Abstract
Three-dimensional (3D) hydrogels made from synthetic polymers have emerged as in vitro cell culture platforms capable of representing the extracellular geometry, modulus, and water content of tissues in a tunable fashion. Hydrogels made from these otherwise non-bioactive polymers can be decorated with short peptides derived from proteins naturally found in tissues to support cell viability and direct phenotype. We identified two key limitations that limit the ability of this class of materials to recapitulate real tissue. First, these environments typically display between 1 to 3 bioactive peptides, which vastly underrepresents the diversity of proteins found in the extracellular matrix (ECM) of real tissues. Second, peptides chosen are ubiquitous in ECM and not derived from proteins found in specific tissues, per se. To overcome this critical limitation in hydrogel design and functionality, we developed an approach to incorporate the complex and specific protein signature of bone marrow into a poly(ethylene glycol) (PEG) hydrogel. This bone marrow hydrogel mimics the elasticity of marrow and has 20 bone marrow-specific and cell-instructive peptides. We propose this tissue-centric approach as the next generation of 3D hydrogel design for applications in tissue engineering and beyond.
Keywords: 3D biomaterial, peptide, stiffness, tissue mimic, mesenchymal stem cell
The vast majority of materials available to study how environmental cues direct cell fate are two-dimensional (2D), ranging from protein-coated surfaces to hydrogels2–4. However, 2D materials restrict cell adhesions to an x-y plane and force an apical-basal polarity6,7. To overcome this, researchers can better recapitulate the in vivo geometry of tissues using hydrogels to culture cells in three-dimensional (3D) environments8. Synthetic hydrogels made from polyethylene glycol (PEG) precursors can be functionalized with peptide motifs that either elicit integrin-binding, allow for cell-mediated matrix degradation9, or bind growth factors and other molecules10. Additionally, PEG hydrogels are independently tunable in both stiffness and ligand density, and they do not have the same degree of batch-to-batch variability inherent to naturally derived protein hydrogels 11–13. For example, Nguyen et al. demonstrated that because synthetic gels were more reproducible they were superior in sensitivity for vascular toxicity screening when compared to Matrigel13.
Despite these advantages, synthetic hydrogels are sometimes considered inferior to more complex, protein-based hydrogels, like Matrigel, which may more accurately mimic the diversity of proteins found in real tissue. Part of this perception is because synthetic gels are decorated with a single cell-binding peptide, RGD, and a degradable crosslinker. Simple RGD-decorated hydrogels do not fully recapitulate the native tissue niche but dominate the synthetic biomaterial literature14. To compete with protein-derived materials, it is imperative to synthesize environments that include the diversity of integrin-binding and protease-sensitive proteins of real tissues. For example, despite clear evidence of the marrow extracellular matrix (ECM) regulating the stem cell niche 15,16, in vitro stem cell culture platforms contain a mere fraction of the biochemical cues typical of bone marrow. In this work, we propose a 3D ECM-inspired hydrogel containing PEG and 20 unique peptides. This tissue-inspired PEG hydrogel aims to capture the protein complexity of the native ECM of bone marrow in a synthetic material that is extremely tunable and can be fabricated with minimal technical expertise. These features enable new avenues for mechanistic research where the protein and mechanical properties of bone marrow can be tuned to understand their role in disease progression.
Our group has previously developed a comprehensive method to determine the ECM of real tissue and apply that knowledge to the design of synthetic tissues. In Galarza et al., we used this approach to develop a brain-mimicking hydrogel 17. However, no such hydrogel system exists for bone marrow, which is critically important given the increasingly appreciated role of the immune system in regulating whole body homeostasis and response to infection. In this work, we synthesized a hydrogel that contains important ECM cues from native bone marrow. Bone marrow is the soft interior tissue between hard compact bone where many immune and stromal stem cells reside. Like every human tissue, bone marrow has unique biophysical features that are critical for cell and organ function. For example, protein composition and tissue stiffness are essential for cellular processes like migration and proliferation1,5,18, as well as regulating stem cell fate and organoid development16,19–21. Thus, it is not surprising that the surrounding ECM plays a crucial role in the proper function of bone marrow, because both hematopoietic and stromal progenitor cells originate from the marrow22. For example, both bone marrow stiffness and fibronectin regulate maintenance of hematopoietic stem cell progenitors23. Additionally, marrow-derived stromal stem cells differentiate into either bone or fat cells in response to mechanical cues24, and the presence or absence of vitronectin in 3D scaffolds can facilitate reversible differentiation into or from osteoblasts25. Therefore, it is critical that in vitro cell culture environments include mechanical (stiffness) and chemical (ECM proteins) cues to study marrow cell biology, as we have done here.
A biomechanics and bioinformatics approach to design a human bone marrow mimicking synthetic hydrogel
We used a top-down engineering approach to identify the physical and chemical properties of bone marrow that could be represented in a synthetic, PEG-based hydrogel (Figure 1a–b). First, we measured the modulus of bone marrow via shear rheology, indentation, and cavitation rheology1. This modulus was then approximated with a PEG hydrogel (a network that is inherently hydrophilic and mimics marrow’s high-water content) by adjusting the crosslinking density. Having our synthetic, PEG-based hydrogel match the modulus is important, because the modulus of the ECM contributes to stem cell fate26. We therefore compared the compressive modulus of porcine marrow and our PEG hydrogel. Both the tissue and synthetic materials closely followed a Hertzian model under low strain regimes (Suppl. Figure 1a–b), suggesting that PEG hydrogels can appropriately model the modulus of marrow under these conditions.
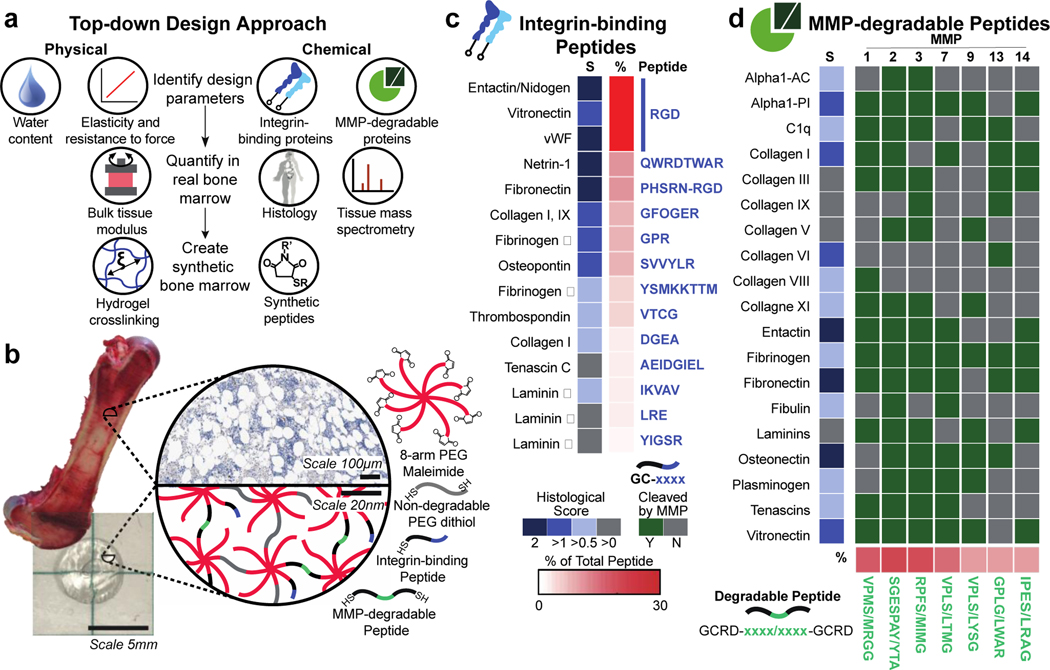
Figure 1. A PEG hydrogel designed to mimic the physical and chemical properties of bone marrow tissue.
a) Tissues have specific physical and chemical properties such as water content, elasticity, integrin-binding, and MMP-degradable proteins. These properties can be quantified in real bone marrow tissue using rheology, mass spectrometry, and tissue histology (Image of human adapted from Protein Atlas5). In PEG hydrogels, these features can be mimicked by tuning the polymer crosslinking density and incorporating peptides (histology from the Protein Atlas5). b) Here, bone marrow tissue (image of porcine bone marrow1) is mimicked with a hydrogel composed of an 8-arm PEG macromer functionalized (image of resulting hydrogel) with c) 13 cysteine-terminated integrin-binding peptides, and crosslinked with d) 7 di-cysteine-terminated MMP-degradable peptides and PEG-dithiol. The known functional sequence for each peptide is depicted in blue for integrin-binding proteins (up to the first 8 amino acids are depicted) and in green for the degradable peptides, where the slash (/) indicates the cleavage location for each enzyme on the matched peptide. Scales for the average histological score and the total percent of each peptide are shown by each peptide/protein pair (Y=yes, N=no, S=Histological Score).
