Abstract
Background:
Development of paired sensory organs is a highly complex and coordinated process. These organs arise from ectodermal thickenings in the cephalic region known as cranial placodes. We have previously shown that Zic1 is a critical regulator for the formation of the pre-placodal region (PPR), the common territory for the development of all cranial placodes in Xenopus laevis.
Results:
In this study, we have analyzed a number of Zic1 targets for their expression during PPR patterning, as well as their regulation by retinoic acid (RA) and one of its major metabolites, 4-oxo-RA. Our findings show that anteriorly Zic1 regulates several transcription factors, Crx, Fezf2, Nkx3–1 and Xanf1 as well as a serine/threonine/tyrosine kinase, Pkdcc.2. These factors are all expressed in the vicinity of the PPR and as such are candidate regulators of placode formation downstream of Zic1. In addition to their differential regulation by RA, we find that 4-oxo-RA is also capable of modulating the expression of these genes, as well as a broad array of RA-regulated genes.
Conclusion:
Our data highlight the complexity of retinoid-mediated regulation required for Zic1-activated anterior structure specification in Xenopus, and the potential physiological role of 4-oxo-RA in cranial placode development.
Keywords: Placode, Retinoic Acid, Patterning, Xenopus
Introduction
Cranial placodes are precursors to specialized sense organs in vertebrates that arise from the head ectoderm during embryonic development. They are ectodermal thickenings that are formed at the neural plate border from the common territory, the pre-placodal region (PPR) and subsequently differentiate into placodes with distinct identities, fates and functions1. Cranial placodes give rise to the adenohypophysis, paired sensory organs (the optic lens, the inner ear and the olfactory epithelium), and sensory cranial ganglia that innervate the orofacial complex2. Disruptions in the formation of these placodes often lead to debilitating disorders and malformations such as deafness, blindness, anosmia, hormonal imbalance, and orofacial sensory deficits.
In order to further our understanding of cranial placode development in Xenopus, our laboratory has established an animal cap explant assay where the activity of the transcription factor Zic1 was found necessary and sufficient for the expression of PPR genes six1 and foxi4.13. The identification of downstream targets of Zic1 via microarray revealed several genes involved in the retinoic acid (RA) signaling pathway, which is essential for the non-cell autonomous specification of PPR by Zic14. Specifically, the RA synthesizing enzyme RALDH2 and the RA-transporter lipocalin-type prostaglandin D2 synthase (LPGDS) that are expressed at the anterior neural plate (ANP) were found to be essential for PPR formation. We further found that the RA catabolizing enzyme, Cyp26c1, and the RA-dependent transcription factor Pitx2c expressed in a domain between ANP and PPR to be critical for PPR formation5. Interestingly, we also discovered that one of the RA metabolites generated by Cyp26c1, 4-oxo-RA, to be present during development at stages critical for the induction and specification of PPR. Consistent with this observation, previous reports have shown that 4-oxo-RA is capable of inducing signaling via retinoic acid receptors (RARs) to modulate the development of the anterior-posterior (A-P) axis in the embryo6,7.
Given the importance of RA in regulating Zic1 targets in the context of PPR formation, we decided to extend our analysis to additional potential targets to assess their regulation by RA, and by its metabolite 4-oxo-RA. The targets selected for further analysis were screened for their expression in the domains relevant for PPR specification, their regulation by Zic1 and their dependence on RA and 4-oxo-RA treatments. Our findings reveal several additional transcription factors, including Crx, Fezf2, Nkx3–1 and Xanf1, as well as a serine/threonine/tyrosine kinase, Pkdcc.2, that are expressed in the domains adjacent to PPR that are regulated by RA. Additionally, we find that 4-oxo-RA is a potent regulator of specification of anterior structures, and of the expression of several Zic1 target genes that are also regulated by RA, suggesting that 4-oxo-RA is capable of playing an important role in the specification of PPR. This is a significant new finding given that RA metabolites are typically thought to be biologically inactive and cleared from the cell in which they are produced. This work has the potential to transform our understanding of RA regulation and function during embryogenesis.
Results
Key anterior patterning genes are expressed during PPR formation
A microarray screen conducted to identify targets of Zic1 yielded several genes known for their patterning functions in other model systems4. We selected for genes highly upregulated by Zic1 and narrowed our choices based on expression profiling data available in Xenbase (xenbase.org), and previously published reports documenting a role for these genes in A-P patterning or development in general. These parameters yielded five candidate genes for further analysis, several of which are transcription factors: crx/otx5b is known to be involved in eye development8, fezf2 is a known regulator of neuronal development9, nkx3–1, has as yet undefined roles in early development but may function in sclerotome development10,11, and xanf1/hesx1, which is important for the development of anterior forebrain across species, including Xenopus 12,13. Interestingly, pkdcc.2, which is a kinase, was also identified as a Zic1 target and regulates the Wnt/planar cell polarity (PCP) pathway in Xenopus14. These genes are collectively referred to as “Zic1 targets” henceforth.
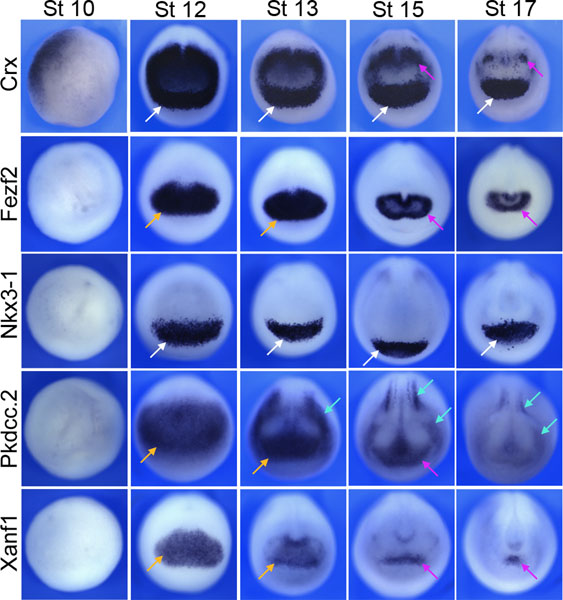
Zic1 has been shown to be a key regulator of the formation of the PPR. Therefore, to further investigate the potential role of these genes in this Zic1-driven pathway, we performed in situ hybridization (ISH) to visualize the spatio-temporal expression of these genes during early embryonic development. Figure 1 shows that these Zic1 targets are expressed during gastrulation (NF stage 10–13) and their domain of expression narrows as development progresses through neurulation (NF stage 15–17). Importantly, fezf2 and xanf1 appear to be expressed in the ANP, whereas nkx3–1 is expressed in the PPR-adjacent domain ventral to the ANP, where it overlaps with the anterior domain of crx. pkdcc.2 shows a broader expression pattern that includes the neural fold as well as the ANP (see Table 1 for a summary of gene expression). Taken together, this analysis reveals that the Zic1 targets are expressed at temporal stages and in spatial domains relevant to PPR formation, either overlapping with or adjacent to the ANP Zic1-expression domain.
Figure 1: Key patterning genes are expressed in the vicinity of the PPR and Zic1 expression domain.
