Abstract
Resistance of cytomegalovirus (CMV) to antiviral agents is a well-recognized phenomenon that has been observed in the laboratory and in the clinical setting. Infections caused by antiviral-resistant CMV have been found exclusively among immunocompromised individuals, including patients with AIDS, bone marrow and solid-organ transplant recipients, and patients with hematologic malignancies, and in individuals with primary immunodeficiencies. The majority of these infections have been described to occur in patients with AIDS receiving prolonged antiviral therapy for CMV end-organ disease. Antiviral agents currently licensed for the treatment of CMV infections include ganciclovir, foscarnet, and cidofovir. Resistance of CMV to ganciclovir is related to mutations in the UL97 region of the viral genome and/or mutations in the viral DNA polymerase. Resistance to foscarnet and cidofovir is associated with mutations in the viral DNA polymerase. Antiviral susceptibility of CMV strains containing DNA polymerase mutations is dependent on the region of the DNA polymerase where the mutations are located. Some DNA polymerase mutant viruses are cross-resistant to ganciclovir, foscarnet, and cidofovir. The recognition that specific UL97 and UL54 mutations are associated with resistance to antiviral agents has led to the development of molecular methods for detection of mutant viruses. This article reviews the mechanisms of resistance of CMV to antiviral agents, the laboratory methods for detection of resistant CMV, and the clinical aspects of infections caused by antiviral-resistant CMV.
Cytomegalovirus (CMV) infections are a major cause of morbidity and mortality among immunocompromised patients, especially recipients of bone marrow and solid-organ transplants and patients with AIDS (40, 51, 53). Specifically, CMV was the most common cause of sight-threatening and life-threatening opportunistic viral infections among patients with AIDS prior to the availability of highly active antiretroviral therapy. Antiviral agents currently licensed for the treatment of CMV infections include ganciclovir, foscarnet, and cidofovir (18, 20, 44, 45). Multiple clinical trials have demonstrated the efficacy of ganciclovir in patients with CMV disease, and published clinical studies suggest that the efficacy of foscarnet is similar to that of ganciclovir in AIDS-related CMV retinitis (41). Although the clinical experience with cidofovir is more limited, recent studies have shown that it is beneficial in the treatment of AIDS patients with CMV retinitis (44, 45).
In severely immunocompromised patients who develop CMV disease, prolonged antiviral therapy is often necessary. A potential risk associated with prolonged use of antiviral compounds is the emergence of resistant viruses. CMV isolates resistant to antiviral agents have been selected in the laboratory (47, 48, 63, 65) and have also been recovered from immunocompromised patients treated with antiviral agents (23, 27, 29, 56, 72). Studies done in the last 5 years have contributed significantly to the current understanding of the mechanisms of resistance of CMV to antiviral drugs. Ganciclovir-resistant CMV strains selected in the laboratory have amino acid deletions or substitutions in conserved regions of the UL97 region and/or point mutations in the DNA polymerase gene of the virus (47, 48, 63, 65). In clinical CMV strains, resistance to ganciclovir has also been associated with mutations in UL97, DNA polymerase, or both viral genes (1–4, 12–15, 30, 31, 36, 37, 49, 57–60, 70–72). The functional consequence of the UL97 mutations is an impaired phosphorylation of ganciclovir in virus-infected cells, with the consequent lack of synthesis of ganciclovir triphosphate, the active form of the drug (48, 61, 63). Characterization of laboratory and clinical CMV strains resistant to foscarnet and cidofovir has demonstrated that resistance to these compounds is associated with amino acid substitutions in conserved regions of the viral DNA polymerase (14, 15, 30, 37, 47, 58, 59, 65). Some of these DNA polymerase mutant isolates are cross-resistant to ganciclovir, foscarnet, and cidofovir.
CMV resistance studies have been hampered by the technical difficulty and lack of standardization of antiviral susceptibility assays. Classically, plaque reduction assays have been considered the “gold standard” method to determine the antiviral susceptibility profile (also termed viral phenotype) of CMV strains (43). Unfortunately, a major problem with plaque reduction assays is the excessive time required to complete the assay and the lack of a consensus method validated in different laboratories. Other methods available for antiviral susceptibility testing include DNA-DNA hybridization, enzyme-linked immunosorbent assay (ELISA), immunofluorescence, and flow cytometry-based assays (33, 50, 67–69). All are limited by lack of standardization and by the length of time required to generate susceptibility results.
The recognition that mutations in the UL97 and/or DNA polymerase region of CMV are associated with antiviral resistance has led to the development of methods for detection of mutant viruses. The UL97 and DNA polymerase sequences (the viral genotype) are generally characterized by combining PCR and sequencing methods. The observation that the majority (94%) of ganciclovir-resistant CMV strains contain specific UL97 mutations prompted the development of screening assays to detect mutant viruses (12, 13, 36). By combining PCR and restriction digestion methods, it has been possible to detect 78% of ganciclovir-resistant CMV isolates containing the most common UL97 mutations (12, 13). These screening methods have been successfully applied to the direct detection of mutant CMV UL97 sequences in clinical specimens (i.e., plasma, blood leukocytes, and cerebrospinal fluid).
From a clinical perspective, the most relevant question is to determine the pathogenic significance of infections caused by resistant CMV strains. Studies by several groups of investigators support the notion that resistance of CMV to antiviral agents is a factor in therapeutic failure and disease progression. Additional studies are needed to fully characterize the genetics of resistance of CMV to different antiviral agents, to determine the importance of individual mutations by assessing their effect on the viral phenotype, and to develop rapid and reliable methods for the timely detection of resistant CMV strains in clinical specimens.
ANTIVIRAL AGENTS FOR TREATMENT OF CMV INFECTIONS
Antiviral agents currently licensed for the treatment of CMV infections include ganciclovir, foscarnet, and cidofovir (5, 18, 20, 44, 45). All three compounds inhibit CMV DNA synthesis by inhibiting the viral DNA polymerase. Ganciclovir (9-[1,3-dyhydroxy-2-propoxymethyl]guanine) is a deoxyguanosine analogue that must be phosphorylated to ganciclovir triphosphate to exert its antiviral activity (20, 32, 54). The UL97 gene of CMV encodes a viral protein kinase that phosphorylates ganciclovir to ganciclovir monophosphate, with cellular kinases carrying out further phosphorylations to ganciclovir di- and triphosphate. Ganciclovir triphosphate is a competitive inhibitor of the natural substrate (deoxyguanosine triphosphate) for the CMV DNA polymerase. In addition, incorporation of ganciclovir triphosphate into viral DNA causes a slowing and subsequent cessation of CMV DNA chain elongation (20, 32, 54).
Foscarnet (trisodium phosphonoformate) is a pyrophosphate analogue that reversibly and noncompetitively inhibits the activity of the CMV DNA polymerase (5, 18). Foscarnet does not require intracellular activation to exert its antiviral activity and is not incorporated into the growing viral DNA chain. Foscarnet reversibly blocks the pyrophosphate binding site of the viral DNA polymerase and inhibits the cleavage of pyrophosphate from deoxynucleoside triphosphates (5, 18).
Cidofovir ([S]-1-[3-hydroxy-2-phosphonylmethoxypropyl]cytosine) is an acyclic nucleoside phosphonate that must be phosphorylated to its diphosphoryl derivative to exert its antiviral activity (22, 44, 45). These phosphorylations are carried out by host cellular enzymes and thus are independent of virus-induced phosphorylating enzymes. Cidofovir diphosphate is a competitive inhibitor of the CMV DNA polymerase. Similarly to ganciclovir, the incorporation of cidofovir diphosphate into viral DNA causes a slowing and subsequent cessation of CMV DNA chain elongation (22).
MECHANISMS OF RESISTANCE
Initial Laboratory and Clinical Studies
The evidence that CMV could become resistant to antiviral compounds was initially obtained in the laboratory by selecting a ganciclovir-resistant CMV strain after passaging a susceptible reference virus (laboratory strain AD169) in the presence of increasing concentrations of ganciclovir (6). This mutant virus, designated B759rD100, was resistant to ganciclovir but susceptible to foscarnet and induced decreased phosphorylation of ganciclovir in infected cells compared with that in cells infected with the control strain AD169. These findings suggested that the resistance of B759rD100 to ganciclovir could be the consequence of a mutation in the viral genome that would reduce the synthesis of a CMV-encoded enzyme contributing to the intracellular phosphorylation of ganciclovir.
