ABSTRACT
Genetic defects in SLC26A3 (DRA), an intestinal Cl−/HCO3− exchanger, result in congenital chloride diarrhea (CLD), marked by lifelong acidic diarrhea and a high risk of inflammatory bowel disease. Slc26a3−/− mice serve as a model to understand the pathophysiology of CLD and search for treatment options. This study investigates the microbiota changes in slc26a3−/− colon, the genotype-related causes for the observed microbiota alterations, its inflammatory potential, as well as the corresponding host responses. The luminal and the mucosa-adherent cecal and colonic microbiota of cohoused slc26a3−/− and wt littermates were analyzed by 16S rRNA gene sequencing. Fecal microbiota transfer from cohoused slc26a3−/− and wt littermates to germ-free wt mice was performed to analyze the stability and the inflammatory potential of the communities.
The cecal and colonic luminal and mucosa-adherent microbiota of slc26a3−/− mice was abnormal from an early age, with a loss of diversity, of short-chain fatty acid producers, and an increase of pathobionts. The transfer of slc26a3−/− microbiota did not result in intestinal inflammation and the microbial diversity in the recipient mice normalized over time. A strong increase in the expression of Il22, Reg3β/γ, Relmβ, and other proteins with antimicrobial functions was observed in slc26a3−/− colon from juvenile age, while the mucosal and systemic inflammatory signature was surprisingly mild. The dysbiotic microbiota, low mucosal pH, and mucus barrier defect in slc26a3−/− colon are accompanied by a stark upregulation of the expression of a panel of antimicrobial proteins. This may explain the low inflammatory burden in the gut of these mice.
KEYWORDS: Anion exchange, intestinal electrolyte transport, inflammatory bowel disease, gut dysbiosis, antimicrobial peptides
Introduction
Mutations in the SLC26A3 (DRA) Cl−/HCO3− exchanger is the underlying mechanism for congenital Cl− diarrhea.1 The disease is characterized by lifelong watery, acidic, and Cl-rich diarrhea, systemic alkalosis, male subfertility, a high propensity for development of inflammatory bowel disease (IBD), and a variable rate of kidney disease, which is likely secondary to fluid loss and electrolyte imbalance.2 SLC26A3 is also strongly downregulated in the ileocolon of patients with inflammatory bowel disease,3,4 and in mice with ileocolonic inflammation,5,6 suggesting that defective SLC26A3 function may play a role in the pathophysiology of IBD.
Slc26a3−/− mice are a suitable animal model to study many aspects of the human disease, because they develop all pathologies described above for CLD patients. During investigations into the molecular nature of the electrolyte transport abnormalities in the colon of the slc26a3−/− mice, we observed a virtual absence of colonic bicarbonate secretion, a lack of the adherent mucus layer, and a high susceptibility for intestinal damage by DSS.7 A further exploration of the sequelae of an absence of Slc26a3-mediated luminal alkalinization revealed that Slc26a3 is responsible for the strong segmental differences of the juxtamucosal pH-microclimate in the murine colon.8 In addition, slc26a3−/− mice developed spontaneous, but mild colonic inflammation, a reduced microbial diversity, and a disturbed mucin biosynthesis.8 Recently, tight junctional defects have also been described in the colon of slc26a3−/− mice.9
The present study was undertaken to study the slc26a3−/− microbiota composition in more detail, its stability over time and its causal relationship to the pathological features observed in the slc26a3−/− colon. In addition, we searched for reasons for the slc26a3−/− microbial colonic phenotype, such as colonic surface pH-microclimate, gastrointestinal transit time, or mucosal anti-microbial peptide expression. Taken together, the findings may explain the observed microbiota shift in the colon of slc26a3−/− mice compared to the colon of their cohoused wt littermates and offer potential treatment strategies for improvement of diarrhea and dysbiosis in CLD patients.
Results
Loss of Slc26a3 shapes fecal microbiota composition already early in life
In animal studies, maternal transmission strongly influence intestinal microbiota composition requiring careful design of studies to address the crosstalk of host genetics and the microbiota.10 Accordingly, feces pellets were collected monthly from cohoused slc26a3−/− and wt littermates, starting post-weaning (approx. 5 weeks of age) to the end of the experimental observation period of 20 weeks. This time point was chosen because slc26a3−/− mice develop diarrhea-related complications during adulthood, mostly anal prolapse, or ulcerations at the tail root near the anus, and have to be sacrificed. Microbiota analysis was performed using 16S rRNA amplicon sequencing of the V4 region. The evaluation of the relative abundance of bacteria on family level in slc26a3−/− mice in comparison to wt littermates exhibited significant increases in Bacteroidaceae, Erysipelotrichaceae and Bifidobacteriaceae, and decreases in Prevotellaceae, Lachnospiraceae, Muribaculaceae ,Helicobacteraceae and Ruminococcaceae (Figure 1a,b). This difference in the composition of the microbiota was present already at weaning and stable over the observation period (Figure 1a). The observed α-diversity metric showed consistent reduction of microbial diversity, with the β-diversity using PCoA revealing a clear separation at each time-point in slc26a3−/− compared to wt mice (Supplementary Figure 1a,b). In addition to the fecal samples, cecal luminal samples were analyzed revealing similar alterations compared to the fecal samples in changes in the α and β diversity (Supplementary Figure 1c,d) and the relative abundance of the families were observed, i.e., significant increases in Bacteroidaceae and Bifidobacteriaceae and decreases in Prevotellaceae, Lachnospiraceae and Lactobacillaceae (Figure 1c,d). In summary, these results demonstrate that slc26a3−/− mice developed an altered but stable luminal microbiota with a dramatically reduced microbial diversity from young age.
Figure 1.
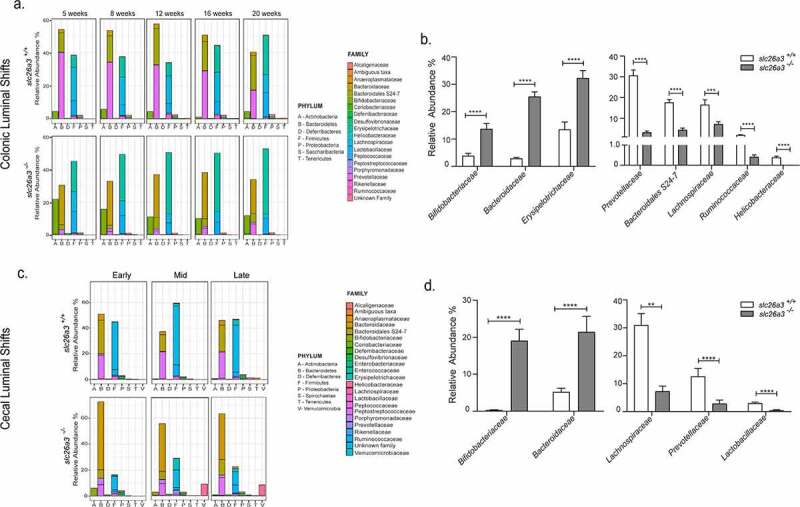
Altered fecal (colonic) and cecal luminal microbiota in the absence of slc26a3 – Relative abundance of the different phyla and bacterial families at different time points of the slc26a3+/+ (wt) and slc26a3−/− mice in the (a) colon and (c) cecum. Overall shifts (combination of all the time points) in the relative abundance of the taxonomic families in the (b) colon (n = 25/group) and in the (d) Cecum (n = 12 for slc26a3+/+and 15 for slc26a3−/−). Colon – n = 5 pairs for all timepoints; Cecum – Early: n = 2–3, Mid: n = 7–8 and Late: n = 3–4.
**p < .005, ***p = .0001, ****p < .0001.
Large differences in the mucosa-associated microbiota of wt and slc26a3−/− mice
Due to the large differences in the surface pH-microclimate and the physicochemical properties of the mucus layer in the cecum and mid-distal colon in slc26a3−/− compared to wt mice, we next focused on the mucosa-associated microbiota. Mice deficient in slc26a3 and wt littermates were sacrificed at different ages and samples from cecum and colon were harvested. From the earliest time point of analysis (4 weeks), significant alterations in the cecal and colonic mucosa-associated microbiota were detected in slc26a3−/− mice compared to cohoused wt (Figure 2a,d). The observed α-diversity and the β-diversity showed a distinct separation between the slc26a3−/− and wt mice, both in the cecum and in the colon (Supplementary Figure 2a-d). Taxonomic breakdown at the family level indicated a decreased abundance in certain populations of the Firmicutes such as the Ruminococcaceae and Lachnospiraceae and an increased abundance of the Muribaculaceae, Bacteroidaceae and Bifidobacteriaceae (Figure 2b,e).
Figure 2.
