Abstract
Background:
Inherited germline TP53 pathogenic and likely pathogenic mutations (gTP53) cause autosomal dominant multicancer predisposition including Li-Fraumeni syndrome (LFS). However, there is no known association of prostate cancer with gTP53.
Objective:
To determine whether gTP53 predisposes to prostate cancer.
Design, setting, and participants:
This multi-institutional retrospective study characterizes prostate cancer incidence in a cohort of LFS males and gTP53 prevalence in a prostate cancer cohort.
Outcome measurements and statistical analysis:
We evaluated the spectrum of gTP53 variants and clinical features associated with prostate cancer.
Results and limitations:
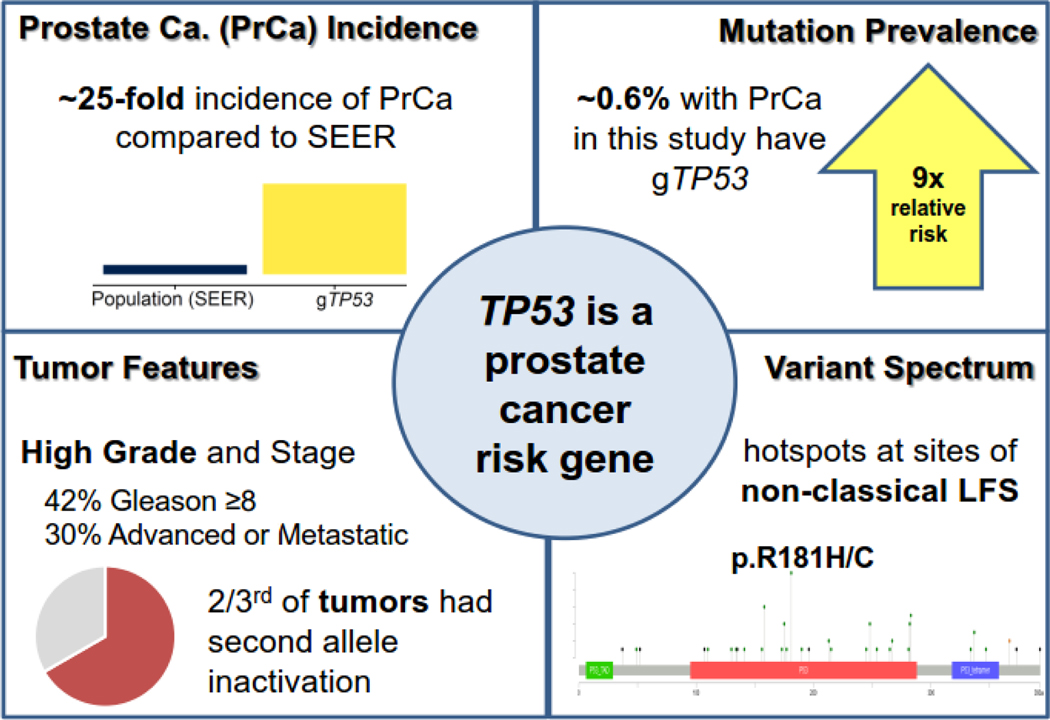
We identified 31 prostate cancer cases among 163 adult LFS males, including 26 of 54 aged ≥50 yr. Among 117 LFS males without prostate cancer at the time of genetic testing, six were diagnosed with prostate cancer over a median (interquartile range [IQR]) of 3.0 (1.3–7.2) yr of follow-up, a 25-fold increased risk (95% confidence interval [CI] 9.2–55; p < 0.0001). We identified gTP53 in 38 of 6850 males (0.6%) in the prostate cancer cohort, a relative risk 9.1-fold higher than that of population controls (95% CI 6.2–14; p < 0.0001; gnomAD). We observed hotspots at the sites of attenuated mutations not associated with classic LFS. Two-thirds of available gTP53 prostate tumors had somatic inactivation of the second TP53 allele. Among gTP53 prostate cancer cases in this study, the median age at diagnosis was 56 (IQR: 51–62) yr, 42% had Gleason ≥8 tumors, and 30% had advanced disease at diagnosis.
Conclusions:
Complementary analyses of prostate cancer incidence in LFS males and gTP53 prevalence in prostate cancer cohorts suggest that gTP53 predisposes to aggressive prostate cancer. Prostate cancer should be considered as part of LFS screening protocols and TP53 considered in germline prostate cancer susceptibility testing.
Patient summary:
Inherited mutations in the TP53 gene are likely to predispose men to aggressive prostate cancer.
Keywords: Attenuated, Genetic testing, Germline, Hypomorphic mutation, Inherited cancer syndrome, Li-Fraumeni syndrome, Pathogenic variant, Prostate cancer, Screening, TP53
1. Introduction
Prostate cancer (PrCa) is a highly heritable disease [1]. Recent data have demonstrated that approximately 5–20% of cases are associated with germline (inherited) mutations in genes associated with recognized cancer predisposition syndromes, such BRCA2 and ATM [2,3]. Germline TP53 pathogenic variants (gTP53) are associated with an autosomal dominant hereditary multicancer predisposition known as Li-Fraumeni syndrome (LFS) [4,5]. Patients with classic LFS develop early-onset, pediatric, and multiple primary cancers with nearly universal development of at least one cancer in their lifetime [6]; presentations with more variable penetrance are increasingly observed [5,7,8]. Panel genetic testing, largely ascertained on individuals with personal and family histories of breast cancers, has demonstrated a more expansive tumor spectrum and more variable penetrance of gTP53 variants than previously appreciated [9]. However, there has been no documented association of PrCa with gTP53, outside of one case report [10]. In a recent review of the TP53 International Agency for Research on Cancer (IARC) database [8], only 1.7% of gTP53 patients were reported to have PrCa, far lower than the 14% lifetime risk of PrCa for average men in the USA [11]. However, this analysis is misleading given that the database is heavily skewed toward men of younger ages than those relevant to typical PrCa risk.
