized equipment and trained personnel [14]. In addition, serology and antigen tests require the production of purified proteins and specific antibodies, a long and expensive process that often leads to batch-to-batch variations [15]. Such problems related to antibodies may be one of the reasons why there is a difference in the detection performance (specificity and sensitivity) observed on rapid antigen tests when these parameters are determined in clinical conditions [16,17].
There are some new detection methods for SARS-CoV-2 infection based on technologies such as Field Effect Transistor, CRISPR-Cas12, Fluorine Doped Tin Oxide electrodes and functionalized gold nanoparticles or magnetic beads; however the equipment and the process to generate the materials needed for their implementation are difficult to obtain in most laboratories [[18], [19], [20], [21]]. As practical COVID-19 detection becomes necessary to save lives and return to a relative normality, there is a pressing need for efficient and affordable diagnosis tools.
Biosensors have been developed for the rapid, sensitive, and stable diagnostic methods that can use novel recognition elements such as nucleic acids aptamers [22,23]. Nucleic acid aptamers are short, single-stranded DNA (ssDNA) or RNA molecules that are selected for binding to a specific target [24]. The high affinity and specificity of aptamers are comparable to those of antibodies, with the advantage of rapid and massive high-quality production by automated synthesizers [25]. Aptamers are obtained through a highly probabilistic process called Systematic Evolution of Ligands by Exponential Enrichment (SELEX) [26]. In SELEX, large single-stranded oligonucleotide combinatorial pools are challenged for binding to a desired target under a defined set of conditions [24,25]. Aptamer selection is attained through iterative steps of target incubation with combinatorial pools, followed by the separation of unbound sequences (partitioning), and the PCR-mediated amplification and purification of the target-bound oligonucleotide species [27,28]. Aptamers with high affinity can be recovered through several partitioning methods although efficiency is often affected by the nonspecific background binding that each SELEX variant may have [29]. SELEX based on capillary electrophoresis (CE-SELEX), where target-bound aptamers are separated in solution, eliminate the possible background against components of the partitioning method resulting in the fast selection of aptamers due to its higher partition efficiency [30].
Aptamers are used in biosensors as biological recognition elements (BRE) for accurate and rapid detection of pathogens through fluorescence, chemiluminescence, electrochemistry and immunoluminescence techniques. These methods often require the chemical modification of the aptamer to produce visual or measurable signals upon interaction with the target molecule. Such modifications include labeling with fluorogenic, electrochemical or chromogenic moieties, addition of catalytic nucleic acids (aptasensors) or even allosteric aptamers (aptazymes) responsive to the target. Recently, new biosensors for COVID-19, including devices targeting the RNA of SARS-CoV-2, and COVID-19 antibodies, as well as immunosensors targeting the S and N proteins have been reported [[31], [32], [33], [34], [35], [36], [37]]. Nonetheless, aptamer-based biosensors offer advantages compared to antibodies such as shorter generation time, lower manufacturing costs, negligible batch-to-batch variability, simple chemical modification, better thermal stability, long shelf-life and higher target selection potential [[38], [39], [40]].
Aptamers against SARS-CoV-2 proteins have been previously isolated, some of them are able to block the interaction of S protein/RBD with ACE2 receptors and capable of inhibiting infection in pseudovirus models demonstrating therapeutic potential [[41], [42], [43], [44]]. Aptamer selection for the same virus using different viral targets and SELEX conditions is a common practice due to the rise of aptamer with different affinity, sequence, structure and binding capabilities [15]. In this regard, a variety of SELEX protocols have been used extensively, such as immobilization-free methods that expose the whole protein surface to the oligonucleotide pool thus making it more accessible to a variety of different aptatopes [45]. This is important when paired (non-competing for the same target site) aptamer screening is sought as different combinations of aptamers can generate a response to a wider range of the analyte concentration [46].
Thus, newly selected aptamers may be necessary for a more versatile response range in novel biosensors. As different BRE are required for biosensor development, aptamers with different interaction properties could be useful, alone or in combination with previously published aptamers. In addition, different aptamers capable of interacting with various epitopes of the S protein may exhibit synergistic effects in the inhibition of the viral entry or the detection of target sequence or structural variants. Therefore, selection and characterization of novel SARS-CoV-2 S protein aptamers are necessary for the development of new theranostic and biotechnology applications.
In the present work, IFCE-SELEX (Ideal Filter Capillary Electrophoresis-SELEX) was used in combination with Next Generation Sequencing (NGS) for the selection and identification of single-stranded DNA (ssDNA) aptamers that specifically bind with high affinity to the SARS-CoV-2 S protein. Furthermore, as proof-of-concept applications, selected paired aptamers were implemented in a electrochemical aptasensor and a simple sandwich-type fluorescent aptasensor capable of detecting and quantifying S protein in diluted human saliva of multiple donors suggesting a novel tool for the rapid and opportune diagnosis of COVID-19.
2. Materials and methods
2.1. Oligonucleotides and proteins
The FWD (5′-CACGACGACAGAGACCACAG-3′) and REV (5′-TGTTCGTCTCTGGCTGCTGG-3′) primers, the 40-nucleotide randomized DNA pool (M2 pool) and the 5′-FAM-C9 aptamer were purchased from T4 oligo® (ADN Artificial S. de R.L. de C.V., Irapuato, México). The 5′-amino-C6-modified C7 and C9 aptamers was purchased from Integrated DNA Technologies Inc. (San Diego, CA). 5′-DTPA2-modified C7 aptamer was purchased from Friz Biochem GmbH (Neuried, Germany). Monofunctional methoxy-polyethylene glycol thiol (PEG, 2 kDa), Bovine serum albumin (BSA), Lysozyme, and Casein were purchased from Merck (Merck KGaA, Darmstadt, Germany). The recombinant baculovirus-produced SARS-CoV-2 S protein (40589-V08B1), the human Angiotensin-Converting Enzyme 2 Protein (ACE2) (10108-H08B), the human Respiratory Syncytial Virus (RSV) glycoprotein G (11070-V08H), the human Coronavirus NL63 (HCoV-NL63) S Protein (40604-V08B), the influenza virus (H1N1) Hemagglutinin protein (40510-V08H), the MERS-CoV virus S protein (40069-V08B), and the SARS-CoV-2 (2019-nCoV) Spike Neutralizing Antibody, Mouse Mab (IgG) were all purchased from Sino Biological (Sino Biological Inc., Beijing, China).
2.2. Ideal-Filter Capillary Electrophoresis SELEX (IFCE-SELEX)
The M2 pool was purified through HPLC and preparative 8% polyacrylamide/7 M Urea gels. Partition by IFCE [30] was achieved using a G7100a Capillary Electrophoresis system (Agilent Technologies, Inc., Santa Clara CA) with standard uncoated fused-silica capillaries (Agilent G1600-60311). Prior to each partition cycle the capillary was preconditioned by flushing 0.1 M NaOH, deionized water and SELEX buffer (50 mM Tris-HCl pH 7.0, 110 mM NaCl, 1 mM MgCl2) for 5 min.
For the initial partition, an interaction mixture containing 100 nM purified S protein and 10 μM M2 ssDNA was prepared in SELEX buffer and incubated for 30 min at room temperature (RT). In all partition cycles the interaction mixture was injected at 34 mbar 10 s pressure pulse and propagated through the non-cooled part of the capillary with a 270 s 20 mbar pressure pulse. The aptamer-S protein complexes were separated applying 10 kV across the capillary for 45 min and collected in SELEX buffer. To increase selection astringency, protein concentration was decreased to 50 nM and 25 nM for the 2nd and 3rd partition cycles, respectively.
