Abstract
Background
Visualizing fibrosis on cardiac magnetic resonance (CMR) imaging with contrast enhancement (late gadolinium enhancement; LGE) is paramount in characterizing disease progression and identifying arrhythmia substrates. Segmentation and fibrosis quantification from LGE-CMR is intensive, manual, and prone to interobserver variability. There is an unmet need for automated LGE-CMR image segmentation that ensures anatomical accuracy and seamless extraction of clinical features.
Objective
This study aimed to develop a novel deep learning solution for analysis of contrast-enhanced CMR images that produces anatomically accurate myocardium and scar/fibrosis segmentations and uses these to calculate features of clinical interest.
Methods
Data sources were 155 2-dimensional LGE-CMR patient scans (1124 slices) and 246 synthetic “LGE-like” scans (1360 slices) obtained from cine CMR using a novel style-transfer algorithm. We trained and tested a 3-stage neural network that identified the left ventricle (LV) region of interest (ROI), segmented ROI into viable myocardium and regions of enhancement, and postprocessed the segmentation results to enforce conforming to anatomical constraints. The segmentations were used to directly compute clinical features, such as LV volume and scar burden.
Results
Predicted LV and scar segmentations achieved 96% and 75% balanced accuracy, respectively, and 0.93 and 0.57 Dice coefficient when compared to trained expert segmentations. The mean scar burden difference between manual and predicted segmentations was 2%.
Conclusion
We developed and validated a deep neural network for automatic, anatomically accurate expert-level LGE- CMR myocardium and scar/fibrosis segmentation, allowing direct calculation of clinical measures. Given the training set heterogeneity, our approach could be extended to multiple imaging modalities and patient pathologies.
Keywords: Segmentation, Contrast-enhanced, CMR, Deep learning, Machine learning
Key Findings.
-
•
An efficient, cost-effective deep learning solution was developed for automatic expert-level segmentation of the left ventricle myocardium, blood pool, and scar/fibrosis regions in contrast-enhanced (late gadolinium enhanced, LGE) cardiac magnetic resonance (CMR) imaging.
-
•
The deep neural network uses a novel convolutional autoencoder postprocessor that guarantees that predicted myocardium segmentations satisfy anatomically inspired geometrical constraints.
-
•
Owing to its high-anatomical-fidelity predictions, the segmentation algorithm can be used to seamlessly compute clinical features, such as left ventricular volume and scar burden.
-
•
A new style-transfer approach was used to augment the training data by transforming nonenhanced CMR imaging into “LGE-like” scans.
Introduction
Many cardiac diseases are associated with structural remodeling of the myocardium. In both ischemic and nonischemic cardiomyopathies, the presence of myocardial fibrosis and scar significantly elevates the risk for lethal heart rhythm disorders and sudden cardiac death (SCD).1, 2, 3 Therefore, assessment of myocardial scar and fibrosis is important for diagnostic and prognostic purposes, in forecasting the trajectory of heart disease,4 evaluating arrhythmia propensity in the heart,5,6 and stratifying patients for SCD risk.7,8 Cardiac magnetic resonance (CMR) imaging with late gadolinium enhancement (LGE) has unparalleled capability in the detection and quantification of scar and fibrosis, visualized as increased brightness in regions with a higher proportion of extracellular space.9 The utility of scar/fibrosis assessment in clinical decision-making has been demonstrated by a large body of clinical research in patients with different cardiomyopathies2,3,6,8,10, 11, 12, 13 and by a number of mechanistic studies of arrhythmogenesis in heart disease.14, 15, 16, 17 However, LGE-CMR image analysis is a laborious task prone to substantial interobserver variability. It requires expert contouring of the epicardial and endocardial borders, the intermediate-intensity peri-infarct zone (gray zone, GZ), and the high-intensity dense scar region. There is an unmet need for an automated method to segment myocardium and scar in LGE-CMR images. Ideally, resulting segmentations should be anatomically accurate, ie, free from nonanatomical artefacts, thus ensuring seamless extraction of important clinical features used in diagnostic and prognostic decisions.
Deep learning (DL)-based image segmentation offers the promise of full automation and output consistency. However, most of the available algorithms require intensive manual interventions, eg, specifying anatomical landmarks18 or labeling boundary slices of the stack at the apex and base of the heart.19 The few DL algorithms developed for LGE-CMR myocardial segmentation20, 21, 22, 23 and the even fewer for LGE-CMR scar/fibrosis segmentation24, 25, 26, 27 all suffer from several limitations. Specifically, these approaches fail to address the presence of resulting poor-performing segmentation outliers, and are not robust to varying image acquisition quality (ie, different scanners and protocols at different centers) or to the varying fibrosis patterns resulting from different heart pathologies, potentially leading to bespoke algorithms, which fail to generalize across populations or produce anatomically plausible heart geometries.24,26,27
Here we develop an anatomically informed DL approach to LGE-CMR image segmentation and clinical feature extraction. We term our technology Anatomical Convolutional Segmentation Network, ACSNet. This fully automated technology applies 3 stages of deep neural networks to segment the left ventricle (LV); contour the LV myocardium, blood pool, and scar/fibrosis regions; and apply geometric constraints to the segmentations to ensure anatomical accuracy. ACSNet is robust to different scar/fibrosis distributions, to inputs from various imaging centers acquired on scanners from different manufacturers, and to multiple CMR modalities. It outperforms interexpert segmentation results and demonstrates consistently accurate performance across often ambiguous regions of the LV (eg, apex and base). Segmentations satisfy anatomical guidelines, allowing for expert-level immediate computation of clinical features, such as scar burden and LV volume.
Methods
Imaging data and processing
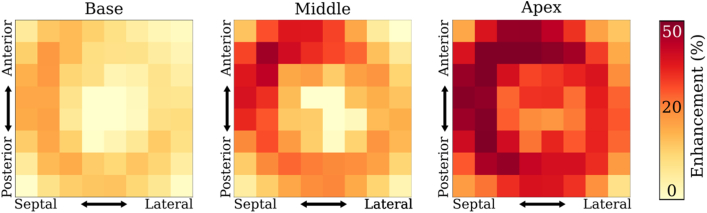
The primary data source for ACSNet was 2-dimensional (2-D) LGE-CMR scans acquired during the Left Ventricular Structural Predictors of Sudden Cardiac Death Study (ClinicalTrials.gov ID NCT01076660) sponsored by Johns Hopkins University. All LGE-CMR images used in this study were acquired using 1.5-T magnetic resonance imaging (MRI) devices (approximately 40% of the scans using Signa; GE Medical Systems, Waukesha, Wisconsin; and roughly 60% using Avanto; Siemens, Erlangen, Germany). The contrast agent used was 0.15−0.20 mmol/kg gadodiamide (Omniscan; GE Healthcare) and the scan was captured 10−30 minutes after injection. The most commonly used sequence was inversion recovery fast-gradient echo pulse, with an inversion recovery time typically starting at 250 ms and adjusted iteratively to achieve maximum nulling of normal myocardium. Typical spatial resolutions ranged from 1.5−2.4 × 1.5−2.4 × 6−8 mm, with 2- to 4-mm gaps. After excluding 7 scans with very poor quality, 1124 2-D LGE-CMR slices were selected from 155 patients with ischemic cardiomyopathy (ICM). Trained experts provided manual segmentations of myocardium and scar/fibrosis as described in previous work.28 The 3-dimensional (3-D) distribution and average amount of contrast enhancement as a proportion of myocardium volume are shown in Figure 1. Regions with high enhancement were predominantly in the apex and anterior septal region for the middle part of the ventricle. Per-patient enhancement region proportion of myocardium volume ranged between 5% and 53%.