That ganciclovir-resistant CMV could emerge in the clinical setting was demonstrated shortly after ganciclovir became available for the treatment of severe CMV infections. During our initial study of ganciclovir for the treatment of CMV disease in immunocompromised hosts, we demonstrated that ganciclovir therapy ablated CMV viremia after a mean of 4.7 days of therapy (28). However, one AIDS patient in that study had CMV persistently isolated from multiple blood cultures despite being treated with multiple courses of ganciclovir. Shortly thereafter, we identified two additional patients (one patient with AIDS, and one patient with chronic lymphocytic leukemia) who also had persistent CMV viremia and progressive CMV disease that were unresponsive to ganciclovir (29). Because of the relentless progression of the CMV disease in these patients and the persistently positive blood cultures during therapy, susceptibilities to ganciclovir of CMV isolates obtained before or shortly after the start of ganciclovir therapy (pretherapy or early isolates) and after several courses of ganciclovir (late isolates) were determined. In all three patients, the late CMV isolates were clearly resistant to ganciclovir, for which the 50% inhibitory concentrations (IC50) were 2 to 12 times higher than those for baseline isolates and 4 to 6 times higher than those for the ganciclovir-susceptible reference strain AD169 (29). Whereas these data demonstrated that ganciclovir therapy could select resistant isolates, the incidence of these infections was not known. The only data available then were from a prospective study of 72 patients with AIDS and CMV infection treated with ganciclovir. In that study, 7.8% of the patients who had been treated with ganciclovir for >3 months eliminated resistant CMV in their urine (23).
To further address the mechanisms of ganciclovir resistance of CMV, laboratory strain 759rD100-1 and 14 clinical isolates (9 of which were resistant to ganciclovir) from the studies cited above were further characterized in a series of experiments aimed to define the antiviral susceptibility pattern of the viruses, the intracellular anabolism of ganciclovir in cells infected with the different strains, and the susceptibility of the viral DNA polymerases to inhibition by antiviral compounds (61). Laboratory strain 759rD100-1 was resistant to ganciclovir and susceptible to foscarnet and had a reduced susceptibility to cidofovir. All nine ganciclovir-resistant clinical isolates were susceptible to foscarnet and cidofovir. Similarly to the ganciclovir-resistant laboratory strain 759rD100-1, the ganciclovir-resistant clinical isolates studied showed a reduced ability to induce intracellular phosphorylation of ganciclovir whereas the ganciclovir-susceptible isolates were able to induce phosphorylation of ganciclovir at levels comparable to those of the ganciclovir-susceptible laboratory strain AD169 (61). The DNA polymerases of a pair of ganciclovir-susceptible and -resistant clinical isolates had comparable sensitivities to ganciclovir triphosphate and foscarnet, with values that were similar to those of the laboratory strain AD169. Based on these results, it was postulated that resistance of clinical CMV isolates to ganciclovir was due to their inability to induce adequate levels of ganciclovir triphosphate in virus-infected cells and that the viral DNA polymerase did not play a role in the resistance of the clinical isolates to ganciclovir. In contrast, the decreased susceptibility to cidofovir of the ganciclovir-resistant laboratory strain 759rD100-1 suggested that the virus could also have an altered DNA polymerase.
Characterization of UL97 Mutations in Ganciclovir-Resistant CMV Laboratory Strains
The initial studies of resistant laboratory and clinical CMV strains related ganciclovir resistance to inadequate intracellular phosphorylation of the compound in cells infected with ganciclovir-resistant viruses. Although experimental evidence at that time suggested that ganciclovir phosphorylation was controlled by a CMV-encoded function, the mechanism by which ganciclovir is phosphorylated in CMV-infected cells remained unresolved until 1992. In that year, two independent groups of investigators provided biochemical, immunological, and genetic evidence that the phosphorylation of ganciclovir in CMV-infected cells is controlled by a protein kinase homologue encoded by the UL97 open reading frame of the virus (46, 63). In one study, a recombinant protein (UL97tr) was obtained by cloning a truncated part of the CMV UL97 open reading frame including sequences homologous to the catalytic domains of protein kinases into an expression vector. This protein was used to produce specific antisera that were shown by Western blotting to react with UL97tr itself and with a protein obtained from CMV-infected cells that had a molecular weight similar to that predicted for UL97. In addition, extracts from bacteria expressing UL97tr efficiently phosphorylated ganciclovir compared with controls. Furthermore, this phosphorylating activity was neutralized by CMV-seropositive serum but not by CMV-seronegative serum (46). In another study, recombinant viruses containing fragments representing the complete UL97 sequence of the ganciclovir-resistant laboratory strain 759rD100 prepared in marker transfer experiments were resistant to ganciclovir and unable to induce phosphorylation of ganciclovir in infected cells. Analysis of the DNA sequence of these recombinant viruses revealed a 12-bp deletion of UL97 codons 590 to 593, which resulted in a 4-amino-acid deletion (Ala-Ala-Cys-Arg) in a conserved region of the UL97 protein possibly implicated in substrate recognition (63). On the basis of these results, it was proposed that UL97 is responsible for the initial phosphorylation of ganciclovir in CMV-infected cells.
These findings prompted the characterization of UL97 sequences in ganciclovir-resistant laboratory and clinical CMV strains. In one study, plaque-purified ganciclovir-resistant strains D1/3/4, D6/3/1, and D10/3/2 were obtained after sequential passages of the susceptible strain, AD169, in increasing concentrations of ganciclovir (48). Cells infected with these ganciclovir-resistant viruses showed reduced levels of ganciclovir phosphorylation, suggesting the presence of mutations in the UL97 region of these viruses. Sequencing studies of this open reading frame demonstrated the same mutation (ATG to ATT), resulting in a methionine-to-isoleucine amino acid change at amino acid residue 460 (M460I mutation) of the UL97 protein in all three resistant strains (48). A recombinant virus (R6HS) prepared after transfection of UL97 sequences from D6/3/1 into cells infected with AD169 was also resistant to ganciclovir and induced ganciclovir phosphorylation levels that were more than 10-fold lower than those in AD169-infected cells (48).
Characterization of UL97 Mutations in Ganciclovir-Resistant Clinical CMV Isolates
Analyses of UL97 sequences in clinical CMV isolates resistant to ganciclovir has also demonstrated the presence of point mutations or deletions in this region of the viral genome. Table 1 summarizes UL97 mutations found in clinical CMV isolates included in studies published in English in the literature between 1995 and 1998. The majority of the 129 ganciclovir-resistant isolates included in Table 1 contained mutations at codon 460 (28 isolates or 22%), 594 (25 isolates or 21%), or 595 (32 isolates or 25%). In most cases, isolates contained single UL97 mutations, with few isolates containing two mutations in this region. Marker transfer experiments have shown that the mutations M460V, M460I, H520Q, A594V, L595S, L595F, deletion of codon 595, deletion of codons 591 to 594, C603W, and C607Y confer ganciclovir resistance when introduced into the genome of recombinant viruses. Similar to the studies done with ganciclovir-resistant laboratory CMV strains, anabolism studies with ganciclovir-resistant clinical CMV isolates have demonstrated that the functional consequence of these UL97 mutations is an impaired intracellular phosphorylation of ganciclovir into ganciclovir monophosphate, with the subsequent lack of synthesis of ganciclovir di- and triphosphate.
TABLE 1.
