Abstract
Background
Systematic case identification is critical to improving population health, but widely used diagnosis code–based approaches for conditions like valvular heart disease are inaccurate and lack specificity.
Objective
To develop and validate natural language processing (NLP) algorithms to identify aortic stenosis (AS) cases and associated parameters from semi-structured echocardiogram reports and compare their accuracy to administrative diagnosis codes.
Methods
Using 1003 physician-adjudicated echocardiogram reports from Kaiser Permanente Northern California, a large, integrated healthcare system (>4.5 million members), NLP algorithms were developed and validated to achieve positive and negative predictive values > 95% for identifying AS and associated echocardiographic parameters. Final NLP algorithms were applied to all adult echocardiography reports performed between 2008 and 2018 and compared to ICD-9/10 diagnosis code–based definitions for AS found from 14 days before to 6 months after the procedure date.
Results
A total of 927,884 eligible echocardiograms were identified during the study period among 519,967 patients. Application of the final NLP algorithm classified 104,090 (11.2%) echocardiograms with any AS (mean age 75.2 years, 52% women), with only 67,297 (64.6%) having a diagnosis code for AS between 14 days before and up to 6 months after the associated echocardiogram. Among those without associated diagnosis codes, 19% of patients had hemodynamically significant AS (ie, greater than mild disease).
Conclusion
A validated NLP algorithm applied to a systemwide echocardiography database was substantially more accurate than diagnosis codes for identifying AS. Leveraging machine learning–based approaches on unstructured electronic health record data can facilitate more effective individual and population management than using administrative data alone.
Keywords: Aortic stenosis, Echocardiography, Machine learning, Population health, Quality and outcomes, Valvular heart disease
Introduction
Systematic, population-level tracking and management of chronic diseases is critical to improving individual and population health. However, for many medical conditions, large-scale identification of affected patient populations is not feasible unless patients have been prospectively entered into clinical registries. For example, valvular heart disease is a common condition affecting millions of persons annually,1 with most cases identified incidentally as part of cardiovascular imaging in asymptomatic patients at various stages of disease severity. Furthermore, until patients meet clinical indications for valvular repair or replacement owing to progression of the valvular disease, they typically undergo serial surveillance by echocardiography. Once disease severity becomes moderate or severe, surveillance is critical to ensure patients receive appropriate, timely medical and/or surgical therapy to prevent heart failure and associated complications.
Unfortunately, in the absence of systematic programs to ensure all patients with valvular heart disease are appropriately followed, it is left to individual physicians to track and manage these patients, which can be challenging without adequate resources or follow-up systems in place. To develop such systems, administrative diagnosis codes are often used to support population management and quality reporting, particularly in large healthcare systems or by payor organizations. However, limitations of diagnosis codes are well documented and include inaccuracy (ie, false-positives and false-negatives) when compared against manual review of medical records, lack of detail (eg, disease severity), and variable patterns of coding across physicians, practices, and health systems.2 Newer methods to overcome these challenges include the application of natural language processing (NLP) algorithms, which are sophisticated machine-learning algorithms that can be applied to large volumes of unstructured free text and semi-structured clinical data (eg, physician progress notes or radiology reports).3 NLP methods have been applied to multiple clinical areas involving unstructured text and big data,4, 5, 6, 7, 8 but less often to identify chronic disease cohorts for population management.9,10 Echocardiography data contain detailed clinical information but are generally semi-structured with unstructured free text sections and not feasible to extract manually on a large scale. NLP methods offer an opportunity to capture the needed clinical data from echocardiograms to develop system-level cohorts of valvular heart disease patients to facilitate more effective individual and population management.
We addressed these challenges by developing and validating detailed NLP algorithms to accurately identify patients with aortic stenosis (AS) and associated parameters from echocardiography reports, and compared the accuracy of the NLP algorithm to the use of administrative diagnostic codes to identify patients with AS in a large, diverse, community-based population.
Methods
Study population
The study population was derived from members in Kaiser Permanente Northern California (KPNC), a large, integrated healthcare delivery system currently providing comprehensive inpatient, emergency department, and outpatient care for >4.5 million members in 21 hospitals and >255 outpatient clinics across Northern California. The KPNC membership is highly representative of the surrounding local and statewide population in terms of age, sex, and race/ethnicity.11
This study was approved by the KPNC institutional review board. Waiver of informed consent was obtained owing to the nature of the study.
Study sample
We identified all patients aged ≥18 years who underwent ≥1 echocardiogram performed as part of usual clinical care between January 1, 2008 and December 31, 2018 within KPNC. We excluded patients who were not active health plan members in KPNC at the time of the echocardiogram or who had unknown sex.
