Abstract
Extracellular vesicles (EVs) have emerged as a promising carrier system for the delivery of therapeutic payloads in multiple disease models, including cancer. However, effective targeting of EVs to cancerous tissue remains a challenge. Here, we show that non-viral transfection of myeloid-derived suppressor cells (MDSCs) can be leveraged to drive targeted release of engineered EVs that can modulate transfer and overexpression of therapeutic anti-cancer genes in tumor cells and tissue. MDSCs are immature immune cells that exhibit enhanced tropism toward tumor tissue and play a role in modulating tumor progression. Current MDSC research has been mostly focused on mitigating immunosuppression in the tumor niche; however, the tumor homing abilities of these cells present untapped potential to deliver EV therapeutics directly to cancerous tissue. In vivo and ex vivo studies with murine models of breast cancer show that non-viral transfection of MDSCs does not hinder their ability to home to cancerous tissue. Moreover, transfected MDSCs can release engineered EVs and mediate anti-tumoral responses via paracrine signaling, including decreased invasion/metastatic activity and increased apoptosis/necrosis. Altogether, these findings indicate that MDSCs could be a powerful tool for the deployment of EV-based therapeutics to tumor tissue.
Keywords: extracellular vesicles, myeloid-derived suppressor cells, tumor tropism, solid tumors, non-viral gene and cell therapies
INTRODUCTION
Extracellular vesicles (EVs) have emerged as a promising carrier system for the delivery of therapeutic payloads for a wide variety of conditions1–10. EVs are cell-derived nanocarriers that mediate the transfer of bioactive cargo (e.g., nucleic acids, proteins) between cells under healthy and pathological conditions11–13. Compared to most nanocarrier systems for therapeutic payload delivery, EVs show improved biocompatibility, reduced immunogenicity, enhanced physicochemical stability in biofluids, and an innate ability to pass through biological barriers1,2,4. As such, a substantial amount of research is currently being devoted to engineering therapeutic EVs for challenging diseases like cancer. EV-based therapies, for example, have shown promise for the treatment of numerous types of cancer, including prostate cancer14, glioblastoma multiforme15, and Lewis lung carcinoma16,17, among others. Additional studies have also shown that EVs naturally derived from tumor cells or certain types of immune cells can drive anti-tumoral activity and could potentially be used as cancer vaccines1.
Strategies to engineer EVs generally come in two forms. The first involves loading the EVs with therapeutic cargo, which could include nucleic acids, proteins, chemotherapeutic drugs, and different types of nanomaterials, among others. The second involves functionalizing the surfaces of the EVs with different types of biomolecules to enhance targeting to specific cell/tissue types, or to reduce clearance and increase circulation time. Surface functionalization is typically done with peptides, antibodies, or aptamers. Despite the promise, however, EV-based therapeutics still face a number of challenges, including difficulties in large-scale EV manufacturing and isolation/purification, and unpredictable or poor cell/tissue targeting efficiencies1. Thus, new approaches are needed to facilitate targeted delivery of EV-based therapeutics to diseased tissues with more scalable manufacturing and isolation/purification procedures.
Here, we explored the use of cell therapies to achieve targeted deployment of EV-based therapies to cancerous tissue by leveraging the tumor-homing abilities of myeloid-derived suppressor cells (MDSCs)18,19. Tumor progression is driven by a complex interplay between different cellular compartments, including cancerous, stromal, and immune cells. Tumor-associated immune cells such as MDSCs have an innate ability to home preferentially to cancerous tissue, where they are known to exert immunosuppressive activity that favors tumor progression. This is achieved by protecting cancerous cells from the host’s immune system and/or exogenous immunotherapies (e.g., CAR-T, CAR-NK cell therapies)20–24. As such, a significant amount of research has gone into developing approaches to counteracting MDSC-driven immunosuppression in the tumor niche25. However, despite their innate tropism toward tumor tissue, there is currently a paucity of research on the use of MDSCs for targeted deployment of EV-based therapeutics to cancerous tissue.
To achieve targeted deployment of EV-based therapies to the tumor niche, we studied the use of MDSCs as in situ delivery vehicles of engineered EVs. MDSCs were engineered to express therapeutic cargo via non-viral electroporation of expression plasmids for Timp3 or Rarres2. These two genes were chosen as model therapeutic cargo because of their role in mediating anti-metastatic and anti-tumoral processes in cancerous tissue26–30. In vitro studies were conducted to evaluate the expression and loading extent and dynamics of Timp3 and Rarres2, both in the transfected MDSCs and in the EVs that were released into the media. We also studied the ability of MDSC-derived EVs to transfect and modulate gene expression and function (e.g., proliferation, invasion) in cancerous cells. Finally, in vivo studies were carried out in a mouse model of breast cancer to evaluate whether engineered MDSCs still exhibited tropism toward tumor tissue, as well as potential changes in gene and protein expression and cell function in the tumor niche. MDSC-driven EV-based therapies for cancer could potentially overcome a number of limitations of current approaches to EV therapies, such as increasing their tumor-targeting abilities without the need for surface functionalization of the EVs with tumor-targeting biomolecules (e.g., simplifying manufacturing), as well as bypassing the need for inefficient EV isolation and purification procedures.
RESULTS
Electroporation of expression plasmids for Timp3 and Rarres2 into MDSCs drives transcript overexpression and transfer into EVs:
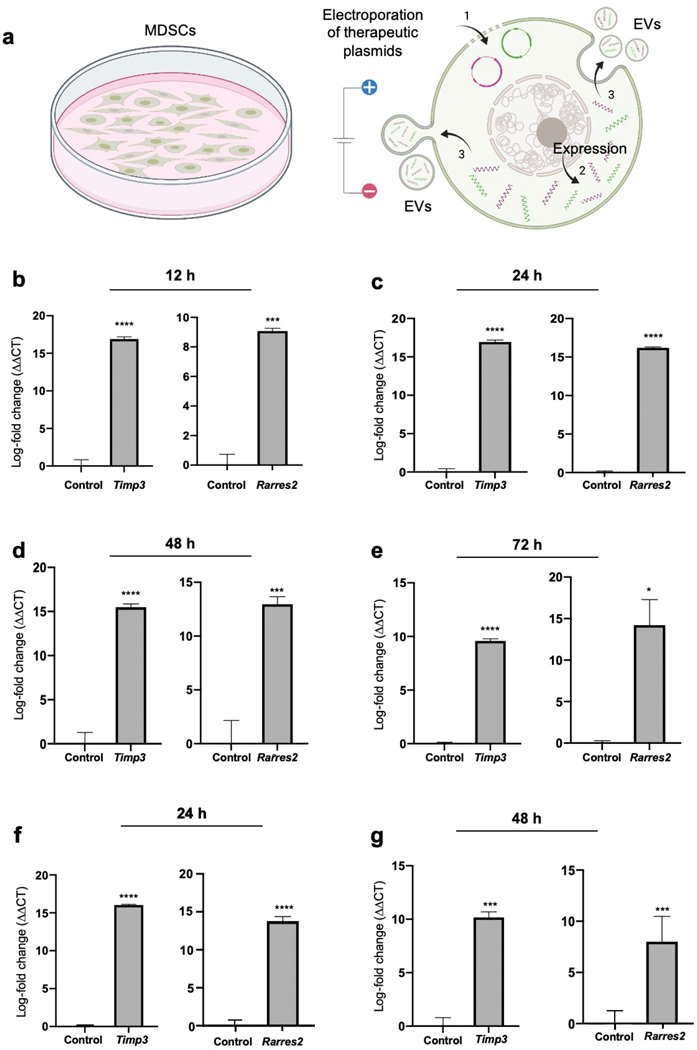
To evaluate if MDSCs can be transfected to drive the release of engineered EVs with desirable cargo (Fig. 1 a), cultures of the murine MDSC cell line, MSC218,19,31,32, were electroporated with expression plasmids for Timp3 and Rarres2, and gene expression was evaluated at 12 – 72 hours post-transfection via qRT-PCR. Sham/empty plasmids with the same backbone were used as controls. qRT-PCR analyses of the electroporated MDSCs showed significantly increased overexpression of Timp3 and Rarres2 for at least 12 – 72 hours post-transfection compared to controls (Fig. 1 b–e). EV isolation from the supernatant and qRT-PCR analysis at 24 – 48 hours post-transfection revealed that the EVs were markedly loaded with Timp3 or Rarres2 transcripts compared to EVs obtained from MDSC cultures that were electroporated with sham plasmids (Fig. 1 f, g). Average EV size was ~180 nm, while EV concentrations ranged from 5×109 – 1010 EVs/ml. To test whether MDSC-derived EVs can be loaded with multiple different transcripts, we co-transfected Timp3 and Rarres2 plasmids into MDSCs and evaluated gene expression and EV loading at 24 – 72 hours post-transfection. qRT-PCR measurements of the cells indicate that co-transfection of the two plasmids led to significantly sustained co-overexpression of Timp3 and Rarres2 in MDSCs for at least 72 hours (Supplementary Fig. 1). Analysis of the EVs released into the supernatant shows that the EVs were successfully co-loaded with Timp3 and Rarres2 transcripts (Supplementary Fig. 1), with levels comparable to those achieved with single-plasmid transfections relative to controls (Fig. 1 f, g). EV size and concentration ranged ~140 – 160 nm, and ~1010 EVs/ml, respectively. Collectively, these findings indicate that MDSCs can be readily transfected via non-viral methods, such as electroporation, and that MDSC transfection can be leveraged to drive the release of engineered EVs loaded with transcripts of the transfected cargo. Moreover, co-transfection experiments indicate that MDSC-derived EVs can also be simultaneously co-loaded with different types of transcripts for therapeutic applications.