To identify the ECM proteins in real marrow, we used a combination of mining published histology data and performing our own mass spectrometry on human marrow (Figure 1a). This search was narrowed to ECM proteins that mediate cell attachment via integrins and are susceptible to proteolysis by matrix metalloproteinases (MMPs). Integrins are the largest class of cell adhesion receptors that mediate attachment to the ECM and activate intracellular signaling27, and collectively the MMP family can degrade most proteins in the ECM28. To design our hydrogel, we annotated the known extracellular integrin-binding and MMP-degradable proteins in human bone marrow using the histology data from the Protein Atlas (Table S3)5. Then, we found the specific peptide sequences within these ECM proteins that are either responsible for high-affinity binding to integrins29, or are highly susceptible to cleavage by MMPs (Tables S1–2). The integrin-binding peptides were synthesized with a single cysteine to attach and be displayed in the hydrogel (Figure 1b–c, and full sequences in Tables S4–5)30–38, and the MMP-degradable peptides were synthesized with cysteines on each end to act as crosslinkers (Figure 1b,d, Table S6).
The histological scores available in the Protein Atlas were used to determine the relative molar peptide concentrations for each ECM protein included in the synthetic bone marrow hydrogel (Figure 1c–d). To validate this approach, ECM proteins were extracted from human bone marrow39 and analyzed via liquid chromatography-mass spectrometry (LC-MS, Figure S1c). The ECM proteins identified with LC-MS in human bone marrow matched the proteins identified using the Protein Atlas better than proteins from two control tissues: lung and brain (Table S4, Fig. S1d). We separately confirmed that human bone marrow tissue is degradable by the MMPs whose cleavage sites we selected for incorporation into the synthetic bone marrow hydrogel (Figure S1e). Together, these data confirmed our approach to identifying the integrin-binding and MMP-degradable protein signature of bone marrow.
Functional validation of bone marrow peptides
Human mesenchymal stem cells (MSCs) were used to test whether stromal cells highly abundant in the marrow could adhere to the integrin-binding peptides in our bone marrow hydrogel. We adapted a competitive cell adhesion assay to measure binding to integrin peptides3,40,41. This involved seeding MSCs in the presence or absence of individual peptides (soluble in the cell culture medium) onto coverslips that had the full integrin-binding peptide cocktail (Figure 1c) covalently attached (Figure 2a). When cells were pre-treated with soluble peptides, we observed a decrease in cell area on the coverslip (Video S1 and S2), which we hypothesized is from the peptides in solution competing for integrin receptors on the cell membrane. We measured cell area two hours after treatment to quantify whether the peptides in solution competed for integrins and therefore blocked adhesion and spreading on the coverslips (Figure 2a–b). We seeded cells onto coverslips that did not have any peptides attached, and quantified protein adsorption (Figure S2a–b), to ensure the cells were binding to the coverslip because of interactions with the BM peptides, and not due to serum from the medium or other non-specific protein binding.
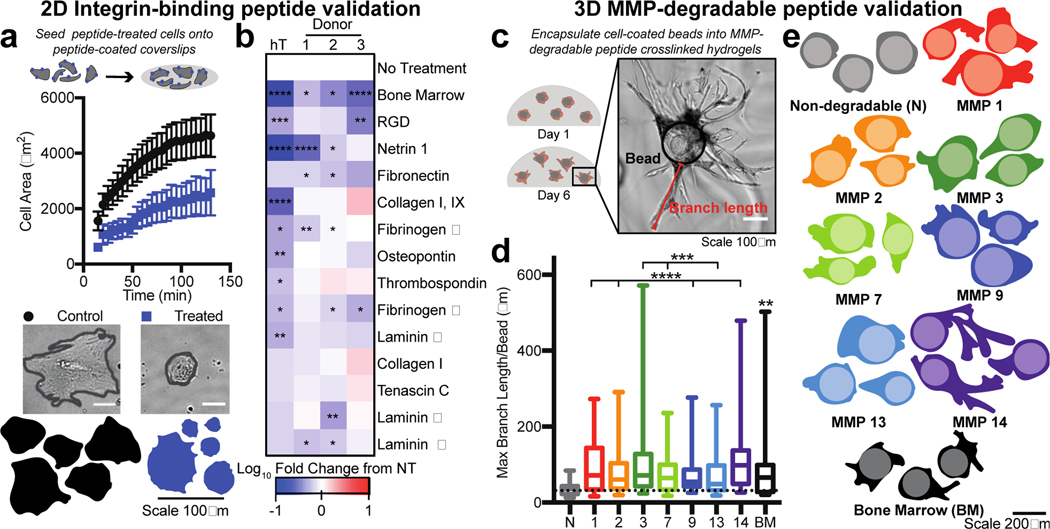
Figure 2. Validation of bone marrow hydrogel peptides.
a) Cells were treated with peptides in solution (medium), and then seeded onto coverslips coated with the bone marrow integrin-binding peptide cocktail. MSC area was measured over approximately 2 hours for cells not treated (control, black) or pre-treated for 30 minutes prior (blue) with soluble integrin-binding peptides and allowed to adhere to a coverslip coupled with all the integrin-binding peptides included in the bone marrow hydrogel design. Representative cell images (scale bar = 50 μm) and traces of MSCs 2 hours after seeding (bottom). Error bars represent SEM. b) Heat map depicting the log10 fold change in cell area at 2 hours compared to no treatment (NT) for each integrinbinding peptide for hTERT MSCs (hT) and three donor MSCs (1–3) (BM=bone marrow peptide cocktail) (N≥2, n≥20 per cell). c) Representative image of MSCs seeded on cytodex beads (black outline) and encapsulated into a hydrogel with MMP degradable crosslinkers (Cell area=red, branch length=green). d) A box and whisker plot for the maximum branch length per bead in each hydrogel condition. e) Representative cell and bead traces in each hydrogel condition, where the lighter colored circle is the bead and the darker color is the cell trace (N=2, n≥15 per cell). Significance is determined using a two-tailed t-test. P-values <0.05 are considered significant, where p<0.05 is denoted with *, ≤0.01 with **, ≤0.001 with ***, and ≤0.0001 with ****.
Three MSC sources from human donors and one immortalized MSC cell line had decreased adhesivity when dosed with the bone marrow integrin-binding peptide cocktail (Figure 2b, Figure S2c). Most individual peptides decreased MSC spreading at the concentration at which they were present in the cocktail (Figure 2b), with the immortalized MSC cell line being more responsive to individual peptides compared to the primary cells. The Collagen I and Tenascin C peptides did not significantly regulate MSC adhesion. We then tested the ability for these peptides to regulate adhesion of human breast cancer cells, which can metastasize to the bone marrow, and found that their adhesion was strongly influenced by the bone marrow peptides (Figure S2c). Overall, this data suggested that each peptide in the mimic could influence cell adhesion to and from our hydrogel matrix.
