Abstract
Background
Cardiorespiratory fitness (CRF) is a potent health marker, the improvement of which is associated with a reduced incidence of non-communicable diseases and all-cause mortality. Identifying metabolic signatures associated with CRF could reveal how CRF fosters human health and lead to the development of novel health-monitoring strategies.
Objective
This article systematically reviewed reported associations between CRF and metabolites measured in human tissues and body fluids.
Methods
PubMed, EMBASE, and Web of Science were searched from database inception to 3 June, 2021. Metabolomics studies reporting metabolites associated with CRF, measured by means of cardiopulmonary exercise test, were deemed eligible. Backward and forward citation tracking on eligible records were used to complement the results of database searching. Risk of bias at the study level was assessed using QUADOMICS.
Results
Twenty-two studies were included and 667 metabolites, measured in plasma (n = 619), serum (n = 18), skeletal muscle (n = 16), urine (n = 11), or sweat (n = 3), were identified. Lipids were the metabolites most commonly positively (n = 174) and negatively (n = 274) associated with CRF. Specific circulating glycerophospholipids (n = 85) and cholesterol esters (n = 17) were positively associated with CRF, while circulating glycerolipids (n = 152), glycerophospholipids (n = 42), acylcarnitines (n = 14), and ceramides (n = 12) were negatively associated with CRF. Interestingly, muscle acylcarnitines were positively correlated with CRF (n = 15).
Conclusions
Cardiorespiratory fitness was associated with circulating and muscle lipidome composition. Causality of the revealed associations at the molecular species level remains to be investigated further. Finally, included studies were heterogeneous in terms of participants’ characteristics and analytical and statistical approaches.
PROSPERO Registration Number
CRD42020214375.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40279-021-01590-y.
Key Points
| A panel of unique lipid species were found to be associated with cardiorespiratory fitness. |
| The majority of circulating glycerolipids, acylcarnitines, and ceramides were negatively associated with cardiorespiratory fitness, highlighting their link to poor cardiometabolic health. |
| Specific glycerophosphocholines and cholesterol esters were found to be positively associated with cardiorespiratory fitness, featuring their roles in health maintenance. |
Introduction
Cardiorespiratory fitness (CRF), defined as the peak oxygen uptake, is a powerful health marker [1]. Importantly, the American Heart Association now recommends assessing it as a vital sign in clinical practice [1]. Indeed, CRF is inversely associated with an incidence of cancer, cardiometabolic diseases, and all-cause mortality [1–6]. Furthermore, improvement in CRF is associated with a reduced incidence of stroke, type 2 diabetes mellitus, dementia, and all-cause mortality [7–12].
Physiologically, CRF reflects the entire oxygen transport chain from its uptake in the lungs to its delivery to the mitochondria for energy production [1]. While the heritability of basal CRF and gains in CRF is around 50%, the mechanisms linking CRF with reduced morbidity and mortality remain largely unknown [1, 13, 14]. As CRF is a better predictor of morbidity and mortality than physical activity itself, physiological adaptations to exercise are likely not sufficient to explain how CRF mitigates morbidity and mortality [15–17]. Identifying metabolic signatures associated with CRF could reveal the metabolic pathways through which CRF acts on morbidity and mortality, lead to the discovery of novel biomarkers of physical fitness, and ultimately pave the way for novel health-monitoring strategies [18, 19].
In the past decade, technological advances in mass spectrometry, nuclear magnetic resonance, and bioinformatics have enabled ‘omics’ scale metabolite phenotyping [20]. Metabolomics is now a powerful tool to investigate at the molecular species level how metabolites relate to the cellular phenotype [21]. While genes encode what may happen, metabolites, influenced by both genome and exposome, provide insights on what has indeed happened [20]. Thus, the metabolome directly reflects cellular activity and is the closest ‘omic’ level to the phenome [20, 21]. Furthermore, metabolites not only constitute building blocks of cell components or fuels in cellular energetics, they also act as driving forces of cellular processes (e.g., cell growth, differentiation, activation, apopotosis) by modulating (through covalent chemical modifications or metabolite-macromolecule interactions) the expression and activity of the other ‘omics’ levels [22, 23]. Conversely, pathological processes can also alter both the metabolome and CRF as well as their mutual associations [24, 25]. Consequently, there are complex interrelated interactions between the genome, the metabolome, the exposome, and disease development, which all influence the phenome (e.g., CRF or health status) [20]. Considering the high clinical relevance of CRF, this work aimed at systematically reviewing the current literature on metabolites in human tissues and body fluids that have been reported to be associated with CRF.
Methods
This systematic review is reported according to the Preferred Reporting Items for Systematic and Meta-Analysis (PRISMA) guidelines [26]. The research question was formulated according to the Population, Exposure, Comparison, Outcome, Study Type framework (Electronic Supplementary Material [ESM]) [27]. The review was registered on PROSPERO (registration number CRD42020214375) on 14 November, 2020 and a protocol was published [28].
Eligibility Criteria
All human studies that (1) were published until the date of the last search, i.e., 3 June, 2021, (2) applied a metabolomics approach, (3) reported metabolites of any tissue, associated with CRF, and (4) measured CRF by means of a cardiopulmonary exercise test (spiroergometry) were eligible. Studies reporting estimated CRF were excluded as estimated CRF correlates only moderately with measured CRF [29]. Studies that were published in languages other than English, German, French, Italian, or Spanish were not included (as the authors have linguistic expertise in these five languages). Finally, non-original articles (i.e., editorials, letters, reviews), meta-analyses, case reports, and conference abstracts were also deemed non-eligible.
Information Sources and Search Strategy
Search strategies were developed in collaboration with an information specialist (CAH) using the Peer Review of Electronic Search Strategies (PRESS) framework [30]. PubMed, Web of Science, and EMBASE were searched. Database-specific subject headings and text word synonyms around the concepts metabolomics and CRF were used. Non-human studies and conference abstracts were excluded. Search results were generated on 20 October, 2020, exported to EndNote X9 (Clarivate, London, UK) and deduplicated. An update search was run on 3 June, 2021. The detailed search strings can be found in the ESM and in the review protocol [28].
Data Management and Extraction
Titles and abstracts of recovered records were reviewed independently by two authors (CG and JC). Articles were deemed as ‘include’, ‘exclude’ or ‘uncertain’ according to the prespecified eligibility criteria. For articles deemed ‘include’ or ‘uncertain,’ full text was retrieved and independently reviewed for eligibility by two authors (CG and JC). Discrepancies during title/abstract or full-text screening were resolved by discussion between the two screening authors. A third party made a final judgment in cases where no resolution was found (LS). To complement the results of direct database searching, bibliographic references of all included articles were screened manually (backward citation tracking), and the citing articles were screened using Scopus (forward citation tracking, on 15 June, 2021). Data were extracted from the full texts and entered in a standardized Excel form. One author extracted the data (CG), and a second author independently checked the extractions (JC). Discrepancies were resolved through discussion (with a third party, if necessary, LS). Corresponding authors were contacted twice by e-mail in cases of missing or unclear data. Information that was extracted can be found in the ESM.
Risk of Bias in Individual Studies
The following key steps of a metabolomics workflow were extracted at the study level: sample collection and storage, sampling time and nutritional protocol, metabolite extraction method, analytical technique, quality control used to assess data quality, data processing, and metabolite annotation. Subsequently, two authors (CG and JC) independently assessed the risk of bias at the study level using the QUADOMICS items applicable to the present work (ESM) [31, 32]. Discrepancies were resolved through discussion (with a third party, if necessary, LS).
Data Synthesis
Qualitative and quantitative data describing associations between metabolites and CRF were synthesized narratively and presented in a tabular and charted format. Metabolites were classified using the chemical taxonomy of the Human Metabolome Database (version 4.0) [33].
Results
Study Selection
The searches yielded 4728 unique records, of which 22 met eligibility criteria as depicted in the PRISMA 2020 flow diagram (Fig. 1) [26]. Authors agreed on all eligibility decisions upon discussion without the need for third-party arbitration. Three additional studies were identified via other methods. One study was identified by a senior author (AST) via handsearching [34], one study was identified through backward citation tracking [35], and another study was identified through forward citation tracking, respectively [36].
Fig. 1.
Preferred Reporting Items for Systematic and Meta-Analysis (PRISMA) 2020 flow diagram. CPET cardiopulmonary exercise test, CRF cardiorespiratory fitness
Characteristics of Included Studies
Included studies were published between 2012 and 2021 (Table 1 and ESM). Fifteen studies included both female and male participants [34–48], and seven studies investigated male participants only [49–55]. One study examined adolescents [38], and the remaining 21 studies enrolled only adults (defined as participants aged > 18 years).
Table 1.