Fig. 1. Non-viral transfection of MDSCs mediates the release of engineered EVs with desirable cargo.
(a) Schematic diagram illustrating the experimental design. (1) MDSCs were electroporated with expression plasmids for Timp3 or Rarres2. Electroporation with sham/empty plasmids served as control. (2) The plasmids are expressed within the MDSCs and (3) transcripts are packed and released within EVs. qRT-PCR analysis of the MDSC cultures at (b) 12, (c) 24, (d) 48, and (e) 72 hours post-electroporation reveals strong Timp3 or Rarres2 overexpression. Analysis of the EVs isolated from the supernatant at (f) 24 and (g) 48 hours post-electroporation indicates successful loading of the EVs with Timp3 or Rarres2. * p< 0.05 (n= 4), *** p< 0.01 (n=3), **** p< 0.0001 (n= 4).
MDSC-derived EVs are internalized by tumor cells and can modulate gene expression:
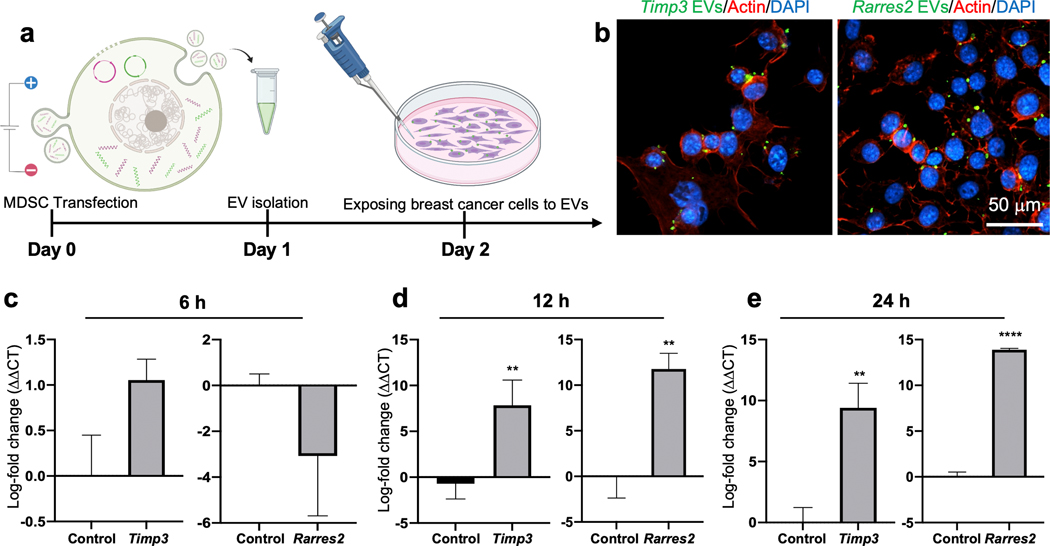
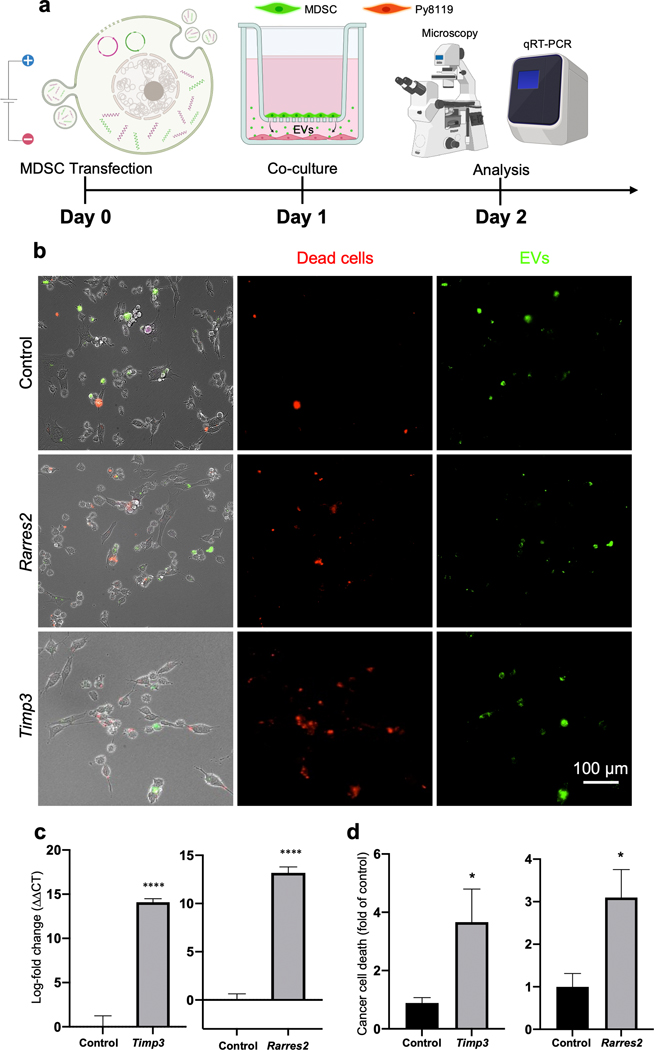
To evaluate if the EVs released by MDSCs following transfection can be internalized and modulate gene expression in tumor cells, we proceeded to electroporate MDSCs with Timp3 or Rarres2 plasmids, and isolated EVs at 24 hours post-transfection, correlating with peak expression of transcripts. MDSCs electroporated with sham plasmids served as control. The isolated EVs were then dyed with a lipophilic PKH probe, and cultures of Py8119 breast cancer cells were exposed to the labeled EVs (~107 EVs/ml) for 6 – 48 hours (Fig. 2 a). Fluorescence microscopy imaging of the Py8119 cells showed successful EV uptake (Fig. 2 b, Supplementary Fig. 2). qRT-PCR analyses of the Py8119 cells exposed to EVs derived from transfected MDSCs indicate that Timp3 and Rarres2 were significantly upregulated after 12 – 24 hours of exposure compared to Py8119 cells exposed to control EVs derived from sham-transfected MDSCs (Fig. 2 c–e). No significant differences were detected after 6 hours of exposure compared to controls, which is likely indicative of insufficient EV internalization and/or transcript upregulation. To verify if MDSCs mediate in situ transfer of engineered EVs and transcripts to Py8119 cells, we labeled the Timp3- and Rarres2-transfected MDSCs with a lipophilic PKH membrane dye used previously by us and others to trace EVs8, and proceeded to co-culture MDSCs with Py8119 cells using a transwell insert. To avoid any potential confounding factors introduced by direct cell-to-cell contact, the transfected MDSCs were plated in the apical chamber, while the Py8119 cells were plated in the basal chamber (Fig. 3 a). Fluorescently labeled EVs were thus expected to be released only by the MDSCs and translocate across the 400 nm pores of the insert membrane into the Py8119 compartment. Fluorescence microscopy imaging of the basal chamber revealed that EVs released by transfected MDSCs from the apical chamber (i.e., labeled green), successfully translocated into the basal chamber, where they were internalized by the Py8119 cells (Fig. 3 b, Supplementary Fig. 3). qRT-PCR analysis of the Py8119 cells showed clear overexpression of Timp3 and Rarres2 compared to Py8119 cells that were co-cultured with MDSCs transfected with sham plasmids (Fig. 3 c). Moreover, dead cell staining with propidium iodine in the Py8119 compartment suggests a cytotoxic effect of Timp3- and Rarres2-loaded EVs, which was absent for sham-loaded EVs. Altogether, these findings indicate that EVs released by transfected MDSCs have the ability to mediate in situ gene transfer and overexpression of therapeutic cargo in cancer cells in a paracrine manner, which could potentially be leveraged for the deployment of EV-based therapies in the tumor niche.
Fig. 2. MDSC-derived EVs can be internalized by cancer cells and modulate gene expression.