2.3. Generation of dsDNA and ssDNA production
Recovered aliquots (8 μL) from each partition were amplified by qPCR using the 0.3 μM primers and the Thermo Scientific™ 1X Maxima SYBR Green master mix (Thermo Fisher Scientific, Inc., Waltham MA), and an initial 95 °C for 7 min incubation followed by the cycling sequence 95 °C for 30s, 58 °C for 30s, 72 °C for 30s until a relatively high level of fluorescence was recorded. Purified amplicons were used as templates for asymmetric emulsion PCR (emPCR) to avoid by-product formation [47]. The oil phase was composed of 4.5% Span 80, 0.4% Tween 80 and 0.05% Triton X-100 in mineral oil. The aqueous phase contained 0.2 mM dNTPs, 0.4 μM FWD primer, 0.03 μM REV primer, 3 mM MgCl2, 1X Taq Buffer (BioTecMol S.A. de C.V., Mexico City, Mexico), 5 pM template and 0.125 U/μL Amplificasa® Taq DNA polymerase (BioTecMol). The 200 μL PCR reaction was combined with 500 μL of oil phase, vortexed for 20 min and cycled 50 times. The PCR water-in-oil emulsion was extracted with water-saturated dichloromethane and centrifuged at 17,200×g for 5 min for aqueous phase recovery. The ssDNA was separated from dsDNA through 10% native polyacrylamide gels followed by crush and soak elution. The ssDNA was quantified using a NanoDrop™ 2000 (Thermo Fisher) and visualized in denaturing 8% polyacrylamide/7 M Urea gels after every purification.
2.4. Next-generation sequencing (NGS)
NSG was performed essentially as previously described [48]. A minimum of 500 ng dsDNA from each cycle was used with the TruSeq DNA PCR-Free LT kit (Illumina Inc., San Diego, CA). The different adapters were added to each partition round following the manufacturer instructions. The partitioned pools with ligated adapters were quantified using the NEBNext® Library Quant Kit for Illumina® and paired-end sequenced in a MiSeq System using a MiSeq Reagent Kit V2 (Illumina) flow cell. NGS data was analyzed with the Galaxy project platform and FASTaptamer software [49].
To further increase the probability of selecting high affinity and specific aptamers, three additional oligodeoxynucleotide pools were included in the sequencing analysis. These pools were obtained from nitrocellulose-bound sequences recovered from three interaction mixtures flowed through a Slot blot apparatus. The first mixture contained only 2R ssDNA (NC), the second (NC-100) and third (NC-25) contained 2R ssDNA and S protein at S protein concentrations of 100 nM and 25 nM, respectively. The ssDNA bound to the nitrocellulose membrane was eluted by incubating at 95 °C in nuclease-free water for 10 min and then amplified by emPCR.
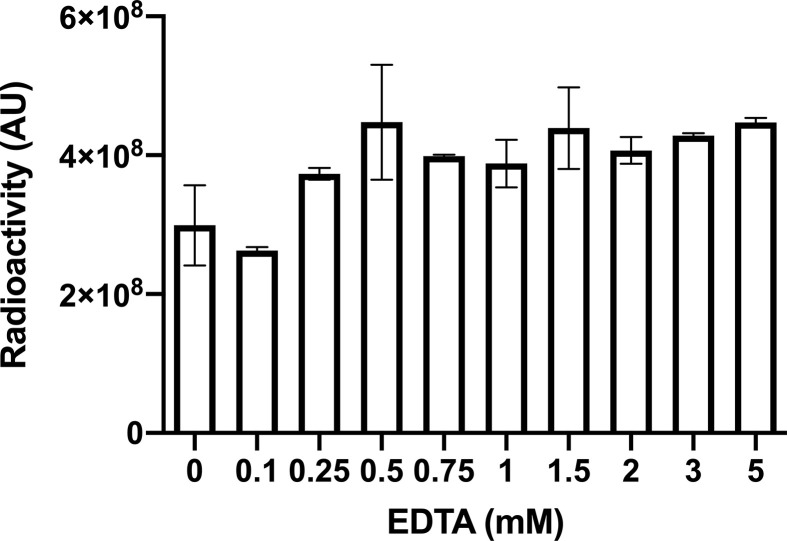
2.5. Binding assays
For Slot blot binding assays, the ssDNA pools and aptamers were radiolabeled using Thermo Scientific™ T4 polynucleotide kinase (PNK) (Thermo Fisher Scientific) and γ-[32P]-ATP (PerkinElmer, Inc., Waltham, MA). For enrichment and dissociation constant (KD) determination, interaction mixtures containing 15 pM of radiolabeled M2 pool, ssDNA pools or aptamers and different S protein concentrations, control proteins or diluted human saliva were incubated for 1 h at RT in either complete SELEX buffer, SELEX buffer without magnesium (TNa7) or with increasing concentrations of EDTA. The interaction mixture was placed in a Slot Blot apparatus (Invitrogen) with a nitrocellulose (NC) membrane (aptamer-S complexes retainer) (Cytiva, Marlborough, MA) layered on a nylon (NY) membrane (unbound aptamer retainer) (Cytiva). Residual radioactivity on the membranes was quantified using an Amersham Typhoon IP Biomolecular Imager (Cytiva). The percentage of bound fraction (%BF) was calculated using the following formula:
| %BF=Residual radioactivity on NC/total radioactivity on NC and NY)*100 |
The K D were calculated using the GraphPad Prism 8.4 software, curve-fitting to one site non-linear regression model:
| B=Bmax∗[M]/(KD+[M]) |
Where B is the bound fraction, Bmax is the maximum binding, [M] is the protein concentration, and K D is the dissociation constant [50].
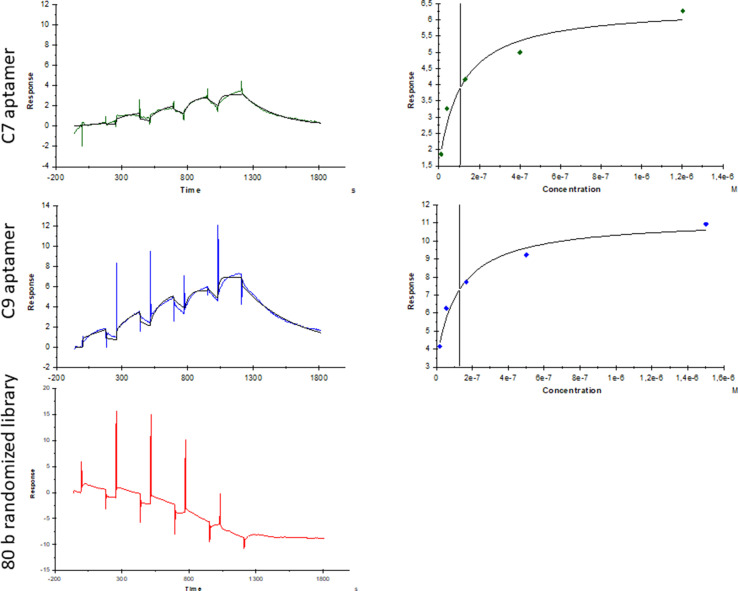
The K D were also determined by single-cycle kinetic analysis using surface plasmon resonance BIAcore T200 instrument (SPR) (GE Healthcare Europe). His-tag Spike protein with a concentration of 148.8 nM (20 ng/μL) was immobilized (lane 2) on a surface sensor (Series S Sensor Chip NTA) (GE Healthcare Europe) with a flow rate of 10 μL/min at 25 °C in TNa7 buffer until values of ∼600 response units were reached. The reference flow cell (lane 1) was left blank. A concentration series of C7, C9 and a randomized 80 nt pool as non-binder control were assessed for binding with S protein at 25 °C. The C7 aptamer was injected at concentrations of 1230, 410, 13, 45, and 15 nM. For C9 aptamer, concentrations were 1523, 507, 169, 56, and 18 nM. The concentrations for the randomized pool were 1335, 445, 148, 49 and 16 nM. The association and dissociation time was 180 s and 600 s, respectively. Data was fitted to a 1:1 binding stoichiometry model using the BIAcore T200 evaluation software 3.2 (Biacore) for K D determination.
2.6. Saliva collection and pretreatment
Whole saliva was collected from consenting healthy volunteer subjects, three male and two female donors, between 08:00 a.m. and 14:00 p.m. to account for the influence of circadian rhythms and food debris. Subjects were asked to rinse their mouths with water and discard this before sample collection. Saliva was allowed to accumulate on the mouth floor. The accumulated saliva was then spit into a polypropylene test tube and this was repeated until enough saliva was collected. During the collection process the sample tubes were kept on ice. Samples were cleared for 30 s in a microfuge. The pellet was discarded, and the supernatant diluted 10-fold in TNa7 buffer. Samples were kept on ice and used immediately.
