Abstract
Phosphocholine molecules decorating bacterial cell wall teichoic acids and outer-membrane lipopolysaccharide have fundamental roles in adhesion to host cells, immune evasion, and persistence. Bacteria carrying the operon that performs phosphocholine decoration synthesize phosphocholine after uptake of the choline precursor by LicB, a conserved transporter among divergent species. Streptococcus pneumoniae is a prominent pathogen where phosphocholine decoration plays a fundamental role in virulence. Here, we present cryo–electron microscopy and crystal structures of S. pneumoniae LicB, revealing distinct conformational states and describing architectural and mechanistic elements essential to choline import. Together with in vitro and in vivo functional characterization, we found that LicB displays proton-coupled import activity and promiscuous selectivity involved in adaptation to choline deprivation conditions, and describe LicB inhibition by synthetic nanobodies (sybodies). Our results provide previously unknown insights into the molecular mechanism of a key transporter involved in bacterial pathogenesis and establish a basis for inhibition of the phosphocholine modification pathway across bacterial phyla.
Cryo-electron microscopy reveals the architecture and mechanism of a key choline transporter contributing to pathogen adaptation.
INTRODUCTION
Teichoic acids (TAs) are fundamental biopolymers that make part of the cell wall of Gram-positive bacteria (1, 2), whereas lipopolysaccharides (LPSs) are exclusively found at the outer membrane of Gram-negatives (3). Although their compositions differ, both TA and LPS function as endotoxins in bacterial pathogens, participate in immune evasion, prevent recognition and opsonization by antibodies, and play important roles in adhesion and colonization (4, 5). Decoration of TA and LPS with phosphocholine moieties is among the most impactful cell wall modifications, conferring virulence advantages to multiple pathogens (6–8). Gram-positive Streptococcus pneumoniae and Gram-negative Haemophilus influenzae, both residing in the human respiratory tract, are well-characterized pathogens that expose phosphocholine epitopes on their cell surface (9, 10). Interactions of phosphocholine with host proteins, such as the platelet activating factor receptor (11, 12), allow adhesion to the surface of host cells followed by cell invasion (13, 14). In addition, S. pneumoniae and commensal streptococci use phosphocholine decorated TA as a platform that anchors a great variety of choline binding proteins, which contribute to adherence, colonization, and virulence (15–17).
Bacteria generally import choline from the extracellular milieu and use it for osmoregulation. However, some bacteria import choline exclusively for modification of TA or LPS (7, 18, 19). The important function of this modification leads to choline auxotrophy in S. pneumoniae (20), although this pathogen can increase the available extracellular natural pool of choline by processing host phospholipids via surface-exposed or secreted phosphodiesterases (21, 22). Thus, choline import catalyzed by LicB is an essential trait of pathogens like S. pneumoniae (Fig. 1A) (20, 23, 24). LicB is a 32-kDa protein, member of the drug/metabolite transporter (DMT) superfamily, which consists of 14 transporter families, harboring more than 300 membrane proteins ubiquitously distributed in eukaryotes, bacteria, and archaea (25). Members of this superfamily display 4 to 10 transmembrane (TM) helices and are derived from primitive proteins of 4 TM helices similar to EmrE, a member of the small multidrug resistance family (26).
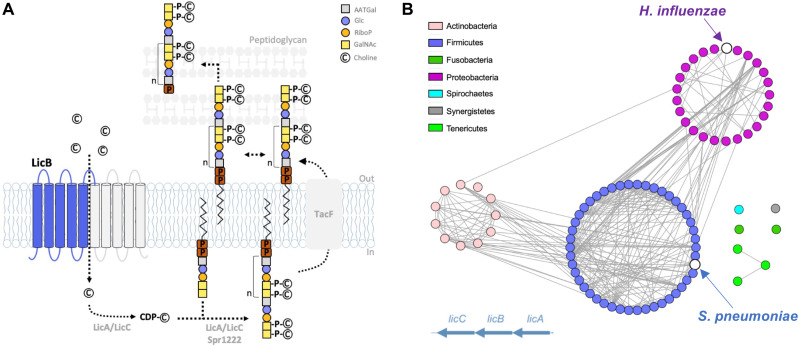
Fig. 1. Phosphocholine decoration of TA and conservation of LicB across divergent bacteria phyla.
(A) Pathway for phosphocholine decoration of TA in S. pneumoniae. Choline is imported by LicB and activated by LicA and LicC before being covalently attached as phosphocholine to TA, which are later exposed at the cell wall. Inverted repeats are shown in blue and gray. (B) Sequence similarity network of bacteria species (nodes) where lic operon genes encoding proteins involved in choline uptake (LicB) and activation (LicA and LicC) are conserved. Edges between nodes correspond to an identity of at least 40% among LicB proteins. Sequences are organized according to bacteria phylum (see fig. S1). S. pneumoniae and H. influenzae are indicated.
The licB gene is located in the lic operon, which encodes the protein machinery that converts choline into phosphocholine and that decorates TA and LPS (23, 27, 28). Inactivation of the licB gene leads to nonviable S. pneumoniae (20), whereas in H. influenzae it leads to choline uptake deficiency under human nasal airway conditions (23). Genes from the lic operon can be found in multiple other Gram-positive and Gram-negative bacterial species, where phosphocholine decoration of TA, LPS, or other structures occurs (24). A sequence similarity network showing licB genes sharing high sequence identity (>40%) among different bacterial phyla demonstrates the important role of this transporter in phosphocholine decoration across Gram-positive and Gram-negative bacteria (Fig. 1B and fig. S1). Despite its clear relevance and potential as a drug target, there are no structural or functional studies aiming to characterize the mechanism of LicB or that provide clues on how to inhibit its function.
To elucidate mechanistic elements essential to LicB function, we determined apo-outward-open and choline-bound-occluded structures of S. pneumoniae LicB solved by cryo–electron microscopy (cryo-EM) and x-ray crystallography and performed in vitro assays in proteoliposomes and in S. pneumoniae cells. We demonstrate that LicB displays promiscuous selectivity involved in adaptation to choline deprivation conditions, and describe architectural elements conserved among divergent bacterial phyla essential for choline binding and proton-coupled import activity. In addition, we describe inhibition of LicB by synthetic nanobodies (sybodies), thereby establishing the basis for selective inhibition of the phosphocholine decoration pathway.
RESULTS
S. pneumoniae LicB is a promiscuous choline transporter
We characterized the transport activity of LicB reconstituted in proteoliposomes using solid supported membrane (SSM) electrophysiology (Fig. 2, A and B) (29). Transport of choline is electrogenic, as evidenced by a positive current consistent with transport of the positively charged choline into proteoliposomes (Fig. 2A). The amplitude of the current measured before reaching electrochemical equilibrium, evidenced by the rapid current decay, is proportional to the initial rate of choline transport. Using this assay, we determined that LicB displays an EC50 (median effective concentration) [apparent Michaelis constant (Km)] for choline of 47 ± 15 μM (Fig. 2B). In contrast, protein-free liposomes exhibit no transport of choline (Fig. 2A). We tested whether LicB can transport alternative compounds with choline-like characteristics. Activation by arsenocholine, a natural compound with a similar composition to choline but containing arsenic instead of nitrogen, can be transported by LicB as evidenced by positive currents arising from transport into proteoliposomes (fig. S2A). LicB displays lower transport affinity for arsenocholine (EC50 = 170 ± 9 μM) (Fig. 2B), likely due to the larger radius of the positively charged trimethyl-arsonium group. These results recapitulate prior observations of arsenocholine transport by a choline-transporting variant of the betaine symporter BetP (30) and by the choline and betaine transporters OpuB and OpuC, respectively (31). In addition, acetylcholine, a positively charged molecule that in contrast to choline carries an ester acetyl instead of a hydroxyl group, is also transported by LicB into proteoliposomes (fig. S2B), albeit with lower affinity (EC50 = 740 ± 84 μM) (Fig. 2B). Together, these results show that LicB is a relatively high-affinity choline transporter that displays promiscuous activity toward other choline-like molecules.
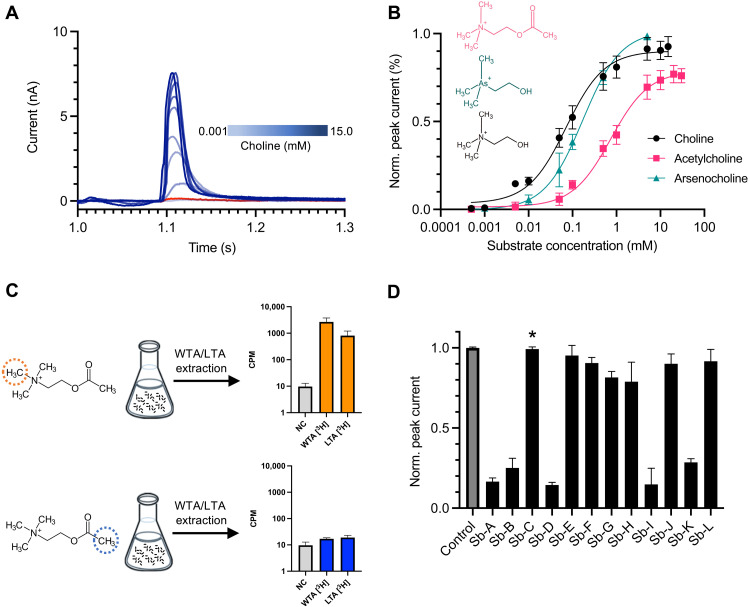
Fig. 2. LicB displays promiscuous activity toward choline-like molecules.
(A) SSM electrophysiology recordings of choline transport in LicB proteoliposomes. Currents recorded between 1 and 1.3 s after addition of choline are shown in blue. Recordings with protein-free liposomes in the presence of 5 mM choline is shown in red. (B) Determination of EC50 values for choline, arsenocholine, and acetylcholine based on SSM recording as shown in (A) and fig. S2. (C) WTA and LTA extraction from S. pneumoniae cells grown in the presence of [3H]-acetylcholine radioactively labeled at different positions indicated by dotted circles. Histograms show the [3H] radioactivity from extracted WTA and LTA. NC, negative control performed with nonradioactive acetylcholine. (D) Normalized amplitudes of SSM currents measured in the presence of sybodies A to L (500 nM), under 5 mM choline transport conditions based on data shown in fig. S3. The asterisk denotes sybody C, which was used for structural studies. n = 3 to 4 biological replicates; n = 2 to 3 technical replicates, for all experiments.