(a) Schematic diagram illustrating the experimental design. MDSCs were electroporated with expression plasmids for Timp3 or Rarres2. Electroporation with sham/empty plasmids served as control. MDSC-derived EVs were isolated and incubated with Py8119 mouse breast cancer cell cultures for 6 – 24 hours. (b) Fluorescence microscopy imaging of the Py8119 cells (labeled red and blue) revealed successful uptake of MDSC-derived EVs (labeled green). The images shown represent EV uptake at 24 h post-exposure. qRT-PCR analysis of the Py8119 cultures at (c) 6, (d) 12, and (d) 24 hours post-EV exposure reveals strong Timp3 or Rarres2 overexpression after 12 hours of exposure. ** p< 0.01 (n= 4), **** p< 0.0001 (n= 4).
Fig. 3. Transfected MDSCs can transfer engineered EVs to breast cancer cells, in situ, and mediate gene expression and cellular responses.
(a) Schematic diagram illustrating the experimental design. MDSCs were electroporated with expression plasmids for Timp3 or Rarres2. Electroporation with sham/empty plasmids served as control. Transfected MDSCs and Py8119 mouse breast cancer cells were co-cultured using a transwell system, with the MDSCs in the apical chamber and Py8119 cells in the basal chamber. The MDSCs were-prelabeled with a membrane dye to trace EV release and uptake. MDSC-derived EVs were thus expected to translocate across the membrane and interact with Py8119 cells in the basal chamber. (b) Fluorescence microscopy imaging of the Py8119 cultures revealed successful translocation and uptake of MDSC-derived EVs (green). Py8119 cells with compromised cell viability/membrane integrity were labeled red. (c) qRT-PCR analyses of the Py8119 cultures showed marked overexpression of Timp3 or Rarres2 after 24 hours. (d) Viability analyses suggest an oncolytic effect for MDC-derived EVs loaded with Timp3 or Rarres2 compared to sham MDSC-derived EVs. * p< 0.05 (n= 10), **** p< 0.0001 (n= 4).
Transfected MDSCs hinder tumor cell proliferation and invasion capabilities in situ:
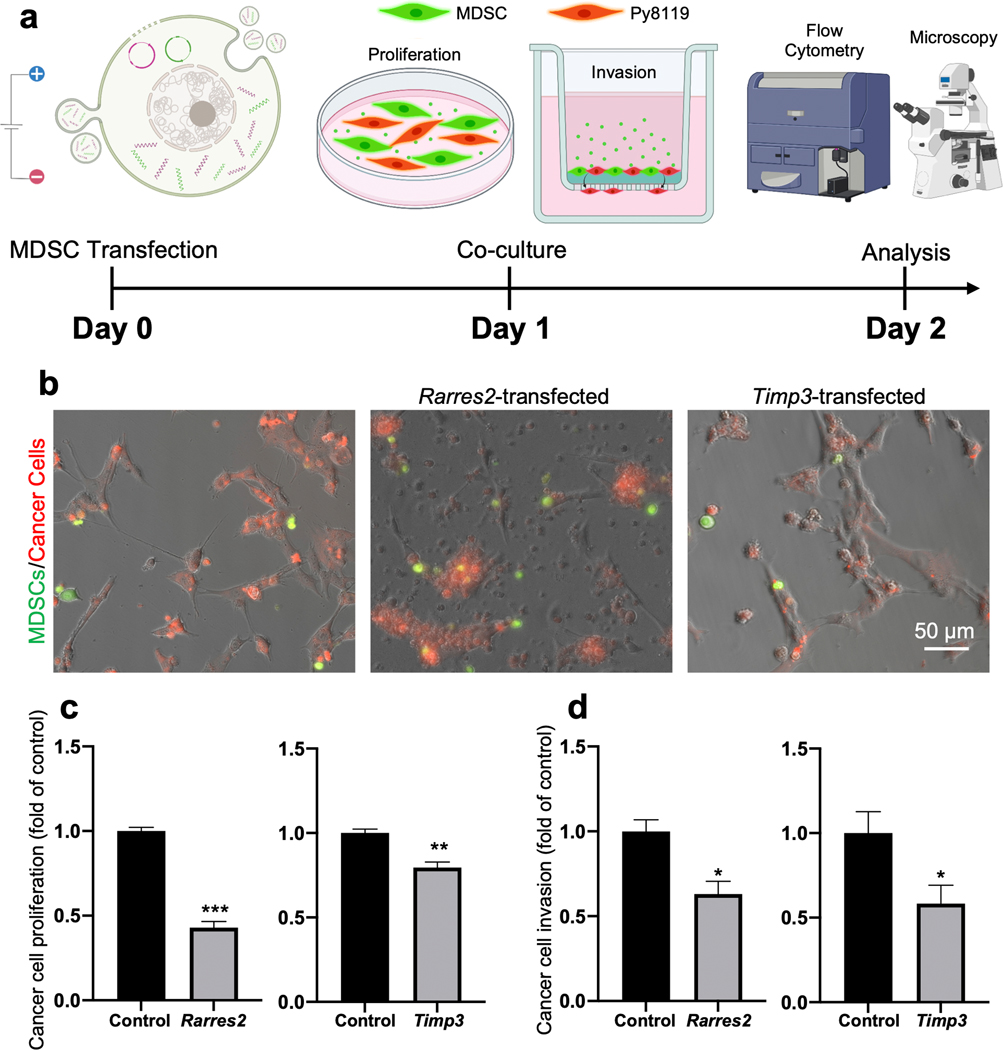
Once we established a paracrine role for MDSC-derived engineered EVs in the modulation of cancer cell responses (e.g., gene expression and viability, Fig. 3), we proceeded to evaluate if MDSCs transfected with Timp3 and Rarres2 plasmids could influence tumor cell proliferation and invasion, in situ. For this, Timp3- or Rarres2-transfected MDSCs and Py8119 cells were mixed and co-cultured in direct contact at a 1:1 ratio to emulate more closely the degree of cell-cell interactions within the tumor niche (Fig. 4 a)19,33–35. Co-cultures with MDSCs transfected with sham plasmids served as control. The cells were pre-labeled with different fluorescent dyes to be able to distinguish them during the analysis phase (i.e., Green: transfected MDSCs; Red: Py8119 breast cancer cells). For proliferation assays, Py8119 cell numbers were quantified by flow cytometry after 24 hours of co-culture. For invasion assays, the cells were plated on Matrigel-coated transwell inserts (Corning BioCoat Matrigel Invasion chambers), and the number of Py8119 cells that invaded through the insert over a period of 24 hours was visualized and quantified with fluorescence microscopy (Fig. 4 b). Flow cytometry analyses revealed a significant decrease in the number of Py8119 cells when co-cultured with Timp3- or Rarres2-transfected MDSCs compared to co-cultures with sham-transfected MDSCs, with a more pronounced oncolytic effect seen for Rarres2 compared to Timp3 (Fig. 4 c). Decreased cancer cell numbers could be potentially driven by EV-mediated cell death, as shown in Fig. 3 c. Moreover, invasion assays revealed that Py8119 cells co-cultured with Timp3- or Rarres2-transfected MDSCs had a tendency to show decreased invasion activity compared to controls (Fig. 4 d). Overall, these findings indicate that transfected MDSCs have the ability to modulate key cancer cell behaviors such as proliferation and invasion, in situ, possibly via the release of engineered EVs loaded with transcripts of the therapeutic cargo, as suggested by the results in Fig. 3.