2.7. Fluorophore-linked aptamer assay (FLAA)
The proof-of-principle FLAA test for SARS-CoV-2 S protein detection was set in 96-well microplates as described with some modifications [51]. Briefly, 100 pmole of 5′-amino-C6-modified C7 aptamer was immobilized on Pierce™ maleic anhydride activated plates (Thermo Fisher Scientific) as capture agent. Plates were blocked using SuperBlock™ reagent (Thermo Fisher Scientific) following the manufacturer's protocol. Different concentrations of S or control proteins were incubated in either TNa7 buffer or TNa7 supplemented with 10% single-donor or multi-donor saliva mix (SDS: 10% Single-donor Saliva; FDS: 10% multi-donor Saliva mix) and washed five times with TNa7 buffer. After washing, 50 pmole of the 5′-FAM-C9 aptamer were added as detection agent and incubated for 1 h at RT and washed five times with TNa7 buffer. To denature and retrieve the detection aptamer, 150 μL of 7 M urea were added and incubated 30 min in agitation at RT. Finally, 25 μL of the urea solution was mixed with 75 μL of TNa7 in 96-well black opaque plates (Corning Inc., Corning NY) and fluorescence measured at 491ex/516em nm using a BioTek® Synergy™ H4 Hybrid Multi-mode microplate reader (Thermo Fisher Scientific). For S protein quantification and spike-and-recovery assays, the background fluorescence signal (SDS or MDS without protein) was subtracted from the sample fluorescence signal (SDS or MDS with added S protein) and then used to determine the S protein concentration with the linear equation y = mx+b.
The maximum theoretical fluorescence intensity was estimated by fitting the calibration data to a nonlinear regression analysis using the formula:
where equals the obtained fluorescence intensity units, is the maximum possible signal obtained and f is the dissociation constant. The limit of detection (LOD) was calculated according to the equation:
where is the constant of random error (which 3 is typically used), is the standard deviation of the blank, and is the slope of the calibration curve [19,27,76].
2.8. Electrochemical assay
The electrochemical detection of the S protein was made utilizing well-established flexible multi-electrode arrays (flex-MEAs) [52]. Briefly, the flex-MEA chip was incubated for 2 h in a 0.5 μM C7 aptamer solution. Afterwards, the chip was rinsed with Tris-HCl 25 mM pH 7.4 and incubated in for 1 h in 5 mg/mL PEG solution, used as a blocking agent, and finally rinse with Tris buffer to remove non-bound molecules. Differential pulse voltammetry (DPV) was used for the characterization of the S protein detection. The protein was spiked in the buffer medium in a concentration range of 1 fg/mL to 100 ng/mL. DPV measurements were conducted in 5 mM ferri-ferrocyanide solution with a potential scan going from 0.0 V to 0.7 V (Ag/AgCl) with increments of 5 mV, an amplitude of 25 mV, a pulse width of 50 ms, a sampling width of 25 ms and a pulse period of 100 ms. In the same way, the control tests with the proteins from the RSV, Influenza and MERS-CoV virus were performed. The ratio between the current signal peak before and after protein addition (|ΔI/I_0 |(%)) was calculated for each tested concentration (c) to produce a calibration curve. The linear part of the obtained calibration curve was fitted by the half-log equation:
| |ΔI/I_0 |(%) = 6.29⋅log(c)+144.72 |
3. Results and discussion
3.1. Binding analysis of IFCE-partitioned pools
SARS-CoV-2 S protein-specific aptamers were selected using IFCE-SELEX to integrate a simple and rapid biosensor for COVID-19. IFCE partitioning was used to obtain high-affinity affinity aptamers with 2 selection cycles using purified recombinant S protein and the randomized oligonucleotide M2 pool (Fig. 1 ). IFCE conditions allowed opposite migration direction of aptamer-target complex and non-binders by decreasing the electroosmotic flow (EOF) in the running buffer. This was confirmed by performing capillary electrophoresis of the M2 pool with low ionic strength (50 mM Tris HCl) and high ionic strength conditions (TNa7) Fig. Multimedia component 5(Supplementary Fig. S1A), as described in the original work [30]. This resulted in suppression of pool migration with TNa7 condition (Fig. Multimedia component 6 Supplementary Fig. S1B). In addition, S protein migration in TNa7 was confirmed using the same conditions resulting in a peak at minute 10 (Fig. Multimedia component 7 Supplementary Fig. S1C). Protein interaction with aptamers changes drastically the mass-to-charge ratio of aptamers resulting in a less negative aptamer-S protein complex whose mobility is more affected by EOF rather than by its electrophoretic mobility. Therefore, a 45-min collection window was established for the partition step [53]. Because the low S protein concentration, the aptamer-complex peak was not detected; however, each electrophoresis condition was amplified by qPCR and aptamer recovery complex was confirmed by direct visualization on polyacrylamide gels (Supplementary Fig. Multimedia component 8Fig. S2A and Fig. Multimedia component 9S2B).
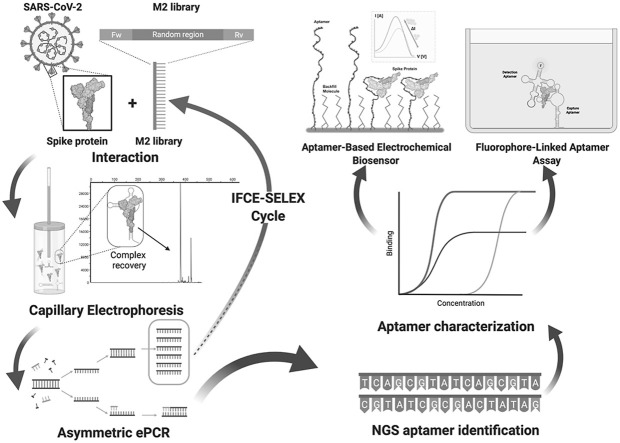
Fig. 1.
Schematics representation of DNA aptamers selection against SARS-CoV-2 Spike (S) protein through IFCE-SELEX. Purified recombinant SARS-CoV-2 S protein was incubated with the M2 ssDNA combinatorial pool containing 40 randomized positions (M2 library). The ssDNA-S protein complexes were partitioned using Ideal-Filter Capillary Electrophoresis (IFCE). The dsDNA generated from the recovered aptamer pools was used as template for asymmetric ePCR in ssDNA production. Each partitioned aptamer pool was allowed to enter in increasingly stringent selection cycles by decreasing the protein concentration by half until 25 nM. Figure created with BioRender.com.
To increase SELEX stringency, the resulting partitioned pool (1R) was used for two subsequent IFCE partition rounds (2R and 3R) with decreasing S protein concentrations (50 and 25 nM, respectively). This method was modified by performing extra selection rounds to increase strong binders, reduce the amount of non-binders and facilitate NGS enrichment analysis [54]. Binding assays showed that the M2 and 2R pools had differential binding affinities for the S protein. The M2 pool showed low binding affinity for S protein at 200 nM concentration, while 2R showed a significantly increased binding affinity (Fig. 2 A). These results suggest that the second IFCE partition round significantly increased the number of high affinity aptamers (HAA) decreasing the amount of protein required for binding. Also, this higher binding affinity appeared specific for S protein as the BSA negative control showed no DNA retention (Fig. 2A). Interestingly, a further selection cycle with decreased S protein concentration produced loss of binding affinity in the third IFCE partition (Fig. Multimedia component 10Supplementary Fig. S3). This approach is useful, however, as seen in some mathematical models, there is a limit in the protein concentration that can be used to improve SELEX. This can be explained because mostly all methods exhibit background binding that can compromise SELEX efficiency because represents a competitive presence [55]. In addition, this is consistent with previous CE-SELEX reports where binding enrichment occurred in early selection rounds and further partition rounds showed no improvement [56] or even the loss of affinity [57,58]. It is unclear why the pool affinity decreases through several partition rounds but other plausible explanations include mutations in aptamer sequences, DNA contamination and even over-amplification of non-aptamer sequences due to Taq DNA polymerase bias, suggesting that decreasing concentrations to a lower limit could be detrimental for the SELEX process [55,59].
