SUMMARY
The endoplasmic reticulum (ER)-to-Golgi transport is critical to protein secretion and intracellular sorting. Here we report a highly elongated tubular ER-Golgi intermediate compartment (t-ERGIC) that selectively expedites the ER-to-Golgi transport for soluble cargoes of the receptor SURF4. Lacking the canonical ERGIC marker ERGIC-53 yet positive for the small GTPases Rab1A/B, the t-ERGIC is further marked by its extraordinarily elongated and thinned shape. With its large surface-to-volume ratio, high intracellular traveling speeds, and ER-Golgi recycling capabilities, the t-ERGIC accelerates the trafficking of SURF4-bound cargoes. The biogenesis and cargo selectivity of t-ERGIC both depend on SURF4, which recognizes the N-terminus of soluble cargoes and co-clusters with the selected cargoes to expand the ER-exit site. In the steady state, the t-ERGIC-mediated fast ER-to-Golgi transport is antagonized by the KDEL-mediated ER retrieval. Together, our results argue that specific cargo-receptor interactions give rise to distinct transport carriers that regulate the trafficking kinetics.
Keywords: Membrane vesicle trafficking, ER-to-Golgi transport, ER-Golgi intermediate compartment, soluble cargo, tubular carrier, SURF4-cargo co-clustering, ER-exit site expansion, SURF4-KDELR antagonism
In brief
Yan et al. unveil an unconventional, highly elongated tubular ERGIC (t-ERGIC), which selectively accelerates the ER-to-Golgi trafficking of soluble cargoes of the receptor SURF4, suggesting that the diversity of ER-to-Golgi transport carriers is attributable to distinct cargo-receptor interactions.
Graphical Abstract
INTRODUCTION
About 6,000 human proteins, after synthesized at the endoplasmic reticulum (ER), are transported to the Golgi apparatus for secretion or sorting to other organelles (Barlowe and Helenius, 2016; Dancourt and Barlowe, 2010; Gomez-Navarro and Miller, 2016; Lee et al., 2004). How cells selectively transport different proteins to fulfill their diverse functions has elicited a wealth of research interest. Under current models, ER-to-Golgi trafficking starts at the ER-exit sites (ERESs), where the cargo receptor or the cargo itself recruits the coat protein complex II (COPII) to generate ER-derived vesicles (Brandizzi and Barlowe, 2013; Kurokawa and Nakano, 2019; Lee et al., 2004; Raote and Malhotra, 2021; Zanetti et al., 2012). Whereas in yeasts these vesicles appear to directly reach the Golgi apparatus via cytoplasmic diffusion (Kurokawa and Nakano, 2019; Lee et al., 2004), in mammalian cells they relay the cargo to the ER-Golgi intermediate compartment (ERGIC) for microtubule-dependent sorting to the Golgi apparatus (Appenzeller-Herzog and Hauri, 2006; Saraste and Marie, 2018). Additionally, proteins lacking affinity to any receptors may be transported via “bulk flow” through nonspecific packaging into the ERES (Barlowe and Helenius, 2016; Lee et al., 2004).
Contrasting with the diversity of protein cargoes and receptors identified over the past decades, the mammalian ERGIC is often simply represented by the presence of the membrane lectin ERGIC-53/LMAN1, which may not define all carriers mediating ER-to-Golgi trafficking (Appenzeller-Herzog and Hauri, 2006; Hauri and Schweizer, 1992; Saraste and Marie, 2018). Whereas electron microscopy often shows ERGIC as vesiculo-tubular clusters (VTCs) <~1 μm in size (Saraste and Svensson, 1991; Schweizer et al., 1988), elongated tubular carriers have also been described (Bannykh et al., 1996; Klumperman et al., 1998; Mironov et al., 2003; Weigel et al., 2021). Live-cell fluorescence microscopy using synchronized cargoes also occasionally notes long (>~1 μm) tubular carriers in ER-to-Golgi trafficking (Ben-Tekaya et al., 2005; Blum et al., 2000; Marra et al., 2001; Presley et al., 1997; Shomron et al., 2021; Simpson et al., 2005; Weigel et al., 2021). Yet, it also appears some cargoes do not enter tubular carriers (Boncompain et al., 2012; Westrate et al., 2020). Whether these morphologically distinct carriers are specific to different protein cargoes, and if so, what determines such selectivity, remain elusive. Moreover, the functional significance of this morphological diversity in regulating transport kinetics remains unexplored. It is further noteworthy that most of the previously examined “model cargoes” are membrane proteins. In contrast, the transport of soluble proteins, which represent ~30% of the anterograde cargoes in mammalian cells (Dancourt and Barlowe, 2010), is less understood.
Here we report SURF4-induced tubular ERGIC (t-ERGIC), a distinct class of ERGIC that specifically accelerates the ER-to-Golgi transport of soluble cargoes of the receptor SURF4, the mammalian homolog of the yeast cargo receptor Erv29p (Belden and Barlowe, 2001; Dancourt and Barlowe, 2010; Mitrovic et al., 2008). The t-ERGIC lacks ERGIC-53 but is marked by the small GTPases Rab1A/B, and further differs from the canonical ERGIC/VTCs by its extremely slender shape. After de novo generation at expanded ERESs via SURF4-cargo interactions, the t-ERGIC travels and recycles in the cell at high speeds to enable efficient ER-to-Golgi transport.
RESULTS
Identification of a Highly Elongated Tubular Organelle through Mislocalized DsRed2-ER-5
We initially attempted to deliver the red fluorescent protein (FP) DsRed2 into the ER lumen of COS-7 cells via an N-terminal signal peptide. Curiously, whereas DsRed2-ER-3 (Figure 1A) correctly localized to the ER (Figure 1B), a construct with slightly varied linkers, DsRed2-ER-5 (Figure 1A), only showed the expected ER localization in ~20% of the transfected cells. A preponderance (~45%) of the cells had fluorescence mainly distributed in peculiar, 2–20 μm long tubular organelles (TOs) (Figures 1B–1D), while another major population (~30%) had most fluorescence in small vesicles (Figures 1D, S1A). Similar TOs and vesicles were also observed for DsRed2-ER-5 expressed in U2OS and HeLa cells (Figures S1B and S1C).
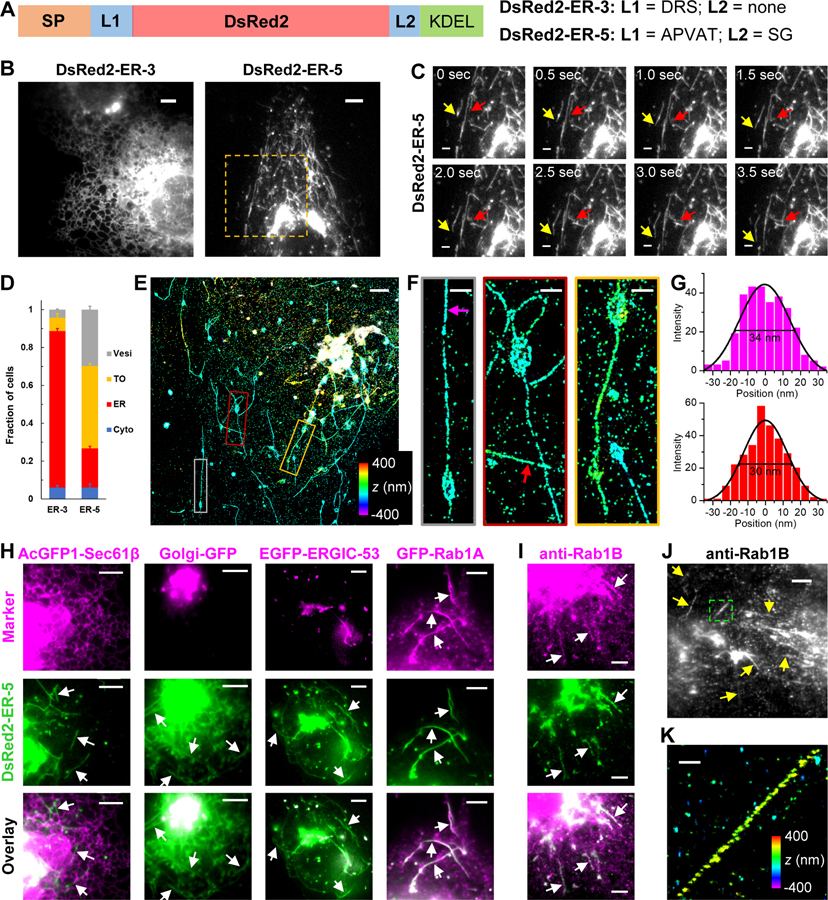
Figure 1. DsRed2-ER-5 Mislocalizes to an Extremely Slender, Rab1-Positive Tubular Organelle.
(A) Schematics of the sequences of DsRed2-ER-3 and DsRed2-ER-5. SP: signal peptide; L: linker.
(B) Representative live-cell images of DsRed2-ER-3 and DsRed2-ER-5 expressed in COS-7 cells.
(C) Time-lapse series of the boxed region in (B), showing the morphology and fast motion of the tubular organelles (TOs) (arrows). See also Video S1.
(D) Classification of the dominating distribution modes of DsRed2-ER-3 and DsRed2-ER-5 in each cell: ER, TO, vesicles (Vesi), and cytoplasm (Cyto). Error bars: SEM (n = 4 with ~50 cells in each replicate).
(E) 3D-STORM image of immunolabeled DsRed2-ER-5 in a COS-7 cell. Color encodes axial position.
(F) Close-ups of the TOs in the three colored boxes in (E).
(G) STORM intensity profiles across the widths of two TOs at the magenta and red arrows in (F). Black curves: Gaussian fits with FWHM (full width at half maximum) of 34 and 30 nm, respectively.
(H) Two-color live-cell images of DsRed2-ER-5 (green) co-expressed with different organelle markers (magenta) in COS-7 cells: the ER marker AcGFP1-Sec61β, the Golgi marker Golgi-GFP, the canonical ERGIC marker EGFP-ERGIC-53, and the small GTPase EGFP-Rab1A.
(I) Two-color image of expressed DsRed2-ER-5 (green) and immunostained endogenous Rab1B (magenta) in a COS-7 cell.
(J) Immunostained endogenous Rab1B in untransfected COS-7 cells.
(K) 3D-STORM image for the boxed region in (J). Color encodes axial position.
Arrows point to TOs. Scale bars: 5 μm (B,H-J); 2 μm (C,E); 500 nm (F,K).
See also Figure S1.
The TOs each had one or a few dilated vacuolar parts and a thin projection, and underwent frequent deformation, fission, and fusion as they moved actively in the cell at typical speeds of 1–2 μm/s (Figure 1C, Video S1). The dilated vacuolar parts were often found at the tubule ends, yet they also frequently slid along the TOs (Video S1). An analogous green FP construct, GCaMP6s-ER-5, behaved similarly and so allowed us to image the TOs with a red live-cell microtubule marker 2xmCherry-EMTB, thus showing their fast sliding along microtubules (Video S2).
Three-dimensional (3D) STORM super-resolution microscopy (Huang et al., 2008) next revealed the unusual ultrastructure of the TOs (Figures 1E and 1F): Whereas the dilated vacuolar parts were spheroids ~500 nm in size, the elongated projections were extremely thin with apparent diameters of ~30 nm (Figure 1G). Given the ~20 nm resolution of STORM and the immunolabeling antibody sizes (Huang et al., 2008), the true diameters should thus be yet smaller.
To identify these TOs, we performed two-color live-cell fluorescence microscopy of DsRed2-ER-5 versus different organelle markers. Co-imaging with ER lumenal and membrane proteins (Figures 1H, S1D, and Video S2) indicated that the TOs and the ER were separate organelles: they were physically unconnected, and the former traveled in the cell at high speeds as the latter exhibited much slower structural dynamics. Similarly, we observed little colocalization or interaction of the TOs with compartments labeled by common markers of the endosomes, autophagosomes, and lysosomes (Figures S1E-S1I).
In contrast, we noticed a substantial accumulation of DsRed2-ER-5 in the Golgi apparatus (Figure 1H). Live-cell recording showed frequent fusion of the DsRed2-ER-5-containing TOs with the Golgi apparatus (green arrows in Video S1), suggesting their involvement in the secretory pathway. Interestingly, whereas the canonical ERGIC marker ERGIC-53 colocalized with DsRed2-ER-5 in vesicle-like structures, it was absent from the TOs (Figure 1H). However, when ERGIC-53 was overexpressed at high levels, it also entered a subset of the TOs (Figure S1J), implying interactions and material exchange between the TOs and the canonical ERGIC. COPII and COPI coats also only partly colocalized with DsRed2-ER-5 in puncta but not in the TOs (Figures S1K and S1L).
We next found Rab1A/B, the major small GTPases in ER-to-Golgi trafficking (Plutner et al., 1991; Stenmark, 2009; Tisdale et al., 1992), are enriched on the surface of the DsRed2-ER-5-containing TOs, a result supported by both expressed EGFP-Rab1A (Figures 1H, S1M) and the immunostaining of endogenous Rab1A (Figure S1N) and Rab1B (Figure 1I). Notably, we further found that in untransfected COS-7 cells, immunostained Rab1B localized to similar, slender and elongated TOs (Figures 1J, 1K, and S1O), suggesting that the TOs naturally exist in the native cell.
The Tubular ERGIC (t-ERGIC) Mediates ER-to-Golgi Trafficking, and Is Formed through Both De Novo Generation and Fusion
Time-lapse imaging captured in some cells the gradual redistribution of DsRed2-ER-5 from the ER to the TOs, Golgi, and vesicles, coupled with a steady reduction of the total fluorescence inside the cell (Figure S2A). Flow cytometry of cells treated by small molecules that respectively inhibited ER-to-Golgi transport (brefeldin A), induced ER stress (thapsigargin and dithiothreitol), and inhibited ER-associated degradation (CB-5083 and MG132) showed that only brefeldin A significantly increased the intracellular DsRed2 fluorescence (Figure S2B). The time-dependent increase of intracellular retention with brefeldin A treatment (Figures 2A and 2B) was accompanied by a redistribution of the DsRed2-ER-5 fluorescence to the ER (Figures 2C, S2C). These results suggest that DsRed2-ER-5 is targeted for ER-to-Golgi transport through the TOs. Together with our results with organelle markers above, we thus identified the TO as a Rab1-coated ERGIC that is not normally enriched with ERGIC-53. Hereafter we refer to it as “tubular ERGIC (t-ERGIC)”.