Figure 1.
Distribution of enhanced myocardium regions for ground truth data. The spatial distribution of regions of enhanced myocardium is shown for 3 regions of the ventricle: basal (left), middle (center), and apical (right). The x-axes capture septal vs lateral location and the y-axes capture anterior vs posterior. The heat map quantifies the proportion of enhanced myocardium located in the respective region, averaged over all patients.
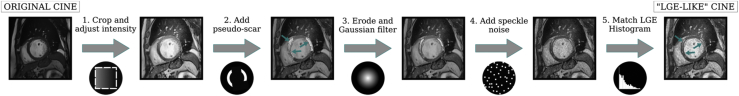
LGE data were supplemented with 1360 2-D short-axis end-diastole cine CMR slices (245 scans) from 2 publicly available sources. The first was MICCAI Automated Cardiac Diagnosis Challenge with scans acquired at University Hospital of Dijon, France, using 2 MRI scanners of different magnetic strengths (1.5 T – Siemens Area; Siemens Medical Solutions, Erlangen, Germany; and 3.0 T – Siemens Trio Tim; Siemens Medical Solutions). Patient pathologies in this set were divided into 5 groups: normal patients, patients with systolic heart failure with infarction, dilated cardiomyopathy patients, hypertrophic cardiomyopathy patients, and patients with abnormal right ventricle.29 The second source was Cardiac MR Left Ventricular Segmentation Challenge with scans acquired at Sunnybrook Hospital, Canada using a 1.5 T scanner (Signa; GE Medical Systems, Waukesha, WI). Patients in this cohort were divided into 4 groups by pathology: normal, heart failure with infarction, heart failure without infarction, and LV hypertrophy.30 Ground truth myocardium segmentations were provided with the scans. The cine CMR data set was converted into “LGE-like” images using a custom style transfer method (Figure 2). First, with a given occurrence probability, a pseudo-enhancement mask was generated independently for each slice by intersecting the myocardium mask with a randomized collection of basic shapes (eg, ellipses, squares, etc) with randomized locations. The probability was selected to match the proportion of slices with enhancement among the LGE scans. The resulting patches were subsequently blurred using Gaussian filters, leading to smoother edges. The resulting mask was overlaid onto the original (dark) myocardium, elevating the signal intensity in the corresponding area. Then, speckle noise was added and, finally, for each cine scan, a histogram match was performed between the newly generated image and randomly sampled scans from the LGE training data set, resulting in a set of “LGE-like” images.
Figure 2.
Conversion process of cine images to “late gadolinium enhancement (LGE)-like” images. (1) The original cine image is cropped/padded to a square and contrast-limited adaptive histogram equalization (CLAHE) is applied. (2) Cine images are further transformed by first generating a pseudo-enhancement (“LGE-like” enhanced myocardium) mask. (3) Pseudo-scar mask is randomly eroded and Gaussian filters are applied to realistically blur the edges. (4) Speckle noise is added to the image to resemble LGE noise. (5) An LGE cardiac magnetic resonance scan is sampled at random and a histogram match is performed.
All LGE and resulting “LGE-like” 2-D slices were preprocessed and stored in a common file format to accommodate multiple medical image file types (eg, DICOM, NIfTI, etc), retaining 3-D ventricular geometry information. Specifically, slices were automatically ordered from apex to base, retaining slice location, image intensities, resolution, and patient orientation information. An average of 1.9 slices per patient, which did not have ground truth myocardial segmentation—most did not contain the LV region of interest (ROI)—were excluded from training to prevent potential bias in the network. The images were standardized in terms of orientation by applying rotations in increments of 90° (90° was chosen to avoid interpolation). If scans originally stored in DICOM had the “WindowCenter,” “WindowLength,” “RescaleSlope,” and “RescaleIntercept” tags populated, the corresponding linear transformation was applied to the raw signal intensities to enhance contrast and brightness.
Finally, to increase the contrast between myocardium and blood pool, contrast-limited adaptive histogram equalization31 was applied. All images were cropped or padded to a square of size 192 × 192 pixels (no aspect ratio distortion), without centering. Finally, resulting images were rescaled in the range [0, 255].
The research protocol used in this study was reviewed and approved by the institutional review board.
Multi-network design
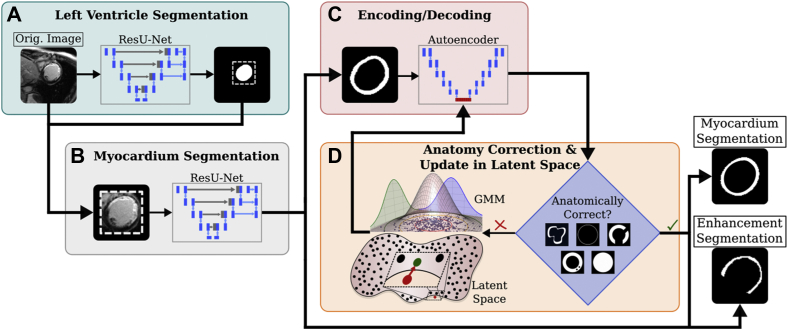
The overall architecture of our DL approach is presented in Figure 3. ACSNet used a multi-network sequential approach in 3 stages to segment viable myocardium, enhanced myocardium (scar/fibrosis), and blood pool. The first network identified the LV ROI, which was then used to zero out signal outside a tight square around the segmented ROI (Figure 3A). The second network differentiated between myocardium and enhancement (Figure 3B). The third was a postprocessing stage, which adjusted the predictions to satisfy anatomical constraints (Figure 3C and 3D).
Figure 3.
ACSNet architecture consisting of 3 interconnected deep learning subnetworks. A: The first residual U-Net (ResU-Net) is used to identify and crop around the left ventricle (LV). B: The second network uses the tightly cropped image from panel A and the LV segmentation to further segment the LV into viable and enhanced myocardium. C: The third network is a convolutional autoencoder trained to encode (compress) and decode myocardial segmentation masks. D: Segmentations from the training set are encoded using the third network to form a latent space. The space is modeled as a Gaussian mixture model (GMM) and conditional resampling is performed to populate the space with anatomically correct samples (black dots). Predicted segmentations are encoded and the nearest-neighbors algorithm is used to return a perturbed, anatomically correct version (green dot) of the original (red dot). GMM image adapted from source.48 Additional details are presented in Methods.
Stage 1: ROI segmentation network
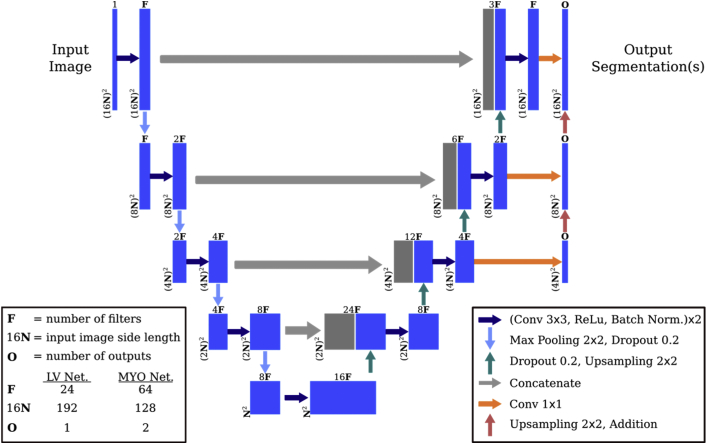
The first network in ACSNet (Figure 3A) was trained to predict a mask of the LV ROI, which included myocardium and blood pool. The goal of this network was to simplify the detailed segmentation problem in the next stage by reducing the very high ratio of background-to-myocardium pixels and limiting the field of view for the second network to mostly myocardium features. The ROI segmentation network (Figure 4) was a U-Net with residuals (ResUNet) of depth 4.19,32 During the downsampling process, each of the 4 depth levels consisted of 2 repetitions of a block made up of a 3 × 3 2-D convolution, followed by a rectified linear unit activation and batch normalization. After the 2 blocks, each was followed by a 2 × 2 max pooling layer and 20% dropout. The upsampling branch had a similar structure using 2 × 2 nearest-neighbor upsampling and identical convolutional layers. ROI predictions were automatically cleaned up by discarding all but 1 connected component, specifically the one closest to the center of the mass of objects in slices located between the 20th and 80th percentiles (higher confidence) of the short-axis height. Lastly, slices close to the base with very large jumps in ROI area were deemed likely above the ventricle and were automatically pruned.
