Abstract
Locus-specific databases are invaluable tools for both basic and clinical research. The extensive information they contain is gathered from the literature and manually curated by experts. Cancer genome sequencing projects generate an immense amount of data, which are stored directly in large repositories (cancer genome databases). The presence of a TP53 defect (17p deletion and/or TP53 mutations) is an independent prognostic factor in chronic lymphocytic leukemia (CLL) and TP53 status analysis has been adopted in routine clinical practice. For that reason, TP53 mutation databases have become essential for the validation of the plethora of TP53 variants detected in tumor samples. TP53 profiles in CLL are characterized by a great number of subclonal TP53 mutations with low variant allelic frequencies and the presence of multiple minor subclones harboring different TP53 mutations. In this review, we describe the various characteristics of the multiple levels of heterogeneity of TP53 variants in CLL through the analysis of TP53 mutation databases and the utility of their diagnosis in the clinic.
Keywords: mutation database, TP53 mutation, chronic lymphocytic leukemia, variant classification guidelines, genetic analysis model
Introduction
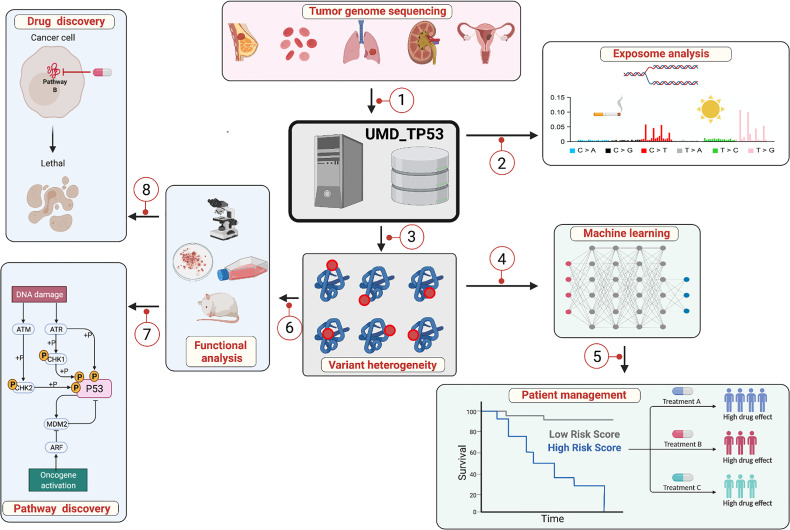
In 1956, Ingram used protein sequencing to provide the first demonstration of a severe disease (human sickle-cell anemia in that work) resulting from a single amino acid substitution (1). Since then, it has been largely demonstrated that gene mutations are the basis for most genetic diseases. The development of DNA sequencing and molecular cloning technologies in the late 1970s contributed greatly to the identification of genes involved in both monogenic and polygenic disorders, including complex diseases like cancer (2). The alterations occurring in those genes are numerous and variable in nature, ranging from point mutations to large deletions or translocations. Moreover, the task of reporting, storing, classifying and analyzing them has been a major challenge (3). To provide a pertinent response to this latter, locus-specific databases (LSDBs) have been developed ( Figure 1 ). Although intended for single genes, LSDBs do offer great accuracy as they are curated manually by experts in the field (4, 5). They also provide information that can be used for large-scale analyses and often include structural, functional or evolutionary data (6). For constitutional mutations associated with a genetic syndrome, several LSDBs also include phenotypic data useful for the study of genotype-phenotype correlation (7).
Figure 1.
The locus-specific database UMD_TP53: a central hub for multifactorial analysis. 1: TP53 variants and patient information are collected and stored in a relational database specifically developed for the storage and the analysis of genetic variants. 2: Exposome analysis: influence of the external and internal environment on the landscape of mutational events to identify the links between exposure to various types of carcinogens, specific mutational events in the TP53 gene and the development of specific cancers. 3: More than 7,000 different TP53 variants have been discovered in various types of cancer with heterogeneous LOF and GOF. 4: Multiple bioinformatics tools, including machine learning, have been developed to predict and classify TP53 variants. 5: Genome-based prognostic biomarkers can be used for several cancer types for potential incorporation into clinical prognostic staging systems or practice guidelines such as TP53 and CLL. 6: Analysis of TP53 variants points to the various functional domains of the protein essential for tumor suppression. 7: Functional analysis has led to the identification of the multiple pathways regulated by TP53. 8: Small molecules have been developed that specifically target missense TP53 variants and restore p53 transcriptional activity, thereby enabling tumor regression. Although this figure describes the TP53 database, the various aspects can be applied to other genes as well.
Genomic studies of tumor samples in the pre-genomic era were focused either on a small number of genes analyzed in large patient cohorts, or on a more significant number of genes but in only a few tumors. Indeed, large-scale analyses combining a multitude of genes and tumors represented a Herculean and costly task. The development of high-throughput methodologies capable of sequencing an entire genome in only a few days (next generation sequencing, NGS) has radically changed the entire field of cancer biology. In the present post-genomic era, whole genome sequencing in a multitude of tumors can be performed in a matter of days. The International Cancer Genome Consortium (ICGC, http://dcc.icgc.org/), the Cancer Genome Atlas Project (TCGA, http://cancergenome.nih.gov/) and the Sanger Institute (http://www.sanger.ac.uk/) have undertaken large-scale cancer genome analyses in different types and subtypes of cancer. That work has led to the creation of large data repositories (cancer genome databases, CGDs) freely available to the entire scientific community (8–10). Both LSDBs and CGDs can be considered as central hubs linking clinical and basic research ( Figure 1 ). They all make important contributions to our knowledge of the intricate pathways regulating cell fate, and our ability to identify new clinical biomarkers and develop novel therapeutic molecules.
TP53 mutation databases are the perfect example of the successful use of these compilations of cancer associated alterations. Indeed, the TP53 suppressor gene is the most frequently mutated gene in human cancer and analyses of these alterations have fueled basic and clinical research, leading in turn to a number of novel therapeutics currently in phase III trials (11).
TP53 Databases and Repositories
Although multiple TP53 LSDBs have been created, only two, UMD_TP53 (Universal Mutation_Database, developed by the present team) and IARC, count 30 years of TP53 mutation analyses in various types of cancer ( Table 1 ) (12, 13). Both have been regularly updated with both TP53 variants and new tools to classify them. The IARC database was updated for the last time in 2019 and is currently awaiting transfer to a new host. The next update to the UMD_TP53 will be performed in March 2022. It will bring a new innovative system to classify TP53 variant pathogenicity and a new version of Seshat to analyze variants (14).
Table 1.