In situ hybridization (ISH) for crx, fezf2, nkx3–1, pkdcc.2 and xanf1 (top to bottom) expression across developmental stages from gastrula to neurula stages. NF stages are indicated on top of each row. In all panels, anterior views, dorsal to top. Anterior neural plate: orange arrows; PPR-adjacent domain: white arrows; Anterior neural fold: purple arrows; Neural crest domain: green arrows. For a summary of the domain of expression, see Table 1.
Table 1:
Summary of the developmental expression of Zic1 targets.
| Gene Name | Anterior Neural Plate | PPR-adjacent Domain | Anterior Neural Fold | Dorsal Neural Fold/Neural Crest Domain |
|---|---|---|---|---|
| crx | − | + | + | − |
| fezf2 | + | − | + | − |
| nkx3–1 | − | + | − | − |
| pkdcc.2 | + | − | + | + |
| xanf1 | + | − | + | − |
(+) present; (−) not detected.
Patterning genes are targets of Zic1
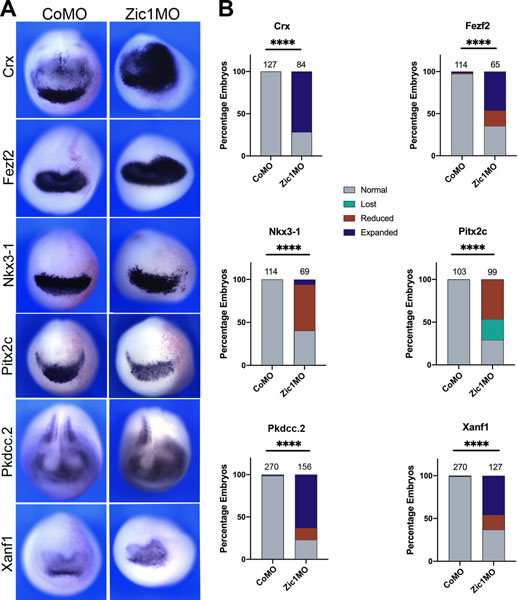
We next sought to verify that Zic1 is required for the expression of these genes. Using a well characterized morpholino antisense oligonucleotides (Zic1MO;15,3,4,5), we knocked down Zic1 in one half of the embryo at the 2-cell stage and assessed the consequences by ISH at NF stage 15 (Figure 2). Our analysis revealed that Zic1 knockdown indeed disrupted the expression of these genes, providing further evidence that they are Zic1 targets. Loss of Zic1 resulted mostly in the expansion of the expression domains for crx, fezf2, pkdcc.2, and xanf1, whereas nkx3–1 expression was significantly reduced. Pitx2c, a previously identified Zic1 target5 was also reduced. These results validate our microarray findings and establish Zic1 as a potent regulator of the above targets.
Figure 2: crx, fezf2, nkx3–1, pkdcc.2 and xanf1 are targets of Zic1.
(A) ISH for crx, fezf2, nkx3–1, pkdcc.2 and xanf1 (top to bottom) in control (CoMO) and Zic1MO-injected NF stage 15 embryos. Representative images are shown, with the injected side on the right. In all panels, anterior views, dorsal to top. (B) Frequency of the phenotypes. The number of embryos analyzed for each condition is on the top of each bar; p-values were calculated using Chi-squared test, **** p<0.0001.
Zic1 induces patterning genes in an RA-dependent manner
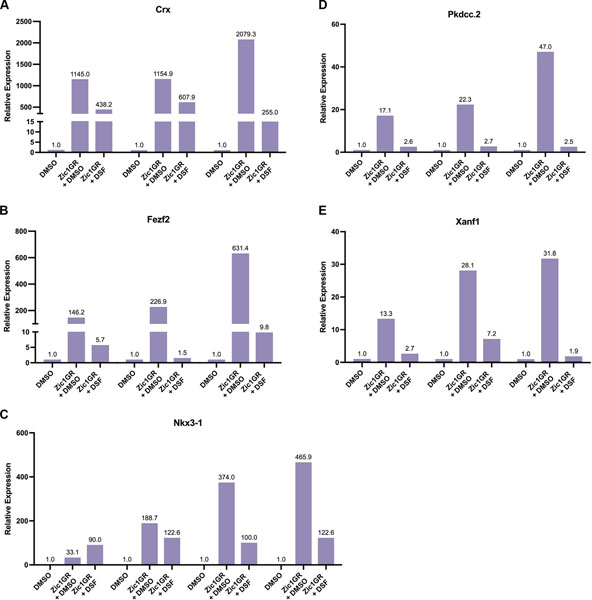
To further validate if the Zic1 targets participate in the PPR formation pathway through RA signaling, we used animal cap explants to investigate their activation under conditions where RA signaling is blocked. As seen in Figure 3, applying disulfiram (DSF), a broad inhibitor of RA synthesis16,17, to animal cap explants expressing Zic1 (Zic1GR;3) abrogated the activation of all Zic1 targets (Figure 3). The result for nkx3–1, was not as consistent, suggesting a more complex regulation mechanism (see Figure 3). Altogether, these data demonstrate that Zic1 regulates these genes indirectly through RA signaling.
Figure 3: Induction of Zic1 targets is dependent on RA synthesis.
qRT-PCR analysis of crx (A), fezf2 (B), nkx3–1 (C), pkdcc.2 (D) and xanf1 (E) expression in Zic1GR injected animal cap explants cultured for 8 hours in the presence of dexamethasone with or without Disulfiram (DSF; 100μM). Three (A, B, D, E) and four (C) biological replicates are shown. Expression levels are normalized to ODC, with fold change in relative expression indicated at the top of each bar.
Patterning Genes respond to both RA and 4-oxo-RA
Further assessment of the impact of RA signaling on the expression of Zic1 targets was performed through dose response experiments. Given our previous findings that Xenopus embryos contain a significant amount of the RA metabolite, 4-oxo-RA5, we wanted to determine if 4-oxo-RA is capable of regulating the expression of Zic1 targets. 4-oxo-RA is one of the polar metabolites generated by the action of Cyp26 enzymes on all-trans RA 18–20, however the extent of its biological activity, if any, has been debated.
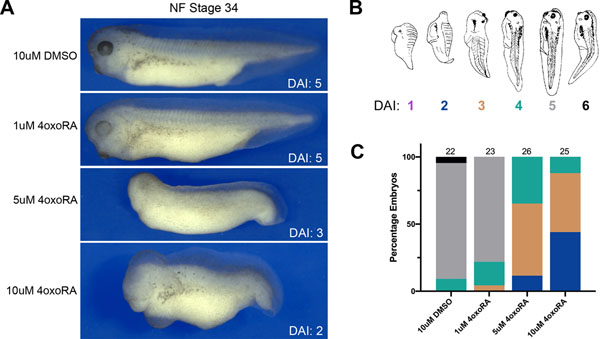
To determine whether 4-oxo-RA induces changes in the overall A-P axis specification in developing Xenopus embryos similar to those seen with RA21–23, NF stage 11 embryos were treated with increasing doses of 4-oxo-RA (Figure 4) and the dorsoanterior index (DAI) was measured at NF stage 34. DAI is a scale developed to assess axis defects in Xenopus embryos, based on whether ventralization or dorsalization has occurred 24. Briefly, a DAI grade between 0–10 is assigned based on the phenotype whereby an embryo with DAI 5 represents a normal embryo, DAI 0 being the most ventroposteriorized with no head or axial structures, and DAI 10 being the most dorsoanteriorized with only head structures that are radially symmetrical (Figure 4B;24). Upon treatment with increasing doses of 4-oxo-RA, embryos display posteriorization with the majority of embryos exhibiting head truncations akin to DAI 2 and 3 (Figure 4A, C). This is consistent with a previous report showing that 4-oxo-RA has axis posteriorizing activity6.