Figure 4.
Left ventricle and myocardium segmentation network architecture. The left ventricle region of interest network (LV Net.) identifies the main region of interest. The second network (MYO Net.) segments the myocardium by differentiating between viable and nonviable tissue represented by each of the 2 outputs. The networks differ by the number of filters, input image size, and number of outputs as indicated.
Stage 2: Left ventricle myocardium segmentation network
The second network (Figure 3B) used the ROI mask predicted by the first network to differentiate LV blood pool, viable myocardium, and regions of enhancement, returning segmentations for the latter 2 tissue types. As preprocessing, the ROI mask center of mass was used to center the LV images in a 128 × 128 pixel square. Next, the predicted LV ROI was used to rescale the intensity values (Supplemental Material, Appendix A).
For the LV myocardium segmentation network, we implemented a modified ResUNet structure (Figure 4), similar to the ROI segmentation network. It differed from the ROI network in that it used twice the number of filters at each of the 4 depth levels owing to the higher complexity of the task. The network outputted 2 masks, 1 representing the entire myocardium and the other identifying only the enhanced tissue. The enhanced regions were minimally cleaned up using an automated series of morphological operations—erosion, opening, and closing—with parameter choices determined by maximizing training data average Dice coefficient using a greedy grid search.
Stage 3: Anatomical autoencoder postprocessing
The final neural network (Figure 3C and 3D) ensured that myocardial segmentation results abided by anatomical guidelines, reducing the performance impact of ambiguous regions (eg, apex and base), where observer ground truth variability was high primarily owing to imaging artifacts. Anatomical corrections were applied on reduced-dimension versions of the myocardial segmentations.
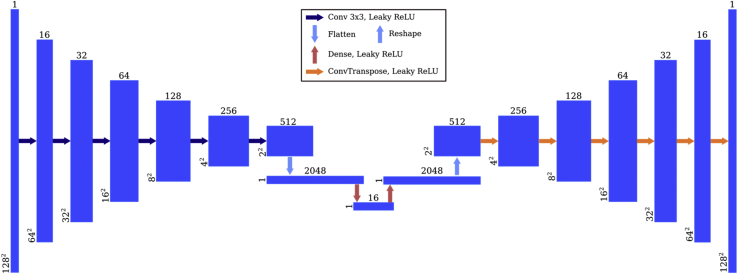
The space of low-dimensional myocardial segmentations was constructed using a convolutional autoencoder network (Figure 5), which consisted of 6 strided 3 × 3-kernel convolutional layers, 2 dense layers, and 6 transpose convolutions. Leaky rectified linear unit activations were used after each layer. On the encoding branch, the number of channels started at 16 and doubled after each convolutional layer to 2048, before being collapsed to the 16-dimensional encoding vector by a dense layer.
Figure 5.
Autoencoder network architecture. The anatomical autoencoder is used as a postprocessing step that takes in myocardial masks and uses a series of convolutions and downsampling layers to create a 16-dimensional latent representation (left side). The decoder piece of the autoencoder (right side) re-creates the original myocardial segmentation from the latent representation.
During training, the autoencoder learned how to encode ground truth myocardial segmentations into the 16-dimensional latent space and use this representation to decode into the original image, effectively resulting in a collection of 16-dimensional vectors representing anatomically correct myocardial segmentations. Next, the latent space was augmented by generating new vectors based on the existing ones. This was done by modeling the existing vectors using a Gaussian mixture model and performing rejection sampling, where the rejection criteria encapsulated the anatomical correctness using a collection of morphological checks. Mathematical details are provided in the Supplemental Material, but a sketch of the model is presented here. The ground truth 16-dimensional representations of the myocardial segmentations were modeled as points coming from a mixture of 5 Gaussian distributions, with weights, means, and covariance matrices estimated using the training data. New samples were drawn from this mixture and kept only if, once decoded, they passed anatomical checks. These checked for convexity defects, holes in myocardium, circularity, number of objects, and myocardial wall thickness. This increased the size of the latent space by sampling an additional 10,000 points.
The 16-dimension representation of a predicted myocardial segmentation was not guaranteed to decode to an anatomically correct image. The “correction” consisted of moving in the direction of the nearest neighbor in the latent space by the smallest distance possible such that the decoding becomes anatomically correct, as defined by the criteria described above. This guarantees anatomical correctness of the predicted myocardium mask.
Finally, the 2-D myocardial segmentations were reconstructed to volumes and additional automatic volumetric checks were applied to remove segmentations from images located below the apex or above the base of the LV. We compared ratios of myocardium to blood pool areas of each slice to identify the longest subsequence of slices in the stack. The threshold used to determine whether to include a slice in the subsequence was approximately a 40% maximum decrease in LV area. Segmented volumes were truncated at the index , where refers to the final index in the subsequence; represents the index of the first “C”-shaped slice (a myocardial segmentation shape that occurs at the boundary of the ventricle and the atrium in the basal region); and represents the index of a large deviation (drop to 60% or increase of 60%) in LV area between successive slices. This check allowed incorporation of at most 1 “C”-shaped slice and excluded slices above the base with no true ROI. The numerical values for the thresholds were determined by ensuring no more than 5% of the ground truth segmented slices would be discarded. Final predicted myocardial segmentations of patient scans therefore passed both per-slice and per-volume anatomical constraints.
Training and evaluation
All train/test splits were performed on the patient level. The training data set consisted of 2484 images from 2 sources: 1124 2-D LGE-CMR slices from 75% of available patients and all 1360 “LGE-like” images. The test set contained only LGE-CMR images from the remaining 25% of patients (269 2-D images). For the myocardium segmentation network, only LGE-CMR scans with enhancement segmentation ground truth were used (roughly 80% of the train and test sets). The autoencoder used ground truth myocardial segmentations from all the available training data. No early stopping or other methods that learn from the validation set were used in training.
To prevent cine-derived “LGE-like” images from dominating the training set, they were weighed less in the loss function.
The loss function used was an equally weighted combination of the balanced cross-entropy loss and the Tversky loss33:
where and are pixel ground truth and predicted values, T/F P/N are true/false positive/negatives, and is a weight on the false positives, which was modulated up to = 0.6 in the first network to avoid over-cropping and down to = 0.4 in the second to limit outliers. The final loss combined per-pixel mean loss (l1) and per-image (l2) loss in equal proportions to incorporate both local and holistic performance. All networks used the Adam optimizer34 with learning rate of 10−3 and trained on NVIDIA Titan RTX graphics processing units using Keras35 and Tensorflow.36
We evaluated ACSNet’s segmentation performance (Table 1) using balanced accuracy (BA), Sørensen-Dice (Dice) coefficient, and Hausdorff distance (HD)37 as metrics. Values were computed by averaging slice values over section of the heart (apex, mid ventricle, base) and over the total heart. Sections of the heart were determined by equipartitioning the short-axis distance between the first and last slice.
