Abstract
Background
Cardiac magnetic resonance (CMR) is the gold standard for left ventricular hypertrophy (LVH) diagnosis. CMR-derived LV mass can be estimated using proprietary algorithms (eg, InlineVF), but their accuracy and availability may be limited.
Objective
To develop an open-source deep learning model to estimate CMR-derived LV mass.
Methods
Within participants of the UK Biobank prospective cohort undergoing CMR, we trained 2 convolutional neural networks to estimate LV mass. The first (ML4Hreg) performed regression informed by manually labeled LV mass (available in 5065 individuals), while the second (ML4Hseg) performed LV segmentation informed by InlineVF (version D13A) contours. We compared ML4Hreg, ML4Hseg, and InlineVF against manually labeled LV mass within an independent holdout set using Pearson correlation and mean absolute error (MAE). We assessed associations between CMR-derived LVH and prevalent cardiovascular disease using logistic regression adjusted for age and sex.
Results
We generated CMR-derived LV mass estimates within 38,574 individuals. Among 891 individuals in the holdout set, ML4Hseg reproduced manually labeled LV mass more accurately (r = 0.864, 95% confidence interval [CI] 0.847–0.880; MAE 10.41 g, 95% CI 9.82–10.99) than ML4Hreg (r = 0.843, 95% CI 0.823–0.861; MAE 10.51, 95% CI 9.86–11.15, P = .01) and InlineVF (r = 0.795, 95% CI 0.770–0.818; MAE 14.30, 95% CI 13.46–11.01, P < .01). LVH defined using ML4Hseg demonstrated the strongest associations with hypertension (odds ratio 2.76, 95% CI 2.51–3.04), atrial fibrillation (1.75, 95% CI 1.37–2.20), and heart failure (4.67, 95% CI 3.28–6.49).
Conclusions
ML4Hseg is an open-source deep learning model providing automated quantification of CMR-derived LV mass. Deep learning models characterizing cardiac structure may facilitate broad cardiovascular discovery.
Keywords: Cardiovascular disease, Convolutional neural network, Deep learning, Left ventricular mass, Machine learning
Key Findings.
-
•
We have developed an open-source deep learning left ventricular (LV) segmentation model that facilitates accurate and automated LV mass estimation using cardiac magnetic resonance (CMR) imaging.
-
•
CMR-derived LV hypertrophy defined using the deep learning model was strongly associated with hypertension, atrial fibrillation, and heart failure.
-
•
By providing accurate cardiac structural measurements at scale, deep learning models have the potential to facilitate broad cardiovascular discovery.
Introduction
Left ventricular hypertrophy (LVH) is defined as pathologically increased LV mass1 and is consistently associated with increased risks of adverse cardiovascular events including heart failure,1, 2, 3 stroke,1 atrial fibrillation,4 and sudden cardiac death.5 The gold standard for LVH diagnosis is cardiac magnetic resonance (CMR) imaging, which provides accurate and reproducible quantification of cardiac structure.6 However, traditional LV mass estimation using CMR requires LV segmentation, which is typically performed manually and requires substantial time and expertise.
The United Kingdom (UK) Biobank is a prospective cohort study composed of over 500,000 individuals designed to facilitate broad-ranging research of diseases affecting middle-aged and older adults. Roughly 40,000 individuals have undergone prospective CMR acquisition, with additional imaging expected in another 60,000 individuals in the near future. However, manually quantified LV mass is available only within roughly 5000 images,7 and additional measurements would be challenging to obtain at scale. Although automated quantification based on proprietary segmentation methods such as InlineVF (Siemens Healthineers, Erlangen, Germany) are accessible, previous work has suggested limited accuracy of resultant LV mass estimates, and the most recent software versions are not available to the majority of researchers.8 Therefore, a freely available method to facilitate accurate and automated quantification of LV mass using raw CMR images could enable impactful cardiovascular discovery research. In particular, deep learning methods may be well suited for estimation of LV mass using CMR.
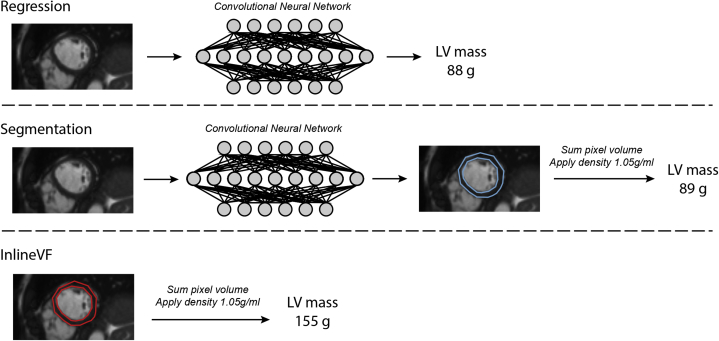
In the current study, we aimed to develop an open-source deep learning model to perform LV mass estimation from CMR images. We compared 2 separate approaches to LV mass estimation: (1) direct estimation trained on manually labeled LV mass values (Machine Learning for Health-Regression [ML4Hreg]), and (2) identification of LV myocardial pixels followed by integration to obtain LV myocardial volume with subsequent conversion to LV mass (Machine Learning for Health-Segmentation [ML4Hseg]) (Figure 1). We then compared the accuracy of both deep learning approaches to LV mass obtained using InlineVF within an independent holdout set using manually labeled LV mass as the gold standard.
Figure 1.