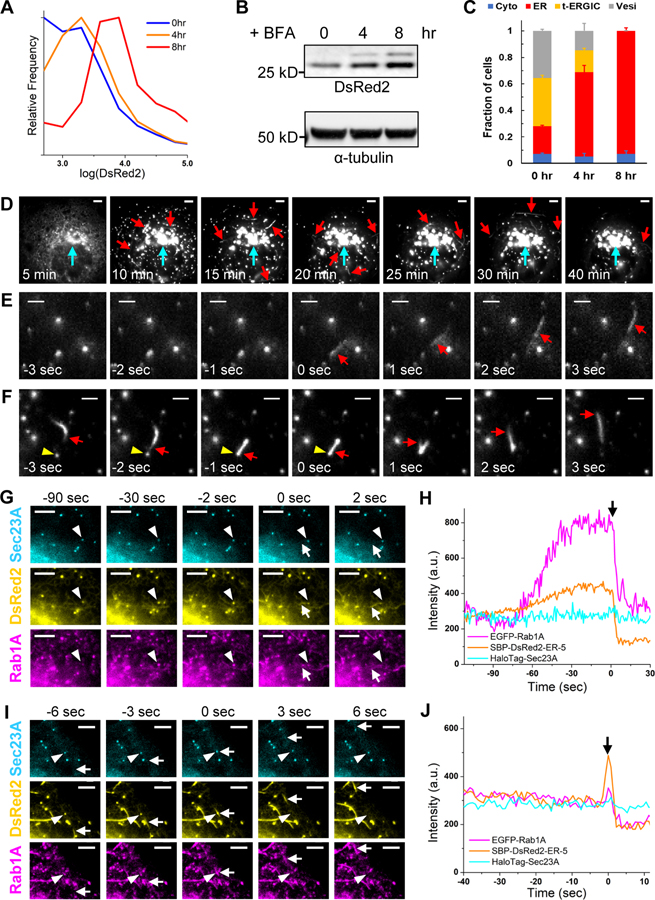
Figure 2. The t-ERGIC Mediates ER-to-Golgi Trafficking, and Is Formed through Both De Novo Generation and Fusion.
(A-C) Flow cytometry histograms (A), lysate immunoblots (B), and subcellular distribution (C) of DsRed2-ER-5 for transfected COS-7 cells treated with 1 μM brefeldin A (BFA) for 0, 4, and 8 hr. Error bars: SEM (n = 3 with ~50 cells in each replicate).
(D) Representative RUSH image sequence of SBP-DsRed2-ER-5 after the addition of 80 μM biotin at time 0. Red and cyan arrows point to t-ERGICs and the Golgi, respectively. See also Video S3.
(E) De novo generation of SBP-DsRed2-ER-5-positive t-ERGIC (arrow) in RUSH. Time 0 corresponds to when budding occurred. Biotin was added at −5 min for cargo release.
(F) Fusion of an SBP-DsRed2-ER-5-positive t-ERGIC (arrow) with an ERES (arrowhead) and subsequent transport of the cargo in the reformed t-ERGIC. Time 0 corresponds to the moment of fusion. Biotin was added at −8 min.
(G) Image sequences of EGFP-Rab1A, SBP-DsRed2-ER-5, and JF635-labeled HaloTag-Sec23A in a RUSH experiment showing the de novo generation of a t-ERGIC (arrow) from the ERES (arrowhead). Time 0 corresponds to when budding occurred. Biotin addition corresponded to −22 min. See also Video S4.
(H) Fluorescence intensity time traces of the three color channels for the ERES indicated by the arrowhead in (G).
(I) Another image sequence of an ERES (arrowhead) in the same RUSH experiment as in (G), showing its fusion with a pre-existing t-ERGIC (arrow). Time 0 corresponds to when fusion occurred. Biotin addition corresponded to −32 min. See also Video S4.
(J) Fluorescence intensity time traces of the three color channels for the ERES indicated by the arrowhead in (I).
Scale bars: 5 μm (D,G,I); 2 μm (E,F).
See also Figure S2.
To elucidate the biogenesis and functions of the t-ERGIC, we employed the Retention Using Selective Hooks (RUSH) assay (Boncompain et al., 2012) to synchronize the release of DsRed2-ER-5 from the ER. In RUSH, the cargo is tagged with a streptavidin binding peptide (SBP) and thus initially retained in the ER by an ER-resident streptavidin “hook”. The addition of biotin outcompetes SBP for streptavidin binding and so enables the synchronized onset of ER-to-Golgi transport of the cargo. Insertion of the SBP tag at the N-terminus of DsRed2-ER-5 (Figure S2D) did not alter the phenotype (Figures S2E and S2F). The SBP-DsRed2-ER-5 cargo was then co-expressed with a streptavidin-KDEL hook, which retained most of the fluorescence signal in the ER. Upon release of the cargo by biotin, we observed that the cargo first concentrated at ERESs and vesicle-like structures throughout the cell (Figure 2D; Video S3). The t-ERGIC emerged right after ER exit (Figures 2D and 2E; Video S3). Single-particle tracking of the vesicles and TOs showed convergence toward the Golgi (Figures S2G and S2H). As the TOs continuously fused with the Golgi (green arrows in Video S3), the fluorescence gradually redistributed from the peripheral ER to the Golgi. These results substantiate that the t-ERGIC mediates anterograde ER-to-Golgi transport.
A closer examination of the RUSH image sequences identified two modes of biogenesis for t-ERGIC: de novo formation (Figure 2E) vs. elongation of existing t-ERGIC through fusion (Figure 2F). Three-color live imaging of the cargo with Rab1A and the ERES marker Sec23A showed that for the first mode, after the cargo was enriched at the ERES, Rab1A gradually accumulated, leveled off, and then budded off together with the cargo into newly formed tubules (Figures 2G and 2H, S2I, Video S4). The budded t-ERGIC then separated from the ERES, leaving behind the Sec23A COPII coat (Figures 2G, S2I), from which another t-ERGIC could bud again (Figure S2J). Accordingly, local fluorescence intensity time traces showed that DsRed2-ER-5 and Rab1A both accumulated at the ERES before they together budded into the t-ERGIC in a single step, whereas Sec23A stayed constant (Figure 2H).
In the second mode, the existing, fast-traveling t-ERGIC tubules fused with the ERES and then budded off as t-ERGIC again (Figure 2F), carrying away the additional cargo while leaving the Sec23A COPII coat behind (Figures 2I and 2J, Video S4). We also often noticed cases in which tubules left the Golgi to travel retrogradely to fuse with the ERES and bring more cargo back to the Golgi (Video S3), a cycling behavior that has been noted previously for ERGIC (Ben-Tekaya et al., 2005; Marra et al., 2001; Sannerud et al., 2006). Thus, the t-ERGIC is a carrier organelle that buds from the ERES and shuttles between the ER and the Golgi to mediate the anterograde transport of cargo proteins.
The accumulation of Rab1A at the ERES before t-ERGIC formation prompted us to test its role in t-ERGIC biogenesis. With a dominant negative Rab1A mutant (N124I) (Moyer et al., 2001; Westrate et al., 2020), we observed that the transfected cells were unable to generate t-ERGIC tubules after the cargo accumulated at the ERES (Figure S2K). Together, our RUSH assays indicate that the Rab1A-dependent t-ERGIC mediates the ER-to-Golgi trafficking of DsRed2-ER-5.
Accelerated ER-to-Golgi Trafficking via the t-ERGIC Is Determined by the N-termini of Soluble Cargoes
Our unexpected discovery of t-ERGIC through DsRed2-ER-5 raises the question of why a similar construct, DsRed2-ER-3, localized predominantly in the ER (Figures 1B, 1D). Given the small dissimilarities between the two constructs (Figure 1A), we wondered whether the property of the N-terminus after the signal-peptide cleavage (P1’) could be important to the fate of the cargo protein. The N-terminus of the cleaved DsRed2-ER-5 begins with the hydrophobic APV tripeptide, whereas that of DsRed2-ER-3 starts with the charged DRS (Figure 1A). Consequently, we constructed a point mutant of DsRed2-ER-5, where the P1’ alanine (A) was substituted by glutamic acid (E). Remarkably, this EPV-DsRed2-ER-5 variant (Figure 3A) was mostly retained in the ER and exhibited few t-ERGICs (Figures 3B and 3C), thus phenocopying DsRed2-ER-3 and serving as a good control for the original DsRed2-ER-5 (hereafter APV-DsRed2-ER-5) for our mechanistic investigations.
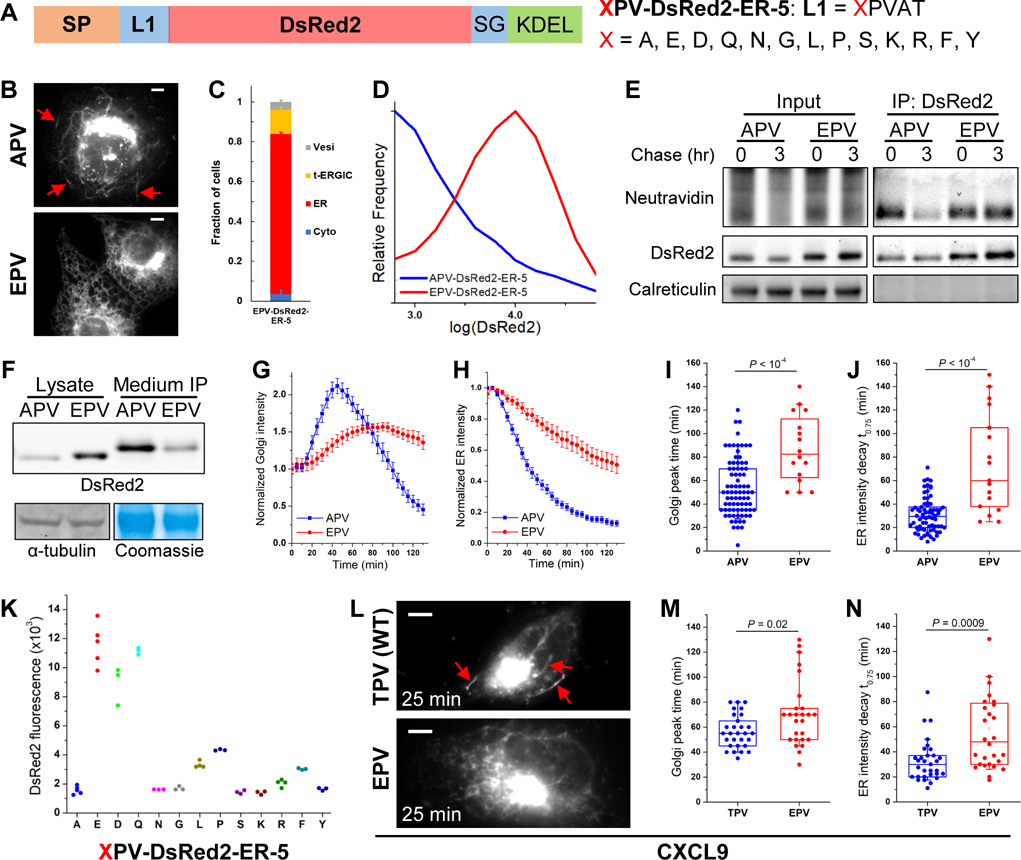
Figure 3. The N-terminus of the Cargo Determines Its Transport with the t-ERGIC and ER-to-Golgi Trafficking Efficiency.
(A) Sequences of the XPV-DsRed2-ER-5 mutations we examined, with varied N-termini after the signal peptide (SP). The original DsRed2-ER-5 has X=A (APV-DsRed2-ER-5).
(B) Representative fluorescence micrographs of APV/EPV-DsRed2-ER-5 in COS-7 cells. Arrows point to t-ERGICs.
(C) Subcellular distribution of EPV-DsRed2-ER-5. Error bars: SEM (n = 3 with ~50 cells in each replicate).
(D) Flow cytometry histograms of APV/EPV-DsRed2-ER-5.
(E) Azidohomoalanine-biotin-alkyne pulse-chase of APV/EPV-DsRed2-ER-5. Newly synthesized proteins were labeled by azidohomoalanine click chemistry and detected by NeutrAvidin (see Methods).
(F) Immunoblots of intracellular (cell lysate) and secreted (anti-FLAG immunoprecipitation from the culture medium) APV/EPV-FLAG-DsRed2-ER-5.
(G,H) Golgi (G) and peripheral ER (H) fluorescence intensity time traces of APV/EPV-SBP-DsRed2-ER-5 in RUSH, pooled from 70 cells from 5 independent runs (APV) or 17 cells from 3 independent runs (EPV). Error bars: SEM. 80 μM biotin was added at time 0.
(I,J) Comparison of the time to the peak fluorescence in the Golgi (I) and the time of fluorescence decay to 75% of the start in the ER (J) of APV/EPV-SBP-DsRed2-ER-5 in RUSH. Each data point is extracted from the RUSH data of one cell in (G,H). Whiskers and boxes show 10%, 25%, 50%, 75%, and 90% quantiles.
(K) Median intracellular fluorescence of different XPV-DsRed2-ER-5 variants expressed in COS-7 cells, as determined by flow cytometry. Each data point corresponds to the median value of ~20,000 positive cells in one biological replicate.
(L) Representative fluorescence micrographs of TPV/EPV-CXCL9-mCherry-SBP in RUSH, 25 min after adding 80 μM biotin. Arrows point to t-ERGICs. See also Figure S3K for image sequences.
(M,N) Comparison of the time to the peak fluorescence in the Golgi (M) and the time of fluorescence decay to 75% of the start in the ER (N) of TPV/EPV-CXCL9-mCherry-SBP in RUSH. Data were from 30 cells (TPV) or 27 cells (EPV) in 3 independent replicates. Whiskers and boxes show 10%, 25%, 50%, 75%, and 90% quantiles.
Scale bars: 5 μm. P values are calculated by two-tailed t tests.
See also Figure S3.
