Abstract
Aluminum-activated malate transporters (ALMTs) form an anion channel family that plays essential roles in diverse functions in plants. Arabidopsis ALMT12, also named QUAC1 (quick anion channel 1), regulates stomatal closure in response to environmental stimuli. However, the molecular basis of ALMT12/QUAC1 activity remains elusive. Here, we describe the cryo-EM structure of ALMT12/QUAC1 from Glycine max at 3.5-Å resolution. GmALMT12/QUAC1 is a symmetrical dimer, forming a single electropositive T-shaped pore across the membrane. The transmembrane and cytoplasmic domains are assembled into a twisted two-layer architecture, with their associated dimeric interfaces nearly perpendicular. GmALMT12/QUAC1-mediated currents display rapid kinetics of activation/deactivation and a bell-shaped voltage dependency, reminiscent of the rapid (R)–type anion currents. Our structural and functional analyses reveal a domain-twisting mechanism for malate-mediated activation. Together, our study uncovers the molecular basis for a previously uncharacterized class of anion channels and provides insights into the gating and modulation of the ALMT12/QUAC1 anion channel.
Structural and functional characterization of ALMT12/QUAC1 reveals domain-twisting mechanism for malate activation.
INTRODUCTION
Anion fluxes in plants play a fundamental role in cell signal transduction and volume regulation. Aluminum-activated malate transporters (ALMTs) form an anion channel family involved in fundamental physiological processes, including stomatal function (1–5), pollen tube growth (6, 7), aluminum (Al3+) resistance (8, 9), mineral nutrition (10), fruit acidity (11), microbe interactions (12), cell signaling (13), and seed development (14). The first ALMT member, Triticum aestivum ALMT1 (TaALMT1), was named because it mediates Al3+-induced malate efflux to confer Al3+ resistance in wheat roots. The Arabidopsis thaliana ALMT12 (AtALMT12), predominantly expressed in guard cells, was the first member in the ALMT family to be implicated in regulating stomatal function in response to abscisic acid, darkness, high CO2 level, and drought.
AtALMT12, also named AtQUAC1 (quick anion channel 1), mediates anion currents with fast kinetics of activation/deactivation and strong voltage dependency (2), similar to that of rapid (R)–type currents of guard cell reported in the patch-clamp study (15–17). The loss of function of almt12/quac1 mutant causes impaired malate-induced anion currents in guard cells for closing stomatal pore (2, 3). Apart from its role in stomatal regulation, AtALMT12/QUAC1 also plays a vital role in cell polarity and the growth of pollen tubes by modulating apical anion efflux (6, 7). AtALMT12/QUAC1 is permeable to various anions, including malate2−, SO42−, NO3−, and Cl−. The activity of AtALMT12/QUAC1 was reported to be modulated by malate (2, 18–20), OST1 phosphorylation (21), and calmodulin (22), but the underlying mechanism is largely enigmatic due to a lack of structural information.
Here, we report a cryo–electron microscopy (cryo-EM) structure of soybean ALMT12/QUAC1 from Glycine max (GmALMT12/QUAC1, ~67% sequence identity to AtALMT12/QUAC1) at the resolution of 3.5 Å in the presence of malate. The structure reveals that GmALMT12/QUAC1 is a symmetrical dimer assembled into twisted two-layer architecture with a central T-shaped pore across the membrane. The pore is lined with highly conserved positively charged residues, rendering the pore surface electropositive, consistent with the GmALMT12/QUAC1 function as an anion channel. Further electrophysiological analyses reveal that GmALMT12/QUAC1 is a malate-regulated inward-rectifying anion channel and displays rapid kinetics in channel gating during activation and deactivation. Further mutagenesis analyses demonstrate that the C-terminal interactions play a vital role in regulating channel function. The comparison of our malate-modified cryo-EM structure with the AlphaFold-predicted structure allows us to propose a mechanism for malate-mediated channel activation. Together, our study uncovers the molecular basis for a previously uncharacterized class of anion channels and provides insights into the gating and modulation of the ALMT12/QUAC1 anion channel.
RESULTS
Bioinformatics analyses of plant ALMTs/QUACs
There are 13 members in the model plant A. thaliana, except for one short sequence of a partial transmembrane domain (TMD) (ALMT11). To better understand how ALMT proteins are represented in the plant kingdom, we clustered ~1700 nonredundant sequences into a superfamily at the Position-Specific Iterative Basic Local Alignment Search Tool (PSI-BLAST) level E ≤ 5 × 10−3, then into two distinct families at a threshold of E ≤ 10−160, and finally into subfamilies at a threshold of E ≤ 10−180, as detailed in table S1. Since more studies have shown that ALMT members function as anion channels rather than transporters, and AtALMT12/QUAC1 is now the best-characterized member, we adopted nomenclature for the QUAC superfamily, which was further divided into families QF1 and QF2. We further found that family QF1 has two subfamilies: QF1A comprises the Arabidopsis ALMT3, ALMT4, ALMT5, ALMT6, and ALMT9 and their homologs, and QF1B has a distinct set of ALMT proteins. Family QF2 has three subfamilies: the Arabidopsis ALMT1, ALMT2, ALMT7, ALMT8, and ALMT10 proteins are in subfamily QF2A, the Arabidopsis ALMT12, ALMT13, and ALMT14 proteins are in QF2B, and another distinct set of ALMT proteins is in QF2C (fig. S1).
The structure-based sequence alignment for GmALMT12/QUAC1 and 13 Arabidopsis ALMT/QUACs is shown in fig. S2. Sequence analyses reveal that the ALMT/QUACs share relatively conserved TM and cytosolic helical regions, suggesting a similar topology and architecture for the ALMT/QUAC1 family. Some conserved positively charged residues are distributed in the TM regions and may be responsible for anion selectivity or voltage sensing. Some fingerprint motifs, including WEP (Trp-Glu-Pro) and PXWXG (Pro-X-Trp-X-Gly), are found in the cytosolic helical region. One of the most divergent regions (~50 to 100 amino acids in length) is found between helices H5 and H6 in the cytosolic portions, harboring potential phosphorylation residues (Ser/Thr) for channel modulation.
Cryo-EM structural determination of GmALMT12/QUAC1
We screened six plant ALMT12/QUAC1s for expression and found that the soybean GmALMT12/QUAC1 was suitable for further structural and functional studies (see Materials and Methods for details). Upon solubilization in 1.0% n-dodecyl-β-d-maltopyranoside (DDM) and 0.02% cholesteryl hemisuccinate (CHS), the GmALMT12/QUAC1 proteins were purified by Ni2+-affinity chromatography, and the resulting peak fractions were pooled and loaded onto a gel filtration column for further purification, buffer and detergent exchange. The final solution contained 0.005% detergent Lauryl Maltose Neopentyl Glycol (LMNG), with either 150 mM NaCl or 75 mM l-malate.
We further determined its cryo-EM structure in the presence of 75 mM l-malate (figs. S3 and S4 and table S2). The dimeric reconstruction at 3.5-Å resolution allowed de novo modeling of 410 of the 537 amino acids (residues 36 to 393 and 455 to 506) per protomer chain. Other regions are not resolved in the density map due to their intrinsic flexibility in the protein. The search in the Dali server (23) for similar structures returned no significant hit. To the best of our knowledge, the GmALMT12/QUAC1 channel has a unique protein fold and architecture, representing a previously uncharacterized class of ion channels.
The twisted two-layer architecture in GmALMT12/QUAC1 channel
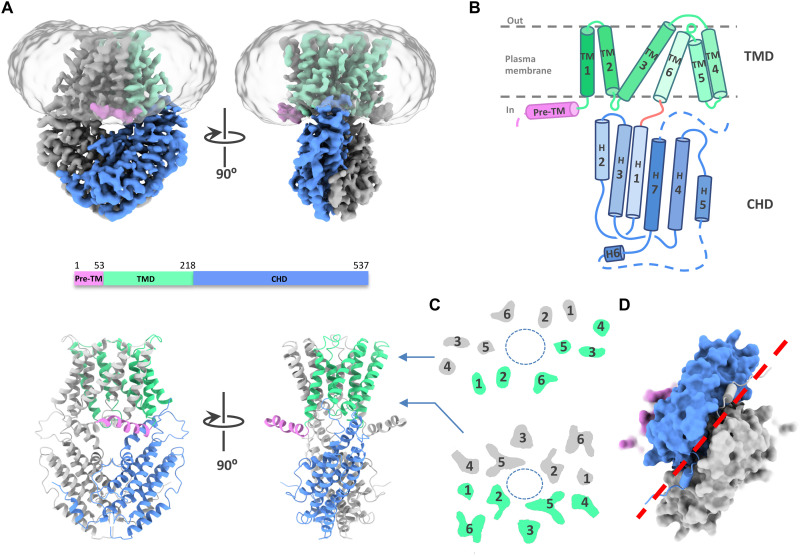
The GmALMT12/QUAC1 channel is a flat vase-shaped homodimer with a twofold axis perpendicular to the plasma membrane (Fig. 1A and movie S1). The overall molecule forms a two-layer architecture, divided into two portions of TMD and the cytoplasmic helical domain (CHD). The domain topology for each protomer is shown in Fig. 1B. TMD comprises six TM helices, arranged as three pairs of V-shaped helical hairpins stacked against each other, whereas the CHD has seven helices, forming a helical bundle with a short domain-swapped finger helix. The N-terminal pre-TM region contains a juxtamembrane helix bent at a conserved proline (P55) before TMD. A disorder region between helices H5 and H6 in the CHD enriches with Ser/Thr residues (fig. S2), raising an intriguing possibility of containing phosphorylation sites for channel regulation.
Fig. 1. Architecture of the GmALMT12/QUAC1 channel.
(A) Overall structure of the GmALMT12/QUAC1 channel. The cryo-EM density maps (top) and ribbon drawings (bottom) are shown, with the pre-TM helix in pink, TMD in green, and CHD in blue in one protomer. Another protomer is colored in gray. (B) Topology of the GmALMT12/QUAC1 protomer. The six helices in TMD are marked by TM1 to TM6, and the seven helices in CHD are marked as H1 to H7. The membrane boundary is shown as gray lines. The disordered regions are indicated as dashed lines. (C) Top view of the cross section of the transmembrane layer at the indicated positions by the arrow lines in (A). The elliptical dashed line marks the pore, and the TMs are indicated. (D) Bottom view of the C-terminal dimeric domains, colored as in (A). The domain-swapped finger helix H6 is shown as cylinder cartoon, and a red dashed line marks the dimeric interface of the CHD.
