Abstract
The history of human sleeping sickness in East Africa is characterized by the appearance of disease epidemics interspersed by long periods of endemicity. Despite the presence of the tsetse fly in large areas of East Africa, these epidemics tend to occur multiply in specific regions or foci rather than spreading over vast areas. Many theories have been proposed to explain this phenomenon, but recent molecular approaches and detailed analyses of epidemics have highlighted the stability of human-infective trypanosome strains within these foci. The new molecular data, taken alongside the history and biology of human sleeping sickness, are beginning to highlight the important factors involved in the generation of epidemics. Specific, human-infective trypanosome strains may be associated with each focus, which, in the presence of the right conditions, can be responsible for the generation of an epidemic. Changes in agricultural practice, favoring the presence of tsetse flies, and the important contribution of domestic animals as a reservoir for the parasite are key factors in the maintenance of such epidemics. This review examines the contribution of molecular and genetic data to our understanding of the epidemiology and history of human sleeping sickness in East Africa.
Although human sleeping sickness may not seem as important on the world stage as diseases such as malaria and AIDS, it is nevertheless an important disease in sub-Saharan Africa and is responsible for a considerable degree of suffering and mortality in countries where it is endemic. Some 55 million people in 36 countries are at risk (79), with an estimated 50,000 new cases reported annually (90). Left untreated, the final outcome of the disease for the individual is death, but equally insidious is the effect on communities and quality of life resulting from the debilitating symptoms. In public health terms, the effects of the disease on community life and, in particular, the contribution of individuals to food production and community support can be measured in terms of disability-adjusted life years lost (DALYS). Human sleeping sickness is responsible for 1.78 million DALYS (90), and the magnitude of this effect can be seen when compared with the related disease leishmaniasis, which has a value of 2.06 million DALYS despite there being 350 million people at risk in 88 countries.
The disease is caused by the protozoan parasite Trypanosoma brucei and is transmitted by the tsetse fly—a link which was established over 100 years ago (17, 135). Despite much research during the intervening years, the disease remains largely unchecked because of the sophisticated biological defense mechanisms adopted by the parasite, the tolerance of the parasite to drug treatment, and the socioeconomic realities of the prevalence of the disease in a region where resource priorities preclude an adequate surveillance and control infrastructure.
One of the characteristic features of the history of African human sleeping sickness in East Africa is the association of specific areas or foci with the disease and a cycling between periodic devastating epidemics interspersed with long periods of low-level endemicity. The key to controlling human sleeping sickness lies with an understanding of the mechanisms responsible for generating these epidemics. To determine the nature of these mechanisms, some specific questions need to be addressed. How does a focus of human sleeping sickness originate? What factors are responsible for maintaining the stability of these foci over long periods? How does a sleeping sickness epidemic develop from low endemic levels within a focus? What factors are responsible for maintaining the epidemic? Clearly, these questions must be addressed in the light of knowledge of the social ecology of the human inhabitants of areas of endemicity and the ecology of the parasite, hosts, and vectors. Although controversy, debate, and conflicting theories have clouded our understanding of these processes, recent detailed analyses of specific human sleeping sickness foci in East Africa have shed more light. In this review, I consider these new data in the light of the history of these sleeping sickness foci; propose an overall model to describe the origins, generation, and dynamics of human sleeping sickness in East Africa; and discuss the implications for disease control in the future.
HUMAN SLEEPING SICKNESS—A DEVASTATING DISEASE WITH FEW CURES
Human sleeping sickness is an extremely debilitating disease and is characterized by two distinct phases: early and late. Following a tsetse fly bite, trypanosomes proliferate in the host bloodstream and undergo antigenic variation to evade the immune system (134). Symptoms of this early phase (nausea, fever, and lethargy) are nonspecific and easily confused with those of other diseases such as influenza and malaria. In the late phase, trypanosomes cross the blood-brain barrier and can be found in neural tissue and cerebrospinal fluid. Subsequent neural damage and host reaction cause the classical symptoms of sleeping sickness: disruption of biological rhythms, inappropriate and irregular sleep patterns, and loss of concentration and coordination. Unless the disease is treated, death will follow. The progress of the disease varies from acute (death, 6 to 12 months) to chronic (death, 5 to 20 years). Few drugs are effective against human sleeping sickness, and little progress has been made in developing new drugs in the last five decades (24, 98). Pentamidine and suramin, developed before the 1920s, remain the drugs of choice for treatment of early-phase disease, while the arsenic-based melarsoprol, developed in 1949, remains the primary drug for treatment of late-phase sleeping sickness. Unfortunately, melarsoprol induces reactive encephalopathy in 10% of patients treated and is fatal in half of those instances (98). More recently, dl-α-difluoromethylornithine (DFMO), an inhibitor of ornithine metabolism, has been used in the treatment of late-phase sleeping sickness, but unfortunately it is effective only for the West African form of the disease (due to Trypanosoma brucei gambiense) and remains ineffectual in the East African form (T. b. rhodesiense). Furthermore, DFMO is no longer commercially available.
The other major strategy used to combat disease, vaccination, has also been thwarted by the ability of the trypanosome to undergo antigenic variation (for reviews, see references 11 and 14). Since there are over 1,000 possible surface coats (132) and many more are generated by recombination (124), the possibility of generating a universal vaccine against human sleeping sickness seems remote. The need for other control strategies, therefore, is paramount.
TRYPANOSOMA BRUCEI—THE CAUSATIVE ORGANISM
Three subspecies of T. brucei are currently recognized (67); (i) T. b. brucei, which is defined as infecting animals but not humans and is present throughout the tsetse region in Africa; (ii) T. b. gambiense, which is defined as human infective and localized to West Africa and results in chronic human sleeping sickness; and (iii) T. b. rhodesiense, which is defined as human infective and localized to East Africa and results in acute sleeping sickness. The three subspecies are morphologically identical, and consequently diagnosis and identification can be difficult. Furthermore, these taxonomic criteria have some loopholes. For example, it is clear that both T. b. gambiense and T. b. rhodesiense have animal reservoir hosts (48, 56, 122); therefore, by implication, an isolate taken from an animal is not necessarily T. b. brucei. Furthermore, the same “subspecies” isolated from different places sometimes has different characteristics, such as isolates taken from the T. b. rhodesiense foci in Uganda and in Zambia (64). Very acute disease is found in the former focus, while low virulence and a high frequency of asymptomatic carriers are a feature of the latter focus (6, 41, 82). Thus, while these taxonomic groups provide a convenient description of the species T. brucei, their use is of limited value in medical and epidemiological studies.
SLEEPING SICKNESS FOCI AND DISEASE EPIDEMICS IN EAST AFRICA
Human sleeping sickness has probably been present in East Africa for many centuries, although we tend to view the disease only after the first descriptions were made by European colonists in the late 19th and early 20th centuries. Atkins in 1742 (quoted in reference 96) described a “sleepy distemper,” while David Livingstone in 1857 (quoted in reference 135) described a “fly disease.” However, it was not until the seminal paper of Sir David Bruce (17) that the tsetse fly was linked to cattle trypanosomiasis and the subsequent reports by Dutton (33) and Stephens and Fantham (115) that the causative agents of, respectively, West and East African sleeping sickness were described.
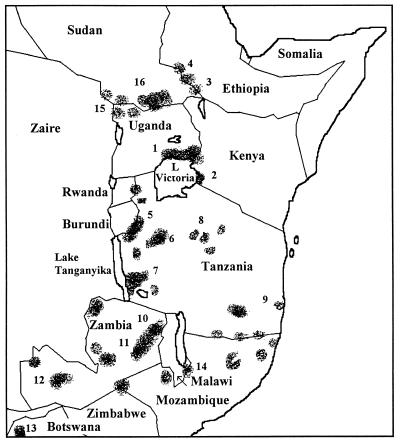
Human sleeping sickness in East Africa has been confined to specific areas (known as foci), and many epidemics have ravaged these foci during recorded history (1900 onwards). Figure 1 shows the current foci of human sleeping sickness and outlines some of the major epidemics associated with each focus.
FIG. 1.
Human sleeping sickness foci in East Africa (data are from references 48, 78, and 79 and other specific reports detailed below. Specific details of epidemics and references are given for numbered foci. Focus 1, Busoga focus (Uganda and Kenya), epidemics, 1900 (18, 21), 1942 (82), 1953 to 1960 (107), and 1964, in Alego, Kenya, (4, 141); 1976 to 1983 (1, 46); and 1988 to 1992 in Tororo District (36, 38, 62, 63, 86). Focus 2, Lambwe Valley focus (Kenya), epidemics, 1960 (2, 8, 40, 137, 141) and 1980 (23, 46, 48, 88, 97). Focus 3, Gilo River focus (Ethiopia), epidemic, 1970 (7, 48, 68). Focus 4, Gambela focus (Ethiopia), epidemic, 1967 (7, 48, 68). Focus 5, Kasulu focus (Tanzania), epidemics, 1930s and 1957 to 1960 (4, 5, 74, 78). Focus 6, Tabora focus (Tanzania), epidemics, 1930s and 1957 to 1960 (4, 5, 74, 78). Focus 7, Rungwa River focus (Tanzania), epidemic, 1920 to 1921 (74, 77, 83). Focus 8, Maswa focus (Tanzania), epidemic, 1920 (29, 39). Focus 9, Matandu River focus (Tanzania), epidemic 1925 (96). Focus 10, North Luangwa Valley focus (Zambia), epidemic 1970 (20). Focus 11, South Luangwa Valley focus (Zambia), epidemic, 1970 (100). Focus 12, Kafue River focus (Zambia), epidemic, 1960 to 1968 (41). Focus 13, Okavango Swamp focus (Botswana), epidemics, 1939 to 1942 and 1957 to 1971 (28, 48, 96). Focus 14, South Malawi focus (Malawi and Mozambique), epidemic, 1912 (43, 96). Focus 15, West Nile (T. b. gambiense) focus (Uganda), epidemic, 1930 (30, 37). Focus 16, Southern Sudan, recent epidemic, 1997 onward (73).
The first reported major epidemic in East Africa was the Great Epidemic of 1900, which devastated the Busoga focus in Uganda and Kenya (18, 21); some half a million people were estimated to be infected during this epidemic (62). T. b. gambiense was thought to be the causative agent, although at that time the more familiar East African subspecies, T. b. rhodesiense, had yet to be described. There were certainly no available diagnostic features which led anyone to suspect that the causative agent was anything other than the previously described T. b. gambiense. Recently, it has been proposed by Koerner et al. that this epidemic was, in fact, caused by T. b. rhodesiense (76). This notion is based on a reexamination of the literature and, in particular, the description of the rapid growth of the parasites in laboratory rodents, a feature not characteristic of T. b. gambiense parasites. The literature describes the presence of T. b. gambiense sleeping sickness in the Busoga focus until the 1950s, but with the unavailability of distinguishing markers and in light of the compelling evidence of Koerner et al. (76), the possibility that T. b. rhodesiense has been there all the time cannot be ruled out. Although this issue may never be resolved, sensitive molecular biological techniques (e.g., PCR) may be able to address this question by using historical collections of microscope slides.
The first reported isolation of T. b. rhodesiense from a sleeping sickness patient was made in 1908 by Stephens and Fantham (115), some 1,200 miles south of the Busoga focus, in the Luangwa Valley in Zambia. Since then, many T. b. rhodesiense epidemics have been reported in East Africa (Figure 1). Based on the timing of the appearance of these epidemics, Ormerod (96) proposed that T. b. rhodesiense spread northward through Rhodesia (now Zimbabwe), Nyasaland (now Malawi), and Tanganyika (now Tanzania) before appearing in the early 1960s in the Busoga focus of Uganda. This theory of the “epidemic spread of Rhodesian sleeping sickness” implies that T. b. rhodesiense is a single strain which has spread throughout East Africa. This is clearly at odds with the proposal of Koerner et al. (76), which suggests that T. b. rhodesiense was already present in the Busoga focus at the turn of the century. At the time when Ormerod (96) proposed his theory, sensitive techniques for the detailed discrimination of T. brucei subsp. were not yet available, and more recent evidence, using molecular biological approaches (62–64), indicates that the T. b. rhodesiense strains responsible from sleeping sickness in the Busoga and Luangwa Valley foci are quite unrelated. It seems unlikely, therefore, that the spread of a single strain can explain the history of epidemics in East Africa.