Guidelines advise extensive cancer screening in patients with LFS, starting in childhood, for example, yearly full-body magnetic resonance imaging (MRI) and brain imaging, targeted ultrasound, assessments for gastrointestinal and skin neoplasms, and breast screening for women [12,13]. Patients with LFS may also have an elevated risk of post-treatment secondary malignancies, for example, after radiation [14,15]. Therefore, LFS status may be considered in cancer treatment planning. For example, in women with LFS, total mastectomy is preferred to partial mastectomy to avoid the need for radiation.
Current cancer screening guidelines for LFS patients do not include PrCa. Moreover, gTP53 status is not currently considered in treatment planning for individuals with PrCa, which often includes decisions between radiation and surgery for patients with early-stage localized disease and regarding adjuvant or salvage radiation in those with later-stage or recurrent disease. Herein, we analyze LFS cohort data alongside laboratory-based clinical genetic testing data in PrCa cohorts, and we demonstrate that gTP53 is associated with the development of aggressive PrCa.
2. Patients and methods
2.1. LFS and PrCa cohorts
The LFS cohort (n = 163 males from 132 families) was created from four datasets of families with a diagnosis of LFS (Supplementary Table 1 and Supplementary material) from Dana Farber Cancer Institute, Huntsman Cancer Institute, Memorial Sloan Kettering Cancer Center, and the University of Pennsylvania. Eligibility criteria for index cases from LFS families were as follows: (1) male sex, (2) age at last follow-up ≥18 yr, and (3) genetic testing confirming heterozygous gTP53 or obligate carrier status within a family. Dates of genetic testing, PrCa diagnosis, or last follow-up and/or death were collected for all LFS men (Supplementary Table 1). The PrCa cohort (n = 6850 individuals) was created from four large series of PrCa patients who had undergone tumor or tumor/germline sequencing (Supplementary material). Pedigrees of patients in both cohorts were collected and examined to ensure that PrCa cases were not double counted due to enrollment in LFS registries or sequencing at more than one institution (Supplementary Fig. 1). Clinical data for both cohorts were collected in accordance with the Declaration of Helsinki guidelines and following approval from the respective institutional review boards at University of Washington, Tulane University, University of Pennsylvania, University of Utah, Memorial Sloan Kettering Cancer Institute, and Dana Farber Cancer Institute.
2.2. TP53 variant review and exclusion of somatic interference
All variants in the LFS and PrCa cohorts were referenced with ClinVar [16] and classified based on TP53-specific American College of Medical Genetics (ACMG) guidelines (Supplementary material) [17]. Subclassification of likely pathogenic or pathogenic variants as full penetrance versus attenuated variants was based on published functional data. Patients in the LFS cohorts were confirmed to have germline variants not due to somatic interference by clonal hematopoiesis of indeterminate potential or other abnormal clonal expansion [18] by testing of ancillary tissues and/or by variant segregation in families. In the PrCa cohorts, somatic interference [18] was excluded by: first, restricting to pathogenic and likely pathogenic variants with >35% variant fraction in blood; second, tumor sequencing when available to confirm presence in the cancer and in the matched normal tissue; and finally, a comparison of variant allele fractions with other observed somatic and germline variants.
2.3. Tumor analyses
Tumor next-generation sequencing data (UW-OncoPlex and MSK-IMPACT) were available for a subset of gTP53 carriers [19,20]. Tumor data were analyzed by an expert molecular pathologist (C.C.P.) for evidence of somatic TP53 second allele inactivation, including assessment of loss of heterozygosity (L.O.H.).
2.4. Statistical analysis
To calculate PrCa risk and incidence rates, the full LFS cohort was restricted to patients without a prior PrCa diagnosis at genetic testing to avoid selection and immortality biases. Risk of PrCa diagnosis relative to the general population was then estimated using a standardized incidence ratio (SIR) comparing the observed numbers with the expected numbers of PrCa diagnoses, with the latter calculated by applying PrCa incidence rates in the population to specific ages and calendar years of follow-up. Follow-up was summarized using reverse Kaplan-Meier estimation [21], which begins at the genetic test, treats death or last follow-up without PrCa diagnosis as events, and censors PrCa diagnoses. This approach gives a robust summary of follow-up for all patients, which is used to calculate the expected number of PrCa diagnoses. The PrCa incidence rates were those observed in the Surveillance, Epidemiology, and End Results (SEER) program from 1997 to 2017 by calendar year and 5-yr age group. Incidence rates over the period of 2018–2020 were assumed to be equal to the rates observed in 2017. Significance of the SIR was evaluated using a two-sided exact Poisson test. Overall and age-specific PrCa incidence rates in the restricted cohort were calculated relative to observed person-years of follow-up, and corresponding rates in SEER were calculated relative to population totals. In the PrCa cohort, the relative risk of carrying gTP53 was compared with frequencies in the gnomAD database using a Fisher’s exact test after excluding individuals with cancer (gnomAD v2.1.1 noncancer, 134 187 samples; Supplementary material).