The dimer formation is mediated by TMDs and CHDs, with buried surface areas of ~3500 Å2 and ~ 3300 Å2, respectively. When viewed from the extracellular side, the TM helices trace the circumference of an ellipse with TM1 to TM6 from one protomer followed by another set of TM1 to TM6 from the second protomer in an antiparallel manner (Fig. 1C). At the membrane, TM1 and TM2 directly interact with TM4 and TM5 from another protomer, respectively. In the cytoplasm, the two CHDs of the dimeric channel unite together at the distal end of the molecule, mediated by extensive interactions. A domain-swapped finger helix (H6) also participates in C-terminal interactions (Fig. 1D). However, the functional role of these interactions remains unclear.
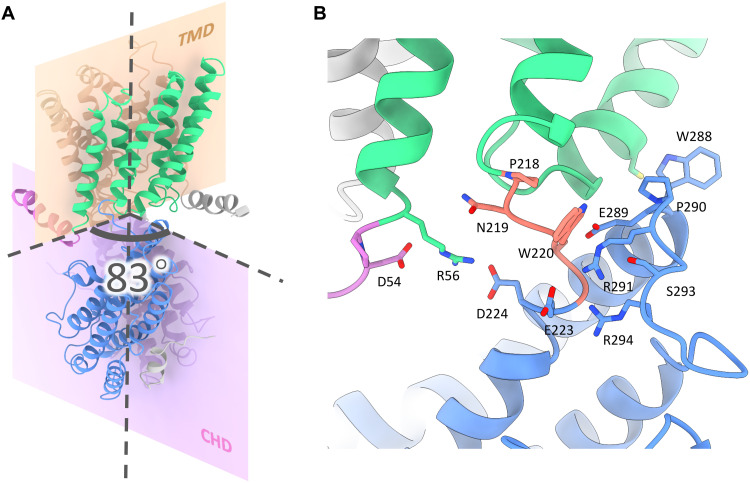
In the two-layer structure of GmALMT12/QUAC1, TMDs connect to the CHDs via a highly conserved PXWXG motif (Pro-X-Trp-X-Gly). They interact with each other in a twisted manner, with their dimeric interfaces nearly perpendicular to each other (forming a dihedral angle of ~83°) (Fig. 2A). Another characteristic motif WEP (Trp-Glu-Pro), located in the connecting loop between helices H2 and H3, is found in the vicinity of the PXWXG motif at the layer interface. These two motifs, together with other surrounding charged residues (Arg/Lys: R56, K63, R291, and R294; Asp/Glu: D54, E223, D224, and E289), form intensive interactions at the layer interface (Fig. 2B). Mutations in the WEP motif (E284Q in TaALMT1 and E276Q in AtALMT12/QUAC1) (18, 24) completely abolish their channel activities, possibly due to impaired coupling between TMD and CHD caused by the altered interaction at the layer interface. Thus, the unique two-layer architecture design allows the CHD and TMD portions to tightly interact with each other, providing a structural basis for coupling channel modulation to gating. These observations also imply that the intramolecular domain rearrangements may occur upon channel activation.
Fig. 2. The twisted two-layer architecture of the GmALMT12/QUAC1 channel.
(A) Dimeric interfaces of TMD and CHD of the GmALMT12/QUAC1 channel. These dimeric interfaces are nearly perpendicular to each other, forming a dihedral angle of ~83°. The ribbon model is colored as in Fig. 1A. (B) Layer interface between TMD and CHD. The TMD portion connects to the CHD portion via a highly conserved PXWXG motif (P218-N219-W220-S221-G222, in salmon), which interacts with another characteristic motif WEP (W288-E289-P290, located between helices H2 and H3, in blue). Other surrounding charged residues at the interface are also shown in sticks, including R56, R291, R294, D54, E223, D224, and E289. The ribbon model is colored as in (A).
Structural features of the ion conduction pathway
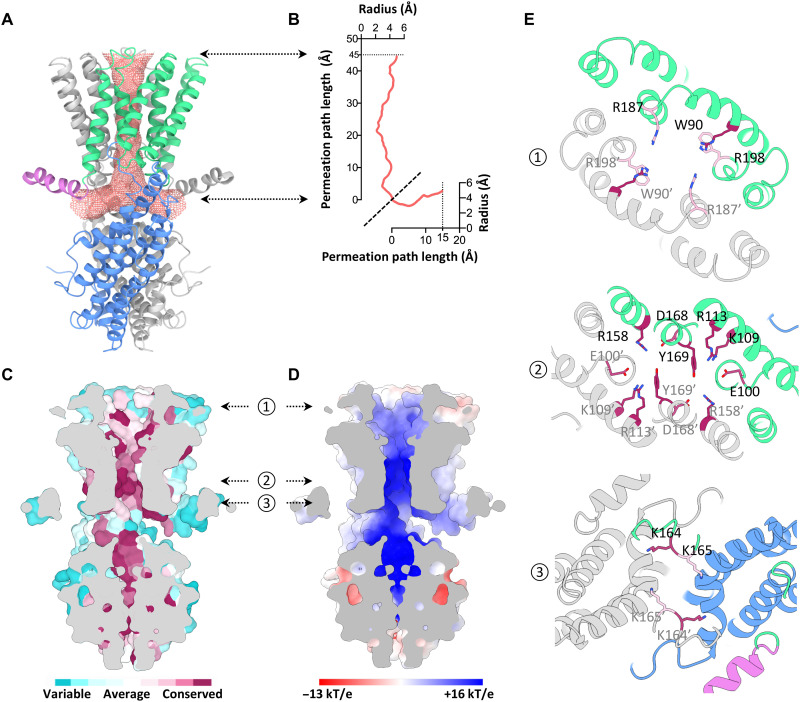
Unlike the dimeric CLC chloride channel with two independent pores (25), GmALMT12/QUAC1 forms a single T-shaped pore with a bifurcated entrance in the cytoplasm (Fig. 3A). The formation of the bifurcated entrance is due to joining the two CHDs in the cytoplasm. The pore is merged into one single pathway within the membrane and eventually forms one exit at the extracellular side. TM2, TM3, TM5, and TM6 from both protomers create a pore with a radius of ~2 to 4 Å across the membrane (Fig. 3B), as estimated by HOLE (26). Overall, the GmALMT12/QUAC1 pore is lined with highly conserved and general hydrophobic residues (Fig. 3C). Nevertheless, some highly conserved positively charged residues are distributed along the pore, thus rendering the surface electropositive (Fig. 3D).
Fig. 3. Structural feature of the GmALMT12/QUAC1 channel pore.
(A) The pore-lining surface was computed by the program HOLE and drawn into a ribbon model of the GmALMT12/QUAC1 pore. We used a simple van der Waals surface for the protein and the program default probe radius of 1.15 Å. The T-shaped pore is shown in salmon dots. (B) Plot of the pore radius as a function of the pore axis. (C and D) Side views of the cross section through the ion-conducting pore. The molecular surface colored with sequence conservation for 137 nonredundant proteins of the QF2B subfamily is shown in (C), and the molecular surface colored with electrostatic potential is shown in (D). Electronegative and electropositive potential in (D) are colored in degrees of red and blue saturation, respectively. (E) Top views of the cross section through the ion-conducting pore at three indicated positions by the arrows in (C) and (D). The ribbon is colored as in (A). The conserved pore-lining charged residues are shown in sticks, colored with the sequence conservation as in (C).
Two arginines (R187 and R198) form a positively charged ring facing the pore entrance at the extracellular side. Three positively charged residues from TM3 and TM4 (K109, R113, and R158) protrude into the pore, making a second positively charged ring within the membrane. They are associated with other conserved residues from TM2 and TM5 (E100, D168, and Y169), mediating an interhelices network within the pore. Another two lysines (K164 and K165) form additional constrictions at the bottom of the T-shaped pore (Fig. 3E). Together, the structural feature of GmALMT12/QUAC1 likely contributes to its function as an anion-conducting channel. The above findings suggest that permeable anions interact with the pore-lining charged residues, thus participating in channel regulation.
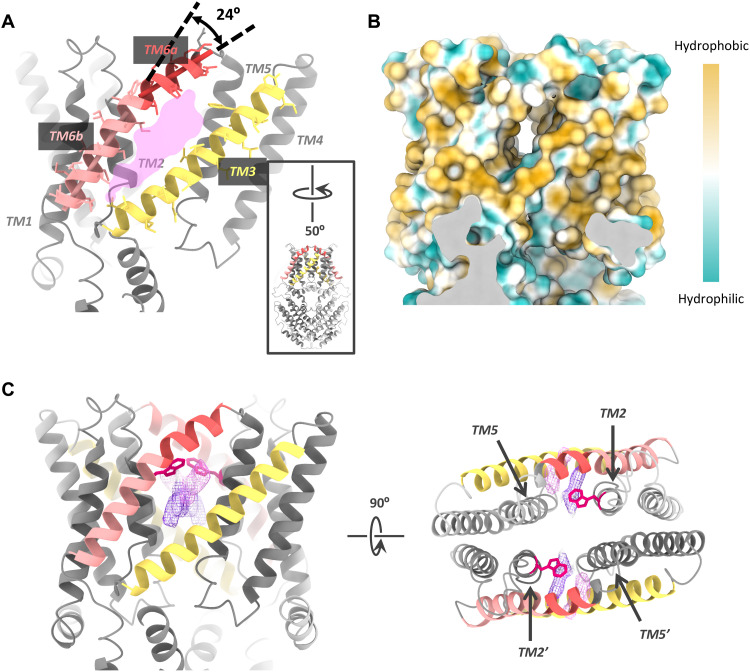
A central kink (~24°) in TM6 causes a lateral fenestration from the membrane to the lumen of the channel pore within each protomer (Fig. 4, A and B, and movie S1). The fenestration measures ~6 Å × 20 Å, and its dimension depends upon the conformation of pore-forming TMs, especially the kinked TM6. The presence of kinked helix and lateral fenestration in the structure implies that the channel in 75 mM malate is likely in a high-energy constrained conformation. Several unmodeled densities and a highly conserved pore-lining W90 residue are found within or near the fenestration (Fig. 4C), raising an interesting question if the bulky W90 residue is involved in the regulation of channel gating.