Acetylcholine can be processed by S. pneumoniae for decoration of TA
Acetylcholine is a neurotransmitter with function in the central and peripheral nervous system, where it is produced in cholinergic neurons and activates acetylcholine receptors before being processed by acetylcholinesterase. S. pneumoniae is one of the major meningitis-causing pathogens in part due to its ability to cross the blood-brain barrier (32). During invasion of the central nervous system, S. pneumoniae can be exposed to acetylcholine. Thus, in light of the promiscuous transport activity of LicB toward this molecule, we tested whether S. pneumoniae can make use of acetylcholine to supply the pathway that carries out decoration of TA. To do this, we grew S. pneumoniae D39V cells in choline-reduced medium containing acetylcholine isotopically labeled with [3H]-methyl at its trimethyl-ammonium group (Fig. 2C). After harvesting the cells, we performed extraction of TA and quantification of their radiolabeled content (Fig. 2C). Our results show prominent [3H] radioactivity in TA extracts, indicating that acetylcholine was used for functionalization of TA.
To demonstrate that acetylcholine has been hydrolyzed by S. pneumoniae, and that only its choline moiety was used for decoration of TA, we performed the same experiment but in the presence of acetylcholine labeled with [3H]-methyl at its acetyl group (Fig. 2C). In this case, we expected to see background levels of radioactivity as the choline moiety will remain unlabeled after acetylcholine hydrolysis. Our results show that extracted TAs display 100-fold lower levels of [3H] radioactivity in comparison to what was observed for extracted TAs, when acetylcholine labeled at its trimethyl-ammonium group was used (Fig. 2C). These results reveal that S. pneumoniae is able to uptake acetylcholine, hydrolyze it, and supply the phosphocholine synthesis pathway for functionalization of TA. Thus, the promiscuous activity of LicB represents an advantage that allows retrieving choline from alternative sources.
Sybodies inhibit LicB activity but do not affect S. pneumoniae growth in vitro
We reasoned that due to the small size of LicB (32 kDa), complex formation with a sybody (~16 kDa) would facilitate structure elucidation by single-particle cryo-EM. Thus, we raised sybodies against biotinylated LicB in the presence of 5 mM choline through a combination of ribosome and phage display (33, 34). Subsequent enzyme-linked immunosorbent assay (ELISA) screening resulted in the identification of multiple sybodies exhibiting good expression levels and behavior on size exclusion chromatography (SEC). To characterize the effect of sybodies on choline transport, we reconstituted LicB in proteoliposomes and performed SSM electrophysiology as described before (35). We measured electrogenic transport after activation with 5 mM choline and evaluated peak current amplitudes in the presence and absence of 12 sybodies (Fig. 2D and fig. S3). Five sybodies displayed strong inhibition of LicB transport activity, as evidenced by a decrease of about 80% of the peak currents, whereas seven sybodies did not affect choline electrogenic transport (Fig. 2D). Whereas the sequences of the complementarity determining regions (CDRs) of inhibitory sybodies shared high similarity, the CDR regions of noninhibitory sybodies were more diverse (fig. S4). We further selected a set of three inhibitory sybodies (sybodies A, B, and D) and one noninhibitory sybody (sybody C), based on their high yields of expression and good behavior during and after purification (fig. S5), and determined their binding kinetics using grating-coupled interferometry (GCI). This showed that all sybodies bind to LicB, with high-affinity displaying binding constants (Kd) in the range of 30 to 40 nM. Binding constants did not differ in the presence of 5 mM choline, indicating that the inhibitory sybodies can bind equally well to choline-bound and substrate-free LicB and are thus not conformationally selective (fig. S6).
Because of their small size, strong antigen affinity, low immunogenicity, and easy production, sybodies are strong candidates for development of therapeutics (36, 37). Thus, we wanted to test whether the inhibitory sybodies A, B, and D were able to affect growth of S. pneumoniae due to inhibition of LicB activity. To test this, we grew unencapsulated S. pneumoniae D39V cells (strain VL567) in liquid medium in the presence of the selected inhibitory sybodies and the noninhibitory sybody C as a control at concentrations ranging from 0.25 to 25 μM (fig. S7). Our results indicate that under these experimental conditions, the three inhibitory sybodies tested did not affect S. pneumoniae growth. We hypothesize that sybodies might be unable to penetrate the cell wall or that the remaining LicB choline transport activity, 14 to 28% based on SSM electrophysiology measurements (Fig. 2D), might suffice to supply the pathway for phosphocholine synthesis.
Structure of outward-open LicB in complex with sybody
We incorporated purified S. pneumoniae LicB in nanodiscs containing a mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) and Escherichia coli polar lipid extract (3:1, w/w) and incubated with the noninhibitory sybody C during reconstitution (fig. S8A). In contrast to the inhibitory sybodies, sybody C led to cryo-EM data of better quality. The LicB:sybody complex structure was solved to a nominal resolution of 3.75 Å (Fig. 3A; figs. S8, B to D, and S9A; and Table 1). Sybody C was found bound via its CDR3 region to cytoplasmic loop 4 located between TM4 and TM5 of LicB (Fig. 3, A and B). The structure revealed a LicB dimer in nanodiscs, with both protomers having the same topological orientation (Fig. 3, A to D). The dimer interface involves hydrophobic interactions between aromatic and aliphatic residues located in TM1, TM8, and TM9, and a putative partially resolved POPG lipid molecule, the most abundant phospholipid in the S. pneumoniae membrane (Fig. 3D and fig. S9, B and C) (38). The polar head group of the lipid faces the extracellular side of the membrane, and the aliphatic tails are surrounded mostly by aromatic residues (fig. S9, B and C).
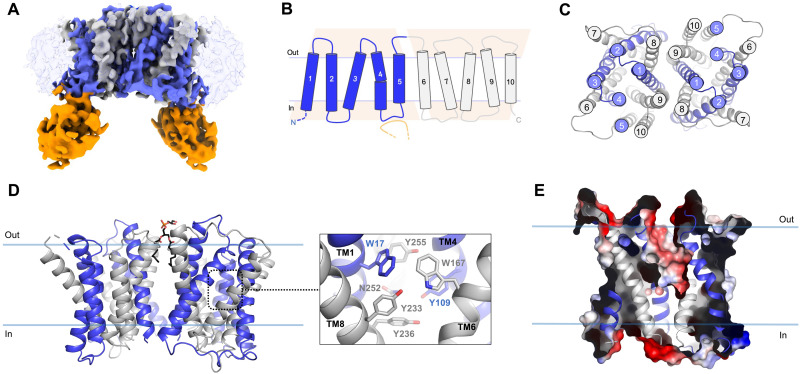
Fig. 3. Cryo-EM structure of outward-open S. pneumoniae LicB in lipid nanodiscs.
(A) Cryo-EM density of LicB homodimer in nanodiscs with bound sybody at 3.75 Å. Inverted repeats, TM1 to TM5 and TM6 to TM10, in each protomer are colored in blue and gray, respectively. Sybody is colored in orange. The nanodisc is shown as transparent light blue. (B) Topology of LicB highlighting the inverted antiparallel repeats shown as pink trapezoids. The CDR3 region of the sybody interacting with the loop connecting TM4 and TM5 is shown in orange. (C) Top view of LicB homodimer. (D) LicB homodimer and central cavity residues. (E) Surface electrostatic potential representation of LicB showing the central cavity opening to the extracellular side of the membrane.
Table 1. Cryo-EM data collection, refinement, and validation statistics.
|
LicB:sybody C in nanodiscs
(PDB ID 7PAF, EMD-13268) |
|
| Data collection and processing | |
| Microscope | Glacios TEM |
| Camera | Gatan K3 GIF |
| Magnification | 46,000 |
| Voltage (kV) | 200 |
| Electron exposure (e−/Å2) | 50 |
| Defocus range (μm) | −0.5 to −3 |
| Pixel size (Å) | 0.878 |
| Symmetry imposed | C2 |
| Initial particle images (no.) | 3,113,507 |
| Final particle images (no.) | 78,649 |
| Map resolution (Å) | 3.75 |
| Fourier shell correlation (FSC) threshold |
0.143 |
| Map resolution range (Å) | 3.0–5.0 |
| 3D-FSC sphericity | 0.989 |
| Refinement | |
| Model resolution (Å) | 3.9 |
| FSC threshold | 0.5 |
| Map sharpening B-factor (Å2) | −75.8 |
| EMRinger score (LicB:sybody) | 2.59 |
| EMRinger score (LicB) | 3.25 |
| Model composition | |
| Nonhydrogen atoms | 6,009 |
| Protein residues | 780 |
| B-factors (Å2) | |
| Protein | 28.24 |
| POPG | 1.65 |
| RMSDs | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.631 |
| Validation | |
| MolProbity score | 1.91 |
| Clashscore | 13.19 |
| Poor rotamers (%) | 0.00 |
| Ramachandran plot | |
| Favored (%) | 95.92 |
| Allowed (%) | 3.82 |
| Disallowed (%) | 0.26 |
| CCmask | 0.73 |
| CCbox | 0.66 |
| CCvolume | 0.68 |
| CCpeaks | 0.55 |
| CCligands | 0.75 |
LicB displays a 10-TM helix topology, with both N- and C-terminal domains facing the cytoplasm (Fig. 3, B and D). LicB exhibits the DMT superfamily fold consisting of two “inverted” structural repeats with antiparallel topology composed of the N-terminal (TM1 to TM5) and the C-terminal (TM6 to TM10) domains (Fig. 3B). TM1-TM4 and TM6-TM9 form a central cavity that opens up toward the extracellular side of the membrane, indicating that the structure is in an outward-open conformation (Fig. 3E). Access to the central cavity from the cytoplasmic face is sealed off by multiple interactions between TM6-TM9 and TM7-TM8 and the cytoplasmic loops connecting TM6-TM7 and TM8-TM9. Multiple aromatic residues including W17 (TM1), Y109 (TM4), W167 (TM6), Y233 (TM9), Y236 (TM9), and Y255 (TM10) participate in the formation of the central cavity (Fig. 3C). The structural fold exhibited by LicB is similar to that of other structurally characterized DMT transporters including the amino acid exporter YddG (39), the Plasmodium falciparum drug transporter PfCRT (40), the triosephosphate/phosphate antiporter TPT (41), and the SLC35 nucleotide sugar antiporters CST (42, 43) and Vrg4 (44, 45).