The peptide sequences for MMP degradation were validated using a cell invasion assay. Cytodex beads were coated with MSCs to standardize cell seeding and simplify the image analysis of branching length. Beads were encapsulated for six days in hydrogels crosslinked with a single MMP-degradable peptide or the full set of degradable crosslinks (Figure 2c). All gels were crosslinked at the same molar ratio of reactive thiols to maleimide groups. In all cases, when degradable peptides were present, MSCs invaded further into the surrounding hydrogel network (Figure 2d–e). MSCs branched the furthest in the bone marrow-cocktail, MMP-3, and −14 crosslinked hydrogels. This suggests that specific individual peptides can be extremely susceptible to degradation and peptide combinations like the combination used here enhance material degradation by bone marrow cells.
Determining the optimal chemical conditions for coupling marrow-specific peptides
We coupled the bone marrow peptides to the hydrogel matrix using a Michael-type addition reaction, which is biocompatible and provides the most efficient incorporation of ligands and the largest range of bulk properties compared to other PEG hydrogels42. Additionally, this reaction can be performed in a biocompatible buffer without UV-crosslinking to maximize cell viability upon encapsulation43. The kinetics of this reaction and the resulting polymer structure in PEG-gels has been studied extensively by our lab and others, so we chose to focus our characterization on the coupling efficiency of peptides44–46. Since the Michael-type donor for this reaction is a thiol, we used a thiol quantification assay to identify uncoupled peptides in solution and ensure that all the peptides reacted with the PEG matrix (Figure S3a). We also found that several parameters regulated the efficiency of peptide incorporation, including polymer wt% and the molar percentage of reactive pairs (Figure S3b–e). While all of these properties also change the effective Young’s modulus of the hydrogel, we determined that an 8-arm PEG gave us the best trade-off: it enabled increased crosslinking without increasing the number of unreacted thiols (Figure S3d).
We exceeded 98% coupling of integrin-binding peptides and 97% coupling of MMP-degradable peptides to an 8-arm PEG-maleimide at 20 wt% (Figure 3a–b). Optimal reaction conditions for integrin-binding peptides occurred in PBS at pH 7.4, but we did note that the peptide cocktail was less soluble in PBS than in DMSO (Figure S3f–g). Separately, we chemically reduced the hydrogel using sodium borohydride to ensure that the disulfide bonds between the thiols were not competing with the Michael-addition reaction. With this assay we found that more than 95% of the hydrogel bonds were from the Michael-type addition reaction (Figure S3h).
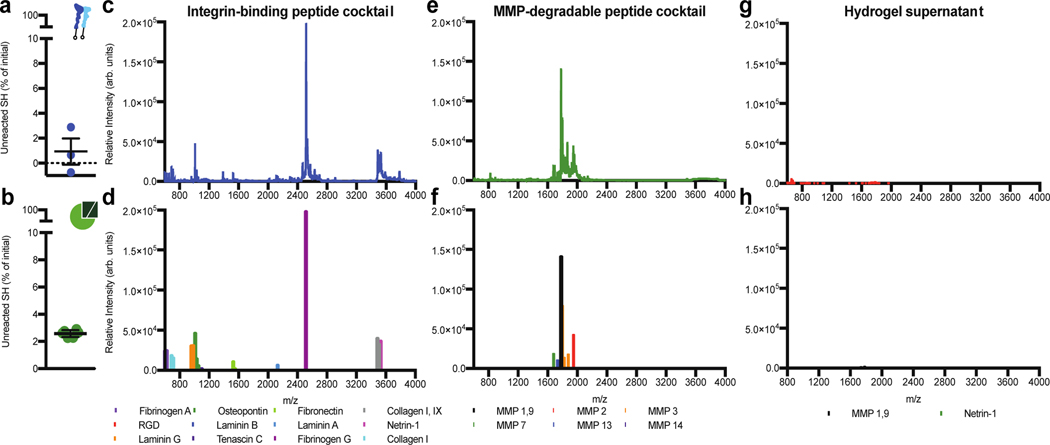
Figure 3. Bone marrow peptides couple to the hydrogel at expected concentrations.
a) The percentage of unreacted thiols when integrin-binding peptides were added to a solution of PEG-maleimide dissolved in PBS at pH 7.4. b) The percentage of unreacted thiols 10 minutes post-crosslinking an 8-arm PEG hydrogel at a 1:1 molar ratio of thiol to maleimide. Error bars represent the SEM (N≥1, n≥3). MALDI-TOF spectrum (top) and identified peptide peaks (bottom) for the c) and d) bone marrow integrin-binding peptides; e) and f) the bone marrow MMP-sensitive peptide crosslinkers, and g) and h) the supernatant of a bone marrow hydrogel swelled for 4 hours in PBS.
We used matrix-assisted laser deposition ionization time of flight (MALDI-TOF) to identify which peptides couple to the hydrogel less efficiently than others. We first made a solution of all the peptides, without PEG present, and identified all except DGEA and AEIDGIEL (Figure 3e–f). These are highly negatively charged peptides, which do not ionize readily, which explains why we could not identify them in a heterogeneous peptide solution. Hydrogels were then formed with all the peptides, swollen in water, and we attempted to identify any unreacted peptides from the lyophilizegd supernatant. Only two peptides were discovered, with a significantly reduced intensity (Figure 3gh). Together, this data shows that our peptides are both crosslinked into the hydrogel and at their expected concentrations.
PEG hydrogels mimic the bulk modulus of bone marrow
The mechanical properties of biomaterials and tissues are known to influence the migration and differentiation of both MSCs and hematopoietic stem cells 24,26,47–51. We have previously shown that porcine bone marrow has a modulus of 4.4±1.0 kPa (Figure 4a)1. We have described the thorough mechanical characterization of intact bone marrow in a prior publication 1. In this prior work, we used indentation, shear rheology, and cavitation rheology to show that the effective modulus (Eeff) of porcine marrow ranges from 0.1 – 24.7kPa, with an average modulus of 4.4 +/− 1.0 kPa. In this previous publication we also learned that bone marrow is an elastic tissue, with similar force responsiveness to the PEG hydrogels developed here. In order to use this heterogeneous real tissue data functionally as a design criterium, we focused on the measurements obtained from indentation, and applied that identical measurement technique to our synthetic hydrogels, focusing on the mean data: 4.4±1.0 kPa. This data has been used to show that hematopoietic progenitor populations can be maintained in the presence of fibronectin at the modulus of bone marrow23, and that scaffolds mimicking this elasticity support megakaryocyte differentiation and platelet release52. This PEG hydrogel can be crosslinked to span the range of stiffness observed in bone marrow (Figure S3c). While many properties can be used to manipulate hydrogel modulus, a 20 wt%, 8-arm, 20kDa PEG hydrogel best matched the reported modulus of porcine bone marrow tissue (Figure 4b).
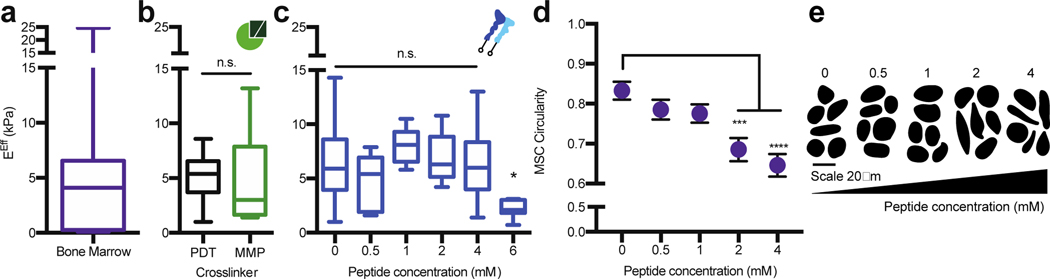
Figure 4. The PEG hydrogel accurately models the bulk compressive properties of bone marrow tissue.
a) Rheology data from Jansen et al., 20151 for the effective Young’s modulus (EEff) of porcine bone marrow at 35°C. b) The EEff for 20 wt%, 8-arm, 20K PEG hydrogels crosslinked at a 1:1 thiol to maleimide molar ratio with 1.5 kDa PEG-dithiol (PDT, black) or with the bone marrow cocktail containing MMP crosslinkers (MMP, green). c) The EEff for 20 wt%, 8-arm, 20K PEG hydrogels crosslinked at a 1:1 thiol to maleimide molar ratio with PDT and coupled with different concentrations of the bone marrow peptide cocktail for 10 minutes before gelation. d) MSCs circularity with respect to peptide concentration and e) representative cell traces for cells encapsulated in a 20 wt%, 8-arm, 20 kDa PEG-crosslinked with the bone marrow cocktail. The significance is determined using a two-tailed t-test where p=0.05, and error bars represent the SEM. (N≥2, n≥3 for mechanical testing; N≥2, n≥10 for cell circularity).
