Abstract
Conservation of large carnivores requires preservation of extensive core habitats and linkages among them. The goal of this study was to identify core habitats and corridors by predicting habitat suitability (an ensemble approach), and calculating resistant kernel and factorial least-cost path modeling for a relatively unknown carnivore, the striped hyaena in Khuzestan area in southwestern Iran. We used the procedure of spatial randomization test to evaluate the coincidence of striped hyaena road crossing with the predicted corridors. The results revealed that elevation, distance to conservation areas, categorical climate and grasslands density were the most influential variables for predicting the occurrence of the striped hyaena in the study area. In the estimated dispersal distance of 70 km, four core habitats were identified. The largest core habitat was located in the northeast of the study area with the highest connectivity contribution. Only about 12% and 1.5% of core habitats and corridors were protected by conservation areas, respectively. Predicted corridors, crossed by roads represented a high risk for striped hyaenas. Adaptive management plan throughout the landscape (conservation of core habitats and corridors, and reducing species mortality on the roads) must be considered by wildlife managers in Iran.
Subject terms: Ecology, Zoology
Introduction
Human activities have threatened large carnivores through habitat loss, fragmentation and isolation at multiple scales1–3. With increasing loss and fragmentation of habitats, there is a crucial need to identify the most important areas for conservation actions4. The conservation areas (CAs) network should protect both core habitats and corridors5. However, several studies report that the existing CAs are primarily small and insufficient to support large carnivores with extensive home range and low population density6–9.
Large carnivores are considered keystone species because they are apex predators and they are umbrella species. Thus conserving carnivores helps regulate prey species and leads to sympatric biota’s conservation10. Large carnivores are also indicator species because of their sensitivity to habitat fragmentation11. Therefore, large carnivores are often selected by researchers as the surrogate species12. However, it is often difficult to identify core habitats and corridors of large carnivores due to their mainly cryptic and nocturnal nature13,14. Species distribution models (SDMs)15 have come to aid researchers in predicting suitable habitats of large carnivores. In addition, SDMs were applied as input data to predict movement corridors used for dispersal and gene flow among core habitats in order to direct management of the species16–18. Identified corridors can be used to direct land managers, for example, managers can prioritize improving wildlife road crossings in areas when roads cross corridors19–21.
Roads have adverse effects on wildlife populations, including large carnivores, and particularly threatened species22–25. Roads fragment continuous habitats and facilitate human access to pristine natural areas26. Furthermore, anthropogenic caused mortalities, road collisions being one, are the main concerns for the conservation of threatened large carnivores24. Several studies have been done in Iran that tested relationship between road collisions of large carnivores and ecological corridors9,19,23; but none focused on the striped hyaena (Hyaena hyaena Linnaeus, 1758).
The striped hyaena occurs in Asia from the Indian subcontinent to the Levant (including 20 countries) and most parts of Africa except the southern part (including 18 countries)27,28. According to the IUCN Red List, the striped hyaena has been categorized as a near threatened species (NT), because of persecution (mainly poisoning the carrions) and decreasing domestic and natural carrions29. The reason for this decrease is the reduction in other sympatric large carnivores’ populations and their prey29,30. The striped hyaena is a classic omnivorous scavenger30,31, which scavenges a variety of foods, including vertebrates, insects and other invertebrates, dried bones, fruits, human organic waste, etc.32,33. Striped hyaenas are one of the least studied large carnivores in Iran and there is limited data available on their habitat needs and spatial distribution. In Iran, the striped hyaena has a widespread distribution; however, its population has decreased severely27. The main causes are habitat loss and anthropogenic activities such as conversion of the natural grasslands to agricultural lands, poaching, poisoning the carrions, using organs for medicine and superstitious beliefs, and road collisions27,34,35. For this regard, identifying striped hyaena habitat suitability, core habitats and connectivity among them are prerequisite steps to delineate management strategies aiming at human-striped hyaena co-existence. This species can be considered as a surrogate species and identifying core habitats and connectivity network can help in locating new CAs and protecting other co-existence species.
This study was carried out in order to (1) assess the habitat suitability of the striped hyaena to predict the core habitats and corridors in Khuzestan area, southwestern Iran, (2) compare identified core habitats and corridors with existing CAs, and (3) overlay the road collisions of the striped hyaena with the predicted corridors.