Structure of choline-bound LicB in occluded state
A choline-bound structure, displaying a different conformational state to that observed by cryo-EM, was obtained after cocrystallization of S. pneumoniae LicB in the presence of choline (Fig. 4A; fig. S10, A and B; and Table 2). An important experimental aspect to solve this structure was to perform dehydration of LicB crystals, which improved diffraction resolution from 6 to 3.8 Å. Phases were determined by molecular replacement using a LicB protomer from the cryo-EM structure as search model. The overall electron density map was of good quality throughout the polypeptide chain, except for loops 2 and 7, which were not included in the final model (Fig. 4A). The crystal structure revealed a conformation occluded from both sides of the membrane (Fig. 4B). Access to the central cavity is sealed by extracellular interactions between helices TM1-TM4 and TM2-TM3, and the loops connecting TM1-TM2 and TM3-TM4, and by cytoplasmic interactions between TM6-TM9 and TM7-TM8, and the cytoplasmic loops connecting TM6-TM7 and TM8-TM9. A clear positive peak in the Fo-Fc map indicated the presence of choline in the central cavity (Fig. 4A and fig. S10B). Residues coordinating the choline molecule include W17 (TM1), Y109 (TM4), W167 (TM6), Y233 (TM8), and Y255 (TM9), which form an “aromatic box” surrounding the trimethylammonium group, whereas Y236 (TM8) and N252 (TM9) coordinate the hydroxyl end of the molecule. Computational docking confirmed that the same binding pocket could accommodate an acetylcholine molecule (fig. S11A). The larger size of acetylcholine likely explains the lower affinity observed for this molecule (Fig. 2B). In agreement with the distribution of substrate binding residues described here for LicB, substrate-bound structures of other DMT transporters, including the triosephosphate/phosphate antiporter TPT (41) and the SLC35 nucleotide sugar antiporters CST (42, 43) and Vrg4 (44, 45), have shown that residues from TM1 to TM4, from the first inverted repeat, and TM6 to TM9, from the second repeat, are involved in substrate coordination. Thus, albeit the notable differences among substrates of DMT transporters, the arrangement of substrate binding residues seems to be conserved.
Fig. 4. Crystal structure of choline-bound occluded S. pneumoniae LicB.
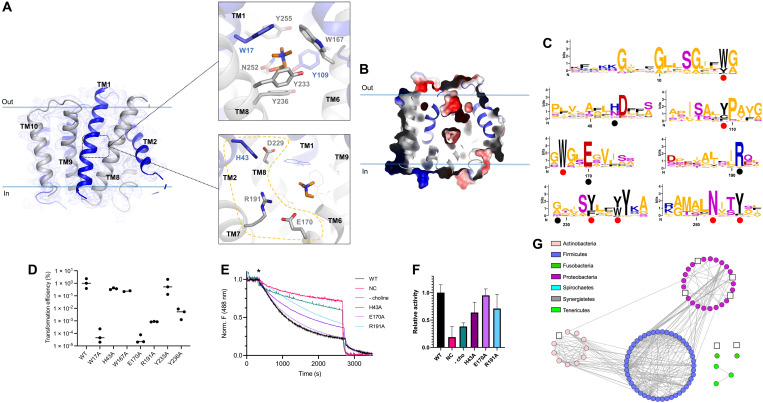
(A) LicB and 2Fo-Fc electron density map contoured at 1.0 σ. Inverted repeats, TM1 to TM5 and TM6 to TM10, are indicated in blue and gray, respectively. Top: Choline binding site. Choline is shown in orange. Bottom: Charged residues in the vicinity of the choline binding site. (B) Surface electrostatic potential representation of LicB showing the central cavity closed to both sides of the membrane. Choline is shown in orange. (C) Sequence logos of the regions containing the choline binding residues (red dots) and charged residues around the central cavity (black dots). Sequences are from LicB proteins analyzed in Fig. 1B (see fig. S1). The y axis denotes positional information in bits. (D) Transformation efficiency assays of S. pneumoniae D39V ∆licB cells complemented with WT Plac-licB or variants (n = 3). (E) Proton transport assay with WT LicB and variants in proteoliposomes. Representative time courses are shown (n = 3). H+ influx was induced by establishing a membrane potential by the addition of the potassium ionophore valinomycin (asterisk). NC indicates negative control (protein-free liposomes). (F) Relative activity of proton transport as in (E). Values after reaching equilibrium at approximately 2700 s were compared (n = 3). Colors correspond to the traces shown in (E). (G) Sequence similarity network as in Fig. 1B, showing bacteria species where residues E170 and R191 are not conserved (white squares, see fig. S1).
Table 2. X-ray data collection and refinement statistics.
Values in brackets are before anisotropic truncation. Values in parentheses are for the last resolution shell. Rmerge = ΣhklΣi |Ii(hkl) − <I(hkl)>|/ΣhklΣi Ii(hkl).
| Data collection | LicB (PDB ID 7B0K) |
| Wavelength (Å) | 1.000031 |
| Space group | C 1 2 1 |
| Unit cell | |
| a/b/c (Å) | 128.90/43.44/126.82 |
| α/β/γ (°) | 90.0/120.43/90.0 |
| Resolution (Å) | 13–3.8 |
| Completeness (%) | 86.8 (37.2) [98.9 (97.2)] |
| No. of measured reflections | 18,226 (690) [20,849 (2,305)] |
| No. of unique reflections | 5,593 (196) [7,167 (694)] |
| I/σI | 4.7 (2.3) [2.05 (1.2)] |
| Rmerge (%) | 14.6 (67.7) [23.1 (144.7)] |
| CC1/2 (%) | 99.6 (85.1) [99.5 (66.3)] |
| Refinement | |
| Rwork/Rfree (%) | 30.91/32.67 |
| RMSD bonds (Å) | 0.003 |
| RMSD angles (°) | 0.564 |
| Ramachandran plot | |
| Outliers (%) | 0.0 |
| Allowed (%) | 6.07 |
| Favored (%) | 93.93 |
| Average B-factor (Å2) | 107.0 |
Conserved choline binding residues are relevant for S. pneumoniae fitness
Sequence conservation analysis revealed that residues W17, Y109, W167, Y233, Y236, N252, and Y255, participating in coordination of choline at the central pocket, are highly conserved among LicB proteins across different bacterial phyla (Fig. 4C and fig. S11B), including the Gram-negative pathogen H. influenzae (fig. S11C). To further show the importance of residues participating in choline binding, we performed complementation assays in S. pneumoniae. First, we constructed strains in which residues participating in the formation of the aromatic box surrounding the trimethylammonium group of choline (Fig. 4A) were exchanged to alanine (W17A, W167A, Y233A, and Y236A) and the genes encoding these variants were cloned under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)–inducible promoter (Plac) and integrated at the ectopic ZIP locus as the second copy of licB (46). Transformation assays were done aiming to replace the native licB gene with an erythromycin resistance cassette, and transformants were plated with and without IPTG. As expected, wild-type (WT) Plac-licB could complement the licB deletion (Fig. 4D). We obtained an average transformation efficiency of approximately 1% in the presence of IPTG, while no colonies appeared in the absence of IPTG. Exchange to alanine of residues Y233 and W167 resulted in transformation efficiencies comparable to WT licB, whereas mutants W17 and Y236 resulted in significantly reduced transformation efficiencies indicating that these are essential residues for LicB function (Fig. 4D). Thus, slight modifications of the binding pocket of LicB are sufficient to perturb fitness of S. pneumoniae.
LicB is a proton-dependent choline symporter
The central cavity of LicB displays multiple charged residues in close proximity to the choline binding site (Fig. 4A), with residues H43, E170, and R191 being conserved among LicB proteins from different bacterial species (Fig. 4C and fig. S11B). The chloroquine resistant transporter PfCRT, which has been described to be a H+-coupled transporter, displays similar charged residues located at its central cavity (40, 47). Charged residues in central cavities of secondary transporters have been frequently associated with proton transport, as these residues are prone to protonation and deprotonation due to fluctuations in their surrounding chemical environment during cycling. We wanted to test whether, similarly to PfCRT, LicB could display H+-coupled activity. To test this, we performed membrane potential–driven H+ transport assays with LicB reconstituted in proteoliposomes in the presence of the fluorophore 9-amino-6-chloro-2-methoxyacridine (ACMA) (Fig. 4, E and F). The robust fluorescence decrease observed upon the addition of valinomycin reflects H+ influx into LicB-WT proteoliposomes, in contrast to the slower quenching of ACMA observed in protein-free liposomes (Fig. 4, E and F). Similar experiments with variants E170A, R191A, and H43A reconstituted in proteoliposomes (Fig. 4, E and F) revealed that H+ influx decreases for variants R191A and H43A, whereas E170A exhibited influx similar to that of WT LicB (Fig. 4, E and F). Complementation assays in S. pneumoniae using the abovementioned transformation assay showed that H43A supports viability, whereas mutants E170A and R191A did not (Fig. 4D). Notably, E170 and R191 are particularly highly conserved residues among bacteria phyla where genes of the lic operon are found (Fig. 4C), with only very few exceptions as indicated in Fig. 4G. Together, these results led us to hypothesize that H+-coupled choline import is a highly conserved trait of LicB proteins across bacteria displaying phosphocholine decoration.
Comparison of LicB outward-open and occluded conformations
Structures of other DMT transporters have been solved in either outward-open or occluded states, and so far, there are no structures available for inward-facing states (39–41). Modeling of different conformations has been possible because of the structural conservation of the DMT fold and the pseudo-symmetrical arrangement of TM helices in two inverted repeats (25). However, until now, LicB is the only DMT transporter with elucidated structures in two distinct structural states. Analysis of the conformational changes taking place during the transition from outward-open to occluded state shows that TM3 and TM6 are the main players in closing the access to the central cavity as they move toward the core of the translocation pathway (Fig. 5, A and B). During this transition, the extracellular halves of TM3 and TM6 tilt about 20°. An inward-facing model based on the pseudo-symmetrical arrangement of the two repeats predicts that the transition from occluded- to inward-open conformation would involve movements of the cytoplasmic halves of TM1 and TM8 (Fig. 5B). Large movements of the extracellular halves of TM3 and TM6 are favored by the flexibility introduced by glycine residues at their midsections. In TM3, this includes residues G83 to G86, G168 in TM6, G14 and G18 to G20 in TM1, and G216 to G220 in TM8. Similar flexible segments have been observed in YddG (39), TPT (41), Vrg4 (45), and CST (42, 43), and glycine residues at this same regions are present in LicB proteins, indicating that similar conformational changes are expected to occur in these proteins.