Table 1.
ACSNet cardiac magnetic resonance imaging with late gadolinium enhancement segmentation performance
| Measure | Apex | Location Middle |
Base | Total | |
|---|---|---|---|---|---|
| LV ROI | |||||
| MYO | BA (%) | 96.3 ± 0.4 | 96.3 ± 0.4 | 96.2 ± 0.5 | 96.3 ± 0.2 |
| Dice | 0.92 ± 0.02 | 0.95 ± 0.01 | 0.94 ± 0.01 | 0.93 ± 0.01 | |
| HD (mm) | 6.9 ± 1.6 | 6.1 ± 0.8 | 6.6 ± 1.5 | 6.5 ± 0.8 | |
| Enhancement region | BA (%) | 93.3 ± 1.1 | 93.1 ± 1.2 | 92.9 ± 1.4 | 93.1 ± 0.7 |
| Dice | 0.75 ± 0.04 | 0.82 ± 0.02 | 0.80 ± 0.04 | 0.79 ± 0.02 | |
| HD (mm) | 6.4 ± 0.7 | 6.6 ± 0.7 | 7.2 ± 1.5 | 6.7 ± 0.6 | |
| Core scar region | BA (%) | 69.9 ± 2.3 | 69.8 ± 2.6 | 70.4 ± 2.6 | 70.0 ± 1.4 |
| Dice | 0.51 ± 0.06 | 0.48 ± 0.07 | 0.59 ± 0.09 | 0.51 ± 0.04 | |
| HD (mm) | 16.8 ± 3.4 | 24.0 ± 6.6 | 19.8 ± 8.7 | 19.9 ± 3.3 | |
| BA (%) | 74.9 ± 2.8 | 74.3 ± 3.1 | 75.5 ± 3.3 | 74.9 ± 1.8 | |
| Dice | 0.57 ± 0.08 | 0.52 ± 0.09 | 0.63 ± 0.11 | 0.57 ± 0.05 | |
| HD (mm) | 14.9 ± 3.7 | 24.4 ± 6.7 | 18.1 ± 8.9 | 18.9 ± 3.5 |
Balanced accuracy (BA), Dice coefficient (Dice), and Hausdorff distance (HD) are shown for 4 regions of interest (ROI) segmented by ACSNet: whole left ventricle (LV ROI), myocardial tissue (MYO), area of enhancement (Enhancement region), and scar tissue (Core scar region).
BA is expressed in percentage terms, Dice is adimensional, and HD is in millimeters. All numbers are averages ± 95% confidence interval size over apex/middle/base/total slices of all patients in the test set.
Additionally, we evaluated LV ROI (myocardium and blood pool) volume, myocardium volume, enhancement region volume, and core scar region volume derived from the segmentations (Table 2). Volumes were calculated by summing voxel volumes and using nearest-neighbor interpolation between slices. Mean absolute errors (MAE) were normalized to the respective ground truth volume. To quantify core scar, the enhanced (scar/fibrosis) region segmented by the network was used to extract the dense core scar region using a modified version of the full width at half maximum38,39 algorithm.
Table 2.
ACSNet cardiac magnetic resonance imaging with late gadolinium enhancement clinical feature performance
| Feature | LV Volume Tertile |
Total | ||
|---|---|---|---|---|
| Lower | Middle | Upper | ||
| LV ROI | ||||
| GT (cc) | 226 (186–259) | 307 (280–327) | 405 (334–573) | 312 (186–573) |
| Pred (cc) | 237 (193–273) | 312 (279–339) | 424 (342–614) | 323 (193–614) |
| Norm. MAE (%) | 10.3 (4.8–18.8) | 4.5 (0.6–8.0) | 4.4 (1.3–10.3) | 6.3 (0.6–18.8) |
| MYO | ||||
| GT (cc) | 121 (85–159) | 171 (110–215) | 180 (114–274) | 158 (85–274) |
| Pred (cc) | 144 (109–186) | 187 (144–226) | 217 (159–351) | 183 (109–351) |
| Norm. MAE (%) | 24.1 (12.6–41.1) | 12.7 (1.8–30.8) | 24.7 (2.2–53.7) | 20.1 (1.8–53.7) |
| Enhancement region | ||||
| GT (cc) | 27 (15–46) | 24 (3–39) | 30 (7–47) | 27 (3–47) |
| Pred (cc) | 21 (7–43) | 19 (0–33) | 26 (3–38) | 22 (0–43) |
| Norm. MAE (%) | 26.6 (5.9–53.4) | 31.1 (3.8–100.0) | 45.4 (15.9–87.5) | 34.2 (3.8–100.0) |
| Core scar region | ||||
| GT (cc) | 13 (6–21) | 11 (1–19) | 17 (4–30) | 13 (1–30) |
| Pred (cc) | 11 (5–20) | 9 (0–17) | 14 (2–22) | 12 (0–22) |
| Norm. MAE (%) | 14.4 (1.2–41.2) | 42.5 (18.2–100.0) | 42.7 (32.1–72.0) | 33.7 (1.2–100.0) |
Ground truth (GT) and predicted (Pred.) volumes and mean absolute error normalized by GT volume (Norm. MAE), together with ranges (parentheses), are shown for 4 regions of interest (ROI) segmented by ACSNet: whole left ventricle (LV ROI), myocardial tissue (MYO), area of enhancement (Enhancement region), and scar tissue (Core scar region). GT and Pred. are expressed in cubic centimeters and Norm. MAE in percentage terms. Numbers represent averages across all patients in the test set (Total) and patients grouped by GT LV volume tertile (lower/middle/upper).
The remote nonenhanced myocardium intensity used by the full width at half maximum algorithm was automatically determined as the median intensity value outside the predicted enhancement region. Differences between ground truth and predictions were reported as the MAE normalized relative to the ground truth value.
Statistical methods
All data analysis in this manuscript was performed using Python 3.4 and open source packages. All results presented without a qualifier represent averages over slices or patients from the 25% of the contrast-enhanced data reserved for testing using a random split. Prediction error was estimated using approximately normal confidence intervals for large n (eg, number of slices) and minimum/maximum ranges for small n (eg, number volumes). Statistically significant difference testing was assessed using Welch’s t test using the Python package scipy.
Results
Segmentation performance
ACSNet segmentations were evaluated using BA, Dice, and HD computed on the test set (see Methods). Table 1 shows that LV ROI identification (first subnetwork) resulted in BA of 96%, Dice coefficient of 0.93, and HD of 6.5 mm. The second subnetwork resulted in 93%, 0.79, and 6.7 mm for the LV myocardium using the same metrics. The same subnetwork evaluated for the identification of the enhancement region led to 70% BA, 0.51 Dice, and 19.9 mm HD. The core scar portion of the enhanced region achieved BA of 74.9%, Dice of 0.57, and HD of 18.9 mm. The anatomical postprocessing (third subnetwork) did not have significant impact on performance metrics.