Materials and methods
Study area
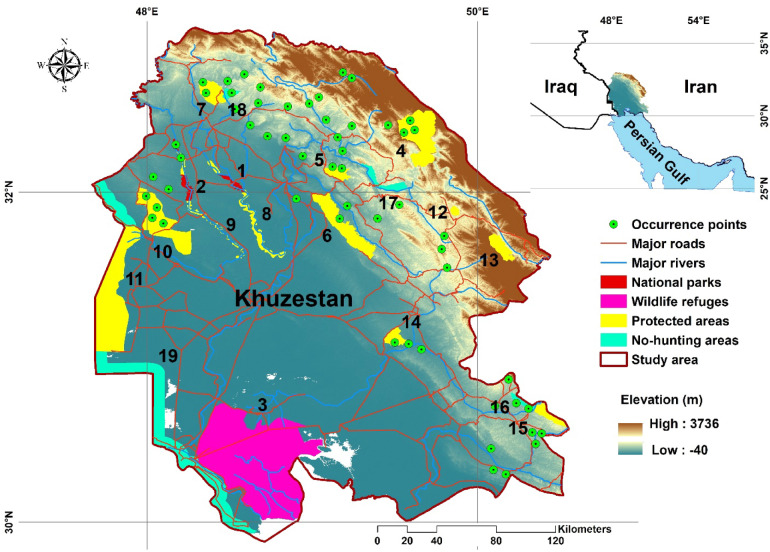
Khuzestan area is a province located the southwest of Iran (area: 64,057 km2) (Fig. 1). Northeast of the study area includes mountainous areas with cold winters (mean 6 °C) and mild summers (mean 25 °C) with the dominant plant species of Hordeum marinum Huds., Onosma rosellatum Lehm. and Ducrosia anethifolia (DC.) Boiss. Other parts of the study area include vast arid plains with mild winters (mean 17 °C) and hot summers (mean 37 °C)36 with dominant plant species of Onopordum heteracanthum C.A.Mey., Chrozophora hierosolymitana Spreng. and Capparis spinosa L. CAs covers about 13% of the study area; includes two national parks (NPs), one wildlife refuge (WR), 12 protected areas (PAs) and four no-hunting areas (NHAs) (Supplementary Information: Table S1, Fig. 1). NPs, WRs and PAs have the highest conservation priorities in Iran, respectively and NHAs were established for poaching control and have the lowest conservation priorities37. NPs, WRs, PAs and NHAs are near to the II, III, IV and IV-VI of the IUCN categories, respectively37. The density of major roads is 70.8 m/km2 in the study area. The study area includes several long rivers (e.g., Karoon, Karkherh and Dez) with a density of 40 m/km2 (Fig. 1). Brown bear (Ursus arctos), Persian leopard (Panthera pardus saxicolor), striped hyaena, grey wolf (Canis lupus), golden jackal (Canis aureus), caracal (Caracal caracal), jungle cat (Felis chaus), wild cat (Felis lybica) and honey badger (Mellivora capensis) are the main carnivores in the study area38,39.
Figure 1.
Study area including Khuzestan area in southwest of Iran, occurrence points and conservation areas (names of conservation areas are available in Table S1). ArcGIS software version 10.1 (https://www.esri.com/en-us/arcgis/products/arcgis-pro/resources) was used to generate the figure.
Occurrence points̕ collection and environmental variables
Occurrence points of the striped hyaena in the study area were collected by Khuzestan provincial office of the Department of Environment (DoE) guards and experts, including the third author, during 2015–2020. A number of 58 occurrence points were obtained for the striped hyaena in the study area. Spatial-autocorrelation was reduced by using the radius of 4 km around each occurrence point according to mean maximum distance moved (MMDM) by the striped hyaena in arid areas of India40 using the Spatially Rarify Occurrence Data tool in the SDMtoolbox41. Only one occurrence point was excluded and 57 occurrence points were used for habitat modeling of the striped hyaena in the study area (Table S2).
All related environmental variables i.e. topographic, climatic, land cover, safety and protection, water resources and human disturbance variables, were considered for habitat modeling of the striped hyaena in the study area (Table S3). Digital Elevation Model (DEM) was download from http://srtm.csi.cgiar.org as the elevation variable with a resolution of 250 m. This data was derived from the 90 m Shuttle Radar Topography Mission (SRTM, http://earthexplorer.usgs.gov). DEM was used to calculate the slope (using Surface Tool) and the topographic roughness index (standard deviation of elevation value of DEM᾽s cells within the radius of 4 km) using ArcGIS software version 10.142 (https://www.esri.com/en-us/arcgis/products/arcgis-pro/resources). A categorical climatic layer created based on De Martonne's classification with eight classes (from very humid to very arid) was used for habitat modeling of the striped hyaena in the study area.
Forests, grasslands and agricultural lands cover-types were derived from the land-cover map of Iran. A circle-moving window with a 4 km radius was used to create density maps of these three cover-types. Normalized Difference Vegetation Index (NDVI) was created by the 16-day composite MODIS data (MODIS MYD 13Q1 V6 map at 250 m cell size; http://earthexplorer.usgs.gov) according to the mean values of the year 2020. Because CAs protect animals from hunting or other human disturbances, distance to CAs was considered. We considered distance to rivers given the importance of water resources for carnivores13 and because the striped hyaena is found in the areas, where water is available within 10 kilometers40,43. Distance to roads was assessed as a predictor. Furthermore, another human disturbance variable, distance to villages was considered because villages attract striped hyaenas to scavenge dead domestic and organic wastes30.
To reduction and choose the optimal variables for habitat modeling of the striped hyaena, the MaxentVariableSelection package44 in R version 3.6.045 (https://www.r-project.org/) was employed by setting a contribution threshold of 1%, regularization multiplier of 1 to 5 with increments of 0.5 and inter-correlation of 0.7. Eight variables with the highest area under the curve (AUC) of receiver operating characteristic (ROC) and the lowest Akaike information criterion (AIC) were chosen by package (Table 1). Variance Inflation Factor (VIF) of selected variables in step 1 was checked using the usdm package46 in R to exclude variables with VIF > 3 (threshold suggested by Zuur et al.47). None of the selected variables was excluded due to low VIFs (Table 1).
Table 1.
Environmental variables using for habitat modeling of the striped hyaena in the study area.