Studies’ and participants’ characteristics
| References | Study design | Participants (n) |
Age (Years ± SD) |
Female sex (%) |
VO2peak (mL/min/kg) |
Health conditions | Tissue samples | Metabolomics approach | Technology used |
|---|---|---|---|---|---|---|---|---|---|
| Stanford et al. [37] | C | 39 |
C1: 58.7 ± 2.5 C2: 29.4 ± 0.6 |
C1: 0 C2: 50 |
N/A |
C1: overweight, obesity, arterial hypertension grade 1 C2: unknown smoking status |
Serum | T (AQ) | LC–MS/MS |
| Duft et al. [38] | RCT | 37 |
CG: 14.72 ± 1.07 TG: 14.44 ± 1.04 |
CG: 52 TG: 50 |
CG: 35.18 ± 6.22 TG: 32.80 ± 6.15 |
Overweight, obesity, unknown smoking status | Serum | T (AQ) | 1H NMR |
| Saleem et al. [36] | L | 100 | 64 ± 6 | 15 | 21.7 ± 5.5 | Overweight, obesity, dyslipidemia, arterial hypertension, CAD, smoking, depression, musculoskeletal issues | Plasma | T (AQ) | HPLC–MS/MS |
| Kujala et al. [49] | CS | 580 | 26.1 ± 6.5 | 0 |
HF: 50.7 ± 4.2 LF: 31.8 ± 3.8 |
Overweight, dyslipidemia, arterial hypertension, diabetes, smoking | Serum | T (AQ) | 1H NMR |
| Fabbri et al. [34] | L | 443 | 68.9 ± 9.4 | 42 | 24.2 ± 6.8 | Dyslipidemia, arterial hypertension, prediabetes, diabetes, smoking | Plasma | T (AQ) | HPLC–MS/MS |
| Monnerat et al. [50] | P | 14 |
HF: 25.8 ± 50.3 LF: 26.0 ± 5.0 |
0 |
HF: 76.3 ± 1.5 LF: 61.0 ± 3.5 |
None | Plasma | U | UHPLC-HRMS |
| Lustgarten et al. [39] | NRT | 77 | 24.4 ± 4.2 | 64 | 44.4 ± 10.8 | Overweight, obesity | Serum | U |
UHPLC-MS/MS GC–MS |
| Harshman et al. [55] | POC | 13 |
C1: 26.67 ± 5.16 C2: 29.43 ± 3.36 |
0 |
C1: 56.57 ± 10.40 C2: 43.03 ± 4.07 |
None | Sweat | T (RQ) | LC–MS/MS |
| Nayor et al. [41] | C | 471 | 53.0 ± 8.0 | 63 | 23.1 ± 7.1 | Overweight, dyslipidemia, arterial hypertension, diabetes, smoking | Plasma | T (AQ), U | LC–MS/MS |
| Contaifer et al. [40] | CS | 49 | 57 | 25 | 14.0 ± 3.4 | Overweight, obesity, dyslipidemia, arterial hypertension, diabetes, heart failure | Plasma | U |
LC–MS/MS GC–MS |
| Mueller-Hennessen et al. [51] | CC | 41 |
P: 51.5 ± 13.7 CG: 47.5 ± 12.8 |
0 |
P: 21.0 ± 8.0 CG: 32.0 ± 6.0 |
Overweight, dyslipidemia, arterial hypertension, diabetes, heart failure, smoking | Plasma | T (RQ) |
LC–MS/MS GC–MS SPE-LC–MS/MS |
| Huffman et al. [42] | RCT | 112 | 18–70 | 48 | 28.0 ± 5.8 | Overweight, obesity, dyslipidemia, arterial hypertension grade 1 | Skeletal muscle | T (AQ) |
MS/MS GC–MS |
| Duft et al. [52] | RCT | 22 |
CG: 47.50 ± 6.20 TG: 48.60 ± 5.50 |
0 |
CG: 29.10 ± 4.70 TG: 28.20 ± 4.70 |
Obesity grade 1 | Serum | T (AQ) | 1H NMR |
| Morris et al. [44] | RCT | 40 | 35.0 ± 14.0 | 50 | 41.1 ± 16.2 | Overweight, obesity | Plasma | T (AQ) | ESI–MS/MS |
| Contrepois et al. [45] | C | 36 | 59.00 ± 8.00 | 42 | 30.60 ± 8.71 | Overweight, arterial hypertension, prediabetes, diabetes, smoking | Plasma | U, ST |
LC–MS/MS Lipidyzer Platform |
| Kistner et al. [46] | CS | 255 | 46.1 ± 16.9 | 42 | 38.8 ± 11.6 | None | Urine | T (AQ) | 1H NMR |
| Chorell et al. [53] | I | 27 |
HF TG: 28.16 ± 2.70 HF CG: 25.58 ± 1.77 LF TG: 26.30 ± 5.30 LF CG: 24.04 ± 1.83 |
0 |
HF TG: 63.20 ± 2.93 HF CG: 63.67 ± 2.80 LF TG: 44.57 ± 5.62 LF CG: 42.71 ± 2.87 |
None | Plasma | U | GC–MS/MS |
| Brennan et al. [47] | RCT | 216 |
CG: 52.3 ± 8.4 TG: 52.4 ± 7.8 |
CG: 66 TG: 64 |
CG: 29.2 ± 6.0 TG: 28.4 ± 5.1 |
Overweight, obesity | Plasma | T (AQ) | LC–MS/MS |
| Morris et al. [43] | CC | 65 |
HF: 28.0 ± 9.0 LF: 36.0 ± 11.0 |
48 |
HF: 54.9 ± 7.5 LF: 30.8 ± 7.2 |
Overweight, obesity | Urine | U | GC–MS |
| Shi et al. [54] | NRT | 20 | 29.42 ± 4.51 | 0 | 59.20 ± 5.90 | None | Serum | U | UHPLC-MS/MS |
| Bye et al. [48] | NRT | 218 |
HF: 49.50 LF: 49.50 |
58 |
HF: 41.43 LF: 31.33 |
None | Serum | U | 1H NMR |
| Michel et al. [35] | CC | 40 |
P: 23.1 ± 5.1 CG: 24.7 ± 6.6 |
35 |
P: 28.8 ± 10.1 CG: 45.7 ± 6.4 |
Fontan patients with systemic left ventricle | Serum | T (AQ) | LC–MS/MS |
AQ absolute quantification, C1 cohort/group 1, C2 cohort/group 2, C cohort, CAD coronary artery disease, CC case–control, CG control groups, CS cross-sectional, ESI–MS/MS electrospray ionization tandem mass spectrometry, GC–MS gas-chromatography tandem mass spectrometry, 1H NMR proton nuclear magnetic resonance, HF high fit, HPLC–MS/MS high-performance liquid-chromatography tandem mass spectrometry, I interventional, L longitudinal, LC–MS/MS liquid-chromatography tandem mass spectrometry, LW low fit, MS/MS tandem mass spectrometry, N/A not applicable, indicates that data were not reported in manuscripts and authors did not respond to our e-mail requests, NRT non-randomized trial, P in the column “age" and “VO2 peak” patients, P in the column study design pilot, POC proof of concept, RCT randomized controlled trial, RQ relative quantification, SD standard deviation, SPE-LC–MS/MS solid-phase extraction liquid-chromatography tandem mass spectrometry, ST semi-targeted, T targeted, TG training/test group, U untargeted, UHPLC-HRMS ultra-high performance liquid-chromatography tandem high-resolution mass spectrometry, VO2peak peak oxygen uptake
Six studies included healthy participants free of any diseases [46, 48, 50, 53–55], of which one enrolled elite long-distance runners [50] and one enrolled amateur marathon runners [54]. In 14 studies, a subfraction or even all participants were overweight or obese [36–45, 47, 49, 51, 52]. Finally, in ten studies, a subfraction or even all participants had a cardiometabolic disease other than overweight or obesity [34–37, 40–42, 45, 49, 51].
Ten studies investigated plasma samples [34, 36, 40, 41, 44, 45, 47, 50, 51, 53], eight analyzed serum samples [35, 37–39, 48, 49, 52, 54], two investigated urine samples [43, 46], one analyzed skeletal muscle [42], and one investigated sweat [55]. Twelve studies applied a targeted approach [34–38, 42, 44, 46, 47, 49, 51, 52], and eight studies applied an untargeted approach [39, 40, 43, 48, 50, 53–55]. Depending on the nature of metabolites, one study used a targeted or untargeted approach [41], whereas another used a targeted or semi-targeted approach [45]. Fifteen studies ran regression analyses to investigate associations between metabolites and CRF [34, 36, 39–41, 43–45, 47–51, 53, 55], and seven studies conducted correlation analyses [35, 37, 38, 42, 46, 52, 54] (ESM).
Risk of Bias in Individual Studies
Risk of bias assessment is summarized in Table 2. Sixteen studies failed to precisely describe the selection process of participants [34, 36, 37, 39, 40, 42–45, 48, 50–55], and 13 studies did not take any actions to avoid overfitting [34–36, 40–47, 50, 55]. While 18 studies collected tissue samples after an overnight fasting [34, 35, 37–39, 41–47, 49–54], Bye et al. [48] collected information on dietary habits without imposing fasting, Harshman et al. [55] collected sweat in a non-fasted state and two studies did not report on the nutritional state (ESM) [36, 40]. Harshman et al. [55] collected sweat during an exercise intervention, while the remaining 21 studies collected tissue samples in a resting state. Mueller-Hennessen et al. [51] did not specify the extraction protocol used. Finally, Lustgarten et al. [39] Huffman et al. [42] Kujala et al. [49] and Mueller-Hennessen et al. [51] did not report on the quality control used.
Table 2.
Risk of bias in individual studies evaluated with QUADOMICS
Green = yes, red = no, orange = unclear
Metabolites Related to CRF
A total of 667 metabolites were reported to be significantly associated (n = 639) or correlated (n = 28) with CRF. These 667 metabolites were measured in plasma (n = 619), serum (n = 18), skeletal muscle (n = 16), urine (n = 11), or sweat (n = 3). Results are presented in a tissue-specific manner with plasma and serum metabolites being grouped together under the term circulating metabolites. Twenty-seven of the metabolites extracted from Nayor et al. [41] were doubly reported as per two different analytical techniques used for their detection (hydrophilic interaction liquid chromatography, positive ion mode analyses of polar and nonpolar plasma lipids using reversed-phase chromatography or negative ion mode analysis of free fatty acids and bile acids using reversed-phase chromatography). Similarly, Contrepois et al. [45] doubly reported four metabolites as they “eluted in multiple peaks”. As all these 31 metabolites were collected from plasma, they were counted only once in this analysis. Furthermore, two studies reported only on metabolites non-significantly associated [50] or correlated [35] with CRF. These metabolites were not considered in the present analysis.
Circulating Metabolites
Circulating metabolites were positively (n = 243) and negatively (n = 394) associated with CRF. As displayed in Fig. 2a, the former consisted of lipids and lipid-like molecules (n = 159), organic acids and derivatives (n = 51), organoheterocyclic compounds (n = 13), organic oxygen compounds (n = 8), benzenoids (n = 6), organic nitrogen compounds (n = 2), nucleosides, nucleotides, and analogs (n = 1), phenylpropanoids and polyketides (n = 1), lignans, neolignans, and related compounds (n = 1), and inorganic compounds (n = 1). As shown in Fig. 2b, metabolites negatively associated with CRF were subdivided into lipids and lipid-like molecules (n = 273), organic acids and derivatives (n = 70), organoheterocyclic compounds (n = 17), organic oxygen compounds (n = 11), nucleosides, nucleotides and analogs (n = 11), organic nitrogen compounds (n = 8), benzenoids (n = 2), phenylpropanoids and polyketides (n = 1), and alkaloids and derivatives (n = 1).