Outcomes
The primary outcomes were the accuracy of NLP algorithms for detecting presence of AS, characterizing severity of AS, and identifying other echocardiographic variables. We also examined the concordance of identifying the presence of AS through an NLP algorithm compared to using ICD-9 or ICD-10 diagnostic codes.
Assembly of development and validation samples of echocardiogram reports
Echocardiogram reports in KPNC are semi-structured free text reports written by physicians, with generally 3 sections: (1) Summary/Impression, (2) Findings, and (3) Measurements. As expected, there is significant variability in the language used in echocardiographic reporting across physician readers, depending on use of free text, health system–wide standard text phrases, or personal templates that are allowed within the echocardiogram software application and electronic health record (EHR). All 21 hospitals used the same electronic reporting system, the McKesson Cardiology Echo reporting system, which underwent several upgrades and updates over the study period, and which offered a high degree of flexibility for the reading physician (Supplemental Material).
To develop and validate our NLP algorithms, we created 1 development and 3 distinct validation datasets from echocardiogram reports between 2008 and 2018. Each dataset consisted of approximately 50% echocardiograms with AS as documented by the interpreting cardiologist of the echocardiogram, which was confirmed by manual review of the report by a board-certified cardiologist (M.D.S.). Presence of AS was defined as the interpreting physician’s written assessment within the echocardiogram. The remaining 50% in each dataset included echocardiogram reports without any AS documented by the interpreting reader, also confirmed by manual physician review for the study. To identify candidate echocardiograms for each dataset, studies from patients with relevant ICD-9 or ICD-10 (International Classification of Diseases, Ninth and Tenth Revision) codes for AS were identified (Supplemental Table 1) for review. All echocardiograms selected for each dataset were randomly sampled from across the study period to ensure temporal variation. For the bicuspid aortic valve (BAV) algorithm, separate development and validation datasets were required to enrich the development and validation datasets with BAV patients, given the low general prevalence of BAV. We identified patients with potential BAV based on relevant ICD-9 (746.4) or ICD-10 codes (Q23.1) and a physician reviewer (M.D.S.) confirmed the presence of BAV, as well as the presence, absence, and severity of AS based upon the reading physician’s determination. For all development and validation datasets (non-BAV and BAV), values of the study’s associated echocardiogram variables (eg, left ventricular ejection fraction [LVEF] and hypertrophy [LVH], mean aortic valve [AV] gradient, etc) were also confirmed via manual review of echocardiogram reports by a cardiologist (M.D.S.).
Development and validation of the natural language processing algorithms
Our goal was to develop a detailed, structured database of echo data from semi-structured echo reports with free text sections using NLP, and to compare AS identification using NLP to using administrative codes alone. The NLP algorithms were developed using I2E software (version 5.4.1; Linguamatics, Inc, Cambridge, UK), which facilitates rapid text mining of unstructured EHR data through the creation of flexible and sophisticated rules-based search algorithms (Supplemental Material). Unique algorithms were developed to abstract data for each of the following variables: (1) presence of native valve AS (ie, interpreting physician’s assessment); (2) severity of AS (ie, physician assessment of mild, mild-to-moderate, moderate, moderate-to-severe, or severe AS); (3) AV maximum velocity (AV max, m/s); (4) peak AV gradient (mm Hg); (5) mean AV gradient (mm Hg); (6) AV velocity-time integral (AV VTI, cm); (7) left ventricular outflow tract (LVOT) VTI (cm); (8) LVOT diameter (cm); (9) AV area (cm2); (10) LVEF (%); (11) end-diastolic volume (mL); (12) ESV (mL); (13) LVH (ie, mild, mild-to-moderate, moderate, moderate-to-severe, or severe); (14) presence of BAV; (15) end-diastolic diameter (cm); and (16) end-systolic diameter (cm). Echo reports with prosthetic AV were identified by the presence of AS query and coded as not having AS (ie, the query is specific for native valve AS, not prosthetic valve AS or text that may describe patient-prosthesis mismatch).
Our target positive predictive value (PPV) and negative predictive value (NPV) values were >95%. For each variable, we refined the initial NLP algorithm within the development dataset until PPV and NPV of >95% were achieved. Each algorithm was then tested on the Validation 1 dataset. If the initial PPV and NPV yielded >95%, the algorithm was considered finalized. If not, the algorithm was iteratively refined until PPV and NPV >95% were achieved, then tested on the next validation dataset, and the process was repeated until the algorithm achieved PPV and NPV >95% on an untested validation dataset. For additional validation of algorithm accuracy, if an algorithm achieved the target >95% PPV and NPV on the first validation test, a second validation was performed on a subsequent validation dataset to further confirm accuracy of the algorithm. For final confirmation of accuracy, all completed algorithms were applied to the full dataset of echocardiograms from the 2008-2018 study period, and >300 echocardiograms were randomly selected to confirm PPV remained >95% based on manual review.