TP53 mutation databases.
| UMD1 | IARC2 | LOVD3 | COSMIC4 | TCGA5 | ICGC6 | MSKSCC7 | GENIE 8 | |
|---|---|---|---|---|---|---|---|---|
| LSDB | LSDB | LSDB | CGD | CGD | CGD | CGD | CGD | |
| Version | 2021R1 | R20, July 2019 | TP53:210617 | v94 | NA | v28 | V10 | V10 |
| Creation date | 1991 | 1991 | 2013 | 2004 | 2008 | 2013 | 2016 | 2016 |
| Last update | 2021 | 2019 | Jun-21 | May-21 | Jun-21 | Mar-21 | Jun-21 | Jun-21 |
| Number of entries | 170,428 | 29,891 | 676 9 | 47,788 | 4,250 | 6557 | 3,249 | 4,813 |
| Unique variant | 8,046 | 4,526 | 400 | 5,705 | 1,961 | 1031 | 11,30 | |
| Cell lines data | Yes | Yes | No | Yes | No | No | No | No |
| Curated publications | 6,704 | 2,273 | 6 | 4,129 | 32 studies | 86 projects | NR | NR |
| Online search | Yes | Yes | No | Yes | Yes | Yes | No | Yes* |
| Publication warning 10 | Yes | No | No | No | No | No | No | No |
| Sex/Age/Ethnicity | No | Yes | No | Partial | Yes | Yes | Yes | Yes |
| Curation for duplicate publications 11 | Yes | Unknown | Unknown | Unknown | NR | NR | NR | NR |
| Sample duplications | No | No | No | Yes | NR | NR | NR | NR |
| SNP curation | Yes | Partial | No | Partial | Partial | Partial | Partial | Partial |
| Availability of functional data | Yes | Yes | No | No | No | No | No | No |
| Availability of predictive data | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes |
| ACMG criteria | Yes | No | No | No | No | No | No | No |
| Data accuracy | Yes | Yes | unknown | Yes | Yes | Yes | Yes | Yes |
| Germline mutation | Yes | Yes | Yes | Yes | No | No | No | No |
| Familial data | No | Yes | No | No | No | No | No | No |
| Availability for Download | Yes | Yes | Yes | Yes | Yes | No* | Yes | Yes |
| Submission for analysis | No | No | Yes | No | No | No | No | No |
| Current status | Alive | on hold | Unknown | Alive | Alive | Alive | Alive | Alive |
| CLL publications/cases | 179 | 31 | 0 | 412 | 0 | 0 | 6 CLL cases | 235 CLL cases |
| Number of TP53 variants in CLL | 4,698 | 187 | 0 | 40 | 0 | 0 | 0 | 13 |
3 https://databases.lovd.nl/shared/genes/TP53; LOVD database includes mostly non-pathogenic SNPs reported in population studies.
5Only the 32 PAN cancer studies (10,967 samples) are included here.
7 https://www.synapse.org/#!Synapse:syn7222066/wiki/405659; MSKSCC data were extracted from GENIE V10.0.
8All GENIE data except MSKSCC study.
9LOVD database includes mostly non-pathogenic SNPs reported in population studies.
10Manuscript known to includes spurious data are flagged.
11Multiple publications report genetic information for the same patient.
*Only via https://genie.cbioportal.org/.
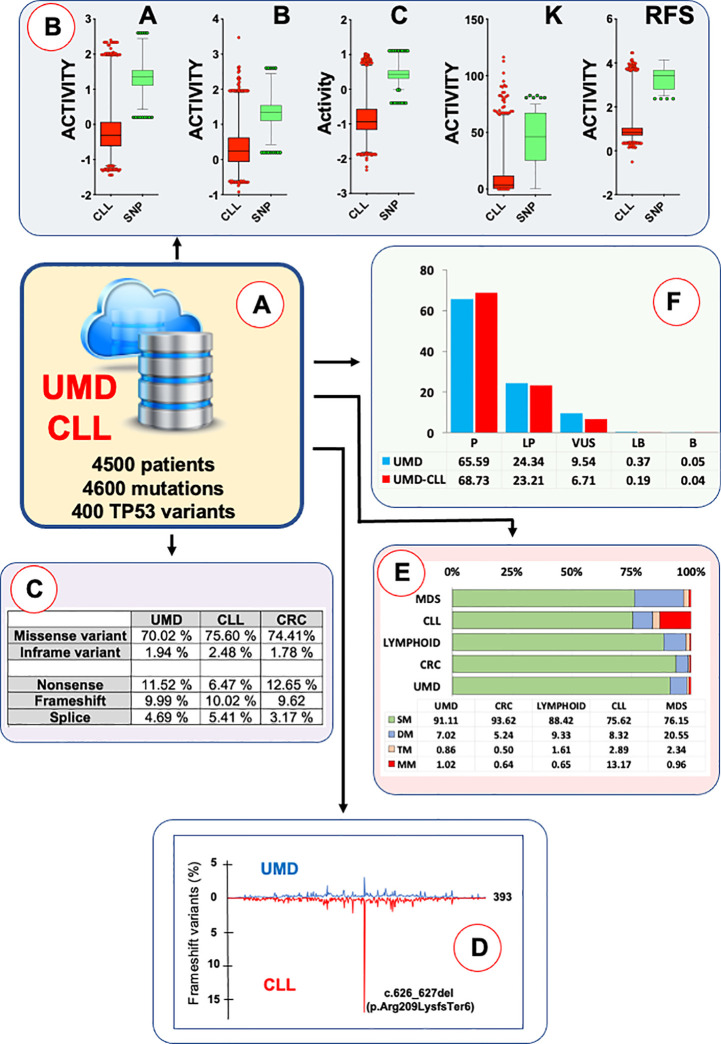
The number of CLL-related TP53 mutations in the various databases is quite low except in UMD_TP53 ( Table 1 ). Because of the clinical importance of TP53 mutations in CLL, a curated subset for that pathology, called UMD_CLL, has been added to the UMD_TP53 database ( Figure 3A ). The latest version of UMD_CLL includes 4,698 mutations, corresponding to 3,419 samples, as patients with multiple TP53 mutations are frequent in this disease. The characteristics of these variants are discussed in the following sections of this review.
Figure 3.
The UMD_CLL database. (A) TP53 mutations from CLL patients included in UMD_TP53 have been manually curated to correct for study duplication. For patients analyzed via Sanger in the nineties and via NGS more recently, only the more recent data were kept in the database as the sensitivity of NGS uncovered less frequent variants. (B) The UMD_CLL database includes three independent sets of functional data used to assess the loss of function of more than 10,000 TP53 variants: A, B and C, data from Giacomelli et al. in mammalian cells; RFS, data from Kotler et al. in mammalian cells; K, data from Kato et al. in yeast cells. Correlation analysis and multidimensional scaling showed excellent agreement between these three sets of data (19). Each dataset has been used to compare the TP53 variants from UMD_CLL (red) to benign TP53 SNPs (green). (C) The landscape of TP53 variants in CLL is similar to that of other types of cancer, with 78% of tumors expressing a mutant TP53 (missense and in-frame variants) and 22% null variants (splice, nonsense and frameshift mutations); (D) Analysis of the distribution of TP53 variants in TP53 protein from CLL patients showed several unusual features, such as a frameshift mutation in codon 209. See text for more details. (E) At least 25% of CLL patients carry at least two pathogenic TP53 variants, and up to 13% carry more than four. This situation is shared only with myelodysplastic syndrome, where up to 20% of patients show two TP53 variants. As half of the CLL data in UMD_TP53 originated from Sanger analyses, it is likely that CLL intratumor heterogeneity is underestimated. (F) All TP53 variants from UMD_TP53 have been classified according to ACMG criteria. For this purpose, all newly discovered, rare, benign SNPs misidentified as pathogenic mutations have been removed from the database.
