Abstract
Strain typing of medically important fungi and fungal population genetics have been stimulated by new methods of tapping DNA variation. The aim of this contribution is to show how awareness of fungal population genetics can increase the utility of strain typing to better serve the interests of medical mycology. Knowing two basic features of fungal population biology, the mode of reproduction and genetic differentiation or isolation, can give medical mycologists information about the intraspecific groups that are worth identifying and the number and type of markers that would be needed to do so. The same evolutionary information can be just as valuable for the selection of fungi for development and testing of pharmaceuticals or vaccines. The many methods of analyzing DNA variation are evaluated in light of the need for polymorphic loci that are well characterized, simple, independent, and stable. Traditional population genetic and new phylogenetic methods for analyzing mode of reproduction, genetic differentiation, and isolation are reviewed. Strain typing and population genetic reports are examined for six medically important species: Coccidioides immitis, Histoplasma capsulatum, Candida albicans, Cryptococcus neoformans, Aspergillus fumigatus, and A. flavus. Research opportunities in the areas of genomics, correlation of clinical variation with genetic variation, amount of recombination, and standardization of approach are suggested.
Strain typing of medically important fungi, i.e., the ability to identify them to the species level and to discriminate among individuals within species, has been galvanized by new methods of tapping the tremendous variation found in fungal DNA. The aim of this review is to show how awareness of fungal population genetics can increase the utility of strain typing to better serve the interests of medical mycology. By paying attention to attributes of the life cycle, such as the mode of reproduction and genetic differentiation or isolation, mycologists can tell which intraspecific groups are worth identifying and can determine the number and type of markers that are needed to do so. The same evolutionary information can be just as valuable to the selection of fungi for development and testing of pharmaceuticals or vaccines. In the process, we should gain a better basic understanding of the behavior of pathogenic fungi in nature.
Initially, most of the application of fungal DNA variation to evolution was directed above the species level (7, 8, 11, 12, 13, 21, 45, 73, 91, 105), but that is no longer the case. Our ability to distinguish individuals in species of systemic and opportunistic pathogenic fungi is approaching the elusive goal of being able to identify every different genotype by a simple laboratory procedure. As discussed below, this goal is within our grasp because of recent reports of studies involving repetitive DNA sequences (33) or microsatellites (19, 25, 39). These advances in detection methods will make studies of nosocomial infections and epidemics much more convincing, and if thought is given to fungal biology, they will take full advantage of the power of DNA variation to identify fungi and track their medically relevant traits. In other words, with a little more effort, strain-typing studies could reveal a number of basic life history features of the fungi and, with that information, could have a greater impact on clinical identification and even research aimed at treatment or prevention.
Two fundamental life history traits that are of interest to medical mycologists are (i) the reproductive mode (i.e., is the fungus asexual and clonal or sexual and recombining?) and (ii) genetic differentiation or isolation (i.e., is the fungus one large interbreeding population, or is it subdivided into discrete populations which are genetically isolated?). Most fungi are sexual, and most are haploid (N), a medical example being Ajellomyces capsulatus (mitosporic name, Histoplasma capsulatum). However, many fungi are thought to be asexual (as many as 25% [58]), medical examples being Aspergillus fumigatus, Trichophyton rubrum and Candida albicans. A few fungi are diploid (2N), C. albicans being a good medical example, and in the Basidiomycota, many species are dikaryotic (N+N), although the medically important basidiomycetes (e.g., Cryptococcus neoformans, anamorph of Filobasidiella neoformans, Trichosporon spp., and Malassezia spp.) are probably haploid when associated with the host (55, 71). Members of sexual species may mate with themselves (selfing) or with other individuals (outcrossing) or may do both. Members of different sexual species may hybridize in laboratory settings, if they are under strong selection for hybrid offspring, but hybridization in nature is rarely encountered in fungi; there are only five good examples of this process. Four of them involve plant-pathogenic fungi that could come in close contact while in the host (Ophiostoma [18], Epichloë [102], Tilletia [41], Typhula [29]), and the fifth, Allomyces (36), is a chytrid with motile gametes that locate each other with the aid of pheromones). Some fungal species have worldwide distributions (e.g., heterothallic Neurospora spp. [93]), but others have populations specific to different continents or hosts (e.g., Armillaria spp. [2, 27] or Pleurotus spp. [115]) and many others are narrowly endemic. The pioneering work on fungal population genetics has been done with plant-pathogenic fungi, and there are good recent reviews of the field (1, 17, 85).
At the outset, it is worth considering the terms “recombination” and “clonality.” Genetic recombination occurs during meiosis, when reassortment and crossing over produce gametes or progeny with different combinations of alleles from those in the parental genomes. In fungi, nonmeiotic recombination, or parasexuality, is also capable of making progeny with new combinations of genes and has been used routinely in laboratory genetic analysis (40). Whether parasexuality is important in nature is controversial, because it is initiated by fusion of vegetative hyphae (anastomosis) of different individuals, a step which appears to be limited to fungi with extremely similar genomes. If it is true that anastomoses are limited to fungi with identical or nearly identical genomes, parasexuality would not be able to make a significant contribution to recombination. Compared with the two mechanisms known for recombination, clonal reproduction is more complicated. Each hyphal apex in a mycelium is clonally related to the others, and fragmentation of a mycelium would qualify as clonal reproduction (for a discussion of this problem in plants, see references 57 and 63]). Production of mitospores (asexual spores, often called conidia) is an improved form of mycelial fragmentation and would seem to be the most common type of fungal clonal reproduction. Yeasts manage to combine mitospore production with vegetative growth, so that every mitotic division leads to mitospore production. However, what about apomictic ascospore production or selfing (homothallism)? The production of ascospores in the absence of recombination seems to point to their other role as resistant propagules. These forms of reproduction would also be considered clonal reproduction by the broad definition that we favor, because the progeny genomes would be identical to the parental genome with no recombination. As a result, if two variable regions of the genome were compared in a collection of individuals, they would appear to be associated in the clonal organism and not in the recombining one. Clonal reproduction, however, is not the only explanation for association between two variable regions. The regions might be in close physical proximity on a chromosome, or they might be more distant but located on a part of a chromosome where crossing over is suppressed. Selection could also lead to their being associated. If the fungal isolates were collected from different, genetically isolated populations, their loci would also appear associated when individuals from the two populations were compared. Similarly, recombination is not the only explanation for a lack of association of loci. Highly mutable loci (hypervariable loci) would mask the association of loci and give the appearance of recombination. Although it would be more accurate to use the terms “recombining” and “nonrecombining” or “associated” and “unassociated,” we will use “recombining” and “clonal” because they are widely understood, if not precisely defined.
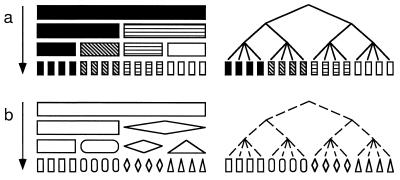
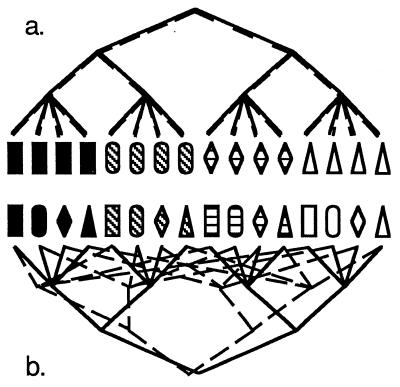
How can knowledge of the reproductive mode and genetic isolation leading to discrete populations or cryptic species make strain-typing data much more valuable to medical mycologists? Imagine an asexual fungus, which must reproduce clonally (Fig. 1 and 2a). There is no recombination in this fungus, and so the entire genome is transmitted intact from generation to generation and every region of the genome has the same evolutionary history (Fig. 2a). Therefore, only one variable region would suffice to identify different genotypes. Because every gene is associated, variation in traits such as pathogenicity can be followed by any variable DNA region, whether or not the two genes are in physical proximity in the genome. Due to clonality, the relatedness of individual fungi can be assessed through any variable region, and the relationship is straightforward. Without mating to keep populations genetically homogeneous or to incorporate foreign genes into local populations, the concepts of gene flow and genetic isolation take on a different meaning. However, the differential success of some clones and the extinctions of others can lead to populations that are distinct from neighboring ones, and gene flow between them certainly can be detected.
FIG. 1.
Relationship between characteristics used for strain typing and their evolutionary history. (a) Increasing diversity of pigmentation through time and the evolutionary relationships of individuals with different pigmentation. (b) Increasing diversity of shape through time and the evolutionary relationships of individuals with different shapes.
FIG. 2.
Effect of reproductive mode on the congruence of evolutionary relationships based on the two characters given in Fig. 1. (a) In a clonal organism, the two features are associated, are found in a few combinations (black rectangles, white triangles, etc.), and have the same evolutionary history. (b) In a recombining organism, the two features are not associated and are found in all possible combinations. As a result, the evolutionary histories of the two features are incongruent.
At the other extreme, imagine a sexual, outbreeding fungus. Here, due to recombination, every region of the genome might have a different evolutionary history, and it would be impossible for a single variable locus to identify genetically different individuals—a collection of loci would be necessary (Fig. 2b). Due to recombination, only loci associated with pathogenicity genes could be used to assay pathogenicity. Genetic distance among isolates could be assayed, but again, the accuracy of the estimate would be proportional to the number of loci assayed. Also, genetic distance would not necessarily have anything to do with clinically relevant characteristics. Gene flow among populations of sexual fungi is an important issue for strain typing. If populations are genetically isolated, a comparison of individual fungi taken from different populations would be like a comparison of clonal organisms, because they do not recombine (Fig. 3). As a consequence, loci with different alleles fixed in different populations could be used to identify individuals to their population of origin; only one such locus is needed, and any one is as good as any other. If differences in pathogenicity were found among populations, any of the loci fixed for different alleles in the populations could be used to track the trait.
FIG. 3.
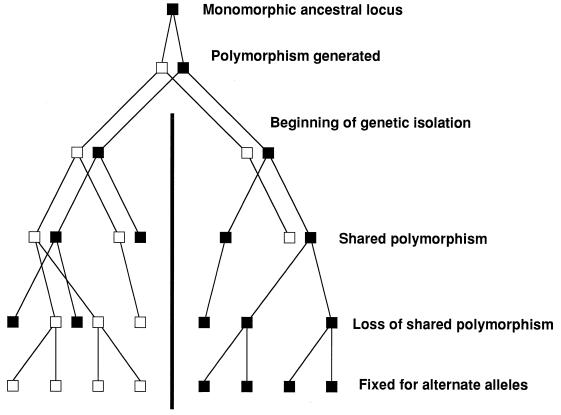
How genetic isolation leads to loss of shared polymorphisms in descendent populations. In the absence of selection to maintain polymorphisms, the fate of all polymorphic loci is fixation through genetic drift. Beginning with two genetically isolated populations that share the polymorphism of the ancestral population, first one and then the other population becomes fixed for one allele. In this case, the two descendent populations have become fixed for alternate alleles, making this locus useful for determining the population of an unknown individual.
Without knowledge of the reproductive mode or genetic differentiation and isolation, strain-typing studies often assume that the fungus is asexual, has clonal reproduction, and is not subdivided into genetically isolated populations. As seen below, these assumptions have not been supported in several recent studies of medically important fungi. Therefore, medical mycologists should be aware of the reproductive mode and population structure of their fungi, because the use of DNA variation to identify fungi and track clinically relevant behavior depends upon it. Researchers working to develop antifungal drugs or vaccines may also be interested in genetically isolated populations or divergent clones, to be sure that all genotypes in the species are affected by the treatment.
USING STRAIN-TYPING DATA TO ANALYZE THE REPRODUCTIVE MODE AND POPULATION STRUCTURE OF FUNGI
Students of charismatic megabiota can study reproductive mode and gene flow with a pair of binoculars or a collection of radio collars, but microbiologists must use genetics. The necessary features are variable traits, in our case polymorphic macromolecules, and methods of analysis that can be used to distinguish clonality from recombination and to detect genetic differentiation and isolation.
GATHERING DATA FOR STRAIN TYPING AND POPULATION GENETICS
Strain typing and population genetics both exploit genetic variation, and almost all studies now focus on genotypic characters instead of phenotypic ones. In the panoply of acronyms that follow, all but the first one analyze DNA variation. To assess the basic features of fungal populations that are essential to interpret strain typing, i.e., reproductive mode and genetic isolation, one needs a series of simple, well-characterized, independent, stable polymorphic loci. Some of the methods below, e.g., electrophoretic karyotyping and fingerprinting methods, cannot provide this type of data. However, once the evolutionary biology of the fungus has been analyzed, methods such as fingerprinting can be useful for distinguishing clones.
Multilocus Enzyme Electrophoresis
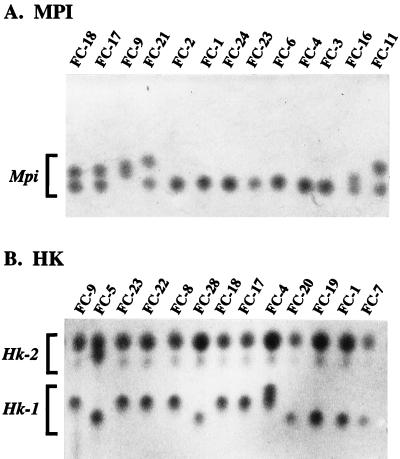
Multilocus enzyme electrophoresis (MLEE) hardly needs an introduction. Any protein (locus) that can be selectively stained can be isolated from a collection of individuals and electrophoresed. If the protein bands are variable, they are considered to be alleles on the basis of mobility (Fig. 4). The technique is robust and continues to be put to good use in studies of medically important fungi, e.g., C. albicans (10) and A. fumigatus (99). An important advantage of this technique is that all alleles are recovered, so that alleles are rarely missing, or null; in other words, the locus is codominant in that both alleles in a diploid can be observed. The criticisms of enzyme electrophoresis are that (i) it assays the genotype indirectly, so that much variation at the nucleotide level may go undetected because nucleotide substitutions do not necessarily change the amino acid composition; (ii) changes in amino acid composition do not necessarily change the electrophoretic mobility of the protein and, as a consequence, alleles that are considered to be the same protein alleles from different individuals may represent different gene alleles; and (iii) selection may be acting on the polymorphisms, so that anonymous DNA markers may give a very different picture from allozyme markers, presumably because the former are neutral and the latter are under some sort of selection (64, 94).