Ascertainment of aortic stenosis by diagnosis codes
Among cohort members, we identified all inpatient, emergency department, and outpatient encounters with an ICD-9 or ICD-10 diagnosis code for AS captured in any diagnosis position during the study period.
Statistical analysis
Analyses were performed using SAS software, version 9.4 (SAS, Cary, NC). We applied the final NLP algorithm for identifying AS (ie, based on the echocardiogram’s reading cardiologist’s assessment of the presence of AS), to all 2008-2018 echocardiogram reports (transthoracic, transesophageal, or stress echo, including full and limited studies) performed in eligible patients during the study period. We then compared results to administrative diagnosis code–based definitions of AS (ICD-9 or ICD-10 codes) found from 14 days before to 6 months after the echocardiogram date. This allowed for a code-based identification of patients who may have had known AS before the echocardiogram, as well as application of new AS diagnosis codes to the patient’s chart if the echocardiogram identified incident AS.
We calculated the PPV and NPV of both NLP algorithms and administrative diagnostic codes to identify AS from the development and validation datasets using the physician-adjudicated echocardiogram result as the “gold standard.” We also calculated and compared the number of echocardiograms with AS identified through NLP compared to those identified using administrative diagnostic codes during the full 2008-2018 study period. Patients with prosthetic valves were identified through the validated NLP algorithms and removed from the analysis of AS ascertainment via NLP vs administrative diagnostic codes. As a sensitivity analysis, we examined this concordance in both the ICD-9 and ICD-10 era, using October 1, 2015 as the transition date to ICD-10 codes. Finally, we calculated the distribution of AS severity, using the validated AS severity NLP algorithm, among those patients with AS identified by NLP but not identified by administrative diagnostic codes, to determine if AS cases without associated diagnosis codes reflected mild through severe disease.
Results
Dataset assembly, accuracy of NLP algorithms, and baseline characteristics
Among eligible echocardiograms between 2008 and 2018, a total of 1003 were selected for the development and validation datasets (210 in the development dataset, 193 in Validation 1 dataset, 300 in Validation 2 dataset, and 300 in Validation 3 dataset, each of which had approximately 50% with AS of any severity by reading physician assessment, and 50% without any identified AS). Development and validation of NLP algorithms for all 16 AS and associated variables were completed, with final PPV and NPV >95% achieved for each variable (Table 1 and Supplemental Table 2) Most variables required iterative testing in 2 validation datasets to ensure that the initial PPV and NPV met our performance criteria. Two queries, AS severity and LVH, required 3 iterations of testing in validation datasets, driven by a higher degree of variability in how physicians reported these measures. The most structured variables from echocardiogram reports, such as AV VTI, LVOT VTI, LVOT diameter, AV area, end-diastolic volume, and end-systolic volume, met our performance criteria after initial development, but we further confirmed that performance characteristics remained at target levels by testing in a second validation dataset.
Table 1.
Performance of aortic stenosis natural language processing algorithms
| Variable | Development dataset results | Final validation dataset results | ||
|---|---|---|---|---|
| PPV | NPV | PPV | NPV | |
| Global aortic stenosis† | 100% | 99% | 99% | 99% |
| Aortic stenosis severity‡ | 100% | 98% | 99% | 96% |
| Aortic valve max velocity† | 100% | 100% | 98% | 100% |
| Left ventricular ejection fraction† | 100% | 100% | 99% | 100% |
| Peak aortic valve gradient† | 100% | 100% | 99% | 98% |
| Mean aortic valve gradient† | 99% | 99% | 100% | 98% |
| Left ventricular hypertrophy‡ | 100% | 100% | 100% | 95% |
| Aortic valve velocity time integral∗ | 100% | 100% | 100% | 100% |
| Left ventricular outflow tract velocity time integral∗ | 100% | 100% | 100% | 100% |
| Left ventricular outflow tract diameter∗ | 100% | 100% | 100% | 99% |
| Aortic valve area∗ | 100% | 100% | 100% | 100% |
| End diastolic volume∗ | 100% | 100% | 100% | 100% |
| End systolic volume∗ | 100% | 100% | 100% | 100% |
| End diastolic diameter† | 100% | 96% | 100% | 96% |
| End systolic diameter† | 100% | 97% | 100% | 96% |
| Bicuspid aortic valve∗ | 100% | 97% | 97% | 100% |
The bicuspid aortic valve natural language processing algorithm used separate development and validation datasets enriched for patients with this condition.