Fig. 4. Transfected MDSCs can also modulate cancer cell proliferation and invasion, in situ.
(a) Schematic diagram illustrating the experimental design. MDSCs were electroporated with expression plasmids for Timp3 or Rarres2. Electroporation with sham/empty plasmids served as control. Transfected MDSCs and Py8119 mouse breast cancer cells were co-cultured in direct contact. Matrigel-coated transwell insets were used for cell invasion studies. The MDSCs were-prelabeled green, and the Py8119 cells were prelabeled red. (b) Fluorescence microscopy imaging of the MDSC/Py8119 co-cultures after 24 hours. (c) Flow cytometry analysis revealed reduced Py8119 cell numbers in co-cultured with Timp3- or Rarres2-transfected MDSCs. (d) Py8119 cell invasion analyses showed reduced Py8119 cell invasion capabilities in co-cultures containing Timp3- or Rarres2-transfected MDSCs. * p<0.05 (n= 4 ), ** p< 0.01 (n= 4), *** p< 0.005 (n= 3).
Transfected MDSCs retain tumor-homing capabilities and modulate gene and protein expression in the tumor niche in vivo:
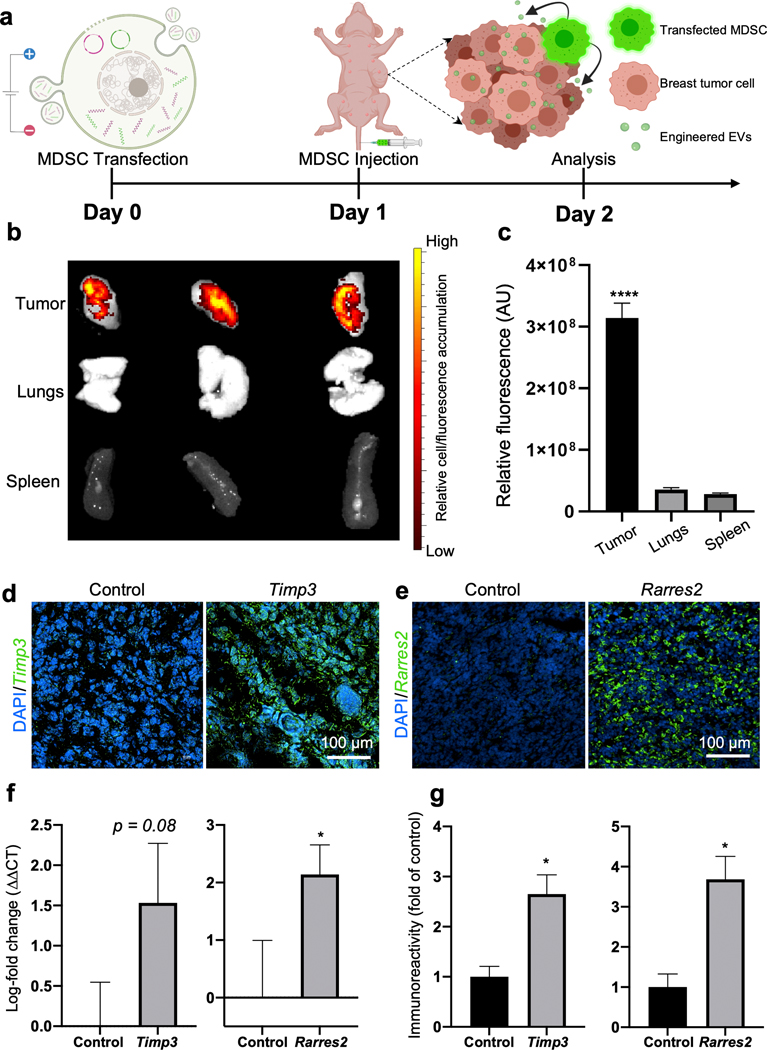
To evaluate whether electroporation with expression plasmids for therapeutic cargo impacted the tropism of MDSCs toward tumor tissue, we proceeded to inject Timp3- and Rarres2-transfected MDSCs into the tail vein of tumor-bearing mice (Fig. 5a). In vivo tumor homing of MDSCs was evaluated via IVIS imaging. For these experiments, we used an orthotopic xenograft model of nude/immunocompromised mice injected with human breast cancer cells (MDA-MB-231 cells) in the mammary gland. The tumors were allowed to reach a size of approximately 5 mm before the transfected MDSCs were injected. Immediately prior to injection, transfected MDSCs were fluorescently labeled with a cell tracker membrane dye to detect the location of the cells and trace the EVs released within the tumor niche. After 24 hours, the mice were sacrificed and IVIS imaging was used to evaluate the accumulation of fluorescence signal stemming from the cell tracker probe in the tumor. IVIS results indicated a strong accumulation of transfected MDSCs in the tumor niche compared to other organs (Fig. 5 b), suggesting that episomal expression of the therapeutic plasmid cargo did not significantly impact the ability of MDSCs to home to cancerous tissue. Ex vivo studies with microfluidic systems incorporating two different types of breast tumor organoids (i.e., aggressive/mesenchymal Py8119 and epithelial Py230)19, and non-tumoral cells (i.e., primary mouse embryonic fibroblasts or pMEFs), further showed that MDSCs transfected with therapeutic or sham cargo/plasmids appear to show similar levels of invasiveness and accumulation in the different organoids (Supplementary Fig. 4). Additional in vivo studies suggest little to no accumulation of MDSCs in clearance organs such as the liver (Supplementary Fig. 5). qRT-PCR analysis and immunostaining of the collected tumor tissue confirmed localized overexpression of Timp3 and Rarres2 at the mRNA and protein levels compared to controls (Fig. 5 d–g). Fluorescence imaging of the cell membrane tracker dye, which in addition to helping localize the injected MDSCs, can also be used to trace MDSC-derived EVs, appears to show MDSC-derived EV uptake by other cells within the tumor niche (Supplementary Fig. 6). Moreover, additional immunostaining revealed a decrease in proliferation activity based on reduced Ki67 immunoreactivity, as well as an increase in pro-apoptotic activity based on enhanced cleaved Caspase-3 immunoreactivity in the tumor niche compared to mice injected with control MDSCs (Supplementary Fig. 7). Altogether, these findings suggest that transfected MDSCs still show remarkable tropism toward tumor tissue, where they can mediate the deployment and transfer of therapeutic EVs into tumor-resident cells, and drive overexpression of therapeutic cargo, abrogating pro-tumoral activity.