TRACKING TRYPANOSOME STRAINS—THE ADVENT OF BIOCHEMICAL AND MOLECULAR APPROACHES
The identification and tracking of strains are crucial to delineating the epidemiology of any parasitic disease. A detailed analysis of the strains responsible for human sleeping sickness was not possible due to morphological identity until the advent of biochemical and molecular approaches. In the early 1980s, the pioneering work of Godfrey and Kilgour (50), Gibson et al. (48), and Tait (120) showed that isoenzyme polymorphism (allozyme analysis) could be used to characterize strains of T. brucei. Two basic questions were addressed at that time: could isoenzyme analysis be used to characterize groups of related strains, and could a genetic system be found in trypanosomes? Gibson et al. (48) demonstrated that a great deal of diversity could be detected in trypanosome strains and that isolates could be grouped on the basis of isoenzyme similarity (zymodemes). Importantly, not only did these groups reflect the geographical location from which the isolates were collected, but also some specific isoenzyme patterns were characteristic of human-infective isolates. Subsequent analyses by this approach addressed specific epidemiological questions in Uganda and Kenya (46, 47). Gibson et al. also recognized that the isoenzyme patterns suggested that a mendelian type of genetic system was operating. Tait (120) took the other approach and used isoenzymes to demonstrate that trypanosomes were diploid and that, by conforming to Hardy-Weinberg equilibria, they were undergoing mating in natural populations. This latter question, of genetic exchange in natural populations, has been under much debate recently and is discussed below. Using the genetic distance measure of Nei (93), Tait demonstrated that T. b. gambiense was a separate subspecies of T. brucei (122) but that T. b. rhodesiense was a host range variant of T. b. brucei (123). To date, T. b. brucei and T. b. rhodesiense remain indistinguishable by isoenzyme methods (51).
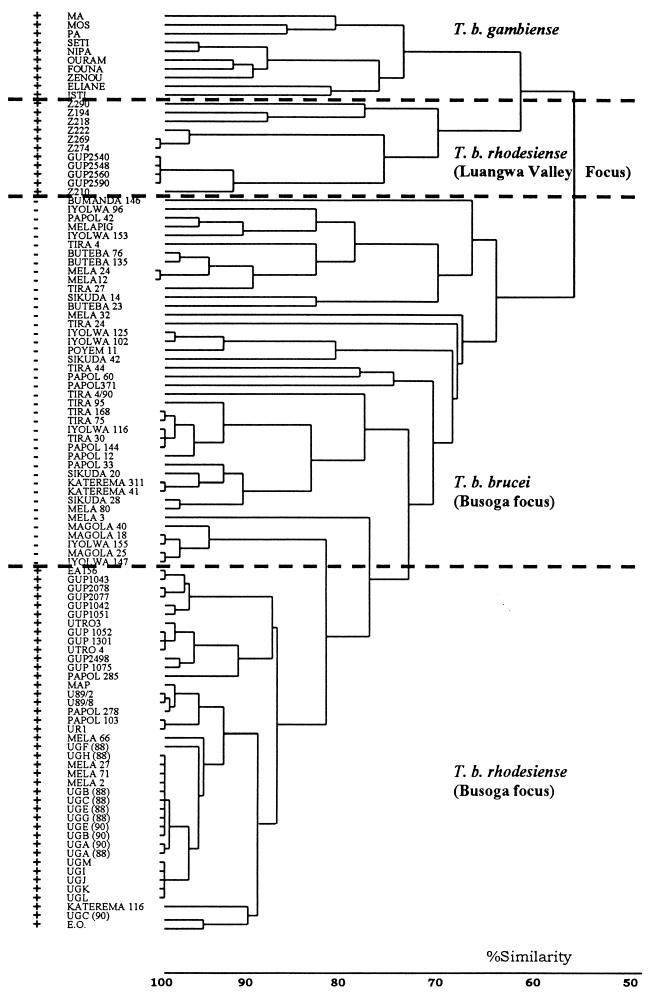
A variety of molecular methods have been used to identify trypanosomes (59, 61). Restriction fragment length polymorphism (RFLP) analysis of kDNA maxicircles (49) distinguished T. brucei strains from East and West Africa based on a variable length polymorphism. RFLP analysis of repetitive DNA sequences (58, 65) has proved to be a very useful technique for the analysis of trypanosome isolates, since banding patterns are generally unique to an isolate but isolates with similar patterns can be grouped by cluster analysis (Fig. 2). By using this approach, it is now possible to distinguish the T. b. rhodesiense (human) isolates from T. b. brucei (nonhuman) isolates in East Africa (63). More recently, other methods of strain typing such as PCR-based randomly amplified polymorphic DNA analysis (85, 136) and analysis of minisatellite sequences (10) have been used. Both of these methods also produce banding patterns (molecular fingerprints) that can be analyzed in a similar way to the RFLP analyses, but as yet the techniques have not been applied directly to epidemiological questions.
FIG. 2.
Relationship between human- and non-human-infective isolates collected during the 1988 to 1990 human sleeping sickness epidemic in the Tororo District of Uganda. A dendrogram depicts the results of a molecular fingerprinting approach based on RFLP analysis with repetitive DNA probes. The banding patterns generated by this molecular fingerprinting approach were compared in pairwise combinations, and a similarity matrix which records levels of similarities and differences in the presence and absence of bands between T. brucei isolates was constructed. The resulting matrix was analyzed by cluster analysis (58) and is depicted in the figure. The names (e.g., TIRA 44) refer to the name of the isolate. Strains identified as + either were isolated from humans or tested positive for human serum resistance (i.e., human-infective strains); strains identified as − were isolated from cattle and were sensitive to human serum (i.e., non-human-infective strains). Details of isolates can be found elsewhere (references 62 to 64). The dendrogram shows that non-human-infective (T. b. brucei) and human-infective (T. b. rhodesiense) isolates, collected during the 1988 to 1990 epidemic in Tororo (Busoga focus), form clearly distinct groups. The strains responsible for human disease in the Luangwa Valley focus, Zambia, are quite unrelated. Human-infective strains from West Africa (T. b. gambiense) are included for comparison.
Some dangers lie in wait for the interpreter of many of these studies. First, all of these studies are based on one sample per individual animal or human. Although it has not yet been fully investigated, considerable genotype selection could occur during the sampling, cloning, and expansion of parasite material. It is likely that future molecular studies will be founded on analyses carried out directly on isolated material by techniques such as PCR. Second, since T. brucei is a diploid organism, care is needed in interpreting heterozygous patterns, since they may appear to be unrelated to their homozygous parental patterns. Third, small sample size predominates in virtually all analyses due to the difficulties of sampling in the field. The continual characterization of trypanosome isolates by using isoenzymes has shown that the more enzyme profiles and isolates are examined, the more zymodemes are discovered (51). An additional problem associated with sample size is the tendency of researchers to pool isolates from different geographical locations and different times. For many epidemiological and population genetic studies, the same time and place must be used for sampling to ensure validity of results (60). The final danger is that the investigator must use appropriate theoretical methods, which take into account the parasite’s genetics and population biology, when analyzing this type of data (125). Many markers, especially isoenzymes, can evolve too fast for use as epidemiological markers, so that results are misinterpreted. For example, increased sampling of isolates from a single geographical location can result in the merging of groupings identified by smaller sample sizes (i.e., a continuum rather than individual groups). Also, identical zymodemes from different populations may be caused by convergent evolution rather than biological or epidemiological similarity. Many of these dangers exist, at least in part, in almost all biochemical and molecular epidemiological studies, and researchers must be aware of the implications.
HUMAN-INFECTIVE STRAINS OF TRYPANOSOMES IN EAST AFRICA—ARE THEY REALLY DISTINCT FROM NON-HUMAN-INFECTIVE STRAINS?
Classical and biochemical taxonomy disagree on the status of human-infective trypanosomes in East Africa. On the one hand, human-infective trypanosomes are given subspecies status as T. b. rhodesiense (67), while on the other hand, they have been classified as host range variants of nonhuman T. b. brucei (123). Further confusion exists because it is possible to select human-infective trypanosomes from their non-human-infective counterparts in the laboratory (52, 71, 101, 133), while in field populations T. b. rhodesiense forms a distinct group (63). Clearly, a paradox exists. The importance of identifying human-infective trypanosomes, determining the relevance of genetic exchange in propagating the human-infective phenotype, and discovering the role of human-infective trypanosomes in human sleeping sickness epidemics is paramount to understanding the history of human sleeping sickness in East Africa.
Biochemical and Molecular Basis of Human Infectivity
As long ago as 1907, Laveran and Mesnil (80) noticed that human serum lysed trypanosomes which had been isolated from animals. Later, Stephens and Fantham (115) first described the human-infective T. b. rhodesiense, which, by implication, must have been resistant to lysis by human serum in order to remain viable in the human host. At the time, little was known about the relationship between trypanosomes isolated from humans and other animals and less still was known about permissive and nonpermissive hosts. Although possibly not realized at the time, the resistance or sensitivity to human serum probably provided the first feature that distinguished human- and non-human-infective trypanosomes. It was not until Rickman and Robson (103, 104) exploited this feature and developed the blood incubation infectivity test (BIIT), which measured resistance to human serum, that T. b. rhodesiense and T. b. brucei could be distinguished without the need for experimental infection of human volunteers. In the BIIT, trypanosome isolates are exposed to human serum and then experimental animals are infected to permit detection of surviving parasites. The test, therefore, is dependent on three factors: the sensitivity of the trypanosome isolate to human serum, the effectiveness of the batch of human serum used, and the ability of the surviving parasites to grow in experimental animals. Although this test represents a considerable improvement over experimental infection of human volunteers, some inconsistencies have been observed in the BIIT performance and in the level of resistance (133). Some of this variability has been removed by the use of a modified test, the in vitro human serum resistance test (19, 69), which has eliminated the need to use infection of experimental animals to detect trypanosomes which have survived lysis by human serum. The likely reason for the remaining variability observed in these tests is that genetic differences in both the host (effectiveness of serum) and parasite (level of resistance) affect the degree of the response. Seed et al. (112) showed that human serum resistance is dependent on both the host and trypanosome genetic backgrounds. Using cloned trypanosome lines that expressed both high and low sensitivity to human serum and different human sera, they showed that both host and trypanosome components caused variation in the trypanolytic activity. Therefore, it seems likely that the variability observed in the human serum resistance test may be due to the combination of different sources of human serum and the trypanosome isolate. In pioneering studies, Rifkin (105) identified the predominant trypanolytic activity as a high-density lipoprotein (HDL)-containing fraction of normal human sera. Using two approaches, she showed that the activity (trypanolytic factor [TLF]) copurified with HDL, and that serum taken from people with Tangier disease—a rare autosomal recessive disorder that is characterized by the absence or severe deficiency of HDL in plasma—lacked TLF activity. These results were confirmed when antibodies against HDL apolipoproteins were found to neutralize the lytic activity of TLF (53). More recent approaches have provided more details of the mechanism of trypanosome killing; these studies have both clouded and clarified this subject. Initially, TLF was reported to be a haptoglobin-related protein which is an apolipoprotein associated with a minor subclass of HDL (53, 113). However, other reports suggested that this might not be the case (12, 55, 81, 99). These conflicts have been resolved by the discovery of a second factor (128), TLF2, which is a non-HDL, unstable high-molecular-mass complex. Evidence suggests that the original HDL-associated TLF activity, TLF1, is inhibited by an endogenous haptoglobin (99) and that TLF2 is probably responsible for the majority of the trypanolytic activity of normal human serum. A model (52) has been proposed which recognizes three phases in the process of TLF-mediated lysis of trypanosomes: (i) binding of TLF to a trypanosome receptor, (ii) internalization by endocytosis, and (iii) lysosomal targeting. Recent evidence indicates that resistance to TLF in human-infective trypanosomes is generated by a mechanism involving decreased internalization (54).
