Abstract
Background.
Determining malaria transmission within regions of low, heterogenous prevalence is difficult. A variety of malaria tests exist and range from identification of diagnostic infection to testing for prior exposure. This study describes the concordance of multiple malaria tests using data from a 2015 household survey conducted in Ethiopia.
Methods.
Blood samples (n = 2279) from 3 regions in northern Ethiopia were assessed for Plasmodium falciparum and Plasmodium vivax by means of microscopy, rapid diagnostic test, multiplex antigen assay, and multiplex assay for immunoglobulin G (IgG) antibodies. Geospatial analysis was conducted with spatial scan statistics and kernel density estimation to identify malaria hot spots by different test results.
Results.
The prevalence of malaria infection was low (1.4% by rapid diagnostic test, 1.0% by microscopy, and 1.8% by laboratory antigen assay). For P. falciparum, overlapping spatial clusters for all tests and an additional 5 unique IgG clusters were identified. For P. vivax, clusters identified with bead antigen assay, microscopy, and IgG partially overlapped.
Conclusions.
Assessing the spatial distribution of malaria exposure using multiple metrics can improve the understanding of malaria transmission dynamics in a region. The relative abundance of antibody clusters indicates that in areas of low transmission, IgG antibodies are a more useful marker to assess malaria exposure.
Keywords: Plasmodium falciparum, Plasmodium vivax, infection test, antibodies, geospatial analysis, GIS, Ethiopia
Over the past decade, malaria cases and deaths have declined worldwide, though malaria still remains a major public health problem in many parts of the world [1]. In 2010, the World Health Organization recommended that all suspected cases of malaria be confirmed by microscopy or rapid diagnostic test (RDT) [2]. RDTs detect malaria antigens such as Plasmodium falciparum histidine-rich protein 2 (HRP2), and/or pan-Plasmodium lactate dehydrogenase (pLDH), and they allow for quick and practical detection of malaria infection [3]. The more traditional method to diagnose malaria is microscopic examination of blood films to identify Plasmodium parasites [4]. The sensitivity of microscopy for diagnosis varies (ranging from 50% to 90%) based on the transmission intensity in a region, expertise of the microscopist, and magnitude of parasitemia in the sample [5-8]. In addition to these diagnostic tests, antibody tests detect prior malaria exposure occurring months to years in the past.
Malaria elimination in a population requires identifying areas where transmission is occurring [9], and neither RDTs nor microscopy are able to detect very low parasite densities (<100/μL), which can be prevalent in low-transmission settings [10-13]. However, more sensitive laboratory-based tests are available to detect malaria infection [14, 15]. The multiplex bead antigen assay simultaneously detects multiple antigens from blood samples with concentrations approximately 200 pg/mL for PfLDH, 100 pg/mL for Plasmodium aldolase, and as low as 1 pg/mL for HRP2 (parasite density for all, <1/μL) [15, 16]. These detection limits provide sensitivity comparable to PCR. The presence of Plasmodium aldolase or pLDH antigens indicates active infection [17, 18], whereas the presence of HRP2 indicates active or recent infection, with HRP2 clearance in systemic circulation occurring 4–7 weeks after treatment [18, 19]. The presence of anti-Plasmodium antibodies can serve as a proxy for prior exposure to malaria parasites, and species-specific immunoglobulin G (IgG) antibodies are known to be produced against P. falciparum and Plasmodium vivax [20-22]. Assessing population-level antibody responses to Plasmodium species can provide an estimate of historical transmission intensity in a region [22-26].
Ethiopia has a very low malaria prevalence nationally (1.2% by RDT) [27] and aims to achieve malaria elimination by 2030 [28]. Even so, about 60% of the population remains at risk, and transmission is highly heterogenous throughout the country [27]. Compared with the rest of the country, the northwest region has a relatively high burden of malaria [29]. Ethiopia is coendemic for P. falciparum and P. vivax malaria [1], with approximately 60% of cases due to P. falciparum and approximately 40% due to P. vivax [30]. To compare the results of multiple tests for malaria infection or exposure, RDT, microscopy, bead antigen assay, and antibody detection assay were conducted on each blood sample. The goal of this study was to assess concordance among the different test results and compare statistically significant hot spots (spatial clusters) of malaria based on the different tests to indicate areas of malaria transmission.
METHODS
Study Design
The study data were obtained from the 2015 Ethiopia Malaria Indicator Survey (MIS), which was conducted from 30 September to 10 December 2015 (coinciding with the high malaria transmission season). The MIS used a 2-stage cluster-randomized sampling technique to select 555 enumeration areas (kebeles/villages) proportional to population size and 25 households randomly selected per enumeration area. Every child <5 years (6–59 months) old in each selected household and all persons in every fourth household were eligible for malaria testing. Survey enumerators recorded the global positioning system (GPS) coordinates of each household [27]. Before enrollment in the MIS, informed consent form was read to each participant in the appropriate local language, and verbal informed consent was obtained. For children <5 years old, the parent’s consent was obtained before any blood sample was collected.
The study protocol was approved by the Ethiopian Public Health Institute Scientific and Ethical Review Committee and the ethical review committees of Malaria Control and Elimination Partnership in Africa/PATH and the Centers for Disease Control and Prevention (CDC) in Atlanta, Georgia (no. 2015-244). Laboratory assays for antigen and IgG antibody detection were conducted at the CDC in Atlanta, and researchers did not have access to identifying information. Data are available on request to the corresponding author and approval from the Ethiopian Public Health Institute.