Fig. 5. Transfected MDSCs retain tumor-homing abilities and drive anti-tumoral gene and protein expression.
(a) Schematic diagram illustrating the experimental design. MDSCs were electroporated with expression plasmids for Timp3 or Rarres2. Electroporation with sham/empty plasmids served as control. Transfected MDSCs were then injected into tumor-bearing mice via the tail vein, and accumulation and gene/protein expression in the tumor was evaluated after 24 hours. (b, c) IVIS imaging after 24 hours revealed strong accumulation of transfected MDSCs in the tumor niche compared to other organs. Transfected MDSCs were fluorescently pre-labeled red for these experiments. (d-g) Tumor tissue analyses indicate marked expression of the therapeutic cargo, Timp3 or Rarres2, both at the (d, e, g) protein and (f) transcript levels. * p< 0.05 (n= 5), **** p< 0.0001 (n= 6).
DISCUSSION
This study reports on a novel approach to deploying EV-based therapeutics into the tumor niche in a targeted manner via the use of MDSC-driven cell therapies. EVs have been shown to offer a number of advantages for therapeutic payload delivery for cancer compared to many other micro- or nano-carrier systems36–38. However, targeted delivery of EVs to cancerous tissue requires complex surface engineering processes that often yield inefficient and/or unpredictable targeting outcomes. Additional challenges to EV-based therapeutics for cancer include difficulties in identifying scalable and efficient EV biomanufacturing and isolation/purification procedures. Thus, there is still a need for novel platform technologies that enable targeted deployment of EV-based therapies to cancerous tissue. MDSCs are immature innate immune cells that are highly expanded in cancer patients20, and are known to exhibit a high degree of tropism toward tumor tissue, where they contribute to the loss of immune effector cell function. We have previously shown in ex vivo and in vivo studies that MDSCs exhibit high dissemination and tumor-tropic capabilities at the single-cell level, as well as contact-guided motility similar to tumor cells18,19,39–42. Thus, pharmacologically counteracting the infiltration of MDSCs into the tumor niche and halting their immunosuppressive activity has emerged as an attractive therapeutic strategy against cancer. However, efforts to effectively stem or reverse MDSC-driven immunosuppression in tumor tissue have been hampered by the lack of druggable targets43. Nevertheless, MDSC homing to tumor tissue could potentially be leveraged to deploy therapies to cancerous tissue in a more targeted manner. While myeloid cells have been recently studied as therapeutic carriers in cancer44,45, to the best of our knowledge, no study has investigated the use of MDSCs to drive engineered EV-based therapies in cancerous tissue to date.
While our results indicate that MDSC-based deployment of therapeutic EVs in breast cancer may be feasible, it is important to point out that, compared to direct delivery of EVs, using cell therapies as an EV delivery vehicle could potentially limit deployment to certain locations protected by biological barriers that are otherwise easily overcome by EVs (e.g., blood-brain barrier)46,47. Although this is clearly an important issue meriting further study, there is evidence that MDSCs can traffic to brain tissue in certain malignancies or in other neurodegenerative conditions48–54. Thus, the use of MDSCs as a platform technology for EV deployment may still be feasible for some applications where biological barriers are present.
Additional challenges potentially stemming from the use of cells to deploy EV therapies could include limited cell availability. However, accumulating evidence suggests that the circulating levels of MDSCs tend to be significantly elevated in cancer patients, and under other non-neoplastic conditions (e.g., stroke, Alzheimer’s disease)33,50,55–57. Thus, isolating and expanding MDSCs from circulation for subsequent use as a therapeutic agent, similar to the methods used for CAR-T or CAR-NK cell therapies, is likely a feasible strategy for the implementation of MDSC-driven EV therapies under multiple conditions (besides cancer). Moreover, additional studies have shown that peripheral blood mononuclear cells (PBMCs) can be differentiated toward MDSCs or MDSC-like cells32,58 in vitro, making PBMCs another potential source of MDSCs for therapeutic applications.
In this study, MDSC-derived EVs were engineered to contain transcripts of Timp3 or Rarress2. The Rarres2 gene encodes for a small protein that is functionally downregulated in various cancers, including adrenocortical carcinomas, melanoma, and breast cancer59, which contributes to dysregulation of the Wnt/β-catenin signaling pathway and tumor progression29,60. The Timp3 gene, on the other hand, falls into the family of tissue inhibitors of metalloproteinases. Timp3 is a well-known inhibitor of cancer cell function, especially invasion, in numerous cancer types, including breast cancer28,61,62. Our results clearly indicate that MDSC-based deployment of Timp3- or Rarres2-loaded EVs results in decreased viability and invasion capabilities in breast cancer cell cultures, as well as reduced proliferation and increased pro-apoptotic activity in vivo. MDSC-derived EVs, however, could conceivably be engineered to contain a wide variety of therapeutic cargo/transcripts besides Timp3 or Rarres2. For example, immunomodulatory transcripts such as IL-12 could potentially be loaded into MDSC-derived EVs to drive more effective immune responses against tumors45. Moreover, in addition to cancer, MDSCs are also known to infiltrate diseased tissue in other conditions, including stroke and Alzheimer’s disease. Therefore, the use of engineered MDSCs as EV delivery vehicles could conceivably go beyond cancer applications, and as such, different genes/transcripts would have to be explored depending on the therapeutic target.
Importantly, our results indicate that, despite potential phenotypic changes following transfection, engineered MDSCs maintain preferential tropism toward the tumor microenvironment, with little-to-no accumulation in off-target tissues such as the liver, which provides crucial evidence for the viability of this immunotherapy. MDSC migration to the tumor microenvironment is mediated by an array of cytokines and chemokines, primarily CCL2, but also other CXC-motif chemokines63. While our data suggest that these recruitment mechanisms are likely maintained in engineered MDSCs, it is possible that the degree of engineered MDSC tropism toward diseased tissue vs. off-target accumulation could be impacted by the method used to transfect the MDSCs (e.g., non-viral vs. viral approaches)64–72, and/or the type of cargo/transcript that is overexpressed in them. Thus, future studies should continue to evaluate how these parameters influence MDSC and MDSC-derived EV targeting to diseased tissues, as well as the potential consequences of off-target secretion of the EVs and corresponding therapeutic payloads in other tissues.