An important breakthrough in our understanding of human sleeping sickness would be to discover the genetic basis of human infectivity. Strain TREU 164, originally isolated from tsetse files in Lugala, Uganda, in 1969, has been shown to be human infective by accidental infection of a laboratory worker (57, 108). By using human serum-resistant and -sensitive trypanosome clones derived from this isolate (ETaT clones) (133), a gene was identified which is expressed (at the RNA level) only in serum-resistant clones (26). The sequence of this gene (SRA) shows that the predicted gene product has features, such as conserved cysteine residues near the C-terminal region, which suggest that it is structurally related to the variant surface glycoproteins (VSGs) (27). (The VSGs are a large family of glycoproteins that coat the entire trypanosome surface and are involved in immune evasion. A single VSG is expressed on the surface of any individual trypanosome, and at relatively frequent intervals a genetic switching event takes place and the VSG is replaced by another family member with different antigenic characteristics.) The predicted amino acid sequence of the SRA gene product suggests that it is intracellular but inserted into a biological membrane. The role of the SRA gene product is unknown, but it is present in very small quantities (106). However, since this gene is not expressed in the procyclic (insect) forms, where endocytosis also does not happen (134), its properties as a membrane-associated, intracellular protein are consistent with a role in endocytosis. If SRA is involved in endocytosis and is the key to human serum resistance, the proposal by Hager and Hajduk (54) would predict that SRA is involved in preventing or reducing the internalization of TLF. As yet, this has not been determined.
In summary, the biochemical evidence shows that the human-infective T. b. rhodesiense differs from the human-infective T. b. brucei by means of its resistance to lysis by components in human serum. Furthermore, this resistance can be acquired by sensitive strains and is caused by the decreased internalization of a TLF. Taken together, the biochemical evidence indicates that T. b. rhodesiense is a host range variant of T. b. brucei.
Identifying Human Trypanosomes in Field Isolates and Detection of Animal Reservoirs for Human Sleeping Sickness
It is important to identify human-infective isolates in the field to determine the relevance of animal reservoirs and the prevalence of human-infective trypanosomes in tsetse flies. As mentioned above, human-infective trypanosomes were identified by experimental infection of human volunteers. By using this approach, it was shown that cattle (95), bushbuck (56), and other wild animals (44) harbored human-infective trypanosomes and therefore could be potential reservoirs. Fortunately for the volunteers, the samples were statistically small, but unfortunately for the epidemiologists, these studies gave little indication of the relative importance of these animal reservoirs. Although the BIIT has been used to identify human-infective trypanosomes in wild animals, domestic animals (48, 111), and tsetse flies (97), equivocal results were obtained in some cases. Isoenzyme analysis has been applied to distinguish field isolates of T. b. brucei and T. b. rhodesiense but has failed to establish unequivocal criteria for distinction (48, 51, 123), leading to the suggestion that T. b. rhodesiense is a host range variant of T. b. brucei (123). Based on identical patterns in human and animal isolates in these isoenzyme studies, animal reservoirs for the human-infective isolates may exist. Such interpretations must be made with caution, but isoenzyme studies on populations from the same locations at the same time (see, e.g., references 46 and 47) clearly implicate cattle as potential reservoirs.
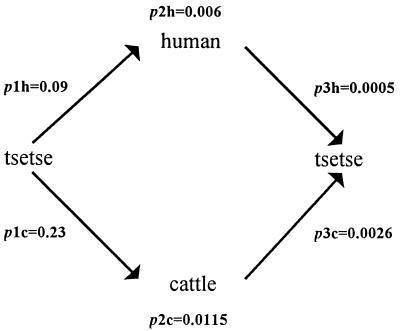
More recently, molecular approaches have been used to identify human-infective T. brucei. Again, molecular markers have failed to provide a clear-cut distinguishing test, probably as a result of the close genetic relationship of T. b. brucei and T. b. rhodesiense. The use of the SRA gene probe (26) as a tool for identifying human-infective strains has potential for use with field isolates, although to date it has not been tested on a large collection of strains. Probably the most successful molecular epidemiological study, in terms of distinguishing human- and non-human-infective trypanosomes, involved a large number of strains circulating during an epidemic in the Tororo District of Uganda in 1988 to 1990 (63). In this study, 88 isolates taken from humans, cattle, and tsetse files in a group of villages within 20 km of each other were analyzed by RFLP analysis of repetitive DNA, isoenzyme electrophoresis, and human serum resistance. The human-infective isolates were clearly distinguishable from the non-human-infective isolates by RFLP analysis (Fig. 2). Furthermore, when Nei’s (93) genetic distance (D) was calculated from the isoenzyme profiles, the genetic distance between the human-infective and non-human-infective populations of trypanosomes collected in the Tororo villages (D = 0.077) (63) was greater than that between two populations of T. b. brucei collected from Lugala, East Africa, and Nigeria, West Africa (D = 0.049) (122). This study also showed that 23% of cattle in these villages carried human-infective T. brucei (63) and that human sleeping sickness is five times more likely to be transmitted by the cattle-fly-human transmission cycle than the human-fly-human cycle (62) (Fig. 3). This showed that the cattle reservoir was clearly important in the generation or maintenance of this epidemic. A comparison with the prevalence of human trypanosome-infected cattle in other epidemics (Table 1) confirmed that this may be a general feature of human sleeping sickness epidemics. In summary, field data suggest that human-infective trypanosomes are distinguishable as a population from their non-human-infective counterparts. These findings contrast with results from other biochemical and molecular studies, indicating that complex mechanisms may be operating in the field which result in the separation of these populations within natural transmission cycles. The crux of the paradox is that while human- and non-human-infective strains are genetically closely related, very little gene flow is apparently occurring between these two host range variants in natural populations. This restricted gene flow could be brought about by lack of genetic exchange between the two groups because of either lack of opportunity or a biological barrier. Lack of opportunity might be promoted by selection against human serum-sensitive strains in human-fly-human or human-fly-animal transmission cycles, thus reducing the opportunity for interaction between human- and non-human-infective strains. Alternatively, the human serum resistance phenotype might be linked to some genetic mechanism (e.g., mating type) that provides a biological barrier to gene flow between these populations. Further research is necessary to elucidate this complex area and understand the interactions of human- and non-human-infective T. brucei in their natural transmission cycles.
FIG. 3.
Dynamics of transmission during the 1988 to 1990 human sleeping sickness epidemic in Tororo, Uganda. p1h and p1c are the probabilities of tsetse flies feeding on, respectively, humans or cattle (calculated from the analysis of tsetse fly blood means). p2h and p2c are the probabilities of either humans or cattle (respectively) being infected with human sleeping sickness trypanosomes. p2h is taken from the frequency of infected humans in the population (0.6%), and p2c is calculated from the frequency of T. brucei-infected cattle (5%) multiplied by the proportion of T. brucei which are human infective (23%). The product of p1 and p2 (p3) is the probability of a fly picking up human-infective parasites from either humans (p3h) or cattle (p3c). In this epidemic, a fly harboring human-infective trypanosomes is five times more likely (p3c = 0.0026) to have picked up that infection from domestic cattle than from an infected person (p3h = 0.0005).
TABLE 1.
Relevance of domestic animal reservoirs in human sleeping sickness foci expressed as the proportion of domestic animals harboring human-infective trypanosomes
| Region and date | No. of domestic animals infected with human-infective isolates | Total no. of domestic animals in study | % of domestic animals harboring human trypanosomes | Reference |
|---|---|---|---|---|
| Lambwe Valley, Kenya (1980) | 14 | 42 | 33 | 47 |
| Busoga, Uganda (1982) | 9 | 43 | 21 | 36 |
| Tororo, Uganda (1988 to 1990) | 3 | 14 | 21 | 36 |
| Tororo, Uganda (1988 to 1990) | 14 | 61 | 23 | 63 |
Is the Human Infectivity Phenotype Spread by Genetic Exchange?
In addition to understanding the transmission of human-infective trypanosomes through animal and vector transmission cycles, another key factor which can contribute to the dispersal of human disease is the spread of the human-infective genotype from human-infective to non-human-infective trypanosomes by genetic exchange. Genetic exchange in trypanosomes was predicted (48, 120) and then demonstrated (70) to occur, but it remains a contentious issue. On the one hand, genetic crosses can be carried out in the laboratory with relative ease (25, 45, 70, 116, 119, 121), but on the other hand, many population genetic studies have indicated that genetic exchange is absent, or at least infrequent, in natural populations (22, 23, 62, 63, 85, 88, 117, 118, 127). A number of factors have confused this debate. First, both clonality and genetic exchange can be detected in different populations (59, 63, 117, 126). The study of the epidemic in Tororo demonstrated that different trypanosome populations (the human-infective and the non-human-infective populations) show different levels of genetic exchange, with the former group being largely clonal (or epidemic) and the latter group showing evidence of frequent genetic exchange within the population (63). Second, different methods of analysis were used, including conformation to Hardy-Weinberg (H-W) equilibria (120), linkage disequilibrium (127), and allele association (63, 87). These statistical analyses all measure different parameters and rely on different assumptions. Conformation to H-W equilibria indicates that a population is undergoing random mating (panmixia), while lack of conformation is explained by either lack of mating or lack of randomness of mating. Linkage disequlibrium statistics measure the variation from linkage equilibrium predicted in a panmictic population. Any barriers to random mating will then create linkage disequilibrium. Allele association measures the breakup of the association of alleles caused by recombination. An index of association of zero is usually taken to indicate frequent genetic exchange but in fact could be generated by any mechanism of recombination. In the light of these inconsistencies, a common approach to analysis might help to standardize interpretations (125). Third, a complex genetic system may exist where polyploidy (45) and selfing (119) may result in disruption to linkage equilibrium. Fourth, pooling of samples from populations, divided by substructuring (60), which have no opportunity for mating has also confused this debate. T. brucei populations clearly have substructuring (63), and this may cause considerable linkage disequilibrium if such subpopulations, which may not genetically recombine, are pooled to enhance sample size for analysis.
Although it is now clear that genetic exchange does play a role in the generation of diversity of trypanosome populations, it is not clear whether genetic exchange occurs between human- and non-human-infective trypanosomes. Further research is needed to establish the role of genetic exchange in human sleeping sickness epidemiology, but clearly complex interactions occur between transmission cycles and genetic cycles in field isolates.
Genetic Diversity in the Tsetse Fly
Different levels of host range specificity seem apparent in T. brucei. Cattle and wild animals appear to be suitable hosts for both human- and non-human-infective T. brucei, while humans are restricted to the former (23, 63). Also some animal hosts can display a resistance to human trypanosomes (92, 103). Little is known about these host restrictions, but it has been proposed that trypanosome strains might be adapted for transmission cycles involving specific hosts (23). Such adaptation may be due to mechanisms such as serum resistance or the recently identified differences in uptake of transferrins (an important trypanosome growth promoter) by the trypanosomes from different host species (14, 15). This latter phenomenon is probably related to antigenic switching and may provide both the flexibility for the trypanosome to adapt to new hosts and the genetic background which enhances transmission through specific host transmission cycles.