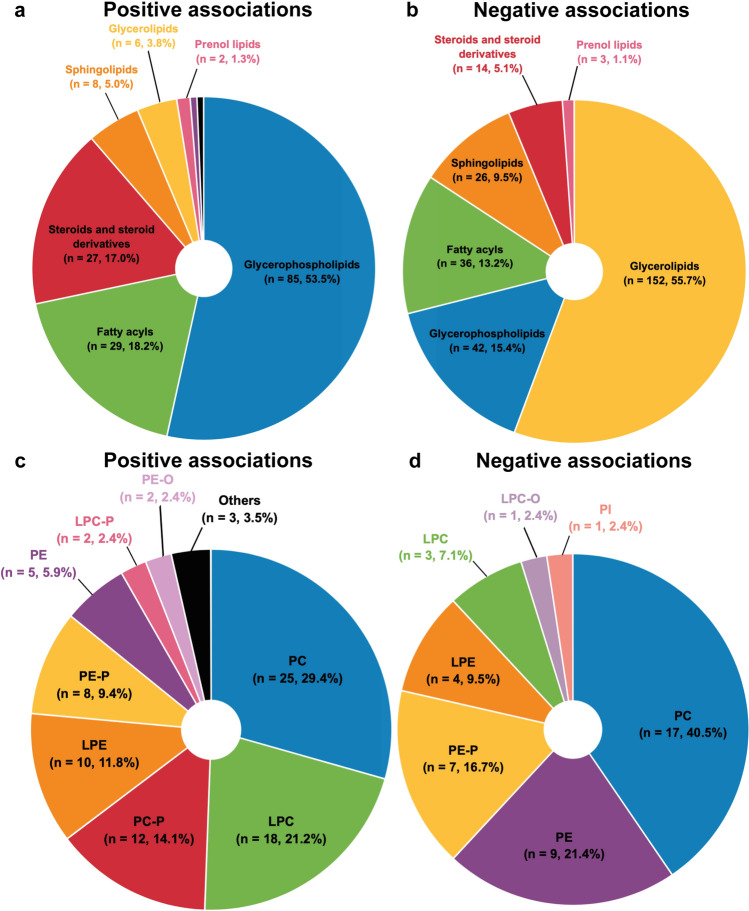
Fig. 2.
Included metabolites on the super class level. a Metabolites super classes positively associated with cardiorespiratory fitness. b Metabolites super classes negatively associated with cardiorespiratory fitness. n number of metabolite species. Figure was created with the Mind the Graph platform (www.mindthegraph.com) and Adobe Illustrator 2021 (Adobe Inc., San Jose, CA, USA)
Among the 159 lipids and lipid-like molecules positively associated with CRF, glycerophospholipids (n = 85), fatty acyls (n = 29), steroids and derivatives (n = 27), sphingolipids (n = 8), and glycerolipids (n = 6) were the most prevalent lipid classes (Fig. 3a). Glycerophospholipids were further subdivided into mainly diacylglycerophosphocholines (PC, n = 25), lyso-acylglycerophosphocholines (LPC, n = 18), alkenyl-acylglycerophosphocholines (n = 12), lyso-acylglycerophosphoethanolamines (n = 10), and alkenyl-acylglycerophosphoethanolamines (n = 8) (Fig. 3c). Fatty acyls comprised very long-chain fatty acids (n = 7), long-chain fatty acids (n = 7), medium-chain fatty acids (n = 5), and acylcarnitines (n = 4). Steroids and derivatives mainly consisted of cholesterol esters (n = 17). Lastly, the sphingolipids consisted of sphingomyelins (n = 5), glycosphingolipids (n = 2), and ceramides (n = 1).
Fig. 3.
Included lipid and lipid-like molecules. a Lipids and lipid-like molecules positively associated with cardiorespiratory fitness, by class level. b Lipids and lipid-like molecules negatively associated with cardiorespiratory fitness, by class level. c Glycerophospholipids positively associated with cardiorespiratory fitness, by direct parent level. d Glycerophospholipids negatively associated with cardiorespiratory fitness, by direct parent level. LPC lyso-acylglycerophosphocholines, LPC-O lyso-alkylglycerophosphocholines, LPC-P lyso-alkenylglycerophosphocholines, LPE lyso-acylglycerophosphoethanolamines, n number of metabolite species, PC diacylglycerophosphocholines, PC-P alkenyl-acylglycerophosphocholines, PE diacylglycerophosphoethanolamines, PE-O alkyl-acylglycerophosphoethanolamines, PE-P alkenyl-acylglycerophosphoethanolamines, PI diacylglycerophosphoinositol. Figure was created with the Mind the Graph platform (www.mindthegraph.com) and Adobe Illustrator 2021 (Adobe Inc., San Jose, CA, USA)
As shown in Fig. 3b, the 273 lipid species negatively associated with CRF consisted of glycerolipids (n = 152), glycerophospholipids (n = 42), fatty acyls (n = 36), sphingolipids (n = 26), steroids and derivatives (n = 14), and prenol lipids (n = 3). Glycerolipids consisted of triacylglycerols (n = 122) and diacylglycerols (n = 30). Glycerophospholipids consisted mainly of PC (n = 17), PE (n = 9), alkenyl-acylglycerophosphoethanolamines (n = 7), and lyso-acylglycerophosphoethanolamines (n = 4) (Fig. 3d). Fatty acyls consisted of acylcarnitines (n = 14), further classified into short-chain (n = 8), medium-chain (n = 3), and long-chain (n = 3) acylcarnitines, and long-chain fatty acids (n = 10). Sphingolipids consisted mainly of ceramides (n = 12), sphingomyelins (n = 7), and glycosphingolipids (n = 6). Last, steroids comprised bile acids and derivatives (n = 7), sulfated steroids (n = 5), and cholesterol esters (n = 2).
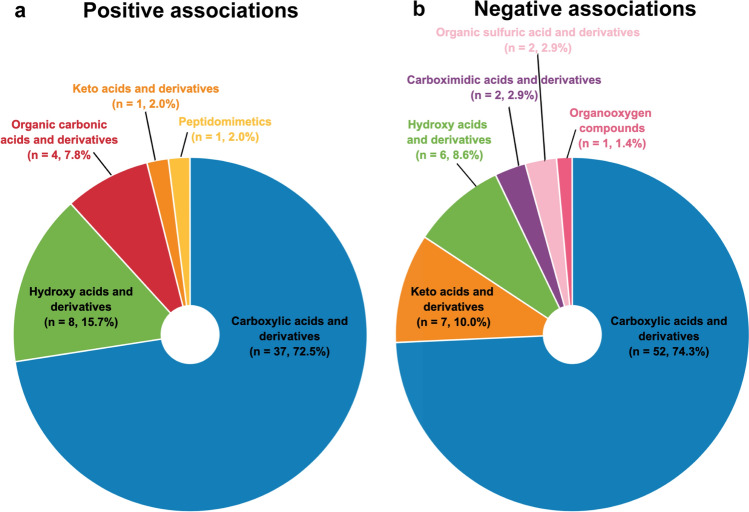
The 51 organic acids and derivatives positively associated with CRF comprised 35 amino acids, peptides, and analogs, of which the most common were alpha amino acids and derivatives (n = 9), N-acyl-alpha amino acids (n = 4), methionine and derivatives (n = 3), prolines and derivatives (n = 3), and dipeptides (n = 3) (Fig. 4a). Seventy organic acids and derivatives were negatively associated or correlated with CRF, of which 51 were amino acids, peptides, and derivatives. The most represented amino acids and analogs were alpha amino acids and derivatives (n = 16), dipeptides (n = 5), arginines and derivatives (n = 4), tyrosine and derivatives (n = 4), prolines and derivatives (n = 3), and phenylalanines and derivatives (n = 3) (Fig. 4b).
Fig. 4.
Included organic acids and derivatives on the class level. a Organic acids and derivatives positively associated with cardiorespiratory fitness. b Organic acids and derivatives negatively associated with cardiorespiratory fitness. n number of metabolite species. Figure was created with the Mind the Graph platform (www.mindthegraph.com) and Adobe Illustrator 2021 (Adobe Inc., San Jose, CA, USA)
The 13 organoheterocyclic compounds, which showed positive associations with CRF, consisted mainly of bilirubins (n = 4). Purines and purine derivates were the most represented metabolites (n = 4) within the 17 organoheterocyclic compounds negatively associated with CRF. Carbohydrates and carbohydrate conjugates were the most common metabolites within the eight organic oxygen compounds positively associated with CRF (n = 5) and in the 11 negatively associated with CRF (n = 8). Eleven nucleosides, nucleotides, and analogs displayed negative associations with CRF, while only one showed a positive association with CRF. Similarly, more organic nitrogen compounds were negatively associated with CRF (n = 8) than positively (n = 2). Finally, six benzenoids species displayed positive associations with CRF, while only two showed negative associations with CRF.
Metabolites Sampled from Skeletal Muscle, Urine, or Sweat
In skeletal muscle, 15 acylcarnitines were positively correlated to CRF, further classified into medium-chain (n = 9), long-chain (n = 5), and short-chain (n = 1) acylcarnitines. In urine, two metabolites (creatinine and uracil) were positively associated with CRF, while eight amino acids and analogs and one straight-chain fatty acid were negatively associated with CRF. Finally, sweat methionine showed a positive association with CRF, while sweat ornithine and phenylalanine displayed negative associations with CRF.
Metabolites Reported Multiple Times in Relation to CRF
Ninety-seven circulating metabolites were reported by two or three distinct studies. Conflicting reports were found for 14 metabolites, which were reported to be either positively or negatively associated or correlated with CRF depending on the studies (Table 3). Fifteen metabolites, including four LPC species, were reported multiple times to be positively associated with CRF (Table 4). Finally, 68 metabolites were consistently negatively associated with CRF (Table 5). The latter included 44 triacylglycerols, nine amino acids, and three ceramides.
