Abstract
The methyltransferase complex (m6A writer), which catalyzes the deposition of N6-methyladenosine (m6A) in mRNAs, is highly conserved across most eukaryotic organisms, but its components and interactions between them are still far from fully understood. Here, using in vivo interaction proteomics, two HAKAI-interacting zinc finger proteins, HIZ1 and HIZ2, are discovered as components of the Arabidopsis m6A writer complex. HAKAI is required for the interaction between HIZ1 and MTA (mRNA adenosine methylase A). Whilst HIZ1 knockout plants have normal levels of m6A, plants in which it is overexpressed show reduced methylation and decreased lateral root formation. Mutant plants lacking HIZ2 are viable but have an 85% reduction in m6A abundance and show severe developmental defects. Our findings suggest that HIZ2 is likely the plant equivalent of ZC3H13 (Flacc) of the metazoan m6A-METTL Associated Complex.
Subject terms: RNA modification, Plant molecular biology, Plant development
The components of m6A writer and their interactions are still far from fully understood. Here, the authors identify two HAKAI-interacting zinc finger proteins, HIZ1 and HIZ2, as components of the Arabidopsis m6A writer complex, and show that hiz2 mutant plants have an 85% reduction in m6A abundance and severe developmental defects.
Introduction
RNA modifications, which collectively constitute the epitranscriptome, can facilitate posttranscriptional regulatory processes1,2. N6-methyladenosine (m6A) is necessary for the regulation of developmental and cell differentiation processes in most eukaryotes, and is one of the most studied internal modifications in eukaryotic messenger RNAs (mRNAs). It is deposited by the methyltransferase complex (m6A writers) predominantly at the 3′ end of transcripts3,4. The methylation can be removed by demethylases (m6A erasers) and recognized by m6A-binding proteins (m6A readers). Identifying and characterizing the components of the m6A writer complex is fundamental to understanding the regulatory roles of this modification.
The m6A writer complex is formed by an assembly of proteins that are conserved between plants and animals. Studies on mammalian and Drosophila systems indicate that the writer complex can be separated into two sub-complexes. Methyltransferase like 3 (METTL3, previously known as MT-A70) equivalent of the Arabidopsis mRNA adenosine methylase A (MTA)5,6, and methyltransferase like 14 (METTL14, plant homolog mRNA adenosine methylase B [MTB])7,8, constitute the m6A-METTL Complex (MAC) and this joins with the m6A-METTL Associated Complex (MACOM)9. MACOM is composed of Wilms’ tumor 1-associating protein (WTAP, plant homolog FKBP12 interacting protein 37 [FIP37])6,10, Vir-like m6A methyltransferase associated (VIRMA, previously called KIAA1429, plant homolog VIRILIZER [VIR])8,11, and HAKAI (also known as Casitas B-lineage lymphoma-transforming sequence-like protein 1 (CBLL1), plant homolog HAKAI)8,11, and in animals it also contains and requires RNA binding motif protein 15 (RBM15) and the zinc finger CCCH domain-containing protein 13 (ZC3H13)12,13. The plant homolog of RBM15, FLOWERING LOCUS PA (FPA), which functions in flowering time control, was recently shown to co-purify with m6A writer proteins but loss of this protein does not influence the global m6A levels14. Despite homology searches, an equivalent of ZC3H13 has not been identified in plants15. In addition to the writer components, a negative regulator C2H2-type zinc finger protein 217 (ZFP217) also associates with the complex in mammalian pluripotent stem cells16.
We previously reported that Arabidopsis HAKAI is required for full m6A methylation, with its knockout leading to a 35% reduction in m6A abundance. Null mutations of any of the other reported m6A writer proteins cause embryo lethality, and hypomorphic partial knockouts lead to ~85% m6A reduction and strong developmental defects3,6,8. Thus, HAKAI might be an accessary protein required for full m6A methylation on all target transcripts or it might be responsible for m6A deposition on just a specific subset of mRNAs. Here, we further characterize the function of Arabidopsis HAKAI and identify HAKAI-interacting zinc finger protein 1 and 2 (HIZ1 and HIZ2) as members of the m6A writer complex. Our findings indicate that HIZ2 could be the long-sought plant homolog of ZC3H13, but in contrast to the other MACOM components, there is limited sequence conservation with their presumed metazoan counterparts.
Results
HAKAI specifically interacts with two zinc finger proteins
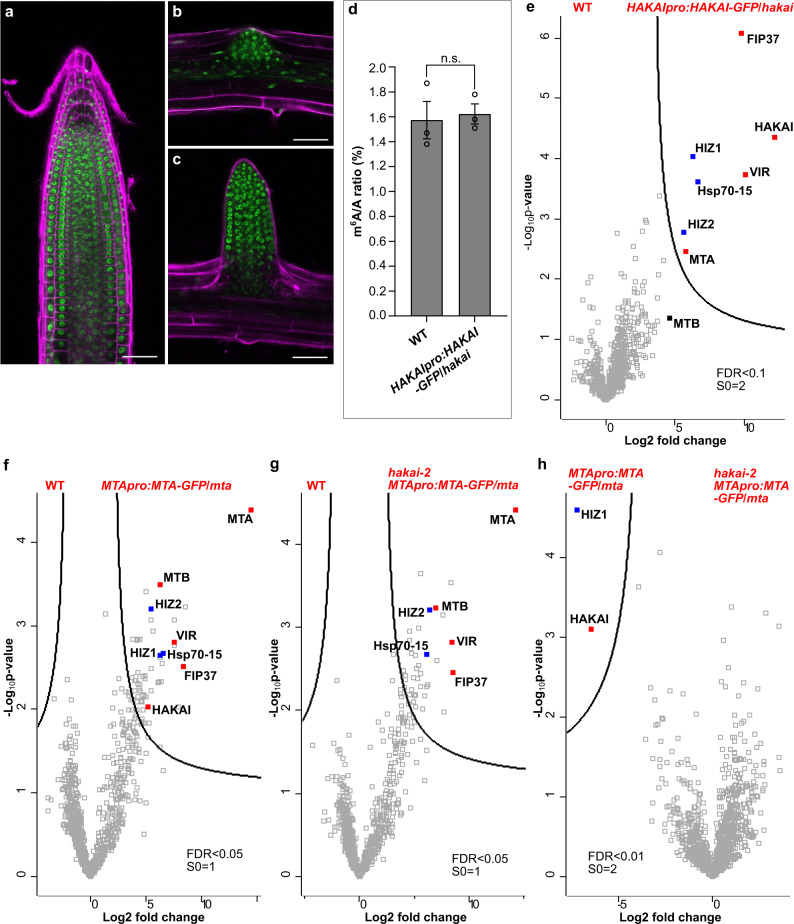
To facilitate the study of proteins interacting with HAKAI in vivo, HAKAI knockout mutant hakai-2 was complemented with HAKAI under its own promoter and with a GFP tag at the carboxy terminus, yielding the homozygous transgenic line—HAKAIpro:HAKAI-GFP/hakai. The GFP-tagged HAKAI protein was primarily localized to the nuclei in the primary root and lateral root initiation sites (Fig. 1a–c), in accordance with the localization of the main m6A methylase MTA6. The m6A level in HAKAIpro:HAKAI-GFP/hakai was restored to that of WT (Fig. 1d), demonstrating that the GFP-tagged HAKAI protein acts normally and complements the hakai-2 reduced m6A phenotype.
Fig. 1. Characterization of the HAKAIpro:HAKAI-GFP/hakai line and in vivo analysis of proteins interacting with HAKAI or MTA using GFP-tagged baits.
a–c The localization of GFP-tagged HAKAI in roots of 5-day-old HAKAIpro:HAKAI-GFP/hakai. a A primary root tip. b A lateral root initiation site. c A formed lateral root. Scale bar = 50 μm. Experiments in a–c were repeated independently at least three times, and representative images are shown. d m6A levels checked by two-dimensional thin layer chromatography (TLC) analysis. Data represent mean ± SE from three biological replicates and statistically significant differences relative to WT were analyzed by two-sided unpaired t-test (n.s. no significance). p = 0.7845. e–h Volcano plots showing proteins co-purified with HAKAI or MTA. The distribution of quantified proteins was plotted according to the Log2 fold change of label free quantification (LFQ) intensities and −Log10p-value obtained from two-sided student t-test of three independent experiments. Significantly enriched proteins were separated from others by a hyperbolic curve. Proteins indicated with filled squares represent known m6A writer complex members (red) and additional components identified in this work (blue). e HAKAIpro:HAKAI-GFP/hakai versus WT. HIZ1 and HIZ2 refer to HAKAI-interacting zinc finger protein 1 and 2 (AT1G32360 and AT5G53440, respectively). MTB is indicated not directly interacting with HAKAI, labeled with a black filled square. f MTApro:MTA-GFP/mta versus WT. g hakai-2 MTApro:MTA-GFP/mta versus WT. h hakai-2 MTApro:MTA-GFP/mta versus MTApro:MTA-GFP/mta. Source data are provided as a Source Data file.
