Abstract
Purpose: Recent studies have found that KRAS mutations predict resistance to monoclonal antibodies targeting the epidermal growth factor receptor in metastatic colorectal cancer (mCRC). A polymorphism in a let-7 microRNA complementary site (lcs6) in the KRAS 3′ untranslated region (UTR) is associated with an increased cancer risk in non-small-cell lung cancer and reduced overall survival (OS) in oral cancers. We tested the hypothesis whether this polymorphism may be associated with clinical outcome in KRAS wild-type (KRASwt) mCRC patients treated with cetuximab monotherapy.
Patients and methods: The presence of KRAS let-7 lcs6 polymorphism was evaluated in 130 mCRC patients who were enrolled in a phase II study of cetuximab monotherapy (IMCL-0144). Genomic DNA was extracted from dissected formalin-fixed paraffin-embedded tumor tissue, KRAS mutation status and polymorphism were assessed using direct sequencing and PCR restriction fragment length polymorphism technique.
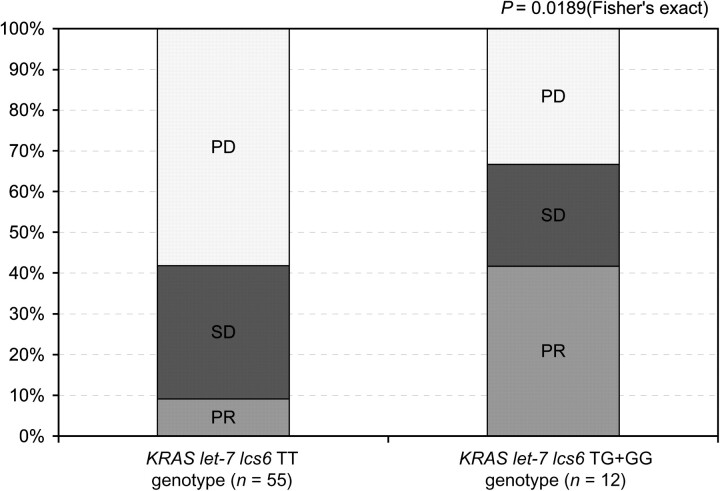
Results: KRAS let-7 lcs6 polymorphism was found to be related to object response rate (ORR) in mCRC patients whose tumors had KRASwt. The 12 KRASwt patients harboring at least a variant G allele (TG or GG) had a 42% ORR compared with a 9% ORR in 55 KRASwt patients with let-7 lcs6 TT genotype (P = 0.02, Fisher’s exact test). KRASwt patients with TG/GG genotypes had trend of longer median progression-free survival (3.9 versus 1.3 months) and OS (10.7 versus 6.4 months) compared to those with TT genotypes.
Conclusions: These results are the first to indicate that the KRAS 3’UTR polymorphism may predict for cetuximab responsiveness in KRASwt mCRC patients, which warrants validation in other clinical trials.
Keywords: cetuximab, KRAS, microRNA polymorphism, metastatic colon cancer
introduction
Colorectal cancer (CRC) remains the second leading cause of cancer deaths in the United States. In 2009, an estimated 146 970 new cases will be diagnosed and 49 920 people will die from this disease [1]. Cetuximab, an immunoglobulin G1 monoclonal antibody to the epidermal growth factor receptor (EGFR), has demonstrated clinical efficacy as monotherapy and when combined with chemotherapy in the treatment of advanced disease [2–7].
Recently, KRAS mutation status has been demonstrated to be a predictive marker of clinical benefit to monoclonal antibodies targeting EGFR [8–11]. Although KRAS mutations, particularly those involving codons 12 and 13 that are found in approximately 30%–40% of patients with metastatic colorectal cancer (mCRC), strongly relate to resistance to monoclonal antibodies targeting EGFR, not all mCRC patients with KRASwt derive benefit from these agents, and there is a need to identify molecular markers that better identify which KRASwt mCRC patients benefit from treatment. Previous studies had identified additional markers in KRASwt patients that can better predict for cetuximab responsiveness. For example, Jacobs et al. found that gene expression levels of two EGFR ligands, epiregulin and amphiregulin (AREG) related to favorable outcome in 220 chemorefractory KRASwt mCRC patients treated with cetuximab and irinotecan [12]. Several studies have demonstrated that high EGFR gene copy number could be a predictive marker in CRC patients treated with cetuximab [13, 14]. Furthermore, Laurent-Puig et al. showed that BRAF status, cytoplasmic expression of PTEN and EGFR amplification were associated with clinical outcome in KRASwt patients treated with a cetuximab-based regimen [15]. Finally, Sartore-Bianchi et al. pointed out that PI3KCA mutations in CRC were associated with clinical resistance to EGFR-targeted monoclonal antibodies including cetuximab and panitumumab [16].