Table 3.
Metabolite species reported to be positively and negatively associated with CRF
| Metabolite super classes | Metabolite species | Studies reporting associations with CRF | |
|---|---|---|---|
| Positive | Negative | ||
| Lipids and lipid-like molecules | Nervonic acid | [41] | [45] |
| Vitamin A | [41] | [45] | |
| CE(20:4) | [41] | [45] | |
| CE(22:4) | [41] | [40] | |
| Organic acids and derivatives | Pyroglutamic acid | [47] | [41] |
| Histidine | [45] | [47] | |
| Methionine | [40] | [47] | |
| Cinnamoylglycine | [41] | [45] | |
| Phenylalanine | [52] | [41] | |
| Serine | [45] | [40] | |
| Tyrosine | [52] | [41, 47] | |
| Organic oxygen compounds | 1,5-Anhydroglucitol | [45] | [47] |
| Kynurenine | [45] | [47] | |
| Organoheterocyclic compounds | C-Glycosyltryptophan | [45] | [41] |
CE cholesterol ester, CRF cardiorespiratory fitness
Table 4.
Metabolites species reported multiple times as positively associated with CRF
| Metabolite super classes | Metabolite species | Studies reporting positive associations with CRF |
|---|---|---|
| Benzenoids | Hippuric acid | [41, 45] |
| Lipids and lipid-like molecules | 12,13-diHOME | [37, 41] |
| Docosahexaenoic acid | [41, 53] | |
| PC(34:2) | [41, 44] | |
| LPC(18:0) | [41, 51] | |
| LPC(18:1) | [41, 51] | |
| LPC(18:2) | [41, 51] | |
| LPC(20:4) | [41, 51] | |
| SM(18:1;2/24:1) | [41, 45] | |
| CE(18:3) | [41, 45] | |
| CE(20:3) | [41, 45] | |
| Organic acids and derivatives | Asparagine | [41, 45] |
| Acetylglycine | [41, 45] | |
| Malic acid | [41, 47] | |
| Organoheterocyclic compounds | Bilirubin | [41, 45, 47] |
CE cholesterol ester, CRF cardiorespiratory fitness, LPC lyso-acylglycerophosphocholines, PC diacylglycerophosphocholines, PC-P alkenyl-acylglycerophosphocholines, SM sphingomyelin
Table 5.
Metabolite species reported multiple times as negatively associated with CRF
| Metabolite super classes | Metabolite species | Studies reporting negative associations with CRF |
|---|---|---|
| Lipids and lipid-like molecules | CAR(3) propionylcarnitine | [41, 45, 47] |
| CAR(6) hexanoylcarnitine | [41, 45] | |
| TAG(46:1), TAG(46:2), TAG(46:3), TAG(47:0), TAG(47:2), TAG(48:1), TAG(48:2), TAG(48:3), TAG(48:4), TAG(48:5), TAG(49:0), TAG(49:1), TAG(49:2), TAG(49:3), TAG(50:1), TAG(50:2), TAG(50:3), TAG(50:4), TAG(50:5), TAG(50:6), TAG(51:0), TAG(51:1), TAG(51:2), TAG(51:3), TAG(52:1), TAG(52:2), TAG(52:3), TAG(52:4), TAG(52:5), TAG(52:6), TAG(52:7), TAG(53:2), TAG(53:3), TAG(54:1), TAG(54:2), TAG(54:3), TAG(54:4), TAG(54:5), TAG(54:6), TAG(54:7), TAG(54:8), TAG(55:2), TAG(55:3), TAG(56:5) | [41, 45] | |
| γ-Tocopherol | [39, 53] | |
| Cer(18:1;2/16:0) | [36, 41, 45] | |
| Cer(18:1;2/18:0) | [34, 36] | |
| Cer(18:1;2/20:0) | [34, 36] | |
| HexCer(18:1;2/18:0) | [36, 45] | |
| SM(18:1;2/18:0) | [41, 45] | |
| SM(18:1;2/18:1) | [36, 41, 45] | |
| Glycocholic acid | [41, 45] | |
| Nucleosides, nucleotides, and analogs | Pseudouridine | [41, 51] |
| Organic acids and derivatives | Alanine | [41, 47] |
| Citrulline | [45, 47] | |
| Creatine | [41, 45, 47] | |
| Lysine | [40, 41, 47] | |
| Ornithine | [41, 45, 47] | |
| Isoleucine | [41, 47] | |
| Leucine | [41, 47] | |
| Thyroxine | [41, 47] | |
| Proline | [41, 47] | |
| Lactic acid | [40, 41] | |
| Pyruvic acid | [41, 47] | |
| Organic nitrogen compounds | Carnitine | [41, 45] |
| Organic oxygen compounds | Gluconic acid | [40, 45] |
CAR carnitine, Cer ceramide, CRF cardiorespiratory fitness, HexCer hexosylceramide, SM sphingomyelin, TAG triacylglycerol
Meta-Analysis
None of the included metabolites fulfilled the criteria, previously described in the review protocol, to be meta-analyzed [28]. Metabolites had (1) to be detected in the same tissue, (2) using the same metabolomics approach (untargeted, semi-targeted, or targeted), (3) in at least three different studies, and (4) to be identified on a level 1 identification according to the Metabolomics Standards Initiative to be meta-analyzed [28, 56].
Discussion
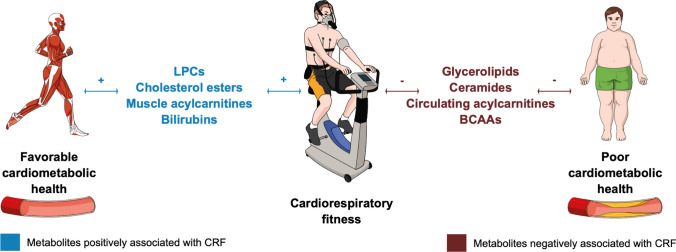
The present work systematically reviewed metabolites that are associated with CRF, a potent marker of human health that should be considered as a vital sign in clinical medicine according to the American Heart Association [1]. Lipids, followed by organic acids, were the metabolites most commonly associated with CRF. Most circulating glycerolipids, acylcarnitines, and ceramides, as known biomarkers of poor cardiometabolic health, showed negative associations with CRF [57–63]. Conversely, most LPC and cholesterol esters were positively associated with CRF (Fig. 5). Branched-chain amino acids (BCAAs) were negatively associated with CRF, while bilirubin displayed positive associations with CRF (Fig. 5). None of the included metabolites fulfilled the qualitative and quantitative criteria defined in the review protocol to be meta-analyzed, which reflects the novelty of the present field. Nevertheless, 83 associations were reported independently by distinct studies, which strengthens the certainty of evidence of these findings. Conversely, conflicting results were found for 14 associations, which reduces their certainty of evidence. The following sections discuss the overall results with a focus on the 83 associations showing the strongest certainty of evidence.
Fig. 5.
Metabolites associated with cardiorespiratory fitness (CRF) as potential driving forces of cardiometabolic health. Figure was created with the Mind the Graph platform (www.mindthegraph.com) and Adobe Illustrator 2021 (Adobe Inc., San Jose, CA, USA). BCAA branched-chain amino acids, LPCs lyso-acylglycerophosphocholines
Lipids and Lipid-Like Molecules
In addition to their well-known functions in energy storage and production, lipids from biological membranes modulate cell surface receptor activities and regulate vesicular trafficking [64–66]. Lipids also act as key signaling molecules, controlling important cellular processes such as cell proliferation, apoptosis, migration, senescence, and inflammation [67, 68]. Alterations in lipid metabolism have been observed in many cardiometabolic, oncological, and neurodegenerative disorders [69, 70]. Remarkably, changes in the lipidome composition have been associated with aging, healthy aging, and age-related disorders [71–74]. Estimated to number in the hundreds of thousands of discrete molecular species, lipids are also the most abundant circulating macromolecules in human plasma [75]. In light of these facts, it is not surprising that lipids were the metabolites most commonly associated with a potent health marker such as CRF.
Specific PC species were previously described as cardiometabolically favorable, while others seem to be metabolically deleterious [62, 63, 76]. For instance, the favorable PC(16:0/22:5) and deleterious PC(16:0/16:0) are part of the ceramide-phospholipid score for the prediction of cardiovascular risk [62, 63], but the exact underlying biological mechanism remains unknown. Interestingly, saturated and monounsaturated PC species were previously positively associated with cardiovascular mortality [76]. The results of the present review support a dual role of PC species with 25 and 17 species being positively or negatively associated with CRF, respectively. No pattern was observed regarding species saturation and their association with CRF. Specifically, PC(34:2) was reported twice to be positively associated with CRF, which contrasts with the fact that this species was previously associated with cardiovascular mortality and aging [76, 77]. However, the annotation PC(34:2) does not unequivocally specify the fatty acyl or alkyl chains in the molecule, which makes data interpretation ambiguous [78]. Thus, PC(34:2) could correspond to different species, such as PC(16:0/18:2) but also PC(14:0/20:2), each of which have potentially different biological roles. Therefore, caution is necessary when interpreting lipidomic data.
Regarding LPC, more species were positively (n = 18) than negatively (n = 3) associated with CRF and four species [LPC(18:0), LPC(18:1), LPC(18:2), and LPC(20:4)] were reported twice as positively associated with CRF [41, 51]. LPC(18:0) and LPC(18:2) were previously associated with reduced cardiovascular mortality [76]. Mechanistically, LPCs are believed to inhibit cholesterol synthesis in macrophages and slow down atherogenesis [79]. LPC(16:0), which was reported once to be positively associated with CRF, is known to be inversely associated with vascular remodeling (intima-media thickness), cardiovascular diseases, and mortality [76, 80]. Finally, circulating LPC levels have been observed to be reduced in rodents with obesity and type 2 diabetes [81].
Twelve alkenyl-acylglycerophosphocholines species were found to be positively associated with CRF. Alkenyl-acylglycerophosphocholines belong to the ether-glycerophospholipid family, which acts, amongst other things, as cellular antioxidants and is therefore considered as metabolically favorable [82, 83]. This could explain why most alkenyl-acylglycerophosphocholines were positively associated with CRF. Additionally, lower circulating levels of ether-glycerophospholipids have been observed in patients with non-alcoholic steatohepatitis and children with type 1 diabetes [84, 85].