Flow cytometry of COS-7 cells transfected with APV-DsRed2-ER-5 and EPV-DsRed2-ER-5 showed markedly higher intracellular fluorescence for the latter (Figure 3D). Pulse-chase experiments indicated that the APV version was selectively removed from the cell (Figure 3E). With FLAG-tagged versions of APV/EPV-DsRed2-ER-5 (Figure S3A), which, without altering the APV/EPV phenotypes (Figures S3B and S3C), facilitated immunoprecipitation from the culture medium, we next found substantially higher extracellular secretion and lower intracellular retention for the APV version (Figure 3F). Immunoblots of ER stress indicators showed no noticeable activation (Figure S3D), indicating that the different fates of the APV and EPV variants were not due to ER-associated degradation. Similar contrasting behavior of APV/EPV-DsRed2-ER-5 was observed in U2OS and HeLa cells (Figure S3E), as well as for APV/EPV variants of the GCaMP6s FP (Figures S3F and S3G).
RUSH experiments showed that after biotin release, contrasting the fast, t-ERGIC-mediated ER-to-Golgi transport of APV-SBP-DsRed2-ER-5 (Figure 2D), EPV-SBP-DsRed2-ER-5 (Figure S3A) did not enter t-ERGIC and was slowly transported to the Golgi (Figure S3H). Quantification of the fluorescence intensity showed substantially faster rises in the Golgi (Figures 3G and 3I) and drops in the ER (Figures 3H and 3J) for the former. These results demonstrate dramatic differences in the ER-to-Golgi transport pathway and efficiency between APV- and EPV-DsRed2-ER-5, which explain their different intracellular retentions.
To further examine the effects of the cargo N-terminus, we compared 13 different amino acid residues at the P1’ position (“XPV-DsRed2-ER-5”; Figure 3A). Interestingly, we found DsRed2 to be strongly retained in the ER when glutamic acid (E), aspartic acid (D), or glutamine (Q) was present at the P1’ position, but often entered the t-ERGIC and got exported out of the ER when the P1’ position was other amino acids, including the structurally similar asparagine (N) (Figures 3K, S3I and S3J).
To test whether this “N-terminus rule” for t-ERGIC-mediated ER export is relevant to endogenous proteins, we examined the normally secreted CXCL9, a soluble chemokine that has been recently implicated in cancer immunotherapy (House et al., 2020; Tokunaga et al., 2018). RUSH showed that whereas the wild-type CXCL9, with a TPV P1’ N-terminus, entered Rab1-positive t-ERGIC tubules upon cargo release, a mutant with an EPV N-terminus did not (Figures 3L, S3K and S3L). Concomitantly, slower ER-to-Golgi transport was found for the latter (Figures 3M and 3N).
Collectively, our data indicate that a group of cargoes with explicit N-terminal features are routed to the t-ERGIC-mediated fast ER-to-Golgi trafficking pathway.
The Biogenesis and Cargo Selectivity of t-ERGIC Both Depend on SURF4
In search of an explanation for how a “D/E/Q but not N” N-terminus could have prevented the DsRed2 cargoes from entering the t-ERGIC, we noticed a recent study that reported an analogous rule for protein secretion (Yin et al., 2018): With a growth hormone cargo, it is found that D/E/Q-containing, but not N-containing, N-terminal tripeptides are unfavored for secretion mediated by the receptor SURF4, whereas hydrophobic-proline-hydrophobic (Φ-P-Φ) tripeptides are the most favored. Accordingly, we examined an APE-DsRed2-ER-5 variant and found it phenocopied EPV-DsRed2-ER-5 (Figures S4A and S4B), thus indicating that the N-terminal tripeptide is also important to cargo sorting into the t-ERGIC. Recent studies on SURF4 and its homologs have generally suggested its preference for hydrophobic N-termini (Belden and Barlowe, 2001; Casler et al., 2019, 2020; Otte and Barlowe, 2004). Therefore, we set out to examine the role of SURF4 in the t-ERGIC pathway.
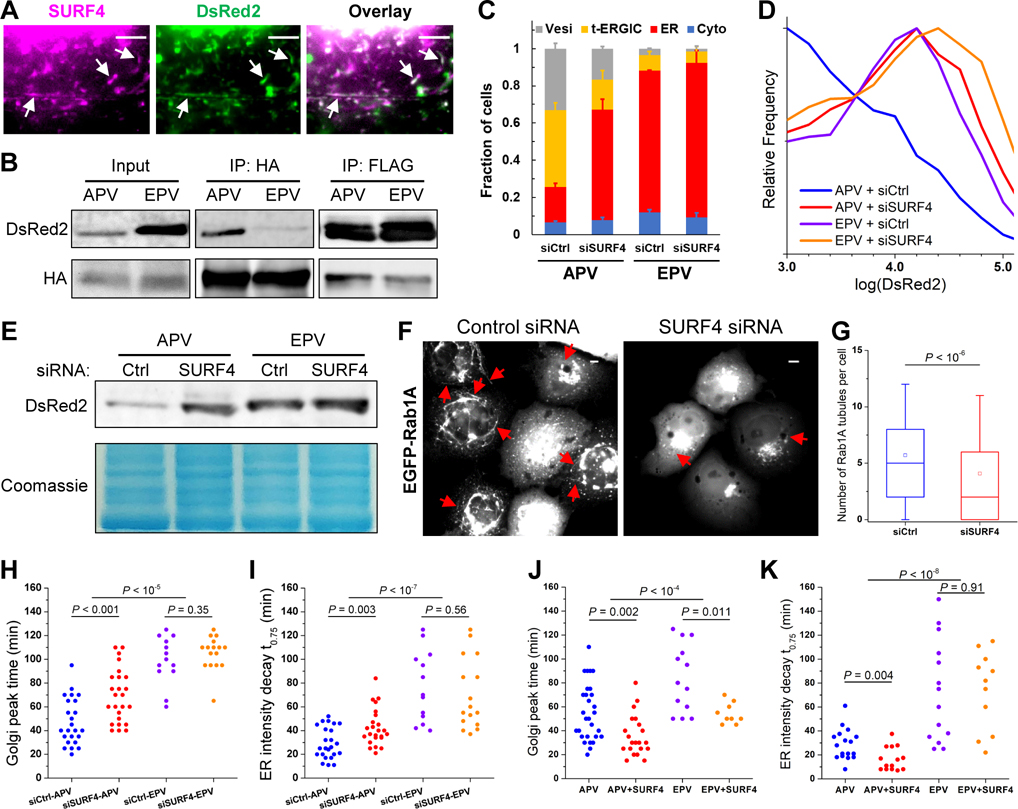
Live-cell imaging showed that AcGFP1-SURF4 colocalized with the t-ERGIC (Figure 4A) and segregated with APV-DsRed2-ER-5 during the de novo generation of t-ERGIC (Figure S4C). Immunolabeled FLAG-SURF4 also colocalized with the t-ERGIC (Figure S4D). These results indicate the co-transport of SURF4 and its cargo (Wang et al., 2021). Notably, SURF4-HA co-immunoprecipitated much more efficiently with APV-FLAG-DsRed2-ER-5 than EPV-FLAG-DsRed2-ER-5, even as the input amount of the former was several-fold lower due to secretion (Figure 4B), thus suggesting that SURF4 selectively sorts the former into the t-ERGIC.
Figure 4. SURF4 Recognizes the N-terminus of the Cargo and Enables t-ERGIC Formation for Expedited ER-to-Golgi Trafficking.
(A) Live-cell images of co-transfected AcGFP1-SURF4 and APV-DsRed2-ER-5 in a COS-7 cell.
(B) Co-immunoprecipitation of APV/EPV-FLAG-DsRed2-ER-5 with co-expressed SURF4-HA.
(C-E) Subcellular distributions (C), flow cytometry histograms (D), and lysate immunoblots (E) of APV/EPV-DsRed2-ER-5 for COS-7 cells co-transfected with control siRNA or SURF4 siRNA. Error bars: SEM (n = 3 with ~50 cells in each replicate).
(F,G) Representative fluorescence micrographs (F) and counts per cell (G) of Rab1A-positive tubules in control or SURF4 siRNA-treated COS-7 cells. Whiskers and boxes show 10%, 25%, 50%, 75%, and 90% quantiles. Open squares indicate means. 287 and 321 cells were quantified for siCtrl and siSURF4, respectively.
(H,I) Comparison of the time to the peak fluorescence in the Golgi (H) and the time of fluorescence decay to 75% of the start in the ER (I) of APV/EPV-SBP-DsRed2-ER-5 with control siRNA or SURF4 siRNA in RUSH. N = 24, 25, 13, and 16 cells for the four conditions, respectively, each from 3 independent runs.
(J,K) Comparison of the time to the peak fluorescence in the Golgi (J) and the time of fluorescence decay to 75% of the start in the ER (K) of APV/EPV-SBP-DsRed2-ER-5 with or without co-expression of FLAG-SURF4 in RUSH. N = 17, 14, 14, and 11 cells for the four conditions, respectively, each from 3 independent runs.
Scale bars: 5 μm. Arrows point to t-ERGICs. P values are calculated by Mann-Whitney test (G), two-way ANOVA (APV vs. EPV in [H-K]), or two-tailed t test (siRNA or SURF4 overexpression in [H-K]).
See also Figure S4.
With small interfering RNA (siRNA) targeting SURF4 (Figure S4E), we next observed a substantial reduction in APV-DsRed2-ER-5 t-ERGICs (Figures 4C, S4F). The intracellular retention of APV-DsRed2-ER-5, as determined by both flow cytometry and immunoblotting, was also significantly enhanced (Figures 4D and 4E). In comparison, intracellular retention of the SURF4-unfavored EPV variant started high and was only mildly affected by the SURF4 siRNA (Figures 4D and 4E). Notably, in cells not expressing DsRed2 cargoes, SURF4 knockdown also markedly reduced the number of EGFP-Rab1A-labeled t-ERGICs (Figures 4F and 4G). RUSH experiments further showed that SURF4 knockdown substantially decreased the trafficking rate of APV- but not EPV-SBP-DsRed2-ER-5 (Figures 4H and 4I). Conversely, when FLAG-SURF4 was overexpressed, the ER-to-Golgi trafficking of APV-SBP-DsRed2-ER-5 was specifically accelerated (Figures 4J and 4K).
Together, our results indicate that the biogenesis and cargo selectivity of t-ERGIC both depend on SURF4, thus explaining the peculiar “N-terminus rule” we identified for t-ERGIC-based ER-to-Golgi transport.
Co-clustering of SURF4 and Cargo Expands the ERES for t-ERGIC Biogenesis
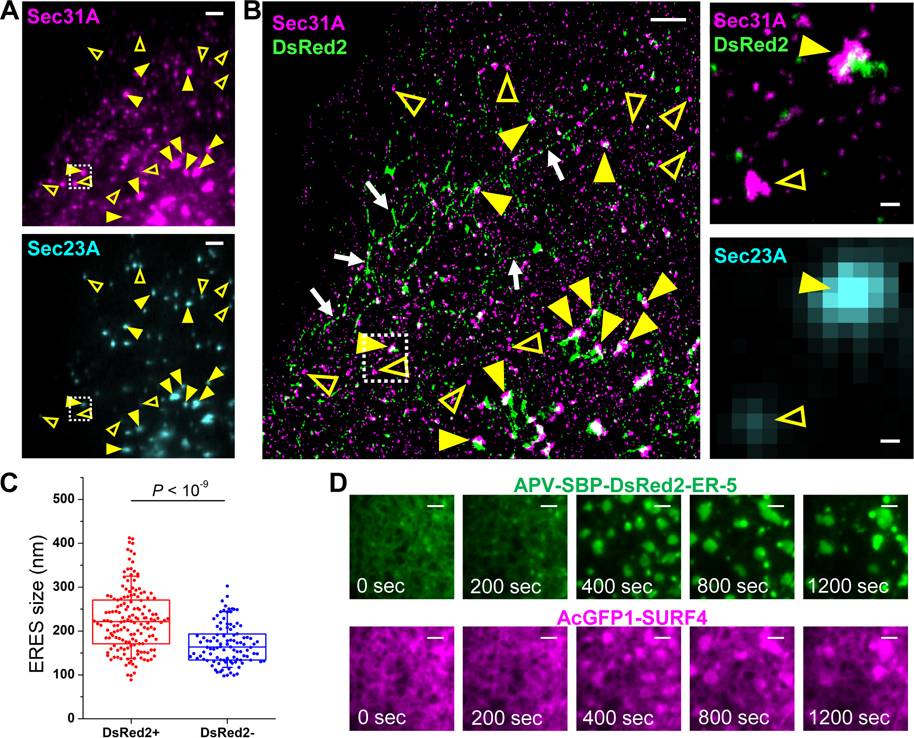
To examine how SURF4 facilitated t-ERGIC biogenesis, we utilized STORM to examine whether SURF4 cargoes were sequestered into a special ERES domain. To this end, we co-transfected cells with APV-DsRed2-ER-5 and EGFP-Sec23A and then immunolabeled Sec31A (Figure 5A). Two-color STORM of Sec31A and APV-DsRed2-ER-5 (Figure 5B) thus showed that at the ERESs, as determined by the colocalization of Sec31A and Sec23A, Sec31A structures that surrounded the APV-DsRed2-ER-5 cargo (e.g., filled arrowheads in Figure 5B) were notably larger than those not loaded with the cargo (e.g., open arrowheads in Figure 5B). As larger ERESs could provide more membrane materials for forming the long t-ERGIC tubules, this observation (statistics in Figure 5C) may explain the specificity of t-ERGIC to SURF4 cargoes. SURF4 siRNA treatment substantially reduced the occurrence of large (>~250 nm) Sec31A cups and hence removed the size difference between DsRed2-loaded and non-loaded ERESs (Figures S5A and S5B), suggesting that SURF4 is necessary for the ERES enlargement in t-ERGIC biogenesis.
Figure 5. SURF4 Co-clusters with Its Cargo to Expand the ERES.
(A) Epifluorescence of immunolabeled Sec31A (top) and EGFP-Sec23A (bottom) in a COS-7 cell co-expressing EGFP-Sec23A and APV-DsRed2-ER-5.
(B) Two-color STORM image of Sec31A and APV-DsRed2-ER-5 for the same view as (A) (left), as well as zoom-ins (right) of the boxed region of the STORM image and the EGFP-Sec23A epifluorescence image. Yellow arrowheads in (A,B) indicate examples of ERES labeled with both EGFP-Sec23A and Sec31A. Filled and open arrowheads indicate DsRed2-loaded and non-loaded ERESs, respectively. Arrows in (B) indicate t-ERGICs.