FIG. 4.
Starch gel electrophoresis of polymorphic enzymatic proteins in C. albicans isolates. (A) Proteins with the activity of mannose-6-phosphate isomerase (MPI) showing several alleles based on electrophoretic mobility in homozygous and heterozygous individuals of this diploid yeast. (B) Proteins with the activity of hexokinase (HK) showing allelic differences at two loci, Hk-1 and Hk-2. Reprinted from reference 95 with permission of the publisher.
Electrophoretic Karyotype Analysis
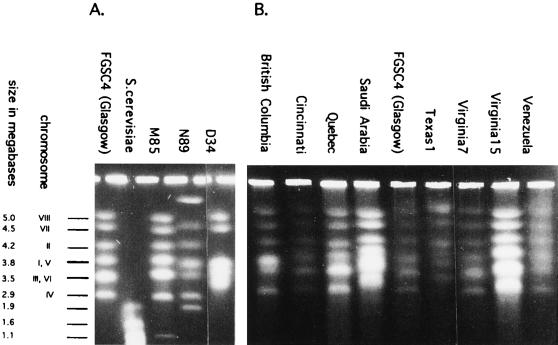
Chromosomal size variation is assayed via an electrophoretic technique, electrophoretic karyotype (EK) analysis, which uses electric fields of alternating orientation to move intact chromosomes through the agarose gel matrix. This biologically interesting and complex topic has recently been reviewed (123). Classic reports for C. albicans pointed out the value of this technique (79, 83), and several other human pathogenic fungi have been studied in this way: Coccidioides immitis (90), Histoplasma capsulatum (107), and Cryptococcus neoformans (119). It may be argued that karyotypes should be less variable for sexual species than for asexual ones, due to the need for pairing at meiosis, but fungal EK are known to display some variation due to loss of dispensable chromosomes (84) and to vary in size and gene arrangement, even in sexual species (51) (Fig. 5).
FIG. 5.
EKs of A. nidulans. (A) Reference isolates of A. nidulans (Glasgow) and Saccharomyces cerevisiae plus three isolates with unusual karyotypes. (B) Isolates from outside Great Britain. In general, the EKs are very similar, as is expected for a meiotic fungus like A. nidulans (Emericella nidulans). However, variation is present. Note the dispensable B-chromosome in M85 (size, ca. 1.1 Mb), the very different pattern in N89 (due to a translocation), a similarly different pattern in D34 (also probably due to translocation), and the relatively small length variation in many chromosomes. Reprinted from reference 49 with permission of the publisher.
Restriction Fragment Length Polymorphism Analysis
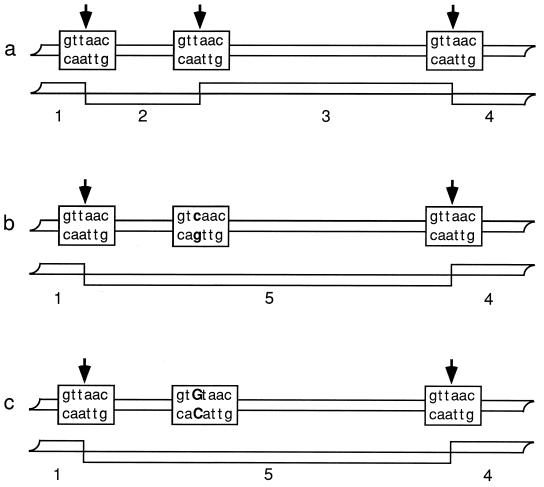
Restriction fragment length polymorphism analysis (RFLP) assays the DNA sequence variation of the genome by using restriction endonucleases to sample short pieces of DNA sequence. Restriction endonucleases recognize specific DNA sequences, usually 4 to 6 nucleotides in length, and cut the DNA in or near the recognition sequence. Alteration of the recognition sequence by nucleotide substitution, insertion or deletion (length mutation), or, for some restriction enzymes, methylation of nucleotides (provided that the DNA is genomic and not amplified or cloned) can prevent the restriction endonuclease from acting and change the fragment pattern (Fig. 6). Length mutations in the region between restriction sites also can change the pattern. Any region of DNA (locus) can be used for RFLP analysis if variation (alleles) can be visualized directly due to multiple copies (mitochondrial DNA [mtDNA], rDNA) or if variation can be visualized indirectly either by hybridization to probe DNA or amplification by PCR. If only one variable restriction endonuclease site is present in the DNA fragment, allele scoring is straightforward. When several variable sites are present, mapping the sites will improve the interpretation. RFLPs were the first DNA markers used for fungal evolutionary biology, and they continue to be put to good use in population genetic studies, e.g., for A. nidulans (48). Critics of RFLP analysis note that while restriction sites in different individuals are most likely to be identical by descent, that is not the case for missing sites, because it is easier to lose a site than to gain one. There are many ways in which a restriction endonuclease site can be lost: any of the several nucleotides in the recognition sequence can be substituted (Fig. 6), or the site can suffer length mutations. Missing sites that have arisen, unknowingly, by different routes confound evolutionary analysis.
FIG. 6.
Multiple origins of the same RFLP fragment due to independent losses of a restriction site. (a) Three sites creating four RFLP fragments. (b) Loss of the middle site through a t-to-c transition, leading to three RFLP fragments through the loss of fragments 2 and 3 and the gain of fragment 5. (c) Loss of the middle site through a t-to-g transversion, leading to the same three RFLP fragments via the same new fragments.
Randomly Amplified Polymorphic DNA or Arbitrarily Primed PCR Analysis
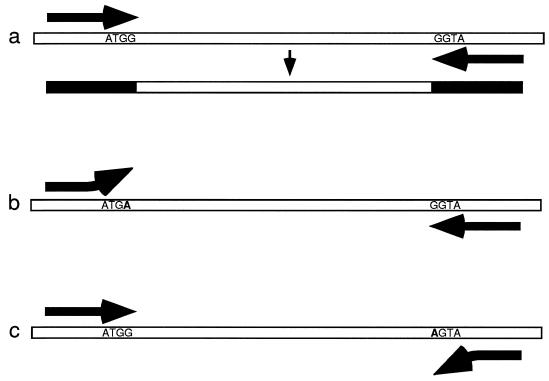
Randomly amplified polymorphic DNA (RAPD) analysis or arbitrarily primed PCR (AP-PCR) analysis is similar to RFLP analysis in that it assays DNA sequence variation in short regions, but instead of analyzing restriction endonuclease recognition sequences, it focuses on PCR priming regions (120). Nucleotide substitutions in the PCR priming regions, particularly the 3′ ends, can prevent primer annealing and PCR amplification. RAPD analysis uses one short PCR primer (ca. 10 bp) and a low annealing temperature to generate several fragments in one amplification. If a comparison of amplifications of several isolates shows a band (locus) that varies, alleles are assigned to the presence [1] and absence [0] of the band. RAPD analysis is technically simple and often detects variation among isolates that are invariant with RFLP analysis, and so it has become quite popular. For example, in comparisons of several DNA-based typing methods for Candida lusitaniae and Aspergillus fumigatus, RAPDs detected variation missed by RFLPs (66, 75). Criticisms of RAPDs initially focused on reproducibility. RAPD analysis succeeds because just one nucleotide substitution can allow or prevent priming, and so it is not surprising that small differences in any aspect of PCR have the same effect. Even if there were no problems with repeatability, there would still be the concern that bands of equal electrophoretic mobility may not be homologous and the related concern that missing bands may not be homologous because they can be lost by several possible nucleotide substitutions in either PCR priming site as well as by length mutations (Fig. 7). There is also the problem of dominant and null alleles; in haploid organisms both the dominant (presence) and null (absence) alleles can be scored, but in diploids it is not possible to distinguish genotypes that are homozygous for the dominant allele from those that are heterozygous. It is also tempting to score more than one variable band per RAPD reaction, although they may not be independent. A recent evaluation of RAPD bands from hybrid plants found that a disturbingly large fraction (13%) of bands with equal mobility were not homologous (97). One solution to this problem is to sequence the RAPD bands to confirm their identity, as has been done in several recent studies (22, 54, 86).
FIG. 7.
Multiple losses of PCR-amplified DNA regions due to independent nucleotide substitutions in the two priming regions. (a) PCR amplification of a DNA sequence showing incorporation of the primer oligonucleotides at each end of the sequence. (b) Failure of PCR amplification due to a G-to-A transition that defeats priming at the left-hand priming region. (c) Failure to amplify DNA due to a second transition that defeats priming at the right-hand priming region.
Sequence-Confirmed Amplified Region Analysis
Starting with variable loci found with RFLP or RAPD analysis, sequence-confirmed amplified region (SCAR) analysis uses DNA sequencing to recover both positive alleles (band present) and null alleles (band absent) and to ensure that alleles from different individuals are homologous. If the RFLP loci are based on DNA hybridization and the variable restriction endonuclease site lies in the probe sequence, it is simple to sequence the probe, design PCR primers to amplify the probe region from every isolate, and use the restriction endonuclease to distinguish the alleles. If, however, the variable restriction endonuclease site lies adjacent to the probe sequence, where it cannot be easily sequenced, or if the variable DNA fragment is a RAPD fragment, where the modified priming site cannot be sequenced, developing the SCAR is more difficult. A robust strategy for finding SCARs involves using RFLPs or RAPDs to generate patterns of bands for a test group of 6 to 10 isolates and then searching for sequence variation among the bands of identical mobility. Sequencing with arbitrary primer pairs (SWAPP [23]) is a refinement of this method which uses two different, ca. 20-bp RAPD primers and low-stringency PCR annealing temperatures to generate PCR bands from every isolate. These bands, which show no variation in agarose gel electrophoresis, are heat denatured, snap cooled to promote single-strand folding, and then electrophoresed on acrylamide under nondenaturing conditions to see if the folded single strands now show variation. If this single-strand conformational polymorphism (SSCP) electrophoresis shows variation in the mobility of the single strands, it is very probably due to conformation changes caused by nucleotide substitutions (88). The variable bands are retrieved from the gel and sequenced. When polymorphic regions are found, new, specific PCR primers are designed and used to amplify the fragment from every isolate in the study. SSCP can be used to score the alleles; alternatively, if the variable nucleotide happens to lie in a restriction endonuclease recognition site, restriction digestion can be used for scoring. Of course, there may be more than one variable nucleotide position in the amplified fragment. However, they are likely to be associated, and only the most informative one is used, i.e., the one in which the allele frequencies are balanced. By this approach, the DNA variation is confined to a single variable nucleotide position or short length mutation, and there are usually just two alleles. Therefore, the problem of ambiguity about the null alleles is overcome because both alleles are recovered; i.e., the locus is codominant. It is still possible that the positive alleles are independently derived, but this is not likely unless the locus is hypervariable (22). This approach has been successfully applied to studies of Coccidioides immitis (22, 23), H. capsulatum (26), and C. albicans (54).
Nucleic Acid Sequencing of Known Genes
Nucleic acid sequencing of known genes has been rendered accessible by inexpensive automated nucleic acid sequencing. Whereas all the preceding techniques used a variety of clever approaches to search for variation without resorting to sequencing, this approach uses brute force. PCR primers that amplify parts of genes from a set of representative isolates are designed by reference to GenBank sequences of the fungus of interest or close relatives. SSCP can be used to screen the PCR products for polymorphism prior to sequencing, and from the sequence all variation in the region is recorded. The technique has all the advantages of SCAR analysis because all the alleles are recovered. If several genes are sequenced, additional opportunities for analysis become available, as discussed below (see, e.g., reference 67). The use of known genes often appeals to a broader scientific audience, but there may be a bias in the evolution of genes that are under strong selection, such as proteins of pathogenic fungi that are recognized as antigens by hosts. While it is true that nucleotides in third codon positions and introns probably would not be subject to selection, their frequency could be affected by proximity to selected regions through genetic hitchhiking (60). The only other drawback comes into play as smaller and younger populations are sampled, because there may be insufficient variation in the genes, or even their introns, to address questions of population genetics. In this event, it is necessary to sample more of the genome to find variation, a task that may involve the use of arbitrary regions as described above.
DNA Fingerprinting with Repetitive DNA Sequences
The techniques mentioned so far are used to describe polymorphic loci one at a time, but both the RFLP and PCR approaches can be used to sample many loci at once if the loci involve repeated DNA sequences. With RFLP fingerprinting, the probe is simply a moderately repeated DNA sequence (ca., 10 to 20 copies per genome) which hybridizes to restriction fragments containing the repeated DNA. With PCR fingerprinting, the primer recognizes a repeated DNA sequence, also producing a large number of fragments with each reaction. Due to the large number of fragments and the mutability of repeated DNA sequences, both the RFLP and PCR fingerprints are quite variable and provide superior discrimination among individual fungal isolates. In comparisons of different strain-typing methods, the fingerprinting methods are usually the most discriminating (33). It is tempting to treat many variable fingerprint fragments as an instant multilocus genotype by assuming that each fragment is an independent locus. Unfortunately, the problems of homology of positive alleles and ambiguity about null alleles are magnified with the complex patterns, and the fragments are not necessarily independent. As a result, fingerprints are useful for identifying individual fungi but problematic for estimating genetic or phylogenetic relationships among individuals.
