Abstract
Purpose
While sampled or short-frame realizations have shown the potential power of deep learning to reduce radiation dose for PET images, evidence in true injected ultra-low-dose cases is lacking. Therefore, we evaluated deep learning enhancement using a significantly reduced injected radiotracer protocol for amyloid PET/MRI.
Methods
Eighteen participants underwent two separate 18F-florbetaben PET/MRI studies in which an ultra-low-dose (6.64 ± 3.57 MBq, 2.2 ± 1.3% of standard) or a standard-dose (300 ± 14 MBq) was injected. The PET counts from the standard-dose list-mode data were also undersampled to approximate an ultra-low-dose session. A pre-trained convolutional neural network was fine-tuned using MR images and either the injected or sampled ultra-low-dose PET as inputs. Image quality of the enhanced images was evaluated using three metrics (peak signal-to-noise ratio, structural similarity, and root mean square error), as well as the coefficient of variation (CV) for regional standard uptake value ratios (SUVRs). Mean cerebral uptake was correlated across image types to assess the validity of the sampled realizations. To judge clinical performance, four trained readers scored image quality on a five-point scale (using 15% non-inferiority limits for proportion of studies rated 3 or better) and classified cases into amyloid-positive and negative studies.
Results
The deep learning–enhanced PET images showed marked improvement on all quality metrics compared with the low-dose images as well as having generally similar regional CVs as the standard-dose. All enhanced images were non-inferior to their standard-dose counterparts. Accuracy for amyloid status was high (97.2% and 91.7% for images enhanced from injected and sampled ultra-low-dose data, respectively) which was similar to intra-reader reproducibility of standard-dose images (98.6%).
Conclusion
Deep learning methods can synthesize diagnostic-quality PET images from ultra-low injected dose simultaneous PET/MRI data, demonstrating the general validity of sampled realizations and the potential to reduce dose significantly for amyloid imaging.
Keywords: PET/MRI, Deep learning, Amyloid PET, Ultra-low-dose PET
Introduction
Alzheimer’s disease (AD) is a severe neurodegenerative disorder, and the number of people with this disease is projected to increase rapidly [1]. One significant biomarker for AD, amyloid plaque buildup in the brain, can be identified with positron emission tomography (PET) imaging [2, 3]. To leverage its exquisite sensitivity [4–6], more frequent PET scans can be used to understand the pathogenesis of the brain proteinopathies involved in dementia [7], to identify at-risk individuals, and to indicate the optimal time point for early intervention in potential anti-amyloid therapies [8]. Advanced hardware such as simultaneous PET/magnetic resonance imaging (MRI) also allows for perfect spatiotemporal correlation of complementary functional (PET) and structural (MRI) information, all of which can contribute to the diagnosis and staging of AD [9].
However, for widespread clinical use and longitudinal imaging studies with large study populations, radioactivity and cost will be limiting factors. The cost from the radiotracers (e.g., amyloid imaging agents) will limit the number of patients to be scanned and reduce the number of potential participants in studies. Furthermore, radioactivity associated with the radiotracers will also present a risk to participants, especially in vulnerable populations, and may discourage patients from enrolling in clinical trials. These factors limit the scalability of PET studies. However, a reduction in injected radiotracer dose implies fewer coincidence events from annihilation photon pairs (“counts”) being collected for image reconstruction. Therefore, reducing the counts collected in PET studies, either by reducing the radiotracer dose or the scan time (to reduce image quality degradation from subject motion as well as to increase machine throughput), but without sacrificing image quality and diagnostic value, will be key to the increased use of this powerful modality.
While researchers have used approximations to investigate dose reduction, or more broadly, low-count imaging in PET, some using machine learning methods (both traditional [10–12] and convolutional neural networks: CNNs [13–19]), we are proposing actual PET/MRI studies with ultra-low-dose injections (~ 2% of the original). In this report, we investigate the use of actual injected ultra-low-dose (AULD) PET images (lower than the ~ 12% previously reported [20]) as inputs rather than list-mode samples (e.g., reconstructing a subset of the original data [13–16, 18, 19, 21] or normal dose with shortened bed time [11, 12, 17, 22–24]). To improve image quality, we will take advantage of the properties of CNNs, where the network could learn important features directly from the data, and possesses translation invariance [25], which allows the network to extract crucial features of the input image regardless of its position in the field of view. This has resulted in multiple medical imaging applications such as image identification [26], generation [27, 28], image segmentation [29], and MR-based attenuation correction [30, 31]. We will specifically build upon our previous study where we trained CNNs to generate an “improved” version of a noisy image created by removing counts from a standard-dose study. Given the intrinsic radiation emission from many modern PET detectors, it is possible that true injected ultra-low-dose images might present a more difficult task than the sampling approach. For this reason, we undertook this study in patients with separate imaging sessions of true injected ultra-low-dose and standard-dose 18F-florbetaben PET/MRI for two reasons: (1) to directly demonstrate the feasibility of true ultra-low-dose acquisition and (2) to further validate our prior undersampling method.
Materials and methods
Fifty total participants (32 for the pre-trained network presented in Chen et al. [13] and 18 as new data) were recruited for this study, approved by the local Institutional Review Board. Written informed consent for imaging was obtained from all participants or an authorized surrogate decision-maker.
