Abstract
Background
PTEN mutations have been reported to be involved in the development and prognosis of endometrial carcinoma (EC). However, a prognostic gene signature associated with PTEN mutational status has not yet been developed. In this study, we generated a PTEN mutation-associated prognostic gene signature for EC.
Methods
We obtained the single-nucleotide variation and transcriptomic profiling data from The Cancer Genome Atlas database as training data and implemented the least absolute shrinkage and selection operator (LASSO) Cox regression algorithm to establish a PTEN mutation-associated prognostic gene signature. The overall survival rates of the high-risk and low-risk groups were determined with the Kaplan-Meier (K-M) method, and the accuracy of risk score prediction was tested by using the receiver operating characteristic (ROC) curve.
Results
The K-M curves revealed that the EC patients with PTEN mutations augured favorable survival outcomes. Differential expression analysis between the EC patients with PTEN mutation and wild-type PTEN identified 224 differentially expressed genes (DEGs). Eighty-four DEGs that manifested prognostic value were fitted into the LASSO-Cox analysis, and a PTEN gene signature with seven mutation-associated prognostic genes that showed robust prognostic ability was constructed; this signature was then successfully validated in the other two datasets from the cBioPortal database as well as with 60 clinical specimens. Furthermore, the PTEN mutation-associated prognostic gene signature proved to be an independent prognostic predictor of EC. Remarkably, the EC patients in the high-risk group were characterized by higher tumor stages and grades as well as lower tumor mutation burden with respect to EC, with a poor survival outcome. Collectively, the PTEN mutation-associated prognostic gene signature that we developed could now be used as a favorable prognostic biomarker for EC.
Conclusion
In summary, we developed and validated a prognostic predictor for EC associated with PTEN mutational status that may be used as a favorable prognostic biomarker and therapeutic target for EC.
1. Background
Endometrial carcinoma (EC) is a common malignancy of the female reproductive system, and its incidence is increasing [1]. EC is a heterogeneous tumor, and the prognosis of patients is closely related to tumor grade and stage; early, accurate, and effective diagnosis is thus helpful in improving the prognosis of the EC patients [2]. Surgery and postoperative radiotherapy are routinely used methods for treating this condition, but there is still a lack of effective treatment for recurrent or progressive EC [3]. Therefore, there is an urgent need to identify additional biological markers for the prognostic prediction of EC.
Phosphatase and tensin homolog on chromosome 10q23 (PTEN), a recognized tumor suppressor gene, is one of the most common mutated genes in human tumors, and it can be detected in a variety of tumor tissues, including EC [4]. Kong et al. [5] reported that the mutation rate of PTEN in EC was higher relative to other tumors and that 37%–61% of their EC patients manifested a PTEN gene mutation. Investigators [6] have also demonstrated that PTEN-deficient endometrial epithelial cells were more likely to convert to complex atypical hyperplasia in response to estrogen stimulation and thus develop into EC; and, accordingly, PTEN deficiency is generally considered to be an early event in EC development. Another study [7] also confirmed that functional loss of PTEN was an early cancerous event that exhibited a higher frequency of PTEN mutations in precancerous or stage I tumors than in advanced or even metastatic EC and that PTEN mutations were associated with more favorable outcomes. Thus, PTEN mutations in endometrial hyperplasia may serve as an early warning indicator for increased cancer risk [8].
In view of the important role played by PTEN mutations in the progression and prognosis of EC, we herein fully revealed the mutational landscape of PTEN in EC and developed a PTEN mutational status-associated prognostic gene signature to predict the prognosis of EC based on The Cancer Genome Atlas (TCGA) database. We also executed external validation using two other datasets from the cBioPortal database as well as 60 clinical specimens so as to prove prognostic ability. Furthermore, we demonstrated the relationships between the PTEN mutational signature and stage/grade as well as tumor mutation burden (TMB) in EC. We posit that the PTEN mutation-associated prognostic gene signature can be used as an acceptable prognostic biomarker for EC.
2. Materials and Methods
2.1. Data Acquisition
Single-nucleotide variation, transcriptomic profiling datasets, and corresponding clinical information of 529 EC patients were downloaded from the TCGA database (https://portal.gdc.cancer.gov/) and considered as the training dataset. The transcriptomic profiling datasets and corresponding clinical information of the validation datasets ucec_tcga_pan_can_atlas_2018 and ucec_tcga_pub included 527 and 331 EC patients, respectively, and were obtained from the cBioPortal database (http://www.cbioportal.org/study/summary?id=ucec_tcga). The clinical information included age, BMI, tumor stage and grade, overall survival time, and survival status (the flowchart of our study is shown in Figure 1).
Figure 1.
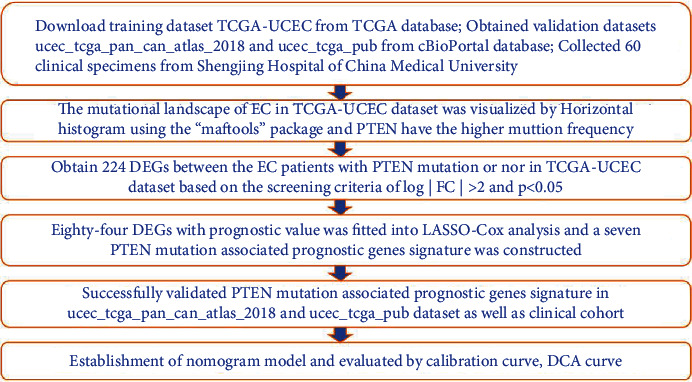
Study workflow. Abbreviations are defined as follows: TCGA: The Cancer Genome Atlas; DEGs: differentially expressed genes; LASSO: least absolute shrinkage and selection operator; UCEC: uterine corpus endometrial carcinoma; DCA: decision curve analysis.