CONCLUSIONS
Our results indicate that a single non-viral transfection of MDSCs with expression plasmids for therapeutic cargo is sufficient to drive the production of engineered EVs loaded with transcripts of the transfected cargo. MDSC-derived EVs were shown to be effectively internalized by tumor cells and to modulate gene expression and tumor cell responses. Notably, when deployed systemically in circulation, transfected MDSCs still exhibited a remarkable ability to home to tumor tissue, where they were shown to mediate engineered EV transfer into the tumor niche and promote overexpression of therapeutic cargo. Using MDSCs to deliver therapeutic EVs to cancerous tissue has the potential to overcome some of the major limitations of current approaches to EV-based therapeutics for cancerous tissue, including bypassing the need for complex EV isolation and purification procedures, as well as the need for surface functionalization with cancer-targeting biomolecules, thus simplifying the manufacturing process. Altogether, our findings indicate that non-virally transfected MDSCs could potentially serve as a powerful platform technology for the deployment of EV-based therapeutics to cancerous tissue.
METHODS
Cell culture:
The mouse mammary cancer cell lines used in this study were derived from MMTV-PyMT transgene-induced mammary tumors in C57BL/6 mice (ATCC, Manassas, VA). The two cell lines used, Py8119 and Py230, were derived from the same tumor model but have distinct mesenchymal (Py8119) or epithelial-like (Py230) features42. The cells were kept in culture with F-12/Kaighn’s medium supplemented with 5% fetal bovine serum (FBS) and 0.1% MITO+ Serum Extender (Corning). The murine MDSC cell line, MSC2 (gift from Gregoire Mignot to Dr. William E. Carson), was cultured in RPMI medium (Gibco, Dublin,IE) containing 10% FBS, 1% sodium pyruvate (Gibco) and 1% antibiotic-antimycotic (Gibco). All the cells were maintained at 37 °C, 5% CO2 and 95% humidity.
Electroporation of MDSCs:
MSC2 cells were transfected via bulk electroporation with either sham or treatment (Timp3, Rarres2) expression plasmids using a Neon Transfection System (ThermoFisher). Transfected MDSCs were cultured in RPMI medium (Gibco, Dublin, IE) containing 10% exosome-depleted FBS and 1% sodium pyruvate. A full list of plasmids used in this study can be found in Table 1.
Table 1.
List of DNA plasmids used in this study.
EV isolation and characterization:
The culture media of transfected MDSCs was collected and centrifuged at 2,000 x g for 20 minutes at 4°C to pellet down and remove dead cells and debris. The supernatant was mixed with Total Exosome Isolation Reagent (Invitrogen, 44–783-59) at a 1:2 ratio and incubated at 4°C overnight. The solution was then centrifuged at 10,000 RPM for 60 minutes at 4°C to precipitate the EVs. To quantify EV size and concentration, EV pellets were re-suspended in 1 mL of serum-free media and analyzed using a Nanosight NS300.
Exposure of breast cancer cell cultures to EVs:
To evaluate the uptake and transfer of transcripts from MDSC-derived EVs to cancer cells, Py8119 cells were exposed to MDSC-derived EVs loaded with Timp3 or Rarres2. EVs were collected and quantified 24 hours post-transfection, as described previously. Py8119 cells were seeded on laminin-coated cover slips (Neuvitro, GG-12–15-Laminin) at a density of 1.5 × 105 cells per replicate and cultured in Ham’s F-12K (Kaighn’s) medium supplemented with 5% EV-free FBS and 1% antibiotic-antimycotic 12h prior to EV exposure. To visualize EV uptake by recipient Py8119 cancer cells, isolated EVs were stained using a PKH67 Green Fluorescent Cell Linker membrane dye kit (Millipore Sigma, MINI67–1KT). Each Py8119 cell replicate was exposed to ~107 EVs. Three independent experiments were run with exposures lasting 6, 12, and 24 hours. Following EVs exposure, the cells were fixed with 10% formalin solution. Phalloidin-iFluor 555 (Abcam, ab176756) was used to stain actin filaments and help visualize Py8119 cells better. The cells were imaged using a Nikon TI2-E fluorescence microscope operating on NIS-Elements AR v5.20.
In situ tracing of EV release and capture:
To evaluate the transfer of EVs from MDSCs to cancer cells, transfected MDSCs and Py8119 cells were co-cultured in 6-well transwell plates (Corning, 3450). MDSCs were stained with a PKH67 green fluorescence dye following transfection, and plated in the apical chamber at a density of 2.5 × 105 cells per well. Py8119 cells were seeded in the basal chamber at the same density. The co-cultures were incubated for 24 hours at 37 °C, 5% CO2 and 95% humidity. Following this, Py8119 cell viability was evaluated using ethidium homodimer (Invitrogen, L3224). The Py8119 cells were then imaged using a Nikon TI2-E fluorescence microscope operating on NIS-Elements AR v5.20. MDSC-derived EVs were imaged under the green fluorescence channel, and Py8119 cells with compromised viability/membrane integrity were imaged under the red fluorescence channel.
Cancer cell proliferation assays:
Cell proliferation studies were performed using co-cultures of transfected MDSCs and Py8119 cells. Py8119 cells were stained with CellTracker red (Invitrogen, 34552) and seeded at a density of 1.5×105 per well in 6-well plates. Transfected MDSCs were stained with CellTracker green (Invitrogen, C7025) and co-plated with the cancer cells at a 1:1 ratio. The co-cultures were incubated at 37 °C, 5% CO2 and 95% humidity for 24 h, and imaged with a Nikon TI2-E fluorescence microscope operating on NIS-Elements AR v5.20. Py8119 cell (red) numbers were quantified via flow cytometry using an LSRII flow cytometer (BD Biosciences).
Cancer cell invasion assays:
Cell invasion studies were performed using the BioCoat Matrigel Invasion Chamber (Corning, 354480) according to the manufacturer’s protocol. Py8119 cells were stained with CellTracker and then co-seeded with transfected MDSCs (1:1 ratio) in each insert. The plates were then incubated for 24 hours at 37°C to allow for cancer cell invasion. Invasion was directed across the inserts by establishing an EV-free FBS gradient. Cancer cells that invaded across the membrane were imaged using a Nikon TI2-E fluorescence microscope and quantified using Image J software (National Institutes of Health, Bethesda, MD).
Orthotopic tumor xenografts:
Immunodeficient nude mice (Jackson Laboratory), 6–8-week-old, were injected with 106 human breast cancer cells MDA-MB-231 (ATCC) suspended in 100 μL of 7 mg/mL basement membrane matrix (Trevigen) in the lower right abdominal mammary fat pad to generate tumors. In some instances, to trace the release and uptake of MDSC-derived EVs, the breast cancer cells were fluorescently pre-labeled with a green MemGlow dye (Cytoskeleton).
In vivo tumor homing studies:
Transfected MDSCs were stained using PKH67 red membrane dye (Millipore Sigma) prior to injection. Tumor-bearing mice were then injected with ~106 MDSCs via the lateral tail vein. Mice were sacrificed 1-day post-injection, and the tumors, lungs and spleens were characterized with an IVIS Imaging System (Xenogen Imaging Technologies). All animal studies were performed in accordance with protocols approved by the Laboratory Animal Care and Use Committee of The Ohio State University.
Immunostaining:
All the antibodies used in this study are listed in Table 2. OCT-embedded tumors were sectioned at 10 μm and mounted in charged microscopy slides. Tissue sections were fixed in cold methanol, blocked with 10% normal goat serum or mouse on mouse (M.O.M.) blocking reagent, and incubated with specific primary antibodies and subsequently with fluorescently tagged secondary antibodies. Tissue sections were imaged using a Nikon TI2-E fluorescence microscope operating on NIS-Elements AR v5.20.