PET/MR data acquisition: pre-trained network
For the network pre-trained with ultra-low-dose list-mode samples, 32 participants (20 female, age 67.7 ± 7.9 years) with MRI and PET data were simultaneously acquired on an integrated PET/MRI scanner with time-of-flight capabilities (SIGNA PET/MR, GE Healthcare, Waukesha, WI, USA). T1-weighted, T2-weighted, and T2 FLAIR morphological MR images were acquired as indicated in Chen et al. [13]. 334 ± 30 MBq of the amyloid radiotracer 18F-florbetaben (Life Molecular Imaging, Berlin, Germany) were injected intravenously and PET data was acquired 90–110 min after injection. The list-mode PET data was reconstructed for the standard-dose ground truth image and every 100th event was sampled and reconstructed (preserving the Poisson nature of the PET acquisition and taking the different randoms levels into account) for a sampled [20]1%-dose PET image. Time-of-flight ordered-subsets expectation-maximization, with two iterations and 28 subsets, and accounting for randoms, scatter, dead-time, and attenuation, was used for all PET image reconstructions. MR attenuation correction was performed using the vendor’s atlas-based method relying on 2-point Dixon imaging [32].
PET/MR data acquisition: ultra-low-dose protocol
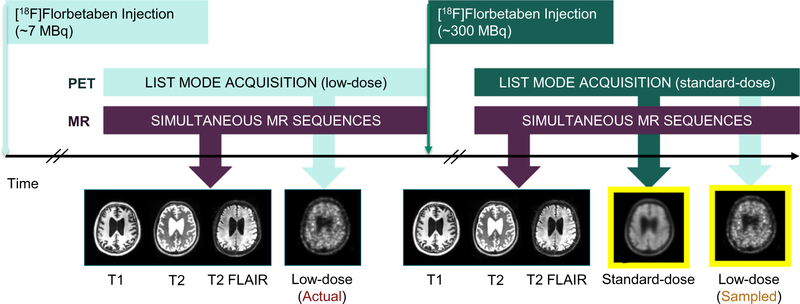
Eighteen (9 female, 72.1 ± 8.6 years) additional participants were scanned with the following injected ultra-low-dose protocol (participant breakdown by diagnosis in Table 1; sample protocol in Fig. 1). These participants were scanned in two PET/MRI sessions (8 on same day: low-dose session followed by full dose; 10 on separate days: 1- to 42-day interval, mean 17.8 days), with 6.64 ± 3.57 and 300 ± 14 MBq 18F-florbetaben injections respectively (2.2 ± 1.3% dose compared to the corresponding standard-dose sessions) representing an approximately 50-fold reduction in radiation dose. For all scans, the T1-, T2-, and T2 FLAIR–weighted MR images were acquired simultaneously with PET (90–110 min after injection; 83–98 min for one participant) on the same scanner. Identical MR acquisitions were performed across the two scanning sessions for all but 10 of the sequences, where the same sequence from the other scan would be used as a substitute. Similar to the previous reconstruction pipeline, time-of-flight ordered-subsets expectation-maximization, with two iterations and 28 subsets, and accounting for randoms, scatter, dead-time, and attenuation (vendor’s zero-TE [ZTE]-based method), was used for all PET image reconstructions. For validation of the sampled ultra-low-dose (SULD) data, the list-mode PET data was reconstructed for the standard-dose ground truth image and was also sampled by a factor determined from the dose reduction between the AULD data and the standard-dose acquisitions and then reconstructed to produce a SULD PET image at the same count level as the AULD image collected in the subject’s other session. Specific dose reduction factors can be found in the Supplementary Materials for all participants.
Table 1.
The participants recruited in this study and their clinical diagnoses
| Diagnosis | Number | ||
|---|---|---|---|
|
| |||
| Pre-trained network | Alzheimer’s disease | 6 | |
| Mild cognitive impairment | 2 | ||
| Dementia with Lewy bodies | 1 | ||
| Parkinson’s disease | 12 | ||
| Healthy control | 11 | ||
| Subtotal | 32 | ||
| Ultra-low-dose protocol | Alzheimer’s disease | 4 | |
| Mild cognitive impairment | 6 | ||
| Healthy control | 8 | ||
| Subtotal | 18 | ||
| Total | 50 | ||
Fig. 1.
Sample scanning protocol for those participants undergoing two injections in the same day. The same MRI sequences are acquired during each session. The standard-dose image is used as the standard space (yellow border) during co-registration
Image pre-processing
To account for any positional offset of the patient during different acquisitions within the same scanning session, all MR images (from both scanning sessions) were co-registered to the reference space of the standard-dose PET images (yellow-bordered images, Fig. 1) using the software FSL [33], with 6 degrees of freedom and correlation ratio as the cost function. All images were resliced to the dimensions of the acquired PET volumes, 89 slices (2.78-mm slice thickness) with 256 × 256 (1.17 × 1.17 mm2) voxels. All images from the low-dose session were also co-registered to the reference space to account for differences between scans. A head mask was made from the T1-weighted image through intensity thresholding and hole filling and applied to the PET and MR images; the normalized volumes were used as inputs to the CNN. The voxel intensities of each volume were normalized by their Frobenius norm.
