Abstract
The increasing mutation frequency of the SARS-CoV-2 virus and the emergence of successive variants have made correct diagnosis hard to perform. Developing efficient and accurate methods to diagnose infected patients is crucial to effectively mitigate the pandemic. Here, we developed an electrochemical immunosensor based on SARS-CoV-2 antibody cocktail-conjugated magnetic nanoparticles for the sensitive and accurate detection of the SARS-CoV-2 virus and its variants in nasopharyngeal swabs. The application of the antibody cocktail was compared with commercially available anti-SARS-CoV-2 S1 (anti-S1) and anti-S2 monoclonal antibodies. After optimization and calibration, the limit of detection (LOD) determination demonstrated a LOD = 0.53–0.75 ng/mL for the antibody cocktail-based sensor compared with 0.93 ng/mL and 0.99 ng/mL for the platforms using anti-S1 and anti-S2, respectively. The platforms were tested with human nasopharyngeal swab samples pre-diagnosed with RT-PCR (10 negatives and 40 positive samples). The positive samples include the original, alpha, beta, and delta variants (n = 10, for each). The polyclonal antibody cocktail performed better than commercial anti-S1 and anti-S2 antibodies for all samples reaching 100% overall sensitivity, specificity, and accuracy. It also showed a wide range of variants detection compared to monoclonal antibody-based platforms. The present work proposes a versatile electrochemical biosensor for the indiscriminate detection of the different variants of SARS-CoV-2 using a polyclonal antibody cocktail. Such diagnostic tools allowing the detection of variants can be of great efficiency and economic value in the fight against the ever-changing SARS-CoV-2 virus.
Keywords: SARS-CoV-2, Electrochemical biosensor, Antigen test, Magnetic nanoparticles, Antibody cocktail, Nasopharyngeal swab specimens
Graphical abstract
1. Introduction
The rapid mutation of the SARS-CoV-2 virus is one of the major problems limiting the quick development of treatments and vaccination. Since its emergence, there have been many variants discovered around the world. The first identified variant was the alpha variant (B.1.1.7) that appeared in the United Kingdom. Later, the beta variant (B.1.351) was identified in South Africa, followed by the delta variant (B.1.617.2) in India, and then other new variants followed everywhere [1]. The emergence of these variants was associated with a high increase in contagiousness, which made controlling the fast spread difficult. From a public health perspective, monitoring SARS-CoV-2 infections need to cover the various variants during screening.
Several approaches have been proposed for SARS-CoV-2 detection and diagnosis [2]. The most used process in routine practice is real-time polymerase chain reaction (RT-PCR) that detect nucleic acids associated with virus particles. This costly method requires lengthy sample preparation and highly trained experts [3]. Taken together, interest in point-of-care (POC) diagnostic tools has increased. Traditional POC tools such as lateral flow assays are based on immunologic analysis to detect viral proteins in samples [4]. They present an excellent choice to tackle the rapid propagation of the pandemic due to their fast turnaround, portability, and simplicity compared with RT-PCR. However, they face significant drawbacks when it comes to sensitivity, especially since some patients have been reported to have a low virus load in the case of SARS-CoV-2.
Electrochemical biosensor platforms are one of the current modalities used as diagnostic tools due to their potential for cost reduction, small size, and portability [5,6]. Screen-printed carbon electrodes (SPCE) are often preferred in sensor platforms due to their small size and suitability for low sample volumes [3,7]. The electrode surface can be modified with various nanoparticles such as gold [8] or carbon nanotubes [9] that increase the surface area of the working electrodes. This increases the number of immobilized detecting agents and increases sensing accuracy [10,11]. Magnetic nanoparticles (MNPs) are among the most interesting nanoparticles used in biomedicine, especially SPCE sensors, due to their easy synthesis process. MNPs have exceptional properties such as high field irreversibility, high saturation field, superparamagnetic and additional anisotropy contributions [4]. These magnetic properties allow them to form a biocompatible disposable recognition surface [12] and increase the sensitivity and stability of the sensing systems [13].
POCs using immunologic interactions (antibodies and antigens) for their sensing efficiency cannot keep up with the increasing number of variants, especially when a new antibody/antigen has to be developed each time. As such, practical tools for the efficient and cost-effective detection of the SARS-CoV-2 virus and its various variants are strongly needed. Indeed, the selection of antibodies is an important consideration in preparing immunosensors. The most accurate result is obtained when the antigen in the sample and the antibody are properly matched [3,14]. In this context, antibody selection is even more critical for platforms designed to detect mutant viruses such as SARS-CoV-2. In this case, it is advantageous to use antibody cocktails with antibodies that recognize multiple viral proteins. According to some reports, antibody cocktails have a higher virus detection rate (including variants) and allow the detection of variants that evade the antibodies used in traditional testing methods (mainly represented by monoclonal antibodies) [15,16]. Even if it is impossible to distinguish which variant is involved, a diagnosis can be made thanks to the interaction of these immobilized antibody cocktails with the antigens in the sample. This is because the detection is mainly achieved through various targets that indiscriminately recognize the virus regardless of its mutation [16].
The present study reports the development of an “All-in-one” electrochemical immunosensor for the indiscriminate diagnosis of SARS-CoV-2 and its variants using a combination of SPCE, MNPs, and an antibody cocktail. For this purpose, three different MNP-based immunosensor platforms were prepared. Two of these platforms were constructed using commercially available SARS-CoV-2-specific anti-S1 and anti-S2 antibodies and compared with the platform using the antibody cocktail to detect the original, alpha, beta, and delta variants of SARS-CoV-2. The analysis performance and data were performed and obtained through electrochemical measurement, including differential pulse voltammetry (DPV), cyclic voltammetry (CV), and electrochemical impedance spectroscopy (EIS).
2. Material and methods
2.1. Chemicals and instruments
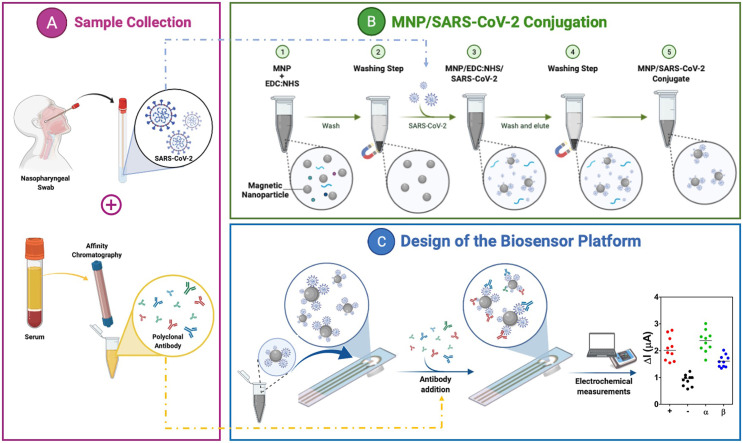
Details of chemicals and instruments can be found in the supplementary information. The experimental steps performed in this study are summarized in Fig. 1 .
Fig. 1.