Microsatellite Analysis
One of the emerging strain-typing techniques exploits the hypervariability of DNA regions made of 10 to 20 or more tandem repeats of nucleotide couplets, triplets, or quadruplets. The ease with which strand slippage during DNA replication can change the number of short repeats makes the level of variability as high as or higher than that of fingerprinting. Use of microsatellites has been demonstrated with C. albicans (39) and H. capsulatum (25). One concern with microsatellites is that alleles may be identical not by descent but by mutation, as is expected with highly mutable sequences (89). Fingerprints of many microsatellite loci can be revealed at once by hybridization of restriction endonuclease-digested fungal DNA to synthetic microsatellite sequences (109). Unfortunately, homology between fragments with the same mobility in different isolates is difficult to establish, as is the case with most fingerprinting techniques.
Amplified Fragment Length Polymorphism Analysis
The polymorphism revealed by amplified fragment length polymorphism (AFLP) analysis depends on restriction endonuclease site differences, just like RFLPs. However, the AFLP process requires that a PCR-amplified library of restriction fragments representative of the entire fungal genome be created (117). The library can be created from very small amounts of DNA, and so it may be useful for medically important fungi that are difficult to cultivate (100). The variable fragments (loci) have two alleles (positive and null), and so the same concerns raised about null alleles with RFLP and RAPD analysis apply. However, many more fragments are produced from each reaction than with RFLP analysis, and the reproducibility is much better than with RAPDs. It should be a good method for prospecting for SCARs and also could provide DNA for multiple gene sequences from pathogens that defy cultivation. Its utility has been demonstrated with several fungi (80, 87, 100).
Once the strain-typing data are in hand, they can be used to analyze genotype diversity among the fungal isolates to address questions about the source of the inoculum, be it reactivation of old infections or newly acquired ones, transfer from other infected humans, or acquisition from environmental sources. However, as discussed in the introduction, to get the most out of the data, they should first be used to analyze two features of the fungus life cycle, i.e., the mode of reproduction and the genetic differentiation or isolation of any groups within the species.
ANALYZING MOLECULAR VARIATION
Reproductive Mode
Almost all mycotic agents make mitospores, and so it is a given that asexual or clonal reproduction is a capability of medically important fungi. This fact can lead to the assumption that asexual reproduction is all that medical fungi can do and that strains of pathogenic fungi are responsible for epidemics. The question for medical mycologists is whether the fungus can also undergo recombination, the hallmark of sexual reproduction. The presence of sexual reproduction under laboratory conditions suggests that the same process is occurring in nature, but selfing or the presence of a preponderance of individuals of one mating type in nature could undermine that assumption.
There are several approaches to distinguishing recombination from clonality (113). Criteria for single-locus data apply to diploid organisms, where fixed heterozygotes and other deviations from Hardy-Weinberg equilibria can be detected. With the exception of C. albicans, medically important fungi are not diploid, and therefore single-locus studies are not informative. However, assaying variation at more than one locus to create a multilocus genotype for haploid fungi is straightforward, as discussed above. Tibayrenc et al. (113) point out that with multilocus genotypes, overrepresented genotypes and association among alleles at different loci or among alleles and phenotypic traits provide evidence of clonality. Overrepresentation of a genotype is certainly a direct consequence of clonal reproduction, but the predominance of one genotype does not rule out recombination in the relationships among the different genotypes. As stated above, it is known that mitosporic fungi can reproduce asexually; the important question is whether they also can recombine. Analyzing representatives of each different multilocus genotype for association among loci is a well-tested method of distinguishing between the null hypothesis of recombination and the alternate hypothesis of clonality (82). A simple way to assess association is to see if all possible combinations of alleles are found for a given pair of loci. This test is best suited for biallelic loci, where the allele frequencies are balanced, and has been used with Coccidioides immitis (22) and C. albicans (54).
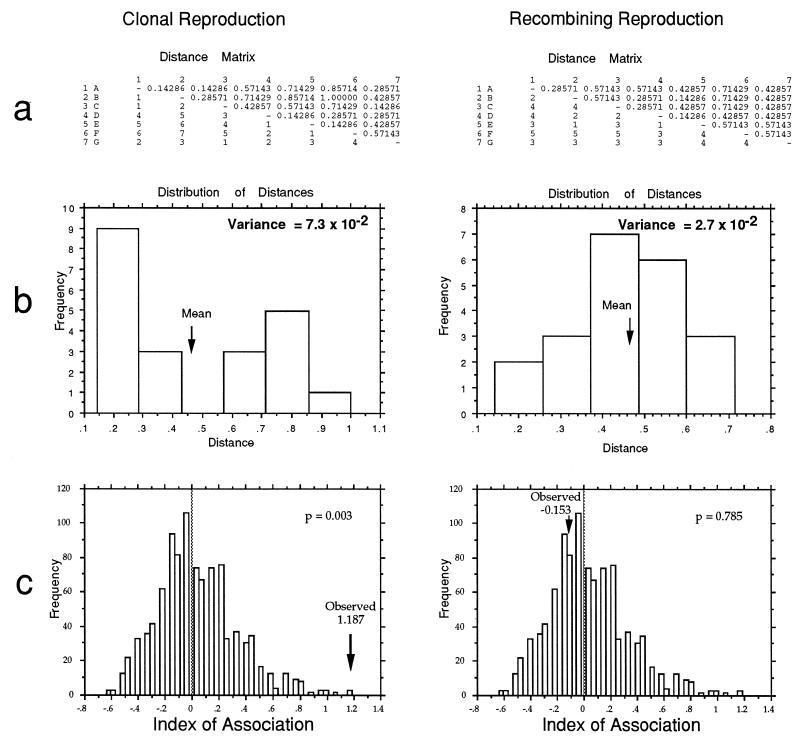
With genotypes made from several discrete loci, association among loci can be measured by a technique developed for barley (20) and adapted for microbes (82). Known as the index of association (IA) test, it is an application of genetic distance analysis. With data sets simulated to represent observed multilocus genotypes for clonal and recombining organisms (Fig. 8), the calculation of IA is demonstrated (Fig. 9). First, distances between multilocus genotypes are determined for all pairs of taxa, and the variance of the distances is calculated (Fig. 9a). The distribution of pairwise distances for recombining organisms should be normal, with a few close relatives, a few distant relatives, and most of the distances near the mean (Fig. 9b); the variance for this distribution is low. For clonal organisms, there should be many close relatives (clone mates), many distant relatives (those on different branches of the tree), and not many at the mean; the variance for this distribution is high (Fig. 9b). The observed data can be used to simulate the type of data expected for a recombining organism by shuffling (resampling without replacement) the alleles at each locus of the observed data (Fig. 10). The variance for the observed data is then compared to the distribution of variances for 1,000 or more independently shuffled data sets, and significance is determined by the fraction of shuffled data sets with higher variances than the observed one. The IA is simply a rescaled variance, with the mean of the distribution of variances for the recombined data sets being defined as an IA of zero. As can be seen in Fig. 9c, the IA of the clonal data is significantly higher than the distribution of IAs for the recombined data (P = 0.003), but not for the recombined data set (P = 0.785).
FIG. 8.
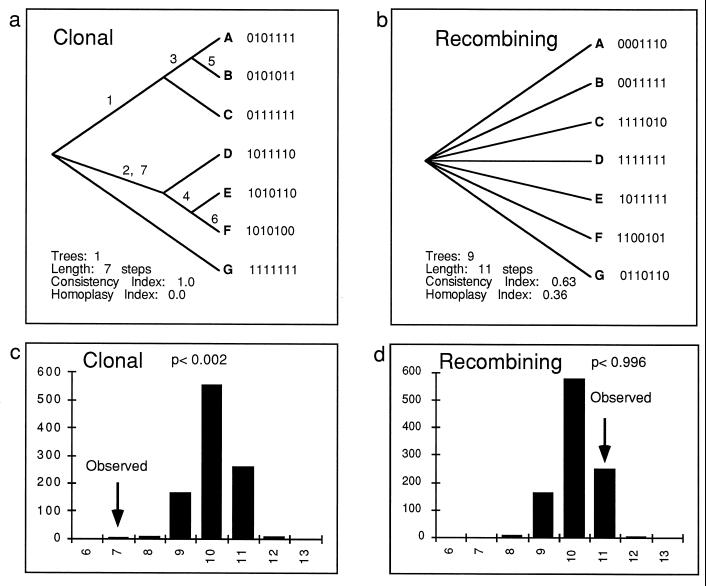
Simulated data sets for a clonal and recombining organism used to explain analytical methods illustrated in Fig. 9 and 11. Each data set consists of seven biallelic loci in seven individuals. The clonal data set was constructed without homoplasy, and the recombined data set was constructed by resampling the alleles at each locus with replacement, as diagrammed in Fig. 10.
FIG. 9.
Method to detect association among loci in individuals characterized by multilocus genotypes, i.e., distinguishing between clonal and recombining organisms by using the distribution of pairwise comparisons of their multilocus genotypes. (a) Distance matrices from all pairwise comparison for clonal and recombining organisms. (b) Bar graph of the frequency distribution of pairwise distances among multilocus genotypes for clonal and recombining organisms. Clonal organisms have an excess of close and distant distances and a high variance. Recombining organisms have a more normal distribution and a low variance. (c) Comparison of the rescaled variance (IA) of clonal and recombining organisms to IAs for 1,000 artificially recombined data sets. Note that the IA of the clonal data is significantly higher than the distribution of IAs for the artificially recombined data sets, while the IA for the recombining organism is close to the mean of the IAs for the artificially recombined data sets.
FIG. 10.
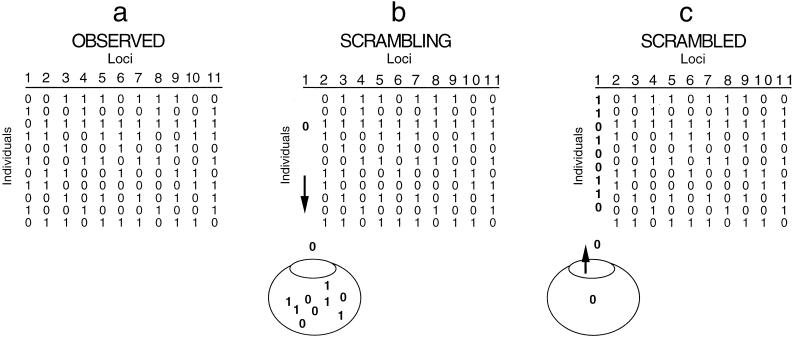
Diagram showing how observed data are shuffled to mimic the effects of recombination. For each locus, the alleles are resampled without replacement to shuffle the alleles among the individuals. The process is repeated for each locus to make a recombined data set. (a) Observed data are shown. (b and c) Alleles at the first locus are resampled without replacement to create an artificially recombined locus.
The aspect of phylogenetics concerned with building evolutionary trees provides another set of useful methods for distinguishing between clonality and recombination, provided that the multilocus genotypes comprise discrete loci. Clonal organisms evolve like phylogenetic trees; each individual has one parent (ancestor), and there is no horizontal transfer of genes or recombination because there is no mating or meiosis. All loci should reflect the same phylogeny, and there will be little homoplasy in a phylogenetic tree based on multilocus genotypes. In a species of recombining organisms, evolution is not tree-like but is more like a net, with each individual having two parents and horizontal gene transfer occurring with each mating (6, 78). Each locus may have a different phylogeny, and a combined phylogenetic analysis of all loci will show more homoplasy than a clonal organism. As can be seen in Fig. 11, it is possible to compare the lengths of phylogenetic trees (which reflects the relative amount of homoplasy) to distinguish between clonal and recombining populations. This type of test was developed to test for evolutionary signal (or, conversely, homoplasy) in phylogenetic data sets (4) and was adapted for population genetics by Burt et al. (22). Here, too, if homoplasy from other sources could be excluded, a high phylogenetic signal would signify high association between alleles at the different loci used to build the tree, while a low signal would indicate low association and hence recombination between the alleles. The same data sets used for the IA test (Fig. 8), one representing a clonal organism and the second representing a recombining one, were used to make parsimony trees (Fig. 11a and b). The most parsimonious tree for the clonal species (Fig. 11a) is as short as possible (each step corresponds to one allele change for each of the seven variable [biallelic] loci), because in clonal species all loci have the same evolutionary history and trees made from different loci will have compatible topologies. In this example, there is no homoplasy in the clonal data set. In a real data set, some homoplasy might be expected, even for strictly clonal organisms. Figure 11b presents the result of parsimony analysis for the recombining population, in this case a consensus tree of the nine most parsimonious trees. Although trees based on each locus alone would be well resolved and short (a tree based on one biallelic locus would have just one branch that partitioned the individuals into two groups), they would not necessarily partition the individuals into the same groups, making trees based on all the loci poorly resolved and longer due to homoplasy. In this case, the tree is four steps longer than the shortest possible tree. As with the IA test, mimicking the effect of recombination by shuffling the alleles for each locus (resampling without replacement) can be used to determine if the tree length for the observed data is significantly shorter than would be expected for recombining populations. The position of the tree length for the observed data in a distribution of tree lengths for 1,000 resampled data sets provides the P value (Fig. 11c and d). The tree length for the clonal data set is significantly shorter than expected for recombining organisms (P = 0.002), allowing us to reject the null hypothesis of recombination; however, this was not so for the recombined data set (P = 0.996).
FIG. 11.