These algorithms met specified performance criteria in the first tested validation set. They were additionally tested in a second validation dataset for further confirmation of accuracy.
Required iteration and testing in 2 validation datasets to meet specified performance criteria.
Required iteration and testing in 3 validation datasets to meet specified performance criteria.
Based on examining accuracy of AS identification from the 1003 physician-adjudicated echocardiogram reports, the developed NLP algorithms were more accurate than using administrative diagnostic codes searched between 14 days prior to and 6 months after the respective echocardiogram date, with PPV and NPV for AS identification with NLP being higher than using diagnostic codes (Table 2). The higher NPV (96%) than PPV (59%) for identifying AS with codes indicates there were more false-positives using codes alone (ie, an ICD-9/10 code for AV disease but no echocardiographic evidence of AS) than false-negatives (ie, no ICD-9/10 code for an associated echo showing AS) among the development and validation datasets.
Table 2.
Positive and negative predictive values of natural language processing algorithm and ICD 9/10 diagnostic codes to identify aortic stenosis compared with physician manual adjudication of medical records in the development and validation datasets
| Total echocardiograms in aortic stenosis development and validation datasets | ||
|---|---|---|
| (N = 1003) | PPV | NPV |
| Identification by ICD 9/10 codes | 59% | 96% |
| Identification by NLP algorithm | 99% | 99% |
ICD 9/10 = International Classification of Diseases, Versions 9 and 10; NLP = natural language processing; NPV = negative predictive value; PPV = positive predictive value.
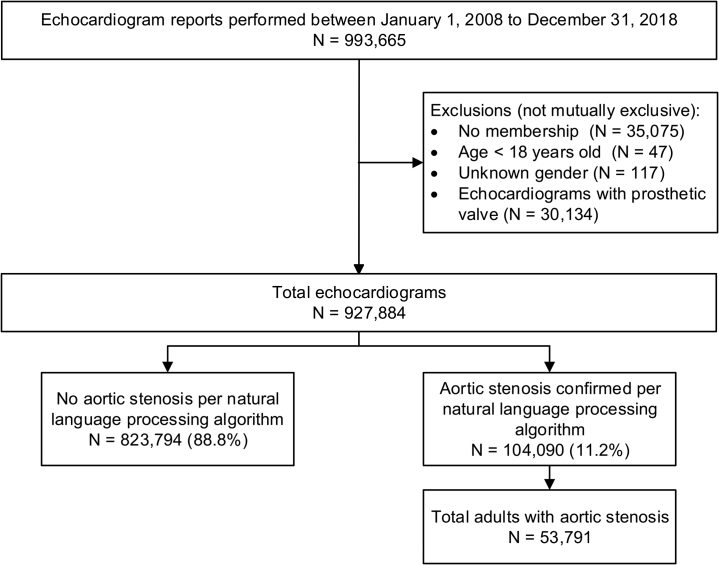
Between 2008 and 2018, we identified 927,884 eligible echocardiograms within KPNC (Figure 1). All 16 echocardiogram algorithms achieved PPV > 95% in over 300 randomly selected echocardiogram reports from the full 2008-2018 dataset. Using our final NLP algorithm for the presence of AS (based upon the reading physician’s interpretation of the echocardiogram), we classified 104,090 echocardiograms (11.2%) from 53,791 unique patients with AS. Patients with AS had mean (standard deviation [SD]) age 75.2 (12.2) years; 52.1% were women, 5.2% Black, 8.4% Asian/Pacific Islander, and 11.1% Hispanic; and had median (interquartile range) 2 (1–4) echocardiograms per patient during the study period. In general, AS patients were significantly older, comprised a higher proportion of white/European patients, and had more echocardiograms per patient than those without AS (Supplemental Table 3).
Figure 1.
Cohort assembly of echocardiograms for adults with aortic stenosis.