Overview of left ventricular (LV) mass algorithms. Depicted is an overview of the 3 approaches to cardiac magnetic resonance–derived LV mass estimation compared in the current study. The top model utilizes deep learning–based regression trained by manually labeled LV mass. The middle model performs deep learning–based segmentation informed by InlineVF contours. The bottom model utilizes the InlineVF automated contours alone. For the deep learning segmentation and InlineVF models, LV segmentations were converted to LV mass by summing pixel volume and multiplying by the density of LV myocardium (1.05 g/mL, see text).
Methods
Study population
The UK Biobank is a population-based prospective cohort of 502,629 participants recruited between 2006 and 2010 in the United Kingdom primarily established to investigate the genetic and lifestyle determinants of disease. The design of the cohort has been described previously.9,10 Briefly, approximately 9.2 million individuals aged 40–69 years living within 25 miles of the 22 assessment centers in England, Wales, and Scotland were invited, and 5.4% participated in the baseline assessment. Extensive questionnaire data, physical measures, and biological samples were collected at recruitment, with ongoing enhanced data collection in large subsets of the cohort, including repeated assessments and multimodal imaging. All participants are followed up for health outcomes through linkage to national health-related datasets. Participants provided written informed consent. The UK Biobank was approved by the UK Biobank Research Ethics Committee (reference number 11/NW/0382). Use of UK Biobank data (application 7089) was approved by the local Mass General Brigham Institutional Review Board.
Cardiac magnetic resonance acquisition
For all analyses, we included individuals who underwent CMR during the UK Biobank imaging assessment and whose bulk CMR data were available for download as of April 30, 2019. The full CMR protocol of the UK Biobank has been described in detail previously.11 Briefly, all CMR examinations were performed in the United Kingdom on a clinical wide-bore 1.5 Tesla scanner (MAGNETOM Aera, Syngo Platform VD13A; Siemens Healthineers, Erlangen, Germany). All acquisitions used balanced steady-state free precession with typical parameters.
The contours extracted from the InlineVF algorithm and stored in each DICOM file’s metadata were further processed into pixel masks that labeled myocardium, LV cavity, and background. The DICOM metadata stores the inner and outer contours of the myocardium as a 1-pixel-wide pixel mask; we converted these contours to polygons, which we processed into a segmentation pixel mask using morphologic image operators. The short-axis CMR sequence in the UK Biobank contained between 6 and 13 short-axis slices extending from base to apex. Height and width of the slices varied by individual but never exceeded 256 in either dimension. All CMR images were zero-padded to be 3-dimensional tensors with shape (256, 256, 13). To facilitate the cross-entropy loss computation the 3 anatomical labels were one-hot encoded to be label masks with shape (256, 256, 13, 3). Each CMR image was normalized on a per-image basis to have mean zero and standard deviation 1.
Left ventricular mass models
We assessed 2 independent deep learning–based approaches to LV mass estimation. The first model was a 3D convolutional neural network regressor trained with the manually annotated LV mass estimates provided by Petersen and colleagues,7 to optimize the log cosh loss function, which behaves like L2 loss for small values and L1 loss for larger values:
Here batch size, , is 4 random samples from the training set of 3178 after excluding testing and validation samples from the total 5065 CMR images with LV mass values included in . The second model, , is a 3D semantic segmenter. To facilitate model development in the absence of hand-labeled segmentations, we trained with the InlineVF contours to minimize the per-pixel cross-entropy between the label and the model’s prediction.
Here the batch size, , was 4 from the total set of 33,071. Height, , and width, , are 256 voxels and there was a maximum of 13 slices along the short axis. There is a channel for each of the 3 labels, which were one-hot encoded in the training data, InlineVF (, and probabilistic values from the softmax layer of . Segmentation architectures used U-Net-style long-range connections between early convolutional layers and deeper layers. Such an approach allows the final segmentation to use high-resolution local information with more abstract contextual features, both of which are critical for semantic segmentation. Since not all CMR images used the same pixel dimensions, we built models to incorporate pixel size values with their fully connected layers before making predictions. An overview of the architectures of both deep learning models is shown in Supplemental Figure 1.
For ML4Hreg, an LV mass estimate was produced directly. To compute an LV mass estimate from ML4Hseg and InlineVF, we calculated LV mass based on the pixels predicted to be myocardium. Specifically, we multiplied the number of pixels corresponding to myocardium by the pixel depth (calculated to be 10 mm using image metadata and visual confirmation; Supplemental Figure 2) to yield total LV myocardial volume in milliliters. We then multiplied the predicted myocardial volume by the tissue density of LV myocardium, which is 1.05 g/cm3, to yield an estimate of LV mass.12 Given evidence of systematic overestimation in raw estimates obtained using both InlineVF and ML4Hseg (Supplemental Figure 3), we centered initial LV mass estimates using the observed mean LV mass of the manually labeled data within strata of sex.
All models were optimized using the Adam variant13 of stochastic gradient descent with initial learning rate 1 × 10-3, exponential learning rate decay, and batch size of 4 on K80 graphical processing units. Additional details regarding model training and evaluation are described in the Supplemental Methods. All models were implemented in tensorflow version 2.1.0 using the ML4H modeling framework.14 Model architectures, trained weights, and more metrics are available at https://github.com/broadinstitute/ml4h/tree/master/model_zoo/cardiac_mri_derived_left_ventricular_mass/.