| Variables category | Variables | Selected by MaxentVariableSelection | VIF |
|---|---|---|---|
| Topography | Elevation | Selected | 1.65 |
| Slope | – | – | |
| Roughness | – | – | |
| Climate | Categorical climate | Selected | 1.13 |
| Land-cover | Forests density | – | – |
| Grasslands density | – | 1.29 | |
| Agricultural lands density | Selected | 1.69 | |
| NDVI | Selected | 1.25 | |
| Prey availability | Distance to CAs | Selected | 1.37 |
| Water resources | Distance to rivers | Selected | 1.2 |
| Human disturbance | Distance to roads | Selected | 1.5 |
| Distance to villages | Selected | 1.18 |
Habitat modeling
Habitat suitability prediction of the striped hyaena was carried out using an R-package biomod248 as an ensemble modeling approach. The predictive accuracy of the habitat suitability model improves by combining different suitability models49,50. Four regression-based models, five machine-learning models and one profile model were implemented for the primary habitat modeling in Biomod2 (Table 2), and four models with AUC > 0.9 and True Statistic Skill (TSS) > 0.75 thresholds were chosen as the best fit51. According to method used by Kaboodvandpour et al.4, six hundred pseudo-absence points were randomly created across the study area (separated by > 4 km from each other) and outside of the 4 km radius circle around each occurrence point. Totally, 658 points (600 pseudo-absence points + 58 occurrence points) and eight environmental variables were used for habitat modeling of the striped hyaena in the study area by using four models of GLM, MaxEnt, GBM and RF (Table 2). Then, map of ensemble habitat suitability was created in biomod2 by weighted-average of models values48. The mean of variables̕ contribution of related models was calculated in Biomod2. In addition, response curves of occurrence points to the variables for the two most accurate models were illustrated in the study area. According to the method of Wan et al.52, the ensemble suitability map was converted into a resistance map. The linear method in rescale by function tool in ArcGIS software and the negative exponential function (R = 1000(−1 × Habitat Suitability))53 were used to create the resistance map in the range of 1 (lowest resistance) to 10 (highest resistance)52.
Table 2.
Different prediction models used for habitat modeling of the striped hyaena in the study area.
| Prediction model category | Prediction model | AUC | TSS |
|---|---|---|---|
| Regression-based models | Generalized linear model (GLM)* | 0.914 | 0.794 |
| Generalized additive model (GAM) | 0.76 | 0.678 | |
| Multivariate adaptive regression splines (MARS) | 0.845 | 0.69 | |
| Flexible discriminant analysis (FDA) | 0.87 | 0.643 | |
| Machine-learning models | Maximum entropy (MaxEnt)* | 0.93 | 0.816 |
| Generalized boosting model (GBM)* | 0.943 | 0.822 | |
| Random forest (RF)* | 0.921 | 0.835 | |
| Classification tree analysis (CTA) | 0.755 | 0.569 | |
| Artificial neural network (ANN) | 0.839 | 0.572 | |
| Profile model | Surface range envelop (SRE) | 0.764 | 0.548 |
*Selected for final habitat modeling.
Core habitats and corridor modeling
Corridor modeling was carried out by using Universal Corridor (UNICOR) software version 1.054 (https://www.fs.usda.gov/treesearch/pubs/40686). The advantage of this software was a dispersal threshold defined by the user to predict core habitats by using resistant kernel9. Connectivity prediction created factorial least-cost path routes with the highest probability of dispersal54,55.
According to the studies of Kruuk33 and Wagner31, the distance threshold of 70,000 (movement abilities of 70 km) was used in resistant kernel analyses. The resistance map was used to identify core habitats of the striped hyaena with the selected scenario. The buffered least-cost paths were then combined through summation to produce the corridor map between all pairs of occurrence points55. The contiguous map of core habitats was converted to a categorical map based on > 10% of the highest records for the species8,37. In other words, contiguous areas with resistance values less than 10 (0–100) were chosen as core habitats. This work was carried out for corridors as well. Only the categorical corridors out of core habitats were considered. The densities of roads and rivers were calculated for each core habitats and corridors. The coverage of CAs with core habitats and categorical corridors of the striped hyaena was calculated separately in the study area.
Contribution of core habitats for connectivity
The Conefor software version 2.656 (http://www.conefor.org/coneforsensinode.html) was used to measure dPC (i) as the reduction of landscape connectivity associated with the loss of core i57, and three subsections of dPCintra, dPCflux and dPCconnector57. dPCintra (i) measures the contribution of core i to landscape connectivity associated with its area and suitability while dPCflux measures the contribution of core i to landscape connectivity associated with dispersal between it and other core areas on the landscape. dPCconnector measures the contribution of core i to landscape connectivity due to its role as a stepping stone, connecting other core areas to each other58,59. To prepare the data for Conefor (i.e., node and distance files), categorical core habitats of the striped hyaena were applied in Conefor Input ArcGIS extension (http://www.jennessent.com/arcgis/conefor_inputs.htm).
Road collisions and predicted corridors
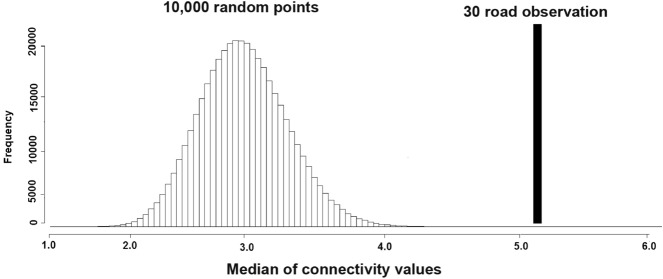
The procedure of spatial randomization test was done to evaluate the occurrence of striped hyaena road crossing within the predicted corridors60,61. A number of 10,000 random points were created along the dangerous roads (roads with record of vehicle collisions, 536 km) in the study area, whereas 30 road observations including 18 successful crossing and 12 collisions records were documented by DoE guards and experts, including the third author with random patrol monitoring during 2015–2020. These 30 road observations were not included in the set of observations used to fit the habitat model. The median value of resistant kernel (predicted connectivity) of road observations was compared with median values of 10,000 random points using a non-parametric test with 107 iterations of 30 locations.