Fig. 2.
IFCE SELEX analysis. A) Binding affinity and specificity of the different aptamer pools partitioned by IFCE-SELEX. Slot blot binding assays were made by incubating purified recombinant SARS-CoV-2 S protein (200 nM) with 10 pM of either the radiolabeled M2 randomized pool or the partitioned pools 2R and 3R. Bovine serum albumin (BSA) (200 nM) was used as negative binding control. The residual radioactivity on the nitrocellulose (NC) and nylon (NY) membranes was used to determine the aptamer fraction bound to the proteins. Error bars represent one standard deviation from triplicate analyses. The results were analyzed using one-way ANOVA with post hoc Tukey's multiple comparisons test (99.9% CI). Asterisks indicate statistical significance (n = 3, p < 0.0001). B) NGS variability analysis. The percentage of unique sequences in each dataset was calculated after sequencing each IFCE-SELEX cycle. The number of unique sequences decreased through the IFCE partition. C) NGS enrichment analysis. R1 data set was matched against 2R. Each black dot represents one sequence obtained from the NGS data. Sequence frequency was plotted as a function of its enrichment-fold from R1 to R2. It was observed that some of the most enriched sequences were also frequent. FASTAptamer toolkit was used for the variability and enrichment analysis. D) Unrooted radial phylogenetic tree of the most enriched sequences in R2 containing the ten selected aptamer sequences. E) Unrooted radial phylogenetic tree of the most enriched sequences against NC membrane. The trees were constructed using the 100 most enriched sequences and analyzed by Molecular Phylogenetic Analysis by Maximum Likelihood method based on the Kimura 2-parameter model conducted in MEGA7. Scale bar indicates 0.5 substitution per site.
3.2. NGS aptamer identification and validation
NGS was used for the identification of individual aptamer sequences from the pools with high affinity and specificity for the S protein. The raw NGS data from unselected M2 pool, all the partitioned pools (1R, 2R and 3R) and the NC, NC-25 and NC-100 nitrocellulose bound pools were pre-processed through the Galaxy project platform before FASTAptamer analysis (Supplementary Table 1).
A reduction in unique sequences was observed as the selection cycles progressed, confirming that the IFCE-SELEX process effectively decreased variability through the partition rounds with the lowest variability in the 3R pool despite the low binding affinity (Fig. 2B). This drop-in variability is also a common event in SELEX processes. Aptamer selection against ibuprofen performed a negative selection step (no target) reducing the number of sequences by 56% [60]. Also, in a SELEX against streptavidin variability decreased through ten selection rounds whereas affinity did not increase after round six [61]. Although the apparent low number of aptamers in 3R, this data was used to discriminate aptamer sequences that are present in both 2R and 3R pools. In addition, bioinformatics motif analysis using Multiple Expectation maximizations for Motif Elicitation Suite (MEME Suite) showed that top enriched sequences motifs are different in 2R and 3R. However, they were not considered for HAA selection due to a high E value (<0.05) in MEME motif analysis (Fig. Multimedia component 11 Supplementary Fig. S4) [62].
To identify HAA, the enrichment-folds (reads per million between selection rounds) were calculated and ranked for every sequence through the partition rounds using the FASTAptamer-Enrich script to observe the most enriched sequences from 2R/1R (Fig. 2C). In addition to this enrichment data, the NC, NC-100 and NC-25 NGS data sets were used to compare NC-100/NC-25 enrichment and identify which aptamer sequences were enriched with the decrement of S protein concentration thus facilitating the HAA discovery.
The arrangement in phylogenetic trees implies the acquisition of new characteristics with respect to their ancestors along the branches. Using Molecular Phylogenetic analysis by Maximum Likelihood method ten candidates were selected from different families in the selection cycle 2 (Table 1 ) and analyzed by binding analysis to find the best HAA (Fig. 2D) [63,64]. All sequences showing high enrichment or frequency in the NC data set were discarded. All ten candidate sequences exhibited no enrichment in the NC data set (Fig. 2E), but some were present in the NC-100 and NC-25 data set.
Table 1.
Sequences of the ten aptamer candidates with higher enrichment/frequency.
| Name | Sequence of selected aptamers (5'→3′) |
|---|---|
| C1 | TCGTAGAGTAGGTGCGGGCTTAACGCTCCAATCCGCTGAC |
| C2 | ATGTAACGTAACATCGTGGATTTGCAGATATTCCCTCAGG |
| C3 | GCTGAACATGCCTTGGAATCATGTTGTCTGCCCAGAGACA |
| C4 | GGGCGCGGGGGACAACGAGATGGGCTTAATGGATAGCAGA |
| C5 | CTCATTAGGCCCATCAAAGTGCCTTAGCTGAATGACCTCA |
| C6 | TGATACGTGACATGGTGCTTGATGAATAGGAATCGTTGT |
| C7 | CACGTGGCCCACGTTAATCCGTTATAAGTCAAGCTCGAT |
| C8 | CTCCCATCGGGATCTTATATCACGAATCGACAACGGTTGA |
| C9 | GGGGGCGTCAAGCGGGGTCACATCGGAGTAGGGAATCTTG |
| C10 | ATCTGAGAGGTCCAAGAGTGCACTGAAGGCATGCCTCTAT |
NGS analysis revealed that the initial pool variability decreased through the SELEX procedure although there were not highly over-represented or predominant sequence motifs as previously described for other CE-SELEX experiments [56,[65], [66], [67], [68], [69]]. Nevertheless, the bioinformatic analysis allowed identification of enriched oligonucleotide sequences. It is also possible that data may be improved if the NGS output is increased by using higher capacity flow cells since other NGS aptamer analyses used a higher number of reads per cycle (>1 million reads) [50].
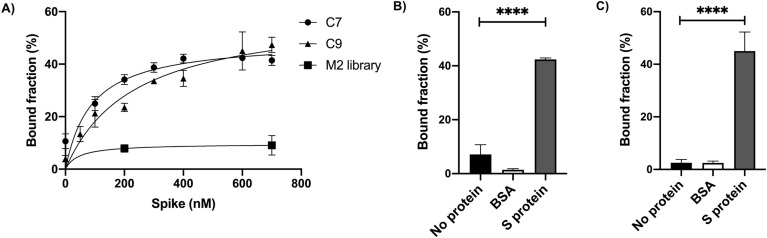
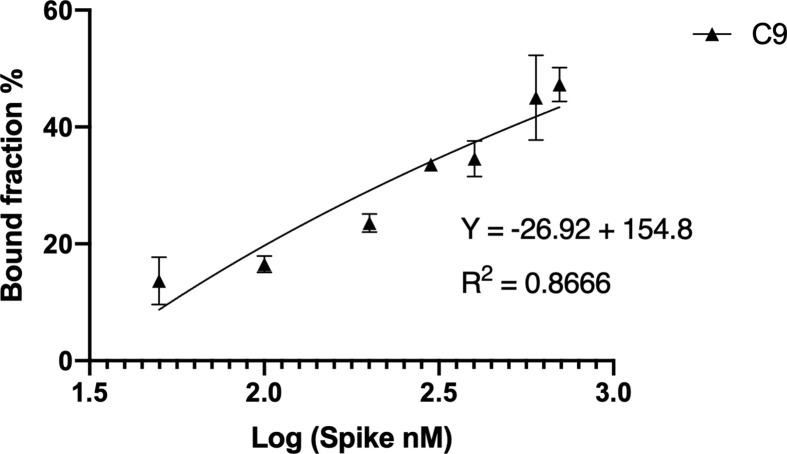
Slot blot binding assays showed candidates C7 and C9 best binding affinities similar to that presented by the whole 2R pool. No significant differences were observed with the BSA negative binding control or without protein, suggesting specific interactions (Fig. 3 B and 3C), as confirmed later by FLAA assay. Candidates C7 and C9 were further analyzed for binding affinity because they showed the lowest dissociation constant K D = 89.41 ± 18 nM and 231.9 ± 15 nM in a linear model, respectively (Fig. 3A and Supplementary Fig. Multimedia component 12Fig. S5A and Fig. Multimedia component 13S5B). In addition, all the K D were also determined by SPR showing nanomolar binding affinities similar to the Slot blot results (Fig. Multimedia component 14 Supplementary Fig. S6).