Illustration of the MNP-based electrochemical immunosensor platform for detecting COVID-19 (and its variants). (A) Nasopharyngeal samples were obtained from patients and identified using RT-PCR for SARS-CoV-2 and its variants. At the same time, antibodies were purified from positive human serums using affinity chromatography and confirmed via specific SARS-CoV-2 ELISA. (B) Conjugation steps of the magnetic nanoparticles (MNPs) for sample preparation. (C) Design and application of the electrochemical sensor in the detection of SARS-CoV-2.
2.2. Collection of clinical specimens
A total of 50 nasopharyngeal swabs were collected at Ege University Hospital (Izmir, Turkey) with informed consent and ethical approval from the Ege University Clinical Research Ethics Committee (No. 20-8T/28). Samples were analyzed using RT-PCR and divided into positive (n = 40) and negative (n = 10) groups. Positive samples were further divided according to the variant (original, alpha, beta, and delta). All samples were coded, de-identified, aliquoted, and stored at −80 °C until use. Swab samples (n = 50) were analyzed with a commercial SARS-CoV-2 nasal antigen rapid test. Serum samples (n = 20) were also collected as before and analyzed by SARS-CoV-2 ELISA assay. Serums were obtained to purify a polyclonal antibody cocktail following the procedure described in our previous work using Protein A affinity chromatography [14].
2.3. MNPs synthesis
MNPs were synthesized by the coprecipitation method according to the procedure of Sanli et al. [12]. Briefly, equal volumes (100 mL) of 2.0 M FeCl3 and 1.0 M FeCl2 solutions were stirred (2300 rpm) at 80 °C under a stream of nitrogen gas. Then, 10 mL of a 25% NH4OH solution was slowly dropped into the solution, followed by a 30 min incubation. The resulting black MNP solution was washed a few times with water and ethanol, and then the MNPs were dried at 80 °C for 6 h. The MNPs were stored at room temperature until use.
2.4. Modification
MNPs were prepared by amino functionalization following Stöber's modification method [17]. MNPs (300 mg) were suspended in 100 mL 80% ethanol and 2.5 mL ammonium solution. After sonication of the solution, 5.0 mL of TEOS (tetraethyl orthosilicate) was added, and the solution was incubated for 4 h at 40 °C. The solution was washed a few times with methanol to remove unreacted TEOS. For the amino-functional MNPs, the obtained silica-coated MNPs were suspended in 100 mL methanol and 5.0 mL of APTES mixture. After sonication for 15 min, the obtained solution was mixed for 6 h at 60 °C. Excess APTES was washed with methanol and water, and the particles were dried at room temperature. The amino-functional MNPs were stored at room temperature until use.
2.5. Bioconjugation
SARS-CoV-2 specific S1 and S2 proteins were attached to the surface of amine-functional MNPs via EDC/NHS chemistry, according to the study of Singh et al. [18]. To this end, 10 mg EDC and 1.7 mg NHS were mixed with 0.5 mg MNP in 150 mL MES buffer (0.1 M, pH 6.0) and allowed to react for 10 min at room temperature with shaking. The MNPs were collected with a neodymium magnet and washed twice with distilled water. The activated MNPs were resuspended in 250 mL PBS buffer (0.01 M, pH 7.4). Subsequently, different concentrations of SARS-CoV-2 specific S1 or S2 proteins (2.5, 10, 25, 50, 75, 100, 150, 200 ng/mL) prepared in PBS buffer were added to 50 mL of MNP solution to a final volume of 300 mL and allowed to react for 2 h at room temperature. The resulting MNP-antigen conjugates were collected with a magnet, washed twice with 1.0% BSA solution prepared in PBS buffer, and finally resuspended in 100 mL of the same solution. All antigen-MNP conjugates were freshly prepared before each use.
2.6. Immunosensor fabrication
To prepare the immunosensor platforms, after performing electrochemical measurements on the bare SPCE, a neodymium magnet was placed in the center of the backside of the SPCE working electrode, and 20 μL of MNP-antigen conjugates were applied to the SPCE surface. Then, 5.0 μL of the 250 μg/mL SARS-CoV-2 specific anti-S1 antibody, anti-S2 antibody, and the antibody cocktail prepared in 1X PBS buffer (pH 7.4) were added to the electrode surface and incubated at room temperature for 30 min. After each modification step, electrochemical measurements were performed. The biosensor responses were calculated based on the difference of the signals before and after the addition of the antibodies, giving the difference of the current values. The electrochemical measurements include differential pulse voltammetry (DPV), cyclic voltammetry (CV), and electrochemical impedance spectroscopy (EIS) (detailed protocol can be found in the supplementary information).
2.7. Optimization and calibration
To optimize the antibodies concentration, the immunosensor platform was designed as previously described, and antibodies were added at different concentrations (25, 50, 125, 250, and 500 μg/mL) and left for 30 min at room temperature. To optimize the incubation time of antibodies, DPV measurements were performed 15, 30, 45 and 60 min after antibodies addition (250 μg/mL) over the surface.
The different immunosensor platforms were calibrated by applying increasing concentrations of the various antigens (S1 and S2) (1.0–200 ng/mL) followed by the addition of the various antibodies at 250 μg/mL. DPV measurements were performed, and the current difference between the antigens and the antibodies addition (ΔμA) was calculated and used for the determination of the analytical features of the immunosensors.
2.8. Sample application
Clinical nasopharyngeal swabs (grouped as original variant (n = 10), alpha variant (n = 10), beta variant (n = 10), delta variant (n = 10), and negative (n = 10)) and vNAT buffer (facilitating extraction) were diluted 100-fold with 1X PBS (pH 7.4) and conjugated with MNPs activated via EDC/NHS according to the conjugation procedure described above. The conjugates (20 μL) were applied to the bare SPCE surface. After washing and drying the SPCE, 5.0 μL of the 250 μg/mL SARS-CoV-2 specific anti-S1 antibody, anti-S2 antibody, and antibody cocktail prepared in 1X PBS buffer (pH 7.4) were dropped onto the electrode surface and incubated at room temperature for 30 min. Electrochemical measurements were performed after each modification step.
2.9. Data and statistical analysis
Results are presented as mean ± standard deviation (SD). The data for the calibration was fit to a nonlinear four-parameter logistic curve, and all parameters were calculated as described previously [19]. The limit of detection (LOD) was calculated as LOD = LOB+(1.645 × SDlow conc. sample). LOB is the limit of blank and is calculated as LOB = meanblank+(1.645xSDblank). Other analytical features such as sensitivity, specificity, and accuracy were calculated according to the formulas described in the Supporting Information. The threshold calculations were performed using the Boxplot analysis method. The data analysis was performed using GraphPad Prism V8.0 software.
Statistical analyses were carried out using one-way analysis of variance (ANOVA), multiple comparisons were performed using a non-parametric test with Kruskal-Wallis and Dunn's posthoc test (GraphPad Prism 8.0). p-values < 0.05 were considered as statistically significant.