Circulating CE levels have been reported to be negatively associated with cardiovascular diseases [86, 87]. Indeed, the formation of cholesterol esters prevents intracellular free cholesterol accumulation [88]. The fact that 17 CE were positively associated with CRF, while only two CE were negatively associated with CRF supports this statement. On the lipid species level, CE(18:3) and CE(20:3) were reported twice as positively associated with CRF [41, 45]. Furthermore, CE(20:4), CE(20:5), CE(22:4), CE(22:5), and CE(22:6), levels of which were previously inversely associated with cardiovascular disease, displayed positive associations with CRF [86]. Most circulating acylcarnitines (regardless of chain length) were negatively associated with CRF, which reflects the fact that an accumulation of circulating acylcarnitines indicates incomplete mitochondrial fatty acid oxidation, and therefore impaired metabolic health [59, 60]. Conversely, muscular acylcarnitines were positively associated with CRF, highlighting improved fatty acid oxidation with a higher fitness level [89]. This is in line with previous data, which showed that exercise improves muscle mitochondrial capacity and the completeness of fatty acid oxidation [90].
Ceramides and their roles in cardiometabolic diseases are receiving growing scrutiny [91, 92]. On a mechanistic level, circulating ceramides are believed to promote foam cell formation, vascular inflammation, and atherosclerosis [93–95]. These findings have progressed to clinical medicine, where ceramides are now used to predict cardiovascular death in patients with and without coronary artery disease [61, 62, 96, 97]. Thus, it is postulated that ceramides act as driving forces of cardiometabolic disorders [98, 99]. In this context, the results of the present review are highly interesting, showing that 12 ceramides were negatively associated with CRF, with only Cer(18:1;2/10:0) being an exception. Furthermore, Cer(18:1;2/16:0) [36, 41, 45], Cer(18:1;2/18:0) [34, 36], and Cer(18:1;2/20:0) [34, 36] were reported several times as negatively associated with CRF. Strikingly, the three cardiometabolically deleterious ceramide species clinically used in the ceramide-phospholipid score (Cer(18:1;2/16:0) [36, 41, 45], Cer(18:1;2/18:0) [34, 36], and Cer(18:1;2/24:1) [36]) were found to be negatively associated with CRF [99]. In light of the these findings, it can be hypothesized that specific CRF-enhancing training could reverse altered ceramide profiles and optimize cardiometabolic health. This needs to be demonstrated in a prospective intervention study.
Organic Acids and Derivatives
Amino acids not only serve as building blocks for proteins but also as signaling molecules, regulators of gene expression, as well as precursors of hormones and neurotransmitters [100]. Circulating levels of the BCAAs isoleucine, leucine, and valine have been associated with obesity, insulin resistance, and type 2 diabetes [101–104]. Initially attributed to an BCAA-mediated activation of the mammalian target of rapamycin pathway [105], these findings are more likely due to an increased ratio of BCAAs to tryptophan and threonine, resulting in central serotonin depletion, hyperphagia, obesity, and a reduced lifespan [106]. Alternatively, it has been suggested that metabolically healthy and cardiorespiratory fit individuals tend to have more efficient BCAA catabolism and fatty acid oxidation, which prevents BCAA accumulation in the circulation [107, 108]. The findings of the present systematic review tend to support the latter as isoleucine [41, 47], leucine [41, 47], and valine [47] were found to be negatively associated with CRF. While asparagine and acetylglycine are known to be inversely associated with the incidence of metabolic syndrome, these two amino acids were reported by distinct studies to be positively associated with CRF [109]. Phenylalanine and tyrosine are known to be elevated in subjects with insulin resistance, diabetes, or coronary artery disease [110]. Both metabolites also displayed higher levels in metabolically unhealthy obese patients, while they were not elevated in metabolically healthy obese subjects [111]. Remarkably, phenylalanine in plasma [41], urine [43], and sweat [55] as well as tyrosine in plasma [41, 47] and urine [46] were negatively associated with CRF. However, the contrary was true for phenylalanine and tyrosine in serum [52].
Other Organic Compounds
Bilirubin, which was positively associated with CRF, is believed to be cardiometabolically favorable. Indeed, elevated bilirubin levels are associated with a reduced incidence of peripheral artery disease and stroke [112]. Conversely, low bilirubin levels have been associated with a higher risk of coronary artery disease, impaired flow-mediated vasodilatation, and increased carotid intima-media thickness [113, 114]. Therefore, elevation of bilirubin levels might be a way through which improvement in CRF mitigates the incidence of cardiometabolic diseases. Hippurate, the levels of which are decreased in patients with metabolic syndrome, was positively associated with CRF [115–117]. Finally, while circulating cell-free nucleic acids are increasingly recognized as potential biomarkers of diseases (‘liquid biopsy’) [118], little is known about the significance of isolated circulating nucleosides, which were mainly negatively associated with CRF [41, 45, 47].
Strengths and Limitations
The present work was the first to systematically review CRF-associated metabolites. In this way, it provides researchers with an objective overview of the current literature and could orientate future research aiming at unraveling metabolic pathways through which CRF mitigates morbidity and mortality. The main limitation of the present systematic review lies in the important heterogeneity of the included studies. Indeed, studies differed in terms of participants’ age, sex, health conditions and medications, body mass index and percentage of body fat, physical activity, and CRF levels. Moreover, fasting protocols, analyzed tissues, sample preparation, extraction methods, analytical techniques, and metabolomics approaches were also heterogeneous. Furthermore, several statistical analyses were used, from simple correlations to multiple linear regressions, adjusting or not for important confounders. Therefore, investigating associations between metabolites and CRF needs to be done in healthy participants before investigating clinical populations. It will then be possible to circumvent the confounding effects of chronic cardiometabolic diseases on lipid metabolism. Second, some associations between metabolites and CRF are likely sex specific and, for female individuals, depend on menopausal status [119]. Unfortunately, data gathered from the included studies and from author contacts did not allow us to analyze associations in a sex-dependent or menopause-dependent manner. Indeed, seven studies investigated male individuals only [49–55], ten publications neither reported sex-specific or menopause-specific results nor provided individual patient data [36–38, 40–42, 44, 46–48], three studies reported sex-specific results without providing individual patient data [34, 39, 43], and two publications provided individual patient data without reporting sex-specific results [35, 45]. Coupled with the important heterogeneity described above, the lack of individual patient data prevented the creation of models predictive of CRF based on a metabolic signature.
Third, this systematic review does not provide information about the cellular origin, destination, or subcellular localization of the circulating pool of metabolites. Thus, potential CRF-promoting nutritional recommendations require further mechanistic studies in model organisms and intervention studies in both model organisms and humans. Consequently, such recommendations cannot be derived at this stage. Fourth, it is essential to harmonize analytical approaches by following recommendations edited by the Metabolomics Society [120, 121]. For instance, analyte concentration in sweat can vary greatly depending on the collection, handling, processing, storage, and skin microbiome [122, 123]. Finally, it is important to conduct regression analyses rather than simple correlation analyses and thereby adjust for relevant confounders [90].
Conclusions
Circulating and muscle lipidome composition was associated with CRF, a clinically highly relevant health parameter. Known biomarkers of poor cardiometabolic health such as circulating glycerolipids, acylcarnitines, and ceramides were negatively associated with CRF. Conversely, circulating LPCs, cholesterol esters, and muscle acylcarnitines were positively associated with CRF, featuring their roles in health maintenance. BCAA and bilirubins showed negative and positive associations with CRF, respectively. It is important to note that the included studies were heterogeneous in terms of participants’ characteristics and analytical and statistical approaches. While causality of the revealed associations remains to be investigated further, lipid metabolism and changes in lipidome composition seem to be tightly related to physical fitness. Deciphering lipid responses to CRF-enhancing interventions could help unravel the metabolic pathways through which CRF mitigates morbidity and mortality.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Funding
Open access funding provided by University of Basel. This review was funded by the Department of Sport, Exercise and Health of the University of Basel, Switzerland.
Conflicts of Interest/Competing Interests
Justin Carrard, Chiara Guerini, Christian Appenzeller-Herzog, Denis Infanger, Karsten Königstein, Lukas Streese, Timo Hinrichs, Henner Hanssen, Hector Gallart-Ayala, Julijana Ivanisevic, and Arno Schmidt-Trucksäss declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Material
All data presented in this review are available within the article and supplementary material.
Code Availability
Not applicable.
Authors’ Contributions
The manuscript underwent several revisions with substantial contributions provided by each co-author. CG, JC, and CAH designed the search string. CAH was responsible for the search strategy. DI and JC designed the data synthesis strategy. CG and JC wrote the protocol, while CAH, LS, KK, TH, HH, HGA, JI, and AST critically revised it. JC registered the protocol in PROSPERO. JC and CG conducted the systematic review, and extracted and summarized the data. JC and CG wrote the present manuscript, while CAH, LS, KK, TH, HH, HGA, JI, and AST critically revised it. JC and CG created the figures. All authors provided critical feedback, and have read and approved the final manuscript.
Footnotes
Justin Carrard and Chiara Guerini: first authors, equally contributed.
Julijana Ivanisevic and Arno Schmidt-Trucksäss: last authors, equally contributed.
Contributor Information
Julijana Ivanisevic, Email: julijana.ivanisevic@unil.ch.
Arno Schmidt-Trucksäss, Email: arno.schmidt-trucksaess@unibas.ch.
