SUMMARY
Wounding is a trigger for both regeneration and defense in plants, but it is not clear whether the two responses are linked by common activation or regulated as trade-offs. Although plant glutamate-receptor-like proteins (GLRs) are known to mediate defense responses, here, we implicate GLRs in regeneration through dynamic changes in chromatin and transcription in reprogramming cells near wound sites. We show that genetic and pharmacological inhibition of GLR activity increases regeneration efficiency in multiple organ repair systems in Arabidopsis and maize. We show that the GLRs work through salicylic acid (SA) signaling in their effects on regeneration, and mutants in the SA receptor NPR1 are hyper-regenerative and partially resistant to GLR perturbation. These findings reveal a conserved mechanism that regulates a trade-off between defense and regeneration, and they also offer a strategy to improve regeneration in agriculture and conservation.
Graphical abstract
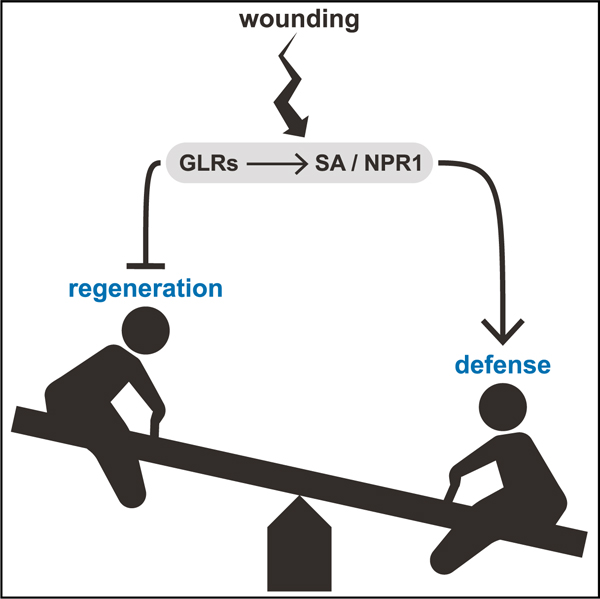
In brief
When attacked, plants initiate defense responses and can also regenerate damaged body parts. It is not known whether the two responses are linked or represent trade-offs. Hernández-Coronado et al. show that plant glutamate receptors mediate a trade-off between defense and regeneration, favoring defense responses in an apparent ‘‘bet-hedging’’ strategy.
INTRODUCTION
Plants have evolved intricate coping mechanisms torespond to biotic and abiotic insults. Two hallmarks of plant’s response to attacks are the rapid initiation of defense responses and the replacement of damaged or lost body parts by whole organ regeneration (Ikeuchi et al., 2019; León et al., 2001). The first steps in both regeneration and defense pathways depend on the stress signals instigated by wounding (Florentin et al., 2013; Savatin et al., 2014), but it is not clear whether their circuitry is wired for differential regulation or their positive activation is linked. For example, the regulation of regenerative processes could require defense responses or rely on the same signals for activation.
There is a well-developed school of thought that growth and defense are trade-offs that emerge from limited resources (Huot et al., 2014; Züst and Agrawal, 2017; Guo et al., 2018). For example, regeneration calls upon many of the same processes as meristematic growth, such as cell division and organogenesis. However, defense and regenerative responses are tightly linked in large part through hormone and calcium signaling and their rapid activation after wounding (Hander et al., 2019; Hoermayer and Friml, 2019; Moeder et al., 2019; Shanmukhan et al., 2020).
In defense responses, the plant hormone jasmonic acid (JA) is a critical signal in the response to necrotrophic pathogens, but JA also has roles in abiotic stress and development (Howe et al., 2018). The primary JA receptor is CORONATINE-INSENSITIVE 1 (COI1), a key regulator of JA signaling (Katsir et al., 2008). Jasmonate promotes an interaction between COI1 (an E3 ligase complex) and JAZ repressor proteins, degrading the repressors and permitting downstream transcriptional responses (Howe et al., 2018). In addition, JA is also a necessary signal for root regeneration in multiple contexts, promoting cell division during repair (Zhang et al., 2019; Zhou et al., 2019).
Another primary defense-response hormone is salicylic acid (SA), which has roles in defense against biotrophic pathogens, mediating both independent and overlapping responses with JA (Yang, 2015). The primary receptor for SA is NONEX-PRESSER OF PR GENES1 (NPR1), with NPR3/NPR4 paralogs acting as repressors of NPR1 (Ding et al., 2018). According to current models, NPR1 is part of a transcription complex interacting with DNA-binding transcriptional co-activators (TGAs) and the mediator/polymerase II complex with SA somehow promoting NPR1 activity to initiate broad transcriptional responses (Peng et al., 2021). Although other pathways mediate defense and regeneration (Ikeuchi et al., 2019; Marhavý et al., 2019), the prominent role of JA and SA in defense and wound responses make them good candidates to regulate interactions between defense and regeneration.
Plant glutamate receptor-like proteins (GLRs) also have a well-documented role in defense downstream of JA (Mousavi et al., 2013; Nguyen et al., 2018). GLRs are non-specific ion channels, with a permeability for calcium Michard et al. (2011), Ortiz-Ramírez et al. (2017). In defense, GLRs have been implicated in calcium signaling that activates JA-dependent systemic signaling to prepare distant organs for a potentially imminent attack (Mousavi et al., 2013; Nguyen et al., 2018). In development, GLRs have been shown to signal through calcium fluxes associated with male gamete function during fertilization (Michard et al., 2011; Ortiz-Ramírez et al., 2017; Wudick et al., 2018b). GLR mutants also show defects in defense against biotrophic pathogens, suggesting that they may also have a role in the SA pathway (Forde and Roberts, 2014). Thus, GLRs mediate some of the plant’s most rapid responses to environmental and developmental inputs. Their role in defense responses has been well established, but their role in regenerative responses remains unclear.
Exploring the regulatory wiring between defense and regeneration has practical implications for human nutrition, as many staple crops, such as potato, cassava, and yam, are grown via regenerative (clonal) propagation (Chin and Tan, 2018; Ikeuchi et al., 2019; Nishimura et al., 2005). Moreover, modern biotechnology approaches commonly employ genetic transformation and regrowth of a regenerative cell mass in plants known as callus (Ikeuchi et al., 2019; Yadava et al., 2016). However, many advanced cultivars in some of the world’s most important crops, such as maize and wheat, are recalcitrant to regeneration (Altpeter et al., 2016; Delporte et al., 2012; Ikeuchi et al., 2019). Current methods to increase regenerative capacity in crops rely on overexpression of plant stem cell/meristem initiation factors, such as Baby Boom and Wuschel (Lowe et al., 2016; Masters et al., 2020; ). However, overexpression of these early growth and meristem regulators can lead to severe developmental effects, and they are impractical for wide use in a variety of plant species. Any mechanisms that could retune the balance between defense and regeneration–preferably limited to the regenerative phases–could be used to improve regeneration for biotechnology, conservation, and propagation of staple food crops.
Here, we uncover mechanisms that regulate defense and regeneration in an antagonistic manner. The suppression of GLR function significantly increased regeneration in Arabidopsis while downregulating defense responses. Transient block experiments with chemical inhibitors of GLR signaling showed that suppression of GLR activity at the time of injury and for several additional hours improved regeneration, implying an immediate and extended role for GLRs in wound responses that mediate regeneration. Although GLRs have been shown to signal through JA, a mutant in the JA receptor, COI1, shows lower regeneration rates in most assays, suggesting the GLR mutant phenotype was not mediated by JA. On the other hand, a mutant in the SA receptor, NPR1, showed dramatically higher regeneration efficiency than wild type. We also found that native SA markers anticorrelate with regeneration efficiency, suggesting that declining regeneration efficiency with age of tissue could be due, in part, to increasing SA levels. Finally, we show that GLRs have a conserved role in maize, as the GLR antagonist CNQX (6-Cyano-7-nitroquinoxaline-2,3-dione) and an SA inhibitor were effective in increasing callus formation in leaf and embryo explants of the recalcitrant B73 maize line. Overall, we conclude that the GLRs regulate the balance between regeneration and defense in an array of contexts and different plant species, favoring defense responses. They now provide a ‘‘druggable target’’ for enhancing plant regeneration and clonal propagation.
RESULTS
Chromatin remodeling is a key step in cellular reprogramming during plant regeneration (Ikeuchi et al., 2019; Rymen et al., 2019), highlighting processes involved in the dramatic alteration of cell fates. To characterize chromatin dynamics during cellular reprogramming, we targeted cells in the microdomain of the root tip undergoing cell-fate transitions in the first 24 h post-injury in Arabidopsis. In these experiments, we used a root regeneration system in which we cut off most of the root meristem, including stem cells and cap, all of which are rapidly replaced within a day during a re-establishment of the proximo-distal (lengthwise) and radial (widthwise) axes (Figure 1A; Efroni et al., 2016; Sena et al., 2009). The expression domain of the functional marker of the root stem cell niche, MONOPTEROS (MP, MP::MP-GFP, Cole et al., 2009), is largely removed by root tip excision but then rapidly reappears in the regenerating stem cell niche region within 2 h after the cuts. We used this highly specific marker to collect reprogramming cells (hereafter, the reprogramming domain) at 2.5, 14, and 16 h after cuts via fluorescence-activated cell sorting (FACS) after rapid digestion of cell walls, followed by isolation of protoplast nuclei (Figure 1B).
Figure 1. The chromatin accessibility landscape changes rapidly in reprogramming cells.
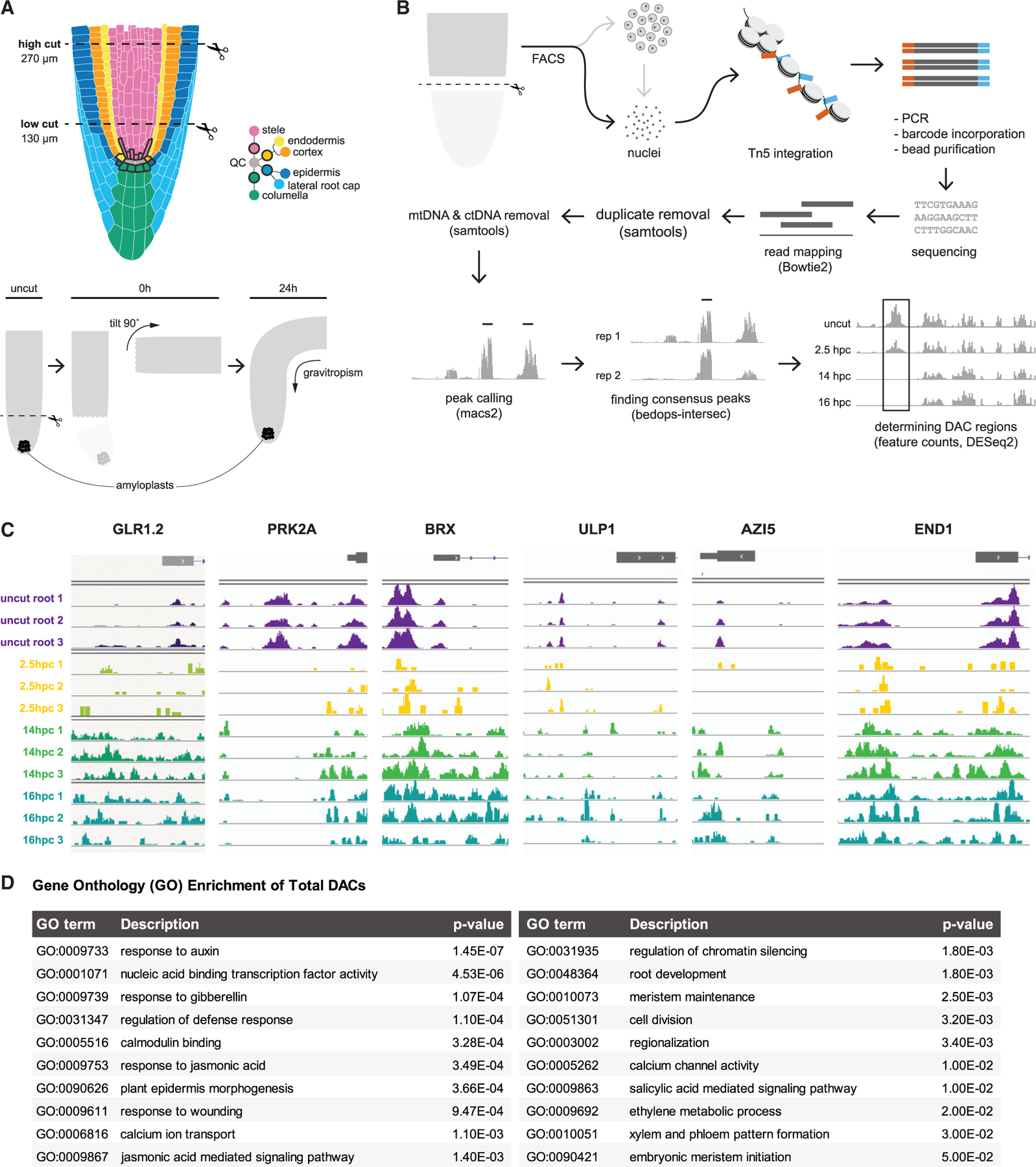
(A) Schematics of the root anatomy and the root regeneration system in which the meristem (top) is excised. Regeneration can be scored by the re-specification of gravity-sensing amyloplasts in newly formed columella cells (below), showing, from left to right, uncut and cut root followed by regeneration of the root tip.
(B) Protocol for collecting nuclei and analyzing open chromatin regions in specific regenerating cells (DAC = differentially accessible chromatin) with arrows indicating workflow.
(C) Examples of changes in chromatin accessibility of the same region before and after cutting at various time points, showing both the gain and loss of chromatin accessibility in regulatory regions and gene bodies. Samples are taken in uncut roots or different hpc (hours post cut).
(D) GO terms associated with genome-wide changes in chromatin accessibility (either opening or closing) during regeneration. GO term analysis was performed using all genes associated with DAC regions versus all genes in the genome, with pvals indicating degree of over-representation by a Fisher’s exact test.