Given these different transmission cycles, it is important to understand the genetic diversity within the tsetse fly, since this is the link between different cycles. This is especially important because genetic exchange between trypanosomes occurs in the tsetse fly (70) and interaction between different transmission cycles could occur here. Very little is known about the genetic diversity of T. brucei strains in tsetse flies, although it is apparent that it is greater than in either cattle or humans (47, 88). When T. brucei samples are taken from tsetse flies collected near human habitation, a high proportion of flies which contain trypanosome zymodemes identical to those found in humans are found (97). These findings suggest that close contact between tsetse flies and humans may enhance the transmission of human strains. Since zymodemes found in tsetse flies differed from those found in humans or cattle, it was proposed that the flies were feeding on wild animals harboring trypanosomes with different zymodemes (88). However, another author suggested that these differences may have been due to comparison of samples taken at different places and times (23). The bulk of the evidence suggests that the genotypes of trypanosomes found in tsetse flies reflect the genotypes found in their hosts, providing that they are sampled at the same time and place. The difficulty in carrying out large-scale analyses of T. brucei genotypes in tsetse flies is compounded by the fact that usually only 0.1% of the flies are infected (86). To adequately resolve the question of genetic diversity in tsetse flies, a large-scale study is required with samples taken from tsetse flies and all possible hosts from the same location and time. Furthermore, efficient PCR-based genetic tools are needed for the rapid screening of tsetse fly and similar field samples.
Genetic Diversity of Trypanosome Strains in the Mammalian Host
The great genotypic diversity found in tsetse flies would be expected to be reflected in the genetic diversity of trypanosomes in the mammalian host. Few studies have addressed this question, since such studies are difficult to conduct and interpret. For example, these studies require large programs of sampling from individual animals, avoidance of selective loss of minor genotypes by amplification of parasites (e.g., in laboratory animals), and the availability of sensitive genetic markers. Some studies (91) have provided evidence for the lack of mixed-species (e.g., T. brucei, T. congolense, and T. vivax) infections in cattle. More recently, a limited study suggested that individual animals may contain only a single trypanosome genotype (66). In this study, single animals were sampled over periods of up to 6 months. DNA was extracted from parasites, and RFLP analysis was carried out. Despite being sampled 6 months apart and, in some cases, from different body tissues (blood or cerebrospinal fluid), the trypanosome genotypes recovered from an individual animal were identical but distinct from those recovered from other animals. This paradoxical finding may be explained by sampling error, a population bottleneck, immune selection, or parasite “social” interactions (66). More work is necessary to investigate this question further, and the ability to amplify DNA from single trypanosomes (84) may be an important tool in such an investigation.
Appearance and Stability of Human-Infective Strains in Sleeping Sickness Foci
Of paramount appearance to the control of human sleeping sickness is a knowledge of the frequency at which human-infective strains appear and the length of time they persist. Although laboratory experiments (see above) show that human-infective strains can be readily obtained from human serum-sensitive ones, the relevance of this to natural populations is not clear. Did human sleeping sickness appear only once in East Africa (the “original” T. b. rhodesiense strain) and spread to other foci, or have human-infective strains arisen independently in different locations in East Africa? Ormerod proposed a theory documenting the spread of a single human sleeping sickness strain from Rhodesia (Zimbabwe) northward to Lake Victoria (96) based on the historical appearance of epidemics. Although consistent with the data available at the time, this theory can now be discounted, since molecular evidence shows that human-infective strains from southeastern Africa (Zambia) are quite unrelated to strains found near Lake Victoria (64). Studies with isoenzymes have suggested that the appearance of new zymodemes correlates with the appearance of an epidemic (46, 47). These data have been interpreted as indicating that the influx of new strains is responsible for new epidemics. In contrast, Hide et al. (62, 63) showed that a single trypanosome strain was responsible for human disease in the Busoga focus from at least the 1960s to the present day. The inconsistency between these conclusions may reside in the fact that isoenzymes are a very “fast” molecular marker, as judged by the almost linear increase in the number of zymodemes with samples examined, and that the underlying strain structure is masked by high levels of genetic variability in isoenzyme profiles. The evidence that a single trypanosome strain is responsible for human disease in the Busoga focus is strongly supported by two observations: first, the similarity among RFLP patterns of human-infective isolates collected in Central Nyanza (Kenya) in 1960 and Tororo (Uganda) in 1990, and second, the difference in RFLP patterns between human- and non-human-infective isolates taken at the same time and place from the Tororo villages in 1990 (63). This observed stability in the human strain lends support to the theory of Koerner et al. (76) that it was in fact the same strain as the one responsible for the great human sleeping sickness epidemic in 1900. Thus, the strain responsible for human sleeping sickness in the Busoga focus appears to have arisen only once and is very stable in this focus. The differences observed between human-infective strains from the Busoga focus and the Zambian focus, with respect to disease prevalence (31, 32), virulence (20, 41, 82, 100), and molecular identity (64), suggest that human infectivity has arisen more than once. This evidence lends support to the notion that each human sleeping sickness focus may be associated with a particular human-infective trypanosome strain which is responsible for the long-term stability of that area as a disease focus (59, 62).
ROLE OF THE TSETSE FLY
The tsetse fly is directly involved in both transmission cycles and genetic cycles, by virtue of playing host to trypanosome genetic exchange (70). Taken together with the well-established observation that destruction of tsetse fly habitats correlates with a reduction in sleeping sickness, the tsetse fly is clearly implicated as a major player in human sleeping sickness epidemics. A number of species of tsetse fly are recognized. Glossina fuscipes (previously known as G. palpalis) is found in riverine and marshland areas and has been implicated as the vector for epidemics in the Lake Victoria area. G. pallidipes, which prefers thicket or forest edge, is less abundant than G. fuscipes in the Lake Victoria area but is the most abundant tsetse fly in the Luangwa Valley focus in Zambia. G. morsitans is found in the Luangwa Valley (as a minority species) and Kafue River foci. Other species associated with human sleeping sickness foci are G. swynnertoni in Tanzania and G. tachinoides in Ethiopia.
One of the main epidemiological concerns has been that different tsetse fly species play different roles in human sleeping sickness. G. pallidipes was the species associated with the T. b. gambiense epidemic in Busoga in the 1940s (82). The discovery of T. b. rhodesiense in G. fuscipes during the Central Nyanza epidemic (114) led to the notion that G. fuscipes had facilitated the influx of T. b. rhodesiense into the area (95). At the time that work was carried out, T. b. gambiense and T. b. rhodesiense were indistinguishable and so, given the controversy over the causative agent of these early epidemics (76), it is difficult to draw conclusions about the involvement of different tsetse fly species in transmission cycles. The situation may be further clouded by the complex ecology of the various species. Coexistence of different tsetse fly species in the same area will create considerable difficulties in unravelling associations between different species and trypanosome populations. For example, in the Luangwa Valley sleeping sickness focus, the savannah (G. morsitans) and forest edge (G. pallidipes) species coexist. Currently, little evidence is available which indicates that unique transmission cycles, such cycles specific to human-infective strains, are associated with particular Glossinia species. The discovery that human-infective trypanosomes (from the Busoga focus) have reduced transmissibility in tsetse flies (89, 140) is, however, of interest in this respect. Human serum-sensitive trypanosome stocks were 1.8 times more likely to mature in the tsetse fly than were their human serum-resistant counterparts. This finding suggests that human-infective trypanosomes are less likely to be transmitted than non-human-infective trypanosomes. Clearly, some other driving force must ensure the survival of human-infective trypanosomes during an epidemic. Two mechanisms could be operating at such times: selection in humans and abundant human-fly-human or human-fly-cattle transmission. These human-infective trypanosomes may also be maintained in the population due to a population bottleneck as a result of the majority of transmission from the tsetse fly occurring at the teneral (first) feed (138, 139).
The destruction of tsetse flies has long been a method of reducing human (and animal) trypanosomiasis, although they are remarkable in their ability to repopulate. For example, G. fuscipes was eradicated from the lakeshore of Lake Victoria between 1954 and 1957 but had to be cleared again by insecticide spraying in 1967 to 1968 and again in 1970 (2, 137). A number of methods have been used to control tsetse fly populations. Aerial or ground spraying, bush clearance, and wildlife eradication have traditionally been used to destroy tsetse flies, although these methods have a significant and nowadays unacceptable environmental impact (72). In the early 1970s, trap and target control methods were first tested in Zimbabwe (129, 130). These methods involved the design of traps with odors, colors, and shapes which attracted tsetse flies. New developments in synthetic pyrethroids (34), which had low toxicity to mammals but were lethal to tsetse flies, combined with the use of these traps offered sample approaches to tsetse fly control. Many studies showed these approaches to be cost-effective (see, e.g., reference 131). In addition, these traps could be used as barriers to tsetse flies by placing traps 100 m apart to form a barrier around a tsetse-free region. By 1990, some 27,000 of these targets were being used as barrier targets in Zimbabwe (9). Despite the success of targets and traps, many regions in East Africa have failed to take up this approach. Generally, this is because of the difficulties in deploying and maintaining traps in inaccessible areas. More recently, pour-on or dipping technologies have been used to control tsetse flies. Specially developed formulations of synthetic pyrethroids, suitable for direct application to cattle, have been tested for their ability to control tsetse flies. Cooper’s Spot On, Ciba Geigy’s Ectopor, and Bayer’s Bayticol are examples of these pour-on formulations, and some success has been achieved with this approach (13). Obviously, this approach is designed to break the tsetse fly transmission cycle, and an initial small incidence of disease must be accepted before control will be achieved.
In summary, a number of new developments in tsetse fly control are emerging, but the success of these approaches depends largely on the implementation of suitable management infrastructures. Such structures must involve national and international surveillance and coordination as well as involvement within communities at a local level (16).
ANATOMY OF A HUMAN SLEEPING SICKNESS EPIDEMIC
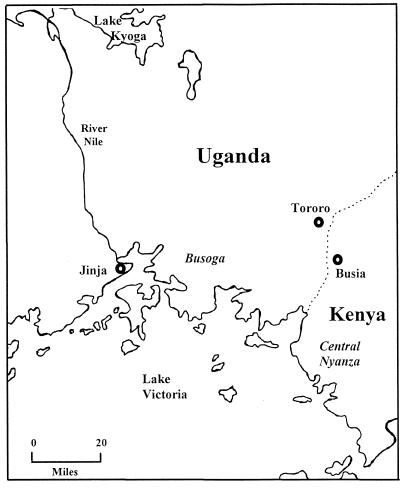
The Busoga focus in southeastern Uganda has long been an important focus of human sleeping sickness. The topography is characterized by a gently undulating slope dropping only 110 m in the 100 km from Lake Victoria to Lake Kyogo (Fig. 4). The terrain is a vast network of slow-moving or stagnant streams overgrown with marsh and dense forest. Wild game are no longer abundant in this region, although bushbusk and antelope are occasionally seen; reptiles such as the monitor lizard are plentiful. Domestic cattle, pigs, goats, and dogs are found near to homesteads. The population density varies between 12 and 300 people per km2 (1), and agriculture is now mainly subsistence despite the presence of a large cotton-growing cash crop industry prior to 1971 (76). Coffee and subsistence crops are grown on small plots close to homesteads. The old cotton plantations, with their heavy use of insecticides, were poor tsetse fly habitats, but the coffee plantations provide a good habitat for tsetse flies. The 1971 takeover by the Amin regime resulted in the destruction of cotton plantations, social turmoil, and poor economic performance. The aftermath was a legacy of abandoned cotton plantations that served as an ideal habitat for tsetse flies and the abandonment of tsetse fly surveillance and control programs (76).
FIG. 4.
Map of the Busoga focus in Uganda and Kenya.