One benefit of synthetic hydrogels is that their moduli can be independently tuned from the concentration of bioactive peptides included. To ensure this was the case for our hydrogel, which includes 20 different peptides, we individually incorporated each peptide cocktail into the hydrogel and tested their effects on modulus. Incorporation of the MMP-sensitive crosslinkers, instead of PEG-dithiol, did not alter the hydrogel modulus (Figure 4b), and the integrin-binding peptides could be incorporated up to a 4 mM total concentration without compromising the bulk modulus (Figure 4c). Through cell tracing experiments, we found that a 2 mM concentration of integrin-binding peptides was needed to achieve significant MSC spreading at 24 hours. We therefore chose a 2 mM integrin-binding peptide concentration; a 20 wt%, 8-arm, 20 kDa PEG-maleimide; crosslinked with a 3:1 ratio of MMP-degradable peptides with PEG-dithiol as the final bone marrow hydrogel formulation (Figure 4d–e).
The synthetic bone marrow hydrogel provides a niche for MSC growth and differentiation
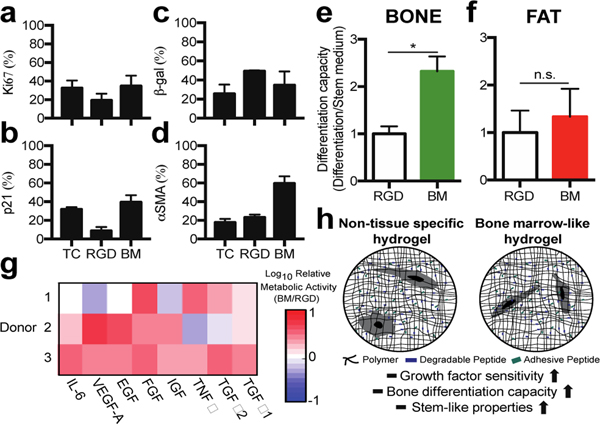
Our results demonstrate an approach to identify the ECM stiffness, integrin-binding proteins, and MMP-degradable sites in real bone marrow, and use that information as design criteria for a synthetic hydrogel. As a proof of concept demonstration, we compared this bone marrow hydrogel to the commonly used RGD-functionalized PEG hydrogel and tissue culture polystyrene (TCPS). We quantified both cell proliferation and differentiation, because these are two phenotypes important for MSCs in the marrow and as they transit to the bone surface or yellow marrow. After one week in culture, the same percentage of MSCs expressed Ki67, a proliferation marker, p21, a cell cycle inhibitor, and β-galactosidase, a marker for senescence, on TCPS as in the bone marrow gel, where cells in the RGD-functionalized PEG hydrogel were less proliferative and had increased senescence (Figure 5a–c).
Figure 5. The bone marrow hydrogel supports MSC growth, and stem-like properties.
Staining for a) Ki67, b) p21, c) beta-galactosidase, and d) α-smooth muscle actin positive cells in a hydrogel with no degradability and 2 mM RGD (RGD) or the bone marrow hydrogel (BM). e) Oil Red O or f) Osteoimage differentiation capacity normalized to the RGD hydrogel. g) Log10 of cell metabolic activity three days after cell encapsulation into the bone marrow hydrogel or an RGD hydrogel for all donor MSCs. Each growth factor was dosed at 20 ng/mL in cell culture medium (n≥3). h) Schematic to compare how the two hydrogels impact observed MSC phenotypes.
We next explored whether MSCs were differentiating toward typical lineages or maintaining their stem-state in the gels compared with control environments. Interestingly, α-smooth muscle actin was highest in the bone marrow hydrogel, suggesting reduced clonogenicity and fat differentiation (Figure 5d)53,54. All donor MSCs were capable of differentiating into bone and fat, shown by staining hydroxyapatite or lipids, respectively (Figure S5). Differentiation capacity was measured by quantifying the ability of cells to differentiate in the presence or absence of differentiation medium. We first performed a control to ensure that the primary cells were responsive to differentiation stimuli on control surfaces (Figure S4). In the bone marrow hydrogel, MSCs had a higher capacity to differentiate into bone compared to RGD-functionalized hydrogels (Figure 5e). In both the RGD-functionalized and bone marrow hydrogels, spontaneous hydroxyapatite formation was observed without the presence of differentiation cues (Figure S5). Adipose differentiation was similar in both materials (Figure 5f). Our results correlate with reports that α-smooth muscle actin positive MSCs filtered from bone marrow have a higher osteogenic differentiation potential54.
We next hypothesized that the bone marrow hydrogel provided a niche for MSCs to differentiate and respond to growth factors typically present in the bone milieu that is responsible for MSC activation, differentiation, proliferation, and trafficking55,56. We treated MSCs encapsulated in hydrogels with a panel of proteins associated with either MSC differentiation or proliferation57. We observed that MSCs encapsulated in the bone marrow hydrogel were more metabolically active when exposed to this panel than when encapsulated in the RGD-functionalized hydrogel (Figure 5g), which agrees with work by others that integrin binding influences sensitivity to soluble factors in the medium 3,58–62. In vivo, the bone marrow niche needs to be able to support progenitor populations and to direct cell differentiation, a feature we demonstrate here in a synthetic biomaterial environment.
Outlook
Here, we combined proteomic-based bioinformatics and biomechanics to make a bone marrow-customized PEG hydrogel. This marrow-mimicking gel is composed of PEG and peptides and polymerizes in 10 seconds under physiological conditions. The novelty of our hydrogel is that it includes 20 different peptides to more fully capture the integrin-binding and MMP sensitive domains of ECM proteins typical of marrow. Both this work and, our recent work to mimic brain tissue17, demonstrate a new approach to hydrogel design when compared to gels that have typically incorporated 1–3 of integrin-binding or MMP-degradable peptides. This approach intentionally changes the motivation of hydrogel design from being application-driven to tissue ECM-mimicking. Another approach to model tissue has been to implant bone-like scaffolds into mice to recruit cells and then use ex vivo culturing to maintain bone marrow cell populations in culture long-term63–65,66. This latter approach is labor intensive and requires technical expertise to fabricate, limiting its throughput. We also argue that these models under-represent the chemical diversity of native tissue, because while they capture the hierarchical structure of bone, they omit the unique ECM protein profile of bone marrow.
Decellularized matrix is currently the only in vitro material capable of including the protein complexity of real tissue67,68,69. It is time-consuming to make and not batch-controlled, making it very useful for some applications, but difficult to use for hypothesis testing based on individual ECM components. Tissue-specific cells can also be made to secrete their own matrix in cell culture, but this matrix is not necessarily representative of the native environment70. As an alternative, we demonstrate an approach to synthetically represent the tissue-specific properties of bone marrow while maintaining control and simplicity. One appeal of this system is that it could be used to co-culture cells or be formed around any cell or organoid of interest20. Additionally, because features can easily be tuned, pseudo-ECM knock-out, -down environments can be used to understand ECM-mediated cell signaling. Future work should focus on a more thorough understanding of how each component of the ECM (and each individual peptide), and how perturbations of these properties, contributes to and changes observed cell phenotypes. For example, work beyond this study could explore whether each peptide is necessary to drive specific bone marrow cell phenotypes. Systems biology-driven experimental design could be applied to perturb individual and groups of peptides to predict the simplest possible hydrogel design needed to achieve the results we present here.