Fig. 3.
Binding Affinity and Specificity of aptamer sequences. A) KD determination. The dissociation constants for aptamers C7 (KD = 89.41 ± 18 nM) (black dots) and C9 (KD = 231.9 ± 15 nM) (black triangles) were calculated fitting the bound fraction curves from slot blot assays to a one binding site non-linear regression model (C7 R2 = 0.83; C9 R2 = 0.92). B) Binding specificity of C7 (left panel) and C9 (right panel) aptamers. Radiolabeled aptamer DNA (15 pM) was incubated with 600 nM of purified S protein. BSA (600 nM) and no protein negative controls were included. The data was analyzed using one-way ANOVA with post hoc Tukey's multiple comparisons test (99% CI). Asterisks indicate statistical significance (n = 3, p < 0.0001). For both aptamers no statistically significant differences were found between BSA and without protein (C7: p > 0.039; C9: p > 0.99).
3.3. FLAA can be used to quantify SARS-CoV-2 S protein
Biosensors can be classified as competitive and sandwich assays depending on the number of BRE that are used. Sandwich-type biosensors are preferred because of its dual recognition mechanism; in this type of biosensors, two different BRE are needed as two spatially distant regions are recognized within the target. This results in higher specificity and selectivity as one BRE is used for capture and the other for signal generation [70].
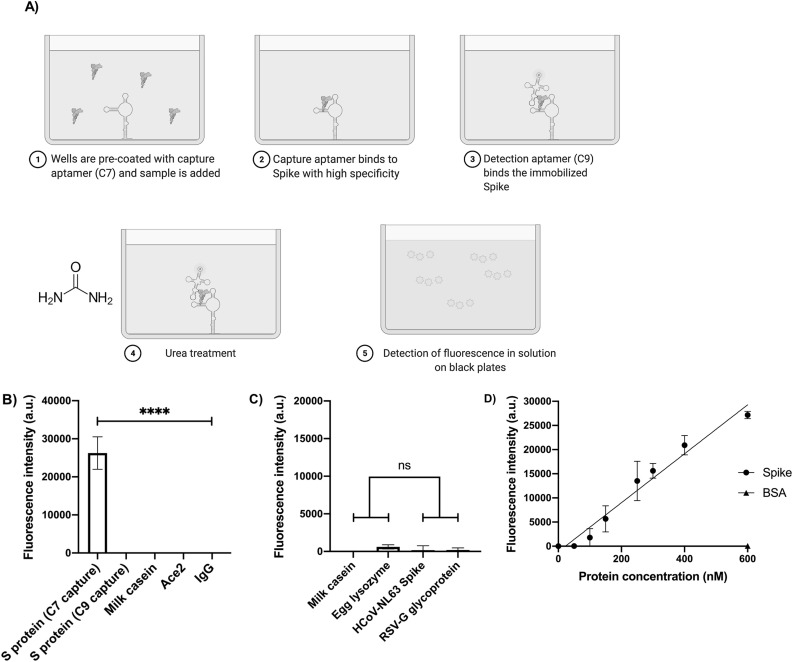
For a FLAA setting, C7 was immobilized on the surface of treated multiwell plates as capture agent and fluorescein-labeled C9 was added as detection agent. Purified recombinant SARS-CoV-2 S protein was added to the C7-containing plates, blocked and incubated in the presence of 10% saliva prior to addition of FAM-labeled C9 (Fig. 4 A). Other non-related proteins (mouse IgG, ACE2 and milk casein) were used as specificity controls (Fig. 4B).
Fig. 4.
FLAA specificity and detection parameters. A) Schematics of the FLAA procedure. Step 1: 5′-amino-C6-modified C7 aptamer was immobilized on the surface of maleic anhydride-activated multiwell plates as capture agent. Step 2: The purified recombinant SARS-CoV-2 S or negative binding control protein were added to the C7-containing plates. Step 3: Fluorescein-labeled C9 was added as detection agent. Step 4: After step 3 the multiwells are washed with TNa buffer. The multiwell plates are incubated with 7 M urea and volume is transferred to black plates. B) FLAA test based on the C7 and C9 aptamers. Purified recombinant S protein (250 nM) was added and incubated before addition of FAM-C9. Milk casein, human ACE2 and mouse IgG, were used as non-related controls to evaluate the FLAA specificity in 10-fold diluted human saliva. The graphs represent the mean and standard deviation from three independent experiments analyzed by one-way ANOVA with post hoc Tukey's multiple comparisons test (95% CI). Asterisks indicate statistical significance (n = 3, p < 0.0001). C) The FLAA test does not detect unrelated proteins and other common cold recombinant surface virus proteins. Milk casein, egg lysozyme, Human Respiratory Syncytial virus (RSV) glycoprotein G and Human Coronavirus (HCoV-NL63) S protein (250 nM) were added and incubated before addition of FAM-C9. The graphs represent the mean and standard deviation from three independent experiments analyzed by one-way ANOVA with post hoc Tukey's multiple comparisons test (95% CI). Asterisks indicate statistical significance (n = 3, p < 0.0001). D) FLAA signal is concentration-dependent of S protein. A FLAA test based was developed in 96-microwell plate format using aptamer C7 as capturing agent and FAM-labeled C9 as detection agent. . The FLAA concentration curve (0 nM–600 nM) of S protein (Black circles) showed a simple linear regression (R2 = 0.94). Background fluorescence from the well without S protein was subtracted from the measurements. BSA (600 nM) was used as negative control (Black triangle). Plotted data represents the mean and standard deviation of three independent experiments (n = 3).
SARS-CoV-2 S protein exhibited the highest fluorescence intensity suggesting a positive recognition and that C7 and C9 bind to different sites within S protein as no signal will be recorded otherwise. Interestingly, when 5′-amino-C6-modified C9 aptamer was used as capture agent and FAM-labeled C7 aptamer as detection agent, no signal was detected when 250 nM of S protein was added in TNa7 buffer. This may be due to the higher K D of C9 (230 nM) compared to C7 (89 nM) suggesting that the capture agent must be the aptamer with the highest affinity for the target if a low limit of detection (LOD) is desired, although the K D may vary depending of the determination method as we observed by SPR analysis (Supplementary Fig. S6). Another explanation may be that the chemical modification itself impaired C9 binding, since such an effect has been observed with other chemical modifications causing the partial or total loss of aptamer binding [71,72]. Also, it has been reported that the negative phosphate backbone of an aptamer can interact with the immobilization surface electrostatically, resulting in denaturation of the aptamer structure. This last may impact more C9 structure (ΔG = −7.4 kcal/mol) since it is less stable than C7 structure (ΔG = −8.9 kcal/mol) [73].
In addition, FLAA was tested against other surface virus proteins, such as human RSV glycoprotein G and HCoV-NL63 S protein, using milk casein and egg lysozyme as negative controls (Fig. 4C). No fluorescence was observed despite other S protein from a human coronavirus was used, thus indicating aptamer specificity for SARS-CoV-2 S glycoprotein. These results showed the suitability of FLAA as a detection test for S protein [45,74,75], using C7 and C9 aptamers as capture and detection agents, respectively.
To further characterize FLAA detection parameters, a calibration curve was generated using TNa buffer. The increase in fluorescence intensity (FI) signal caused by the formation of a tertiary complex (C7–S protein-C9) was determined by fluorometry. Background fluorescence was subtracted from all data points and plotted against increasing S protein concentrations to establish the dynamic detection range. The resulting standard curve was fitted to a simple linear regression analysis where the following equation was obtained:
The calculated LOD of the FLAA assay was 41.87 nM with a sensitivity (S) extracted from the slope of the obtained equation of 51 nM. The calculated was 70585 which corresponds to a protein concentration greater than the tested in this assay (600 nM); this result implies that the dynamic detection range of this assay using these BREs is wider and may be used with higher protein concentrations.