(C) Statistics of the sizes of DsRed2-loaded and non-loaded, Sec23A-positive ERESs, based on the STORM-determined sizes of the Sec31A clusters. Whiskers and boxes show 10%, 25%, 50%, 75%, and 90% quantiles. P value is calculated by a two-tailed t test. n = 5 STORM images from 2 independent replicates were quantified.
(D) Representative RUSH image sequence showing the formation and fusion of co-clustered large domains of AcGFP1-SURF4 and APV-SBP-DsRed2-ER-5. Biotin was added at time 0 for cargo release. See also Video S6.
Scale bars: 2 μm (A,B,D), 200 nm (zoom-ins of B).
See also Figure S5.
Live-cell RUSH assay provided additional insights. Upon the release of APV-SBP-DsRed2-ER-5 by biotin, we observed the co-clustering of this cargo with AcGFP1-SURF4 at the ERES, as well as their co-translocation into the t-ERGIC (Video S5). Interestingly, in a fraction (~20%) of the cells characterized by high expression levels, we further observed the co-expansion of SURF4 and cargo into large, coalescing domains (Figure 5D, Video S6). 3D-STORM of fixed cells showed that SURF4 and the cargo were both membrane-associated at the expanded clusters (Figures S5C and S5D).
Antagonism between SURF4 and KDEL Receptors Regulates the Steady-State Location of Cargo Proteins
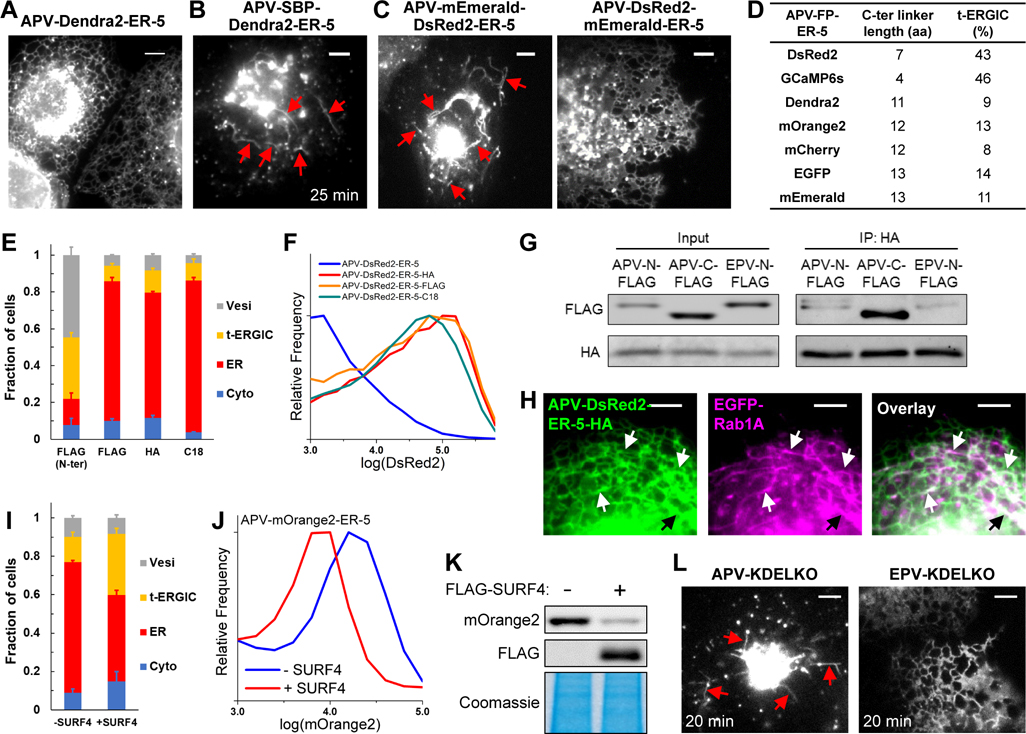
While we have elucidated how soluble proteins of different N-termini were differentially selected by SURF4 for entering the t-ERGIC secretion pathway, further experiments indicated another layer of complexity. Specifically, whereas we showed above that APV-DsRed2-ER-5 and APV-GCaMP6s-ER-5 both mainly localized to t-ERGIC tubules in the steady state, analogous constructs of other FPs, including Dendra2, mOrange2, mCherry, EGFP, and mEmerald, mainly localized to the ER and had only ~10% cells dominated by fluorescence in the t-ERGIC (Figures 6A, S6A and S6B).
Figure 6. Antagonism between SURF4 and KDELR Determines the Fates of Secretory Cargoes.
(A) Representative fluorescence micrograph of APV-Dendra2-ER-5 in COS-7 cells.
(B) Representative fluorescence micrograph of APV-SBP-Dendra2-ER-5 in RUSH, 25 min after the addition of 80 μM biotin.
(C) Representative fluorescence micrographs of APV-DsRed2-mEmerald-ER-5 and APV-mEmerald-DsRed2-ER-5 in COS-7 cells.
(D) Table summarizing the C-terminal linker lengths and the fractions of t-ERGIC-predominant cells for different APV-FP-ER-5 constructs. See Methods for detail.
(E,F) Subcellular distributions (E) and flow cytometry histograms (F) of APV-(FLAG-)DsRed2-ER-5 and its C-terminal inserted derivatives. The “FLAG (N-ter)” data in (E) duplicates “APV” in Figure S3B.
(G) Co-immunoprecipitation of APV-FLAG-DsRed2-ER-5, APV-DsRed2-ER-5-FLAG, and EPV-FLAG-DsRed2-ER-5 with KDELR3-HA.
(H) Two-color live-cell fluorescence micrographs of APV-DsRed2-ER-5-HA and EGFP-Rab1A in a co-transfected cell.
(I-K) Subcellular distribution (I), flow cytometry histograms (J), and lysate immunoblots (K) of APV-mOrange2-ER5 in COS-7 cells with or without the co-expression of FLAG-SURF4.
(L) Representative live-cell fluorescence micrographs of APV/EPV-DsRed2-ER-5-KDELKO in RUSH, 20 min after biotin-induced cargo release.
Scale bars: 5 μm. Error bars: SEM (n = 3 with ~50 cells in each replicate). Arrows indicate t-ERGICs.
See also Figures S6 and S7.
Curiously, RUSH experiments on APV-SBP-FP-ER-5 showed that upon cargo release, all FPs were efficiently trafficked to the Golgi via t-ERGIC (Figures 6B, S6D and S6E). Thus, although all FP cargoes entered the t-ERGIC pathway, other factors modulated their steady-state location. One possibility is that the cargoes were differentially retrieved back from the Golgi to the ER. As the Golgi-to-ER transport is often mediated by KDEL receptors (KDELRs), which recognize KDEL-like motifs at the cargo C-termini (Munro and Pelham, 1987; Wilson et al., 1993), we compared APV-mEmerald-DsRed2-ER-5 and APV-DsRed2-mEmerald-ER-5, which respectively had C-termini identical to that of APV-DsRed2-ER-5 and APV-mEmerald-ER-5 (Figure S6C). Remarkably, in the steady state, we found the former was often in the t-ERGIC (Figure 6C), whereas the latter was mainly in the ER (Figure 6C) and was better retained in the cell (Figure S6C).
To rationalize how the C-termini of APV-DsRed2-ER-5 and APV-mEmerald-ER-5, which both ended with KDEL, could differently interact with KDELRs, we noted that recent structural analysis indicates that the KDEL-binding pocket of the KDELR is largely buried inside the membrane (Bräuer et al., 2019). It is thus possible that the binding efficiency of KDELRs may depend on how well the C-terminus KDEL motifs are exposed. Indeed, as we examined the linker between the folded FP core and the C-terminus KDEL motif, we found that the DsRed2 and GCaMP6s constructs had much shorter linkers when compared to the other FPs (Figure 6D, Methods).
To test whether this linker length could be significant, we inserted into APV-DsRed2-ER-5 three different sequences (FLAG-tag of 8 aa, HA-tag of 9 aa, and a random 18-aa linker) between the DsRed2 C-terminus and the KDEL motif (Figure S7A). Remarkably, these constructs of extended pre-KDEL linkers were all mainly localized to the ER (Figures 6E, S7B) and showed substantially increased intracellular retention (Figure 6F). Co-immunoprecipitation showed that the C-terminally extended cargo indeed interacted with the KDELR much more strongly when compared to control constructs in which the same extension was added to the N-terminus (Figure 6G). Conversely, as we truncated 10 C-terminal residues before the KDEL motif in APV-EGFP-ER-5, increased t-ERGIC presence of the cargo was observed (Figures S7C and S7D) together with reduced intracellular retention (Figure S7E). Together, our results suggest that the efficacy of KDELR-mediated Golgi-to-ER transport, and hence the steady-state localization of cargoes, depend on how well the C-terminus KDEL motif is exposed.
Notably, although the C-terminally extended APV-DsRed2-ER-5-HA mainly localized to the ER, co-imaging with EGFP-Rab1A showed that it also populated Rab1A-positive t-ERGICs (Figure 6H), whereas EPV-DsRed2-ER-5-HA did not (Figure S7F). Moreover, as we overexpressed FLAG-SURF4 in cells expressing APV-mOrange2-ER-5, more cells were characterized by fluorescence in the t-ERGICs (Figure 6I), and the intracellular retention decreased (Figures 6J and 6K).
We also examined APV/EPV-DsRed2-ER-5 variants with the C-terminal KDEL motif removed (“KDELKO”). Whereas low intracellular retention was found in the steady state for both the APV and EPV variants, which precluded the direct visualization of DsRed2 fluorescence in live cells, immunofluorescence against DsRed2 showed the presence of the former, but not the latter, in the t-ERGIC (Figure S7G). Meanwhile, RUSH of APV/EPV-SBP-DsRed2-ER-5-KDELKO visualized the entering of the APV variant, but not the EPV variant, into t-ERGIC for accelerated anterograde transport (Figures 6L, S7H). We further note that our consistent RUSH results above on the TPV and EPV versions of CXCL9 (Figure 3L-N) were also without KDEL-like C-terminal motifs. Thus, the KDEL signal did not affect whether the cargoes utilized the t-ERGIC pathway.
Together, our results indicate that the N-terminal SURF4 signal and the C-terminal KDELR signal independently promote the anterograde and retrograde trafficking of soluble proteins, and hence antagonistically regulate the steady-state localization and retention of the cargo. Proteins with both signals may be enriched in the intermediate organelles along secretory pathways: This strategy appears to be utilized by the cell (below), while also facilitating the mechanistic investigations in this work.
DISCUSSION
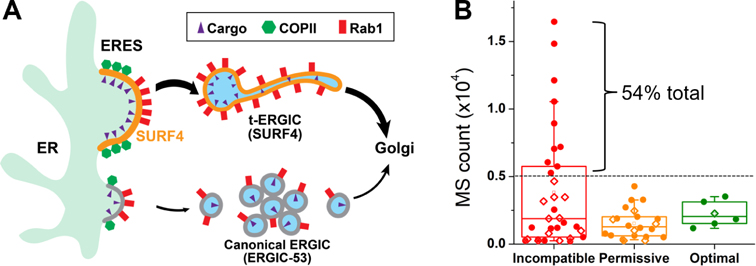
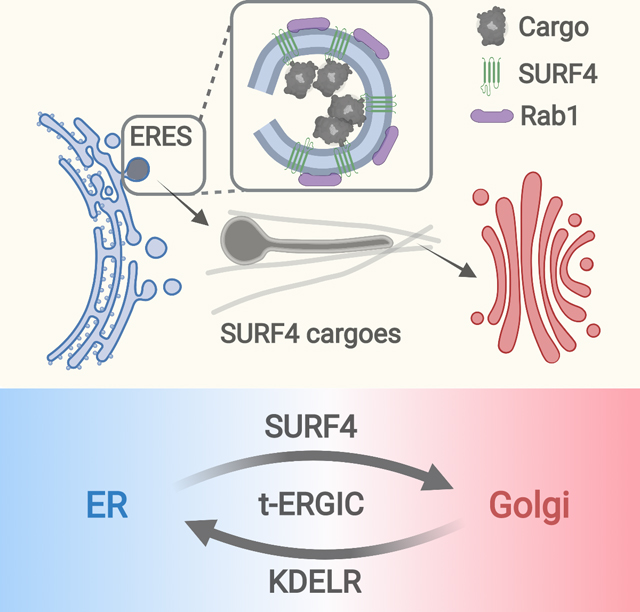
Although the molecular diversity of cargo-receptor interactions in the early secretory pathways have been extensively characterized biochemically, their cell biological consequences, including the diversity of cargo carriers and the differential transport kinetics, are less understood (Barlowe and Helenius, 2016; Dancourt and Barlowe, 2010; Gomez-Navarro and Miller, 2016). Our results showed that SURF4-cargo interactions give rise to a morphologically and functionally distinct compartment that specifically expedites the ER-to-Golgi transport of SURF4 cargoes (Figure 7A).
Figure 7. SURF4-Mediated t-ERGIC Transport Leads to Differential Protein Trafficking Rates and Steady-State Localizations.
(A) Working model of differential ER-to-Golgi trafficking via the SURF4-mediated t-ERGIC vs. the canonical ERGIC.
(B) Categorization of the mass spectrometry (MS) counts of ER-lumen proteins in the HeLa cell (Itzhak et al., 2016), based on the N-terminal tripeptide, into SURF4-incompatible (D/E/Q-containing), optimal (X-P-Y without D/E/Q), and permissive (others). Proteins with and without KDEL-like motifs (Raykhel et al., 2007) are marked by filled circles and open diamonds, respectively.
See also Table S1.
Early electron microscopy studies depict ERGICs as VTCs with ~100 nm tubular buds (Saraste and Svensson, 1991; Schweizer et al., 1988). The abundance of ERGIC-53 in the VTCs has since made it a canonical marker for ERGIC (Appenzeller-Herzog and Hauri, 2006; Hauri and Schweizer, 1992; Saraste and Marie, 2018). Although tubular carriers of >~1 μm lengths have been observed for certain cargoes (Ben-Tekaya et al., 2005; Blum et al., 2000; Marra et al., 2001; Presley et al., 1997; Sannerud et al., 2006; Shomron et al., 2021; Simpson et al., 2005; Weigel et al., 2021), it remains unclear what cargo features and/or their molecular interactions lead to these phenotypes. Of note, recent work (Shomron et al., 2021; Weigel et al., 2021) has observed tubular ER-to-Golgi intermediates in the synchronized trafficking of several membrane-protein cargoes, yet the tubule-generation mechanisms and functional implications are not addressed. In particular, in such synchronized-release experiments, the ERES expands >~2-fold in diameter due to cargo accumulation (Weigel et al., 2021), which may inadvertently contribute to tubular carrier generation.