In sum, we have captured the ECM of real bone marrow using simple chemistry in a widely-used material that is adaptable to high throughput, systems-level screens71. We propose this approach could be applied to any tissue or organ, creating a new class of designer biomaterials that can be employed to elucidate ECM-driven mechanisms in cells not easily achieved by other systems.
Materials and methods
Cell culture
All cell culture supplies were purchased from Thermo Fisher Scientific (Waltham, MA) unless otherwise noted. Human mesenchymal stem cells (MSCs) were received through a material transfer agreement with Texas A&M University College of Medicine Institute for Regenerative Medicine at the Scott & White Hospital funded by the National Institute of Health (NIH). MSCs were isolated from the bone marrow of three healthy donors, two females (29 and 24 years old) and one male (24 years old). MSCs were cultured in alpha minimum essential medium (αMEM), supplemented with 16.5% fetal bovine serum (FBS) and 1% L-glutamine, and used between the 2nd and 6th passage. The ability for cells to differentiate was confirmed at the end of passage 6 (or later) for each cell source. The hTERT MSCs were provided from Dr. Junya Toguchida and the human breast cancer cell line MDA-MB-231 was provided by Dr. Shannon Hughes. These were cultured in Dulbecco’s modified eagle’s medium (DMEM), supplemented with 1% L-glutamine, 1% penicillin– streptomycin, 10% FBS, 1% non-essential amino acids, and 1% sodium pyruvate.
Identifying integrin-binding and MMP-degradable proteins in bone marrow
Manual data mining was used to identify 48 integrin-binding proteins and 44 MMP-degradable proteins (Tables S1 and S2). These proteins were quantified in human bone marrow using the Protein Atlas (Table S3)5. The histological score was annotated for each protein. The value of the histological score for the hematopoietic cells was averaged across all the patients scored. This list was used to identify which proteins or protein substrates would be represented by integrin-binding moieties or degradable peptide sequences for the majority of the proteins identified in bone marrow tissue. The histological value was used to determine the percentage of each integrin-binding peptide and MMP-degradable crosslinker to use for proteins in bone marrow.
Solid-phase peptide synthesis
All peptides were synthesized on a CEM’s Liberty Blue automated solid phase peptide synthesizer (CEM, Mathews, NC) using Fmoc protected amino acids (Iris Biotech GMBH, Germany). Peptide was cleaved from the resin by sparging-nitrogen gas through a solution of peptide-resin and trifluoroacetic acid (TFA), triisopropylsilane, water, and 2,2′(Ethylenedioxy)diethanethiol at a ratio of 92.5:2.5:2.5:2.5 % by volume, respectively (Sigma-Aldrich, St. Louis, MO) for 3 hours at room temperature in a peptide synthesis vessel (ChemGlass, Vineland, NJ). The peptide solution was filtered to remove the resin and the peptide was precipitated out using diethyl ether at −80°C. Molecular mass was validated using a MicroFlex MALDI-TOF (Bruker, Billerica, MA) using α-cyano-4-hydroxycinnamic acid as the matrix (Sigma-Aldrich). Peptides were purified to ≥95% on a VYDAC reversed-phase c18 column attached to a Waters 2487 dual λ adsorbable detector and 1525 binary HPLC pump (Waters, Milford, MA).
The following sequences were synthesized: GCGDDEA, GPRGGC, CSRARKQAASIKVAVADR, CSVTCG, CGGYSMKKTTMKIIPFNRLTIG, GCKQLREQ, GCDPGYIGSR, GRGDSPCG, GCRDRPFSMIMGDRCG, GCRDGPLGLWARDRCG, GCRDVPLSLTMGDRCG, and GCRDGPQGIWGQDRCG. The following sequences were purchased from GenScript (Piscataway, NJ) at >96% purity: CGGSVVYGLR, CGPHSRNGGGGGGRGDS, CGP(GPP)5GFOGER(GPP)5, CGGAEIDGIEL, GCRDIPESLRAGDRCG, GCGGQWRDTWARRLRKFQQREKKGKCRKA, GCRDVPLSLYSGDRCG, GCRDSGESPAYYTADRCG, and GCRDVPMSMRGGDRCG.
Polymerization of 3D bone marrow and RGD hydrogels
A 20 kDa 8-arm PEG-maleimide (Jenkem Technology, Plano, TX) was reacted with 2mM of the bone marrow integrin-binding peptide cocktail (Table S4) for 10 minutes in serum-free medium at pH 7.4, forming a solution of PEG with integrin-binding peptides attached. This solution was crosslinked with di-thiol peptides and PEGs to form a gel. The reaction was performed at a 1:1 molar ratio of thiol to maleimide in PBS at pH 7.4, and the crosslinker cocktail was composed of 75 mole% of 1.5 kDa linear PEG-dithiol (Jenkem) and 25 mole% of the MMP-degradable cocktail (Table S5). Gels were polymerized in 10 μL volumes with 1,000 cells/μL. The gel is formed by mixing the two solutions together via pipet, and the reaction occurs within seconds. Cell culture medium was added after 5 minutes to swell the material for at least 18 hours before use. Other hydrogel combinations were made with a 2, 10, and 20 kDa 4-arm PEG-maleimide, all crosslinked at a 1:1 molar ratio of thiol to maleimide with 1.5 kDa linear PEG-dithiol. The RGD-functionalized hydrogel was synthesized in the same way, but 2mM of the peptide GRGDSPCG was replaced for the BM cocktail and the 1.5kDa linear PEG-dithiol was used for the crosslinker at a 1:1 molar ratio of thiol to maleimide.
ECM protein enrichment from tissues
Tissue samples from healthy women between ages 45–60 were obtained from Cooperative Human Tissue Network funded by the National Cancer Institute (NCI) under IRB exempt status. Insoluble ECM proteins were extracted from 500 mg of tissue using the CNMCS compartmental protein extraction kit according to the manufacturer’s instructions (Millipore, Billerica, MA). This resulted in an insoluble ECM pellet.
Mass spectrometry
Two biological replicates were analyzed for human bone marrow, brain, and lung tissues. The ECM-rich pellet remaining from the CNCMS kit was solubilized and reduced in 8 M urea, 100 mM of ammonium bicarbonate, and 10 mM dithiothreitol (DTT) for 30 minutes at pH 8 and 37°C. Samples were alkylated with 25 mM iodoacetamide (Sigma-Aldrich) in the dark at room temperature for 30 minutes before the solution was quenched with 5 mM DTT. Prior to cleavage, the solution was diluted to 2 M urea with 100 mM ammonium bicarbonate at pH 8. Proteins were cleaved via trypsin and Lys-C endoproteinase (Promega, Madison, WI), at a ratio of 1:50 enzyme to protein overnight (12–16 hours) at 37°C. Samples were cleaned and concentrated using a C18 column. A reverse phase LC gradient was used to separate peptides prior to mass analysis. Mass spectrometry analysis was performed in an Orbitrap Fusion Tribrid. Peptides were aligned against the Matrisome using the Thermo Proteome Discoverer 1.41.1429. Parameters used trypsin as a protease, with 4 missed cleavage per peptide, a precursor mass tolerance of 10 ppm, and fragment tolerance of 0.6 Da.
MMP degradation of bone marrow tissue
The MMP degradation assay was adapted from a protocol by Skjøt-Arkil et al. 72. The ECM-rich pellet from the CNMCS kit was solubilized in 8 M urea at pH 8 and lyophilized in 200 μg aliquots. The lyophilized ECM was resuspended in 100 mM Tris-HCl, 100 mM NaCl, 10 mM CaCl2, and 2 mM ZnOAc at pH 8.0. (Sigma-Aldrich) MMP-1, MMP-3 (901-MP, 513-MP, R&D Systems, Minneapolis, MN) MMP-2, MMP-9, MMP-13, MMP-14 (ab125181, ab168863, ab134452, ab168081, Abcam, Cambridge, MA), and MMP-7 (CC1059, Millipore) were activated according to the manufacturer’s instructions and mixed individually with 200 μg of tissue per 1 μg of either active enzyme, or MMP buffer was used as a control. Samples were mixed for 18 hours at 37°C, at which point the reaction was terminated with 25 μM of GM6001 (Millipore). Digested protein was run on a Novex 12% Tris-glycine polyacrylamide gel, stained using silver stain (Thermo) and imaged using the IN Genius Syngene Bioimaging platform (Frederick, MD).