Results
Habitat modeling and variables contribution
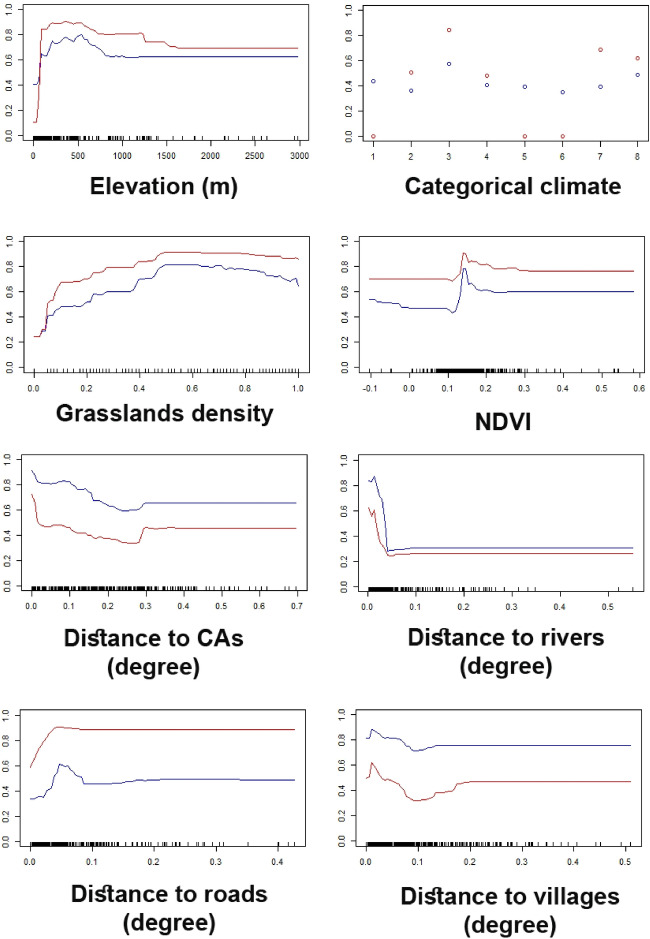
Habitat suitability prediction revealed that elevation, distance to CAs, categorical climate and grasslands density were the most influential variables for predicting the occurrence of the striped hyaena in the study area (Table S4). The optimal range of elevation for the striped hyaena occurrence was 500–1500 m in the study area, and stabilized at 2000 m. The striped hyaena occurred mainly in semi-arid, arid and Mediterranean areas, respectively. As NDVI in natural grasslands increased, the probability of the striped hyaena occurrence increased and then stabilized at 0.2 (from − 1 to 1). As distance to CAs and distance to rivers increased, the probability of striped hyaena occurrence decreased. By increasing distance to roads, the probability of the striped hyaena occurrence increased gradually and then stabilized at about 13 km (Fig. 2). Finally, probability of striped hyaena occurrence increased with increasing distance to villages and then stabilized at about 26 km. Ensemble suitability map showed that hills and hillsides of northeast, east and southeast of the study area had the highest suitability for the striped hyaena (Fig. 3). Habitat suitability models of GLM, MaxEnt, GBM and RF are shown at Supplementary Information (Figure S1).
Figure 2.
Response curves of occurrence points of the striped hyaena to the environmental variables (the two most accurate models of RF [red] and GBM [blue] were considered). Y-axis represents the probability of the striped hyaena occurrence. X-axis of categorical climate variable represents: (1) sea and lake, (2) semi-humid, (3) semi-arid, (4) humid, (5) very arid, (6) very humid, (7) Arid and (8) Mediterranean (each 0.1 geographical degree in the study area is approximately equal to 13.2 km).
Figure 3.
Ensemble habitat suitability map for the striped hyaena in the study area based on the four optimal models of GLM, MaxEnt, RF, and GBM. ArcGIS software version 10.1 (https://www.esri.com/en-us/arcgis/products/arcgis-pro/resources) was used to generate the figure.
Core habitats and corridors
Four core habitats were identified, covering 25% of the study area (Fig. 4; Table 3). The largest habitat patch was Core1, located northeast of the study area (about 11,400 km2) (Fig. 4). The second-largest habitat patch was Core4, located southeast of the study area (about 2700 km2) (Fig. 4, Table 3). One NP, eight PAs and three NHAs were located within identified core habitats. About 11% of the predicted core habitats were covered by CAs (Table 3). Core2 had the highest percentage of coverage with CAs (33%). Core2 and Core3 had the highest density of roads (81.1 m/km2) and rivers (83.95 m/km2), respectively (Table 3).
Figure 4.
Core habitat and corridors for the striped hyaena in the study area (a: Categorical core habitats and corridor paths, b: Contiguous core habitats and c: Contiguous corridor paths). ArcGIS software version 10.1 (https://www.esri.com/en-us/arcgis/products/arcgis-pro/resources) and UNICOR software version 1.0 (https://www.fs.usda.gov/treesearch/pubs/ 40,686) were used to generate the figure.
Table 3.
Properties of predicted core habitats and corridors for the striped hyaena in the study area (number of core habitats and corridors are
available at Fig. 4).
| Number | Area (km2) | Protected | Road density (m/km2) | River density (m/km2) | Number of CAs inside cores and corridors | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Area (km2) | % | NP | WR | PA | NHA | |||||
| Core habitats | 1 | 11,392 | 1129.69 | 9.92 | 64.5 | 59.08 | – | – | 4 | 2 |
| 2 | 1195.4 | 394.74 | 33.02 | 81.1 | 12.39 | 1 | – | 2 | – | |
| 3 | 512.89 | 98.06 | 19.12 | 50 | 83.95 | – | – | 1 | – | |
| 4 | 2726.07 | 140.61 | 5.16 | 65.56 | 55.64 | – | – | 1 | 1 | |
| Total | 15,826.36 | 1763.1 | 11.14 | 69.16 | 55.87 | 1 | – | 8 | 3 | |
| Corridors | 1 | 209.29 | 16.62 | 7.94 | 231.97 | 102.84 | 1 | – | – | – |
| 2 | 1043.41 | – | – | 81.52 | 71.58 | – | – | – | – | |
| Total | 1253.67 | 16.62 | 1.33 | 106.55 | 76.79 | 1 | – | – | – | |
The connectivity for the striped hyaena in the study area was maintained between core habitats from northwest to southeast (Fig. 4). Two main corridors were detected among core habitats. Corridor1 had moderate connectivity between Core1 and Core2 (Fig. 4). Corridor2 among Core1, Core3 and Core4 had high connectivity between the northeast and southeast of the study area. This corridor had two branches: one from Core1 to Core3 and another from Core1 to Core4. Only one NP was located within corridors. Overall, less than 2% of corridors were covered by CAs (Table 3) and in Corridor2 was outside of a CA. Corridor1 had the highest density of roads (231.97 m/km2) and rivers (102.84 m/km2) (Table 3).