The chromatin accessibility landscape changes rapidly in reprogramming cells
Up to 2,000 nuclei from the highly localized reprogramming domain were isolated from protoplasts and subjected to small-sample assay for transposase accessible chromatin (ATAC)-seq protocols (Buenrostro et al., 2013, 2015, STAR Methods). As a control for protoplast effects, open chromatin regions (ATAC-seq peaks) from nuclei of whole, uncut root tips were compared with nuclei taken from the same region but subjected to cell wall digestion before harvest, showing minimal differences. We then used MACS2 (Zhang et al., 2008) to call peaks and then DESeq2 (Love et al., 2014) to find regions of differential chromatin accessibility between control cell replicates and each of the post-cut time points (Figure 1B; STAR Methods).
Replicates for the 14-h cut time points had the strongest signals, showing about 750 promoter and 500 coding regions with a significant decrease in accessibility, whereas nearly 7,000 peaks in the promoter and 3,000 peaks in coding regions gained accessibility (examples in Figure 1C). A gene ontology analysis of loci associated with significantly changing ATAC peaks at 14 h revealed processes, such as auxin and wounding response (Figure 1D), providing validation that chromatin accessibility reflected categories of genes known to be upregulated in root regeneration.
Interestingly, among the highly significant gene ontology (GO) terms were calcium ion transport and calcium channel activity (p < 10‒3 and <10‒2, respectively, Figure 1D), which included genes from several GLR ion channels. In particular, the majority of the 20 annotated GLRs in Arabidopsis showed significant changes in open chromatin in their promoter coding regions, with many showing increases in accessibility at 14 h (Figure 2A). At the transcriptional level, many GLRs were upregulated during the early stages of regeneration potentially influenced by changes in chromatin accessibility (Figure 2B). For example, GLR1.2 showed a significant increase in chromatin accessibility in its promoter region at 14 h (Figure 2A) and an increase in mRNA abundance at later time points in the RNA-seq time course analysis of regenerating roots (Figure 2B). The potential involvement of the GLRs was intriguing because they could mediate early signals that trigger regenerative processes. Although chromatin accessibility was not always correlated with gene expression, we focused on GLRs that showed a trend of increased accessibility during regeneration together with either increasing expression during regeneration or high expression levels in the reprogramming zone (e.g., GLR1.2, 1.4, 2.2, 3.1, 3.2, 3.3; Figures 2A and 2B).
Figure 2. The GLR family undergoes rapid chromatin modification and transcriptional regulation.
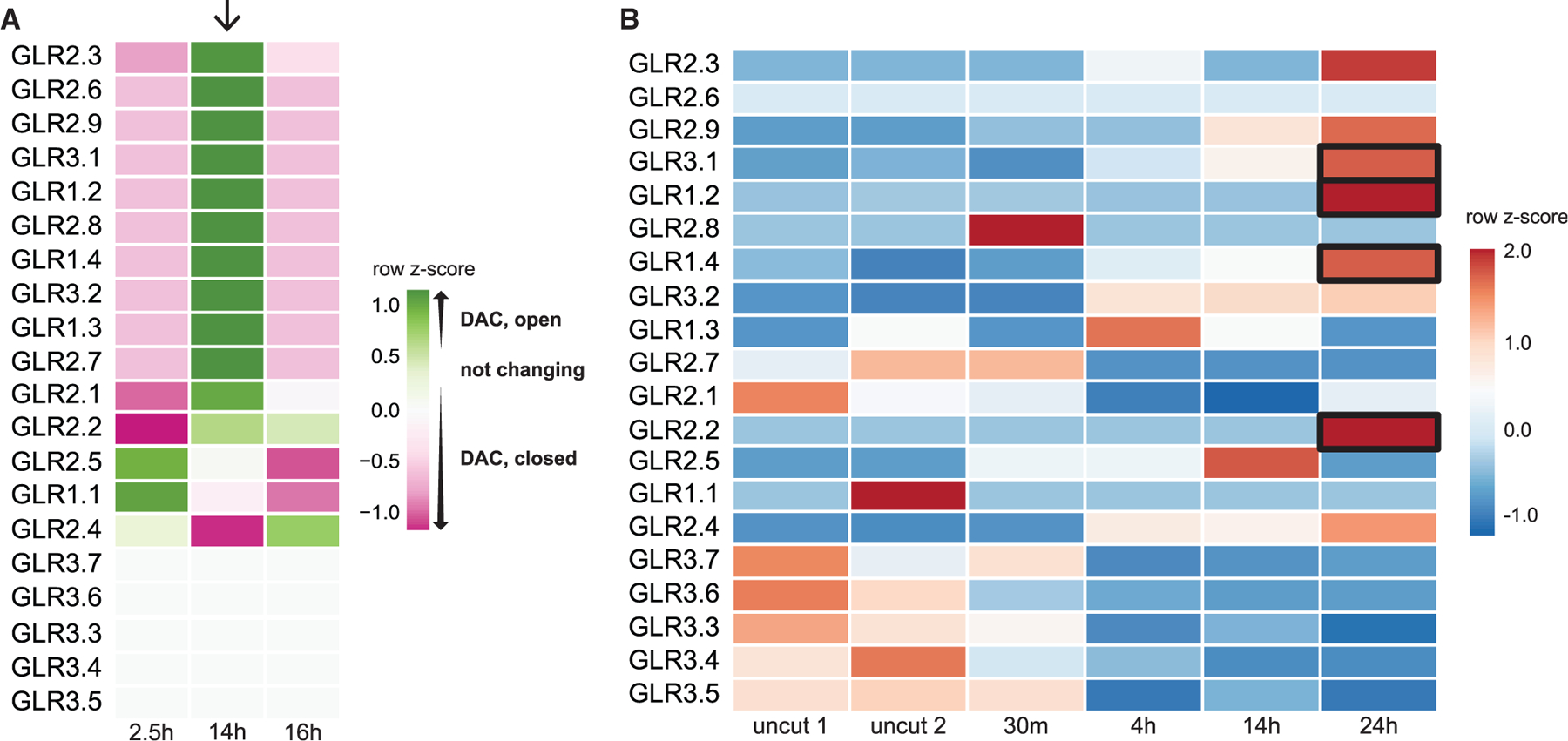
(A) Heatmap showing trends in chromatin accessibility among GLRs during regeneration at three time points representing hours post cut. Color represents the relative accessibility (per ATAC-seq analysis in Figure 1B) as a row Z score, which is standard deviations from the row mean. Arrow points to a trend of opening chromatin at 14 h among a subset of GLRs.
(B) Heatmap depicting GLR transcriptional analysis from RNA-seq profiles in a time course of uncut and regenerating root stumps at four time points after cutting (row Z score followed by log2 normalization). Boxes highlight several GLRs that increase in expression during the first 24 h of regeneration and whose chromatin becomes more accessible.
Perturbation of GLR signaling improves regeneration
We initially hypothesized that GLRs, along with their well-characterized role in defense, could mediate ion signals that trigger cellular dedifferentiation and regeneration. In the root regeneration system (hereafter root-from-stump), regeneration can be screened quantitatively by the reprogramming of vascular cells to columella cap cells, which sense gravity and quickly restore downward growth (Sena et al., 2009). However, single mutants in several candidate genes, including GLR1.2 (At5g48400), GLR1.4 (At3g07520), and GLR3.3 (At1g42540; Table S1), showed no difference in regeneration compared with wild type.
To address potential redundancy, we focused on the higher order glr1.2/1.4/2.2/3.3 mutant, which included several of our candidates and was previously shown to be severely impaired in calcium flux in pollen tubes (Wudick et al., 2018b). However, counter to expectations, the quadruple glr1.2/1.4/2.2/3.3 mutant showed twice the regeneration efficiency as wild type in cuts in the more differentiated regions of the root (270 µm from the tip, Figure 3A). In addition, other higher order GLR mutants also showed improvement in regeneration, although not as dramatic as the glr1.2/1.4/2.2/3.3 combination (Figure 3A). None of the mutants, including the quadruple combinations, showed root growth phenotypes, with root length and meristem size indistinguishable from wild-type Columbia-0 (Col-0) plants (Figures S1A and S1B), providing evidence that enhanced regeneration was not trivially due to a larger meristem before cutting. We note that none of the mutants showed a regeneration phenotype (increased or decreased regeneration) in the more competent, younger region of the meristem (within ~130 µm from the tip, Figure S2A). This age effect of GLR mutations on regeneration was a consistent phenomenon in our subsequent experiments that we pursue further on.
Figure 3. Perturbation of GLR signaling improves regeneration.
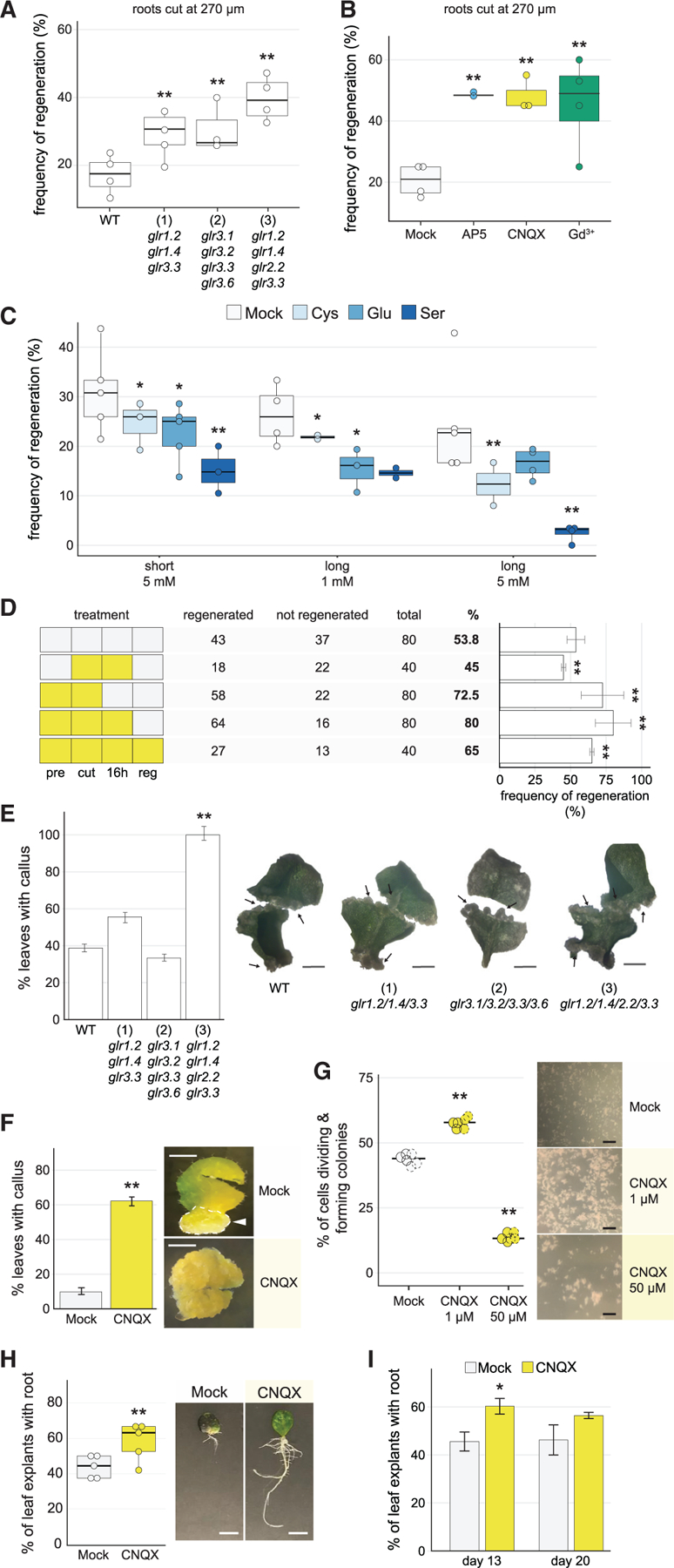
(A) Regeneration frequency of wild-type plants (Col-0) compared with triple and quadruple glr mutants: (1) glr1.2/1.4/3.3, (2) glr3.1/3.2/3.3/3.6, and (3) glr1.2/1.4/2.2/3.3 (chi-square test, Fisher’s combined probability test; n > 19 for each genotype in each of 4 trials; **p < 10–7).
(B) Constitutive post-injury treatment (cut on treatment plates and kept on treatment plates for the duration of the experiment) of cut roots on various calcium signaling antagonists, showing enhanced regeneration on all treatments. Roots were grown on normal media, cut, and transferred to treatment plates (chi-square test, Fisher’s combined probability test; n > 20 for each treatment in each of 4 trials, Gd3+ n > 12 one trial, **p < 10–11).
(C) Regeneration frequency in Col-0 plants treated with different amino acid agonists for GLRs. Regeneration frequencies were diminished in all treatments (chi-square test, Fisher’s combined probability test; n > 25 for each agonist treatment in each condition, 6 trials short treatment, 4 trials long treatment, *p < 0.02, **p < 10–6). Roots were grown on standard media, cut, and transferred to agonists. For ‘‘short’’ treatment, plants were exposed to 5 mM of L-glutamate (Glu), L-cysteine (Cys), or L-serine (Ser) for <2 min and transferred back to 0.53 MS plates to assess regeneration. For ‘‘long’’ treatment, plants were maintained on plates with treatment until regeneration was assessed.
(D) CNQX transient ‘‘window’’ treatments, showing that inhibiting GLRs at the time of injury is critical for enhancing regeneration. Pattern at left shows treatment scheme with white boxes indicating mock and yellow boxes indicating CNQX treatment. Pre, 1 h pretreatment; Cut, cutting plate; 16 h, begins immediately after cutting and ends 16 h later; Reg, begins 16 h after cutting and continues for the duration of the experiment. (Chi-square, n ≥ 40 for each treatment, **p < 0.0008).
(E) The glr quadruple mutant (3) shows enhanced callus formation, with bar graph showing percent of leaves that formed callus for each genotype, scored at 2 weeks post callus initiation. Right, representative images of callus formation in leaves from wild type, triple and quadruple GLR mutants, showing more extensive callus formation in mutants versus. Wild-type leaves that did form callus. In all cases, formation of callus occurred only in some regions of the leaf (partial). The error bars are proportional to the standard error of the pooled percentage computed using binomial distribution (chi-square test; n R 32 for each genotype; **p < 10‒9). Arrows indicate regions of callus formation. Scale bars, 0.5 cm.