Use of parsimony tree length to distinguish between clonal reproduction and recombination. (a) Parsimony tree for a clonal organism based on data in Fig. 8. Note that it is fully resolved and as short as possible (seven loci, seven steps), because the data set was constructed without homoplasy. Of course, not all clonal data sets would be free of homoplasy, but methods have been proposed to estimate the amount of homoplasy expected for a clonal organism (81). (b) Parsimony tree for a recombining organism based on data in Fig. 7a. Note that this tree is a consensus of nine most parsimonious trees, all of which are four steps longer than the shortest possible tree. The lack of resolution and the longer tree length are the result of each locus having a different evolutionary history. (c) Tree length of a clonal organism compared to the tree lengths for 1,000 artificially recombined data sets (cf. Fig. 10). Note that only 1 of 1,000 data sets had a tree length as short as the observed data. (d) Tree length of a recombining organism compared to the tree lengths for 1,000 artificially recombined data sets. Note that all but 4 of 1,000 data sets had a tree length as short as the observed data.
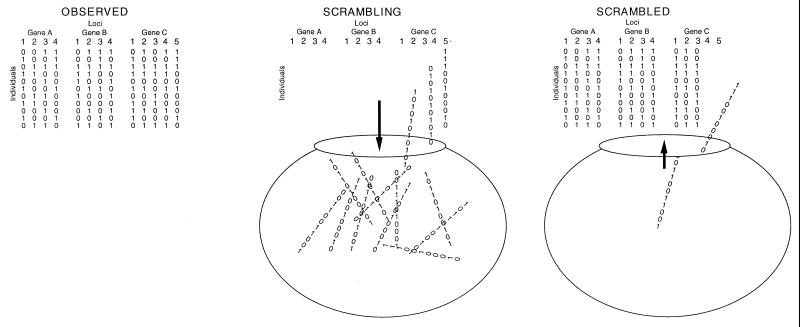
In the phylogenetic analysis described above, each locus was defined as one variable nucleotide position, or site. If gene sequences for several genes were available, so that the gene was the locus with several variable sites, the same principle of concordance of evolutionary histories could be used to distinguish clonality from recombination. Detection of microbial recombination by comparing phylogenetic trees built for different genes was first used with Escherichia coli (34). If the trees were concordant, there was strong evidence for clonality, but if the trees were in conflict, recombination was a likely explanation. As with all tests of the mode of reproduction, the sample of individuals must be from one population and the loci cannot be hypervariable. Violating the former will bias the analysis toward clonality, and violating the latter will bias the analysis toward recombination. A test developed to evaluate the congruence of phylogenetic trees of taxa above the species level, the partition homogeneity test (PHT), is ideal for this intraspecific comparison (37, 62). For congruent gene trees, i.e., those from a clonal species, the sum of the lengths of the most parsimonious trees for each gene should not change significantly if the different sites (polymorphic nucleotides in each gene) are swapped among genes. In other words, whether the polymorphic sites are contiguous in the same gene or are drawn from other genes throughout the genome, the sum of gene tree lengths should remain the same because the entire genome evolves as a unit. For incongruent gene trees, as would be expected for a recombining fungus, the sum of the trees for the observed data should be shorter than the sum of the trees made after the sites have been swapped among genes. This disparity would result from bringing together sites from distant regions of the genome into one resampled gene. Under recombination, the disparate regions would have different evolutionary histories, and making a tree with these loci would require extra steps due to homoplasy. As before, significance is established by making many randomly swapped data sets and comparing the observed sum of lengths to the distribution for 1,000 or more swapped data sets (Fig. 12 and 13) (50, 67).
FIG. 12.
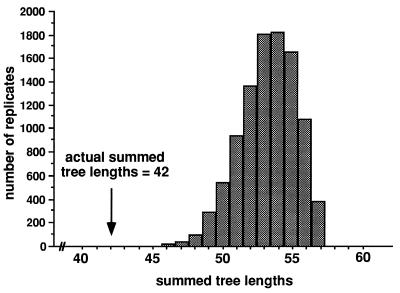
Partition homogeneity test of A. flavus (group 1), showing that the sum of observed parsimony tree lengths for five gene genealogies is significantly (P < 0.0001) shorter than the distribution of summed tree lengths following shuffling of polymorphic sites among the five genes (Fig. 13). Because shuffling of sites should not affect tree length for clonal organisms, the inference is that A. flavus is recombining. Reprinted from reference 50 with permission of the publisher.
FIG. 13.
Shuffling of polymorphic sites among three genes. Each scrambled data set is made by sampling sites without replacement while maintaining the number of sites for each gene. Note that this shuffling differs from that in Fig. 9, where alleles are shuffled among loci. Here, the alleles are maintained in their original order but the sites (loci) are shuffled.
A likelihood method to distinguish between clonality and recombination, one that has clonality as the null hypothesis, has also been demonstrated (22). For all possible phylogenetic trees, the likelihood of the data for each locus is calculated with and without the constraint that each locus must fit the same phylogenetic tree topology. The sum of the highest likelihoods for all of the loci, without the constraint of having the same phylogenetic tree topology, then is compared to the highest sum of likelihoods for all the loci, given the constraint of having to conform to the same tree topology. Similar likelihoods would be expected for clonal organisms because all loci evolve together; much higher likelihoods would be expected for the sum of individual loci without the constraint of a common tree topology if the organism was recombining (22). Unfortunately, the number of possible trees grows much faster than the number of individuals (6 individuals have 105 possible trees, 7 have 945, 10 have a bit over 2 × 106 [38]). At present, this method is limited to a small number of individuals and loci due to computational demands.
Genetic Differentiation and Isolation
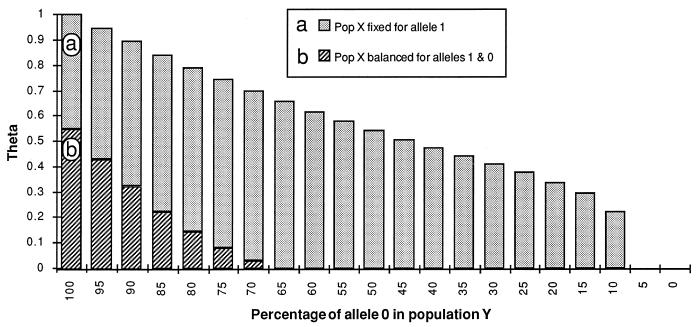
Wright’s work on F statistics (121) provided the foundation for studies of gene flow by pointing out that subdivision of a large population into smaller ones could be detected by comparing allele frequencies at polymorphic loci. He compared the probability of selecting identical alleles from two individuals in a single subgroup to the probability of selecting identical alleles from one individual in each of two subgroups. If individuals in both subgroups freely interbreed, so that gene flow is high, the probabilities of retrieving identical alleles in each sampling should be identical or nearly so; if the subgroups are isolated and gene flow is low, the probability of selecting the same allele from one subgroup should be much higher than that for individuals from the two groups. His statistic for this comparison, Fst, varies from 0 (no isolation) to 1 (complete isolation) and has been estimated in a variety of ways. A convenient method of estimating Wright’s Fst is theta (31). Theta behaves similarly to Fst and has been used to detect genetic differentiation and isolation in human pathogenic fungi (22). The full range of theta, from 0 for free gene flow to 1 for no gene flow, assumes that loci have been sampled randomly, with no bias for polymorphic loci. However, it is often the case that only polymorphic loci are sampled in the first population, because monomorphic loci are not useful for tests of reproductive mode. In this case, theta cannot show the full range of values from 0 to 1, and lower values (e.g., 0.5) may indicate the absence of gene flow (Fig. 14).
FIG. 14.
Simulation of the behavior of Wright’s Fst, estimated by using theta (31), a statistic used to infer genetic isolation. Theta is defined as (Q − q)/(1 − q), where Q is the probability that two randomly sampled genes within a population are the same allele and q is the probability that two genes randomly selected from different populations are the same allele. (a) Behavior of theta when the locus may be fixed in both populations. Theta ranges from 0 when each population is fixed for the same allele to 1 when each population is fixed for a different allele. (b) Behavior of theta when the locus may be fixed in one population but is balanced for the two alleles in the other (50:50). Theta ranges from 0 when allele frequencies are identical (50:50) to ca. 0.5 when one population is fixed for either allele and the other population remains balanced. Populations for which data are biased toward polymorphic loci result in overestimates of gene flow compared to populations for which both polymorphic and monomorphic loci are sampled.
The availability of gene sequence data from several genes allows for phylogenetic tests of genetic isolation based on the same logic as was presented above for detecting recombination. With sexual fungi, when sequence data for several different genes are combined to make one phylogenetic tree, the expectation is that the combined gene tree will be poorly resolved compared to individual gene trees because the different genes have different evolutionary histories and their trees will have different topologies. However, if a branch in the combined gene tree is found and is strongly supported, there is an indication of genetic isolation. This partition in the tree reflects the fate of polymorphic loci when a population is divided by some event. At the time the population is divided, both progeny populations share the polymorphisms. However, as time goes on in the absence of selection to maintain the polymorphism, the polymorphism is lost, first from one population and then from the other, resulting in the two populations becoming fixed for different alleles (Fig. 3). Only when allele frequencies are significantly different, culminating when they have become fixed for different alleles, can they be detected. For every ancestral polymorphic locus that becomes fixed for alternate alleles in the two descendent populations, a greater number become fixed for the same allele (only those with a 50:50 balance of alleles in the ancestral population would have an equal chance of becoming fixed for alternate alleles). This approach has been used to demonstrate possible cryptic species for Coccidioides immitis (67).
As mentioned in the Introduction, there is a relationship between the detection of reproductive mode and genetic isolation. To search for recombining groups, it is essential to sample individuals from a small geographic area so that the fungi have the opportunity to interbreed, whether or not they are capable of doing so. If isolates are inadvertently taken from more than one genetically isolated population, their relationships will appear clonal because recombination between the populations is prevented. Therefore, if the goal of the study is to analyze the mode of reproduction, it is important to first test for genetic isolation.
SPECIFIC FUNGAL EXAMPLES
Cases in which the aforementioned methods of data acquisition and analysis have been used with pathogenic fungi are reviewed below. Highlighted are studies of strain typing, reproductive mode, genetic differentiation and isolation, and the uncovering of cryptic species.
Coccidioides immitis
Coccidioides immitis is a haploid, filamentous ascomycete that makes mitospores (arthroconidia) in the environment and spherules filled with endospores in patients (92). Neither mating nor meiosis has ever been found in this fungus.
Strain typing.
C. immitis recently entered the molecular age with an RFLP study of repeated DNA (probably mtDNA and rDNA) from 1 Venezuelan and 14 Californian isolates (122). The technique sorted the isolates into two groups, one with two Californian clinical isolates, including the often studied Silveira isolate, and the other with the rest. Other studies shortly followed that provided information for strain typing set in the context of fungal evolutionary biology.
Reproductive mode.
Clonality and recombination were examined in 25 isolates of C. immitis from Tucson, Az., through analysis of 14 polymorphic loci (22). Each locus was discovered via SWAPP (23), and the variation was localized to a single variable nucleotide position or small length mutation (22). The study found that isolates from different patients were genetically distinct, i.e., that there were no overrepresented genotypes. In addition to the 25 isolates, another 5 came from repeated isolations from the same patients, and these fungi showed no genetic changes over a maximum of 8 years. Comparison of the observed genotypes to artificially recombined datasets by the IA test failed to reject the null hypothesis of recombination, and the tree length test showed a much longer tree than the number of biallelic loci (38:14), although there was support for association at P = 0.04 (Table 1). The tree length test appears to be a more sensitive method of detecting association among loci than the IA test. C. immitis, with its abundant mitospores, should be capable of clonal reproduction. However, none of the isolates in this study were identical, and so other causes of association among loci may be responsible for the tree length result. Of the 14 loci, 3 had more than one variable nucleotide position within ca. 500 bp, but there was no evidence of recombination over this short distance. Among several pairs of loci, however, all possible combinations of alleles were found, as would be expected with recombination. Finally, using a subset of the data, a likelihood-ratio test with and without the constraint of associated loci allowed rejection of the clonal hypothesis. A criticism of this type of study concerns the possibility of hypervariable loci, which would give the observed result regardless of the reproductive mode. In this case, the authors presented several arguments against hypervariability in the loci, including the occurrence of only two of four possible alleles (nucleotides) at each of the 14 loci, the lack of recombination among variable nucleotide positions separated by 500 bp or less, and the lack of variation at the same loci in individual fungi collected from geographically distant locations. Another criticism concerns the relatively small number of isolates sampled. Given that C. immitis produces mitospores, it would be reassuring to find isolates with identical genomes; the fact that they have not been found suggests that the extent of genetic variation in C. immitis has not yet been thoroughly sampled.
TABLE 1.
Reproductive mode assayed by IA and tree length tests
| Species | No. of loci | No. of isolates | No. of genotypes | IAb | Lb | Locic | Reference |
|---|---|---|---|---|---|---|---|
| Coccidioides immitis | 14 | 25 | 25 | 0.041ns | 38* | D | 22 |
| Histoplasma capsulatum | 11 | 30 | 30 | 0.043ns | 45* | D | 26 |
| Candida albicans | 10a | 26 | 0.517* | 53ns | D | 54 | |
| Candida albicans | 12 | 41 | 0.661*** | 96*** | P | 96 | |
| Candida albicans | 9 | 48 | 0.524*** | 150*** | P | 10 | |
| Aspergillus fumigatus | 8 | 24 | −0.002ns | 25ns | P | 99 | |
| Cryptococcus neoformansd | |||||||
| var. neoformans and gattii | 8 | 233 | 13 | 1.619*** | 29*** | P | 16 |
| var. neoformans | 7 | 230 | 10 | 1.247*** | 16* | P | 16 |
| serotype A | 5 | 222 | 7 | 0.393ns | 9ns | P | 16 |
10 nucleotide sites grouped into 5 independent loci.