2.2. Specimen Collection
We selected a total of 60 patients with primary endometrial cancer who were admitted to the Department of Gynecology and Obstetrics of Shengjing Hospital between January of 2016 and January of 2017. The patients in the primary endometrial cancer group were 35–60 years of age, with a mean age of 54.5 years. There were 30 cases of patients with FIGO stage I, 15 with stage II, and 15 with stages III–IV. There were 15 patients graded G1, 17 graded G2, and 28 graded G3. All patients underwent staging surgery for endometrial cancer, with removal of pelvic lymph nodes and abdominal para-aortic lymph nodes. A patient was confirmed as having endometrial cancer by pathologists, and none of the patients had received chemotherapy or radiotherapy prior to surgery. This study was approved by the Ethics Committee of Shengjing Hospital of the China Medical University, and informed consent was obtained from all patients. In addition, all methods were performed in accordance with the relevant guidelines and regulations.
2.3. Identification of Differentially Expressed Genes
We employed the “limma” package to identify the differentially expressed genes (DEGs) between the EC patients with PTEN mutation and those with wild-type PTEN. Our screening criteria for DEGs were log | FC | >2 and P < 0.05, and the results were visualized as heatmaps and volcano maps.
2.4. Construction and Validation of a PTEN Mutational Status-Associated Prognostic Signature
The “survival” package was used to screen the DEGs with prognostic value by univariate Cox regression analysis based on the screening criterion of P < 0.05. Next, the key DEGs with prognostic value were further selected by least absolute shrinkage and selection operator (LASSO) regression and stepwise regression analyses. We used multivariate Cox regression to calculate the regression coefficients of the key DEGs with prognostic value and generated a PTEN mutational status-associated prognostic signature. The risk score for each EC patient was calculated using the following formula: risk score = exp1 × β1 + exp2 × β2 ⋯ +expn × βn (expn represents the expression value of each key DEG with prognostic value, and βn represents the regression coefficient) [9]. The EC patients were classified into the high- and low-risk groups based on the median risk score, and the Kaplan-Meier (K-M) method and log-rank test were applied to evaluate the survival between the high- and low-risk groups. Time-dependent and time-independent receiver operating characteristic (ROC) curves were constructed to evaluate the prognostic ability of the PTEN mutational status-associated prognostic signature, and we validated the signature using the ucec_tcga_pan_can_atlas_2018 and ucec_tcga_pub. Finally, the univariate and multivariate Cox regression analyses were used to determine whether the PTEN-associated signature possessed independent prognostic value in both testing and validation datasets.
2.5. Real-Time qPCR
Real-time qPCR was used to detect the relative expression levels of PTEN-associated genes in 60 EC tissues. Total RNA from EC samples was extracted by using TRIzol reagent (Invitrogen, USA) and reverse-transcribed to cDNA, and the RT-qPCR was performed using SYBR Premix Ex Taq (Takara, Japan). GAPDH was selected as an internal reference to detect the relative expression levels of PTEN-associated genes in EC tissues based on the 2-ΔΔCt method, and the primer sequences for RT-qPCR are presented in Supplementary Table 1. Next, we established a PTEN-associated signature based on the relative expression levels of PTEN-associated genes to verify the results of our bioinformatics analysis. A K-M curve was used to evaluate the survival between the high- and low-risk groups, and we constructed ROC curves to evaluate the prognostic ability of the PTEN-associated signature.
2.6. Construction and Validation of a Nomogram Model Based on the PTEN-Associated Signature and Clinical Factors
The “rms” and “survival” packages were used to construct a nomogram model based on the PTEN-associated signature and clinical factors, and consistency between actual and predicted survival rates was assessed with calibration curves. We employed decision curve analysis (DCA) to evaluate the accuracy of the prognostic prediction model [10, 11].
2.7. Statistical Analysis
We used the “maftools” package to reveal PTEN mutational status in EC. Perl was used to calculate the TMB of the patients with EC from TCGA database, and the Wilcoxon rank-sum test was employed for comparative analysis between the two groups. The DEGs related to overall survival (OS) were screened out by univariate Cox regression analysis, and the LASSO-Cox regression algorithm was used to establish the risk-prognosis model. The OS rates of the high-risk and low-risk groups were determined by the K-M method, and the accuracy of risk score prediction was assessed by the ROC curve. The univariate and multivariate Cox regression analyses were used to assess whether the PTEN-associated signature displayed independent prognostic value. The above statistics were analyzed with R (version 3.6.3) software, and a test level of P < 0.05 was considered statistically significant.
3. Results
3.1. PTEN Mutational Status in Endometrial Carcinoma
The mutational landscape of EC in TCGA database was visualized by horizontal histogram using the “maftools” package, and PTEN depicted a high mutation frequency (64%; Figure 2(a)). The K-M curves revealed that the EC patients with PTEN mutation (PTEN mut) exhibited a longer survival time than the patients with wild-type PTEN (PTEN wild; P < 0.001; Figure 2(b)), and the percentage chart shows that the PTEN mutation occurred more frequently in the younger patients with a lower stage and grade of cancer (P < 0.001; Figures 2(c)–2(e)).
Figure 2.
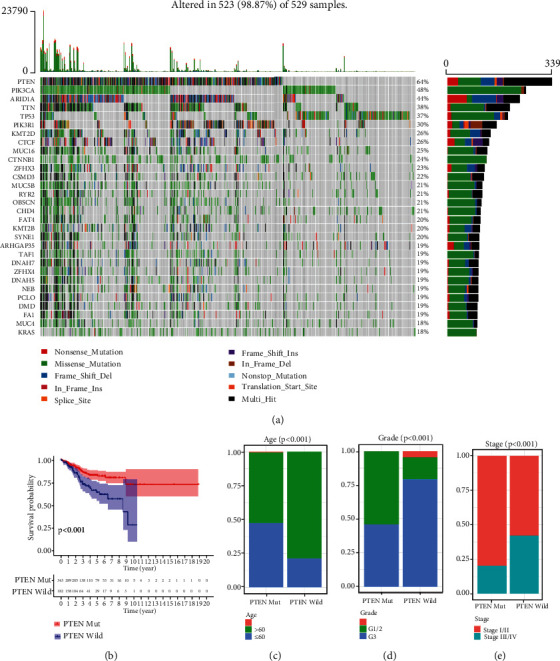
PTEN mutational status of endometrial carcinoma in The Cancer Genome Atlas (TCGA) dataset. (a) Mutational landscape of endometrial carcinoma (EC) in TCGA dataset. (b) Kaplan-Meier curves revealed that the EC patients with PTEN mutation showed a favorable survival outcome. (c–e) Percentage chart showing that the PTEN mutation occurred more frequently at a younger age and with a lower tumor stage and grade in EC patients.