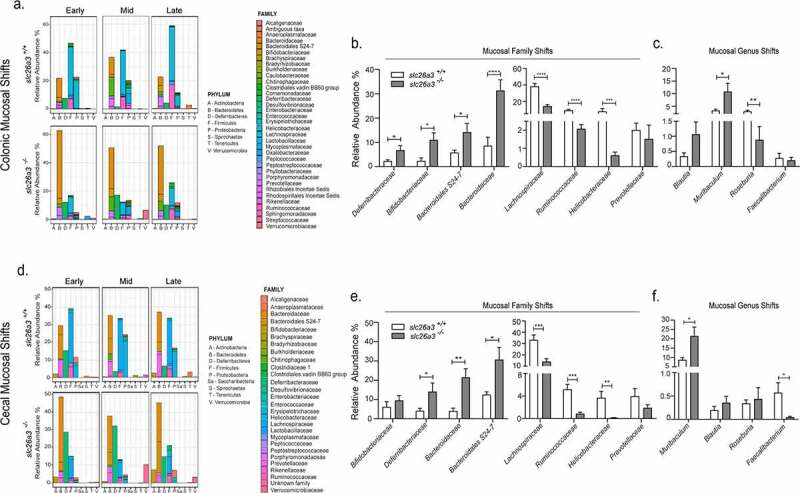
Altered colonic mucosal adherent microbiota in the absence of slc26a3 – Relative abundance of the different phyla and bacterial families at different time points of the slc26a3+/+ (wt) and slc26a3−/− mice in the (a) colon and (d) cecum. Overall shifts (combination of all the time points) in the relative abundance of the taxonomic families and genus in the (b, c) colon (n= 12 for slc26a3+/+and 17 for slc26a3−/−) and in the (e,f) cecum (n = 11 for slc26a3+/+and 14 for slc26a3−/−). Colon Early: n = 5–7, Mid: n = 3–4 and Late: n = 4–6.; Cecum – Early: n = 4–5, Mid: n = 4–5 and Late: n = 3–4.
*p < .05, **p < .005, ***p = .0001, ****p < .0001.
Several interesting observations were made that may explain the pathophysiological aspects of the colonic slc26a3−/− phenotype: Firstly, the phylum Deferribacteres containing mucolytic bacteria,11 was significantly increased in the absence of slc26a3 (Figure 2b,e). This may in part explain the strong reduction in mucosa-adherent mucus in slc26a3−/− colon.7,8 Secondly, there was a significant and strong decrease in short-chain fatty acid (SCFA) producing genera Roseburia12(Figure 2c,f), and Faecalibacterium13 (Figure 2f), as well as family Lachnospiraceae14 (Figure 2b,e). The potential consequences are outlined in the discussion.
pH-dependency of invitro growth of individual bacteria
One possible reason for the changes in the luminal as well as the mucosa-associated microbiota of slc26a3−/− compared to cohoused wt littermates may be the low pH in the lumen as well as directly above the epithelial cells.8 We therefore assessed the growth rate of a panel of commensal bacteria in a pH-dependent manner. One such panel of growth curves is shown in Figure 3. Notably, some bacterial families that are strongly overrepresented in the fecal microbiota of the slc26a3−/− colon, namely Bifidobacteriaceae (Figure 3a) were particularly acid resistant. Other bacterial species and families, such as Muribaculaceae (Figure 3b) and Lachnospiraceae displayed a dramatic reduction of growth in acidic conditions, which is in line with their decreased abundance in the lumen and mucosa of slc26a3−/− mice. However, Prevotellaceae were similarly resistant to mildly acidic growth conditions, yet were underrepresented in the slc26a3−/− microbiota (Table 1). Thus, additional host or community-dependent factors are likely to shape the microbiota in the slc26a3−/− intestine.
Figure 3.
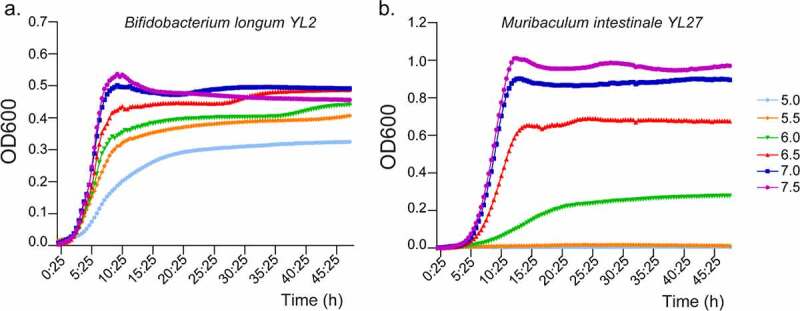
Growth curves of different bacterial strains at different pH. Growth curve of (a) Bifidobacterium longum YL2 and (b) Muribaculum intestinale YL27. n = 3.
Table 1.
Panel of growth curves of different bacterial strains at different pH
| Bacteria | Family | Peak growth (h:min) |
pH |
|||||
|---|---|---|---|---|---|---|---|---|
| 7.5 | 7.0 | 6.5 | 6.0 | 5.5 | 5.0 | |||
| Prevotella rodentium | Prevotellaceae | 10:25 | 100 | 100 | 80.83 | 71.87 | 60.53 | 37.03 |
| Muribaculum intestinale YL27 | Muribaculaceae | 12:55 | 100 | 89.09 | 60.27 | 10.35 | 1.09 | 0.49 |
| Bifidobacterium longum YL2 | Bifidobacteriaceae | 9:35 | 100 | 93.47 | 79.06 | 64.08 | 57.99 | 34.87 |
| Bacteroides cecimuris I48 | Bacteroidaceae | 17:55 | 100 | 97.53 | 62.72 | 1.51 | 0.56 | 1.18 |
| Blautia coccoides YL58 | Lachnospiraceae | 12:05 | 100 | 86.17 | 66.47 | 48.74 | 29.40 | 1.61 |
Growth is depicted as percentage with the peak growth at 7.5 set at 100%.
Gastrointestinal transit time in slc26a3−/− and wt littermates
Another possible reason for the different microbiota composition could be a different gastrointestinal transit time.15 We therefore assessed the total gastrointestinal (GI) transit time in slc26a3−/− and wt littermates aged 9–15 weeks. The mice were subjected to the procedure on 3 d, with 3 d intermission, so that the stool was definitively free of any red dye, when the next GI transit time measurement was performed. The results of the three measurements were averaged and were 146 ± 13 min for the wt mice, and 168 ± 17 min for the slc26a3−/− mice (n = 5, ns) (Table 2a), with the overall stool water content during the entire measurement period remaining consistently and significantly higher in the slc26a3−/− mice (Table 2b).
Table 2.
Total Gastrointestinal (GI) transit time and stool water content.
| a. | |||||||
|---|---|---|---|---|---|---|---|
| Genotype | GI transit time measurements (min) |
||||||
| I | II | III | IV | V | Average | SEM | |
| Slc26a3+/+ | 122.33 | 153 | 168 | 165.66 | 125.33 | 146. 86 | 13.38 |
| Slc26a3−/− | 166.33 | 185 | 116.33 | 136 | 241 | 168.93 | 17.59 |
Transfer of slc26a3-deficient microbiota into germ-free mice
In order to investigate the inflammatory potential of the slc26a3−/− colonic microbiota, a fecal microbiota transfer (FMT) experiment was performed. Four-week-old germ-free mice received either stool samples from SPF slc26a3+/+ mice (Donor wt) or from the slc26a3−/− mice (Donor ko). Analysis was then performed 8 weeks post-transplant (Details of the experimental setup and subsequent weight/growth parameters are shown in Supplementary Figure 3). No histological or molecular signs of inflammation were found in the recipient mice receiving either an FMT from wt or slc26a3−/− mice. The small but significant increases of the inflammatory markers Ly6g, Cd3 and Tnfα that was observed in the slc26a3−/− colon was not transferred to the Recipient ko group, both at the mRNA level (Figure 4a) and at the protein level for CD3 (Figure 4b). The spleens of the donors and recipients were analyzed in search for a potential systemic inflammatory reaction. All mice that were raised in a germ-free environment had significantly lower mRNA levels of the neutrophil marker Ly6g than the slc26a3−/− and the donor wt mice, whereas the mRNA expression of the Cd3 T-cell receptor, and of Tnfα, was not different in any of the mice (Figure 4c).
Figure 4.
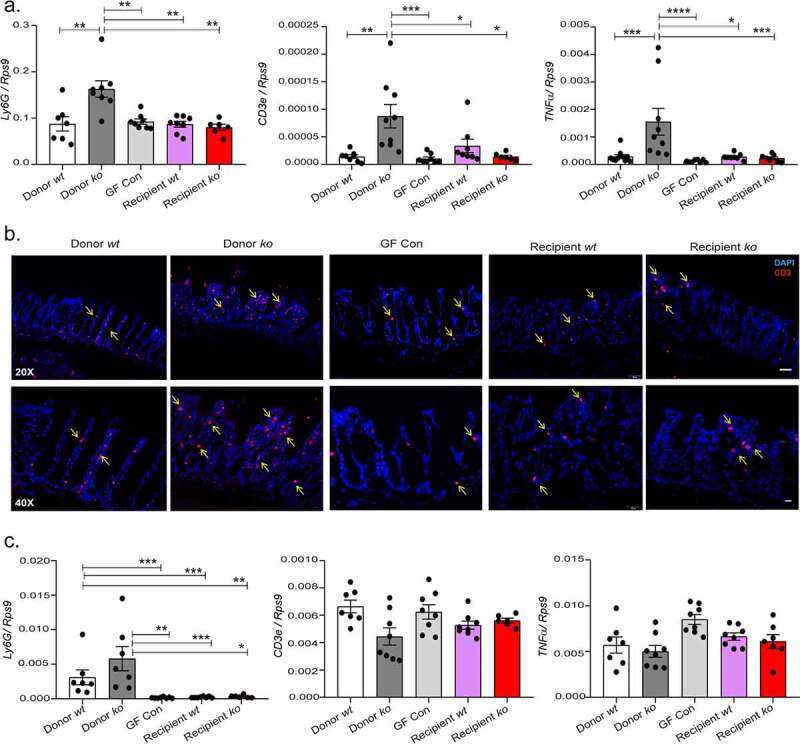
Non transfer of mucosal inflammation into the germ-free mice (a) Lack of development of mucosal inflammation as seen by the gene expression analysis of Ly6g, Cd3e and Tnfα in the mid distal colon. (b) Immunohistochemistry of CD3 T cells in the mid distal colon. (n = 5). (c) No systemic inflammation observed in all groups as seen by gene expression analysis of Ly6g, Cd3e and Tnfα in the spleen.