Fig. 4. The lateral fenestration in the GmALMT12/QUAC1 channel.
(A) Lateral fenestration with a dimension of ~6 Å × 20 Å. The fenestration is formed by TM2, TM3, TM5, and TM6 within one protomer. A kink of ~24° in TM6 is indicated. For a better view of the fenestration, the molecule is rotated by 50°, as shown in the inset. (B) The molecular lipophilicity potential map is shown on the surface of the channel, as orientated in (A). Hydrophobic and hydrophilic potentials are colored in degrees of gold and cyan saturation, respectively. (C) Two unmodeled densities (in purple and magenta) within the fenestration. The side view (right) and top view (left) are shown. The W90 residues are shown as red sticks.
Functional characterization of GmALMT12/QUAC1 by TEVC
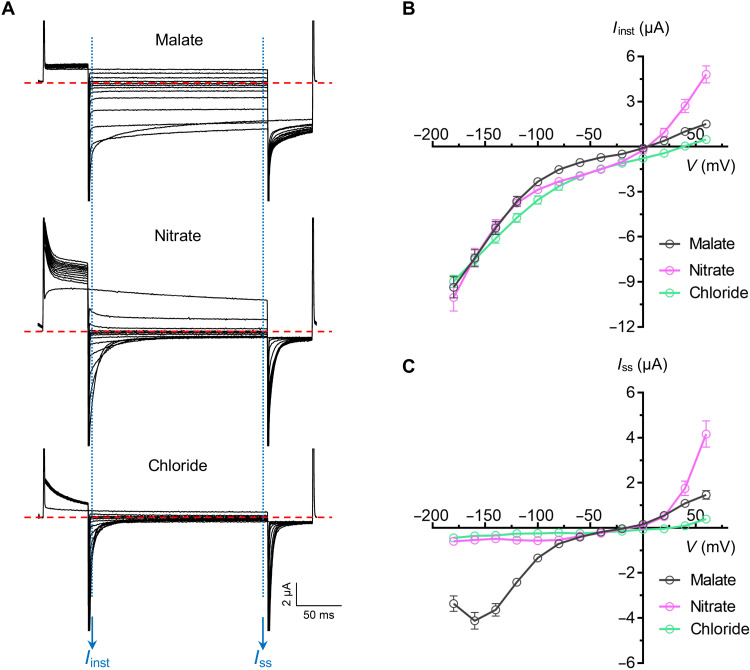
A total of 34 ALMTs were found in soybean (27), but none of them has yet been functionally characterized by electrophysiology. We expressed GmALMT12/QUAC1 in Xenopus laevis oocytes and measured channel conductance by applying the two-electrode voltage clamp (TEVC) technique in different anionic bath solutions, including malate, nitrate, or chloride (Fig. 5A). In the recordings, the application of voltage pulses elicited rapidly activating currents, resembling AtALMT12/QUAC1 (2). They share fast kinetics in the channel activation and deactivation, as evidenced in the analyses of the instant and steady-state current-voltage relationships (Fig. 5). In contrast to substantial instantaneous currents, only small steady-state currents can be seen in the recordings at negative membrane potentials. We found that the steady-state currents mediated by GmALMT12/QUAC1 displayed a bell-shaped current-voltage curve (Fig. 5C). This electrophysiological behavior points to a strong voltage dependency for the GmALMT12/QUAC1 channel activation, a hallmark feature of the R-type anion current in guard cells (2, 16, 17).
Fig. 5. TEVC recording of the GmALMT12/QUAC1 channel in X. laevis oocytes.
(A) Representative current traces recorded at different voltages (from +60 to −180 mV in 20-mV decrement) in various external solutions: 30 mM l-malate, 30 mM NaNO3, or 30 mM NaCl. (B) Instant current–voltage (Iinst-V) relations of GmALMT12/QUAC1 (data are means ± SEM, n ≥ 8). (C) Steady-state current–voltage (Iss-V) relations of GmALMT12/QUAC1 (data are means ± SEM, n ≥ 8).
In comparison, the activation/deactivation kinetics and voltage dependency of GmALMT12/QUAC1 in NaNO3 or NaCl solutions are distinguished from those in malate solutions. Unexpectedly, the steady-state currents at negative membrane potentials are much smaller in NaNO3 or NaCl solutions (Fig. 5, A and C). Overall, our TEVC recordings demonstrate that external anions play a crucial role in regulating GmALMT12/QUAC1 channel properties, especially the activation/deactivation kinetics and steady-state currents. We propose that the anions interplay with the pore-lining residues, such as the interhelix charged residues interacting network within the pore (Fig. 3E), and further investigation is needed to understand the underlying mechanism regulated by different anions.
Single-channel analysis of GmALMT12/QUAC1 channel in planar lipid bilayer
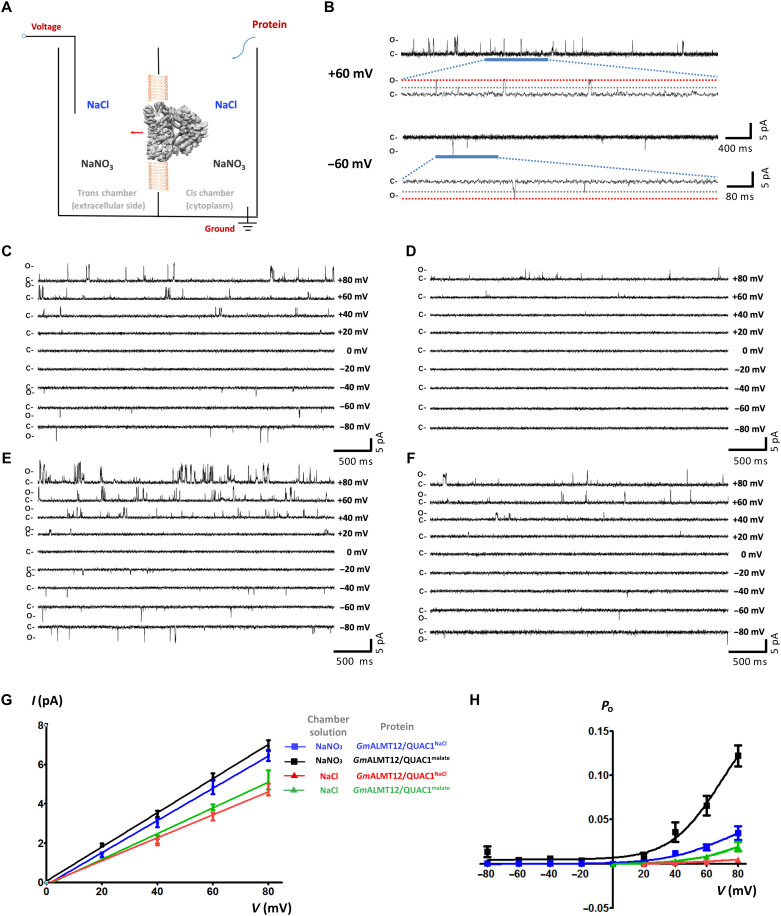
To further understand the single-channel properties, we applied the planar lipid bilayer (PLB) technique to perform single-channel recording using purified proteins. We first fused the GmALMT12/QUAC1NaCl (purified in 150 mM NaCl) proteins into a lipid bilayer and measured channel conductance under different symmetrical solutions (Fig. 6A). When using symmetrical NaNO3 solutions, the application of a series of TM voltages to the trans chamber resulted in frequent openings and closings of the GmALMT12/QUAC1 channel, as evidenced by the current fluctuation in the recording traces at different applied voltages (Fig. 6, B and C). We also found that the channel gating transits between two predominant states of fully open and closed, and sometimes to a subconductance that is ~50% level of the full conductance (Fig. 6B). This observation is consistent with the two-entrance T-shaped tunnel presented in the GmALMT12/QUAC1 channel (Fig. 3A). The intermediate subconductance (at ~50% full level) suggests that one of two entrances is blocked or closed. In addition, the GmALMT12/QUAC1 channel gating displays a flickering feature that represents the rapid character of the activation/deactivation kinetics for R-type anion channels.
Fig. 6. Single-channel analysis of the GmALMT12/QUAC1 channel in planar lipid bilayer.
(A) Schematic illustration of single-channel recording of GmALMT12/QUAC1 channel in a PLB. The chambers were filled with 1.0 ml of symmetrical solutions of 150 mM NaCl or NaNO3, and GmALMT12/QUAC1NaCl or GmALMT12/QUAC1malate (purified in 150 mM NaCl or 75 mM l-malate, respectively) was added to the cis side. The trans chamber, representing the extracellular compartment, was connected to the head stage input of a bilayer voltage-clamp amplifier. The cis chamber, representing the cytoplasmic compartment, was held at virtual ground. (B) Representative current traces for single-channel analysis of GmALMT12/QUAC1NaCl at +60- or −60-mV holding potentials in the NaNO3 solution. The closed (C) and full-open (O) states are indicated. (C and D) Representative current traces for single-channel analysis of GmALMT12/QUAC1NaCl at different holding potentials, as indicated. The chamber solutions are 150 mM NaNO3 (C) or NaCl (D). (E and F) Representative current traces for single-channel analysis of GmALMT12/QUAC1malate at different holding potentials, as indicated. The chamber solutions are 150 mM NaNO3 (E) or NaCl (F). (G and H) Current-voltage relationships (G) and open probabilities (H) for the single-channel recordings of GmALMT12/QUAC1. The chamber solutions (NaNO3 or NaCl) and proteins (GmALMT12/QUAC1NaCl or GmALMT12/QUAC1malate) are indicated in the inset (data are means ± SEM, n ≥ 4). Note: Due to the opposite virtual grounding, the positive currents recorded in PLB correspond to the negative currents observed in TEVC.
In contrast, we observed much reduced channel activity in the measurements in the solution of 150 mM NaCl (Fig. 6D). The mean single-channel conductance for GmALMT12/QUAC1NaCl is 75.5 ± 4.5 pS (n = 5) and 58.5 ± 7.0 pS (n = 5) in the solutions of NaNO3 and NaCl, respectively (Fig. 6G). The channel open probability displays a strong voltage dependency, much higher at positive voltage than at negative one (Fig. 6H). As in our experimental setup, positive currents mean anions flow from the cis chamber (representing cytoplasm) to the trans chamber (representing extracellular side), and vice versa. Therefore, these results demonstrate that GmALMT12/QUAC1 functions as an anion efflux channel (corresponding to anions flow from cis to trans chamber). In addition to these, we tried to perform single-channel recording in 75 mM malate solution but failed due to the instability of the bilayer in high malate concentration. Whether malate acts as a permeable anion or a gating modifier for GmALMT12/QUAC1 remains elusive.