We identified HAKAI interacting proteins by crosslinking of the nuclear fraction from HAKAIpro:HAKAI-GFP/hakai followed by anti-GFP co-immunoprecipitation and mass spectrometry14. Major proteins co-purifying with HAKAI were FIP37, VIR, MTA, two zinc finger proteins (AT1G32360 and AT5G53440, hereafter referred to as HAKAI-interacting zinc finger protein 1 and 2, HIZ1 and HIZ2, respectively) and a heat shock protein 70 family member (Hsp70-15, protein ID: A0A178W9Z4), whereas MTB was not among the directly-interacting proteins (Fig. 1e and Supplementary Data 1). To further investigate the role of HAKAI in the physical interaction between Arabidopsis m6A writer complex members, we utilized the MTApro:MTA-GFP/mta line where the embryo lethal phenotype of MTA null mutant (Salk_114710) is complemented using the MTA coding sequence translationally fused to GFP at its carboxy terminus under the control of the MTA promoter (Supplementary Fig. 1a). As we were interested in finding out how HAKAI influences the composition of the writer complex, we crossed hakai-2 with the MTApro:MTA-GFP/mta line and from the F3 progeny we selected plants homozygous for both hakai-2 and the MTA null mutant together with the complementing MTApro:MTA-GFP transgene (hakai-2 MTApro:MTA-GFP/mta). The localization and amount of GFP-tagged MTA protein in hakai-2 MTApro:MTA-GFP/mta was similar to MTApro:MTA-GFP/mta (Supplementary Fig. 1b, c). The m6A level in MTApro:MTA-GFP/mta was at the WT level, and that in hakai-2 MTApro:MTA-GFP/mta was, as expected, similar to hakai-2 measurements (Supplementary Fig. 1d), confirming the functional complementation by the GFP-tagged MTA under its own promoter.
We next carried out in vivo crosslinking and GFP pull-down experiments using both hakai-2 MTApro:MTA-GFP/mta and MTApro:MTA-GFP/mta lines. All known plant m6A writer proteins, including VIR, FIP37, MTB and HAKAI, were found interacting with MTA (Fig. 1f and Supplementary Data 1). The two zinc finger proteins which were pulled out with HAKAI were also found as interacting partners with MTA (Fig. 1f). However, the knockout of HAKAI led to the disappearance of the interaction between MTA and HIZ1 whereas HIZ2 remained as an interacting partner with MTA (Fig. 1g, h). Therefore, we conclude that HAKAI is a bona fide m6A writer protein in Arabidopsis, that HIZ1 and HIZ2 appear to be subunits of Arabidopsis m6A writer complex, and that HAKAI is essential for the interaction between HIZ1 and MTA.
HIZ1 levels are regulated by HAKAI, and HIZ1 ectopic expression reduces m6A abundance
Both HIZ1 and HIZ2 are annotated as zinc finger CCCH-domain containing proteins (TAIR database, https://www.arabidopsis.org/), however, their sequences share little homology. HIZ1 is composed of 384 amino acid residues (aa) while HIZ2 is more than three times as large (1181 aa or 1169 aa dependent on splice variant) (Supplementary Fig. 2). There are three zinc finger CCCH domains in HIZ1 protein sequence but only partial match is found in HIZ2 (Supplementary Fig. 2).
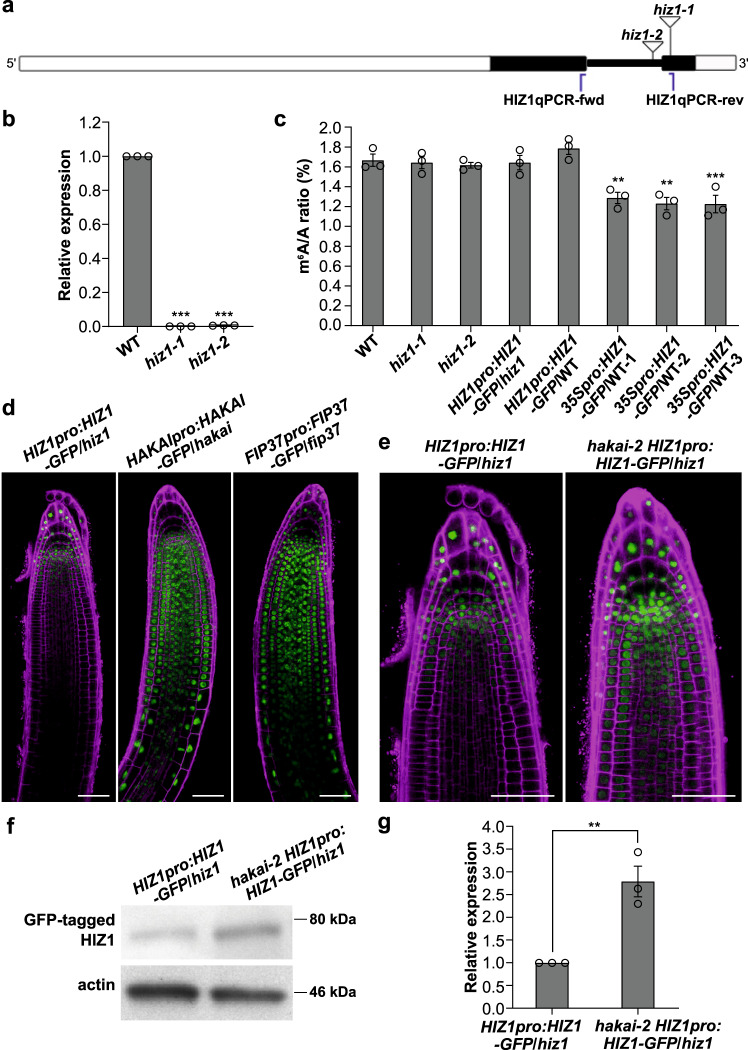
To address the function of HIZ1 in writing m6A methylation, mutant Arabidopsis lines homozygous for gene-disruptive T-DNA insertions—Salk_045882 (hiz1-1) and Salk_000717 (hiz1-2) were identified (Fig. 2a). RT-qPCR analysis confirmed that HIZ1 transcripts were absent in these two lines (Fig. 2b), but they displayed WT phenotypes, indicating that the loss-of-function of HIZ1 has negligible impact on overall growth and development. Recombinant constructs encoding carboxy terminus GFP-tagged HIZ1 either under its own promoter or the constitutive cauliflower mosaic virus (CaMV) 35S promoter, were constructed and transformed into hiz1-1 and wild-type (WT) respectively to generate a complementation line (HIZ1pro:HIZ1-GFP/hiz1) and an ectopic overexpressor line (35Spro:HIZ1-GFP/WT) (Supplementary Fig. 3a). Assays on 2-week-old seedlings showed that the m6A amount in hiz1 mutants and the complementation line stayed unchanged relative to WT, while that in the overexpressor lines driven by 35S promoter decreased by 24% (Fig. 2c). When the HIZ1 promoter rather than the constitutive 35S promoter was driving HIZ1-GFP expression (HIZ1pro:HIZ1-GFP/WT), the m6A to A ratio remained at the WT level (Fig. 2c). As expected, the expression of 35Spro:HIZ1-GFP was observed mainly in the nuclei and was strongly expressed along the whole root (Supplementary Figs. 3b and 4). The localization of GFP-tagged HIZ1 under its own promoter was also nuclear, but within root structures, it was found predominantly in cells at the root tips and in root hairs (Fig. 2d, e and Supplementary Fig. 4). In the root tips, HIZ1 localized to the columella and stem cell niche (including the quiescent center and stem cells surrounding it) of the primary roots (Fig. 2d, e). This stem cell niche localization is somewhat different from that of HAKAI and FIP37, which show reduced expression in these cells and higher expression in the apical meristem and basal meristem zones of the primary root tip where HIZ1-GFP is lower (Fig. 2d). The localization of GFP-tagged HIZ1 in HIZ1pro:HIZ1-GFP/WT was similar but the protein amount was slightly higher (Supplementary Fig. 3b, c). During lateral root development, the expression of HIZ1pro:HIZ1-GFP was maintained at a low level prior to lateral root emergence from the primary root, and afterwards its expression increased and concentrated in the lateral root tips (Supplementary Fig. 3d). In contrast, the expression of GFP-tagged HAKAI or FIP37 was much stronger during lateral root initiation and development (Supplementary Fig. 3d).
Fig. 2. Characterizing Arabidopsis HIZ1.
a Schematic of HIZ1 (AT1G32360) genomic DNA sequence with T-DNA insertion sites and the primer pair used for RT-qPCR. White rectangles denote UTRs of HIZ1 genomic DNA, black rectangles denote exons and thick black lines represent introns. hiz1-1 refers to Salk_045882 and hiz1-2 refers to Salk_000717. Blue lines represent the locations of primers. b The transcript levels of HIZ1 analyzed by RT-qPCR using the primer pair labeled in a. CBP20 was used as a reference gene. Data here and in c represent mean ± SE from three biological replicates and statistically significant differences relative to WT were analyzed by One-Way ANOVA (one-sided test) and marked with asterisks (**p < 0.01; ***p < 0.001). p < 0.0001 (hiz1-1); p < 0.0001 (hiz1-2). c m6A levels checked by two-dimensional thin layer chromatography (TLC) analysis. In one-way ANOVA test, p = 0.9996 (hiz1-1); p = 0.9907 (hiz1-2); p = 0.9996 (HIZ1pro:HIZ1-GFP/hiz1); p = 0.6393 (HIZ1pro:HIZ1-GFP/WT); p = 0.0036 (35Spro:HIZ1-GFP/WT-1); p = 0.001 (35Spro:HIZ1-GFP/WT-2); p = 0.0009 (35Spro:HIZ1-GFP/WT-3). d, e Localization of GFP-tagged proteins in primary root tips of 3-day-old seedlings. Scale bar = 50 μm. Experiments in d, e were repeated independently at least three times, and representative images are shown. f Western blot demonstrating the increased protein level of GFP-tagged HIZ1 in the hakai-2 background. Experiments in f were repeated independently three times with similar results. g Transcript levels of HIZ1pro:HIZ1-GFP analyzed by RT-qPCR using the same primer pair as that in b. CBP20 was used as a reference gene. Data represent mean ± SE from three biological replicates and the statistically significant difference was analyzed by two-sided unpaired t-test and marked with asterisks (**p < 0.01). p = 0.0061. Source data are provided as a Source Data file.