Contribution of core habitats for connectivity
Based on dPC index at the estimated dispersal distance scenario, Core1 had the highest contribution to habitat connectivity (Table 4). Based on the results of dPCintra and dPCflux, Core1 had the highest intrapatch connectivity and the highest flux according to patch area and the position within the landscape. Core4 had the highest second contribution. Core1 had the highest contribution as the stepping-stone (Table 4).
Table 4.
Values of dPC index and its three fractions (intra, flux and connector) calculated for predicted four core habitats at the dispersal scenario of 70 km (number of core habitats are available in Fig. 4).
| dPC | dPCintra | dPCflux | dPCconnector | |
|---|---|---|---|---|
| Core1 | 93.87 | 64.01 | 29.02 | 0.84 |
| Core2 | 13.06 | 0.7 | 12.36 | 0 |
| Core3 | 5.29 | 0.13 | 5.16 | 0 |
| Core4 | 20.11 | 3.67 | 16.44 | 0 |
Road collisions and predicted corridors
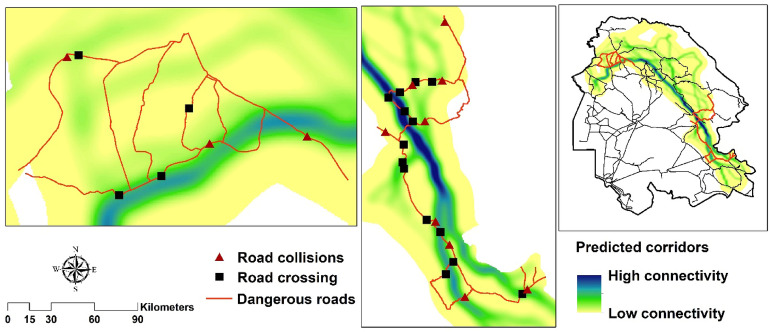
Out of 12 road collisions for the striped hyaena, four were males, seven were females and one was a cub. Six road collisions occurred during winter (from January to March), four during spring (from April to June) and two during summer (from July to September) (Table S5). Predicted corridors, crossed by roads represented a high risk for striped hyaenas (Fig. 5). The spatial randomization test revealed that observations points (crossing + collisions) were more likely to be within corridors than random points (Fig. 6). Observation points had a significantly higher connectivity score than the randomly selected locations (P < 0.001).
Figure 5.
Predicted corridors, striped hyaena road observations (18 crossing + 12 collisions, data collected during 2015–2020) and roads in the study area. ArcGIS software version 10.1 (https://www.esri.com/en-us/arcgis/products/arcgis-pro/resources) and UNICOR software version 1.0 (https://www.fs.usda.gov/treesearch/pubs/40686) were used to generate the figure.
Figure 6.
Spatial randomization test: 30 road observations (18 crossing + 12 collisions) of the striped hyaena and 10,000 random points along the dangerous roads of the study area.
Discussion
We found four variables of elevation, distance to CAs, categorical climate and grasslands density to be significant predictors of striped hyaena occurrence in Khuzestan area, southwestern Iran. We identified four core habitats and two corridors that have the potential to maintain connectivity. The largest core habitat (Core 1) had the highest priority for conservation. Only about one tenth of core habitats was protected by CAs.
Rieger43 reported that the striped hyaena occurred in Iran at elevations up to 2250 m. Here, we predicted slightly lower value of 2000 m. Our habitat modeling predicted that striped hyaenas are limited by higher elevation, however, that is disagreed with the results of Shamoon and Idan62. In our study area, the striped hyaena preferred mainly semi-arid and arid areas with a moderate density of grasslands, and this finding is supported by Leakey et al.63.
The striped hyaena were more likely to occur near rivers, and this finding was supported by Rieger43, and Singh et al.40, and near the villages, which was mentioned earlier by Singh et al.40, Akay et al.28 and Farhadinia et al.64. In addition, Bhandari et al.65 found that the striped hyaena prefers open landscapes along rivers and human settlements, because of suitable cover and access to resources. Other studies found presence of domestic animals in the striped hyaena scats, which was indicative of frequent near human settlements30. In Iran, preying upon livestock by the striped hyaena is rare, and this species approaches the villages for feeding on carrions of domestic animals64.
Core habitats, corridors and CAs
We identified four core habitats for striped hyaenas; however, all had about 10% protection status. Core1 is the largest patch of suitable habitat and occupies a central location relative to other habitat in the study area, which has made the core habitat with the highest flux and as a connector (stepping-stone) within the landscape. Therefore, Core1 had the highest contribution for connectivity in the study area. Only about 12% of core habitats were protected by CAs, which was less than the amount set for near threatened and threatened mammals in Iran66 (i.e., 20%). In addition, Farashi et al.67 reported 66% coverage of CAs with suitable areas of the striped hyaena in Iran, which is remarkably higher compared to the obtain value in this study (11.4%). That is why in their study, occurrence points (centroid of the area of occupancy) of the Atlas of Mammals of Iran39 were used for habitat modeling of the striped hyaena with insufficient occurrence points (just one point) in Khuzestan area (our study area). In this study, by applying sufficient occurrence points for habitat modeling of the striped hyaena, predicted large core habitats in this area needs more CAs to cover unprotected core habitats. Furthermore, establishing more strictly conservation areas is politically challenging. Therefore, we strongly recommend establishing new less strictly conservation areas, such as NHAs. In addition, a small proportion of corridors was protected by CAs. This means that more CAs are needed for conservation of corridors of the striped hyaena in the study area.