Figure 4: 4-oxo-RA causes posteriorization of the embryo axis.
(A) Brightfield images of Xenopus NF stage 34 embryos treated at NF stage 11 with either 10μM DMSO or increasing doses of 4-oxo-RA as shown. In all panels, lateral views, dorsal to top, anterior to left. (B) Schematic representation of the dorso-anterior index (DAI) assignments is as defined previously 24. (C) Frequency of DAI phenotypes of 4-oxo-RA treated embryos. Total embryos analyzed per condition are indicated at the top of the bar.
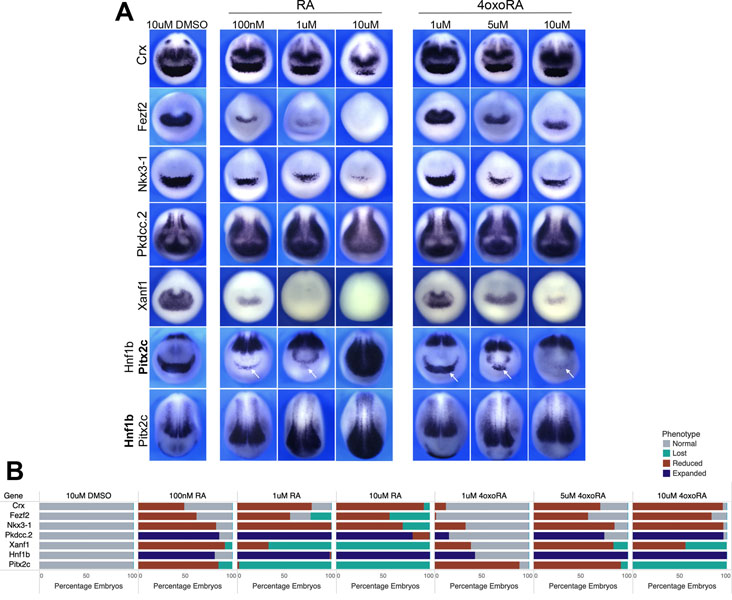
Next, we assessed the gene expression changes in Zic1 targets induced by RA and 4-oxo-RA in parallel. Figure 5 shows the summary findings of changes in gene expression of crx, fezf2, nkx3–1, pkdcc.2 and xanf1. We have previously shown that pitx2c and hnf1b show decreasing and increasing expression, respectively, upon treatment with increasing RA doses5 and they serve as positive controls for the treatments. Our analysis reveals 4-oxo-RA as not only capable of regulating the expression of Zic1 targets, but that the effect of 4-oxo-RA is very similar to that of RA, albeit at a higher dose (Figure 4, compare 1μM RA and 10μM 4-oxo-RA). These findings suggest that Zic1 targets are sensitive to RA levels, and demonstrate that the RA metabolite, 4-oxo-RA has biological signaling activity and regulates the expression of Zic1 targets in a manner similar to RA.
Figure 5: Zic1 targets show sensitivity to both RA and 4-oxo-RA.
(A) ISH for crx, fezf2, nkx3–1, pkdcc.2 and xanf1 expression on stage 15 embryos treated with either 10μM DMSO or increasing doses of RA (left) or 4-oxo-RA (right) as shown. Bottom two rows show pitx2c and hnf1b as controls in a double ISH where the gene considered is indicated in bold pitx2c (middle row) and hnf1b (bottom row), respectively. White arrows indicate position of the PPR-adjacent domain of pitx2c. All images show anterior views with dorsal to top, except for hnf1b, where dorsal views are presented with anterior to bottom. (B) Frequency of phenotypes with n ≥ 50 embryos per condition from two biological replicates.
RNA-seq reveals broad similarities in gene activation between RA and 4-oxo-RA
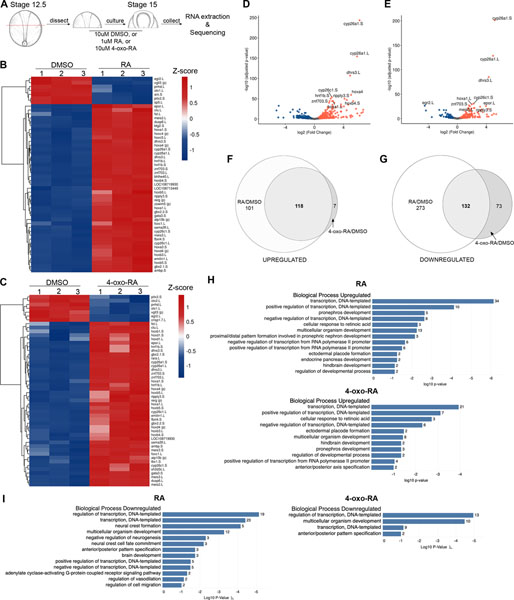
The discovery that 4-oxo-RA regulates the expression of patterning genes similarly to RA prompted us to use next generation sequencing (NGS) to determine the extent to which the two retinoids bring about similar changes in gene expression. We subjected anterior halves of Xenopus embryos to RA or 4-oxo-RA treatment mid-gastrulation (NF stage 12.5) and analyzed the global changes in RNA expression at NF stage 15 (Figure 6A). Heat map analysis of the top 50 differentially regulated genes by RA (Figure 6B) and 4-oxo-RA (Figure 6C) showed gene expression changes in known RA-target genes, as well as several novel genes. Further examination revealed that several of the highly upregulated genes between RA and 4-oxoRA were identical, such as cyp26a1, cyp26c1, dhrs3, hoxa1, ripply3 and znf703 (Figure 6D, E). Zic1 targets were also identified in these data (Table 2), with the magnitude of gene expression change (represented by log2 fold change) correlating with loss or expansion of expression observed in Figure 5. The only exception was nkx3–1, which was not detected in the 4-oxo-RA vs DMSO dataset (see discussion). Altogether, these data further validate that Zic1 targets are regulated similarly by both RA and 4-oxoRA, consistent with the whole embryo immersion experiments above (Figure 5).
Figure 6: RNA-Seq analysis reveals similarities in genes regulated by RA and 4-oxo-RA.
(A) Schematic of experimental design for RNA-Seq analysis. (B) Heatmap analysis showing the top 50 differentially expressed genes based on pairwise comparison of normalized count matrix between 10μM DMSO and 1μM RA treated anterior embryo halves. (C) Heatmap analysis of the top 50 differentially expressed genes between 10μM DMSO and 10μM 4-oxo-RA treated anterior embryo halves, similarly analyzed as (B). Heatmaps were colored by z-score. (D) Volcano plots showing significance of differential gene expression on y-axis (log10 adjusted p-value) vs. magnitude of differential gene expression on x-axis (log2 fold change) for 10μM DMSO versus 1μM RA treated anterior halves, with the top 10 genes labeled with their Xenbase ID. (E) Volcano plot similarly analyzed as (D) showing top 10 differentially regulated genes for 10μM DMSO versus 10μM 4-oxo-RA treated anterior halves, with top 10 genes with their Xenbase ID. Venn Diagram analysis for genes upregulated (F) or downregulated (G) upon RA and 4-oxoRA treatments. GO analysis using DAVID Bioinformatics Resources was performed for genes upregulated by RA (H, top) and 4-oxo-RA (H, bottom). GO analysis for genes downregulated by RA (I, left) and 4-oxo-RA (I, right).