Concordance of administrative diagnosis codes with NLP-based classification of AS and severity
Among 104,090 echocardiograms classified by NLP as having AS, only 64.6% (n = 67,297) had a diagnosis code for AS between 14 days before and 6 months after the echocardiogram date (Table 3). Findings were similar when stratified between the ICD-9 and ICD-10 eras (Supplemental Table 4). In addition, among the patients with 101,811 echocardiograms associated with a diagnosis code for AS within the specified time period before and after the echocardiogram, the PPV for a diagnosis code–based identification of AS was only 66.1% using the NLP-based classification as the gold standard, indicating that a codes-based approach did not capture 33.9% of patients with AS noted on echocardiogram. Of note, among the 826,073 echocardiograms without a diagnosis code for AS within the specified time period, the NPV was 95.5% based on the NLP-based classification for the absence of AS, indicating that over our decade-long study period, 4.5% (n = 36,793) of all echocardiograms without an associated AS diagnosis code actually do have AS based upon the NLP-based classification. Although among the 823,794 echocardiograms classified by NLP as not having AS, 4.1% (n = 34,514) had a diagnosis code for AS between 14 days before and 6 months after the echocardiogram date, this rate dropped to 1.1% if the most nonspecific ICD-9 code (424.1), which codes for both aortic stenosis and aortic regurgitation, is excluded, suggesting a substantial degree of misclassification from administrative diagnosis codes.
Table 3.
Application of validated natural language processing algorithm vs administrative diagnosis codes to identify aortic stenosis among all adult echocardiograms, 2008–2018
| Validated NLP algorithm |
Total echocardiograms, n (column %) | |||
|---|---|---|---|---|
| Positive for AS | Negative for AS | |||
| AS ICD 9/10 codes† | Positive for AS | 67,297 | 34,514 | 101,811 (11.0) |
| Negative for AS | 36,793 | 789,280 | 826,073 (89.0) | |
| Total echocardiograms, n (row %) | 104,090 (11.2) | 823,794 (88.8) | 927,884 | |
AS = aortic stenosis.
ICD-9 codes included 395.0, 746.3, 396.2, and 424.1; ICD-10 codes included I06.0, I06.2, I35.0, and Q23.0.
Further, in the 104,090 echocardiograms with AS by NLP, there was a broad distribution of AS severity using the NLP algorithm, and only 4.2% did not have a specific physician-designated AS severity level (Table 4). The NLP-derived hemodynamic parameters matched well to the physician-designated AS severity level. The mean (SD) AV max for physician-designated mild, mild-moderate, moderate, moderate-severe, and severe AS were 2.4 m/s (0.4), 2.8 m/s (0.5), 3.2 m/s (0.5), 3.6 m/s (0.6), and 4.2 m/s (0.8), respectively; the mean (SD) AV mean gradient for physician-designated mild, mild-moderate, moderate, moderate-severe, and severe AS were 12.9 mm Hg (4.9), 18.3 mm Hg (5.8), 24.5 mm Hg (7.9), 31.5 mm Hg (9.4), and 43.8 mm Hg (16.0), respectively.
Table 4.
Distribution of severity of aortic stenosis based on natural language processing algorithm applied to echocardiogram reports between 2008 and 2018, overall and stratified by the presence or absence of administrative diagnosis codes
| Severity of aortic stenosis | Overall N = 104,090 |
Diagnostic code for aortic stenosis N = 67,297 |
No diagnostic code for aortic stenosis N = 36,793 |
|---|---|---|---|
| Mild | 44,767 (43.0) | 17,290 (25.7) | 27,477 (74.7) |
| Mild-to-moderate | 7130 (6.9) | 4958 (7.4) | 2172 (5.9) |
| Moderate | 22,888 (22.0) | 19,049 (28.3) | 3839 (10.4) |
| Moderate-to-severe | 6987 (6.7) | 6624 (9.8) | 363 (1.0) |
| Severe | 17,916 (17.2) | 17,457 (25.9) | 459 (1.3) |
| No severity found | 4402 (4.2) | 1919 (2.9) | 2483 (6.7) |
Severity of aortic stenosis was based upon interpreting physician assessment of the echocardiogram identified through validated natural language processing algorithm.
We next evaluated the distribution of AS severity in echocardiograms that did or did not have an associated diagnosis code for AS. In the subset of 67,297 echocardiograms that had both NLP-classified AS and an associated AS diagnosis code, disease severity was fairly evenly distributed across the spectrum of severity (ie, mild, moderate, and severe), but with a rightward shift toward more severe disease compared to the overall cohort of echocardiograms with AS. Among the 36,793 echocardiograms with NLP-classified AS but without any associated AS diagnosis code, slightly less than three-quarters had mild AS, and 18.6% had severity that was greater than mild. Of all the echocardiograms with moderate or greater disease severity, 9.8% had no associated diagnostic code for AS.
Discussion
In our study, we successfully developed and validated algorithms using NLP to extract detailed data elements from semi-structured echocardiogram reports with free text sections within a large, integrated healthcare delivery system. We applied the validated NLP algorithms to nearly 1 million echocardiogram reports to identify nearly 54,000 patients with AS between 2008 and 2018. To understand if case identification of AS was superior using validated NLP algorithms compared to traditional case identification using administrative diagnostic codes, we examined the ability of each method to identify cases of AS in a large, diverse, community-based population. We found our validated NLP algorithms were significantly more accurate than using administrative diagnosis codes for identifying AS.