3.2. Identification of Differentially Expressed Genes and Construction of the PTEN Mutational Status-Associated Prognostic Signature
Considering the robust prognostic value of PTEN mutational status, we developed a PTEN mutational status-associated prognostic signature to predict the prognosis of EC. First, the “limma” package was used to identify 224 DEGs (37 upregulated genes and 187 downregulated genes) between the EC patients with PTEN mutation and those without, based on the screening criteria of log | FC | >2 and P < 0.05 (Figures 3(a) and 3(b); Supplementary Table 2). Eighty-four DEGs with prognostic value were selected using univariate Cox regression analysis based on the screening criterion of P < 0.05 (Supplementary Table 3). We performed LASSO analysis based on the 84 DEGs and obtained nine genes (Figures 3(c) and 3(d)). Stepwise regression analyses then further reduced the nine genes to seven, and a PTEN mutation-associated prognostic signature was constructed. The risk score for each EC patient was calculated using the following formula: risk score = 0.2023 × expGDPD2 + 0.0029 × expGRB7 + 0.2875 × expKCNK9 + 0.1244 × expMUC3A + 0.4495 × expMYT1 + 0.2650 × expRPS6KA6 + 0.0359 × expTSPYL5 (Table 1). Differential analysis revealed that all seven genes were more highly expressed in the PTEN-wild group than in the PTEN-mut group (Figures 3(e)–3(k)).
Figure 3.
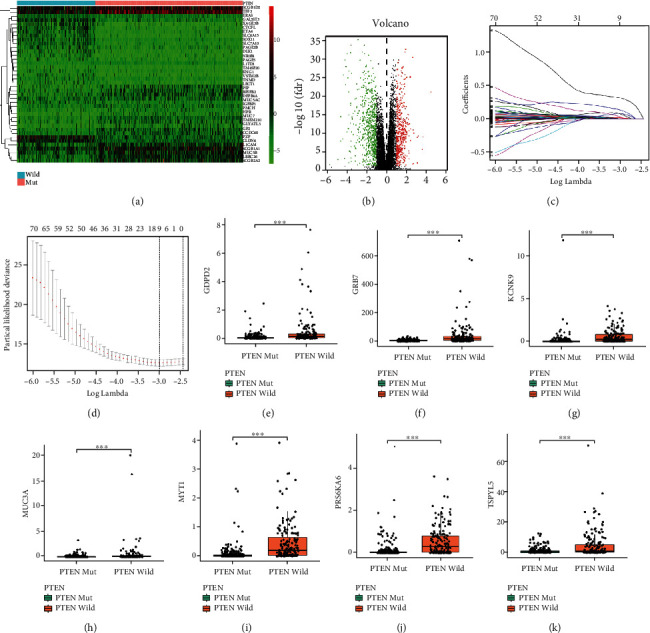
Identification of differentially expressed genes (DEGs) and construction of the PTEN mutational status-associated prognostic signature. (a) Heatmap showing the top 40 DEGs between the EC patients with and without PTEN mutation. (b) Volcano plot showing the DEGs between the EC patients with and without PTEN mutation. (c) LASSO coefficient profiles of the 84 DEGs with prognostic value. (d) Nine PTEN mutation prognostic genes obtained from LASSO regression based on 10-fold cross-validation and minimal criteria. (e–k) The relative expression levels of the seven prognostic genes (GDPD2, GRB7, KCNK9, MUC3A, MYT1, RPS6KA6, and TSPYL5) between the EC patients with and without PTEN mutation.
Table 1.
Regression coefficients of the seven PTEN mutational status-associated prognostic genes.
| Id | Coef | HR | HR.95 L | HR.95H | P value |
|---|---|---|---|---|---|
| GDPD2 | 0.202281 | 1.224192 | 1.0066 | 1.48882 | 0.042781 |
| GRB7 | 0.002907 | 1.002911 | 1.000964 | 1.004862 | 0.003377 |
| KCNK9 | 0.287543 | 1.333148 | 1.113335 | 1.596361 | 0.001761 |
| MUC3A | 0.124442 | 1.132517 | 1.051765 | 1.219468 | 0.000976 |
| MYT1 | 0.449528 | 1.567573 | 1.155313 | 2.126944 | 0.003887 |
| RPS6KA6 | 0.265031 | 1.303471 | 0.941477 | 1.804652 | 0.110342 |
| TSPYL5 | 0.035858 | 1.036509 | 1.013534 | 1.060005 | 0.001716 |
3.3. Evaluation and Validation of the PTEN Mutational Status-Associated Prognostic Signature
We then evaluated and validated the prognostic ability of the PTEN mutational status-associated prognostic signature in both the training and validation datasets. The risk score and survival status of the EC patients are shown in Figures 4(a), 4(d), 4(g), and 4(j); and the EC patients were classified into high- and low-risk groups based on the median risk score. The K-M curve analysis indicated that the patients in the high-risk group evinced a poor survival outcome (Figures 4(b), 4(e), 4(h), and 4(k)), and the values for the areas under the ROC curve (AUC) at 1, 3, and 5 years in TCGA dataset were 0.706, 0.694, and 0.662, respectively (Figure 4(c)). The AUC values at 1, 3, and 5 years in the ucec_tcga_pan_can_atlas_2018 dataset were 0.753, 0.804, and 0.853, respectively (Figure 4(f)); the respective values at 1, 3, and 5 years in the ucec_tcga_pub dataset were 0.888, 0.862, and 0.859 (Figure 4(i)); and the AUC values at 1, 3, and 5 years in our clinical specimens were 0.910, 0.806, and 0.782, respectively (Figure 4(l)). All of the above results highlighted the robust predictive potential of our PTEN mutational status-associated prognostic signature.
Figure 4.