3. Results
3.1. Incidence of PrCa in LFS
Among a cohort of LFS individuals created from datasets at four academic institutions, we identified 163 males aged ≥18 yr with a confirmed gTP53 mutation or obligate carrier status from 132 families (Supplementary Tables 1 and 2, and Supplementary Fig. 1). We identified PrCa in 31 of 163 (19%) adult males in this cohort, including 26 of 54 males (48%) aged ≥50 yr (Table 1). The frequency of PrCa within age deciles was similar in all four datasets (Supplementary Table 3). In a restricted analysis of 117 men who did not have a PrCa diagnosis at the time of genetic testing (Supplementary Table 1), six were diagnosed with PrCa over a median (interquartile range [IQR]) of 3.0 (1.3–7.2) yr of follow-up. The risk of a PrCa diagnosis in this subgroup of LFS men was 25 times that in the general population (95% confidence interval [CI] 9.2–55; p < 0.0001), with incidence rates significantly elevated at all ages (Fig. 1, Table 2, and Supplementary Fig. 2).
Table 1 –
Age distribution of men with prostate cancer (PrCa) in Li-Fraumeni syndrome (LFS) cohorts
| Age group | Combined LFS/LFL cohorts | ||
|---|---|---|---|
|
|
|||
| n | PrCa | % | |
| 18–29 | 32 | 0 | 0 |
| 30–39 | 35 | 1 | 2.9 |
| 40–49 | 42 | 4 | 10 |
| 50–59 | 32 | 15 | 47 |
| 60–69 | 15 | 7 | 47 |
| 70–79 | 7 | 4 | 57 |
| ≥80 | 0 | 0 | NA |
| Total (w/ages) | 163 | 31 | 19 |
LFL = Li-Fraumeni–like syndrome; NA = not available.
Age based on cancer diagnosis age or current age if unaffected.
Fig. 1 –
Summary of results (graphical abstract). LFS = Li-Fraumeni syndrome; PrCa = prostate cancer; SEER = Surveillance, Epidemiology, and End Results.
Table 2 –
Annual prostate cancer incidence rates per 1000 in the Li-Fraumeni syndrome (LFS) and in the Surveillance, Epidemiology, and End Results (SEER) registry
| Age (yr) | Restricted LFS cohort a | SEER | ||
|---|---|---|---|---|
|
|
||||
| Rate per 1000 | 95% CI | Rate per 1000 | 95% CI | |
| 0–49 | 3.9 | 0.48–14 | 0.050 | 0.049–0.050 |
| 50–59 | 40 | 4.8–144 | 2.03 | 2.02–2.04 |
| 60–74 | 90 | 11–326 | 7.10 | 7.08–7.13 |
| 0–74 | 10 | 3.8–23 | 1.07 | 1.07–1.07 |
CI = confidence interval.
Results are by post hoc age at risk and all ages combined.
LFS cohort restricted to men with no prostate cancer diagnosis at the time of their genetic test.
3.2. Prevalence of gTP53 in individuals with prostate cancer
To determine the prevalence of gTP53 in PrCa patients, we analyzed four large germline or tumor sequencing datasets comprising 6850 PrCa patients (Table 3 and Supplementary Table 2), excluding cases suggestive of somatic interference [18]. We identified gTP53 in 38 patients overall (0.55% prevalence), with prevalence rates ranging from 0.27% to 0.84% across the four independent cohorts (Table 3). The relative risk of having gTP53 was significantly elevated at 9.1 (95% CI 6.2–14, p < 0.0001) compared with the gnomAD noncancer population database (Fig. 1). Restricting both the sequencing cohorts and the gnomAD to TP53 variants classified as likely pathogenic or pathogenic in ClinVar, the relative risk remained statistically significantly elevated at 8.7 (95% CI 4.8–16, p < 0.0001; Table 3).
Table 3 –
Germline TP53 mutations in prostate cancer (PrCa) cohort compared with the general population
| Cohort | Total | LP/P + VUS/LP | ClinVar LP/P | ||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| n (%) | RR a (95% CI) | p value | n (%) | RR a (95% CI) | p value | ||
| RL | 3329 | 23 (0.69) | 11 (7.1–18) | <0.0001 | 10 (0.30) | 11 (5.5–23) | <0.0001 |
| AL-1 | 831 | 7 (0.84) | 14 (6.4–30) | <0.0001 | 5 (0.60) | 22 (8.7–57) | 0.002 |
| AL-2 | 2191 | 6 (0.27) | 4.5 (2.0–10) | 0.0004 | 1 (0.05) | NA | NA |
| TCGA | 499 | 2 (0.40) | 6.6 (1.6–27) | 0.009 | 0 (NA) | NA | NA |
| Combined | 6850 | 38 (0.55) | 9.1 (6.2–14) | <0.0001 | 16 (0.23) | 8.7 (4.8–16) | <0.0001 |
AL = Academic Laboratory; CI = confidence interval; RL = reference laboratory; LP/P = likely pathogenic and pathogenic mutations; RR = relative risk; TCGA = The Cancer Genome Atlas; VUS = variants of uncertain significance.
gnomAD v2.1.1 noncancer, 134 187 sample data for TP53 were downloaded and TP53 variants classified as LP/P mutations as per ClinVar or VUS/LP in ClinVar, classified as likely pathogenic in this study. The rate of LP/P and VUS/LP TP53 variants in gnomAD was 0.059%, and the rate of ClinVar LP/P TP53 variants in gnomAD was 0.026%.