UL97 mutations in clinical CMV isolates resistant to ganciclovir
| UL97 mutation | Amino acid change | No. of isolates | Reference(s) |
|---|---|---|---|
| M460V | Methionine to valine | 21 | 1, 12, 13, 27, 57, 58, 72 |
| M460I | Methionine to isoleucine | 7 | 3, 13, 58, 59, 71 |
| N510S | Aspartic acid to serine | 5 | 13, 31 |
| H520Q | Histidine to glutamine | 6 | 13, 30, 36 |
| A590T | Alanine to threonine | 1 | 72 |
| A591D | Alanine to aspartic acid | 1 | 72 |
| A591V | Alanine to valine | 2 | 13, 58 |
| C592G | Cysteine to glysine | 5 | 13, 30, 58 |
| A594V | Alanine to valine | 24 | 3, 12, 13, 30, 31, 58, 70 |
| A594T | Alanine to threonine | 1 | 30 |
| del591–594 | Deletion of alanine-alanine-cysteine-arginine | 1 | 13 |
| L595S | Leucine to serine | 23 | 3, 12, 13, 30, 49, 58, 70, 72 |
| L595F | Leucine to phenylalanine | 5 | 12, 58, 70 |
| L595T | Leucine to threonine | 1 | 58 |
| del595 | Deletion of leucine | 2 | 2 |
| L595W | Leucine to tryptophan | 1 | 13 |
| E596G | Glutamic acid to glysine | 1 | 13 |
| E596D | Glutamic acid to aspartic acid | 1 | 72 |
| N597I | Asparagine to isoleucine | 1 | 72 |
| G598V | Glysine to valine | 1 | 72 |
| K599M | Lysine to methionine | 1 | 72 |
| del600 | Deletion of leucine | 1 | 16 |
| C603W | Cysteine to tryptophan | 5 | 14, 58 |
| C607Y | Cysteine to tyrosine | 6 | 4, 59 |
| C603Y | Cysteine to tyrosine | 1 | 72 |
| A606D | Alanine to aspartic acid | 1 | 72 |
| V665I | Valine to isoleucine | 4 | 70 |
In addition to the mutations cited above, other UL97 mutations (N510S, A590T, A591V, A591D, C592G, L595W, L595T, E596G, E596D, N597I, G598V, K599M, C603Y, A606D, and V665I) have been found in ganciclovir-resistant clinical CMV isolates (Table 1). N510S has been reported to occur in one clinical CMV isolate resistant to ganciclovir that also contained a deletion of UL97 codons 591 to 594. Because this deletion confers resistance to ganciclovir, the role of N510S as a marker of resistance is unclear. V665I has been found in resistant isolates containing other UL97 mutations related to ganciclovir resistance. Therefore, the significance of this mutation is not clear. Although the effect of A591V, C592G, L595W, L595T, E596G, N597I, G598V, K599M, C603Y, A606D, and V665I in conferring resistance to ganciclovir has not been studied with recombinant viruses or in ganciclovir anabolism studies, these mutations are located at or next to UL97 codons shown to be involved in ganciclovir resistance. Further work is necessary to determine the role of these amino acid substitutions in ganciclovir resistance.
The biological role of the UL97 protein is not known. It has sequence homology to bacterial phosphotransferases as well as other protein kinases of human herpesviruses, some of which play a crucial regulatory role in gene expression (7). It has been suggested that mutations related to resistance to ganciclovir alter the substrate binding sites of UL97 so that it can no longer efficiently phosphorylate ganciclovir. However, these mutations do not inactivate UL97, as evidenced by the ability of the mutated protein to phosphorylate itself and other protein substrates (7, 38).
Characterization of DNA Polymerase (UL54) Mutations in Drug-Resistant CMV Laboratory Strains
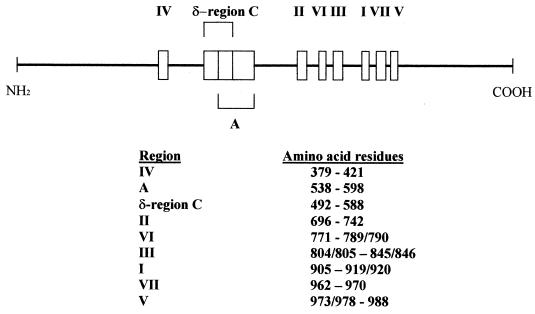
The CMV DNA polymerase is the target of the antiviral agents currently available to treat CMV infections. Therefore, mutations in the viral gene that encodes the DNA polymerase can result in resistance of the virus to these drugs. Sequence analysis of the DNA polymerase of ganciclovir-resistant CMV strains selected in the laboratory has shown the presence of mutations in this region of the viral genome (47, 65). For instance, each one of the ganciclovir-resistant strains D1/3/4, D6/3/1, and D10/3/2 (obtained after sequential passages of the susceptible strain, AD169, in increasing concentrations of ganciclovir) contained mutations in the DNA polymerase gene (47). D1/3/4 and D6/3/1 had a single-base change (A to C) at position 2160, which results in an amino acid change from leucine to isoleucine at residue 501 (L501I mutation). The mutation in D10/3/2 consists of a single-base change at position 1893 (G to T), which results in a change from phenylalanine to valine at residue 412 (F412V mutation) (47). Antiviral susceptibility studies revealed that these isolates were resistant to ganciclovir, susceptible to foscarnet, and resistant to cidofovir (Table 2). Because these mutants also contained UL97 mutations, the role of the DNA polymerase mutations in conferring antiviral resistance was further examined in recombinant viruses. Transfer of the L501I mutation found in D1/3/4 to the wild-type, AD169, resulted in a recombinant virus (HP/A1B/4) that was also resistant to ganciclovir, sensitive to foscarnet, and resistant to cidofovir. However, in contrast to the parental strain D1/3/4, cells infected with the recombinant strain HP/A1B/4 had normal levels of ganciclovir phosphorylation. Similarly, analysis of the sequence of the DNA polymerase of ganciclovir-resistant laboratory strain 759rD100 revealed a C-to-G change at position 3619 that results in a replacement of alanine by glycine at position 987 (A987G mutation) (65). Transfer of the A987G to the wild-type, AD169, resulted in a recombinant virus (GDGrP53) that was resistant to ganciclovir, sensitive to foscarnet, resistant to cidofovir, and able to induce phosphorylation of ganciclovir in infected cells as efficiently as did the wild-type, AD169. The mutations found in the drug-resistant laboratory mutants described above are localized in highly conserved regions of the CMV DNA polymerase (Fig. 1). F412V lies within conserved region IV, L501I is located in ∂-region C, and A987G is located in conserved region V. It has been postulated that these DNA polymerase mutations could be involved in substrate recognition and could cause drug resistance by decreasing the affinity of the viral polymerase for antiviral compounds (47, 65).
TABLE 2.
Effect of UL97 and UL54 mutations in antiviral susceptibilities of laboratory CMV isolates
| Strain | Amino acid change (residue) in:
|
Fold change in IC50 of:
|
Reference(s) | |||
|---|---|---|---|---|---|---|
| UL97 sequence | UL54 sequence | Ganciclovir | Foscarnet | Cidofovir | ||
| R6HS | Methionine to isoleucine (460) | Wild type | 5 | NCa | NC | 48 |
| HP/A1B/4 | Wild type | Leucine to isoleucine (501) | 5 | NC | 8–15 | 47 |
| GDGrH5 | Wild type | Alanine to glysine (987) | 6 | NAa | NA | 65 |
| GDGrP53 | Wild type | Alanine to glysine (987) | 4 | NA | 10 | 65 |
| D6/3/1 | Methionine to isoleucine (460) | Leucine to isoleucine (501) | 20 | 1.2 | 8–16 | 47, 48 |
| D1/3/4 | Methionine to isoleucine (460) | Leucine to isoleucine (501) | 20 | 1.2 | 8–16 | 47, 48 |
| D10/3/2 | Methionine to isoleucine (460) | Phenylalanine to valine (412) | 20 | 1.2–1.5 | 8–16 | 47, 48 |
| 759rD100 | Deletion (590–593) | Alanine to glysine (987) | 20 | NC | 7 | 63, 65 |
NC, no change; NA, not available.
FIG. 1.
Schematic representation of the CMV DNA polymerase (UL54). Boxes represent regions of the DNA polymerase highly conserved among different herpesviruses. Data are from references 15, 16, 58, and 65.
Characterization of UL54 Mutations in Drug-Resistant Clinical CMV Isolates
The finding of mutations in the UL54 region of drug-resistant laboratory strains prompted the determination of the sequence of this region in clinical CMV isolates. UL54 mutations in clinical CMV isolates that have been reported in the literature in English between 1995 and 1998 are summarized in Table 3. These data should be interpreted with caution because the role of the UL54 mutations listed in conferring resistance has been analyzed in recombinant viruses in only few of the studies. In recombinant viruses, F412C and L501I cause resistance to both ganciclovir and cidofovir (14, 58); T700A and V715M cause resistance to foscarnet (3); L802M has been associated with resistance to both ganciclovir and foscarnet or only to foscarnet (14, 16); and A809V causes resistance to both ganciclovir and cidofovir (15). Of note is that the T700A mutation has also been found in a foscarnet-susceptible clinical CMV isolate obtained from a patient who had not been previously treated with foscarnet (30). The reason for the discrepant effect of L802M in antiviral susceptibilities has not been fully clarified. In addition, all the isolates listed in Table 3 were resistant to ganciclovir and the majority contained one or two ganciclovir resistance UL97 mutations. Therefore, the contribution of UL54 mutations to the ganciclovir resistance of the isolates listed in Table 3 cannot be differentiated from that of the UL97 mutations present in the same isolates. Clinical CMV isolates that are resistant to ganciclovir, foscarnet, and cidofovir have also been reported. In addition to ganciclovir resistance mutations in UL97, these multiresistant isolates have two or more of the following mutations in UL54: N408D, F412C, K513E, D588E, V781I, L802M, K805Q, T821I, S897P, and R1052C (58).