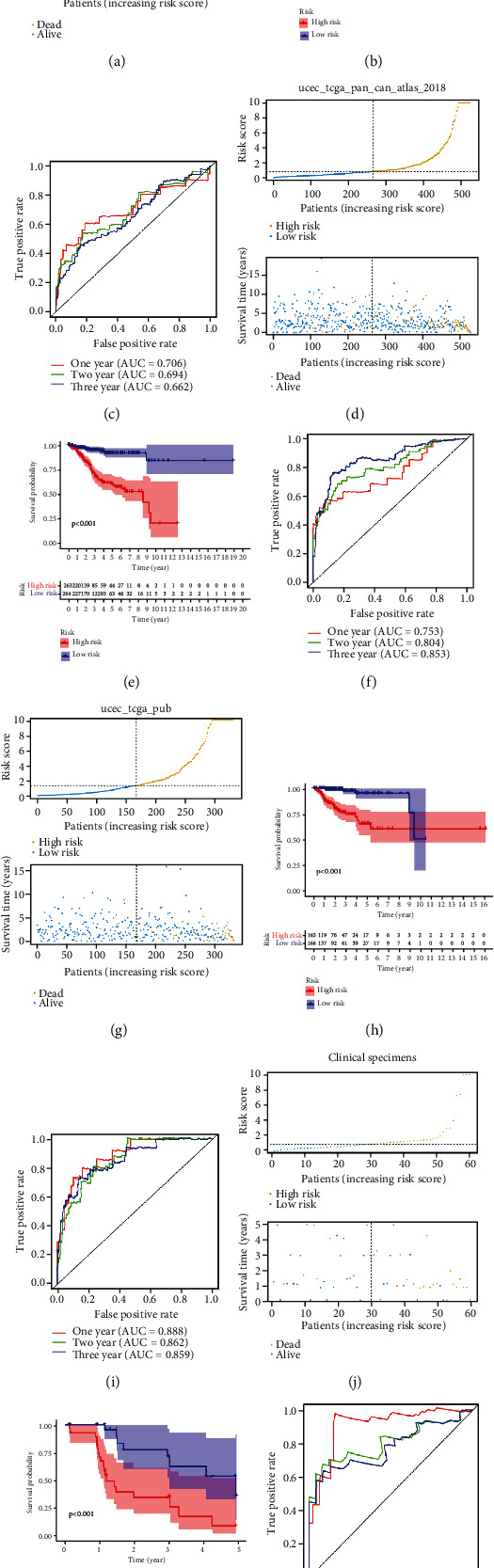
Evaluation and validation of the PTEN mutational status-associated prognostic signature in the training and validation datasets. Risk score, survival status, K–M curve, and ROC curve in TCGA (a–c), ucec_tcga_pan_can_atlas_2018 (d–f), ucec_tcga_pub dataset (g–i), and clinical specimens (j–l).
3.4. Independent Prognostic Value of the PTEN Mutational Status-Associated Prognostic Signature and Its Relationship with Clinicopathological Characteristics
To determine whether the PTEN-associated signature possessed independent prognostic value, the univariate and multivariate Cox regression analyses were performed in TCGA dataset and the clinical cohort, and both indicated that stage, grade, and risk score were related to the prognosis of the EC patients in TCGA dataset (Figures 5(a) and 5(b)). The univariate Cox regression analysis revealed that age, tumor stage and grade, and risk score were related to the prognosis of the EC patients in the clinical cohort, while the multivariate Cox regression analysis suggested that age, stage, and risk score were related to the prognosis of the EC patients (Figures 5(c) and 5(d)). Finally, in TCGA dataset, the percentage chart revealed that younger age and higher stage and grade of tumors in the EC patients were associated with high risk for EC (P < 0.05; Figures 5(e)–5(g)).
Figure 5.
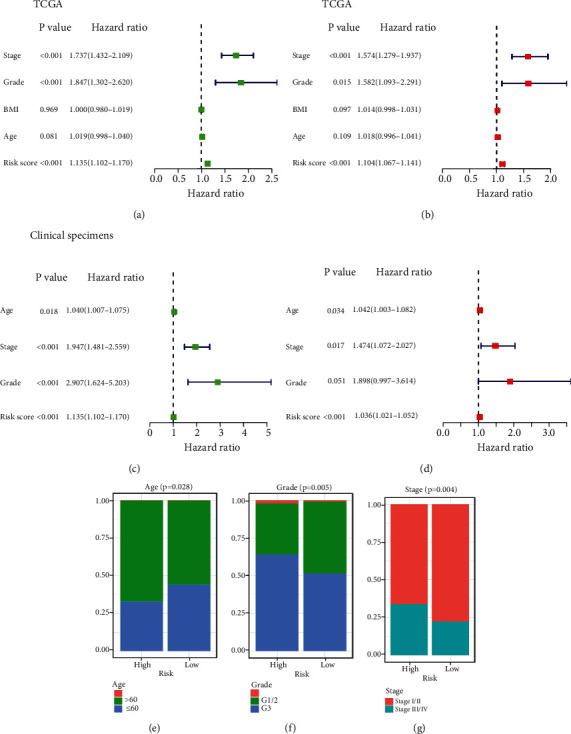
Independent prognostic value of the PTEN mutational status-associated prognostic signature and its relationship to clinicopathological characteristics. (a) Univariate Cox regression analysis of TCGA dataset. (b) Multivariate Cox regression analysis of TCGA dataset. (c) Univariate Cox regression analysis in the clinical cohort. (d) Multivariate Cox regression analysis in the clinical cohort. (e–g) Percentage chart showing that high-risk was associated with younger age and higher tumor stage and grade in EC patients.
3.5. Construction and Validation of a Nomogram Model Based on the PTEN-Associated Signature and Clinical Factors
The “rms” and “survival” packages were employed to construct a nomogram model based on the seven genes of the PTEN-associated signature and the clinical factors to predict the survival rates of EC patients at 1, 3, and 5 years (Figure 6(a)). The calibration curves for these periods revealed high consistency between the actual and predicted survival rates, suggesting the powerful predictive performance of the nomogram model (Figures 6(b)–6(d)); and the DCA curve indicated that the prognostic ability of the model was accurate (Figure 6(e)). Thus, we successfully validated our nomogram model in a clinical cohort (Supplementary Figure 1).
Figure 6.