In addition to tumor characteristics playing an important role in determining responsiveness to cetuximab, the genetic makeup of patients may also contribute to determining cetuximab sensitivity. Several studies have found that FcγRIIa–FcγRIIIa polymorphisms, as well as COX-2 and EGFR germline polymorphisms, are associated with clinical outcome in mCRC patients treated with single-agent cetuximab independent of KRAS status [17–19]. MicroRNAs are small, noncoding RNAs that regulate gene expression by degrading and/or suppressing the translation of target messenger RNA (mRNA) by base pairing in the 3′-untranslated region (UTR) of mRNA [20]. Very recently, microRNA polymorphisms were discovered and are becoming increasingly important in the fast growing field of personalized medicine. MicroRNA polymorphisms could present at or near a microRNA-binding site of functional genes. MicroRNA polymorphisms can affect gene expression by interfering with microRNA function. They have been shown to affect drug response and have the potential to confer drug resistance [21, 22]. The let-7 family of microRNAs were found to regulate KRAS activity by binding to the 3′-UTR of human KRAS gene [23]. Previous study demonstrated that a microRNA polymorphism in the let-7 microRNA complementary-binding site (lcs6) of the 3′UTR of KRAS gene was associated with increased KRAS expression in in vitro model [24]. Furthermore, this polymorphism was found to be associated with increased cancer risk in non-small-cell lung cancer (NSCLC) patients and reduced overall survival (OS) in oral cancers [24, 25], suggesting functional and clinical significance.
Due to the important role of KRAS mutation status in predicting cetuximab efficacy in CRC, we hypothesized that this KRAS let-7 lcs6 polymorphism may predict efficacy of cetuximab in KRASwt mCRC patients. We studied this polymorphism in 130 mCRC patients who were refractory to fluoropyrimidine, irinotecan, and oxaliplatin, and treated with cetuximab as monotherapy in a phase II study (IMCL-0144).
patients and methods
patient characteristics and statistical analysis
One hundred and thirty (38%) of the 346 patients enrolled in IMCL-0144 had tumor tissues available and amenable for analysis of KRAS let-7 lcs6 polymorphism (Table 1). IMCL-0144 involved patients with histopathologically confirmed mCRC, who were treated with cetuximab monotherapy following failure of therapeutic regimens that included fluoropyrimidine, irinotecan and oxaliplatin [26]. All 130 patients who had available tumor tissue samples were included in the present pharmacogenetic study, irrespective of clinical outcome and KRAS mutation status. This study was performed at the University of Southern California/Norris Comprehensive Cancer Center (USC/NCCC) following approval by the Institutional Review Board of the University of Southern California for Medical Sciences. All patients provided their written informed consent for tissue collection to allow study of molecular correlates.
Table 1.
Pretreatment characteristics among patients whose specimens available for genotyping in IMCL-0144
| Single-agent cetuximab (n = 130) | ||
| Frequency | % | |
| Median age, year (range) | 60 (29–85) | |
| Sex | ||
| Female | 66 | 51 |
| Male | 64 | 49 |
| Race | ||
| Caucasian | 121 | 93 |
| African-American | 3 | 2 |
| Asian | 3 | 2 |
| Other | 3 | 2 |
| ECOG performance status | ||
| 0 | 52 | 41 |
| 1 | 76 | 59 |
| 2 | 0 | 0 |
| KRAS mutation status* (*Codons 12 and 13) | ||
| Wild-type | 88 | 68 |
| Mutant | 42 | 32 |
ECOG, Eastern Cooperative Oncology Group.
The primary objective of this pharmacogenetic study was to evaluate relationships between KRAS let-7 lcs6 polymorphism and tumor response in KRASwt mCRC patients treated with single-agent cetuximab, whereas secondary objectives included evaluations of relationships of the polymorphism to progression-free survival (PFS) and OS. The PFS was calculated from the time of the first date of cetuximab treatment until the first observation of disease progression or death from any cause. If a patient had not progressed or died, PFS was censored at the time of the last follow-up. The OS time was calculated as the period from the first day of cetuximab infusion or until death from any cause, at which the point data were censored.