The most recent epidemic in the Busoga focus began in 1971, with 169 cases being detected and treated. Heightened medical surveillance and tsetse fly control terminated this outbreak at the end of that year, and no further cases were reported. In 1972, further breakdown of tsetse fly control measures led to the spread of G. fuscipes fuscipes (94), and changes in agricultural practices encouraged bush growth followed by expansion of tsetse fly habitats. By 1976, the epidemic had flared up again, and it peaked in 1980 with 8,465 cases (1.2% of the population of the affected region being infected) (1, 62, 76). Since analysis of trypanosome zymodemes (46) showed that six zymodemes were found in human-infective isolates, it was proposed that either new strains had entered the region or genetic recombination had generated the isoenzyme variation. The similarity of RFLP patterns among these isolates and those from a later epidemic in Tororo (63) and the observation of hybrid isoenzyme patterns in human trypanosome zymodemes (46) suggest the latter interpretation. By 1984, the epidemic had spread eastward (35), and the first few cases were observed in the Tororo District of southeastern Uganda (Fig. 4). Before this date, there had been no recorded cases of sleeping sickness in this District. However, 297 cases were recorded in 1988, and a peak of 352 cases was reached in 1990. This latter phase of the epidemic offered the opportunity to carry out a detailed analysis of a very localized area, where detailed epidemiological data and trypanosome isolates could be collected alongside each other (86). Samples were taken from tsetse flies, cattle, pigs, and humans from a series of villages within a 10-km radius in the Tororo District in 1988 to 1992. The infection rate in this region was found to be 0.6% of people, 5% of cattle, and 0.1% of tsetse flies (86). Since the percentage for tsetse flies is surprisingly low, a very high biting rate must occur to achieve reasonable levels of disease transmission. Analysis of tsetse fly blood meals indicated a “preference” for tsetse fly feeding behavior, with 59% having fed on monitor lizards, 23% having fed on cattle, and only 9% having fed on humans (140). These figures probably reflect the abundance and availability of hosts rather than on actual feeding preference. Since it is unknown if T. brucei can survive or be detected in cold-blooded monitor lizards, future studies should include sampling of these animals to rule out any possible epidemiological involvement.
Trypanosomes from this area were analyzed to determine the identity of the causative agent of this disease. Isoenzyme analysis of 47 isolates from Tororo (36) demonstrated a large diversity of zymodemes that were broadly similar to those found in a collection of 104 isolates from the “old” Busoga sleeping sickness area. Another study carried out on samples from the Buvuma Islands in Lake Victoria, an area which had not been subjected to the tsetse fly control measures carried out on the mainland, also revealed a range of zymodemes broadly similar to that on the mainland, although a greater diversity of zymodemes was observed in the tsetse flies (38). This large diversity of zymodemes observed in these studies does not necessarily imply that a large number of strains are present. Many similarities can be seen between the banding patterns in different zymodemes, suggesting that some of these zymodemes may be related or derived from genetic hybrids. Thus, the great diversity could merely reflect allelic variation within the population of isolates. Furthermore, the broad overlap in the zymodemes compared with those of the Busoga area probably suggests that the trypanosome samples are all derived from a single strain. In another study, a detailed analysis of 88 isolates by isoenzyme variation, human serum resistance, and RFLP analysis showed that a single strain was responsible for human sleeping sickness in this epidemic; moreover, this strain was also responsible for the human disease in Busoga in the 1980s and also in the Central Nyanza region of Kenya in the 1960s (63). These findings provided strong evidence that a single strain is responsible for human disease in the whole Busoga focus. Furthermore, this study showed that human-infective strains (as defined by isolation from humans or resistance to human serum) could be clearly distinguished from non-human-infective strains. This had an important impact on the ability to identify and quantify the importance of animal reservoirs. While studies in Tororo (36) and earlier studies (46) had implicated cattle as a human reservoir in this region, RFLP analysis demonstrated that 23% of cattle harbored human-infective trypanosomes in Tororo (63). Taking into account transmission parameters, it was calculated that the cattle-fly-human transmission cycle is five times more probable than the human-fly-human cycle (62) (Fig. 3). Thus, cattle were very important as an animal reservoir in this epidemic.
Studies on trypanosome genetic exchange during this epidemic generated conflicting results. Considerable linkage disequilibrium was observed when 44 stocks from Tororo were examined (118), suggesting that the trypanosomes in this area were largely clonal. In contrast, another study of 88 stocks demonstrated that the human-infective isolates were epidemic (i.e., genetic exchange was masked by the epidemic spread of a single strain) and the non-human-infective strains were undergoing genetic exchange (62, 63). It was proposed that the difference in levels of genetic exchange between the human- and non-human-infective trypanosomes is due to a reduced transmissibility of the human-infective trypanosomes in tsetse flies (89, 140), leading to a reduction in the opportunity for mating in this population. It is likely that the lower levels of genetic exchange in the human-infective trypanosomes contributed to the stability of the strain responsible for sleeping sickness in the Busoga focus.
In summary, the data from this human sleeping sickness focus indicate that the crucial features of an epidemic are the presence of a human-infective trypanosome strain, a good tsetse fly habitat, a cattle reservoir, and the opportunity for all of these components to interact by close contact.
An Area of a Human Sleeping Sickness Focus Where the Disease is Endemic—What Is the Risk of an Epidemic?
One of the paradoxes of the history of human sleeping sickness is that in some areas disease is endemic with the occurrence of occasional epidemics while other areas apparently have no recorded cases of the disease. A crucial requirement for disease control is to define those regions with a high risk of epidemic formation and those at lower risk. The ability to predict the likelihood of an epidemic would enable control measures and surveillance infrastructures to be put into place in advance. Quite naturally, most epidemiological studies have focused on the devastating epidemics which have ravaged East Africa but few studies have investigated the areas of endemicity with few or no reported cases of human disease. One of the requirements for such a study is to choose a location where epidemiological parameters can be directly compared during epidemic and endemic periods. The Busia District of western Kenya is located within the Busoga focus some 10 to 20 miles from the location of the 1988 to 1992 human sleeping sickness epidemic at Tororo. This region currently has endemic human sleeping sickness, with only seven reported cases in 1991, and thus offers a good opportunity to compare areas of endemic and epidemic infection in a sleeping sickness focus. Infected people in the Busia District were found to be harboring the same strain of T. brucei as were those infected in Tororo (66), and, albeit in a limited study, human-infective trypanosomes were also found in domestic cattle (66). One cow infected with human-infective trypanosomes was only 3.5 years old, and by virtue of being born after any cases of human sleeping sickness in the area, must have contracted the parasite from tsetse flies. Although more studies are required to define the parameters more accurately, the evidence suggest that human-infective trypanosomes are present in cattle and circulating in tsetse flies in this area of endemicity.
MODEL FOR THE ORIGINS AND MAINTENANCE OF HUMAN SLEEPING SICKNESS FOCI IN EAST AFRICA
Three basic features define the model proposed here for the origins and maintenance of human sleeping sickness foci in East Africa: (i) each sleeping sickness focus has a single strain of human-infective trypanosome associated with it; (ii) this strain is maintained within the ecosystem of the focus, even in times of endemicity; and (iii) this strain generates an epidemic when certain favorable epidemiological conditions prevail.
A number of stages may be involved in this process. (i) Infrequently, throughout East Africa, the human-infective genotype arises in the background trypanosome population either by general mechanisms such as mutation and genetic recombination or by specific mechanisms such as selection in human serum and selection by host-specific transferrin uptake. These mutant genotypes are then either lost or maintained in the population by genetic drift or genetic recombination. If maintained, these genotypes may create the genetic environment for the generation of a human-infective strain, i.e., the strain responsible for that given focus. (ii) Once a transmission cycle which has the occasional involvement of humans is established, selection in humans acts to maintain a single or small number of human-infective strains in the transmission cycle. The low frequency of infected tsetse flies (0.1%) and infrequent encounters with humans create population bottleneck, resulting in restriction of genetic diversity and lack of frequent genetic exchange in the human-infective strain. (iii) Epidemic selection of the human strain follows when ecological conditions change such that the tsetse fly population (or, more strictly, the biting rate) increases. Close interaction among humans, tsetse, and animal reservoirs and lack of treatment of human disease all serve to increase the density of the infected human population. A full-blown epidemic caused by single trypanosome strain occurs. A sufficient density of infected people is necessary to overcome the disadvantaged transmissibility of human-infective strains. (iv) A period of endemicity follows as population immunity, tsetse fly control, and drug treatment decrease the number of cases. The human-infective strains are left circulating in the tsetse fly and animal reservoirs until such a time as changes in the ecological situation again trigger another epidemic.
From the point of view of control, this model proposes that the key factors in an epidemic are the presence of a human-infective strain; a high tsetse fly infestation or biting rate; close contact between tsetse fly, animal reservoirs, and humans; and the density of the infected human population. It may not be necessary to tackle all of these key factors to prevent or control an epidemic, since a reduction in some or only one factor(s) may be all that is required. An understanding of the quantitative effects of each factor may allow us to select the most appropriate strategy for a given situation.
This model has a number of suggestions of key strategies for controlling an epidemic: (i) prevention of transmission from tsetse flies to humans by tsetse fly control or changes to sociological and agricultural practices which reduce tsetse fly-human contact; (ii) prevention of transmission from domestic animals to humans by changes in agricultural practice which separate animals from domestic dwellings or the treatment of cattle harboring human trypanosomes; and (iii) treatment of people or thinning out of population density to reduce the selection for human-infective trypanosomes.
This model also predicts some additional preventive measures which can be taken to reduce the risk of the generation of an epidemic in areas of endemicity: (i) surveillance to determine the levels of human-infective trypanosomes in the human, tsetse fly, and domestic animal population, and (ii) development of longer-term agricultural and social practices to minimize contact between humans, tsetse flies, and domestic animals (e.g., barrier trapping around the larger settlements).
To achieve these aims, a number of additional tools are required. First, a sensitive detection system, specific to the human-infective strain in a given focus, is needed for surveillance and monitoring. Second, the use of environmentally friendly and effective tsetse fly control and detection methods is needed to reduce transmission. Third, effective drug treatment for both human and cattle is needed to reduce the concentration of infected hosts.
CONCLUSIONS AND FUTURE PERSPECTIVES
Human sleeping sickness in East Africa has been well documented for over 100 years, and yet we seem no closer to eradicating this devastating disease. The continued vulnerability of Africa to this disease is highlighted by the recent epidemic in southern Sudan (73) in which some 10 million people are at risk and in which, in some areas, 20% of the population are infected. Multifaceted approaches to controlling the disease are still being developed along both traditional and more novel lines, such as new approaches to drug discovery based on the anti-trypanosomal activity of plants used in traditional medicine in Uganda (42); the release of sterile male tsetse fly on the island of Zanzibar and the consequent eradication of tsetse flies there (3); and the monitoring of tsetse fly habitats by using satellite imagery (75, 109, 110). Despite these approaches, the disease remains largely intractable. The trypanosome has evolved a niche by clever manipulation of its environment—hosts and vectors—and, perhaps very significantly, by existing in a location where financial and political constraints prevent implementation of control measures. The history of human sleeping sickness in East Africa is inextricably linked with political unrest, scarcity of resources, and lack of the necessary administrative infrastructure to take advantage of recent developments in technology. If these issues are addressed, the current developments in technologies should allow us to consign East African human sleeping sickness to history.
ACKNOWLEDGMENTS
I thank Sue Welburn, Ian Maudlin and Andy Tait, at Glasgow University, for discussion of ideas and for their contributions to our fruitful collaborations in this subject.
I thank the Wellcome Trust and the University of Salford for financial support.