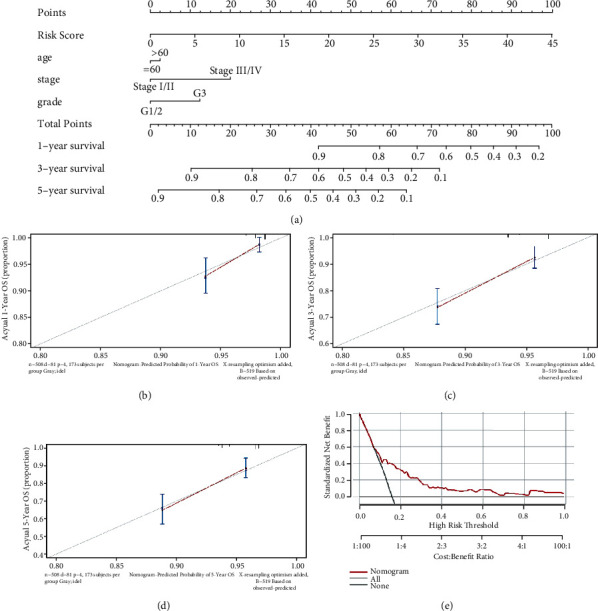
Construction of a nomogram model in TCGA dataset. (a) A nomogram for predicting the 1-, 3-, and 5-year overall survival rates of EC patients. (b–d) The calibration curve at 1, 3, and 5 years. (e) A DCA curve was used to evaluate the accuracy of the nomogram model.
3.6. Mutational Landscape Associated with the PTEN Mutational Status-Associated Prognostic Signature
TMB refers to the total number of replacement and insertion/deletion mutations in each group of nucleobases in the coding region of the evaluated gene exon within the tumor cell genome [12]. Figures 7(a)–7(c) show that the patients in the high-risk group and those with the PTEN mutation reflected a higher TMB value, while the Sankey diagram shows the relationships among risk score, PTEN mutational status, TMB, and survival status (Figure 7(d)). Finally, we investigated the mutational landscape associated with the PTEN mutational status-associated prognostic signature and observed a higher PTEN mutation frequency in the high-risk group (Figure 7(e)).
Figure 7.
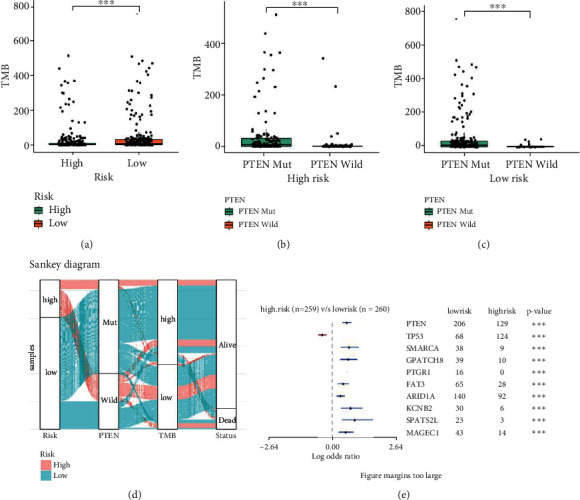
Mutational landscape associated with the PTEN mutational status-associated prognostic signature. (a) Differences in tumor mutation burden (TMB) between high- and low-risk groups. (b) Differences in TMB between patients with and without PTEN mutation in the high-risk group. (c) Differences in TMB between patients with and without PTEN mutation in the low-risk group. (d) Sankey diagram shows the relationships among risk score, PTEN mutational status, TMB, and survival status. (e) Mutational landscape associated with the PTEN-associated signature.
4. Discussion
Mutations in the tumor suppressor PTEN constitute the most frequent type of mutation observed in EC [13], and PTEN has been shown to interact with cell adhesion complexes and to stabilize cell junctions, thereby reducing invasion and metastasis of a range of cancer cells that include EC [14–16]. Previous studies have scrutinized the influence of PTEN mutation on the progression and prognosis of endometrial cancer, but only few have focused on the development of a PTEN mutational status-associated prognostic signature [17–19]. In the present study, based on our analysis of the downloaded single-nucleotide variation and transcriptomic profiling datasets, we found that PTEN embodied a higher mutation frequency in EC patients and that PTEN mutation was associated with younger age and a lower stage and grade of tumors, as well as a favorable survival outcome for EC patients. We then explored and verified a PTEN mutational status-associated prognostic signature that was associated with the malignant progression and prognosis of EC patients. We posit that this PTEN mutation-associated signature constitutes a novel means to predict prognosis and to evaluate efficacy in EC and may thus become a new target for the treatment of EC patients in the future.
In this study, we developed a PTEN mutation-associated signature that included seven genes (GDPD2, GRB7, KCNK9, MUC3A, MYT1, RPS6KA6, and TSPYL5) that were selected by LASSO-Cox analysis, and by reviewing previous studies, we found that RPS6KA6 and TSPYL5 had been reported to be associated with the occurrence and development of EC. Ribosomal S6 kinase 4 (RSK4) is a tumor suppressor gene product (also known as RPS6KA6) that has been shown to be significantly downregulated in multiple malignancies—including those of the breast, colon, kidney, ovarian, and acute myeloid leukemia [20–27]; and it can inhibit tumor cell proliferation, invasion, and the epithelial mesenchymal transformation [28, 29]. One study [30] showed that hypermethylation of RSK4 in EC resulted in a lowered expression level of RSK4 in EC relative to normal endometrial tissues and that reduced RSK4 methylation was associated with higher EC grade. The testis-specific protein Y-encoded-like 5 (TSPYL5) is a member of the TSPYL family, and according to the current studies, the TSPYL5 expression is deleted or downregulated in many tumors [31]. As a new tumor suppressor molecule, the TSPYL5 is closely related to the malignant progression and prognosis of tumors [32–34] and has been reported to be associated with tumor differentiation, cell cycle, and survival in EC [35, 36]. Although the roles of GRB7, KCNK9, MUC3A, and MYT1 in EC have not been exposed, their actions in other tumors have been investigated. For example, growth factor receptor-bound protein 7 (GRB7) is an important bridging protein that is involved in the physiological and pathological processes such as embryonic development, angiogenesis, metabolic regulation, and tumorigenesis by binding to tyrosine kinase receptors (RTKs). GRB7 has also been described as being involved in cellular proliferation, migration, and invasion, cancer prognosis, and tumor-associated angiogenesis of a variety of tumors [37–39]. TASK-3 (also called KCNK9) is a member of the K2P potassium channel family, is overexpressed in a variety of tumor tissues such as breast cancer, gastric adenocarcinoma, ovarian cancer, and lung adenocarcinoma, and is closely related to tumor progression [40–43]. The MUC3A gene is mapped to a mucin cluster located on chromosome 7q22, is a tumor suppressor gene found to be expressed at low levels in a variety of tumors, and is involved in the malignant progression of tumors and in their prognoses [44, 45]. Myelin transcription factor 1 (MyT1) is principally expressed in developing central nervous system cells and mediates the proliferation and differentiation of oligodendrocytes and the formation of the myelin sheath of nerve cells [46]. Recent studies have shown that MYT1 is also involved in the malignant progression of gastric cancer, liver cancer, and glioblastomas [47–49]. Although glycerophosphodiester phosphodiesterase 2 (GDPD2) is primarily involved in lipid metabolism, its actions in EC and other tumors remain unreported [50].