As early as 2005, in collaboration with C. Ishioka’s group, UMD_TP53 was updated with TP53 functional data to improve the curation of the database and develop the first tools to assess TP53 variant loss of function (LOF) (15, 16). These tools have shown tremendous value for distinguishing true oncogenic TP53 variants from passenger or artifactual mutations. Data from two recent large-scale studies analyzing TP53 LOF via multiple assays in mammalian cells have also been included in UMD_TP53 to refine TP53 variant classification (17–19). Version 1 of Seshat was released in 2018. Seshat is a web service for annotating TP53 information derived from sequencing data. It allows the use of mutation annotation format (MAF) or variant call format (VCF) files. Seshat performs accurate variant annotations using the nomenclature of the Human Genome Variation Society and the stable TP53 genomic reference provided by Locus Reference Genomic (14).
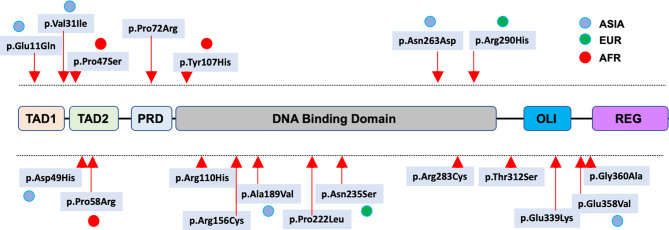
Several single nucleotide polymorphisms (SNPs) in the coding region of the TP53 gene have been identified and extensively characterized. Among the missense SNPs, rs1042522 (p.Pro72Arg) is common in all populations across the globe. Contrastingly, rs1800371 (p.Pro47Ser) has been shown to be specific to the African population (20). Both SNPs are included in ClinVar and considered benign according to American College of Medical Genetics and Genomics (ACMG) criteria. In a recent survey, new TP53 missense SNPs, including five variants specific to the Asian population, were identified and characterized ( Figure 2 ) (21). None of these variants were found to display LOF compared to the normal TP53 gene ( Figure 3B ) and they are now defined as bona fide benign SNPs (21). In UMD_TP53, these variants are specifically flagged as germline SNPs. However, other LSDBs and CGDs define several of them as somatic and potentially pathogenic variants.
Figure 2.
Distribution of benign missense TP53 SNPs in the p53 protein. SNPs specific for an ethnic population are indicated by colored dots.
The three major CGDs (ICGC, TCGA and GENIE) include data from both whole exome and whole genome sequencing of multiple tumors ( Table 1 ). CGDs list fewer TP53 variants than LSDBs do. However, the former are able to show the full pattern of mutations in a single tumor, which enables analyses that are not possible with the latter. For example, CGDs enable the identification of mutual exclusivity of genomic alterations to identify genes belonging to a same functional pathway, as they do not mutate simultaneously in a same patient (22).
Shaping the Landscape of TP53 Mutations in CLL
Although most DNA damage resulting from endogenous and/or exogenous insults is successfully managed by the various DNA repair mechanisms, some does escape those processes and transform into stable mutations. Of these latter, only a few will target cancer genes and thus confer a growth advantage (driver mutations). The remaining mutations will be co-selected during the neoplastic process (passenger mutations). The number of driver mutations is very low (less than 20). However, that of passenger mutations is several orders of magnitude higher, ranging from 0.8 substitutions per megabase for hematological neoplasms such as CLL to 9 or 11 for lung cancer or melanoma respectively (23).
As a result, due to the specificity of the damage caused by such insults and the specific repair mechanisms used by the cell to correct the damage, mutagenic processes generate characteristic point mutation rate spectra, which are called mutational signatures. These signatures point to the mutagenic processes active in a tumor and reveal the high tissue specificity of these mutagenic mechanisms. For these analyses, passenger mutations are preferred as they are not subject to any selective process. In contrast, mutations in driver genes are highly biased as only those able to drive a cancer phenotype will be selected, whether it is via the LOF of a tumor suppressor gene or the gain of function of an oncogene. For the latter, mutations (predominantly missense variants) are restricted to a few codons in the gene targeting key functional residues. For tumor suppressor genes, mutations (predominantly nonsense or frameshift) will lead to a null phenotype or the synthesis of an inactive truncated protein. The mode of inactivation of TP53 is unique compared to other tumor suppressor genes, with more than 80% of somatic and germline TP53 alterations being missense mutations that lead to the synthesis of a stable mutant protein that accumulates in the nucleus of tumor cells (24). The classification of TP53 as a tumor suppressor gene led to a general belief wherein the loss of TP53 function is the sole mechanism associated with TP53 mutations. In fact, this strong selection to maintain expression of mutants in tumors is known to have a vital role in transformation, including dominant activity (DN) and/or a gain of function (GOF), making TP53 variants oncogenic. The distribution of mutations in the p53 protein is also unique among oncogenes and tumor suppressor genes as nearly all of the protein’s 393 amino acid residues have been the target of at least one mutation in human cancer. Each residue in the core domain (containing the DNA-binding region) has been found to be mutated at least five times in independent tumors, and up to 6,000 times for hotspot mutants.
Nevertheless, the distribution of these mutations, and therefore the landscape of TP53 variants observed in a number of types of cancer, is very heterogenous. This aspect may result from the specificity of the insults that generate the mutations (5-methylcytosine deamination at CpG dinucleotide, UV, tobacco carcinogens or chemotherapy) or from the tissue-specific selection of TP53 variants with a special growth advantage (25). At first glance, an analysis of the 3,914 cases of CLL in UMD_TP53 shows a mutation profile similar to those of other cancers ( Figures 3C, D ), with 76% of missense mutations mostly localized in the DNA-binding domain of TP53.
The unusual feature of TP53 mutation in CLL is the presence of a specific hot spot variant: a deletion of two nucleotides at codon 209 (c.626_627del) leading to premature termination (p.Arg209LysfsTer6) ( Figure 3E ). Frameshift variants are found all along the TP53 gene in every type of cancer, but variant c.626_627del is highly predominant in CLL (15% of frameshift mutations in CLL compared to 1 to 2% in other cancer types). The sequence around codon 209 contains an inverted repeat that could explain its specific mutability. Furthermore, the observation of this variant in both untreated and treated patients indicates that it originates from an unknown endogenous mechanism. Although frameshift variants are usually not expressed due to NMD (nonsense-mediated mRNA decay) and protein instability, a specific selection for a truncated TP53 cannot be formally excluded.
Subclonality of TP53 Mutations
Whole exome and whole genome sequencing have provided new insights into the heterogeneity and evolution of tumors, with, importantly, the detection of a high number of subclones in a single tumor (26, 27). This knowledge on the subclonality of TP53 mutations is likely to have implications for biomarker discovery and/or cancer therapy, particularly in the era of targeted treatments. Furthermore, indications of a relationship between this heterogeneity and clinical outcomes are emerging.
TP53 mutated subclones with variant allele frequencies (VAFs) lower than 10% (range 0.3% to 10%, depending on the study), undetectable by conventional Sanger sequencing, have been reported in multiple studies (28–33). Subclonal TP53 variants and high VAF variants have the same profile, including similar hot spot variants. Longitudinal studies have shown that some of these clones can become more prevalent during the development of the tumor, regardless of whether the patient was treated or not. These small mutated subclones have been shown to be associated with unfavorable prognoses in some studies. However, this issue remains controversial, and there is currently no use of mutated subclones in the clinic. TP53 classifications and the methods and cut-offs used to define low VAF clones must be harmonized to enable consensus.