We found that over 35% of echocardiograms with AS identified by NLP were performed on patients who did not have an associated AS diagnosis code up to 14 days before or 6 months after the procedure. Further, nearly 20% of the echocardiograms had hemodynamically significant AS (ie, greater than mild disease by the reading physician’s assessment). Although the majority of echocardiograms without administrative diagnosis codes were for mild AS, even mild AS warrants follow-up with an echocardiogram every 3–5 years, per recent clinical practice guidelines.12 The significant portion of hemodynamically significant AS cases lacking codes suggests that codes are inconsistently applied in AS cases.
In addition, the suboptimal PPV of 66% for administrative diagnostic codes being associated with an NLP-confirmed case of AS in the full 2008–2018 dataset may also reflect that AV disease diagnosis codes are not highly specific for AS, and instead may be applied in cases of aortic regurgitation, may be mistakenly applied in cases of aortic sclerosis without stenosis, or may reflect older codes in patients who previously received AV surgery. While the poor specificity was worse in the ICD-9 era, PPV was still suboptimal at 76%—indicating a lot of false-positive cases—when examined in the ICD-10 era (Supplemental Table 4). This lack of both specificity and sensitivity of AS administrative diagnosis codes, coupled with the lack of information on disease severity, which determines clinical follow-up, makes diagnosis codes an inferior method for case ascertainment and as a tool to guide population management compared to NLP methods. Incorporating NLP tools into traditional quality improvement programs could facilitate more effective individual and population management than relying on administrative and diagnosis codes alone.
NLP has been used for various clinical applications, including identification of postoperative surgical13,14 and critical care complications,15 identification of specific radiographic findings (eg, pulmonary nodules from radiology reports),16 medication extraction from clinical notes,17 improvement of clinical prediction models,18 and identification of clinical conditions (eg, pneumonia,19,20 heart failure21). However, the use of NLP to identify cases of valvular heart disease and assess its severity is rare and has been infrequently done.22,23 No published study has demonstrated the superiority of an NLP-based approach compared to using traditionally used administrative diagnostic codes across multiple coding eras.23
Use of NLP methods for improving both research and clinical quality of care has been discussed for decades,9,24 but despite the widespread implementation of EHR systems and rapid expansion in availability of unstructured data, it has a limited role within most healthcare delivery systems. Despite the maturation and integration of NLP-based methods in other industries, integration of NLP has been slow in healthcare. However, recent initiatives by several large technology companies attempting to expand their footprint into healthcare may accelerate this integration.25,26
Although some routine tracking of conditions27 or procedures28,29 exists for quality or regulatory reporting purposes using various approaches, most medical conditions are not routinely identified and assessed from the clinically rich data in EHRs using validated methods. In a systematic review to identify examples where NLP or other text-mining methods were used to conduct more precise case detection beyond structured codes, 67 studies published between 2000 and 2015 for 41 different clinical conditions were found, none of which included valvular heart disease,10 though proof-of-concept data exist30 and more recent efforts have successfully queried and abstracted echocardiography data on a larger scale.22 One possible explanation is that echocardiography reports have been less well integrated into EHRs, making them difficult to query.
In our study, we developed and validated machine learning–based methods applied to semi-structured echocardiogram report data to identify patients across the spectrum of disease severity for the most common adult cardiac valvular condition. Using NLP methods, we comprehensively identified AS patients from echocardiography reports. Systematically incorporating NLP tools into quality improvement programs could facilitate more cost-effective individual and population management for AS rather than relying on administrative diagnosis codes alone,31 which can misclassify individuals and do not provide detailed information on disease severity.
Our study has certain limitations. While we studied echocardiogram reports derived from a large, diverse population receiving care within an integrated healthcare delivery system, the operating characteristics of our NLP algorithms and administrative diagnosis codes for identification of AS, and discrepancies between the 2 methods, may vary in other health systems. However, given the lack of detail for valvular conditions among current administrative diagnosis codes, NLP-based case identification has many advantages, such as identifying disease severity, which is not noted in the current diagnosis code classification scheme, as well as identifying the accompanying echo parameters that contribute to a patient’s overall course and prognosis (ie. AV velocity, mean AV gradient, LVEF). In addition, given the broad time period examined and the large number of reading cardiologists within our healthcare system, our NLP algorithms likely capture a diversity of text combinations to describe AS that would be applicable to other health systems. All patients were members of KPNC at the time of echocardiogram, and KPNC has low overall annual attrition (loss of membership) rates of 2%–3%, and lower rates among older persons (age >65 years), who comprise the majority of AS patients. Despite this, some patients identified with AS by NLP may not have had the opportunity to have an AS diagnosis code applied to their medical record owing to membership cancellation, no further follow-up visits, etc; but given our low attrition rates and high rates of access to care, these possibilities likely did not meaningfully bias the results.