Study Area and Population
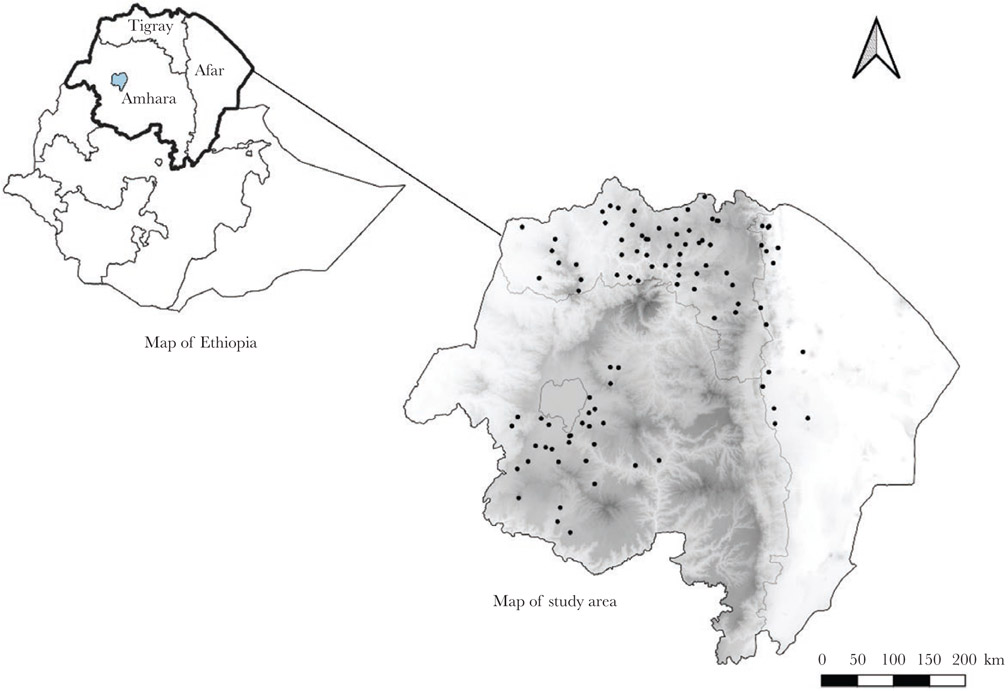
The study involved the 3 northern administrative regions of Ethiopia (Amhara, Tigray, Afar; Figure 1). The physical environment is heterogenous, including both mountains and desert and altitude ranges from below sea level to >2500 m above sea level [31]. The sample data set for analysis (n = 2279) consisted of persons who had results for all 4 tests (RDT, microscopy, multiplex bead antigen assay, and IgG antibody detection).
Figure 1.
Map of Ethiopia, highlighting the study regions (Amhara, Afar, and Tigray). Map on the right shows the distribution of locations where dried blood spot samples were collected in the 3 study regions. Each black dot represents 1 cluster of households in the enumeration area (kebele). Shading indicates elevation, with darker areas at a higher elevation (elevation range, from 131 m below to 4092 m above sea level).
Microscopy and RDTs
Whole-blood samples were tested using the Carestart (AccessBio) multispecies RDT, testing for the presence of HRP2 and pLDH antigens. Participants with a positive RDT result were immediately treated for malaria according to the national malaria treatment guidelines [27]. Thick and thin blood smears for microscopy were made on one slide and stained with 3% Giemsa, and the slides were read by microscopists at the Ethiopian Public Health Institute. A slide was considered negative if no Plasmodium asexual forms or gametocytes were identified after reading 100 fields. All positive slides and 5% of negative slides from each region were reread at the Adama Malaria Control Reference Center.
Multiplex Antigen and IgG Detection Assays
Reagent preparation and multiplex bead-based antigen detection [16] and IgG detection assay [29] were performed as described elsewhere. Briefly, a 6-mm punch equivalent to 10 μL of whole blood was used from each dried blood spot, with blood eluted overnight to a 1:20 concentration in blocking buffer (phosphate-buffered saline containing 0.5% bovine serum albumin, 0.05% Tween 20, 0.02% sodium azide, 0.5% polyvinyl alcohol, 0.8% polyvinylpyrrolidone, and 3-μg/mL Escherichia coli extract).
The 1:20 whole-blood dilution was used for the multiplex bead assay to detect HRP2, pLDH, and Plasmodium aldolase antigens. A 1:200 dilution of serum was used for the multiplex IgG detection assay to detect antibodies to a panel of 10 Plasmodium antigens. For P. falciparum, the following were tested: circumsporozoite protein (PfCSP), early transcribed membrane protein 5 antigen 1 (PfEtramp5Ag1), glutamate-rich protein (PfGLURP-R0), liver-stage antigen 1 (PfLSA1), schizont egress antigen-1 (PfSEA1), apical membrane antigen 1 N terminal region (PfAMA1), and merozoite surface protein 1 19-kD region (PfMSP1); for P. vivax: PvAMA1, PvMSP1, and a chimeric form of the PvMSP1 antigen with additional T- and B-cell epitopes (chPvMSP1). Threshold assay signal was dichot-omized into antigen or IgG positive or negative as appropriate, based on the log-normal mean plus 3 standard deviations of a panel of 92 known negative blood samples from US residents for all assays. For both antigen detection and IgG assays, each assay plate included a buffer blank and positive and negative controls to ensure appropriate assay data collection.