CNN implementation, pre-trained network
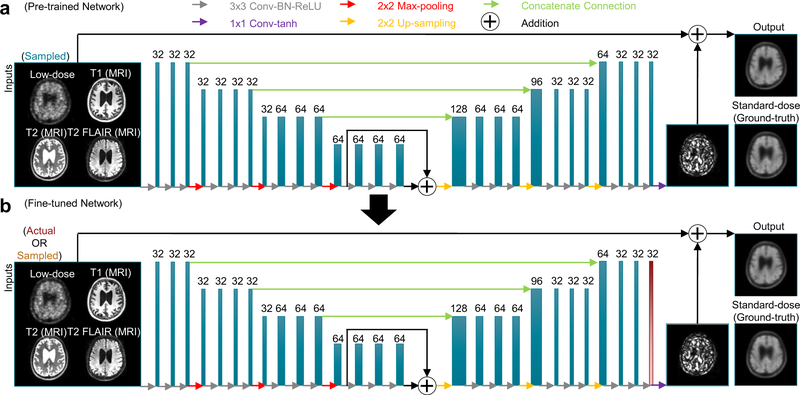
A U-Net CNN was trained in the work from Chen et al. [13], with 2848 inputs (32 datasets with 89 slices each) and an eightfold data augmentation. Briefly, the inputs of the network are the multi-contrast MR images (T1-, T2-, and T2 FLAIR–weighted) and, during the pre-training phase, the sampled 1%-dose PET image. The standard-dose PET image was used as the ground truth (Fig. 2).
Fig. 2.
A schematic of the U-Net used in this work. Initially, the ultra-low-dose PET and the 3 MR contrasts are used as inputs and pre-trained on the standard-dose PET image (a). The weights from all the layers are then saved as the starting point for the 2nd fine-tuning step (b). Only the last layer (red) is trained with data from the ultra-low-dose injection protocol. The arrows denote computational operations and the tensors are denoted by boxes with the number of channels above each box. Conv: convolution; BN: batch normalization; ReLU: rectified linear unit activation; tanh: hyperbolic tangent
The encoder portion is composed of layers which perform two-dimensional convolutions (using 3 × 3 filters), batch normalization, and rectified linear unit activation operations. 2-by-2 max pooling is used to reduce the dimensionality of the data. A residual connection was used in the central layers to connect its input and output. In the decoder portion, the data in the encoder layers are concatenated with those in the decoder layers. Linear interpolation is performed to restore the data to its original dimensions. 1 × 1 convolutions and hyperbolic tangent activation were used in the final layer to obtain the output, which is then added with the input low-dose image to obtain the enhanced PET image. The network was trained with an initial learning rate of 0.0002 and a batch size of four over 100 epochs. The L1 norm was selected as the loss function and adaptive moment estimation as the optimization method [34].
CNN implementation, ultra-low-dose protocol
In the participants with both actual low-dose and standard-dose imaging sessions, to adjust for the potential subtle differences between the datasets and to prevent overfitting due to the number of the additional participants, the weights from the last layer of this pre-trained model were fine-tuned separately with either the AULD or SULD dataset as inputs. A larger head mask was made by dilating the previous head mask by 8 voxels and applied to all PET and MR images. Ninefold cross-validation was used to use all data efficiently and to prevent training and testing on the same subjects (16 subjects for training, 2 subjects for testing per network trained). The network was fine-tuned with a learning rate of 0.0001 and a batch size of four over 100 epochs. For testing, images acquired during the actual injected ultralow-dose session were used as inputs.
Assessment of image quality
Using the software FreeSurfer [35, 36], a brain mask derived from the T1-weighted images of each subject was used for voxel-based analyses. The mean cerebral uptake values were calculated for all image types (standard-dose images were multiplied by the low-dose percentage) and correlation coefficients were computed. The mean cerebral uptake values for the two ultra-low-dose images (AULD vs. SULD) were also correlated to test the validity of the undersampling method. For each axial slice, the image quality of the deep learning-enhanced PET images and the original low-dose PET images within the brain mask were compared to the standard-dose image using several metrics: peak signal-to-noise ratio (PSNR),structural similarity (SSIM)[37],and root mean square error (RMSE). The metrics for each subject were obtained by a weighted average (by voxel number) of the slices. Paired t tests were conducted (95% confidence interval, Bonferroni-corrected for three comparisons) to compare the three metrics between the methods (actual injected vs. sampled) as well as between the ultra-low-dose and their CNN-enhanced counterparts.
Clinical readings
The enhanced PET images, low-dose PET images, and the standard-dose PET image of each subject were anonymized and presented in random order to four clinicians (G.D., M.E.I.K., G.Z., M.Z.), all of whom had been certified to read amyloid PET imaging. The amyloid uptake status (positive, negative, uninterpretable) of each image was determined. The standard-dose image readings were treated as the ground truth; the accuracy, sensitivity, and specificity were calculated for the readings of the low-dose (two versions) and the enhanced images (two versions).
For each PET image, the clinicians assigned an image quality score on a five-point scale: 1 = uninterpretable, 2 = poor, 3 = adequate, 4 = good, 5 = excellent. The scores were dichotomized into 1–2/3–5, and the percentage of images deemed adequate or better was calculated for each method. The 95% confidence interval for the difference in the proportions of high scores was constructed and compared to a predetermined non-inferiority threshold of −15%.
The agreement of the four readers was assessed using Gwet’s agreement coefficient 1 (AC1) [38]. For intra-reader agreement, the standard-dose PET images were clinically read by the same clinicians a second time for amyloid status in a separate reading session (at least 6 weeks apart).