(F) Callus formation in leaves treated with the GLR antagonist CNQX is enhanced, showing percent of leaves with callus. Images show roots in both conditions that formed callus. Arrow indicates region of callus formation in mock, while the treated sample is completely transformed to callus, indicating accelerated callus formation. The error bars are proportional to the standard error of the pooled percentage (chi-square test; n > 48 for each treatment; *p < 10‒29) Scale bars, 0.5 cm.
(G) Callus microcolony formation from single cells is enhanced by addition of 1-µM CNQX to the liquid media (chi-square test; n = 8.0 × 104 cells, where frequency of callus formation was estimated from hema-cytometer readings of 3 samples from each of two biological replicates and extrapolated; **p < 10‒30). At right, representative images of microcolony formation. Scale bar, 2 mm.
(H) Root-from-leaf regeneration under CNQX treatment, showing more leaves with roots in CNQX treatment. (Chi-square test; Fisher’s combined probability test, n ≥ 18 for each treatment in each of 6 trials, trial 3, n = 12; **p < 0.0005) Image shows more extensive root system of CNQX-treated leaves. Scale bars, 65 mm.
(I) Root-from-leaf regeneration under CNQX treatment assessed at 13 and 20 days. The difference in de novo root formation between treated and untreated leaves diminished over 20-day incubation (chi-square test; n = 40 for each treatment at each time point; *p < 0.03).
To explore the effect of the strong mutant combination, we examined the expression domains of the GLR using an atlas of tissue-specific expression (Li et al., 2016), observing that GLRs 1.2, 1.4, 2.2, and 3.3 had largely complementary patterns in root tissues (ground tissue, pericycle, epidermis, and vascular tissue, respectively, Figure S2B). This indicates that our mutant combination covered almost all domains of the meristem. We surmise the glr1.2/1.4/2.2/3.3 combination had an additive effect in which the regeneration phenotype was not detectable until signaling from a critical number of tissues was blocked.
To test whether the GLRs function pre or post-wounding, we took advantage of GLR channel inhibitors used in animals and plants by conditionally blocking GLR-mediated signaling after wounding (Krystal et al., 1999; Michard et al., 2011; Wudick et al., 2018a). Gadolinium (Gd3+) is a broad inhibitor of calcium channels, whereas AP5 and CNQX were originally described as antagonist of mammalian NMDA- and AMPA-type glutamate receptors, respectively (Krystal et al., 1999; Michard et al., 2011) and later found to efficiently inhibit various GLR members in Physcomitrium and Arabidopsis (Michard et al., 2011; Ortiz-Ramírez et al., 2017; Wudick et al., 2018a; Mou et al., 2020).
Using these inhibitors, plants were grown on standard plates, their meristems excised, and then transferred to plates containing calcium channel antagonists or to standard media (mock) for the remainder of the experiment. All three pharmacological treatments dramatically enhanced regeneration efficiency (Figure 3B), reaching a nearly 3-fold increase in regeneration in the challenging proximal wound cuts and exceeding the effects of the glr quadruple mutant (Figure 3A versus 3B). The fact that the glutamate receptor antagonists had the same effect as the broad calcium signaling antagonist Gd3+ suggests that GLRs–and not other calcium channels–mediate the effect on regeneration. We surmise that chemical treatments had a more dramatic effect on regeneration than mutants due to even higher order redundancy among the GLRs, many of which are expressed in the root (Figure S2B).
In addition, even brief post-injury treatment of roots with the canonical GLR agonists L-cysteine, L-serine, and L-glutamate, which has been shown to induce defense responses (Goto et al., 2020), resulted in the opposite effect, dramatically inhibiting regeneration (Figure 3C). Thus, GLR signaling strength has a direct effect on regeneration both above and below native levels.
Using the GLR inhibitor CNQX, we tested the time window in which glutamate receptor signaling affects regeneration. A 1-h pretreatment alone followed by transfer of cut roots onto mock plates improved regeneration significantly (Figure 3D), suggesting that the GLRs function, in large part, immediately after wounding. We could not observe enhancement of regeneration with a 16- or a 4-h CNQX treatment post-cutting alone without pretreatment (Figure 1D, compare rows 1 and 2, Figure S2C). However, when a pretreatment was combined with a 16-h incubation on CNQX post-cutting, regeneration increased to even higher levels than the maximal treatment (1-h pretreatment + transfer to CNQX plates for 3 days, Figure 3D). Thus, CNQX also improves regeneration in the hours after injury. Taken together, these results suggest that blocking the function of a redundant set of GLR acts immediately upon injury and continuously for several hours to suppress regeneration after wounding.
Plants can regenerate many different organs in varied developmental contexts (Ikeuchi et al., 2019; Sinnott, 1960). Thus, to ask if the GLRs function broadly in regeneration, we tested GLR mutants and chemical treatments in other regeneration systems in Arabidopsis. Many types of regeneration occur through callus, a root-like mass of cells analogous to blastema in animals, which gives rise to shoots and roots (Birnbaum and Sánchez Alvarado, 2008; Sugimoto et al., 2010, 2011). To test the role of GLR-mediated signaling in callus formation (hereafter, callus-from-leaves), early-life-stage (8 days post germination, dpg.) or later-life-stage (15 dpg.) leaves were excised at the base of the petiole, cut in half, and cultured on callus inducing media (CIM) plates for 2 weeks. Although no difference in callus formation efficiency was observed in early-stage leaves (Figure S2D), the glr1.2/1.4/2.2/3.3 mutant showed an increase in callus formation in older leaves, with more than a 2-fold higher efficiency than wild type for the quadruple mutant (Figure 3E). We again draw attention to the fact that the more dramatic effect on later-stage leaves is reminiscent of the glr quadruple mutant’s more pronounced effect on older cells in the root meristem (previous section)–a repeat of the so-called the age effect.
We repeated the callus-from-leaves experiments using chemical treatments, plating young (8 dpg) and later-stage (15 dpg) leaves on CIM plates containing CNQX or mock treatments (STAR Methods). Consistently, GLR inhibitors increased callus formation in normally older recalcitrant leaves (Figure 3F). After 15 days on CIM, untreated leaves formed callus at a frequency of 10%, whereas callus formation in CNQX-treated leaves was up to six times higher, reaching about ~60% regeneration in repeated trials (Figure 3F). In other regeneration contexts, CNQX improved the formation of microcalli from single cells (protoplasts, Figure 3G) and also improved root formation from the base of Arabidopsis leaves (hereafter, root-from-leaf, Chen et al., 2014; Figure 3H). The difference in root formation between treated and untreated leaves in the root-from-leaf system diminished over longer-term incubation (20 days, Figure 3I). However, we note that the effects of treatment were still critical for overall regeneration success in other systems (e.g., Figure 3B). Overall, the results show that GLRs inhibit the regeneration response in a broad set of regeneration contexts in Arabidopsis.
CNQX affects regeneration through the salicylic acid pathway
To examine the downstream effects of blocking GLR signaling after wounding, we generated an RNA-seq time course of control versus CNQX-treated roots after cutting (uncut [2×], 30 min, 4 h, 14 h, and 24 h). We dissected root tissues to harvest only the distal root tip or regenerating root stumps containing the reprogramming domain. We filtered the dataset for the top 10% most variable genes and performed a differential time series analysis on control versus CNQX-treated samples using ImpulseDE (Sander et al., 2017), identifying about 1,400 differentially expressed (DE) transcripts categorized into 11 different patterns (STAR Methods).
Most DE genes fell into a pattern of repression by CNQX throughout the time points as illustrated for cluster 9 (Figures 4A and S3A). These clusters were overwhelmingly represented by stress- and defense-responsive GO terms (response to abiotic stimulus p < 10‒8, response to stress p < 10‒19, response to other organisms p < 10‒7, Table S2). We then specifically examined genes annotated with a role in ‘‘defense.’’ Many of these genes were induced in mock but failed to show upregulation in the CNQX or in the glr quadruple mutant at 24 h post cut (hpc). (Figures 4B and 4C; Table S3). Thus, CNQX treatment appears to downregulate many stress responses in reprogramming cells (Figure 4B), showing that the GLRs mediate a response to root meristem damage next to the wound site.
Figure 4. A time course shows dramatic changes in stress responses with CNQX treatment.
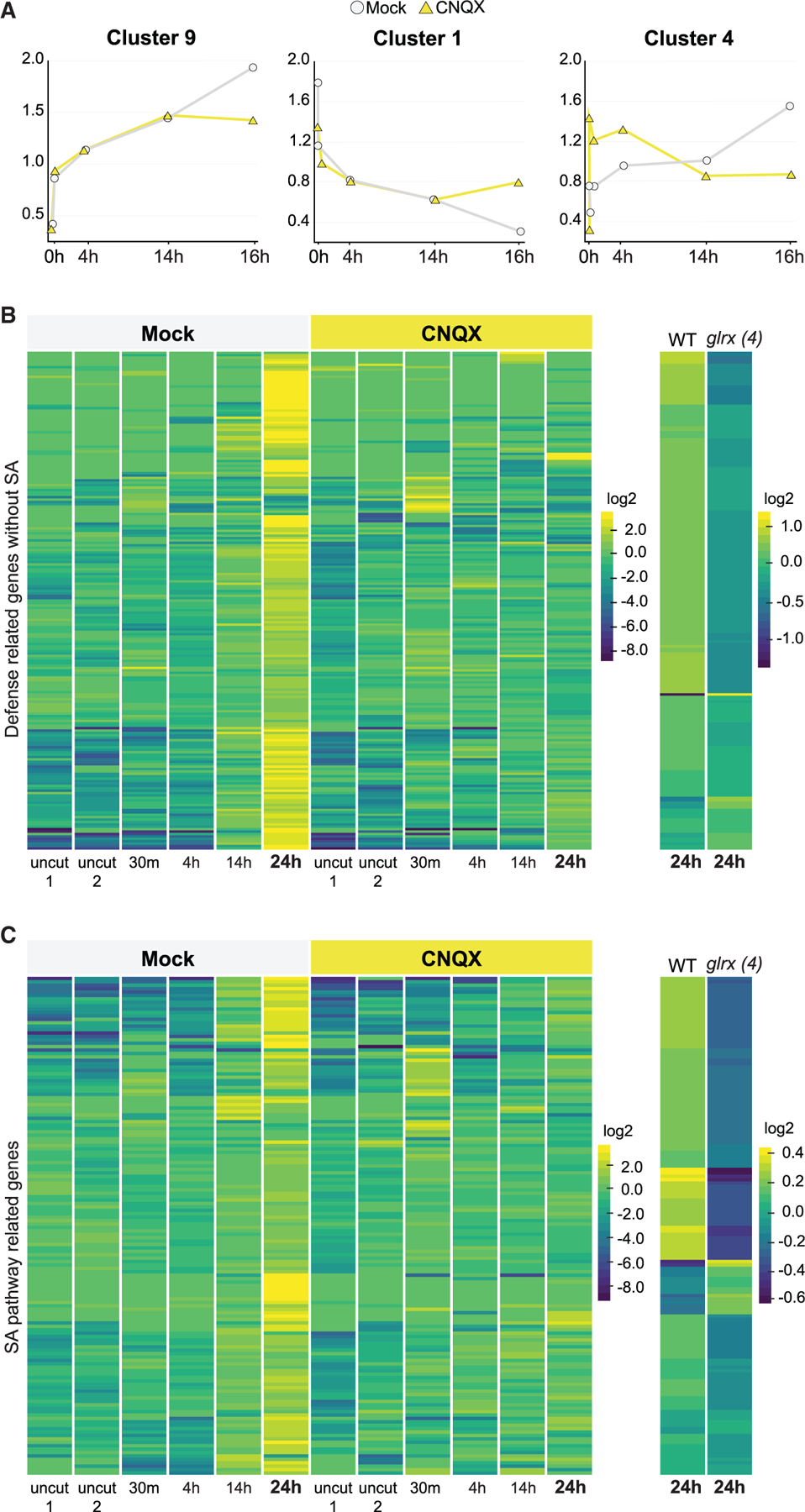
(A) Plots summarizing 3 of the 11 different patterns found among approximately 1,400 differentially regulated transcripts along a regeneration time course (0 min, 30 min, 4 h, 14 h, and 24 h), using ImpulseDE (see experimental model and subject details).
(B and C) Heatmaps of defense-annotated genes (B) and SA responsive genes (C) that are upregulated in regeneration at 24 h in untreated roots, showing a failure to upregulate the same genes in CNQX treatments and in the glrx4 mutant (glr1.2/1.4/2.2/3.3). For treatments, plants were grown on standard plates and then transferred to mock or CNQX plates (50 µM). Read counts were row normalized and log2 transformed. Col-0 versus glr1.2/1.4/2.2/3.3 mutant plants were profiled at 24 h after injury only and normalized separately. Genes in heatmaps are listed in Table S3.
Given that JA is involved in both root regeneration (Zhang et al., 2019; Zhou et al., 2019) and GLR-mediated defense (Mousavi et al., 2013; Nguyen et al., 2018), we tested a fertile hypomorph mutant of COI1 (Katsir et al., 2008). If the GLR-mediated ion signaling inhibits regeneration through JA signaling, we reasoned that loss-of-function mutants in COI1 would phenocopy GLR mutants.
However, the coi1–2 hypomorph showed lower regeneration in callus formation (Figures 5A and 5B) and root-from-leaf regeneration (Figures 5C and 5D), the latter in agreement with previous reports (Zhang et al., 2019). In the root-from-stump assay, the coi1–2 allele did not show consistent results (Figures S3B and S3C). Nonetheless, overall, the largely lower regeneration efficiency of the coi1 mutant fails to implicate JA in mediating the GLR effect on regeneration.
Figure 5. CNQX affects regeneration through the SA pathway.