*p < .05, **p < .008, ***p ≤ .0007, ****p < .0001. slc26a3+/+ (wt) and slc26a3−/− (ko). Each dot represents one mouse.
Improved species richness post FMT
Microbiota analysis demonstrated that wt donor and recipients had a similar microbiota, while in contrast, slc26a3−/− donor and germ-free wt mice receiving the stool from slc26a3−/− mice exhibited strong deviation (Figure 5a). Specifically, the microbiota in recipient mice was characterized by higher and distinct diversity compared to the Donor ko microbiota, whereas the wt donor and recipient microbiota was very similar (Figure 5b,c). These shifts were also seen in the cecal recipient microbiota (Supplementary Figure 4a-c). In addition to its improved species richness, the colonic and cecal luminal microbiota of the recipient ko mice also showed decreases in the relative abundance of the families Bacteroidaceae and Erysipelotrichaceae, and an expansion in protective Muribaculaceae .15,16 An increase in Prevotellaceae was also observed in the recipient mice. Only Lachnospiraceae had comparable trends between the donor and recipient mice (Figure 5d-h and Supplementary Figure 4d-h), possibly because some species were reduced to undetectable levels in the slc26a3−/− microbiota and could not be recovered despite the “normal” gut milieu of the recipients.
Figure 5.
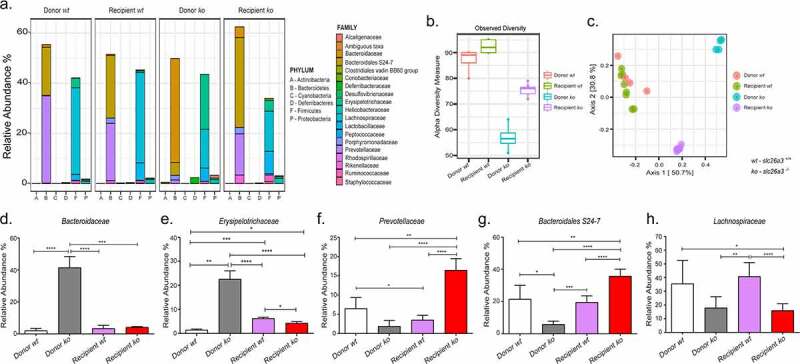
Improved and altered abundance of colonic taxonomic families post FMT (a) Relative abundance of the different phyla and families in the Donor wt, Recipient wt, Donor ko and Recipient ko. (b) Increase in the species richness post gavage transfer in the Recipient ko group. (c) Principal coordinate analysis depicting distinct microbial signatures between Donor ko and Recipient ko groups. (d,e.) Decrease in pro-inflammatory families Bacteroidaceae and Erysipelotrichaceae. (f) Increase in the relative abundance of Prevotellaceae and (g) protective Muribaculaceae . (h) Continued lowered abundance of Lachnospiraceae. n = 4 pairs for Donor wt, and Donor ko, n = 8 for Recipient wt and n = 7 for Recipient ko.
*p < .05, **p < .005, ***p = .0001, ****p < .0001.
Strong upregulation of antimicrobial proteins in slc26a3−/− colonic mucosa
The microbiota dysbiosis, in combination with a reduced Muc2 synthesis,8 an absent firm mucus layer,7 and tight junctional defects9 poses a strong threat to the colonic mucosal integrity. Nevertheless, the observed spontaneous inflammation in the slc26a3−/− mice was very mild and occurred only late in their lifespan,8 and this study. We therefore studied the expression of a panel of antimicrobial peptides and other protective proteins that have shown to be essential for the maintenance of microbial homeostasis and mucosal integrity. No significant difference was observed in the expression of the cathelicidin Cramp,17 in the barrier-protective trefoil factor Tff318 in the murine beta-defensin 4 (Defb4)19 and murine beta-defensin 14 (Defb14), the murine orthologue for HBD4 (Figure 6a-d) between wt and slc26a3−/− groups. Surprisingly, however, a very strong mRNA upregulation of the colon-expressed antimicrobial Reg3 lectins,20 possibly mediated by the strong increase in Il22 mRNA, was observed in the colonic mucosa (Figure 6e-g). This was accompanied by concomitant increases in the transcript levels for antibacterial-secreted phospholipase A2 (Pla2g2a), goblet cell-derived resistin like molecule beta (Relmβ)21 and Angiogenin 4 (Ang4).22 Intestinal alkaline phosphatase (iAlp) expression was also found upregulated in slc26a3−/− colonic mucosa (Figure 6h-k).
Figure 6.
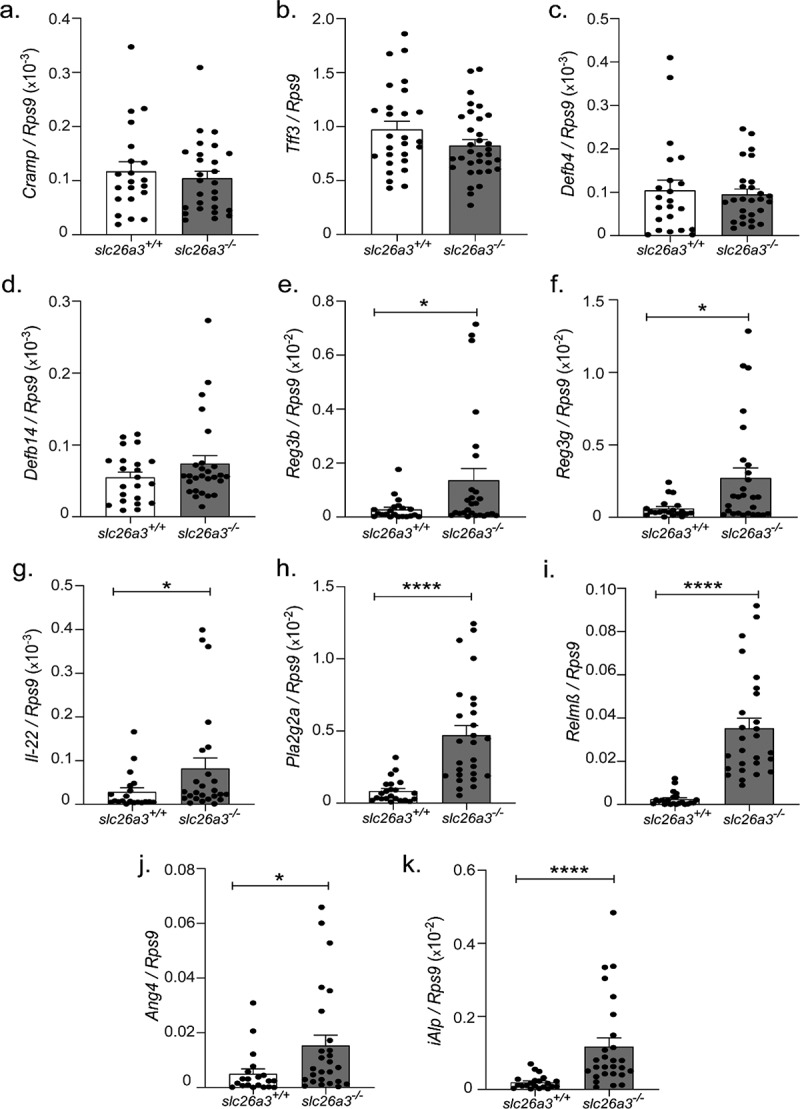
Shifts in the gene expression of antimicrobial peptides in the absence of slc26a3−/−. Unaltered (a-d) Cramp, Tff3, Defb4 and Defb14. Upregulated. (e-k) Reg3b, Reg3g, Il-22, Pla2g2a, Relmβ, Ang4, and iAlp.
*p < .05, ****p < .0001. Each dot represents one mouse.
Discussion
The human autosomal recessive disease congenital chloride diarrhea (CLD) was originally thought to be excessively rare and restricted to a few geographic locations with high levels of consanguinity.23 Recently, cases are being described more frequently from all continents. This is likely due to prenatal tentative diagnosis by abdominal ultrasound, and confirmation by sequencing techniques after birth that have become widely available in many areas of the world. While the disease need not be lethal any more, the substitution of electrolytes and fluid, which is the current mainstay of therapy, permits survival but does not attenuate the diarrhea. Moreover, many centers report a high percentage of grave complications, such as inflammatory bowel disease2,24 or chronic renal failure,25 both likely direct sequelae of the intestinal manifestations of CLD. It is therefore important to find causes that may aggravate the diarrheal, and/or the inflammatory intestinal phenotype in CLD patients, and to design treatment and preventive strategies.
The genetic deletion of Slc26a3 created a mouse model that recapitulates many aspects of the human CLD disease.26 In addition to the virtual absence of colonic bicarbonate secretion and a lack of firm mucus layer, we noticed in a past study an extraordinarily high susceptibility of the slc26a3−/− mice to DSS.7 Even a 2% DSS concentration in the drinking fluid resulted in rapid death, with an enormous increase in the number of large lymphoid aggregates in the intestine, whereas wt littermate mice showed only very mild inflammation under the same conditions.7 Because a dysbiotic microbiome has been linked to the severity of DSS colitis,27,28 we had also analyzed the fecal microbiota of slc26a3−/− and wt littermates at the phylum level and found a strongly reduced diversity and an altered ratio of Firmicutes/Bacteroidetes in the slc26a3−/− colon.8 These findings prompted us to explore the slc26a3−/− microbiota in more detail.