Table 2: Magnitude of gene expression change.
Fold change (Log2 fold change) observed for Zic1 targets in the RNA-Seq analysis. Positive fold changes represent upregulated genes, whereas downregulated genes show negative log2 fold change.
| Zic1 Target | Log2 Fold Change RA vs. DMSO | Log2 Fold Change 4-oxoRA vs. DMSO |
|---|---|---|
| Crx | −1.24 | −0.53 |
| Fezf2 | −1.43 | −0.64 |
| Nkx3–1 | −0.56 | not identified |
| Pkdcc.2 | 0.62 | 0.63 |
| Xanf1 | −2.09 | −1.13 |
| Hnf1b | 2.44 | 1.96 |
| Pitx2c | −2.04 | −1.62 |
To determine the extent to which the gene expression changes induced by the two retinoids overlap, Venn Diagram comparisons between RA vs. DMSO and 4-oxo-RA vs. DMSO were made (Figure 6F, G). Interestingly, the majority of the genes upregulated by 4-oxo-RA were also upregulated by RA, whereas the majority of genes downregulated by RA were not downregulated by 4-oxo-RA. Despite these differences, a significant overlap in common genes was observed in both the upregulated and downregulated datasets (see Table 3 for the top 20 genes). GO analyses identified several terms pertaining to A-P axis specification, placode formation and neural crest development were enriched in the embryos exposed to RA and 4-oxo-RA (Figure 6H, I). Particularly, ectodermal placode formation was enriched in genes upregulated by RA and 4-oxo-RA and A-P pattern specification was enriched in genes downregulated in both conditions. Taken together, these results demonstrate that globally, the genes expression changes induced by 4-oxo-RA are highly similar to those induced by RA, lending credence to 4-oxo-RA’s ability to act as a morphogen during development.
Table 3: Top 20 common genes upregulated and downregulated by RA and 4-oxo-RA.
The genes are ranked by fold change from high to low. (p), provisional name.
| Common Genes Upregulated | Common Genes Downregulated |
|---|---|
| hoxb5.S | vgll3 (p) |
| cyp26b1.L | egr2.L |
| LOC494794 | arx.S |
| hoxb6.L | pnhd.L |
| cyp26a1.L | trpm1 (p) |
| hoxd4.L | barhl2.S |
| hoxa4 (p) | otx1.L |
| akr1c2.S | crisp1.7.L |
| dhrs3.L | wnt8b (p) |
| epor.L | sp5.L |
| hoxb4.S | dmrta1.S |
| clu.L | Xelaev18032865m |
| ripply3.S | sst.1.S |
| rerg (p) | nog2.S |
| cd69 (p) | hesx1.L |
| LOC108701043 | nrp1.S |
| LOC108697667 | XB5848002.L |
| tlx3 (p) | rspo2.L |
| bhlhe40.L | twist1.S |
| hoxb3.L | wnt8b.S |
Discussion
This study highlights the complexity of morphogens that operate during cranial placode development, and broadly, during the development of anterior structures in the Xenopus embryo. In our previous work4,5, we elucidated an RA-dependent program of cranial placode specification executed by Zic1. In this study, we have further defined the extent of this regulation by characterizing several Zic1 targets that are expressed in the ANP or the PPR-adjacent domain. In addition, we followed up on our previous observation that the RA metabolite, 4-oxo-RA, is present at high levels in the developing Xenopus embryo5 to show that 4-oxo-RA is in fact capable of extensive gene expression regulation in a manner similar to RA. Our findings point to 4-oxo-RA as an active morphogen during embryonic development, further suggesting that gradients established during embryonic development cannot be fully explained by a “source and sink” model25,26.
Zic1 targets are RA dependent patterning genes
Several Zic1 targets expressed in these domains are genes with predicted or defined roles in embryonic development, further validating Zic1’s role as a key regulator of developmental processes involved in head formation through modulation of RA. Crx (also known as otx5b) has been shown to be critical for photoreceptor development in the retina 27,28. Fezf2 is an important regulator of forebrain development, and more specifically, of projection neurons in the mammalian neocortex, as well as monoaminergic neurons29, 30.
On the other hand, while several members of the NKX gene family have been shown to play key roles during development31, the role of nkx3–1 is less clear. Reports in mice have shown nkx3–1 to be involved in sclerotome development32, minor salivary glands and prostate duct morphogenesis33. Interestingly, in Xenopus, crx, nkx3–1 and pitx2c were strongly induced by pitx1, a gene that is also expressed in the PPR-adjacent domain34. The study also suggested that while the induction of crx and nkx3–1 appears direct, the induction of pitx2c is likely indirect. This finding is especially intriguing in light of our previous work showing pitx2c as a Zic1 target5 and our microarray analysis that identified pitx1 as a Zic1 target, however at levels approximately half that of pitx2c (data not shown). In future work, it will be interesting to elucidate the relationship between these genes, and we speculate that these interactions might contribute to the variability of nkx3–1 induction levels in response to Zic1GR expression (Figure 3 and Figure 3).
Xanf1 (also known as hesx1) is perhaps the best-characterized gene in embryonic development, though its regulation by Zic1 has not been previously described. Xanf1 functions during forebrain development to regulate formation of the anterior neuroectoderm in mice12 and Xenopus35, where it acts as a transcriptional repressor. Mutations in human Hesx1 gene have been reported to cause septo-optic dysplasia (SOD)36, an early forebrain development disorder presenting with hypoplasia of the optic nerves, midline brain defects, as well as pituitary-related deficiencies37.
Out of the five Zic1 targets presented in this study, pkdcc2 is the only gene that is not a transcription factor14. pkdcc.2 is one of two Xenopus homologs of the mouse and human Pkdcc gene, encoding a secreted tyrosine kinase38. Pkdcc knockdown in mice has been shown to result in a host of defects, including short limbs and cleft palate39. In Xenopus, pkdcc.2 and pkdcc.1 have been shown to regulate blastopore and neural tube closure through regulation of the Wnt-PCP signaling pathway14, highlighting pkdcc.2’s role as a patterning gene during embryonic development. With the exception of nkx3–1, blocking RA signaling through DSF causes reduction in gene expression of these Zic1 target genes, indicating that they are involved in the Zic1-activated pathway that is critical for cranial placode specification5. Future studies will examine the functional role of these genes in cranial placode specification.
4-oxo-RA is an active morphogen during Xenopus development
Another important finding of this study is the demonstration that 4-oxo-RA is an important morphogen capable of inducing gene expression changes in Xenopus embryos similar to RA. Based on genetic evidence, it has been proposed that retinoid metabolites do not regulate development40, however this study did not specifically assess the effect of any metabolite on gene expression of RA-targets. Our approach involved directly treating the embryo with 4-oxo-RA during the “RA sensitivity” window 41 to assess if any morphological defects or gene expression changes could be observed. Indeed, we observe A-P defects in 4-oxo-RA treated embryos (Figure 4) consistent with previous results6. In addition, we observed quantifiable changes in known RA target genes, both through ISH experiments and RNA-seq analyses (as detailed below).