Our results showed that for soluble cargoes, the receptor SURF4 defines an ERGIC-53-negative but Rab1-positive ERGIC domain that is morphologically distinct from VTCs, being ~10 μm long and <30 nm in diameter and traveling throughout the cell over long distances before fusing with the Golgi. As we reexamined the limited known previous examples of tubular carriers for soluble cargoes, we noticed a model cargo “lumenal GFP” that inexplicably generated long tubular carriers (Blum et al., 2000). Analysis of its sequence showed a SURF4-optimal LPV tripeptide after the signal peptide cleavage site, thus well explained by our model.
The biogenesis of the very long t-ERGIC tubules demands a large amount of membrane materials. STORM showed that in the steady state, ERESs loaded with SURF4 cargoes were considerably larger. As we depleted SURF4, the enlarged ERESs disappeared and the t-ERGIC diminished. A drop in ERES size has been previously noticed with SURF4 knockdown in Caenorhabditis elegans (Saegusa et al., 2018). With RUSH, we visualized the co-clustering of SURF4 and cargo at the ERES and further noted their co-expansion into large, coalescing domains in some cells. Whereas ERES expansion via SURF4-cargo co-clustering provides a potential mechanism for t-ERGIC generation, additional machineries await to be identified to explain the recruitment of Rab1, which we showed to enable both the ERES budding of new t-ERGICs and the ERES fusion with pre-existing t-ERGICs. The recruitment of ERGIC to ERES has been discussed in ER-export models for bulky cargoes such as procollagens (Lujan et al., 2021; Raote and Malhotra, 2021; Santos et al., 2015). In our system, as Rab1 is present on both membranes, the t-ERGIC-ERES fusion may be considered homotypic.
Though the SURF4-mediated transport is known to be substantially faster than the bulk flow (Belden and Barlowe, 2001; Casler et al., 2019, 2020; Dancourt and Barlowe, 2010; Emmer et al., 2018; Malkus et al., 2002; Otte and Barlowe, 2004; Saegusa et al., 2018; Yin et al., 2018), our results unveiled that SURF4 establishes a distinct ERGIC form for this process. By virtue of its en bloc cargo packaging, high moving speed, and fast recycling capability, the t-ERGIC provides an efficient trafficking pathway. With its extremely elongated shape and hence high surface-to-volume ratio, the t-ERGIC may be particularly efficient for the transport of receptor-bound cargoes at the membrane while minimizing the nonspecific trafficking of other soluble proteins in the lumen (Saraste and Marie, 2018).
While our RUSH results showed that SURF4 cargoes consistently entered the t-ERGICs for rapid ER-to-Golgi transport, in the steady state some cargoes localized more strongly to the ER, even though Rab1 co-labeling showed that they entered t-ERGICs. Whereas KDELRs provide a well-studied mechanism for retrograde trafficking (Gomez-Navarro and Miller, 2016), we unveiled an interesting effect, in which the C-terminal KDEL motif was less accessed by the KDELRs when closely linked to a well-folded core. Extending this linker substantially increased the KDEL-KDELR affinity, under which condition the cargo became more localized to the ER. Overexpressing SURF4 tipped this balance again and led to increased cargo localization to the t-ERGIC and decreased intracellular retention. Together, we thus showed that the N-terminus-selective, SURF4-mediated t-ERGIC fast route for ER-to-Golgi transport was counterbalanced by the C-terminal ER-retrieval signal.
For ER-resident soluble proteins, one may expect that the SURF4 signal to be negatively selected. We examined the P1’ N-terminal tripeptides for the 61 proteins annotated as “ER-lumen” in a subcellular fractionation-mass spectrometry dataset of HeLa cells (Itzhak et al., 2016). Notably, the 10 most abundant proteins, making up 54% of the total, all have SURF4-incompatible (D/E/Q-containing) N-termini together with KDEL-like C-terminal motifs (Figure 7B, Table S1). Low SURF4-binding affinity may thus have been evolutionarily selected for the abundant ER-resident proteins (Yin et al., 2018).
Intriguingly, among the 61 “ER-lumen” proteins, we also identified six candidates with SURF4-optimal P1’ N-termini (Figure 7B, Table S1). Here, we included all proteins with X-P-Y N-terminal tripeptides in which neither X nor Y was D/E/Q, which appears to be a sufficient but not necessary SURF4 signal (Casler et al., 2019, 2020; Huang et al., 2021; Wang et al., 2021; Yin et al., 2018). Although five out of these six proteins have KDEL-like ER retrieval motifs (Raykhel et al., 2007), a survey of the literature and our immunofluorescence images both indicated the substantial presence of these proteins outside the ER in the Golgi, vesicles, and the extracellular space (Honoré, 2009; Tsukumo et al., 2009; Vorum et al., 1999). Among them, immunolabeled calumenin, which is shown as a SURF4 client in a recent in vitro assay (Huang et al., 2021), as well as RCN1 and RCN3 in the same CREC protein family (Honoré, 2009), colocalized with EGFP-Rab1A-marked t-ERGIC (Figure S7I-K). The assay (Huang et al., 2021) further identifies SDF4 (Cab45), yet another member of the CREC family with both a SURF4-optimal N-terminus and a good ER-retrieval motif, as a notable SURF4 client. We found immunolabeled endogenous SDF4 exhibited little ER localization but colocalized well with the EGFP-Rab1A-marked t-ERGIC (Figure S7L). Together, these results suggest that antagonistic trafficking may be utilized by the cell to enrich proteins with both good SURF4 and KDELR signals in the intermediate organelles along secretory pathways.
In summary, by identifying t-ERGIC as a SURF4-mediated, morphologically and functionally distinct compartment that specifically expedites the ER-to-Golgi transport of SURF4 cargoes, our results argue that specific cargo-receptor interactions give rise to distinct transport carriers, which in turn regulate the ER-to-Golgi trafficking kinetics. Meanwhile, the antagonism between the N-terminal ER export and C-terminal ER retrieval signals unveiled in this work demonstrates how the cargo primary structure may be utilized to achieve exquisite, hierarchical controls of protein trafficking and localization.
Limitations of the study
In this work, we have mostly worked with FPs, in particular DsRed2, as the model cargoes, and validated results with the naturally secreted protein CXCL9. Through immunofluorescence, we further showed the localization of several endogenous SURF4 clients to the t-ERGIC. Future studies should examine additional native proteins to test generalizability. In a similar vein, we have focused on COS-7 cells to facilitate microscopy and transfection, with additional consistent results shown for U2OS and HeLa cells. It would be helpful to examine real secretory cells in future work. Finally, as we have focused on the interactions of soluble cargoes with SURF4, it remains open whether other receptor-cargo interactions may produce related or distinct ERGIC forms. Two recent studies have noted tubular ER-to-Golgi intermediates in the synchronized trafficking of several membrane-protein cargoes (Shomron et al., 2021; Weigel et al., 2021), but effects due to the substantial ERES expansion before cargo release await to be clarified, as discussed above. To what degrees these carriers are related to or dissimilar from the t-ERGIC we reported here for SURF4 clients, which we showed robustly in both synchronized and unsynchronized experiments and for immunolabeled endogenous proteins, presents a pressing question for future efforts.
STAR METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ke Xu (xuk@berkeley.edu).
Materials availability
The plasmids generated in this study have been deposited to Addgene.
Data and code availability
The raw data and custom codes for data analysis are available upon request to the Lead Contact.
Experimental model and subject details
Cell culture
COS-7, U2OS, and HeLa cells were obtained from the Cell Culture Facility at University of California Berkeley. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS, 1× GlutaMax, and 1× non-essential amino acids at 37°C, 5% CO2, and ambient oxygen. Lipofectamine 3000 was used for transient transfection according to the manufacturer’s protocol. In general, cells were plated 20–24 hr before transfection to reach 60%−70% confluency. A total of 1 μg plasmid was used for each well of a 12-well plate. A mass ratio of 4:1 was used for co-transfection of the cargo and one organelle marker, and a mass ratio of 3:1:1 was used for co-transfection of the cargo and two markers. Experiments were performed 20–24 hr post-transfection, except for RUSH, where the plasmids were expressed for 40–48 hr to ensure adequate expression.
Plasmids
pDsRed2-ER-5 was constructed from pDendra2-ER-5 by replacing Dendra2 with DsRed2 using the AgeI and Kpn2I sites. The EPV construct pEPV-DsRed2-ER-5 (A18E mutation of APV-DsRed2-ER-5) was generated by changing the alanine codon (GCA) to a glutamate codon (GAA) using the BmtI and AgeI sites of the ER-5 plasmids. N-terminal tags (SBP, FLAG) were inserted using the AgeI site, duplicating the APVAT/EPVAT linker. Other A18X mutations were generated in a similar manner. C-terminal tags (HA, FLAG, C18) were inserted using the Kpn2I site, duplicating the SG linker. Other pFP-ER-5 plasmids were generated by replacing DsRed2 with corresponding FPs. pDsRed2-ER-3 was constructed from pmEmerald-ER-3 by replacing mEmerald-KDEL with DsRed2-KDEL using the AgeI and EcoRI sites. pmEmerald-ERGIC-53 (high expression) was constructed by appending the sequence of ERGIC-53 after its signal peptide to pEPV-mEmerald-ER-5 using the Kpn2I and EcoRI sites. pAcGFP1-SURF4 was constructed by replacing Sec61β of pAcGFP1-Sec61β with PCR-amplified SURF4 cDNA from COS-7 cells using the Kpn2I and SalI sites. pStr-KDEL_APV/EPV-SBP-DsRed2-ER-5 was constructed by replacing SBP-EGFP-Ecadherin of pStr-KDEL_SBP-EGFP-Ecadherin with APV/EPV-SBP-DsRed2-ER-5 using the AscI and XbaI sites. pHaloTag-Sec23A was constructed by replacing EGFP of pEGFP-Sec23A with HaloTag using AgeI and Kpn2I sites. pStr-KDEL_TPV-CXCL9-mCherry-SBP was constructed by inserting the synthesized human CXCL9-mCherry-SBP (Twist Bioscience) in between the AscI and XbaI sites. The TPV-to-EPV mutation was generated by PCR between the intrinsic BsrGI site and the XbaI site. pStr-KDEL_APV-Dendra2/mCherry/EGFP-ER-5 was constructed by the Gibson assembly (New England BioLabs E2611) of pStr-KDEL_SBP-EGFP-Ecadherin linearized by AscI and XbaI, PCR-amplified calreticulin signal peptide-SBP tag, and PCR-amplified Dendra2/mCherry/EGFP-SGKDEL. pFLAG-SURF4 was constructed by replacing the AcGFP1 in pAcGFP1-SURF4 by the FLAG tag using the AgeI and Kpn2I sites. pAPV-DsRed2-mEmerald-ER-5 and pAPV-mEmerald-DsRed2-ER-5 were made by inserting mEmerald into pDsRed2-ER-5 using the Kpn2I site and the AgeI site, respectively. pAPV-EGFP(1–228)-ER-5 was constructed by replacing the full-length EGFP of pAPV-EGFP-ER-5 with EGFP(1–228) using the AgeI and Kpn2I sites. pAPV/EPV-DsRed2-ER-5-KDELKO and pStr-KDEL_APV/EPV-SBP-DsRed2-ER-5-KDELKO were made by removing the KDEL motif in the corresponding plasmids using the Kpn2I and EcoRI sites.
All constructed plasmids were prepared from DH5α, XL1-Blue, or Stbl3 cells using the QIAprep Spin Miniprep Kit (QIAGEN). Protein-coding sequences were verified by Sanger sequencing at UC Berkeley DNA Sequencing Facility.
Antibodies
The following secondary antibodies were conjugated in house using previously described protocol (Dempsey et al., 2011): donkey anti-mouse IgG-CF568, goat anti-mouse IgG2b-Alexa Fluor 647, goat anti-mouse IgG1-CF568, donkey anti-chicken IgY-Alexa Fluor 488, donkey anti-rat IgG-Alexa Fluor 488. These antibodies (0.3–0.4 mg/mL) were used at 1:60 dilution for immunofluorescence and 1:300 for immunoblotting.
The following antibodies and dilutions were used for immunofluorescence: rabbit anti-β-COP (1:100), rabbit anti-Rab1A (1:50), rabbit anti-Rab1B (1:40), mouse anti-DsRed (1:100), rabbit anti-FLAG (1:500), rabbit anti-Sec31A (1:200), mouse anti-calumenin (1:30), rabbit anti-SDF4 (1:100), rabbit anti-RCN1 (1:100), rabbit anti-RCN3 (1:100), rabbit anti-GFP-Alexa Fluor 647 (1:300), goat anti-rabbit-Alexa Fluor 647 (1:400), goat anti-mouse-Alexa Fluor 647 (1:400).
For immunoblotting, the following antibodies and dilutions were used: mouse anti-DsRed (1:300), mouse anti-α-tubulin (1:3000), NeutrAvidin-Alexa Fluor 647 (1:600), chicken anti-α-tubulin (1:600), mouse anti-PERK (1:300), rat anti-GRP94 (1:600), rabbit anti-GRP78 (1:1000), rabbit anti-FLAG (1:1000), mouse anti-HA (1:2000), rabbit anti-GFP-Alexa Fluor 647 (1:1500), goat anti-rabbit-Alexa Fluor647 (1:2000).
Method details
Drug treatments
Cells were transfected for 20–24 hr before the addition of the drug for the indicated time. The following chemicals were used: brefeldin A, dithiothreitol, MG132, CB-5083, thapsigargin, dimethyl sulfoxide.
Live-cell fluorescence microscopy
Cells were plated in Lab-Tek II chambered coverglass and transfected as described above. For cells transfected with HaloTag-Sec23A, 0.2 μM of JF635 HaloTag ligand was added to the cell culture medium 1 hr before imaging. After incubation at 37°C for 30 min, the cells were rinsed with the normal cell culture medium for 5 min × 6 times. Prior to imaging, 25 mM HEPES was added to the cell culture medium to maintain the pH in the ambient environment.