Competitive binding assay
Glass coverslips were prepared with 1 ug/cm2 of the bone marrow peptide coupled to the surface using silane chemistry described by Barney et al3. Cells were seeded at 4,000 cells/cm2 in their normal growth medium after 30 minutes of pretreatment with individual peptides or the complete bone marrow cocktail. Bone marrow was dosed at a molar amount of 25 nmol/mL of medium and the molar amount dosed for each individual peptide was as follows: GRGDSPCG at 600 pmol/mL, CGPHSRNGGGGGGRGDS and GCGGQWRDTWARRLRKFQQREKKGKCRKA at 220 pmol/mL, CGP(GPP)5GFOGER(GPP)5, CGGSVVYGLR, and GPRGGCG at 160 pmol/mL, CSVTCG and CGGYSMKKTTMKIIPFNRLTIG at 100 pmol/mL, GCGDDEA, SRARKQAASIKVAVADRGCG, GCKQLREQ, and CGGAEIDGIEL at 60 pmol/mL, and GCDPGYIGSR at 40 pmol/mL. Cells were imaged beginning 10 minutes after seeding in an environment-controlled Zeiss Axio Observer Z1 microscope (Carl Zeiss, Oberkochen, Germany) using an AxioCam MRm camera and an EC Plan-Neofluar 20X 0.4 NA air objective. Images were taken using Zeiss Axio Observer Z1 (Zeiss) at five-minute intervals for 2 hours, and cell areas were manually traced in ImageJ (NIH, Bethesda, MD).
Cell invasion into MMP-degradable hydrogels
Cytodex1 microcarrier beads (Sigma-Aldrich) were swollen in sterile 1X PBS (1 g beads/50 mL PBS) and autoclaved for 30 minutes at 121°C. Flasks were coated with poly (2-hydroxyethy methacrylate) suspended in ethanol at 20 mg/mL and allowed to evaporate in a biosafety cabinet for 30 minutes to make them non-adherent. Cells were seeded at 10–50 cells/bead in non-adherent flasks at a 0.1 mL of beads/mL of media. The flask was shaken every hour for 4 hours to ensure coating onto beads, and cells were allowed to grow on beads for 48 hours post-seeding. Hydrogels were prepared with 4-arm 20kDa PEG-maleimide at a 20wt% cross-linked at a 1:1 molar ratio with 50% 1.5 kDa linear PEG-dithiol and 50% of each individual MMP-degradable peptide sequence (Table S5). Hydrogels were imaged at days 1, 3, and 6 and all image analysis was performed in ImageJ.
Validation of peptide incorporation
The Measure-iT thiol kit was used to quantify unreacted thiols. Buffers were prepared according to the manufacturer’s guidelines. Mono-functional peptides were incorporated at 1 mM in a 100 μL volume of 8-arm, 20kDa PEG-maleimide at 20wt% for 10 minutes before reacting with 100 μL of the Measure-iT thiol working solution. MMP-degradable peptides were reacted with an 8-arm, 20kDa PEG-maleimide at 20wt% in 10 μL volumes for 10 minutes before reacting with 100 μL of the Measure-iT thiol working solution. The hydrogel was reduced by immersing hydrogels in sodium borohydride (NaBH, Sigma-Aldrich) in water at a molar ratio of 4:1 NaBH to thiol for 4 hours before adding Measure-iT thiol working solution. All solutions or hydrogel supernatants were read at an excitation of 494 nm and emission of 517 nm, according to manufacturer’s instructions. To quantify which peptides did not react, the supernatant from a hydrogel swollen in water for 2 hours was lyophilized, resuspended in 1:1 acetonitrile and ultrapure water with 0.1% TFA at a theoretical concentration 100 pmol/μL, assuming 0% of the peptides coupled to the hydrogel. Peptides were identified using a MicroFlex MALDI-TOF (Buker) with either saturated α-cyano-4-hydroxy cinnamic acid or 10 mg/mL 2,5-dihydroxybenzoic acid as our matrix (Sigma-Aldrich).
Hydrogel mechanical and structural characterization
The effective Young’s modulus was measured using indentation testing on 10 μL volumes of the 3D hydrogels. A custom-built instrument was used as previously described73. Bone marrow mechanical data was taken from Jansen et al.1 For this application, a flat punch probe was applied to samples at a fixed displacement rate of 10 μm/s, for a maximum displacement of 100 μm. The first 10% of the linear region of the force-indentation curves were analyzed using a Hertzian model modified by Hutchens et al. to account for dimensional confinement described by the ratio between the contact radius (a) and the sample height (h) (0.5<a/h<2)74.
MSC spreading with varying peptide concentrations
hTERT MSCs were encapsulated into the 3D bone marrow hydrogels with peptide concentrations varying from 0 to 4 mM of the bone marrow peptide cocktail. After 24 hours, hydrogels were fixed in 10% formalin for 10 minutes and stained with AlexaFluor 555 phalloidin (A34055, 1:40) and DAPI (1:10,000). Cells were imaged Zeiss Spinning Disc Observer Z1 microscope (Zeiss) using an HRm AxioCam and an EC Plan-Neofluar 20X 0.5 NA air objective. Images were taken using Zen (Zeiss) and cell areas were traced in ImageJ.
Differentiation of MSCs across biomaterials
Differentiation of cells was assayed across 5 different biomaterial platforms: tissue culture polystyrene, glass coverslips, 2D PEG hydrogels, and 3D PEG hydrogels with either the bone marrow cocktail or the RGD peptide functionality. Glass coverslips were prepared the same way as for the competitive binding assay. 2D PEG-phosphorylcholine (PEG-PC, Sigma-Aldrich) hydrogels were prepared with bone marrow peptides coupled to the surface at 1 ug/cm2 as described by Herrick et al.4 PC was kept at 17 wt% (0.6 M) and PEG-dimethacrylate (Mn 750) was added at 1.1 wt% (0.015 M) for a ~4 kPa hydrogel. Cells were seeded at a density of 15,000 cells/cm2 on plastic and coverslips, 30,000 cells/cm2 for 2D hydrogels, and 2,000 cells/μL in 3D hydrogels. For osteoblast differentiation, cells were provided cell culture medium supplemented with 10 mM glycerol phosphate (Santa Cruz Biotechnology, Dallas, TX), 1 nM dexamethasone, and 50 μM L-ascorbic acid 2-phosphate (Sigma-Aldrich). For adipose cell differentiation, cells were provided cell culture medium supplemented with 0.5 μM isobutylmethylxanthine, 0.5 μM dexamethasone, and 50 μM indomethacin (Sigma-Aldrich). Cells were maintained for 21 days with medium changes every 3–4 days. After 21 days, cells and materials were fixed in 10% formalin prior to staining. Oil Red O staining was used to identify lipid formation and hydroxyapatite formation was identified using an OsteoImage mineralization assay (Lonza, Basel, Switzerland). Both staining procedures were performed according to the manufacturer’s instructions. Differentiation capacity was determined by dividing the percentage of cells that differentiated in differentiation medium by the percentage that differentiated in stem cell medium. This number for both conditions was normalized to the RGD hydrogel.
Cell proliferation in response to growth factors
MSCs were encapsulated at 1,000 cells/μL in the bone marrow hydrogel or a 20wt%, 8-arm, 20kDa PEG-maleimide functionalized with 1 mM GRGDSPC (Genscript) crosslinked 100% with 1.5 kDa PEG-dithiol. Gels were individually dosed with 20 ng/mL of select growth factors: transforming growth factor-β1 (Millipore), transforming growth factor-β2 (Sigma-Aldrich), transforming growth factor-α, insulin-like growth factor, fibroblast growth factor-1, epidermal growth factor (R&D Systems), vascular endothelial growth factor-A, and interleukin-6 (Abcam). After 5 days in culture, with media changes every 2 days, cell proliferation was measured with CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega) at 490 nm (BioTek ELx800 microplate reader, Winooski, VT). Final results were normalized to a proliferation reading of cells grown in hydrogels for 24 hours in the normal cell culture medium.