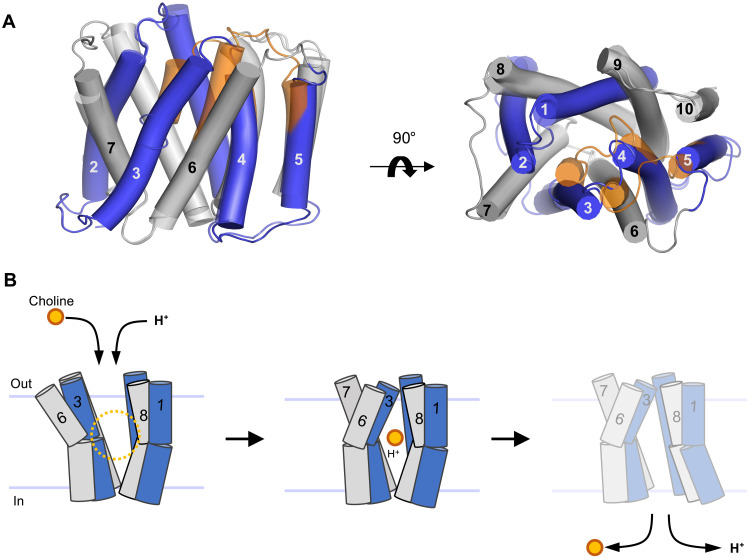
Fig. 5. Alternating access model of choline import.
(A) Superposition of outward-open and occluded structures of LicB. Inverted repeats, TM1 to TM5 and TM6 to TM10, are indicated in blue and gray, respectively. Helices that undergo large conformational changes are shown in orange in the occluded state. (B) Schematic of the proposed transport cycle depicting the movements of TM3 and TM6 that close the extracellular entry pathway, whereas the predicted movements of TM1 and TM8 open the cytoplasmic exit pathway. Choline (orange) and protons enter the binding cavity in the outward-open state. LicB changes conformation to the occluded state before opening up to the cytoplasm for choline and proton release.
DISCUSSION
Phosphocholine synthesis is an essential part of the pathways that decorate TA and LPS in pathogenic bacteria. Both biopolymers are important for adhesion to host cells, immune evasion, and bacterial persistence (4, 5). Pathogens like S. pneumoniae and H. influenzae are unable to synthesize choline; thus, uptake of this molecule is a fundamental step in exploiting the advantages that TA or LPS phosphocholine decoration provides. The essential role of LicB in supplying with choline the protein machinery encoded by the lic operon to perform TA and LPS decoration makes LicB and other proteins related to this process promising targets for drug development. In this context, inhibitory sybodies targeting LicB display great potential to counteract bacterial pathogens. However, further strategies aiming to facilitate sybodies diffusion across the bacterial capsule and cell wall need to be developed.
Multiple architectural and mechanistic elements described here report previously unknown findings on LicB function. The role of catalytic residues involved in choline binding and proton coupling in S. pneumoniae LicB and their high conservation across divergent bacteria phyla support the essential role of these mechanistic elements in supplying the phosphocholine synthesis pathway for decoration of TA and LPS. A homology model of H. influenzae LicB reveals a central cavity with an aromatic box and charged residues at the same positions as in S. pneumoniae LicB (fig. S11C).
The promiscuous selectivity of LicB is a striking finding that point toward an adaptation mechanism of S. pneumoniae under conditions with limited accessibility to free choline. We demonstrated that S. pneumoniae can catabolize acetylcholine and use the choline product for TA functionalization. However, because acetylcholinesterase rapidly processes free acetylcholine at synaptic clefts, it is unlikely for S. pneumoniae to find high concentrations of this molecule during invasion of the central nervous system. Furthermore, physiological concentrations of choline oscillate between 10 and 50 μM in the plasma and 5 and 7 μM in the cerebrospinal fluid (48), whereas only approximately 33 nM of acetylcholine is present in the cerebrospinal fluid (49). It is not known whether the concentration of these two molecules would change during meningitis progression. The nature of the protein(s) that performs acetylcholine hydrolysis in S. pneumoniae and how conserved is this mechanism among other bacteria remain to be shown. Docking analysis of acetylcholine indicates that the highly conserved catalytic residues involved in choline binding suffice to coordinate acetylcholine (fig. S11A). This, together with the high sequence conservation of the residues participating in the formation of the central binding pocket (Fig. 4C and fig. S11B) and neighbor residues located along the translocation pathway (fig. S11D), led us to hypothesize that LicB proteins in other bacteria would be able to import acetylcholine as well.
From the structurally characterized DMT transporters, PfCRT is the most similar to LicB in terms of function as it performs symport toward the cytoplasm (40, 47). PfCRT exports drugs from the acidic digestive vacuole of intra-erythrocytic P. falciparum parasites (47). Comparison of outward-open LicB and outward-open PfCRT [Protein Data Bank (PDB): 6UKJ], which superpose with a root mean square deviation (RMSD) of 2.6 Å (Cα atoms), reveals similarities in the location of charged residues in their central cavities: H43LicB/H97PfCRT, E170LicB/D137PfCRT, R191LicB/R231PfCRT, and D229LicB/D326PfCRT. Like LicB, PfCRT also makes use of the electrochemical proton gradient (50). Proton-coupled transport by LicB implicates that acidification of the external medium would result in faster choline uptake. This might be relevant at the natural niche of pathogens like S. pneumoniae and H. influenzae, the human nasopharynx, which displays mild acidic conditions (5.0 < pH < 6.5) (51, 52). In the absence of a proton gradient or neutral pH or in the presence of a mutation that disrupts proton transport (H43A), transport of choline is driven by the membrane potential and likely by a choline concentration gradient because the phosphocholine synthesis pathway quickly diminishes the concentration of free choline in the cytoplasm (27, 53, 54).
The cryo-EM structure of LicB in nanodiscs revealed a homodimer arrangement. Dimerization has been observed in crystal structures of DMT transporters including the triosephosphate/phosphate antiporter TPT (41), the guanosine diphosphate (GDP)–mannose transporter Vrg4 (44, 45), and the cytidine monophosphate (CMP)–sialic acid transporter CST (42, 43). Notably, the dimer arrangement displayed by LicB is very different to that observed in the crystal structures of these DMT transporters. While the LicB interprotomer interactions involve TM1, TM8, and TM9 (Fig. 3C), the interactions observed in the structures of TPT, Vrg4, and CST involve TM5 and TM10. The packing of the crystal structure of LicB shows a symmetry partner that interacts through TM5 and TM10, thus displaying the same dimer topology as that described for TPT, Vrg4, and CST (fig. S10C). Although for TPT it is not known whether the dimer is functionally relevant, it has been shown that dimerization of Vrg4 and CST allows faster transport and enhances protein stability (42, 44). Residues at the interface of the LicB dimer observed in the cryo-EM structure and between symmetry partners present in the crystal structure are not highly conserved (fig. S12, A and B). Although LicB is mainly monomeric after purification in detergent (fig. S13), dimerization in lipid nanodiscs, which represent a native-like environment, might point toward a functional dimer. Furthermore, the presence of interfacial lipids in oligomeric assemblies has been shown to play fundamental roles in stabilization and regulation of protomer association (55). Thus, the putative POPG lipid molecule at the interface between LicB protomers in nanodiscs might contribute to its stabilization (Fig. 3D and fig. S9, B and C), expected to also occur in the S. pneumoniae membrane where POPG is the most abundant phospholipid (38). However, the impact of dimerization and interfacial lipids in LicB function remains to be shown.
More broadly, we showed a comprehensive mechanistic analysis of a DMT transporter essential for bacterial cell wall modification. Because of the widespread importance of phosphocholine decoration of TA and LPS in bacterial pathogens, our results constitute a substantial advance that is likely to contribute to the development of new strategies to modulate interactions with the host. Sybody-like molecules represent promising chemical structures that will facilitate the engineering of new molecules targeting LicB proteins.
MATERIALS AND METHODS
LicB expression, purification, and nanodisc reconstitution
The gene encoding S. pneumoniae LicB with an N-terminal 10× His affinity tag in a modified pET-19b vector (Novagen) was overexpressed in E. coli BL21-Gold (DE3) (Stratagene) cells. The cells were grown at 37°C in Terrific broth medium supplemented with 1% (w/v) glucose and ampicillin (100 μg/ml). Protein overexpression was induced with 0.2 mM IPTG, and cells were incubated for 1 hour before harvesting. Frozen cell pellets were resuspended in 50 mM tris-HCl (pH 8.0), 500 mM NaCl, 5 mM β-mercaptoethanol, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). Cells were disrupted; the membrane fraction was separated by ultracentrifugation and subsequently flash-frozen with liquid nitrogen until further use. Frozen membranes were solubilized for 2 hours at 4°C in 50 mM tris-HCl (pH 8.0), 200 mM NaCl, 15% (v/v) glycerol, 20 mM imidazole (pH 8.0), 1% (w/v) lauryl maltose neopentyl glycol (LMNG) (Anatrace), and 2 mM β-mercaptoethanol. After centrifugation, the supernatant was loaded onto a Ni-NTA superflow affinity column (Qiagen); preequilibrated on 50 mM tris-HCl (pH 8.0), 500 mM NaCl, 10% glycerol, 20 mM imidazole (pH 8.0), 0.02% LMNG, and 2 mM β-mercaptoethanol; and subsequently washed with buffer containing 50 mM tris-HCl (pH 8.0), 500 mM NaCl, 10% (v/v) glycerol, 50 mM imidazole (pH 8.0), 0.02% LMNG, and 2 mM β-mercaptoethanol. The protein was eluted with buffer containing 50 mM tris-HCl (pH 8.0), 200 mM NaCl, 10% (v/v) glycerol, 500 mM imidazole (pH 8.0), 0.02% LMNG, and 2 mM β-mercaptoethanol. The buffer was exchanged to 10 mM tris-HCl (pH 8.0), 150 mM NaCl, and 0.012% LMNG with PD-10 columns (GE Healthcare) for the removal of imidazole. The 10× His affinity tag was removed by overnight incubation with tobacco etch virus protease, which was later removed by passing through a Ni-NTA affinity column. LicB was further purified by SEC with running buffer containing 10 mM tris-HCl (pH 8.0), 150 mM NaCl, and 0.012% LMNG using a Superdex 200 Increase 10/300 GL column (GE Healthcare). Purified LicB was reconstituted in MSP1D1 nanodiscs using a ratio of 3:9:7:175 (LicB:sybody:MSP1D1:lipids) in a buffer containing 50 mM tris-HCl (pH 8.0), 50 mM NaCl, and 10% (v/v) glycerol. The lipid mixture used consists of 16:0 to 18:1 POPG:E. coli polar lipid extract (Avanti) in a 3:1 (w/w) ratio. Detergent was removed by adding Bio-Beads SM2 (Bio-Rad). After Bio-Beads removal, the mixture was centrifuged and loaded on a Superdex 200 Increase 10/300 GL column (GE Healthcare) equilibrated with buffer containing 50 mM tris-HCl (pH 8.0) and 50 mM NaCl. The peak corresponding to LicB:sybody in nanodiscs was collected and used for single-particle cryo-EM studies.