TABLE 3.
UL54 mutations in clinical CMV isolates resistant to antiviral agents
| Region | UL54 mutationa | Amino acid change | Viral phenotypeb
|
UL97 mutationsc | Reference(s) | ||
|---|---|---|---|---|---|---|---|
| Ganciclovir | Foscarnet | Cidofovir | |||||
| IV | N408D | Asparagine to aspartic acid | R | S | R | No | 30, 58 |
| F412C | Phenylalanine to cysteine | R | S | R | Yes | 14, 58 | |
| D413E | Aspartic acid to glutamic acid | R | S | R | Yes | 30 | |
| ∂-region C | L501F | Leucine to phenylalanine | R | S | R | Yes | 30, 37 |
| L501I | Leucine to isoleucine | R | S | R | Yes | 58 | |
| T503I | Threonine to isoleucine | R | S | R | Yes | 58 | |
| K513R | Lysine to arginine | R | S | R | Yes | 58 | |
| K513E | Lysine to glutamic acid | R | S | R | Yes | 59 | |
| P522A | Proline to alanine | R | S | ND | Yes | 30 | |
| T700A | Threonine to alanine | R | S | R | No | 30 | |
| II | T700A | Threonine to alanine | S | R | S | No | 3 |
| M715V | Valine to methionine | S | R | S | No | 3 | |
| I722V | Isoleucine to valine | R | S | R | Yes | 58 | |
| VI | V781I | Valine to isoleucine | R | R | S | Yes | 58 |
| III | L802M | Leucine to methionine | R | R | S | Yes | 14 |
| A809V | Alanine to valine | R | R | S | Yes | 15 | |
| G841A | Glysine to alanine | R | S | ND | Yes | 30 | |
| Other | S676G | Serine to glysine | R | S | R | Yes | 58 |
| G678S | Glysine to serine | R | S | R | Yes | 59 | |
| Y751H | Tyrosine to histidine | R | S | R | Yes | 58 | |
Mutations transferring antiviral resistance to recombinant viruses are shown in boldface type.
R, S, and ND denote resistant, sensitive, and not done, respectively.
Refers to ganciclovir resistance UL97 mutations.
A more detailed analysis of the effect of specific UL54 mutations in the susceptibility of CMV to different antiviral agents has been performed recently by constructing UL54 mutant recombinants by a cosmid cotransfection approach (16, 42). In this method, recombinant UL54 mutant viruses were generated by cotransfection of nine overlapping CMV DNA fragments into fibroblasts, and their drug susceptibility profiles were determined in a plaque reduction assay and compared with drug susceptibilities of a wild-type recombinant virus (16). A total of 17 recombinant viruses expressing various point mutations in UL54 were generated, and their antiviral susceptibilities were characterized. The in vitro susceptibility data of the recombinant viruses to ganciclovir, foscarnet, and cidofovir are summarized in Table 4. From these data, it has been concluded that (i) mutations located in UL54 regions IV and ∂-region C (N terminus of UL54) and mutations in region V (C-terminus of UL54) cause resistance to ganciclovir and cidofovir, (ii) mutations in conserved regions II and VI (located in the central part of the UL54 polypeptide) cause resistance to foscarnet, (iii) mutations in conserved region III are associated with various antiviral susceptibility profiles, (iv) mutations located outside conserved regions of UL54 are not associated with significant changes in antiviral susceptibilities of CMV, and (v) multiple UL54 mutations are additive and cause resistance to multiple antiviral agents.
TABLE 4.
Effect of UL54 mutations in antiviral susceptibilities of recombinant CMV isolatesa
| Region | Mutation | Amino acid change | Viral phenotypeb
|
||
|---|---|---|---|---|---|
| Ganciclovir | Foscarnet | Cidofovir | |||
| IV | N408D | Asparagine to aspartic acid | R | S | R |
| F412C | Phenylalanine to cysteine | R | S | R | |
| F412V | Phenylalanine to valine | R | S | R | |
| ∂-region C | L501I | Leucine to isoleucine | R | S | R |
| K513E | Lysine to glutamic acid | R | S | R | |
| P522S | Proline to serine | R | S | R | |
| L545S | Leucine to serine | R | S | R | |
| D588E | Aspartic acid to glutamic acid | S | R | S | |
| II | T700A | Threonine to alanine | S | R | S |
| V715M | Valine to methionine | S | R | S | |
| VI | V781I | Valine to isoleucine | S | R | S |
| III | L802M | Leucine to methionine | S | R | S |
| K805Q | Lysine to glutamine | S | S | R | |
| A809V | |||||
| T821I | Threonine to isoleucine | R | S | R | |
| V | A987G | Alanine to glysine | R | S | R |
| Other | S676G | Serine to glysine | S | S | S |
| V759M | Valine to methionine | S | S | S | |
| Multiple | F412C L802M | R | R | R | |
| K513E D588E | R | R | R | ||
| K805Q T821I | R | R | R | ||
Data are from reference 16.
R and S denote resistant and sensitive, respectively.
Effect of UL97 and UL54 Mutations on Antiviral Susceptibilities of CMV
From published data, it is possible to address the effect of UL97 and UL54 mutations on the susceptibility of CMV to antiviral agents. A summary of results of antiviral susceptibility testing of well-characterized CMV laboratory strains is presented in Table 2. Although limited, these data indicate different antiviral susceptibility patterns depending upon whether the CMV strain contains mutations only in UL97, only in UL54, or in both genes. As illustrated in Table 2, CMV strains with only UL97 mutations are resistant to ganciclovir but susceptible to foscarnet and cidofovir, CMV strains with certain UL54 mutations are cross-resistant to ganciclovir and cidofovir, and CMV strains containing UL97 and UL54 mutations (double-mutant strains) are highly resistant to ganciclovir.
Similar antiviral susceptibility patterns have been found among clinical CMV isolates. In one study, characterization of 28 ganciclovir-resistant clinical CMV isolates revealed that low-level resistance to ganciclovir (defined as a ganciclovir IC50 of ≥8 μM and ≤30 μM) was associated with UL97 mutations while high-level resistance to ganciclovir (IC50 of ≥30 μM) and cross-resistance to cidofovir and/or foscarnet was associated with the presence of both UL97 and UL54 mutations (58). In addition, the study found a significant correlation between the level of resistance and the duration of prior ganciclovir therapy. For instance, the mean ganciclovir IC50 for ganciclovir-resistant isolates obtained during the first 9 months of ganciclovir treatment in that study was statistically significantly lower than that for ganciclovir-resistant isolates obtained after at least 9 months of treatment (25 and 54 μM, respectively). The frequency of CMV isolates highly resistant to ganciclovir also increased with the length of therapy, with 19% of the ganciclovir-resistant isolates obtained during the first 9 months being highly resistant to ganciclovir versus 64% of the isolates obtained after at least 9 months of therapy. It has been suggested that these findings support a model in which treatment with ganciclovir initially would select for CMV variants with low-level ganciclovir resistance and UL97 mutations and then would select for a subpopulation of virus acquiring UL54 mutations (in addition to UL97 mutations) and high-level resistance to ganciclovir (58).
LABORATORY METHODS FOR ANTIVIRAL SUSCEPTIBILITY TESTING OF CMV ISOLATES
The recognition that virulent drug-resistant CMV strains could emerge in the clinical setting has important implications, including the need to develop laboratory methods for determining antiviral susceptibilities that can be used in diagnostic laboratories for the detection of resistant viruses. Methods used in the laboratory to determine susceptibilities of CMV isolates to antiviral compounds may be classified as phenotypic or genotypic. Phenotypic methods are designed to determine the concentration of an antiviral agent that would inhibit the virus in culture. Generally, these are culture-based methods in which a known amount of infectious virus is grown in the presence of different concentrations of the antiviral agent. Viral production is then plotted against antiviral concentration to determine the IC50 of the agent for the virus which is the concentration of antiviral agent (expressed as micromolar or micrograms per milliliter) producing 50% inhibition of the virus in culture. On the basis of the results of phenotypic assays, viruses are classified as susceptible or resistant to a given antiviral compound.