Although changes in gene expression upon exogenous treatment of embryos with RA or 4-oxo-RA may not necessarily reflect an endogenous role for these molecules, this approach still provides meaningful information on their potential activity. Prior to this work, two studies have examined 4-oxo-RA in Xenopus embryonic development, but this work presents the first investigation of 4-oxo-RA activity at a global level in the embryo. It was previously shown that 4-oxo-RA is capable of interacting with retinoic acid receptors (RARs) with almost similar affinity as RA for RAR⍺, but with significantly higher affinity for RARβ and RARγ42. This work along with our prior study5 demonstrated that 4-oxo-RA levels could be measured in Xenopus embryos at consistently higher levels than RA, suggesting that it may be the major retinoid species in frogs. However, it is unclear whether these levels of 4-oxo-RA are due to maternal accumulation, or generated specifically for signaling during embryonic development as a consequence of Cyp26 enzymes RA catabolizing activity5. In addition, we have previously shown that cranial placode formation via Zic1 does not require RARs4. It will be interesting to see which receptors are responsible for retinoid signaling for placode specification, and subsequently, if 4-oxo-RA has a higher affinity for this receptor and can compete with RA.
The genes identified in our RNA-seq analysis were specifically enriched for anterior structures by only analyzing the anterior half of the embryo. This also allowed us to exclude gene expression changes resulting from interactions with the second major source of RA in the embryo, the paraxial mesoderm43. As seen in Figure 6, the majority of genes upregulated by 4-oxo-RA are also upregulated by RA, albeit at a lower dose. These genes, including dhrs3, cyp26a1, hoxd4, hoxa1 and hnf1b etc. are known RA-responsive genes and have previously been identified in screens for RA-targets44, demonstrating that 4-oxo-RA is an active retinoid in the Xenopus embryo. Interestingly, with the exception of nkx3–1, all Zic1-targets characterized in this study were also identified in the RNA-seq (see Table 2) with similar up or downregulation trends as observed in Figure 5. In this analysis, nkx3–1 did not reach the threshold of adjusted p-value < 0.05. This could be due to the fact that the embryos analyzed via RNA-seq were treated with the respective retinoids for a shorter period of time (from NF stage 12.5 to NF stage 15; Figure 6) as compared to the whole embryo treatment (from NF stage 11 to 15; Figure 5). It is worth noting that despite the similarities in the pathways up or downregulated by RA and 4-oxo-RA, the impact of RA is greater, with over 600 differentially regulated genes that span a greater range of biological processes than 4-oxo-RA (Figure 6). Based on these data, we conclude that while 4-oxo-RA is capable of inducing similar gene expression changes, 4-oxo-RA affects fewer targets than RA.
Overall, our results demonstrate that the presence of Cyp26 enzymes in a particular domain adjacent to an RA synthesizing domain does not necessarily serve as a “sink”, as the metabolites generated have biological activity. We speculate that this might indicate a role of 4-oxo-RA as a “backup” mechanism, where in the event of excess RA degradation, crucial genes may still be activated. Alternatively, it may be a remnant of maternal RA deposits in the yolk that accumulates over time and can perform essential gene activation in the absence of sufficient RA. Consistent with previous reports45,46 we also find that 4-oxo-RA induces cyp26a1 and cyp26c1 expression, raising the interesting possibility that these enzymes are also needed to further oxidize the metabolites 4-oxo-RA, 18-OH-RA and 4-OH-RA, as seen for human Cyp26c147. While it has been shown that 4-oxo-RA can bind to RARs42 and alter the expression of genes known that contain RAREs, it is also possible that it is regulating gene expression in an RARs independent manner, based on cellular and tissue context. These are important questions that require further investigation to understand the physiological function for 4-oxo-RA during development.
Experimental Procedures
Plasmids and oligonucleotides
Xenopus laevis Zic1GR, and the tracer β-galactosidase mRNAs were synthesized using the Message mMachine SP6 and T7 Transcription kits (Ambion, SP6 Cat# AM1340; T7 Cat# AM1344) as described previously5. A well characterized Zic1 morpholino antisense oligonucleotide (Zic1MO;15,3,4,5) was purchased from GeneTools (Philomath, OR). A standard control morpholino (CoMO) was used as control.
Embryos, injections, treatments and animal cap explant culture
As described in Dubey et. al., 2021, Xenopus laevis embryos were staged according to Nieuwkoop and Faber (NF)48 and raised in 0.1X Normal Amphibian Medium (NAM)49. All experiments in this study were conducted in accordance with the guidelines of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All protocols requiring Xenopus were approved by the Institutional Animal Care and Use Committee of New York University under animal protocol #IA16–00052. In microinjection experiments, embryos were injected in one blastomere at the 2-cell stage (NF stage 2) and collected at NF stage 15 for in situ hybridization (ISH). 40–60ng of MO or CoMO and 0.5 ng of β-galactosidase mRNA as a lineage tracer were used in injections. CoMO-injected embryos were used as controls. Retinoic acid (all-trans RA, Sigma-Aldrich, Cat# R2625) and 4-oxo-RA (Sigma-Aldrich, Cat#07558) treatments were performed using stock solutions prepared in DMSO (dimethylsulfoxide; Sigma-Aldrich Cat# D8418). NF stage 11 embryos were incubated in RA diluted in 0.1X NAM (100nM, 1μM, 10μM) or 4-oxo-RA diluted in 0.1X NAM (1μM, 5μM, 10μM) and collected at NF stage 15 for ISH (Figure 5) or NF stage 34 (Figure 4). Anterior halves of the embryos used in Figure 6 were treated at NF stage 12.5 and collected at NF stage 15. Sibling embryos treated with 10μM DMSO served as controls.
For Animal cap explant assays, one blastomere of a 2-cell stage embryo was injected in the animal pole region with 250pg Zic1GR mRNA, as previously described4. The explants were dissected at the blastula stage (NF stage 9) and culture in 0.5X NAM with 10μM dexamethasone (Sigma-Aldrich, Cat# D1756) for 8 hours at 24°C unless otherwise noted, then collected and immediately stored at −80°C. 100μM Disulfiram (DSF, tetraethylthiuram disulfide, Sigma-Aldrich, Cat# 86720) was added to the incubation medium for DSF-treated caps. All experiments were performed on at least three independent batches of embryos (obtained from three separate females). For whole embryo treatment and injection experiments, 50 embryos per injection and per marker were used, and 12–15 animal caps per condition were used in animal cap explant assays.
In situ hybridization
In situ hybridization was performed as previously described5,50–51. Briefly, NF stage 15 embryos were fixed in MEMFA (0.1 M MOPS, 2 mM EGTA, 1 mM MgSO4 and 3.7% formaldehyde) for 1 hour. Red-Gal (Research Organics, Cat#C40040) staining was performed prior to in situ hybridization (ISH) when β-galactosidase mRNA was used as tracer. No tracers were used in RA and 4-oxoRA immersion experiments.