3. Results and discussion
Managing SARS-CoV-2 has been mostly revolving around the efficient diagnosis of infections. Therefore, many efforts were spent developing specific tools targeting functionally active structures of the virus [20]. The main proteins targeted for SARS-CoV-2 diagnostic are the spike glycoprotein (S), envelope protein (E), matrix protein (M), and nucleocapsid protein (N) [21]. Although RT-PCR is currently the standard method for diagnosing SARS-CoV-2, it has some inconveniences such as the cost, need of trained personnel, and centralization [22]. As such, researchers turned toward developing point-of-care (POC) tools and portable biosensors to circumvent the difficulties seen in PCR. Most of the developed biosensors are designed to detect the S and N proteins of the virus using commercial antibodies [[23], [24], [25], [26]]. The S protein is the preferred target for diagnosing SARS-CoV-2 as it is required for viral entry into host cells and, therefore, directly contributes to the virulence of SARS-CoV-2. The S1 subunit has low evolutionary protein homologies within the coronavirus family, suggesting that it may potentially exhibit less cross-reactivity among endemic coronaviruses [27]. The N protein plays an essential role in the transcription and replication of viral RNA, packaging the encapsulated genome into virions and inhibiting the cell cycle process of the host cells. The N protein is abundantly expressed during infections and has high immunogenic activity. Therefore, it is crucial for antibody-based detection of SARS-CoV-2. However, the N protein homology between SARS-CoV-2 and SARS-CoV-1 is estimated at 90% compared with S protein (77%), specifically the S1 subunit including the RBD (66%). This makes the N protein unfavorable in terms of cross-reactivity [28]. Recent studies have shown that N protein-based antibody assays can exhibit a higher false-negative rate than the S1 subunit. The S1 subunit purified from mammalian cells shows the highest performance distinguishing COVID-19 patients from controls [29,30]. Despite all this, the mutation occurrences on the S protein that continuously produce newly emerging variants make this target difficult and less trustworthy.
In this work, an electrochemical immunosensor system based on antigen-antibody affinity using magnetic nanoparticles and an antibody cocktail was developed for the indiscriminate detection of SARS-CoV-2, including its variants. The platform's performance was compared with the same system using commercial SARS-CoV-2-specific anti-S1 and anti-S2 monoclonal antibodies. These platforms are designed as antigen tests. Typically, monoclonal antibodies (mAbs) are used for such antigen tests due to their high specificity. Yet, polyclonal antibodies have a high capacity to detect different epitopes, which is of great importance due to the continuous emergence of new SARS-CoV-2 variants [16,31,32].
Studies suggest the use of mAb cocktails for the treatment of COVID-19 [16,33]. In these studies, it was observed that the viral load decreased when synthetically prepared mAbs were mixed and administered to the patient to mimic the immune system. In another study, mAbs were also effective against variants [16]. These therapeutic approaches are also known as passive vaccination. In addition to therapeutic approaches and vaccines, evidence is accumulating for the use of antibody cocktails in diagnosis. In our previous study, cocktail antibodies were used to develop paper-based lateral flow platforms with dye-loaded polymersomes to detect the original variant of SARS-CoV-2 antigens [14]. The current study aimed to take further advantage of the polyclonal antibody cocktail advantages in detecting the different variants of SARS-CoV-2 following our latest work demonstrating that the polyclonal antibody cocktail can identify specific regions from the English variant through mass spectrometry analysis [34].
3.1. Conjugate characterization
MNPs were synthesized and functionalized with amino groups to be used for the development of immunosensors. Amino functional MNPs were conjugated with target proteins (S1, S2, and nasopharyngeal swabs) to generate different immunosensors via EDC:NHS crosslinkers. SEM images were performed to characterize the MNPs and conjugates. An increase in size was observed after conjugating the SARS-CoV-2 protein with the amino-functional MNP (Fig. S1).
3.2. Surface modification of immunosensors
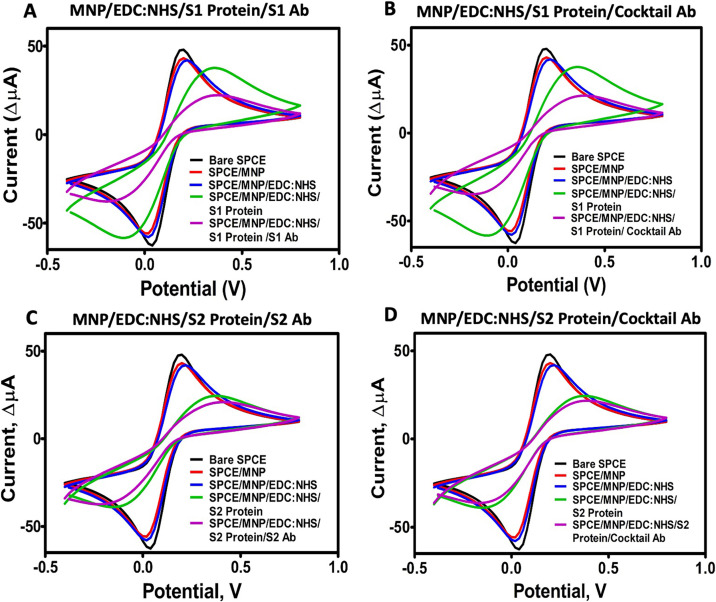
The electrochemical behavior of the developed immunosensors was investigated by electroanalytical techniques (cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS)). When the non-conjugated amino-functional MNPs, MNPs with EDC:NHS crosslinkers, and target proteins-conjugated MNPs were dropped onto the empty SPCE, a typical decrease in both oxidation and reduction of CV peak heights were observed [[35], [36], [37]]. The reduction of peaks can be explained because large groups of bulky materials added to the surface prevented electrons from entering the electrode surface. Although there was no significant difference between the peaks of EDC:NHS-treated MNP and amino-functional MNP, the peak decreased significantly after the MNP surface was conjugated with target proteins (Fig. 2 ). This decrease indicates the successful conjugation between the amino group of the MNP and the carboxyl groups of target proteins via the EDC:NHS chemistry.
Fig. 2.
Cyclic voltammetry (CV) analysis of the modification steps of the immunosensor systems. (A) S1 Protein/S1 Ab pair, (B) S1 Protein/Ab cocktail pair, (C) S2 Protein/S2 Ab pair, (D) S2 Protein/Ab cocktail pair.