The proteomics data suggest that HAKAI has an effect on the interaction between MTA and HIZ1. Therefore, HIZ1pro:HIZ1-GFP/hiz1 was crossed with hakai-2 to check whether the expression and localization of GFP-tagged HIZ1 would change in a hakai-2 background. The homozygous line, hakai-2 HIZ1pro:HIZ1-GFP/hiz1, was used for imaging the localization of GFP-tagged HIZ1. We found that the disruption of HAKAI led to an increase of HIZ1pro:HIZ1-GFP expression in the primary root tip (Fig. 2e). We observed a similar increase in expression in the lateral roots of hakai-2 HIZ1pro:HIZ1-GFP/hiz1, and this was noticeable before lateral root emergence (Supplementary Fig. 3d). Western blotting confirmed the higher level of GFP-tagged HIZ1 upon the knockout of HAKAI (approximately a 3-fold increase) (Fig. 2f). To test whether this increase of protein level is due to an increase in HIZ1 transcript levels, we performed RT-qPCR to check the HIZ1pro:HIZ1-GFP transcript abundance in HIZ1pro:HIZ1-GFP/hiz1 and hakai-2 HIZ1pro:HIZ1-GFP/hiz1. The transcript levels for HIZ1pro:HIZ1-GFP were 2.8-fold increased in the absence of HAKAI (Fig. 2g). Thus, HAKAI does not seem to act as a posttranslational regulator of HIZ1, but rather affects promoter activity or transcript stability.
Reducing m6A modulates auxin responses and root development
Plants with decreased m6A levels show reduced apical dominance and partial agravitropic root growth as a result of impaired auxin responses3,6,17. Although the root morphology of hiz1-1 is similar to that of WT, the overexpressor line, 35Spro:HIZ1-GFP/WT-1, exhibited similar root phenotypes to known m6A writer mutants, including shorter primary roots and a decreased number of lateral roots (Supplementary Fig. 5).
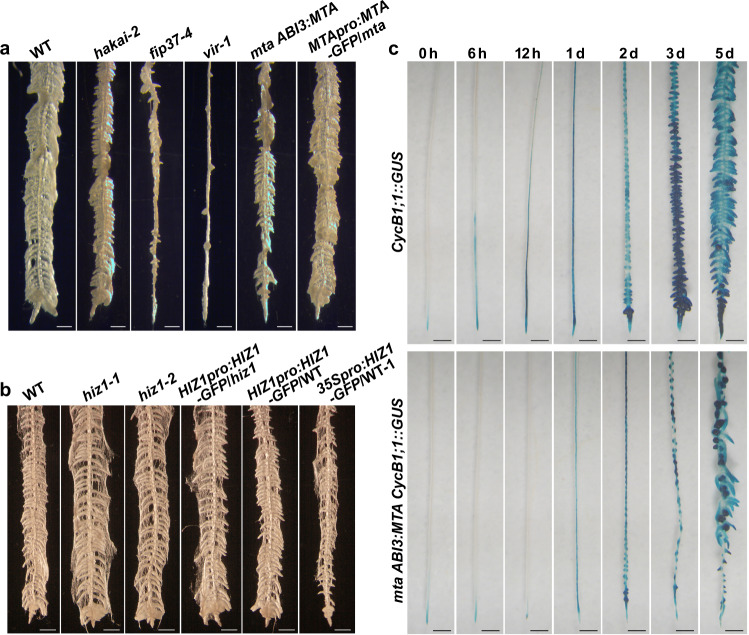
Both primary and lateral root development and growth are controlled by the plant hormone, auxin, and an auxin gradient, established via polar transport, is critical for lateral root initiation18. Thus, in WT Arabidopsis plants, lateral roots can be induced along the entire length of the primary root by first treating seedlings with N-1-naphthylphthalamic acid (NPA), which inhibits the major auxin efflux carriers19, then treating with the synthetic auxin 1-naphthaleneacetic acid (NAA). Because NAA can freely enter cells by passive diffusion, its intracellular level is normally controlled by the efflux carriers20. Thus, the sequential treatment with NPA and NAA results in an auxin maxima and potential activation of all pericycle cells for lateral root initiation21. We wished to test if the low m6A lines were impaired in their ability to respond to this treatment. Four-day-old Arabidopsis seedlings were first placed on medium containing NPA for 3 days and then transferred onto medium containing NAA. On the 5th day of the NAA treatment, induced lateral roots were clearly visible along the whole primary root of WT plants (Fig. 3a). However, fewer induced lateral roots were observed in all mutants for m6A writer complex members apart from HIZ1 (Fig. 3a, b). This reduction was most severe in vir-1 plants (Fig. 3a). In mta mutant plants, the reduction of lateral root number was restored to the WT levels by introducing the WT MTA transgene (Fig. 3a). We suspected that early cell division may be influenced by m6A deficiency. To further study the influence of m6A on early cell division in lateral root induction, the mta ABI3:MTA plant (where the ABI3:MTA construct complements the embryo lethal phenotype of the MTA knockout mutant but exhibits mutant phenotype in a mature plant)3,6 was crossed with a mitotic marker line CycB1;1::GUS18, and the GUS expression was compared between the MTA, CycB1;1::GUS and mta, CycB1;1::GUS plants (mta ABI3:MTA CycB1;1::GUS). The GUS expression in the mta ABI3:MTA background was markedly decreased during lateral root induction compared with that in the WT background, indicating reduced initiation of cell division (required for lateral root initiation and development) (Fig. 3c). In addition, the GFP expression analysis of MTApro:MTA-GFP/mta treated with NPA and NAA demonstrated that the MTApro:MTA-GFP transgene was strongly expressed during the whole lateral root induction process, primarily at root tips and lateral root initiation sites (Supplementary Fig. 6). Altogether, this is consistent with a reduced auxin response in the Arabidopsis root upon m6A deficiency. When the same NPA and NAA treatments were performed on hiz1 mutants and HIZ1-GFP lines (HIZ1pro:HIZ1-GFP/hiz1, HIZ1pro:HIZ1-GFP/WT and 35Spro:HIZ1-GFP/WT-1), the overexpressor line where HIZ1 was driven by the constitutive 35S promoter showed decreased lateral root induction whilst other lines displayed WT levels of lateral root induction (Fig. 3b).
Fig. 3. Root morphology under the pericycle activation for lateral root induction.
a, b Root morphology of seedlings after being treated with 1-naphthaleneacetic acid (NAA, 10 μM) for 5 days following N-1-naphthylphthalamic acid (NPA) treatment. Roots were soaked in 100% ethanol for a few seconds to minimize the effect of root hairs in photographing lateral roots. Scale bar = 500 μm. c GUS staining showing CycB1;1::GUS activities in WT and mta ABI3:MTA backgrounds during a time-course growth on NAA after NPA treatment. Scale bar = 500 μm.
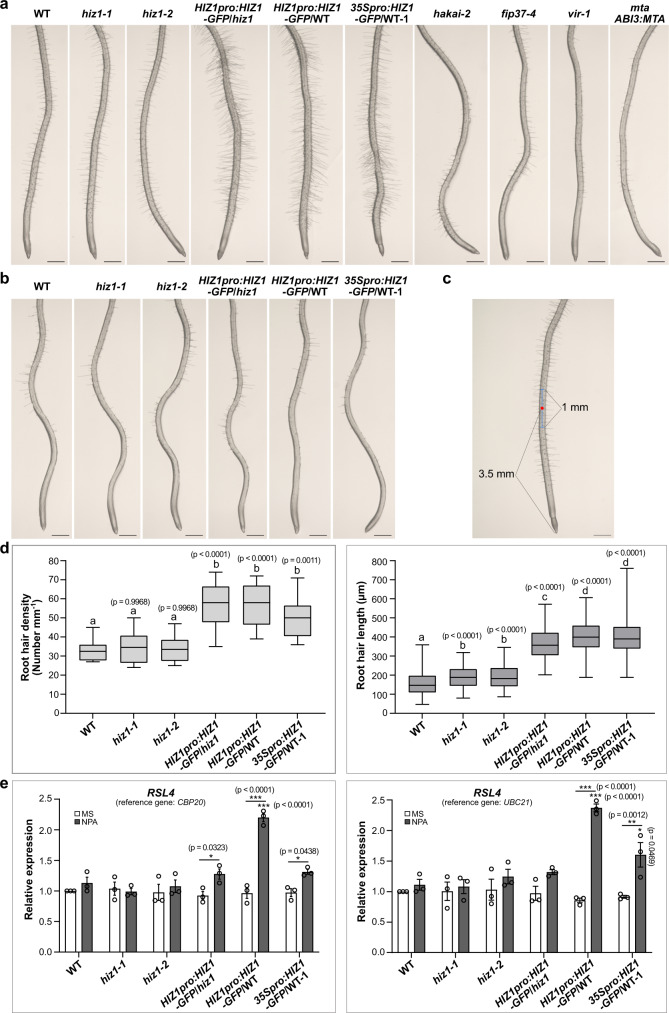
Whilst conducting these experiments, we observed that in all HIZ1-GFP lines (HIZ1pro:HIZ1-GFP/hiz1, HIZ1pro:HIZ1-GFP/WT and 35Spro:HIZ1-GFP/WT-1), there were significantly increased root hair lengths and density following NPA treatment compared to WT (Fig. 4a–d). This morphological change was observed only in HIZ1-GFP lines (most significant in HIZ1pro:HIZ1-GFP/WT) and not in any other m6A writer mutants (Fig. 4a).