Since the strong segmental differences in the surface pH-microclimate in the murine colon are caused by the differences in expression and function of Slc26a3, with alkaline surface pH in the cecum and mid-distal colon and a fairly low surface pH in the proximal colon, where Slc26a3 expression is low,8 we focused on the mucosa-associated microbiota in this study. Specifically, we speculated the microbiota to be particularly influenced by the acidic surface pH in the cecum and mid-distal colon of the slc26a3−/− mice. For each of these locations, the slc26a3−/− microbiota showed a dramatic reduction in diversity. In addition, dramatic shifts were observed in the microbiota composition by the individual families, along with clear differences in the relative OTU abundances. Families with a known inflammatory potential, such as Bacteroidaceae and Erysipelotrichaceae, or with a mucolytic potential, such as the Deferribacteraceae (Mucispirillum), were strongly overrepresented, while the butyrate producers Lachnospiraceae (Roseburia) and Ruminococcaceae (Faecalibacterium) were strongly underrepresented in the mucosa-attached microbiota.
For some families, there were also significant differences in either the direction of change or the magnitude of change between wt and slc26a3−/− mice. One prominent example is the Muribaculaceae family , i.e., compare Figure 1b with Figure 2b,e. While changes in the magnitude can be more easily result simply as consequence of the different communities present in the lumen and the mucosal layer, changes in direction are likely the result of combinations of physiological and ecological factors. Previous studies in mice already suggested that the abundance of Muribaculaceae family members varies in association with different environmental conditions, including diet and transit time.29 Moreover, metagenomic analysis of various representatives from this diverse family revealed the presence of enzymatic clusters, for example the presence of trophic guilds for carbohydrate metabolism from various sources (i.e. plant glycans, host glycans), for breakdown of antimicrobial peptides, for evading immune detection, for fimbria development, dealing with oxidative stress, etc.30 This suggests that the members of the family can adjust to environmental changes, and partition into environmental niches. Hence, we assume that specific members of the Muribaculaceae family find suboptimal growth conditions in the lumen of slc26a3−/− colon, but that adaptive strategies increasing their fitness may result in relatively better conditions when they attach to the slc26a3−/− mucosa, than for other species. Importantly, these are likely not the same strains or species yet, the resolution of the 16S rRNA amplicon sequencing does not permit a reliable species identification. Further supporting the particular observations around this family is the significantly larger relative abundance in the microbiota in the GF mice when gavaged with microbiota from slc26a3−/− colon than from wt colon (Figure 5g). Of note, a similar situation after transfer into GF mice (without a relative enrichment at the slc26a3−/− mucosa) is seen for the genus Prevotella (Figure 5f).
Together, these findings were interesting, because they may help explain the slc26a3−/− colonic phenotype (and guide the investigations necessary to improve the clinical outcome in CLD patients). First, the increased abundance of mucolytic Deferribacteres, i.e., Mucispirillum schaedleri, may be related to the mucus layer defect in the slc26a3−/− colon. In addition, this species has been found increased in inflammation and expresses genes related to host chemokine and cytokine release.31 Several other families underrepresented in slc26a3−/− colon are also known to stimulate mucin biosynthesis, secretion and/or mucus layer quality.32,33 Second, the strongly decreased abundance of SCFA producers may explain why the slc26a3−/− mice cannot compensate their colonic fluid and base loss. Both in wt and slc26a3−/− isolated colonic mucosa, a luminal addition of the SCFA anion propionate was able to increase the luminal alkalinization rate.34 This suggests that the exchange of luminal SCFA against HCO3−, previously described in the rodent and human colon,35 albeit by an as yet molecularly undefined pathway, is intact in slc26a3−/− mucosa. A low abundance of SCFA producers may add to the diarrheal severity, and improving SCFA availability in the CLD colon a way to improve the diarrheal state. The reduction in SCFA producers in the microbiota of slc26a3−/− feces was also seen in a microbiota analysis by Kumar et al.,9 and may explain the slc26a3−/− increased DSS susceptibility, because a loss of SCFA producers is associated with an increased DSS susceptibility in mice.28 Although their methodological approach differed from ours, the group also found an increased abundance of families with a known inflammatory potential, such as Bacteroidaceae and Erysipelotrichaceae.9
To examine whether the strikingly different surface pH in the cecum and mid-distal colon of slc263−/− and wt mice may be the major determinant for the enormous differences in both the relative composition and the absolute abundance of members of the different families, we assessed the growth curves of a number of gut bacteria at different pH values in vitro. All studied bacteria replicated more slowly at acidic pH, although the relative acid resistance differed greatly between families. The high abundance of the Bifidobacteriaceae in the feces may be related to their acid-resistance and the low abundance of Helicobacter to their acid sensitivity,36 as well as to a described antagonistic function of Bifidobacteriaceae on Helicobacter growth.37 However, Prevotella rodentium was also fairly acid-resistent, yet the Prevotellacae family was underrepresented in the slc26a3−/− microbiota. Evidently, other factors are also important in facilitating growth and persistence of bacteria in relatively acidic pH surroundings in vivo. The method, which studies the growth of a single species in vitro, does not allow to pick up on the important interactions of the different microbiota with each other nor the important host–microbiota interactions that shape the microbiota.
Several groups have studied the inflammatory response and/or the microbiota in mice, which are defective for an intestinal acid/base transporter. The nhe3−/− mouse strongly upregulates INFγ in the small intestine,38 and Kiela et al. showed that this was part of a response against bacteria, which renders the mice extremely sensitive to small intestinal damage by DSS.39 The pH mileau was alkaline in the small and large intestine of nhe3−/− mice,40,41 but interestingly, while the small intestinal mucosa is hypoabsorptive,40 the large intestinal mucosa is hyperabsorptive, due to a very strong upregulation of ENaC and associated transporters (Junhua Li, unpublished). Thus, the nhe3−/− mice form fecal pellets in the colon, whereas the slc26a3−/− mice do not. Interestingly, the hyperabsorptive nhe3−/− distal colon displays a firmly adherent mucus layer, but an increased bacterial content in the mucosa.42 The nhe3−/− mouse displays a reduced colonic microbial diversity and a microbiota-dependent distal colitis in some animal houses.43,44 Interestingly, Engevik et al., did not find inflammation but showed a correlation between an increased sodium content in the nhe3−/− ileum and in the proliferation of B. thetaiotaomicron, which in turn increases fucosylation and thus shapes its own niche for optimal growth,41 and may enhance host-commensal symbiosis.45 An increased fucosylation, as well as dysbiosis, was also observed in the small intestine of CFTR-deficient mice,46,47 but in these mice, the small intestine has an acidic luminal mileau and is hyperabsorptive.48 A recent review summarizes the data about microbiota alterations in different mouse models that are deficient in intestinal ion transport protein, and it becomes clear that there is yet a lot to be learned about the causal relationships.49
We also measured the GI transit time, which affects the microbiota composition and diversity, because species with longer replication times are underrepresented if transit is very rapid.50 Surprisingly, both wt and slc26a3−/− mice displayed similar transit times, despite the difference in stool water. Current dogma states that a higher gut fluidity stimulates intestinal motility,51 but this appears not to be the case if the underlying cause for the increased fluid content is solely due to a decrease in absorption. The difference in stool water, observed in wt and slc26a3−/− mice,11 also does not explain the observed microbiota changes, because an increase in stool water was associated with increased abundance of Roseburia, Faecalibacterium and Lachnospira in another study, while these species were strongly decreased in slc26a3−/− microbiota.52 Therefore, we assume that the enormous upregulation of antimicrobial peptides, discussed further below, with their recognized potential to modulate microbiota composition, may also play a dominant role.
In order to study the inflammatory potential of the slc26a3−/− microbiota in an immunologically intact host, the luminal colonic content of slc26a3−/− mice and cohoused littermates was transferred to germ-free mice. The germ-free mice gavaged with either slc26a3−/− or wt microbiota did not develop intestinal inflammation 8 weeks later and the microbial diversity had markedly increased in the germ-free mice that had been gavaged with the slc26a3−/− microbiota, compared to that of its donor. No differences in α-diversity was seen between the donor and recipient wt microbiota. In addition, the absolute abundance of several families were in the range of the donor wt microbiota, with the exception of Lachnospiraceae, which remained at a lower level, possibly because selected members of that family were not present in the slc26a3−/− microbiota already before the gavage. Our findings demonstrate that the slc26a3−/− microbiota, while being highly abnormal and clearly harvesting high numbers of facultative pathobionts, is not capable to induce inflammation in a genetically healthy, intestinal electrolyte transport-competent, germ-free mouse.
Of course, we could have now proceeded to gavage the slc26a3−/− microbiota into germ-free mice that are immunologically incompetent, such as IL-10-deficient mice.53,54 However, we were more intrigued by the fact that the slc26a3−/− mice, even at the end of their expected lifetime, had only developed mild distal colonic and no systemic inflammation with the exception of an increased serum IL-6 level, a known disease activity marker for Crohn’s disease55 (Figure 4c and Supplementary Figure 6). Immunohistochemically, a mild increase in CD3e-positive cells in the slc26a3−/− colonic mucosa/submucosa was revealed. We had reported similar findings before and had provided evidence that the mice do not die of the intestinal inflammation.8 This lack of more prominent inflammation in slc26a3−/− mice is highly surprising, given the severe disturbances in three important protective entities in the colon: the microbiota (this study), the firm mucus layer7 and mucus production,8 and the tight junctional structures.9 In addition, the mild spontaneous inflammation in these mice was in stark contrast with the severe disease even after mild DSS challenge. Since it is now known that DSS causes direct toxicity to enterocytes even in the complete absence of other cell types,56 inactivating their defensive strategies, we speculated that the slc26a3−/− colonocytes may have activated defense strategies to counteract the negative effects of the dysbiotic microbiota on mucosal homeostasis.