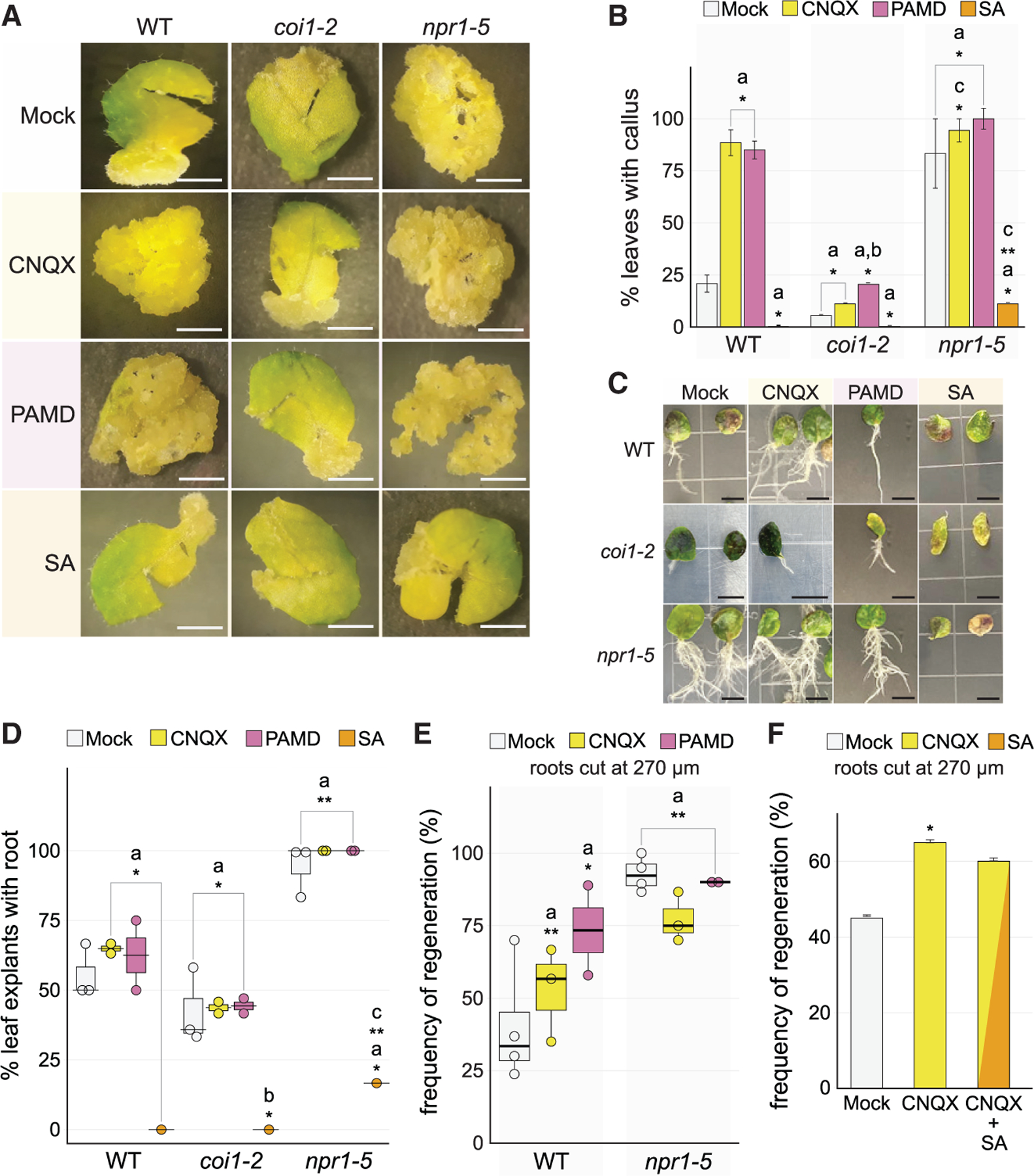
(A) Representative images of callus formation in leaves of wild type (Col-0), coi1–2, and npr1–5 on Mock, CNQX, PAMD, or SA, all under constitutive post-injury treatment. Productive callus appears as clusters of amorphous, pale (yellow) cell masses. Scale bars, 0.5 cm.
(B) Quantification of callus formation shown in (A) (chi-square test; n => 18 for each genotype in each treatment; asignificant difference with respect to Col-0 Mock, *p < 10‒6, **p < 10‒11; bsignificant difference with respect to coi1–2 Mock, *p = 0.003; csignificant difference with respect to npr1 Mock, *p < 0.04, **p < 10–9).
(C) Root-from-leaf regeneration in wild type (Col-0), coi1–2, and npr1–5 treated on Mock, CNQX, SA or PAMD under constitutive post-injury treatments. Representative images show differences in responses based on genotype and treatment. Scale bars, 65 mm.
(D) Quantification of root-from-leaf regeneration shown in (C) (chi-square test, Fisher’s combined probability test; n ≥ 12 for each treatment in each genotype in each of 3 trials; asignificant difference with respect to Col-0 Mock, *p < 0.03, **p < 10‒7; bsignificant difference with respect to coi1–2 Mock, *p < 0.02; csignificant difference with respect to npr1 Mock, *p < 10‒20).
(E) Regeneration frequency of Col-0 and npr1–5 mutant on different constitutive post-injury treatments. The error bars are proportional to the standard error of the pooled percentage computed using binomial distribution (chi-square test, Fisher’s combined probability test; n ≥ 18 for each treatment in each genotype in each of 4 trials; asignificant difference with respect to Col-0 Mock, *p < 0.05, **p < 10‒6).
(F) Root regeneration frequency under combined constitutive post-injury treatment of CNQX and SA, showing that SA addition does not fully counteract GLR blockage by CNQX (chi-square test; n = 40; *p < 0.02).
On the other hand, the RNA-seq time course showed that many SA responsive genes were upregulated in regeneration under mock conditions but failed to show that upregulation on CNQX treatment and in glr1.2/1.4/2.2/3.3 mutant (Figure 4C). In addition, we observed that SA responsive markers increased along the longitudinal axis of the root as cells matured away from the stem cell niche (Figure S4). The increase in SA response is inversely correlated with the competence of cells to regenerate along the longitudinal axis of the root (Sena et al., 2009). The observation is analogous to a previous report showing that callus formation in garlic leaf decreased as leaf tissues matured and SA levels rose (Mostafa et al., 2020). These trends provided an intriguing connection between SA and the ‘‘age effect’’ we observed in several contexts in which perturbing GLRs improved regeneration only in older tissues. This led to the hypothesis that SA mediates GLR signaling in older tissues that normally inhibits regeneration.
To test the role of SA in GLR-mediated regeneration response, we assayed several mutants of the SA signal transduction pathway for regeneration phenotypes, with the receptor, NPR1, showing the most dramatic effect (Ding et al., 2018; Figures S5A–S5D). Compared with wild type, callus formation in the npr1–5 mutant showed more than a 3-fold increase, including more complete reprogramming of explants to callus (Figures 5A and 5B). In root-from-leaf regeneration, the npr1 mutant showed more than a 2-fold increase in efficiency (Figures 5C and 5D) and, similarly, in the root-from-stump assay, npr1 roots showed an approximate 2-fold increase in regeneration in high cuts (Figure 5E). Consistently, transient treatment with the SA inhibitor 4-phenyl-2-{[3-(trifluoromethyl)-aniline]methylidene}cyclohexane-1,3-dione (PAMD) showed higher regeneration than mock-treated wild-type roots in all three regeneration systems, although PAMD has a strong inhibitory effect on long-term growth (Figures 5A–5E; Seo et al., 2012). In contrast, treatment of roots with SA greatly inhibited regeneration in all the systems tested (Figures 5A–5D, S5E, and S5F). These strong effects with transient treatments support a role for SA signaling in the inhibition of regeneration in the post-injury environment.
Examining epistatic effects between the GLR and SA pathway, we observed at least partial insensitivity of npr1 to regeneration enhancement by CNQX treatment in most assays (Figures 5A–5D). The insensitivity of npr1 to CNQX was most apparent in the more challenging regeneration assays, such as high cuts in root-from-stump regeneration (Figures S5A and S5D). In addition, although wild-type plants showed increased regeneration in SA inhibitor PAMD, the same treatment had no effect on regeneration in the glr1.2/1.4/2.2/3.3 mutant combination, showing resistance to the SA inhibitor (Figure S5E). The results are consistent with GLR perturbations already suppressing downstream SA signaling. Simultaneous treatment with CNQX and SA in wildtype plants showed only a small, non-significant decrease in root regeneration levels compared with CNQX treatment alone (Figure 5F). However, treatment of the glr1.2/1.4/2.2/3.3 mutant with SA still inhibits regeneration as it does in wild type, a partial rescue of the wild-type response by apparent restoration of downstream SA signaling (Figure S4E). These results support a role for SA downstream of GLR signaling. Together, the results suggest that the GLR-mediated effects on regeneration work in large part through the SA signaling pathway.
Blocking GLRs severely disrupts ion flux, inhibits defense, and augments repair
The involvement of the GLR calcium channels in our regeneration assays implies a role for rapid ion signaling near the injury site. To test whether GLRs mediate ion signaling in our root-from-stump assay, we used a calcium-specific vibrating probe to measure extracellular calcium fluxes in wild type and glr1.2/1.4/2.2/3.3 root stumps for several minutes after wounding. Without wounding, glr1.2/1.4/2.2/3.3 roots did not show a significant difference in calcium flux when compared with wild type (Figure S6A). Upon wounding, wild-type roots showed high Ca2+ influx at the injury site for 10 min after which the influx decreased over the recovery time (Figure 6A, gray markers and gray trend line). In contrast, the glr quadruple mutant best-fit curve showed lower Ca2+ influx immediately after wounding and a faster recovery to stable levels (Figure 6A, blue markers and blue trend line).
Figure 6. Blocking GLRs severely disrupts ion flux, inhibits defense, and augments repair.
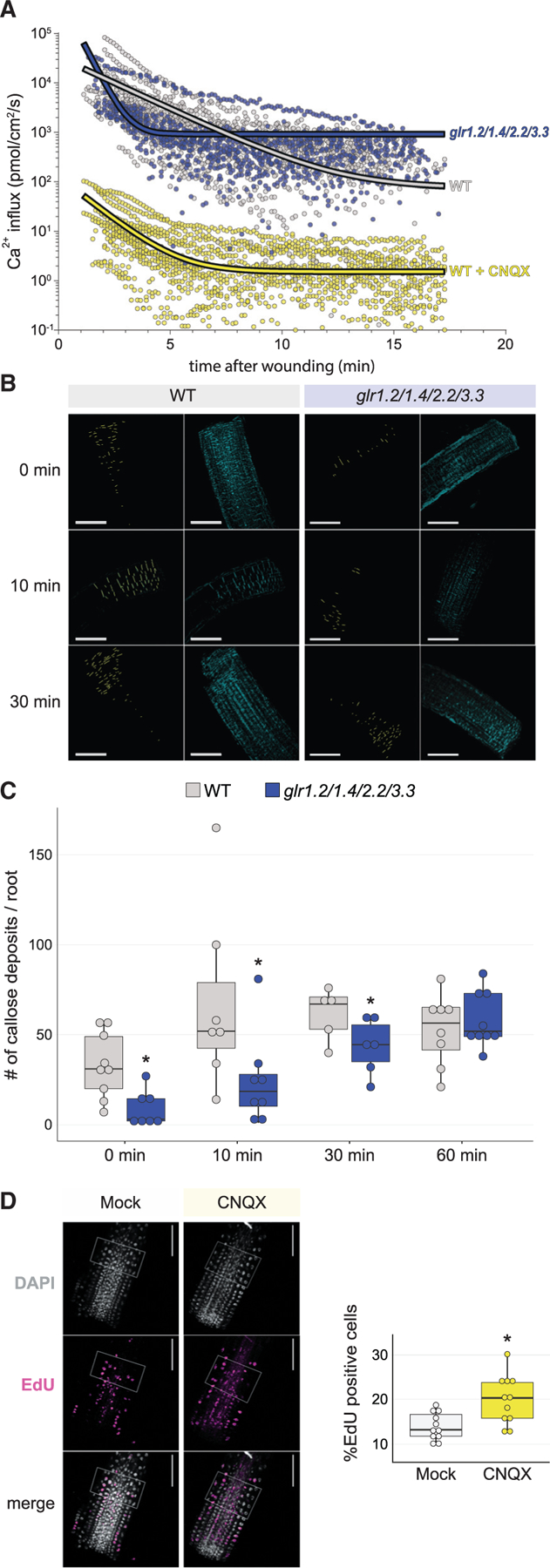
(A) Vibrating probe measurements over time after root tip cutting show an increase in the rate of positive ion (Ca2+) flux in the glr1.2/1.4.2.2/3.3 quadruple mutant compared with wild type (top), showing a severe disruption in ion flux (wild type, n = 13; glrx4, n = 11). In wild type (bottom), treatment with CNQX decreases calcium influx 4-fold overall, showing a similar pattern of calcium influx over time compared with the mutant trend above.
(B and C) Representative images (B) and quantification (C) of callose deposition in the minutes after injury, showing a decrease in callose accumulation in the glrx4 mutant compared with wild type (t test; n ≥ 5 for each genotype at each time point; *p = 0.003 for 0 min, which is effectively 3–5 min after wounding due to experimental limitations, *p = 0.035 for 10 min, *p = 0.03 for 30 min & n.s. for 60 min) with images over time. Callose deposition was analyzed using the DAPI filter (cyan) after aniline blue staining and particle analysis (e.g., yellow pixels in first and third column of B). Scale bars, 100 mm.
(D) Representative images and quantification of EdU staining for cells that have entered or undergone S phase, showing increased division rates in Col-0 roots treated with CNQX (t test; mock, n =18; CNQX, n = 16; *p = 0.003).
The steeper decline in influx over time in the mutant curve amounts to a higher recovery constant compared with wild type (K = 2.041 versus K = 0.485, p < 0.0001), exhibiting lower levels of calcium influx within the first 5 min after wounding. At recovery phase, however, the glr quadruple mutant plateaued at an influx rate one order of magnitude higher than wild type (920.2 versus 74.6 pmol/cm2/s, p = 0.0002; Figure 6A, compare fitted curves), indicating that a larger calcium influx is sustained in the mutant.