Another characteristic is the high number of CLL patients with multiple TP53 variants ( Figure 3F ). This feature appears specific to CLL; it has not been observed in other types of cancer (22). Bi-allelic TP53 inactivation could explain two TP53 variants but not a higher number of them (range 3 to 10) (34). This high intratumor heterogeneity has been detected in multiple independent studies and validated by specific analyses such as FASAY (functional analysis of separated alleles in yeast) and SMRT (single-molecule real-time sequencing) that confirm different allelic locations for these TP53 variants. Like for minor subclones, most TP53 variants identified in tumors bearing multiple TP53 variants are truly pathogenic. The basis of this specific selection for multiple TP53 variants during the course of CLL is currently unknown.
TP53 Mutation Heterogeneity and Pathogenicity
As early as the nineties, it was obvious that TP53 mutant LOF was heterogenous. Variants were classified as “contact” or “structural,” depending on whether the substituted amino-acid acted directly on DNA interactions (p.Arg273His) or caused a general effect on the protein structure (p.Arg175His). Several classifications for variants based on TP53 aspects have been suggested to stratify patients with TP53 mutations but none have reached the clinic due to the high heterogeneity of the variants and the specificity of the variants among cancer types. A number of predictive tools have been developed, exploiting such information as sequence phylogenetic conservation, amino acid physicochemical properties, functional domains and structural attributes. Commonly used variant effect prediction methods include SIFT (35, 36), PolyPhen (37), GERP++ (38), Condel (39), CADD (40), fathmm (41), MutationTaster (42), MutationAssessor (43), GESPA (44) and, more recently, REVEL (45) and ENVISION (46). Several of these methods, such as fathmm, Condel, CADD and REVEL, integrate data from multiple tools to improve classification accuracy. Recent methods have used machine learning processes. Their training and validation were conducted using datasets of classified variants taken from either pathogenic (COSMIC, TCGA, GENIE, HGMD) or benign (dbSNP, gnomAD or Clinvar) variant databases. Nonetheless, for TP53 and other genes, these various classifiers have heterogenous outcomes and no consensus for their use has been reached. GENIE uses SIFT and PolyPhen, whereas TCGA uses SIFT, PolyPhen and MutationAssessor, and COSMIC uses fathmm. When employing predicting methods based on phylogenetic conservation, tools based on amino acid physicochemical properties such as SIFT or PolyPhen should be used with great precaution as the relation of the deleteriousness of the protein predicted by these tools and any association with disease is far from being straightforward.
To solve this issue, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) have published standards and guidelines for the interpretation of sequence variants (47). These guidelines describe a proposition for classifying variants as “pathogenic,” “likely pathogenic”, “uncertain significance”, “likely benign” or “benign” according to a series of criteria with levels of evidence defined as “very strong”, “strong”, “moderate” or “supporting”. They have been widely adopted by clinical laboratories around the world. However, these recommendations were primarily designed for constitutional variants. Thus, their use for somatic variants requires some adjustment (48). The two main criteria used for the levels of evidence were based on population (BA1, BS1 and BS2) and functional (BS3) data, which are now fully available in UMD_TP53.
One of the main advantages for TP53 over other tumor suppressor genes is the availability of a range of functional data for all possible missense mutations occurring in the coding region for the large isoform of the protein. Data from three independent large-scale saturation mutagenesis screening studies carried out in different settings (yeast or mammalian) and with different readouts (transcription, growth arrest or apoptosis) are currently available (17, 18, 49). A correlation analysis showed excellent agreement between LOF for the protein and the occurrence of these variants in different cancer types, making this criterion suitable for defining PS3 for ACMG classification ( Figure 3B ) (19). An analysis of the UMD_CLL database indicates that 91.9% of the TP53 variants identified in CLL, whether clonal or subclonal, are classified as pathogenic and 6.7% as VUS ( Figure 3F ).
A Snapshot of TP53 Mutation Status in CLL
TP53 status in tumors is complex as multiple mechanisms can impair TP53 tumor suppression pathways. Furthermore, it is quite likely that cancer specificity plays an important role in this process due to the large diversity of TP53 function and regulation among the various tissues. Although MDM2 expression is upregulated in numerous cancers, resulting in a loss of p53-dependent activities, its frequency in CLL is quite low. Other mechanisms, such as the dysregulation of the microRNA network that controls TP53, are also possible but their importance in CLL needs further investigation.
In contrast, CGDs have made it possible to identify the co-occurrence or mutual exclusivity of specific genetic events. In the former, alterations of certain combinations of genes tend to co-exist in a same tumor, whereas in the latter, mostly only one out of a group of genes is altered. Individual alterations targeting similar biological processes are believed to be mutually redundant, with one alteration being sufficient to deregulate the affected process. Identifying mutual exclusivity can therefore help to identify unknown functional interactions. In CLL, this type of analysis is averted by the important genetic heterogeneity of the tumors, showing multiple subclones with different genetic alterations. Because NGS gives a global picture of these events, defining whether or not they occur in the same cells is difficult. This problem will likely be resolved once sufficient single-cell genomic analyses have been performed.
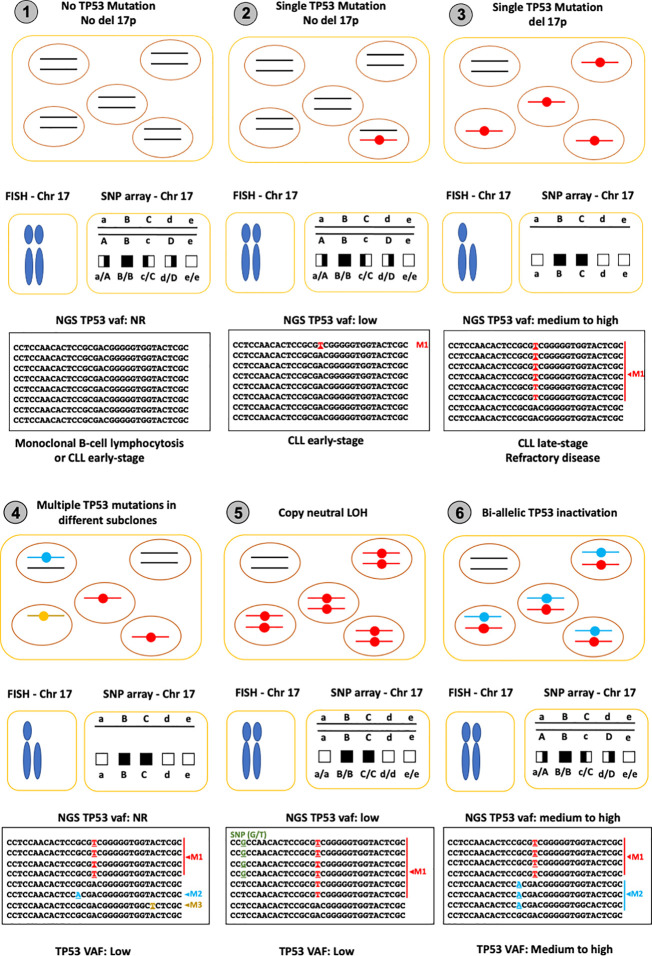
As shown in Figure 4 , TP53 status in CLL can be very heterogeneous, as the prevalence of TP53 abnormalities, including 17p deletion and TP53 mutations, varies across the different phases of the disease (26). Furthermore, the subclonal heterogeneity of the tumors can sometimes be misleading. Indeed, bulk NGS analyses generate an averaged picture of a given population of cells, which may result in an underestimation of their true heterogeneity. Nevertheless, a general picture emerges from the various studies. TP53 mutations are not the prime event in CLL. In the early phase of the disease (stage 0), TP53 mutations appear to be either absent or infrequent, but this issue needs to be carefully reevaluated using NGS assays validated for limits of detection (LOD) ranging from 0.05% to 1% ( Figure 4 , panel 1). Furthermore, because these variants are usually not associated with a deletion of the second allele, FISH or SNP arrays are not suitable for early detection analyses ( Figure 4 , panel 2). 17p deletion and complex karyotypes occur during disease progression, leading to the conventional view of CLL with a single TP53 mutation associated with TP53 loss of heterozygosity (LOH) ( Figure 4 , panel 3). Targeted, high-depth, NGS of TP53 coupled with an adequate pipeline able to reach a LOD of at least 1% has led to the discovery of multiple subclones expressing different pathogenic TP53 variants ( Figure 4 , panel 4). Why CLL has such a propensity for TP53 mutations is currently unknown. However, it is clear that CLL depends on signals from the microenvironment and that its cells cycle between lymphoid tissue sites such as lymph nodes and peripheral blood. It is possible that the strong proliferation signals provided by the microenvironment in lymph nodes require a loss of several anti-proliferative signals such as that provided by TP53.