Region-based analyses
FreeSurfer-derived cortical parcellations and cerebral segmentations based on the Desikan-Killiany Atlas [39] were grouped into 10 larger regions for analysis (groupings in the Supplementary Materials). The whole cerebellum was used as a reference region for calculating SUVR values of all PET images. The mean SUVR and the voxel-wise standard deviation (SD) were calculated for the large regions in each subject. To assess tracer uptake agreement between images, the coefficient of variation (CV=SD/mean) was calculated and compared between methods with paired t tests.
Results
Qualitative assessment of image quality
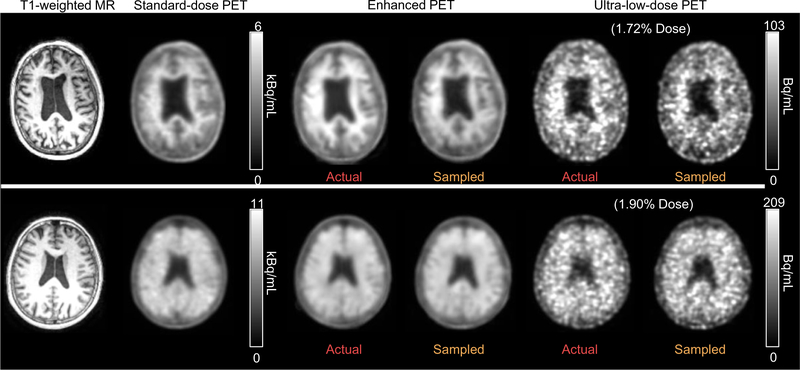
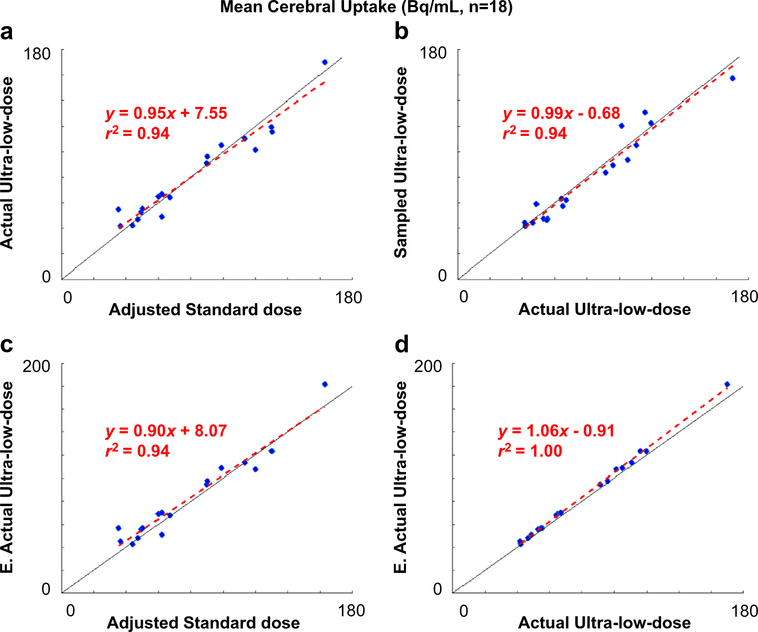
Qualitatively, the enhanced images show marked improvement in image quality compared with the low-dose images and visually resemble the ground truth image (Fig. 3). The mean radiotracer uptake within the brain correlated strongly across image types (standard-dose vs. AULD, AULD vs. enhanced AULD, standard-dose vs. enhanced AULD, AULD vs. SULD) and the line of best fit is close to the line of identity (Fig. 4).
Fig. 3.
Representative amyloid PET images (top: amyloid negative, bottom: amyloid positive). The enhanced PET images show significantly reduced noise compared to both the actual injected and sampled ultra-low-dose PET images, and bear resemblance in uptake pattern to the standard-dose images
Fig. 4.
Correlation of mean cerebral uptake between image types. The uptake values of the standard-dose images were multiplied by each dataset’sdose reduction factor (ratio between the injected ultra-low-dose and the injected standard-dose). E.: enhanced
Quantitative assessment of image quality
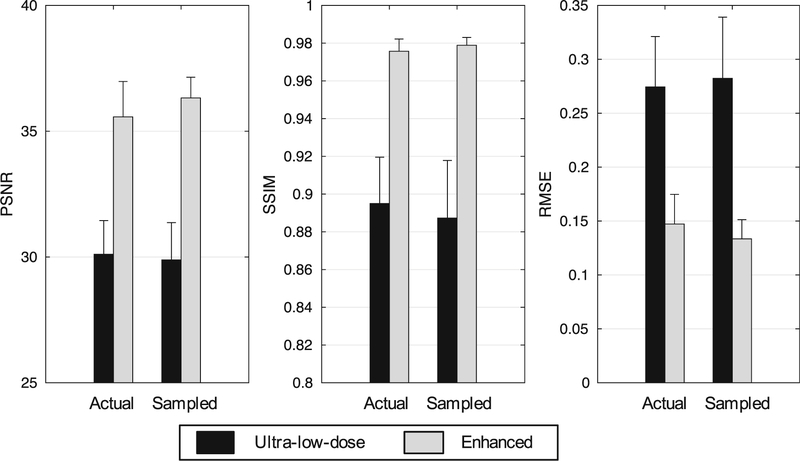
The metrics showed that the enhanced images vastly outperformed their ultra-low-dose counterparts (p < 0.001 for all comparisons). Between the AULD and SULD images, the metrics did not show significant differences (p > 0.05/3for all comparisons), although the enhanced SULD images showed higher PSNR, SSIM, and lower RMSE than the enhanced AULD images (Fig. 5).