Sybody selection
LicB carrying an N-terminal Avi-tag, followed by a SSGTSS linker sequence to warrant efficient biotinylation, was expressed and purified as described above. Enzymatic biotinylation was performed using recombinant BirA in the presence of 0.5 mM biotin, 20 mM magnesium acetate, 20 mM adenosine 5′-triphosphate, and 5% glycerol (56). The reaction was incubated for 16 hours at 4°C. Protein biotinylation was confirmed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) after incubation of the reaction product with streptavidin. Biotinylated LicB was desalted with buffer containing 10 mM tris-HCl (pH 8.0), 150 mM NaCl, and 0.012% LMNG, and His-tagged BirA was removed using a Ni-NTA superflow affinity column. The protein was aliquoted at a concentration of 5 μM, flash-frozen in liquid nitrogen, and stored at −80°C until used for nanobody selection. Sybody selection was performed as previously described (33). In short, sybodies were selected against biotinylated LicB in the presence of 5 mM choline using the three sybody libraries concave, loop, and convex. After one round of ribosome display, two rounds of phage display were performed, switching the panning surface in each round. During the second round of phage display, an off-rate selection was carried out using nonbiotinylated LicB at a concentration of 5 μM. The enrichment was monitored throughout the selection process by quantitative polymerase chain reaction (qPCR), which looked ideal with values of 4.6 (concave), 2.5 (loop), and 3.3 (convex) after the first round of phage display and 1922 (concave), 107 (loop), and 70 (convex) after the second round of phage display. Ninety-five single clones of each library were screened by ELISA, resulting in 40 hits. Sanger sequencing revealed 29 unique sybodies, of which 26 showed a monodisperse peak at the expected elution volume on a Sepax SRT-10C SEC-100 column. From these, the best behaved sybodies during expression and purification (12 sybodies, all belonging to the convex sybody library; fig. S4) were selected for further functional characterization.
Sybody expression and purification
Sybodies were expressed and purified as previously described (33, 34), with minor modifications. Briefly, the sybody-encoding gene in the pSBinit vector (Addgene, #110100) was transformed into E. coli MC1061-competent cells, which were then grown at 37°C in Terrific broth medium supplemented with 0.004% glycerol, ampicillin (100 μg/ml), and streptomycin (100 μg/ml). Overexpression of the protein was induced with 0.02% l-arabinose at an OD600 (optical density at 600 nm) of 0.7, followed by 15-hour incubation at 22°C before harvesting. The cell pellet was resuspended in 40 mM tris-HCl (pH 7.4), 500 mM NaCl, and 0.5 mM PMSF, and cells were lysed using a tip sonicator. The cell debris was discarded by centrifugation, and the supernatant was loaded on a Ni-NTA affinity chromatography column equilibrated with 40 mM tris-HCl (pH 7.4), 150 mM NaCl, and 50 mM imidazole. The column was washed with 40 mM tris-HCl (pH 7.4), 150 mM NaCl, and 50 mM imidazole, and the protein was eluted with 40 mM tris-HCl (pH 7.4), 150 mM NaCl, and 300 mM imidazole. Imidazole was then removed using a PD-10 column (GE Healthcare). The concentrations of sybody preparations were determined by measuring A280, and the quality of the purified sybodies was assessed by SDS-PAGE and SEC.
Sybody binding constant determination
Kinetic characterization of sybodies binding to biotinylated LicB was performed using GCI on a WAVEsystem instrument (Creoptix AG). Biotinylated LicB was captured onto a Streptavidin PCP-STA WAVEchip (polycarboxylate quasi-planar surface, Creoptix AG) to a density of 1700 pg/mm2. Sybodies were injected at increasing concentrations ranging from 5 to 405 nM using a threefold serial dilution and five different concentrations in buffer containing 10 mM tris-HCl (pH 8.0), 150 mM NaCl, and 20 mM imidazole supplemented with 0.02% LMNG with or without 5 mM choline. Sybodies were injected for 200 s at a flow rate of 50 μl/min. Dissociation recording was set to 900 s to allow the return to baseline. Blanks were injected after every second analyte injection. All sensorgrams were recorded at 25°C, and the data were analyzed on the WAVEcontrol software (Creoptix AG). Data were double-referenced by subtracting the signals of buffer injections (blanks) and by subtracting the signals of a reference channel. A Langmuir 1:1 model was used for data fitting. For sybody C, the two highest concentrations were excluded from data fitting, due to unspecific interaction with the flow channels at high concentrations.
Sample preparation and cryo-EM data acquisition
A sample of nanodisc reconstituted LicB:sybody C complex was concentrated between 1 and 1.5 mg/ml using a Vivaspin concentrator with a 30,000-Da cutoff (GE Healthcare). Cryogenic samples were prepared using Mark IV Vitrobot (Thermo Fisher Scientific) at 95% humidity at 4°C. Five microliters of the sample was applied to glow-discharged Quantifoil R1.2/1.3 300-mesh copper holey carbon grids, blotted for 4 s using a blotting force of 3. The grids were flash-frozen in a mixture of propane and ethane and then cooled with liquid nitrogen. Movies were recorded with SerialEM on a Glacios microscope (Thermo Fisher Scientific) operated at 200 kV, equipped with a K3 direct electron detector (Gatan). Images were recorded with a defocus range of 0.5 and 3 μm and a pixel size of 0.878 Å/pixel at a nominal magnification of 46,000×. Each micrograph was dose-fractionated to 25 frames under a dose rate of 12.5 electrons per pixel per second, with a total exposure time of 4 s, resulting in a total dose of about 50 e−/Å2.
Cryo-EM data processing
Data processing was carried out entirely in cryoSPARC v3.2.0 (57). Beam-induced drift of 8229 raw movies was corrected, and the movies were aligned using patch motion correction. The contrast transfer function (CTF) of each aligned micrograph was determined using patch CTF estimation. Micrographs were Fourier-cropped once during patch motion correction to adjust the pixel size to 0.878 Å per pixel (0.439 Å per pixel in super-resolution mode). Micrographs were classified and some discarded, based on CTF fit resolution, relative ice thickness, and total full-frame motion, resulting in 8008 micrographs. A small subset of micrographs was used to adjust parameters for optimal picking of particles, which was performed using blob picker. A total of 3,113,507 particles were extracted with a box size of 384 pixel and Fourier-cropped to 192 pixels, resulting in a pixel size of 1.756 Å per pixel. Multiple rounds of two-dimensional (2D) classification with a batch size of 200 particles per class were performed to discard bad particles. The selected particles were used to perform sequential ab initio reconstructions with three or four classes, no similarity, and C1 symmetry. The best-resolved class was used as reference for multiple rounds of heterogeneous refinement with the particle sets selected from the preceding round of 3D classification. After heterogeneous refinement, 148,064 good particles were re-extracted, resulting in a pixel size of 0.878 Å per pixel, and further refined in a nonuniform (NU) refinement using C2 symmetry, input reconstruction map filtered to 12 Å, and per-group CTF parameters. NU refinement resulted in a map with a 4.2-Å resolution, according to the 0.143 cutoff criterion. This reconstruction map was used to generate templates for automated template-based picking from all good micrographs. Particles were re-extracted, and a similar processing procedure as that described above was applied. After NU refinement, a set of 78,649 particles were locally CTF refined before local resolution refinement, which resulted in a map with a 3.75-Å resolution, according to the 0.143 cutoff criterion. The local resolution was estimated with cryoSPARC v3.2.0 (57), and the directional resolution estimation was done using the 3DFSC server, which calculated a sphericity of 0.989 (58).
Model building
Model building was performed in Coot (59). The cryo-EM density of the LicB dimer was of sufficiently high resolution to unambiguously build LicB polypeptide chain into the cryo-EM density. The sybody was only partially resolved, with the part interacting with LicB being its best-resolved region. Thus, a homology model of the sybody (based on PDB: 5m14) was positioned by rigid body in the density map, accompanied by manual building of the CDR3 region. Models were improved by iterative cycles of real-space refinement in Phenix (60) with application of secondary structure constraints. Validations of models were performed in Phenix. The final refined structure has 95.43% of residues in the Ramachandran favored region, 4.32% in Ramachandran allowed, and 0.25% as Ramachandran outliers. Figures of models and densities were made in PyMOL (The PyMOL, Molecular Graphics Systems, Schödinger LLC) and ChimeraX (61). The average Q-score value of the final model was 0.498, whereas the expected value at 3.75-Å resolution was 0.445 (62). The EMRinger score for the final model is 2.59, with the LicB chains displaying an EMRinger score of 3.25 (63).
LicB crystallization
LicB in buffer containing 10 mM tris-HCl (pH 8.0), 150 mM NaCl, and 0.012% LMNG was concentrated up to 8 mg/ml using a 30-kDa molecular weight cutoff Vivaspin 20 concentrator (GE Healthcare). Extensive optimization of crystal conditions using sitting-drop and hanging-drop vapor diffusion and other postcrystallization treatments yielded plate-shaped crystals after 3 to 4 days of incubation at 20°C. The reservoir conditions contained 1.5 mM choline, 100 mM Hepes (pH 7.5), and 33% polyethylene glycol 400 (PEG 400). Optimized crystals were dehydrated and cryoprotected by gently increasing PEG 400 concentration in the drop to a final concentration at 30%. The best diffracting crystals were obtained by air dehydration for 20 min in the drop. Crystals were scooped from the drop at different time points followed by flash-freezing by immersion in liquid nitrogen.