To determine FLAA detection performance in biofluids, spike-and-recovery assays were performed using diluted human saliva as matrix. As biological matrices may contain components that affect the response to the analyte more than the standard diluent (TNa), a spike-and-recovery assay is appropriate to assess the difference response between the standard diluent and the biological matrix [77]. A specific amount of purified S protein (spike) in 10-fold diluted human saliva (from multiple or single donors) was added to the microplate wells in TNa buffer and the fluorescence response (recovery) read after incubation in comparison with the response without saliva. BSA was used as negative control (Fig. 4D). It was established that the FLAA assay determines S protein concentration with an average recovery well within the 80–110% acceptable range (Table 2 ) [73]. These results also showed that other components found in saliva had low detrimental effect in the capacity of C7 and C9 aptamers to detect and quantify S protein, indicating that the FLAA may be suitable for COVID-19 detection in diluted saliva samples (Supplementary Fig. Multimedia component 15Fig. S7A andFig. Multimedia component 16 S7B).
Table 2.
Spike-and-recovery assessment for FLAA in diluted human saliva.
| Sample | Spike added (nM) | Expected (nM) | Observed (nM) | Average recovery (%) |
|---|---|---|---|---|
| SDS + Spike | 500 | 500 | 516 ± 24.2 | 103.27 |
| MDS + Spike | 250 | 250 | 235 ± 27.9 | 94.30 |
| MDS + Spike | 500 | 500 | 439 ± 13 | 87.80 |
| SDS + BSA | 500 | 0 | −7 ± 70 | – |
SDS: 10% Single Donor Saliva; MDS: 10% Multiple Donor Saliva.
As a second proof-of-principle test, the C7 aptamer was implanted in an electrochemical aptasensor configuration. A 5′-end thiol-terminal group was added to the C7 aptamer for its immobilization on a gold electrode through a sulfur-gold bond (Fig. 5 A). Furthermore, thiol terminated PEG molecules were used as anti-fouling backfill to suppress unspecific binding [78]. The binding of the target to the receptor layer induced conformational changes within the aptamer film. These modulations of the receptor layer caused alterations of the ferri/ferrocyanide charge transfer characteristics which were registered by differential pulse voltammetry (DPV). Firstly, the starting DPV current signal of the sensor electrodes was measured without analyte exposure. Subsequently, the sensor responses were recorded after 30-min incubation for different S protein concentrations covering a range from 1 fg/mL to 100 ng/mL. An increase of the peak current signal was observed as the concentration of the protein rose. The current increase can be understood as a result of a reduced charge transfer resistance due to conformational rearrangements within the receptor layer (Fig. 5A). The sensitivity of the sensor was calculated to 6.29 ± 0.98/decade while the LOD was calculated to 8.85 fg/mL (0.07 fM). Correspondingly, a dynamic detection was feasible in the range from 8.85 fg/mL to 100 pg/mL. Hereby, the calculated K D for 5′-thio-C7 was 141 fg/mL (1.04 fM) (Fig. 5B).
Fig. 5.
Electrochemical detection. A) Schematic representation of the surface of a gold electrode functionalized with C7 aptamer by a sulfur-gold binding through thiol groups at the 5′-end (5′-thio-C7). The formation of the aptamer-S protein complex changes the aptamer conformation resulting in modification of the electrochemical signal, shortening the charge transfer distance of the redox molecules and thereby increasing the current as the concentration of the protein increases (inlet). B) Current signal I(Ipeak-I0)/I0I (%) of the differential potential voltammetry response versus increasing analyte concentration (1 fg/mL – 100 ng/mL). Inlet: same relation but in a semi-log representation. A linear fit was calculated over the linear dynamic detection range (Data points) by excluding the data points in saturation region (Excluded). The selectivity is demonstrated by the sensor signals obtained for Glycoprotein G of the RSV, the Hemagglutinin protein of the influenza (H1N1) virus, and the S protein of the MERS-CoV virus.
A high selectivity of the C7 aptamer was obtained for S protein of the SARS-CoV-2 virus over proteins from other viruses such as the Glycoprotein G of the RSV, the Hemagglutinin protein of the influenza (H1N1) virus, or the S protein of the MERS-CoV virus even at high concentrations (Fig. 5B).
4. Conclusion
In the present work, C7 and C9 aptamers specific for the SARS-CoV-2 S protein were selected by combining IFCE partition with an optimized SELEX protocol. The high partition efficiency coupled with the use of emPCR for efficient aptamer amplification allowed the enrichment of HHA in only two selection cycles. Aptamer sequence identification was facilitated by using Slot-blot assays with the generated pools and the use of NGS data combined with phylogenetic analysis. This combination of methods can be easily applied to different targets for rapid HAA discovery.
The newly developed FLAA test for SARS-CoV-2 S protein detection is presented as a proof-of-concept to validate the potential of DNA aptamer applications in the fight against COVID-19. The C7 and C9 aptamers were deemed suitable for the design of FLAA detection methods as both bind S in a variety of conditions and appear to have different binding sites within the protein as the observed K D for each aptamer suggests that no signal would be retained in case of intramolecular competition. This indicated a potential application in clinical samples, which was further validated by spike-and-recovery assays. Furthermore, a second validation approach was demonstrated for the implementation of the C7 aptamer in an aptasensor system. A flexMEA chip was utilized to electrochemically determine the aptamer-S protein complex formation with high sensitivity and selectivity. In addition, C7 and C9 binding capacities and biofluid resilience are useful for application in places where minimal clinical facilities are difficult to meet as is the case for developing countries. As a next stage, we are currently evaluating the FLAA test with clinical samples to establish clinical applicability and final costs.
Author contributions
MAMR, PAFU and VMGV contributed equally to this work by performing experiments, bioinformatics analysis, manuscript writing and data interpretation.
MC and GM provided the design of the M2 pool.
SRMR, GFM and DM performed the electrochemical experiments and results.
LMAS was the project coordinator providing funding, data analysis, manuscript writing and revision.
Acknowledgements
This project was funded by CONACyT grants 311385 and 300073 and SRE-AMEXCID grant 2020-6. We thank to Ian Gering and María Fernanda Pérez y Pérez for excellent technical assistance during SPR and binding experiments, respectively.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ab.2022.114633.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. S1A.
Supplementary Fig. S1B.
Supplementary Fig. S1C.
Supplementary Fig. S2A.
Supplementary Fig. S2B.
Supplementary Fig. S3.
Supplementary Fig. S4.
Supplementary Fig. S5A.
Supplementary Fig. S5B.
Supplementary Fig. S6.
Supplementary Fig. S7A.
Supplementary Fig. S7B.