Fig. 5.
Image quality metrics comparing the ultra-low-dose PET images to their counterparts enhanced from the network. For the three metrics, comparison is to the ground truth standard-dose PET images. PSNR: peak signal-to-noise ratio; SSIM: structural similarity; RMSE: root mean square error
Clinical readings
Gwet’s AC1 was used to evaluate inter-rater agreement. The readers had generally high agreement for each method (Table 2), with the standard-dose images having the highest agreement followed by the enhanced AULD images.
Table 2.
Agreement between the readers and assigned quality score across image types.
| Metric | Standard-dose | AULD | SULD | Enhanced AULD | Enhanced SULD |
|---|---|---|---|---|---|
|
| |||||
| Gwet’s AC1, uptake status (95% CI) | 0.89 (0.73, 1.00) | 0.75 (0.10, 1.00) | 0.85 (0.34, 1.00) | 0.81 (0.60, 1.00) | 0.71 (0.42, 0.99) |
| Gwet’s AC1, quality score (95% CI) | 0.57 (0.39, 0.74) | 0.27 (−0.02, 0.55) | 0.27 (−0.03, 0.57) | 0.70 (0.58, 0.82) | 0.69 (0.51, 0.86) |
| Quality score (mean ± SD) | 4.13 ± 0.64 | 1.40 ± 0.49 | 1.40 ± 0.49 | 3.33 ± 0.63 | 3.38 ± 0.64 |
AC, agreement coefficient; AULD, actual injected ultra-low-dose; CI, confidence interval; SD, standard deviation; SULD, sampled ultra-low-dose
Of the 72 total reads of the standard-dose ground truth amyloid images, 34 (47%) were amyloid positive. As expected, the AULD and SULD PET images were inadequate with a majority of them uninterpretable (43/72 reads, 60%).
In terms of accuracy, sensitivity, and specificity of the clinical assessments between the enhanced images and the standard-dose images, readings of the enhanced images had high values in general, with the enhanced AULD images slightly outperforming the enhanced SULD images (Table 3; confusion matrices in Table 4). The accuracy of the enhanced AULD images was close to the reader reproducibility (98.6%, 95% confidence interval: 92.5%, 100.0%).
Table 3.
The accuracy, sensitivity, and specificity of the uptake status readings between the enhanced images and the standard-dose images.
| Metric (95% CI) | Enhanced from AULD | Enhanced from SULD |
|---|---|---|
|
| ||
| Accuracy | 97.2% (90.3–99.7%) | 91.7% (82.7–96.9%) |
| Sensitivity | 100% (89.7–100%) | 88.2% (72.5–96.7%) |
| Specificity | 94.7% (82.3–99.4%) | 94.7% (82.3–99.4%) |
AULD, injected ultra-low-dose; CI, confidence interval; SULD, sampled ultra-low-dose
Table 4.
Confusion matrices between the standard-dose and the images enhanced from actual injected ultra-low-dose (AULD) and sampled ultra-low-dose (SULD) data
| Enhanced from AULD data |
||||
|---|---|---|---|---|
| Negative | Positive | Total | ||
|
| ||||
| Standard-dose | Negative | 36 | 2 | 38 |
| Positive | 0 | 34 | 34 | |
| Total | 36 | 36 | 72 | |
| Enhanced from SULD data | ||||
| Negative | Positive | Total | ||
| Standard-dose | Negative | 36 | 2 | 38 |
| Positive | 4 | 30 | 34 | |
| Total | 40 | 32 | 72 | |
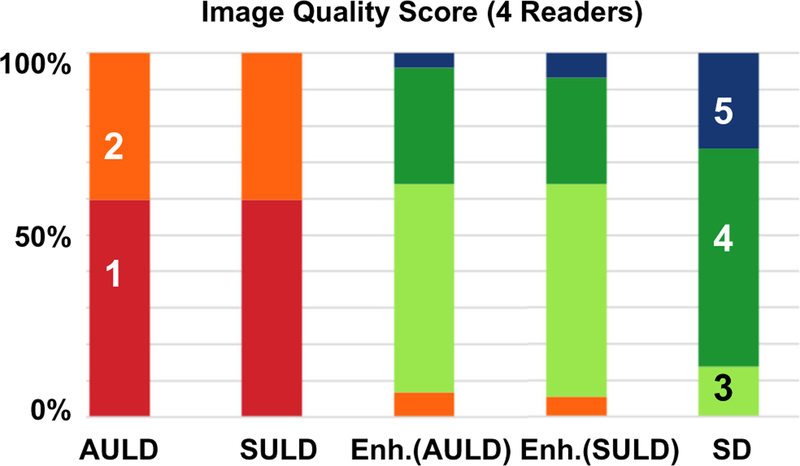
Compared to the ratings, the readers had less agreement with each other in assigning the quality scores (Table 2). The readers had the most agreement in the enhanced AULD images followed by the enhanced SULD images. The image quality scores assigned by each reader to each of the reconstructed PET volumes are shown in Fig. 6, along with mean scores for each group. All of the standard-dose images were scored as 3 or above, while both sets of ultra-low-dose images scored 2 or below. The enhanced images had a slightly lower proportion of high-scoring images than the standard-dose images, but still fell within the non-inferiority threshold of - 15% (confidence intervals between enhanced AULD and standard-dose: [− 13%, − 1%]; between enhanced SULD and standard-dose: [− 11%, 0%]). Detailed confusion matrices of the readers are provided in the Supplementary Materials.