Disease associations
Given established associations between increased LV mass and the presence of LVH with cardiovascular disease, we assessed for associations between CMR-derived LV mass (using each method) and prevalent hypertension, atrial fibrillation, and heart failure. For these analyses, LVH was defined as LV mass index >72 g/m2 in men and >55 g/m2 in women,7 and alternatively as the sex-specific 90th percentile of LV mass.1 Indexing for body surface area was performed using the DuBois formula.15 Diseases were defined using self-report and inpatient ICD-9/10 codes (updated through March 31, 2020; Supplemental Table 1).
Statistical analysis
The primary measure of LV mass estimation accuracy was the Pearson correlation between model-estimated LV mass values and hand-labeled LV mass within a holdout set independent of model training. We also calculated the mean absolute error (MAE) and analyzed agreement using Bland-Altman plots16 as secondary measures. Correlation coefficients were compared using Dunn and Clark’s z statistic17 for overlapping dependent correlations. Confidence intervals for MAE were obtained using 1000-iteration bootstrapping. A linear recalibration formula to correct for bias in InlineVF-based measurements was obtained by regressing manually labeled LV mass on LV mass estimates obtained using InlineVF. Associations between LV mass and LVH with prevalent disease were assessed using logistic regression models with adjustment for age and sex.
Although our primary aim was to obtain accurate LV mass estimates, given that previous deep learning segmentation models have reported model performance in terms of pixel-wise agreement,18 we performed a secondary model evaluation in which we assessed pixel-wise agreement of regions segmented as LV blood pool and LV myocardium by calculating Dice scores. In these analyses, ML4Hseg was compared against its training data (ie, InlineVF) as a measure of internal validity, and both ML4Hseg and InlineVF segmentations were then compared against 73 gold-standard segmentations of short-axis images manually produced by a cardiologist (J.P.).
To assess the behavior of the deep learning segmentation model, we generated saliency maps (maps denoting CMR regions identified as myocardium). Statistical analyses were performed using R v3.5 (packages “data.table,” “ggplot2,” “epiR,” “pROC,” “nricens”).19,20 All 2-tailed P values < .05 were considered statistically significant.
Results
Within 33,071 individuals who underwent CMR, we trained models to derive CMR-based LV mass using deep learning regression (ML4Hreg) and segmentation (ML4Hseg). The mean age was 64 ± 8 years and 52% were female. Other baseline characteristics of the training and test sets are shown in Table 1.
Table 1.
Baseline characteristics
| Training set (N = 33,071) | Holdout set (N = 5393) | |
|---|---|---|
| Age | 64.2 ± 7.5 | 63.6 ± 7.7 |
| Female | 17,183 (52.0%) | 2847 (52.8%) |
| Race/Ethnicity | - | - |
| White | 32,013 (96.8%) | 5235 (97.1%) |
| Asian or Pacific Islander | 446 (1.3%) | 61 (1.1%) |
| Black | 207 (0.6%) | 29 (0.5%) |
| Mixed | 151 (0.5%) | 23 (0.4%) |
| Other | 159 (0.5%) | 27 (0.5%) |
| Unknown | 95 (0.3%) | 18 (0.3%) |
| Systolic blood pressure (mm Hg) | 138 ± 18 | 137 ± 18 |
| Diastolic blood pressure (mm Hg) | 79 ± 10 | 79 ± 10 |
| HTN | 10,122 (30.6%) | 1572 (29.1%) |
| Diabetes | 1288 (3.9%) | 186 (3.4%) |
| Heart failure | 191 (0.6%) | 26 (0.5%) |
| Myocardial infarction | 686 (2.1%) | 101 (1.9%) |
| CMR-derived LV mass (InlineVF, g) | 154.9 ± 38.5 | 154.9 ± 38.2 |
| CMR-derived LV mass (InlineVF centered, g) | 90.0 ± 33.1 | 90.1 ± 32.8 |
| CMR-derived LV mass (regression, g) | 88.2 ± 16.2 | 87.6 ± 15.6 |
| CMR-derived LV mass (segmentation, g) | 88.9 ± 28.4 | 89.1 ± 27.8 |
CMR = cardiac magnetic resonance; HTN = hypertension; LV = left ventricular.
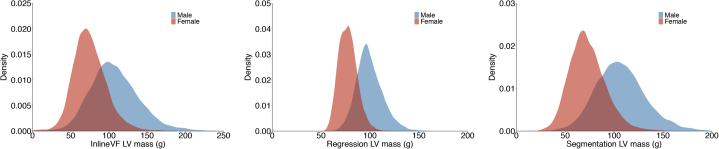
Initial LV mass estimates obtained using InlineVF and ML4Hseg demonstrated evidence of systematic overestimation, which was corrected after centering each distribution upon the mean observed manually labeled LV mass. An example of a uniform overestimation error is depicted in Supplemental Figure 4. The distributions of LV mass stratified by sex using each method are shown in Figure 2.
Figure 2.
Distributions of cardiac magnetic resonance (CMR)-derived left ventricular (LV) mass obtained using each estimation method. Depicted are density plots showing the distribution of CMR-derived LV mass (x-axis) using mean-centered InlineVF (left panel), the deep learning regression model (middle panel), and the deep learning segmentation model (right panel). Results are shown for the full sample with available CMR imaging (N=38,464).