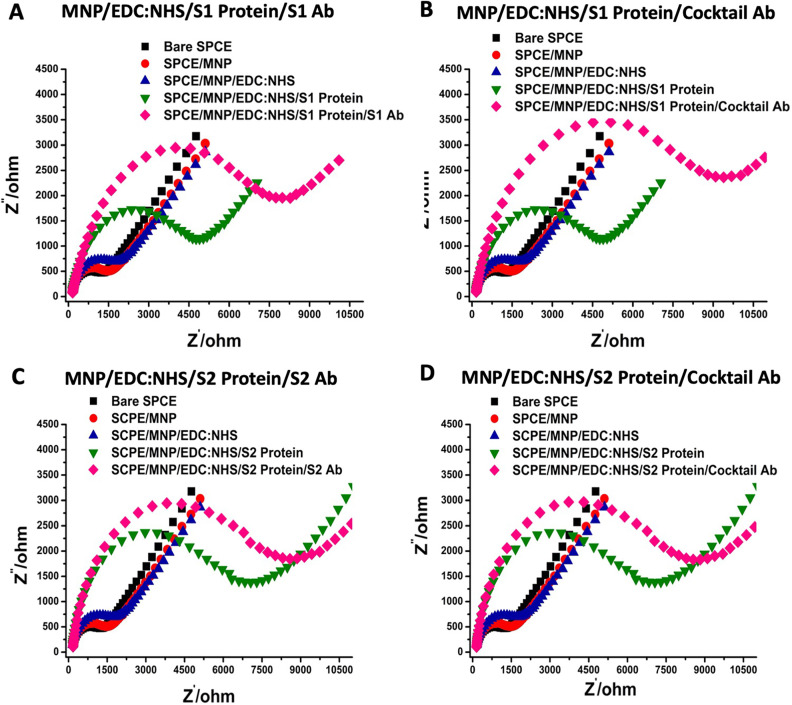
In the next step, commercial SARS-CoV-2-specific anti-S1, anti-S2 mAbs, and the antibody cocktail were applied to the surface. Based on the successful protein-antibody interactions, similar electrochemical behaviors were observed in this step. The recognition of the S1 and S2 proteins by their respectively specific antibodies happened as expected. Furthermore, a polyclonal antibody cocktail could identify both S1 and S2 antigens, as demonstrated by the CV measurements (Fig. 2). Additionally, EIS measurements were also performed and analyzed by the Nyquist plot. This plot consists of two parts, including lower and upper frequencies. At lower frequencies, the diagonal lines observed indicate the typical diffusion of the redox behavior on the electrode surface, while at higher frequencies, the observed semicircular peaks indicate the change in electron transfer resistance. To annotate the EIS measurements, an electrical circuit was designed including Rs (solution resistance), Zw (Warburg impedance), Cdl (double layer capacitance), and Rct (change in electron transfer resistance). As expected, the resistance values increased and continued to grow after each modification step of the immunosensor platform (Fig. 3 ). The data of the CV and EIS measurements are summarized in Table S1-S2.
Fig. 3.
Electrochemical impedance spectroscopy (EIS) analysis of the immunosensor systems at each modification step. (A) S1 Protein/S1 Ab pair, (B) S1 Protein/Ab cocktail pair, (C) S2 Protein/S2 Ab pair, (D) S2 Protein/Ab cocktail pair.
3.3. Optimization studies for antibody immobilization
The immobilization time of the antibodies, which significantly affects the performance of the MNP-based immunosensor platforms was optimized using a 250 μg/mL antibody concentration and incubated for different periods. The incubation of 30 min demonstrated the best optimal condition for the signal analysis (Fig. S2 A-C). The optimization steps also addressed the concentration of the antibody immobilized over the biosensor. The MNP-based immunosensor prepared by incubating 250 μg/mL antibody showed the best oxidative current response for each antibody and was considered as optimal (Fig. S2 D-F).
3.4. Analytical features
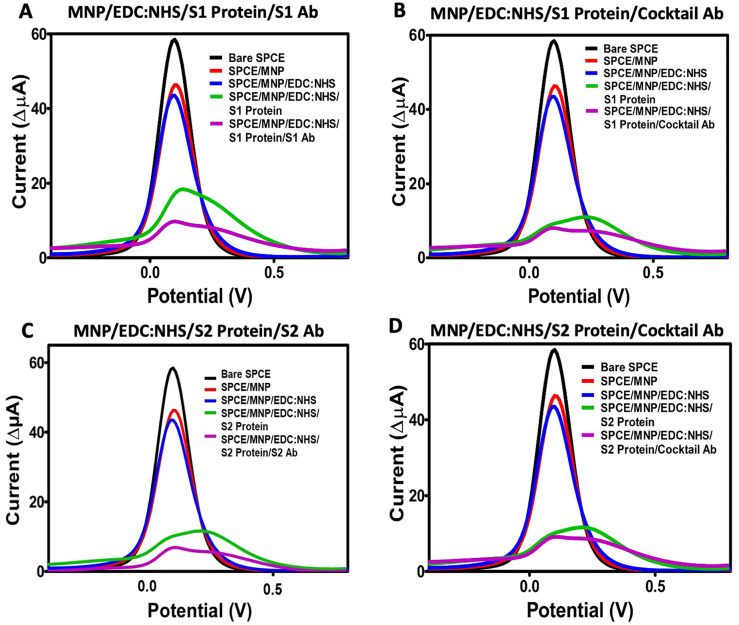
Differential pulse voltammetry (DPV) measurements were used to determine other analytical parameters of the developed immunosensors. Similar to the results obtained from CV, a decrease in oxidation peaks was observed after each modification step (Fig. 4 ).
Fig. 4.
Differential pulse voltammetry (DPV) measurements of all modification steps of the immunosensor systems. (A) MNP/EDC:NHS/S1 Protein/S1 Ab; (B) MNP/EDC:NHS/S1 Protein/Cocktail Ab; (C) MNP/EDC:NHS/S2 Protein/S2 Ab; (D) MNP/EDC:NHS/S2 Protein/Cocktail Ab.
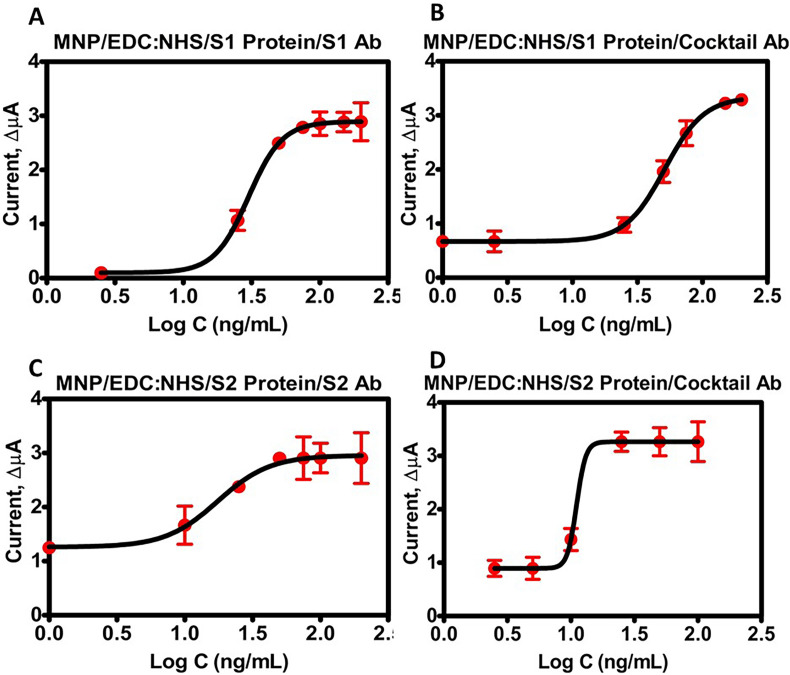
Calibration graphs were made using the current difference of DPV peaks obtained by adding antibodies and MNP-conjugated to target proteins over the electrode surface (Fig. 5 ). The data was fit onto a nonlinear four-parameter logistic curve. The LOD of the MNP-S1 protein conjugate-based immunosensor was calculated to be 0.93 ng/mL after the addition of the S1 antibody. In contrast, the use of the antibody cocktail yielded 0.53 ng/mL (Fig. 5A and B, Table 1 ). For the MNP-S2 protein conjugate, the LOD was estimated as 0.99 ng/mL with anti-S2 antibody and 0.75 ng/mL for the antibody cocktail (Fig. 5C and D, Table 1).