REFERENCES
- 1.Abaru D E. Sleeping sickness in Busoga, Uganda, 1976–1983. Trop Med Parasitol. 1985;36:72–76. [PubMed] [Google Scholar]
- 2.Allsopp R, Baldry D A T. A general description of the Lambwe Valley area of South Nyanza District, Kenya, Bull. W H O. 1972;47:691–698. [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. Sterile sex conquers sleeping sickness. New Sci. 1997;156:12. [Google Scholar]
- 4.Apted F I C. The epidemiology of Rhodesian sleeping sickness. In: Mulligan H W, editor. The African trypanosomiases. London, United Kingdom: George Allen and Unwin; 1970. pp. 645–660. [Google Scholar]
- 5.Apted F I C, Ormerod W E, Smyly D P, Stronach B W. A comparative study of the epidemiology of endemic rhodesian sleeping sickness in different parts of Africa. J Trop Med Hyg. 1963;66:1–16. [PubMed] [Google Scholar]
- 6.Bajyana-Songa E, Hamers R, Rickman L R, Nantulya V M, Mulla A F, Magnus E. Evidence for widespread asymptomatic Trypanosoma rhodesiense human infection in the Luangwa Valley (Zambia) Trop Med Parasitol. 1991;42:389–393. [PubMed] [Google Scholar]
- 7.Baker J R, McConnell E, Kent D C, Hady J. Human trypanosomiasis in Ethiopia. Ecology of Illubabor Province and epidemiology in the Baro River area. Trans R Soc Trop Med Hyg. 1970;64:523–530. doi: 10.1016/0035-9203(70)90074-x. [DOI] [PubMed] [Google Scholar]
- 8.Baldry D A T. A history of Rhodesian sleeping sickness in the Lambwe Valley. Bull W H O. 1972;47:699–718. [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett J. Control strategies for African trypanosomiases: their sustainability and effectiveness. In: Hide G, Mottram J C, Coombs G H, Holmes P H, editors. Trypanosomiasis and leishmaniasis: biology and control. Wallingford, United Kingdom: CAB International; 1997. pp. 347–359. [Google Scholar]
- 10.Barrett M P, MacLeod A, Tovar J, Sweetman J P, Tait A, Le Page R W F, Melville S E. A single locus minisatellite sequence which distinguishes between Trypanosoma brucei isolates. Mol Biochem Parasitol. 1997;86:95–99. doi: 10.1016/s0166-6851(97)90009-8. [DOI] [PubMed] [Google Scholar]
- 11.Barry J D B. The biology of antigenic variation in African trypanosomes. In: Hide G, Mottram J C, Coombs G H, Holmes P H, editors. Trypanosomiasis and leishmaniasis: biology and control. Wallingford, United Kingdom: CAB International; 1997. pp. 89–108. [Google Scholar]
- 12.Barth P. A new method for the isolation of the trypanocidal factor from normal human serum. Acta Trop. 1989;46:71–73. doi: 10.1016/0001-706x(89)90019-3. [DOI] [PubMed] [Google Scholar]
- 13.Baylis M, Stevenson P. Trypanosomiasis and tsetse control with insecticidal pour-ons—fact and fiction? Parasitol Today. 1998;14:77–82. doi: 10.1016/s0169-4758(97)01170-8. [DOI] [PubMed] [Google Scholar]
- 14.Bitter W, Gerrits H, Kieft R, Borst P. The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature. 1998;391:499–502. doi: 10.1038/35166. [DOI] [PubMed] [Google Scholar]
- 15.Borst P, Bitter W, Blundell P, Cross M, McCulloch R, Rudenko G, Taylor M C, van Leeuwen F. The expression sites for variant surface glycoproteins of Trypanosoma brucei. In: Hide G, Mottram J C, Coombs G H, Holmes P H, editors. Trypanosomiasis and leishmaniasis: biology and control. Wallingford, United Kingdom: CAB International; 1997. pp. 109–132. [Google Scholar]
- 16.Brightwell R, Dransfield R D, Stevenson P, Williams B. Changes over twelve years in populations of Glossina pallidipes and Glossina longipennis (Diptera: Glossinidae) subject to varying trapping pressure at Nguruman, south-west Kenya. Bull Entomol Res. 1997;87:349–370. [Google Scholar]
- 17.Bruce D. Preliminary report on the tsetse fly disease or Nagana in Zululand. Durban, South Africa: Bennett and David; 1895. [Google Scholar]
- 18.Bruce D. Further report on sleeping sickness in Uganda. Rep Sleeping Sickness Comm R Soc. 1903;4:3–6. [Google Scholar]
- 19.Brun R, Jenni L. Human serum resistance of metacyclic forms of Trypanosoma brucei brucei, T. brucei rhodesiense and T. brucei gambiense. Parasitol Res. 1987;73:218–223. doi: 10.1007/BF00578507. [DOI] [PubMed] [Google Scholar]
- 20.Buyst H. The epidemiology, clinical features, treatment and history of sleeping sickness in the northern edge of the Luangwa fly belt. Med J Zambia. 1974;8:2–12. [Google Scholar]
- 21.Christy C. Sleeping Sickness. Comm R Soc. 1903;4:3–6. [Google Scholar]
- 22.Cibulskis R E. Origins and genetic diversity in natural populations of Trypanosoma brucei. Parasitology. 1988;96:303–322. doi: 10.1017/s0031182000058315. [DOI] [PubMed] [Google Scholar]
- 23.Cibulskis R E. Genetic variation in Trypanosoma brucei and the epidemiology of sleeping sickness in the Lambwe Valley, Kenya. Parasitology. 1992;104:99–109. doi: 10.1017/s0031182000060844. [DOI] [PubMed] [Google Scholar]
- 24.Croft S L, Urbina J A, Brun R. Chemotherapy of human leishmaniasis and trypanosomiasis. In: Hide G, Mottram J C, Coombs G H, Holmes P H, editors. Trypanosomiasis and leishmaniasis: biology and control. Wallingford, United Kingdom: CAB International; 1997. pp. 245–258. [Google Scholar]
- 25.Degen R, Pospichal H, Enyaru J C K, Jenni L. Sexual compatibility among Trypanosoma brucei isolates from an epidemic area in Southeastern Uganda. Parasit Res. 1995;81:253–257. doi: 10.1007/BF00937118. [DOI] [PubMed] [Google Scholar]
- 26.De Greef C, Imbrechts H, Matthyssens G, Van Meirvenne N, Hamers R. A gene expressed only in serum-resistant variants of Trypanosoma brucei rhodesiense. Mol Biochem Parasitol. 1989;36:169–176. doi: 10.1016/0166-6851(89)90189-8. [DOI] [PubMed] [Google Scholar]
- 27.De Greef C, Hamers R. The serum resistance-associated (SRA) gene of Trypanosoma brucei rhodesiense encodes a variant surface glycoprotein-like protein. Mol Biochem Parasitol. 1994;68:277–284. doi: 10.1016/0166-6851(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 28.De Raadt P. African sleeping sickness today. Trans R Soc Trop Med Hyg. 1976;70:114–116. doi: 10.1016/0035-9203(76)90165-6. [DOI] [PubMed] [Google Scholar]
- 29.Duke H L. An enquiry into an outbreak of human trypanosomiasis in a “G. morsitans” belt, to the east of Mwanza, Tanganyika Territory. Proc R Soc London Ser B. 1923;94:250. [Google Scholar]
- 30.Duke H L. On the occurrence in main of strains of T. gambiense non-transmissible cyclically by G. palpalis. Parasitology. 1930;22:491–504. [Google Scholar]
- 31.Dukes P, Scott C M, Rickman L R, Wupara F. Sleeping sickness in the Luangwa Valley of Zambia. A preliminary report of the 1982 outbreak at Kasyasya Village. Bull Soc Pathol Exot. 1983;76:605–613. [PubMed] [Google Scholar]
- 32.Dukes P, Rickman L R, Killick-Kendrick R, Kakoma I, Wurapa F K, De Raadt P, Morrow R. A field comparison of seven diagnostic tests for human trypanosomiasis in the Luangwa Valley, Zambia. Tropenmed Parasitol. 1984;35:141–147. [PubMed] [Google Scholar]
- 33.Dutton J E. Preliminary note on the trypanosomes occurring in the blood of man. Br Med J. 1902;2:881. [Google Scholar]
- 34.Elliot M. A photostable pyrethroid. Nature. 1973;246:169–170. doi: 10.1038/246169a0. [DOI] [PubMed] [Google Scholar]
- 35.Enyaru J C K, Odiit M, Gashumba J K, Carasco J F, Rwendeire A J J. Characterisation by isoenzyme electrophoresis of Trypanozoon stocks from sleeping sickness endemic areas of Southeast Uganda. Bull W H O. 1992;70:631–636. [PMC free article] [PubMed] [Google Scholar]
- 36.Enyaru J C K, Stevens J R, Odiit M, Okuna N M, Carasco J F. Isoenzyme comparison of Trypanozoon isolates from two sleeping sickness areas of south-eastern Uganda. Acta Trop. 1993;55:97–115. doi: 10.1016/0001-706x(93)90072-j. [DOI] [PubMed] [Google Scholar]
- 37.Enyaru J C K, Allingham R, Bromidge T, Kanmogue G D, Carasco J F. The isolation and genetic heterogeneity of Trypanosoma brucei gambiense from north-west Uganda. Acta Trop. 1993;54:31–39. doi: 10.1016/0001-706x(93)90066-k. [DOI] [PubMed] [Google Scholar]
- 38.Enyaru J C K, Matovu E, Odiit M, Okedi L A, Rwendeire A J J, Stevens J R. Genetic diversity in Trypanosoma (Trypanozoon) brucei isolates from mainland and Lake Victoria island populations in south-eastern Ugands: epidemiological and control implications. Ann Trop Med Parasitol. 1997;91:107–113. doi: 10.1080/00034983.1997.11813118. [DOI] [PubMed] [Google Scholar]
- 39.Fairbairn H. Sleeping sickness in Tanganika Territory, 1922–1946. Trop Dis Bull. 1948;45:1–17. [Google Scholar]
- 40.Ford J. The role of the trypanosomiases in African ecology. A study of the tsetse fly problem. Oxford, United Kingdom: Clarendon Press; 1971. [Google Scholar]
- 41.Foulkes J. Human trypanosomiasis in Zambia. Med J Zambia. 1970;4:167–177. [Google Scholar]
- 42.Freiburghaus F, Ogwal E N, Nkunya M H H, Kaminsky R, Brun R. In vitro antitrypanosomal activity of African plants used in traditional medicine in Uganda to treat sleeping sickness. Trop Med Int Health. 1996;1:765–771. doi: 10.1111/j.1365-3156.1996.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 43.Gashumba J K, Komba E K, Truc P, Allingham R M, Ferris V, Godfrey D G. The persistence of genetic homogeneity among Trypanosoma brucei rhodesiense isolates from patients in north-west Tanzania. Acta Trop. 1994;56:341–348. doi: 10.1016/0001-706x(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 44.Geigy R, Mwambu P M, Kaufmann M. Sleeping sickness survey in Musoma District, Tanzania. IV. Examination of wild mammals as a potential reservoir for T. rhodesiense. Acta Trop. 1971;30:49–56. [PubMed] [Google Scholar]