Statistical Analysis
Summary statistics were calculated using SAS software, version 9.4 (SAS Institute). Any RDT result positive for HRP2 was considered a P. falciparum infection, and any result positive for pLDH only was considered a P. vivax infection. Likewise, any multiplex bead antigen assay positive for HRP2 was considered a P. falciparum infection, and any test positive for pLDH or Plasmodium aldolase alone (and not HRP2) was considered a P. vivax infection. These assumptions were made owing to the low prevalence of P. malariae and Plasmodium ovale in Ethiopia [32, 33]. The 10 antigens assessed for IgG antibody detection were categorized into 3 groups: “short-term” P. falciparum, “long-term” P. falciparum, and P. vivax antibodies. Short-term antibodies were more likely to have been acquired in the past year and included antibodies to PfCSP, PfEtramp5Ag1, PfGLURP-R0, PfLSA1, and PfSEA1 [34, 35]. Long-term P. falciparum antibodies (acquired at any time in life) included antibodies to PfAMA1 and PfMSP1 [24, 35]. P. vivax antibodies to these antigens (PvAMA1, PvMSP1, and chPvMSP1) were all considered long-term. Although the long-term antibodies generally persist longer than the short-term antibodies, some of the short-term antibodies may last for years, especially in adults [34].
Geospatial Analysis and Mapping
Cluster analysis within each region was performed using the Kulldorff spatial scan statistic in SaTScan software (version 9.7). Statistically significant clusters were identified using circular and elliptical windows of varying sizes, from zero to a maximum of 40% of the total population at risk, discrete purely spatial Poisson modeling, and 999 Monte Carlo simulations. The window with the highest log-likelihood ratio was defined as the most likely cluster if the P value was <.05 [36]. A criterion of “no geographic overlap” was specified for identifying any secondary clusters [37]. The spatial clusters were mapped using QGIS 3.16.3 software [38].
In addition, kernel density estimation (KDE) was conducted to identify clusters of high malaria prevalence from smoothed prevalence surfaces. Two spatial intensity surfaces were compared: one surface for testing positive for the test of interest and another for all survey participants. The ratio of the 2 surfaces provided the kernel-smoothed prevalence surface map. The maximum bandwidth (approximately 35 km) was calculated using the maximal smoothing principle [39]. Only areas within 35 km of a kebele were considered for the kernel-smoothing procedure, to prevent oversmoothing or estimating prevalence very far from where data were collected. An adaptive bandwidth and the Diggle edge correction was used [40]. To identify areas of significantly higher prevalence, 1-sided asymptotic tolerance contours (α = .05) were calculated [41]. The kernel-smoothed maps were created using R software, version 4.03.3 (R Foundation for Statistical Computing) [42].
RESULTS
Since enumeration areas were randomly selected, in proportion to population size, the number of participants varied between the 3 regions (Table 1). Tigray had the most participants included in this analysis, at 1133 (49.7%), followed by Amhara (742 [32.6%]) and Afar (404 [17.7%]). Nearly half of the samples were from children <5 years old (45.3%) and the second-largest age group was persons aged ≥25 years (24.6%). The prevalence of malaria infection (by RDT, microscopy, or multiplex bead antigen assay) was low (<2.0% overall), though evidence of prior infection via IgG detection was much higher. For P. falciparum, 30.8% of all participants were seropositive for short-term IgG antibodies, and 38.1% for long-term antibodies. For P. vivax, 39.9% of participants were seropositive for long-term antibodies. In this sample, for both P. falciparum and P. vivax, Amhara had a higher antibody prevalence than the other 2 regions.
Table 1.
Characteristics of the Study Population and Malaria Prevalence Estimates for Amhara, Afar, and Tigray Regions in 2015
| Variable | All | Afar Region | Amhara Region | Tigray Region |
|---|---|---|---|---|
| Samples, no. (%) | 2279 (100) | 404 (17.7) | 742 (32.6) | 1133 (49.7) |
| Female sex, no. (%) | 1188 (52.1) | 201 (49.8) | 367 (49.5) | 620 (54.7) |
| Age group, no. (%) | ||||
| <5 y | 1032 (45.3) | 259 (64.1) | 292 (39.4) | 481 (42.5) |
| 5–14 y | 419 (18.4) | 56 (13.9) | 161 (21.7) | 202 (17.8) |
| 15–24 y | 267 (11.7) | 31 (7.7) | 86 (11.6) | 150 (13.2) |
| ≥25 y | 561 (24.6) | 58 (14.4) | 203 (27.4) | 300 (26.5) |
| Result, no. (% [95% CI]) | ||||
| RDT positivea | 31 (1.4 [.9–1.8]) | 1 (0.3 [.0–.7]) | 10 (1.4 [.5–2.2]) | 20 (1.8 [1.–2.5]) |
| Microscopy positivea | 23 (1.0 [.6–1.4]) | 4 (1.0 [.0–2.0) | 7 (.9 [.3–1.6]) | 12 (1.1 [.5–1.7]) |
| Bead antigen assay positivea | 40 (1.8 [1.2–2.3]) | 4 (1.0 [.0–2.0]) | 12 (1.6 [.7–2.5]) | 24 (2.1 [1.3– 3.0]) |
| Short–term Plasmodium falciparum IgG positive | 701 (30.8 [28.9–32.7]) | 82 (20.3 [16.4–24.2]) | 372 (50.1 [46.5–53.7]) | 247 (21.8 [19.4–24.2]) |
| Long–term P. falciparum IgG positive | 868 (38.1 [36.1–40.1]) | 137 (33.9 [29.3–38.5]) | 349 (47.0 [43.4–50.6]) | 382 (33.7 [31.0–36.6]) |
| Plasmodium vivax IgG positive | 910 (39.9 [37.9–41.9]) | 110 (27.2 [22.9–31.6]) | 403 (54.3 [50.7–57.9]) | 397 (35.0 [32.3–37.8]) |
| Any IgG positive | 1327 (58.2 [56.2–60.3]) | 195 (48.3 [43.4–53.1]) | 559 (75.3 [72.2–78.4]) | 573 (50.6 [47.7–53.5]) |
| Negative for all tests | 943 (41.4 [39.4–43.4]) | 205 (50.7 [45.9–55.6]) | 182 (24.5 [21.4–27.6]) | 556 (49.1 [46.2–52.0]) |
Abbreviations: CI, confidence interval; IgG, immunoglobulin G; RDT, rapid diagnostic test.