Genotypic assays are designed to determine whether mutations known to confer antiviral resistance are present in the genome of the viruses being studied. Generally, genotypic methods involve the use of molecular technologies to obtain sequence information concerning target regions of the viral genome. Genotypic assays are useful when the genetic basis for antiviral resistance has been identified and when there is a predictable relationship between the presence or absence of a genetic variant and measurable antiviral resistance. The significance of mutations found in genotypic studies must be further evaluated by assessing the effect of each mutation on the phenotype of recombinant viruses and on the structure and function of mutated viral proteins. Whereas these latter studies are required for understanding the mechanisms underlying the development of antiviral resistance, antiviral susceptibility methods used in diagnostic laboratories are usually limited to phenotypic and genotypic assays. While currently available methods have some utility for patient management, further work is necessary to develop rapid and reproducible assays that will permit the timely detection of resistant viral isolates in clinical specimens for the purpose of guiding therapeutic decisions.
Phenotypic Methods
A variety of phenotypic assays have been used in different studies to determine antiviral susceptibilities of clinical and laboratory CMV strains. These methods include the plaque reduction assay, assays based on DNA hybridization, ELISA-based assays, and assays based on flow cytometry (33, 43, 50, 67–69). Each of these methods measures the inhibition of viral growth in the presence of antiviral agents. The difference is that the plaque reduction assay measures the inhibition of replication of infectious virus, assays based on DNA hybridization measure the inhibition of viral DNA synthesis, and ELISA- and flow cytometry-based assays measure the inhibition of synthesis of one or more viral proteins. The results of these studies are difficult to compare because a standardized phenotypic assay is not used and different criteria are used to define CMV resistance.
Plaque reduction assay.
The plaque reduction assay has been considered the gold standard for antiviral susceptibility testing of CMV and other viruses (6). In this assay, a standardized inoculum of a stock virus (previously subjected to titer determination in cell culture) is inoculated into cultures and incubated in the presence of the antiviral agent. The cultures are then observed for the presence of viral plaques. The IC50 of the agent for the isolate is defined as the concentration of agent causing a 50% reduction in the number of plaques produced. Plaque reduction assays are labor-intensive, involving the following steps: (i) isolation of the virus in culture, (ii) preparation of viral stocks (by sequential passaging of the viral isolate in culture until an adequate amount of virus is present to perform the assay), (iii) titer determination of viral stocks, and (iv) the actual antiviral susceptibility testing. Therefore, plaque reduction assays are limited by the excessive time required to complete the assay (4 to 6 weeks) and the lack of a standardized method validated across different laboratories. In addition, the continuous passaging of the isolates needed to prepare viral stocks may influence the results of plaque reduction assays by selecting CMV strains that are not representative of the original population of viruses.
DNA hybridization assay.
In the DNA hybridization assay, IC50s of agents for CMV isolates are determined by measuring the effect of the agents on CMV DNA synthesis in culture (21). Cell cultures are inoculated with a standardized amount of CMV and incubated in the presence of different concentrations of antiviral agent until control wells (without drug) show 60 to 80% cytopathic effect (typically after 6 to 8 days in culture). Then the cells are lysed, and whole genomic DNA is extracted and transferred by capillary action onto negatively charged nylon membranes. The membranes are hybridized to an 125I-labelled human CMV probe (Diagnostic Hybrids, Athens, Ohio), rinsed, washed, and counted in a gamma counter for 2 min. Mean hybridization values (in counts per minute [cpm]) for each concentration of antiviral agent are calculated and expressed as a percentage of the cpm in control cultures. The IC50 is defined as the concentration of antiviral agent resulting in a 50% reduction in viral nucleic acid hybridization values compared with the hybridization values of controls. Similarly to plaque reduction assays, DNA hybridization assays require the isolation of CMV strains, sequential passaging of the isolates in culture for preparation of viral stocks, titer determination of viral stocks, and susceptibility testing. There is good correlation between results obtained by DNA hybridization assays and plaque reduction assays (21). One disadvantage of DNA hybridization assays is that they require the use of radiolabelled probes. However, DNA hybridization assays have the advantage over plaque reduction assays of eliminating the variation due to subjective errors resulting from plaque counting by different individuals.
Other phenotypic methods.
A variety of culture-based methods have also been used in different laboratories to determine antiviral susceptibilities of CMV isolates. The principle behind these methods is the same as in plaque reduction assays, but viral production is measured by using immunofluorescence-, immunoperoxidase-, ELISA-, or flow cytometry-based methods for detection and quantitation of cells expressing CMV antigens (immediate-early, early, or late) (33, 50, 67–69). Dose-inhibition curves are then constructed to determine the IC50s of the agents for the isolates studied. These assays are also laborious and have the same limitations as the plaque reduction assays.
To reduce the turnaround time involved in identifying drug-resistant CMV isolates, considerable efforts have been made to develop rapid screening assays as well as methods that can be used to determine susceptibilities directly in clinical specimens or in primary cultures. A modified plaque reduction assay has been used for susceptibility testing of CMV isolates directly in primary cultures of clinical specimens after a single replication cycle of the virus. This method provided susceptibility results in 5 days, clearly differentiated between susceptible and resistant CMV isolates, and compared favorably with a conventional plaque reduction assay (52). In another study, blood leukocytes from patients with documented CMV viremia were used as the inoculum in a modified plaque reduction assay (34). This method provided results within 4 to 6 days and reliably detected ganciclovir- and foscarnet-resistant CMV isolates. A practical limitation of this method is that the level of CMV viremia in patients is often too low to allow it to be used. Additional studies are needed to evaluate the applicability and reproducibility of these rapid phenotypic assays across different laboratories.
Practical considerations.
Interpretation of the results of antiviral resistance studies is difficult because the different susceptibility assay systems for viruses are not well standardized and disparate results can be obtained when different methods are used. Table 5 summarizes published IC50s of drugs for well-characterized reference CMV strains in experiments performed in different laboratories by using a variety of phenotypic assays. Table 6 summarizes published IC50s used in different studies to define antiviral resistance of clinical CMV isolates. These results clearly illustrate that there is significant variability among methods and laboratories. Variables that affect the results of susceptibility testing include the type of cell culture used, the size of the viral inoculum, the method used, and the laboratory performing the assay. When feasible, a baseline viral isolate from the same patient should be tested in parallel with the isolate suspected of being resistant. Unfortunately, baseline isolates are often not available. Antiviral susceptibility assays should also include well-characterized susceptible and resistant reference isolates.
TABLE 5.
Antiviral susceptibilities of reference CMV strains
| Methoda | Ganciclovir IC50 (μM) to:
|
Foscarnet IC50 (μM) to:
|
Cidofovir IC50 (μM) to:
|
Reference | |||
|---|---|---|---|---|---|---|---|
| AD169b | 759rD100c | AD169 | 759rD100 | AD169 | 759rD100 | ||
| PRA | 3.1 | 17 | |||||
| 1 | 12 | 150 | 150 | 6 | |||
| 4.9 | 35 | 64 | 24 | 0.6 | 61 | ||
| 4 | 210 | 0.54 | 64 | ||||
| 10 | 250 | 1 | 47 | ||||
| 5.3 | 58.9d | 52 | |||||
| 2.5 | 55 | 10 | |||||
| 10 | 200 | 0.5 | 48 | ||||
| 0.6 | 10 | ||||||
| DNA | 1.7 | 39 | 50 | 35 | 0.49 | 1.5 | 61 |
| 0.7 | 19 | 8 | |||||
| LARA | 6.5 | 31.6d | 52 | ||||
| ELISA | 5.4 | 150 | 67 | ||||
| 6.25 | 210 | 1 | 47 | ||||
PRA, plaque reduction assay; DNA, DNA hybridization assay; LARA, late-antigen reduction assay.
Susceptible CMV strain that contains UL97 and DNA polymerase wild-type sequences.
Ganciclovir-resistant CMV strain that contains a UL97 deletion and a DNA polymerase mutation.
Ganciclovir-resistant strain RCL1 was used.
TABLE 6.