Malate regulation on GmALMT12/QUAC1 channel gating
To investigate the effect of malate regulation on channel activity, we reproduced the same experiments with GmALMT12/QUAC1malate (purified in 75 mM l-malate) as above. We observed that GmALMT12/QUAC1malate exhibits a similar current amplitude but a much higher open probability than those with GmALMT12/QUAC1NaCl in both solutions of NaNO3 and NaCl (Fig. 6, E and F). The mean single-channel conductance for GmALMT12/QUAC1malate is 88.5 ± 2.4 pS (n = 4) and 62.7 ± 4.1 pS (n = 4) in the solutions of NaNO3 and NaCl, respectively (Fig. 6G). The open probabilities of GmALMT12/QUAC1malate also display a similar voltage dependency as observed for GmALMT12/QUAC1NaCl (Fig. 6H). We found that the proteins purified in the presence of 75 mM malate or 150 mM NaCl resulted in distinct channel behaviors in the PLB recordings. This observation suggests that malate interacts with GmALMT12/QUAC1 and modulates gating by up-regulating channel open probability.
To further examine malate regulation on the channel gating, 2 mM l-malate was added to the solutions upon GmALMT12/QUAC1NaCl fused into a lipid bilayer. The channels became more active after adding 2 mM l-malate to either the trans or cis chamber (fig. S5, A and B). The current amplitude slightly increases, whereas the open probability becomes much higher (fig. S5, C and D). This observation confirms that malate can stimulate channel activity via enhanced open probability. Our findings also suggest that the malate regulation site may be located within the pore and can be accessed from either the extracellular or intracellular side of the membrane. Together, we propose that permeable anions may interact with charged residues within the channel pore and regulate gating. We also speculate that the GmALMT12/QUAC1 structure with an open pore in the presence of 75 mM l-malate may represent a malate-activated state, although the malate-binding site remained unclear due to insufficient information.
Essential role of the juxtamembrane pre-TM helix in channel function
The sequence analysis shows that the N-terminal segment before TMD is one of the most divergent regions in the ALMT/QUAC family (fig. S2). The GmALMT12/QUAC1 structure reveals that this region contains a juxtamembrane pre-TM helix (Fig. 1B), enriching positively charged residues. To examine its functional role, we designed a truncation construct (∆1–53). Unexpectedly, we found that the ∆1–53 mutant showed nearly null currents in the TEVC recordings (fig. S6). The observation demonstrates that the intracellular pre-TM helix region plays a vital role in channel function. Considering its adjacent association with the inner leaflet of the membrane, we propose that the pre-TM helix region may serve as a lever to regulate the proper conformation and function of the GmALMT12/QUAC1 channel.
The crucial function of C-terminal dimeric interaction
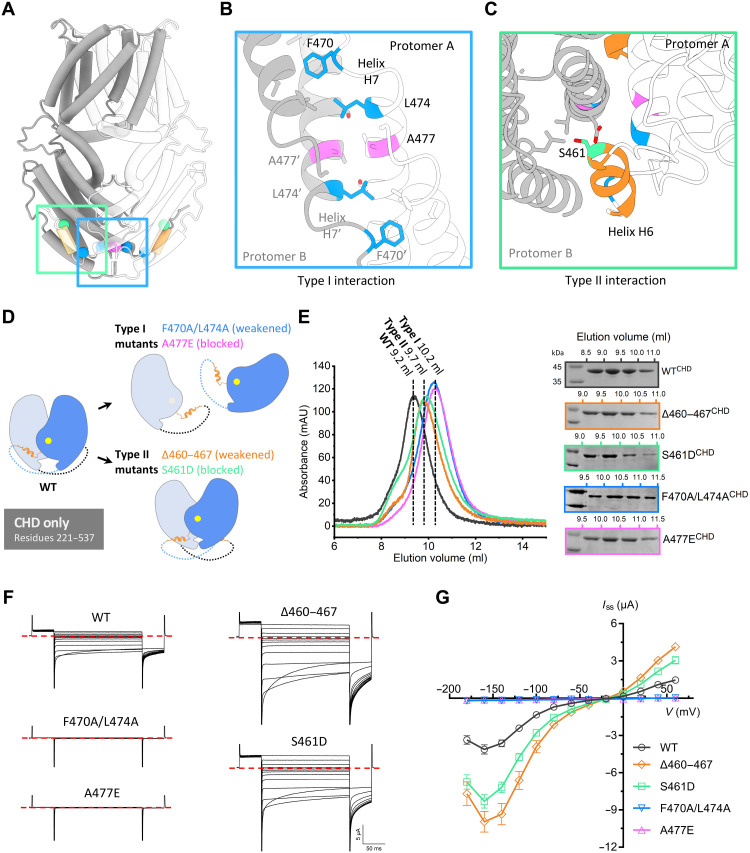
The cytosolic C-terminal portion is mainly formed by α helices, except for a large divergent region between helices H5 and H6 that is disordered or missing in the ALMT/QUAC family (fig. S2). The two CHDs in GmALMT12/QUAC1 are not isolated, but tightly interact at the distal end of the molecule, resulting in a domain-twisted architecture (Fig. 2A). As the CHD-mediated dimeric interactions are mainly mediated by helices H6 and H7, we simply classify them into two types of interactions (Fig. 7A). Type I zipper-like interaction is mediated by helix H7 from each protomer, arranged in an antiparallel manner. Two highly conserved hydrophobic residues (F470 and L474) protrude into the hydrophobic pocket formed by another protomer, forming a zipper-like interaction at the dimer interface (Fig. 7B). Type II domain-swapped finger helix interaction is mediated by a short finger helix H6, which is bound to the helical bundle formed by another subunit (Fig. 7C).
Fig. 7. Effect of dimeric interactions on channel activity.
(A) Cartoon of the GmALMT12/QUAC1 channel, with one protomer in gray and one in white. The domain-swapped helix H6 (residues 460 to 467) is colored in orange, and key residues at the dimeric interface are highlighted: S461 in green, A477 in magenta, and F470/L474 in blue. (B and C) Close-up views of the type I interaction (B) and type II interaction (C) at the cytoplasmic dimer interface. Residues at the dimeric interface are shown as sticks and colored as in (A). (D) Mutagenesis design for disrupting the interaction at the interface of dimeric CHDs. (E) Purification of the CHD proteins (residues 221 to 537, wild type or mutants) on a Superdex 75 (10/300) column. Compared to the wild type (peak at ~9.2 ml), the eluted peaks for the type I mutants (A477E and F470A/L474A) shift backward to ~10.2 ml, suggesting a disassociation of dimer into monomer. The elution peaks at ~9.7 ml for the type II mutants (∆460–467 and S461D) suggest altered conformations upon the removal of finger helix interaction. The eluted fractions were analyzed by SDS-PAGE, as indicated. (F and G) TEVC recording of GmALMT12/QUAC1 channel (wild type and mutants) in X. laevis oocytes. The same set of mutations, as in (D), was generated into full-length constructs for conductance measurement in the bath solution of 30 mM l-malate (data are means ± SEM, n ≥ 8). Representative current traces recorded at different voltages (from +60 to −180 mV in 20-mV decrement) are shown in (F). The steady-state current–voltage (Iss-V) relations are shown in (G). Data are means ± SEM, n ≥ 8.
To investigate the functional role of these dimeric interactions in channel regulation, we generated mutations, aiming to disrupt the type I zipper-like interaction (A477E and F470A/L474A) or to break the type II finger helix interaction (∆460–467 and S461D) at the dimeric interface (Fig. 7D). For the type I mutations, the substitution of a bulky acidic side chain in the mutant A477E is expected to block the CHD dimeric interface, or the removal of bulky side chains in the double mutant F470A/L474A will likely weaken the zipper-like interaction at the CHD dimeric interface. Similarly, the substitution of a bulky acidic side chain of the buried serine residue in the S461D mutant is expected to block the swapped-helix interaction, or the deletion of the helix H6 region in ∆460–467 mutant will abolish the finger helix–mediated type II interaction.
The above mutations were generated into the CHD (residues 221 to 537) bacteria-expression constructs and the full-length oocyte-expression constructs. We first characterized the oligomeric state of the purified CHD proteins by gel filtration chromatography. As seen from the gel filtration profile of the Superdex 75 (10/30) column, the elution volume of wild-type CHD protein (9.2 ml) was slightly smaller than that of bovine serum albumin (~67 kDa, 9.5 ml), suggesting that its apparent molecular weight may be slightly greater than 67 kDa. Thus, the wild-type CHD (monomer, ~38 kDa) likely maintains a dimeric form in solution. Both type I mutants (A477E and F470A/L474A) have an elution volume (10.2 ml) similar to that of ovalbumin (~43 kDa, 10.1 ml), suggesting that they are likely dissociated into monomeric forms (Fig. 7E). We also analyzed the purified CHD proteins (wild type and A477E mutant) by two different cross-linking experiments using glutaraldehyde. They both revealed that the wild-type CHD results in more cross-linked dimer than the A477E mutant under parallel conditions, as evidenced by the SDS–polyacrylamide gel electrophoresis (PAGE) analyses (fig. S7). This observation implies that type I mutations destabilize the interaction at the dimeric interface between CHDs. In comparison, both type II mutants (S461D or ∆460–467) are eluted at ~9.7 ml on the same Superdex 75 column, implying a distinct conformation from those of wild-type and type I mutants (Fig. 7E). This finding suggests that the removal or release of finger helix interactions within two CHDs may lead to altered dimeric structure with rearranged CHDs.