Includes a positive result for any malaria antigen, including histidine-rich protein 2, pan-Plasmodium lactate dehydrogenase, and Plasmodium aldolase.
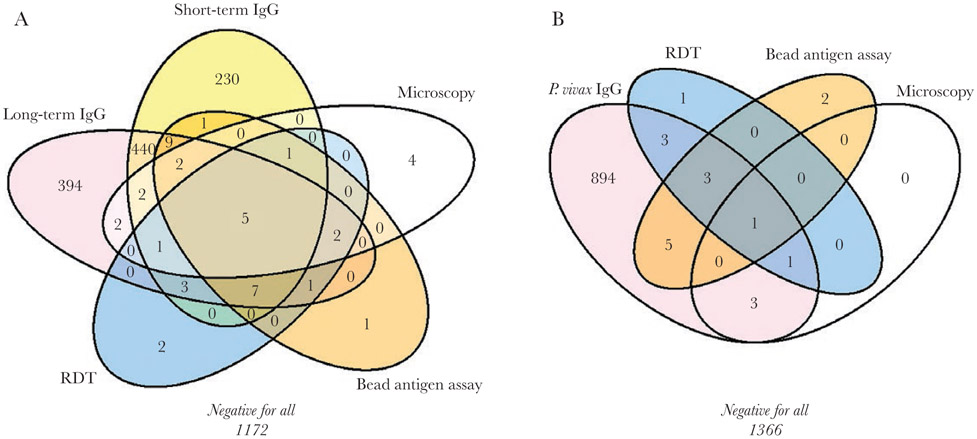
Overall, concordance among the different malaria tests was varied (Figure 2A). For P. falciparum, only 5 participants (0.2%) tested positive with all tests: RDT, bead-antigen assay, microscopy, and short- and long-term antibodies. Among the 22 samples positive for P. falciparum with RDT, 8 (36.4%) also had a positive result with both bead antigen assay and microscopy. A high concordance was observed between short- and long-term P. falciparum antibodies; however, 394 people (17.3%) tested positive for long-term P. falciparum antibodies alone. Among those 394 persons, 51.0% were aged ≥15 years. About half of all participants (51.4%) tested negative for P. falciparum infection or exposure.
Figure 2.
Concordance of different tests for malaria infection or prior exposure to malaria, Plasmodium falciparum (A) or Plasmodium vivax (B). Results reflect field and laboratory samples from 2279 participants in the Malaria Indicator Survey, 2015. Abbreviations: IgG, immunoglobulin G; RDT, rapid diagnostic test.
For P. vivax, only 1 sample (0.04%) tested positive with RDT, bead-antigen assay, microscopy, and P. vivax antibodies (Figure 2B). Among the 9 samples positive for P. vivax with RDT, only 1 (11.1%) also tested positive with bead antigen assay and microscopy. For IgG detection, 894 samples (39.2%) tested positive for anti–P. vivax antibodies alone. Most participants (59.9%) tested negative for any P. vivax infection or prior exposure.
When the 3 diagnostic tests for malaria infection (RDT, microscopy, and bead-based assay) were compared, the bead antigen assay provided the highest number of positive results. For all Plasmodium species, the bead antigen assay detected 40 positive samples (1.8%), compared with 31 (1.4%) for RDT and 23 (1.0%) for microscopy. For P. falciparum infection, the bead antigen assay identified 29 positive samples (1.3%), compared with 22 (0.97%) for RDT and 18 (0.79%) for microscopy, and for P. vivax, the bead antigen assay identified 11 positive samples (0.48%), compared with 9 (0.39%) and 4 (0.18%), respectively.
For seroprevalence of anti-Plasmodium IgG antibodies, higher prevalence estimates were found for the long-term antibodies than for the short-term antibodies (Supplementary Table 1). For all ages, the highest overall seroprevalence was found for the P. vivax chPvMSP1 antigen (35.8%) followed by P. falciparum MSP1 (34.4%). In addition, the levels of IgG antibodies against each antigen reliably increased with age among the 4 age groups (0–4, 5–14, 15–24, and ≥25 years) (Supplementary Figure 1).
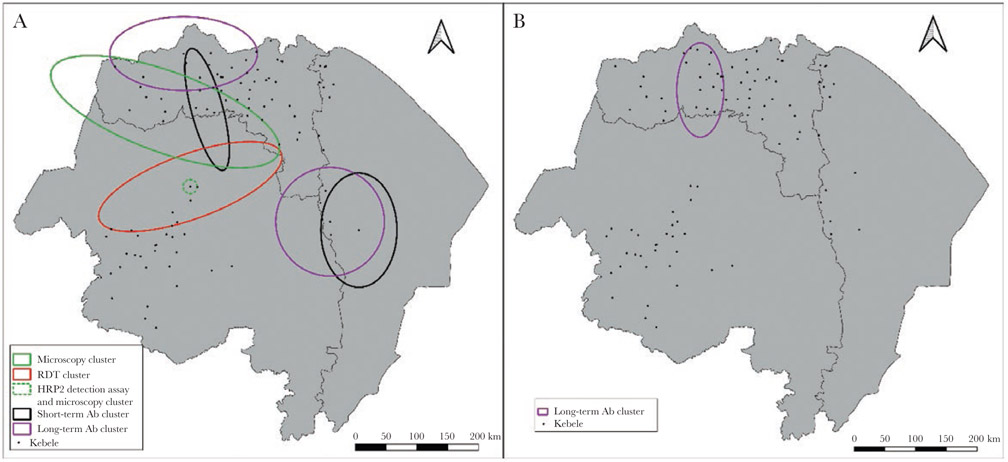
Clusters of malaria cases were first assessed using the Kulldorff spatial scan statistic within each region independently to avoid large clusters between regions that would include large areas without data. Overlapping clusters of P. falciparum infection (RDT, microscopy, and antigen assay) were found in Amhara only (Figure 3). In Tigray and Afar, partially overlapping clusters of short- and long-term anti–P. falciparum antibodies were found. Also in Tigray, a cluster of anti–P. vivax antibodies was identified. Five of the 9 clusters identified from the spatial scan statistic were antibody clusters. Overall, there was clear spatial heterogeneity in malaria prevalence, particularly with the P. falciparum RDT, bead antigen assay, and microscopy (Supplementary Figure 2). Spatial heterogeneity was also found for P. vivax malaria infection and exposure (Supplementary Figure 3).