Live-cell fluorescence microscopy was performed on an Olympus IX73 inverted epifluorescence microscope with a water-immersion objective (Olympus, UPLSAPO60XW, NA 1.2) and a mercury lamp, or a Nikon Eclipse Ti-E inverted fluorescence microscope with an oil-immersion objective (Nikon CFI Plan Apochromat λ 100×, NA 1.45) with 488-nm, 560-nm, and 647-nm lasers modulated by an acousto-optic tunable fiber (AOTF, Gooch & Housego, 97–03151-01). Cells were imaged at 2–20 frames per second (fps) at room temperature to moderately slow down the motion of the fast-moving t-ERGIC. Concurrent multi-color imaging was achieved by modulating the AOTF to allow frame-synchronized alternating excitation at 488, 560, and 647 nm (Yan et al., 2020) with a multi-bandpass filter cube (Semrock Di01-R405/488/561/635 and Chroma ZET405/488/561/640m).
RUSH assay
Constructs for the RUSH assay (pStr-KDEL plasmids with a co-expressed streptavidin-KDEL hook) were modified from previous work (Boncompain et al., 2012). Cells were plated in 8-well Lab-Tek chambered coverglass and transfected. The cells were imaged in the cell culture medium with 25 mM HEPES on the abovementioned Nikon Ti-E microscope. 80 μM D-biotin was added to the imaging medium to release the cargo. For quantification of the ER-to-Golgi transport kinetics, images were acquired every 5 min for ~10 predetermined positions using Micro-Manager (Edelstein et al., 2014). For high temporal resolution imaging, images were acquired continuously at 2–20 fps.
Cell fixation, immunolabeling, and epifluorescence microscopy
Cells were plated in Lab-Tek II chambered coverglass or on 12-mm #1.5 coverslips in 24-well plates and transfected as described above. Cells were fixed with 3% paraformaldehyde (PFA) with 0.02%−0.1% glutaraldehyde (GA) in DPBS for 30 min at room temperature. We found that the t-ERGIC morphology was best preserved in the presence of GA, yet a high concentration of GA impeded epitope immunolabeling, especially for antibodies against β-COP and Rab1A. The sample was then reduced with 0.1% NaBH4 in DPBS for 5 min, and rinsed with DPBS for 10 min × 3 times.
For immunolabeling, the cells were blocked with the blocking buffer (3% bovine serum albumin [BSA] and 0.1% saponin dissolved in DPBS) for 1 hr at room temperature. Primary antibodies were diluted in the blocking buffer at the abovementioned ratios. Cells were incubated with the primary antibodies for 1 hr at room temperature or overnight at 4°C. Cells were then rinsed with the washing buffer (0.1× blocking buffer diluted in DPBS) for 10 min × 3 times before incubation with the secondary antibodies diluted in the blocking buffer for 1 hr at room temperature. After the secondary labeling, cells were rinsed with the washing buffer for 10 min × 3 times and finally with DPBS for 10 min.
Conventional epifluorescence microscopy was performed in DPBS on the same setups for live-cell fluorescence microscopy.
STORM super-resolution microscopy
STORM experiments were conducted as previously described (Gorur et al., 2017; Hauser et al., 2018). Briefly, the sample was immersed in a photoswitching buffer (5% D-(+)-glucose, 100 mM cysteamine, 0.8 mg/mL glucose oxidase, and 40 μg/mL catalase in 100 mM Tris-HCl pH 7.5) and mounted on a custom-built STORM microscope with a cylindrical lens for 3D-STORM (Huang et al., 2008). Single-molecule images were collected at 110 fps for 50,000–80,000 frames for the construction of each super-resolution image. For two-color STORM, Alexa Fluor 647 and CF568 were sequentially imaged as described above with the 647-nm laser and the 560-nm laser, respectively.
Flow cytometry
Flow cytometry was carried out on an Attune NxT flow cytometer (Thermo Fisher) per the manufacturer’s protocols. Transfected cells were trypsinized using TrypLE, neutralized with DMEM, and transferred to a 1.7 mL microcentrifuge tube. ~50,000 cells were analyzed. At 30%−60% transfection efficiency, non-transfected cells in the sample were thresholded by the fluorescence signal and not shown in the graphs.
Protein gel electrophoresis, Coomassie Blue staining, and immunoblotting
Cells were plated and transfected in 12-well plates. 20–24 hr after transfection, the cell culture medium was centrifuged at 2,000 g for 2 min and collected for the analysis of secreted proteins (see Immunoprecipitation [IP] and Co-IP below). Cells on the surface of the plate were then lysed in the Triton lysis buffer (1% TritonX-100, 137 mM NaCl, 50 mM Tris-HCl pH 7.5, 1× protease inhibitor cocktail, and 1× phosphatase inhibitor cocktail in water) for 30 min on ice. The lysate was then centrifuged at 16,000 g for 15 min at 4°C. The supernatant was added to 1× LDS sample buffer with 300 mM DTT and incubated for 10 min at 75°C.
Samples were run in NuPAGE Bis-Tris gels (4%−12% for small volumes of samples, 10% for larger amounts) in 1× MOPS SDS running buffer at 90 V for 1–2 hr.
For Coomassie Blue staining, the gel was rinsed in water for 5 min, incubated with 50 mL Bio-Safe Coomassie Blue Stain for 2 hr, and rinsed with water for 30 min × 3 times.
For immunoblotting, the sample in the gel was transferred to a low-fluorescence PVDF membrane in the transfer buffer (25 mM Tris base, 192 mM glycine, and 10% v/v methanol in water) at 18 V for 50–70 min at room temperature using the Mini Gel system. The membrane was blocked in the blocking buffer of 5% BSA in TBST (137 mM NaCl, 2.7 mM KCl, 19 mM Tris-HCl pH 7.5, and 0.1% v/v Tween 20 in water) for 1 hr at room temperature. Primary and dye-conjugated secondary antibodies were diluted in the blocking buffer and incubated with the membrane for 1 hr each. After each round of labeling, the membrane was washed in TBST for 10 min × 3 times. A laboratory rocker (Bellco Biotechnology 7740–10010) was used for all steps. The fluorescently labeled membrane was imaged by the Typhoon FLA 9500 scanner (GE Healthcare Life Sciences) per the manufacturer’s protocol.
Pulse-chase assay
Azidohomoalanine-based pulse-chase assay was performed according to a previous protocol (Wang et al., 2017). Cells were plated and transfected in 6-cm dishes. Pulse-chase was performed by incubating the cells in methionine/cysteine/glutamine-free DMEM with 10% dialyzed FBS, 1× GlutaMax, 0.2 mM L-cysteine, and 50 μM L-azidohomoalanine for 1 hr at 37°C. The medium was then replaced by the normal cell culture medium supplemented with 2 mM L-methionine for 0–3 hr at 37°C for the chase. The cells were then lysed in the sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 100 mM Tris-HCl pH 8.0 and 1× protease inhibitor cocktail in water) for 30 min at 4°C, and then centrifuged at 16,000 g for 15 min at 4°C.
Labeling of the incorporated AHA by biotin-alkyne was conducted using the Click-&-Go Protein Reaction Buffer Kit (Click Chemistry Tools) according to the manufacturer’s protocol. The reacted mixture was dialyzed in PBST buffer (DPBS with 0.05% v/v Tween 20) by a centrifugal filter with a 3 KDa molecular weight cut-off. The supernatant was immunoprecipitated by custom-made anti-RFP beads (see Immunoprecipitation [IP] and Co-IP below). Biotinylated proteins were detected by western blotting using NeutrAvidin-Alexa Fluor 647.
Immunoprecipitation (IP) and co-IP
IP was used to enrich target proteins from the cell culture medium and pulse-chased samples. For anti-FLAG IP of the secreted APV/EPV-FLAG-DsRed2-ER-5, 10–20 μL rat anti-FLAG magnetic beads were added to 1 mL of the centrifuged cell culture medium (above). For anti-DsRed IP of the AHA pulse-chased samples (above), rabbit anti-RFP magnetic beads were prepared by conjugating 5 μg rabbit anti-RFP antibody to 50 μL Protein G magnetic beads according to the manufacturer’s protocol, and 20–50 μL conjugated beads was used for IP.
Co-IP was performed for cell lysates from 10-cm dishes. Cells were lysed in the digitonin lysis buffer (1% digitonin, 137 mM NaCl, 10% w/v glycerol, and 1× protease inhibitor cocktail in water) with pH controlled at 7.0 (for co-IP of APV/EPV-FLAG-DsRed2-ER-5 and SURF4-HA, 17 mM Na2HPO4, 13 mM NaH2PO4) or 6.5 (for co-IP with KDELR3-HA, 18 mM Na2HPO4, 32 mM NaH2PO4) for 30 min at 4°C. The lysate was centrifuged at 16,000 g for 20 min at 4°C, and the supernatant was diluted 2× with 200 mM NaCl with 1× protease inhibitor cocktail for co-IP. The rat anti-FLAG magnetic beads and the mouse anti-HA magnetic beads were used for co-IP.
The sample-loaded beads in 1.7 mL tubes were incubated on a tube rotator (VWR 10136–084) for 3 hr at 4°C. The beads were then washed in the washing buffer (corresponding lysis buffer without digitonin and glycerol supplemented with 0.03% v/v Tween 20) on ice for 5 min × 3 times. The IP-ed proteins were eluted by 20 μL 1.5× LDS sample buffer diluted in the washing buffer for 10 min × 2 times at 80°C, with intermittent vortex mixing.
RNA interference
Silencer Select siRNA against SURF4 was from Thermo Fisher (Ambion 4427037-s13651). Scrambled Silencer Select control siRNA (GUACCAAUUCGUAAGUGUUTT; AACACUUACGAAUUGGUACTT) was synthesized by Thermo Fisher. Cells were plated in 6-well plates. siRNA transfection was conducted using Lipofectamine RNAiMAX (Invitrogen) per the manufacturer’s protocol. Cells were replated on day 3 to ~70% confluency and transfected with plasmids using Lipofectamine 3000 on day 4.
RT-PCR assay
siRNA-transfected cells were harvested on Day 5 by trypsinization. Total RNA was extracted with the RNeasy Mini Kit (QIAGEN). Reverse transcription was performed using the GoScript Reverse Transcription Kit (Promega) per the manufacturer’s protocol. 20 ng cDNA was used for PCR amplification with the iProof High-Fidelity PCR Kit (Bio-Rad) for 24 cycles. The PCR product was analyzed in 1.5% agarose gel stained by SYBR Safe and imaged by the Typhoon TLA 9500 scanner. Primers used for PCR are listed in the Key resources table.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-β-COP | Invitrogen | Cat# PA1-061 |
| Rabbit anti-Rab1A | Cell Signaling Technology | Cat# 13075 |
| Rabbit anti-Rab1B | Proteintech | Cat# 17824-1-AP |
| Mouse anti-DsRed | Santa Cruz | Cat# sc-390909 |
| Rabbit anti-FLAG | Cell Signaling Technology | Cat# 14793 |
| Rabbit anti-Sec31A | Proteintech | Cat# 17913-1-AP |
| Mouse anti-calumenin | Santa Cruz | Cat# sc-271357 |
| Mouse anti-α-tubulin | Sigma-Aldrich | Cat# T9026 |
| Chicken anti-α-tubulin | Abcam | Cat# ab89984 |
| Mouse anti-PERK | Santa Cruz | Cat# sc-377400 |
| Rat anti-GRP94 | Santa Cruz | Cat# sc-32249 |
| Rabbit anti-GRP78 | Invitrogen | Cat# PA5-29705 |
| Mouse anti-HA | Invitrogen | Cat# 26183 |
| Rabbit anti-RFP | Rockland | Cat# 600-401-379 |
| Rabbit anti-SDF4 | Sigma-Aldrich | Cat# HPA011249 |
| Rabbit anti-RCN1 | Sigma-Aldrich | Cat# HPA038474 |
| Rabbit anti-RCN3 | Sigma-Aldrich | Cat# HPA050402 |
| Rabbit anti-GFP-Alexa Fluor 647 | Invitrogen | Cat# A31852 |
| Goat anti-rabbit-Alexa Fluor 647 | Invitrogen | Cat# A21245 |
| Goat anti-mouse-Alexa Fluor 647 | Invitrogen | Cat# A21236 |
| Donkey anti-mouse IgG-CF568 | Jackson ImmunoResearch; dye conjugated in house | Cat# 715-005-151 |
| Goat anti-mouse IgG2b-Alexa Fluor 647 | Jackson ImmunoResearch; dye conjugated in house | Cat# 115-005-207 |
| Goat anti-mouse IgG1-CF568 | Jackson ImmunoResearch; dye conjugated in house | Cat# 115-005-205 |
| Donkey anti-chicken IgY-Alexa Fluor 488 | Jackson ImmunoResearch; dye conjugated in house | Cat# 703-005-155 |
| Donkey anti-rat IgG-Alexa Fluor 488 | Jackson ImmunoResearch; dye conjugated in house | Cat# 712-005-153 |
| Bacterial and virus strains | ||
| DH5α | NEB | Cat# C2987H |
| XL1-Blue | Agilent | Cat# 200249 |
| Stbl3 | Invitrogen | Cat# C737303 |
| Golgi-GFP BacMam | Invitrogen | Cat# C10592 |
| Chemicals, peptides, and recombinant proteins | ||
| CF568 NHS ester | Biotium | Cat# 92131 |
| Alexa Fluor 647 NHS ester | Invitrogen | Cat# A37573 |
| Alexa Fluor 488 NHS ester | Invitrogen | Cat# A20000 |