Fig. 6.
Clinical image quality scores as assigned by the four readers. AULD: actual injected ultra-low-dose; Enh.: Enhanced; SD: standard-dose; SULD: sampled ultra-low-dose
Region-based analyses
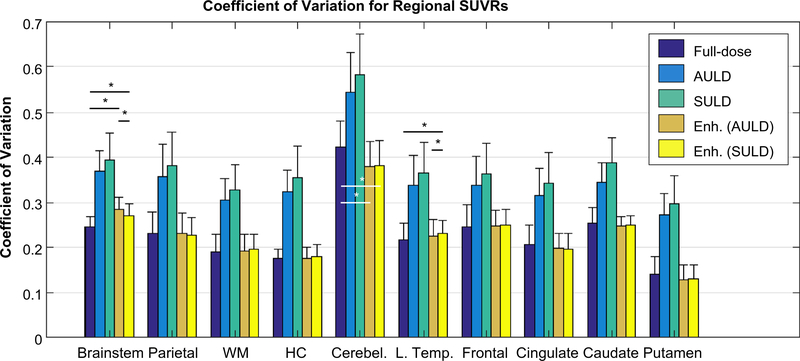
The CV of the regional SUVRs was higher in the AULD and SULD images than the standard-dose or the enhanced images. Performing paired t tests between the standard-dose and enhanced image types in each region (three tests per region) showed that a majority of comparisons were not significantly different (p > 0.05/3) (Fig. 7).
Fig. 7.
Coefficient of variation (mean/standard deviation) of regional standard uptake value ratios (SUVRs) compared between image types across all subjects. Pair-wise paired t tests were only performed between the standard-dose and enhanced images (three image types with similar coefficients of variation) at the p = 0.05/3 (Bonferroni corrected) level, with asterisks denoting significance. AULD: actual injected ultra-low-dose; Enh.: enhanced; HC: hippocampus; L. Temp.: lateral temporal; SULD: sampled ultra-low-dose; WM: white matter
Discussion
In this study, we demonstrated the feasibility of ultra-low-dose deep learning–enhanced amyloid PET/MRI imaging, using a small percentage (~ 2.2%) of the traditional injected dose. Dramatically lowering injected dose will enable more frequent scanning under current radiation safety thresholds and has a wide range of potential applications, such as for use in studies requiring multiple follow-ups (e.g., clinical trials of amyloid-clearing drugs), multi-tracer studies, and longitudinal studies of younger populations (e.g., dementia in Down syndrome patients or those with familial AD) to evaluate their disease progression. In addition, we validated the list-mode undersampling method used to approximate ultra-low-dose studies; this supports the idea that large-scale retrospective ultra-low-dose PET studies can be performed.
Metrics-wise, the enhanced images vastly outperformed their ultra-low-dose counterparts; comparing the enhanced AULD vs. enhanced SULD images showed slightly (but not significant) improved metrics for the SULD images. This may be due to the use of the pre-trained network, which was trained with sampled list-mode reconstructions. The SULD images were also in the same space as the standard-dose images; it is possible that the co-registration/interpolation step necessary for the AULD inputs might affect CNN performance. On the other hand, the CV of the regional SUVRs provides another metric to show the improved image quality of the enhanced images compared to their ultra-low-dose counterparts.
Qualitatively, the enhanced images are smoother in appearance than the standard-dose images; this difference resulted in the readers preferring the standard-dose images over the enhanced images. However, the enhanced images still produced a similar proportion of “high quality” images compared to the standard-dose. On the other hand, both sets of ultra-low-dose images were very noisy and had the same proportion of uninterpretable images.
The correlations between AULD and SULD images showed that the intrinsic radiation of the PET detector ring did not affect the AULD data, though this might become a factor as we extend our research into lower dose reduction regimes. Between the SULD and AULD images, the high correlation coefficient (almost 1) validated the use of undersampling the list-mode data and strongly suggests that results obtained from such data would translate to actual injected low-dose studies.
Similar studies on various tracers have also shown that readers can also read images reconstructed with reduced counts, albeit with less certainty [23] and the equivalent dose reduction factor is not as extreme (25%) [24]as that used in this work (~ 2%). With deep learning enhancement, the images achieve similar diagnostic value in accuracy and are much preferred by the readers compared to the ultra-low-dose images. This shows the potential of employing scan protocols that use massively lower injected dose than current convention.