Table 2.
List of antibodies used in this study.
| Target | Primary Antibody | Raised | Cat. No. | Company | Conc. | Secondary antibody | Conc. |
|---|---|---|---|---|---|---|---|
| Timp3 | Timp3 | Rabbit | ab39184 | Abcam | 1:250 | Goat pAb to rabbit IgG 488 (H+L) | 1:200 |
| Chemerin | Rarres2 | Rabbit | ab103153 | Abcam | 1:500 | Goat pAb to rabbit IgG 488 (H+L) | 1:200 |
| Ki67 | Ki67 | Rabbit | ab15580 | Abcam | 1:200 | Goat pAb to rabbit IgG 488 (H+L) | 1:200 |
| Cleaved Capsase-3 | Caspase-3 (mouse) | Rabbit | ab449 | Abcam | 1:50 | Goat pAb to rabbit IgG 647 (H+L) | 1:200 |
Gene expression analyses:
Total RNA was extracted using TRIzol reagent (ThermoFisher). Reverse transcription reactions were performed using 500–1000 ng RNA in a 20 μl reaction with the superscript VILO cDNA synthesis kit (ThermoFisher). cDNA was used as a template to measure expression levels by quantitative real-time PCR using predesigned primers. Real-time PCR reactions were performed using the QuantStudio 3 Real-Time PCR System with TaqMan fast advance chemistry (Thermo Scientific) with the following conditions: 95 °C 10 min, 40 cycles of 95 °C 1 min, 60 °C 1 min, and 72 °C 1 min. Gene expression was normalized against the house keeping genes GAPDH and ATP-6. A full description of primers can be found in Table 3.
Table 3.
List of primers used for gene expression analysis.
| Gene Symbol | Gene name | Gene aliases | Species | Company | Cat. No. |
|---|---|---|---|---|---|
| Gapdh | glyceraldehyde-3-phosphate dehydrogenase | Gapdh | Mouse | Thermofisher Scientific | Mm99999915_g1 |
| Timp3 | Metallopeptidase inhibitor | TIMP-3 | Mouse | Thermofisher Scientific | Mm033403204_m1 |
| Rarres2 | Retinoic acid receptor | AI303516 | Mouse | Thermofisher Scientific | Mm00503579_m1 |
| ATP6 | ATP synthase F0 subunit 6 | Gm10925 | Mouse | ThermoFisher Scientific | Mm03649417_g1 |
Statistical analysis:
All data are reported as the mean and standard error. Statistical analyses were completed using SigmaPlot v14.0 and Prism v10. Comparisons between groups were performed based on 3 – 10 biological replicates. Statistical outliers (i.e., >3 studentized standard deviations) were excluded from the analyses.
Supplementary Material
ACKNOWLEDGEMENTS
Funding for this work was partly provided by a New Innovator Award (NIBIB/NIH, DP2EB028110) and DP1DK126199 to DGP. Most illustrations were created using BioRender.com.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Zhang X. et al. Engineered extracellular vesicles for Cancer therapy. Advanced Materials 33, 2005709 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Elsharkasy OM et al. Extracellular vesicles as drug delivery systems: Why and how? Advanced drug delivery reviews 159, 332–343 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Kamerkar S. et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 546, 498–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez-Erviti L. et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature biotechnology 29, 341–345 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Duarte-Sanmiguel S, Higuita-Castro N, Gallego-Perez D, in Electroporation Protocols 79–84 (Humana, 2020). [DOI] [PubMed] [Google Scholar]
- 6.Lemmerman LR et al. Nanotransfection-based vasculogenic cell reprogramming drives functional recovery in a mouse model of ischemic stroke. Science Advances 7, eabd4735, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang S. et al. Non-viral reprogramming of human nucleus pulposus cells with FOXF1 via extracellular vesicle delivery: an in vitro and in vivo study. European Cells & Materials 41, 90–107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallego-Perez D. et al. Topical tissue nano-transfection mediates non-viral stroma reprogramming and rescue. Nature nanotechnology 12, 974 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson SW & Nguyen J. Exosomes as therapeutics: the implications of molecular composition and exosomal heterogeneity. Journal of Controlled Release 228, 179–190 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Fuhrmann G, Herrmann IK & Stevens MM. Cell-derived vesicles for drug therapy and diagnostics: opportunities and challenges. Nano today 10, 397–409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalluri R. & LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maas SL, Breakefield XO & Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends in cell biology 27, 172–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tkach M. & Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell 164, 1226–1232 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Saari H. et al. Microvesicle- and exosome-mediated drug delivery enhances the cytotoxicity of Paclitaxel in autologous prostate cancer cells. Journal of Controlled Release 220, 727–737, (2015). [DOI] [PubMed] [Google Scholar]
- 15.Setti M. et al. Extracellular vesicle-mediated transfer of CLIC1 protein is a novel mechanism for the regulation of glioblastoma growth. Oncotarget 6, 31413–31427, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Z. et al. Immunoregulatory Roles of Extracellular Vesicles and Associated Therapeutic Applications in Lung Cancer. Frontiers in Immunology 11, 5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahlgren J. et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Research 40, e130, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duarte-Sanmiguel S. et al. Guided migration analyses at the single-clone level uncover cellular targets of interest in tumor-associated myeloid-derived suppressor cell populations. Scientific reports 10, 1–8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla VC et al. Reciprocal Signaling between Myeloid Derived Suppressor and Tumor Cells Enhances Cellular Motility and is Mediated by Structural Cues in the Microenvironment. Advanced Biosystems 4, 2000049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wesolowski R, Markowitz J. & Carson WE. Myeloid derived suppressor cells - a new therapeutic target in the treatment of cancer. Journal for immunotherapy of cancer 1, 10, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stiff A. et al. Myeloid-derived suppressor cells express Bruton’s tyrosine kinase and can be depleted in tumor bearing hosts by ibrutinib treatment. Cancer Res 76, 2125–2136, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stiff A. et al. MDSC inhibit FcR-mediated natural killer cell function via nitric oxide. Nitric Oxide Production by Myeloid Derived Suppressor Cells Plays a Role in Impairing Fc Receptor-Mediated Natural Killer Cell Function. Clin CA Res 24, 1891 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubovsky JA et al. Ibrutinib is an irreversible molecular inhibitor of ITK driving a Th1-selective pressure in T lymphocytes. Blood 122, 2539–2549, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mundy-Bosse BL et al. Myeloid-derived suppressor cell inhibition of the IFN response in tumor-bearing mice. Cancer Res 71, 5101–5110, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He W. et al. Re-polarizing Myeloid-derived Suppressor Cells (MDSCs) with Cationic Polymers for Cancer Immunotherapy. Scientific reports 6, 24506, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H-L et al. TIMP3 expression associates with prognosis in colorectal cancer and its novel arylsulfonamide inducer, MPT0B390, inhibits tumor growth, metastasis and angiogenesis. Theranostics 9, 6676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celebiler A. et al. Predicting invasive phenotype with CDH1, CDH13, CD44, and TIMP3 gene expression in primary breast cancer. Cancer science 100, 2341–2345 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anania M. et al. TIMP3 regulates migration, invasion and in vivo tumorigenicity of thyroid tumor cells. Oncogene 30, 3011–3023 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Pachynski RK et al. Chemerin suppresses breast cancer growth by recruiting immune effector cells into the tumor microenvironment. Frontiers in immunology 10, 983 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu-Chittenden Y. et al. RARRES2 functions as a tumor suppressor by promoting β-catenin phosphorylation/degradation and inhibiting p38 phosphorylation in adrenocortical carcinoma. Oncogene 36, 3541–3552 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stiff A. et al. Myeloid-derived suppressor cells express Bruton’s tyrosine kinase and can be depleted in tumor-bearing hosts by ibrutinib treatment. Cancer research 76, 2125–2136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trikha P. et al. Targeting myeloid-derived suppressor cells using a novel adenosine monophosphate-activated protein kinase (AMPK) activator. Oncoimmunology 5, e1214787 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markowitz J, Wesolowski R, Papenfuss T, Brooks TR & Carson WE. Myeloid-derived suppressor cells in breast cancer. Breast cancer research and treatment 140, 13–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beury DW et al. Cross‐talk among myeloid‐derived suppressor cells, macrophages, and tumor cells impacts the inflammatory milieu of solid tumors. Journal of leukocyte biology 96, 1109–1118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nam S, Lee A, Lim J. & Lim J-S. Analysis of the expression and regulation of PD-1 protein on the surface of myeloid-derived suppressor cells (MDSCs). Biomolecules & therapeutics 27, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higuita-Castro N. et al. Soft lithography-based fabrication of biopolymer microparticles for nutrient microencapsulation. Industrial Biotechnology 8, 365–371 (2012). [Google Scholar]