*P < 0.05; ***, P < 0.001; ns, not significant.
Loci: D, multiple codominant DNA loci; P, multiple codominant protein loci.
Excluding isolates with serotype AD.
Genetic differentiation and isolation.
The same loci used in the Arizona study of C. immitis were characterized for collections of clinical isolates in California and Texas to study gene flow among the populations (24). At four of the loci, allele frequencies between California on the one hand and Arizona or Texas on the other were very different and F statistics (estimated as theta) showed little or no gene flow. Gene flow between Arizona and Texas was also significantly reduced, but not as much as between either of these states and California. A criticism of this study is that the test was biased because the Arizona loci were selected to be polymorphic, preventing any loci from being fixed for either allele in Arizona (cf. Fig. 14); in this case, the effect of the bias was to overestimate gene flow. It seems safe to say that genetically isolated populations of C. immitis are present, although sampling at locations between Tucson, Az., and San Antonio, Tex., might show a gradient of changing gene frequencies instead of a sharp boundary.
Cryptic species.
Seventeen C. immitis isolates were separated into two groups, those inside and those outside California, in phylogenetic trees made from partial gene sequence of five protein-coding loci (chitin synthase, chitinase, orotidine monophosphate decarboxylase, serine proteinase, and a T-cell reactive protein with similarity to a mammalian dioxygenase) (67, 68). There were no shared polymorphisms between the two groups; i.e., polymorphisms in the ancestral population had been eliminated in at least one of the two groups over time, so that none remained. Assuming a mutation rate of 10−9 per nucleotide per year, the two groups have been genetically isolated for ca. 11 × 106 years, considerably longer than for sibling Drosophila species (108). The Silveira isolate, collected in California in 1951, fell in the non-California group. This isolate was different from most California isolates in the RFLP study (122). It is possible that the patient acquired coccidioidomycosis outside California, in a manner similar to that of a Texas patient who had acquired C. immitis in California (24). It is also possible that isolates with non-California genotypes are found in California, necessitating a renaming of the groups. Because C. immitis is not transmitted from host to host and must be acquired from the environment, examining environmental isolates may provide the best data on which genotypes are endemic in any given locale.
Surprisingly, recombination seems to be part of the C. immitis life cycle. The evidence for recombination in Arizona is strong, but there are not enough polymorphic loci in the California population to test for recombination. Although no genetic evidence for clonal reproduction was found, it seems almost certain that this type of reproduction also is operating, given that mycelia of this fungus produce abundant mitospores. The discovery of strong and old genetic isolation, as would be expected for different species, was also a surprise. The processes that lead to speciation seem to be operating today, as shown by the reduction of gene flow between Arizona and Texas. Typing of isolates to the California and non-California cryptic species can be accomplished by examining any of 17 sites fixed for alternate alleles (67, 68), and typing of non-California isolates to Arizona or Texas can be accomplished by examining no more than 4 loci fixed in Texas. Identification of individual, recombined C. immitis isolates would require the use of quite a few biallelic loci, gene sequences with several sites, or hypervariable loci with many alleles such as microsatellites.
Histoplasma capsulatum
Histoplasma capsulatum (anamorph or mitosporic state of Ajellomyces capsulatus) is a haploid, filamentous ascomycete that grows as a yeast in patients. It makes two types of mitospores as well as meiospores. Mating is regulated by two alleles at one locus and results in meiotic progeny (ascospores), as observed in cultivation (70).
Strain typing.
H. capsulatum was an early subject of studies of molecular variation. Two RFLP studies involving mtDNA and rDNA probes (106, 116) revealed polymorphisms in H. capsulatum isolates, and the availability of a yeast phase-specific nuclear gene (yps-3) provided an additional tool to type strains (65). Types based on mtDNA variation or yps-3 variation are congruent for U.S. H. capsulatum isolates from individuals without AIDS. Central American isolates, or U.S. isolates from AIDS patients, which may have originated in Central America or the Caribbean, have a different yps-3 genotype and harbor several mtDNA subtypes (65). Recent characterization of 16 biallelic loci characterized as single-nucleotide substitutions have produced unique multilocus genotypes for each of 30 H. capsulatum isolates from Indianapolis (26). The more recent discovery of multiallelic microsatellite loci indicates that individual isolates may be characterized with just a few loci (25).
Cryptic species.
H. capsulatum has three varieties, capsulatum, duboisii, and farciminosum. H. capsulatum var. capsulatum and H. capsulatum var. duboisii have the same teleomorph, Ajellomyces capsulatus. Although H. capsulatum var. farciminosum has not reproduced meiotically in cultivation, it does respond to pairings with mating tester strains of var. capsulatum, but not to the point of making meiospores. H. capsulatum var. capsulatum is endemic in North America and South America; H. capsulatum var. duboisii is endemic in equatorial west Africa, and the yeast cells that it makes in vivo are reported to be larger than those of H. capsulatum var. capsulatum. H. capsulatum var. farciminosum is said to be found only in animals, whereas the two other varieties can be found in both humans and animals. H. capsulatum var. capsulatum is most commonly associated with lung disease, which H. capsulatum var. duboisii is not. Judging from their clinical behavior, they seem to be overt species, but morphologically there are few differences. Comparison of nucleic acid sequences from the large-subunit rDNA and DNA-DNA hybridization of single isolates of all three H. capsulatum varieties showed that they are closely related, but the relationships among them were not resolved (56).
Reproductive mode.
Carter et al. (26) used the aforementioned biallelic nucleotide substitution loci to address the question of reproductive mode in 30 clinical H. capsulatum isolates. With the IA test, the null hypothesis of recombination could not be rejected, and, as with C. immitis, the tree length test showed a tree much longer than a strictly clonal tree (45:11), but one that was significantly (P = 0.05) shorter than those of most artificially recombined data (Table 1). This result is consistent with the knowledge that H. capsulatum mates in culture (70). Unlike the mtDNA and yps-3 data, the nucleotide substitution loci did not place isolates from AIDS patients into a group separate from the isolates from individuals without AIDS. However, the segregation of isolates from AIDS patients in previous studies may be related to their suspected Central American or Caribbean origin (65), and isolates from these locations were not included in the study by Carter et al. (26).
Genetic differentiation and isolation.
A pioneering isoenzyme study of H. capsulatum surveyed 339 isolates for eight polymorphic enzyme loci, at least one of which probably contained more than one locus (46, 47). Based on allele frequencies reported for each of four sites (two in Missouri and one each in Kentucky and Michigan), there was as much variation within populations as between them, and the strongest genetic differentiation occurred between Michigan and the southern sites. Unfortunately, the data were not presented as multilocus genotypes for each isolate, and so tests of reproductive mode are not possible.
H. capsulatum var. capsulatum mates in cultivation and recombines in nature. The abundant production of mitospores also suggests that clonal reproduction is common in the fungus, although identical genotypes were not found in an 11-locus study of 30 clinical isolates (26). The three varieties based on phenotypic and clinical differences may represent species, but the necessary genetic studies have not been performed. If the varieties are as genetically distinct as species and they can still mate, the scarcity of fungal hybridization might be explained. That is, the fungal species that can hybridize are those that morphologically are so similar that they are not recognized as distinct species, letting their hybridization go undetected. Populations within varieties have not been identified, and isozyme data from North America do not support genetically isolated populations. Given the New World distribution of H. capsulatum var. capsulatum, it might be useful to compare isolates from North and South America.
Candida albicans
The ascomycete Candida albicans, a relative of Saccharomyces cerevisiae (7, 69), is apparently diploid and has never been observed to mate in the laboratory or in nature. It grows as a yeast and as a hypha.
Strain typing.
The full complement of methods has been tried on C. albicans. A recent comparison of techniques (33) showed that RFLPs of middle repeated sequences (103) and PCR fingerprinting with M13 provided the best discrimination. Pujol et al. (95) took the comparison among techniques a step further by making comparisons among four methods, three that generate many bands in one procedure and one that analyzes loci singly, MLEE. They made the comparison with similarity coefficients and phenetic trees, which carry the assumption of clonal reproduction. Two of the multiband methods (RAPDs and hybridization of restriction digestions to the Ca3 repeat) showed agreement with MLEE at the deep branches of the phenograms but not in the fine relationships among closely related isolates. The third method, hybridization of CARE2 repeated DNA to restriction digests, was at odds with the other three. While it is clear that the multiband methods allow many different genotypes to be recognized, use of the data to infer genetic relationships is compromised by problems with homology of bands or fragments. Previous analysis of the Ca3 repeated sequence (101) showed that the sequence evolves extremely rapidly, even between transfers in the laboratory. This hypervariability is probably due to transposition of the repeated sequence used to type the strains (76). Another type of hypervariable locus, the microsatellites, has also been assayed in C. albicans, showing that single loci can be extremely polymorphic, with as many as 11 alleles in this diploid ascomycete (19, 39). A key question for the use of repeated sequences or microsatellites to do more than strain typing is, “Are the alleles identical by descent or by convergence?” A study comparing results from these extremely variable loci to more sedate, biallelic loci could answer the question about hypervariability and identity and provide a useful service to the field. Such a study in horseshoe crabs (Limulus polyphemus) found incongruence between genealogies based on the microsatellites or the flanking regions (89). However, these hypervariable loci may be just what is needed when the question concerns the origin of nosocomial infections, because they have so many alleles.
Cryptic species.
Sullivan et al. (109) uncovered a group of C. albicans isolates that were genetically isolated and that could be distinguished from other Candida species by a combination of characters: chlamydospore aggregation, serotype, and carbohydrate utilization. Their discovery employed RFLPs in DNA hybridizing to microsatellite sequences and was corroborated by using RAPDs, isoenzyme analysis, and comparison of sequence of the V3 region of large-subunit rDNA. The genetically isolated group is now known as C. dubliniensis (110).
Reproductive mode.
Pujol et al. (96) used isoenzyme variation to study the reproductive mode of oral C. albicans isolates from human immunodeficiency virus (HIV)-positive patients. Analysis of 55 isolates at 13 polymorphic loci found 2 significantly overrepresented genotypes and between 3 and 6 of 13 loci with genotypic frequencies that differed significantly from Hardy-Weinberg expectation; both results provide evidence of clonality. However, genotypic frequencies for most of the loci were not significantly different from the Hardy-Weinberg expectation, indicating that recombination may also be operating (95). Our analysis of their isozyme data (12 loci and 41 genotypes) by both the IA and the tree length test allowed rejection of the null hypothesis of recombination at P < 0.001 (Table 1); i.e., by these tests, the population is clonal.
Boerlin et al. (10) characterized 10 polymorphic isoenzyme loci in a collection of 189 C. albicans isolates from oral cavities of both HIV-positive and HIV-negative patients, as well as some isolates from patients with invasive disease. They found 52 different genotypes, 1 of which was overrepresented. Of the 10 loci, 8 showed genotypic frequencies significantly different from Hardy-Weinberg expectations, and only 1 of 28 pairwise comparisons loci showed a lack of association. Clearly, there is good evidence for clonality. The two loci with genotypic frequencies expected under Hardy-Weinberg equilibrium may argue for recombination or may represent hypervariable loci. The authors used a distance method (unweighted pair group method with arithmetic mean [UPGMA]) to make a phylogenetic tree based on the isoenzyme data, and it failed to show grouping of isolates from HIV-positive or HIV-negative patients or of invasive isolates. This type of analysis makes the assumption of purely clonal reproduction, which may be justified in this case.
Gräser et al. (54) used 12 nucleic acid loci, characterized as single nucleotide substitutions or small length mutations on six PCR fragments, to study 52 C. albicans isolates taken from 52 asymptomatic subjects. These codominant loci were discovered by using SWAPP (23), and the isolates showed overrepresentation of some multilocus genotypes and significant deviation from Hardy-Weinberg expectations in 8 of the 12 loci; again, this provides evidence for clonality. However, genotypic frequencies were within Hardy-Weinberg expectations for 4 of the loci, and most of 66 pairs of loci were not associated (48 to 53, depending on the method of determining significance). Our analysis of their data (Table 1, conservatively grouping 10 of their nucleotide sites into 5 independent loci for 26 genotypes) by IA test supported rejection of recombination (P < 0.05), while use of the tree length test did not support rejection of recombination (P < 0.1). As with the Pujol et al. (96) and Boerlin et al. (10) studies, there is clear evidence of clonal reproduction in the Gräser et al. (54) study, but there is also evidence for recombination, including finding all possible genotypes for two pairs of loci (54). How can one reconcile the disagreement regarding reproductive mode in the several studies? As both groups of Candida researchers noted in a recent exchange on this topic (112, 114), the data from both groups show that Candida is not freely recombining (as also seen in our test of data from both groups [Table 1]), and that Candida is not completely clonal (as evidenced by Gräser et al. [54] finding all possible genotypes for pairs of diploid, biallelic loci). It seems likely that the next step will be to quantify the amount of recombination, perhaps by the method recently proposed by Maynard Smith and Smith (81) or one like it. Another reason for differences between studies performed by the two groups may lie in the type of loci or in the sample of individuals. Because the sample of individuals is different in each study, the reproductive modes and population structures for the two samples could be different. C. albicans is transmitted from host to host, and clonal spread could be easier in this fungus than in a species that must be acquired from the environment. A thorough investigation of genetic isolation in C. albicans would also be worthwhile, remembering that different methods of sampling nucleic acid variation demonstrated the same three deeply branching groups of isolates while showing variation in the shallow branches (95). One recent search for genetic isolation by using RFLPs of rDNA and RAPDs failed to find differences among Asian, North American, and European isolates, but new combinations of RFLP bands found in Asian isolates could be explained by hybridization between North American and European isolates (30).