Figure 3.
All statistically significant spatial clusters for Plasmodium falciparum (A) and Plasmodium vivax (B) malaria. Clusters were assessed independently by region. Each cluster indicates a significant geospatial cluster of prevalence (P < .05). Abbreviations: Ab, antibody; HRP2, histidine-rich protein 2; RDT, rapid diagnostic test.
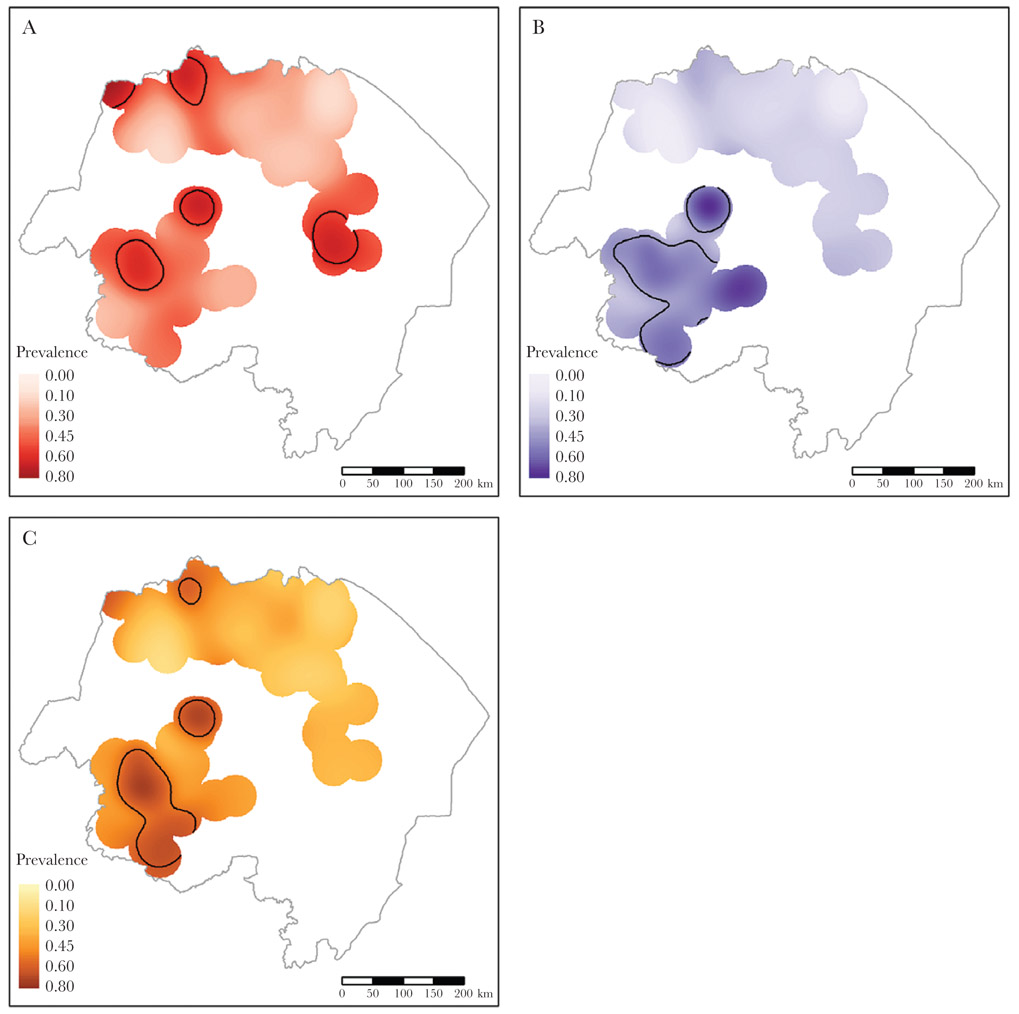
The kernel-smoothed spatial prevalence identified more clusters of smaller size than the SaTScan analysis (Figure 4). For P. falciparum, overlapping clusters were identified in northern Amhara for all tests (RDT, bead HRP2 antigen assay, microscopy, and short- and long-term antibodies) (Supplementary Figure 4 and Figure 4). Five unique clusters were identified for long-term P. falciparum antibodies, 2 of which overlap either completely or partially with the 2 short-term P. falciparum antibody clusters. The remaining long-term P. falciparum antibody clusters (in Tigray and Afar) coincide with the clusters identified by SaTScan within those regions. For the anti–P. vivax IgG, 3 clusters were identified by kernel smoothing (Figure 4). The antibody cluster in Tigray partially overlapped with the P. vivax bead antigen assay and microscopy clusters (Supplementary Figure 5, and Figure 4). Although no clusters of P. vivax infection were identified in any region by SaTScan, clusters for all 3 tests of active infection were identified in northwest Tigray by kernel smoothing.
Figure 4.
Kernel-smoothed prevalence surfaces for long-term anti–Plasmodium falciparum antibodies (A), short-term anti-P. falciparum antibodies (B), and anti–Plasmodium vivax antibodies (C). Areas encircled by black lines represent areas of significantly higher prevalence (asymptotic P < .05).