| NeutrAvidin | Thermo Fisher | Cat# 31000 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat# 276855 |
| Brefeldin A | Abcam | Cat# 120299 |
| Dithiothreitol (DTT) | Thermo Fisher | Cat# R0861 |
| MG132 | Tocris | Cat# 1748 |
| CB-5083 | Cayman Chemical | Cat# 19311 |
| Thapsigargin | Invitrogen | Cat# T7459 |
| JF635 HaloTag ligand | Lavis Lab | Promega Cat# GA1120 |
| D-biotin | J&K | Cat# 322564 |
| Paraformaldehyde (PFA) | Electron Microscopy Sciences | Cat# 15714 |
| Glutaraldehyde (GA) | Electron Microscopy Sciences | Cat# 16120 |
| NaBH4 | Sigma-Aldrich | Cat# 213462 |
| Bovine serum albumin (BSA) | Sigma-Aldrich | Cat# A3059 |
| Saponin | Sigma-Aldrich | Cat# S4521 |
| D-(+)-glucose | Sigma-Aldrich | Cat# G7528 |
| Cysteamine | TCI | Cat# A0648 |
| Glucose oxidase | Sigma-Aldrich | Cat# G2133 |
| Catalase | Sigma-Aldrich | Cat# C30 |
| TritonX-100 | Sigma-Aldrich | Cat# T8787 |
| NaCl | Sigma-Aldrich | Cat# S9888 |
| Protease inhibitor cocktail | Thermo Fisher | Cat# 87786 |
| Phosphatase inhibitor cocktail | Sigma-Aldrich | Cat# P0044 |
| Bio-Safe Coomassie Blue Stain | Bio-Rad | Cat# 1610786 |
| Tris base | Acros Organics | Cat# 42457-1000 |
| Glycine | Sigma-Aldrich | Cat# G8898 |
| Methanol | VWR | Cat# BDH1135 |
| KCl | Sigma-Aldrich | Cat# P9541 |
| Tween 20 | Sigma-Aldrich | Cat# P7949 |
| L-cysteine | Alfa Aesar | Cat# J63745 |
| L-azidohomoalanine (AHA) | Click Chemistry Tools | Cat# 1066 |
| L-methionine | Alfa Aesar | Cat# J61904 |
| Sodium dodecyl sulfate (SDS) | Sigma-Aldrich | Cat# L6026 |
| Biotin-alkyne | Click Chemistry Tools | Cat# 1266 |
| Digitonin | Sigma-Aldrich | Cat# D141 |
| Glycerol | Alfa Aesar | Cat# 38988 |
| Na2HPO4 | Macron Fine Chemicals | Cat# 7917-04 |
| NaH2PO4 | Fisher Chemical | Cat# S369 |
| Agarose | Lonza | Cat# 50002 |
| SYBR Safe | Invitrogen | Cat# S33102 |
| Critical commercial assays | ||
| Lipofectamine 3000 | Invitrogen | Cat# L3000008 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat# 27106 |
| Click-&-Go Protein Reaction Buffer kit | Click Chemistry Tools | Cat# 1262 |
| Lipofectamine RNAiMAX | Invitrogen | Cat# 13778 |
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| GoScript Reverse Transcription Kit | Promega | Cat# A50001 |
| iProof High-Fidelity PCR Kit | Bio-Rad | Cat# 1725330 |
| Experimental models: Cell lines | ||
| COS-7 | UC Berkeley Cell Culture Facility | ATCC CRL-1651 |
| U2OS | UC Berkeley Cell Culture Facility | ATCC HTB-96 |
| HeLa | UC Berkeley Cell Culture Facility | ATCC CCL-2 |
| Oligonucleotides | ||
| Silencer Select siRNA against SURF4 | Thermo Fisher | Ambion 4427037-s13651 |
| Select control siRNA | Thermo Fisher | GUACCAAUUCGUAAGUGUU(TT) |
| SURF4 RT-PCR F | This work | CTGCTCCTAGCAGAATCCC |
| SURF4 RT-PCR R | This work | TGCATGGGCTTGTAGACTG |
| GAPDH RT-PCR F | This work | CATCACCATCTTCCAGGAGC |
| GAPDH RT-PCR R | This work | GGATGATGTTCTGGAGAGCC |
| Recombinant DNA | ||
| pDendra2-ER-5 | Addgene | 57716 |
| pmEmerald-ER-3 | Addgene | 54082 |
| pAcGFP1-Sec61β | Addgene | 15108 |
| pRTN4A-AcGFP1 | Addgene | 61807 |
| pEGFP-ERGIC-53 (low expression) | Addgene | 38270 |
| pEGFP-Rab1A | Addgene | 49467 |
| pEGFP-Rab5B | Addgene | 61802 |
| pEGFP-Rab11A | Addgene | 12674 |
| pEGFP-Rab7A | Addgene | 12605 |
| pClover-LAMP1 | Addgene | 56528 |
| pEGFP-p62 | Addgene | 38277 |
| pEGFP-Sec23A | Addgene | 66609 |
| pStr-KDE L_S BP-EGFP-Ecadherin | Addgene | 65286 |
| pcDNA3-FLAG-Rab1A-N124I | Addgene | 46778 |
| pTol2-elavl3-H2B-GCaMP6s | Addgene | 59530 |
| pTwist-CMV BetaGlobin-KDELR3-HA (HA tag inserted between E143 and A144 of human KDELR3) | Twist Bioscience | This work |
| pTwist-CMV BetaGlobin-SURF4-HA (HA tag inserted between D263 and K265 of human SURF4) | Twist Bioscience | This work |
| pDsRed2-ER-5 (pAPV-DsRed2-ER-5) | Generated here | This work |
| pEPV-DsRed2-ER-5 | Generated here | This work |
| pDsRed2-ER-3 | Generated here | This work |
| pAPV-SBP-DsRed2-ER-5 | Generated here | This work |
| pEPV-SBP-DsRed2-ER-5 | Generated here | This work |
| pmEmerald-ERGIC-53 (high expression) | Generated here | This work |
| pAcGFP1-SURF4 | Generated here | This work |
| pFLAG-SURF4 | Generated here | This work |
| pStr-KDEL_APV-SBP-DsRed2-ER-5 | Generated here | This work |
| pStr-KDEL_EPV-SBP-DsRed2-ER-5 | Generated here | This work |
| pHaloTag-Sec23A | Generated here | This work |
| pStr-KDEL_TPV-CXCL9-mCherry-SBP | Generated here | This work |
| pStr-KDEL_EPV-CXCL9-mCherry-SBP | Generated here | This work |
| pAPV-DsRed2-mEmerald-ER-5 | Generated here | This work |
| pAPV-mEmerald-DsRed2-ER-5 | Generated here | This work |
| pAPV-EGFP(1-228)-ER-5 | Generated here | This work |
| pAPV -DsRed2-ER-5-KDELKO | Generated here | This work |
| pEPV -DsRed2-ER-5-KDELKO | Generated here | This work |
| Software and algorithms | ||
| ImageJ (Fiji) | NIH | https://imagej.net/software/fiji/ |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html |
| Micro-Manager | (Edelstein et al., 2014) | https://micro-manager.org/ |
| Other | ||
| Dulbecco’s Modified Eagle Medium (DMEM) | Gibco | Cat# 31053-028 |
| Fetal bovine serum (FBS) | Gibco | Cat# A3160401 |
| GlutaMax | Gibco | Cat# 35050061 |
| Non-essential amino acids | Gibco | Cat# 11140050 |
| TrypLE | Gibco | Cat# 12604013 |
| 1M HEPES | Gibco | Cat# 15630080 |
| 1M Tris-HCl pH 7.5 | Corning | Cat# 46-030-CM |
| 1M Tris-HCl pH 8.0 | Corning | Cat# 46-031-CM |
| DPBS | Corning | Cat# 21-030-CV |
| Methionine/cysteine/glutamine-free DMEM | Gibco | Cat# 21013024 |
| Dialyzed FBS | Gibco | Cat# A3382001 |
| 6-well plates | Corning | Cat# 3516 |
| 12-well plate | Corning | Cat# 3513 |
| 6-cm dishes | Falcon | Cat# 353004 |
| 10-cm dishes | Corning | Cat# 430167 |
| Lab-Tek II chambered coverglass | Thermo Fisher | Cat# 155409 |
| Acousto-optic tunable fiber (AOTF) | Gooch & Housego | Cat# 97-03151-01 |
| NuPAGE Bis-Tris gels (4%–12%) | Invitrogen | Cat# NP0321 |
| NuPAGE Bis-Tris gels (10%) | Invitrogen | Cat# NP0315 |
| MOPS SDS running buffer | Invitrogen | Cat# NP0001 |
| LDS sample buffer | Invitrogen | Cat# NP0007 |
| Low-fluorescence PVDF membrane | Thermo Fisher | Cat# 22860 |
| Mini Gel system | Invitrogen | Cat# NW2000 |
| Centrifugal filters (3 KDa molecular weight cut-off) | Millipore | Cat# UFC500324 |
| Rat anti-FLAG magnetic beads | Thermo Fisher | Cat# A36797 |
| Mouse anti-HA magnetic beads | Thermo Fisher | Cat# 88836 |
| Protein G magnetic beads | Invitrogen | Cat# 10003D |
Estimation of C-terminal linker lengths of FPs
Crystal structures of relevant FPs were identified in the Protein Data Bank (PDB). We defined the free C-terminal linker as the C-terminal tail extending from the folded core of the protein in the structure, plus the unresolved sequence of the FP and the “SG” linker in the ER-5 constructs. The number of amino acid residues was counted as the length of the C-terminal linker. PDB structures used: 1G7K (DsRed, for DsRed2), 3WLC (GCaMP6m, for GCaMP6s), 2VZX (Dendra2, for Dendra2), 2H5Q (mCherry, for mOrange2 and mCherry), and 2Y0G (EGFP, for EGFP and mEmerald).
Quantification and statistical analysis
Single-particle tracking analysis
The time-sorted image sequence was imported into Fiji (Schindelin et al., 2012) and analyzed through the TrackMate plugin (Tinevez et al., 2017) with the LoG detector and the simple LAP tracker. For t-ERGICs, the enlarged bodies were tracked as single particles, and the tubules were not separately tracked. The tracked trajectories were outputted to MATLAB for plotting into scaled displacements of trajectories. The Golgi territory was manually defined.
RUSH analysis
To analyze the ER-to-Golgi trafficking rate, the Golgi area and the ER area were manually defined in the image before biotin addition using Fiji. The background-subtracted intensities in the two regions were plotted as a function of time and normalized to the initial values. Golgi peak time was defined as the time corresponding to the highest intensity in the Golgi region. ER intensity decay t0.75 was defined as the time when the ER intensity dropped to 0.75 of the initial value. For the very slow ER intensity decay of EPV-DsRed2-ER-5 samples, extrapolation was used to estimate the t0.75.
ERES size measurement
STORM single-molecule coordinates of each Sec31A cluster were plotted in the imaging plane, whose long and short axes were defined by a principal direction algorithm as described (Yan et al., 2020). Distributions along the long axis and the short axis were respectively fitted by Gaussian curves. The average of the FWHMs of the two Gaussians was taken as the estimated size of the ERES.
Statistical analysis
The sample size was not predetermined by statistical methods. Fluorescence images and immunoblots were representative of at least three biological replicates. The sample size and significance test of statistical analyses were indicated in the figure legends. The value of “n” corresponds to the number of biological replicates. A P value lower than 0.05 was considered statistically significant. Graphs were generated using Origin 8.5 (OriginLab).
Supplementary Material
Video S1. DsRed2-ER-5 in fast-moving tubular organelles (TOs) in a COS-7 cell, related to Figure 1C. Time 117.0 sec in the video corresponds to “0 sec” in Figure 1C. Cyan arrows point to the Golgi. Green arrows mark examples of TO fusion with the Golgi. Playback speed is 3× real time.
Video S2. Two-color live-cell imaging of the TOs versus microtubules and the ER in the COS-7 cell, related to Figure 1. Top: GCaMP6s-ER-5-containing TOs (green) moving along microtubules labeled by 2xmCherry-EMTB (magenta). Middle: DsRed2-ER-5-containing TOs (green) moving independently of the ER marker AcGFP1-Sec61β (magenta). Bottom: DsRed2-ER-5-containing TOs (green) moving independently of the ER marker Rtn4a-AcGFP1 (magenta). Playback speed is 8× real time.
Video S3. Live-cell imaging for RUSH of SBP-DsRed2-ER-5 in COS-7 cells, related to Figure 2. Left: Same data as presented in Figure 2D. Right: data from another run. 80 μM biotin was added at time 0. The fluorescent cargo was redistributed from the ER to the Golgi via the t-ERGIC. Green arrows mark examples of TO fusion with the Golgi. Timestamp: min:sec. Playback speed is 8× real time.
Video S4. Three-color live-cell imaging for RUSH of APV-SBP-DsRed2-ER-5 (yellow) with the co-transfection of EGFP-Rab1A (magenta) and HaloTag-Sec23A (labeled by JF635, cyan) in a COS-7 cell, related to Figure 2. 80 μM biotin was added at −20 min. White arrowheads point to the same ERESs as in Figure 2G and Figure 2I for the two t-ERGIC biogenesis modes. Timestamps of 118.5 s and 717 s in the video correspond to the “0 sec” time points in Figure 2G and Figure 2I, respectively. Playback speed is 9× real time.
Video S5. RUSH of APV-SBP-DsRed2-ER-5 (yellow) with co-transfection of AcGFP1-SURF4 (magenta) and HaloTag-Sec23A (labeled by JF635, cyan) in a COS-7 cell, related to Figure 5. 80 μM biotin was added at time 0. Timestamp: min:sec. Playback speed is 9× real time.
Video S6. Co-clustering of APV-SBP-DsRed2-ER-5 (green) and AcGFP1-SURF4 (magenta) into gradually expanding domains upon cargo release in RUSH in a COS-7 cell, related to Figure 5D. 80 μM biotin was added at time 0. Timestamp: min:sec. Playback speed is 12× real time.
Highlights.
SURF4 cargoes are selectively exported from the ER into an elongated tubular ERGIC
Tubular ERGIC specifically accelerates the ER-to-Golgi trafficking of SURF4 cargoes
SURF4-dependent expansion of the ERES induces the biogenesis of tubular ERGIC
Competitive SURF4 export and KDELR retrieval signals determine cargo localization
ACKNOWLEDGEMENTS
We thank Dr. Mary West of QB3 Cell and Tissue Analysis Facility for technical support with flow cytometry. We acknowledge support from the National Institute of General Medical Sciences of the National Institutes of Health (DP2GM132681). K.X. is a Chan Zuckerberg Biohub investigator and acknowledges additional support by the Packard Fellowships for Science and Engineering.