Figure 4.
TP53 status in CLL patients, a snapshot. The top panel displays a schematic view of the tumor with the two TP53 alleles. The middle panel shows cytogenetic analysis performed by FISH (left) or by SNP arrays (right). The lower panel displays an example of the read alignments from NGS. 1: No TP53 mutation: In monoclonal B-cell lymphocytosis, TP53 mutation and 17p deletion are very rare, leading to negative results for both FISH and genetic analysis. 2: TP53 mutation without LOH: In early stages of CLL, the frequency of TP53 mutation is low (less than 10%) with many cases showing no LOH. Sensitive sequencing analysis with NGS is able to identify low VAF TP53 variants (variant M1 in the lower panel). 3: TP53 mutation with LOH: In late-stage or relapsing disease, TP53 mutations associated with 17p deletion can be found in 30 to 50% of CLL patients. In the majority of cases, VAF is greater than 50% due to the loss of the second allele. This situation is commonly seen in CLL. 4: Multiple TP53 and LOH: in both early and late-stage disease, FASAY (functional analysis of separated alleles of p53 on yeast) or SMRT (single-molecule, real-time sequencing) has demonstrated a high level of intratumoral heterogeneity in CLL with the presence of multiple independent subclones expressing different pathogenic TP53 variants (M1, M2 and M3 in the lower panel). Although 17p deletion is often observed in these patients, it is difficult to determine if subclones expressing different TP53 variants are associated with it, and even more so if the VAF of the variant is low. 5: Copy neutral LOH: Following the initial mutational inactivation of one allele, the remaining wild-type allele is deleted concurrently with the duplication of the mutated allele, leading to copy neutral LOH (cnLOH). Detecting cnLOH is difficult and thus the frequency of the event is currently unknown. Without SNP array analysis and if the VAF of the variant is lower than 50%, this situation can be misidentified as a tumor without LOH. Tumors with VAF greater than 50% without obvious 17p deletion should be checked for cnLOH. 6: Bi-allelic mutations: Inactivation of the TP53 gene via different mutations in the two alleles is possible but difficult to distinguish from intratumoral heterogeneity. Although this situation is often described as plausible in many reviews, it has never been formally identified, as only single-cell sequencing would be able to validate bi-allelic TP53 inactivation.
Another genetic configuration observed in CLL is copy neutral LOH (cnLOH), with the same mutation in both alleles of a given cell ( Figure 4 , panel 5). This genetic event is attributed to mitotic recombination in tumor cells where the wild-type allele is replaced by the mutant allele leading to a large region of homozygosity that can be detected early by SNP-arrays. Inversely, this situation cannot be detected by any karyotyping analyses and could be misinterpreted as heterozygous mutation if the sequencing VAF is below 50%. The situation described in panel 6 of Figure 4 (two different mutations in the two alleles of a single cell) is theoretically possible and often described as a potential status in CLL and other tumors. However, such a situation appears to be very uncommon and has never been formally observed in CLL.
TP53 Clinical Considerations
TP53 mutations have been described in CLL since the early 1990s (50, 51). An association between TP53 mutations, drug resistance and poor clinical outcomes was first demonstrated in 1993 by El Rouby et al. and thereafter confirmed in further studies (52–54). In 2000, using FISH analysis for multiple chromosomal markers, Dohner et al. showed that 17p deletion, where the TP53 gene is located, was an independent predictor of disease progression and survival (55). Using either 17p deletion or TP53 mutation as a biomarker, subsequent studies confirmed this finding and resulted in TP53’s classification as a well-established prognostic marker furthermore able to provide pertinent information for establishing an appropriate course of treatment for patients.
The therapeutic approach for CLL carrying TP53 mutations will be addressed in detail in another review in this series. There are, however, some biological aspects and some issues related to methodological/technical details that can be discussed here.
There is no longer a place for chemo-based regimens in patients with CLLs presenting TP53 mutations. The introduction of novel targeted agents has greatly altered the clinical course of these patients, who now benefit from responses that were never observed during the chemo(immunotherapy) era (56). That said, even with the use of novel agents, CLL remains incurable. Patients with TP53 disruption (TP53 mutation/17p deletion) exhibit worse clinical outcomes compared to those without it, indicating that the management of the former is still an unmet challenge (57). This is more evident for relapsed/refractory (R/R) CLL (58–62) where data on front-line therapies are still scarce because the follow-up of clinical trials at the front line is still short (63, 64). Moreover, little is known on R/R CLL response to novel agents, a setting wherein TP53 disruption seems to be an unfavorable prognostic/predictive factor (65, 66).
Another parameter that needs to be taken into consideration is that in both clinical trials and clinical care, TP53 disruptions are considered equal whatever their nature. There is thus no differentiation being made between patients with monoallelic or bi-allelic aberrations, despite data suggesting that the latter may exhibit more aggressive clinical courses (33, 60). Similarly, the number or type of mutations receives no consideration as a specific clinical feature either.
Moreover, in untreated CLL with TP53 mutations, there is a subset of patients with indolent clinical courses, which suggests that other disease- and/or patient-related parameters may alter the impact of TP53 disruption (67, 68). Also, genomic instabilities at the chromosomal and molecular level, as well as the immunogenetic features of the clonotypic B-cell receptor, namely the somatic hypermutation status of the immunoglobulin heavy variable gene, have been proposed as factors that may aggravate or alleviate the impact of TP53 mutations (69–72).