With the understanding of biochemical analyses for these mutations in CHD, we further investigated their functional effects on the full-length channel using oocyte-expression constructs by electrophysiology. We performed all experiments parallel to minimize variations in expression levels due to experimental factors, such as complementary RNA (cRNA) dose and expression time. As a result of disrupting the zipper-like interaction at the dimeric interface, the type I mutations cause nearly null currents in the conductance measurement with bath solution of 30 mM l-malate (Fig. 7, F and G). It is worth mentioning that the CHD dimer-interface disrupting mutations do not necessarily cause disassociation of full-length channel into monomeric subunit, as the dimeric interaction in TMD portion is intact. These observations demonstrate that the disruption of the CHD dimeric interface completely abolished the channel activity. Therefore, type I interaction joins two CHDs together, and likely serves as an anchor point allowing torsional force transduction between CHD and TMD. Unexpectedly, type II mutations stimulate channel activities, evidenced by the substantially enhanced currents in TEVC recordings (Fig. 7, F and G). These findings demonstrate that the removal or release of finger helix interaction up-regulates channel activity. To examine the surface expression of the above mutants in oocytes, we fused green fluorescent proteins (GFPs) to the C terminus of the following proteins, including wild type, ∆460–467, S461D, and A477E (fig. S8). The fluorescence and confocal imaging show that their surface expressions were at similar levels. Therefore, the observed conductance changes in TEVC measurement are likely caused by altered channel properties.
Gating of the GmALMT12/QUAC1 channel
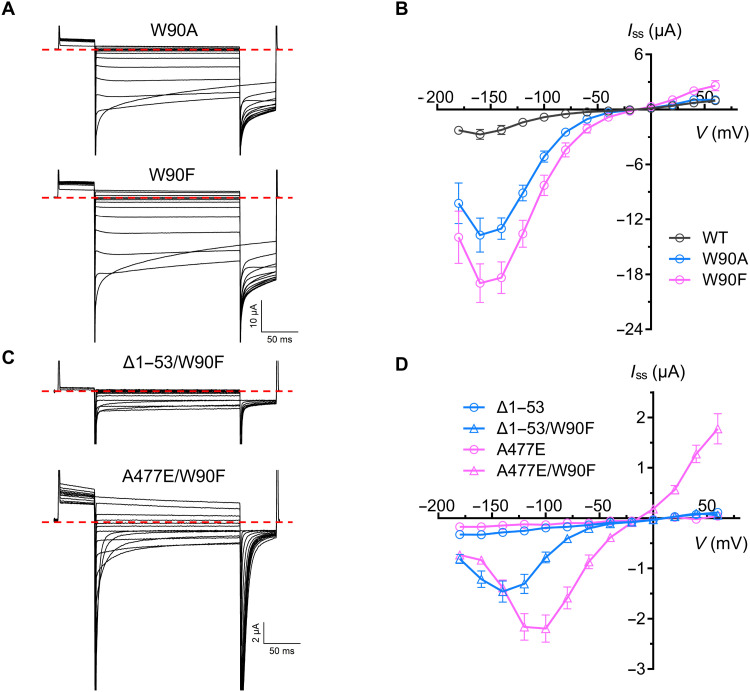
Our electrophysiological experiments have demonstrated that malate plays a vital role in modulating GmALMT12/QUAC1 activity; however, we wonder how malate regulation is coupled to channel activation. The GmALMT12/QUAC1 structure reveals that the conserved pore-lining tryptophan W90, located in the middle of the membrane, is in close contact with an unmodeled density within the fenestration (Fig. 4C and movie S1), raising an intriguing question if W90 is involved in channel activation. To investigate its functional role in channel regulation, we made mutations of W90A and W90F. Compared to wild type, the removal of indole ring in W90A mutant causes ~5 times larger conductance in the presence of external malate. A similar aromatic amino acid substitution in the W90F mutant unexpectedly results in even larger channel currents (Fig. 8, A and B). It seems that larger side-chain substitution (W90F versus W90A) leads to even larger enhanced channel activity. There is no simple explanation for the results from these mutations; however, this distinctive mutational effect suggests that W90 may interact with pore-lining residues and probably undergoes conformational changes during channel activation. Thus, we propose that the conserved W90 functions as a toggle switch to regulate channel gating.
Fig. 8. Analyses of GmALMT12/QUAC1 W90 mutants by TEVC recording.
(A and B) Representative current traces (A) and steady-state current–voltage (Iss-V) relations (B) of GmALMT12/QUAC1 mutants, W90A and W90F, at different applied voltages (from +60 to −180 mV in 20-mV decrement) in the external solutions of 30 mM l-malate (data are means ± SEM, n ≥ 6). (C and D) Representative current traces (C) and steady-state current–voltage (Iss-V) relations (D) of GmALMT12/QUAC1 mutants, Δ1–53/W90F and A477E/W90F, at different applied voltages (from +60 to −180 mV in 20-mV decrement) in the external solutions of 30 mM l-malate (data are means ± SEM, n ≥ 6).
This notion is confirmed in the conductance measurement of combined mutants of W90F with other two loss-of-function mutants, ∆1–53 and A477E. The null mutant ∆1–53 lacks its N-terminal pre-TM helix region but maintains a dimeric structure. Another null mutant A477E likely forms a loosened dimeric channel with disassociated CHDs. However, both mutants completely abolish channel activities, probably due to impaired coupling between malate regulation to channel gating. We found that the channel activity of both null mutants was partially restored to moderate levels after the addition of the W90F mutation (Fig. 8, C and D). The confocal imaging analysis indicated that the C-terminal GFP fusion of these mutants (W90F, A477E, and A477E/W90F) showed a similar expression level in oocytes (fig. S8). Therefore, the recovery of channel activity in the null mutants confirms that W90 can directly regulate channel gating.
Comparison of the cryo-EM structure with AlphaFold-predicted structure
Recently, AlphaFold has made significant progress in three-dimensional (3D) structure prediction from protein sequence (28). By taking advantage of this advanced technology, we generated an AlphaFold-predicted model for the GmALMT12/QUAC1 protein that is the focus of our current study. Unexpectedly, the AlphaFold-predicted GmALMT12/QUAC1 structure (AF-structure) is similar to that determined by cryo-EM (cryo-EM structure), with a root mean square deviation (RMSD) of 2.1 Å/160 Cα and 1.4 Å/207 Cα for TMD and CHD, respectively. However, further superimposition reveals a noticeable difference in the orientation of TMD versus CHD in GmALMT12/QUAC1, with an RMSD of 3.1 Å/388 Cα and 3.2 Å/776 Cα for the monomer and dimer, respectively (fig. S9A).
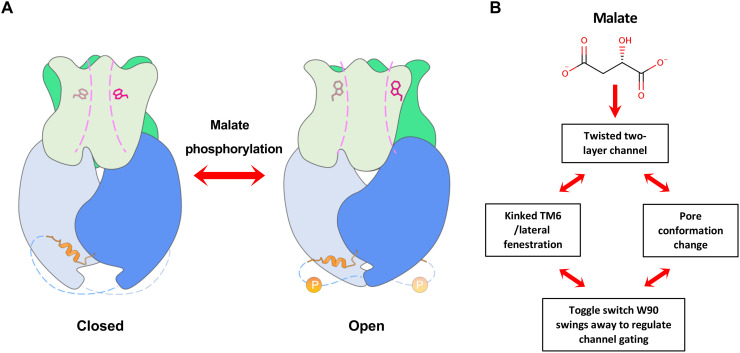
Unlike the kinked TM6 observed in the cryo-EM structure, TM6 in the AF-structure adopts a straight conformation, eliminating the lateral fenestration in TMD (fig. S9B). As the malate regulation effect on the channel is not taken into account, the resulting predicted model may represent an apo-structure in a resting state. The cryo-EM structure determined in the presence of malate reveals a high-energy twisted conformation with an open pore, thus representing a malate-activated structure. These two structures allow us to make comparisons and gain insight into the gating mechanism of the GmALMT12/QUAC1 channel. It seems that the pore-forming helices move slightly outward, as suggested by the porcupine plot on the Cα comparison between the AF-structure and the cryo-EM structure (fig. S9C). Another noticeable difference is the dihedral angle between the TMD and CHD interfaces, which is 93.5° and 83.0° for the AF-structure and cryo-EM structure, respectively (fig. S9D and movie S2). This observation shows a rotation (~10.5°) between these two states, suggesting that domain reorganization occurs during the conformational conversion. The AF-structure shows a similar pore, except a narrow constriction near W90. Further investigation is needed to understand the functional role of the pore-lining residues. We also speculate that the domain reorganization is associated with malate regulation and subsequently triggers conformational changes of the toggle switch within the pore to activate the channel (fig. S9E). On the basis of our data, we propose the following mechanism for GmALMT12/QUAC1 activation: Upon malate binding, the twisted two-layer architecture in GmALMT12/QUAC1 undergoes intramolecular domain reorganization, which subsequently induces the conformational change of the toggle switch W90. These changes promote the channel conversion from the basal to the activated state (Fig. 9 and movie S3).
Fig. 9. The proposed model for GmALMT12/QUAC1 regulation.
(A) Model for the GmALMT12/QUAC1 channel regulation. (B) Upon malate binding, GmALMT12/QUAC1 undergoes intramolecular domain reorganization within the twisted two-layer architecture and subsequently induces the conformational change of the toggle switch W90. These changes promote the channel conversion from closed to open state. Other modulations, such as phosphorylation or calmodulin binding, can further enhance the malate-mediated activation, probably via the release of its inhibitory domain-swapped helix from the interprotomer interaction.
DISCUSSION
GmALMT12/QUAC1 is a candidate for R-type anion channel
Our work provides a glimpse of the molecular structure of the ALMT12/QUAC1 anion channel. The structure of GmALMT12/QUAC1 reveals a twisted two-layer (TMD and CHD) architecture (Fig. 2A). Two interacting CHDs in the channel are tightly bound together in the cytoplasm, forming a T-shaped bifurcated pore across the membrane (Fig. 3A). Highly conserved charged residues line within the pore, rendering the pore electropositive for conducting anions (Fig. 3, C to E). The TMD and CHD portions interact in a twisted manner, rendering the channel in a high-energy state. Under such a constrained conformation, the channel will frequently switch between the open and closed states, resulting in rapid kinetics in channel gating (movie S2). Therefore, this unique architecture design provides a structural basis for ALMT12/QUAC1 function as an R-type channel.