Fig. 5.
Calibration graphs of the immunosensor systems. (A) MNP/EDC:NHS/S1 Protein/S1 Ab, (B) MNP/EDC:NHS/S1 Protein/Cocktail Ab, (C) MNP/EDC:NHS/S2 Protein/S2 Ab, (D) MNP/EDC:NHS/S2 Protein/Cocktail Ab.
Table 1.
Analytical parameters.
| Analytical Parameters | Values |
|||
|---|---|---|---|---|
| MNP-S1/S1 Ab | MNP-S1/Cocktail Ab | MNP-S2/S2 Ab | MNP-S2/Cocktail Ab | |
| Detection Range (ng/mL) | 2.5–200 | 1–200 | 1–200 | 2.5–100 |
| a (zero concentration) | 0.099 | 0.493 | 1.249 | 0.894 |
| b (slope factor) | 3.502 | 2.861 | 2.715 | 12.029 |
| c (mid-range concentration) | 29.977 | 51.209 | 16.504 | 11.067 |
| d (infinite concentration) | 2.894 | 3.343 | 2.913 | 3.265 |
| Correlation coefficient | 0.952 | 0.994 | 0.771 | 0.921 |
| LOB (ng/mL) | 0.828 | 0.510 | 0.835 | 0.510 |
| LOD (ng/mL) | 0.930 | 0.538 | 0.994 | 0.756 |
| Repeatability (±SD) | 0.03 | 0.008 | 0.08 | 0.051 |
| Coefficient of variation (%) | 2.630 | 3.870 | 3.340 | 3.360 |
The repeatability (±SD) was tested by measuring the current differential at the same concentration on different electrodes (n = 3) by selecting the midpoint of the calibration graph for each sensor platform (Table 1). The coefficient of variation was calculated accordingly and was less than 10% for the four different sensors. The low coefficients of variation obtained for each sensor confirmed that the immunosensors have high repeatability [38]. The various analytical parameters analyzed for the current electrochemical immunosensor are summarized in Table 1.
3.5. SARS-CoV-2 analysis in clinical samples
The RT-PCR confirmed clinical samples were used to investigate the detection performance of the developed SARS-CoV-2 immunosensors using different antibodies. A total of 50 nasopharyngeal swabs were collected from patients, of which ten were negative, and the rest were positive (n = 40). Moreover, the positive samples were classified as original, alpha, beta, and delta variants according to RT-PCR data.
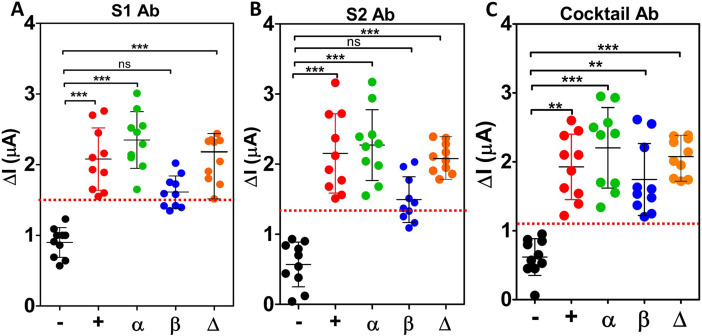
In the designed immunosensor platforms, MNP was conjugated to the swab samples and vNAT buffer. DPV measurements were performed before and after immobilization of the antibodies on the electrode surface, and the oxidation current difference was calculated. Fig. 6 shows the electrochemical immunosensor response obtained for the anti-S1 antibody, anti-S2 antibody, and antibody cocktail for swab samples from patients with negative, original, alpha, beta, and delta variants. In addition, Tables S3 and S4 provide the current differential values obtained for each sample and vNAT buffer. Differential values for the vNAT buffer were similar to the current differential values for the negative samples. These results indicate that the vNAT buffer did not cause interference on the sensor platform. The response of the sensor platforms to clinical samples was classified by calculating the threshold for each immunosensor, with samples that responded below the threshold classified as negative and values that responded above the threshold as positive. The red lines in Fig. 6 demonstrate the threshold values: 1.5 μA for the anti-S1 antibody, 1.4 μA for the anti-S2 antibody, and 1.1 μA for the antibody cocktail. The results of the MNP-based immunosensor platform developed for each antibody to detect the SARS-CoV-2 and its different variants were compared with the Ct values data and a commercially available rapid antigen test (lateral flow assay) (Table S4). Using these results, the sensitivity, specificity, and accuracy were calculated for the three different immunosensors (Table 2 ). The antibody cocktail provided the best analytical features reaching 100% accuracy, specificity, and sensitivity compared with the other two platforms (Table 2).
Fig. 6.
Electrochemical signals of COVID-19 patients swab specimens (+(original), α, β, and Δ variants) and non-infected individuals (negative “-”). Each bar represents the mean ± SD of three separate measurements. The red lines represent the threshold lines. Below this line indicates negative results, and above indicates positive results. (A) MNP/EDC:NHS/Clinical samples/anti-S1 antibody platform, (B) MNP/EDC:NHS/Clinical samples/anti-S2 antibody platform, (C) MNP/EDC:NHS/Clinical sample/Antibody Cocktail sensor platform. ns: non-significant; *: p > 0.05, **: p < 0.01, and ***: p < 0.001 compared with the negative group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Clinical performance of three different immunosensor platforms.
| RT-PCR |
Sensitivity | Specificity | Accuracy | |||
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| Anti-S1 antibody immunosensor | Positive | 36 | 0 | 90% | 100% | 92% |
| Negative | 4 | 10 | ||||
| Total | 40 | 10 | ||||
| Anti-S2 antibody immunosensor | Positive | 35 | 0 | 87.5% | 100% | 90% |
| Negative | 5 | 10 | ||||
| Total | 40 | 10 | ||||
| Antibody cocktail immunosensor | Positive | 40 | 0 | 100% | 100% | 100% |
| Negative | 0 | 10 | ||||
| Total | 40 | 10 | ||||
Statistical analysis of the different immunosensors responses to SAR-CoV-2 variants showed that the anti-S1 antibody-based platform had a significant difference (p < 0.001) between the negative samples and the original variant, alpha variant, and delta variant (Fig. 6A). However, no difference was found between the negative and beta variant for this immunosensor (p > 0.05). These results are similar for the anti-S2 antibody-based immunosensor (Fig. 6B). This suggests that the beta variant may contain mutations and structural changes that limit the recognition ability of the monoclonal antibodies targeted against the original variant. It further confirms the reported information that the variants are resistant to neutralization by antibodies erected against the original variant [16]. The statistical analysis of the antibody cocktail-based sensor resulted in a significant difference between negative samples and all the variants regardless of their type (p < 0.05) (Fig. 6C).