Colonic epithelial cells express a number of antimicrobial peptides (AMP).19,57 The endogenous upregulation or the exogenous delivery of antimicrobial peptides protects the host against invasion of pathogens or damage by pathogen-associated molecular patterns.58,59 We measured the mRNA expression levels for a large panel of antimicrobial peptides and other substances with a known protective function (Figure 6). The results were striking: while no changes in expression levels was observed in the defensins, in the cathelicidin Cramp and in the trefoil factor Tff3, there was a very strong increase in the expression of the Reg3 lectins, Reg3β and Reg3γ (Figure 6e,f). These intestinal Reg3 proteins have recently been shown to be upregulated in mice that lack a firm mucus layer22 and to be protective against both colitis59,60 and diet- or alcohol-induced liver disease61 by reducing bacterial translocation in the gut in the presence of a dysfunctional epithelial barrier.62,63 Because the Reg3 proteins are discussed as IL-22 target genes,64,65 we also investigated the mucosal expression of Il22, and it was upregulated already at juvenile age, long before signs of inflammation are observed (Figure 6g and Supplementary Figure 5d). IL-22 is produced by several populations of immune cells including mucosa-resident dendritic and innate lymphoid cells (ILC) type 3 in response to bacterial stimuli,66 or other cytokines such as IL23,67 and mediates mucosal protective and regenerative functions.
The strong upregulation of Reg3β as well as Reg3γ and Relmβ in the slc26a3−/− colon suggests a protection against both gram-negative and gram-positive bacteria. REG3β binds to carbohydrate moieties on lipopolysaccharides (LPS) and thus kills Gram-negative bacteria.68,69 RELMβ also directly kills gram-negative bacteria.70 Initial interactions between Reg3γ, the murine orthologue of human REG3α, and its bacterial targets are mediated by binding to peptidoglycan, which is freely accessible only in gram-positive bacteria.71 The bactericidal mechanism of the Reg3 lectins and RELM involves permeabilization of the bacterial membrane by pore formation.70,72 The Reg3 proteins, as well as RELMβ, have been shown to be particularly important for enforcing the physical separation of microbiota and host and for limiting microbiota activation of adaptive immunity.60,70,73 As shown previously, the protective effect by the Reg3 lectins is functional also in the absence of a firm mucus layer.
Other proteins with important antibacterial properties that were upregulated are the enzymes such as the secreted phospholipase A2 type II (PlpA2IIA),74 and the intestinal alkaline phosphatase (iALP), which is also involved in preventing bacterial translocation in the gut.75,76 The findings may explain why both the slc26a3−/− mice, and the Muc2-deficient mice display spontaneous inflammation only in the distal parts of the colon, where the expression of Reg3β and Reg3γ, as well as iALP activity, are much lower than in the proximal colon.22 They also help to understand why we did not see bacteria in the colonic cryptal lumen in an earlier study, despite the complete lack of an adherent mucus layer.7 Finally, the high Relmβ expression, which is produced in goblet cells and promotes mucus secretion,77,78 may help explain why the size of many of the goblet thecae appeared less rounded, as if emptied, in the slc26a3−/− colonic mucosa.8
In summary, this study demonstrates that cohoused slc26a3−/− and wt littermates on the same diet until analysis, develop a very different colonic microbiota, with a dramatic loss of diversity, of short-chain fatty acid producers, and a strong increase of several facultative pathobionts. Most likely because of the dramatic upregulation of a panel of antimicrobial proteins in the slc26a3−/− colon of these immunocompetent mice, colonic inflammation was found only distally, and was mild, even in the presence of a very marked barrier defect as well as a pro-inflammatory microbiota. CLD patients have a dramatically increased risk of developing IBD,2 and it will be a task for the future to study whether the risk of CLD patients to develop IBD may be related to their genetically or environmentally determined ability to mount an antimicrobial defense.
Materials and methods
Animals
slc26a3−/− mice26 were bred and maintained at Hannover Medical School under standard temperature and light conditions, as previously described8 and were allowed free access to food and a half maximal Pedialyte drinking solution to prevent dehydration and enable increased survival post weaning. All mice in the experiments were age matched and used between 4 and 20 weeks of age. The wild-type and knockout littermates were co-housed from birth and monitored daily. Four-week-old male and female germ-free C57BL/6 J mice were housed in static micro-isolators (gnotocages) with autoclaved food, water and bedding at the Hannover Medical School. All experiments involving animals were approved by the Hannover Medical School committee on investigations involving animals and an independent committee assembled by the local authorities (Authorization number: 33.14–42502-04-14/1549 and 33.19–42502-04-20/3561).
Microbiota transfer
Cecal and colonic luminal content from the co-housed slc26a3 +/+ (wt) and slc26a3 −/− (ko) mice were collected and immediately suspended in sterile phosphate buffered saline (PBS)/glycerol (30%), snap-frozen and stored at −80°C until further use. Aliquots were then thawed and homogenized to get an even suspension. Each germ-free mouse was intragastrically gavaged with 50 µl cecal-colonic fecal suspensions from either the wt or ko donor, for over a period of 5 days, with a gap between D 2 and D 4. Germ-free mice gavaged with neither of the suspensions served as internal controls. Mice were then housed in microbiota – specific microisolators and sacrificed 8 weeks post colonization. Weight of the mice was recorded weekly once.
Histology and immunohistochemistry
Colonic tissues from each group, were harvested and fixed in 4% paraformaldehyde (PFA). 3 µm paraffin-embedded sections were stained with hematoxylin and eosin. To identify CD3e-positive cells, immunohistochemistry was performed on frozen sections of the colon. Mid- distal colonic tissues from the slc26a3 +/+, slc26a3 −/− and gavaged mice was frozen in Tissue Tek OCT and 10 µm cryosections were made. The sections were then fixed for 10 min in ice-cold acetone. Prior to staining, the sections were rehydrated with TBS-T for 10 min and then blocked with 10% rat serum in TBS-T for 30 min at room temperature. The slides were then stained with the diluted primary antibody, AntiCD3- Cy3 clone 17A2. After staining the nuclei with DAPI, the slides were mounted and examined under the Olympus FluoView™ FV1000 confocal microscope.
Quantitative PCR protocol
Gene expressions of a variety of pro-inflammatory cytokines and antimicrobial peptides were analyzed in the mid-distal colon by qPCR using ribosomal protein S9 (RPS9) as reference gene. Additionally, the spleen was analyzed for pro-inflammatory cytokines. RNA extraction, cDNA transcription and qPCR analysis were performed as per manufacturer’s instructions. Briefly, the total RNA was extracted using RNeasy® Mini Kit (Qiagen GmbH) and the quality was assessed using QIAxcel RNA QC Kit v2.0 (Qiagen GmbH). 1 µg RNA was then reverse transcribed with the QuantiTect® Reverse Transcription Kit (Qiagen GmbH). cDNA was diluted 1:40 with DNase free water, and 4 µL of the dilution was used as a template for PCR. Each reaction additionally contained 5 µL 2X qPCRBIO SyGreen Mix Lo-ROX (PCR Biosystems) and an appropriate amount of primers. The panel of primers are given in supplementary Table 1.
Microbiota analysis
Sample preparation – luminal microbiota
Stool/fecal pellets were collected from gavaged mice and donor mice post sacrifice and immediately stored at −80°C. Stool pellets were similarly collected from the slc26a3 wt and ko mice at multiple time points (Weeks 5, 8, 12, 16 and 20 for colon and Early: 4–9 weeks, Mid: 10–15 weeks and Late: 16–20 weeks for cecum) and stored at −80°C. DNA was extracted using a method as described previously.79 Briefly, samples were first suspended in a solution containing 200 µL 0.1 mm zirconia/silica beads, 500 µL extraction buffer (200 mM NaCl, 200 mM Tris [pH 8.0] and 20 mM Sodium EDTA), 200 µL 20% SDS and 500 µL phenol:chloroform:isoamyl alcohol mixture (pH 7.9, 25:24:1). The samples were subsequently lysed twice by mechanical disruption using bead beater for 2 min. It was then followed by extraction with phenol:chloroform:isoamyl alcohol (pH 7.9, 25:24:1), and precipitation with ice-cold isopropanol. The DNA extracts were resuspended in Tris-EDTA (TE) buffer.
Sample preparation – mucosa adherent microbiota
Mucosal scrapings from slc26a3 wt and ko were collected at different time points and stored in TriZol at −80°C. For the analysis, samples were once again clustered into three groups, namely “Early” (4–9 weeks), “Mid” (10–15 weeks) and “Late” (16–20 weeks). The scrapings were first homogenized thoroughly in 1 ml TriZol and 200 µL 1 mm beads using a bead beater. To ensure complete dissociation of nucleoprotein complexes, the homogenized samples were incubated at room temperature for 5 min followed by spinning it down at 300 g for 5 min at 4°C. Phase separation was performed by adding 200 µL chloroform per 1 ml TriZol to the supernatant. It was vortexed vigorously and then spun down at ≤12,000 g for 15 min at 2–8°C. The upper aqueous phase was then transferred into a tube and RNA was precipitated with ice-cold isopropanol. After washing the RNA extract, it was resuspended in fresh pure water and stored immediately at −80°C. The RNA was then converted to bacterial cDNA using the RevertAid First Strand cDNA Synthesis Kit according to manufacturer’s instructions. The presence of bacterial cDNA was confirmed by performing a PCR.