Evolutionary studies of C. albicans have found a cryptic species, C. dubliniensis, and have provided data consistent with both clonal and recombining modes of reproduction in this yeast. Hypervariable microsatellite loci, useful for identification of individuals, are being characterized and promise to make it possible to distinguish even very similar genotypes by one technique. It would be worthwhile to search for genetically isolated populations within larger samples of C. albicans and, as with many fungi, to attempt to estimate the amount of recombination.
Cryptococcus neoformans
Cryptococcus neoformans (the anamorph of the basidiomycete Filobasidiella neoformans) is a haploid yeast with close relatives in the mycoparasitic genus Tremella (111). There are two varieties, neoformans and gattii. Environmentally, C. neoformans var. neoformans is found in bird excreta and C. neoformans var. gattii is associated with Eucalyptus trees (35). Each variety has two serotypes, A and D in C. neoformans var. neoformans and B and C in C. neoformans var. gattii. Mating is controlled by two alleles (a and α) at a single mating locus, but in C. neoformans var. neoformans serotype A, no isolates with mating type a have been recovered. In cultivation, C. neoformans can reproduce via mitospores (yeast cells) and mate to produce meiospores (basidiospores) (71).
Strain typing.
Brandt et al. (14) developed 10 isoenzyme loci to characterize 34 C. neoformans isolates and then expanded the study to include ca. 350 isolates (15, 16). For the 99% that were C. neoformans var. neoformans, they found 16 electrophoretic types (multilocus genotypes), and for the 3 C. neoformans var. gattii isolates, they found another 3 genotypes. The most common genotypes correlated with the geographic area of origin for the isolates, with California isolates being distinct from those from the southeastern United States. There was no correlation between genotype and isolates from AIDS patients. RAPDs were then used to subtype isolates in the largest genotype, resulting in 19 RAPD subtypes, which no longer showed a correlation with the area of geographic origin (15). Isoenzyme and RAPD analysis of repeated isolates from the same patient showed no variation, although EK instability has been seen in C. neoformans in cultivation and during animal passage (42, 44).
Reproductive mode.
In the isoenzyme studies mentioned above, the presence of overrepresented genotypes provided evidence for clonality. We reanalyzed the data (Table 1), using the eight simplest loci and 16 C. neoformans var. neoformans genotypes from 234 isolates excluding the AD serotype. Both the IA and tree length tests allowed significant rejection of the null hypothesis of recombination when all isolates were assumed to be members of a single species. If it is assumed that there are two species that correlate with the A and D serotypes and that the shuffling of alleles among loci (Fig. 10) is limited to members of the same serotype (A or D), the significance of the IA test was reduced (P < 0.05) and that of the tree length test was lost entirely. An attractive hypothesis is that serotypes of C. neoformans var. neoformans are indicators of cryptic speciation (42) and that recombination is occurring within the A and D serotype groups. Consistent with this explanation is the deep divergences between clusters of isolates of these two serotypes in distance analyses (14, 16).
Chen et al. (28) and Franzot et al. (43) studied variation in isolates from New York and Brazil by using two types of data, RFLPs hybridizing to the moderately repeated DNA sequence CNRE-1, and DNA sequencing of the URA5 gene, respectively. In the combined analysis, Franzot et al. (43) argued for clonal reproduction, noting that some genotypes were overrepresented, that EKs were heterogeneous (heterogeneous chromosomes might be incompatible with pairing at meiosis), and that the mating type of the isolates was always α instead of a mix of α and a. As with Candida, it seems certain that clonal reproduction is operating in natural populations of C. neoformans, but there is evidence that some recombination is also present. For example, parsimony analysis of the DNA sequence data for the URA5 gene returned 1,276 most parsimonious trees, an unexpected result for a clonal organism, and it would be surprising if a fungus that displayed the ability to mate in the laboratory (71) never made use of it in nature. On the other hand, haploid C. neoformans var. neoformans isolates of mating type α can produce basidiospores without mating, which would be another method of clonal reproduction (118).
Genetic differentiation and cryptic species.
Boekhout et al. (9) compared the two C. neoformans varieties, neoformans and gattii (anamorphs of F. neoformans var. neoformans and F. neoformans var. bacillispora, respectively), in terms of EKs and RAPDs. With both methods, there were consistent differences between the varieties, leading the authors to suggest that they should be considered separate species. Although it is known from laboratory studies that matings between members of the two varieties demonstrate recombination (72, 104), it seems likely that gene flow between them is greatly reduced in nature. Early comparisons of the genomes of these two taxa also showed them to be distinct (5).
In the same study, Boekhout et al. (9) addressed variation within the varieties. In C. neoformans var. neoformans, EKs were quite variable and there was no correlation with the pattern of bands and the source of the isolates (saprobic, clinical, or veterinary). Neither was there a correlation between source and EK in C. neoformans var. gattii. In addition, there was evidence from comparisons of the same strain kept under different culture conditions that the EK could change over time. With RAPD analysis, again there was no correlation with the source of the isolates, but in C. neoformans var. gattii only there was a correlation with geographic origin. The isozyme studies mentioned above (14, 16) showed strong separation between isolates in serotypes A and D, and analysis of the reproductive mode was strongly influenced by assuming that individuals in the two serotypes were reproductively isolated. It may be that serotypes A and D have become fixed in cryptic species. Serotype AD is more closely related to serotype A in the isozyme studies and has far less genetic diversity than serotype A.
Evolutionary studies of C. neoformans have provided good evidence for clonality, but some of the data also are consistent with recombination if species are defined narrowly. The varieties are genetically isolated, as would be expected for species, and analysis of isozyme data suggests that cryptic species exist within a single variety, C. neoformans var. neoformans. As with H. capsulatum, matings between varieties may represent another example of overlooked fungal hybridization.
Aspergillus fumigatus and A. flavus
Aspergillus fumigatus and A. flavus are haploid filamentous ascomycete fungi that make abundant mitospores but never have been observed to make meiospores in cultivation or in nature.
Strain typing.
A. fumigatus , an opportunistic human pathogen, has been a frequent subject of epidemiological studies because it strikes immunosuppressed patients while they are in the hospital, generating suspicion about nosocomial infection. Comparative studies have shown that RAPDs provided better discrimination than immunoblot fingerprinting or RFLPs of repeated DNAs (most probably mtDNA or rDNA [77]). A more recent RAPD study found different patterns for five isolates, each from a different patient, but identical patterns for one clinical isolate and several isolates from the hospital environment (74). The patient isolates were collected over a 4-month period, and the matching environmental isolates were taken within as few as 4 days after the collection of the patient isolate. Comparing the results of three typing methods, isoenzymes, RAPDs and RFLPs, for genomic DNA, Lin et al. (75) found that RAPD and RFLP showed the most variability. Girardin et al. (53) used RFLPs combined with DNA-DNA hybridization to a moderately repeated, inactive, retrotransposon-like DNA, Afut1, to examine isolates taken over a 16-month period from five leukemia patients, one asthma patient, and the environment. Each patient had a genetically distinct isolate, and in three cases an environmental isolate was found with the same genotype as a clinical isolate, but in only one case was the fungus obtained from the environment at a time when the patient could have become infected by it. In other cases, the predominant genotype in a hospital room was not the one recovered from patients housed in the room. The points that A. fumigatus isolates are genetically variable, that many of them are capable of causing disease, and that they are all found in the environment were dramatically made in a huge study of Afut1 variation in over 800 clinical and environmental isolates (52). The number of different genotypes was about half the number of isolates, but cases where two isolates had the same genotype were explained by repeated isolations from the same patient or environmental location. Identical genotypes from different geographic locations were rare (ca 3%), showing that Afut1 variation is very effective for strain typing.
Cryptic species.
Rinyu et al. (98) examined 61 clinical and environmental A. fumigatus isolates for colony and microscopic morphology by a number of molecular techniques, i.e., analysis of isoenzymes, RFLPs of mtDNA and rDNA, and RAPDs. Although there was no correlation between the molecular markers and either pathogenicity or geographic origin, one isolate appeared quite distinct by both isoenzymes and RAPDs, leading to speculation that it might represent a distinct species. A similar result was found in another study in which 2 of 35 isolates appeared quite different by isoenzyme, RFLP, and RAPD analyses, sharing no “alleles” with the other isolates or with each other (75). A study of agricultural isolates of the second most common agent of aspergillosis, A. flavus, using partial DNA sequence for five genes and compatibility of gene genealogies, showed two genetically isolated groups in what had been considered to be a good morphological species (50).
Reproductive mode.
The strain-typing studies mentioned above certainly found repeated genotypes characteristic of clonal reproduction. In the largest study, which used similar typing methods, over 400 distinct genotypes were found, suggesting a role for recombination (32). Rinyu et al. (98) used isoenzyme and RAPD data to show some overrepresented genotypes, but the absence of a strict correlation between phenetic trees based on the two data sets and the presence of all six possible two-locus genotypes for the two most common isoenzyme loci (PHO, alleles a and b; EST, alleles a, b, and c) also provided evidence for recombination. Rodriguez et al. (99) characterized 12 polymorphic isoenzyme loci in 63 A. fumigatus isolates. They found two alleles per individual at four loci: three peptidases and one hexokinase. They interpreted the extra alleles as evidence that A. fumigatus was diploid and used the excess and absence of heterozygosity to argue that A. fumigatus was clonal. However, under the alternative hypothesis that A. fumigatus is haploid and that the extra alleles are due to gene duplication, our analysis of the 8 unduplicated loci and 24 unique genotypes by both IA and tree length tests suggests that A. fumigatus is recombining (Table 1). For A. flavus, Geiser et al. (50) used nucleotide sequence for five genes to demonstrate recombination in the larger of the two genetically isolated groups in this species. Their use of the PHT gave a significant result (P < 0.0001) for incongruence among the five gene genealogies, consistent with recombination. Taken together, the data make A. flavus a very good candidate for a recombining fungus, in spite of the absence of sexual morphology. Although the medically important species lack the morphology of sex, other Aspergillus species have it, including Neosartorya fischeri, a very close relative of A. fumigatus and Petromyces alliaceus, a close relative of A. flavus. However, the presence of sexual morphology does not necessarily mean the absence of clonal reproduction. For A. nidulans, a genetic model organism, Geiser et al. (48) showed that recombination explained the relationships among at least 20 British genotypes but that the 20 genotypes were overrepresented and their prevalence was a product of clonal propagation.
For the medically important Aspergillus species, A. fumigatus and A. flavus, clonality seems a given, but recombination is playing a part in A. flavus and may be doing so in A. fumigatus. Cryptic species have been found in A. flavus, and they could be present in A. fumigatus. Genetically isolated populations within the species have not been identified. Identical genotypes have been found in locations that are geographically well separated; therefore, isolated populations may not exist.
FUTURE DIRECTIONS
Genomics
Fungal evolutionary biologists use genomics of a sort every time they use nucleotide sequences from well-studied fungi to design amplification primers for the more poorly known relatives of these fungi. With the completion of the Saccharomyces cerevisiae genomic sequence, a strong start on the Candida albicans genome project, and the expectation that Aspergillus nidulans and Neurospora crassa will be sequenced in the near future, fungal evolutionary biologists can begin to think about more ambitious use of genomics. Strain typing could be a prime beneficiary, because every region of the genome (exons, introns, intergenic regions, selfish elements, microsatellites, promoters, enhancers, and categories to be discovered) can be characterized and compared to find regions with the right amount of variation to distinguish evolutionarily meaningful groups. In the process, genome projects would gain information on the diversity in genomic regions, something that is absolutely lacking in the genomic sequence of a single isolate.
Clinical Variation
The clinical relevance of species identification has been shown again and again in the differences among pathogens and their closest relatives. Now that genetically isolated groups are being found within morphological species of medically important fungi, one cannot help wondering if similar clinical differences might be found among the newly recognized cryptic species. Due to genetic isolation and fixation of alleles at previously polymorphic loci, identification of pathogens to one cryptic species is straightforward. If this information could be included in case studies, correlations between genotype and clinical behavior might emerge. In the absence of these data, it might be prudent to perform tests of new pharmaceuticals or vaccines on members of all genetically isolated groups of the pathogenic fungus of choice. The strategy of testing control measures against a battery of genetically different pathogenic fungi is an established practice in agriculture, where new plant varieties are evaluated for resistance to fungi with different avirulence genes (3).
Amount of Recombination
All the fungi discussed above make mitotic spores; therefore, it is almost certain that clonal or mitotic reproduction is occurring in all of them, even if it has not been shown. For all of them, there is also evidence for recombination, whether or not they can be mated in the laboratory. For those that cannot be mated in the laboratory yet show recombination in nature, it is not clear how they recombine or how often they do so. With Coccidioides immitis, an estimate of the number of recombination events needed to account for the observed recombination implied that a frequency of sex far too low to be directly observed would suffice (22). Studies that assess not only the presence of recombination but also its level are needed next (59, 60, 61, 81).
Standardization of Approach
For each pathogenic fungus, there are typically several groups of researchers studying epidemiological and evolutionary questions, and each group often has a different approach. A similar situation existed in fungal phylogenetics before sequencing of ribosomal genes and spacers became the norm. The advantage of sequencing over previous methods was that sequence data obtained by one group could easily be added to by another group, which propelled the field at a much higher rate. Use of a common set of sequence-confirmed loci for each pathogenic fungus could help do the same for the field of strain typing.
ACKNOWLEDGMENTS
We thank Arturo Casadevall for providing data for analysis in Table 1, David Soll for providing us with Fig. 4, and Matthew Fisher, Arturo Casadevall, Rytas Vilgalys, and Martin Bidartondo for criticizing draft manuscripts.
Writing of this review was supported by grants from the National Institutes of Health to J.W.T.