References
- 1.Ross R, Blair SN, Arena R, Church TS, Despres JP, Franklin BA, et al. Importance of assessing cardiorespiratory fitness in clinical practice: a case for fitness as a clinical vital sign: a scientific statement from the american heart association. Circulation. 2016;134:e653–e699. doi: 10.1161/cir.0000000000000461. [DOI] [PubMed] [Google Scholar]
- 2.Robsahm TE, Falk RS, Heir T, Sandvik L, Vos L, Erikssen JE, et al. Measured cardiorespiratory fitness and self-reported physical activity: associations with cancer risk and death in a long-term prospective cohort study. Cancer Med. 2016;5:2136–2144. doi: 10.1002/cam4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gander JC, Sui X, Hébert JR, Hazlett LJ, Cai B, Lavie CJ, et al. Association of cardiorespiratory fitness with coronary heart disease in asymptomatic men. Mayo Clin Proc. 2015;90:1372–1379. doi: 10.1016/j.mayocp.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrell SW, Finley CE, Radford NB, Haskell WL. Cardiorespiratory fitness, body mass index, and heart failure mortality in men. Circ Heart Fail. 2013;6:898–905. doi: 10.1161/CIRCHEARTFAILURE.112.000088. [DOI] [PubMed] [Google Scholar]
- 5.Juraschek SP, Blaha MJ, Whelton SP, Blumenthal R, Jones SR, Keteyian SJ, et al. Physical fitness and hypertension in a population at risk for cardiovascular disease: the Henry Ford Exercise Testing (Fit) Project. J Am Heart Assoc. 2014;3:e001268. doi: 10.1161/JAHA.114.001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizzo NS, Ruiz JR, Hurtig-Wennlöf A, Ortega FB, Sjöström M. Relationship of physical activity, fitness, and fatness with clustered metabolic risk in children and adolescents: the European Youth Heart Study. J Pediatr. 2007;150:388–394. doi: 10.1016/j.jpeds.2006.12.039. [DOI] [PubMed] [Google Scholar]
- 7.Prestgaard E, Mariampillai J, Engeseth K, Erikssen J, Bodegård J, Liestøl K, et al. Change in cardiorespiratory fitness and risk of stroke and death. Stroke. 2018 doi: 10.1161/strokeaha.118.021798. [DOI] [PubMed] [Google Scholar]
- 8.de Lannoy L, Sui X, Lavie CJ, Blair SN, Ross R. Change in submaximal cardiorespiratory fitness and all-cause mortality. Mayo Clin Proc. 2018;93:184–190. doi: 10.1016/j.mayocp.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Tarp J, Støle AP, Blond K, Grøntved A. Cardiorespiratory fitness, muscular strength and risk of type 2 diabetes: a systematic review and meta-analysis. Diabetologia. 2019;62:1129–1142. doi: 10.1007/s00125-019-4867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrman JK, Brawner CA, Al-Mallah MH, Qureshi WT, Blaha MJ, Keteyian SJ. Cardiorespiratory fitness change and mortality risk among black and white patients: Henry Ford Exercise Testing (Fit) Project. Am J Med. 2017;130:1177–1183. doi: 10.1016/j.amjmed.2017.02.036. [DOI] [PubMed] [Google Scholar]
- 11.Tari AR, Nauman J, Zisko N, Skjellegrind HK, Bosnes I, Bergh S, et al. Temporal changes in cardiorespiratory fitness and risk of dementia incidence and mortality: a population-based prospective cohort study. Lancet Public Health. 2019;4:e565–e574. doi: 10.1016/s2468-2667(19)30183-5. [DOI] [PubMed] [Google Scholar]
- 12.Imboden MT, Harber MP, Whaley MH, Finch WH, Bishop DL, Fleenor BS, et al. The association between the change in directly measured cardiorespiratory fitness across time and mortality risk. Prog Cardiovasc Dis. 2019;62:157–162. doi: 10.1016/j.pcad.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Bouchard C, Sarzynski MA, Rice TK, Kraus WE, Church TS, Sung YJ, et al. Genomic predictors of the maximal O2 uptake response to standardized exercise training programs. J Appl Physiol. 1985;2011(110):1160–1170. doi: 10.1152/japplphysiol.00973.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouchard C, Daw EW, Rice T, Perusse L, Gagnon J, Province MA, et al. Familial resemblance for VO2max in the sedentary state: the heritage family study. Med Sci Sports Exerc. 1998;30:252–258. doi: 10.1097/00005768-199802000-00013. [DOI] [PubMed] [Google Scholar]
- 15.Myers J, Kaykha A, George S, Abella J, Zaheer N, Lear S, et al. Fitness versus physical activity patterns in predicting mortality in men. Am J Med. 2004;117:912–918. doi: 10.1016/j.amjmed.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 16.Lee D-C, Sui X, Ortega FB, Kim Y-S, Church TS, Winett RA, et al. Comparisons of leisure-time physical activity and cardiorespiratory fitness as predictors of all-cause mortality in men and women. Br J Sports Med. 2011;45:504–510. doi: 10.1136/bjsm.2009.066209. [DOI] [PubMed] [Google Scholar]
- 17.Williams PT. Physical fitness and activity as separate heart disease risk factors: a meta-analysis. Med Sci Sports Exerc. 2001;33:754–761. doi: 10.1097/00005768-200105000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanford JA, Nogiec CD, Lindholm ME, Adkins JN, Amar D, Dasari S, et al. Molecular transducers of physical activity consortium (motrpac): mapping the dynamic responses to exercise. Cell. 2020;181:1464–1474. doi: 10.1016/j.cell.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Després J-P. Predicting longevity using metabolomics: a novel tool for precision lifestyle medicine? Nat Rev Cardiol. 2020;17:67–68. doi: 10.1038/s41569-019-0310-2. [DOI] [PubMed] [Google Scholar]
- 20.Ivanisevic J, Thomas A. Metabolomics as a tool to understand pathophysiological processes. In: Giera M, editor. Clinical metabolomics: methods and protocols. New York: Springer; 2018. pp. 3–28. [DOI] [PubMed] [Google Scholar]
- 21.Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinschen MM, Ivanisevic J, Giera M, Siuzdak G. Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol. 2019;20:353–367. doi: 10.1038/s41580-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart DS. Metabolomics for investigating physiological and pathophysiological processes. Physiol Rev. 2019;99:1819–1875. doi: 10.1152/physrev.00035.2018. [DOI] [PubMed] [Google Scholar]
- 24.Porcu E, Gilardi F, Darrous L, Yengo L, Bararpour N, Gasser M, et al. Triangulating evidence from longitudinal and mendelian randomization studies of metabolomic biomarkers for type 2 diabetes. Sci Rep. 2021;11:6197. doi: 10.1038/s41598-021-85684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeiher J, Ombrellaro KJ, Perumal N, Keil T, Mensink GBM, Finger JD. Correlates and determinants of cardiorespiratory fitness in adults: a systematic review. Sports Med Open. 2019;5:39. doi: 10.1186/s40798-019-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Methley AM, Campbell S, Chew-Graham C, McNally R, Cheraghi-Sohi S. PICO, PICOS and SPIDER: a comparison study of specificity and sensitivity in three search tools for qualitative systematic reviews. BMC Health Serv Res. 2014;14:579. doi: 10.1186/s12913-014-0579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrard J, Guerini C, Appenzeller-Herzog C, Infanger D, Königstein K, Streese L, et al. The metabolic signature of cardiorespiratory fitness: a protocol for a systematic review and meta-analysis. BMJ Open Sport Exerc Med. 2021;7:e001008. doi: 10.1136/bmjsem-2020-001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loprinzi PD, Edwards MK, Addoh O, Bentley JP. Evaluation of the convergent validity of an estimated cardiorespiratory fitness algorithm. Eur J Appl Physiol. 2018;118:629–636. doi: 10.1007/s00421-018-3803-5. [DOI] [PubMed] [Google Scholar]
- 30.McGowan J, Sampson M, Salzwedel DM, Cogo E, Foerster V, Lefebvre C. Press peer review of electronic search strategies: 2015 guideline statement. J Clin Epidemiol. 2016;75:40–46. doi: 10.1016/j.jclinepi.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 31.Parker LA, Gómez Saez N, Lumbreras B, Porta M, Hernández-Aguado I. Methodological deficits in diagnostic research using ‘-omics’ technologies: evaluation of the quadomics tool and quality of recently published studies. PLoS ONE. 2010;5:e11419. doi: 10.1371/journal.pone.0011419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lumbreras B, Porta M, Márquez S, Pollán M, Parker LA, Hernández-Aguado I. Quadomics: an adaptation of the quality assessment of diagnostic accuracy assessment (quadas) for the evaluation of the methodological quality of studies on the diagnostic accuracy of ‘-omics’-based technologies. Clin Biochem. 2008;41:1316–1325. doi: 10.1016/j.clinbiochem.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 33.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, et al. Hmdb 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabbri E, Yang A, Simonsick EM, Chia CW, Zoli M, Haughey NJ, et al. Circulating ceramides are inversely associated with cardiorespiratory fitness in participants aged 54–96 years from the baltimore longitudinal study of aging. Aging Cell. 2016;15:825–831. doi: 10.1111/acel.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michel M, Dubowy K-O, Entenmann A, Karall D, Adam MG, Zlamy M, et al. Targeted metabolomic analysis of serum amino acids in the adult fontan patient with a dominant left ventricle. Sci Rep. 2020;10:8930. doi: 10.1038/s41598-020-65852-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saleem M, Herrmann N, Dinoff A, Marzolini S, Mielke MM, Andreazza A, et al. Association between sphingolipids and cardiopulmonary fitness in coronary artery disease patients undertaking cardiac rehabilitation. J Gerontol A Biol Sci Med Sci. 2018;75:671–679. doi: 10.1093/gerona/gly273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanford KI, Lynes MD, Takahashi H, Baer LA, Arts PJ, May FJ, et al. 12,13-Dihome: an exercise-induced lipokine that increases skeletal muscle fatty acid uptake. Cell Metab. 2018;27:1111–20.e1113. doi: 10.1016/j.cmet.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duft RG, Castro A, Bonfante ILP, Lopes WA, da Silva LR, Chacon-Mikahil MP, et al. Altered metabolomic profiling of overweight and obese adolescents after combined training is associated with reduced insulin resistance. Sci Rep. 2020;10:16880. doi: 10.1038/s41598-020-73943-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lustgarten MS, Price LL, Logvinenko T, Hatzis C, Padukone N, Reo NV, et al. Identification of serum analytes and metabolites associated with aerobic capacity. Eur J Appl Physiol. 2013;113:1311–1320. doi: 10.1007/s00421-012-2555-x. [DOI] [PubMed] [Google Scholar]