Conclusion
In conclusion, our study demonstrates the ability of NLP to accurately identify and characterize the severity of AS from semi-structured and unstructured echocardiogram reports and supports the potential value of NLP to enhance quality improvement and research efforts for this condition. Future studies leveraging NLP-derived data to evaluate the association between severity of AS and clinical outcomes, along with identifying predictors of AS progression, will further advance personalized and population-based care strategies to optimize surveillance and treatment of adults with this common valvular heart condition.
Acknowledgments
The authors wish to thank Kathleen Nogueria, MHA, for technical assistance with NLP queries.
Funding Sources
This work was supported by grants from The Permanente Medical Group Delivery Science and Applied Research and Physician Researcher Programs.
Disclosures
The authors have no conflicts to disclose.
Authorship
All authors attest they meet the current ICMJE criteria for authorship.
Patient Consent
Waiver of informed consent was obtained owing to the nature of the study.
Ethics Statement
The research protocol used in this study was reviewed and approved by the Kaiser Permanente Northern California institutional review board.
Guidelines Statement
The research reported in this paper adhered to the STROBE guidelines for observational research.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.cvdhj.2021.03.003.
Appendix. Supplementary data
References
- 1.Nkomo V.T., Gardin J.M., Skelton T.N., Gottdiener J.S., Scott C.G., Enriquez-Sarano M. Burden of valvular heart diseases: a population-based study. Lancet. 2006;368:1005–1011. doi: 10.1016/S0140-6736(06)69208-8. [DOI] [PubMed] [Google Scholar]
- 2.Liebovitz D.M., Fahrenbach J. COUNTERPOINT: Is ICD-10 diagnosis coding important in the era of big data? No. Chest. 2018;153:1095–1098. doi: 10.1016/j.chest.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 3.Sheikhalishahi S., Miotto R., Dudley J.T., Lavelli A., Rinaldi F., Osmani V. Natural language processing of clinical notes on chronic diseases: systematic review. JMIR Med Inform. 2019;7:e12239. doi: 10.2196/12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ly T., Pamer C., Dang O., et al. Evaluation of Natural Language Processing (NLP) systems to annotate drug product labeling with MedDRA terminology. J Biomed Inform. 2018;83:73–86. doi: 10.1016/j.jbi.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 5.McEntire R., Szalkowski D., Butler J., et al. Application of an automated natural language processing (NLP) workflow to enable federated search of external biomedical content in drug discovery and development. Drug Discov Today. 2016;21:826–835. doi: 10.1016/j.drudis.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Jonnagaddala J., Liaw S.T., Ray P., Kumar M., Chang N.W., Dai H.J. Coronary artery disease risk assessment from unstructured electronic health records using text mining. J Biomed Inform. 2015;58(Suppl):S203–S210. doi: 10.1016/j.jbi.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang M., Chang M., Reed J.Z., Milward D., Xu J.J., Cornell W.D. Developing timely insights into comparative effectiveness research with a text-mining pipeline. Drug Discov Today. 2016;21:473–480. doi: 10.1016/j.drudis.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Chang H.M., Huang E.W., Hou I.C., Liu H.Y., Li F.S., Chiou S.F. Using a text mining approach to explore the recording quality of a nursing record system. J Nurs Res. 2019;27:e27. doi: 10.1097/jnr.0000000000000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velupillai S., Suominen H., Liakata M., et al. Using clinical Natural Language Processing for health outcomes research: overview and actionable suggestions for future advances. J Biomed Inform. 2018;88:11–19. doi: 10.1016/j.jbi.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ford E., Carroll J.A., Smith H.E., Scott D., Cassell J.A. Extracting information from the text of electronic medical records to improve case detection: a systematic review. J Am Med Inform Assoc. 2016;23:1007–1015. doi: 10.1093/jamia/ocv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon N.P. Division of Research, Kaiser Permanente Medical Care Program; Oakland, CA: 2013. Characteristics of Adult Health Plan Members in Kaiser Permanente’s Northern California Region, as Estimated from the 2011 Member Health Survey. April 2013. [Google Scholar]
- 12.Otto C.M., Nishimura R.A., Bonow R.O., et al. 2020 ACC/AHA Guideline for the Management of Patients With Valvular Heart Disease: Executive Summary: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation. 2021;143:e35–e71. doi: 10.1161/CIR.0000000000000932. [DOI] [PubMed] [Google Scholar]