3.3. Spectrum of gTP53 variants reveals hotspots associated with attenuated mutations
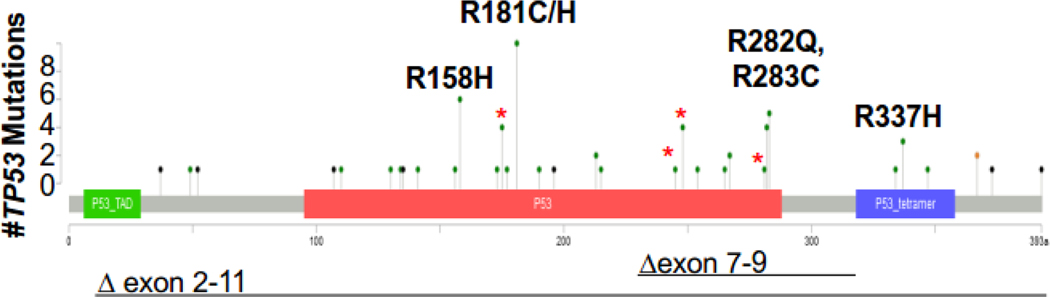
To study the spectrum of gTP53 variants and clinicopathological PrCa features, we combined gTP53 PrCa cases from the LFS and PrCa cohorts as no significant differences were observed for any variable between cohorts (Table 4). Of 67 individuals with PrCa and gTP53, 20 (30%) had classic LFS variants (Table 4). Thirty-two individuals (48%) had gTP53 variants with published evidence of being attenuated (or hypomorphic) mutations, and 15 (22%) had mutations suggestive of being attenuated (Table 4). Five codons associated with attenuated mutations (158, 181, 282, 283, and 337) accounted for 28 of the 67 cases (42%; Fig. 1 and 2, and Supplementary Table 2). Moreover, rare attenuated mutations at codon 181 (n = 10) were found in all series (except TCGA), and codons 158, 283, and 337 were enriched in four or more series (Supplementary Table 2). Specific TP53 variants, p.R158H, p.R181H, p.R282Q, p.R283C, and p.R337H, were each significantly enriched in PrCa cohorts versus gnomAD noncancer controls, with relative risks ranging from 6.1 (for p.R283C) to 92 (for p.R158H; Supplementary Table 4).
Table 4 –
Characteristics of 67 PrCa patients with confirmed gTP53
| LFS | PrCa | |||
|---|---|---|---|---|
|
|
||||
| n | % | n | % | |
| Site | n = 31 | n = 36 | ||
| LFS-1 | 10 | 32 | - | - |
| LFS-2 | 3 | 10 | - | - |
| LFS-3 | 6 | 19 | - | |
| LFS-4 | 12 | 39 | - | - |
| RL-1 | - | - | 7 | 19 |
| RL-2 | - | - | 23 | 64 |
| RL-3 | - | - | 4 | 11 |
| TCGA | - | - | 2 | 5.6 |
| Age of PrCa diagnosis | n = 31 | n = 27 | ||
| Median (IQR) | 56 (50–64) | 56 (51–60) | ||
| Mutation class | n = 31 | n = 36 | ||
| Classic | 12 | 39 | 8 | 22 |
| Reduced penetrance | 16 | 52 | 16 | 44 |
| Unknown | 3 | 10 | 12 | 33 |
| Tumor LOH | n = 6 | n = 9 | ||
| LOH present | 6 | 100 | 4 | 44 |
| Personal cancer history | n = 31 | n = 12 | ||
| None | 11 | 35 | 5 | 42 |
| Sarcoma | 8 | 26 | 1 | 8.3 |
| Adrenal cortical tumor | 0 | - | 0 | - |
| Brain | 2 | 6.5 | 0 | - |
| Leukemia | 0 | - | 0 | - |
| ≥1 LFS-core cancer a | 10 | 32 | 1 | 8.3 |
| Cancer timing for MP | n = 19 | n = 4 | ||
| Other cancer after PrCa | 7 | 37 | 3 | 75 |
| Other cancer before PrCa | 7 | 37 | 1 | 25 |
| Both before and after PrCa | 5 | 26 | 0 | - |
| Cancer family history | n = 31 | n = 13 | ||
| None | 1 | 3.2 | 1 | 7.7 |
| Sarcoma | 9 | 29 | 2 | 15 |
| Adrenal cortical tumor | 3 | 10 | 1 | 7.7 |
| Brain | 8 | 26 | 5 | 38 |
| Leukemia | 3 | 10 | 2 | 15 |
| Breast age <31 | 4 | 13 | 0 | - |
| Any LFS core cancer a | 16 | 52 | 8 | 62 |
| Breast (any age) | 21 | 68 | 6 | 46 |
| Prostate | 9 | 29 | 4 | 31 |
| Clinical LFS criteria | n = 28 | n = 7 | ||
| Classic | 4 | 14 | 1 | 14 |
| Chompret | 13 | 46 | 4 | 57 |
| Birch and/or Eeles | 19 | 68 | 4 | 57 |
| No criteria met | 8 | 29 | 3 | 43 |
| PrCa Gleason score | n = 20 | n = 14 | ||
| GS 6–7 | 12 | 60 | 7 | 50 |
| GS 8–10 | 8 | 40 | 7 | 50 |
| PSA at diagnosis | n = 14 | n = 9 | ||
| Median (IQR) | 6 (4–12) | 6 (3–23) | ||
| >20 | 3 | 21 | 3 | 33 |
| PrCa diagnosis | n = 11 | n = 7 | ||
| Screening | 8 | 73 | 4 | 57 |
| Incidental | 1 | 9.1 | 1 | 14 |
| Symptoms | 2 | 18 | 2 | 29 |
| Stage at diagnosis | n = 21 | n = 13 | ||
| Localized (Nx or N0) | 17 | 81 | 7 | 54 |
| N1 or de novo M1 | 4 | 19 | 6 | 46 |
ACT = adrenal cortical tumor; GS = Gleason score; IQR = interquartile range; LFS = Li-Fraumeni syndrome; LOH = loss of heterozygosity; MP = multiple primary; PrCa = prostate cancer; PSA = prostate-specific antigen; RL = reference laboratory; TCGA = The Cancer Genome Atlas.