References
- 1.Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO . 1–23. World Heal. Organ; 2021. https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-update (COVID-19 Weekly Epidemiological Update). [Google Scholar]
- 4.Abduljalil J.M., Abduljalil B.M. Epidemiology, genome, and clinical features of the pandemic SARS-CoV-2: a recent view. New Microbes New Infect. 2020;35:100672. doi: 10.1016/j.nmni.2020.100672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar A., Prasoon P., Kumari C., Pareek V., Faiq M.A., Narayan R.K., Kulandhasamy M., Kant K. SARS-CoV-2-specific virulence factors in COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.26615. [DOI] [PubMed] [Google Scholar]
- 8.Coutard B., Valle C., De Lamballerie X., Canard B., Seidah N.G., Decroly E. 2020. The Spike Glycoprotein of the New Coronavirus 2019-nCoV Contains a Furin- like Cleavage Site Absent in CoV of the Same Clade. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsueh P.R., Huang L.M., Chen P.J., Kao C.L., Yang P.C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woo P.C.Y., Lau S.K.P., Chu C., Chan K., Tsoi H., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W., Poon L.L.M., Wong S.S.Y., Guan Y., Peiris J.S.M., Yuen K. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/jvi.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang T., Wu Q., Zhang Z. Probable Pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1578. doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pang J., Wang M.X., Ang I.Y.H., Tan S.H.X., Lewis R.F., Chen J.I.-P., Gutierrez R.A., Gwee S.X.W., Chua P.E.Y., Yang Q., Ng X.Y., Yap R.K., Tan H.Y., Teo Y.Y., Tan C.C., Cook A.R., Yap J.C.-H., Hsu L.Y. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J. Clin. Med. 2020;9 doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sánchez-Báscones E., Parra F., Lobo-Castañón M.J. Aptamers against viruses: selection strategies and bioanalytical applications. TrAC Trends Anal. Chem. (Reference Ed.) 2021;143 doi: 10.1016/j.trac.2021.116349. [DOI] [Google Scholar]
- 16.Hayer J., Kasapic D., Zemmrich C. Real-world clinical performance of commercial SARS-CoV-2 rapid antigen tests in suspected COVID-19: a systematic meta-analysis of available data as of November 20, 2020. Int. J. Infect. Dis. 2021;108:592–602. doi: 10.1016/j.ijid.2021.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alghounaim M., Bastaki H., Bin Essa F., Motlagh H., Al-Sabah S. The performance of two rapid antigen tests during population-level screening for SARS-CoV-2 infection. Front. Med. 2021;8:1–5. doi: 10.3389/fmed.2021.797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahari S., Roberts A., Shahdeo D., Gandhi S. eCovSens-ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCovid-19 antigen, a spike protein domain 1 of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.24.059204. [DOI] [Google Scholar]
- 20.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Il Kim S. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal Swab Specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 21.Singh N.K., Ray P., Carlin A.F., Magallanes C., Morgan S.C., Laurent L.C., Aronoff-Spencer E.S., Hall D.A. Hitting the diagnostic sweet spot: point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens. Bioelectron. 2021;180:113111. doi: 10.1016/j.bios.2021.113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song K.M., Lee S., Ban C. Aptamers and their biological applications. Sensors. 2012;12:612–631. doi: 10.3390/s120100612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Z. Chen, Q. Wu, J. Chen, • Xiaohua Ni, J. Dai, A DNA aptamer based method for detection of SARS-CoV-2 nucleocapsid protein, (n.d.). 10.1007/s12250-020-00236-z. [DOI] [PMC free article] [PubMed]
- 24.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2016;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun H., Zu Y. A Highlight of recent advances in aptamer technology and its application. Molecules. 2015;20:11959–11980. doi: 10.3390/molecules200711959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 27.Dembowski S.K., Bowser M.T. Microfluidic methods for aptamer selection and characterization. Analyst. 2018;143:21–32. doi: 10.1039/c7an01046j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoltenburg R., Reinemann C., Strehlitz B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 2005;383:83–91. doi: 10.1007/s00216-005-3388-9. [DOI] [PubMed] [Google Scholar]
- 29.Ruscito A., DeRosa M.C. Small-molecule binding aptamers: selection strategies, characterization, and applications. Front. Chem. 2016;4:14. doi: 10.3389/fchem.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le A.T.H., Krylova S.M., Kanoatov M., Desai S., Krylov S.N. Ideal-filter capillary electrophoresis (IFCE) facilitates the one-step selection of aptamers. Angew. Chem. Int. Ed. 2019;58:2739–2743. doi: 10.1002/anie.201812974. [DOI] [PubMed] [Google Scholar]
- 31.Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J., Zhang Y.-P., Li C.-P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ali M.A., Hu C., Jahan S., Yuan B., Saleh M.S., Ju E., Gao S.J., Panat R. Sensing of COVID-19 antibodies in seconds via aerosol jet nanoprinted reduced-graphene-oxide-coated 3D electrodes. Adv. Mater. 2021;33:1–15. doi: 10.1002/adma.202006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elledge S.K., Zhou X.X., Byrnes J.R., Martinko A.J., Lui I., Pance K., Lim S.A., Glasgow J.E., Glasgow A.A., Turcios K., Iyer N.S., Torres L., Peluso M.J., Henrich T.J., Wang T.T., Tato C.M., Leung K.K., Greenhouse B., Wells J.A. Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. Nat. Biotechnol. 2021;39:928–935. doi: 10.1038/s41587-021-00878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen, Biosens. Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerutti F., Burdino E., Milia M.G., Allice T., Gregori G., Bruzzone B., Ghisetti V. Urgent need of rapid tests for SARS CoV-2 antigen detection: evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J. Clin. Virol. 2020;132:104654. doi: 10.1016/j.jcv.2020.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eissa S., Zourob M. Development of a low-cost cotton-tipped electrochemical immunosensor for the detection of SARS-CoV-2. Anal. Chem. 2021;93:1826–1833. doi: 10.1021/acs.analchem.0c04719. [DOI] [PubMed] [Google Scholar]
- 37.Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh N.K., Jain P., Das S., Goswami P. Dye coupled aptamer-captured enzyme catalyzed reaction for detection of pan malaria and p. Falciparum species in laboratory settings and instrument-free paper-based platform. Anal. Chem. 2019;91:4213–4221. doi: 10.1021/acs.analchem.9b00670. [DOI] [PubMed] [Google Scholar]
- 39.Vargas-Montes M., Cardona N., Moncada D.M., Molina D.A., Zhang Y., Gómez-Marín J.E. Enzyme-linked aptamer assay (ELAA) for detection of toxoplasma ROP18 protein in human serum. Front. Cell. Infect. Microbiol. 2019;9:1–13. doi: 10.3389/fcimb.2019.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wędrowska E., Wandtke T., Piskorska E., Kopiński P. The latest achievements in the construction of influenza virus detection aptasensors. Viruses. 2020;12:1–14. doi: 10.3390/v12121365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun M., Liu S., Wei X., Wan S., Huang M., Song T., Lu Y., Weng X., Lin Z., Chen H., Song Y., Yang C. Aptamer blocking Strategy inhibits SARS-CoV-2 virus infection. Angew. Chem. Int. Ed. 2021;60:10266–10272. doi: 10.1002/anie.202100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X., ling Wang Y., Wu J., Qi J., Zeng Z., Wan Q., Chen Z., Manandhar P., Cavener V.S., Boyle N.R., Fu X., Salazar E., Kuchipudi S.V., Kapur V., Zhang X., Umetani M., Sen M., Willson R.C., hsia Chen S., Zu Y. Neutralizing aptamers block S/RBD-ACE2 interactions and prevent host cell infection. Angew. Chem. Int. Ed. 2021;60:10273–10278. doi: 10.1002/anie.202100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 44.Schmitz A., Weber A., Bayin M., Breuers S., Fieberg V., Famulok M., Mayer G. A SARS-CoV-2 spike binding DNA aptamer that inhibits pseudovirus infection by an RBD-independent mechanism. Angew. Chem. Int. Ed. 2021;60:10279–10285. doi: 10.1002/anie.202100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bin Seo H., Gu M.B. Aptamer-based sandwich-type biosensors. J. Biol. Eng. 2017;11:1–7. doi: 10.1186/s13036-017-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morales M.A., Halpern J.M. Guide to selecting a biorecognition element for biosensors. Bioconjugate Chem. 2018;29:3231–3239. doi: 10.1021/acs.bioconjchem.8b00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao K., Ding W., Wang F., Li H., Ma D., Wang H. Emulsion PCR: a high efficient way of PCR amplification of random DNA libraries in aptamer selection. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel M., Suess B. Nucleic acid aptamers : selection, characterization, and application. Methods Mol. Biol. 2016;1380:113–125. doi: 10.1007/978-1-4939-3197-2. [DOI] [PubMed] [Google Scholar]