Data collection
LicB crystals diffracted x-rays up to about 6- to 7-Å resolution in general. Drop dehydration through air exposure over 30 min led to crystals that diffracted x-rays to higher resolution. A dataset collected from one LicB crystal that showed anisotropic diffraction and belonged to space group C121 was used to determine the structure. The unit cell constants of this crystal were a = 128.9 Å, b = 43.44 Å, c = 126.82 Å and α = 90°, β = 120.43°, γ = 90° (Table 2). Data were processed and merged with XDS (64), and anisotropic scaling/ellipsoid truncation was performed (65). Resolution limits after ellipsoid truncation were a* = 4.0 Å, b* = 3.5 Å, and c* = 4.2 Å. Karplus CC* (Pearson correlation coefficient)–based data cutoff approach was used to determine the usable resolution of the datasets (66). The resolution limit was set considering a CC1/2 > ∼40% based on data merging statistics and a CC* analysis against unmerged intensities in Phenix package (60) satisfying Karplus CC* against CCwork and CCfree criteria, as well as Rfree of the highest-resolution shell against the refined structure being less than or equal to ∼50%. A second criterion for limiting the resolution was the overall completeness percentage observed after anisotropic ellipsoid truncation, which was kept above 85%. Diffraction data were collected at the beamline X06SA at the Swiss Light Source (SLS) (Villigen).
LicB crystal structure determination
The crystal structure of LicB was solved by molecular replacement using the program PHASER (67). The model generated by single-particle cryo-EM was used as reference. Multiple rounds of refinement in Phenix (60) and model building using Coot (59) were performed. Map sharpening was used to facilitate model building. X-ray data and refinement statistics are given in Table 2. The final refined structure has Rwork = 29.69% and Rfree = 31.97%, with 92.16% of residues in the Ramachandran favored region, 7.84% in Ramachandran allowed, and 0.0% as Ramachandran outliers. Molecular graphics were created in PyMOL (The PyMOL, Molecular Graphics Systems, Schödinger LLC). Surface electrostatics were calculated with the APBS PyMOL plugin.
Reconstitution in proteoliposomes
Lipid mixtures of 1-palmitoyl-2-oleoyl-phosphatidylethanolamine (POPE):POPG 3:1 or E. coli polar lipid extract (ECPL):Phosphatidylcholine (PC) 3:1 (w/w) (Avanti) were mixed, dried from chloroform, and solubilized in buffer containing 20 mM tris (pH 8.0), 150 mM NaCl, and 2 mM β-mercaptoethanol to a final concentration of 20 mg/ml. The lipids were flash-frozen with liquid nitrogen and stored at −80°C. Unilamellar liposomes were formed after a 1:1 dilution in buffer and extrusion through a polycarbonate filter (400-nm pore size). After addition of 0.2% n-Dodecyl-b-D-Maltopyranoside (DDM), liposomes were mixed with LicB to have a 1:50 (w/w) TM protein:lipid ratio. Detergent was removed with Bio-Beads SM2 (Bio-Rad). Proteoliposomes at a final concentration of 20 mg/ml of lipids were resuspended in 10 mM Hepes (pH 7.3), 100 mM KCl, and 5 mM choline for the proton transport assay and in 20 mM tris (pH 8.0), 150 mM KCl, and 2 mM β-mercaptoethanol for the SSM-based electrophysiology assays. The proteoliposomes were aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C until further use.
SSM-based electrophysiology
SSM-based electrophysiology was conducted using a SURFE2R N1 instrument (Nanion Technologies) following published protocols (35). The 3-mm SURFE2R N1 single sensors (Nanion Technologies) were alkylated by adding 100 μl of 0.5 mM thiol solution (1-octadecanethiol in isopropanol), subsequently incubating for 1 hour at room temperature in a closed petri dish. The sensor was then washed with isopropanol and Milli-Q water. A 1.5-μl lipid solution (7.5 μg/μl 1,2-diphytanoyl-sn-glycero-3-phosphocholine in n-decane) was applied on the surface of the gold electrode followed by immediate addition of 100 μl of SSM buffer containing 50 mM tris-HCl (pH 8.0), 5 mM MgCl2, and 150 mM KCl. The proteoliposome suspension at a lipid concentration (20 mg/ml) was diluted 1:20 in SSM buffer, and the mixture was sonicated for 20 s. This treatment yields small liposomes (<100 nm). Ten microliters of the diluted proteoliposomes was added to the chip before centrifugation at 2000g for 30 min. The conductance and capacitance were measured to ensure the quality of the SSM before each measurement. A conductance value below 5 nS and capacitance between 15 and 35 nF were considered acceptable. Activating buffers (buffer A) were prepared from a large stock of nonactivating buffer (buffer B: SSM buffer) by the addition of the substrate choline or other compounds such as arsenocholine and acetylcholine at different concentrations ranging between 1 μM and 30 mM. Currents corresponding to different substrate concentrations were measured three times and on three different chips. Peak current values were determined and normalized according to the maximum peak current at 5 mM choline for each individual chip. The normalized peak currents were plotted to calculate EC50 values. Assays with sybodies were performed at a constant concentration of choline at 5 mM in buffer A and 500 nM sybody in buffers A and B, as previously described (35). Before the measurements in the presence of sybody, the maximal peak current was determined with 5 mM choline in buffer A and used to normalize the peak currents in the presence of sybody. Data for each sybody were collected from three independent sensor preparations and measured in triplicates. Analysis of the data was performed in OriginPro (OriginLab Corporation) and GraphPad Prism 9 (GraphPad Software).
Proton transport assay
POPE:POPG (3:1) proteoliposomes were thawed, and 20 μl of the proteoliposomes was sonicated and diluted to 500 μl with buffer containing 10 mM Hepes (pH 7.3), 10 mM KCl, 90 mM NaCl, 5 mM choline, and 0.75 μM ACMA before the assay. Time-course fluorescence was measured at 20°C using a Jasco fluorimeter. The change in ACMA fluorescence was detected using an excitation wavelength of 410 nm and an emission wavelength of 480 nm. After equilibration of the system, H+ and choline influx was initiated by establishing a membrane potential by the addition of the K+-selective ionophore valinomycin (5 nM). The activity was assessed for 1200 s. The proton gradient was collapsed by the addition of 0.5 μM carbonyl cyanide m-chlorophenyl hydrazone. Time courses were repeated at least three times for each individual experiment.
SEC coupled to multiangle light scattering
SEC–multiangle light scattering (MALS) measurements of LicB were performed at 18°C in 10 mM tris-HCl (pH 8.0), 150 NaCl, and 0.012% LMNG using a GE Healthcare Superdex 200 Increase 10/300 GL column on an Agilent 1260 high-performance liquid chromatography. The column was equilibrated overnight for a stable baseline before data collection. Monitoring of the elution was carried out with a multiwavelength absorbance detector at 280 and 254 nm, a Wyatt Heleos II 8+ MALS detector, and a Wyatt Optilab rEX differential refractive index detector. Interdetector delay volumes, light scattering detector normalization, and broadening corrections were calibrated using a bovine serum albumin solution (2 mg/ml; ThermoPierce) and standard protocols in ASTRA 6 (Wyatt Technologies). Weight-averaged molar mass, elution concentration, and mass distribution of the samples were calculated using the ASTRA 6 software (Wyatt Technologies). The specific refractive index increment for the detergent LMNG was assumed to be 0.146 mg/ml according to experimental data.
Mutagenesis
Point mutations H43A and R191A were introduced by site-directed mutagenesis using primers (table S1). Point mutations Y233A, Y236A, N252A, W17A, W167A, and E170A were introduced by cloning using gBlock gene fragments (Integrated DNA Technologies) (table S1).
Sequence similarity network
A sequence similarity network of LicB proteins from multiple bacteria species (nodes) was generated using the EFI-EST webserver (68). Bacteria species were selected considering that lic operon genes encoding LicB and proteins that perform choline activation (LicA and LicC) are present (69). Sequences were visualized with 40% identity and organized by phylum in Cytoscape (70). The phylogenetic tree showing the evolutionary relationships among the selected bacteria species was generated with iTOL (71).
Structural sequence conservation analysis
The sequence conservation analysis shown in figs. S11 (B and D) and S12 was computed using the ConSurf server (72). Briefly, LicB homologs displaying at least 35% identity were selected from a protein sequence BLAST search on the National Center for Biotechnology Information (NCBI) database using S. pneumoniae LicB protein sequence as a query. We then generated a multiple-sequence alignment using the HHMER algorithm provided by ConSurf, with conservation scores plotted in PyMOL (The PyMOL, Molecular Graphics Systems, Schödinger LLC).
Docking of acetylcholine
Docking of acetylcholine to the LicB structures was done with Autodock Vina (73). The molecule was downloaded in the SDF format from the ZINC database (74) and converted into a PDBQT format using Open Babel (75). Docking was carried out over a search space of 28 Å × 56 Å × 30 Å in the outward-open state and 38 Å × 34 Å × 42 Å covering the entire entry pathway to the central cavity.
Construction of mutants in S. pneumoniae
Strain VL4243 [S. pneumoniae D39V, prsI::PF6-lacI-tetR (gen), bgaA::Plac-licB (tet)] was made as follows. A second copy of licB under control of the IPTG-inducible Plac promoter (76) was integrated in the genome of parental strain D39V at the bgaA locus. Golden Gate assembly of three parts was used. The first part is the amplification by PCR using chromosomal DNA of strain VL1998 (laboratory collection) as template with primers OVL5139 and OVL5623 (table S1). The resulting product contained the upstream homologous region to integrate at the bgaA locus, the tetracycline marker (tet), Plac, and the Ribosome binding site. The second part is the amplification by PCR of licB, without its own RBS, from D39V with primers OVL5624 and OVL5625. The third part is the amplification by PCR using chromosomal DNA of strain VL1998 as template with primers OVL5626 and OVL2082, resulting in the downstream homologous region to integrate at the bgaA locus. After purification, the three parts were digested and ligated together during an assembly reaction with T4 DNA ligase buffer (Vazyme), T4 DNA ligase (Vazyme), and Esp3I [New England Biolabs (NEB)]. The assembly mixture was incubated in a thermocycler (PCR Max) by cycling 25 × the series 1.5 min at 37°C and 3 min at 16°C. The sample was then incubated for 5 min at 37°C and subsequently incubated for 10 min at 80°C. Strain VL333 [prsI::PF6-lacI-tetR (gen) (77)] was transformed with the assembly mixture, and transformants were selected using tetracycline. The bgaA locus of the resulting strain, VL4243, was confirmed by sequencing. To create seven different point mutations in licB, the desired amino acids were changed for alanine in the second copy of licB at bgaA. Golden Gate assembly was used to amplify two parts per point mutation. The first part is the amplification by PCR of the upstream region of bgaA::Plac-licB from VL4243 until the desired point mutation with forward primer OVL2077 and reverse primer for each point mutation, OVL6035 (Y233A), OVL6037 (Y236A), OVL6039 (W17A), OVL6041 (W167A), OVL6043 (E170A), OVL6045 (R191A), or OVL6047 (H43A). The second part is the amplification by PCR of bgaA::Plac-licB from VL4243, started at the point mutation location until the downstream homologous region to integrate at the bgaA locus. Different forward primers were used for each point mutation: OVL6036 (Y233A), OVL6038 (Y236A), OVL6040 (W17A), OVL6042 (W167A), OVL6044 (E170A), OVL6046 (R191A), and OVL6048 (H43A); the reverse primer was identical, OVL2082. After purification, the two parts were digested with Esp3I and ligated together during an assembly reaction with T4 DNA ligase buffer (Vazyme), T4 DNA ligase (Vazyme), and Esp3I (NEB). Strain VL333 was transformed with the assembly mixture with tetracycline selection. The bgaA region containing the Plac-licB genes of the resulting strains, VL4250 (Y233A), VL4251 (Y236A), VL4252 (W17A), VL4253 (W167A), VL4254 (E170A), VL4255 (R191A), VL4256, and (H43A), was confirmed by sequencing.