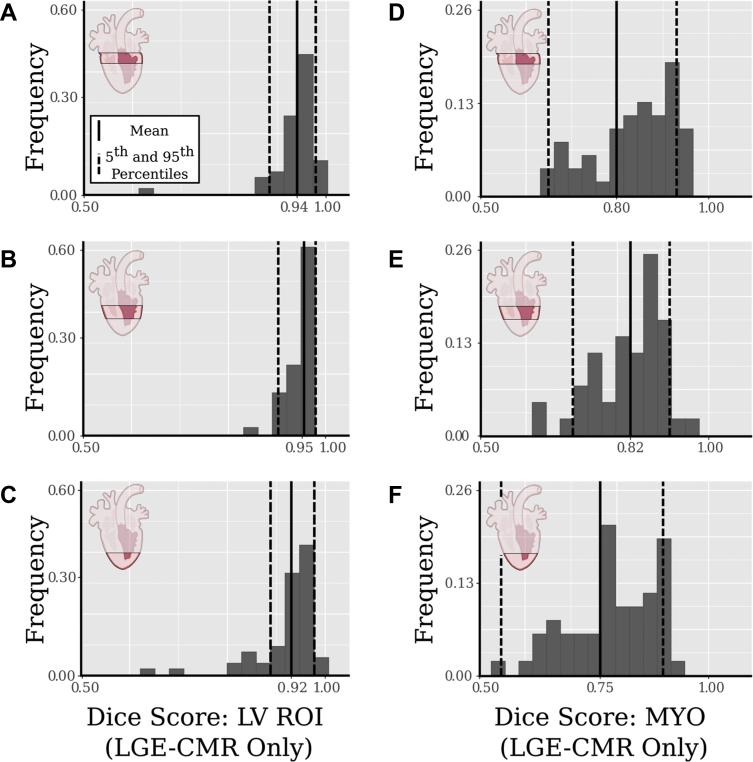
Figure 6 illustrates the consistency of ACSNet results across 3 regions of the LV (apex, middle, and base) through histograms of per-slice Dice scores. Dice scores are shown for the LV ROI (Figure 6A–6C) and LV myocardial segmentations (Figure 6D–6F). The average Dice scores for each region are 0.94 (Figure 6A) and 0.80 (Figure 6D) for basal slices, 0.95 (Figure 6B) and 0.82 (Figure 6E) for middle slices, and 0.92 (Figure 6C) and 0.75 (Figure 6F) for apical slices. Further details of LV ROI and myocardial segmentations for the 3 regions of the ventricle are in Table 1.
Figure 6.
Left ventricle (LV) region of interest (ROI) and myocardium segmentation results by region. Histograms of per-slice Dice scores are shown for 3 regions of the heart (rows from top to bottom: basal, middle, and apical). Columns represent the left ventricle (LV ROI) segmentation (A–C) and myocardium (MYO) segmentation (D–F). The averages are shown as solid vertical lines, and the dotted lines represent the 5th and 95th percentiles. LGE-CMR = cardiac magnetic resonance imaging with late gadolinium enhancement.
Table 3 presents a comparison of Dice scores and HD for previously published LV myocardial segmentation methods, showing that ACSNet achieved the lowest HD among those LGE-CMR myocardium segmentation methods. The Dice score is similar to the other methods’ results. ACSNet improved upon the interobserver Dice score of 0.76 as well as the interobserver HD (10.6 ± 4.65 mm endocardial HD and 12.5 ± 5.38 mm epicardial HD) achieved in the Multi-Sequence Cardiac MRI Segmentation Challenge.40,41
Table 3.
Comparison of cardiac magnetic resonance imaging with late gadolinium enhancement segmentation results for the left ventricle myocardium
| Method | MYO Dice score | MYO Hausdorff distance (mm) |
|---|---|---|
| ACSNet | 0.79 ± 0.02 | 6.70 ± 0.53 |
| Interobserver40,41 | 0.76 ± 0.08 | 12.50 ± 5.38 |
| Zabihollahy et al24 | 0.85 ± 0.03 | 19.21 ± 4.74 |
| Yue et al20 | 0.76 ± 0.23 | 11.04 ± 5.82 |
| Roth et al21 | 0.78 | 16.30 |
| Mean result of MS-CMRSeg MICCAI Challenge45 | 0.77 ± 0.10 | 18.06 ± 12.18 |
| Chen et al22 | 0.83 ± 0.04 | 12.45 ± 3.14 |
All entries were rounded from the provided values to the nearest tenths place. Note: These sources use different data sets or additional cardiac magnetic resonance (CMR) scans; data for Interobserver,40,41 Yue et al,20 Roth et al,21 and Chen et al22 are based on the 2019 CMRSeg MICCAI challenge45 consisting of 2-D late gadolinium enhancement (LGE)-CMR and corresponding balanced steady-state free precision from 45 patients, various subsets of whom were used as test sets. Zabihollahy et al24 used 3 orthogonal views of 34 subjects with 3-D LGE-CMR scans.
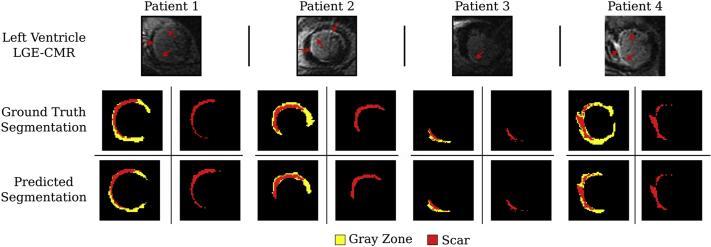
Figure 7 illustrates examples of ACSNet’s performance in terms of scar segmentation. The first row shows the original scan, the middle row presents the ground truth scar and GZ segmentations, and the bottom row shows the predicted segmentations. Results for patients 1–3 are representative examples of scar and GZ segmentations. Patient 4 was included to show an example of an outlier for which GZ segmentation has low accuracy. Of note, the low GZ accuracy did not hinder the scar segmentation performance. Balanced accuracy, Dice score, and HD for all enhancement and core scar for the apex, middle, and base of the LV are shown in Table 1.
Figure 7.
Scar segmentation results. Segmentations of enhancement regions from the myocardium segmentation network represent gray zone (yellow) and scar (red). The first row shows the original scan, the middle row shows the ground truth scar and gray zone segmentations, and the bottom row shows the predicted segmentations. LGE-CMR = cardiac magnetic resonance imaging with late gadolinium enhancement.
Clinical feature calculation
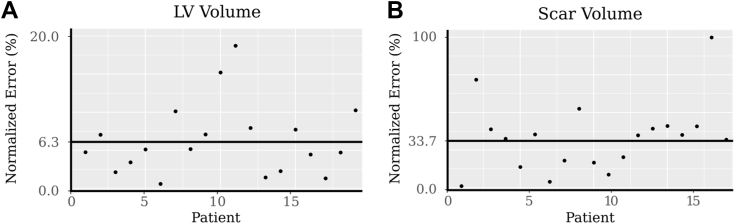
ACSNet was used to seamlessly calculate clinical features, such as scar burden and LV volume. Our results demonstrate no statistically significant difference between features computed using automatic vs manual (expert-level) myocardium and regions of enhancement segmentations (P value = .71 for LV volume and P value = .46 for scar volume). Figure 8A shows the normalized absolute error of each LV volume in the test set (dots), together with the MAE at 6.3% (line). Similarly, Figure 8B shows scar volume comparison to ground truth, which resulted in an MAE of 33.7%. Also shown are each patient’s scar volume absolute errors (dots). Table 2 presents ground truth and predicted volumes of the entire LV, LV myocardium, enhancement region, and core scar volumes in cubic centimeters (cc), as well as MAE in patients grouped by volume of the LV. Scar burden, calculated as the mean scar-to-myocardium volume fraction, differed by 2% when comparing automatic and manual segmentations (Figure 8).
Figure 8.
Scar and left ventricle (LV) volumes. LV (A) and scar (B) volume error is computed as the absolute error normalized by each respective volume. Each point represents the error in LV volume of a single segmented patient scan. The solid black line shows the mean.