The association of KRAS let-7 lcs6 polymorphism with tumor response was determined by contingency table and the Fisher’s exact test. The association between this polymorphism with OS and PFS was analyzed using Kaplan–Meier plots and the log-rank test. The level of significance was set to a P value of <0.05, and P values are given for -two-sided testing. All statistical tests were performed using the SAS statistical package version 9.1 (SAS Institute Inc. Cary, NC), and Epilog Plus Version 1.0 (Epicentre Software, Pasadena, CAc).
clinical evaluation of response criteria
Objective tumor response was assessed every 6 weeks during the course of the study and criteria were based on modified World Health Organisation guidelines [26]. Response to cetuximab was determined by an independent response assessment committee that was blinded to the investigator-reported measurements and assessments were reported in the study. A partial response required at least a 50% reduction in the sum of the bidimensional products of all measurable lesions documented at least 4 weeks apart. Treatment was continued in the absence of intolerable toxicity or progressive disease, defined as at least a 25% increase in measurable disease, unequivocal growth of existing nonmeasurable disease, the appearance of one or more new lesions or reappearance of old lesions.
KRAS mutation status and KRAS let-7 lcs6 genotyping
Tissue specimens from primary tumors were collected and genomic DNA was extracted using the QIAamp kit (Qiagen, CA). KRAS mutation status was determined by direct sequencing as previously described [17]. Briefly, microdissected tumor DNA was amplified using following primer set: forward: 5′-TGA CTG AAT ATA AAC TTG TGG TAG TTG -3′, and reverse: 5′-TCG TCC ACA AAA TGA TTC TGA A-3′. PCR fragments were sequenced on an ABI 3100A Capillary Genetic Analyzer (Applied Biosystems), and analyzed in both sense and antisense directions for the presence of heterozygous mutations. Analysis of the DNA sequence was performed using ABI Sequencing Scanner v1.0 (Applied Biosystems). KRAS let-7 lcs6 polymorphism (rs61764370) was tested using PCR restriction fragment length polymorphism (PCR-RFLP) technique. Briefly, forward primer 5′-TTA GGA GAG ACG GGG TTT CA-3′ and reverse primer 5′-AAA TGA GTT CTG CAA AAC AGG-3′ were used for PCR amplification, PCR products were digested by restriction enzyme TfiI (New England Biolab, MA), and alleles were separated on 4% NuSieve ethidium bromide-stained agarose gel.
results
The 130 patients whose tissues samples were available for pharmacogenomic and molecular analyses in the present study had a similar median PFS (1.3 months), OS (6.3 months), and ORR (9.2%) values compared with patients whose tissues samples were not available for assessment in the present study (n = 216); the respective median values were PFS 1.5 months, OS 6.8 months, and ORR 13%, respectively [26].
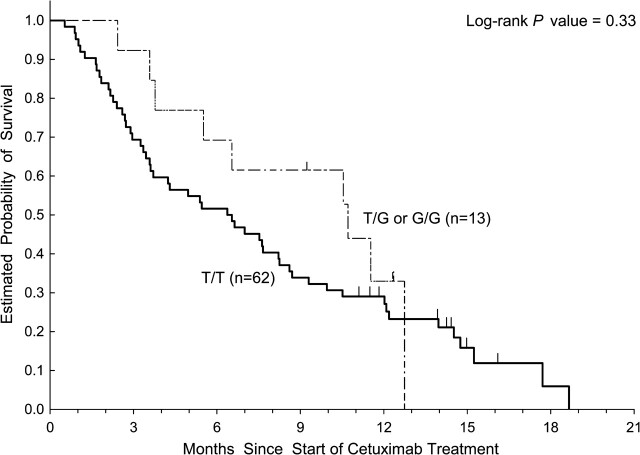