Road collisions and predicted corridors
Road collisions were mainly observed in the predicted corridors and a few in the edge of core habitats. However, crossings and collisions occurred even in areas of low predicted connectivity. Corridor1 had a relatively high density of roads and it was bisected about 10 times by roads. Consequently, some of road collisions observed here. In return, Corridor2 had lower density of roads and was bisected similar times with Corridor1. However, a higher number of road collisions were observed here, because higher connectivity caused more individuals movement of striped hyaenas in Corridor2 (from Core1 to Core3 and Core4 and vice versa). Areas that were predicted to be corridors had more road kill observations and that fits out hypothesis that striped hyaenas will be in greater risk of road collisions when moving between core habitats. Our results support previous findings on the use of resistant kernel and factorial least-cost path analyses for effective prioritization of dangerous roads23,61.
Conservation implications for the striped hyaena
With increasing human population and habitat loss, the pressure on the large carnivores, including the striped hyaena has increased2. Large core habitats could help the striped hyaena meet its ecological requirements11. Increasing the amount of CAs is necessary for the conservation of the large carnivores4, and in particularly for the striped hyaena as demonstrated in this study. In addition, maintaining landscape connectivity is necessary for carnivores68, consequently, population gene diversity is conserved16. We urge decision makers to take into account the results of this study when planning corridors between core habitats.
Striped hyaenas movements between core habitats may result in more human-hyaena interactions and therefore additional mitigation efforts is necessary to ensure the safety of the species. For example, increasing local knowledge about the behavior of the striped hyaena (feeding on domestic carrions) and low probability of attacks on domestic animals could be effective for conservation of the species64,69. Facilitating safe wildlife crossing of roads e.g. use of multiple warning signs in dangerous roads in high risk road sections could mitigate the number of road collisions19. Actually, adaptive management plan throughout the landscape (conservation of core habitats and corridors, and reducing species mortality on the roads) must be considered by DoE managers23.
Conclusions
This study was carried out in Khuzestan area in southwestern Iran (mainly arid and semi-arid areas). Four core habitats were detected in this study. The largest one is located in the northeast of the study area. The connectivity was maintained from northwest to southeast of the study area with two main corridors. The result of this study can help direct future conservation plans for the striped hyaena.
Supplementary Information
Acknowledgements
We would like to thank guards and experts of Khuzestan provincial office of the Department of Environment of Iran who provided occurrence points for this study. This study was supported by Research and Technology of Agricultural Sciences and Natural Resources University of Khuzestan (project number 991/22).
Author contributions
K.A. conceptualized and designed the project. R.A. collected the data. K.A. and A.M analyzed the data and interpreted results. K.A. and A.M wrote the manuscript with support from R.A. All authors discussed the results and commented on the manuscript
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-07386-y.
References
- 1.Bennett, A. F. Linkages in the Landscape The Role of Corridors and Connectivity in Wildlife Conservation. (IUCN, Gland, Switzerland and Cambridge, UK).
- 2.Berger J, Young JK, Berger KM. Protecting migration corridors: Challenges and optimism for Mongolian Saiga. PLOS Biol. 2008;6:e165. doi: 10.1371/journal.pbio.0060165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy SM, et al. Consequences of severe habitat fragmentation on density, genetics, and spatial capture-recapture analysis of a small bear population. PLOS ONE. 2017;12:1–20. doi: 10.1371/journal.pone.0181849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaboodvandpour S, Almasieh K, Zamani N. Habitat suitability and connectivity implications for the conservation of the Persian leopard along the Iran-Iraq border. Ecol. Evol. 2021;11:13464–13474. doi: 10.1002/ece3.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilty JA, Lidicker WZ, Jr, Merenlender AM. Corridor Ecology: The Science and Practice of Linking Landscapes for Biodiversity Conservation. Washington DC: Island Press; 2012. [Google Scholar]
- 6.Noss RF, Quigley HB, Hornocker MG, Merrill T, Paquet PC. Conservation biology and carnivore conservation in the rocky mountains. Conserv. Biol. 1996;10:949–963. [Google Scholar]
- 7.Terraube J, Van Doninck J, Helle P, Cabeza M. Assessing the effectiveness of a national protected area network for carnivore conservation. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-16792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashrafzadeh MR, et al. A multi-scale, multi-species approach for assessing effectiveness of habitat and connectivity conservation for endangered felids. Biol. Conserv. 2020;245:108523. [Google Scholar]