There are several limitations to our study, one of them being the registration (and interpolation) of the AULD and MRI images discussed previously. Further investigation on the effect of simultaneity as well as training PET-only networks (e.g., using the network structure in [15]; preliminary results shown in the Supplementary Materials) will elucidate the extent of this effect on the low-dose image enhancement. The rounding error that occurs when converting the DRF into an undersampling factor will also contribute to inaccuracies during the correlation study. The parameters used for reconstructing the AULD and SULD datasets are also a potential source of bias in this study. Ideally, the PET images at different count levels should be individually optimized during reconstruction. However, we did not opt to perform this optimization due to the varying methods implemented in literature [23, 24] as well as the large parameter search space. The bias due to image quality between the two ultra-low-dose datasets should also be minimized due to the two image types having the same reconstruction parameters. The difference in attenuation correction methods as well as dose reduction levels between the two datasets used (for the pre-trained network vs. the ultra-low-dose protocol) is also a potential source of bias, although we believe this issue has been resolved through fine-tuning the network with datasets coming from the ultra-low-dose protocol. Finally, other potential sources of bias include the time between the two scanning sessions, participant population, and the difference in brain physiology and function at the time of scanning due to these differences, though studies have shown that the change in physiology will be minimal given our time frame [40]. Due to the challenges in scheduling and acquiring multiple PET sessions in these patients at risk of dementia, the size of the test set is also necessarily small. This may result in a non-representative sample and larger studies could be considered (particularly using sampled list-mode data now that they have been validated).
Conclusion
This work has shown that high-quality amyloid PET images can be generated using deep learning methods starting from simultaneously acquired MR images and actual ultra-low-dose PET injections. Moreover, we have validated the method of undersampling list-mode data for ultra-low-dose imaging. The enhanced images demonstrated diagnostic value with high accuracy, sensitivity, and specificity, as well as quantitative value through SUVR analyses. The results of this work can potentially contribute to the implementation of lower-count (shorter time and/or reduced dose) amyloid PET imaging and its increased utility.
Supplementary Material
Acknowledgments
The authors would like to thank Tie Liang, EdD for the statistical analyses and Jiahong Ouyang, MS, for the discussions on network structure and training.
Funding This project was made possible by the NIH grants P41-EB015891 and P50-AG047366 (Stanford Alzheimer’s Disease Research Center), GE Healthcare, the Michael J. Fox Foundation for Parkinson’s Disease Research, the Foundation of the ASNR, and Life Molecular Imaging.
Footnotes
Conflict of interest Outside submitted work: GZ-Subtle Medical Inc., co-founder and equity relationship. No other potential conflicts of interest relevant to this article exist.
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00259-020-05151-9.
Code availability Custom code was used for this project.
Data availability Data was collected at the authors’ institutions and is available when requested for review.
Compliance with ethical standards
Ethics approval All procedures involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent to participate Written informed consent for imaging was obtained from all participants or an authorized surrogate decision-maker.
Consent for publication N/A
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alzheimer’s A. 2019 Alzheimer’s disease facts and figures. Alzheimers Dement. 2019;15:321–87. [Google Scholar]
- 2.Berti V, Pupi A, Mosconi L. PET/CT in diagnosis of dementia. Arm N Y Acad Sci. 2011;1228:81–92. 10.1111/j.17496632.2011.06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishii K. PET approaches for diagnosis of dementia. AJNR Am J Neuroradiol. 2014;35:2030–8. 10.3174/ajnr.A3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Judenhofer MS, Wehrl HF, Newport DF, Catana C, Siegel SB, Becker M, et al. Simultaneous PET-MRI: a new approach for functional and morphological imaging. Nat Med. 2008;14:459–65. 10.1038/nml700. [DOI] [PubMed] [Google Scholar]
- 5.Phelps ME. PET: physics, instrumentation, and scanners. New York: Springer; 2006. [Google Scholar]
- 6.Wemick MN, Aarsvold JN. Emission tomography: the fundamentals of PET and SPECT. Amsterdam. Boston: Elsevier Academic Press; 2004. [Google Scholar]
- 7.Sperling RA, Momrino EC, Schultz AP, Betensky RA, Papp KV, Amariglio RE, et al. The impact of amyloid-beta and tau on prospective cognitive decline in older individuals. Ann Neurol. 2019;85:181–93. 10.1002/ana.25395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sevigny J, Chiao P, Bussiere T, Weinreb PFI, Williams L, Maier M, et al. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature. 2016;537:50–6. 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 9.Catana C, Drzezga A, Fleiss WD, Rosen BR. PET/MRI for neurologic applications. J Nucl Med. 2012;53:1916–25. 10.2967/jnumed.112.105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bland J, Mehranian A, Belzunce MA, Ellis S, McGinnity CJ, Hammers A, et al. MR-guided kernel EM reconstruction for reduced dose PET imaging. IEEE Trans Radiat Plasma Med Sci. 2018;2?235–43. 10.1109/TRPMS.20l7.2771490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang J, Gao Y, Shi F, Lalush DS, Lin W, Shen D. Prediction of standard-dose brain PET image by using MRI and low-dose brain [18F]FDG PET images. Med Phys. 2015;42:5301–9. 