Microbial 16S rRNA gene analysis
Prior to sequencing, the crude DNA extracts were resuspended in TRIS-EDTA (TE) buffer containing 100 mg/ml RNase and column purified. Amplification of the V4 region (F515/R806) of the 16S rRNA gene was performed as described previously.80 Using the Illumina MiSeq platform (PE250), the samples were sequenced. Sequences were then filtered for low quality reads (q ≥ 30) and bar code binning was performed using the QIIME v1.8.0.81 The obtained reads were then clustered into operational taxonomical units (OTUs) based in the 97% nucleotide identity of the amplicon sequences using UCLUST reference OTU picking. This was followed by further taxonomic classification using the Ribosomal Database Project (RDP) classifier executed at 80% bootstrap confidence cutoff.82,83 The sequences without a matching reference dataset were then grouped as de novo using UCLUST. The OTU absolute abundance table and mapping file were used for statistical analyses and data visualization in the R statistical programming environment package phyloseq.84
pH-dependency of in vitro growth of various microbiota components
In brief, all bacteria were grown in BHI medium. For each media, pH was decreased with hydrochloric acid. All cultures were grown in 200 µl in 96-well plates on 37°C in anaerobic conditions, using a biotek absorbance reader.
Total gastrointestinal transit time
The total Gastrointestinal transit time was measured as previously described with a few modifications.85 Briefly, a solution of carmine red (6% in 0.5% methycellulose solution) was mixed with the powdered normal chow/food, reshaped into pellets, dried and sterilized prior to feeding. Both the slc26a3−/− and wt were fasted for approximately 5 h and then placed in clean transparent empty cages for observation. Post fasting, the mice were fed the red pellets and the interval between the first food intake to the appearance of the first red stool pellet was considered as the total GI transit time.
Statistics
Statistical analysis was performed using GraphPad Prism Version 8.00. Unless otherwise indicated, comparisons of two groups that passed the Shapiro–Wilk normality test were compared with the two-tailed Student t-test. In case one or both groups did not pass the normality test, analysis was performed using the nonparametric Mann–Whitney U-test. All results were expressed as the mean ± SEM.
| b. | |||||
|---|---|---|---|---|---|
| Genotype | Stool water content (%) |
||||
| 0 h | 1 h | 2 h | 3 h | 4 h | |
| Slc26a3+/+ | 61.38 | 63.13 | 63.32 | 53.60 | 56.86 |
| Slc26a3−/− | 88.86 | 85.68 | 85.52 | 85.79 | 83.34 |
| P value | 0.0016 | 0.0211 | 0.0053 | 0.0024 | 0.0002 |
(a.) The transit time for each mice (n = 5/genotype) is depicted as an average of three measurements. The overall average and SEM indicate that there is no significant difference between slc26a3−/− and the wt littermates. p = 0.3786. (b.) The stool water content, however, remained significantly higher in the slc26a3−/− throughout the measurement. Zero hour indicates the timepoint right at the end of the fasting period; 1, 2, 3, 4 h indicate the timepoint since the mice began eating. Each value is an average of a total of three measurements in five mice.
Supplementary Material
Acknowledgments
We gratefully thank and acknowledge Brigitte Rausch and Anne Knoll-Schluch for their help with the animal breeding and genotyping; Anna Smozcek for brilliant technical assistance with gnotobiotic experiments; Achim Gronow for excellent technical assistance with 16S rRNA sequencing; Dr Joana Martins and Prof. Dr rer.nat Immo Prinz for the antibody anti-CD3- Cy3 clone 17A2 and guidance with the immunohistochemistry staining.
Funding Statement
This work was supported by the China Scholarship Council [201704910936, 202006160035]; Deutsche Forschungsge-meinschaft [SFB900/B8, Project ID 158989968; SE260/13-4,19-1,22-1]; DFG Priority Program [SPP1656 BL953/5-28]; Sonderforchungsbereiche (SFB) [621/C9]; and Volkswagen Foundation [ZN1953].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. Raw data for the 16S rRNA sequencing were generated at Helmholtz Center for Infection Research, Braunschweig, Germany. Derived data supporting the findings of this study are available from the corresponding author, Ursula Seidler, on request.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Seidler U, Nikolovska K.. slc26 family of anion transporters in the gastrointestinal tract: expression, function, regulation, and role in disease. Compr Physiol. 2019;9(2):839–18. doi: 10.1002/cphy.c180027. [DOI] [PubMed] [Google Scholar]
- 2.Norsa L, Berni Canani R, Duclaux- Loras R, Bequet E, Köglmeier J, Russell RK, Uhlig HH, Travis S, Hollis J, Koletzko S, et al. Inflammatory bowel disease in patients with congenital chloride diarrhoea. J Crohn’s Colitis. 2021;15(10):1679–1685. doi: 10.1093/ecco-jcc/jjab056. [DOI] [PubMed] [Google Scholar]
- 3.Camarillo GF, Goyon EI, Zuñiga RB, Salas LAS, Escárcega AEP, Yamamoto-Furusho JK. Gene expression profiling of mediators associated with the inflammatory pathways in the intestinal tissue from patients with ulcerative colitis. Mediators Inflamm. 2020;2020:9238970. doi: 10.1155/2020/9238970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farkas K, Yeruva S, Rakonczay Z, Ludolph L, Molnár T, Nagy F, Szepes Z, Schnúr A, Wittmann T, Hubricht J, et al. New therapeutic targets in ulcerative colitis: the importance of ion transporters in the human colon. Inflamm Bowel Dis. 2011;17(4):884–898. doi: 10.1002/ibd.21432. [DOI] [PubMed] [Google Scholar]
- 5.Yang H, Jiang W, Furth EE, Wen X, Katz JP, Sellon RK, Silberg DG, Antalis TM, Schweinfest CW, Wu GD. Intestinal inflammation reduces expression of DRA, a transporter responsible for congenital chloride diarrhea. Am J Physiol Liver Physiol. 1998;275(6):G1445–1453. doi: 10.1152/ajpgi.1998.275.6.G1445. [DOI] [PubMed] [Google Scholar]
- 6.Xiao F, Juric M, Li J, Riederer B, Yeruva S, Singh AK, Zheng L, Glage S, Kollias G, Dudeja P, et al. Loss of downregulated in adenoma (DRA) impairs mucosal HCO3− secretion in murine ileocolonic inflammation. Inflamm Bowel Dis. 2012;18(1):101–111. doi: 10.1002/ibd.21744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao F, Yu Q, Li J, V JME, Singh AK, Xia W, Riederer B, Engelhardt R, Montrose M, Soleimani M, et al. Slc26a3 deficiency is associated with loss of colonic HCO 3 − secretion, absence of a firm mucus layer and barrier impairment in mice. Acta Physiol. 2014;211(1):161–175. doi: 10.1111/apha.12220. [DOI] [PubMed] [Google Scholar]
- 8.Kini A, Singh AK, Riederer B, Yang I, Tan Q, Di Stefano G, Tan Q, Xiao F, Xia W, Suerbaum S, et al. Slc26a3 deletion alters pH-microclimate, mucin biosynthesis, microbiome composition and increases the TNFα expression in murine colon. Acta Physiol. 2020;230(2):e13498. doi: 10.1111/apha.13498. [DOI] [PubMed] [Google Scholar]
- 9.Kumar A, Priyamvada S, Ge Y, Jayawardena D, Singhal M, Anbazhagan AN, Chatterjee I, Dayal A, Patel M, Zadeh K, et al. A novel role of SLC26A3 in the maintenance of intestinal epithelial barrier integrity. Gastroenterology. 2021;160(4):1240–1255. doi: 10.1053/j.gastro.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grönlund MM, Grzeskowiak Ł, Isolauri E, Salminen S. Influence of mother’s intestinal microbiota on gut colonization in the infant. Gut Microbes. 2011;2(4):227–233. doi: 10.4161/gmic.2.4.16799. [DOI] [PubMed] [Google Scholar]
- 11.Huang EY, Inoue T, Leone VA, Dalal S, Touw K, Wang Y, Musch MW, Theriault B, Higuchi K, Donovan S, et al. Using corticosteroids to reshape the gut microbiome: implications for inflammatory bowel diseases. Inflamm Bowel Dis. 2015;21(5):963–972. doi: 10.1097/MIB.0000000000000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, Bonnaure-Mallet M, Jolivet-Gougeon A. Roseburia spp.: a marker of health? Future Microbiol. 2017;12(2):157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11(4):841–852. doi: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vital M, Karch A, Pieper DH, Shade A. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems. 2017;2(6):e00130–17. doi: 10.1128/msystems.00130-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, Gallini CA, Beal C, Van Hylckama-Vlieg JET, Ballal SA, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8(7):1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker KD, Albeke SE, Gigley JP, Goldstein AM, Ward NL. Microbiome composition in both wild-type and disease model mice is heavily influenced by mouse facility. Front Microbiol. 2018;9:1598. doi: 10.3389/fmicb.2018.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’huigin C, Gong W, Zhou J, Xiang Y, Tessarollo L, Yao X, Huang J, Trinchieri G, Badger J H, Bian X, et al. The antimicrobial peptide CRAMP is essential for colon homeostasis by maintaining microbiota balance. J Immunol. 