The results show that the immunosensor designed with the antibody cocktail has a higher ability to discriminate between positive and negative samples and a higher variant detection rate. Our information on SARS-CoV-2 and its variants suggest that they contain mutations that can cause functional changes, especially in the spike glycoprotein, unlike the original virus [39]. The alpha variant has amino acid changes, deletions, insertions, and synonymous mutations. It contains several biologically important mutations, including E484K and N501Y. Similarly, the E484K mutation was also found in the beta variant [40], whereas in the delta variant, the L452R and P681R mutations in the S protein were most prominent [41]. Due to these alterations, the efficacy of antigen tests that detect the S protein is limited against the variants. On the other hand, tests developed with the polyclonal antibody cocktail that recognize SARS-CoV-2 proteins are more effective in detecting variants. The results of our study also showed that the assay platform designed with the antibody cocktail could detect spike or nucleocapsid-specific immunoglobulins in swap samples for all variants. This is related to the advantages of the antibody cocktail mentioned earlier. In addition, it is possible to detect not only one but all immunoglobulins whose levels change at different times after infection. Furthermore, the antibody cocktail sensor proved to be more suitable for future studies considering the increasing number of mutations of SARS-CoV-2, as it provided significant results for the different variants.
All swab samples were applied to a commercially available rapid antigen test kit, and the results' representative images are shown in Fig. S3. According to the rapid antigen test results, it reacted directly proportional to the viral load in all variants. The increase in Ct values (lower viral load) was associated with an increased frequency of false negatives. This demonstrates the success of the systems developed in this study compared to an existing rapid antigen test. We also assessed the developed biosensor with other sensor platforms described in the literature (Table 3 ). The main advantage of our proposed system compared with other platforms resides in using an antibody cocktail-MNP combination for the indiscriminate detection of various variants. Although our immunosensor platforms have a similar working performance to other systems, they are superior because most of the other platforms are focused on a single target that can be affected if a new mutation touches the targeted structures whereas, our system can be seen as an all-in-one system in terms of variants detection.
Table 3.
Overview of reported electrochemical biosensor platforms for SARS-CoV-2 diagnosis.
| Electrochemical Method | Target Biomarker | Linear Range | LOD | Biological Sample | Ref. |
|---|---|---|---|---|---|
| DPV measurement | S1 spike protein | 5.0–500 ng/mL | 2.9 ng/mL | Human serum | [42] |
| Paper based electrochemical measurement | RNA of the N gene | 585.4–5.854 × 107 copies/μL | 6.9 copies/μL | Nasopharyngeal swab | [43] |
| DPV measurement | Spike protein | 0.04–10 μg/mL | 19 ng/mL | Untreated saliva | [44] |
| Nucleocapsid protein | 0.01–0.6 μg/mL | 8.0 ng/mL | |||
| DPV measurement | ORF1ab | 10−2 fM-1.0 pM | 200 copies/mL | Throat swab, Urine, Feces, Plasma, Whole blood, Saliva | [45] |
| DPV measurement | Nucleocapsid protein | 0.22–333 fM | 27 fM | Nasopharyngeal swab | [46] |
| EIS measurement | S protein | 1.0 fM-1.0 nM | 2.8 fM | Fetal bovine Serum, Rabbit Serum | [47] |
| RBD | 16.9 fM | ||||
| SWV measurement | SP-RBD | 1.0–1000 ng/mL | 0.11 ng/mL | Human sera | [48] |
| SWV measurement | SARS-CoV-2 spike antibody | 0.1–1000 ag/mL | 0.01 ag/mL | Pretreated saliva and oropharyngeal swab | [49] |
| DPV measurement | MNP-S1/anti-S1 Ab | 2.5–200 ng/mL | 0.93 ng/mL | Nasopharyngeal swab (original, alpha, beta, and delta variants) | This work |
| MNP-S1/Ab cocktail | 1.0–200 ng/mL | 0.53 ng/mL | |||
| MNP-S2/anti-S2 Ab | 1.0–200 ng/mL | 0.99 ng/mL | |||
| MNP-S2/Ab cocktail | 2.5–100 ng/mL | 0.75 ng/mL |
AuNP: Gold nanoparticle, Cys: Cysteamine, DPV: Differential pulse voltammetry, EIS: Electrochemical impedance spectroscopy, FET: Field-effect transistor, LSG: Laser-scribed graphene, MNP: Magnetic nanoparticles, ORF: Open reading frame, PBASE: 1-pyrenebutyric acid N-hydroxysuccinimide ester, RBD: Receptor binding domain, SP-RBD: Spike protein receptor-binding domain, SWV: Square-wave voltammetry.
3.6. Limitations and perspectives
This study can be considered an “emergency case technology” in the context of new information, product development, and rapid adaptation strategies to help control the current pandemic. Because the pandemic is an unusual and unpredictable global problem, knowledge is growing daily. As new variants emerge, there is a need for adaptive technologies that can adapt quickly. Clearly, the antibody cocktails used in this study represent a way to increase sensitivity in detecting viral antigens by recognizing different targets [34]. However, it is unavoidable that using these antibodies derived from human samples is not a sustainable resource. Other alternative approaches are needed, both to establish a stable production source and to be able to recognize new variants. In this context, further studies should be conducted to use aptamer-based biorecognition molecules or to produce recombinant antibodies as an alternative for systems that can rapidly adapt to the current and next possible pandemic processes, which will include current and potential mutations.
4. Conclusion
As long as the pandemic COVID-19 continues, there is a need for sensitive and accurate alternative diagnostic methods against its variants. We propose the concept of an “all-in-one” biosensor as a versatile sensor that can detect various variants simultaneously. In this study, three different immunosensor platforms prepared with different antibodies were investigated using the same series of experiments and clinical samples. The literature search revealed that this study is the first to compare magnetic nanoparticle-based immunosensor systems prepared with an antibody cocktail purified by affinity chromatography from patient serum samples, as well as commercially available anti-S1 and anti-S2 antibodies. The design proposed with an antibody cocktail proved to be more successful in terms of sensitivity, accuracy, and specificity in detecting variants. This study contributes to the current debate on SARS-CoV-2 variants detection. We believe our method can be used as a potential diagnostic tool during this global crisis and other COVID-19 like pandemics.