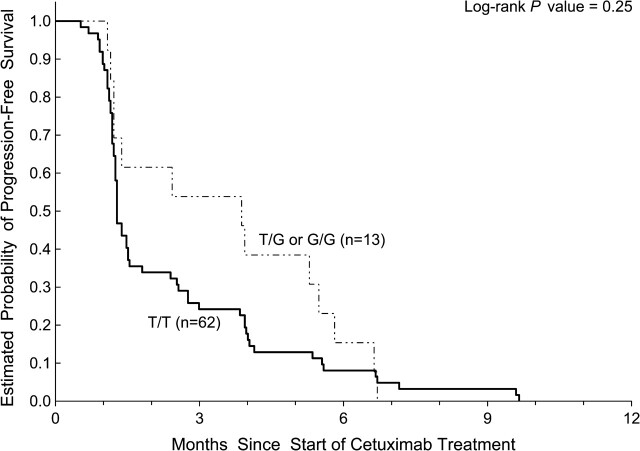
Analysis of KRAS let-7 lcs6 polymorphism was available in 111 patients due to exhaustion of available DNA from previous diagnostic testing. Thirteen of 111 patients (12%) were not assessable for tumor response. In 98 patients assessable for tumor response, 67 patients had wild-type KRAS and 31 patients had mutant KRAS. None of the 31 patients with a documented KRAS mutation responded to cetuximab, whereas 10 of the 67 KRASwt patients responded (0% versus 15%, respectively; P = 0.01, Fisher’s exact test). In the 111 patients assessable for PFS and OS, KRASwt patients had significantly longer PFS (P = 0.023) and OS (P = 0.02) values compared with KRAS-mutated patients. Fifty-five (82%) of the 67 KRASwt patients had the KRAS let-7 lcs6 TT genotype. There are 12 KRASwt patients harboring at least one let-7 lcs6 variant G allele (TG or GG). These 12 patients had a 42% (5/12) ORR compared with 55 KRASwt patients with wild-type TT genotype who had a 9% (5/55) ORR (P = 0.02, Fisher’s exact test; Figure 1). None of the 31 KRAS mutant patients had an objective response to cetuximab regardless of KRAS let-7 lcs6 polymorphism. Of the 28 KRAS mutant patients with KRAS let-7 lcs6 TT genotype, 10 had stable disease and 18 had progressive disease as their best response, whereas all 3 patients whose tumors had KRAS mutations and had a heterozygous TG genotype had progressive disease as their best response. There were no statistically significant associations between KRAS let-7 lcs6 polymorphism and OS, PFS either in wild-type or in mutant KRAS patients (Table 2). KRASwt patients who possessed a TT genotype (n = 62) had a median PFS of 1.3 months (95%CI: 1.2–1.5 months) compared with patients with at least one variant allele (TG or GG; n = 13) who had a median PFS of 3.9 months (95% CI: 1.2–5.5 months; P = 0.25, log-rank test). The median OS for KRASwt patients with a TT genotype was 6.4 months (95%CI: 3.6–8.2 months) compared with 10.7 months (95% CI: 5.5–12.7 months) for those harboring at least one variant allele (P = 0.33, log-rank test; Figures 2 and 3).
Figure 1.
Tumor response by KRAS let-7 lcs6 polymorphism in KRASwt patients enrolled in IMCL-0144.
Table 2.
PFS and OS by KRAS let-7 lcs6 polymorphism in mCRC patients treated with cetuximab monotherapy
| PFS | OS | |||||
| KRAS mutation status | KRAS wild type | KRAS mutant | KRAS wild type | KRAS mutant | ||
| Polymorphism | N | Median, m (95% CI) | N | Median, m (95% CI) | Median, m (95% CI) | Median, m (95% CI) |
| KRAS let-7 lcs6 | ||||||
| TT | 62 | 1.3 (1.2–1.5) | 32 | 1.3 (1.2–2.3) | 6.4 (3.6–8.2) | 5.9 (2.5–7.9) |
| TG+GG | 13 | 3.9 (1.5–5.5) | 4 | 1.2 (1.2–2.8) | 10.7 (5.5–12.7) | 2.8 (2.3–12.4) |
| P value | 0.25 | 0.71 | 0.33 | 0.84 | ||
PFS, progression-free survival; OS, overall survival; m, months; CI, confidence interval.
Figure 2.
Overall survival by KRAS let-7 lcs6 polymorphism in KRASwt patients treated with cetuximab monotherapy.
Figure 3.