Fig. 4. Root hair morphology under the N-1-naphthylphthalamic acid treatment.
a Root morphology after being treated with N-1-naphthylphthalamic acid (NPA, 10 μM) for 6 days. Scale bar = 500 μm. b Roots from 0.5× MS control of the same age as those in a. Scale bar = 500 μm. Experiments in a, b were repeated independently at least three times, and representative images are shown. c A schematic illustrating how root hair density and lengths were measured. The number of root hairs in the middle 1 mm segment (shown using a blue segment with a red dot in the middle) of the imaged root represents the root hair density. The lengths of 10 root hairs on each side of the primary root in the middle 1 mm segment were measured to indicate its representative root hair length. Scale bar = 500 μm. d Statistical data showing root hair density and lengths. Ten seedlings were used for the measurement for each line. Data represent mean ± SE and statistically significant differences were analyzed by One-Way ANOVA (one-sided test). p values labeled in the diagram represent the comparisons with WT and letters represent significance at p < 0.01. In box plots, the center line in each box indicates the median. The lower and upper bounds of each box represent the first quartile (25%) and the third quartile (75%), respectively. The bottom and top of whiskers denote the minimum and maximum, respectively. e Transcript levels of RSL4 analyzed by RT-qPCR. Samples were roots grown on NPA or the control (0.5× MS medium without NPA) for 6 days after germinating on 0.5× MS for 4 days. Data represent mean ± SE from three biological replicates. Statistically significant differences were analyzed by two-way ANOVA (one-sided test) and marked with asterisks (*p < 0.05; ***p < 0.001). Asterisks labeled just above the column are for comparing with WT grown under the same condition and those labeled above both columns for each line are for the comparison between two growth conditions (with or without NPA). Source data are provided as a Source Data file.
HIZ1pro:HIZ1-GFP is expressed in hair cells and this expression is strongly elevated following NPA treatment. In contrast, 35Spro:HIZ1-GFP is expressed in all root cells (including root hairs) (Supplementary Fig. 4). Consistent with the root hair phenotypes, HIZ1-GFP transgene abundance in HIZ1pro:HIZ1-GFP/hiz1 increased by 2-fold relative to the WT HIZ1 transcript level (Supplementary Fig. 7). This suggests that the HIZ1-GFP transgene not only complements the loss of HIZ1 expression in hiz1-1, but also leads to the local overexpression of HIZ1 to some extent, possibly due to the lack of regulatory sequences from the endogenous 3′ UTR (Supplementary Fig. 3a), which is itself methylated and is absent in the carboxy-terminal GFP tagged lines. In all of the HIZ1-GFP lines, the root hair physiological changes were also mirrored at the RNA level by the increased expression of the bHLH transcription factor ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4) following NPA treatment (Fig. 4e). RSL4 is a key regulator of root hair genes, the constitutive expression of which promotes root hair formation22. The induction of RSL4 is indicative of the initiation of root hair specific pathways in the NPA-treated HIZ1-GFP lines. However, this root hair promoting activity of HIZ1 appears to require the presence of HAKAI, as the positive effect on root hair development was lost when the HIZ1-GFP lines were crossed into genetic backgrounds in which HAKAI was knocked out (Supplementary Fig. 8).
Transcriptome-wide analysis of m6A topology in WT, hakai-2, fip37-4, and vir-1
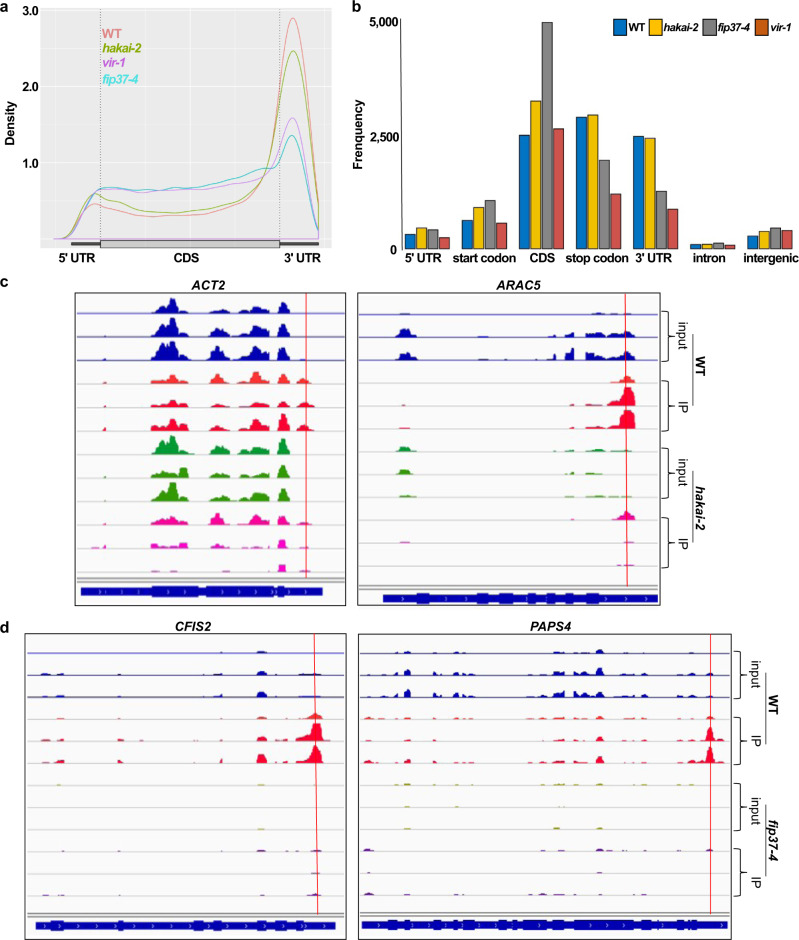
HAKAI knockout lines have a 35% lower m6A/A ratio compared with WT. However, the m6A levels are higher than in hypomorphic mutants of other key complex members, and phenotypically hakai mutants resemble WT. The double mutants between hakai and core complex members exhibit a much-enhanced phenotype, characteristic of more severe m6A deficiency, than the single core complex mutants alone8, indicating an additive or synergistic effect of the mutations. The reduced level of m6A seen in the hakai lines could be due to a general reduction of methylation across all target transcripts, or it could result from loss of methylation from just a specific subset. To address this question, we carried out methylated RNA immunoprecipitation with next-generation sequencing (MeRIP-Seq) on mRNAs extracted from 2-week-old seedlings of WT and writer mutants and compared results for hakai-2 with those of WT and of the two other complex member mutants, vir-1 and fip37-4. MeRIP-Seq enriches for m6A-containing mRNA fragments, which are defined as peaks relative to the input sample. We identified 12,715 peaks for WT, 13,546 for hakai-2, 8883 for vir-1, and 9809 for fip37-4. We did not expect peaks to disappear completely in vir-1 and fip37-4, as both mutants are hypomorphic (~85% reduction in m6A), their complete knockout being lethal6,8. The topology and number of m6A peaks at the transcript level was very similar between hakai-2 and WT, and significantly different in the fip37-4 and vir-1 mutants (Fig. 5a). All three mutants had a decrease in m6A methylation at the 3′ ends of transcripts (Fig. 5a), as well as a relative increase at the 5′ ends and across the coding sequence (CDS). Similar results for fip37 were previously observed by Shen et al.23. At the genomic level, most of the m6A sites were found in protein coding transcripts and in long non-coding RNAs (lncRNAs) (Supplementary Fig. 9). There was an increase in m6A deposition in the intergenic regions for all three mutants, which was most pronounced in the fip37-4 immunoprecipitation (IP) samples (Fig. 5b).
Fig. 5. MeRIP-Seq analysis of three key components of the m6A writer complex mutants.
a Metagene profile of the global distribution of m6A in WT, hakai-2, fip37-4 and vir-1. b Frequency of m6A sites in the different transcript parts in relation to their genomic position. c m6A topology differences between hakai-2 and WT in two key transcripts involved in root hair development (images from Integrative Genomics Viewer [IGV]). IP: immunoprecipitation. y axis scale for ACT2: 0–4000 reads; y axis scale for ARAC5 inputs: 0–1000 reads and for IPs: 0–4000 reads. Red lines here and in d represent the positions of predicted m6A peaks. d m6A topology differences between fip37-4 and WT in two examples of the GOBP mRNA polyadenylation group (images from IGV). y axis scale for both CFIS2 and PAPS4: 0–1000 reads.
We wanted to test whether the topological differences in m6A deposition can be biologically meaningful, globally. Thus, we carried out a GO enrichment analysis on the protein coding methylated transcript lists which were present in WT and absent in hakai-2 (798), fip37-4 (1566) or vir-1 (3079) (Supplementary Data 2–4). For hakai-2, we created a GOBP list containing those GO terms that were at least 2-fold enriched and had a p value <0.05 (Fisher test) (Supplementary Data 5). Some of the highest (50.2-fold) enriched processes were RNA localization to chromatin (GO: 1990280), regulation of antisense RNA transcription (GO: 0060194), circadian regulation of translation (GO: 0097167) and regulation of transcription by RNA polymerase V (GO: 1904279). We were particularly interested in processes that are involved in root hair initiation or elongation, as our data showed that HIZ1 is involved in root hair development in a HAKAI-dependent manner. We identified six transcripts that were 2.49-fold enriched in association with root hair cell differentiation (GO: 0048765), trichoblast maturation (GO: 0048764), and 2.41-fold enriched in trichoblast differentiation (GO: 0010054) in WT versus hakai-2 mutant (Supplementary Data 5). All of these six transcripts were methylated at their 3′ ends in WT plants, and this was missing from the hakai-2 samples. MeRIP-Seq profiles of actin 2 (ACT2, AT3G18780) and Arabidopsis Rac-like GTP binding protein 5 (ARAC5, AT1G75840) belonging to this GO term enriched set are presented in Fig. 5c. The 1566 differential peaks in WT versus fip37-4 mutant and 3079 for WT versus vir-1 were also tested for GOBP enrichment. We expected terms associated with embryo development and root development to be represented as we already have phenotypic evidences for these processes being influenced by the lack of mRNA methylation6,8. For fip37-4, one of the top categories was embryo development (GO: 0009790) (2.06-fold enrichment), as well as root development (GO: 0048364) (2-fold enrichment) (Supplementary Data 5). The embryo development was only 1.64-fold enriched in the vir-1 samples, however, lateral root formation (GO: 0010311) was enriched 3.19-fold (Supplementary Data 5). The root hair development associated transcripts were also enriched in fip37-4, however different sets of transcripts are losing methylation in this mutant compared to hakai-2. One of the highest enriched terms in the fip37-4 list was mRNA polyadenylation (GO: 0006378), with a 6.21-fold enrichment (Supplementary Data 5). Association between mRNA methylation and polyadenylation site choice was already established by Parker et al. in the vir-1 mutant line24. We found that all six members of the group including nuclear poly(A) polymerase 4 (PAPS4, AT4G25550) and CFIS2 (AT4G25550) are missing their m6A peaks in the 3′ UTR (Fig. 5d) or significantly reduced (FY, AT5G13480) in the fip37-4 mutant.