Ganciclovir, foscarnet, and cidofovir IC50s used in clinical studies to define resistant CMV isolates
In an effort to reduce assay variability and to reach a consensus on the definition of antiviral resistance in CMV, the AIDS Clinical Trials Group (ACTG) has evaluated a consensus plaque reduction assay by testing a panel of well-characterized clinical and laboratory CMV strains in 13 laboratories across the United States. Preliminary results indicate that susceptible and resistant CMV isolates can be readily distinguished by the consensus assay (19). It has been proposed to use a ganciclovir IC50 of >12 μM, a foscarnet IC50 of >400 μM, and a cidofovir IC50 of >2 μM as the cutoff values that define CMV resistance to these compounds (19, 24). More recently, a more accurate cutoff of >4 μM has been set to define resistance to cidofovir (11). CMV strains for which the ganciclovir IC50 is >6 μM and <12 μM are considered to be of an “intermediate-resistance” phenotype and have decreased susceptibility to this compound.
Genotypic Methods
The recognition that specific mutations in the UL97 and UL54 genes of CMV are associated with resistance of the virus to antiviral compounds has led to the development of genotypic methods for the detection of mutant CMV isolates.
The strong clustering of mutations at specific UL97 codons in ganciclovir-resistant CMV isolates has led to the development of diagnostic screening assays based on restriction enzyme analysis of selected PCR products amplified from CMV-infected cell cultures (12, 36). Mutations at codons 460 and 594 result in the loss of naturally occurring restriction enzyme sites, whereas mutations at codons 520 and 595 result in additional restriction sites. The presence of mutations is determined by distinctive restriction patterns visualized by gel electrophoresis (Table 7). This screening method can detect mutants when they reach 10% of the viral population in a clinical isolate; in one study, it identified 78% of the isolates resistant to ganciclovir in phenotypic assays (13). A major advantage of this restriction digestion analysis is that provisional susceptibility information can be obtained in less than 2 days and that it can be used directly with clinical samples (i.e., blood leukocytes, plasma, cerebrospinal fluid, aqueous humor). A disadvantage is that this method may not detect specific mutations involving base changes that do not result in a change in the restriction enzyme pattern. In addition, it will “miss” ganciclovir-resistant CMV isolates containing DNA polymerase mutations but wild-type UL97 sequences (30). Therefore, while a positive result is virtually diagnostic of ganciclovir resistance, a negative result does not necessarily indicate that a virus is susceptible to ganciclovir. DNA polymerase mutations are detected by sequencing the relevant portions of the polymerase-encoding gene. This procedure is currently performed only in a few research laboratories. Restriction enzyme analysis of PCR fragments has been recently used to detect the ganciclovir resistance mutation L501F and to detect the ganciclovir and foscarnet resistance mutation A809V in the DNA polymerase region of clinical CMV isolates (15, 37).
TABLE 7.
Restriction enzyme digestion screening for UL97 mutations related to ganciclovir resistance
| Enzyme | UL97 Codon | Fragment size (bp)a
|
|
|---|---|---|---|
| Wild-type sequence | Mutant sequence | ||
| NlaIII | 460 | 198, 168, 126, 9 | 324, 168, 9 |
| AluI | 520 | 295, 189, 17 | 268, 189, 27, 17 |
| HhaI | 594 | 50, 38, 18, 12 | 62, 38, 18 |
| MseI | 595 | 76, 42 | 46, 42, 30 |
| TaqI | 595 | 99, 19 | 71, 28, 19 |
Underlined fragments are normally seen in polyacrylamide gels after ethidium bromide staining.
Studies combining both genotypic and phenotypic assays to characterize selected CMV strains are crucial to further our understanding of CMV resistance. However, it should be noted that viruses containing UL97 or UL54 mutations causing antiviral resistance can be susceptible in phenotypic assays. For instance, a clinical CMV isolate with a A594V mutation in UL97 was susceptible to ganciclovir by the plaque reduction assay in one study (70). In another study, a CMV isolate, obtained from a bone marrow transplant recipient, that contained an A594V mutation in UL97 was susceptible to ganciclovir in a DNA hybridization assay (31). Similarly, the foscarnet resistance T700A mutation in UL54 has been found in a foscarnet-susceptible isolate (30). Because the T700A mutation has been associated with a slow growth of CMV in culture, it was proposed that the presence of this mutation could introduce a bias in the evaluation of antiviral susceptibility assays since susceptible viruses could mask the slow-growing resistant mutants in culture (3). The discrepancy between a resistant genotype (i.e., with a mutant UL97 or UL54 sequence) and a susceptible phenotype could indicate that the sensitivity of susceptibility assays for the detection of isolates containing antiviral resistance mutations is lower than that of PCR sequencing methods. Because immunocompromised patients can harbor multiple isolates, it is also possible that culture-based susceptibility assays will select a different virus population from that selected by PCR and characterized in sequencing studies (31). Nevertheless, phenotypic testing of viruses with known UL97 and UL54 mutations is important to define cross-resistance patterns and to determine the importance of individual mutations in the CMV genome by assessing their effect on the phenotype of mutant viruses. Identification of mutations conferring high-level drug resistance (in phenotypic assays) would help in the design of rapid assays targeted to detect highly resistant CMV isolates.
INCIDENCE AND CLINICAL SIGNIFICANCE OF INFECTIONS CAUSED BY DRUG-RESISTANT CMV
The vast majority of infections caused by drug-resistant CMV have been found in patients with AIDS. CMV resistant to antiviral agents has also been recovered from other immunocompromised hosts, including patients with primary immunodeficiencies or hematologic disorders and recipients of bone marrow, heart, and lung allografts (1, 27, 29, 37, 49, 56, 72). Because of the underlying disorders, it has been difficult to determine whether the outcome of these infections is due to the poor immune status of the host, the presence of CMV resistant to antiviral therapy, or both. Nevertheless, abundant data in the literature suggest that resistance of CMV to ganciclovir is associated with lack of therapeutic response and progression of CMV disease. Isolation of ganciclovir-resistant CMV or detection of ganciclovir resistance UL97 and/or UL54 mutations in viral isolates or directly in clinical samples from patients with AIDS has been associated with recurrent or progressive CMV retinitis despite therapy (2, 14, 15, 50, 59, 70), occurrence of contralateral eye disease among patients with unilateral CMV retinitis (39), progressive extraocular CMV disease, development of CMV disease of the central nervous system while on therapy for CMV retinitis (57, 71), persistent isolation or detection of CMV in blood leukocyte fractions, persistent detection of CMV DNA in plasma fractions (57, 70), and increase in the CMV DNA burden in blood leukocyte and plasma fractions (9, 35, 57).
That ganciclovir-resistant CMV strains are pathogenic is further supported by the detection of ganciclovir resistance UL97 and/or UL54 mutations in CMV isolates from gastrointestinal tissue (36), cerebrospinal fluid (71), and bronchoalveolar lavage fluid (1, 49). The ganciclovir resistance UL97 and UL54 mutations have also been found by direct analysis in samples of vitreous fluid and brain tissue from patients with end-organ CMV disease (37, 59). Although the information on the clinical outcome of infections caused by foscarnet- and cidofovir-resistant CMV is limited, available data suggest that these infections are also associated with progressive CMV disease and therapeutic failure. In one study, failure of intravitreal cidofovir therapy in patients with AIDS and CMV retinitis previously treated with systemic ganciclovir or intravenous cidofovir was associated with the isolation of cidofovir- and ganciclovir-resistant isolates containing mutations in the UL97 and UL54 genes (59).
There is limited information about the in vivo persistence of CMV strains containing UL97 or DNA polymerase mutations. In one study (70), a previously present M460V UL97 mutation was absent from a plasma sample obtained 6 months after cessation of ganciclovir therapy. In contrast, a previously detected L595F UL97 mutation was still present in a plasma sample obtained from another patient 9 months after cessation of ganciclovir therapy (70). In another study (30), a CMV strain with a ganciclovir-resistant phenotype and an M460V mutation in UL97 persisted for a prolonged period, even when the patient was no longer being treated with ganciclovir. These findings suggest that CMV strains with certain UL97 (or DNA polymerase mutations) may be stable in vivo even in the absence of selective antiviral pressure.