Two types of interactions between dimeric CHDs play distinctive roles in regulating channel function (Fig. 7, B and C). Type I interaction, mediated by helix H7 in a zipper-like fashion, not only joins the two CHDs together to form the unique architecture with a T-shape bifurcated pore but also plays an essential role in the regulation of channel gating, including activation/deactivation kinetics and voltage dependency. Biochemical and functional analyses on type I interaction suggest that CHDs interact together and form an anchoring point at the distal end of the molecule, thus providing a structural basis for the coupling of mechanical force transduction between channel regulation and gating. Type II interaction, mediated by finger helix H6 in a domain-swapped manner, is not essential but involved in modulating channel activity. Our analysis suggests that the finger helix interaction regulates channel conformation and may be related to high-energy state transitions in the domain-twisted architecture. Type II interaction plays an inhibitory role in channel modulation, as its release enhances the channel activity. The sequence corresponding to the finger helix H6 is highly conserved in the QF2B subfamily (fig. S2), suggesting that type II interactions and their associated regulation may be subfamily specific. The upstream region of helix H6 represents one of the most divergent regions of the ALMT/QUAC family (fig. S2), harboring potential phosphorylation residues (Ser/Thr) for channel modulation. Thus, finger helix H6 and its associated type II interaction may offer additional elements for other modulations, such as kinase phosphorylation or calmodulin binding (Fig. 9A).
Tryptophan W90, a bulky residue in the middle of the channel pore, may act as a toggle switch to regulate gating. Removal of the indole ring (W90A) or replacement with a similar phenyl ring (W90F) results in up-regulation of channel activity (Fig. 8, A and B). The direct regulation of channel gating by W90 was further confirmed by the restoration of channel activity by W90F in null mutants (∆1–53 and A477E). The W90F-mediated currents share similar voltage dependency and kinetics of deactivation with those of the wild-type channel; however, the A477E/W90F mutant–mediated currents exhibit altered voltage dependency and quicker kinetics of deactivation (Fig. 8). These observations suggest that the CHD-mediated dimeric interaction plays a vital role in regulating voltage dependency of the channel conductance.
Similar to AtALMT12/QUAC1, TEVC measurement demonstrates that GmALMT12/QUAC1-mediated currents display rapid kinetics of activation/deactivation and a bell-shaped voltage dependency, reminiscent of the R-type anion currents (Fig. 5). Furthermore, the gating behaviors of GmALMT12/QUAC1 channel in PLB recordings exhibit flickering features that may represent the rapid characters of single-channel opening and closing for R-type channels (Fig. 6, C to F). This is consistent with our observation of the high-energy domain-twisting architecture of the GmALMT12/QUAC1 channel. Together, our work provides structural and functional evidence supporting ALMT12/QUAC1 as an R-type anion channel. However, further studies are needed to confirm that ALMT12/QUAC1 forms the R-type anion channel, especially genetic evidence.
Malate binding and channel regulation
The highly conserved charged residues within the pore form a network of interhelix interaction for interplaying with permeable anions (Fig. 3E). In TEVC measurements, the GmALMT12/QUAC1-mediated currents show rapid kinetics of activation/deactivation, but different external anions (malate2−, NO3−, and Cl−) in the bath solution lead to distinctive steady-state currents and voltage dependency (Fig. 5). Similar phenomena have been reported for AtALMT12/QUAC1 (29). We suggest that the distinction among different anions is likely caused by their interaction with charged residues within the channel pore. Larger anions (e.g., malate2− and other organic anions) may form more interactions than smaller anions (e.g., Cl−), leading to different channel activity and voltage dependency. Therefore, we speculate that anions with different sizes/charges may act as door wedges to regulate channel gating.
Since TEVC and PLB establish their virtual grounding oppositely (see Materials and Methods for details), the positive currents recorded in PLB correspond to the negative currents observed in TEVC, both representing anion efflux from the cytoplasm (Figs. 5 and 6). The GmALMT12/QUAC1 proteins reconstituted in lipid bilayer exclusively mediate outward currents, consistent with its function as an anion efflux channel. However, discrepancies are also found here, as we also observed noticeable outward currents in TEVC measurements, indicating external anion influx into the cell (Fig. 5). Similar outward currents were also described for AtALMT12/QUAC1 (29). This may be due to the passive movement of the anion across the open channel driven by the membrane potential and ionic gradient in TEVC recordings.
TEVC measurements indicate that malate up-regulates GmALMT12/QUAC1 channel activity. To further investigate the modulatory effect of malate, we performed single-channel recording in lipid bilayers with proteins purified in 75 mM malate or 150 mM NaCl. We elucidated in the PLB recordings that the malate regulatory effect is mainly through increasing the open probability (Fig. 6 and fig. S5), possibly via direct binding to the channel. This notion is further confirmed in the experiments of adding 2 mM malate to either cis or trans chamber in PLB recording with protein purified in NaCl (fig. S5). Furthermore, this finding also suggests that malate can act from either the extracellular or intracellular side and that the site of action is likely located inside the pore.
The anion channels: SLAC1 versus ALMT12/QUAC1
Tight regulation of stomatal pores is crucial to the land plants, as excessive water loss is devastating, and insufficient CO2 intake is also harmful. It is well established that guard cell anion effluxes are essential events to initiate stomatal closure in response to environmental stimuli (30). Previous studies showed that the guard cells harbor two distinct types of anion channels: rapid (R)–type and slow (S)–type (15–17, 31, 32). The R-type channel is voltage-dependent; it is activated by depolarization within milliseconds, inactivated during prolonged stimulation, and deactivated rapidly at hyperpolarized potentials (17). Extracellular malate can slow down the deactivation process (18). In contrast, the S-type channel shows weak voltage dependence and takes up to several tens of seconds to be activated, followed by slow inactivation kinetics (17).
Recent work of slow anion channel 1 (SLAC1), an S-type anion channel found in Arabidopsis (33–35), has uncovered detailed mechanisms underlying S-type channel activity (36–41). Our current work suggests that ALMT12/QUAC1 is the candidate for an R-type channel. SLAC1 and ALMT12/QUAC1 differ in structure, apparently, in the mechanism for activation and regulation. SLAC1 has a trimeric architecture with three independent pores, gated by two interplayed phenylalanines. In contrast, ALMT12/QUAC1 forms an entirely different dimeric assembly with a single T-shaped bifurcated pore. Distinct from the SLAC1 regulation controlled by phosphorylation and dephosphorylation via various protein kinases and phosphatase (36–40), the ALMT12/QUAC1 gating is directly regulated by malate, which is much faster. These findings may explain the fast kinetics observed in the ALMT12/QUAC1 channel, compared with the slow activity found in the S-type channel.
As both SLAC1 and ALMT12/QUAC1 represent two essential distinct channels that mediate osmotic active anion efflux from guard cell, one remaining puzzle is how they interplay to initiate stomatal closure in response to various environmental stimuli. Further investigation to address this issue will shed light on the understanding of guard cell stomatal signaling and provide critical information for engineering drought-resistant or water-use efficiency crops.
MATERIALS AND METHODS
Bioinformatics analysis of ALMT/QUAC proteins
Sequences related to ALMT/QUACs were searched and analyzed by using PSI-BLAST (42). Searches at E < 5 × 10−3 starting with 13 individual Arabidopsis ALMT/QUACs identified a common pool of over 1700 plant ALMT/QUAC-related protein sequences, which were pooled together and used for subclassification into families and subfamilies. Detailed information is reported in the footnotes in table S1.
Cloning and expression of ALMT12/QUAC1 in yeast Schizosaccharomyces pombe
Full-length coding sequences of six plant ALMT12/QUAC1s from rice, wheat, corn, soybean, tomato, and cotton were cloned into a modified pREP1 vector with a C-terminal 10× His tag. The resulting constructs were transformed into a leucine auxotrophic Schizosaccharomyces pombe strain, and the transformants were selected on the standard Edinburgh minimal medium (EMM) plates without leucine, as previously described (43). To prevent protein expression during strain growth, 25 μM thiamine was added to inhibit the promoter.
Scaled-up production and purification of GmALMT12/QUAC1
To prepare the seed, transformed cells were inoculated into 100 ml of EMM culture medium supplemented with 25 μM thiamine and were shaken at 200 rpm and 30°C for 24 hours. For protein expression and scaled-up production, the seed cells were collected by centrifugation and washed with sterile water twice before inoculation to a culture of 500 ml. After 12-hour growth, 500 ml of fresh medium was supplemented, and the culture continued to grow for an additional 24 hours. Cells were harvested through centrifugation for 20 min at 4500 rpm.
For protein purification, cells were resuspended in lysis buffer [50 mM tris-HCl (pH 8.0), 200 mM NaCl, 2.5% glycerol, aprotinin (1 μg/ml), leupeptin (1 μg/ml), pepstatin (1 μg/ml), 2 mM phenylmethylsulfonyl fluoride, and 2 mM dithiothreitol] and lysed using a high-pressure cell disrupter (JNBIO) with four passes at ~18,000 psi. Cell debris was removed by centrifugation at 12,000 rpm for 15 min, and the supernatant was subjected to a further ultracentrifugation at 41,000 rpm for 1 hour. The membrane was collected and homogenized in a solubilization buffer [50 mM tris-HCl (pH 8.0), 200 mM NaCl, and 2.5% glycerol] and incubated with a final concentration of 1.0% DDM and 0.02% CHS by gentle stirring for 1 hour at 4°C. After ultracentrifugation at 35,000 rpm for 50 min, the resulting supernatant was purified by Ni2+-affinity column pre-equilibrated with the same solubilization buffer supplemented with 0.05% DDM and 2 mM tris(2-carboxyethyl)phosphine. After 20 column volumes of buffer wash, the protein was eluted with 350 mM imidazole in the solubilization buffer. The resulting GmALMT12/QUAC1 protein, without removal of His tag, was concentrated to ~10 mg/ml and loaded onto a Superose 6 (10/300) gel filtration column for further purification and detergent exchange. The gel filtration buffer contained 20 mM tris, 150 mM NaCl (pH 8.0, adjusted with HCl) or 75 mM l-malate (pH 8.0, adjusted with NaOH), and 0.005% LMNG. The elution fractions were analyzed by SDS-PAGE, and the peak fractions were collected and concentrated for functional analysis (~2 mg/ml) or making cryo-EM grids (~5 mg/ml).
Cryo-EM grid preparation and data acquisition
Four microliters of GmALMT12/QUAC1malate proteins (purified in 75 mM l-malate) was applied to newly glow-discharged holy carbon film grids (Au R1.2/1.3, 300 meshes, Quantifoil, Germany). The grids were blotted with force 3 and blotting time of 9.0 s at 100% humidity and 4°C and were vitrified by plunge freezing into liquid ethane using Vitrobot Mark IV (Thermo Fisher Scientific, USA). All movies were collected on a Titan Krios G2 transmission electron microscope (Thermo Fisher Scientific, USA) operated at 300 kV, equipped with a Gatan K2 Summit direct detection camera (Gatan Company, USA) and a post-column GIF energy filter.