We also wanted to see if there was any connection between the methylation levels of any transcript and their abundance. We carried out a transcriptomic analysis of all three mutants where we identified differentially expressed genes (1.5-fold change cut off). We then looked at the proportional representation of the transcripts that lost their methylation in these gene lists. We did not find any over representation of lack of methylation in the up or down regulated sets in any of the mutants (as an example, vir-1 analysis is in Supplementary Data 6). Next, we used the dataset from Lavenus et al.25, where the authors specifically identified genes that are regulated by auxin response factor 7 (ARF7), a transcription factor which is required together with ARF19 for lateral root initiation in response to auxin. We extracted all of the methylated genes from this dataset, which gave us a list of 90 m6A methylated transcripts, more than half (52%) of the verified ARF7 responsive gene dataset (173 genes) (Supplementary Data 7). We again wanted to know whether the loss of methylation in ARF7 responsive transcripts would affect their expression at the transcriptional level. Therefore, we extracted those genes that lose methylation in the vir-1 mutant (a list of 19 genes, Supplementary Data 7). Out of the 19 transcripts that lost methylation, only two were found in the vir-1 differentially expressed gene list. Nevertheless, these two transcripts ARF19 and auxin resistant 1 (AUX1) are both instrumental in lateral root development and have decreased expression in vir-1. This finding is in agreement with defective lateral root development phenotypes we observe in m6A deficient plants8. Next we looked at how methylation affects gene expression during root hair development. From a gene expression dataset of Lan et al.26 that reports a root hair dataset compared with data obtained from analyzing all root tissues, we identified 365 methylated transcripts out of 635 upregulated genes. This equates to a 57% representation (representation 1, p < 0.152) of the mRNA methylation in those transcript that increase abundance during root hair development (Supplementary Data 8). Next, we looked at the representation of methylated transcripts in the downregulated list of genes, which was 72.5%, a statistically significant increase (representation 1.3, p < 1.042e−29, http://nemates.org/MA/progs/overlap_stats.html) (Supplementary Data 8). This suggests that during root hair development, the presence of m6A is associated with a decrease in abundance of the relevant transcripts.
HIZ2 is required for full m6A methylation and normal growth and development
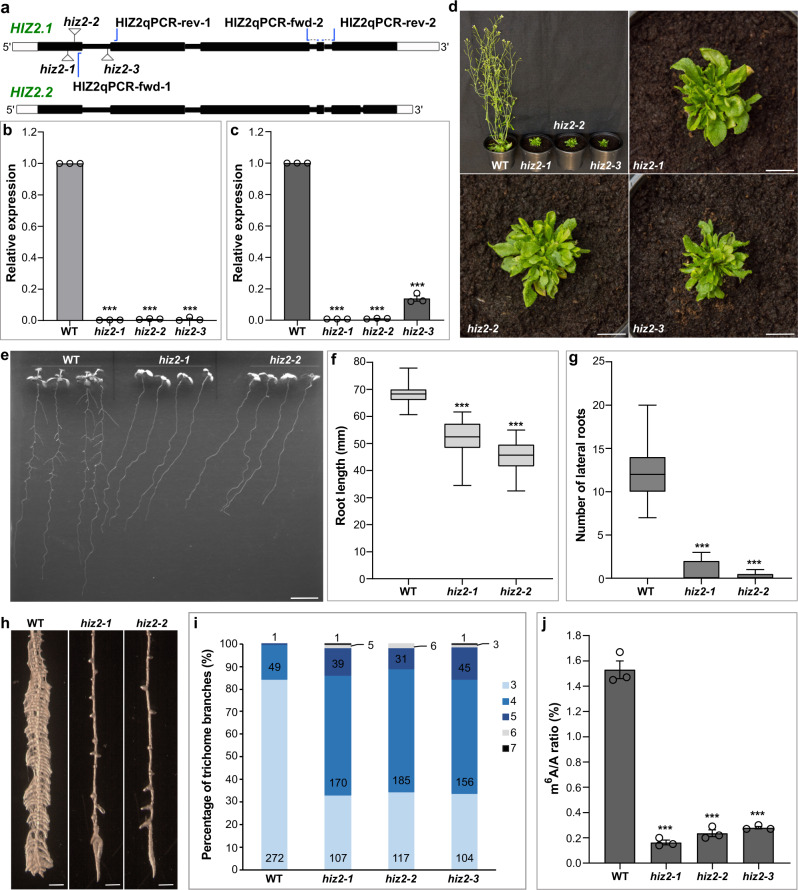
From the co-immunoprecipitation experiments we identified two zinc finger proteins (HIZ1 and HIZ2) as HAKAI interacting partners. While HIZ1 is a writer complex member only by its interaction with HAKAI, HIZ2 was shown to stay bound to MTA even when HAKAI was absent. Thus, we wanted to know whether HIZ2 is required for m6A deposition. We identified three homozygous T-DNA insertion mutants disrupting the expression of HIZ2, referred to as hiz2-1 (Salk_120590), hiz2-2 (Salk_020625), and hiz2-3 (Salk_126486) (Fig. 6a). RT-qPCR analysis confirmed that the correct transcripts of HIZ2 in hiz2-1 and hiz2-2 were absent and in hiz2-3 they were markedly reduced and truncated (Fig. 6b, c). Unlike the major hypomorphic m6A writer mutants (mta ABI3:MTA, fip37-4 and vir-1) where a full knockout appears to be lethal3,6,8, the three hiz2 mutants are viable but demonstrate strong developmental defects, including retarded growth, reduced apical dominance and small and bushy rosette leaves, which are similar to, but more severe than, the phenotypes of the major hypomorphic m6A writer mutants (Fig. 6d and Supplementary Fig. 10). In addition, 11-day-old hiz2 mutants grown on 0.5× MS medium displayed significantly shorter primary roots and nearly no lateral roots compared with WT (Fig. 6e–g). Under NPA and NAA treatment, hiz2 mutants showed only a few very short induced lateral roots (Fig. 6h), reminiscent of that in vir-1 plants (Fig. 3a). Trichome branching is known to be influenced by mRNA methylation3,27,28. The hypomorphic mta mutant has an increased trichome branching phenotype3, which is also observed in the hiz2 mutants (Supplementary Fig. 11). The percentage of trichomes in hiz2 mutants with more than three branches was 65% compared to 15% in WT, and 14% had five and more branches whereas WT had only 0.3% (Fig. 6i). Measurement of m6A levels in hiz2 mutants revealed a decrease of 85% (Fig. 6j), which is comparable to the decrease of m6A levels in the major hypomorphic m6A writer mutants3,8. Thus, we conclude that HIZ2 is an important component of the Arabidopsis m6A writer complex. It is potentially a core component of the finely tuned mechanism of mRNA m6A modification involved in Arabidopsis growth and development.
Fig. 6. Characterizing Arabidopsis HIZ2.
a Schematic of HIZ2 (AT5G53440) genomic DNA sequence with T-DNA insertion sites. HIZ2.1 and HIZ2.2 refer to two splice variants. White rectangles denote UTRs, black rectangles denote exons and thick black lines represent introns. hiz2-1 to hiz2-3 refer to Salk_120590, Salk_020625 and Salk_126486, respectively. HIZ2qPCR-fwd-1 and HIZ2qPCR-rev-1 were used as the first primer pair while HIZ2qPCR-fwd-2 and HIZ2qPCR-rev-2 were the second primer pair in RT-qPCR. b, c HIZ2 transcript levels checked by RT-qPCR using the first primer pair (b) and the second primer pair (c) labeled in a. CBP20 was used as a reference gene. Data represent mean ± SE from three biological replicates. Statistically significant differences relative to WT here and in f, g, j were analyzed by One-Way ANOVA (one-sided test) and marked with asterisks (***p < 0.001). d Five-week-old hiz2 mutants and WT planted in compost. Scale bar = 1 cm. e Eleven-day-old seedlings vertically cultured on 0.5× MS plus 1% sucrose. Scale bar = 1 cm. f, g Statistical data showing the root phenotypes of seedlings in e. Data represent mean ± SE (n = 25 individual seedlings). In box plots, the center line in each box indicates the median. The lower and upper bounds of each box represent the first quartile (25%) and the third quartile (75%), respectively. The bottom and top of whiskers denote the minimum and maximum, respectively. h Roots under the Stereo Dissecting Microscope after the treatment with N-1-naphthylphthalamic acid (NPA, 10 μM) and then transferred onto 0.5× MS with 1-naphthaleneacetic acid (NAA, 10 μM) for 5 days. Roots were soaked in 100% ethanol for a few seconds to minimize the effect of root hairs in photographing lateral roots. Scale bar = 500 μm. i Percentage of trichome branches on the first pair of rosette leaves from 2-week-old plants cultured in compost. Twenty leaves were used for counting the trichome branching in each line. j m6A levels checked by two-dimensional thin layer chromatography (TLC) analysis. Data represent mean ± SE from three biological replicates. Source data are provided as a Source Data file.