- 37.Wu Y. et al. Surface‐Mediated Nucleic Acid Delivery by Lipoplexes Prepared in Microwell Arrays. Small 9, 2358–2367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X. et al. Nanochannel electroporation as a platform for living cell interrogation in acute myeloid leukemia. Advanced Science 2, 1500111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallego-Perez D. et al. Microfabricated mimics of in vivo structural cues for the study of guided tumor cell migration. Lab on a Chip 12, 4424–4432 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Gallego-Perez D. et al. On-chip clonal analysis of glioma-stem-cell motility and therapy resistance. Nano letters 16, 5326–5332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu S-Q et al. The human PMR1 endonuclease stimulates cell motility by down regulating miR-200 family microRNAs. Nucleic acids research 44, 5811–5819 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shukla VC et al. Lab-on-a-chip platforms for biophysical studies of cancer with single-cell resolution. Trends in biotechnology 36, 549–561 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Kerkar SP et al. IL-12 triggers a programmatic change in dysfunctional myeloid-derived cells within mouse tumors. The Journal of clinical investigation 121, 4746–4757, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shields CW et al. Cellular backpacks for macrophage immunotherapy. Science Advances 6, eaaz6579 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaczanowska S. et al. Genetically engineered myeloid cells rebalance the core immune suppression program in metastasis. Cell 184, 2033–2052. e2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banks WA et al. Transport of Extracellular Vesicles across the Blood-Brain Barrier: Brain Pharmacokinetics and Effects of Inflammation. International Journal of Molecular Sciences 21, 4407, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto J. et al. Transmission of α-synuclein-containing erythrocyte-derived extracellular vesicles across the blood-brain barrier via adsorptive mediated transcytosis: another mechanism for initiation and progression of Parkinson’s disease? Acta Neuropathologica Communications 5, 71, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X. et al. BATF2 prevents glioblastoma multiforme progression by inhibiting recruitment of myeloid-derived suppressor cells. Oncogene 40, 1516–1530 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mirghorbani M, Van Gool S. & Rezaei N. Myeloid-derived suppressor cells in glioma. Expert review of neurotherapeutics 13, 1395–1406 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Pawelec G, Verschoor CP. & Ostrand-Rosenberg S. Myeloid-derived suppressor cells: not only in tumor immunity. Frontiers in immunology 10, 1099 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salminen A, Kaarniranta K. & Kauppinen A. The potential importance of myeloid-derived suppressor cells (MDSCs) in the pathogenesis of Alzheimer’s disease. Cellular and Molecular Life Sciences 75, 3099–3120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sendo S, Saegusa J. & Morinobu A. Myeloid-derived suppressor cells in non-neoplastic inflamed organs. Inflammation and regeneration 38, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawano T. et al. Temporal and spatial profile of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) in ischemic stroke in mice. PloS one 14, e0215482 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Leo A, Ugolini A. & Veglia F. Myeloid cells in glioblastoma microenvironment. Cells 10, 18, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wesolowski R. et al. Circulating myeloid-derived suppressor cells increase in patients undergoing neo-adjuvant chemotherapy for breast cancer. Cancer Immunology, Immunotherapy 66, 1437–1447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khaled YS, Ammori BJ & Elkord E. Increased Levels of Granulocytic Myeloid-Derived Suppressor Cells in Peripheral Blood and Tumour Tissue of Pancreatic Cancer Patients. Journal of Immunology Research 2014, 879897, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma P. et al. Circulating Myeloid Derived Suppressor Cells (MDSC) That Accumulate in Premalignancy Share Phenotypic and Functional Characteristics With MDSC in Cancer. Frontiers in Immunology 2014, Article ID 879897 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benner B. et al. Generation of monocyte-derived tumor-associated macrophages using tumor-conditioned media provides a novel method to study tumor-associated macrophages in vitro. Journal for immunotherapy of cancer 7, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu-Chittenden Y. et al. Serum RARRES2 Is a Prognostic Marker in Patients With Adrenocortical Carcinoma. Journal of Clinical Endocrinology and Metabolism 101, 3345–3352, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu-Chittenden Y. et al. RARRES2 functions as a tumor suppressor by promoting β-catenin phosphorylation/degradation and inhibiting p38 phosphorylation in adrenocortical carcinoma. Oncogene 36, 3541–3552, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anand-Apte B. et al. A review of tissue inhibitor of metalloproteinases-3 (TIMP-3) and experimental analysis of its effect on primary tumor growth. Biochemistry and Cell Biology 74, (1996). [DOI] [PubMed] [Google Scholar]
- 62.Qi JH et al. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nature Medicine 9, 401–415, (2003). [DOI] [PubMed] [Google Scholar]
- 63.Cha YJ & Koo JS. Role of tumor-associated myeloid cells in breast cancer. Cells 9, 1785, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi J. et al. A review of electroporation-based intracellular delivery. Molecules 23, 3044, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Yan Z, Xia X. & Lin Y. A Novel Electroporation System for Living Cell Staining and Membrane Dynamics Interrogation. Micromachines 11, 767, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng C-X et al. Lentiviral Vectors and Adeno-Associated Virus Vectors: Useful Tools for Gene Transfer in Pain Research. The Anatomical Review 301, 825–836, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nayerossadat N, Maedeh T. & Ali PA. Viral and nonviral delivery systems for gene delivery. Advanced Biomedical Research 1, 27, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang L. et al. Magnetic tweezers‐based 3D microchannel electroporation for high‐throughput gene transfection in living cells. Small 11, 1818–1828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang L. et al. Dielectrophoresis-assisted 3D nanoelectroporation for non-viral cell transfection in adoptive immunotherapy. Lab on a Chip 15, 3147–3153 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Chang L. et al. 3D nanochannel electroporation for high-throughput cell transfection with high uniformity and dosage control. Nanoscale 8, 243–252 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Chang L. et al. Micro-/nanoscale electroporation. Lab on a Chip 16, 4047–4062 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Bertani P. et al. Bosch etching for the creation of a 3D nanoelectroporation system for high throughput gene delivery. Journal of Vacuum Science & Technology B, Nanotechnology and Microelectronics: Materials, Processing, Measurement, and Phenomena 33, 06F903 (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.