Discussion
In this study we present a DL approach for automatic and anatomically accurate segmentation of myocardium and scar/fibrosis on LGE-CMR images and for extraction of anatomical features, such as scar burden and ventricular volume. The complex learning process involves 3 subnetworks, each having distinct tasks: the first reduces class imbalance between the ROI and background, the second delineates the endocardium and epicardium, and the third ensures anatomical correctness for both slices and volumes. We show that ACSNet outperforms interexpert (ie, manual) scores and performs well on inputs with various scar distribution patterns acquired from numerous imaging centers and MRI machines. Our technology is seamlessly employed to extract clinical anatomical covariate data, potentially enhancing the prognostic utility of LGE-CMR.
Advantages of ACSNet
ACSNet fully automates the segmentation of LV LGE-CMR images. The high number of manual steps and the interobserver variability associated with this task have hindered implementing LGE-CMR image analysis as part of routine patient assessment and prognostication. For instance, scar burden and LV volume computed from myocardial and scar/fibrosis segmentations have been associated with risk of SCD42 but are seldom used in practice to guide primary prevention. ACSNet can produce accurate segmentations within seconds from raw medical images, making it possible to more easily incorporate LGE-CMR image analysis in clinical decision-making.
ACSNet achieves good performance despite the complexity of LGE-CMR images. Contouring of LGE-CMR images is complicated by the presence of both low- (viable) and high- (scar/fibrosis) signal-intensity myocardium regions. As a result, manual segmentations can be variable even across experts, potentially affecting estimated features of clinical interest. The same complications also affect computer-aided segmentation algorithms, which can struggle with visually similar, but distinct anatomical entities (eg, myocardium and blood pool). ACSNet’s results demonstrate robust learning, leading to reliable segmentations, despite inherent noise present in ground truth data. There were no clear common characteristics of the cases that had lower performance. The network maintains consistently high performance across all regions of the heart. This is prioritized by design in favor of higher average Dice scores with poor-performing outlier slices. Despite ACSNet’s success with whole-ventricle segmentation, some outliers were present when segmenting the area of enhancement (Figure 7, patient 4). However, the proposed method to tease out the core scar does not seem sensitive to the overestimation of GZ, leading to accurate clinical feature estimation.
In this study, we use a novel method to ensure anatomical accuracy of segmentations: we integrate, in the DL approach, an additional deep neural network (ACSNet’s third subnetwork) encompassing a number of per-slice and per-volume morphological checks. The distribution-based model of the latent space allows for complex anatomical segmentations such as “C”-shaped myocardium, which can occur in the ventricle’s base, while also smoothing out potentially erroneous myocardial segmentation predictions. This low number of large deviations from ground truth is illustrated by ACSNet’s performance in terms of HD (Table 3). Moreover, ACSNet uses volumetric checks that standardize and automate the identification of apical and basal (beginning and end) slices, a time-consuming and often error-prone process when performed manually. Importantly, these checks also establish consistency and reliability in the calculation of clinical features (eg, LV volume and scar burden).
ACSNet takes advantage of the more widely available cine data with ground truth segmentation labels and addresses the scarcity of available segmented LGE-CMR data. Importantly, ACSNet performs well despite data scarcity owing to the innovative style transfer process to augment the training data presented here. This process generates pseudo-enhancement for nonenhanced cine using a low-cost cine-to-LGE conversion algorithm. The method tripled the available training data and added heterogeneity to the learning process in terms of patient cohorts and health centers. By training ACSNet with both LGE and “LGE-like” cine CMR images from a broad range of cohorts, the technology holds the promise to fully automate segmentation of short-axis cardiac images across multiple medical imaging modalities. For example, since style-transferred cine images were already part of training, ACSNet would be expected to segment cine scans with high accuracy. Similarly, given that signal intensity preprocessing was minimal, our approach could generalize to computed tomography images, which, like CMR, display a high-intensity blood pool and low-intensity myocardium. Finally, ACSNet performance was validated on ICM patient data, though ACSNet could be easily applied to non-ICM patient scans as well.
Lastly, we envision ACSNet to be an important and necessary first step in a number of fields related to cardiac imaging. For example, in machine learning or radiomics applied to CMR, having an efficient way to discard information outside the ROI can greatly enhance models’ abilities to learn without getting bogged down with extraneous information. Furthermore, personalized computational heart modeling simulating cardiac electrophysiology to identify arrhythmogenic pathways and arrhythmia dynamics14, 15, 16 or the targets for ablation therapy43,44 often require segmentations to capture heart geometry and scar distribution. Their efficiency and robustness could therefore be drastically improved by ACSNet.
Comparison to other work
Segmentation algorithms for the LV myocardium have predominantly focused on cine CMR images. Despite promising advances, most cine segmentation algorithms still require manual steps. For example, the method by Zheng and colleagues19 requires a preprocessing step to discard apical and basal slices and a manual curation of “difficult cases.” An attempt by Bello and colleagues18 at segmenting cine images relies on ground truth landmark annotations to prevent anatomically inconsistent outliers. The current limitations in cine segmentation have demonstrated that LGE-CMR require a tailored segmentation algorithm, rather than a reimplementation of methods developed for cine scans.
Some DL methods have been proposed specifically for LGE-CMR myocardial or scar segmentation; however, these solutions also have a number of limitations. The approach by Campello and colleagues23 for segmenting the myocardium in LGE-CMR images attempted to address LGE-CMR data scarcity by using a costly DL cine-to-LGE style transfer approach. However, in the process, the style-transferred cine images lost the salient aspect of LGE-CMR, the scar/fibrosis features. A recent attempt by Zabihollahy and colleagues24 at myocardial and scar/fibrosis segmentation on 3-D LGE-CMR resulted in artifacts, such as disjoint pieces of the myocardium, despite the benefit of a 10-fold increase in the number of slices per patient furnished by the 3-D acquisition. Additionally, 3-D LGE-CMR data are relatively scarce, as most hospitals and healthcare centers acquire in 2-D. The 2019 CMRSeg MICCAI challenge for myocardial segmentation20, 21, 22, 49,45 and a study focused on scar segmentation27 both required LGE-CMR and corresponding cine scans for each patient. Furthermore, Fahmy and colleagues27 exclusively utilized images of patients with hypertrophic cardiomyopathy and did not present overall myocardial segmentation performance, which could have been traded off for better scar segmentation. An attempt by Moccia and colleagues25 at predicting enhancement segmentations required manually segmented ground truth myocardium as an additional network input; this requirement limited their data set to only 30 patients, all from a single center.
A few recent methods have proposed postprocessing steps to improve the anatomical accuracy of myocardial segmentations from cine images.46, 47 Although these algorithms smooth out resulting segmentations, they have a number of limitations: they use generic techniques unable to capture nuances of heart anatomy47; they require an already highly accurate segmentation as input to function well46; or they do not incorporate 3-D constraints.46
Study limitations
As with any DL algorithms, ACSNet could benefit from a larger cohort with more diverse image data, including different imaging centers, machines, scar patterns based on different cardiomyopathies, etc. The LGE-CMR images in this study originated exclusively from ICM patients, which could affect the algorithm’s ability to generalize to non-ICM. However, while the pseudo-enhancement generated for the synthetic data was done through a random process and not based on established scar distributions, its addition improved the network’s ability to identify regions of enhancement.