Finally, there is a discrepancy regarding the threshold for reporting TP53 mutations detected by NGS in the clinical setting versus the official guidelines that merits discussion. According to the latest versions of these latter, only mutations with VAF ≥10% should be reported and used for directing treatment choice (73). This conservative approach within the official guidelines is based mainly on the fact that the clinical impact of small TP53 clones, especially those below 5%, has not been demonstrated to date in prospective clinical trials. However, diagnostic laboratories are becoming more experienced in NGS data output management, and resultantly, clones down to 2-5% are being reported in the clinical setting and, in the majority of cases, taken into consideration for clinical decision-making.
Remaining Challenges and Perspectives
Compared to other cancer types, the clinical value of TP53 status in CLL has always been uncontested and it is now a required biomarker for patient stratification. It is therefore essential that TP53 analyses be performed in a standardized manner to provide consistent data across the various clinical laboratories. For this purpose, the TP53 Network of the European Research Initiative on Chronic Lymphocytic Leukemia (ERIC) had released a first recommendation in 2012 and updated it recently to take into account the emergence of NGS (73, 74). Nevertheless, considering the quick evolution of methodologies and the discovery of the subclonal heterogeneity of TP53 variants with low VAF clones, a new consensus must be reached for the controversial issue of the limit of detection used to report TP53 variants in clinical laboratories. Although conventional Sanger sequencing has been widely used in the past, it is now clear that NGS-based analysis should become mandatory for the clinical detection of low VAF clones. The current situation is unclear, with several studies suggesting that patients with low VAF TP53 clones have the same clinical prognosis as patients with high VAF ones, and other studies unable to confirm that finding. Reaching a consensus to define a robust, clinically justified LOD will be essential for improving patient stratification. Furthermore, despite their relative infrequency, it will be important to evaluate the real incidence of multiple TP53 subclonal mutations using adequate methodologies as well as their evolution during the course of disease and with different types of treatment. Whether or not this reservoir of heterogenous oncogenic TP53 variants is an essential component of the plasticity of CLL remains to be addressed.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
TS is supported by Cancéropole Ille de France (convention n°2019-1-EMERG-22-INSERM 6-1). PG is supported by the Lion’s Cancer Research Foundation, Uppsala and the ALF funds. Figures were created with BioRender.com.
References
- 1. Ingram VM. A Specific Chemical Difference Between the Globins of Normal Human and Sickle-Cell Anaemia Haemoglobin. Nature (1956) 178:792–4. doi: 10.1038/178792a0 [DOI] [PubMed] [Google Scholar]
- 2. Collins FS. Positional Cloning Moves From Perditional to Traditional. Nat Genet (1995) 9:347–50. doi: 10.1038/ng0495-347 [DOI] [PubMed] [Google Scholar]
- 3. Horaitis O, Cotton RG. The Challenge of Documenting Mutation Across the Genome: The Human Genome Variation Society Approach. Hum Mutat (2004) 23:447–52. doi: 10.1002/humu.20038 [DOI] [PubMed] [Google Scholar]
- 4. Auerbach AD, Burn J, Cassiman JJ, Claustres M, Cotton RG, Cutting G, et al. Mutation (Variation) Databases and Registries: A Rationale for Coordination of Efforts. Nat Rev Genet (2011) 12:881; discussion 881. doi: 10.1038/nrg3011-c1 [DOI] [PubMed] [Google Scholar]
- 5. Dalgleish R. LSDBs and How They Have Evolved. Hum Mutat (2016) 37:532–9. doi: 10.1002/humu.22979 [DOI] [PubMed] [Google Scholar]
- 6. Soussi T. Locus-Specific Databases in Cancer: What Future in a Post-Genomic Era? The TP53 LSDB Paradigm. Hum Mutat (2014) 35:643–53. doi: 10.1002/humu.22518 [DOI] [PubMed] [Google Scholar]
- 7. Pinard A, Miltgen M, Blanchard A, Mathieu H, Desvignes JP, Salgado D, et al. Actionable Genes, Core Databases, and Locus-Specific Databases. Hum Mutat (2016) 37:1299–307. doi: 10.1002/humu.23112 [DOI] [PubMed] [Google Scholar]
- 8. Zhang J, Baran J, Cros A, Guberman JM, Haider S, Hsu J, et al. International Cancer Genome Consortium Data Portal–A One-Stop Shop for Cancer Genomics Data. Database (Oxford) (2011) 2011:bar026. doi: 10.1093/database/bar026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res (2018) 47:D941–7. doi: 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Network CGAR, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat Genet (2013) 45:1113–20. doi: 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheok CF, Verma CS, Baselga J, Lane DP. Translating P53 Into the Clinic. Nat Rev Clin Oncol (2011) 8:25–37. doi: 10.1038/nrclinonc.2010.174 [DOI] [PubMed] [Google Scholar]
- 12. Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 Variations in Human Cancers: New Lessons From the IARC TP53 Database and Genomics Data. Hum Mutat (2016) 37:865–76. doi: 10.1002/humu.23035 [DOI] [PubMed] [Google Scholar]
- 13. Leroy B, Ballinger ML, Baran-Marszak F, Bond GL, Braithwaite A, Concin N, et al. Recommended Guidelines for Validation, Quality Control, and Reporting of TP53 Variants in Clinical Practice. Cancer Res (2017) 77:1250–60. doi: 10.1158/0008-5472.CAN-16-2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tikkanen T, Leroy B, Fournier JL, Risques RA, Malcikova J, Soussi T. Seshat: A Web Service for Accurate Annotation, Validation, and Analysis of TP53 Variants Generated by Conventional and Next-Generation Sequencing. Hum Mutat (2018) 39:925–33. doi: 10.1002/humu.23543 [DOI] [PubMed] [Google Scholar]
- 15. Soussi T, Kato S, Levy PP, Ishioka C. Reassessment of the TP53 Mutation Database in Human Disease by Data Mining With a Library of TP53 Missense Mutations. Hum Mutat (2005) 25:6–17. doi: 10.1002/humu.20114 [DOI] [PubMed] [Google Scholar]
- 16. Soussi T, Asselain B, Hamroun D, Kato S, Ishioka C, Claustres M, et al. Meta-Analysis of the P53 Mutation Database for Mutant P53 Biological Activity Reveals a Methodologic Bias in Mutation Detection. Clin Cancer Res (2006) 12:62–9. doi: 10.1158/1078-0432.CCR-05-0413 [DOI] [PubMed] [Google Scholar]
- 17. Kotler E, Shani O, Goldfeld G, Lotan-Pompan M, Tarcic O, Gershoni A, et al. A Systematic P53 Mutation Library Links Differential Functional Impact to Cancer Mutation Pattern and Evolutionary Conservation. Mol Cell (2018) 71:178–190.e8. doi: 10.1016/j.molcel.2018.06.012 [DOI] [PubMed] [Google Scholar]