- 9.Mohammadi A, et al. Identifying priority core habitats and corridors for effective conservation of brown bears in Iran. Sci. Rep. 2021;11:1–13. doi: 10.1038/s41598-020-79970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beier P, Majka DR, Spencer WD. Forks in the road: Choices in procedures for designing wildland linkages. Conserv. Biol. 2008;22:836–851. doi: 10.1111/j.1523-1739.2008.00942.x. [DOI] [PubMed] [Google Scholar]
- 11.Calvignac S, Hughes S, Hänni C. Genetic diversity of endangered brown bear (ursus arctos) populations at the crossroads of Europe, Asia and Africa. Divers. Distrib. 2009;15:742–750. [Google Scholar]
- 12.Khosravi R, Hemami MR, Cushman SA. Multi-scale niche modeling of three sympatric felids of conservation importance in central Iran. Landsc. Ecol. 2019;34:2451–2467. [Google Scholar]
- 13.Almasieh K, Rouhi H, Kaboodvandpour S. Habitat suitability and connectivity for the brown bear (Ursus arctos) along the Iran-Iraq border. Eur. J. Wildl. Res. 2019;65:1–12. [Google Scholar]
- 14.Balme GA, Hunter LTB, Slotow R. Evaluating methods for counting cryptic carnivores. J. Wildl. Manage. 2009;73:433–441. [Google Scholar]
- 15.Guisan A, Zimmermann NE. Predictive habitat distribution models in ecology. Ecol. Modell. 2000;135:147–186. [Google Scholar]
- 16.McRae BH, Beier P. Circuit theory predicts gene flow in plant and animal populations. Proc. Natl. Acad. Sci. USA. 2007;104:19885–19890. doi: 10.1073/pnas.0706568104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farhadinia MS, et al. Leveraging trans-boundary conservation partnerships: Persistence of Persian leopard (Panthera pardus saxicolor) in the Iranian Caucasus. Biol. Conserv. 2015;191:770–778. [Google Scholar]
- 18.Almasieh K, Mirghazanfari SM, Mahmoodi S. Biodiversity hotspots for modeled habitat patches and corridors of species richness and threatened species of reptiles in central Iran. Eur. J. Wildl. Res. 2019;65:1–13. [Google Scholar]
- 19.Mohammadi A, et al. Road expansion: A challenge to conservation of mammals, with particular emphasis on the endangered Asiatic cheetah in Iran. J. Nat. Conserv. 2018;43:8–18. [Google Scholar]
- 20.Cushman SA, Lewis JS, Landguth EL. Evaluating the intersection of a regional wildlife connectivity network with highways. Mov. Ecol. 2013;1:1–11. doi: 10.1186/2051-3933-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammadi A, Fatemizadeh F. Quantifying landscape degradation following construction of a highway using landscape metrics in Southern Iran. Front. Ecol. Evol. 2021;9:836. [Google Scholar]
- 22.Crooks KR. Relative sensitivities of mammalian carnivores to habitat fragmentation. Conserv. Biol. 2002;16:488–502. [Google Scholar]
- 23.Moqanaki EM, Cushman SA. All roads lead to Iran: Predicting landscape connectivity of the last stronghold for the critically endangered Asiatic cheetah. Anim. Conserv. 2017;20:29–41. [Google Scholar]
- 24.Neumann W, et al. Difference in spatiotemporal patterns of wildlife road-crossings and wildlife-vehicle collisions. Biol. Conserv. 2012;145:70–78. [Google Scholar]
- 25.Mohammadi A, Kaboli M. Evaluating wildlife-vehicle collision hotspots using kernel-based estimation: A focus on the endangered Asiatic cheetah in central Iran. Human-Wildlife Interact. 2016;10:103–109. [Google Scholar]
- 26.Benítez-López A, Alkemade R, Verweij PA. The impacts of roads and other infrastructure on mammal and bird populations: A meta-analysis. Biol. Conserv. 2010;143:1307–1316. [Google Scholar]
- 27.Dadashi-Jourdehi A, Shams-Esfandabad B, Ahmadi A, Rezaei HR, Toranj-Zar H. Predicting the potential distribution of striped hyena Hyaena hyaena in Iran. Belgian J. Zool. 2020;150:185–195. [Google Scholar]
- 28.Akay AE, Inac S, Yildirim IC. Monitoring the local distribution of striped hyenas (Hyaena hyaena L.) in the Eastern Mediterranean Region of Turkey (Hatay) by using GIS and remote sensing technologies. Environ. Monit. Assess. 2011;181:445–455. doi: 10.1007/s10661-010-1840-6. [DOI] [PubMed] [Google Scholar]
- 29.AbiSaid, M. & Dloniak, S. M. D. Hyaena hyaena. The IUCN Red List of Threatened Species 2015. (2015).
- 30.Alam MS, Khan JA. Food habits of striped hyena (Hyaena hyaena) in a semi-arid conservation area of India. J. Arid Land. 2015;7:860–866. [Google Scholar]
- 31.Wagner, A. P. Behavioral ecology of the striped hyena (Hyaena hyaena). ProQuest Diss. Theses 195–195 (2006).
- 32.Hofer H. Species Accounts, Status Survey and Conservation Action Plan of Hyaena. Information Press; 1998. [Google Scholar]
- 33.Kruuk H. Feeding and social behaviour of the striped hyaena (Hyaena vulgaris Desmarest) East African Wildl. J. 1976;14:91–111. [Google Scholar]
- 34.Tourani M, Moqanaki EM, Kiabi BH. Vulnerability of striped hyaenas, hyaena hyaena, in a human-dominated landscape of central Iran. Zool. Middle East. 2012;56:133–136. [Google Scholar]
- 35.Parchizadeh J, Belant JL. Human-caused mortality of large carnivores in Iran during 1980–2021. Glob. Ecol. Conserv. 2021;27:e01618. [Google Scholar]
- 36.Almasieh K, Zoratipour A, Negaresh K, Delfan-Hasanzadeh K. Habitat quality modelling and effect of climate change on the distribution of Centaurea pabotii in Iran. Spanish J. Agric. Res. 2018;16:e0304. [Google Scholar]
- 37.Ahmadi M, et al. Species and space: A combined gap analysis to guide management planning of conservation areas. Landsc. Ecol. 2020;35:1505–1517. [Google Scholar]
- 38.Yusefi GH, Faizolahi K, Darvish J, Safi K, Brito JC. The species diversity, distribution, and conservation status of the terrestrial mammals of Iran. J. Mammal. 2019;100:55–71. [Google Scholar]
- 39.Karami, M., Ghadirian, T. & Faizolahi, K. The atlas of mammals of Iran ; Jahad daneshgahi, kharazmi Branch (Department of the Environment of Iran, 2016).