Immunofluorescence and senescence stains
After 7 days, cells were fixed, permeabilized, and stained. The following antibodies were used for immunofluorescence: Ki67 (ab16667, 1:200, Abcam), p21 (ab7903, 1:200, Abcam), alpha smooth muscle actin (ab7817, 1:200, Abcam). Beta-galactosidase activity was determined using the Senescence Cell Histochemical Staining Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Cell nuclei were stained with DAPI at a 1:10,000 dilution. Samples were imaged on a Zeiss Cell Observer SD.
Statistical Analysis
Statistical analysis was accomplished using GraphPad’s Prism v7.0a. Data is reported as the mean ± standard error. The term “N” indicates the number of biological replicates performed and “n” indicates the number of technical replicates used per biological replicate. Unless otherwise noted, a two-tailed t-test was performed on the biological replicates. P-values <0.05 are considered significant, where p<0.05 is denoted with *, ≤0.01 with **, ≤0.001 with ***, and ≤0.0001 with ****.
Supplementary Material
Acknowledgments
We would like to thank Dr. Sarah Perry, Dr. Peter Chien, and Dr. Lila Gierasch for technical assistance and use of equipment. Research reported in this publication was supported by the Office of the Director, National Institutes of Health of the National Institutes of Health under Award Number S10OD010645. SRP is a Pew Biomedical Scholar supported by the Pew Charitable Trusts. SRP was supported by a faculty development award from Barry and Afsaneh Siadat. This work was funded by an NIH New Innovator award (1DP2CA186573–01) and a National Science Foundation (NSF) CAREER grant (DMR-1454806) to SRP.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Data Availability Statement
The raw/processed data required to reproduce these findings cannot be shared at this time due to technical or time limitations.
Lauren Jansen: Conceptualization, Methodology, Validation, Formal Analysis, Investigation, Data Curation, Writing – Original Draft, Writing – Review & Editing, Visualization Hyuna Kim: Data curation, Validation, Formal Analysis, Investigation, Writing – Review & Editing, Visualization Christopher Hall: Methodology, Investigation, Writing – Review & Editing Thomas McCarthy: Investigation, Writing – Review & Editing: Michael Lee: Methodology, Investigation, Writing – Original Draft, Writing – Review & Editing Shelly Peyton: Conceptualization, Writing- Reviewing and Editing, Resources, Data Curation, Writing – Original Draft, Writing – Review & Editing, Supervision, Funding Acquisition
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jansen LE, Birch NP, Schiffman JD, Crosby AJ & Peyton SR Mechanics of intact bone marrow. J Mech Behav Biomed Mater 50, 299–307, doi: 10.1016/j.jmbbm.2015.06.023 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen TV, Sleiman M, Moriarty T, Herrick WG & Peyton SR Sorafenib resistance and JNK signaling in carcinoma during extracellular matrix stiffening. Biomaterials 35, 5749–5759, doi: 10.1016/j.biomaterials.2014.03.058 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Barney LE et al. A cell-ECM screening method to predict breast cancer metastasis. Integr Biol (Camb) 7, 198–212, doi: 10.1039/c4ib00218k (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrick WG et al. PEG-phosphorylcholine hydrogels as tunable and versatile platforms for mechanobiology. Biomacromolecules 14, 2294–2304, doi: 10.1021/bm400418g (2013). [DOI] [PubMed] [Google Scholar]
- 5.Uhlén M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Baker EL, Bonnecaze RT & Zaman MH Extracellular matrix stiffness and architecture govern intracellular rheology in cancer. Biophys J 97, 1013–1021, doi: 10.1016/j.bpj.2009.05.054 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaman MH et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. PNAS 103, 15–16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kloxin AM, Kloxin CJ, Bowman CN & Anseth KS Mechanical properties of cellularly responsive hydrogels and their experimental determination. Adv Mater 22, 3484–3494, doi: 10.1002/adma.200904179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeForest CA & Anseth KS Advances in bioactive hydrogels to probe and direct cell fate. Annual review of chemical and biomolecular engineering 3, 421–444, doi: 10.1146/annurevchembioeng-062011-080945 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Xie AW & Murphy WL Engineered biomaterials to mitigate growth factor cost in cell biomanufacturing. Current Opinion in Biomedical Engineering 10, 1–10 (2019). [Google Scholar]
- 11.Brooks EA, Jansen LE, Gencoglu MF, Yurkevicz AM & Peyton SR Complementary, Semiautomated Methods for Creating Multidimensional PEG-Based Biomaterials. Acs Biomaterials Science & Engineering 4, 707–718, doi: 10.1021/acsbiomaterials.7b00737 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Cruz-Acuna R. et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat Cell Biol 19, 1326–1335, doi: 10.1038/ncb3632 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen EH et al. Versatile synthetic alternatives to Matrigel for vascular toxicity screening and stem cell expansion. Nature Biomedical Engineering 1, 0096, doi: 10.1038/s41551-017-0096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huettner N, Dargaville TR & Forget A. Discovering Cell-Adhesion Peptides in Tissue Engineering: Beyond RGD. Trends Biotechnol 36, 372–383, doi: 10.1016/j.tibtech.2018.01.008 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Watt FM & Huck WT Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol 14, 467–473, doi: 10.1038/nrm3620 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Vining KH & Mooney DJ Mechanical forces direct stem cell behaviour in development and regeneration. Nat Rev Mol Cell Biol 18, 728–742, doi: 10.1038/nrm.2017.108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galarza S, Crosby AJ, Pak C. & Peyton SR Control of Astrocyte Quiescence and Activation in a Synthetic Brain Hydrogel. Advanced healthcare materials, e1901419, doi: 10.1002/adhm.201901419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peyton SR et al. Marrow-derived stem cell motility in 3D synthetic scaffold is governed by geometry along with adhesivity and stiffness. Biotechnol Bioeng 108, 1181–1193, doi: 10.1002/bit.23027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert PM et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329, 1078–1081, doi: 10.1126/science.1191035 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gjorevski N. et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Caiazzo M. et al. Defined three-dimensional microenvironments boost induction of pluripotency. Nat Mater 15, 344–352, doi: 10.1038/nmat4536 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Choi JS & Harley BA Challenges and Opportunities to Harnessing the (Hematopoietic) Stem Cell Niche. Curr Stem Cell Rep 2, 85–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi JS & Harley B. Marrow-inspired matrix cues rapidly affect early fate decisions of hematopoietic stem and progenitor cells. Science Advances 3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen JH et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. 2014, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeForest CA & Tirrell DA A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nature Materials 14, 523–531 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Chaudhuri O. et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nature MAterials 15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hynes RO Integrins: Bidirectional, Allosteric Signaling Machines. Cell 110, 673–687 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Löffek S, Schilling O. & Franzke CW Biological role of matrix metalloproteinases: A critical balance. European Respiratory Journal 38, 191–208 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Naba a. et al. The Matrisome: In Silico Definition and In Vivo Characterization by Proteomics of Normal and Tumor Extracellular Matrices. Molecular and Cellular Proteomics 11, M111.014647 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knight CG et al. The Collagen-binding Adomains of Integrins 1 1 and 2 1Recognize the Same Specific Amino Acid Sequence, GFOGER, in Native (Triple-helical) Collagens. Journal of Biological Chemistry 275, 35–40, doi: 10.1074/jbc.275.1.35 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Patterson J. & Hubbell JA Enhanced proteolytic degradation of molecularly engineered PEG hydrogels in response to MMP-1 and MMP-2. Biomaterials 31, 7836–7845, doi: 10.1016/j.biomaterials.2010.06.061 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Plow EF, Haas TA, Zhang L, Loftus J. & Smith JW Ligand binding to integrins. J Biol Chem 275, 21785–21788, doi: 10.1074/jbc.R000003200 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Lishko VK et al. Multiple binding sites in fibrinogen for integrin alphaMbeta2 (Mac-1). J Biol Chem 279, 44897–44906, doi: 10.1074/jbc.M408012200 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Staatz WD et al. Identification of a Tetrapeptide Recognition Sequence for the alpha2beta1 Integrin in Collagen. The Journal of Biological Chemistry 266, 7363–7367 (1991). [PubMed] [Google Scholar]
