Figure 5.
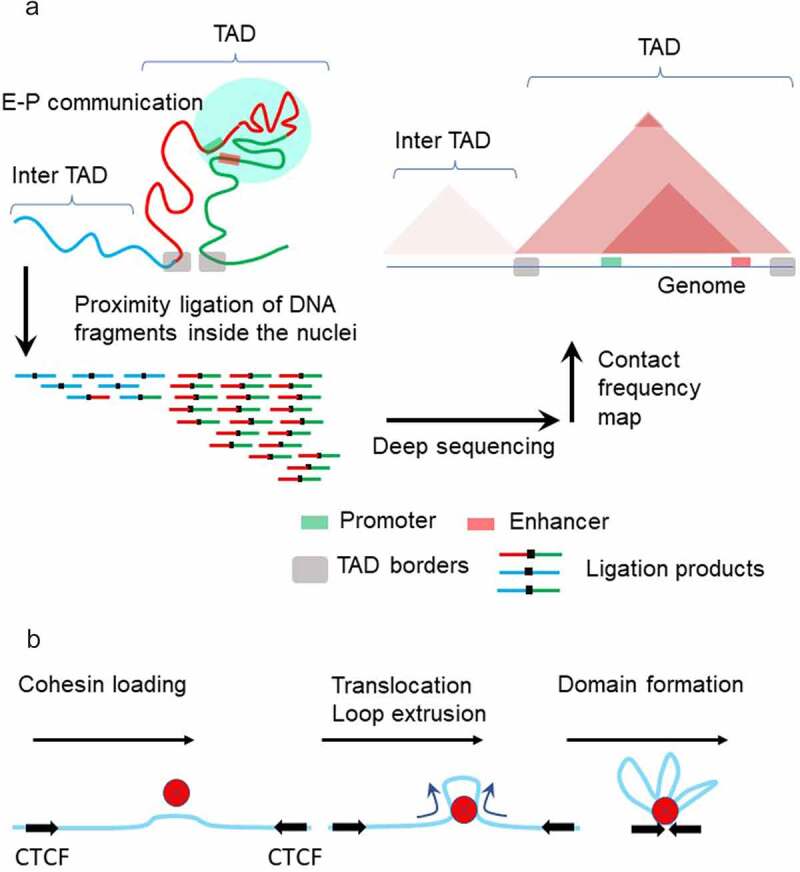
Model of chromatin topology Chromatin fibers are partitioned into topologically associating domains (TADs) (A, left). TADs in the genome are detected by Hi-C method. First, cells are fixed by formaldehyde treatment, which crosslinks chromatin segments that are in proximity. After digestion with restriction enzyme(s), DNA fragments are re-ligated. Proximity between chromatin segments results in the higher incidence that fragments are ligated together (A, left). Deep sequencing of the ligated fragments and mapping sequencing reads on the genome enables genome-wide identification of contact frequencies among different genomic loci (A, right). TADs preferentially self-associate to create discrete structural blocks which appear as triangles in the Hi-C map. Within TADs, sub-domains with higher contact frequencies are formed, often representing the confinement of region between enhancer and promoter. Chromatin loop extrusion is mediated by cohesin and CTCF proteins (b). Cohesin loads on DNA and begins translocating along DNA, resulting in loop extrusion. This process halts when cohesin encounters CTCF molecules, forming a chromatin loop with cohesin and CTCF present at its base.