To further evaluate the prognostic ability of the PTEN-associated signature in both training and validation datasets as well as in clinical specimens, an ROC curve was plotted and the AUC was calculated. We found a mean AUC of over 0.78, suggesting that the prognostic ability of the PTEN-associated signature was robust. Moreover, univariate and multivariate Cox regression analyses revealed that the PTEN-associated signature was an independent prognostic predictor for EC. Therefore, we posit that the PTEN-associated signature has the potential to be a promising clinical prognostic tool for EC.
5. Conclusions
In summary, we developed and validated a prognostic predictor for EC associated with PTEN mutational status. The PTEN mutation-associated prognostic gene signature may therefore be used as a set of favorable prognostic biomarkers and therapeutic targets for EC.
Acknowledgments
We wish to thank the authors who provided TCGA and cBioPortal public datasets.
Data Availability
All data generated or analyzed during this study are included in this published article.
Ethical Approval
This study was approved by the ethics committee of the Shengjing Hospital of China Medical University. In addition, all methods were executed in accordance with the relevant guidelines and regulations.
Consent
Informed consent was obtained from all patients.
Disclosure
This manuscript was submitted as a preprint in the link https://www.researchsquare.com/article/rs-724760/v1 [51].
Conflicts of Interest
The authors declare that they have no competing interests.
Authors' Contributions
Ying Wu, Jun Wang, Lina Ge, and Qing Hu conceived and designed the study. Ying Wu, Jun Wang, Lina Ge, and Qing Hu developed the methodology. Ying Wu, Jun Wang, Lina Ge, and Qing Hu analyzed and interpreted the data. Ying Wu and Qing Hu wrote, reviewed, and/or revised the manuscript.
Supplementary Materials
Supplementary Figure 1: validation of a nomogram model in the clinical cohort. (A) A nomogram for predicting the 1-, 3-, and 5-year overall survival rates of EC patients. (B–D) The calibration curve at 1, 3, and 5 years. (E) A DCA curve was used to evaluate the accuracy of the nomogram model. Supplementary Table 1: the sequences of primers used for RT-qPCR. Supplementary Table 2: two hundred and twenty-four DEGs (37 upregulated genes and 187 downregulated genes) between the EC patients with PTEN mutation or not. Supplementary Table 3: eighty-four DEGs with prognostic value were selected by univariate Cox regression analysis.
References
- 1.Hou R., Wang L., Wu Y. J. Predicting ATP-binding cassette transporters using the random forest method. Frontiers in Genetics . 2020;11:p. 156. doi: 10.3389/fgene.2020.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuo K., Machida H., Frimer M., et al. Prognosis of women with stage I endometrioid endometrial cancer and synchronous stage I endometrioid ovarian cancer. Gynecologic Oncology . 2017;147(3):558–564. doi: 10.1016/j.ygyno.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu K. H., Broaddus R. R. Endometrial Cancer. The New England Journal of Medicine . 2020;383(21):2053–2064. doi: 10.1056/NEJMra1514010. [DOI] [PubMed] [Google Scholar]
- 4.Leslie N. R., Downes C. P. PTEN function: how normal cells control it and tumour cells lose it. The Biochemical Journal . 2004;382(1):1–11. doi: 10.1042/BJ20040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong D., Suzuki A., Zou T. T., et al. PTEN1 is frequently mutated in primary endometrial carcinomas. Nature Genetics . 1997;17(2):143–144. doi: 10.1038/ng1097-143. [DOI] [PubMed] [Google Scholar]
- 6.Malentacchi F., Turrini I., Sorbi F., et al. Pilot investigation of the mutation profile of PIK3CA/PTEN genes (PI3K pathway) in grade 3 endometrial cancer. Oncology Reports . 2019;41(3):1560–1574. doi: 10.3892/or.2018.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Risinger J. I., Hayes K., Maxwell G. L., et al. PTEN mutation in endometrial cancers is associated with favorable clinical and pathologic characteristics. Clinical Cancer Research . 1998;4(12):3005–3010. [PubMed] [Google Scholar]
- 8.Abd El-Maqsoud N. M., El-Gelany S. Differential expression patterns of PTEN in cyclic, hyperplastic and malignant endometrium: its relation with ER, PR and clinicopathological parameters. Journal of the Egyptian National Cancer Institute . 2009;21(4):323–331. [PubMed] [Google Scholar]
- 9.Li X., Meng Y. Survival analysis of immune-related lncRNA in low-grade glioma. BMC Cancer . 2019;19(1):p. 813. doi: 10.1186/s12885-019-6032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu C., Zhang Y. Development and validation of prognostic nomogram for young patients with gastric cancer. Annals of Translational Medicine . 2019;7(22):p. 641. doi: 10.21037/atm.2019.10.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou X., Wang D., Zuo J., et al. Development and validation of a prognostic nomogram for HIV/AIDS patients who underwent antiretroviral therapy: data from a China population-based cohort. Ebiomedicine . 2019;48:414–424. doi: 10.1016/j.ebiom.2019.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bi F., Chen Y., Yang Q. Significance of tumor mutation burden combined with immune infiltrates in the progression and prognosis of ovarian cancer. Cancer Cell International . 2020;20:p. 373. doi: 10.1186/s12935-020-01472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Y. S., Huang H. D., Yeh K. T., Chang J. G. Identification of novel mutations in endometrial cancer patients by whole-exome sequencing. International Journal of Oncology . 2017;50(5):1778–1784. doi: 10.3892/ijo.2017.3919. [DOI] [PubMed] [Google Scholar]