Credit author statement
Ceren Durmus: Methodology, Validation, Formal analysis, Investigation, Writing - Original Draft, Writing - Review & Editing, Visualization. Simge Balaban Hanoglu: Methodology, Validation, Formal analysis, Investigation, Writing - Original Draft, Writing - Review & Editing, Visualization. Duygu Harmanci: Methodology, Validation, Formal analysis, Investigation, Writing - Original Draft, Writing - Review & Editing, Visualization. Hichem Moulahoum: Methodology, Validation, Writing - Original Draft, Writing - Review & Editing, Project administration. Kerem Tok: Validation, Investigation. Faezeh Ghorbanizamani: Methodology, Validation, Writing - Original Draft, Writing - Review & Editing, Project administration. Serdar Sanli: Investigation. Figen Zihnioglu: Conceptualization, Methodology, Resources, Supervision, Project administration. Serap Evran: Resources, Project administration. Candan Cicek: Resources. Ruchan Sertoz: Resources. Bilgin Arda: Resources. Tuncay Goksel: Resources. Kutsal Turhan: Resources. Suna Timur: Conceptualization, Methodology, Validation, Resources, Writing - Review & Editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This study was supported by The Scientific and Technological Research Council of Turkey (TUBITAK, Project Grant No: 120R021) and Republic of Turkey, Ministry of Development (Project Grant No: 2016K121190). The authors are thankful to the EGE-MATAL for providing its facilities for all experimental stages.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2022.123356.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bernal J.L., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G. New England Journal of Medicine; 2021. Effectiveness of Covid-19 Vaccines against the B. 1.617. 2 (Delta) Variant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranjan P., Singhal A., Yadav S., Kumar N., Murali S., Sanghi S.K., Khan R. Rapid diagnosis of SARS-CoV-2 using potential point-of-care electrochemical immunosensor: toward the future prospects. Int. Rev. Immunol. 2021;40(1–2):126–142. doi: 10.1080/08830185.2021.1872566. [DOI] [PubMed] [Google Scholar]
- 3.Rahmati Z., Roushani M., Hosseini H., Choobin H. Electrochemical immunosensor with Cu2O nanocube coating for detection of SARS-CoV-2 spike protein. Mikrochim. Acta. 2021;188(3):105. doi: 10.1007/s00604-021-04762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akbarzadeh A., Samiei M., Davaran S. Magnetic nanoparticles: preparation, physical properties, and applications in biomedicine. Nanoscale Res. Lett. 2012;7(1):144. doi: 10.1186/1556-276X-7-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan R.S., Khurshid Z., Yahya Ibrahim Asiri F. Advancing point-of-care (PoC) testing using human saliva as liquid biopsy. Diagnostics. 2017;7(3):39. doi: 10.3390/diagnostics7030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei Q., Qi H., Luo W., Tseng D., Ki S.J., Wan Z., Gorocs Z., Bentolila L.A., Wu T.T., Sun R., Ozcan A. Fluorescent imaging of single nanoparticles and viruses on a smart phone. ACS Nano. 2013;7(10):9147–9155. doi: 10.1021/nn4037706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyszczuk-Rotko K., Kozak J., Sztanke M., Sztanke K., Sadok I. A screen-printed sensor coupled with flow system for quantitative determination of a novel promising anticancer agent candidate. Sensors. 2020;20(18):5217. doi: 10.3390/s20185217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pingarrón J.M., Yanez-Sedeno P., González-Cortés A. Gold nanoparticle-based electrochemical biosensors. Electrochim. Acta. 2008;53(19):5848–5866. [Google Scholar]
- 9.Wang J. Carbon‐nanotube based electrochemical biosensors: a review. Electroanalysis: Int. J. Devoted Fundament. Pract. Aspects Electroanal. 2005;17(1):7–14. [Google Scholar]
- 10.Mabey D., Peeling R.W., Ustianowski A., Perkins M.D. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004;2(3):231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 11.Urdea M., Penny L.A., Olmsted S.S., Giovanni M.Y., Kaspar P., Shepherd A., Wilson P., Dahl C.A., Buchsbaum S., Moeller G., Hay Burgess D.C. Requirements for high impact diagnostics in the developing world. Nature. 2006;444(1):73–79. doi: 10.1038/nature05448. Suppl 1. [DOI] [PubMed] [Google Scholar]
- 12.Sanli S., Ghorbani-Zamani F., Moulahoum H., Gumus Z.P., Coskunol H., Odaci Demirkol D., Timur S. Application of biofunctionalized magnetic nanoparticles based-sensing in abused drugs diagnostics. Anal. Chem. 2020;92(1):1033–1040. doi: 10.1021/acs.analchem.9b04025. [DOI] [PubMed] [Google Scholar]
- 13.Rocha-Santos T.A.P. Sensors and biosensors based on magnetic nanoparticles. Trac. Trends Anal. Chem. 2014;62:28–36. [Google Scholar]
- 14.Ghorbanizamani F., Tok K., Moulahoum H., Harmanci D., Hanoglu S.B., Durmus C., Zihnioglu F., Evran S., Cicek C., Sertoz R., Arda B., Goksel T., Turhan K., Timur S. Dye-loaded polymersome-based lateral flow assay: rational design of a COVID-19 testing platform by repurposing SARS-CoV-2 antibody cocktail and antigens obtained from positive human samples. ACS Sens. 2021;6(8):2988–2997. doi: 10.1021/acssensors.1c00854. [DOI] [PubMed] [Google Scholar]
- 15.Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., Logue J., Zuiani A., McCulloch D., Lelis F.J.N., Henson S., Monaco D.R., Travers M., Habibi S., Clarke W.A., Caturegli P., Laeyendecker O., Piechocka-Trocha A., Li J.Z., Khatri A., Chu H.Y., Collection M.C., Processing T., Villani A.C., Kays K., Goldberg M.B., Hacohen N., Filbin M.R., Yu X.G., Walker B.D., Wesemann D.R., Larman H.B., Lederer J.A., Elledge S.J. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370(6520) doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., Gilchuk P., Zost S.J., Tahan S., Droit L., Turner J.S., Kim W., Schmitz A.J., Thapa M., Wang D., Boon A.C.M., Presti R.M., O'Halloran J.A., Kim A.H.J., Deepak P., Pinto D., Fremont D.H., Crowe J.E., Jr., Corti D., Virgin H.W., Ellebedy A.H., Shi P.Y., Diamond M.S. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27(4):717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stöber W., Fink A., Bohn E. Controlled growth of monodisperse silica spheres in the micron size range. J. Colloid Interface Sci. 1968;26(1):62–69. [Google Scholar]
- 18.Singh V., Krishnan S. An electrochemical mass sensor for diagnosing diabetes in human serum. Analyst. 2014;139(4):724–728. doi: 10.1039/c3an01542d. [DOI] [PubMed] [Google Scholar]
- 19.Armbruster D.A., Pry T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008;29(Suppl 1):S49–S52. Suppl 1. [PMC free article] [PubMed] [Google Scholar]