2018;200(6):2174–2185. doi: 10.4049/jimmunol.1602073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braga Emidio N, Brierley SM, Schroeder CI, Muttenthaler M. Structure, function, and therapeutic potential of the trefoil factor family in the gastrointestinal tract. ACS Pharmacol Transl Sci. 2020;3(4):583–597. doi: 10.1021/acsptsci.0c00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blyth GAD, Connors L, Fodor C, Cobo ER. The network of colonic host defense peptides as an innate immune defense against enteropathogenic bacteria. Front Immunol. 2020;11:965. doi: 10.3389/fimmu.2020.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin JH, Seeley RJ. REG3 proteins as gut hormones? Endocrinology. 2019;160(7):1677–1678. doi: 10.1210/en.2019-00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8(6):411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 22.Paassen N B-V, Loonen LMP, Witte-Bouma J, Male AM K-V, de Bruijn ACJM, van der Sluis M, Lu P, van Goudoever JB, Wells JM, Dekker J, et al. Mucin Muc2 deficiency and weaning influences the expression of the innate defense genes Reg3β, Reg3γ and Angiogenin-4. PLoS One. 2012;7(6):e38798. doi: 10.1371/journal.pone.0038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kere J, Lohi H, Höglund P. Genetic disorders of membrane transport. III. Congenital chloride diarrhea. Am J Physiol - Gastrointest Liver Physiol. 1999;276(1):G7–G13. doi: 10.1152/ajpgi.1999.276.1.g7. [DOI] [PubMed] [Google Scholar]
- 24.Hihnala S, Höglund P, Lammi L, Kokkonen J, Ormälä T, Holmberg C. Long-term clinical outcome in patients with congenital chloride diarrhea. J Pediatr Gastroenterol Nutr. 2006;42(4):369–375. doi: 10.1097/01.mpg.0000214161.37574.9a. [DOI] [PubMed] [Google Scholar]
- 25.Wedenoja S, Örmälä T, Berg UB, Halling SFE, Jalanko H, Karikoski R, Kere J, Holmberg C, Höglund P. The impact of sodium chloride and volume depletion in the chronic kidney disease of congenital chloride diarrhea. Kidney Int. 2008;74(8):1085–1093. doi: 10.1038/ki.2008.401. [DOI] [PubMed] [Google Scholar]
- 26.Schweinfest CW, Spyropoulos DD, Henderson KW, Kim J-H, Chapman JM, Barone S, Worrell RT, Wang Z, Soleimani M. slc26a3 (dra)-deficient mice display chloride-losing diarrhea, enhanced colonic proliferation, and distinct up-regulation of ion transporters in the colon. J Biol Chem. 2006;281(49):37962–37971. doi: 10.1074/jbc.M607527200. [DOI] [PubMed] [Google Scholar]
- 27.Ozkul C, Ruiz VE, Battaglia T, Xu J, Roubaud-Baudron C, Cadwell K, Perez-Perez GI, Blaser MJ. A single early-in-life antibiotic course increases susceptibility to DSS-induced colitis. Genome Med. 2020;12(1):65. doi: 10.1186/s13073-020-00764-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laffin M, Fedorak R, Zalasky A, Park H, Gill A, Agrawal A, Keshteli A, Hotte N, Madsen KL. A high-sugar diet rapidly enhances susceptibility to colitis via depletion of luminal short-chain fatty acids in mice. Sci Rep. 2019;9(1):12294. doi: 10.1038/s41598-019-48749-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagkouvardos I, Lesker TR, Hitch TCA, Gálvez EJC, Smit N, Neuhaus K, Wang J, Baines JF, Abt B, Stecher B, et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome. 2019;7(1):28. doi: 10.1186/s40168-019-0637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ormerod KL, Wood DLA, Lachner N, Gellatly SL, Daly JN, Parsons JD, Dal’Molin CGO, Palfreyman RW, Nielsen LK, Cooper MA, et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome. 2016;4(1):36. doi: 10.1186/s40168-016-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loy A, Pfann C, Steinberger M, Hanson B, Herp S, Brugiroux S, Gomes Neto JC, Boekschoten MV, Schwab C, Urich T, et al. Lifestyle and horizontal gene transfer-mediated evolution of mucispirillum schaedleri, a core member of the murine gut microbiota. mSystems. 2017;2(1):e00171–16. doi: 10.1128/msystems.00171-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graziani F, Pujol A, Nicoletti C, Dou S, Maresca M, Giardina T, Fons M, Perrier J. Ruminococcus gnavus E1 modulates mucin expression and intestinal glycosylation. J Appl Microbiol. 2016;120(5):1403–1417. doi: 10.1111/jam.13095. [DOI] [PubMed] [Google Scholar]
- 33.Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut. 2020;69(12):2232–2243. doi: 10.1136/gutjnl-2020-322260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi H, Nagai H, Ichiro OK, Soleimani M, Suzuki Y. Segmental differences in Slc26a3-dependent Cl− absorption and HCO3− secretion in the mouse large intestine in vitro in ussing chambers. J Physiol Sci. 2021;71(1):5. doi: 10.1186/s12576-020-00784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dohgen M, Hayahshi H, Yajima T, Suzuki Y. Stimulation of bicarbonate secretion by luminal short-chain fatty acid in the rat and human colon in vitro. Jpn J Physiol. 1994;44(5):519–531. doi: 10.2170/jjphysiol.44.519. [DOI] [PubMed] [Google Scholar]
- 36.Sjöström JE, Larsson H. Factors affecting growth and antibiotic susceptibility of Helicobacter pylori: effect of pH and urea on the survival of a wild-type strain and a urease-deficient mutant. J Med Microbiol. 1996;44(6):425–433. doi: 10.1099/00222615-44-6-425. [DOI] [PubMed] [Google Scholar]
- 37.Kim JE, Kim MS, Yoon YS, Chung MJ, Yum DY. Use of selected lactic acid bacteria in the eradication of Helicobacter pylori infection. J Microbiol. 2014;52(11):955–962. doi: 10.1007/s12275-014-4355-y. [DOI] [PubMed] [Google Scholar]
- 38.Woo AL, Gildea LA, Tack LM, Miller ML, Spicer Z, Millhorn DE, Finkelman FD, Hassett DJ, Shull GE. In vivo evidence for interferon-γ-mediated homeostatic mechanisms in small intestine of the NHE3 Na+/H+ exchanger knockout model of congenital diarrhea. J Biol Chem. 2002;277(50):49036–49046. doi: 10.1074/jbc.M205288200. [DOI] [PubMed] [Google Scholar]
- 39.Kiela PR, Laubitz D, Larmonier CB, Midura-Kiela MT, Lipko MA, Janikashvili N, Bai A, Thurston R, Ghishan FK. Changes in mucosal homeostasis predispose NHE3 knockout mice to increased susceptibility to DSS-induced epithelial injury. Gastroenterology. 2009;137(3):965–975. doi: 10.1053/j.gastro.2009.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia W, Yu Q, Riederer B, Singh AK, Engelhardt R, Yeruva S, Song P, Tian D-A, Soleimani M, Seidler U. The distinct roles of anion transporters Slc26a3 (DRA) and Slc26a6 (PAT-1) in fluid and electrolyte absorption in the murine small intestine. Pflügers Arch. 2014;466(8):1541–1556. doi: 10.1007/s00424-013-1381-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engevik MA, Aihara E, Montrose MH, Shull GE, Hassett DJ, Worrell RT. Loss of NHE3 alters gut microbiota composition and influences Bacteroides thetaiotaomicron growth. Am J Physiol Gastrointest Liver Physiol. 2013;305(10):G697–711. doi: 10.1152/ajpgi.00184.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.V JME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63(2):281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Midura-Kiela MT, Ramalingam R, Hassan KA, Besselsen DG, Golebiewski M, Ghishan FK, Hill FM, Laubitz D, McFadden R-MT, Shehab KW, et al. Reduced colonic microbial diversity is associated with colitis in NHE3-deficient mice. Am J Physiol Liver Physiol. 2013;305(10):G667–677. doi: 10.1152/ajpgi.00189.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrison CA, Laubitz D, Ohland CL, Midura-Kiela MT, Patil K, Besselsen DG, Jamwal DR, Jobin C, Ghishan FK, Kiela PR. Microbial dysbiosis associated with impaired intestinal Na + /H + exchange accelerates and exacerbates colitis in ex-germ free mice. Mucosal Immunol. 2018;11(5):1329–1341. doi: 10.1038/s41385-018-0035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nat. 2014;514(7524):638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiru S, Devereux G, King A. Abnormal fucosylation of ileal mucus in cystic fibrosis: i A histochemical study using peroxidase labelled lectins. J Clin Pathol. 1990;43(12):1014–1018. doi: 10.1136/jcp.43.12.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomsson KA, Hinojosa-Kurtzberg M, Axelsson KA, Domino SE, Lowe JB, Gendler SJ, Hansson GC. Intestinal mucins from cystic fibrosis mice show increased fucosylation due to an induced Fucα1-2 glycosyltransferase. Biochem J. 2002;367(Pt 3):609–616. doi: 10.1042/BJ20020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan Q, Di Stefano G, Tan X, Renjie X, Römermann D, Talbot SR, Seidler UE. Inhibition of Na+/H+ exchanger isoform 3 improves gut fluidity and alkalinity in cystic fibrosis transmembrane conductance regulator-deficient and F508del mutant mice. Br J Pharmacol. 2021;178(5):1018–1036. doi: 10.1111/bph.15323. [DOI] [PubMed] [Google Scholar]