- 14.Dorr C., Janik C., Weg M., et al. Transposon mutagenesis screen identifies potential lung cancer drivers and CUL3 as a tumor suppressor. Molecular Cancer Research . 2015;13(8):1238–1247. doi: 10.1158/1541-7786.MCR-14-0674-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q. A., Yang Y., Liang X. Retraction note: lncRNA CTBP1-AS2 sponges miR-216a to upregulate PTEN and suppress endometrial cancer cell invasion and migration. Journal of Ovarian Research . 2020;13(1):p. 147. doi: 10.1186/s13048-020-00754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xin W., Gao X., Zhao S., et al. LncRNA RP11-395G23.3 suppresses the endometrial cancer progression via regulating microRNA-205-5p/PTEN axis. American Journal of Translational Research . 2020;12(8):4422–4433. [PMC free article] [PubMed] [Google Scholar]
- 17.Che Y., Yao Q., Dai S., Luo B., Wang Y. Study of the mutation and expression of PTEN gene in endometrial carcinoma and epithelial ovarian cancer. Zhonghua Fu Chan Ke Za Zhi . 2002;37(10):608–611. [PubMed] [Google Scholar]
- 18.Zhang H., Wang S., Cacalano N., et al. Oncogenic Y68 frame shift mutation of PTEN represents a mechanism of docetaxel resistance in endometrial cancer cell lines. Scientific Reports . 2019;9(1):p. 2111. doi: 10.1038/s41598-019-38585-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao Y., Liang B. PTEN mutation: a potential prognostic factor associated with immune infiltration in endometrial carcinoma. Pathology, Research and Practice . 2020;216(6):p. 152943. doi: 10.1016/j.prp.2020.152943. [DOI] [PubMed] [Google Scholar]
- 20.López-Vicente L., Armengol G., Pons B., et al. Regulation of replicative and stress-induced senescence by RSK4, which is down-regulated in human tumors. Clinical Cancer Research . 2009;15(14):4546–4553. doi: 10.1158/1078-0432.CCR-08-3159. [DOI] [PubMed] [Google Scholar]
- 21.Su X., Li Z., He C., Chen W., Fu X., Yang A. Radiation exposure, young age, and female gender are associated with high prevalence of RET/PTC1 and RET/PTC3 in papillary thyroid cancer: a meta-analysis. Oncotarget . 2016;7(13):16716–16730. doi: 10.18632/oncotarget.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thakur A., Rahman K. W., Wu J., et al. Aberrant expression of X-linked genes RbAp46, Rsk4, and Cldn2 in breast cancer. Molecular Cancer Research . 2007;5(2):171–181. doi: 10.1158/1541-7786.MCR-06-0071. [DOI] [PubMed] [Google Scholar]
- 23.Thakur A., Xu H., Wang Y., Bollig A., Biliran H., Liao J. D. The role of X-linked genes in breast cancer. Breast Cancer Research and Treatment . 2005;93(2):135–143. doi: 10.1007/s10549-005-4516-0. [DOI] [PubMed] [Google Scholar]
- 24.Arechavaleta-Velasco F., Zeferino-Toquero M., Estrada-Moscoso I., et al. Ribosomal S6 kinase 4 (RSK4) expression in ovarian tumors and its regulation by antineoplastic drugs in ovarian cancer cell lines. Medical Oncology . 2016;33(2):p. 11. doi: 10.1007/s12032-015-0724-6. [DOI] [PubMed] [Google Scholar]
- 25.Rafiee M., Keramati M. R., Ayatollahi H., et al. Down-regulation of ribosomal S6 kinase RPS6KA6 in acute myeloid leukemia patients. Cell Journal . 2016;18(2):159–164. doi: 10.22074/cellj.2016.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai J., Ma H., Huang F., et al. Low expression of RSK4 predicts poor prognosis in patients with colorectal cancer. International Journal of Clinical and Experimental Pathology . 2014;7(8):4959–4970. [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J., Jia Q., Zhang Y., et al. Prominent roles of ribosomal S6 kinase 4 (RSK4) in cancer. Pathology, Research and Practice . 2021;219:p. 153374. doi: 10.1016/j.prp.2021.153374. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J., Li Q. Y., Liu J. L., Wei W., Yang H. W., Tang W. RSK4 knockdown promotes proliferation, migration and metastasis of human breast adenocarcinoma cells. Oncology Reports . 2015;34(6):3156–3162. doi: 10.3892/or.2015.4291. [DOI] [PubMed] [Google Scholar]
- 29.Ye Q., Wang X., Jin M., et al. Effect of RSK4 on biological characteristics of colorectal cancer. World Journal of Surgical Oncology . 2018;16(1):p. 240. doi: 10.1186/s12957-018-1474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dewdney S. B., Rimel B. J., Thaker P. H., et al. Aberrant methylation of the X-linked ribosomal S6 kinase RPS6KA6 (RSK4) in endometrial cancers. Clinical Cancer Research . 2011;17(8):2120–2129. doi: 10.1158/1078-0432.CCR-10-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M., Ingle J. N., Fridley B. L., et al. TSPYL5 SNPs: association with plasma estradiol concentrations and aromatase expression. Molecular Endocrinology . 2013;27(4):657–670. doi: 10.1210/me.2012-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang C., He C., Ruan P., Zhou R. TSPYL5 activates endoplasmic reticulum stress to inhibit cell proliferation, migration and invasion in colorectal cancer. Oncology Reports . 2020;44(2):449–456. doi: 10.3892/or.2020.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim E. J., Lee S. Y., Kim T. R., et al. TSPYL5 is involved in cell growth and the resistance to radiation in A549 cells via the regulation of p21(WAF1/Cip1) and PTEN/AKT pathway. Biochemical and Biophysical Research Communications . 2010;392(3):448–453. doi: 10.1016/j.bbrc.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 34.Sun Q., Guo D., Li S., et al. Combining gene expression signature with clinical features for survival stratification of gastric cancer. Genomics . 2021;113(4):2683–2694. doi: 10.1016/j.ygeno.2021.06.018. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Nan F., Lu K., et al. Identification of key genes in endometrioid endometrial adenocarcinoma via TCGA database. Cancer Biomarkers . 2017;21(1):11–21. doi: 10.3233/CBM-170164. [DOI] [PubMed] [Google Scholar]