Transformation efficiency assay
Transformation efficiencies were determined to test whether mutated licB alleles would support growth as the only copy of licB in the pneumococcal genome. Transformation assays were performed by transforming DNA of a licB::ery cassette to the strains with both copies of licB (without and with point mutation). DNA of licB::ery was obtained by amplification by PCR of strain VL4249 (laboratory collection) with primers OVL5635 and OVL5636. To transform S. pneumoniae, cells were grown in C+Y medium (pH 6.8) at 37°C to OD595 of 0.1. Subsequently, cells were treated for 12 min at 37°C with synthetic CSP-1 (100 ng/ml) and incubated for 20 min at 30°C with the transforming DNA (licB::ery). After incubation with the transforming DNA, cells were grown in C+Y medium (pH 6.8) at 37°C for 60 min. S. pneumoniae transformants were selected by plating, in triplicate, inside Columbia agar supplemented with 4% of defibrinated sheep blood (Thermo ScientificTM OxoidTM) with erythromycin (0.5 μg/ml) and 0 or 0.1 mM IPTG. To obtain the viable count, the transformant mix was diluted 1000 or 10,000 times and plated without induction and selection, in triplicate, inside Columbia agar supplemented with 4% of defibrinated sheep blood. Transformation efficiency in percentage is calculated as the number of transformants divided by the number of total colony-forming units (viable count).
S. pneumoniae sybodies susceptibility assays
Growth assays of unencapsulated S. pneumoniae strain VL567 (cps::chl) were performed in the presence or absence of sybodies A, B, C, and D at three different concentrations. Bacteria were grown in microtiter plates (CytoOne, CC 7672-7596) and incubated at 37°C, without shaking, in Tecan i-Control infinite 200 PRO. Bacterial growth was monitored by measuring OD595 every 10 min during 8 hours. Measurements were performed using three replicates per condition. Strain VL567 was grown in C+Y (pH 6.8) until OD595 reached 0.1. Cells were spun down to remove the medium and replaced by fresh C+Y medium. These washed precultures were diluted 20-fold in C+Y without sybody or in the presence of the four sybodies at different final concentrations per well.
S. pneumoniae TA extraction and analysis
Cells were grown in 5 ml of modified C+Y medium without choline or yeast containing radiolabeled [3H]-acetylcholine at either the acetyl group or the amino group at 37°C until OD600 reached 0.5. The cell solution was split in two equal halves to lipoteichoic acid (LTA) and wall teichoic acid (WTA) extraction. For LTA extraction, cells were centrifuged at 5000g for 5 min and the pellet was washed twice with 1 ml of buffer containing 20 mM MES (pH 6.5), 0.5 M sucrose, and 20 mM MgCl2 before resuspending it in 1 ml of the same buffer. Proteoplasts were generated by the addition of lysozyme (20 mg/ml) and 100 U of mutanolysin. The mixture was incubated for 30 min at 37°C. The protoplasts were pelleted at 5000g for 5 min and lysed in ice cold-buffer containing 20 mM Hepes (pH 8.0), 100 mM NaCl, 1 mM dithiothreitol, 1 mM MgCl2, 1 mM CaCl2, 2× cOmplete protease inhibitors, ribonuclease (RNase) A (6 μg/ml), and deoxyribonuclease (DNase) I (6 μg/ml). Unbroken spheroplasts were removed by centrifugation at 5000g for 10 min. The membrane fraction was collected by ultracentrifugation for 1 hour at 100,000g and at 4°C. The pellet was resuspended in 100 μl of buffer containing 50 mM MES (pH 6.5) and 150 mM NaCl, and the solution was flash-frozen and stored at −80°C for further use. For WTA extraction, cells were pelleted by centrifugation at 5000g for 5 min and resuspended in 1 ml of buffer containing 50 mM MES (pH 6.5) before a second centrifugation at 5000g for 5 min and resuspension in 1 ml of buffer containing 50 mM MES (pH 6.5) and 4% (w/v) SDS. The solution was incubated at 100°C for 1 hour. The sacculi were collected with a 5000g centrifugation for 5 min and washed with 1 ml of buffer containing 50 mM MES (pH 6.5). The sample was centrifuged in a clean tube at 14,000g for 5 min. The pellet was washed with 1 ml of buffer containing 50 mM MES (pH 6.5) and 4% (w/v) SDS, twice with 1 ml of buffer containing 50 mM MES (pH 6.5) and 2% (w/v) NaCl, and once in buffer containing 50 mM MES (pH 6.5). The sample was centrifuged at 14,000g for 5 min and resuspended in 1 ml of buffer containing 20 mM tris-HCl (pH 8.0), 0.5% (w/v) SDS, and 20 μg of proteinase K. The sample was incubated for 4 hours at 50°C while shaking at 1000 rpm. The pellet was collected by centrifugation at 14,000g for 5 min and washed with 1 ml of buffer containing 50 mM MES (pH 6.5) and 2% (w/v) NaCl. The sample was then washed three times with distilled water. The pellet was collected by centrifugation and hydrolyzed in 0.5 ml of alkaline solution of 1 N sodium hydroxide. The mixture was incubated for 16 hours at 25°C while shaking at 1000 rpm. Insoluble material was pelleted by centrifugation at 140,000g for 5 min, and the supernatant was transferred into a clean tube. Tris-HCl (125 μl) (pH 7.8) was added to neutralize the reaction, and the sample was flash-frozen until further use. The samples were quantified by liquid scintillation counting (TRI-CARB, PerkinElmer).
Acknowledgments
We thank Nanion Technologies for technical assistance. We thank the staff at the electron microscopy facility at Biozentrum (BioEM laboratory) and at the PX beamline of the SLS. We thank T. Sharpe from the Biophysics facility at Biozentrum for SEC-MALS data analysis and X. L. Blatter for assistance in cell expressions.
Funding: Work in the Perez laboratory was supported by the Swiss National Science Foundation (SNSF) (PP00P3_170607), the Helmut Horten Stiftung (HHS), and Nanion Research Grant Initiative. Work in the Veening laboratory was supported by the SNSF (project grants 31003A_172861 and 310030_192517), SNSF JPIAMR grant (40AR40_185533), SNSF NCCR “AntiResist” (51NF40_180541), and ERC consolidator grant 771534-PneumoCaTChER. Work in the Seeger laboratory was supported by the SNSF (project grant 310030_188817) and an ERC consolidator grant (no. 772190).
Author contributions: N.B. performed purifications, crystallization experiments, and functional assays. N.B. and C.P. prepared samples for cryo-EM, processed x-ray and cryo-EM data, and built and validated the models. A.-S.R. performed complementation and growth assays under supervision from J.-W.V. G.C. produced samples for sybody selection. C.A.J.H. and G.C. performed sybody selection under supervision from M.A.S. G.C. established WTA and LTA extraction assays, and N.B. performed extraction experiments. C.P. and N.B. wrote the manuscript with input from all authors. C.P. conceived the project.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Electron microscopy density maps and atomic models have been deposited in the EMDB and PDB, respectively, with accession codes EMD-13268 and PDB 7PAF. Atomic coordinates for the reported crystal structure have been deposited in the PDB under accession code 7B0K.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S13
Table S1
REFERENCES AND NOTES
- 1.Weidenmaier C., Peschel A., Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6, 276–287 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Brown S., Santa Maria J. P. Jr., Walker S., Wall teichoic acids of Gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silhavy T. J., Kahne D., Walker S., The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raetz C. R. H., Whitfield C., Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swoboda J. G., Campbell J., Meredith T. C., Walker S., Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 11, 35–45 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swiatlo E., Champlin F. R., Holman S. C., Wilson W. W., Watt J. M., Contribution of choline-binding proteins to cell surface properties of Streptococcus pneumoniae. Infect. Immun. 70, 412–415 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiser J. N., Shchepetov M., Chong S. T., Decoration of lipopolysaccharide with phosphorylcholine: A phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65, 943–950 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiser J. N., Pan N., McGowan K. L., Musher D., Martin A., Richards J., Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187, 631–640 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillespie S. H., Ainscough S., Dickens A., Lewin J., Phosphorylcholine-containing antigens in bacteria from the mouth and respiratory tract. J. Med. Microbiol. 44, 35–40 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Barbier M., Oliver A., Rao J., Hanna S. L., Goldberg J. B., Albertí S., Novel phosphorylcholine-containing protein of Pseudomonas aeruginosa chronic infection isolates interacts with airway epithelial cells. J. Infect. Dis. 197, 465–473 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Cundell D. R., Gerard N. P., Gerard C., Idanpaan-Heikkila I., Tuomanen E. I., Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377, 435–438 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Iuchi H., Ohori J., Kyutoku T., Ito K., Kurono Y., Role of phosphorylcholine in Streptococcus pneumoniae and nontypeable Haemophilus influenzae adherence to epithelial cells. Auris Nasus Larynx 46, 513–519 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Pang B., Winn D., Johnson R., Hong W., West-Barnette S., Kock N., Swords W. E., Lipooligosaccharides containing phosphorylcholine delay pulmonary clearance of nontypeable Haemophilus influenzae. Infect. Immun. 76, 2037–2043 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ring A., Weiser J. N., Tuomanen E. I., Pneumococcal trafficking across the blood-brain barrier. Molecular analysis of a novel bidirectional pathway. J. Clin. Invest. 102, 347–360 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenow C., Ryan P., Weiser J. N., Johnson S., Fontan P., Ortqvist A., Masure H. R., Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25, 819–829 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Garcia P., Gonzalez M. P., Garcia E., Lopez R., Garcia J. L., LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 31, 1275–1277 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Gosink K. K., Mann E. R., Guglielmo C., Tuomanen E. I., Masure H. R., Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 68, 5690–5695 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomasz A., Choline in the cell wall of a bacterium: Novel type of polymer-linked choline in Pneumococcus. Science 157, 694–697 (1967). [DOI] [PubMed] [Google Scholar]