As indicated above, the majority of infections caused by drug-resistant CMV have been found in patients with AIDS. The frequency of these infections is not well known and has varied among different studies. In one study that analyzed the prevalence of infections caused by ganciclovir-resistant CMV strains in 31 patients with AIDS and CMV retinitis, 5 (38%) of 13 patients who were treated with intravenous ganciclovir for more than 3 months were excreting ganciclovir-resistant CMV strains in their urine (23). This represented an overall incidence of ganciclovir resistance of 7.8% in this patient population. In a more recent study of 207 patients with AIDS and newly diagnosed CMV retinitis who were enrolled in a randomized trial comparing intravenous ganciclovir and foscarnet, ganciclovir-resistant CMV was found in 5.3% of the patients who remained viremic after being treated with ganciclovir for a minimum of 2.5 months (62). In a prospective study of 76 patients with AIDS and CMV retinitis treated initially with intravenous ganciclovir as the initial induction and maintenance therapy, 11.4% of the patients had ganciclovir-resistant CMV isolated from blood or urine after 6 months of treatment and 27.5% of the patients had ganciclovir-resistant CMV in their blood or urine after 9 months of therapy (39). In another study, ganciclovir-resistance UL97 mutations were found directly in blood leukocytes of 31% of patients with AIDS and CMV retinitis treated with ganciclovir for at least 3 months (35).
The incidence of ganciclovir-resistant CMV infections in patients treated with oral ganciclovir has been analyzed in several studies. In a randomized trial comparing oral ganciclovir to placebo for prevention of CMV disease in 725 human immunodeficiency virus- and CMV-seropositive individuals, the prevalence of isolates resistant to ganciclovir among individuals who remained culture positive after being treated for a minimum of 3 months was <1% (19, 26). In another study, ganciclovir resistance UL97 mutations were found in plasma samples from 4.8% of 117 AIDS patients who had received prophylaxis or preemptive therapy with oral ganciclovir for 12 months (60). In that study, the detection of these UL97 mutations in plasma was associated with clinical progression (60).
Data on the incidence of foscarnet- or cidofovir-resistant CMV infections in AIDS patients are more limited, but available information suggests that the incidence of infections caused by CMV strains resistant to these compounds is low. For instance, foscarnet-resistant isolates were not found in patients with AIDS and CMV retinitis with persistent viremia who had been treated with foscarnet for a median of 5.1 months (62). In one study, cidofovir-resistant isolates were obtained from 2 (3%) of 64 patients with AIDS and CMV retinitis who received intraocular cidofovir injections for the treatment of recurrent CMV retinitis (59). However, the frequency of cidofovir-resistant CMV infections might be higher among patients harboring ganciclovir-resistant CMV strains. In a retrospective analysis of 28 ganciclovir-resistant CMV isolates, resistance to cidofovir was found in 11 (39%) of the isolates (58). All these cidofovir-resistant isolates were highly resistant to ganciclovir and had been recovered from patients after extended treatment with ganciclovir. Although these data do not predict the incidence of cidofovir resistance in the clinical setting, they help to identify the appropriate background (high level of ganciclovir resistance; prolonged prior therapy with ganciclovir) where cidofovir resistance should be considered.
Although the frequency of infections caused by drug-resistant CMV in solid-organ transplant recipients is not known, it is probably low. In one study, none of 33 blood or tissue CMV isolates recovered from solid-organ transplant recipients who developed CMV viremia after receiving prophylaxis and/or treatment with intravenous ganciclovir were resistant to this compound (8). Reports of drug-resistant CMV infections in this patient population remain anecdotal (1, 49).
The limited information available on the incidence of drug-resistant CMV infections in the context of bone marrow transplantation suggests that it is not a frequent problem. In one study, ganciclovir-resistant CMV isolates were recovered from the blood and bronchoalveolar lavage fluid of a patient treated with ganciclovir for more than 2 months (27). In a survey of the European Group for Blood and Bone Marrow Transplantation, 23 patients with ganciclovir-resistant CMV infections were reported from 19 of the 68 participating centers (55). However, resistance to ganciclovir was documented in only 2 cases. In another study (56), a ganciclovir-resistant CMV isolate was recovered from a lung autopsy sample from a bone marrow transplant recipient with a second episode of CMV pneumonitis who had received acyclovir prophylaxis and two courses of ganciclovir therapy for a total of 55 days. In a recent study, CMV isolates containing UL97 mutations associated with resistance to ganciclovir were recovered after 41 and 73 days of ganciclovir therapy in two patients who had previously received prophylaxis with acyclovir (31). Although the incidence of drug-resistant CMV infections in patients with primary immunodeficiencies is not known, a recent report suggest that ganciclovir-resistant CMV strains can emerge rapidly (within 3 weeks of therapy initiation) in patients with primary combined immunodeficiency and CMV infection who are treated with ganciclovir (72).
Because acyclovir has the same mechanism of activation and action as ganciclovir against CMV (66), it is theoretically possible that CMV isolates resistant to ganciclovir will emerge under selective pressure during exposure to acyclovir. Whether prior exposure to acyclovir contributes to the selection of ganciclovir-resistant CMV in the clinical setting is not clear. In one study, a ganciclovir-resistant isolate was recovered prior to ganciclovir therapy in a patient with chronic leukemia who had been treated intermittently with intravenous acyclovir. This isolate contained a V594 mutation in UL97 (30). In another study, a CMV isolate with reduced susceptibility to ganciclovir was recovered from a solid-organ transplant recipient after a 14-day course of treatment with acyclovir (17). Other studies have suggested that antiviral prophylaxis does not result in the selection of resistant CMV isolates. For instance, none of six CMV isolates recovered from the urine of six patients with AIDS treated with acyclovir for an average of 15 months were resistant to ganciclovir (25). In another study, none of 17 CMV isolates from solid-organ transplant recipients who developed active CMV infections after receiving prophylaxis with acyclovir were resistant to ganciclovir (8). Confirmation of whether drug-resistant CMV strains are selected for during prior prophylaxis with acyclovir will require additional studies.
CONCLUSIONS
Resistance of CMV to antiviral agents is a well-documented complication of long-term antiviral therapy. This problem has been observed mostly in patients with AIDS and CMV retinits (before the advent of highly active antiretroviral therapy), in whom drug-resistant CMV infections have been associated with clinical progression and therapeutic failure. Current data suggest that the incidence of infections caused by drug-resistant CMV in bone marrow or solid-organ transplant recipients is low. Because of the routine use of antiviral agents for prophylaxis or preemptive therapy for CMV infections in bone marrow or solid-organ transplant recipients, surveillance antiviral susceptibility studies of CMV isolates from patients developing active CMV infections would be appropriate. CMV resistance studies have been limited by the difficulty and lack of standardization of antiviral susceptibility assays. In contrast, significant advances have been made in recent years in understanding the mechanisms by which CMV becomes resistant to antiviral agents. The recognition that specific mutations in the UL97 and UL54 genes of CMV are associated with different antiviral susceptibility patterns has prompted the development of molecular laboratory methods for detection of mutant viral sequences in viral isolates and directly in clinical specimens. The incorporation of these methods in the routine of diagnostic virology laboratories in the future should contribute to a more rapid and sensitive detection of drug-resistant CMV.
ACKNOWLEDGMENTS
This study was supported by the NIH (grant AI-27761), Minnesota Medical Foundation, and International Center for Antiviral Research and Epidemiology (ICARE).