- 40.Contaifer D, Buckley LF, Wohlford G, Kumar NG, Morriss JM, Ranasinghe AD, et al. Metabolic modulation predicts heart failure tests performance. PLoS ONE. 2019 doi: 10.1371/journal.pone.0218153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nayor M, Shah RV, Miller PE, Blodgett JB, Tanguay M, Pico AR, et al. Metabolic architecture of acute exercise response in middle-aged adults in the community. Circulation. 2020 doi: 10.1161/circulationaha.120.050281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huffman KM, Koves TR, Hubal MJ, Abouassi H, Beri N, Bateman LA, et al. Metabolite signatures of exercise training in human skeletal muscle relate to mitochondrial remodelling and cardiometabolic fitness. Diabetologia. 2014;57:2282–2295. doi: 10.1007/s00125-014-3343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris C, Grada CO, Ryan M, Roche HM, De Vito G, Gibney MJ, et al. The relationship between aerobic fitness level and metabolic profiles in healthy adults. Mol Nutr Food Res. 2013;57:1246–1254. doi: 10.1002/mnfr.201200629. [DOI] [PubMed] [Google Scholar]
- 44.Morris C, O'Grada CM, Ryan MF, Gibney MJ, Roche HM, Gibney ER, et al. Modulation of the lipidomic profile due to a lipid challenge and fitness level: a postprandial study. Lipids Health Dis. 2015;14:65. doi: 10.1186/s12944-015-0062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Contrepois K, Wu S, Moneghetti KJ, Hornburg D, Ahadi S, Tsai MS, et al. Molecular choreography of acute exercise. Cell. 2020;181:1112–30.e1116. doi: 10.1016/j.cell.2020.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kistner S, Rist MJ, Döring M, Dörr C, Neumann R, Härtel S, et al. An nmr-based approach to identify urinary metabolites associated with acute physical exercise and cardiorespiratory fitness in healthy humans: results of the KarMeN study. Metabolites. 2020 doi: 10.3390/metabo10050212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennan AM, Benson M, Morningstar J, Herzig M, Robbins J, Gerszten RE, et al. Plasma metabolite profiles in response to chronic exercise. Med Sci Sports Exerc. 2018;50:1480–1486. doi: 10.1249/mss.0000000000001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bye A, Vettukattil R, Aspenes ST, Giskeødegård GF, Gribbestad IS, Wisløff U, et al. Serum levels of choline-containing compounds are associated with aerobic fitness level: the HUNT-study. PLoS ONE. 2012;7:e42330. doi: 10.1371/journal.pone.0042330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kujala UM, Vaara JP, Kainulainen H, Vasankari T, Vaara E, Kyröläinen H. Associations of aerobic fitness and maximal muscular strength with metabolites in young men. JAMA Netw Open. 2019;2:e198265. doi: 10.1001/jamanetworkopen.2019.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monnerat G, Sánchez CAR, Santos CGM, Paulucio D, Velasque R, Evaristo GPC, et al. Different signatures of high cardiorespiratory capacity revealed with metabolomic profiling in elite athletes. Int J Sports Physiol Perform. 2020 doi: 10.1123/ijspp.2019-0267. [DOI] [PubMed] [Google Scholar]
- 51.Mueller-Hennessen M, Sigl J, Fuhrmann JC, Witt H, Reszka R, Schmitz O, et al. Metabolic profiles in heart failure due to non-ischemic cardiomyopathy at rest and under exercise. ESC Heart Fail. 2017;4:178–189. doi: 10.1002/ehf2.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duft RG, Castro A, Bonfante ILP, Brunelli DT, Chacon-Mikahil MPT, Cavaglieri CR. Metabolomics approach in the investigation of metabolic changes in obese men after 24 weeks of combined training. J Proteome Res. 2017;16:2151–2159. doi: 10.1021/acs.jproteome.6b00967. [DOI] [PubMed] [Google Scholar]
- 53.Chorell E, Svensson MB, Moritz T, Antti H. Physical fitness level is reflected by alterations in the human plasma metabolome. Mol Biosyst. 2012;8:1187–1196. doi: 10.1039/c2mb05428k. [DOI] [PubMed] [Google Scholar]
- 54.Shi R, Zhang J, Fang B, Tian X, Feng Y, Cheng Z, et al. Runners’ metabolomic changes following marathon. Nutr Metab (Lond) 2020;17:19. doi: 10.1186/s12986-020-00436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harshman SW, Browder AB, Davidson CN, Pitsch RL, Strayer KE, Schaeublin NM, et al. The impact of nutritional supplementation on sweat metabolomic content: a proof-of-concept study. Front Chem. 2021;9:659583. doi: 10.3389/fchem.2021.659583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller M, Stone NJ, Ballantyne C, Bittner V, Criqui MH, Ginsberg HN, et al. Triglycerides and cardiovascular disease. Circulation. 2011;123:2292–2293. doi: 10.1161/CIR.0b013e3182160726. [DOI] [PubMed] [Google Scholar]
- 58.Mach F, Baigent C, Catapano AL, Koskinas KC, Casula M, Badimon L, et al. 2019 ESV/EAS guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur Heart J. 2020;41:111–188. doi: 10.1093/eurheartj/ehz455. [DOI] [PubMed] [Google Scholar]
- 59.McCoin CS, Knotts TA, Adams SH. Acylcarnitines: old actors auditioning for new roles in metabolic physiology. Nat Rev Endocrinol. 2015;11:617–625. doi: 10.1038/nrendo.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, Light AR, Hoppel CL, Campbell C, Chandler CJ, Burnett DJ, et al. Acylcarnitines as markers of exercise-associated fuel partitioning, xenometabolism, and potential signals to muscle afferent neurons. Exp Physiol. 2017;102:48–69. doi: 10.1113/ep086019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laaksonen R, Ekroos K, Sysi-Aho M, Hilvo M, Vihervaara T, Kauhanen D, et al. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond ldl-cholesterol. Eur Heart J. 2016;37:1967–1976. doi: 10.1093/eurheartj/ehw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hilvo M, Meikle PJ, Pedersen ER, Tell GS, Dhar I, Brenner H, et al. Development and validation of a ceramide- and phospholipid-based cardiovascular risk estimation score for coronary artery disease patients. Eur Heart J. 2019;41:371–380. doi: 10.1093/eurheartj/ehz387. [DOI] [PubMed] [Google Scholar]
- 63.Hilvo M, Wallentin L, Lakic TG, Held C, Kauhanen D, Jylhä A, et al. Prediction of residual risk by ceramide-phospholipid score in patients with stable coronary heart disease on optimal medical therapy. J Am Heart Assoc. 2020;9:e015258. doi: 10.1161/JAHA.119.015258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arish M, Husein A, Kashif M, Sandhu P, Hasnain SE, Akhter Y, et al. Orchestration of membrane receptor signaling by membrane lipids. Biochimie. 2015;113:111–124. doi: 10.1016/j.biochi.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 65.Meer GV, Sprong H. Membrane lipids and vesicular traffic. Curr Opin Cell Biol. 2004;16:373–378. doi: 10.1016/j.ceb.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 66.Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 67.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 68.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 69.Chen L, Chen X-W, Huang X, Song B-L, Wang Y, Wang Y. Regulation of glucose and lipid metabolism in health and disease. Sci China Life Sci. 2019;62:1420–1458. doi: 10.1007/s11427-019-1563-3. [DOI] [PubMed] [Google Scholar]
- 70.Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol. 2018;19:175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carrard J, Gallart-Ayala H, Infanger D, Teav T, Wagner J, Knaier R, et al. Metabolic view on human healthspan: a lipidome-wide association study. Metabolites. 2021;11:287. doi: 10.3390/metabo11050287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beyene HB, Olshansky G, Smith AAT, Giles C, Huynh K, Cinel M, et al. High-coverage plasma lipidomics reveals novel sex-specific lipidomic fingerprints of age and BMI: evidence from two large population cohort studies. PloS Biol. 2020;18:e3000870. doi: 10.1371/journal.pbio.3000870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonzalez-Covarrubias V, Beekman M, Uh HW, Dane A, Troost J, Paliukhovich I, et al. Lipidomics of familial longevity. Aging Cell. 2013;12:426–434. doi: 10.1111/acel.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson LC, Parker K, Aguirre BF, Nemkov TG, D’Alessandro A, Johnson SA, et al. The plasma metabolome as a predictor of biological aging in humans. Geroscience. 2019;41:895–906. doi: 10.1007/s11357-019-00123-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quehenberger O, Dennis EA. The human plasma lipidome. N Engl J Med. 2011;365:1812–1823. doi: 10.1056/NEJMra1104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sigruener A, Kleber ME, Heimerl S, Liebisch G, Schmitz G, Maerz W. Glycerophospholipid and sphingolipid species and mortality: the Ludwigshafen Risk and Cardiovascular Health (LURIC) Study. PLoS ONE. 2014;9:e85724. doi: 10.1371/journal.pone.0085724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kawanishi N, Kato Y, Yokozeki K, Sawada S, Sakurai R, Fujiwara Y, et al. Effects of aging on serum levels of lipid molecular species as determined by lipidomics analysis in japanese men and women. Lipids Health Dis. 2018;17:135. doi: 10.1186/s12944-018-0785-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liebisch G, Vizcaino JA, Kofeler H, Trotzmuller M, Griffiths WJ, Schmitz G, et al. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54:1523–1530. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rozenberg O, Shih DM, Aviram M. Human serum paraoxonase 1 decreases macrophage cholesterol biosynthesis. Arterioscler Thromb Vasc Biol. 2003;23:461–467. doi: 10.1161/01.ATV.0000060462.35946.B3. [DOI] [PubMed] [Google Scholar]