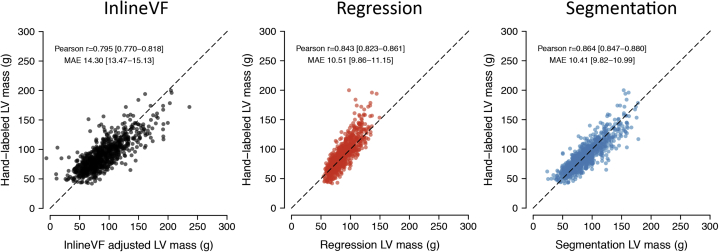
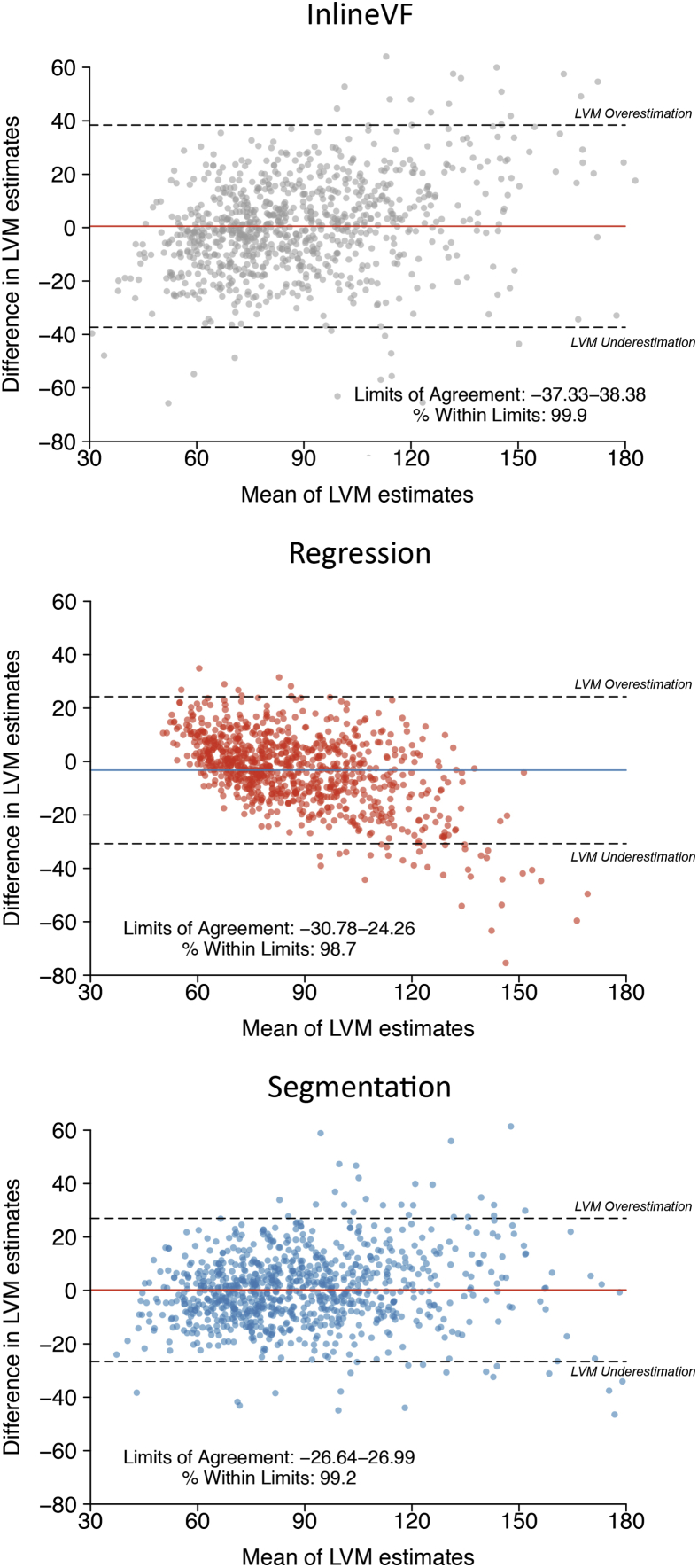
In an independent holdout set of 891 individuals with manually labeled LV mass estimates available, ML4Hseg had favorable correlation with manually labeled LV mass (r = 0.864, 95% confidence interval [CI] 0.847–0.880; MAE 10.41 g, 95% CI 9.82–10.99) as compared to ML4Hreg (r = 0.843, 95% CI 0.823–0.861; MAE 10.51, 95% CI 9.86–11.15, P = .01) and centered InlineVF (r = 0.795, 95% CI 0.770–0.818; MAE 14.30, 95% CI 13.46–11.01, P < .01, Figure 3). Bland-Altman plots demonstrated reasonable agreement between both ML4Hseg and ML4Hreg and manually labeled LV mass, although ML4Hreg tended to progressively underestimate greater LV mass values (Figure 4). Saliency maps suggested that the models appropriately identified areas of LV myocardium for LV mass estimation (Supplemental Figure 5 and Supplemental Methods). Correlations between manually labeled LV mass with InlineVF and ML4Hseg prior to mean centering are shown in Supplemental Figures 3 and 6. Correlation between manually labeled LV mass and InlineVF additionally adjusted using linear recalibration was slightly improved (r = 0.838, 95% CI 0.817–0.856) and is shown in Supplemental Figure 6. A bias-corrected InlineVF LV mass can be calculated using the following equation: .
Figure 3.