Lower calcium influx observed in the mutant immediately after injury could result from a direct contribution of the GLRs or an indirect consequence of an altered cellular physiology in the mutant. To distinguish these possibilities, we performed the same measurements on wild-type roots treated with CNQX after wounding. Upon the chemical treatment, calcium influx is severely affected decreasing by four orders of magnitude. Nonetheless, similar dynamics were observed with the lowest calcium influx occurring at 6 min after wounding (K = 0.8218) followed by sustained calcium levels in the recovery phase (Figure 6A, yellow markers and yellow trend line). The comparable dynamics of glr mutants and the transient CNQX-blocking of GLR function confirms an overall impairment in the kinetics of homeostatic re-establishment of calcium after wounding further supporting that GLR channels mediate Ca2+ influxes in response to injury.
Earlier studies of injury in soybean showed that callose synthase, which deposits callose on the cell cortex in response to injury (Ellinger and Voigt, 2014), is directly activated by a calcium flux (Kauss, 1985; Wissemeier and Horst, 1995). Using this evidence, we hypothesized that GLRs activate callose synthase in response to wounding through their calcium permeability. In this scenario, callose buildup then seals the injury helping to re-establish normal homeostasis. The model implies that perturbation of GLRs, which we showed earlier as decreasing calcium fluxes upon wounding, should prevent callose buildup.
To test the model, we measured callose buildup in cells at 0, 10, 30, and 60 min after wounding by quantitative confocal imaging of aniline blue stained roots in wild type and the glr1.2/1.4/2.2/3.3 mutant. Indeed, the glr mutants had significantly less callose deposition (Figures 6B and 6C), showing that perturbation of GLR signaling reduced a defense response in the immediate vicinity of the wound. Thus, the glr mutant’s impaired Ca2+ influx response immediately after wounding might not suffice to activate callose deposition to the same extent as in wild type. However, after 10 min, when influxes plateau, the sustained higher Ca2+ influx seems to compensate for the early low influx, and the differences in callose deposition become less evident until no significant difference was observed at 60 min (Figures 6C and S6B).
Focusing on regenerative responses, we also asked whether perturbation of GLRs lead to cellular changes that could enhance regeneration. We first examined cell division rates, a well-documented regeneration response in the root injury model (Heyman et al., 2016). To measure cell-cycle activity, we used the nucleoside analog 5-ethynyl-2′-deoxyuridine (EdU), which is incorporated into DNA during synthesis and labels cells in S phase. Quantitative measurements of EdU-stained roots showed an increase in the frequency of S-phase-labeled cells in the root stump of wild-type CNQX-treated plants where cells reprogram to reform the distal meristem, suggesting an increase in division rates (Figure 6D). In addition, a quantitative index of cell identity (ICI, Efroni et al., 2015; Efroni et al., 2016) showed that CNQX-treated cells had a more ‘‘youthful’’ stem cell-like character at 24 h after cutting compared with the mock cells (Figure S6C). Together, these results show the physiological consequences of GLR signaling near the wound site favor defense responses in wild type, such as the buildup of callose, while inhibiting regenerative responses, such as cell division and cellular reprogramming. Thus, the GLRs manage a regeneration versus defense balance in the aftermath of wounding.
GLR-mediated signaling affects regeneration in flowering plants broadly
To determine if the GLRs’ role in regeneration is conserved among plants, we took advantage of GLR antagonists to conditionally perturb GLR signaling in other species. Maize is a monocot crop species separated from Arabidopsis, a dicot, by an estimated 150 million years (Chaw et al., 2004). Due to the recalcitrance of callus formation from vegetative tissues in maize, biotechnology protocols employ a callus from embryo protocol for transformation (Green and Phillips, 1975). We cultured maize embryo cells on CIM adjusted for maize using the regeneration competent Hi-II line, which is commonly used for transformation, and the recalcitrant B73 line, which is a parental line used in advanced hybrid seed production, scoring callus formation after 15 days (STAR Methods).
Both lines showed an increase in callus formation when incubated on CNQX after embryo isolation (Figure 7A). In the recalcitrant B73 line, CNQX treatment increased the formation of productive calli (>5 mm) by 50% (Figure 7A). In addition, the productive calli were 30% larger in CNQX treatment (Figure 7B). CNQX-treated B73 maize embryos even reached similar callus formation rates of Hi-II on mock treatments (Figure 7A). Moreover, B73 embryos incubated on CNQX had smaller, cytoplasmically clear cells that typically give rise to regenerative organs (Figures 7B and S7A).
Figure 7. Blocking calcium channels improves regeneration in the maize recalcitrant line B73.
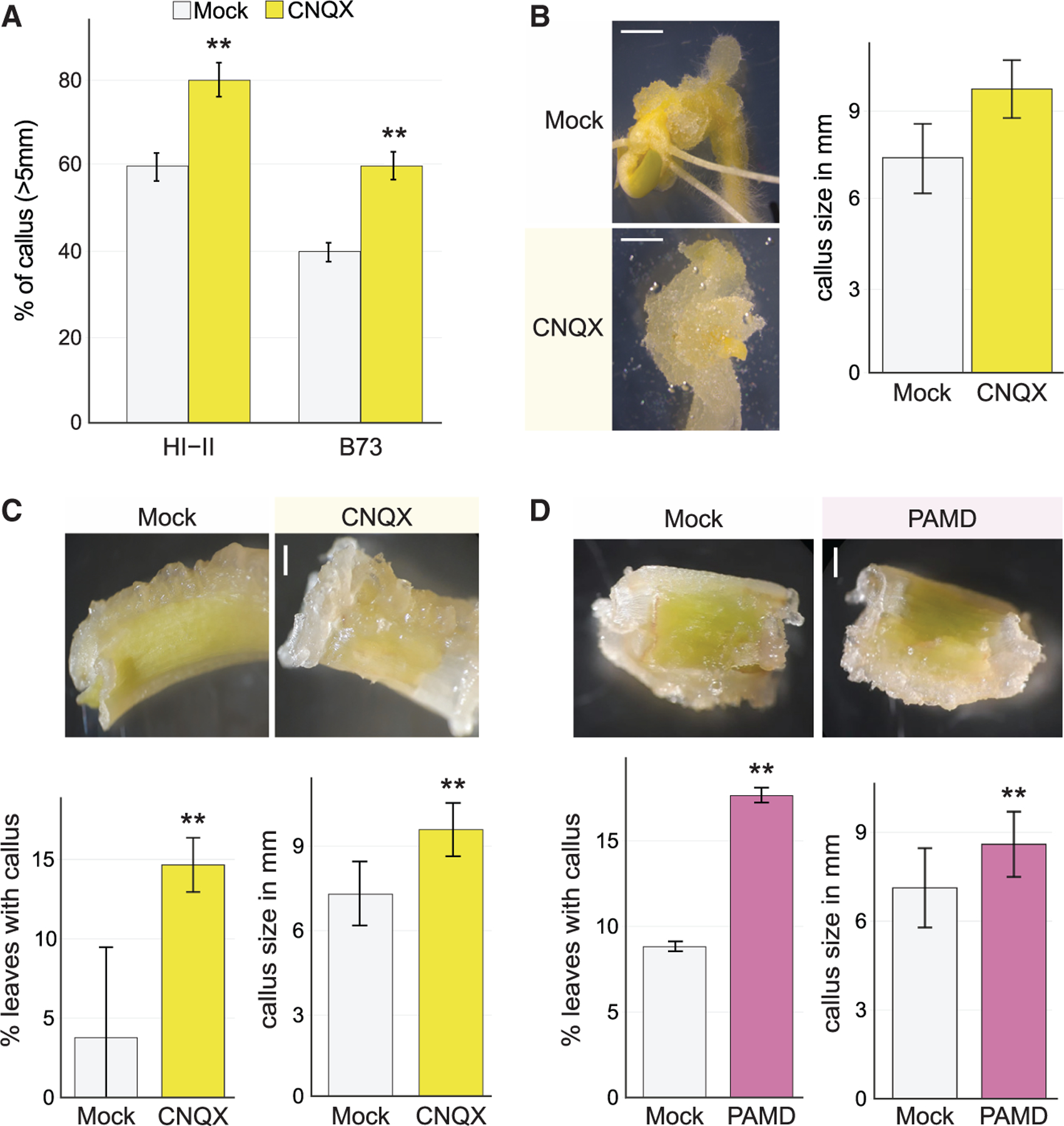
(A) CNQX constitutive post embryo harvest treatment increases the percent of callus formation from embryos in the regeneration competent Hi-II line (chi-square test; n = 48; **p < 0.0001) and the regeneration recalcitrant B73 line (chi-square test; n = 48 for each genotype in each treatment; **p < 0.0001).
(B) Representative images and quantification of callus formation from embryo in the B73 line and quantification. Under CNQX constitutive post embryo harvest treatment, a larger mass of callus was obtained (t test; n = 24 for each treatment; **p <10‒8). Scale bars, 2 mm.
(C) Regeneration of the maize seedling’s first leaf and coleoptile in the recalcitrant B73 line comparing mock and CNQX constitutive post-leaf harvest treated samples with quantitative analysis showing more than 3× higher frequency of callus formation in CNQX-treated cells (chi-square test; n ≥ 34 for each treatment; **p < 0.0001) and improvement in callus size (t-test; n = 24; **p < 0.0001). Scale bars, 1 mm.
(D) Callus formation from B73 leaves with PAMD constitutive post leaf harvest treatment, showing an improvement in callus formation frequency (chi-square test; n ≥ 34 for each treatment; **p < 0.0001) and callus size due to treatment (t test; n = 34 for each treatment; **p < 0.001). Scale bars, 1 mm.
To determine whether calcium signaling antagonists could enhance regeneration in highly recalcitrant tissue, we incubated the first leaves and the sheath (coleoptile) of B73 on mock or CNQX plates for 15 days. Callus formed on around 4.4% of mock-treated B73 leaf explants, whereas callus formation on CNQX-treated B73 leaf explants more than tripled to about 15% (Figure 7C). Callus microcolony formation from Hi-II protoplasts also increased on CNQX (Figure S7B). In addition, treatment with PAMD increased coleoptile to callus formation rate and size to similar levels as CNQX for the coleoptile (Figure 7D). The result suggests that the role of GLRs and the SA pathway in regeneration is conserved in maize. Thus, GLR-mediated signaling inhibits regeneration in a range of contexts across flowering plant species.
DISCUSSION
Upon wounding, GLRs balance defense and regeneration
The plant’s two primary strategies to defend against attacks– defense and regeneration–share many of the same rapid wound responsive triggers, such as calcium, reactive oxygen species, and electrical signals (Chen et al., 2016; Efroni et al., 2016; Florentin et al., 2013; Hoermayer and Friml, 2019; Moeder et al., 2019; Savatin et al., 2014). The key finding here is that GLR-mediated signals negatively regulate regeneration but positively regulate defense response. The result shows that the activation of the two processes can be decoupled. GLR-mediated signaling tunes responses in favor of defense, enhancing callose deposition and the expression of defense-related genes. At the same time, GLR signaling dampens the regeneration response, slowing division rates, cellular reprogramming, and meristem recovery. The effects all appear to take place in the immediate vicinity of the wound without long-distance systemic signaling, as conditional GLR inhibition improved regeneration in isolated explants of leaves or embryos where no systemic signaling is possible. Thus, the plant’s wound response appears to be, at least to some degree, a trade-off between efficient repair and a robust defense response and the plant does not necessarily optimize wound responses for either process.
GLR signaling acts mainly through the SA pathway
The concept of a trade-off between growth and defense is well established (Huot et al., 2014; van Butselaar and Van den Ackerveken, 2020; Züst and Agrawal, 2017, but also see Kliebenstein [2016]). Both SA and JA have been implicated in boosting defense while slowing growth (Huot et al., 2014; van Butselaar and Van den Ackerveken, 2020); therefore, the increase in regeneration we observed by blocking GLRs or the SA pathway may reflect a growth versus defense trade-off. However, perturbations of defense pathways did not always affect regeneration and growth in the same way. For example, the coi1–2 mutant also shows increased growth but lower regeneration in most of the assays. In addition, the quadruple glr mutant, which showed increased regeneration, did not show enhanced root growth. (In fact, other groups found that a glr3.6 knockdown had root growth defects, Singh et al., 2016.) Furthermore, our conditional treatments showed that the effects of GLR signaling on regeneration and defense responses could be entirely recapitulated by blocking the channels transiently upon wounding, and many of the effects of the glr mutants, such as decreased callose deposition, were apparent only after wounding. Thus, we posit that GLR-mediated signaling represents a different type of tradeoff–not in balancing growth versus defense–but routing the complex responses to an injury. The local GLR-mediated response appears to allow a ‘‘bet-hedging’’ strategy between defense and regeneration.
Both SA and JA have been shown previously to mediate GLR responses in wounding (Forde and Roberts, 2014). Therefore, why were the regeneration effects mediated by SA and not JA? Although both hormone pathways have overlapping function, they also mediate response to different types of pathogens independently (Yang, 2015). Still, it is not apparent how those independent roles would lead to their differential effects on regeneration. Another explanation centers on JA’s competing roles in wound response. That is, in addition to its well-documented roles in defense (Howe et al., 2018), recent reports have implicated JA with a positive role in regeneration (Zhang et al., 2019; Zhou et al., 2019). Thus, in disturbing JA signaling, any gain in regenerative capacity resulting from a down regulation of defense is offset by also inhibiting the developmental responses needed for repair. Indeed, whereas our transcriptional analysis showed that the GLRs activated SA responses, the effect on JA response was more complex, with both up- and downregulation of JA-responsive genes. Overall, SA may provide a less ‘‘conflicted’’ pathway to downregulate regeneration and upregulate defense. Thus, SA may be a better pathway to mediate the trade-off between the two.
Calcium fluxes are severely disturbed around the wound in GLR mutants
GLR channels are known to mediate calcium signaling (Michard et al., 2017; Wudick et al., 2018a), whereas other work has documented the complexity of ion-mediated signaling during wounding in plants (Meyer and Weisenseel, 1997). In the wounding experiments, maize roots wounded by deep incisions showed very fast membrane depolarizations, followed by periods of gradual re- or hyper-polarization. Similarly, our results show high Ca2+ influxes at the injury site in Arabidopsis wild-type roots followed by gradual recovery of fluxes. Maize root wounding also showed that the observed re-polarization was composed of major effluxes of potassium and chloride and smaller influxes of calcium and protons, which could be expected given the concentration gradients of these metabolites and ions.