- 18. Giacomelli AO, Yang X, Lintner RE, McFarland JM, Duby M, Kim J, et al. Mutational Processes Shape the Landscape of TP53 Mutations in Human Cancer. Nat Genet (2018) 50:1381–7. doi: 10.1038/s41588-018-0204-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carbonnier V, Leroy B, Rosenberg S, Soussi T. Comprehensive Assessment of TP53 Loss of Function Using Multiple Combinatorial Mutagenesis Libraries. Sci Rep (2020) 10:20368. doi: 10.1038/s41598-020-74892-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X, Dumont P, Della Pietra A, Shetler C, Murphy ME. The Codon 47 Polymorphism in P53 Is Functionally Significant. J Biol Chem (2005) 280:24245–51. doi: 10.1074/jbc.M414637200 [DOI] [PubMed] [Google Scholar]
- 21. Doffe F, Carbonnier V, Tissier M, Leroy B, Martins I, Mattsson JSM, et al. Identification and Functional Characterization of New Missense SNPs in the Coding Region of the TP53 Gene. Cell Death Differ (2021) 28:1477–92. doi: 10.1038/s41418-020-00672-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep (2019) 28:1370–84.e5. doi: 10.1016/j.celrep.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ding L, Bailey MH, Porta-Pardo E, Thorsson V, Colaprico A, Bertrand D, et al. Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell (2018) 173:305–320.e10. doi: 10.1016/j.cell.2018.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soussi T, Wiman KG. TP53: An Oncogene in Disguise. Cell Death Differ (2015) 22:1239–49. doi: 10.1038/cdd.2015.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leroy B, Anderson M, Soussi T. TP53 Mutations in Human Cancer: Database Reassessment and Prospects for the Next Decade. Hum Mutat (2014) 35:672–88. doi: 10.1002/humu.22552 [DOI] [PubMed] [Google Scholar]
- 26. Lazarian G, Guièze R, Wu CJ. Clinical Implications of Novel Genomic Discoveries in Chronic Lymphocytic Leukemia. J Clin Oncol (2017) 35:984–93. doi: 10.1200/JCO.2016.71.0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fabbri G, Dalla-Favera R. The Molecular Pathogenesis of Chronic Lymphocytic Leukaemia. Nat Rev Cancer (2016) 16:145–62. doi: 10.1038/nrc.2016.8 [DOI] [PubMed] [Google Scholar]
- 28. Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Famà R, et al. Clinical Impact of Small TP53 Mutated Subclones in Chronic Lymphocytic Leukemia. Blood (2014) 123:2139–47. doi: 10.1182/blood-2013-11-539726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bomben R, Rossi FM, Vit F, Bittolo T, D’Agaro T, Zucchetto A, et al. TP53 Mutations With Low Variant Allele Frequency Predict Short Survival in Chronic Lymphocytic Leukemia. Clin Cancer Res (2021) 27:5566–75. doi: 10.1158/1078-0432.CCR-21-0701 [DOI] [PubMed] [Google Scholar]
- 30. Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M, et al. Clinical Impact of Clonal and Subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM Mutations in Chronic Lymphocytic Leukemia. Blood (2016) 127:2122–30. doi: 10.1182/blood-2015-07-659144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brieghel C, Kinalis S, Yde CW, Schmidt AY, Jønson L, Andersen MA, et al. Deep Targeted Sequencing of TP53 in Chronic Lymphocytic Leukemia: Clinical Impact at Diagnosis and at Time of Treatment. Haematologica (2019) 104:789–96. doi: 10.3324/haematol.2018.195818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stengel A, Kern W, Haferlach T, Meggendorfer M, Fasan A, Haferlach C. The Impact of TP53 Mutations and TP53 Deletions on Survival Varies Between AML, ALL, MDS and CLL: An Analysis of 3307 Cases. Leukemia (2017) 31:705–11. doi: 10.1038/leu.2016.263 [DOI] [PubMed] [Google Scholar]
- 33. Blakemore SJ, Clifford R, Parker H, Antoniou P, Stec-Dziedzic E, Larrayoz M, et al. Clinical Significance of TP53, BIRC3, ATM and MAPK-ERK Genes in Chronic Lymphocytic Leukaemia: Data From the Randomised UK LRF CLL4 Trial. Leukemia (2020) 34:1760–74. doi: 10.1038/s41375-020-0723-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malcikova J, Stano-Kozubik K, Tichy B, Kantorova B, Pavlova S, Tom N, et al. Detailed Analysis of Therapy-Driven Clonal Evolution of TP53 Mutations in Chronic Lymphocytic Leukemia. Leukemia (2015) 29:877–85. doi: 10.1038/leu.2014.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kumar P, Henikoff S, Ng PC. Predicting the Effects of Coding non-Synonymous Variants on Protein Function Using the SIFT Algorithm. Nat Protoc (2009) 4:1073–81. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 36. Ng PC, Henikoff S. Predicting Deleterious Amino Acid Substitutions. Genome Res (2001) 11:863–74. doi: 10.1101/gr.176601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A Method and Server for Predicting Damaging Missense Mutations. Nat Methods (2010) 7:248–9. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a High Fraction of the Human Genome to be Under Selective Constraint Using GERP++. PloS Comput Biol (2010) 6:e1001025. doi: 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. González-Pérez A, López-Bigas N. Improving the Assessment of the Outcome of Nonsynonymous SNVs With a Consensus Deleteriousness Score, Condel. Am J Hum Genet (2011) 88:440–9. doi: 10.1016/j.ajhg.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A General Framework for Estimating the Relative Pathogenicity of Human Genetic Variants. Nat Genet (2014) 46:310–5. doi: 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day IN, et al. An Integrative Approach to Predicting the Functional Effects of Non-Coding and Coding Sequence Variation. Bioinformatics (2015) 31:1536–43. doi: 10.1093/bioinformatics/btv009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster Evaluates Disease-Causing Potential of Sequence Alterations. Nat Methods (2010) 7:575–6. doi: 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 43. Reva B, Antipin Y, Sander C. Predicting the Functional Impact of Protein Mutations: Application to Cancer Genomics. Nucleic Acids Res (2011) 39:e118. doi: 10.1093/nar/gkr407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Khurana JK, Reeder JE, Shrimpton AE, Thakar J. GESPA: Classifying nsSNPs to Predict Disease Association. BMC Bioinf (2015) 16:228. doi: 10.1186/s12859-015-0673-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet (2016) 99:877–85. doi: 10.1016/j.ajhg.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gray VE, Hause RJ, Luebeck J, Shendure J, Fowler DM. Quantitative Missense Variant Effect Prediction Using Large-Scale Mutagenesis Data. Cell Syst (2018) 6:116–124.e3. doi: 10.1016/j.cels.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med (2015) 17:405–24. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn (2017) 19:4–23. doi: 10.1016/j.jmoldx.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, et al. Understanding the Function-Structure and Function-Mutation Relationships of P53 Tumor Suppressor Protein by High-Resolution Missense Mutation Analysis. Proc Natl Acad Sci USA (2003) 100:8424–9. doi: 10.1073/pnas.1431692100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gaidano G, Ballerini P, Gong JZ, Inghirami G, Neri A, Newcomb EW, et al. P53 Mutations in Human Lymphoid Malignancies: Association With Burkitt Lymphoma and Chronic Lymphocytic Leukemia. Proc Natl Acad Sci USA (1991) 88:5413–7. doi: 10.1073/pnas.88.12.5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fenaux P, Preudhomme C, Lai JL, Quiquandon I, Jonveaux P, Vanrumbeke M, et al. Mutations of the P53 Gene in B-Cell Chronic Lymphocytic Leukemia: A Report on 39 Cases With Cytogenetic Analysis. Leukemia (1992) 6:246–50. [PubMed] [Google Scholar]