Whole-mount ISH was performed in 4 ml glass vials on a standard bench top nutator. crx, fezf2 and nkx3–1 (purchased form OpenBiosystems; ThermoFisher Scientific, MA), hnf1b52 and pitx2c53 digoxygenin (DIG)-labeled antisense RNA probes were synthesized using the Genius Kit (Roche, Cat#1175025). To generate pkdcc.2 and Xanf1 probes, pkdcc.2 and xanf1 sequences were amplified from Xenopus laevis NF Stage 12 cDNA and cloned into pGEMT-Easy (Promega, Cat#A1360), and linearized templates were used to synthesize the corresponding DIG-labeled probes.
Hybridization was performed overnight at 60°C. For detection of RNA probes, the embryos were incubated with an anti-DIG antibody conjugated to alkaline phosphatase (Roche, Cat#11093274910; 1:2000 dilution) overnight at 4°C. Several successive washes were used for excess antibody removal and the chromogenic reaction performed overnight at room temperature in BM purple (Roche, Cat#11442074001). The embryos were subsequently bleached in 10% hydrogen peroxide in methanol for 48 h and subsequently imaged in 1× PBS.
RNA Sequencing
Extracted Total RNA from anterior halves of NF stage 15 embryos was verified on the Agilent Bioanalyzer system using a nanochip to verify the RNA integrity number (RIN). High quality total RNA (500ng) was input into the Illumina Truseq v2 stranded mRNA library prep kit (Cat#20020595) according to the manufacturer’s protocol. Generated cDNA was amplified with 12 PCR cycles. Final generated libraries were verified on the Agilent Tapestation 2200 system using high sensitivity DNA Screentape. Sample concentration was checked on the Invitrogen qubit system using the high sensitivity DNA reagent. Each sample was normalized to a specific nanomolar concentration and pooled equally into a final combined pool. The pool was run on 1 lane of Novaseq SP100 flow cell (run length was 50 paired end bases) on the Illumina Novaseq 6000 sequencing system.
Bioinformatics Analysis
RNA-seq data were analyzed by sns rna-star pipeline (https://igordot.github.io/sns/routes/rna-star.html). Adapters and low quality bases were trimmed using Trimmomatic (v0.36)54. Sequencing reads were mapped to the reference genome (XENLA_9.2, http://www.xenbase.org/common/displayJBrowse.do?data=data/xl9_2) using the STAR aligner (v2.7.3)55. Alignments were guided by a Gene Transfer Format (GTF) file, which was converted from a General Feature Format (GFF3) file. The mean read insert sizes and their standard deviations were calculated using Picard tools (v.2.18.20) (http://broadinstitute.github.io/picard). The genes-samples counts matrix was generated using featureCounts (v1.6.3)56, normalized based on their library size factors using DEseq257, and differential expression analysis was performed.
The Read Per Million (RPM) normalized BigWig files were generated using deepTools(v.3.1.0)58. To compare the level of similarity among the samples and their replicates, we used two methods: principal-component analysis and Euclidean distance-based sample clustering. All the downstream statistical analyses and plot generation were performed in R environment (4.0.3) (https://www.r-project.org/). Significant testing was done using Wald Chi-squared test and the Benjamini-Hochberg (BH) procedure was used to generate adjusted p-values that were used as inputs for Volcano plot analysis of differential gene expression. Gene ontology (GO) enrichment analyses were performed using DAVID Bioinformatics Resources 6.859,60.
qRT-PCR analysis
Total RNA was extracted from 12–15 animal cap explants using the RNeasy Micro kit (Qiagen, Cat# 74004) and quantified using a Nanodrop spectrophotometer (Nanodrop Technologies). Genomic DNA was removed by on-column digestion with RNase-free DNase I for 15 mins at room temp. 10ng total RNA was used per reaction. qRT-PCR was performed using the One Step RT-PCR kit (Applied Bioystems, Cat# 4388869, 4389988). The mean Ct value for each biological replicate was calculated from 4 technical replicates. The primers used for qRT-PCR are listed below.
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| pkdcc.2 | 5’ GGACAGTTCCCAATGACTTCT 3’ | 5’ GTGACGCTAGAGGGCTTATTT 3’ |
| xanf1 | 5’ AGAGAGAACTGAGCTGGTATAGG 3’ | 5’ GGATAGGAATTCACTCGGAACAC 3’ |
| nkx3–1 | 5’ AGGTCATAGAGCTGGAAAGAAAG 3’ | 5’ AGGCTCTTGGCAAGCTG 3’ |
| crx | 5’ CAAACCCACGGAAACAAAGAAG 3’ | 5’ GCACTCGGGATTCTGGTAAAT 3’ |
| fezf2 | 5’ GTCTGCGGAAAGGTGTTTAATG 3’ | 5’ CGTGTGGATGATCTTGTGTCT 3’ |
| ODC | 5’ ACATGGCATTCTCCCTGAAG 3’ | 5’ TGGTCCCAAGGCTAAAGTTG 3’ |
Imaging
Embryos processed for in situ hybridization were imaged using a dual light-fluorescence Leica M165 Stereomicroscope, with the same exposure and magnification used for each marker in the experiment. Representative images of the dominant phenotype were captured and the image panels were composed using Adobe Photoshop.
Statistical Analysis
Unless otherwise stated, experiments were conducted in embryos from three or more female Xenopus for biological replicates. No prior analysis was done to calculate the sample size. As previously described5, to assess changes in expression of the genes of interest, Zic1MO-injected embryos were compared to CoMO injected embryos, and for each unilaterally injected embryo the injected side was compared to the uninjected side as well, for an additional internal control. Each embryo was assigned one of the following phenotypes: normal, lost or reduced, expanded, or ectopic. For RA/4-oxo-RA treated embryos, the treated embryos were compared with DMSO-treated controls at an equivalent dose and assigned phenotypes as above. For whole-mount ISH experiments, the data from all biological replicates was pooled for statistical analysis, and significance testing was performed using the Chi-squared test using GraphPad Prism 9 (GraphPad Software). p<0.05 was considered significant, unless otherwise stated. For animal cap qRT-PCR experiments, a representative graph is shown in the main results, and the remaining two or three biological replicates are provided in Figure 3. Data in Figure 5 was visualized using Tableau (Tableau Software).
Acknowledgements
We thank the members of Saint-Jeannet laboratory for technical assistance and helpful discussions. We would like to thank Paul Zappile at the Genome Technology Center (GTC) for expert library preparation and RNA sequencing, and Ziyan Lin at the Applied Bioinformatics Laboratories (ABL) for providing bioinformatics support and helping with the analysis and interpretation of the data. This work has used computing resources at the NYU School of Medicine High Performance Computing Facility. This work also benefited from the support of Xenbase (http://www.xenbase.org/ - RRID:SCR_003280) and the National Xenopus Resource (http://mbl.edu/xenopus/ - RRID:SCR_013731).
Funding:
This work was supported by grants from the National Institutes of Health to J-P.S-J. (R01-DE025806) and to A.D (F32-DE027599). GTC and ABL are shared resources partially supported by the Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center.
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
All data supporting the results are available from the corresponding author upon request. The RNA-seq data have been deposited onto GEO under accession number: GSE180269.