- 19.Whiting G. C., Gillespie S. H., Investigation of a choline phosphate synthesis pathway in Streptococcus pneumoniae: Evidence for choline phosphate cytidylyltransferase activity. FEMS Microbiol. Lett. 143, 279–284 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Kharat A. S., Tomasz A., Drastic reduction in the virulence of Streptococcus pneumoniae expressing type 2 capsular polysaccharide but lacking choline residues in the cell wall. Mol. Microbiol. 60, 93–107 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Fan X., Goldfine H., Lysenko E., Weiser J. N., The transfer of choline from the host to the bacterial cell surface requires glpQ in Haemophilus influenzae. Mol. Microbiol. 41, 1029–1036 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Vollmer W., Tomasz A., Identification of the teichoic acid phosphorylcholine esterase in Streptococcus pneumoniae. Mol. Microbiol. 39, 1610–1622 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Fan X., Pericone C. D., Lysenko E., Goldfine H., Weiser J. N., Multiple mechanisms for choline transport and utilization in Haemophilus influenzae. Mol. Microbiol. 50, 537–548 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Clark S. E., Weiser J. N., Microbial modulation of host immunity with the small molecule phosphorylcholine. Infect. Immun. 81, 392–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jack D. L., Yang N. M., Saier M. H. Jr., The drug/metabolite transporter superfamily. Eur. J. Biochem. 268, 3620–3639 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Schuldiner S., Granot D., Mordoch S. S., Ninio S., Rotem D., Soskin M., Tate C. G., Yerushalmi H., Small is mighty: EmrE, a multidrug transporter as an experimental paradigm. News Physiol. Sci. 16, 130–134 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Zhang J. R., Idanpaan-Heikkila I., Fischer W., Tuomanen E. I., Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 31, 1477–1488 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Bondy J., Osharovich S., Storm J., Durning G., McAuliffe T., Fan X., Haemophilus influenzae LicB contributes to lung damage in an aged mice co-infection model. Microb. Pathog. 90, 1–6 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Bazzone A., Barthmes M., Fendler K., SSM-based electrophysiology for transporter research. Methods Enzymol. 594, 31–83 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Perez C., Faust B., Mehdipour A. R., Francesconi K. A., Forrest L. R., Ziegler C., Substrate-bound outward-open state of the betaine transporter BetP provides insights into Na+ coupling. Nat. Commun. 5, 4231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann T., Warmbold B., Smits S. H. J., Tschapek B., Ronzheimer S., Bashir A., Chen C., Rolbetzki A., Pittelkow M., Jebbar M., Seubert A., Schmitt L., Bremer E., Arsenobetaine: An ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes. Environ. Microbiol. 20, 305–323 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Yau B., Hunt N. H., Mitchell A. J., Too L. K., Blood–brain barrier pathology and CNS outcomes in Streptococcus pneumoniae meningitis. Int. J. Mol. Sci. 19, 3555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimmermann I., Egloff P., Hutter C. A. J., Arnold F. M., Stohler P., Bocquet N., Hug M. N., Huber S., Siegrist M., Hetemann L., Gera J., Gmür S., Spies P., Gygax D., Geertsma E. R., Dawson R. J. P., Seeger M. A., Synthetic single domain antibodies for the conformational trapping of membrane proteins. eLife 7, e34317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmermann I., Egloff P., Hutter C. A. J., Kuhn B. T., Bräuer P., Newstead S., Dawson R. J. P., Geertsma E. R., Seeger M. A., Generation of synthetic nanobodies against delicate proteins. Nat. Protoc. 15, 1707–1741 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Barland N., Perez C., Selection of transporter-targeted inhibitory nanobodies by solid-supported-membrane (SSM)-based electrophysiology. J. Vis. Exp. (2021). [DOI] [PubMed] [Google Scholar]
- 36.Dumoulin M., Conrath K., van Meirhaeghe A., Meersman F., Heremans K., Frenken L. G. J., Muyldermans S., Wyns L., Matagne A., Single-domain antibody fragments with high conformational stability. Protein Sci. 11, 500–515 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iezzi M. E., Policastro L., Werbajh S., Podhajcer O., Canziani G. A., Single-domain antibodies and the promise of modular targeting in cancer imaging and treatment. Front. Immunol. 9, 273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trombe M. C., Laneelle M. A., Laneelle G., Lipid composition of aminopterin-resistant and sensitive strains of Streptococcus pneumoniae. Effect of aminopterin inhibition. Biochim. Biophys. Acta 574, 290–300 (1979). [DOI] [PubMed] [Google Scholar]
- 39.Tsuchiya H., Doki S., Takemoto M., Ikuta T., Higuchi T., Fukui K., Usuda Y., Tabuchi E., Nagatoishi S., Tsumoto K., Nishizawa T., Ito K., Dohmae N., Ishitani R., Nureki O., Structural basis for amino acid export by DMT superfamily transporter YddG. Nature 534, 417–420 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Kim J., Tan Y. Z., Wicht K. J., Erramilli S. K., Dhingra S. K., Okombo J., Vendome J., Hagenah L. M., Giacometti S. I., Warren A. L., Nosol K., Roepe P. D., Potter C. S., Carragher B., Kossiakoff A. A., Quick M., Fidock D. A., Mancia F., Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature 576, 315–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y., Nishizawa T., Takemoto M., Kumazaki K., Yamashita K., Hirata K., Minoda A., Nagatoishi S., Tsumoto K., Ishitani R., Nureki O., Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity. Nat. Plants 3, 825–832 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Nji E., Gulati A., Qureshi A. A., Coincon M., Drew D., Structural basis for the delivery of activated sialic acid into Golgi for sialyation. Nat. Struct. Mol. Biol. 26, 415–423 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Ahuja S., Whorton M. R., Structural basis for mammalian nucleotide sugar transport. eLife 8, e45221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker J. L., Corey R. A., Stansfeld P. J., Newstead S., Structural basis for substrate specificity and regulation of nucleotide sugar transporters in the lipid bilayer. Nat. Commun. 10, 4657 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker J. L., Newstead S., Structural basis of nucleotide sugar transport across the Golgi membrane. Nature 551, 521–524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keller L. E., Rueff A. S., Kurushima J., Veening J. W., Three new integration vectors and fluorescent proteins for use in the opportunistic human pathogen Streptococcus pneumoniae. Genes 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fidock D. A., Nomura T., Talley A. K., Cooper R. A., Dzekunov S. M., Ferdig M. T., Ursos L. M. B., Sidhu A. b. S., Naudé B., Deitsch K. W., Su X.-z., Wootton J. C., Roepe P. D., Wellems T. E., Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6, 861–871 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sweet D. H., Miller D. S., Pritchard J. B., Ventricular choline transport: A role for organic cation transporter 2 expressed in choroid plexus. J. Biol. Chem. 276, 41611–41619 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Tohgi H., Abe T., Kimura M., Saheki M., Takahashi S., Cerebrospinal fluid acetylcholine and choline in vascular dementia of Binswanger and multiple small infarct types as compared with Alzheimer-type dementia. J. Neural Transm. 103, 1211–1220 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Juge N., Moriyama S., Miyaji T., Kawakami M., Iwai H., Fukui T., Nelson N., Omote H., Moriyama Y., Plasmodium falciparum chloroquine resistance transporter is a H+-coupled polyspecific nutrient and drug exporter. Proc. Natl. Acad. Sci. U.S.A. 112, 3356–3361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.du Plessis J. L., Stefaniak A. B., Wilhelm K. P., Measurement of skin surface pH. Curr. Probl. Dermatol. 54, 19–25 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Harell M., Mover-Lev H., Levy D., Sade J., Gas composition of the human nose and nasopharyngeal space. Acta Otolaryngol. 116, 82–84 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Kwak B. Y., Zhang Y.-M., Yun M., Heath R. J., Rock C. O., Jackowski S., Park H.-W., Structure and mechanism of CTP:phosphocholine cytidylyltransferase (LicC) from Streptococcus pneumoniae. J. Biol. Chem. 277, 4343–4350 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Wang L., Jiang Y. L., Zhang J. R., Zhou C. Z., Chen Y., Structural and enzymatic characterization of the choline kinase LicA from Streptococcus pneumoniae. PLOS ONE 10, e0120467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta K., Donlan J. A. C., Hopper J. T. S., Uzdavinys P., Landreh M., Struwe W. B., Drew D., Baldwin A. J., Stansfeld P. J., Robinson C. V., The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 541, 421–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuhn B. T., Zimmermann I., Egloff P., Hürlimann L. M., Hutter C. A. J., Miscenic C., Dawson R. J. P., Seeger M. A., Geertsma E. R., Biotinylation of membrane proteins for binder selections. Methods Mol. Biol. 2127, 151–165 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Tan Y. Z., Baldwin P. R., Davis J. H., Williamson J. R., Potter C. S., Carragher B., Lyumkis D., Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L. W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using x-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pintilie G., Zhang K., Su Z., Li S., Schmid M. F., Chiu W., Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat. Methods 17, 328–334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barad B. A., Echols N., Wang R. Y. R., Cheng Y., DiMaio F., Adams P. D., Fraser J. S., EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D., Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karplus P. A., Diederichs K., Linking crystallographic model and data quality. Science 336, 1030–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zallot R., Oberg N., Gerlt J. A., The EFI web resource for genomic enzymology tools: Leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 58, 4169–4182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., Mering C., STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ben Chorin A., Masrati G., Kessel A., Narunsky A., Sprinzak J., Lahav S., Ashkenazy H., Ben-Tal N., ConSurf-DB: An accessible repository for the evolutionary conservation patterns of the majority of PDB proteins. Protein Sci. 29, 258–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trott O., Olson A. J., AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sterling T., Irwin J. J., ZINC 15—Ligand discovery for everyone. J. Chem. Inf. Model. 55, 2324–2337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., Hutchison G. R., Open Babel: An open chemical toolbox. J. Chem. 3, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sorg R. A., Gallay C., Van Maele L., Sirard J. C., Veening J. W., Synthetic gene-regulatory networks in the opportunistic human pathogen Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 117, 27608–27619 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu X., Gallay C., Kjos M., Domenech A., Slager J., Kessel S. P., Knoops K., Sorg R. A., Zhang J.-R., Veening J.-W., High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol. Syst. Biol. 13, 931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S13
Table S1