Correlation between manually labeled left ventricular (LV) mass and derived left ventricular mass estimated using each model. Depicted are plots illustrating the correlation between manually labeled LV mass (y-axis) and cardiac magnetic resonance–derived LV mass using InlineVF (left panel), deep learning regression (middle panel), and deep learning segmentation (right panel). Results are shown among individuals within the test set independent of model training. Estimates for InlineVF and the segmentation model are displayed after centering the distribution upon the observed sex-stratified mean manually labeled LV mass (see text).
Figure 4.
Bland-Altman plots comparing manually labeled left ventricular (LV) mass and derived LV mass using each model. Depicted are Bland-Altman plots16 showing agreement between manually labeled LV mass and LV mass estimated using InlineVF (top), ML4Hreg (middle), and ML4Hseg (bottom). Estimates using InlineVF and ML4Hseg are depicted after mean centering (see text). In each plot, each point represents a paired observation (ie, the manually labeled LV mass estimate and the model predicted LV mass estimate). The x-axis depicts increasing mean of the paired observations. The y-axis depicts the difference between the paired observations, with negative values representing pairs in which manually labeled LV mass was larger than model-predicted LV mass (underestimation using the model). The colored horizontal line shows the overall mean difference within each sample, and the hashed horizontal lines show the upper and lower bounds of the mean difference (defined as ±1.96 standard deviations of the difference). The corresponding bounds (a surrogate for level of agreement) and the proportion of observations within those bounds are depicted on each plot. A total of 13 (InlineVF), 1 (ML4Hreg), and 1 (ML4Hseg) outlying observations are not depicted for graphical purposes.
Associations between CMR-based LV mass and prevalent disease
We assessed for associations between CMR-derived LV mass and prevalent cardiovascular disease. At the time of CMR acquisition, there were 11,271 prevalent hypertension, 1053 atrial fibrillation, and 241 heart failure events. When compared to the other approaches, LVH defined using ML4Hseg consistently demonstrated the strongest associations with hypertension (odds ratio [OR] 2.76, 95% CI 2.51–3.04), atrial fibrillation (OR 1.75, 95% CI 1.37–2.20), and heart failure (OR 4.67, 95% CI 3.28–6.49, Table 2).
Table 2.
Associations between deep learning segmentation–derived left ventricular mass index and prevalent disease
| N events† | Odds ratio with covariate (95% CI) |
|||
|---|---|---|---|---|
| LVMI (per 1 SD) | LVH | LVH (90th percentile) | ||
| Hypertension | ||||
| InlineVF | 11,271 | 1.43 (1.39–1.47) | 2.30 (2.15–2.46) | 2.33 (2.17–2.50) |
| Regression | 11,271 | 1.27 (1.24–1.30) | 1.67 (1.38–2.01) | 1.64 (1.53–1.76) |
| Segmentation | 11,271 | 1.55 (1.51–1.59) | 2.76 (2.51–3.04) | 2.39 (2.23–2.57) |
| Atrial fibrillation | ||||
| InlineVF | 1053 | 0.99 (0.93–1.05) | 1.19 (0.99–1.44) | 1.27 (1.04–1.53) |
| Regression | 1053 | 1.00 (0.93–1.07) | 1.13 (0.59–1.93) | 0.99 (0.80–1.21) |
| Segmentation | 1053 | 1.13 (1.06–1.21) | 1.75 (1.37–2.20) | 1.61 (1.34–1.93) |
| Heart failure | ||||
| InlineVF | 241 | 1.45 (1.29–1.63) | 2.92 (2.16–3.89) | 3.02 (2.23–4.04) |
| Regression | 241 | 1.39 (1.23–1.57) | 3.94 (1.75–7.67) | 2.36 (1.71–3.20) |
| Segmentation | 241 | 1.71 (1.51–1.93) | 4.67 (3.28–6.49) | 3.73 (2.78–4.95) |
LVH = left ventricular hypertrophy; LVMI = left ventricular mass index.
Total N = 37,261 with available phenotypic data and cardiac magnetic resonance–derived left ventricular mass estimates obtained using each method.
Pixel-wise agreement
We then assessed agreement between ML4Hseg and its training data (ie, InlineVF) within a held-out test set of InlineVF examples (n = 300, 15,507 voxels). Agreement was very high for LV blood pool segmentation, with a mean dice score of 0.955. Although slightly lower, agreement for myocardial segmentation was also high (n = 300, 17,766 voxels), with a mean Dice score of 0.900. We then compared segmentations from the 2 segmentation-based models (ML4Hseg and InlineVF) to hand-labeled contours. Dice scores for ML4Hseg (0.903 for LV; 0.631 for myocardium) were consistently greater than those for InlineVF (0.890 for LV; 0.601 for myocardium), although overall agreement for myocardial segmentations was only moderate. The standard deviation of the study-level Dice coefficient was lower using ML4Hseg (0.083) as opposed to InlineVF (0.098), suggesting more consistent performance. Visual examination of segmentations obtained using ML4Hseg vs InlineVF suggest that ML4Hseg may be more accurate owing to a reduction in gross segmentation errors (Supplemental Figure 4). A comparison of ML4Hseg and InlineVF with previously published segmentation models is shown in Supplemental Table 2.
Discussion
Within more than 30,000 individuals with CMR imaging performed as part of the UK Biobank prospective cohort study, we developed and tested 2 deep learning–based approaches to automated LV mass estimation. When compared to a second deep learning approach and the proprietary D13A InlineVF automated segmentation software, ML4Hseg demonstrated the greatest correlation (86%) and lowest estimation error (approximately 10 g) when compared against manually labeled LV mass in a test set independent of model training. Importantly, greater CMR-derived LV mass obtained using our segmentation model had the strongest associations with prevalent hypertension, atrial fibrillation, and heart failure. We have shared our model architecture publicly and aim to return CMR-derived LV mass values to the UK Biobank for use by other researchers in order to facilitate future cardiovascular discovery utilizing rich cardiac structural imaging features.