Progression-free survival by KRAS let-7 lcs6 polymorphism in KRASwt patients treated with cetuximab monotherapy.
discussion
Our data have shown for the first time that a KRAS let-7 microRNA-binding site polymorphism (lcs6) in the 3′-UTR of the KRAS gene relates to tumor response in KRASwt mCRC patients treated with cetuximab monotherapy. Although patient numbers are small, our preliminary data demonstrated that KRASwt patients with KRAS let-7 lcs6 TT genotype had worse ORR compared with those with TG/GG genotype. The 12 KRASwt patients harboring at least a variant G allele (TG or GG) had a 42% ORR compared with 55 KRASwt patients who possessed wild-type TT genotype with only 9% ORR (P = 0.02, Fisher’s exact test). Patients with TG/GG allele had a longer PFS and OS but did not reach statistical significance due to the small sample size.
MicroRNA polymorphisms are emerging as significant molecular markers in the field of personalized medicine. They have been shown to affect drug response and are reported to be associated with many diseases including cancer. KRAS let-7 lcs6 microRNA polymorphism has been shown to be associated with oral cancer survival and lung cancer risk in different studies [24]. Christensen et al. examined the let-7 lcs6 polymorphism and the association with disease occurrence and OS in a population-based case–control study of patients with squamous cell carcinoma of the head and neck (HNSCC). Their results suggest that HNSCC patients who carry the KRAS let-7 lcs6 variant allele have a significantly reduced OS compared with patients with a let-7 lcs6 wild-type TT genotype [25]. Meanwhile, Chin et al. evaluated the same let-7 lcs6 polymorphism and its relationship with the risk of developing NSCLC. Their data have shown that the lcs6 variant allele in a KRAS microRNA complementary site is significantly associated with increased risk for NSCLC among those with a moderate smoking history [24].
Recent studies have shown that KRAS is regulated by the let-7 microRNA family. The 3′-UTR region of the human KRAS genes contains multiple let-7 complementary sites (lcs), allowing let-7 to regulate KRAS activity [23]. In CRC, transfection of cell lines with a let-7a-1 precursor microRNA resulted in growth suppression and a decrease in KRAS protein levels, suggesting that let-7 microRNA may play a role in suppressing colon cancer growth [27]. Moreover, it have been demonstrated that let-7 microRNA is not only involved in the growth of colon cancer cells, but can also modulate tumor sensitivity to chemotherapeutic agents [28]. Nakajima et al. have shown an association for members of the let-7 family of microRNAs with responsiveness to the oral fluoroyprimidine S-1. They found the level of expression of let-7g strongly related to responsiveness to S-1 treatment in 46 patients with recurrent or refractory advanced CRC [28].
These findings suggest that patients with KRASwt CRCs who also carry a variant KRAS let-7 lcs6 allele have a higher probability of responding to cetuximab monotherapy compared with those with wild-type TT genotype. However, these results were derived retrospectively and involve a relatively small number of patients, and therefore should be considered hypothesis generating and subject to confirmation in prospective and randomized controlled studies. Surprisingly, these findings that are opposite to our hypothesis that the variant let-7 lcs6 allele has been associated with lower let-7 levels and increased KRAS expression in NSCLC patients compared with the wild-type allele leads to greater activation of Ras/MAPK pathway, which is a known mechanism of resistance to anti-EGFR monoclonal antibodies [24]. One possible explanation for our finding is colon cancer patients whose tumors harbor the variant lcs6 allele may also have increased KRAS expression and activity, resulting in increased tumoral oncogenic addiction in the presence of EGFR signaling. As KRAS is a key mediator of the EGFR signal transduction primarily via the MAPK pathway, it seems plausible that patients with increased tumoral KRAS expression as a result of the lcs6 variant allele, may demonstrate increased sensitivity to the suppression of this increased oncogenic signaling by cetuximab treatment. As these tumors are KRASwt, cetuximab treatment will suppress EGFR tyrosine kinase phosphorylation reverting KRAS to its inactive GDP state, and the rapid loss of prosurvival signals and the simultaneous increase in proapoptotic signals may commit the cell to apoptotic death. This provides a plausible explanation as to why patients harboring the variant KRAS let-7 lcs6 polymorphism demonstrate a superior response to cetuximab. Similar observations were found in NSCLC patients treated with the EGFR tyrosine kinase inhibitor (TKI) gefitinib [29]. Weiss et al. showed microRNA-128b negatively regulates EGFR by binding to the EGFR 3′-UTR. In their lung cancer cell line and clinical specimen analyses, they found a loss of microRNA-128b related to increased EGFR expression and responsiveness to treatment with EGFR TKIs [29]. Furthermore, in vitro and in vivo studies are needed to explore the possible mechanism between this let-7 lcs6 polymorphism and cetuximab efficacy.