LFS core cancers: ACT, brain, leukemia, sarcoma, and breast age <31 yr.
Fig. 2 –
Distribution of TP53 pathogenic and likely pathogenic variants. Lollipop plot depicts 67 germline P/LP variants in patients with prostate cancer. Green color represents missense variants, black represents truncating variants (frameshift, nonsense, and splice site), and black lines represent large deletion variants. Hotspots for prostate cancer were observed at sites of reduced penetrance variants at codons 158, 181, 282, 283, and 337. Classic LFS hotspot mutations (R175H, G245S, R248Q, and R282W) are indicated with red asterisks. LFS = Li-Fraumeni syndrome; P/LP = pathogenic and likely pathogenic.
3.4. Tumor analysis in gTP53 carriers supports role in tumorigenesis
As a biallelic loss suggests that the gTP53 variant contributed to cancer formation, we analyzed available prostate tumors from gTP53 carriers who had adequate tumor for evaluation. Ten of 15 available tumors (67%), including nine with likely or known attenuated mutations, had evidence of somatic inactivation of the second TP53 allele (Table 4 and Supplementary Table 2). The tumors without evidence of somatic inactivation had either p.R282Q or p.R283C alleles.
3.5. Clinicopathological features of gTP53 carriers with PrCa
The median age of PrCa diagnosis in males with gTP53 in this study was 56 (IQR: 51–60) yr (Table 4). PrCa incidence was increased compared with the SEER data in each group analyzed (Table 2 and Supplementary Fig. 2). Of individuals with gTP53 PrCa in the combined cohorts, 26% had a personal diagnosis and 55% had a family history of an LFS core cancer. Only 26% of the families in the combined cohort did not meet any LFS diagnostic criteria. Nearly all individuals had a family history of cancer, and 30% of patients had a family history of PrCa. A family history of breast cancer was reported in 61% of the combined cohort. Of men with LFS, PrCa was the only or the most recent cancer diagnosis in 58% (18 of 31).
Of PrCa cases, 27% were diagnosed as locally advanced (N1) or de novo metastatic disease. Similarly, 41% of gTP53 individuals had high-grade disease (Gleason ≥8; Fig. 1 and Table 4). Prostate-specific antigen (PSA) at diagnosis ranged from 1.1 to 171 ng/dl, and 68% of individuals were diagnosed by screening. Three individuals were diagnosed due to symptoms of advanced Gleason 9–10 PrCa at ages 50, 51, and 63 yr (Supplementary Table 2).
4. Discussion
We estimate the risk of PrCa diagnosis in individuals with LFS, and the frequency of rare germline pathogenic and likely pathogenic TP53 variants in a PrCa cohort. We found that LFS men have a 25-fold increased risk of PrCa compared with the general population. Further, we found that 0.55% (range 0.27–0.84%) of individuals with PrCa in a large sequencing cohort have gTP53. Tumor sequencing detected second allele inactivation in a majority of available tumor cases, supporting the biological relevance of the gTP53 variants to tumorigenesis. Collectively, these data demonstrate for the first time that gTP53 variants contribute to PrCa risk.
There is a paucity of data on PrCa risk in individuals with gTP53 [8,10]. While an analysis of LFS males in IARC [8] suggested that PrCa was enriched among cancers diagnosed in late adulthood, comparable average ages of cancer onset in LFS patients and SEER population suggested that the contribution of gTP53 variants was minimal. More recently, Shin et al [6] reported that the incidence of non–LFS-related tumors was higher in male gTP53 carriers between ages 35 and 65 yr, but PrCa was not analyzed individually. Our findings may differ from prior literature because a larger cohort of older LFS males, including families with attenuated LFS, was included.
Over half of the gTP53 variants we reported are considered attenuated or hypomorphic variants not typically associated with classic LFS. Individuals with attenuated gTP53 likely still have a multicancer predisposition syndrome of clinical significance [22,23]. We hypothesize that these individuals are more likely to live into adulthood and be at risk for developing prostate and other adult cancers, compared with those with full-penetrance gTP53 variants who typically have a severe early-onset cancer phenotype with high mortality rates at young ages.