Data collection was performed on energy-filtered transmission electron microscopy mode with a silt width of 20 e−·V. The magnification was set to a nominal ×130,000, corresponding to a calibrated pixel size of 1.04 Å/pixel at the specimen level (0.52 Å/pixel in superresolution mode). Images were recorded using SerialEM (version 3.8.4) (44) with a beam-image shift method (45). During the 8-s exposure, 32 frames were collected with a dose of around 60 e−/Å2. A total of 6189 movie stacks were collected, with defocus ranging from −1.2 to −2.2 μm.
Image processing and model building
For cryo-EM image processing, all main steps were performed using RELION 3.0 (46) or cryoSPARC 3.1 (47). Pyem (48) and UCSF Chimera (49) were used for format conversion of data files and reconstructions analysis, respectively.
Briefly, 6189 movie stacks were aligned by 5 × 5 patches with dose weighting and binned to 1.04 Å using MotionCor2 (50). The contrast transfer function parameters were estimated using Gctf (51). Micrographs with defects in the thon rings were discarded. A total of 2,987,283 candidate particles were initially picked using Gautomatch without template (52), and followed by several rounds of 2D classification in cryoSPARC, giving a set of 2,136,754 high-quality particles. A rough initial model was generated by a subset of 100,000 particles. Subsequentially, six rounds of 3D classification were performed at C1 symmetry. Three parallel 3D classifications were performed within each round, and the resulting good particles were combined for sequential round classification. After that, a set of 428,433 particles that generated a map of 4.0 Å using NU-refinement in cryoSPARC was subjected to Bayesian polishing. Three rounds of 3D classification were further performed as described below. Each round contains an NU-refinement at C2 symmetry in cryoSPARC followed by an alignment-free 3D classification with a micelle-free mask in RELION. Last, NU-refinement of 169,576 particles yielded a reconstruction with a resolution of 3.5 Å, based on the gold-standard Fourier shell correlation using the 0.143 criterion. Density modification was performed using PHENIX (53). Local resolution was determined using ResMap (54).
A de novo atomic model was built manually in Coot (55) using the predicted secondary structure from MPI quick2d (56). The model was further refined using PHENIX (53). The final model was validated using EMRinger (57). All structure figures were prepared in UCSF ChimeraX (58).
TEVC recordings
The TEVC recordings were conducted as previously described (40). Briefly, all complementary DNAs (cDNAs) for GmALMT12/QUAC1, including wild type or mutants, were cloned into plasmid pGHME2 for expression in X. laevis oocyte. Linearized plasmids were used to generate cRNAs using T7 polymerase. Thirty-six nanograms of cRNA of each construct was injected into isolated oocytes. Oocytes were then incubated at 18°C for ~48 hours in ND96 buffer [96 mM NaCl, 1.8 mM CaCl2, 1 mM MgCl2, 2 mM KCl, and 5 mM Hepes-Na (pH 7.5)].
Using a Oocyte Clamp OC-725C amplifier (Warner Instruments) and a Digidata 1550 B low-noise data acquisition system with pClamp software (Molecular Devices), TEVC recordings were performed in the bath solutions: 10 mM MES/tris (pH 5.6), 1 mM Ca(gluconate)2, 1 mM Mg(gluconate)2, 70 mM Na(gluconate), 1 mM LaCl3, 30 mM NaCl or NaNO3, or 30 mM l-malate (pH 5.6, prepared from l-malic acid with NaOH). Osmolarity was adjusted to ~220 mOsmol kg−1 with d-sorbitol. The microelectrode solutions contained 3 M KCl, and the bath electrode was a 3 M KCl agar bridge. Voltage-clamp currents were measured in response to 200-ms-long voltage steps to test potentials that ranged from +60 to −180 mV in 20-mV decrement. Before each voltage step, the membrane potential was held at +60 mV for 50 ms, and following each voltage step, the membrane potential was returned to −180 mV for 50 ms. I-V relations for GmALMT12/QUAC1 channels were generated from currents measured at 1.5 ms after the beginning (instantaneous currents, Iinst) or 5 ms before the end (steady-state currents, Iss) of each test voltage step. Three independent batches of oocytes were investigated and showed consistent findings. Data from one representative oocyte batch were shown. The recordings were analyzed using Clampfit 10.6 (Molecular Devices) and Prism (version 5.0, GraphPad).
PLB recordings
Lipid bilayer experiments were conducted as previously described in our study of the ZAR1 channel (59). The purified GmALMT12/QUAC1 (in 150 mM NaCl or 75 mM l-malate), at a concentration of 1.5 to 2.5 μg/ml, was fused into PLBs formed by painting a lipid mixture of phosphatidylethanolamine and phosphatidylcholine (Avanti Polar Lipids) in a 5:3 ratio in decane across a 200-μm hole in a polystyrene partition separating the internal and external solutions in polysulfonate cups (Warner Instruments). The trans chamber (1.0 ml), representing the extracellular compartment, was connected to the head stage input of a bilayer voltage-clamp amplifier. The cis chamber (1.0 ml), representing the cytoplasmic compartment, was held at virtual ground. Solutions used for I-V relationship analysis were as follows: 150 mM NaCl or 150 mM NaNO3. All solutions were buffered with 20 mM tris (pH 8.0). Purified proteins were added to the cis side and fused with the lipid bilayer.
Current traces were recorded every 1 to 2 min after application of the voltage to the trans side. Single-channel currents were recorded using Bilayer Clamp BC-525D (Warner Instruments), filtered at 800 Hz using Low-Pass Bessel Filter 8 Pole (Warner Instruments), and digitized at 4 kHz. All experiments were performed at room temperature (23 ± 2°C). The recordings were analyzed using Clampfit 10.6 (Molecular Devices) and Prism (version 5.0, GraphPad).
Biochemical characterization of CHDs
cDNAs encoding the C-terminal CHD (residues 221 to 537) of GmALMT12/QUAC1 (wild type or mutants) were subcloned into a modified bacterial expression vector pET-24a, with a C-terminal 10× His tag. Transformed Escherichia coli BL21 (DE3) cells were grown in a Terrific Broth medium containing kanamycin (50 μg/ml). Protein expression was induced in cells grown to an OD600 (optical density at 600 nm) of ~0.8 with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and followed by overnight cell growth at 16°C. Cells were collected and subsequently lysed using a high-pressure cell disrupter (JNBIO) with three passes at ~15,000 psi. The target protein was purified by using Ni2+ affinity chromatography and further analyzed by Superdex 75 (10/300) gel filtration chromatography and SDS-PAGE.
Chemical cross-linking experiments for CHDs
For chemical cross-linking experiments, purified CHD proteins (wild type and A477E mutant) were concentrated to ~0.2 mg/ml. Equal amounts of purified CHD proteins were incubated with 0.01% glutaraldehyde at 4°C for 3 hours. Then, a final concentration of 50 mM tris-HCl (pH 8.0) was added to quench the reactions. Samples were further analyzed by 10% SDS-PAGE. For parallel comparison, the protein amount and cross-linking conditions were the same for wild type and A477E CHD at each step.
The glycerol gradient fixation (GraFix) was generated by an upper buffer containing 5% (v/v) glycerol, 20 mM Hepes-Na (pH 7.5), and 200 mM NaCl, and a lower buffer containing 20% (v/v) glycerol, 20 mM Hepes-Na (pH 7.5), 200 mM NaCl, and 0.15% (v/v) glutaraldehyde. Purified CHD proteins were concentrated to ~0.4 mg/ml, and a 0.2-ml sample was loaded onto the top of the gradient. Ultracentrifugation was performed at 40,000 rpm in a Beckman SW41Ti rotor for 18 hours at 4°C. The fractions of 0.5 ml were collected and quenched with a final concentration of 80 mM glycine. Fractions were further analyzed by 10% SDS-PAGE.
Fluorescence and confocal imaging
To examine the surface expression of GmALMT12/QUAC1 wild type and mutants on the oocyte plasma membrane, we fused GFP to the C terminus of the following proteins, including wild type, ∆460–467, S461D, and A477E. After incubation at 18°C for ~48 hours in ND96 buffer (see the “TEVC recordings” section for details), the oocytes expressed with GFP-tagged proteins were analyzed with a confocal laser microscope. The fluorescence intensities were measured with ImageJ software. We performed all experiments parallel to minimize variations in expression levels due to experimental factors, such as cRNA dose and expression time.
Three-dimensional structure modeling using AlphaFold
Briefly, the sequence for AlphaFold prediction was generated by two individual GmALMT12/QUAC1 sequences, connected by a linker of tandemly repeated sequence of (GSGS)50. Running with the local installed AlphaFold2 program and the CASP14 preset and databases (as of 23 July 2021), five dimer models were generated, with their pLDDT (predicted lDDT-Cα) scores ranging from 64.6 to 68.8. All the AlphaFold-predicted structures (AF-structure) are nearly identical, with apparent twofold symmetry. The two protomers in the top-rank dimer model (scored 68.8) display an RMSD of 0.92 Å. One protomer was selected for further structural comparison with the cryo-EM structure. For a better view, the disordered (residues 1 to 35, 394 to 454, and 507 to 537) regions were removed in the final model.
Acknowledgments
We thank the staff at the Center for Biological Imaging (CBI), Institute of Biophysics, Chinese Academy of Sciences.
Funding: This project is financially supported by the National Key Research and Development Program of China (2020YFA0509903 and 2016YFA0500503 to Y.-h.C., 2017YFA0504703 and 2020YFA0509901 to Y. Zhai, and 2021YFA1300702 to M.S.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24020305 to Y.-h.C. and XDB37040102 to F.S.), the National Natural Science Foundation of China (31872721 to Y.-h.C. and 31771566 to Y. Zhai), the International Partnership Program of Chinese Academy of Sciences (153E11KYSB20190029 to Y.-h.C. and F.L.), and NIH grant (1R35GM134920-01 to F.L.).