Our study supports and extends previous work demonstrating the potential for deep learning to provide automated quantification of imaging phenotypes. Over the last few years, several deep learning–based approaches to LV segmentation have been proposed, primarily utilizing variants of a convolutional neural network architecture.18 Within the UK Biobank, Aung and colleagues21 utilized a combination of deep learning and manual segmentation to extend LV mass estimates to approximately 16,000 images, in order to facilitate genetic analyses. Similarly, Bai and colleagues22 utilized a neural network to estimate several cardiac structural features to enable broad phenotypic association testing. In keeping with most previous models,22 we utilized a convolutional neural network architecture with U-net style connections between convolutional layers and deeper layers, allowing models to learn abstract contextual features to inform semantic segmentation. For training, we utilized contours extracted from the InlineVF proprietary algorithm, allowing us to leverage a comparatively large training set. Furthermore, we explicitly compared several approaches to automated LV mass estimation, observing that an image segmentation model demonstrated favorable performance when compared to deep learning–based regression and a recalibrated InlineVF-based method. Importantly, we validated the performance of our best-performing algorithm by assessing correlation and agreement against manually labeled estimates, and by testing for expected associations with prevalent disease.
Our results provide insight into the comparative accuracy of potential methods to estimate cardiac structural features, suggesting that automated segmentation may provide superior performance. Specifically, we compared 3 approaches to automated LV mass estimation: deep learning–based segmentation (ML4Hseg), deep learning–based regression on hand-labeled LV mass estimates (ML4Hreg), and use of the automated contours provided by the InlineVF D13A proprietary software. Even when compared to the deep learning regressor trained to estimate LV mass directly, we observed favorable accuracy using ML4Hseg, which more closely mirrors the manual process of clinical LV mass estimation, in which cardiac radiologists manually label pixels as LV myocardium. Of note, even though we trained ML4Hseg using InlineVF contours, ML4Hseg resulted in substantially more accurate LV mass estimates than InlineVF alone. Improvement upon the InlineVF training data may be related to a lower likelihood of committing gross segmentation errors or an intrinsic robustness to noise present in training labels, as described previously with deep learning architectures.23,24 Although ML4Hseg also outperformed ML4Hreg, our results demonstrate that a regression approach can achieve reasonable accuracy, which may be improved as more gold-standard LV mass estimates become available. Future work is needed to better understand the relative strengths and weaknesses of various approaches to deep learning–based cardiac structural characterization, as well as to assess the comparative generalizability of such approaches when transferred to external datasets.
The current work highlights the potential for deep learning to derive clinically relevant imaging phenotypes in an efficient and automated manner. Increased LV mass and LVH have long been implicated as important risk factors for adverse cardiovascular events.1, 2, 3, 4, 5 The detection of LVH is clinically relevant, since the majority of cases are related to hypertension, for which treatment can lead to regression of hypertrophy and improvement in cardiovascular risk profile.25,26 The associations and effect sizes we observed between CMR-derived LVH and prevalent hypertension, atrial fibrillation, and heart failure were consistently strongest using ML4Hseg and are broadly consistent with prior studies.2,4 On balance, our findings suggest that deep learning may facilitate recognition of clinically relevant degrees of increased LV mass in a manner deployable at scale.
We submit that our findings may directly enable future cardiovascular research focused on CMR-derived LV mass, and potentially additional imaging phenotypes. The UK Biobank has performed CMR in more than 35,000 individuals, with imaging expected to extend to nearly 100,000 individuals in the near future. Although LV mass and LVH reflect clinically important aspects of cardiac structure, quantification of LV mass using gold-standard CMR is traditionally performed manually, which is time-consuming and requires specialized expertise. Although UK Biobank images include the InlineVF D13A automated contours that may be used to estimate LV mass, our findings support previous studies demonstrating substantial overestimation.8 Furthermore, InlineVF is a proprietary algorithm whose latest versions are not accessible to all investigators.8 To this end, our deep learning model ML4Hseg provides more accurate and substantially less biased estimates, and the code underlying the model is available for public use. For investigators opting not to deploy our deep learning model, we also provide a formula to obtain linearly adjusted LV mass using InlineVF D13A, which demonstrated considerable bias correction and only moderately lower correlation as compared to our segmentation model.
Our study should be interpreted in the context of design. First, although the correlation between our deep learning model and manually labeled LV mass was very good (86%), it was not perfect, which may result in some misclassification of LV mass. Nevertheless, it was the best-performing model of the approaches tested in terms of correlation, agreement, and absolute error. Second, the number of manually labeled LV mass values available to train ML4Hreg and evaluate each model was relatively limited. Future models trained on a greater number of ground truth examples may enable the development of more accurate deep learning–based LV mass estimates. Third, although pixel-wise agreement with hand-labeled segmentations was favorable using ML4Hseg as opposed to InlineVF, overall agreement using ML4Hseg was only modest, and initial LV mass estimates using both ML4Hseg and InlineVF showed evidence of systematic overestimation. Therefore, it is possible that InlineVF (and secondarily ML4Hseg) tends to result in contours that are systematically too large, with simple mean centering resulting in correction of the resulting distributional shift. Since InlineVF is proprietary, we are unable to fully evaluate the mechanism by which such overestimations may occur, although approaches to modify or exclude inaccurate labels may further improve the performance of future models. Fourth, although higher pixel-wise agreement has been reported using previous deep learning approaches,18,27, 28, 29, 30, 31 we note that the majority of such models were trained using hand-labeled segmentations provided on standardized image sets, which may not be directly comparable to our UK Biobank test set images. Owing to absence of pretrained weights or incompatibility of older models with our codebase, we were unable to directly compare the performance of ML4Hseg with previous models within our test set. In light of these limitations, we acknowledge that external validation of ML4Hseg would be needed prior to deployment in datasets outside of the UK Biobank.