- 80.Fernandez C, Sandin M, Sampaio JL, Almgren P, Narkiewicz K, Hoffmann M, et al. Plasma lipid composition and risk of developing cardiovascular disease. PLoS ONE. 2013;8:e71846. doi: 10.1371/journal.pone.0071846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barber MN, Risis S, Yang C, Meikle PJ, Staples M, Febbraio MA, et al. Plasma lysophosphatidylcholine levels are reduced in obesity and type 2 diabetes. PLoS ONE. 2012;7:e41456. doi: 10.1371/journal.pone.0041456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dean JM, Lodhi IJ. Structural and functional roles of ether lipids. Protein Cell. 2018;9:196–206. doi: 10.1007/s13238-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watschinger K, Werner ER. Orphan enzymes in ether lipid metabolism. Biochimie. 2013;95:59–65. doi: 10.1016/j.biochi.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, Min H-K, et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 2009;50:1827–1838. doi: 10.1002/hep.23229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Orešič M, Simell S, Sysi-Aho M, Näntö-Salonen K, Seppänen-Laakso T, Parikka V, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. 2008;205:2975–2984. doi: 10.1084/jem.20081800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Toledo E, Wang DD, Ruiz-Canela M, Clish CB, Razquin C, Zheng Y, et al. Plasma lipidomic profiles and cardiovascular events in a randomized intervention trial with the Mediterranean diet. Am J Clin Nutr. 2017;106:973–983. doi: 10.3945/ajcn.116.151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gerl MJ, Vaz WLC, Domingues N, Klose C, Surma MA, Sampaio JL, et al. Cholesterol is inefficiently converted to cholesteryl esters in the blood of cardiovascular disease patients. Sci Rep. 2018;8:14764. doi: 10.1038/s41598-018-33116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo J, Yang H, Song B-L. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21:225–245. doi: 10.1038/s41580-019-0190-7. [DOI] [PubMed] [Google Scholar]
- 89.Purdom T, Kravitz L, Dokladny K, Mermier C. Understanding the factors that effect maximal fat oxidation. J Int Soc Sports Nutr. 2018;15:3. doi: 10.1186/s12970-018-0207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bewick V, Cheek L, Ball J. Statistics review 7: correlation and regression. Crit Care. 2003;7:451–459. doi: 10.1186/cc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Summers SA. Could ceramides become the new cholesterol? Cell Metab. 2018;27:276–280. doi: 10.1016/j.cmet.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 92.Poss AM, Holland WL, Summers SA. Risky lipids: refining the ceramide score that measures cardiovascular health. Eur Heart J. 2019;41:381–382. doi: 10.1093/eurheartj/ehz525. [DOI] [PubMed] [Google Scholar]
- 93.Li W, Yang X, Xing S, Bian F, Yao W, Bai X, et al. Endogenous ceramide contributes to the transcytosis of oxldl across endothelial cells and promotes its subendothelial retention in vascular wall. Oxid Med Cell Longev. 2014;2014:823071. doi: 10.1155/2014/823071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang X, Zhang Y, Wang P, Zhang S-Y, Dong Y, Zeng G, et al. Adipocyte hypoxia-inducible factor 2α suppresses atherosclerosis by promoting adipose ceramide catabolism. Cell Metab. 2019;30:937–51.e935. doi: 10.1016/j.cmet.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 95.Chaurasia B, Summers SA. Ceramides: lipotoxic inducers of metabolic disorders. Trends Endocrinol Metab. 2015;26:538–550. doi: 10.1016/j.tem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 96.Havulinna AS, Sysi-Aho M, Hilvo M, Kauhanen D, Hurme R, Ekroos K, et al. Circulating ceramides predict cardiovascular outcomes in the population-based finrisk 2002 cohort. Arterioscler Thromb Vasc Biol. 2016;36:2424–2430. doi: 10.1161/ATVBAHA.116.307497. [DOI] [PubMed] [Google Scholar]
- 97.Meeusen JW, Donato LJ, Bryant SC, Baudhuin LM, Berger PB, Jaffe AS. Plasma ceramides. Arterioscler Thromb Vasc Biol. 2018;38:1933–1939. doi: 10.1161/ATVBAHA.118.311199. [DOI] [PubMed] [Google Scholar]
- 98.Choi RH, Tatum SM, Symons JD, Summers SA, Holland WL. Ceramides and other sphingolipids as drivers of cardiovascular disease. Nat Rev Cardiol. 2021 doi: 10.1038/s41569-021-00536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hilvo M, Vasile VC, Donato LJ, Hurme R, Laaksonen R. Ceramides and ceramide scores: clinical applications for cardiometabolic risk stratification. Front Endocrinol (Lausanne). 2020 doi: 10.3389/fendo.2020.570628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. 2009;37:1–17. doi: 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- 101.Lackey DE, Lynch CJ, Olson KC, Mostaedi R, Ali M, Smith WH, et al. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. Am J Physiol Endocrinol Metab. 2013;304:E1175–E1187. doi: 10.1152/ajpendo.00630.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Piccolo BD, Comerford KB, Karakas SE, Knotts TA, Fiehn O, Adams SH. Whey protein supplementation does not alter plasma branched-chained amino acid profiles but results in unique metabolomics patterns in obese women enrolled in an 8-week weight loss trial. J Nutr. 2015;145:691–700. doi: 10.3945/jn.114.203943. [DOI] [PubMed] [Google Scholar]
- 103.Fiehn O, Garvey WT, Newman JW, Lok KH, Hoppel CL, Adams SH. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese african-american women. PLoS ONE. 2010;5:e15234. doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huffman KM, Shah SH, Stevens RD, Bain JR, Muehlbauer M, Slentz CA, et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care. 2009;32:1678–1683. doi: 10.2337/dc08-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014;19:418–430. doi: 10.1016/j.cmet.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Solon-Biet SM, Cogger VC, Pulpitel T, Wahl D, Clark X, Bagley E, et al. Branched chain amino acids impact health and lifespan indirectly via amino acid balance and appetite control. Nat Metab. 2019;1:532–545. doi: 10.1038/s42255-019-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kujala UM, Peltonen M, Laine MK, Kaprio J, Heinonen OJ, Sundvall J, et al. Branched-chain amino acid levels are related with surrogates of disturbed lipid metabolism among older men. Front Med. 2016;3:57. doi: 10.3389/fmed.2016.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kainulainen H, Hulmi JJ, Kujala UM. Potential role of branched-chain amino acid catabolism in regulating fat oxidation. Exerc Sport Sci Rev. 2013;41:194–200. doi: 10.1097/JES.0b013e3182a4e6b6. [DOI] [PubMed] [Google Scholar]
- 109.Ntzouvani A, Nomikos T, Panagiotakos D, Fragopoulou E, Pitsavos C, McCann A, et al. Amino acid profile and metabolic syndrome in a male mediterranean population: a cross-sectional study. Nutr Metab Cardiovasc Dis. 2017;27:1021–1030. doi: 10.1016/j.numecd.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 110.Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125:2222–2231. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Batch BC, Shah SH, Newgard CB, Turer CB, Haynes C, Bain JR, et al. Branched chain amino acids are novel biomarkers for discrimination of metabolic wellness. Metabolism. 2013;62:961–969. doi: 10.1016/j.metabol.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Perlstein TS, Pande RL, Creager MA, Weuve J, Beckman JA. Serum total bilirubin level, prevalent stroke, and stroke outcomes: NHANES 1999–2004. Am J Med. 2008;121:781–8.e781. doi: 10.1016/j.amjmed.2008.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schwertner H, Jackson WG, Tolan G. Association of low serum concentration of bilirubin with increased risk of coronary artery disease. Clin Chem. 1994;40(1):18–23. doi: 10.1093/clinchem/40.1.18. [DOI] [PubMed] [Google Scholar]
- 114.Erdogan D, Gullu H, Yildirim E, Tok D, Kirbas I, Ciftci O, et al. Low serum bilirubin levels are independently and inversely related to impaired flow-mediated vasodilation and increased carotid intima-media thickness in both men and women. Atherosclerosis. 2006;184:431–437. doi: 10.1016/j.atherosclerosis.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 115.Shearer J, Duggan G, Weljie A, Hittel DS, Wasserman DH, Vogel HJ. Metabolomic profiling of dietary-induced insulin resistance in the high fat–fed c57bl/6j mouse. Diabetes Obes Metab. 2008;10:950–958. doi: 10.1111/j.1463-1326.2007.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Calvani R, Miccheli A, Capuani G, Tomassini Miccheli A, Puccetti C, Delfini M, et al. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes (Lond) 2010;34:1095–1098. doi: 10.1038/ijo.2010.44. [DOI] [PubMed] [Google Scholar]
- 117.Waldram A, Holmes E, Wang Y, Rantalainen M, Wilson ID, Tuohy KM, et al. Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J Proteome Res. 2009;8:2361–2375. doi: 10.1021/pr8009885. [DOI] [PubMed] [Google Scholar]
- 118.Pös O, Biró O, Szemes T, Nagy B. Circulating cell-free nucleic acids: characteristics and applications. Eur J Hum Genet. 2018;26:937–945. doi: 10.1038/s41431-018-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tokarz J, Adamski J. Chapter 2. Confounders in metabolomics. In: Adamski J, editor. Metabolomics for biomedical research. Academic Press; 2020. p. 17–32. 10.1016/B978-0-12-812784-1.00002-5pp, https://www.sciencedirect.com/science/article/pii/B9780128127841000025?via%3Dihub.
- 120.Beger RD, Dunn W, Schmidt MA, Gross SS, Kirwan JA, Cascante M, et al. Metabolomics enables precision medicine: “a white paper, community perspective”. Metabolomics. 2016;12:149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sansone S-A, Fan T, Goodacre R, Griffin JL, Hardy NW, Kaddurah-Daouk R, et al. The metabolomics standards initiative. Nat Biotechnol. 2007;25:846–848. doi: 10.1038/nbt0807-846b. [DOI] [PubMed] [Google Scholar]
- 122.Hussain JN, Mantri N, Cohen MM. Working up a good sweat: the challenges of standardising sweat collection for metabolomics analysis. Clin Biochem Rev. 2017;38:13–34. [PMC free article] [PubMed] [Google Scholar]
- 123.Kabashima K, Honda T, Ginhoux F, Egawa G. The immunological anatomy of the skin. Nat Rev Immunol. 2019;19:19–30. doi: 10.1038/s41577-018-0084-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.