10.1118/1.4928400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le A, Pei Z, Adeli E, Yan W, Guangkai M, Feng S, et al. Multilevel canonical correlation analysis for standard-dose PET image estimation. IEEE Trans Image Process. 2016;25:3303–15. 10.1109/TIP.2016.2567072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen KT, Gong E, de Carvalho Macruz FB, Xu J, Bounds A, Khalighi M, et al. Ultra-low-dose (18)F-Florbetaben amyloid PET imaging using deep learning with multi-contrast MRI inputs. Radiology. 2019;290:649–56. 10.1148/radiol.2018180940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K, Wu D, Gong K, Dutta J, Kim JH, Son YD, et al. Penalized PET reconstruction using deep learning prior and local linear fitting. IEEE Transactions on Medical Imaging. 2018;37:1478–87. 10.1109/TMI.2018.2832613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouyang J, Chen KT, Gong E, Pauly J, Zaharchuk G. Ultra-low-dose PET reconstruction using generative adversarial network with feature matching and task-specific perceptual loss. Med Phys. 2019;46:3555–64. 10.1002/mp.13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Zhou L, Yu B, Wang L, Zu C, Lalush DS, et al. 3D auto-context-based locality adaptive multi-modality GANs for PET synthesis. IEEE Trans Med Imaging. 2019;38:1328–39. 10.1109/TMI.2018.2884053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang L, Qiao Y, Nie D, An L, Wang Q, Shen D. Deep autocontext convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI. Neurocomputing. 2017;267:406–16. 10.1016/j.neucom.2017.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Gong E, Ouyang J, Pauly J, Zaharchuk G. Ultra-low-dose 18F-FDG brain PET/MR denoising using deep learning and multi-contrast information. Houston, TX: SPIE Medical Imaging; 2020. [Google Scholar]
- 19.Sanaat A, Arabi H, Mainta I, Garibotto V, Zaidi H. Projection-space implementation of deep learning-guided low-dose brain PET imaging improves performance over implementation in image-space. J Nucl Med. 2020. 10.2967/jnumed.119.239327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefferkoetter J, Nai YH, Reilhac A, Townsend DW, Eriksson L, Conti M. Low dose positron emission tomography emulation from decimated high statistics: a clinical validation study. Med Phys. 2019;46:2638–45. 10.1002/mp.13517. [DOI] [PubMed] [Google Scholar]
- 21.Herholz K, Evans R, Anton-Rodriguez J, Hinz R, Matthews JC. The effect of 18F-florbetapir dose reduction on region-based classification of cortical amyloid deposition. Eur J Nucl Med Mol Imaging. 2014;41:2144–9. 10.1007/s00259-014-2842-3. [DOI] [PubMed] [Google Scholar]
- 22.Chen KT, Schürer M, Ouyang J, Koran MEI, Davidzon G, Mormino E, et al. Generalization of deep learning models for ultra-low-count amyloid PET/MRI using transfer learning. Eur J Nucl Med Mol Imaging. 2020. 10.1007/s00259-020-04897-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiller F, Frings L, Thurow J, Meyer PT, Mix M. Limits for reduction of acquisition time and administered activity in (18)F-FDG PET studies of Alzheimer dementia and frontotemporal dementia. J Nucl Med. 2019;60:1764–70. 10.2967/jnumed.119.227132. [DOI] [PubMed] [Google Scholar]
- 24.Tiepolt S, Barthel H, Butzke D, Hesse S, Patt M, Gertz HJ, et al. Influence of scan duration on the accuracy of beta-amyloid PET with florbetaben in patients with Alzheimer’s disease and healthy volunteers. Eur J Nucl Med Mol Imaging. 2013;40:238–44. 10.1007/s00259-012-2268-8. [DOI] [PubMed] [Google Scholar]
- 25.Gens R, Domingos P. Deep Symmetry Networks. Advances in neural information processing systems. 2014.
- 26.He KM, Zhang XY, Ren SQ, Sun J. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. Ieee I Conf Comp Vis. 2015:1026–34. 10.1109/Iccv.2015.123. [DOI] [Google Scholar]
- 27.Chen H Low-dose CT with a residual encoder-decoder convolutional neural network (RED-CNN). arXiv: arXiv; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Gong E, Fan AP, Goubran M, Khalighi MM, Zaharchuk G. Predicting (15)O-Water PET cerebral blood flow maps from multi-contrast MRI using a deep convolutional neural network with evaluation of training cohort bias. J Cereb Blood Flow Metab. 2019: 271678X19888123. doi: 10.1177/0271678X19888123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–48. 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging-based attenuation correction for PET/MR imaging. Radiology. 2018;286:676–84. 10.1148/radiol.2017170700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torrado-Carvajal A, Vera-Olmos J, Izquierdo-Garcia D, Catalano OA, Morales MA, Margolin J, et al. Dixon-VIBE deep learning (DIVIDE) pseudo-CT synthesis for pelvis PET/MR attenuation correction. J Nucl Med. 2018. 10.2967/jnumed.118.209288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iagaru A, Mittra E, Minamimoto R, Jamali M, Levin C, Quon A, et al. Simultaneous whole-body time-of-flight 18F-FDG PET/MRI: a pilot study comparing SUVmax with PET/CT and assessment of MR image quality. Clin Nucl Med. 2015;40:1–8. 10.1097/RLU.0000000000000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW. Smith SM. Fsl. Neuroimage. 2012;62:782–90. 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Kingma DP. Ba J. A Method for Stochastic Optimization. arXiv: Adam; 2014. [Google Scholar]
- 35.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 36.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9: 179–94. 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13:600–12. [DOI] [PubMed] [Google Scholar]
- 38.Gwet KL. Computing inter-rater reliability and its variance in the presence of high agreement. Br J Math Stat Psychol. 2008;61:29–48. 10.1348/000711006X126600. [DOI] [PubMed] [Google Scholar]
- 39.Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–80. 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 40.Hatashita S, Wakebe D, Kikuchi Y, Ichijo A. Longitudinal assessment of amyloid-ß deposition by [18F]-Flutemetamol PET imaging compared with [11C]-PIB across the spectrum of Alzheimer’s disease. Front Aging Neurosci. 2019;11. 10.3389/fnagi.2019.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.