In the PrCa sequencing cohorts, we found the relative risk of carrying gTP53 variants to be comparable with that of BRCA2 (relative risk 4.7–8.6) [24,25], a gene for which PrCa screening recommendations are modified [26,27]. This argues for consideration of PrCa screening in LFS guidelines. Moreover, since gTP53 carriers may be at an increased risk for radiation-induced sarcomas [5,14], our data may have treatment implications for gTP53 individuals with PrCa, for example, consideration of MRI versus computed tomography screening and avoidance of therapeutic radiation. The high rate of aggressive disease in our series (30% locally advanced or de novo metastatic) is consistent with prior observations that somatic TP53 mutations are associated with advanced PrCa, and supports the consideration of earlier and/or more frequent screening with PSA and attention to prostate pathology on full-body MRI screening to identify disease at a stage amenable to surgical resection in male gTP53 carriers from LFS families. For example, one LFS patient in this series had a glioblastoma at age 24 yr, developed symptomatic hematospermia at age 50 yr, and was diagnosed with Gleason score 9 de novo metastatic PrCa, a situation that might have been avoided with screening. In addition, the high rate of other LFS core cancers (61%) and breast cancer at any age (61%) in the family history of the cohort highlights the potential value of cascade familial testing to reduce cancer risk in relatives of individuals with PrCa identified to have gTP53.
Our study has limitations—the patients represented in all series were ascertained by either clinical testing of PrCa patients or LFS status, and presumably reflect enrichment of family or personal history and younger age. Ascertainment bias is particularly important to consider for the referral laboratory series, which also had very limited clinical data. Part of the increased risk of PrCa diagnosis in the LFS cohort may be due to more frequent testing and biopsy in these men; however, it seems unlikely that this bias could explain the 25-fold increased risk relative to the general US population. In addition, the LFS cohort size is small. However, LFS is an extremely rare cancer syndrome, afflicting approximately one in 10 000–30 000 individuals [28]; therefore, centralized efforts, such as through the LiFE consortium [29], will be required to confirm the risk estimates and clinical correlations presented. It is possible that TP53-mutant clonal hematopoiesis of indeterminate potential could contaminate our series [18]; however, paired tumor/normal testing in sequencing cohorts was analyzed, variant allele fraction cutoffs were employed, and LFS males had ancillary tissue or familial testing, proving that the mutation was inherited. Although tumor sequencing results are supportive of mechanistic causality, additional studies would be needed to prove the role in tumorigenesis. Lastly, most individuals did not find out that they were a gTP53 carrier until after their PrCa diagnosis and could not be utilized in SIR calculations.
Despite these limitations, our complimentary analyses showing an increased incidence of PrCa in an LFS cohort, far higher prevalence of gTP53 variants in a PrCa cohort than in population controls, tumor data supporting a role in tumorigenesis, and biological plausibility of observing more PrCa in attenuated gTP53 are collectively compelling. Confirmation in larger numbers of PrCa patients with paired testing of blood and tumor will be helpful, as will better understanding of modifying factors in the context of gTP53 variants. Exact PrCa risk estimates will likely be specific to variant and population.
5. Conclusions
This study contributes to a greater understanding of TP53-associated cancer risk, demonstrating that adult cancers such as PrCa in LFS are understudied and merit further attention. Attenuated or hypomorphic gTP53 alleles provide a plausible hypothesis as to why some gTP53 patients develop late adulthood cancers that would not have been appreciated previously in individuals with more severe gTP53 variants that predispose to high cancer burden and earlier mortality. Screening guidelines for adults with attenuated LFS phenotypes are needed urgently. We suggest that current LFS screening guidelines be updated to consider annual PrCa screening in men with at least 10 yr of life expectancy. While gTP53 mutation rates are low in PrCa cohorts, the clinical importance of the finding for the patient and family is significant. We suggest evaluation of TP53 in germline genetic testing in PrCa patients, ideally by paired tumor-normal testing, to rule out somatic interference.
Supplementary Material
Acknowledgments:
We thank the UW Genetics and Solid Tumors Laboratory and NGS Analytics Lab for support with genetic testing and data analysis.
Financial disclosures: Colin C. Pritchard certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Kara N. Maxwell: no disclosures reported. Heather H. Cheng: research funding to institution from Clovis, Janssen, Sanofi, Medivation/Astellas, and Color Foundation; consultancy to AstraZeneca. Jacquelyn Powers, Roman Gulati, Elisa M. Ledet, Casey Morrison, Anh Le, and Ryan Hausler: no disclosures reported. Jill Stopfer: consultancy to AstraZeneca. Sophie Hyman, Wendy Kohlmann, Anne Naumer, Jennie Vagher, Samantha Greenberg, Lorraine Naylor, and Mercy Laurino: no disclosures reported. Eric Q. Konnick: travel and honoraria from Roche, Ventana, Medscape, and Clinical Care Options (CCO). Brian H. Shirts and Saud H. Al-Dubayan: no disclosures reported. Eliezer M. Van Allen: consultant/advisory roles from Genome Medical, Invitae, Tango Therapeutics, Manifold Bio, Monte Rosa Therapeutics, and Janssen; research support from BMS and Novartis. Bastien Nguyen and Joseph Vijai: no disclosures reported. Wassim Abida: honoraria from CARET, Roche, Medscape, Aptitude Health, GlaxoSmithKline, Clovis Oncology, and ORIC Pharmaceuticals; consultant for Clovis Oncology, Janssen, MORE Health, ORIC Pharmaceuticals, and Daiichi Sankyo; research funding from AstraZeneca, Zenith Epigenetics, Clovis Oncology, GlaxoSmithKline, ORIC Pharmaceuticals, and Epizyme. Maria Carlo, Marianne Dubard-Gault, and Daniel J. Lee: no disclosures reported. Luke D. Maese: consultant for Jazz Pharmaceuticals. Diana Mandelker, Bruce Montgomery, and Michael J. Morris: no disclosures reported. Piper Nicolosi and Robert L. Nussbaum: employee and shareholder of Invitae Corporation. Lauren E. Schwartz, Zsofia Stadler, Judy E. Garber, and Kenneth Offit: no disclosures reported. Joshua D. Schiffman: employee and shareholder of PEEL Therapeutics. Peter S. Nelson: consultant for Astellas, Janssen, and Bristol Myers Squibb. Oliver Sartor: consultant for Advanced Accelerator Applications, Astellas, AstraZeneca, Bayer Blue Earth Diagnostics Inc., Bavarian, Nordic, Bristol, Myers, Squibb, Clarity, Pharmaceuticals, Clovis, Constellation, Dendreon, EMD, Serono, Fusion, Janssen, Myovant, Myriad, Noria, Therapeutics, Inc., Novartis, Noxopharm, Progenics, POINT, Biopharma, Pfizer, Sanofi, Tenebio, Telix, Theragnostics, Dendreon, Endocyte, Innocrin, Invitae, Merck, and SOTIO; research funding from Advanced Accelerator Applications, AstraZeneca, Bayer, Invitae, and Merck. Michael F. Walsh: no disclosures reported. Colin C. Pritchard: consultant for AstraZeneca.