- 40.Singh P, Gopalaswamy AM, Karanth KU. Factors influencing densities of striped hyenas (Hyaena hyaena) in arid regions of India. J. Mammal. 2010;91:1152–1159. [Google Scholar]
- 41.Brown JL. SDMtoolbox: A python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. Methods Ecol. Evol. 2014;5:694–700. doi: 10.7717/peerj.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esri. ArcGIS 10.1. Environ. Syst. Res. Institute, Redlands, CA, USA (2012).
- 43.Rieger I. A review of the biology of striped hyaenas, Hyaena hyaena (Linne, 1758) Saugetierkund. Mitt. 1979;27:81–95. [Google Scholar]
- 44.Jueterbock, A. ‘ MaxentVariableSelection ’ vignette (2015).
- 45.Team, R. C. R: A language and environment for statistical computing. R Foundation for Statistical Computing. (2019).
- 46.Naimi B, Hamm NAS, Groen TA, Skidmore AK, Toxopeus AG. Where is positional uncertainty a problem for species distribution modelling? Ecography (Cop.) 2014;37:191–203. [Google Scholar]
- 47.Zuur AF, Ieno EN, Elphick CS. A protocol for data exploration to avoid common statistical problems. Methods Ecol. Evol. 2010;1:3–14. [Google Scholar]
- 48.Thuiller W, Lafourcade B, Engler R, Araújo MB. BIOMOD: A platform for ensemble forecasting of species distributions. Ecography (Cop.) 2009;32:369–373. [Google Scholar]
- 49.Shahnaseri G, et al. Contrasting use of habitat, landscape elements, and corridors by grey wolf and golden jackal in central Iran. Landsc. Ecol. 2019;34:1263–1277. [Google Scholar]
- 50.Araújo MB, New M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007;22:42–47. doi: 10.1016/j.tree.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 51.Eskildsen A, et al. Testing species distribution models across space and time: high latitude butterflies and recent warming. Glob. Ecol. Biogeogr. 2013;22:1293–1303. [Google Scholar]
- 52.Wan HY, Cushman SA, Ganey JL. Improving habitat and connectivity model predictions with multi-scale resource selection functions from two geographic areas. Landsc. Ecol. 2019;34:503–519. [Google Scholar]
- 53.Mateo-Sánchez MC, et al. A comparative framework to infer landscape effects on population genetic structure: Are habitat suitability models effective in explaining gene flow? Landsc. Ecol. 2015;30:1405–1420. [Google Scholar]
- 54.Landguth EL, Hand BK, Glassy J, Cushman SA, Sawaya MA. UNICOR: A species connectivity and corridor network simulator. Ecography (Cop.) 2012;35:9–14. [Google Scholar]
- 55.Cushman SA, McKelvey KS, Schwartz MK. Use of empirically derived source-destination models to map regional conservation corridors. Conserv. Biol. 2009;23(2):368–376. doi: 10.1111/j.1523-1739.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 56.Saura S, Torné J. Conefor Sensinode 2.2: A software package for quantifying the importance of habitat patches for landscape connectivity. Environ. Model. Softw. 2009;24:135–139. [Google Scholar]
- 57.Saura S, Pascual-Hortal L. A new habitat availability index to integrate connectivity in landscape conservation planning: Comparison with existing indices and application to a case study. Landsc. Urban Plan. 2007;83:91–103. [Google Scholar]
- 58.Saura S, Rubio L. A common currency for the different ways in which patches and links can contribute to habitat availability and connectivity in the landscape. Ecography (Cop.) 2010;33:523–537. [Google Scholar]
- 59.Avon C, Bergès L. Prioritization of habitat patches for landscape connectivity conservation differs between least-cost and resistance distances. Landsc. Ecol. 2016;31:1551–1565. [Google Scholar]
- 60.Cushman SA, Lewis JS, Landguth EL. Why did the bear cross the road? Comparing the performance of multiple resistance surfaces and connectivity modeling methods. Diversity. 2014;6:844–854. [Google Scholar]
- 61.Mohammadi A, et al. Integrating spatial analysis and questionnaire survey to better understand human-onager conflict in Southern Iran. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-021-91921-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shamoon H, Shapira I. Limiting factors of Striped Hyaena, Hyaena hyaena, distribution and densities across climatic and geographical gradients (Mammalia: Carnivora) Zool. Middle East. 2019;65:189–200. [Google Scholar]
- 63.Leakey LN, et al. Diet of striped hyaena in northern Kenya. Afr. J. Ecol. 1999;37:314–326. [Google Scholar]
- 64.Farhadinia MS, Johnson PJ, Hunter LTB, Macdonald DW. Wolves can suppress goodwill for leopards: Patterns of human-predator coexistence in northeastern Iran. Biol. Conserv. 2017;213:210–217. [Google Scholar]
- 65.Bhandari S, Bhusal DR, Psaralexi M, Sgardelis S. Habitat preference indicators for striped hyena (Hyaena hyaena) in Nepal. Glob. Ecol. Conserv. 2021;27:e01619. [Google Scholar]
- 66.Farashi A, Shariati M. Biodiversity hotspots and conservation gaps in Iran. J. Nat. Conserv. 2017;39:37–57. [Google Scholar]
- 67.Farashi A, Shariati M, Hosseini M. Identifying biodiversity hotspots for threatened mammal species in Iran. Mamm. Biol. 2017;87:71–88. [Google Scholar]
- 68.Boitani L, Ciucci P, Corsi F, Dupre E. Range and corridors for brown bears in the eastern potential. Ursus. 1999;11:123–130. [Google Scholar]
- 69.Bhandari S, Morley C, Aryal A, Shrestha UB. The diet of the striped hyena in Nepal’s lowland regions. Ecol. Evol. 2020;10:7953–7962. doi: 10.1002/ece3.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.