- 36.Witek Ł., Janikowski T., Bodzek P., Olejek A., Mazurek U. Expression of tumor suppressor genes related to the cell cycle in endometrial cancer patients. Advances in Medical Sciences . 2016;61(2):317–324. doi: 10.1016/j.advms.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Y., Pei Y., Yang L., et al. Upregulated GRB7 promotes proliferation and tumorigenesis of bladder cancer via phospho-AKT pathway. International Journal of Biological Sciences . 2020;16(16):3221–3230. doi: 10.7150/ijbs.49410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vermehren-Schmaedick A., Mhawech-Fauceglia P., Park B. S., Pejovic T., Luoh S. W. The prognostic significance of GRB7 protein expression and localization in human breast and ovarian cancers. Oncotarget . 2020;11(24):2273–2289. doi: 10.18632/oncotarget.27593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villalobo A., Berchtold M. W. The role of calmodulin in tumor cell migration, invasiveness, and metastasis. International Journal of Molecular Sciences . 2020;21(3) doi: 10.3390/ijms21030765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zúñiga R., Concha G., Cayo A., et al. Withaferin a suppresses breast cancer cell proliferation by inhibition of the two-pore domain potassium (K2P9) channel TASK-3. Biomedicine & Pharmacotherapy . 2020;129:p. 110383. doi: 10.1016/j.biopha.2020.110383. [DOI] [PubMed] [Google Scholar]
- 41.Cikutović-Molina R., Herrada A. A., González W., Brown N., Zúñiga L. TASK-3 gene knockdown dampens invasion and migration and promotes apoptosis in KATO III and MKN-45 human gastric adenocarcinoma cell lines. International Journal of Molecular Sciences . 2019;20(23) doi: 10.3390/ijms20236077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Innamaa A., Jackson L., Asher V., et al. Expression and prognostic significance of the oncogenic K2P potassium channel KCNK9 (TASK-3) in ovarian carcinoma. Anticancer Research . 2013;33(4):1401–1408. [PubMed] [Google Scholar]
- 43.Ko J. H., Gu W., Lim I., Bang H., Ko E. A., Zhou T. Ion channel gene expression in lung adenocarcinoma: potential role in prognosis and diagnosis. PLoS One . 2014;9(1):p. e86569. doi: 10.1371/journal.pone.0086569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu T., Liu Y., Zhang Y., et al. Increased expression of MUC3A is associated with poor prognosis in localized clear-cell renal cell carcinoma. Oncotarget . 2016;7(31):50017–50026. doi: 10.18632/oncotarget.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao W., Xie Y. KIAA1429 promotes the progression of lung adenocarcinoma by regulating the m6A level of MUC3A. Pathology, Research and Practice . 2021;217:p. 153284. doi: 10.1016/j.prp.2020.153284. [DOI] [PubMed] [Google Scholar]
- 46.Pandya P., Pasvolsky R., Babichev Y., et al. PICOT binding to the polycomb group protein, EED, alters H3K27 methylation at the MYT1 PRC2 target gene. Biochemical and Biophysical Research Communications . 2019;509(2):469–475. doi: 10.1016/j.bbrc.2018.12.153. [DOI] [PubMed] [Google Scholar]
- 47.Toledo C. M., Ding Y., Hoellerbauer P., et al. Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in glioblastoma stem-like cells. Cell Reports . 2015;13(11):2425–2439. doi: 10.1016/j.celrep.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le A. P., Zhang L. L., Liu W., Shi Y. F. Cantharidin inhibits cell proliferation and induces apoptosis through G2/M phase cell cycle arrest in hepatocellular carcinoma stem cells. Oncology Reports . 2016;35(5):2970–2976. doi: 10.3892/or.2016.4684. [DOI] [PubMed] [Google Scholar]
- 49.Lee N. R., Kim D. Y., Jin H., et al. Inactivation of the Akt/FOXM1 signaling pathway by Panobinostat suppresses the proliferation and metastasis of gastric cancer cells. International Journal of Molecular Sciences . 2021;22(11) doi: 10.3390/ijms22115955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsutsumi T., Matsuda R., Morito K., et al. Identification of human glycerophosphodiesterase 3 as an ecto phospholipase C that converts the G protein-coupled receptor 55 agonist lysophosphatidylinositol to bioactive monoacylglycerols in cultured mammalian cells. Biochimica et Biophysica Acta - Molecular and Cell Biology of Lipids . 2020;1865(9):p. 158761. doi: 10.1016/j.bbalip.2020.158761. [DOI] [PubMed] [Google Scholar]
- 51.Qing H., Jun W., Lina G., Ying W. Significance of PTEN Mutational Status Associated Gene Signature in the Progression and Prognosis of Endometrial Carcinoma. Research Square . 2021 doi: 10.21203/rs.3.rs-724760/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: validation of a nomogram model in the clinical cohort. (A) A nomogram for predicting the 1-, 3-, and 5-year overall survival rates of EC patients. (B–D) The calibration curve at 1, 3, and 5 years. (E) A DCA curve was used to evaluate the accuracy of the nomogram model. Supplementary Table 1: the sequences of primers used for RT-qPCR. Supplementary Table 2: two hundred and twenty-four DEGs (37 upregulated genes and 187 downregulated genes) between the EC patients with PTEN mutation or not. Supplementary Table 3: eighty-four DEGs with prognostic value were selected by univariate Cox regression analysis.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