- 20.Chang M.C., Hur J., Park D. Interpreting the COVID-19 test results: a guide for physiatrists. Am. J. Phys. Med. Rehabil. 2020;99(7):583–585. doi: 10.1097/PHM.0000000000001471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moulahoum H., Ghorbanizamani F., Zihnioglu F., Turhan K., Timur S. How should diagnostic kits development adapt quickly in COVID 19-like pandemic models? Pros and cons of sensory platforms used in COVID-19 sensing. Talanta. 2021;222:121534. doi: 10.1016/j.talanta.2020.121534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barlev-Gross M., Weiss S., Ben-Shmuel A., Sittner A., Eden K., Mazuz N., Glinert I., Bar-David E., Puni R., Amit S., Kriger O., Schuster O., Alcalay R., Makdasi E., Epstein E., Noy-Porat T., Rosenfeld R., Achdout H., Mazor O., Israely T., Levy H., Mechaly A. Spike vs nucleocapsid SARS-CoV-2 antigen detection: application in nasopharyngeal swab specimens. Anal. Bioanal. Chem. 2021;413(13):3501–3510. doi: 10.1007/s00216-021-03298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Lillehoj P.B. Microfluidic magneto immunosensor for rapid, high sensitivity measurements of SARS-CoV-2 nucleocapsid protein in serum. ACS Sens. 2021;6(3):1270–1278. doi: 10.1021/acssensors.0c02561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eissa S., Alhadrami H.A., Al-Mozaini M., Hassan A.M., Zourob M. Voltammetric-based immunosensor for the detection of SARS-CoV-2 nucleocapsid antigen. Mikrochim. Acta. 2021;188(6):199. doi: 10.1007/s00604-021-04867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng T., Sui Z., Huang Z., Xie J., Wen K., Zhang Y., Huang W., Mi W., Peng K., Dai X., Fang X. Point-of-care test system for detection of immunoglobulin-G and -M against nucleocapsid protein and spike glycoprotein of SARS-CoV-2. Sensor. Actuator. B Chem. 2021;331:129415. doi: 10.1016/j.snb.2020.129415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501 e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tilocca B., Soggiu A., Sanguinetti M., Musella V., Britti D., Bonizzi L., Urbani A., Roncada P. Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses. Microb. Infect. 2020;22(4–5):188–194. doi: 10.1016/j.micinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan X., Chen D., Xia Y., Wu X., Li T., Ou X., Zhou L., Liu J. Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect. Dis. 2020;20(4):410–411. doi: 10.1016/S1473-3099(20)30114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu R., Han H., Liu F., Lv Z., Wu K., Liu Y., Feng Y., Zhu C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin. Chim. Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Read A.J., Gauci C.G., Lightowlers M.W. Purification of polyclonal anti-conformational antibodies for use in affinity selection from random peptide phage display libraries: a study using the hydatid vaccine EG95. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009;877(14–15):1516–1522. doi: 10.1016/j.jchromb.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuravsky M.L., Schmalhausen E.V., Pozdnyakova N.V., Muronetz V.I. Isolation of antibodies against different protein conformations using immunoaffinity chromatography. Anal. Biochem. 2012;426(1):47–53. doi: 10.1016/j.ab.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 33.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., Perry C., Pan C., Hosain R., Mahmood A., Davis J.D., Turner K.C., Hooper A.T., Hamilton J.D., Baum A., Kyratsous C.A., Kim Y., Cook A., Kampman W., Kohli A., Sachdeva Y., Graber X., Kowal B., DiCioccio T., Stahl N., Lipsich L., Braunstein N., Herman G., Yancopoulos G.D., Trial I. REGN-COV2, a neutralizing antibody cocktail, in outpatients with covid-19. N. Engl. J. Med. 2021;384(3):238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tok K., Moulahoum H., Ghorbanizamani F., Harmanci D., Balaban Hanoglu S., Durmus C., Evran S., Cicek C., Sertoz R., Arda B., Goksel T., Turhan K., Timur S., Zihnioglu F. Simple workflow to repurpose SARS-CoV-2 swab/serum samples for the isolation of cost-effective antibody/antigens for proteotyping applications and diagnosis. Anal. Bioanal. Chem. 2021;413:7251–7263. doi: 10.1007/s00216-021-03654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durmus C., Aydindogan E., Gumus Z.P., Endo T., Yamada S., Coskunol H., Timur S., Yagci Y. Catechol-attached polypeptide with functional groups as electrochemical sensing platform for synthetic cannabinoids. ACS Appl. Polym. Mater. 2019;2(2):172–177. [Google Scholar]
- 36.Balaban S., Man E., Durmus C., Bor G., Ceylan A.E., Pinar Gumus Z., Evran S., Coskunol H., Timur S. Sensor platform with a custom‐tailored aptamer for diagnosis of synthetic cannabinoids. Electroanalysis. 2020;32(3):656–665. [Google Scholar]
- 37.Balaban S., Beduk T., Durmus C., Aydindogan E., Salama K.N., Timur S. Laser‐scribed graphene electrodes as an electrochemical immunosensing platform for cancer biomarker ‘eIF3d. Electroanalysis. 2021;33(4):1072–1080. [Google Scholar]
- 38.Chen L.C., Wang E., Tai C.S., Chiu Y.C., Li C.W., Lin Y.R., Lee T.H., Huang C.W., Chen J.C., Chen W.L. Improving the reproducibility, accuracy, and stability of an electrochemical biosensor platform for point-of-care use. Biosens. Bioelectron. 2020;155:112111. doi: 10.1016/j.bios.2020.112111. [DOI] [PubMed] [Google Scholar]
- 39.Darby A.C., Hiscox J.A. Covid-19: variants and vaccination. BMJ. 2021;372:n771. doi: 10.1136/bmj.n771. [DOI] [PubMed] [Google Scholar]
- 40.Mahase E. Covid-19: what new variants are emerging and how are they being investigated? BMJ. 2021;372:n158. doi: 10.1136/bmj.n158. [DOI] [PubMed] [Google Scholar]
- 41.Shiehzadegan S., Alaghemand N., Fox M., Venketaraman V. Analysis of the delta variant B.1.617.2 COVID-19. Clin. Pract. 2021;11(4):778–784. doi: 10.3390/clinpract11040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beduk T., Beduk D., de Oliveira Filho J.I., Zihnioglu F., Cicek C., Sertoz R., Arda B., Goksel T., Turhan K., Salama K.N., Timur S. Rapid point-of-care COVID-19 diagnosis with a gold-nanoarchitecture-assisted laser-scribed graphene biosensor. Anal. Chem. 2021;93(24):8585–8594. doi: 10.1021/acs.analchem.1c01444. [DOI] [PubMed] [Google Scholar]
- 43.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14(12):17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabiani L., Saroglia M., Galata G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L., Li H., Zhao C.Y., Zhang L., Wei J., Zhang Y.P., Li C.P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raziq A., Kidakova A., Boroznjak R., Reut J., Opik A., Syritski V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021;178:113029. doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ali M.A., Hu C., Jahan S., Yuan B., Saleh M.S., Ju E., Gao S.J., Panat R. Sensing of COVID-19 antibodies in seconds via aerosol jet nanoprinted reduced-graphene-oxide-coated 3D electrodes. Adv. Mater. 2021;33(7) doi: 10.1002/adma.202006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liv L. Electrochemical immunosensor platform based on gold-clusters, cysteamine and glutaraldehyde modified electrode for diagnosing COVID-19. Microchem. J. 2021;168:106445. doi: 10.1016/j.microc.2021.106445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.