- 49.Alam K.K., Chang J.L., Burke D.H. FASTAptamer: a bioinformatic toolkit for high-throughput sequence analysis of combinatorial selections. Mol. Ther. Nucleic Acids. 2015;4:e230. doi: 10.1038/mtna.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho M., Xiao Y., Nie J., Stewart R., Csordas A.T., Oh S.S., Thomson J.A., Soh H.T. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee K.H., Zeng H. Aptamer-based ELISA assay for highly specific and sensitive detection of Zika NS1 protein. Anal. Chem. 2017;89:12743–12748. doi: 10.1021/acs.analchem.7b02862. [DOI] [PubMed] [Google Scholar]
- 52.Figueroa-Miranda G., Chen S., Neis M., Zhou L., Zhang Y., Lo Y., Tanner J.A., Kreidenweiss A., Offenhäusser A., Mayer D. Multi-target electrochemical malaria aptasensor on flexible multielectrode arrays for detection in malaria parasite blood samples. Sensor. Actuator. B Chem. 2021;349 doi: 10.1016/j.snb.2021.130812. [DOI] [Google Scholar]
- 53.Zhang H., Li X.-F., Le X.C. Tunable aptamer capillary electrophoresis and its application to protein analysis. J. Am. Chem. Soc. 2008;130:34–35. doi: 10.1021/ja0778747. [DOI] [PubMed] [Google Scholar]
- 54.Le A.T.H., Krylova S.M., Beloborodov S.S., Wang T.Y., Hili R., Johnson P.E., Li F., Veedu R.N., Belyanskaya S., Krylov S.N. How to Develop and prove high-efficiency selection of ligands from oligonucleotide libraries: a universal framework for aptamers and DNA-encoded Small-molecule ligands. Anal. Chem. 2021;93:5343–5354. doi: 10.1021/acs.analchem.1c00601. [DOI] [PubMed] [Google Scholar]
- 55.Wang J., Rudzinski J.F., Gong Q., Soh H.T., Atzberger P.J. Influence of target concentration and background binding on in vitro selection of affinity reagents. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mendonsa S.D., Bowser M.T. In vitro selection of high-affinity DNA ligands for human IgE using capillary electrophoresis. Anal. Chem. 2004;76:5387–5392. doi: 10.1021/ac049857v. [DOI] [PubMed] [Google Scholar]
- 57.Jing M., Bowser M.T. Tracking the emergence of high affinity aptamers for rhVEGF165 during capillary electrophoresis-systematic evolution of ligands by exponential enrichment using high throughput sequencing. Anal. Chem. 2013;85:10761–10770. doi: 10.1021/ac401875h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J., Bowser M.T. Capillary electrophoresis–SELEX selection of catalytic DNA aptamers for a Small-molecule porphyrin target. Anal. Chem. 2013;85:1525–1530. doi: 10.1021/ac302721j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu C., Yang G., Ghulam M., Li L., Qu F. Evolution of multi-functional capillary electrophoresis for high-efficiency selection of aptamers. Biotechnol. Adv. 2019;37:107432. doi: 10.1016/j.biotechadv.2019.107432. [DOI] [PubMed] [Google Scholar]
- 60.Kim Y.S., Hyun C.J., Kim I.A., Gu M.B. Isolation and characterization of enantioselective DNA aptamers for ibuprofen. Bioorg. Med. Chem. 2010;18:3467–3473. doi: 10.1016/j.bmc.2010.03.074. [DOI] [PubMed] [Google Scholar]
- 61.Schütze T., Wilhelm B., Greiner N., Braun H., Peter F., Mörl M., Erdmann V.A., Lehrach H., Konthur Z., Menger M., Arndt P.F., Glökler J. Probing the SELEX process with next-generation sequencing. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ajawatanawong P. Molecular phylogenetics: concepts for a newcomer. Adv. Biochem. Eng. Biotechnol. 2017;160:185–196. doi: 10.1007/10_2016_49. [DOI] [PubMed] [Google Scholar]
- 64.Setlem K., Mondal B., Ramlal S., Kingston J. Immuno affinity SELEX for simple, rapid, and cost-effective aptamer enrichment and identification against aflatoxin B1. Front. Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.01909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu C., Yang G., Ghulam M., Li L., Qu F. Evolution of multi-functional capillary electrophoresis for high-efficiency selection of aptamers. Biotechnol. Adv. 2019;37:107432. doi: 10.1016/j.biotechadv.2019.107432. [DOI] [PubMed] [Google Scholar]
- 66.Mendonsa S.D., Bowser M.T. In vitro selection of aptamers with affinity for neuropeptide Y using capillary electrophoresis. J. Am. Chem. Soc. 2005;127:9382–9383. doi: 10.1021/ja052406n. [DOI] [PubMed] [Google Scholar]
- 67.Mosing R.K., Mendonsa S.D., Bowser M.T. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal. Chem. 2005;77:6107–6112. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- 68.Tok J., Lai J., Leung T., Li S.F.Y. Selection of aptamers for signal transduction proteins by capillary electrophoresis. Electrophoresis. 2010;31:2055–2062. doi: 10.1002/elps.200900543. [DOI] [PubMed] [Google Scholar]
- 69.Williams B.A.R., Lin L., Lindsay S.M., Chaput J.C. Evolution of a histone H4-K16 acetyl-specific DNA aptamer. J. Am. Chem. Soc. 2009;131:6330–6331. doi: 10.1021/ja900916p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen A., Yan M., Yang S. Split aptamers and their applications in sandwich aptasensors. TrAC Trends Anal. Chem. (Reference Ed.) 2016;80:581–593. doi: 10.1016/j.trac.2016.04.006. [DOI] [Google Scholar]
- 71.Elskens J.P., Elskens J.M., Madder A. Chemical modification of aptamers for increased binding affinity in diagnostic applications: current status and future prospects. Int. J. Mol. Sci. 2020;21:1–31. doi: 10.3390/ijms21124522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Odeh F., Nsairat H., Alshaer W., Ismail M.A., Esawi E., Qaqish B., Al Bawab A., Ismail S.I. Aptamers chemistry: chemical modifications and conjugation strategies. Molecules. 2020;25 doi: 10.3390/molecules25010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walter J.G., Kökpinar Ö., Friehs K., Stahl F., Scheper T. Systematic investigation of optimal aptamer immobilization for protein-microarray applications. Anal. Chem. 2008;80:7372–7378. doi: 10.1021/ac801081v. [DOI] [PubMed] [Google Scholar]
- 74.Higuchi A., Siao Y.-D., Yang S.-T., Hsieh P.-V., Fukushima H., Chang Y., Ruaan R.-C., Chen W.-Y. Preparation of a DNA Aptamer−Pt complex and its use in the colorimetric sensing of thrombin and anti-thrombin antibodies. Anal. Chem. 2008;80:6580–6586. doi: 10.1021/ac8006957. [DOI] [PubMed] [Google Scholar]
- 75.Schlecht U., Malavé A., Gronewold T., Tewes M., Löhndorf M. Comparison of antibody and aptamer receptors for the specific detection of thrombin with a nanometer gap-sized impedance biosensor. Anal. Chim. Acta. 2006:65–68. doi: 10.1016/j.aca.2006.01.016. 573–574. [DOI] [PubMed] [Google Scholar]
- 76.Macdougall D., Crummett W.B. Guidelines for data acquisition and data quality evaluation in environmental chemistry. Anal. Chem. 1980;52:2242–2249. [Google Scholar]
- 77.Andreasson U., Perret-Liaudet A., van Waalwijk van Doorn L.J.C., Blennow K., Chiasserini D., Engelborghs S., Fladby T., Genc S., Kruse N., Kuiperij H.B., Kulic L., Lewczuk P., Mollenhauer B., Mroczko B., Parnetti L., Vanmechelen E., Verbeek M.M., Winblad B., Zetterberg H., Koel-Simmelink M., Teunissen C.E. A practical guide to immunoassay method validation. Front. Neurol. 2015;6:1–8. doi: 10.3389/fneur.2015.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Figueroa-Miranda G., Wu C., Zhang Y., Nörbel L., Lo Y., Tanner J.A., Elling L., Offenhäusser A., Mayer D. Polyethylene glycol-mediated blocking and monolayer morphology of an electrochemical aptasensor for malaria biomarker detection in human serum. Bioelectrochemistry. 2020;136:107589. doi: 10.1016/j.bioelechem.2020.107589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.