Funding/Support and role of the sponsor: Research support for this study was generously provided by the National Institutes of Health (K08CA215312, Kara N. Maxwell; R50CA221836, Roman Gulati; R01CA242218, Judy E. Garber; P30CA016520, Abramson Cancer Center; P30CA008748, Memorial Sloan Kettering Cancer Center; P50CA097186, Pacific Northwest Prostate Cancer SPORE, Colin C. Pritchard, Heather H. Cheng, Peter S. Nelson; P30CA015704 FredHutch/UW Cancer Consortium), the Department of Defense (W81XWH-18–1-0756, PC170510, Colin C. Pritchard; W81XWH-18–1-0356, PC170503P2, PC200262P1, Colin C. Pritchard; W81XWH-17–2-0043, Heather H. Cheng), Prostate Cancer Foundation (20YOUN02, Kara N. Maxwell, Colin C. Pritchard, Heather H. Cheng, Peter S. Nelson), Burroughs Wellcome Foundation (1017184, Kara N. Maxwell), Basser Center for BRCA at the University of Pennsylvania (Kara N. Maxwell), Breast Cancer Research Foundation (Judy E. Garber, Kenneth Offit), Brotman Baty Institute for Precision Medicine (Brian H. Shirts, Colin C. Pritchard), UW/FHCRC Institute for Prostate Cancer Research (Colin C. Pritchard, Heather H. Cheng, Peter S. Nelson), Niehaus Center for Inherited Cancer Genomics (Michael F. Walsh), Marie-Josée and Henry R. Kravis Center for Molecular Oncology (Michael F. Walsh), Elephant p53 (EP53) Program and its generous funding provided to Huntsman Cancer Institute by the State of Utah (Joshua D. Schiffman), 5 For The Fight (Joshua D. Schiffman), Soccer for Hope Foundation (Joshua D. Schiffman), and Li-Fraumeni Syndrome Association (Joshua D. Schiffman).
APPENDIX
K.N. Maxwell, H.H. Cheng, J. Powers, R. Gulati, E.M. Ledet, C. Morrison, A. Le, R. Hausler, J. Stopfer, S. Hyman, W. Kohlmann, A. Naumer, J. Vagher, S. Greenberg, L. Naylor, M. Laurino, E.Q. Konnick, B.H. Shirts, S.H. Al-Dubayan, E.M. Van Allen, B. Nguyen, J. Vijai, W. Abida, M. Carlo, M. Dubard-Gault, D.J. Lee, L.D. Maese, D. Mandelker, B. Montgomery, M.J. Morris, P. Nicolosi, R.L. Nussbaum, L.E. Schwartz, Z. Stadler, J.E. Garber, K. Offit, J.D. Schiffman, P.S. Nelson, O. Sartor, M.F. Walsh, C.C. Pritchard
This study demonstrates that germline mutations in the TP53 gene are likely to predispose men to prostate cancer. Current Li-Fraumeni syndrome screening guidelines should be updated to consider annual prostate cancer screening, and TP53 should be considered in germline prostate cancer susceptibility testing.
Footnotes
Study concept and design: Maxwell, Cheng, Powers, Gulati, Walsh, Pritchard.
Acquisition of data: Maxwell, Cheng, Powers, Ledet, Morrison, Le, Hausler, Stopfer, Hyman, Kohlmann, Naumer, Vagher, Greenberg, Naylor, Laurino, Al-Dubayan, Van Allen, Abida, Carlo, Dubard-Gault, Lee, Maese, Mandelker, Montgomery, Morris, Nicolosi, Nussbaum, Schwartz, Stadler, Garber, Offit, Schiffman, Nelson, Sartor, Walsh, Pritchard.
Analysis and interpretation of data: Maxwell, Cheng, Powers, Gulati, Hausler, Konnick, Shirts, Al-Dubayan, Van Allen, Nguyen, Vijai, Garber, Offit, Walsh, Pritchard.
Drafting of the manuscript: Maxwell, Cheng, Powers, Gulati, Walsh, Pritchard.
Critical revision of the manuscript for important intellectual content: Maxwell, Cheng, Powers, Gulati, Ledet, Morrison, Le, Hausler, Stopfer, Hyman, Kohlmann, Naumer, Vagher, Greenberg, Naylor, Laurino, Konnick, Shirts, Al-Dubayan, Van Allen, Nguyen, Vijai, Abida, Carlo, Dubard-Gault, Lee, Maese, Mandelker, Montgomery, Morris, Nicolosi, Nussbaum, Schwartz, Stadler, Garber, Offit, Schiffman, Nelson, Sartor, Walsh, Pritchard.