Conclusion
Utilizing a unique resource of CMR images obtained within more than 35,000 individuals, we developed ML4Hseg—a deep learning segmentation model that provides automated LV mass estimation with favorable accuracy as compared to deep learning regression or the InlineVF proprietary algorithm. Importantly, model-derived LV mass estimates demonstrated expected associations with cardiovascular disease. We have made our algorithm publicly available for future use, and submit that such deep learning approaches may facilitate broad cardiovascular discovery by enabling future analyses of CMR-derived cardiac structural phenotypes available at scale.
Acknowledgments
FundingSources: Dr Khurshid is supported by NIH T32HL007208. Dr Pirruccello is supported by a John S. LaDue Memorial Fellowship. Dr Ho is supported by NIH R01HL134893, R01HL140224, and K24HL153669. Dr Lubitz is supported by NIH 1R01HL139731 and American Heart Association (AHA) 18SFRN34250007. Dr Ellinor is supported by NIH 1R01HL092577, R01HL128914, K24HL105780, American Heart Association 18SFRN34110082, and Foundation Leducq 14CVD01. Dr Anderson is supported by NIH R01NS103924 and AHA 18SFRN34250007.
Disclosures: Dr Pirruccello has served as a consultant for Maze Therapeutics. Dr Philippakis receives sponsored research support from Bayer AG, IBM, Intel, and Verily. He has also received consulting fees from Novartis and Rakuten. Dr Ho receives sponsored research support from Bayer AG and Gilead Sciences. Dr Ho has received research supplies from EcoNugenics. Dr Friedman receives sponsored research support from Bayer AG and IBM. Dr Anderson receives sponsored research support from Bayer AG and has consulted for ApoPharma, Inc. Dr Batra receives sponsored research support from Bayer AG and IBM, and consults for Novartis. Dr Lubitz receives sponsored research support from Bristol Myers Squibb / Pfizer, Bayer AG, Boehringer Ingelheim, and Fitbit, and has consulted for Bristol Myers Squibb / Pfizer and Bayer AG, and participates in a research collaboration with IBM. Dr Ellinor receives sponsored research support from Bayer AG. Dr Ellinor has consulted for Bayer AG, Novartis, MyoKardia, and Quest Diagnostics.
Ethics Statement
The UK Biobank was approved by the UK Biobank Research Ethics Committee (reference number 11/NW/0382). Use of UK Biobank data (application 7089) was approved by the local Mass General Brigham Institutional Review Board.
Patient Consent
Participants provided written informed consent.
Authorship
All authors attest they meet the current ICMJE criteria for authorship.
Disclaimer
Given his role as Editor-in-Chief, David McManus had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to the Deputy Editor, Hamid Ghanbari.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.cvdhj.2021.03.001.
Appendix. Supplementary data
References
- 1.Bluemke D.A., Kronmal R.A., Lima J.A.C., et al. The relationship of left ventricular mass and geometry to incident cardiovascular events: the MESA (Multi-Ethnic Study of Atherosclerosis) study. J Am Coll Cardiol. 2008;52:2148–2155. doi: 10.1016/j.jacc.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawel-Boehm N., Kronmal R., Eng J., et al. Left ventricular mass at MRI and long-term risk of cardiovascular events: the Multi-Ethnic Study of Atherosclerosis (MESA) Radiology. 2019;293:107–114. doi: 10.1148/radiol.2019182871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazzeroni D., Rimoldi O., Camici P.G. From left ventricular hypertrophy to dysfunction and failure. Circ J. 2016;80:555–564. doi: 10.1253/circj.CJ-16-0062. [DOI] [PubMed] [Google Scholar]
- 4.Chrispin J., Jain A., Soliman E.Z., et al. Association of electrocardiographic and imaging surrogates of left ventricular hypertrophy with incident atrial fibrillation: MESA (Multi-Ethnic Study of Atherosclerosis) J Am Coll Cardiol. 2014;63:2007–2013. doi: 10.1016/j.jacc.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haider A.W., Larson M.G., Benjamin E.J., Levy D. Increased left ventricular mass and hypertrophy are associated with increased risk for sudden death. J Am Coll Cardiol. 1998;32:1454–1459. doi: 10.1016/s0735-1097(98)00407-0. [DOI] [PubMed] [Google Scholar]
- 6.Lenstrup M., Kjaergaard J., Petersen C.L., Kjaer A., Hassager C. Evaluation of left ventricular mass measured by 3D echocardiography using magnetic resonance imaging as gold standard. Scand J Clin Lab Invest. 2006;66:647–657. doi: 10.1080/00365510600892233. [DOI] [PubMed] [Google Scholar]
- 7.Petersen S.E., Aung N., Sanghvi M.M., et al. Reference ranges for cardiac structure and function using cardiovascular magnetic resonance (CMR) in Caucasians from the UK Biobank population cohort. J Cardiovasc Magn Reson. 2017;19:18. doi: 10.1186/s12968-017-0327-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suinesiaputra A., Sanghvi M.M., Aung N., et al. Fully-automated left ventricular mass and volume MRI analysis in the UK Biobank population cohort: evaluation of initial results. Int J Cardiovasc Imaging. 2018;34:281–291. doi: 10.1007/s10554-017-1225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudlow C., Gallacher J., Allen N., et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Littlejohns T.J., Sudlow C., Allen N.E., Collins R. UK Biobank: opportunities for cardiovascular research. Eur Heart J. 2019;40:1158–1166. doi: 10.1093/eurheartj/ehx254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen S.E., Matthews P.M., Francis J.M., et al. UK Biobank’s cardiovascular magnetic resonance protocol. J Cardiovasc Magn Reson. 2016;18:8. doi: 10.1186/s12968-016-0227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myerson S.G., Bellenger N.G., Pennell D.J. Assessment of left ventricular mass by cardiovascular magnetic resonance. Hypertension. 2002;39:750–755. doi: 10.1161/hy0302.104674. [DOI] [PubMed] [Google Scholar]
- 13.Kingma D.P., Ba J. Adam: A Method for Stochastic Optimization. arXiv:14126980 [cs] [Internet] 2017 [cited 2021 Feb 13] http://arxiv.org/abs/1412.6980 Available at.