Discussion
Both the plant and the mammalian HAKAI are members of the m6A writer complex8,11. The mammalian HAKAI was originally characterized as an E3 ubiquitin ligase29, thus we thought that the plant homolog may carry out a similar function, and might regulate the m6A writer complex members at a posttranslational level. The proteomics experiments revealed that HAKAI binds to two writer complex members, which we name HIZ1 and HIZ2. HAKAI’s presence is essential for the HIZ1 binding to the core member MTA (METTL3), thus we suspected that the presence of HIZ1 in the complex may be regulated by posttranslational modification carried out by HAKAI. We did observe an increased amount of GFP-tagged HIZ1 in the absence of HAKAI but at the same time the transcript level of the HIZ1pro:HIZ1-GFP transgene was also increased, indicating HIZ1 is regulated by HAKAI, but not necessarily at a posttranslational level. Thus, whether the plant HAKAI possesses an E3 ligase function and if so, what the significance of this is, remains an open question.
To test whether the reduced level of m6A seen in the hakai lines is due to a general reduction of methylation across all target transcripts, or results from loss of methylation from just a specific subset, we determined the topology of m6A sites globally in WT, hakai-2, and two other complex member mutants, fip37-4 and vir-1. The distribution of m6A peaks was very similar between hakai-2 and WT, and the number of peaks that were methylated in WT but which were absent or substantially reduced in hakai-2 were fewer than those that were missing in the fip37-4 and vir-1 mutants. Thus, the majority of the 35% reduction in m6A abundance seen in hakai mutants is likely due to a global reduction in methylation. However, a small number of transcripts appear to be disproportionally affected, and we found that those transcripts which are methylated in WT but not in hakai-2 were enriched in association with root hair development processes. Although hakai-2 does not show root hair phenotypic changes, the methylation changes in relation to root hair development may be significant, as the HIZ1 overexpressor shows increased root hair length and density in response to NPA treatment in a HAKAI-dependent manner. When we compared m6A topology between the three mutants, it was apparent that both vir-1 and fip37-4 lost most of the 3′ end m6A peaks and had increase in 5′ end and middle peaks. As far as biological processes are concerned, transcripts that lost m6A peaks in the 3′ UTRs were highly associated with embryo development and lateral root development, which was common between fip37-4 and vir-1. One of the highest enrichment terms was mRNA polyadenylation and this was only found in the fip37-4 dataset. An association between mRNA methylation and polyadenylation site choice was previously shown in the vir-1 mutant line24. Parker et al. propose that for certain transcripts, the proper sites for polyadenylation are directly labeled by a nearby m6A24. We found that six transcripts that are involved in mRNA polyadenylation were missing m6A peaks in the 3′ UTR (PAPS2, PAPS4, CPSF160, CFIS2, CPSF73-I) or the peaks were significantly reduced (FY) in the fip37-4 mutants. Thus, we would like to suggest that the poly(A) synthesis and the cleavage site may also be altered by the m6A dependent expression changes of the enzymes directly involved in this process.
Our study identified two Arabidopsis zinc finger proteins that co-purified with HAKAI, HIZ1 and HIZ2. Similar to the core complex member mutants, the knockout of HIZ2 results in decreased global m6A levels, and very severe developmental defects. The mammalian CCCH-type zinc finger protein, ZC3H13, and its Drosophila homolog Female lethal 2 (Fl(2)d)-associated complex component (Flacc), have been identified as writer complex members9,13. The Arabidopsis HIZ2 is composed of a similar number of amino acid residues and possesses low-complexity regions, similar to Flacc (Supplementary Fig. 2). In addition, the lack of full CCCH domains in both Flacc and HIZ2 (Supplementary Fig. 2), suggests that HIZ2 is structurally more similar to the Drosophila Flacc than to the mammalian ZC3H13 protein. Disruption of Flacc function results in reduced m6A level and defects in sex determination due to aberrant splicing of Sex lethal (Sxl)9, similar to the knockout of the Drosophila METTL3 homolog Inducer of meiosis 4 (dIme4)30,31. The Drosophila Flacc is specifically involved in stabilizing the methylase complex, serving as a bridge between the RNA binding protein Spenito (Nito, homologous to Arabidopsis FPA) and Fl(2)d (FIP37 homolog)9. Depletion of mouse ZC3H13 impairs embryonic stem cell self-renewal and triggers differentiation13. In human, the functional role of ZC3H13 other than interacting with WTAP is poorly understood, although a recent report suggests that mutant ZC3H13 may facilitate glioblastoma progression32,33. However, whilst the Drosophila and the mammalian zinc finger homologs may seem to be involved in different processes and are structurally somewhat divergent, human ZC3H13 can restore the interaction between Nito and Fl(2)d upon deletion of endogenous Drosophila Flacc9. Although rather divergent in sequence and structure, the requirement for HIZ2, ZC3H13 and Flacc in the Arabidopsis, mouse and Drosophila methylation writer complex indicates a conserved role for a zinc finger protein. In Arabidopsis, HIZ2 is likely to be a functional homolog of Drosophila Flacc, and it interacts with MTA (METTL3) and HAKAI. However, the interaction of HIZ2 with either FIP37 or FPA is yet to be explored.
Another mammalian zinc finger protein which has been implicated in m6A writing is mouse ZFP217, which belongs to the C2H2-type zinc finger protein family, and acts as a multi-functional oncogene16,34. ZFP217 transcriptionally activates Nanog and Sox2, as well as other genes linked to the undifferentiated state of embryonic pluripotent stem cells16. In addition, through an interaction with the writer complex, ZFP217 inhibits m6A deposition in the same mRNA molecules. When ZFP217 is depleted, the expression of pluripotency factors decreases in parallel with a global increase of m6A methylation, which triggers the degradation of core stem cell transcripts16. Thus, ZFP217 regulates the balance between stem cell regeneration and differentiation. We found that the Arabidopsis HIZ1 overexpression driven by the constitutive 35S promoter led to a 24% reduction in global m6A abundance. Thus, we propose that the Arabidopsis HIZ1, via its binding to HAKAI, may be an inhibitor of mRNA methylation, similar to ZFP217. In addition, the GFP-tagged HIZ1 under its own promoter demonstrated the highest expression in stem cell niches in both primary and lateral root tips. Thus, the plant HIZ1 and the mouse ZFP217 show some degree of functional similarities in spite of their lack of a common structural or sequence relationship. However, it should be noted that we don’t have evidences from knockout mutants to support the proposed function of HIZ1.
When HIZ1 was overexpressed under the strong and constitutive 35S promoter the plants showed some characteristics of the core writer complex mutants, for instance, decreased level of m6A and reduced lateral root induction in response to NPA and NAA treatment. When HIZ1 was overexpressed under the control of its own promoter, the m6A inhibitory effect was not observed, neither was the decrease in lateral root induction. However, in the HIZ1 overexpressor under 35S promoter, and in plants transformed with HIZ1 under its own promoter, we observed increased root hair density and lengths upon NPA treatment. This effect was strongest when the HIZ1 native promoter was used, and under NPA treatment, this promoter drives strong GFP expression in root hairs. The increase in root hair length and density is likely acting through the key regulator RSL4, the expression of which increases in these plants. However, RSL4 itself is not methylated in our data sets, thus, the mRNA methylation process probably indirectly regulates its expression under auxin efflux inhibition. Interestingly, a previous study using a panel of 166 geographically diverse Arabidopsis accessions to look at natural variation in root hair response to low phosphate conditions, identified a SNP which the authors attributed as causative of the root hair phenotype. This SNP resides between HIZ1 and an upstream gene and they went on to show that root hair density (though not length) was altered in response to low phosphate in a HIZ1 insertion mutant35. Neither root hair density nor length were altered in the mutant under phosphate replete conditions35. These results do however, indicate a natural function for HIZ1 in root hair formation, and as the root hair phosphate response is auxin-mediated36, it may explain why the effects following NPA treatment are observed in the plants in which HIZ1-GFP is under the control of either the constitutive 35S or its native promoter.
We propose a model where HIZ1 interacts with the methylation writer complex (via HAKAI) to fine tune methylation by down regulating methylase activity in those cells in which it is expressed, similar to the methylase suppressing activity of mouse ZFP217. In this scenario, strong and constitutive expression of HIZ1 would be expected to result in a global (and HAKAI dependent) reduction in m6A, as we observe. However, it should be noted that the HIZ1 knockout lines do not show obvious root meristem defects that might be predicted to result if they contain higher m6A levels in the stem cell niche. An alternative possibility is that overexpression of HIZ1 disrupts the structure and stoichiometry of the writer complex. This seems less likely, as the HIZ1 knockout mutant has normal m6A levels (in whole seedlings), and it is not strongly expressed outside of the meristematic and root hair cells, suggesting that it is not ubiquitously required as a key structural component.
Based on our findings, we propose that HAKAI acts as a “bridge”, connecting HIZ1 with the core m6A writer components. In addition, HIZ2 is an essential member of the m6A writer complex required for full methylation of transcripts and is likely the plant equivalent of ZC3H13 (Flacc) of the mammalian (Drosophila) m6A-METTL Associated Complex.
Methods
Plant materials and growth conditions
Arabidopsis lines (ecotype Colombia-0) used include WT, m6A writer mutants and transgenic lines containing sequences encoding m6A writer proteins. Specifically, mta ABI3:MTA, fip37-4 and vir-1 are hypomorphic alleles of MTA, FIP37 and VIR, respectively8. hakai-2 is a HAKAI knockout line generated via CRISPR-Cas9 mutagenesis8. T-DNA insertion mutants, including Salk_045882 (hiz1-1), Salk_000717 (hiz1-2), Salk_120590 (hiz2-1), Salk_020625 (hiz2-2) and Salk_126486 (hiz2-3), were obtained from Nottingham Arabidopsis Stock Center (NASC) (Nottingham, UK).