References
- 1.Saint-Jeannet J-P, Moody SA. Establishing the pre-placodal region and breaking it into placodes with distinct identities. Developmental biology. 2014;389(1):13–27. doi: 10.1016/j.ydbio.2014.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlosser G Chapter Four - Making Senses: Development of Vertebrate Cranial Placodes. In: Jeon K, ed. International Review of Cell and Molecular Biology. Vol 283. Academic Press; 2010:129–234. doi: 10.1016/S1937-6448(10)83004-7 [DOI] [PubMed] [Google Scholar]
- 3.Hong C-S, Saint-Jeannet J-P. The Activity of Pax3 and Zic1 Regulates Three Distinct Cell Fates at the Neural Plate Border. Molecular Biology of the Cell. 2007;18(6):2192–2202. doi: 10.1091/mbc.e06-11-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaurena MB, Juraver-Geslin H, Devotta A, Saint-Jeannet J-P. Zic1 controls placode progenitor formation non-cell autonomously by regulating retinoic acid production and transport. Nature Communications. 2015;6:7476. doi: 10.1038/ncomms8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubey A, Yu J, Liu T, Kane MA, Saint-Jeannet J-P. Retinoic acid production, regulation and containment through Zic1, Pitx2c and Cyp26c1 control cranial placode specification. Development. 2021;148(4). doi: 10.1242/dev.193227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pijnappel WWM, Hendriks HFJ, Folkers GE, et al. The retinoid ligand 4-oxo-retinoic acid is a highly active modulator of positional specification. Nature. 1993;366:340. doi: 10.1038/366340a0 [DOI] [PubMed] [Google Scholar]
- 7.Achkar CC, Derguini F, Blumberg B, et al. 4-Oxoretinol, a new natural ligand and transactivator of the retinoic acid receptors. Proceedings of the National Academy of Sciences. 1996;93(10):4879–4884. doi: 10.1073/pnas.93.10.4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitaker SL, Knox BE. Conserved Transcriptional Activators of the Xenopus Rhodopsin Gene. Journal of Biological Chemistry. 2004;279(47):49010–49018. doi: 10.1074/jbc.M406080200 [DOI] [PubMed] [Google Scholar]
- 9.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(47):17184–17189. doi: 10.1073/pnas.0508732102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka M, Lyons GE, Izumo S. Expression of the Nkx3.1 homobox gene during pre and postnatal development. Mechanisms of Development. 1999;85(1):179–182. doi: 10.1016/s0925-4773(99)00084-2 [DOI] [PubMed] [Google Scholar]
- 11.Herbrand H, Pabst O, Hill R, Arnold H-H. Transcription factors Nkx3.1 and Nkx3.2 (Bapx1) play an overlapping role in sclerotomal development of the mouse. Mechanisms of Development. 2002;117(1):217–224. doi: 10.1016/s0925-4773(02)00207-1 [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Barbera JP, Rodriguez TA, Beddington RS. The homeobox gene Hesx1 is required in the anterior neural ectoderm for normal forebrain formation. Dev Biol. 2000;223(2):422–430. doi: 10.1006/dbio.2000.9757 [DOI] [PubMed] [Google Scholar]
- 13.Andoniadou CL, Signore M, Young RM, et al. HESX1- and TCF3-mediated repression of Wnt/β-catenin targets is required for normal development of the anterior forebrain. Development. 2011;138(22):4931–4942. doi: 10.1242/dev.066597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vitorino M, Silva AC, Inácio JM, et al. Xenopus Pkdcc1 and Pkdcc2 Are Two New Tyrosine Kinases Involved in the Regulation of JNK Dependent Wnt/PCP Signaling Pathway. PLOS ONE. 2015;10(8):e0135504. doi: 10.1371/journal.pone.0135504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132(10):2355–2363. doi: 10.1242/dev.01823 [DOI] [PubMed] [Google Scholar]
- 16.Kitson TM. The effect of disulfiram on the aldehyde dehydrogenases of sheep liver. Biochem J. 1975;151(2):407–412. doi: 10.1042/bj1510407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veverka KA, Johnson KL, Mays DC, Lipsky JJ, Naylor S. Inhibition of aldehyde dehydrogenase by disulfiram and its metabolite methyl diethylthiocarbamoyl-sulfoxide. Biochemical Pharmacology. 1997;53(4):511–518. doi: 10.1016/s0006-2952(96)00767-8 [DOI] [PubMed] [Google Scholar]
- 18.White JA, Beckett-Jones B, Guo YD, et al. cDNA cloning of human retinoic acid-metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J Biol Chem. 1997;272(30):18538–18541. doi: 10.1074/jbc.272.30.18538 [DOI] [PubMed] [Google Scholar]
- 19.White JA, Ramshaw H, Taimi M, et al. Identification of the human cytochrome P450, P450RAI-2, which is predominantly expressed in the adult cerebellum and is responsible for all-trans-retinoic acid metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6403–6408. doi: 10.1073/pnas.120161397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taimi M, Helvig C, Wisniewski J, et al. A Novel Human Cytochrome P450, CYP26C1, Involved in Metabolism of 9-cis and All-trans Isomers of Retinoic Acid. Journal of Biological Chemistry. 2004;279(1):77–85. doi: 10.1074/jbc.M308337200 [DOI] [PubMed] [Google Scholar]
- 21.Katsuyama Y, Saiga H. Retinoic acid affects patterning along the anterior-posterior axis of the ascidian embryo. Dev Growth Differ. 1998;40(4):413–422. doi: 10.1046/j.1440-169x.1998.t01-2-00006.x [DOI] [PubMed] [Google Scholar]
- 22.Grandel H, Lun K, Rauch G-J, et al. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129(12):2851–2865. doi: 10.1242/dev.129.12.2851 [DOI] [PubMed] [Google Scholar]
- 23.Dubey A, Rose RE, Jones DR, Saint-Jeannet J-P. Generating retinoic acid gradients by local degradation during craniofacial development: One cell’s cue is another cell’s poison. genesis. 2018;56(2):e23091. doi: 10.1002/dvg.23091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao KR, Elinson RP. The entire mesodermal mantle behaves as Spemann’s organizer in dorsoanterior enhanced Xenopus laevis embryos. Developmental Biology. 1988;127(1):64–77. doi: 10.1016/0012-1606(88)90189-3 [DOI] [PubMed] [Google Scholar]
- 25.Crick F Diffusion in Embryogenesis. Nature. 1970;225(5231):420–422. doi: 10.1038/225420a0 [DOI] [PubMed] [Google Scholar]
- 26.Shimozono S, Iimura T, Kitaguchi T, Higashijima S-i, Miyawaki A. Visualization of an endogenous retinoic acid gradient across embryonic development. Nature. 2013;496(7445):363–366. doi: 10.1038/nature12037 [DOI] [PubMed] [Google Scholar]
- 27.Vignali R, Colombetti S, Lupo G, et al. Xotx5b, a new member of the Otx gene family, may be involved in anterior and eye development in Xenopus laevis. Mechanisms of Development. 2000;96(1):3–13. doi: 10.1038/nature12037 [DOI] [PubMed] [Google Scholar]
- 28.Shen Y-c, Raymond PA. Zebrafish cone-rod (crx) homeobox gene promotes retinogenesis. Developmental Biology. 2004;269(1):237–251. doi: 10.1016/j.ydbio.2004.01.037 [DOI] [PubMed] [Google Scholar]