DISCUSSION
Spatial clustering of P. falciparum and P. vivax was identified in northern Ethiopia by using data from independent assays for parasite exposure, providing evidence that malaria infection (or exposure) was not randomly distributed within the 3 regions. Overlapping clusters were identified for P. falciparum by all 4 tests, whereas for P. vivax, only 3 clusters (antibody, bead antigen assay, and microscopy) overlapped. Many antibody clusters were identified for both P. falciparum and P. vivax, indicating that in areas of low transmission, antibody data provide more information to assess malaria exposure in a population. Importantly, the combination of results from multiple test types provides more information—this is required in areas of high malaria heterogeneity.
Owing to the unique nature of the targets detected by each test, it was not surprising that some discordance was found among test results for malaria infection and exposure. The overall malaria infection prevalence with the bead antigen assay was 1.8%, which was 25% higher than with RDT and 57% higher than with microscopy. Evidence for malaria exposure in this part of Ethiopia appears to be low to moderate as measured by antibody prevalence (38.1% and 39.9% sero-positivity for long-lived IgG against P. falciparum and P. vivax antigens, respectively). As expected, overall seroprevalence was higher for long-lived than for short-lived IgGs [24, 35, 43], and by age 5 years, 4.2%–17.0% of children have IgG seroconverted to the P. falciparum and P. vivax targets on our panel.
Although overall seroprevalence was higher for the long-term IgG, in Amhara the prevalences of long-term and short-term IgG were very similar (47.0% and 50.1%, respectively). This is likely a result of more recent exposure in Amhara, as there was an increase in malaria cases presenting to health facilities in late 2014 and 2015 [44]. Seroprevalences to long-term P. falciparum and P. vivax antibodies were generally equivalent, in contrast with tests for infection, which found a predominance of P. falciparum. This pattern of slightly higher seroprevalence for P. falciparum compared with P. vivax has been found by previous studies in Ethiopia [29, 45, 46]. These contrasts may point to better diagnostic tests for P. falciparum than for P. vivax, as the HRP2 antigen target has a higher sensitivity than pLDH [47]; lower P. vivax infection densities that may still induce an antibody response; or the potential ability of P. vivax to induce IgG with longer half-lives.
Although overall concordance among the different tests was low, geospatial overlap of several different clusters of malaria was identified, providing evidence for a genuine finding of relatively higher malaria transmission in specific areas among these 3 Ethiopian regions. The geospatial overlap of P. falciparum clusters detected by RDT, bead antigen assay, microscopy, and antibodies in western Amhara indicate that relatively more P. falciparum transmission was occurring there. In Amhara, 4 kebeles accounted for all of the P. falciparum cases detected by RDT and 3 of those kebeles accounted for all of the P. falciparum cases detected by bead antigen assay. Such findings can assist programmatic decision making by highlighting the geographically heterogenous distribution of P. falciparum infection, which is common in low-endemic settings [29, 45].
For P. vivax, overlapping clusters for the bead antigen assay, microscopy, and antibody detection were found with KDE, but not with SaTScan. This is likely because clustering was assessed within each region with SaTScan, instead of comparing all 3 regions, as was done with KDE. In addition, SaTScan relies on circular or elliptical windows to identify clusters, which may miss more subtle clusters of varying shapes that can be identified by KDE. Within Tigray, the SaTScan analysis identified 1 large cluster of P. vivax antibody prevalence, coinciding in the same area identified by KDE. Almost no P. vivax infection was detected in Amhara, but there was a high prevalence of previous P. vivax exposure, as indicated by the presence of anti–P. vivax antibodies. This may indicate that P. vivax transmission has decreased in Amhara over time.
For both P. falciparum and P. vivax, antibody data provided more positive results and a more detailed representation of malaria transmission compared with other indicators. Five hot spots of long-term antibody prevalence for P. falciparum and 3 for P. vivax were identified with the kernel-smoothed maps (compared with 2 hot spots and 1 hot spot, respectively, with SaTScan analysis). In studies with similar age distributions, these findings accentuate the utility of using antibody data to estimate transmission intensity in settings of low endemicity [23-26]. As infections become more rare in a population, they are harder to identify because a higher proportion are asymptomatic [48]. In addition, antibody data are a more sensitive indicator of malaria transmission compared with indicators of active infection, because antibodies linger within a person, and there is therefore more time to measure exposure. Many studies have used spatial scan statistics to identify disease clusters [49, 50], but in areas with sparse data or disperse population density, KDE is useful for identifying more refined hot spots. However, more dense sampling of the population would inherently provide tighter clusters of positivity. Concordance between both spatial models would provide the highest confidence that malaria exposure in an area is relatively high.
Our analyses are subject to several limitations. The study samples were collected from a cross-sectional survey conducted in 2015. As malaria is dynamic by place and time [45, 50], the results presented here indicate malaria prevalence during the study period and likely do not indicate current malaria prevalence in northern Ethiopia. Future studies could estimate malaria prevalence using the tests described here at multiple time points during the year to assess how transmission varies. In Ethiopia, the MIS is conducted periodically (approximately every 4–5 years) during the peak malaria transmission season; therefore, these data could be compared with data from a future MIS, controlling for seasonality. In addition, the samples analyzed here were a subset of the original MIS; therefore, the sample may not be a statistically representative sample of the 3 administrative regions. Not all dried blood spots collected were shipped to the CDC, and not all could be assessed with the multiplex antigen detection or antibody detection assays. Samples are assumed to reflect malaria infection and exposure at their respective locations, and we did not account for population movement or individual travel history, although the reported rate of recent travel in this sample was very low (2.1%). A person was considered seropositive to short- or long-term IgG antibodies if their dried blood spot was IgG positive to any of the multiple targets designated as inducing a short- or long-term IgG response. For this reason, absolute seroprevalence estimates in this study would be more sensitive but potentially less specific.