- 49.Engevik AC, Engevik MA. Exploring the impact of intestinal ion transport on the gut microbiota. Comput Struct Biotechnol J. 2021;19:134–144. doi: 10.1016/j.csbj.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asnicar F, Leeming ER, Dimidi E, Mazidi M, Franks P, Al Khatib H, Valdes AM, Davies R, Bakker E, Francis L, et al. Blue poo: impact of gut transit time on the gut microbiome using a novel marker. Gut. 2021;70(9):1665–1674. doi: 10.1136/gutjnl-2020-323877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Read NW. Colon: relationship between epithelial transport and motility. Pharmacology. 1988;36(1):120–125. doi: 10.1159/000138430. [DOI] [PubMed] [Google Scholar]
- 52.Jalanka J, Major G, Murray K, Singh G, Nowak A, Kurtz C, Silos-Santiago I, Johnston JM, de Vos WM, Spiller R. The effect of psyllium husk on intestinal microbiota in constipated patients and healthy controls. Int J Mol Sci. 2019;20(2):433. doi: 10.3390/ijms20020433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Büchler G, Wos-Oxley ML, Smoczek A, Zschemisch NH, Neumann D, Pieper DH, Hedrich HJ, Bleich A. Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota-host interactions. Inflamm Bowel Dis. 2012;18(5):943–954. doi: 10.1002/ibd.21895. [DOI] [PubMed] [Google Scholar]
- 54.Bruesch I, Meier P, Vital M, Pieper DH, Selke K, Böhlen S, Basic M, Meier M, Glage S, Hundrieser J, et al. Analysis of Cdcs1 colitogenic effects in the hematopoietic compartment reveals distinct microbiome interaction and a new subcongenic interval active in T cells. Mucosal Immunol. 2019;12(3):691–702. doi: 10.1038/s41385-019-0133-9. [DOI] [PubMed] [Google Scholar]
- 55.Mavropoulou E, Mechie NC, Knoop R, Petzold G, Ellenrieder V, Kunsch S, Pilavakis Y, Amanzada A, Bonaz B. Association of serum interleukin-6 and soluble interleukin-2-receptor levels with disease activity status in patients with inflammatory bowel disease: a prospective observational study. PLoS One. 2020;15(5):e0233811. doi: 10.1371/journal.pone.0233811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, Elci B, Brandenberg N, Kolotuev I, Gjorevski N, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nat. 2020;585(7826):574–578. doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- 57.Mukherjee S, Hooper LV. Antimicrobial defense of the intestine. Immunity. 2015;42(1):28–39. doi: 10.1016/j.immuni.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H, Xia X, Han F, Jiang Q, Rong Y, Song D, Wang Y. Cathelicidin-BF, a novel antimicrobial peptide from bungarus fasciatus, attenuates disease in a dextran sulfate sodium model of colitis. Mol Pharm. 2015;12(5):1648–1661. doi: 10.1021/acs.molpharmaceut.5b00069. [DOI] [PubMed] [Google Scholar]
- 59.Bajic D, Niemann A, Hillmer AK, Mejias-Luque R, Bluemel S, Docampo M, Funk MC, Tonin E, Boutros M, Schnabl B, et al. Gut microbiota-derived propionate regulates the expression of Reg3 mucosal lectins and ameliorates experimental colitis in mice. J Crohn’s Colitis. 2020;14(10):1462–1472. doi: 10.1093/ecco-jcc/jjaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loonen LMP, Stolte EH, Jaklofsky MTJ, Meijerink M, Dekker J, Van Baarlen P, Wells JM. REG3γ-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014;7(4):939–947. doi: 10.1038/mi.2013.109. [DOI] [PubMed] [Google Scholar]
- 61.Wang L, Fouts DE, Stärkel P, Hartmann P, Chen P, Llorente C, DePew J, Moncera K, Ho SB, Brenner DA, et al. Intestinal reg3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe. 2016;19(2):227–239. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hartmann P, Seebauer CT, Mazagova M, Horvath A, Wang L, Llorente C, Varki NM, Brandl K, Ho SB, Schnabl B. Deficiency of intestinal mucin-2 protects mice from diet-induced fatty liver disease and obesity. Am J Physiol Gastrointest Liver Physiol. 2016;310(5):G310–322. doi: 10.1152/ajpgi.00094.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendrikx T, Schnabl B. Antimicrobial proteins: intestinal guards to protect against liver disease. J Gastroenterol. 2019;54(3):209–217. doi: 10.1007/s00535-018-1521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zindl CL, Lai JF, Lee YK, Maynard CL, Harbour SN, Ouyang W, Chaplin DD, Weaver CT. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci U S A. 2013;110(31):12768–12773. doi: 10.1073/pnas.1300318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hammer AM, Morris NL, Cannon AR, Khan OM, Gagnon RC, Movtchan NV, van Langeveld I, Li X, Gao B, Choudhry MA. Interleukin-22 prevents microbial dysbiosis and promotes intestinal barrier regeneration following acute injury. Shock. 2017;48(6):657–665. doi: 10.1097/SHK.0000000000000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Castleman MJ, Dillon SM, Purba CM, Cogswell AC, Kibbie JJ, McCarter MD, Santiago ML, Barker E, Wilson CC. Commensal and pathogenic bacteria indirectly induce IL-22 but not IFNγ production from human colonic ILC3s via multiple mechanisms. Front Immunol. 2019;10:649. doi: 10.3389/fimmu.2019.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aden K, Rehman A, Falk-Paulsen M, Secher T, Kuiper J, Tran F, Pfeuffer S, Sheibani-Tezerji R, Breuer A, Luzius A, et al. Epithelial IL-23R signaling licenses protective IL-22 responses in intestinal inflammation. Cell Rep. 2016;16(8):2208–2218. doi: 10.1016/j.celrep.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stelter C, Käppeli R, König C, Krah A, Hardt WD, Stecher B, Bumann D, May RC. Salmonella-induced mucosal lectin RegIIIβ kills competing gut microbiota. PLoS One. 2011;6(6):e20749. doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miki T, Holsts O, Hardt WD. The bactericidal activity of the C-type lectin regIIIβ against gram-negative bacteria involves binding to lipid A. J Biol Chem. 2012;287(41):34844–34855. doi: 10.1074/jbc.M112.399998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Propheter DC, Chara AL, Harris TA, Ruhn KA, Hooper LV. Resistin-like molecule β is a bactericidal protein that promotes spatial segregation of the microbiota and the colonic epithelium. Proc Natl Acad Sci U S A. 2017;114(42):11027–11033. doi: 10.1073/pnas.1711395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lehotzkya RE, Partchb CL, Mukherjeea S, Casha HL, Goldman WE, Gardner KH, Hooper LV. Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc Natl Acad Sci U S A. 2010;107(17):7722–7727. doi: 10.1073/pnas.0909449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mukherjee S, Zheng H, Derebe MG, Callenberg KM, Partch CL, Rollins D, Propheter DC, Rizo J, Grabe M, Jiang QX, et al. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nat. 2014;505(7481):103–107. doi: 10.1038/nature12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Hensbergen VP, Wu Y, van Sorge NM, Touqui L. Type IIA secreted phospholipase A2 in host defense against bacterial infections. Trends Immunol. 2020;41(4):313–326. doi: 10.1016/j.it.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 75.Lallès JP. Intestinal alkaline phosphatase: novel functions and protective effects. Nutr Rev. 2014;72(2):82–94. doi: 10.1111/nure.12082. [DOI] [PubMed] [Google Scholar]
- 76.Fawley J, Koehler S, Cabrera S, Lam V, Fredrich K, Hessner M, Salzman N, Gourlay D. Intestinal alkaline phosphatase deficiency leads to dysbiosis and bacterial translocation in the newborn intestine. J Surg Res. 2017;218:35–42. doi: 10.1016/j.jss.2017.03.049. [DOI] [PubMed] [Google Scholar]
- 77.Krimi RB, Kotelevets L, Dubuquoy L, Plaisancié P, Walker F, Lehy T, Desreumaux P, Van Seuningen I, Chastre E, Forgue-Lafitte ME, et al. Resistin-like molecule β regulates intestinal mucous secretion and curtails TNBS-induced colitis in mice. Inflamm Bowel Dis. 2008;14(7):931–941. doi: 10.1002/ibd.20420. [DOI] [PubMed] [Google Scholar]
- 78.Bergstrom KSB, Morampudi V, Chan JM, Bhinder G, Lau J, Yang H, Ma C, Huang T, Ryz N, Sham HP, et al. Goblet cell derived RELM-β recruits CD4+ T cells during infectious colitis to promote protective intestinal epithelial cell proliferation. PLoS Pathog. 2015;11(8):e1005108. doi: 10.1371/journal.ppat.1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 84.McMurdie PJ, Holmes S, Watson M. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Z, Chalazonitis A, Huang YY, Mann JJ, Margolis KG, Yang QM, Kim DO, Côté F, Mallet J, Gershon MD. Essential roles of enteric neuronal serotonin in gastrointestinal motility and the development/survival of enteric dopaminergic neurons. J Neurosci. 2011;31(24):8998–9009. doi: 10.1523/JNEUROSCI.6684-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. Raw data for the 16S rRNA sequencing were generated at Helmholtz Center for Infection Research, Braunschweig, Germany. Derived data supporting the findings of this study are available from the corresponding author, Ursula Seidler, on request.