- 45.Gibson W C. Analysis of a genetic cross between Trypanosoma brucei rhodesiense and T. b. brucei. Parasitology. 1989;99:391–402. doi: 10.1017/s0031182000059114. [DOI] [PubMed] [Google Scholar]
- 46.Gibson W C, Gashumba J K. Isoenzyme characterization of some Trypanozoon stocks from a recent trypanosomiasis epidemic in Uganda. Trans R Soc Trop Med Hyg. 1983;77:114–118. doi: 10.1016/0035-9203(83)90033-0. [DOI] [PubMed] [Google Scholar]
- 47.Gibson W C, Wellde B T. Characterization of Trypanozoon stocks from the South Nyanza sleeping sickness focus in Western Kenya. Trans R Soc Trop Med Hyg. 1985;79:671–676. doi: 10.1016/0035-9203(85)90187-7. [DOI] [PubMed] [Google Scholar]
- 48.Gibson W C, Marshall T F C, Godfrey D G. Numerical analysis of enzyme polymorphism. A new approach to the epidemiology and taxonomy of trypanosomes of the genus Trypanozoon. Adv Parasitol. 1980;18:175–245. doi: 10.1016/s0065-308x(08)60400-5. [DOI] [PubMed] [Google Scholar]
- 49.Gibson W C, Borst P, Fase-Fower F. Further analysis of intraspecific variation in Trypanosoma brucei using restriction site polymorphisms in the maxi-circle kinetoplast DNA. Mol Biochem Parasitol. 1985;51:21–366. doi: 10.1016/0166-6851(85)90026-x. [DOI] [PubMed] [Google Scholar]
- 50.Godfrey D G, Kilgour V. Enzyme electrophoresis in characterising the causative organism of gambian trypanosomiasis. Trans R Soc Trop Med Hyg. 1976;70:219–224. doi: 10.1016/0035-9203(76)90043-2. [DOI] [PubMed] [Google Scholar]
- 51.Godfrey D G, Baker R D, Rickman L R, Mehlitz D. The distribution, relationships and identification of enzymic variants within the subgenus Trypanozoon. Adv Parasitol. 1990;29:1–74. doi: 10.1016/s0065-308x(08)60104-9. [DOI] [PubMed] [Google Scholar]
- 52.Hajduk S L, Hager K, Esko J D. High-density lipoprotein-mediated lysis of trypanosomes. Parasitol Today. 1992;8:95–98. doi: 10.1016/0169-4758(92)90247-y. [DOI] [PubMed] [Google Scholar]
- 53.Hajduk S L, Moore D R, Vasudevacharya J, Siqueira H, Torri A F, Tytler E M, Esko J D. Lysis of Trypanosoma brucei by a toxic subspecies of human high density lipoprotein. J Biol Chem. 1989;264:5210–5217. [PubMed] [Google Scholar]
- 54.Hager K M, Hajduk S L. Mechanism of resistance in African trypanosomes to cytotoxic human HDL. Nature. 1997;385:823–826. doi: 10.1038/385823a0. [DOI] [PubMed] [Google Scholar]
- 55.Hawking F. The resistance to human plasma of Trypanosoma brucei, Trypanosome rhodesiense and Trypanosoma gambiense. I. Analysis of the composition of trypanosome strains. Trans R Soc Trop Med Hyg. 1976;70:504–512. doi: 10.1016/0035-9203(76)90138-3. [DOI] [PubMed] [Google Scholar]
- 56.Heisch R B, McMahon J P, Manson-Bahr P E C. The isolation of Trypanosoma rhodesiense from bushbuck. Br Med J. 1958;2:1203. doi: 10.1136/bmj.2.5106.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herbert W J, Parratt D, van Meirvenne N, Lennox B. An accidental laboratory infection with trypanosomes of a defined stock. II. Studies on the serological response of the patient and the identity of the infecting organism. J Infect. 1980;2:113–124. doi: 10.1016/s0163-4453(80)91109-3. [DOI] [PubMed] [Google Scholar]
- 58.Hide G. The molecular identification of trypanosomes. Methods Mol Biol. 1996;50:243–263. doi: 10.1385/0-89603-323-6:243. [DOI] [PubMed] [Google Scholar]
- 59.Hide G. The molecular epidemiology of trypanosomatids. In: Hide G, Mottram J C, Coombs G H, Holmes P H, editors. Trypanosomiasis and leishmaniasis: biology and control. Wallingford, United Kingdom: CAB International; 1997. pp. 289–304. [Google Scholar]
- 60.Hide G. Comment on Dr Tibayrenc’s lecture. In: Silva R A M S, Davila A M R, editors. : Proceedings of the First Internet Conference on Salivarian Trypanosomes. FAO Animal Production and Health paper 136. 1997. pp. 47–51. [Google Scholar]
- 61.Hide G, Tait A. The molecular epidemiology of parasites. Experientia. 1991;47:128–142. doi: 10.1007/BF01945413. [DOI] [PubMed] [Google Scholar]
- 62.Hide G, Tait A, Maudlin I, Welburn S C. The origins, dynamics and generation of Trypanosoma brucei rhodesiense epidemics in East Africa. Parasitol Today. 1996;12:50–55. doi: 10.1016/0169-4758(96)80654-5. [DOI] [PubMed] [Google Scholar]
- 63.Hide G, Welburn S C, Tait A, Maudlin I. Epidemiological relationships of Trypanosoma brucei stocks from South East Uganda: evidence for different population structures in human and non human trypanosomes. Parsitology. 1994;109:95–111. doi: 10.1017/s0031182000077805. [DOI] [PubMed] [Google Scholar]
- 64.Hide G, Buchanan N, Welburn S C, Maudlin I, Barry J D, Tait A. Trypanosoma brucei rhodesiense: characterization of stocks from Zambia, Kenya and Uganda using repetitive DNA probes. Exp Parasitol. 1991;72:430–439. doi: 10.1016/0014-4894(91)90089-f. [DOI] [PubMed] [Google Scholar]
- 65.Hide G, Cattand P, Le Ray D, Barry J D, Tait A. The identification of T. brucei subspecies using repetitive DNA sequences. Mol Biochem Parasitol. 1990;39:213–226. doi: 10.1016/0166-6851(90)90060-y. [DOI] [PubMed] [Google Scholar]
- 66.Hide G, Angus S, Holmes P H, Maudlin I, Welburn S C. Comparison of Trypanosoma brucei strains circulating in an endemic and an epidemic area of a sleeping sickness focus. Exp Parasitol. 1998;89:21–29. doi: 10.1006/expr.1998.4265. [DOI] [PubMed] [Google Scholar]
- 67.Hoare C A. The trypanosomes of mammals. Oxford, United Kingdom: Blackwell Scientific Publications Ltd.; 1972. [Google Scholar]
- 68.Hutchinson M P. Human trypanosomiasis in south west Ethiopia (March 1967 to March 1970) Ethiop Med J. 1971;9:3–69. [PubMed] [Google Scholar]
- 69.Jenni L, Brun R. A new in vitro test for human serum resistance of Trypanosoma (Trypanozoon) brucei. Acta Trop. 1982;39:281–284. [PubMed] [Google Scholar]
- 70.Jenni L, Marti S, Schweizer J, Betschart B, Le Page R W F, Wells J M, Tait A, Paindavoine P, Pays E, Steinert M. Hybrid formation between African trypanosomes during cyclical transmission. Nature. 1986;322:173–175. doi: 10.1038/322173a0. [DOI] [PubMed] [Google Scholar]
- 71.Jennings F W, Urquhart G M. Induction of human serum-sensitive Trypanosoma brucei stabilates into human serum-resistant “T. rhodesiense.”. Trans R Soc Trop Med Hyg. 1985;79:80–85. doi: 10.1016/0035-9203(85)90243-3. [DOI] [PubMed] [Google Scholar]
- 72.Jordan A M. Trypanosomiasis control and african rural development. New York, N.Y: Longman; 1986. [Google Scholar]
- 73.Kigotho A W. Southern Sudan hit by epidemic of sleeping sickness. Lancet. 1997;350:502. [Google Scholar]
- 74.Kilhamia C M, Komba E, Mella P N P, Mbwanbo H A. Trypanosomiasis. In: Mwaluko G M P, Kilama W L, Mandara M P, Murru M, MacPherson C N L, editors. Health and disease in Tanzania. London, United Kingdom: Harper Collins Academic; 1991. pp. 133–143. [Google Scholar]
- 75.Kitron U, Otieno L H, Hungerford L L, Odulaja A, Brigham W U, Okello O O, Joselyn M, Mohamedahmed M M, Cook E. Spatial analysis of the distribution of tsetse flies in the Lambwe Valley, Kenya, using LANDSAT™ satellite imagery and GIS. J Anim Ecol. 1996;65:371–380. [Google Scholar]
- 76.Koerner T, de Raadt P, Maudlin I. The 1901 Uganda sleeping sickness epidemic revisited—a case of mistaken identity? Parasitol Today. 1995;11:303–306. doi: 10.1016/0169-4758(95)80046-8. [DOI] [PubMed] [Google Scholar]
- 77.Komba E K, Odiit M, Mbulamberi D B, Chimfwembe E C, Nantuyla V M. Multicentre evaluation of an antigen-detection ELISA for the diagnosis of Trypanosoma brucei rhodesiense sleeping sickness. Bull W H O. 1992;70:57–61. [PMC free article] [PubMed] [Google Scholar]
- 78.Komba E K, Kibona S N, Ambwene A K, Stevens J R, Gibson W C. Genetic diversity among Trypanosoma brucei rhodesiense isolates from Tanzania. Parasitology. 1998;115:571–579. doi: 10.1017/s0031182097001856. [DOI] [PubMed] [Google Scholar]
- 79.Kuzoe F A S. Current situation of African trypanosomiasis. Acta Trop. 1993;54:153–162. doi: 10.1016/0001-706x(93)90089-t. [DOI] [PubMed] [Google Scholar]
- 80.Laveran A, Mesnil F. Trypanosomes and trypanosomiasis. London, United Kingdom: Balliere, Tindall and Cox; 1907. [Google Scholar]
- 81.Lorenz P, James R W, Owen J S, Betschart B. Heterogeneity in the properties of the trypanolytic factor in normal human serum. Mol Biochem Parasitol. 1994;64:153–164. doi: 10.1016/0166-6851(94)90143-0. [DOI] [PubMed] [Google Scholar]
- 82.MacKichan I W. Rhodesian sleeping sickness in eastern Uganda. Trans R Soc Trop Med Hyg. 1944;38:49–60. [Google Scholar]
- 83.MacLean G. Ann. Trop Med Parasitol. 1926;20:329. [Google Scholar]
- 84.MacLeod A, Turner C M R, Tait A. Detection of single copy gene sequences from single trypanosomes. Mol Biochem Parasitol. 1997;84:267–270. doi: 10.1016/s0166-6851(96)02810-1. [DOI] [PubMed] [Google Scholar]
- 85.Mathieu-Daude F, Stevens J, Welsh J, Tibayrenc M, McLelland M. Genetic diversity and population structure of Trypanosoma brucei: clonality versus sexuality. Mol Biochem Parasitol. 1995;72:89–101. doi: 10.1016/0166-6851(95)00083-d. [DOI] [PubMed] [Google Scholar]
- 86.Maudlin, I., S. C. Welburn, J. K. Gashumba, N. Okuna, and M. Kalunda. 1990. The role of cattle in the epidemiology of sleeping sickness in Uganda. Bull. Soc. Fr. Parasitol. 8(Suppl. 2):788.