Overlapping spatial clusters of different metrics provide more confidence that malaria transmission in a setting is truly higher than in surrounding areas. Antibody clusters, especially where short-term and long-term IgG clusters overlap, may be used to narrow down areas that warrant follow-up activities, including further surveys to investigate malaria transmission. For regions of low malaria endemicity and coendemic Plasmodium species, combining multiple measures of infection or exposure can provide a detailed assessment of transmission.
Supplementary Material
Acknowledgments.
We are grateful to the study participants and the research team in Ethiopia, including the microscopists and survey enumerators for their commitment in conducing the Ethiopia National Malaria Indicator Survey 2015. The research was supported in part by an appointment to the Research Participation Program at the Centers for Disease Control and Prevention, administered by the Oak Ridge Institute for Science and Education.
Disclaimer.
The findings and conclusions of this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the US President’s Malaria Initiative.
Financial support.
This work was supported by the US President’s Malaria Initiative and the Global Fund to fight AIDS, Tuberculosis and Malaria, via the Ethiopian Federal Ministry of Health.
Footnotes
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. World malaria report 2019. Geneva, Switzerland: World Health Organization, 2019. [Google Scholar]
- 2.World Health Organization. Guidelines for the treatment of malaria. 3rd ed. Geneva, Switzerland: World Health Organization, 2015. https://www.ncbi.nlm.nih.gov/books/NBK294440/?report=reader. Accessed 16 August 2021. [Google Scholar]
- 3.Moody A Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev 2002; 15:66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Payne D Use and limitations of light microscopy for diagnosing malaria at the primary health care level. Bull World Health Organ 1988; 66:621–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Barat L, Chipipa J, Kolczak M, Sukwa T. Does the availability of blood slide microscopy for malaria at health centers improve the management of persons with fever in Zambia? Am J Trop Med Hyg 1999; 60:1024–30. [DOI] [PubMed] [Google Scholar]
- 6.Reyburn H, Mbatia R, Drakeley C, et al. Overdiagnosis of malaria in patients with severe febrile illness in Tanzania: a prospective study. BMJ 2004; 329:1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyburn H, Ruanda J, Mwerinde O, Drakeley C. The contribution of microscopy to targeting antimalarial treatment in a low transmission area of Tanzania. Malar J 2006; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Assefa A, Ahmed AA, Deressa W, et al. Assessment of subpatent Plasmodium infection in northwestern Ethiopia. Malar J 2020; 19:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bousema T, Griffin JT, Sauerwein RW, et al. Hitting hotspots: spatial targeting of malaria for control and elimination. PLoS Med 2012; 9:e1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. Malaria rapid diagnostic test performance: summary results of WHO product testing of malaria RDTs: round 1-8 (2008-2018). Geneva, Switzerland: World Health Organization, 2018. https://apps.who.int/iris/bitstream/handle/10665/276193/9789241514958-eng.pdf?ua=1/. Accessed 17 March 2021. [Google Scholar]
- 11.Bousema T, Okell L, Felger I, Drakeley C. Asymptomatic malaria infections: detectability, transmissibility and public health relevance. Nat Rev Microbiol 2014; 12:833–40. [DOI] [PubMed] [Google Scholar]
- 12.Okell LC, Ghani AC, Lyons E, Drakeley CJ. Submicroscopic infection in Plasmodium falciparum-endemic populations: a systematic review and meta-analysis. J Infect Dis 2009; 200:1509–17. [DOI] [PubMed] [Google Scholar]
- 13.McMorrow ML, Aidoo M, Kachur SP. Malaria rapid diagnostic tests in elimination settings—can they find the last parasite? Clin Microbiol Infect 2011; 17:1624–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berry A, Benoit-Vical F, Fabre R, Cassaing S, Magnaval JF. PCR-based methods to the diagnosis of imported malaria. Parasite 2008; 15:484–8. [DOI] [PubMed] [Google Scholar]
- 15.Rogier E, Plucinski M, Lucchi N, et al. Bead-based immunoassay allows sub-picogram detection of histidine-rich protein 2 from Plasmodium falciparum and estimates reliability of malaria rapid diagnostic tests. PLoS One 2017; 12:e0172139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plucinski MM, Herman C, Jones S, et al. Screening for Pfhrp2/3-deleted Plasmodium falciparum, non-falciparum, and low-density malaria infections by a multiplex antigen assay. J Infect Dis 2019; 219:437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plucinski MM, McElroy PD, Dimbu PR, et al. Clearance dynamics of lactate dehydrogenase and aldolase following antimalarial treatment for Plasmodium falciparum infection. Parasit Vectors 2019; 12:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalrymple U, Arambepola R, Gething PW, Cameron E. How long do rapid diagnostic tests remain positive after anti-malarial treatment? Malar J 2018; 17:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plucinski MM, Dimbu PR, Fortes F, et al. Posttreatment HRP2 clearance in patients with uncomplicated Plasmodium falciparum malaria. J Infect Dis 2017; 217:685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plucinski MM, Candrinho B, Chambe G, et al. Multiplex serology for impact evaluation of bed net distribution on burden of lymphatic filariasis and four species of human malaria in northern Mozambique. PLoS Negl Trop Dis 2018; 12:e0006278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Priest JW, Plucinski MM, Huber CS, et al. Specificity of the IgG antibody response to Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale MSP119 subunit proteins in multiplexed serologic assays. Malar J 2018; 17:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogier E, Moss DM, Chard AN, et al. Evaluation of immunoglobulin g responses to Plasmodium falciparum and Plasmodium vivax in Malian school children using multiplex bead assay. Am J Trop Med Hyg 2017; 96:312–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corran P, Coleman P, Riley E, Drakeley C. Serology: a robust indicator of malaria transmission intensity? Trends Parasitol 2007; 23:575–82. [DOI] [PubMed] [Google Scholar]