Arabidopsis seeds were cultured in plates or compost depending on the subsequent use. Seeds planted in plates were sterilized by soaking in 5% NaClO for 4 min and then washed with sterile water for five times before being planted onto 0.5× MS medium plus 1% sucrose. Plates were placed in the cold room (4 °C, dark) for 2 days and then kept in the tissue culture room (16 h light: 8 h dark, 22 °C) until used. Plants grown directly in compost were maintained in the phytotron (16 h light: 8 h dark, 22 °C day: 18 °C night).
Generation of transgenic lines
Recombinant plasmid constructs generated in this study include HAKAIpro:HAKAI-GFP, MTApro:MTA-GFP, FIP37pro:FIP37-GFP, HIZ1pro:HIZ1-GFP, and 35Spro:HIZ1-GFP. These constructs were generated by cloning the respective genomic DNA sequence (coding sequence for MTA) without the stop codon and 3′ UTR to Gateway entry vector pCR™8/GW/TOPO® (Invitrogen), then recombining into the appropriate pGreen-based GFP Gateway destination vector37 using Gateway® LR Clonase™ II Enzyme Mix (Invitrogen). Transgenic lines were obtained by floral dip transformation38 with the above recombinant constructs. Homozygous transgenic lines were selected and GFP-tagged protein localization in roots was determined via confocal microscopy (Leica TCS SP5). Primers used for cloning and screening were listed in Supplementary Data 9.
Poly(A) RNA preparation and m6A measurement
Total RNA was isolated using hot phenol extration39. Poly(A) RNA was prepared according to the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, NEB). m6A levels were measured by two-dimensional thin layer chromatography (TLC) analysis as described in Zhong et al.6. Fifty nanogram of poly(A) RNA was digested by 1 μL of Ribonuclease T1 (1000 U μL−1; Thermo Fisher Scientific) in 1× polynucleotide kinase (PNK) buffer A at 37 °C for 1 h. 5′ ends of digested RNA fragments were labeled with 0.5 μL of [γ-32P]ATP (6000 Ci mmol−1; PerkinElmer) using 10 U of T4 PNK. After overnight ethanol precipitation, the labeled RNA pellet was resuspended in 10 μL of 50 mM sodium acetate (pH 5.5) and digested by nuclease P1 (Sigma-Aldrich) at 37 °C for at least 1 h to produce mononucleotides. One microliter of the digested sample was loaded onto the cellulose F TLC plate (20 × 20 cm; Merck) and developed in a solvent system, with isobutyric acid:0.5 M NH4OH (5:3, v/v) as the first dimension buffer and isopropanol:HCl:water (70:15:15, v/v/v) as the second dimension buffer. The labeled nucleotides were identified and quantified by using a storage phosphor screen (Fuji-Screen) and Bio-Rad Molecular Imager FX system in combination with Quantity One software (v4.6.2.70).
In vivo interaction proteomics-mass spectrometry analysis
The in vivo interaction proteomics-mass spectrometry (IVI-MS) libraries were prepared according to the method described in Parker et al.14. Freshly harvested 2-week-old seedlings were crosslinked using 1% formaldehyde and the crosslinking was quenched by 0.125 M glycine solution. Nuclei were isolated from ground frozen sample powder using HONDA buffer (20 mM Hepes KOH pH 7.4, 10 mM MgCl2, 440 mM sucrose, 1.25% Ficoll, 2.5% Dextran T40, 0.5% Triton X-100, 5 mM DTT, 5 mM phenylmethylsulfonyl fluoride [PMSF] and 1% Protease Inhibitor Cocktail [Sigma-Aldrich]). After centrifuging at 2000 × g for 17 min at 4 °C, the pellet was washed twice with HONDA buffer. The isolated nuclei samples were lysed using nuclei lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 1% SDS, 1 mM PMSF and 1% Protease Inhibitor Cocktail [Sigma-Aldrich]) via sonication using a water bath sonicator (Diagenode Bioruptor 200) on low power with four cycles of 30 s ON and 60 s OFF. After centrifugation at 16,100 × g for 15 min at 4 °C, the supernatant was diluted using IP dilution buffer (16.7 mM Tris-HCl pH 8.0, 1.2 mM EDTA pH 8.0, 167 mM NaCl, 1.1% Triton X-100 and 1% Protease Inhibitor Cocktail [Sigma-Aldrich]) and then pre-washed GFP-Trap_A agarose beads (ChromoTek) was added to pull down target protein complex. The mixture was incubated at 4 °C with rotation for 5 h. Afterwards, beads were washed three times using beads washing buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA pH 8.0, 1% Triton X-100, 0.1% SDS and 1 mM PMSF). Samples were incubated at 90 °C for 30 min to reverse formaldehyde crosslinking. After running samples on SDS-PAGE gel, the gel for the same lane was cut into five slices. Protein samples were digested into peptides by trypsin and then analyzed on LTQ Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific). WT was used as a control and three biological replicates were performed for each line.
After mass spectrometry, raw files corresponding to five slices from the same lane were merged and analyzed via MaxQuant (v1.6.0.16) using label free quantification (LFQ)40. Unique and razor peptides were used for protein quantification and the protein and peptide false discovery rates (FDRs) were set to 0.01. MaxQuant output files containing proteins that were identified in each sample were subsequently analyzed using Perseus (v1.6.15.0)41. Potential interacting proteins were determined using two-sided student t-test and visualized by volcano plots. The significance was analyzed based on the negative logarithm of the p-value derived from the t-test (−log10p-value). Threshold values (FDR) were set between 0.01–0.1 and S0 (curve bend) at 1 or 2. In volcano plots, significantly-enriched proteins were separated from others by a hyperbolic curve.
Gene expression analysis by RT-qPCR
Total RNAs were extracted from 2-week-old seedlings (for confirming T-DNA inserted mutations and the expression levels of transgenes) or from root samples under different treatments. Total RNAs were treated with Ambion® TURBOTM DNase (Invitrogen) and then purified via GeneJET RNA Cleanup and Concentration Micro Kit (Thermo Fisher Scientific). First-strand cDNAs were synthesized using ProtoScript II Reverse Transcriptase (NEB). qPCR was carried out on LightCycler® 480 instrument (Roche) using SensiMix™ SYBR® Low-ROX (Bioline). Three biological replicates were analyzed for each sample. Relative expression levels were determined using the 2−ΔΔCT method42. Arabidopsis CBP20 (AT5G44200) and UBC21 (AT5G25760) were used as internal controls. qPCR primer pairs were listed in Supplementary Data 9.
Western blot
Proteins were extracted from 2-week-old seedlings cultured on 0.5× MS medium using the nuclei lysis buffer as mentioned above. Protein samples were separated on Bio-Rad Mini-PROTEAN® TGX™ Precast Gel and transferred onto Amersham™ Protran® western blotting membrane (nitrocellulose, pore size: 0.2 μm). Subsequent steps were performed following instructions for ‘WesternBreeze® Chemiluminescent Western Blot Immunodetection’ Kit (Invitrogen). The GFP antibody used is a mixture of two monoclonal antibodies from mouse IgG1κ (clone 7.1 and 13.1, Sigma-Aldrich) at a 1:1000 dilution. An anti-actin antibody, a rabbit polyclonal antibody targeting a set of actins in Arabidopsis (Agrisera) was used as a control (at 1:2500 dilution). The protein amounts were quantified using Quantity One software (v4.6.2.70). Uncropped scans for western blot membranes were included in the Source Data file or Supplementary Information file.
Analysis of root phenotypes
Arabidopsis primary root lengths and the number of lateral roots were recorded from 10-day-old seedlings vertically cultured on 0.5× MS with 1% sucrose. Primary root lengths were measured using ImageJ software (v1.8.0_172). Pericycle activation for lateral root initiation was performed according to the method described in Himanen et al.21. Specifically, 4-day-old Arabidopsis seedlings vertically cultured on 0.5× MS were transferred to vertical square plates containing 0.5× MS and 10 μM NPA. On the 3rd day of culturing on NPA, they were transferred onto 0.5× MS plus 10 μM NAA. To observe root hair morphology on NPA, seedlings grown on NPA remained growing on the same medium after the 3rd day and images of root hairs were taken on the 6th day of culturing on NPA. Roots were photographed under the Stereo Dissecting Microscope (Zeiss Stemi SV6). The number and lengths of root hairs were measured using ImageJ software (v1.8.0_172). Statistical analysis was performed using GraphPad Prism (v9.2.0).
GUS expression assay
Harvested roots were immersed in GUS staining reagent mix (50 mM phosphate buffer, 10 mM EDTA, 20% methanol, 0.1% TritonX-100 and 1 mg mL−1 X-Gluc) while shaking for 12 h. Afterwards, stained roots were soaked in 100% ethanol for destaining. Photos were taken under the Stereo Dissecting Microscope (Zeiss Stemi SV6).
MeRIP-Seq and differential expression analysis
Total RNA from three replicates of 2-week-old seedling grown on 0.5× MS was isolated using hot phenol extraction39. This was followed by one-round poly(A) purification using oligo d(T) magnetic beads (NEB). 1.5–2 µg of mRNA was fragmented to 100–150 nts using RNA Fragmentation Reagent (Thermo Fisher Scientific) followed by overnight ethanol precipitation. After centrifugation and washing, the pellets were resuspended in 10 µL of H2O. Nine microliter of the solution was used for immunoprecipitation, and one microliter for preparing the input libraries. The fragmented RNA was mixed with Protein G magnetic bead prebound monoclonal anti-m6A antibody (1 µL) from the EpiMark N6-Methyladenosine Enrichment Kit (NEB), resuspended in 300 µL EpiMark IP buffer supplemented with murine RNase inhibitor (NEB). All following steps were as described by the manufacturer. After the last wash we carried out an extra washing step using H2O. We omitted the final elution step as we carried out the cDNA synthesis on the magnetic beads using ScriptSeq v2 RNA-Seq Library Preparation Kit (Illumina). The libraries for both IP and input were size selected using E-Gel SizeSelect II (Invitrogen), and quality checked on Agilent High Sensitivity DNA Chips (Agilent). The pooled libraries were sequenced on Nextseq 500 (Illumina) at the Nottingham University’s Sequencing Service.