- 52. el Rouby S, Thomas A, Costin D, Rosenberg CR, Potmesil M, Silber R, et al. P53 Gene Mutation in B-Cell Chronic Lymphocytic Leukemia Is Associated With Drug Resistance and is Independent of MDR1/MDR3 Gene Expression. Blood (1993) 82:3452–9. doi: 10.1182/blood.V82.11.3452.3452 [DOI] [PubMed] [Google Scholar]
- 53. Dohner H, Fischer K, Bentz M, Hansen K, Benner A, Cabot G, et al. P53 Gene Deletion Predicts for Poor Survival and Non-Response to Therapy With Purine Analogs in Chronic B-Cell Leukemias. Blood (1995) 85:1580–9. doi: 10.1182/blood.V85.6.1580.bloodjournal8561580 [DOI] [PubMed] [Google Scholar]
- 54. Wattel E, Preudhomme C, Hecquet B, Vanrumbeke M, Quesnel B, Dervite I, et al. P53 Mutations Are Associated With Resistance to Chemotherapy and Short Survival in Hematologic Malignancies. Blood (1994) 84:3148–57. doi: 10.1182/blood.V84.9.3148.3148 [DOI] [PubMed] [Google Scholar]
- 55. Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic Aberrations and Survival in Chronic Lymphocytic Leukemia. N Engl J Med (2000) 343:1910–6. doi: 10.1056/NEJM200012283432602 [DOI] [PubMed] [Google Scholar]
- 56. Burger JA. Treatment of Chronic Lymphocytic Leukemia. N Engl J Med (2020) 383:460–73. doi: 10.1056/NEJMra1908213 [DOI] [PubMed] [Google Scholar]
- 57. Ding W. The Ongoing Unmet Needs in Chronic Lymphocytic Leukemia: TP53 Disruption, Richter, and Beyond. Hematol Oncol Clin North Am (2021) 35:739–59. doi: 10.1016/j.hoc.2021.04.001 [DOI] [PubMed] [Google Scholar]
- 58. Stilgenbauer S, Eichhorst B, Schetelig J, Hillmen P, Seymour JF, Coutre S, et al. Venetoclax for Patients With Chronic Lymphocytic Leukemia With 17p Deletion: Results From the Full Population of a Phase II Pivotal Trial. J Clin Oncol (2018) 36:1973–80. doi: 10.1200/JCO.2017.76.6840 [DOI] [PubMed] [Google Scholar]
- 59. Stilgenbauer S, Eichhorst B, Schetelig J, Coutre S, Seymour JF, Munir T, et al. Venetoclax in Relapsed or Refractory Chronic Lymphocytic Leukaemia With 17p Deletion: A Multicentre, Open-Label, Phase 2 Study. Lancet Oncol (2016) 17:768–78. doi: 10.1016/S1470-2045(16)30019-5 [DOI] [PubMed] [Google Scholar]
- 60. Brown JR, Hillmen P, O’Brien S, Barrientos JC, Reddy NM, Coutre SE, et al. Extended Follow-Up and Impact of High-Risk Prognostic Factors From the Phase 3 RESONATE Study in Patients With Previously Treated CLL/SLL. Leukemia (2018) 32:83–91. doi: 10.1038/leu.2017.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, et al. Targeting BTK With Ibrutinib in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med (2013) 369:32–42. doi: 10.1056/NEJMoa1215637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum K, et al. Ibrutinib Treatment for First-Line and Relapsed/Refractory Chronic Lymphocytic Leukemia: Final Analysis of the Pivotal Phase Ib/II PCYC-1102 Study. Clin Cancer Res (2020) 26:3918–27. doi: 10.1158/1078-0432.CCR-19-2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fischer K, Al-Sawaf O, Bahlo J, Fink AM, Tandon M, Dixon M, et al. Venetoclax and Obinutuzumab in Patients With CLL and Coexisting Conditions. N Engl J Med (2019) 380:2225–36. doi: 10.1056/NEJMoa1815281 [DOI] [PubMed] [Google Scholar]
- 64. Burger JA, Sivina M, Jain N, Kim E, Kadia T, Estrov Z, et al. Randomized Trial of Ibrutinib vs Ibrutinib Plus Rituximab in Patients With Chronic Lymphocytic Leukemia. Blood (2019) 133:1011–9. doi: 10.1182/blood-2018-10-879429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, et al. Targeting BCL2 With Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med (2016) 374:311–22. doi: 10.1056/NEJMoa1513257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roberts AW, Ma S, Kipps TJ, Coutre SE, Davids MS, Eichhorst B, et al. Efficacy of Venetoclax in Relapsed Chronic Lymphocytic Leukemia Is Influenced by Disease and Response Variables. Blood (2019) 134:111–22. doi: 10.1182/blood.2018882555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Baliakas P, Jeromin S, Iskas M, Puiggros A, Plevova K, Nguyen-Khac F, et al. Cytogenetic Complexity in Chronic Lymphocytic Leukemia: Definitions, Associations, and Clinical Impact. Blood (2019) 133:1205–16. doi: 10.1182/blood-2018-09-873083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Leeksma AC, Baliakas P, Moysiadis T, Puiggros A, Plevova K, Van Der Kevie-Kersemaekers AM, et al. Genomic Arrays Identify High-Risk Chronic Lymphocytic Leukemia With Genomic Complexity: A Multi-Center Study. Haematologica (2021) 106:87–97. doi: 10.3324/haematol.2019.239947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Baliakas P, Moysiadis T, Hadzidimitriou A, Xochelli A, Jeromin S, Agathangelidis A, et al. Tailored Approaches Grounded on Immunogenetic Features for Refined Prognostication in Chronic Lymphocytic Leukemia. Haematologica (2019) 104:360–9. doi: 10.3324/haematol.2018.195032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sutton LA, Hadzidimitriou A, Baliakas P, Agathangelidis A, Langerak AW, Stilgenbauer S, et al. Immunoglobulin Genes in Chronic Lymphocytic Leukemia: Key to Understanding the Disease and Improving Risk Stratification. Haematologica (2017) 102:968–71. doi: 10.3324/haematol.2017.165605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ljungström V, Cortese D, Young E, Pandzic T, Mansouri L, Plevova K, et al. Whole-Exome Sequencing in Relapsing Chronic Lymphocytic Leukemia: Clinical Impact of Recurrent RPS15 Mutations. Blood (2016) 127:1007–16. doi: 10.1182/blood-2015-10-674572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yu L, Kim HT, Kasar S, Benien P, Du W, Hoang K, et al. Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation. Clin Cancer Res (2017) 23:735–45. doi: 10.1158/1078-0432.CCR-16-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Malcikova J, Tausch E, Rossi D, Sutton LA, Soussi T, Zenz T, et al. ERIC Recommendations for TP53 Mutation Analysis in Chronic Lymphocytic Leukemia-Update on Methodological Approaches and Results Interpretation. Leukemia (2018) 32:1070–80. doi: 10.1038/s41375-017-0007-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pospisilova S, Gonzalez D, Malcikova J, Trbusek M, Rossi D, Kater AP, et al. ERIC Recommendations on TP53 Mutation Analysis in Chronic Lymphocytic Leukemia. Leukemia (2012) 26:1458–61. doi: 10.1038/leu.2012.25 [DOI] [PubMed] [Google Scholar]