REFERENCES
- 1.Anderson J B, Kohn L M. Clonality in soilborne, plant-pathogenic fungi. Annu Rev Phytopathol. 1995;33:369–391. doi: 10.1146/annurev.py.33.090195.002101. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J B, Korhonen K, Ullrich R C. Relationships between European and North American biological species of Armillaria mellea. Exp Mycol. 1980;4:87–95. [Google Scholar]
- 3.Appel D J, Gordon T R. Relationships among pathogenic and nonpathogenic isolates of Fusarium oxysporum based on the partial sequence of the intergenic spacer region of the ribosomal DNA. Mol Plant-Microbe Interact. 1996;9:125–138. doi: 10.1094/mpmi-9-0125. [DOI] [PubMed] [Google Scholar]
- 4.Archie J W. A randomization test for phylogenetic information in systematic data. Syst Zool. 1989;38:239–252. [Google Scholar]
- 5.Aulakh H S, Strauss S E, Kwon-Chung K J. Genetic relatedness of Filobasidiella neoformans (Cryptococcus neoformans) and Filobasidiella bacillispora (Cryptococcus bacillosporus) as determined by deoxyribonucleic acid base composition and sequence homology studies. Int J Syst Bacteriol. 1981;31:97–103. [Google Scholar]
- 6.Avise J C. Molecular markers, natural history and evolution. New York, N.Y: Chapman & Hall; 1994. [Google Scholar]
- 7.Barns S M, Lane D J, Sogin M L, Bibeau C, Weisburg W G. Evolutionary relationships among pathogenic Candida species and relatives. J Bacteriol. 1991;173:2250–2255. doi: 10.1128/jb.173.7.2250-2255.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berbee M L, Taylor J W. 18S ribosomal RNA gene sequence characters place the human pathogen Sporothrix schenckii in the genus Ophiostoma. Exp Mycol. 1991;16:87–91. [Google Scholar]
- 9.Boekhout T, van Belkum A, Leenders A C, Verbrugh H A, Mukamurangwa P, Swinne D, Scheffers W A. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int J Syst Bacteriol. 1997;47:432–442. doi: 10.1099/00207713-47-2-432. [DOI] [PubMed] [Google Scholar]
- 10.Boerlin P, Boerlin-Petzold F, Goudet J, Durussel C, Pagani J-L, Chave J-P, Bille J. Typing Candida albicans oral isolates from human immunodeficiency virus-infected patients by multilocus enzyme electrophoresis and DNA fingerprinting. J Clin Microbiol. 1996;34:1235–1248. doi: 10.1128/jcm.34.5.1235-1248.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowen A R, Chen-Wu J L, Momany M, Young R, Szaniszlo P J, Robbins P W. Classification of fungal chitin synthases. Proc Natl Acad Sci USA. 1992;89:519–523. doi: 10.1073/pnas.89.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowman B H, Taylor J W. Molecular phylogeny of pathogenic and non-pathogenic Onygenales. In: Reynolds D R, Taylor J W, editors. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. Wallingford, United Kingdom: CAB International; 1993. pp. 169–178. [Google Scholar]
- 13.Bowman B H, White T J, Taylor J W. Evolutionary relationships of human pathogenic fungi: multiple origins of pathogenicity in the fungal order Onygenales. Mol Phylogenet Evol. 1996;6:89–96. doi: 10.1006/mpev.1996.0061. [DOI] [PubMed] [Google Scholar]
- 14.Brandt M E, Bragg S L, Pinner R W. Multilocus enzyme typing of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2819–2823. doi: 10.1128/jcm.31.10.2819-2823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandt M E, Hutwagner L C, Klug L A, Baughman W S, Rimland D, Graviss E A, Hamill R J, Thomas C, Pappas P G, Reingold A L, Pinner R W. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. J Clin Microbiol. 1996;34:912–917. doi: 10.1128/jcm.34.4.912-917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandt M E, Hutwagner L C, Kuykendall R J, Pinner R W. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. J Clin Microbiol. 1995;33:1890–1895. doi: 10.1128/jcm.33.7.1890-1895.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brasier C M. Episodic selection as a force in fungal microevolution, with special reference to clonal speciation and hybrid introgression. Can J Bot. 1995;73:S1213–S1221. [Google Scholar]
- 18.Brasier C M, Kirk S A, Pipe N D, Buck K W. Rare interspecific hybrids in natural populations of the Dutch elm disease pathogens Ophiostoma ulmi and Ophiostoma novo-ulmi. Mycol Res. 1997;102:45–57. [Google Scholar]
- 19.Bretagne S, Costa J-M, Besmond C, Carsique R, Calderone R. Microsatellite polymorphism in the promoter sequence of the elongation factor 3 gene of Candida albicans as the basis for a typing system. J Clin Microbiol. 1997;35:1777–1780. doi: 10.1128/jcm.35.7.1777-1780.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown A H D, Feldman M W, Nevo E. Multilocus structure of natural populations of Hordeum spontaneum. Genetics. 1980;96:523–536. doi: 10.1093/genetics/96.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruns T D, Vilgalys R, Barns S M, Gonzalez D, Hibbett D S, Lane D J, Simon L, Stickel S, Szaro T M, Weisburg W G, Sogin M L. Evolutionary relationships within the fungi: analyses of nuclear small subunit rRNA sequences. Mol Phylogenet Evol. 1992;1:231–241. doi: 10.1016/1055-7903(92)90020-h. [DOI] [PubMed] [Google Scholar]
- 22.Burt A, Carter D A, Koenig G L, White T J, Taylor J W. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc Natl Acad Sci USA. 1996;93:770–773. doi: 10.1073/pnas.93.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burt A, Carter D A, White T J, Taylor J W. DNA sequencing with arbitrary primer pairs. Mol Ecol. 1994;3:523–525. doi: 10.1111/j.1365-294x.1994.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 24.Burt A, Dechairo B M, Koenig G L, Carter D A, White T J, Taylor J W. Molecular markers reveal differentiation among isolates of Coccidioides immitis from California, Arizona and Texas. Mol Ecol. 1997;6:781–786. doi: 10.1046/j.1365-294x.1997.00245.x. [DOI] [PubMed] [Google Scholar]
- 25.Carter D A, Burt A, Taylor J W, Koenig G L, Dechairio B, White T J. A set of electrophoretic molecular markers for strain typing and population genetic studies of Histoplasma capsulatum. Electrophoresis. 1997;18:1047–1053. doi: 10.1002/elps.1150180703. [DOI] [PubMed] [Google Scholar]
- 26.Carter D A, Burt A, Taylor J W, Koenig G L, White T J. Clinical isolates of Histoplasma capsulatum from Indianapolis have a recombining population structure. J Clin Microbiol. 1996;34:2577–2584. doi: 10.1128/jcm.34.10.2577-2584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chase T E, Ullrich R C. Genetic basis of biological species in Heterobasidion annosum: Mendelian determinants. Mycologia. 1990;82:67–72. [Google Scholar]
- 28.Chen F, Currie B P, Chen L-C, Spitzer E D, Casadevall A. Genetic relatedness of Cryptococcus neoformans clinical isolates grouped with the repetitive DNA probe CNRE-1. J Clin Microbiol. 1995;33:2818–2822. doi: 10.1128/jcm.33.11.2818-2822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christen A A, Bruehl G W. Hybridization of Typhula ishikariensis and T. idahoensis. Phytopathology. 1979;69:263–266. [Google Scholar]
- 30.Clemons K V, Feroze F, Holmberg K, Stevens D A. Comparative analysis of genetic variability among Candida albicans isolates from different geographic locales by three genotypic methods. J Clin Microbiol. 1997;35:1332–1336. doi: 10.1128/jcm.35.6.1332-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cockerham C C, Weir B S. Estimation of gene flow from F-statistics. Evolution. 1993;47:855–863. doi: 10.1111/j.1558-5646.1993.tb01239.x. [DOI] [PubMed] [Google Scholar]
- 32.Debeaupuis J-P, Sarfati J, Chazalet V, Latgé J-P. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect Immun. 1997;65:3080–3085. doi: 10.1128/iai.65.8.3080-3085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-Guerra T M, Martinez-Suarez J V, Laguna F, Rodriguez-Tudela J L. Comparison of four molecular typing methods for evaluating genetic diversity among Candida albicans isolates from human immunodeficiency virus-positive patients with oral candidiasis. J Clin Microbiol. 1997;35:856–861. doi: 10.1128/jcm.35.4.856-861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dykhuizen D E, Green L. Recombination in Escherichia coli and the definition of biological species. J Bacteriol. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellis D H, Pfeiffer T J. Natural habitat of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1990;28:1642–1644. doi: 10.1128/jcm.28.7.1642-1644.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emerson R, Wilson C M. Interspecific hybrids and the cytogenetics and cytotaxonomy of Euallomyces. Mycologia. 1954;46:393–434. [Google Scholar]
- 37.Farris J S, Kallersjo M, Kluge A G, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–319. [Google Scholar]
- 38.Felsenstein J. The number of evolutionary trees. Syst Zool. 1978;27:27–33. [Google Scholar]
- 39.Field D, Eggert L, Metzger D, Rose R, Wills C. Use of polymorphic short and clustered coding-region microsatellite to distinguish strains of Candida albicans. FEMS Immunol Med Microbiol. 1996;15:73–79. doi: 10.1111/j.1574-695X.1996.tb00056.x. [DOI] [PubMed] [Google Scholar]
- 40.Fincham J R S, Day P R. Fungal genetics. 4th ed. Berkeley: University of California Press; 1979. [Google Scholar]
- 41.Fischer G W, Holton C S. Biology and control of the smut fungi. New York, N.Y: Ronald Press; 1957. [Google Scholar]
- 42.Franzot S P, Fries B C, Casadevall A. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Genetic relationship between Cryptococcus neoformans var. neoformans strains of serotype A and D, abstr. R-24; p. 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franzot S P, Hamdan J S, Currie B P, Casadevall A. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J Clin Microbiol. 1997;35:2243–2251. doi: 10.1128/jcm.35.9.2243-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fries B C, Chen F, Currie B P, Casadevall A. Karyotype instability in Cryptococcus neoformans infection. J Clin Microbiol. 1996;34:1531–1534. doi: 10.1128/jcm.34.6.1531-1534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gargas A, DePriest P T, Grube M, Tehler A. Multiple origins of lichen symbioses in fungi suggested by SSU rDNA phylogeny. Science. 1995;268:1492–1495. doi: 10.1126/science.7770775. [DOI] [PubMed] [Google Scholar]
- 46.Gaur P K, Lichtwardt R W. Mating types and convertibility among soil isolates of Histoplasma capsulatum. Mycologia. 1980;72:404–406. [PubMed] [Google Scholar]
- 47.Gaur P K, Lichtwardt R W, Hamrick J L. Isozyme variation among soil isolates of Histoplasma capsulatum. Exp Mycol. 1981;5:69–77. [Google Scholar]
- 48.Geiser D M, Arnold M L, Timberlake W E. Sexual origins of British Aspergillus nidulans isolates. Proc Natl Acad Sci USA. 1994;91:2349–2352. doi: 10.1073/pnas.91.6.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geiser D M, Arnold M L, Timberlake W E. Wild chromosomal variants in Aspergillus nidulans. Curr Genet. 1996;29:293–300. doi: 10.1007/BF02221561. [DOI] [PubMed] [Google Scholar]
- 50.Geiser D M, Pitt J I, Taylor J W. Cryptic speciation and recombination in the aflatoxin producing fungus Aspergillus flavus. Proc Natl Acad Sci USA. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geiser D M, Timberlake W E, Arnold M L. Loss of meiosis in Aspergillus. Mol Biol Evol. 1996;13:809–817. doi: 10.1093/oxfordjournals.molbev.a025641. [DOI] [PubMed] [Google Scholar]
- 52.Girardin H, Sarfati J, Kobayashi H, Bouchara J P, Latgé J P. Use of DNA moderately repetitive sequence to type Aspergillus fumigatus isolates from aspergilloma patients. J Infect Dis. 1994;169:683–685. doi: 10.1093/infdis/169.3.683. [DOI] [PubMed] [Google Scholar]
- 53.Girardin H, Sarfati J, Traore F, Camet J D, Derouin F, Latgé J P. Molecular epidemiology of nosocomial invasive Aspergillosis. J Clin Microbiol. 1994;32:684–690. doi: 10.1128/jcm.32.3.684-690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gräser Y, Volovsek M, Arrington J, Schonian G, Presber W, Mitchell T G, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guého, E., J. Faergemann, C. Lyman, and E. J. Anaissie. 1994. Malassezia and Trichosporon: two emerging pathogenic basidiomycetous yeast-like fungi. J. Med. Vet. Mycol. 32(Suppl. 1):367–378. [PubMed]
- 56.Gueho E, Leclerc M C, de Hoog G S, Dupont B. Molecular taxonomy and epidemiology of Blastomyces and Histoplasma species. Mycoses. 1997;40:69–81. doi: 10.1111/j.1439-0507.1997.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 57.Harper J L. Population biology of plants. London, United Kingdom: Academic Press, Ltd.; 1977. [Google Scholar]
- 58.Hawksworth D, Kirk P, Sutton B, Pegler D. Ainsworth’s and Bisby’s dictionary of the Fungi. 8th ed. Wallingford, United Kingdom: CAB International; 1996. [Google Scholar]
- 59.Hein J. A heuristic method to reconstruct the history of sequences subject to recombination. J Mol Evol. 1993;36:396–405. [Google Scholar]
- 60.Hudson R R. How can the low levels of DNA sequence variation in regions of the Drosophila genome with low recombination rates be explained? Proc Natl Acad Sci USA. 1994;91:6815–6818. doi: 10.1073/pnas.91.15.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hudson R R, Saez A G, Ayala F J. DNA variation at the Sod locus of Drosophila melanogaster: an unfolding story of natural selection. Proc Natl Acad Sci USA. 1997;94:7725–7729. doi: 10.1073/pnas.94.15.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huelsenbeck J P, Bull J J, Cunningham C W. Combining data in phylogenetic analysis. Trends Ecol Evol. 1996;11:152–158. doi: 10.1016/0169-5347(96)10006-9. [DOI] [PubMed] [Google Scholar]