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Appenzeller-Herzog C, and Hauri HP (2006). The ER-Golgi intermediate compartment (ERGIC): In search of its identity and function. J. Cell Sci. 119, 2173–2183. [DOI] [PubMed] [Google Scholar]
- Bannykh SI, Rowe T, and Balch WE (1996). The organization of endoplasmic reticulum export complexes. J. Cell Biol. 135, 19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, and Helenius A (2016). Cargo capture and bulk flow in the early secretory pathway. Annu. Rev. Cell Dev. Biol. 32, 197–222. [DOI] [PubMed] [Google Scholar]
- Belden WJ, and Barlowe C (2001). Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science 294, 1528–1531. [DOI] [PubMed] [Google Scholar]
- Ben-Tekaya H, Miura K, Pepperkok R, and Hauri H-P (2005). Live imaging of bidirectional traffic from the ERGIC. J. Cell Sci. 118, 357–367. [DOI] [PubMed] [Google Scholar]
- Blum R, Stephens DJ, and Schulz I (2000). Lumenal targeted GFP, used as a marker of soluble cargo, visualises rapid ERGIC to Golgi traffic by a tubulo-vesicular network. J. Cell Sci. 113, 3151–3159. [DOI] [PubMed] [Google Scholar]
- Boncompain G, Divoux S, Gareil N, De Forges H, Lescure A, Latreche L, Mercanti V, Jollivet F, Raposo G, and Perez F (2012). Synchronization of secretory protein traffic in populations of cells. Nat. Methods 9, 493–498. [DOI] [PubMed] [Google Scholar]
- Brandizzi F, and Barlowe C (2013). Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuer P, Parker JL, Gerondopoulos A, Zimmermann I, Seeger MA, Barr FA, and Newstead S (2019). Structural basis for pH-dependent retrieval of ER proteins from the Golgi by the KDEL receptor. Science 363, 1103–1107. [DOI] [PubMed] [Google Scholar]
- Casler JC, Papanikou E, Barrero JJ, and Glick BS (2019). Maturation-driven transport and AP-1–dependent recycling of a secretory cargo in the Golgi. J. Cell Biol. 218, 1582–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casler JC, Zajac AL, Valbuena FM, Sparvoli D, Jeyifous O, Turkewitz AP, Horne-Badovinac S, Green WN, and Glick BS (2020). ESCargo: a regulatable fluorescent secretory cargo for diverse model organisms. Mol. Biol. Cell 31, 2892–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancourt J, and Barlowe C (2010). Protein sorting receptors in the early secretory pathway. Annu. Rev. Biochem. 79, 777–802. [DOI] [PubMed] [Google Scholar]
- Dempsey GT, Vaughan JC, Chen KH, Bates M, and Zhuang X (2011). Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 8, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, and Stuurman N (2014). Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmer BT, Hesketh GG, Kotnik E, Tang VT, Lascuna PJ, Xiang J, Gingras AC, Chen XW, and Ginsburg D (2018). The cargo receptor SURF4 promotes the efficient cellular secretion of PCSK9. Elife 7, e38839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Navarro N, and Miller E (2016). Protein sorting at the ER-Golgi interface. J. Cell Biol. 215, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorur A, Yuan L, Kenny SJ, Baba S, Xu K, and Schekman R (2017). COPII-coated membranes function as transport carriers of intracellular procollagen I. J. Cell Biol. 216, 1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri HP, and Schweizer A (1992). The endoplasmic reticulum-Golgi intermediate compartment. Curr. Opin. Cell Biol. 4, 600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M, Yan R, Li W, Repina NA, Schaffer DV, and Xu K (2018). The spectrin-actin-based periodic cytoskeleton as a conserved nanoscale scaffold and ruler of the neural stem cell lineage. Cell Rep. 24, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honoré B (2009). The rapidly expanding CREC protein family: Members, localization, function, and role in disease. Bioessays 31, 262–277. [DOI] [PubMed] [Google Scholar]
- House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL, Todd KL, Henderson MA, Giuffrida L, Petley EV, et al. (2020). Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin. Cancer Res. 26, 487–504. [DOI] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, and Zhuang X (2008). Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yin H, Li B, Wu Q, Liu Y, Poljak K, Maldutyte J, Tang X, Wang M, Wu Z, et al. (2021). An in vitro vesicle formation assay reveals cargo clients and factors that mediate vesicular trafficking. Proc. Natl. Acad. Sci. 118, e2101287118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhak DN, Tyanova S, Cox J, and Borner GHH (2016). Global, quantitative and dynamic mapping of protein subcellular localization. Elife 5, e16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J, Schweizer A, Clausen H, Tang BL, Hong W, Oorschot V, and Hauri HP (1998). The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J. Cell Sci. 111, 3411–3425. [DOI] [PubMed] [Google Scholar]
- Kurokawa K, and Nakano A (2019). The ER exit sites are specialized ER zones for the transport of cargo proteins from the ER to the Golgi apparatus. J. Biochem. 165, 109–114. [DOI] [PubMed] [Google Scholar]
- Lee MCS, Miller EA, Goldberg J, Orci L, and Schekman R (2004). Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20, 87–123. [DOI] [PubMed] [Google Scholar]
- Lujan P, Angulo-Capel J, Chabanon M, and Campelo F (2021). Interorganelle communication and membrane shaping in the early secretory pathway. Curr. Opin. Cell Biol. 71, 95–102. [DOI] [PubMed] [Google Scholar]
- Malkus P, Jiang F, and Schekman R (2002). Concentrative sorting of secretory cargo proteins into COPII-coated vesicles. J. Cell Biol. 159, 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P, Maffucci T, Daniele T, Tullio G. Di, Ikehara Y, Chan EKL, Luini A, Beznoussenko G, Mironov A, and De Matteis MA (2001). The GM130 and GRASP65 golgi proteins cycle through and define a subdomain of the intermediate compartment. Nat. Cell Biol. 3, 1101–1113. [DOI] [PubMed] [Google Scholar]
- Mironov A, Beznoussenko GV, Trucco A, Lupetti P, Smith JD, Geerts WJC, Koster AJ, Burger KNJ, Martone ME, Deerinck TJ, et al. (2003). ER-to-Golgi carriers arise through direct en bloc protrusion and multistage maturation of specialized ER exit domains. Dev. Cell 5, 583–594. [DOI] [PubMed] [Google Scholar]
- Mitrovic S, Ben-Tekaya H, Koegler E, Gruenberg J, and Hauri H-P (2008). The cargo receptors Surf4, endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-53, and p25 are required to maintain the architecture of ERGIC and Golgi. Mol. Biol. Cell 19, 1976–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer BD, Allan BB, and Balch WE (2001). Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic 2, 268–276. [DOI] [PubMed] [Google Scholar]
- Munro S, and Pelham HRB (1987). A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907. [DOI] [PubMed] [Google Scholar]
- Otte S, and Barlowe C (2004). Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nat. Cell Biol. 6, 1189–1194. [DOI] [PubMed] [Google Scholar]
- Plutner H, Cox AD, Pind S, Khosravi-Far R, Bourne JR, Schwaninger R, Der CJ, and Balch WE (1991). Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J. Cell Biol. 115, 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJM, and Lippincott-Schwartz J (1997). ER-to-Golgi transport visualized in living cells. Nature 389, 81–85. [DOI] [PubMed] [Google Scholar]
- Raote I, and Malhotra V (2021). Tunnels for protein export from the endoplasmic reticulum. Annu. Rev. Biochem. 90, 605–630. [DOI] [PubMed] [Google Scholar]
- Raykhel I, Alanen H, Salo K, Jurvansuu J, Van DN, Latva-Ranta M, and Ruddock L (2007). A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol. 179, 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saegusa K, Sato M, Morooka N, Hara T, and Sato K (2018). SFT-4/Surf4 control ER export of soluble cargo proteins and participate in ER exit site organization. J. Cell Biol. 217, 2073–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R, Marie M, Nizak C, Dale HA, Pernet-Gallay K, Perez F, Goud B, and Saraste J (2006). Rab1 defines a novel pathway connecting the pre-Golgi intermediate compartment with the cell periphery. Mol. Biol. Cell 17, 1514–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AJM, Raote I, Scarpa M, Brouwers N, and Malhotra V (2015). TANGO1 recruits ERGIC membranes to the endoplasmic reticulum for procollagen export. Elife 4, e10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J, and Marie M (2018). Intermediate compartment (IC): from pre-Golgi vacuoles to a semi-autonomous membrane system. Histochem. Cell Biol. 150, 407–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J, and Svensson K (1991). Distribution of the intermediate elements operating in ER to Golgi transport. J. Cell Sci. 100, 415–430. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, and Schmid B (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A, Fransen JAM, Bachi T, Ginsel L, and Hauri HP (1988). Identification, by a monoclonal antibody, of a 53-kD protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J. Cell Biol. 107, 1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomron O, Nevo-Yassaf I, Aviad T, Yaffe Y, Zahavi EE, Dukhovny A, Perlson E, Brodsky I, Yeheskel A, Pasmanik-Chor M, et al. (2021). COPII collar defines the boundary between ER and ER exit site and does not coat cargo containers. J. Cell Biol. 220, e201907224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JC, Nilsson T, and Pepperkok R (2005). Biogenesis of tubular ER-to-Golgi transport intermediates. Mol. Biol. Cell 17, 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H (2009). Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525. [DOI] [PubMed] [Google Scholar]
- Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, and Eliceiri KW (2017). TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90. [DOI] [PubMed] [Google Scholar]
- Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, and Balch WE (1992). GTP-binding mutants of Rab1 and Rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the golgi complex. J. Cell Biol. 119, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga R, Zhang WU, Naseem M, Puccini A, Berger MD, Soni S, McSkane M, Baba H, and Lenz H-J (2018). CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation–a target for novel cancer therapy. Cancer Treat. Rev. 63, 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukumo Y, Tsukahara S, Saito S, Tsuruo T, and Tomida A (2009). A novel endoplasmic reticulum export signal: Proline at the +2-position from the signal peptide cleavage site. J. Biol. Chem. 284, 27500–27510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorum H, Hager H, Christensen BM, Nielsen S, and Honoré B (1999). Human calumenin localizes to the secretory pathway and is secreted to the medium. Exp. Cell Res. 248, 473–481. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang J, Lee YM, Ng S, Shi Y, Hua Z-C, Lin Q, and Shen H-M (2017). Nonradioactive quantification of autophagic protein degradation with L-azidohomoalanine labeling. Nat. Protoc. 12, 279–288. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Xu B, Huang D, Nie C, Pu L, Zajac GJM, Yan H, Zhao J, Shi F, et al. (2021). Receptor-mediated ER export of lipoproteins controls lipid homeostasis in mice and humans. Cell Metab. 33, 350–366. [DOI] [PubMed] [Google Scholar]
- Weigel AV, Chang C-L, Shtengel G, Xu CS, Hoffman DP, Freeman M, Iyer N, Aaron J, Khuon S, Bogovic J, et al. (2021). ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell 184, 2412–2429. [DOI] [PubMed] [Google Scholar]
- Westrate LM, Hoyer MJ, Nash MJ, and Voeltz GK (2020). Vesicular and uncoated Rab1-dependent cargo carriers facilitate ER to Golgi transport. J. Cell Sci. 133, jcs239814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DW, Lewis MJ, and Pelham HRB (1993). pH-dependent binding of KDEL to its receptor in vitro. J. Biol. Chem. 268, 7465–7466. [PubMed] [Google Scholar]
- Yan R, Chen K, and Xu K (2020). Probing nanoscale diffusional heterogeneities in cellular membranes through multidimensional single-molecule and super-resolution microscopy. J. Am. Chem. Soc. 142, 18866–18873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Garcia MR, Novak AJ, Saunders AM, Ank RS, Nam AS, and Fisher LW (2018). Surf4 (Erv29p) binds amino-terminal tripeptide motifs of soluble cargo proteins with different affinities, enabling prioritization of their exit from the endoplasmic reticulum. PLoS Biol. 16, e2005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Pahuja KB, Studer S, Shim S, and Schekman R (2012). COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 14, 20–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. DsRed2-ER-5 in fast-moving tubular organelles (TOs) in a COS-7 cell, related to Figure 1C. Time 117.0 sec in the video corresponds to “0 sec” in Figure 1C. Cyan arrows point to the Golgi. Green arrows mark examples of TO fusion with the Golgi. Playback speed is 3× real time.
Video S2. Two-color live-cell imaging of the TOs versus microtubules and the ER in the COS-7 cell, related to Figure 1. Top: GCaMP6s-ER-5-containing TOs (green) moving along microtubules labeled by 2xmCherry-EMTB (magenta). Middle: DsRed2-ER-5-containing TOs (green) moving independently of the ER marker AcGFP1-Sec61β (magenta). Bottom: DsRed2-ER-5-containing TOs (green) moving independently of the ER marker Rtn4a-AcGFP1 (magenta). Playback speed is 8× real time.
Video S3. Live-cell imaging for RUSH of SBP-DsRed2-ER-5 in COS-7 cells, related to Figure 2. Left: Same data as presented in Figure 2D. Right: data from another run. 80 μM biotin was added at time 0. The fluorescent cargo was redistributed from the ER to the Golgi via the t-ERGIC. Green arrows mark examples of TO fusion with the Golgi. Timestamp: min:sec. Playback speed is 8× real time.
Video S4. Three-color live-cell imaging for RUSH of APV-SBP-DsRed2-ER-5 (yellow) with the co-transfection of EGFP-Rab1A (magenta) and HaloTag-Sec23A (labeled by JF635, cyan) in a COS-7 cell, related to Figure 2. 80 μM biotin was added at −20 min. White arrowheads point to the same ERESs as in Figure 2G and Figure 2I for the two t-ERGIC biogenesis modes. Timestamps of 118.5 s and 717 s in the video correspond to the “0 sec” time points in Figure 2G and Figure 2I, respectively. Playback speed is 9× real time.
Video S5. RUSH of APV-SBP-DsRed2-ER-5 (yellow) with co-transfection of AcGFP1-SURF4 (magenta) and HaloTag-Sec23A (labeled by JF635, cyan) in a COS-7 cell, related to Figure 5. 80 μM biotin was added at time 0. Timestamp: min:sec. Playback speed is 9× real time.
Video S6. Co-clustering of APV-SBP-DsRed2-ER-5 (green) and AcGFP1-SURF4 (magenta) into gradually expanding domains upon cargo release in RUSH in a COS-7 cell, related to Figure 5D. 80 μM biotin was added at time 0. Timestamp: min:sec. Playback speed is 12× real time.
Data Availability Statement
The raw data and custom codes for data analysis are available upon request to the Lead Contact.