Our experiments with the glr quadruple mutant demonstrate an impairment of calcium influxes that are reduced even further to almost background levels by pretreatment with CNQX. These results suggest that, without Ca2+ signaling, the default pathway reverts to favor regeneration. Although we cannot rule out that the observed effects mediated by the GLRs were caused by ion fluxes other than calcium, the equivalent calcium flux dynamics obtained from CNQX-exposed roots and the observed phenotype in callose deposition further support a role in calcium signaling. The lower levels of calcium influx observed in the mutant 2–8 min after wounding could account for a lower callose synthase activity, resulting in less callose deposition (observed at time 0 after wounding). Following the observations of the maize results described earlier, the subsequent disturbances we observed in Ca2+ flux levels could be due to lower callose deposits that allow a leakage of ions and other metabolites, such as charged macromolecules.
In this model, the GLRs represent a rapid response mechanism to injury; therefore, a key question is when after wounding do they exert their effects? Ion signaling acts fast in most contexts, which leads to the expectation that GLR signaling affects regeneration in the seconds or minutes after wounding. Indeed, our experiments showed that a 1-h pretreatment ending right after injury led to a significant increase in regeneration. This shows that GLRs mediate rapid signaling after wounding. However, we could increase regeneration further with prolonged treatment on CNQX, showing the GLRs also mediate a longer-term response. One explanation is that certain responses, such as callose deposition, represent early outcomes of GLR signaling, whereas GLRs also mediate responses that act on a longer time frame, such as the SA responses we observed in our expression analysis around 24 h after wounding. This leads to a scenario in which defense responses can inhibit regeneration for some time after wounding. To augment regeneration, defense responses need to be inhibited continually for some time after wounding, such as the clinical use of fever suppressants to prevent temperature spiking during an infection. In the context of normal function, the GLRs may signal over an extended time to sustain the defense response and inhibit regeneration.
SA contributes to age-dependent regeneration
Interestingly, the involvement of the SA pathway in regeneration may also provide a clue to the longstanding observation that regeneration efficiency decreases with tissue age (Binding, 1975; Chen et al., 2014; Hoque and Mansfield, 2004; Prakash and Gurumurthi, 2010; Sinnott, 1960; Vasil and Vasil, 1974). SA levels were previously shown to be anticorrelated with callus formation in garlic leaves (Mostafa et al., 2020), and our own analysis showed that SA-response markers were anticorrelated with zones of regeneration efficiency in roots. Furthermore, our data showed that perturbing GLRs, which signal to SA, had stronger effects in enhancing regeneration in older tissues. A consistent explanation to these observations is that mature tissues exhibit higher levels of SA signaling that prime them for defense responses at the cost of regenerative processes. The model implies that inhibiting SA response in these older tissues reverts regenerative competence–in part–to a younger developmental state.
Finally, plants deal with a wide array of insults and attacks, including pathogens, herbivores, and mechanical damage. In many contexts, the strategic response is a combination of defense and regeneration. However, neither response is apparently optimized. Early screens established the prevalence of mutants for enhanced defense responses (Glazebrook et al., 1996), whereas we show here perturbations that can enhance regeneration. Inhibition of the GLR-SA pathway provides evidence of a trade-off between the two responses and exposes one mechanism that the plant uses to balance its complex response to the environment.
Limitations of the study
Regeneration can vary by ambient conditions, time of year, and subtle growth conditions; therefore, mock control or WT plants are needed for each experimental trial. Nonetheless, although starting regeneration rates may vary by trial, effects are generally consistent over multiple trials. Repeats of the same experiment need to be treated as independent trials. Here, we used Fisher’s combined p value method. The net result is that absolute regeneration levels for a given experimental set should be considered in a range rather than an absolute value.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be made available upon reasonable request by the Lead Contact, Kenneth D. Birnbaum, ken.birnbaum@nyu.edu.
Materials availability
Arabidopsis mutant lines used in this study are available from the authors or from the sources cited in the text or methods. Chemical reagents can be obtained by suppliers, as listed in the reagent table below.
Data and code availability
Raw and processed data files have been submitted to GEO and are publicly available as of the date of publication under accessions numbers GSE168381 (ATAC-seq data) and GSE168385 (RNA-seq data).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The ATACseq and RNAseq analysis, as well as all wild type controls, were performed using Arabidopsis Columbia (Col-0) ecotype. The mutant lines used in this study are: glr1.2 (SALK_114822), glr3.3–1 (SALK_077608), glr3.3–2 (SALK_082194), glr1.2/1.4–1/3.3–1, glr1.2/1.4–1/2.2/3.3–1 (Wudick et al., 2018b), coi1–2 (Xu et al., 2002) and npr1–5 (Zipfel et al., 2004), glr3.1/3.2/3.3/3.6 (Mou et al., 2020). Other SA pathway related mutants tested were: cbp60g-1 (SALK_023199C), cbp60a-1/cbp60g-1 (CS72190), npr3–2/npr4–2 (CS72351), cpk11–2/cpk5/cpk6 ( CS69906), cpk6(a) (CS65905), cpk6(b) (SALK_025460). All in the Col-0 genetic background. All plants were grown on regular agar plates (0.8% agar, ½ MS, 0.5% sucrose) in Percival chambers at 23°C under 16 hr light/8 hr darkness.
METHOD DETAILS
Root stump regeneration assay
Seeds were stratified at 4°C for two days, placed on regular agar plates, and then grown vertically for 7 days. Plants were transferred to a ‘‘cutting board’’ (3% agar, ½ MS, 0.5% sucrose), upon which root meristems were cut at ~130um (low cuts) or 270um (high cuts), which removed most of the meristem. Cuts were made by using a 30G sterile dental needle (ExelInt) by hand under a dissecting microscope. Plates were turned 90° degrees immediately after cutting. Root regeneration was scored for gravitropism as in Figure 1A at 1,2, and 3 days post cut (dpc). A clear gravitropic response determined regeneration, where regenerated roots were also checked under a dissecting microscope to ensure the primary root attained morphological characteristics of a regenerated meristem.
ATAC-seq analysis
Seeds of the pMP::MP-GFP background were stratified and grown in standard growth conditions on standard plates for 7 days. Plants were then transferred to cutting board plates, as above, and cut at ~130µm. The excised roots were then harvested at 2.5h, 14h, or 16 h after cutting, with triplicate samples collected. pMP::MP-GFP is typically induced within 2 h, and localized mainly in the nucleus but also in the cytoplasm. However, due to the low signal observed in the nucleus at early time points, the excised roots were subjected to rapid enzymatic digestion of cell walls as described for maize (Ortiz-Ramirez et al., 2017), with a shorter enzymatic incubation of 1 hr. GFP+ cells were collected by FACS and protoplasts were lysed in deionized water. For the time points after root tip cutting, about 1000–2000 GFP-expressing protoplasts were used per sample. After protoplasts were lysed, the low-input Tn5 tagmentation protocol was used, decreasing the concentration of both Tn5 and primers (Buenrostro et al., 2015). A final library purification was done using Ampure XP beads to selectively avoid fragments of less than 100 bp that correspond to primers and primer dimers. For uncut controls, about 50K protoplasts from distal meristem dissections were used. Nuclei were either mechanically released or obtained after root protoplasting, stained with DAPI (1µM) and isolated by FACS. The standard Tn5 tagmentation protocol was used according to the described protocols (Buenrostro et al., 2015; Lu et al., 2017). In brief, a purification step of the transposed DNA fragments was done using Qiagen MinElute PCR Purification Kit prior library amplification (see key resources table for primers used). The appropriate number of cycles for library amplification were experimentally determined by qPCR.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| none | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| (2E)-4-phenyl-2-[[[3-(trifluoromethyl)phenyl]amino]methylidene]cyclohexane-1,3-dione | Aurora Fine Chemicals | 180.584.762 |
| SULFO-CYANINE5 AZIDE 1MG | LUMIPROBE | A3330 |
| CNQX | Santa Cruz | sc-203003A |
| Gadolinium(III) chloride | Santa Cruz | sc-224004 |
| D(−)-2-Amino-5-phosphonovaleric acid (D-AP5) | Santa Cruz | sc-20043 |
| ABA | Sigma | 862169 |
| Aniline Blue | Sigma | 415049 |
| L-ASCORBIC ACID | Sigma | A4544 |
| 6-Benzylaminopurine | Sigma | B3408 |
| Copper (II) Sulfate | Sigma | 451657 |
| DAPI | Sigma | D9542 |
| 2,4-Dichlorophenoxyacetic acid | Sigma | D7299 |
| L-Glutamic acid potassium salt monohydrate | Sigma | G1501 |
| L-Cysteine (Cys) | Sigma | C7352 |
| L-Serine (Ser) | Sigma | S4311 |
| Inositol | Sigma | I3011 |
| 1-Naphthaleneacetic Acid | Sigma | N0640 |
| Nicotinic acid | Sigma | N0671 |
| Salicylic Acid | Sigma | 247588 |
| Vitamin B1 | Sigma | T4625 |
| Vitamin B6 | Sigma | P5669 |
| ZEATIN | Sigma | Z0164 |
| Critical commercial assays | ||
| Nugen OvationRNA Amplification System V2 | Tecan Genomics | 3100-A01 |
| Nugen Ovation Ultralow DR Multiplex System | Tecan Genomics | 0344NB-08 |
| Deposited data | ||
| Raw and processed ATAC-seq data from root reprogramming cells | This paper | GSE168381 |
| Raw and processed RNA-seq data from root reprogramming cells | This paper | GSE168385 |
| Experimental models: Organisms/strains | ||
| Arabidopsis thaliana Columbia ecotype | ABRC | N/A |
| Arabidopsis thaliana glr1.2 mutant | ABRC | SALK_114822 |
| Arabidopsis thaliana glr3.3–1 mutant | ABRC | SALK_077608 |
| Arabidopsis thaliana glr3.3–2 mutant | ABRC | SALK_082194 |
| Arabidopsis thaliana glr1.2/1.4–1/3.3–1 mutant | Wudick et al., 2018b | N/A |
| Arabidopsis thaliana glr1.2/1.4–1/2.2/3.3–1 mutant | Wudick et al., 2018b | N/A |
| Arabidopsis thaliana glr3.1/3.2/3.3/3.6 mutant | Mou et al., 2020 | N/A |
| Arabidopsis thaliana coi1–2 mutant | Xu et al., 2002 | N/A |
| Arabidopsis thaliana npr1–5 mutant | Zipfel et al., 2004 | N/A |
| Arabidopsis thaliana cbp60g-1 mutant | ABRC | SALK_023199C |
| Arabidopsis thaliana cbp60a-1/cbp60g-1 mutant | ABRC | CS72190 |
| Arabidopsis thaliana npr3–2/npr4–2 mutant | ABRC | CS72351 |
| Arabidopsis thaliana cpk11–2/cpk5/cpk6 mutant | ABRC | CS69906 |
| Arabidopsis thaliana cpk6(a) mutant | ABRC | CS65905 |
| Arabidopsis thaliana cpk6(b) mutant | ABRC | SALK_025460 |
| Zea mays B73 | Jackson Lab CSHL | N/A |
| Zea mays HiII | Jackson Lab CSHL | N/A |
| Oligonucleotides | ||
| Ad1_noMX | Buenrostro, et al., 2013 | AATGATACGGCGACCACCGAGATCTACACTCGTCGGCAGCGTCAGATGTG |
| Ad2.1_TAAGGCGA | Buenrostro, et al., 2013 | CAAGCAGAAGACGGCATACGAGATTCGCCTTAGTCTCGTGGGCTCGGAGATGT |
| Ad2.2_CGTACTAG | Buenrostro, et al., 2013 | CAAGCAGAAGACGGCATACGAGATCTAGTACGGTCTCGTGGGCTCGGAGATGT |
| Ad2.3_AGGCAGAA | Buenrostro, et al., 2013 | CAAGCAGAAGACGGCATACGAGATTTCTGCCTGTCTCGTGGGCTCGGAGATG |
| Ad2.4_TCCTGAGC | Buenrostro, et al., 2015 | CAAGCAGAAGACGGCATACGAGATGCTCAGGAGTCTCGTGGGCTCGGAGATGT |
| Software and algorithms | ||
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| macs2 version 2.1.1 | Zhang et al., 2008 | https://pypi.org/project/MACS2/2.1.1.20160309/ |
| DEseq2 |
Love et al., 2014
|
https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| HISAT2 | Kim, et al., 2015 | http://daehwankimlab.github.io/hisat2/manual/ |
| Image J software Fiji version 1.52p | Schindelin, et al., 2019 | http://imagej.nih.gov/ij/ |
| VirtualPlant 1.3 | Katari et al., 2010 | http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/ |
All sequenced libraries were aligned using Bowtie2 and the peaks corresponding to accessible chromatin regions were identified with MACS2. Consensus peaks among replicates were obtained and the counts were determined. The Differential Accessible Chromatin (DAC) regions between samples were analyzed for significance using DEseq2 comparing each time point vs. uncut roots. To eliminate false-positives and to increase the confidence in our results, we 1) only considered accessible regions present in all replicates, and 2) used a padj value below 0.01 in our DAC analysis.