Author contributions: L.Q. performed protein purification, cryo-EM data collection, TEVC experiments, and data analysis. L.-h.T., J.-s.X., and Y. Zhu performed cryo-EM data collection and structural determination. X.-h.Z. performed mutagenesis and protein purification. M.S. performed PLB experiments and data analysis. C.r.Z., M.-h.W., and X.-l.L. performed experiments. F.L., F.S., and Y. Zhai analyzed data. Y.-h.C. initiated the project, planned and analyzed experiments, supervised the research, and wrote the manuscript with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The accession number for the 3D cryo-EM density map reported in this paper is EMD-32328, and the Protein Data Bank (PDB) accession code for the coordinate is 7W6K.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
REFERENCES AND NOTES
- 1.Meyer S., Scholz-Starke J., De Angeli A., Kovermann P., Burla B., Gambale F., Martinoia E., Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J. 67, 247–257 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Meyer S., Mumm P., Imes D., Endler A., Weder B., Al-Rasheid K. A. S., Geiger D., Marten I., Martinoia E., Hedrich R., AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 63, 1054–1062 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Sasaki T., Mori I. C., Furuichi T., Munemasa S., Toyooka K., Matsuoka K., Murata Y., Yamamoto Y., Closing plant stomata requires a homolog of an aluminum-activated malate transporter. Plant Cell Physiol. 51, 354–365 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Angeli A., Zhang J., Meyer S., Martinoia E., AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nat. Commun. 4, 1804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovermann P., Meyer S., Hörtensteiner S., Picco C., Scholz-Starke J., Ravera S., Lee Y., Martinoia E., The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 52, 1169–1180 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Gutermuth T., Herbell S., Lassig R., Brosché M., Romeis T., Feijó J. A., Hedrich R., Konrad K. R., Tip-localized Ca2+-permeable channels control pollen tube growth via kinase-dependent R- and S-type anion channel regulation. New Phytol. 218, 1089–1105 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Herbell S., Gutermuth T., Konrad K. R., An interconnection between tip-focused Ca2+ and anion homeostasis controls pollen tube growth. Plant Signal. Behav. 13, e1529521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki T., Yamamoto Y., Ezaki B., Katsuhara M., Ahn S. J., Ryan P. R., Delhaize E., Matsumoto H., A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37, 645–653 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Hoekenga O. A., Maron L. G., Piñeros M. A., Cançado G. M. A., Shaff J., Kobayashi Y., Ryan P. R., Dong B., Delhaize E., Sasaki T., Matsumoto H., Yamamoto Y., Koyama H., Kochian L. V., AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 9738–9743 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ligaba A., Maron L., Shaff J., Kochian L., Piñeros M., Maize ZmALMT2 is a root anion transporter that mediates constitutive root malate efflux. Plant Cell Environ. 35, 1185–1200 (2012). [DOI] [PubMed] [Google Scholar]
- 11.De Angeli A., Baetz U., Francisco R., Zhang J., Chaves M. M., Regalado A., The vacuolar channel VvALMT9 mediates malate and tartrate accumulation in berries of Vitis vinifera. Planta 238, 283–291 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Rudrappa T., Czymmek K. J., Paré P. W., Bais H. P., Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramesh S. A., Tyerman S. D., Xu B., Bose J., Kaur S., Conn V., Domingos P., Ullah S., Wege S., Shabala S., Feijó J. A., Ryan P. R., Gillham M., GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat. Commun. 6, 7879 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H., Li Y., Zhu J. K., Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants. 4, 989–996 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Keller B. U., Hedrich R., Raschke K., Voltage-dependent anion channels in the plasma membrane of guard cells. Nature 341, 450–453 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietrich P., Hedrich R., Interconversion of fast and slow gating modes of GCAC1, a Guard Cell Anion Channel. Planta 195, 301–304 (1994). [Google Scholar]
- 17.Schroeder J. I., Keller B. U., Two types of anion channel currents in guard cells with distinct voltage regulation. Proc. Natl. Acad. Sci. U.S.A. 89, 5025–5029 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mumm P., Imes D., Martinoia E., Al-Rasheid K. A. S., Geiger D., Marten I., Hedrich R., C-terminus-mediated voltage gating of Arabidopsis guard cell anion channel QUAC1. Mol. Plant 6, 1550–1563 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Hedrich R., Marten I., Lohse G., Dietrich P., Winter H., Lohaus G., Heldt H.-W., Malate-sensitive anion channels enable guard cells to sense changes in the ambient CO2 concentration. Plant J. 6, 741–748 (1994). [Google Scholar]
- 20.Raschke K., Alternation of the slow with the quick anion conductance in whole guard cells effected by external malate. Planta 217, 651–657 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Imes D., Mumm P., Böhm J., Al-Rasheid K. A. S., Marten I., Geiger D., Hedrich R., Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 74, 372–382 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Luu K., Rajagopalan N., Ching J. C. H., Loewen M. C., Loewen M. E., The malate-activated ALMT12 anion channel in the grass Brachypodium distachyon is co-activated by Ca2+/calmodulin. J. Biol. Chem. 294, 6142–6156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm L., DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuichi T., Sasaki T., Tsuchiya Y., Ryan P. R., Delhaize E., Yamamoto Y., An extracellular hydrophilic carboxy-terminal domain regulates the activity of TaALMT1, the aluminum-activated malate transport protein of wheat. Plant J. 64, 47–55 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Dutzler R., Campbell E. B., Cadene M., Chait B. T., MacKinnon R., X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415, 287–294 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S. P., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Peng W., Wu W., Peng J., Li J., Lin Y., Wang Y., Tian J., Sun L., Liang C., Liao H., Characterization of the soybean GmALMT family genes and the function of GmALMT5 in response to phosphate starvation. J. Integr. Plant Biol. 60, 216–231 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malcheska F., Ahmad A., Batool S., Müller H. M., Ludwig-Müller J., Kreuzwieser J., Randewig D., Hänsch R., Mendel R. R., Hell R., Wirtz M., Geiger D., Ache P., Hedrich R., Herschbach C., Rennenberg H., Drought-enhanced xylem sap sulfate closes stomata by affecting ALMT12 and guard cell ABA synthesis. Plant Physiol. 174, 798–814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pantoja O., Recent advances in the physiology of ion channels in plants. Annu. Rev. Plant Biol. 72, 463–495 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Schroeder J. I., Hagiwara S., Cytosolic calcium regulates ion channels in the plasma membrane of Vicia faba guard cells. Nature 338, 427–430 (1989). [Google Scholar]
- 32.Hedrich R., Busch H., Raschke K., Ca2+ and nucleotide dependent regulation of voltage dependent anion channels in the plasma membrane of guard cells. EMBO J. 9, 3889–3892 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vahisalu T., Kollist H., Wang Y. F., Nishimura N., Chan W. Y., Valerio G., Lamminmäki A., Brosché M., Moldau H., Desikan R., Schroeder J. I., Kangasjärvi J., SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452, 487–491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Negi J., Matsuda O., Nagasawa T., Oba Y., Takahashi H., Kawai-Yamada M., Uchimiya H., Hashimoto M., Iba K., CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452, 483–486 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Saji S., Bathula S., Kubo A., Tamaoki M., Kanna M., Aono M., Nakajima N., Nakaji T., Takeda T., Asayama M., Saji H., Disruption of a gene encoding C4-dicarboxylate transporter-like protein increases ozone sensitivity through deregulation of the stomatal response in Arabidopsis thaliana. Plant Cell Physiol. 49, 2–10 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K. A. S. S., Romeis T., Hedrich R., Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U.S.A. 106, 21425–21430 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S. C., Lan W., Buchanan B. B., Luan S., A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. U.S.A. 106, 21419–21424 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandt B., Brodsky D. E., Xue S., Negi J., Iba K., Kangasjärvi J., Ghassemian M., Stephan A. B., Hu H., Schroeder J. I., Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. U.S.A. 109, 10593–10598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maierhofer T., Diekmann M., Offenborn J. N., Lind C., Bauer H., Hashimoto K., Al-Rasheid K. A. S., Luan S., Kudla J., Geiger D., Hedrich R., Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci. Signal. 7, ra86 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Deng Y.-N., Kashtoh H., Wang Q., Zhen G.-X., Li Q., Tang L.-H., Gao H.-L., Zhang C.-R., Qin L., Su M., Li F., Huang X.-H., Wang Y.-C., Xie Q., Clarke O. B., Hendrickson W. A., Chen Y.-H., Structure and activity of SLAC1 channels for stomatal signaling in leaves. Proc. Natl. Acad. Sci. U.S.A. 118, e2015151118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y.-H., Hu L., Punta M., Bruni R., Hillerich B., Kloss B., Rost B., Love J., Siegelbaum S. A., Hendrickson W. A., Homologue structure of the SLAC1 anion channel for closing stomata in leaves. Nature 467, 1074–1080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J., Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X.-H., Su M., Gao F., Xie W., Zeng Y., Li D., Liu X., Zhao H., Qin L., Li F., Liu Q., Clarke O. B., Lam S. M., Shui G., Hendrickson W. A., Chen Y.-H., Structural basis for activity of TRIC counter-ion channels in calcium release. Proc. Natl. Acad. Sci. U.S.A. 116, 4238–4243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Wu C., Huang X., Cheng J., Zhu D., Zhang X., High-quality, high-throughput cryo-electron microscopy data collection via beam tilt and astigmatism-free beam-image shift. J. Struct. Biol. 208, 107396 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 48.D. Asarnow, E. Palovcak, Y. F. Cheng, UCSF pyem v0.5 (2019); doi: 10.5281/zenodo.3576630. [DOI]
- 49.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.K. Zhang, F. Sun, Gautomatch: An efficient and convenient gpu-based automatic particle selection program (2011); https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/.
- 53.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L.-W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in PHENIX. Acta Crystallogr. D. Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gabler F., Nam S.-Z., Till S., Mirdita M., Steinegger M., Söding J., Lupas A. N., Alva V., Protein sequence analysis using the MPI bioinformatics toolkit. Curr. Protoc. Bioinformatics 72, e108 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Barad B. A., Echols N., Wang R. Y. R., Cheng Y., Dimaio F., Adams P. D., Fraser J. S., EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bi G., Su M., Li N., Liang Y., Dang S., Xu J., Hu M., Wang J., Zou M., Deng Y., Li Q., Huang S., Li J., Chai J., He K., Chen Y.-H., Zhou J.-M., The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528–3541.e12 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Dreyer I., Gomez-Porras J. L., Riaño-Pachón D. M., Hedrich R., Geiger D., Molecular evolution of slow and quick anion channels (SLACs and QUACs/ALMTs). Front. Plant Sci. 3, 263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 and S2
References
Movies S1 to S3