- 35.Ruoslahti E. RGD and other recognition sequences for integrins. Annual Review of cell and developmental biology 12, 697–715 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Yamada KM Adhesive Recognition Sequence. The Journal of Biological Chemistry 266, 12809–12812 (1991). [PubMed] [Google Scholar]
- 37.Yebra M. et al. Recognition of the neural chemoattractant Netrin-1 by integrins alpha6beta4 and alpha3beta1 regulates epithelial cell adhesion and migration. Developmental Cell 5, 695–707 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Prater CA, Plotkin J, Jaye D. & Frazier WA The properdin-like type I repeats of human thrombospondin contain a cell attachment site. Journal of Cell Biology 112, 1031–1040 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naba A, Clauser KR, Lamar JM, Carr SA & Hynes RO Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. Elife 3, e01308, doi: 10.7554/eLife.01308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu YJ et al. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 160, 659–672, doi: 10.1016/j.cell.2015.01.007 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Makarem R. et al. Competitive binding of vascular cell adhesion molecule-1 and the HepII/IIICS domain of fibronectin to the integrin alpha 4 beta 1. J Biol Chem 269, 4005–4011 (1994). [PubMed] [Google Scholar]
- 42.Phelps EA et al. Maleimide cross-linked bioactive PEG hydrogel exhibits improved reaction kinetics and cross-linking for cell encapsulation and in situ delivery. Adv Mater 24, 64–70, 62, doi: 10.1002/adma.201103574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jansen LE, Negrón-Piñeiro LJ, Galarza S. & Peyton SR Control of Thiol-Maleimide Reaction Kinetics in PEG Hydrogel Networks. Acta Biomaterialia 70, 120–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J. et al. Characterization of the crosslinking kinetics of multi-arm poly(ethylene glycol) hydrogels formed via Michael-type addition. Soft Matter 12, 2076–2085, doi: 10.1039/c5sm02668g (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darling NJ, Hung YS, Sharma S. & Segura T. Controlling the kinetics of thiol-maleimide Michael-type addition gelation kinetics for the generation of homogenous poly(ethylene glycol) hydrogels. Biomaterials 101, 199–206, doi: 10.1016/j.biomaterials.2016.05.053 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Jansen LE, Negron-Pineiro LJ, Galarza S. & Peyton SR Control of thiol-maleimide reaction kinetics in PEG hydrogel networks. Acta Biomater 70, 120–128, doi: 10.1016/j.actbio.2018.01.043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi JS & Harley BA Marrow-inspired matrix cues rapidly affect early fate decisions of hematopoietic stem and progenitor cells. Sci Adv 3, e1600455, doi: 10.1126/sciadv.1600455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peyton SR & Putnam AJ Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol 204, 198–209, doi: 10.1002/jcp.20274 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Engler AJ, Sen S, Sweeney HL & Discher DE Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689, doi: 10.1016/j.cell.2006.06.044 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Yang C, Tibbitt MW, Basta L. & Anseth KS Mechanical memory and dosing influence stem cell fate. Nat Mater 13, 645–652, doi: 10.1038/nmat3889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rape AD, Zibinsky M, Murthy N. & Kumar S. A synthetic hydrogel for the high-throughput study of cell-ECM interactions. Nature Communications 6, 8129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tozzi L. et al. Multi-channel silk sponge mimicking bone marrow vacular niche for platelet production. Biomaterials 178, 122–133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talele NP, Fradette J, Davies JE, Kapus A. & Hinz B. Expression of alpha-Smooth Muscle Actin Determines the Fate of Mesenchymal Stromal Cells. Stem Cell Reports 4, 1016–1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grcevic D. et al. In vivo fate mapping identifies mesenchymal progenitor cells. Stem Cells 30, 187–196, doi: 10.1002/stem.780 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zachar L, Bacenkova D. & Rosocha J. Activation, homing, and role of the mesenchymal stem cells in the inflammatory environment. Journal of Inflammation Research 9, 231–240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cornelissen AS, Maijenburg MW, Nolte MA & Voermans C. Organ-specific migration of mesenchymal stromal cells: Who, when, where and why? Immunol Lett 168, 159–169, doi: 10.1016/j.imlet.2015.06.019 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Rodrigues M, Griffith LG & Wells A. Growth factor regulation of proliferation and survival of multipotential stromal cells. Stem cell Research and therapy 1, 32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balanis N, Yoshigi M, Wendt MK, Schiemann WP & Carlin CR β3 Integrin-EGF receptor cross-talk activates p190RhoGAP in mouse mammary gland epithelial cells. Molecular biology of the cell 22, 4288–4301, doi: 10.1091/mbc.E10-080700 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang F. et al. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci U S A 95, 14821–14826 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moro L. et al. Integrin-induced epidermal growth factor (EGF) receptor activation requires c-Src and p130Cas and leads to phosphorylation of specific EGF receptor tyrosines. The Journal of biological chemistry 277, 9405–9414, doi: 10.1074/jbc.M109101200 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Mercurio AM, Bachelder RE, Bates RC & Chung J. Autocrine signaling in carcinoma: VEGF and the alpha6beta4 integrin. Semin Cancer Biol 14, 115–122, doi: 10.1016/j.semcancer.2003.09.016 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Schwartz AD, Hall CL, Barney LE, Babbitt CC & Peyton SR Integrin alpha6 and EGFR signaling converge at mechanosensitive calpain 2. Biomaterials 178, 73–82, doi: 10.1016/j.biomaterials.2018.05.056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Torisawa YS et al. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro. Nat Methods 11, 663–669, doi: 10.1038/nmeth.2938 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Lee J. et al. Implantable microenvironments to attract hematopoietic stem / cancer cells. PNAS 109, 19638–19643 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Villasante A, Marturano-Kruik A. & Vunjak-Novakovic G. Bioengineered human tumor within a bone niche. Biomaterials 35, 5785–5794, doi: 10.1016/j.biomaterials.2014.03.081 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marturano-Kruik A. et al. Human bone perivascular niche-on-a-chip for studying mestastatic colonization. PNAS (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bersini S. et al. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials 35, 2454–2461, doi: 10.1016/j.biomaterials.2013.11.050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marinkovic M. et al. One size does not fit all: 72 developing a cell-specific niche for in vitro study of cell behavior. Matrix Biol, doi: 10.1016/j.matbio.2016.01.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rijal G. & Li W. A versatile 3D tissue matrix 73 scaffold system for tumor modeling and drug screening. Science Advances 3, e1700764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ragelle H. et al. Comprehensive proteomic characterization of stem cell-derived extracellular 74 matrices. Biomaterials 128, 147–159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ranga A. et al. 3D niche microarrays for systems-level analyses of cell fate. Nature Communications 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skjot-Arkil H. et al. Measurement of MMP-9 and 12 degraded elastin (ELM) provides unique information on lung tissue degradation. BMC Pulm Med 12, 34, doi: 10.1186/1471-2466-12-34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan EP, Smith EJ, Hayward RC & Crosby AJ Surface Wrinkles for Smart Adhesion. Advanced Materials 20, 711–716, doi: 10.1002/adma.200701530 (2008). [DOI] [Google Scholar]
- 74.Hutchens SB & Crosby AJ Soft-solid deformation mechanics at the tip of an embedded needle. Soft Matter 10, 3679–3684, doi: 10.1039/c3sm52689e (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.