Regeneration assays in calcium antagonist treatments or SA treatments
For treatments, seeds were grown and meristems were cut as detailed above for standard growth on standard plates. For the different treatments, plates were prepared as followed: mock plates (0.8% agar, ½ MS, 0.5% sucrose, water); mock cutting board (3% agar, ½ MS, 0.5% sucrose, water); CNQX, GD3+ or AP5 plates (0.8% agar, ½ MS, 0.5% sucrose and either 50µM CNQX, 1µM GD3+ or 200µM AP5); CNQX, GD3+ or AP5 cutting board (3% agar, ½ MS, 0.5% sucrose and 50µM CNQX,1µM GD3+ or 200µM AP5 respectively); Mock plates for SA treatments (0.8% agar, ½ MS, 0.5% sucrose, DMSO); mock cutting board for SA treatments (3% agar, ½ MS, 0.5% sucrose, DMSO); SA or PAMD plates (0.8% agar, ½ MS, 0.5% sucrose and either 1µM SA or 1µM PAMD); CNQX and SA plates (0.8% agar, ½ MS, 0.5% sucrose, 50µM CNQX, 1µM SA). Seven days after plating, seeds from standard plates were transferred to either the CNQX or mock cutting board for rapid resection and then immediately placed back on either CNQX plates or control plates. In the experiment shown in the main Figure, plants were pre-incubated for 1 h on either mock or CNQX plates to ensure cells were exposed to CNQX at the time of cutting.
Regeneration assays in GLR agonist treatments
Arabidopsis Col-0 seeds were grown and meristems were cut as detailed above for standard growth on standard plates. For the different concentrations used in the agonist treatments, regular plates (0.8% agar, ½ MS, 0.5% sucrose) were supplemented with 1mM or 5mM of either L-Glutamate (Glu), L-Cysteine (Cys) or L-Serine (Ser). In short treatment plants were exposed to the amino acid tested (5mM) for <2 min during root tip cut and transfer to half strength MS plates to assess regeneration. For long treatment, plants were cut and maintained on plates with the treatment until regeneration was assessed.
Root stump regeneration assays with transient CNQX window treatments
For transient treatments, seeds were grown and meristems were cut as detailed for CNQX treatments with regeneration. For the short early window treatments, plants were cut on the CNQX cutting board and transferred to CNQX plates for 4 h then transferred to mock plates for the remainder of the period. For the later treatment, plants were transferred to the mock cutting board for resection, transferred again to a mock plate after cutting for 4 h and then transferred to a CNQX plate for the remainder of the experiment. For the 16 h window treatments, plants were exposed to a 1h pretreatment by placing them on mock or CNQX plates and then transferred to the cutting board without the treatment. After root tip excision, plants were placed on or off CNQX for 16 h followed by a second transfer to either mock or CNQX plates for regeneration. Regeneration was scored as above. Controls included all the same transfers where one set of plates was transferred on and off mock plates (mock) and a second set of plates was transferred on and off only CNQX plates (positive control).
RNA-seq time course
Plants were grown on standard plates in standard conditions and transferred to CNQX or mock cutting board plates at 7 days after plating. Immediately after transfer, plants were cut at 270 µm (high cuts) and then transferred to CNQX plates or mock plates. Approximately 20 stump tips consisting of about 100 µm of the stump were collected at each time point (30 min, 4 h, 14 h, 24 h). For controls, two separate sets of uncut roots were transferred to CNQX plates or mock plates and incubated for 14 h before collecting root tip tissue, which also consisted of about 100 µm of the distal root tip. RNA was extracted using the Qiagen MicroRNAeasy Kit. We performed cDNA construction and library amplification with Nugen OvationRNA Amplification System V2. Libraries were made using the Nugen Ovation Ultralow DR Multiplex System (Tecan). DNA fragmenting was performed using the Covaris S220. Short read sequences were generated on an Illumina NextSeq 500 and were aligned using HISAT2. Counts were normalized between samples using DESeq2’s median of ratios method before analyzing for differentially expressed genes.
Analysis of RNA-seq time course
The time series included six time points for mock and CNQX treated plants (2 uncut controls, 30 min post cut, 4h post cut,14 h post cut, 24 h post cut). First, of the 32,833 annotated transcripts, the top 10th percentile most variable genes were selected [(genevarfilter(data,’Percentile’,90)) in Matlab], resulting in a list of 3,283 genes. Any gene with a zero value in any sample was removed (leaving 3131 genes). The resulting matrix was then subject to time series analysis using ImpulseDE version 1.12.0 in R (Chechik and Koller, 2009; Sander et al., 2017). All default values were used (n_iter = 100,n_randoms = 50000, Q_value = 0.01) in a case vs. control analysis with the 6 mock samples vs. 6 CNQX-treated samples used for comparative differential expression. Genes classified as differentially expressed ‘DE’ in ‘at least 2 TPs for case vs. control were selected for further analysis (1436 genes). Impulse categorized the DE genes into 19 different clusters. Each cluster was then subjected to GO term analysis using the biomaps function in VirtualPlant 1.3 (Katari et al., 2010). Overall trends for each cluster (control vs. treat) were derived by taking the row sum and normalizing by the row mean.
Extracellular Ca2+ flux measurements after wounding
The ion-selective vibrating probe was used to estimate extracellular calcium flux at the wounding site (as per Wudick et al., 2018b) between 1–2 min after the cut due to the constrains in the technique. Arabidopsis roots were hydrated approximately 1 h before the measurements to equilibrate with the Basic Measuring Solution (BMS): 0.5 mM KCl, 0.1 mM CaCl2, pH 5.5 (unbuffered) (Shabala et al, 2005) or BMS+CNQX (50 µM). All the roots were cut and measured in BMS. The vibrating probe was moved with an excursion of 10mm, completing the cycle of acquisition in 10 s. Each of the roots was measured until the fluxes stabilized at approximately 10–15 min after starting the measures. All the points were plotted, and an exponential decay fit was employed to extract the asymptote (plateau) and the decay constant (k) that were then compared with the extra sum of squares F-test. The statistics and fitting of the data were run on Graphpad Prism.
EdU staining
Cell proliferation analysis was performed with the EdU click reaction containing 2Mm CuSO4.5H2O (Sigma), 8 µM Sulfo-Cyanine5 azide (Lumiprobe) and 20mg/ml Ascorbic acid (Sigma). For EdU incorporation, Col-0 plants were grown under standard conditions described above, and, at 7 days after plating, were transferred to ½ MS media supplemented with CNQX (50 mM) or mock. After 60 min pre-incubation, plants were placed on CNQX or mock cutting board, cut at 270 µm (high cuts) and immediately transferred to ½ MS medium containing CNQX (50 µM)/5 µM EdU or mock/5 µM EdU for 30min. Plants were fixed and permeabilized according to published protocols (Kazda et al., 2016). The next day, samples were washed with PBS twice and incubated in the EdU click reaction cocktail for 30 min in the dark. After two washes with PBS, plants were placed on 0.1mM DAPI in PBS and incubated for 30 min at 22 °C in the dark. Two final washes with PBS were performed. Samples were observed under an inverted fluorescence microscope. Images were analyzed with Image J software Fiji 1.52p version (http://imagej.nih.gov/ij/). Using images from a median optical section, labeled cells were quantified for EdU and DAPI in the meristematic zone. The boundary between the meristematic and the elongation zones is defined by a twofold or more increase in the distance between nuclei in the cortical cell layer.
Callose staining
Plants grown under standard conditions on standard plates, as above, were transferred to CNQX or mock cutting board 7 days after plating and cut at 270 µm (high cuts). Plants were fixed on a 1:3 acetic acid/ethanol solution overnight after harvesting either immediately (0 min), after 10 min, or after 30 min. Note that 0 min is effectively 3–5 min after wounding accounting for the time it takes to fix the roots. The next day plants were stained in aniline blue solution (750 µl of 67 mM K3PO4 pH 9.5, 240 µl of distilled water, 1 µl of Silwet-77, and 10 µl of 1% aniline blue) in the dark for 1 h prior to imaging as described previously (Cui and Lee, 2016). Images were analyzed with Image J software Fiji 1.52p version (http://imagej.nih.gov/ij/).
Arabidopsis tissue culture and callus induction
Plants were grown under standard conditions in standard plates for each genotype shown in the main text. Detached leaf explants from 8-day-old (young leaves) and 15-day-old (old leaves) seedlings were cultured on media for callus induction: ½ MS + 3% sucrose + 1% Agar + 2 mg/L 2,4 D + 0.5 mg/l AIA + 0.5 mg/L Zeatin, pH 5,7. For callus induction, two different calcium channel antagonists were tested in the media: Gd3+ 1µM or CNQX at 1µM or 50 µM. SA treatment was tested using 50 µM SA or 10 µM PAMD on CIM. Sterile water was used instead of calcium channel antagonists as a mock control. Detached leaves were cut in half. Each plate contained either nine 15-day-old leaves or sixteen 8-day-old leaves. The plates were left at 23 °C under dark photoperiod. Callus formation was scored at day 15 after plating.
Root from leaf regeneration
For root-from-leaf regeneration, seeds were stratified at 4°C for two days, placed on regular agar plates (0.7% agar, ½ MS, 0.5% sucrose) and grown vertically in Percival chambers at 23°C under 16 hr light/8 hr darkness for 15 days. Different chemicals were tested to improve root regeneration, supplementing either with 50 µM CNQX, 50 µM SA or 10 µM PAMD. Detached leaf explants from 15-day seedlings were cultured on regular agar plates grown under the above conditions. Each plate contained twelve leaves. Root from leaf regeneration was scored at day 15 after plating.
Maize tissue culture and callus induction
Mature dry seeds of maize inbred lines HI-II and B73 were surface sterilized in 50ml falcon tube with 70% ethanol for 10 min and then in 4% sodium hypochlorite (NaOCl) supplemented with 0.1% Tween20 for 30 min, washed 3 times in sterile distilled water and incubated in 4% chloramine for 20min under shaking. The seeds were rinsed 4 times with sterilized water and plated on ½ MS basal media with 0.7% agar and pH 5.7, and incubated at 28 °C in darkness. Immature embryos were excised to use as explants after 3 days on ½ MS medium. The swollen mature embryos were removed from seeds with a scalpel and cultured, scutellum side up, on induction medium. The first leaf and coleoptile were taken for explants at 6 days after germination on ½ MS medium. The coleoptiles were longitudinally split with a sharp scalpel to expose shoot meristem and simultaneously cultured on callus induction media with the split side facing the media. Callus induction media contained N6 basal media supplemented with 3% sucrose, 2 mg/l glycine, 1 mg/l vitamin B1, 0.5 mg/l vitamin B6, 0.5 nicotinic acid, 120 mg/l inositol, 7 g/l powdered agar, 2 mg/l 2,4-dichlorophenoxyacetic acid (2,4-D), 1 mg/l 1-naphthaleneacetic acid (NAA), CNQX (50 µM for treated samples) and pH 5.7 (Shen et al., 2012). The embryos and first leaf/coleoptile were incubated for 15 days in darkness at 28 °C. After 3 weeks of culture on induction medium, the percentages and size of primary calli were determined.
Protoplast culture
Plants were grown under standard conditions and approximately 0.6 g of the aerial parts of 1-week-old sterile plantlets was soaked in 5 mL of maceration medium (MGG) in a Petri dish. Five milliliters of MGG were added, bringing the total volume to 10 mL. Maceration was performed in the dark, at 24°C, overnight (12 h). After cell wall digestion, 20 mL of protoplast suspension in MGG was filtered through a sterile 80-mm mesh filter into 10 mL washing solution (2.5% KCl and 0.2% CaCl2) already added to a 30-mL tube. After centrifugation (70g, 6 min), protoplast pellets were gently resuspended in 25 mL washing solution and centrifuged again (70g, 6 min). Washing was performed two more times. Protoplast numbers were estimated before the last centrifugation. After a 1 h incubation at 4°C, the protoplast suspension was diluted in protoplast induction media (PIM) to a concentration of 8.3 × 105 protoplasts per milliliter. Protoplasts were diluted 1:10 in PIM. The effect of CNQX on cell division was evaluated by adding 1µM or 50 µM CNQX to the liquid media. The plates were subsequently placed in large plastic containers to limit evaporation and cell suspensions were cultured in the dark at 20°C (Chupeau et al, 2013). On day 11, cell colonies formed in a liquid culture dish were visualized and counted.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical data of all experiments, as the statistical tests used, p values and the number of individuals tested (n) can be found in the figure legends and in the results section. All detailed raw data on phenotypes, their statistical analysis, and multiple testing results are contained in Table S4. For the analysis of mutants and treatments for regeneration, standard Chi-square (e.g., regeneration proportions) or t tests (e.g., root length) were used for statistical analysis of phenotypes and traits. In many cases, experiments were repeated up to 4 times for rigor and/or to test more subtle differences between phenotypic classes. If repeat experiments were performed under the same condition at the approximate same time, the results were pooled for statistical tests. In some cases, experiments were performed months or years apart and batch effects were apparent. For example, background control (WT or mock) regeneration rates can vary by season and ambient humidity levels. In these cases, we combined separate tests for a given hypothesis using Fisher’s Method for Combining P Values, using the formula pchisq((sum(log(p))*−2), df=length(p)*2, lower.tail=F) in R, where p is a row vector of all probability values for a given hypothesis. The multiple regeneration systems tested led to 127 tests in total, so, to account for multiple testing, we applied the Benjamini-Hochberg correction for all pvals reported for each test as the false discovery rate, FDR. None of the p values reported as significant lost their significance (fell above the significance cutoff of 5% FDR).
Supplementary Material
Highlights.
Perturbation of plant glutamate receptors improves regeneration in monocots and dicots
The role of glutamate receptors in regeneration is mediated by salicylic acid signaling
Increased salicylic acid response in older tissue is anticorrelated with regeneration
Perturbing glutamate receptors improves regeneration of recalcitrant maize B73 lines
ACKNOWLEDGMENTS
We thank Carlos Ortiz-Ramıŕez for helpful input on GLRs. K.D.B. is supported by the National Institutes of Health (R35GM136362) and the National Science Foundation (1934388). J.A.F. was supported by the NIH (R01-GM131043) and the NSF (MCB-1930165).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2022.01.013.
DECLARATION OF INTERESTS
A patent (application #63120640) has been filed related to this work on compositions and methods to enhance plant regeneration.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
SUPPORTING CITATIONS
The following references appear in the supplemental information: Brady et al. (2007).