- 87.Maynard Smith J, Smith N H, O’Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mihok S, Otieno L H, Darji N. Population genetics of Trypanosoma brucei and the epidemiology of human sleeping sickness in the Lambwe Valley. Parasitology. 1990;100:219–233. doi: 10.1017/s0031182000061229. [DOI] [PubMed] [Google Scholar]
- 89.Milligan P J M, Maudlin I, Welburn S C. Trypanozoon: infectivity to humans is linked to reduced transmissibility in tsetse. II. Genetic mechanisms. Exp Parasitol. 1995;81:409–415. doi: 10.1006/expr.1995.1132. [DOI] [PubMed] [Google Scholar]
- 90.Molyneux D H. Current public health status of the trypanosomiases and leishmaniases. In: Hide G, Mottram J C, Coombs G H, Holmes P H, editors. Trypanosomiasis and leishmaniasis: biology and control. Wallingford, United Kingdom: CAB International; 1997. pp. 39–50. [Google Scholar]
- 91.Morrison W I, Wells P W, Moloo S K, Paris J, Murray M. Interference in the establishment of superinfections with Trypanosoma congolense in cattle. J Parasitol. 1982;68:755–764. [PubMed] [Google Scholar]
- 92.Mulla A F, Rickman L R. Evidence for the presence of an innate trypanosomicidal factor in the serum of a non immune African waterbuck (Kobus ellipsiprymmus) Trans R Soc Trop Med Hyg. 1988;82:97–98. doi: 10.1016/0035-9203(88)90274-x. [DOI] [PubMed] [Google Scholar]
- 93.Nei M. Genetic distance between populations. Am Nat. 1972;106:283–292. [Google Scholar]
- 94.Okoth J O, Kapaata R. Trypanosome infection rates in Glossina fuscipes fuscipes Newst. in the Busoga sleeping sickness focus, Uganda. Ann Trop Med Parasitol. 1986;80:459–461. doi: 10.1080/00034983.1986.11812048. [DOI] [PubMed] [Google Scholar]
- 95.Onyango R J, van Hoeve K, de Raadt P. The epidemiology of Trypanosoma rhodesiense sleeping sickness in Alego location, Central Nyanza, Kenya. Evidence that cattle may act as a reservoir host of trypanosomes infectious to man. Trans R Soc Trop Med Hyg. 1966;60:175–182. doi: 10.1016/0035-9203(66)90024-1. [DOI] [PubMed] [Google Scholar]
- 96.Ormerod W E. The epidemic spread of Rhodesian sleeping sickness 1908–1960. Trans R Soc Trop Med Hyg. 1961;55:525–538. doi: 10.1016/0035-9203(60)90110-3. [DOI] [PubMed] [Google Scholar]
- 97.Otieno L H, Darji N. Characterization of potentially man-infective Trypanosoma brucei from endemic areas of sleeping sickness in Kenya. Trop Med Parasitol. 1985;36:123–126. [PubMed] [Google Scholar]
- 98.Pepin J, Milord F. The treatment of African trypanosomiasis. Adv Parasitol. 1994;33:1–47. doi: 10.1016/s0065-308x(08)60410-8. [DOI] [PubMed] [Google Scholar]
- 99.Raper J, Nussenzweig V, Tomlinson S. The main lytic factor of Trypanosoma brucei brucei in normal human serum is not high density lipoprotein. J Exp Med. 1996;183:1023–1029. doi: 10.1084/jem.183.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rickman L R. Investigations into an outbreak of human trypanosomiasis in the lower Luangwa Valley, Eastern Province, Zambia. E Afr Med J. 1974;51:467–487. [PubMed] [Google Scholar]
- 101.Rickman L R, Kolala F. Relationship between Trypanosoma brucei and T. rhodesiense. Trans R Soc Trop Med Hyg. 1980;74:817–819. doi: 10.1016/0035-9203(80)90214-x. [DOI] [PubMed] [Google Scholar]
- 102.Rickman L R, Kolala F. Effects of some animal sera on Trypanosoma brucei rhodesiense and T. b. brucei clones. Trop Med Parasitol. 1982;33:129–135. [PubMed] [Google Scholar]
- 103.Rickman L R, Robson J. The blood incubation infectivity tests: a simple test which may serve to distinguish Trypanosoma brucei from T. rhodesiense. Bull W H O. 1970;42:650–651. [PMC free article] [PubMed] [Google Scholar]
- 104.Rickman L R, Robson J. The testing of proven T. brucei and T. rhodesiense strains by the blood incubation infectivity test. Bull W H O. 1970;42:911–916. [PMC free article] [PubMed] [Google Scholar]
- 105.Rifkin M R. Identification of the trypanocidal factor in normal human serum: high density lipoprotein. Proc Natl Acad Sci USA. 1978;75:3450–3454. doi: 10.1073/pnas.75.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rifkin M R, De Greef C, Jiwa A, Landsberger F R, Shapiro S Z. Human serum-sensitive Trypanosoma brucei rhodesiense: a comparison with seriologically identical human serum-resistant clones. Mol Biochem Parasitol. 1994;66:211–220. doi: 10.1016/0166-6851(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 107.Robertson D H H, Baker J R. Human trypanosomiasis in south-east Uganda. 1. A study of the epidemiology and the present virulence of the disease. Trans R Soc Trop Med Hyg. 1958;52:337–348. doi: 10.1016/0035-9203(58)90047-6. [DOI] [PubMed] [Google Scholar]
- 108.Robertson D H H, Pickens S, Lawson J H, Lennox B. An accidental laboratory infection with African trypanosomes of a defined stock. I. The clinical course of the infection. J Infect. 1980;2:105–112. doi: 10.1016/s0163-4453(80)91084-1. [DOI] [PubMed] [Google Scholar]
- 109.Robinson T, Rogers D J, Williams B. Univariate analysis of tsetse habitat in the commonly fly belt of Southern Africa using climate and remotely sensed vegetation data. Med Vet Entomol. 1997;11:223–234. doi: 10.1111/j.1365-2915.1997.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 110.Robinson T, Rogers D J, Williams B. Mapping tsetse habitat suitability in the common fly belt of Southern Africa using multivariant analysis of climate and remotely sensed vegetation data. Med Vet Entomol. 1997;11:235–245. doi: 10.1111/j.1365-2915.1997.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 111.Robson J, Rickman L R, Allsopp R, Scott D. The composition of the Trypanosoma brucei subgroup in non human reservoirs in the Lambwe Valley of Kenya, with particular reference to the distribution of T. rhodesiense. Bull W H O. 1972;46:765–770. [PMC free article] [PubMed] [Google Scholar]
- 112.Seed J R, Sechelski J B, Ortiz J O, Chapman J F. Relationship between human serum trypanocidal activity and host resistance to the african trypanosomes. J Parasitol. 1993;79:226–232. [PubMed] [Google Scholar]
- 113.Smith A B, Esko J D, Hajduk S L. Killing of trypanosomes by the human haptoglobin-related protein. Science. 1995;268:284–286. doi: 10.1126/science.7716520. [DOI] [PubMed] [Google Scholar]
- 114.Southon H A, Robertson D H H. isolation of Trypanosoma rhodesiense from wild Glossina palpalis. Nature. 1961;189:411–412. [Google Scholar]
- 115.Stephens J W W, Fantham H B. On the peculiar morphology of a trypanosome from a case of sleeping sickness and the possibility of its being a new species (T. rhodesiense) Ann. Trop Med Parasitol. 1910;4:343–350. [Google Scholar]
- 116.Sternberg J, Turner C M R, Wells J M, Ranford-Cartwright L C, Le Page R W F, Tait A. Gene exchange in African trypanosomes: frequency and allelic segregation. Mol Biochem Parasitol. 1989;34:269–280. doi: 10.1016/0166-6851(89)90056-x. [DOI] [PubMed] [Google Scholar]
- 117.Stevens J R, Tibayrenc M. Trypanosoma brucei s.l.: evolution, linkage and the clonality debate. Parasitology. 1996;112:481–488. doi: 10.1017/s0031182000076940. [DOI] [PubMed] [Google Scholar]
- 118.Stevens J R, Welburn S C. Genetic processes within an epidemic of sleeping sickness. Parasitol Res. 1993;79:421–427. doi: 10.1007/BF00931833. [DOI] [PubMed] [Google Scholar]
- 119.Tait A, Buchanan N, Hide G, Turner C M R. Evidence for self-fertilisation in T. brucei. Mol Biochem Parasitol. 1996;76:31–42. doi: 10.1016/0166-6851(95)02528-6. [DOI] [PubMed] [Google Scholar]
- 120.Tait A. Evidence for diploidy and mating in trypanosomes. Nature. 1980;287:536–538. doi: 10.1038/287536a0. [DOI] [PubMed] [Google Scholar]
- 121.Tait A, Turner C M R. Genetic exchange in Trypanosoma brucei. Parasitol Today. 1990;6:70–75. doi: 10.1016/0169-4758(90)90212-m. [DOI] [PubMed] [Google Scholar]
- 122.Tait A, Babiker E A, Le Ray D. Enzyme variation in Trypanosoma brucei spp. I. Evidence for the subspeciation of Trypanosoma brucei gambiense. Parasitology. 1984;89:311–326. doi: 10.1017/s0031182000001335. [DOI] [PubMed] [Google Scholar]
- 123.Tait A, Barry J D, Wink R, Sanderson A, Crowe J S. Enzyme variation in Trypanosoma brucei ssp. II. Evidence for T. b. rhodesiense being a subset of variants of T. b. brucei. Parasitology. 1985;90:89–100. doi: 10.1017/s0031182000049040. [DOI] [PubMed] [Google Scholar]
- 124.Thon G, Baltz T, Eisen H. Antigenic diversity by the recombination of pseudogenes. Genes Dev. 1989;3:1247–1254. doi: 10.1101/gad.3.8.1247. [DOI] [PubMed] [Google Scholar]
- 125.Tibayrenc M. Population genetics of parasitic protozoa and other microorganisms. Adv Parasitol. 1995;36:47–115. doi: 10.1016/s0065-308x(08)60490-x. [DOI] [PubMed] [Google Scholar]
- 126.Tibayrenc M. Evolutionary genetics of Trypanosoma, Leishmania and other micro-organisms: epidemiological, taxonomical and medical implications. In: Hide G, Mottram J C, Coombs G H, Holmes P H, editors. Trypanosomiasis and leishmaniasis: biology and control. Wallingford, United Kingdom: CAB International; 1997. pp. 305–315. [Google Scholar]
- 127.Tibayrenc M, Kejllberg F, Ayala F J. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, and Leishmania, Naegleria, Plasmodium, Trichomonas and Trypanosoma and their medical and taxonomic consequences. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tomlinson S, Raper J. The lysis of Trypanosoma brucei brucei by human serum. Nat Biotechnol. 1996;14:717–721. doi: 10.1038/nbt0696-717. [DOI] [PubMed] [Google Scholar]
- 129.Vale G A. The responses tsetse flies (Diptera: Glossinidae) to mobile and stationary baits. Bull Entomol Res. 1974;64:545–588. [Google Scholar]
- 130.Vale G A, Hargrove J W. Field attraction of tsetse flies (Diptera: Glossinidae) to ox odour: effects of dose. Trans Rhod Sci Assoc. 1975;56:46–50. [Google Scholar]
- 131.Vale G A, Hargrove J W, Cockbill G F, Phelps R J. Field trials of baits to control populations of Glossinia morsitans morsitans Westwood and G. pallidipes Austen (Diptera: Glossinidae) Bull Entomol Res. 1986;76:179–193. [Google Scholar]
- 132.Van der Ploeg L H T, Valerio D, De Lange T, Bernards A, Borst P, Grosveld F G. An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustered in the genome. Nucleic Acids Res. 1982;10:5905–5923. doi: 10.1093/nar/10.19.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Van Meirvenne N, Magnus E, Janssens P G. The effect of normal human serum on trypanosomes of distinct antigenic type (Etat 1-12) isolated from a strain of Trypanosoma brucei rhodesiense. Ann Soc Belge Med Trop. 1976;56:55–63. [PubMed] [Google Scholar]
- 134.Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Br Med J. 1985;42:105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- 135.Vickerman K. Landmarks in trypanosome research. In: Hide G, Mottram J C, Coombs G H, Holmes P H, editors. Trypanosomiasis and leishmaniasis: biology and control. Wallingford, United Kingdom: CAB International; 1997. pp. 1–38. [Google Scholar]
- 136.Waitumbi J N, Murphy N B. Inter-and intra-species differentiation of trypanosomes by genomic fingerprinting with arbitrary primers. Mol Biochem Parasitol. 1993;58:181–186. doi: 10.1016/0166-6851(93)90105-7. [DOI] [PubMed] [Google Scholar]
- 137.Watson H J C. The epidemiology of human sleeping sickness in the Lambwe Valley, South Nyanza, Kenya. Bull W H O. 1972;47:719–726. [PMC free article] [PubMed] [Google Scholar]
- 138.Welburn S C, Maudlin I. The nature of the teneral state in Glossina and its role in the acquisition of trypanosome infection in tsetse. Ann Trop Med Parasitol. 1992;86:529–536. doi: 10.1080/00034983.1992.11812703. [DOI] [PubMed] [Google Scholar]
- 139.Welburn S C, Maudlin I. Current trends in parasite vector interactions. In: Hide G, Mottram J C, Coombs G H, Holmes P H, editors. Trypanosomiasis and leishmaniasis: biology and control. Wallingford, United Kingdom: CAB International; 1997. pp. 315–326. [Google Scholar]
- 140.Welburn S C, Maudlin I, Milligan P J M. Trypanozoon: infectivity to humans is linked to reduced transmissibility in tsetse. I. Comparison of human serum-resistant and human serum-sensitive field isolates. Exp Parasitol. 1995;81:404–408. doi: 10.1006/expr.1995.1131. [DOI] [PubMed] [Google Scholar]
- 141.Willett K C. Some observations on the recent epidemiology of sleeping sickness in Nyanza region, Kenya, and its relation to the general epidemiology of Gambian and Rhodesian sleeping sickness in Africa. Trans R Soc Trop Med Hyg. 1965;59:374–394. doi: 10.1016/0035-9203(65)90055-6. [DOI] [PubMed] [Google Scholar]