In summary, this study supports the role of let-7 lcs6 polymorphisms as a predictive marker of cetuximab efficacy in patients with KRASwt mCRC treated with cetuximab as monotherapy. Due to the retrospective nature of this study, these results should be interpreted carefully. Prospective, randomized controlled and biomarker embedded clinical trials are needed to confirm and validate our findings.
funding
This work was funded by the NIH grant 5 P30CA14089-27I and the Dhont Family Foundation
disclosure
H-JL has received honoraria from Merck KG and Bristol-Myers Squibb; EKR is employed by Imclone Systems, Inc.; DJM is employed by Merck Co., Inc; CL is employed by and has an ownership interest in Bristol-Myers Squibb.
Acknowledgments
The study was performed in the Sharon A. Carpenter Laboratory at USC/Norris Comprehensive Cancer Center and in memory of David Donaldson. The study was previously presented at American Society of Clinical Oncology 2009 (Abstract ID 4061).
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer Statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 3.Saltz LB, Meropol NJ, Loehrer PJ, Sr., et al. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 4.Bokemeyer C, Bondarenko I, Makhson A, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27:663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 5.Jonker DJ, O'Callaghan CJ, Karapetis CS, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 6.Sobrero AF, Maurel J, Fehrenbacher L, et al. EPIC: phase III trial of cetuximab plus irinotecan after fluoropyrimidine and oxaliplatin failure in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:2311–2319. doi: 10.1200/JCO.2007.13.1193. [DOI] [PubMed] [Google Scholar]
- 7.Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 8.Di Fiore F, Blanchard F, Charbonnier F, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by cetuximab plus chemotherapy. Br J Cancer. 2007;96:1166–1169. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lievre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 10.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 11.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs B, De Roock W, Piessevaux H, et al. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol. 2009;27:5068–5074. doi: 10.1200/JCO.2008.21.3744. [DOI] [PubMed] [Google Scholar]
- 13.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 14.Personeni N, Fieuws S, Piessevaux H, et al. Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: a fluorescent in situ hybridization study. Clin Cancer Res. 2008;14:5869–5876. doi: 10.1158/1078-0432.CCR-08-0449. [DOI] [PubMed] [Google Scholar]
- 15.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 16.Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 17.Lurje G, Nagashima F, Zhang W, et al. Polymorphisms in cyclooxygenase-2 and epidermal growth factor receptor are associated with progression-free survival independent of K-ras in metastatic colorectal cancer patients treated with single-agent cetuximab. Clin Cancer Res. 2008;14:7884–7895. doi: 10.1158/1078-0432.CCR-07-5165. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, Gordon M, Schultheis AM, et al. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J Clin Oncol. 2007;25:3712–3718. doi: 10.1200/JCO.2006.08.8021. [DOI] [PubMed] [Google Scholar]
- 19.Bibeau F, Lopez-Crapez E, Di Fiore F, et al. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol. 2009;27:1122–1129. doi: 10.1200/JCO.2008.18.0463. [DOI] [PubMed] [Google Scholar]
- 20.Reddy SD, Gajula RP, Pakala SB, Kumar R. MicroRNAs and cancer therapy: the next wave or here to stay? Cancer Biol Ther. 2010 Apr 4;9(7) doi: 10.4161/cbt.9.7.11402. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416. doi: 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24:489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen BC, Moyer BJ, Avissar M, et al. A let-7 microRNA-binding site polymorphism in the KRAS 3' UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30:1003–1007. doi: 10.1093/carcin/bgp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenz HJ, Van Cutsem E, Khambata-Ford S, et al. Multicenter phase II and translational study of cetuximab in metastatic colorectal carcinoma refractory to irinotecan, oxaliplatin, and fluoropyrimidines. J Clin Oncol. 2006;24:4914–4921. doi: 10.1200/JCO.2006.06.7595. [DOI] [PubMed] [Google Scholar]
- 27.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima G, Hayashi K, Xi Y, et al. Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in Colon Cancer. Cancer Genomics Proteomics. 2006;3:317–324. [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss GJ, Bemis LT, Nakajima E, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19:1053–1059. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]