- 29.Shimizu T, Hibi M. Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Development, Growth & Differentiation. 2009;51(3):221–231. doi: 10.1111/j.1440-169X.2009.01088.x [DOI] [PubMed] [Google Scholar]
- 30.Zhang S, Li J, Lea R, Vleminckx K, Amaya E. Fezf2 promotes neuronal differentiation through localised activation of Wnt/β-catenin signalling during forebrain development. Development (Cambridge, England). 2014;141(24):4794–4805. doi: 10.1242/dev.115691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanfel MN, Moses KA, Schwartz RJ, Zimmer WE. Regulation of organ development by the NKX-homeodomain factors: an NKX code. Cell Mol Biol (Noisy-le-grand). 2005;Suppl 51:Ol785–799. [PubMed] [Google Scholar]
- 32.Herbrand H, Pabst O, Hill R, Arnold HH. Transcription factors Nkx3.1 and Nkx3.2 (Bapx1) play an overlapping role in sclerotomal development of the mouse. Mech Dev. 2002;117(1–2):217–224. doi: 10.1016/s0925-4773(02)00207-1 [DOI] [PubMed] [Google Scholar]
- 33.Schneider A, Brand T, Zweigerdt R, Arnold H-H. Targeted disruption of the Nkx3.1 gene in mice results in morphogenetic defects of minor salivary glands: parallels to glandular duct morphogenesis in prostate. Mechanisms of Development. 2000;95(1):163–174. doi: 10.1016/s0925-4773(00)00355-5 [DOI] [PubMed] [Google Scholar]
- 34.Jin Y, Weinstein DC. Pitx1 regulates cement gland development in Xenopus laevis through activation of transcriptional targets and inhibition of BMP signaling. Developmental biology. 2018;437(1):41–49. doi: 10.1016/j.ydbio.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ermakova GV, Alexandrova EM, Kazanskaya OV, Vasiliev OL, Smith MW, Zaraisky AG. The homeobox gene, Xanf-1, can control both neural differentiation and patterning in the presumptive anterior neurectoderm of the Xenopus laevis embryo. Development. 1999;126(20):4513–4523. doi: 10.1242/dev.126.20.4513 [DOI] [PubMed] [Google Scholar]
- 36.Dattani MT, Martinez-Barbera J-P, Thomas PQ, et al. Mutations in the homeobox gene HESX1/Hesx1 associated with septo-optic dysplasia in human and mouse. Nature Genetics. 1998;19(2):125–133. doi: 10.1038/477 [DOI] [PubMed] [Google Scholar]
- 37.Webb EA, Dattani MT. Septo-optic dysplasia. European journal of human genetics: EJHG. 2010;18(4):393–397. doi: 10.1038/ejhg.2009.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bordoli MR, Yum J, Breitkopf SB, et al. A secreted tyrosine kinase acts in the extracellular environment. Cell. 2014;158(5):1033–1044. doi: 10.1016/j.cell.2014.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imuta Y, Nishioka N, Kiyonari H, Sasaki H. Short limbs, cleft palate, and delayed formation of flat proliferative chondrocytes in mice with targeted disruption of a putative protein kinase gene, Pkdcc (AW548124). Developmental Dynamics. 2009;238(1):210–222. doi: 10.1002/dvdy.21822 [DOI] [PubMed] [Google Scholar]
- 40.Niederreither K, Abu-Abed S, Schuhbaur B, Petkovich M, Chambon P, Dollé P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nature Genetics. 2002;31:84. doi: 10.1038/ng876 [DOI] [PubMed] [Google Scholar]
- 41.Cunningham ML, Mac Auley A, Mirkes PE. From gastrulation to neurulation: Transition in retinoic acid sensitivity identifies distinct stages of neural patterning in the rat. Developmental Dynamics. 1994;200(3):227–241. doi: 10.1002/aja.1002000305 [DOI] [PubMed] [Google Scholar]
- 42.Blumberg B, Bolado J, Derguini F, et al. Novel retinoic acid receptor ligands in Xenopus embryos. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(10):4873–4878. doi: 10.1073/pnas.93.10.4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molotkova N, Molotkov A, Sirbu IO, Duester G. Requirement of mesodermal retinoic acid generated by Raldh2 for posterior neural transformation. Mechanisms of development. 2005;122(2):145–155. doi: 10.1016/j.mod.2004.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arima K, Shiotsugu J, Niu R, et al. Global analysis of RAR-responsive genes in the Xenopus neurula using cDNA microarrays. Developmental Dynamics. 2005;232(2):414–431. doi: 10.1002/dvdy.20231 [DOI] [PubMed] [Google Scholar]
- 45.Topletz AR, Tripathy S, Foti RS, Shimshoni JA, Nelson WL, Isoherranen N. Induction of CYP26A1 by metabolites of retinoic acid: evidence that CYP26A1 is an important enzyme in the elimination of active retinoids. Mol Pharmacol. 2015;87(3):430–441. doi: 10.1124/mol.114.096784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Idres N, Marill J, Flexor MA, Chabot GG. Activation of retinoic acid receptor-dependent transcription by all-trans-retinoic acid metabolites and isomers. J Biol Chem. 2002;277(35):31491–31498. doi: 10.1074/jbc.M205016200 [DOI] [PubMed] [Google Scholar]
- 47.Zhong G, Ortiz D, Zelter A, Nath A, Isoherranen N. CYP26C1 Is a Hydroxylase of Multiple Active Retinoids and Interacts with Cellular Retinoic Acid Binding Proteins. Molecular pharmacology. 2018;93(5):489–503. doi: 10.1124/mol.117.111039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin). A systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. 1956. Amsterdam: North-Holland Publishing Co. [Google Scholar]
- 49.Slack JM, Forman D. An interaction between dorsal and ventral regions of the marginal zone in early amphibian embryos. J Embryol Exp Morphol. 1980;56:283–299. [PubMed] [Google Scholar]
- 50.Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods in Cell Biology. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6 [DOI] [PubMed] [Google Scholar]
- 51.Saint-Jeannet J-P. Whole-Mount In Situ Hybridization of Xenopus Embryos. Cold Spring Harbor Protocols. 2017;2017(12):pdb.prot097287. doi: 10.1101/pdb.prot097287 [DOI] [PubMed] [Google Scholar]
- 52.Demartis A, Maffei M, Vignali R, Barsacchi G, De Simone V. Cloning and developmental expression of LFB3/HNF1 beta transcription factor in Xenopus laevis. Mech Dev. 1994;47(1):19–28. doi: 10.1016/0925-4773(94)90092-2 [DOI] [PubMed] [Google Scholar]
- 53.Jeong Y-HP, Park B-K; Saint-Jeannet J-P and Lee Y-H. Developmental expression of Pitx2c in Xenopus trigeminal and profundal placodes. The International Journal of Developmental Biology. 2014;58:701–704. doi: 10.1387/ijdb.140254js [DOI] [PubMed] [Google Scholar]
- 54.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29(1):15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30(7):923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 57.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramírez F, Ryan DP, Grüning B, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Research. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 60.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the results are available from the corresponding author upon request. The RNA-seq data have been deposited onto GEO under accession number: GSE180269.