- 24.Drakeley CJ, Corran PH, Coleman PG, et al. Estimating medium- and long-term trends in malaria transmission by using serological markers of malaria exposure. Proc Natl Acad Sci U S A 2005; 102:5108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart L, Gosling R, Griffin J, et al. Rapid assessment of malaria transmission using age-specific sero-conversion rates. PLoS One 2009; 4:e6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bousema T, Youssef RM, Cook J, et al. Serologic markers for detecting malaria in areas of low endemicity, Somalia, 2008. Emerg Infect Dis 2010; 16:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ethiopian Public Health Institute. Ethiopia national malaria indicator survey 2015. Addis Ababa, Ethiopia: Ethiopian Public Health Institute, 2016. [Google Scholar]
- 28.Ethiopian Ministry of Health. National malaria elimination road map. Addis Ababa, Ethiopia: Federal Ministry of Health, 2017. [Google Scholar]
- 29.Assefa A, Ali Ahmed A, Deressa W, et al. Multiplex serology demonstrate cumulative prevalence and spatial distribution of malaria in Ethiopia. Malar J 2019; 18:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taffese HS, Hemming-Schroeder E, Koepfli C, et al. Malaria epidemiology and interventions in Ethiopia from 2001 to 2016. Infect Dis Poverty 2018; 7:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asefa M, Cao M, He Y, Mekonnen E, Song X, Yang J. Ethiopian vegetation types, climate and topography. Plant Divers 2020; 42:302–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alemu A, Fuehrer HP, Getnet G, Tessema B, Noedl H. Plasmodium ovale curtisi and Plasmodium ovale wallikeri in North-West Ethiopia. Malar J 2013; 12:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feleke SM, Brhane BG, Mamo H, et al. Sero-identification of the aetiologies of human malaria exposure (Plasmodium spp.) in the Limu Kossa District of Jimma Zone, South western Ethiopia. Malar J 2019; 18:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helb DA, Tetteh KK, Felgner PL, et al. Novel serologic biomarkers provide accurate estimates of recent Plasmodium falciparum exposure for individuals and communities. Proc Natl Acad Sci U S A 2015; 112:E4438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ondigo BN, Hodges JS, Ireland KF, et al. Estimation of recent and long-term malaria transmission in a population by antibody testing to multiple Plasmodium falciparum antigens. J Infect Dis 2014; 210:1123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waller L, Gotway C. Applied spatial statistics for public health data. Hoboken, New Jersey: John Wiley & Sons, 2004. [Google Scholar]
- 37.Kulldorff M SaTScan user guide for version 9.7, 2017. http://www.satscan.org/. Accessed 16 February 2021. [Google Scholar]
- 38.QGIS Development Team. QGIS geographic information system. open source geospatial foundation project, 2021. http://qgis.osgeo.org. Accessed 12 February 2021.
- 39.Terrell G The maximal smoothing principle in density estimation. J Am Stat Assoc 1990; 85:410–77. [Google Scholar]
- 40.Diggle P A kernel-method for smoothing point process data. Appl Stat-J Roy St C 1985; 34:138–47. [Google Scholar]
- 41.Davies TM, Hazelton ML. Adaptive kernel estimation of spatial relative risk. Stat Med 2010; 29:2423–37. [DOI] [PubMed] [Google Scholar]
- 42.Davies TM, Marshall JC, Hazelton ML. Tutorial on kernel estimation of continuous spatial and spatiotemporal relative risk. Stat Med 2018; 37:1191–221. [DOI] [PubMed] [Google Scholar]
- 43.Ondigo BN, Hamre KES, Frosch AEP, Ayodo G, White MT, John CC. Antibody profiles to P. falciparum antigens over time characterize acute and long-term malaria exposure in an area of low and unstable transmission. Am J Trop Med Hyg 2020; 103:2189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lankir D, Solomon S, Gize A. A five-year trend analysis of malaria surveillance data in selected zones of Amhara region, Northwest Ethiopia. BMC Public Health 2020; 20:1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tadesse FG, van den Hoogen L, Lanke K, et al. The shape of the iceberg: quantification of submicroscopic Plasmodium falciparum and Plasmodium vivax parasitaemia and gametocytaemia in five low endemic settings in Ethiopia. Malar J 2017; 16:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yalew WG, Pal S, Bansil P, et al. Current and cumulative malaria infections in a setting embarking on elimination: Amhara, Ethiopia. Malar J 2017; 16:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B, Sun Z, Li X, et al. Performance of pfHRP2 versus pLDH antigen rapid diagnostic tests for the detection of Plasmodium falciparum: a systematic review and meta-analysis. Arch Med Sci 2017; 13:541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris I, Sharrock WW, Bain LM, et al. A large proportion of asymptomatic Plasmodium infections with low and sub-microscopic parasite densities in the low transmission setting of Temotu Province, Solomon Islands: challenges for malaria diagnostics in an elimination setting. Malar J 2010; 9:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oviedo A, Knipes A, Worrell C, et al. Combination of serological, antigen detection, and DNA data for Plasmodium falciparum provides robust geospatial estimates for malaria transmission in Haiti. Sci Rep 2020; 10:8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seyoum D, Yewhalaw D, Duchateau L, Brandt P, Rosas-Aguirre A, Speybroeck N. Household level spatio-temporal analysis of Plasmodium falciparum and Plasmodium vivax malaria in Ethiopia. Parasit Vectors 2017; 10:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.