Another limitation inherent to supervised learning tasks in medical imaging is the potential subjectivity of ground truth. Considering that myocardial segmentation interobserver Dice can be as low as 0.76,40,41 one can expect even lower concordance for the more complicated regions of scar/fibrosis. A supervised DL algorithm is trained to best replicate whatever ground truth it is provided, the quality of which, therefore, sets an upper bound on the algorithm’s segmentation performance. Given the significant interobserver disagreement on ground truth, exceedingly high concordance of an algorithm with 1 expert should be seen as a sign of poor generalization, rather than strong performance. In ACSNet, small potential increases in performance metrics are explicitly traded off for reasonable segmentations that preserve anatomical cardiac features and avoid artifacts.
Additionally, we did not perform a broad hyperparameter sweep when developing the structure of our networks. However, manual exploration of different hyperparameter implementations resulted in similar Dice scores for myocardium segmentation, which indicates the robustness of the core ACSNet model. This is suggested, for example, in Table 3, where ACSNet myocardial segmentations have significantly lower HD when compared to other methods. However, it is important to stress that the comparison in Table 3 should be interpreted more as a general guidepost rather than a precise comparison, given that either the presented segmentation methods require additional inputs unavailable for our dataset (eg, corresponding/matching cine MRI for each LGE scan), or the details of the methodology were not made publicly available for us to replicate.
Conclusion
We demonstrated an efficient, cost-effective DL solution for automatic expert-level segmentation of LV LGE-CMR images and seamless, accurate derivation of scar burden and LV volume. ACSNet segmentations uphold anatomical constraints and have been trained on scans from heterogeneous cohorts, thus offering the potential for much broader application across imaging modalities and patient pathologies.
Acknowledgments
Funding Sources
NT acknowledges support from NIH (grants R01HL142496, R01HL124893 and U01HL126273), a grant from the Leducq Foundation, and a grant for the Lowenstein Foundation. MM is grateful for support from the Simons Foundation, from NSF 2031985, 1837991, and AFOSR FA9550-20-1-0288. KCW is grateful for support from NIH grant HL103812 and HL132181.
Disclosures
The authors have no conflict to disclose.
Authorship
All authors attest they meet the current ICMJE criteria for authorship.
Patient Consent
All patients provided written informed consent.
Ethics Statement
The research protocol used in this study was reviewed and approved by the Johns Hopkins University Institutional Review Board.
Appendix Supplementary data
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.cvdhj.2021.11.007.
Supplemental Material
References
- 1.Zipes D.P., Wellens H.J.J. Sudden cardiac death. Circulation. 1998;98:2334–2351. doi: 10.1161/01.cir.98.21.2334. [DOI] [PubMed] [Google Scholar]
- 2.Kuruvilla S., Adenaw N., Katwal A.B., Lipinski M.J., Kramer C.M., Salerno M. Late gadolinium enhancement on cardiac magnetic resonance predicts adverse cardiovascular outcomes in nonischemic cardiomyopathy: a systematic review and meta-analysis. Circ Cardiovasc Imaging. 2014;7:250–258. doi: 10.1161/CIRCIMAGING.113.001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marco A.D., Anguera I., Schmitt M., et al. Late gadolinium enhancement and the risk for ventricular arrhythmias or sudden death in dilated cardiomyopathy: systematic review and meta-analysis. JACC Heart Fail. 2017;5:28–38. doi: 10.1016/j.jchf.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 4.van der Burg A.E.B., Bax J.J., Boersma E., Pauwels E.K.J., Wall E.E., Schalij M.J. Impact of viability, ischemia, scar tissue, and revascularization on outcome after aborted sudden death. Circulation. 2003;108:1954–1959. doi: 10.1161/01.CIR.0000091410.19963.9A. [DOI] [PubMed] [Google Scholar]
- 5.Wu K.C. Sudden cardiac death substrate imaged by MRI: from investigational tool to clinical applications. Circ Cardiovasc Imaging. 2017;10:7. doi: 10.1161/CIRCIMAGING.116.005461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roes S.D., Borleffs C.J.W., Geest R.J., et al. Infarct tissue heterogeneity assessed with contrast-enhanced MRI predicts spontaneous ventricular arrhythmia in patients with ischemic cardiomyopathy and implantable cardioverter-defibrillator. Circ Cardiovasc Imaging. 2009;2:183–190. doi: 10.1161/CIRCIMAGING.108.826529. [DOI] [PubMed] [Google Scholar]
- 7.Scott P.A., Morgan J.M., Carroll N., et al. The extent of left ventricular scar quantified by late gadolinium enhancement MRI is associated with spontaneous ventricular arrhythmias in patients with coronary artery disease and implantable cardioverter-defibrillators. Circ Arrhythm Electrophysiol. 2011;4:324–330. doi: 10.1161/CIRCEP.110.959544. [DOI] [PubMed] [Google Scholar]
- 8.AlJaroudi W.A., Flamm S.D., Saliba W., Wilkoff B.L., Kwon D. Role of CMR imaging in risk stratification for sudden cardiac death. JACC Cardiovasc Imaging. 2013;6:392–406. doi: 10.1016/j.jcmg.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Kim R.J., Fieno D.S., Parrish T.B., et al. Relationship of MRI delayed contrast enhancement to irreversible injury, infarct age, and contractile function. Circulation. 1999;100:1992–2002. doi: 10.1161/01.cir.100.19.1992. [DOI] [PubMed] [Google Scholar]
- 10.Disertori M., Rigoni M., Pace N., et al. Myocardial fibrosis assessment by LGE is a powerful predictor of ventricular tachyarrhythmias in ischemic and nonischemic LV dysfunction: a meta-analysis. JACC Cardiovasc Imaging. 2016;9:1046–1055. doi: 10.1016/j.jcmg.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 11.Mordi I., Jhund P.S., Gardner R.S., et al. LGE and NT-ProBNP identify low risk of death or arrhythmic events in patients with primary prevention ICDs. JACC Cardiovasc Imaging. 2014;7:561–569. doi: 10.1016/j.jcmg.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Cain M.A., Metzl M.D., Patel A.R., et al. Cardiac sarcoidosis detected by late gadolinium enhancement and prevalence of atrial arrhythmias. Am J Cardiol. 2014;113:1556–1560. doi: 10.1016/j.amjcard.2014.01.434. [DOI] [PubMed] [Google Scholar]
- 13.Marra M.P., De Lazzari M., Zorzi A., et al. Impact of the presence and amount of myocardial fibrosis by cardiac magnetic resonance on arrhythmic outcome and sudden cardiac death in nonischemic dilated cardiomyopathy. Heart Rhythm. 2014;11:856–863. doi: 10.1016/j.hrthm.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Balaban G., Halliday B.P., Porter B., et al. Late-gadolinium enhancement interface area and electrophysiological simulations predict arrhythmic events in patients with nonischemic dilated cardiomyopathy. JACC Clin Electrophysiol. 2021;7:238–249. doi: 10.1016/j.jacep.2020.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kucukseymen S., Yavin H., Barkagan M., et al. Discordance in scar detection between electroanatomical mapping and cardiac MRI in an infarct swine model. JACC Clin Electrophysiol. 2020;6:1452–1464. doi: 10.1016/j.jacep.2020.08.033. [DOI] [PubMed] [Google Scholar]