For the MeRIP analysis, sequencing reads contaminating adapter sequences and low quality reads (phred scores <30) were removed using Trimgalore (v0.4.4, https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The processed fastq reads were aligned to the Ensembl annotated TAIR10.49 reference genome using STAR (v2.5.0)43. The resultant bam files were indexed using SAMtools (v1.10, PMID: 19505943)44 and m6A enriched regions identified in m6A immunoprecipitated samples over inputs using m6aViewer (v1.6.1) (PMID: 28724534)45. BEDtools (v2.27.1, PMID: 20110278)46 was used to extend peaks by 100 bp upstream and downstream. Only those peaks represented in at least two replicates, and 4-fold enriched were taken forward for further analysis using the RNAmod, interactive web-based platform (http://61.147.117.195/RNAmod/) with default settings47. Using the Peak matrix dataset created by the RNAmod platform, we produced lists of peaks which were present in WT samples and missing from the mutant data. The GO enrichment analysis on these gene lists was carried out using PANTHER (v16.) Classification System website (http://pantherdb.org).
For differential expression the MeRIP-Seq input files qualities were assessed by fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) prior to trimming with Trimmomatic (v0.38)48. Trimmed reads were aligned to the TAIR10 Arabidopsis genome (TAIR10.48) using Bowtie2 (v2.4.2)49 with local alignment enabled. Aligned bam files were indexed with SAMtools (v1.10)44 prior to gene counting with HTSeq (v0.9.1)50. A matrix of gene counts was generated from the read counts and imported into R-studio (v1.4.1717-3, https://www.rstudio.com/). Differential expression analysis was performed using DESeq2 (v2.11.40.6)51. To generate a list of differentially expressed genes, we chose genes with a log2 fold change above 1.5 (enriched) or below −1.5 (depleted) and adjusted p-value of less than 0.05. The default tests for calculating p-value (Wald) and correcting for multiple testing (Benjamini and Hochberg) were used. We identified gene list overlaps between our methylation datasets and differential expression data from our input samples as well as from published expression data25,26. To calculate the statistical significance, the online tool (http://nemates.org/MA/progs/overlap_stats.html) was used.
Analysis of trichome branching
The number of trichome branches were counted from the first pair of rosette leaves of 2-week-old Arabidopsis plants cultured in compost. Trichomes with different number of branches were photographed under the Stereo Dissecting Microscope (Zeiss Stemi SV6).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by Biotechnology and Biological Sciences Research Council (BBSRC) grants (grants BB/M008606/1 and BB/S006478/1) awarded to R.G.F. and by BBSRC grant (BB/M010066/1) awarded to G.G.S. The PhD program of M.Z. was supported by China Scholarship Council Research Excellence Scholarship and the University of Nottingham Future Food Beacon Doctoral/Post-Doctoral Prize.
Source data
Author contributions
M.Z., Z.B., K.M., and S.Z. conducted experiments, Z.B., N.P.M., and N.A. carried out bioinformatics analyses. R.G.F. conceived research and designed experiments. M.Z., Z.B., and R.G.F. wrote the manuscript. G.G.S. and N.P.M. edited manuscript. All authors read and commented on the final version of the manuscript.
Peer review
Peer review information
Nature Communications thanks Brian Gregory and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
Sequence details for Arabidopsis genes used in this study can be found in TAIR database (https://www.arabidopsis.org/) with the following accession numbers: AT5G01160 (HAKAI), AT1G32360 (HIZ1), AT5G53440 (HIZ2), AT4G10760 (MTA), AT3G54170 (FIP37) and AT3G05680 (VIR). Protein sequences for possible HIZ2 homologs in other organisms used in this study can be found in the UniProt Knowledgebase (UniProtKB, https://www.uniprot.org/) with the following identifiers: Q5T200-1 (Human ZC3H13), E9Q784-1 (Mouse ZC3H13), and Q9VWN4-1 (Drosophila Flacc). The mass spectrometry proteomics data generated in this study have been deposited in the ProteomeXchange Consortium via the PRIDE52 partner repository under the dataset identifier PXD026287. The MeRIP-Seq data generated in this study have been deposited at NCBI’s Gene Expression Omnibus (GEO) under the accession number GSE174573. The transcriptomics data generated in this study have been deposited at GEO under the accession number GSE188610. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-28753-3.
References
- 1.Arribas-Hernández L, Brodersen P. Occurrence and functions of m6A and other covalent modifications in plant mRNA. Plant Physiol. 2020;182:79–96. doi: 10.1104/pp.19.01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang H, Weng H, Chen J. The biogenesis and precise control of RNA m6A methylation. Trends Genet. 2020;36:46–52. doi: 10.1016/j.tig.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodi Z, et al. Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front. Plant Sci. 2012;3:48. doi: 10.3389/fpls.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- 6.Zhong S, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Růžička K, et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. N. Phytol. 2017;215:157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knuckles P, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz S, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue Y, et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:1–17. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patil DP, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen J, et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 2018;69:1028–1038. e1026. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker MT, et al. Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA. eLife. 2021;10:e65537. doi: 10.7554/eLife.65537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balacco DL, Soller M. The m6A writer: rise of a machine for growing tasks. Biochemistry. 2019;58:363–378. doi: 10.1021/acs.biochem.8b01166. [DOI] [PubMed] [Google Scholar]
- 16.Aguilo F, et al. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell. 2015;17:689–704. doi: 10.1016/j.stem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhat SS, et al. mRNA adenosine methylase (MTA) deposits m6A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. PNAS. 2020;117:21785–21795. doi: 10.1073/pnas.2003733117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casimiro I, et al. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell. 2001;13:843–852. doi: 10.1105/tpc.13.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abas, L. et al. Naphthylphthalamic acid associates with and inhibits pin auxin transporters. PNAS118, e2020857118 (2021). [DOI] [PMC free article] [PubMed]
- 20.Delbarre A, Muller P, Imhoff V, Guern J. Comparison of mechanisms controlling uptake and accumulation of 2, 4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta. 1996;198:532–541. doi: 10.1007/BF00262639. [DOI] [PubMed] [Google Scholar]
- 21.Himanen K, et al. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell. 2002;14:2339–2351. doi: 10.1105/tpc.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi K, Menand B, Bell E, Dolan L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 2010;42:264. doi: 10.1038/ng.529. [DOI] [PubMed] [Google Scholar]
- 23.Shen L, et al. N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev. Cell. 2016;38:186–200. doi: 10.1016/j.devcel.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker MT, et al. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. eLife. 2020;9:e49658. doi: 10.7554/eLife.49658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavenus J, et al. Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. Plant Cell. 2015;27:1368–1388. doi: 10.1105/tpc.114.132993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan P, Li W, Lin WD, Santi S, Schmidt W. Mapping gene activity of Arabidopsis root hairs. Genome Biol. 2013;14:R67. doi: 10.1186/gb-2013-14-6-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scutenaire J, et al. The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell. 2018;30:986–1005. doi: 10.1105/tpc.17.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei LH, et al. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell. 2018;30:968–985. doi: 10.1105/tpc.17.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita Y, et al. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 2002;4:222–231. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- 30.Haussmann IU, et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 31.Lence T, et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242–247. doi: 10.1038/nature20568. [DOI] [PubMed] [Google Scholar]
- 32.Horiuchi K, et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow RD, et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat. Neurosci. 2017;20:1329. doi: 10.1038/nn.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee DF, Walsh MJ, Aguiló F. ZNF217/ZFP217 meets chromatin and RNA. Trends Biochem. Sci. 2016;41:986–988. doi: 10.1016/j.tibs.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stetter MG, Schmid K, Ludewig U. Uncovering genes and ploidy involved in the high diversity in root hair density, length and response to local scarce phosphate in Arabidopsis thaliana. PLoS ONE. 2015;10:e0120604. doi: 10.1371/journal.pone.0120604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhosale R, et al. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat. Commun. 2018;9:1409. doi: 10.1038/s41467-018-03851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong S, Lin Z, Fray RG, Grierson D. Improved plant transformation vectors for fluorescent protein tagging. Transgenic Res. 2008;17:985. doi: 10.1007/s11248-008-9199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox J, Mann M. Maxquant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 41.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antanaviciute A, et al. m6aViewer: software for the detection, analysis, and visualization of N6-methyladenosine peaks from m6A-seq/ME-RIP sequencing data. RNA. 2017;23:1493–1501. doi: 10.1261/rna.058206.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Q, Gregory RI. RNAmod: an integrated system for the annotation of mRNA modifications. Nucleic Acids Res. 2019;47:W548–W555. doi: 10.1093/nar/gkz479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Sequence details for Arabidopsis genes used in this study can be found in TAIR database (https://www.arabidopsis.org/) with the following accession numbers: AT5G01160 (HAKAI), AT1G32360 (HIZ1), AT5G53440 (HIZ2), AT4G10760 (MTA), AT3G54170 (FIP37) and AT3G05680 (VIR). Protein sequences for possible HIZ2 homologs in other organisms used in this study can be found in the UniProt Knowledgebase (UniProtKB, https://www.uniprot.org/) with the following identifiers: Q5T200-1 (Human ZC3H13), E9Q784-1 (Mouse ZC3H13), and Q9VWN4-1 (Drosophila Flacc). The mass spectrometry proteomics data generated in this study have been deposited in the ProteomeXchange Consortium via the PRIDE52 partner repository under the dataset identifier PXD026287. The MeRIP-Seq data generated in this study have been deposited at NCBI’s Gene Expression Omnibus (GEO) under the accession number GSE174573. The transcriptomics data generated in this study have been deposited at GEO under the accession number GSE188610. Source data are provided with this paper.