- 63.Janzen D H. What are dandelions and aphids? Am Nat. 1977;111:586–589. [Google Scholar]
- 64.Karl S A, Avise J C. Balancing selection at allozyme loci in oysters: implications from nuclear RFLPs. Science. 1992;256:100–102. doi: 10.1126/science.1348870. [DOI] [PubMed] [Google Scholar]
- 65.Keath E J, Kobayashi G S, Medoff G. Typing of Histoplasma capsulatum by restriction fragment length polymorphisms in a nuclear gene. J Clin Microbiol. 1992;30:2104–2107. doi: 10.1128/jcm.30.8.2104-2107.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.King D, Rhine-Chalberg J, Pfaller M A, Moser S A, Merz W G. Comparison of four DNA-based methods for strain deliniation of Candida lusitaniae. J Clin Microbiol. 1995;33:1467–1470. doi: 10.1128/jcm.33.6.1467-1470.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koufopanou V, Burt A, Taylor J W. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci USA. 1997;94:5478–5482. doi: 10.1073/pnas.94.10.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koufopanou V, Burt A, Taylor J W. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci USA. 1998;95:8414. doi: 10.1073/pnas.94.10.5478. . (Correction.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kurtzman C P, Robnett C J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35:1216–1233. doi: 10.1128/jcm.35.5.1216-1223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwon-Chung K J. Sexual stage of Histoplasma capsulatum. Science. 1972;175:326. doi: 10.1126/science.175.4019.326. [DOI] [PubMed] [Google Scholar]
- 71.Kwon-Chung K J. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- 72.Kwon-Chung K J, Bennett J E, Rhodes J C. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Leeuwenhoek. 1982;48:25–38. doi: 10.1007/BF00399484. [DOI] [PubMed] [Google Scholar]
- 73.Leclerc M C, Philippe H, Guého E. Phylogeny of dermatophytes and dimorphic fungi based on large subunit ribosomal RNA sequence comparisons. J Med Vet Mycol. 1994;32:331–341. doi: 10.1080/02681219480000451. [DOI] [PubMed] [Google Scholar]
- 74.Leenders A, van Belkum A, Janssen S, de Marie S, Kluytmans J, Wielenga J, Löwenberg B, Verbrugh H. Molecular epidemiology of an apparent outbreak of invasive aspergillosis in a hematology ward. J Clin Microbiol. 1996;34:345–351. doi: 10.1128/jcm.34.2.345-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin D, Lehmann P F, Hamory B H, Padhye A A, Durry E, Pinner R W, Lasker B A. Comparison of three typing methods for clinical and environmental isolates of Aspergillus fumigatus. J Clin Microbiol. 1995;33:1596–1601. doi: 10.1128/jcm.33.6.1596-1601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lockhart S R, Fritch J J, Meier A S, Schroppel K, Srikantha T, Galask R, Soll D R. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J Clin Microbiol. 1995;33:1501–1509. doi: 10.1128/jcm.33.6.1501-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loudon K W, Burnie J P, Coke A P, Matthews R C. Application of polymerase chain reaction to fingerprinting Aspergillus fumigatus by random amplification of Polymorphic DNA. J Clin Microbiol. 1993;31:1117–1121. doi: 10.1128/jcm.31.5.1117-1121.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maddison W. Phylogenetic histories within and among species. In: Hoch P C, Stephenson A G, editors. Experimental and molecular approaches to plant biosystematics. St. Louis: Missouri Botanical Garden; 1995. pp. 273–287. [Google Scholar]
- 79.Magee B B, Magee P T. Electrophoretic karyotypes and chromosome numbers in Candida species. J Gen Microbiol. 1987;133:425–430. doi: 10.1099/00221287-133-2-425. [DOI] [PubMed] [Google Scholar]
- 80.Majer D, Mithen R, Lewis B G, Vos P, Oliver R P. The use of AFLP fingerprinting for the detection of genetic variation in fungi. Mycol Res. 1996;100:1107–1111. [Google Scholar]
- 81.Maynard Smith J, Smith N H. Detecting recombination from gene trees. Mol Biol Evol. 1998;15:590–599. doi: 10.1093/oxfordjournals.molbev.a025960. [DOI] [PubMed] [Google Scholar]
- 82.Maynard Smith J, Smith N H, O’Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Merz W G, Connelly C, Hieter P. Variation of electrophoretic karyotype among clinical isolates of Candida albicans. J Clin Microbiol. 1988;26:842–845. doi: 10.1128/jcm.26.5.842-845.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miao V P, Covert S F, VanEtten H D. A fungal gene for antibiotic resistance on a dispensable (“B”) chromosome. Science. 1991;254:1773–1776. doi: 10.1126/science.1763326. [DOI] [PubMed] [Google Scholar]
- 85.Milgroom M G. Recombination and the multilocus structure of fungal populations. Annu Rev Phytopathol. 1996;34:457–477. doi: 10.1146/annurev.phyto.34.1.457. [DOI] [PubMed] [Google Scholar]
- 86.Mondon P, Brenier M P, Symoens F, Rodriguez E, Coursange E, Chaib F, Lebeau B, Piens M A, Tortorano A-M, Mallie M, Chapuis F, Carlotti A, Villard J, Viviani M A, Nolard N, Bastide J-M, Ambroise-Thomas P, Grillot R. Molecular typing of Aspergillus fumigatus strains by sequence-specific DNA primer (SSDP) analysis. FEMS Immunol Med Microbiology. 1997;17:95–102. doi: 10.1111/j.1574-695X.1997.tb01001.x. . (Erratum, 18:217, 1997.) [DOI] [PubMed] [Google Scholar]
- 87.Mueller U G, Lipari S E, Milgroom M G. Amplified fragment length polymorphism (AFLP) fingerprinting of symbiotic fungi cultured by the fungus-growing ant Cyphomyrmex minutus. Mol Ecol. 1996;5:119–122. doi: 10.1111/j.1365-294x.1996.tb00297.x. [DOI] [PubMed] [Google Scholar]
- 88.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Orti G, Pearse D E, Avise J C. Phylogenetic assessment of length variation at a microsatellite locus. Proc Natl Acad Sci (USA) 1997;20:10745–10749. doi: 10.1073/pnas.94.20.10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pan S, Cole G T. Electrophoretic karyotypes of clinical isolates of Coccidioides immitis. Infect Immun. 1992;60:4872–4880. doi: 10.1128/iai.60.11.4872-4880.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan S, Sigler L, Cole G T. Evidence for a phylogenetic connection between Coccidioides immitis and Uncinocarpus reesei (Onygenaceae) Microbiology. 1994;140:1481–1494. doi: 10.1099/00221287-140-6-1481. [DOI] [PubMed] [Google Scholar]
- 92.Pappagianis D. Epidemiology of coccidioidomycosis. Curr Top Med Mycol. 1988;2:199–238. doi: 10.1007/978-1-4612-3730-3_6. [DOI] [PubMed] [Google Scholar]
- 93.Perkins D D, Turner B C, Barry E G. Strains of Neurospora collected from nature. Evolution. 1976;30:281–313. doi: 10.1111/j.1558-5646.1976.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 94.Pogson G H, Mesa K A, Boutilier R G. Genetic population structure and gene flow in the Atlantic Cod Gadus morhua: a comparison of allozyme and nuclear RFLP loci. Genetics. 1995;139:375–385. doi: 10.1093/genetics/139.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pujol C, Joly S, Lockhart S R, Noel S, Tibayrenc M, Soll D R. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J Clin Microbiol. 1997;35:2348–2358. doi: 10.1128/jcm.35.9.2348-2358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pujol C, Reynes J, Renaud F, Raymond M, Tibayrenc M, Ayala F J, Janbon F, Mallie M, Bastide J-M. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc Natl Acad Sci USA. 1993;90:9456–9459. doi: 10.1073/pnas.90.20.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rieseberg L H. Homology among RAPD fragments in interspecific comparisons. Mol Ecol. 1996;5:99–105. [Google Scholar]
- 98.Rinyu E, Varga J, Ferenczy L. Phenotypic and genotypic analysis of variability in Aspergillus fumigatus. J Clin Microbiol. 1995;33:2567–2575. doi: 10.1128/jcm.33.10.2567-2575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rodriguez E, De Meeus T, Mallie M, Renaud F, Symoens F, Mondon P, Piens M-A, Lebeau B, Viviani M A, Grillot R, Nolard N, Chapuis F, Tortorano A-M, Bastide J-M. Multicentric epidemiological study of Aspergillus fumigatus isolated by multilocus enzyme electrophoresis. J Clin Microbiol. 1996;34:2559–2568. doi: 10.1128/jcm.34.10.2559-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rosendahl S, Taylor J W. Development of multiple genetic markers for studies of genetic variation in arbuscular mycorrhizal fungi using AFLP. Mol Ecol. 1997;6:821–830. [Google Scholar]
- 101.Sadhu C, McEachern M J, Rustchenko-Bulgac E P, Schmid J, Soll D R, Hicks J B. Telomeric and dispersed repeat sequences in Candida yeasts and their use in strain identification. J Bacteriol. 1991;173:842–850. doi: 10.1128/jb.173.2.842-850.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schardl C L, Leuchtmann A, Tsai H, Collett M A, Watt D M, Scott D B. Origin of a fungal symbiont of perennial ryegrass by interspecific hybridization of a mutualist with the ryegrass choke pathogen, Epichloë typhina. Genetics. 1994;136:1307–1317. doi: 10.1093/genetics/136.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scherer S, Stevens D A. A Candida albicans dispersed, repeated gene family and its epidemiologic applications. Proc Natl Acad Sci USA. 1988;85:1452–1456. doi: 10.1073/pnas.85.5.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schmeding K A, Jong S C, Hugh R. Sexual compatability between serotypes of Filobasidiella neoformans (Cryptococcus neoformans) Curr Microbiol. 1981;5:133–138. [Google Scholar]
- 105.Spatafora J, Blackwell M. Molecular systematics of unitunicate perithecial Ascomycetes. The Clavicipitales-Hypocreales connection. Mycologia. 1993;85:912–922. [Google Scholar]
- 106.Spitzer E D, Lasker B A, Travis S J, Kobayashi G, Medoff G. Use of mitochondrial and ribosomal DNA polymorphisms to classify clinical and soil isolates of Histoplasma capsulatum. Infect Immun. 1989;57:1409–1412. doi: 10.1128/iai.57.5.1409-1412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Steele P E, Carle G F, Kobayashi G S, Medoff G. Electrophoretic analysis of Histoplasma capsulatum chromosomal DNA. Mol Cell Biol. 1989;9:983–987. doi: 10.1128/mcb.9.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stephens J C, Nei M. Phylogenetic analysis of polymorphic DNA sequences in the Adh locus in Drosophila melanogaster and its sibling species. J Mol Evol. 1985;22:289–300. doi: 10.1007/BF02115684. [DOI] [PubMed] [Google Scholar]
- 109.Sullivan D, Bennett D, Henman M, Harwood P, Flint S, Mulcahy F, Shanley D, Coleman D. Oligonucleotide fingerprinting of isolates of Candida species other than C. albicans and of atypical Candida species from human immunodeficiency virus-positive and AIDS patients. J Clin Microbiol. 1993;31:2124–2133. doi: 10.1128/jcm.31.8.2124-2133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sullivan D J, Westerneng T J, Haynes K A, Bennett D E, Coleman D C. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 111.Swann E C, Taylor J W. Phylogenetic diversity of yeast-producing basidiomycetes. Mycol Res. 1995;99:205–210. [Google Scholar]
- 112.Tibayrenc M. Are Candida albicans natural populations subdivided? Trends Microbiol. 1997;5:253–254. doi: 10.1016/S0966-842X(97)01068-8. [DOI] [PubMed] [Google Scholar]
- 113.Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Breniere S F, Darde M, Ayala F J. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci USA. 1991;88:5129–5133. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vilgalys R, Gräser Y, Schönian G, Presber W, Mitchell T. Are Candida albicans natural populations subdivided? Trends Microbiol. 1997;5:254–257. doi: 10.1016/S0966-842X(97)01068-8. . (Response.) [DOI] [PubMed] [Google Scholar]
- 115.Vilgalys R, Sun B L. Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proc Natl Acad Sci USA. 1994;91:4599–4603. doi: 10.1073/pnas.91.10.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vincent R D, Goewert R, Goldman W E, Kobayashi G S, Lambowitz A M, Medoff G. Classification of Histoplasma capsulatum isolates by restriction fragment polymorphisms. J Bacteriol. 1986;165:813–818. doi: 10.1128/jb.165.3.813-818.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wickes B L, Mayorga M E, Edman U, Edman J C. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc Natl Acad Sci USA. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wickes B L, Moore T D E, Kwon-Chung K J. Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology. 1994;140:543–550. doi: 10.1099/00221287-140-3-543. [DOI] [PubMed] [Google Scholar]
- 120.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zimmermann C R, Snedker C J, Pappagianis D. Characterization of Coccidioides immitis isolates by restriction fragment length polymorphisms. J Clin Microbiol. 1994;32:3040–3042. doi: 10.1128/jcm.32.12.3040-3042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zolan M E. Chromosome length polymorphisms in fungi. Microbiol Rev. 1995;59:686–698. doi: 10.1128/mr.59.4.686-698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]