- 16.Arevalo H.J., Vadakkumpadan F., Guallar E., et al. Arrhythmia risk stratification of patients after myocardial infarction using personalized heart models. Nat Commun. 2016;7:11437. doi: 10.1038/ncomms11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashikaga H., Arevalo H., Vadakkumpadan F., et al. Feasibility of image-based simulation to estimate ablation target in human ventricular arrhythmia. Heart Rhythm. 2013;8:1109–1116. doi: 10.1016/j.hrthm.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bello G.A., Dawes T.J.W., Duan J., et al. Deep-learning cardiac motion analysis for human survival prediction. Nat Mach Intell. 2019;1:95–104. doi: 10.1038/s42256-019-0019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Q., Delingette H., Duchateau N., Ayache N. 3D consistent and robust segmentation of cardiac images by deep learning with spatial propagation. IEEE Trans Med Imaging. 2018:1. doi: 10.1109/TMI.2018.2820742. [DOI] [PubMed] [Google Scholar]
- 20.Yue Q., Luo X., Ye Q., Xu L., Zhuang X. Cardiac segmentation from LGE MRI using deep neural network incorporating shape and spatial priors. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. 2019;11765 LNCS:559–567. [Google Scholar]
- 21.Roth H., Zhu W., Yang D., Xu Z., Xu D. In: Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges. Pop M., Sermesant M., Camara O., et al., editors. Springer International Publishing; 2020. Cardiac segmentation of LGE MRI with noisy labels; pp. 228–236. [Google Scholar]
- 22.Chen C., Ouyang C., Tarroni G., et al. International Workshop on Statistical Atlases and Computational Models of the Heart. Springer; 2019. Unsupervised multi-modal style transfer for cardiac MR segmentation; pp. 209–219. [Google Scholar]
- 23.Campello V., Martín-Isla C., Izquierdo C., Petersen S.E., Ballester M.A.G., Lekadir K. International Workshop on Statistical Atlases and Computational Models of the Heart. Springer; 2019. Combining multi-sequence and synthetic images for improved segmentation of late gadolinium enhancement cardiac MRI; pp. 290–299. [Google Scholar]
- 24.Zabihollahy F., Rajchl M., White J.A., Ukwatta E. Fully automated segmentation of left ventricular scar from 3D late gadolinium enhancement magnetic resonance imaging using a cascaded multi-planar U-net (CMPU-net) Med Phys. 2020;47:1645–1655. doi: 10.1002/mp.14022. [DOI] [PubMed] [Google Scholar]
- 25.Moccia S., Banali R., Martini C., et al. Development and testing of a deep learning-based strategy for scar segmentation on CMR-LGE images. MAGMA. 2019;32:187–195. doi: 10.1007/s10334-018-0718-4. [DOI] [PubMed] [Google Scholar]
- 26.Fahmy A.S., Neisius U., Chan R.H., et al. Three-dimensional deep convolutional neural networks for automated myocardial scar quantification in hypertrophic cardiomyopathy: a multicenter multivendor study. Radiology. 2019;294:190737. doi: 10.1148/radiol.2019190737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahmy A.S., Rowin E., Chan R.H., Manning W.J., Maron M.S., Nezafat R. Improved quantification of myocardium scar in late gadolinium enhancement images: deep learning based image fusion approach. J Magn Reson Imaging. 2021;54:303–312. doi: 10.1002/jmri.27555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu K.C., Gerstenblith G., Guallar E., et al. Combined cardiac magnetic resonance imaging and C-reactive protein levels identify a cohort at low risk for defibrillator firings and death. Circ Cardiovasc Imaging. 2012;5:178–186. doi: 10.1161/CIRCIMAGING.111.968024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernard O., Lalande A., Zotti C., et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Trans Med Imaging. 2018;37:2514–2525. doi: 10.1109/TMI.2018.2837502. [DOI] [PubMed] [Google Scholar]
- 30.Radau P., Lu Y., Connelly K., Paul G., Dick A., Wright G. Evaluation framework for algorithms segmenting short axis cardiac MRI. The MIDAS Journal. 2009 [Google Scholar]
- 31.Pizer S.M., Amburn E.P., Austin J.D., et al. Adaptive histogram equalization and its variations. Computer Vision, Graphics, and Image Processing. 1987;39:355–368. [Google Scholar]
- 32.Ronneberger O., Fischer P., Brox T. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Navab N., Hornegger J., Wells W., Frangi A., editors. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 33.Salehi S.S.M., Erdogmus D., Gholipour A. International Workshop on Machine Learning in Medical Imaging. Springer; 2017. Tversky loss function for image segmentation using 3D fully convolutional deep networks; pp. 379–387. [Google Scholar]
- 34.Kingma D., Ba J. Adam: a method for stochastic optimization. Int. Conf. on Learn. Represent. 2014 [Google Scholar]
- 35.Chollet François, others. Keras 2015. https://keras.io Retrieved from.
- 36.Abadi M., Agarwal A., Barham P., et al. TensorFlow: large-scale machine learning on heterogeneous systems. 2015. tensorflow.org Software available from.
- 37.Birsan T., Tiba D. IFIP Conference on System Modeling and Optimization. Springer; 2005. One hundred years since the introduction of the set distance by Dimitrie Pompeiu; pp. 35–39. [Google Scholar]
- 38.Schmidt A., Azevedo C.F., Cheng A., et al. Infarct tissue heterogeneity by magnetic resonance imaging identifies enhanced cardiac arrhythmia susceptibility in patients with left ventricular dysfunction. Circulation. 2007;115:2006–2014. doi: 10.1161/CIRCULATIONAHA.106.653568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jablonowski R., Chaudhry U., Pals J., et al. Cardiovascular magnetic resonance to predict appropriate implantable cardioverter defibrillator therapy in ischemic and nonischemic cardiomyopathy patients using late gadolinium enhancement border zone: comparison of four analysis methods. Circ Cardiovasc Imaging. 2017;10 doi: 10.1161/CIRCIMAGING.116.006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhuang X. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2016. Multivariate mixture model for cardiac segmentation from multi-sequence MRI; pp. 581–588. [Google Scholar]
- 41.Zhuang X. Multivariate mixture model for myocardial segmentation combining multi-source images. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2019;41:2933–2946. doi: 10.1109/TPAMI.2018.2869576. [DOI] [PubMed] [Google Scholar]
- 42.Klem I., Weinsaft J.W., Bahnson T.D., et al. Assessment of myocardial scarring improves risk stratification in patients evaluated for cardiac defibrillator implantation. J Am Coll Cardiol. 2012;60:408–420. doi: 10.1016/j.jacc.2012.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prakosa A., Arevalo H.J., Deng D., et al. Personalized virtual-heart technology for guiding the ablation of infarct-related ventricular tachycardia. Nat Biomed Eng. 2018;2:732–740. doi: 10.1038/s41551-018-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ballistipedia Bivariate. 2020. http://ballistipedia.com/index.php?title=File:Bivariate.png Retrieved from.
- 45.Boyle P.M., Zghaib T., Zahid S., et al. Computationally guided personalized targeted ablation of persistent atrial fibrillation. Nat Biomed Eng. 2019;3:870–879. doi: 10.1038/s41551-019-0437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhuang X., Xu J., Luo X., et al. Cardiac segmentation on late gadolinium enhancement MRI: a benchmarkstudy from multi-sequence cardiac MR segmentation challenge. arXiv preprint arXiv. 2006 doi: 10.1016/j.media.2022.102528. [DOI] [PubMed] [Google Scholar]
- 47.Painchaud N., Skandarani Y., Judge T., Olivier B., Lalande A., Jodoin P.M. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2019. CardiacMRI segmentation with strong anatomical guarantees; pp. 632–640. [Google Scholar]
- 48.Larrazabal A.J., Martínez C., Glocker B., Ferrante E. Post-DAE: Anatomically plausible segmentation viapost-processing with denoising autoencoders. IEEE Trans Med Imaging. 2020;39:3813–3820. doi: 10.1109/TMI.2020.3005297. [DOI] [PubMed] [Google Scholar]
- 49.Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19:716–723. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.