REFERENCES
- Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binding H (1975). Reproducibly high planting efficiencies of isolated protoplasts from shoot cultures of tobacco. Plant Physiol 35, 325–327. [Google Scholar]
- Birnbaum KD, and Sánchez Alvarado AS (2008). Slicing across kingdoms: regeneration in plants and animals. Cell 132, 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee J-Y, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, and Benfey PN (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol 109, 21–29, 1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw S-M, Chang C-C, Chen H-L, and Li W-H (2004). Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol 58, 424–441. [DOI] [PubMed] [Google Scholar]
- Chechik G, and Koller D (2009). Timing of gene expression responses to environmental changes. J. Comput. Biol 16, 279–290. [DOI] [PubMed] [Google Scholar]
- Chen L, Sun B, Xu L, and Liu W (2016). Wound signaling: the missing link in plant regeneration. Plant Signal. Behav 11, e1238548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Qu Y, Sheng L, Liu J, Huang H, and Xu L (2014). A simple method suitable to study de novo root organogenesis. Front. Plant Sci 5, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CF, and Tan HS (2018). The use of proteomic tools to address challenges faced in clonal propagation of tropical crops through somatic embryogenesis. Proteomes 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupeau M-C, Granier F, Pichon O, Renou J-P, Gaudin V, and Chupeau Y (2013). Characterization of the early events leading to totipotency in an Arabidopsis protoplast liquid culture by temporal transcript profiling. Plant Cell 25, 2444–2463. 10.1105/tpc.113.109538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, and Werr W (2009). DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136, 1643–1651. [DOI] [PubMed] [Google Scholar]
- Cui W, and Lee JY (2016). Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat. Plants 2, 16034. [DOI] [PubMed] [Google Scholar]
- Delporte F, Jacquemin JM, Masson P, and Watillon B (2012). Insights into the regenerative property of plant cells and their receptivity to transgenesis: wheat as a research case study. Plant Signal. Behav 7, 1608–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, and Zhang Y (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173, 1454–1467.e15. [DOI] [PubMed] [Google Scholar]
- Efroni I, Ip P-L, Nawy T, Mello A, and Birnbaum KD (2015). Quantification of cell identity from single-cell gene expression profiles. Genome Biol 16, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip P-L, Rahni R, DelRose N, Powers A, Satija R, and Birnbaum KD (2016). Root regeneration triggers an embryolike sequence guided by hormonal interactions. Cell 165, 1721–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D, and Voigt CA (2014). Callose biosynthesis in Arabidopsis with a focus on pathogen response: what we have learned within the last decade. Ann. Bot 114, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentin A, Damri M, and Grafi G (2013). Stress induces plant somatic cells to acquire some features of stem cells accompanied by selective chromatin reorganization. Dev. Dyn 242, 1121–1133. [DOI] [PubMed] [Google Scholar]
- Forde BG, and Roberts MR (2014). Glutamate receptor-like channels in plants: a role as amino acid sensors in plant defence? F1000Prime Rep 6, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, and Ausubel FM (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Maki N, Ichihashi Y, Kitazawa D, Igarashi D, Kadota Y, and Shirasu K (2020). Exogenous treatment with glutamate induces immune responses in Arabidopsis. Mol. Plant. Microbe Interact 33, 474–487. [DOI] [PubMed] [Google Scholar]
- Green CE, and Phillips RL (1975). Plant regeneration from tissue cultures of Maize1. Crop Sci 15, 417–421. [Google Scholar]
- Guo Q, Yoshida Y, Major IT, Wang K, Sugimoto K, Kapali G, Havko NE, Benning C, and Howe GA (2018). JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 115, E10768–E10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hander T, Fernández-Fernández ÁD, Kumpf RP, Willems P, Schatowitz H, Rombaut D, Staes A, Nolf J, Pottie R, Yao P, et al. (2019). Damage on plants activates Ca2+-dependent metacaspases for release of immunomodulatory peptides. Science 363, eaar7486. [DOI] [PubMed] [Google Scholar]
- Heyman J, Cools T, Canher B, Shavialenka S, Traas J, Vercauteren I, Van den Daele H, Persiau G, De Jaeger G, Sugimoto K, and De Veylder L (2016). The heterodimeric transcription factor complex ERF115-PAT1 grants regeneration competence. Nat. Plants 2, 16165. [DOI] [PubMed] [Google Scholar]
- Hoermayer L, and Friml J (2019). Targeted cell ablation-based insights into wound healing and restorative patterning. Curr. Opin. Plant Biol 52, 124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque ME, and Mansfield JW (2004). Effect of genotype and explant age on callus induction and subsequent plant regeneration from root-derived callus of Indica rice genotypes. Plant Cell Tissue Organ Cult 78, 217–223. [Google Scholar]
- Howe GA, Major IT, and Koo AJ (2018). Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol 69, 387–415. [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, and He SY (2014). Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Favero DS, Sakamoto Y, Iwase A, Coleman D, Rymen B, and Sugimoto K (2019). Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol 70, 377–406. [DOI] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, et al. (2010). VirtualPlant: a software platform to support systems biology research. Plant Physiol 152, 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, and Howe GA (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105, 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H (1985). Callose biosynthesis as a Ca2+-regulated process and possible relations to the induction of other metabolic changes. J. Cell Sci. Suppl 2, 89–103. [DOI] [PubMed] [Google Scholar]
- Kazda A, Akimcheva S, Watson JM, and Riha K (2016). Cell proliferation analysis using EdU labeling in whole plant and histological samples of Arabidopsis. In Plant Cell Division: Methods and Protocols, Caillaud M-C, ed. (Springer; ), pp. 169–182. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ (2016). False idolatry of the mythical growth versus immunity tradeoff in molecular systems plant pathology. Physiol. Mol. Plant Pathol 95, 55–59. [Google Scholar]
- Krystal JH, D’Souza DC, Petrakis IL, Belger A, Berman RM, Charney DS, Abi-Saab W, and Madonick S (1999). NMDA agonists and antagonists as probes of glutamatergic dysfunction and pharmacotherapies in neuropsychiatric disorders. Harv. Rev. Psychiatry 7, 125–143. [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León J, Rojo E, and Sánchez-Serrano JJ (2001). Wound signalling in plants. J. Exp. Bot 52, 1–9. [DOI] [PubMed] [Google Scholar]
- Li S, Yamada M, Han X, Ohler U, and Benfey PN (2016). High-resolution expression map of the Arabidopsis Root reveals alternative splicing and lincRNA regulation. Dev. Cell 39, 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho M-J, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, et al. (2016). Morphogenic regulators Baby Boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hofmeister BT, Vollmers C, DuBois RM, and Schmitz RJ (2017). Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res 45, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Kurenda A, Siddique S, Dénervaud Tendon V, Zhou F, Holbein J, Hasan MS, Grundler FM, Farmer EE, and Geldner N (2019). Single-cell damage elicits regional, nematode-restricting ethylene responses in roots. EMBO J 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters A, Kang M, McCaw M, Zobrist JD, Gordon-Kamm W, Jones T, and Wang K (2020). Agrobacterium-mediated immature embryo transformation of recalcitrant maize inbred lines using morphogenic genes. J. Vis. Exp 156. 10.3791/60782. [DOI] [PubMed] [Google Scholar]
- Meyer AJ, and Weisenseel MH (1997). Wound-induced changes of membrane voltage, endogenous currents, and ion fluxes in primary roots of maize. Plant Physiol 114, 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham M, Liu LH, Obermeyer G, and Feijó JA (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332, 434–437. [DOI] [PubMed] [Google Scholar]
- Michard E, Simon AA, Tavares B, Wudick MM, and Feijó JA (2017). Signaling with ions: the keystone for apical cell growth and morphogenesis in pollen tubes. Plant Physiol 173, 91–111. 10.1104/pp.16.01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder W, Phan V, and Yoshioka K (2019). Ca(2+) to the rescue - Ca(2+) channels and signaling in plant immunity. Plant Sci 279, 19–26. [DOI] [PubMed] [Google Scholar]
- Mostafa HHA, Wang H, Song J, and Li X (2020). Effects of genotypes and explants on garlic callus production and endogenous hormones. Sci. Rep 10, 4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou W, Kao Y-T, Michard E, Simon AA, Li D, Wudick MM, Lizzio MA, Feijó JA, and Chang C (2020). Ethylene-independent signaling by the ethylene precursor ACC in Arabidopsis ovular pollen tube attraction. Nat. Commun 11, 4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi SA, Chauvin A, Pascaud F, Kellenberger S, and Farmer EE (2013). Glutamate RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500, 422–426. [DOI] [PubMed] [Google Scholar]
- Nguyen CT, Kurenda A, Stolz S, Chételat A, and Farmer EE (2018). Identification of cell populations necessary for leaf-to-leaf electrical signaling in a wounded plant. Proc. Natl. Acad. Sci. USA 115, 10178–10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A, Ashikari M, Lin S, Takashi T, Angeles ER, Yamamoto T, and Matsuoka M (2005). Isolation of a rice regeneration quantitative trait loci gene and its application to transformation systems. Proc. Natl. Acad. Sci. USA 102, 11940–11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ramírez C, Michard E, Simon AA, Damineli DSC, Hernández-Coronado M, Becker JD, and Feijó JA (2017). Glutamate RECEPTOR-LIKE channels are essential for chemotaxis and reproduction in mosses. Nature 549, 91–95. [DOI] [PubMed] [Google Scholar]
- Peng Y, Yang J, Li X, and Zhang Y (2021). Salicylic acid: biosynthesis and signaling. Annu. Rev. Plant Biol 72, 761–791. [DOI] [PubMed] [Google Scholar]
- Prakash MG, and Gurumurthi K (2010). Effects of type of explant and age, plant growth regulators and medium strength on somatic embryogenesis and plant regeneration in Eucalyptus amaldulensis. Plant Cell Tissue Organ Cult 100, 13–20. [Google Scholar]
- Rymen B, Kawamura A, Lambolez A, Inagaki S, Takebayashi A, Iwase A, Sakamoto Y, Sako K, Favero DS, Ikeuchi M, et al. (2019). Histone acetylation orchestrates wound-induced transcriptional activation and cellular reprogramming in Arabidopsis. Commun. Biol 2, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J, Schultze JL, and Yosef N (2017). ImpulseDE: detection of differentially expressed genes in time series data using impulse models. Bioinformatics 33, 757–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatin DV, Gramegna G, Modesti V, and Cervone F (2014). Wounding in the plant tissue: the defense of a dangerous passage. Front. Plant Sci 5, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena G, Wang X, Liu H-Y, Hofhuis H, and Birnbaum KD (2009). Organ regeneration does not require a functional stem cell niche in plants. Nature 457, 1150–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo EK, Nakamura H, Mori M, and Asami T (2012). Screening and characterization of a chemical regulator for plant disease resistance. Bioorg. Med. Chem. Lett 22, 1761–1765. [DOI] [PubMed] [Google Scholar]
- Shabala L, Cuin TA, Newman IA, and Shabala S (2005). Salinity-induced ion flux patterns from the excised roots of Arabidopsis SOS mutants. Planta 222, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Shanmukhan AP, Mathew MM, Radhakrishnan D, Aiyaz M, and Prasad K (2020). Regrowing the damaged or lost body parts. Curr. Opin. Plant Biol 53, 117–127. [DOI] [PubMed] [Google Scholar]
- Shen Y, Jiang Z, Yao X, Zhang Z, Lin H, Zhao M, Liu H, Peng H, Li S, and Pan G (2012). Genome expression profile analysis of the immature maize embryo during dedifferentiation. PLoS One 7, e32237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Chien CT, and Chang IF (2016). The Arabidopsis glutamate receptor-like gene GLR3.6 controls root development by repressing the Kip-related protein gene KRP4. J. Exp. Bot 67, 1853–1869. [DOI] [PubMed] [Google Scholar]
- Sinnott EW (1960). Plant Morphogenesis (McGraw-Hill; ), p. 244. [Google Scholar]
- Sugimoto K, Gordon SP, and Meyerowitz EM (2011). Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol 21, 212–218. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, and Meyerowitz EM (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18, 463–471. [DOI] [PubMed] [Google Scholar]
- van Butselaar T, and Van den Ackerveken G (2020). Salicylic acid steers the growth-immunity tradeoff. Trends Plant Sci 25, 566–576. [DOI] [PubMed] [Google Scholar]
- Vasil V, and Vasil IK (1974). Regeneration of tobacco and Petunia plants from protoplasts and culture of corn protoplasts. In Vitro 10, pp. 83–96. [DOI] [PubMed] [Google Scholar]
- Wissemeier AH, and Horst WJ (1995). Effect of calcium supply on aluminium-induced callose formation, its distribution and persistence in roots of soybean (Glycine max (L.) Merr.). J. Plant Physiol 145, 470–476. [Google Scholar]
- Wudick MM, Michard E, Oliveira Nunes C, and Feijó JA (2018a). Comparing plant and animal glutamate receptors: common traits but different fates? J. Exp. Bot 69, 4151–4163. [DOI] [PubMed] [Google Scholar]
- Wudick MM, Portes MT, Michard E, Rosas-Santiago P, Lizzio MA, Nunes CO, Campos C, Damineli DSC, Carvalho JC, Lima PT, et al. (2018b). CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca(2+) homeostasis. Science 360, 533–536. [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, and Xie D (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadava P, Abhishek A, Singh R, Singh I, Kaul T, Pattanayak A, and Agrawal PK (2016). Advances in maize transformation technologies and development of transgenic maize. Front. Plant Sci 7, 1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-X, Ahammed GJ, Wu C, Fan S. y., and Zhou Y-H (2015). Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Curr. Protein Pept. Sci 16, 450–461. [DOI] [PubMed] [Google Scholar]
- Zhang G, Zhao F, Chen L, Pan Y, Sun L, Bao N, Zhang T, Cui C-X, Qiu Z, Zhang Y, et al. (2019). Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 5, 491–497. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Lozano-Torres JL, Blilou I, Zhang X, Zhai Q, Smant G, Li C, and Scheres B (2019). A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177, 942–956.e14. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, and Boller T (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. [DOI] [PubMed] [Google Scholar]
- Züst T, and Agrawal AA (2017). Trade-offs Between plant growth and defense Against insect herbivory: an emerging mechanistic synthesis. Annu. Rev. Plant Biol 68, 513–534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data files have been submitted to GEO and are publicly available as of the date of publication under accessions numbers GSE168381 (ATAC-seq data) and GSE168385 (RNA-seq data).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