- 13.Murff H.J., FitzHenry F., Matheny M.E., et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA. 2011;306:848–855. doi: 10.1001/jama.2011.1204. [DOI] [PubMed] [Google Scholar]
- 14.FitzHenry F., Murff H.J., Matheny M.E., et al. Exploring the frontier of electronic health record surveillance: the case of postoperative complications. Med Care. 2013;51:509–516. doi: 10.1097/MLR.0b013e31828d1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taggart M., Chapman W.W., Steinberg B.A., et al. Comparison of 2 natural language processing methods for identification of bleeding among critically ill patients. JAMA Netw Open. 2018;1 doi: 10.1001/jamanetworkopen.2018.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang S.K., Garry K., Chung R., et al. Natural language processing for identification of incidental pulmonary nodules in radiology reports. J Am Coll Radiol. 2019;16:1587–1594. doi: 10.1016/j.jacr.2019.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng C., Rashid N., Koblick R., An J. Medication extraction from electronic clinical notes in an integrated health system: a study on aspirin use in patients with nonvalvular atrial fibrillation. Clin Ther. 2015;37:2048–2058 e2042. doi: 10.1016/j.clinthera.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Marafino B.J., Park M., Davies J.M., et al. Validation of prediction models for critical care outcomes using natural language processing of electronic health record data. JAMA Netw Open. 2018;1 doi: 10.1001/jamanetworkopen.2018.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkin P.L., Froehling D., Wahner-Roedler D., et al. NLP-based identification of pneumonia cases from free-text radiological reports. AMIA Annu Symp Proc. 2008:172–176. [PMC free article] [PubMed] [Google Scholar]
- 20.Jones B.E., South B.R., Shao Y., et al. Development and validation of a natural language processing tool to identify patients treated for pneumonia across VA emergency departments. Appl Clin Inform. 2018;9:122–128. doi: 10.1055/s-0038-1626725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Luo J., Hao S., et al. NLP based congestive heart failure case finding: a prospective analysis on statewide electronic medical records. Int J Med Inform. 2015;84:1039–1047. doi: 10.1016/j.ijmedinf.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Nath C., Albaghdadi M.S., Jonnalagadda S.R. A natural language processing tool for large-scale data extraction from echocardiography reports. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Small A.M., Kiss D.H., Zlatsin Y., et al. Text mining applied to electronic cardiovascular procedure reports to identify patients with trileaflet aortic stenosis and coronary artery disease. J Biomed Inform. 2017;72:77–84. doi: 10.1016/j.jbi.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Jha A.K. The promise of electronic records: around the corner or down the road? JAMA. 2011;306:880–881. doi: 10.1001/jama.2011.1219. [DOI] [PubMed] [Google Scholar]
- 25.Evans M., Stevents L. Big Tech Expands Footprint in Health. Wall Street Journal. November 27, 2018;2018 [Google Scholar]
- 26.Singer N., Wakabayashi D. New York Times; 2019. Google to Store and Analyze Millions of Health Records. [cited 2019 Nov 11]. [Google Scholar]
- 27.Bagai A., Chen A.Y., Udell J.A., et al. Association of cognitive impairment with treatment and outcomes in older myocardial infarction patients: a report from the NCDR Chest Pain-MI Registry. J Am Heart Assoc. 2019;8 doi: 10.1161/JAHA.119.012929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moussa I., Hermann A., Messenger J.C., et al. The NCDR CathPCI Registry: a US national perspective on care and outcomes for percutaneous coronary intervention. Heart. 2013;99:297–303. doi: 10.1136/heartjnl-2012-303379. [DOI] [PubMed] [Google Scholar]
- 29.Holmes D.R., Jr., Nishimura R.A., Grover F.L., et al. Annual outcomes with transcatheter valve therapy: from the STS/ACC TVT Registry. Ann Thorac Surg. 2016;101:789–800. doi: 10.1016/j.athoracsur.2015.10.049. [DOI] [PubMed] [Google Scholar]
- 30.Chung J., Murphy S. Concept-value pair extraction from semi-structured clinical narrative: a case study using echocardiogram reports. AMIA Annu Symp Proc. 2005:131–135. [PMC free article] [PubMed] [Google Scholar]
- 31.Manuel D.G., Rosella L.C., Stukel T.A. Importance of accurately identifying disease in studies using electronic health records. BMJ. 2010;341:c4226. doi: 10.1136/bmj.c4226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.