Statistical analysis: Maxwell, Cheng, Powers, Gulati, Pritchard.
Obtaining funding: Maxwell, Cheng, Gulati, Garber, Offit, Schiffman, Nelson, Walsh, Pritchard.
Administrative, technical, or material support: None.
Supervision: Garber, Offit, Schiffman, Nelson, Sartor, Walsh, Pritchard.
Other: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Eeles R, Goh C, Castro E, et al. The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol 2014;11:18–31. [DOI] [PubMed] [Google Scholar]
- [2].Nicolosi P, Ledet E, Yang S, et al. Prevalence of germline variants in prostate cancer and implications for current genetic testing guidelines. JAMA Oncol 2019;5:523–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 2016;375:443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Malkin D, Li FP, Strong LC, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 1990;250:1233–8. [DOI] [PubMed] [Google Scholar]
- [5].Bougeard G, Renaux-Petel M, Flaman JM, et al. Revisiting Li-Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol 2015;33:2345–52. [DOI] [PubMed] [Google Scholar]
- [6].Shin SJ, Dodd-Eaton EB, Peng G, et al. Penetrance of different cancer types in families with Li-Fraumeni syndrome: a validation study using multicenter cohorts. Cancer Res 2020;80:354–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mai PL, Best AF, Peters JA, et al. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer 2016;122:3673–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Amadou A, Waddington Achatz MI, Hainaut P. Revisiting tumor patterns and penetrance in germline TP53 mutation carriers: temporal phases of Li-Fraumeni syndrome. Curr Opin Oncol 2018;30:23–9. [DOI] [PubMed] [Google Scholar]
- [9].Rana HQ, Gelman R, LaDuca H, et al. Differences in TP53 mutation carrier phenotypes emerge from panel-based testing. J Natl Cancer Inst 2018;110:863–70. [DOI] [PubMed] [Google Scholar]
- [10].Spees CK, Kelleher KJ, Abaza R, Clinton SK. Prostate cancer and Li-Fraumeni syndrome: implications for screening and therapy. Urol Case Rep 2015;3:21–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Siegel DA, O’Neil ME, Richards TB, Dowling NF, Weir HK. Prostate cancer incidence and survival, by stage and race/ethnicity—United States, 2001–2017. MMWR Morb Mortal Wkly Rep 2020;69:1473–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kratz CP, Achatz MI, Brugieres L, et al. Cancer screening recommendations for individuals with Li-Fraumeni syndrome. Clin Cancer Res 2017;23:e38–45. [DOI] [PubMed] [Google Scholar]
- [13].Frebourg T, Bajalica Lagercrantz S, Oliveira C, Magenheim R, Evans DG, European Reference Network GENTURIS. Guidelines for the Li-Fraumeni and heritable TP53-related cancer syndromes. Eur J Hum Genet 2020;28:1379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Heymann S, Delaloge S, Rahal A, et al. Radio-induced malignancies after breast cancer postoperative radiotherapy in patients with Li-Fraumeni syndrome. Radiat Oncol 2010;5:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hendrickson PG, Luo Y, Kohlmann W, et al. Radiation therapy and secondary malignancy in Li-Fraumeni syndrome: a hereditary cancer registry study. Cancer Med 2020;9:7954–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fortuno C, Lee K, Olivier M, et al. Specifications of the ACMG/AMP variant interpretation guidelines for germline TP53 variants. Hum Mutat 2021;42:223–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Weitzel JN, Caho EC, Nehoray B, et al. Somatic TP53 variants frequently confound germline testing results. Genet Med 2018;20:809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pritchard CC, Salipante SJ, Koehler K, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn 2014;16:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials 1996;17:343–6. [DOI] [PubMed] [Google Scholar]
- [22].Powers J, Pinto EM, Barnoud T, et al. A rare TP53 mutation predominant in Ashkenazi Jews confers risk of multiple cancers. Cancer Res 2020;80:3732–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mastellaro MJ, Seidinger AL, Kang G, et al. Contribution of the TP53 R337H mutation to the cancer burden in southern Brazil: insights from the study of 55 families of children with adrenocortical tumors. Cancer 2017;123:3150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst 1999;91:1310–6. [DOI] [PubMed] [Google Scholar]
- [25].Lecarpentier J, Silvestri V, Kuchenbaecker KB, et al. Prediction of breast and prostate cancer risks in male BRCA1 and BRCA2 mutation carriers using polygenic risk scores. J Clin Oncol 2017;35:2240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].NCCN. NCCN guidelines version 1.2019. Prostate cancer early detection. [Google Scholar]
- [27].Giri VN, Knudsen KE, Kelly WK, et al. Implementation of germline testing for prostate cancer: Philadelphia Prostate Cancer Consensus Conference 2019. J Clin Oncol 2020;38:2798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].MacFarland SP, Zelley K, Long JM, et al. Earlier colorectal cancer screening may be necessary in patients with Li-Fraumeni syndrome. Gastroenterology 2019;156:273–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mai PL, Sand SR, Saha N, et al. Li-Fraumeni Exploration Consortium Data Coordinating Center: building an interactive web-based resource for collaborative international cancer epidemiology research for a rare condition. Cancer Epidemiol Biomarkers Prev 2020;29:927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.