- 14.ML4CVD Group Machine Learning for Health (ML4H). GitHub 2020. https://github.com/broadinstitute/ml
- 15.Du Bois D., Du Bois E.F. A formula to estimate the approximate surface area if height and weight be known. 1916. Nutrition. 1989;5:303–311. discussion 312–313. [PubMed] [Google Scholar]
- 16.Bland J.M., Altman D.G. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8:135–160. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- 17.Dunn O.J., Clark V. Correlation coefficients measured on the same individuals. Journal of the American Statistical Association. 1969;64:366–377. [Google Scholar]
- 18.Chen C., Qin C., Qiu H., et al. Deep learning for cardiac image segmentation: a review. Front Cardiovasc Med. 2020;7:25. doi: 10.3389/fcvm.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2015. R: A language and environment for statistical computing.https://www.R-project.org/ [Google Scholar]
- 20.Dowle M., Srinivasan A., Gorecki J., et al. data.table: Extension of “data.frame.” Version 1.12.6. https://CRAN.R-project.org/package=data.table
- 21.Aung N., Vargas J.D., Yang C., et al. Genome-wide analysis of left ventricular image-derived phenotypes identifies fourteen loci associated with cardiac morphogenesis and heart failure development. Circulation. 2019;140:1318–1330. doi: 10.1161/CIRCULATIONAHA.119.041161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai W., Suzuki H., Huang J., et al. A population-based phenome-wide association study of cardiac and aortic structure and function. Nat Med. 2020;26:1654–1662. doi: 10.1038/s41591-020-1009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen P, Liao B, Chen G, Zhang S. Understanding and utilizing deep neural networks trained with noisy labels. arXiv:1905.05040.
- 24.Rolnick D, Veit A, Belongie S, Shavit N. Deep learning is robust to massive label noise. arXiv:1705.10694.
- 25.Dahlöf B., Pennert K., Hansson L. Reversal of left ventricular hypertrophy in hypertensive patients. Am J Hypertens. 1992;5:95–110. doi: 10.1093/ajh/5.2.95. [DOI] [PubMed] [Google Scholar]
- 26.Okin P.M., Devereux R.B., Jern S., et al. Regression of electrocardiographic left ventricular hypertrophy during antihypertensive treatment and the prediction of major cardiovascular events. JAMA. 2004;292:2343–2349. doi: 10.1001/jama.292.19.2343. [DOI] [PubMed] [Google Scholar]
- 27.Li C., Tong Q., Liao X., et al. APCP-NET: Aggregated Parallel Cross-Scale Pyramid Network for CMR Segmentation. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) [Internet] Venice, Italy: IEEE, 2019 [cited 2021 Feb 3], pp. 784–788. https://ieeexplore.ieee.org/document/8759147/ Available from.
- 28.Painchaud N, Skandarani Y, Judge T, Bernard O, Lalande A, Jodoin P-M. Cardiac MRI Segmentation with Strong Anatomical Guarantees. arXiv:1907.02865 [cs, eess] 2019; 11765:632–640. [DOI] [PubMed]
- 29.Zotti C., Luo Z., Lalande A., Jodoin P.-M. Convolutional neural network with shape prior applied to cardiac MRI segmentation. IEEE J Biomed Health Inform. 2019;23:1119–1128. doi: 10.1109/JBHI.2018.2865450. [DOI] [PubMed] [Google Scholar]
- 30.Pop M., Sermesant M., Jodoin P.-M., et al., editors. Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges [Internet] Springer International Publishing; Cham: 2018. http://link.springer.com/10.1007/978-3-319-75541-0 [cited 2021 Feb 3]. Available from. [Google Scholar]
- 31.Khened M., Kollerathu V.A., Krishnamurthi G. Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. arXiv:1801.05173 [cs] [Internet] 2018 [cited 2021 Feb 3] http://arxiv.org/abs/1801.05173 Available from. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.