REFERENCES
- 1.Alain S, Honderlick P, Grenet D, Stern M, Vadam C, Sanson-Le Pors M, Mazeron M. Failure of ganciclovir treatment associated with selection of a ganciclovir-resistant cytomegalovirus strain in a lung transplant recipient. Transplantation. 1997;63:1533–1536. doi: 10.1097/00007890-199705270-00031. [DOI] [PubMed] [Google Scholar]
- 2.Baldanti F, Silini E, Sarasini A, Talarico C L, Stanat S C, Biron K K, Furione M, Bono F, Palú G, Gerna G. A three-nucleotide deletion in the UL97 open reading frame is responsible for the ganciclovir resistance of a human cytomegalovirus clinical isolate. J Virol. 1995;69:796–800. doi: 10.1128/jvi.69.2.796-800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldanti F, Underwood M R, Stanat S C, Biron K K, Chou S, Sarasini A, Silini E, Gerna G. Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype, while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double-resistant human cytomegalovirus strains recovered from patients with AIDS. J Virol. 1996;70:1390–1395. doi: 10.1128/jvi.70.3.1390-1395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldanti F, Underwood M R, Talarico C L, Simoncini L, Sarasini A, Biron K K, Gerna G. The Cys607Tyr change in the UL97 phosphotransferse confers ganciclovir resistance to two human cytomegalovirus strains recovered from two immunocompromised individuals. Antimicrob Agents Chemother. 1998;42:444–446. doi: 10.1128/aac.42.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balfour H H., Jr Management of cytomegalovirus disease with antiviral drugs. Rev Infect Dis. 1990;12(Suppl. 7):S849–S860. doi: 10.1093/clinids/12.supplement_7.s849. [DOI] [PubMed] [Google Scholar]
- 6.Biron K K, Fyfe J A, Stanat S C, Leslie L K, Sorrell J B, Lambe C U, Coen D M. A human cytomegalovirus mutant resistant to the nucleoside analog 9-{[2-hydroxy-1-(hydroxymethyl)ethoxy]methyl}guanine (BW B759U) induces reduced levels of BW B759U triphosphate. Proc Natl Acad Sci USA. 1986;83:8769–8773. doi: 10.1073/pnas.83.22.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biron K K. Cytomegalovirus: genetics of drug resistance. In: Mills J, editor. Antiviral chemotherapy. Vol. 4. New York, N.Y: Plenum Press; 1996. pp. 135–143. [DOI] [PubMed] [Google Scholar]
- 8.Boivin G, Erice A, Crane D D, Dunn D L, Balfour H H., Jr Ganciclovir susceptibilities of cytomegalovirus (CMV) isolates from solid organ transplant recipients with CMV viremia after antiviral prophylaxis. J Infect Dis. 1993;168:332–335. doi: 10.1093/infdis/168.2.332. [DOI] [PubMed] [Google Scholar]
- 9.Boivin G, Chou S, Quirk M R, Erice A, Jordan M C. Detection of ganciclovir resistance mutations and quantitation of cytomegalovirus (CMV) DNA in leukocytes of patients with fatal disseminated CMV disease. J Infect Dis. 1996;173:523–528. doi: 10.1093/infdis/173.3.523. [DOI] [PubMed] [Google Scholar]
- 10.Cherrington J M, Miner R, Hitchcock M J M, Lalezari J P, Drew W L. Susceptibility of human cytomegalovirus to cidofovir is unchanged after limited in vivo exposure to various clinical regimens of drug. J Infect Dis. 1996;173:987–992. doi: 10.1093/infdis/173.4.987. [DOI] [PubMed] [Google Scholar]
- 11.Cherrington J M, Fuller M D, Lamy P D, Miner R, Lalezari J P, Nuessle S, Drew W L. In vitro antiviral susceptibilities of isolates from CMV retinitis patients receiving first or second line cidofovir therapy: relationship to clinical outcome. J Infect Dis. 1998;178:1821–1825. doi: 10.1086/314487. [DOI] [PubMed] [Google Scholar]
- 12.Chou S, Erice A, Jordan M C, Vercellotti G M, Michels K R, Talarico C L, Stanat S C, Biron K K. Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J Infect Dis. 1995;171:576–583. doi: 10.1093/infdis/171.3.576. [DOI] [PubMed] [Google Scholar]
- 13.Chou S, Guentzel S, Michels K R, Miner R C, Drew W L. Frequency of UL97 phosphotransferase mutations related to ganciclovir resistance in clinical cytomegalovirus isolates. J Infect Dis. 1995;172:239–242. doi: 10.1093/infdis/172.1.239. [DOI] [PubMed] [Google Scholar]
- 14.Chou S, Marousek G, Guentzel S, Follansbee S E, Poscher M E, Lalezari J P, Miner R C, Drew W L. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J Infect Dis. 1997;176:786–789. doi: 10.1086/517302. [DOI] [PubMed] [Google Scholar]
- 15.Chou S, Marousek G, Parenti D M, Gordon S M, LaVoy A G, Ross J G, Miner R C, Drew W L. Mutation in region II of the DNA polymerase gene conferring foscarnet resistance in cytomegalovirus isolates from 3 subjects receiving prolonged antiviral therapy. J Infect Dis. 1998;178:526–530. doi: 10.1086/515648. [DOI] [PubMed] [Google Scholar]
- 16.Cihlar T, Fuller M D, Cherrington J M. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J Virol. 1998;72:5927–5936. doi: 10.1128/jvi.72.7.5927-5936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole N L, Balfour H H., Jr In vitro susceptibility of cytomegalovirus isolates from immunocompromised patients to acyclovir and ganciclovir. Diagn Microbiol Infect Dis. 1987;6:255–261. doi: 10.1016/0732-8893(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 18.Chrisp P, Clissold S P. Foscarnet: a review of its antiviral activity, pharmacokinetic properties, and therapeutic use in immunocompromised patients with CMV retinitis. Drugs. 1991;41:104–129. doi: 10.2165/00003495-199141010-00009. [DOI] [PubMed] [Google Scholar]
- 19.Crumpacker C S, Baldanti F, Boeck M, Chou S, Danner S, Drew W L, Emanuel D, Erice A, Hardy W D, Spector S A. Drug resistance in cytomegalovirus: current knowledge and implications for patient management. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;12(Suppl. 1):S1–S22. [PubMed] [Google Scholar]
- 20.Crumpacker C S. Ganciclovir. N Engl J Med. 1996;335:721–729. doi: 10.1056/NEJM199609053351007. [DOI] [PubMed] [Google Scholar]
- 21.Dankner W M, Scholl D, Stanat S C, Martin M, Sonke R L, Spector S A. Rapid antiviral DNA-DNA hybridization assay for human cytomegalovirus. J Virol Methods. 1990;28:293–298. doi: 10.1016/0166-0934(90)90122-v. [DOI] [PubMed] [Google Scholar]
- 22.De Clercq E. Therapeutic potential of HPMPC as an antiviral drug. Rev Med Virol. 1993;3:85–96. [Google Scholar]
- 23.Drew W L, Miner R C, Busch D F, Follansbee S E, Gullett J, Mehalko S G, Gordon S M, Owen W F, Jr, Matthews T R, Buhles W C, DeArmond B. Prevalence of resistance in patients receiving ganciclovir for serious cytomegalovirus infections. J Infect Dis. 1991;163:716–719. doi: 10.1093/infdis/163.4.716. [DOI] [PubMed] [Google Scholar]
- 24.Drew W L, Miner R, Saleh E. Antiviral susceptibility testing of cytomegalovirus: criteria for detecting resistance to antivirals. Clin Diagn Virol. 1993;1:179–185. doi: 10.1016/0928-0197(93)90012-t. [DOI] [PubMed] [Google Scholar]
- 25.Drew W L, Anderson R, Lang W, Miner R C, Davis G, Lalezari J. Failure of high-dose oral acyclovir to suppress CMV viruria or induce ganciclovir-resistant CMV in HIV antibody positive patients. J Acquired Immune Defic Syndr Hum Retrovirol. 1995;8:289–291. doi: 10.1097/00042560-199503010-00010. [DOI] [PubMed] [Google Scholar]
- 26.Drew W L, Miner R C, Crager M, Stempien M J. Program and Abstracts of the 35th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1995. Does resistance to ganciclovir develop in patients receiving prophylactic drug? abstr. H135; p. 203. [Google Scholar]
- 27.Drobyski W R, Knox K K, Carrigan D R, Ash R C. Foscarnet therapy for ganciclovir-resistant cytomegalovirus in marrow transplantation. Transplantation. 1991;52:155–157. [PubMed] [Google Scholar]
- 28.Erice A, Jordan M C, Chace B A, Fletcher C, Chinnok B J, Balfour H H., Jr Ganciclovir treatment of cytomegalovirus disease in transplant recipients and other immunocompromised hosts. JAMA. 1987;257:3082–3087. [PubMed] [Google Scholar]
- 29.Erice A, Chou S, Biron K K, Stanat S C, Balfour H H, Jr, Jordan M C. Progressive disease due to ganciclovir-resistant cytomegalovirus in immunocompromised patients. N Engl J Med. 1989;320:289–293. doi: 10.1056/NEJM198902023200505. [DOI] [PubMed] [Google Scholar]
- 30.Erice A, Gil-Roda C, Pérez J, Balfour H H, Jr, Sannerud K J, Hanson M N, Boivin G, Chou S. Antiviral susceptibilities and analysis of UL97 and DNA polymerase sequences of clinical cytomegalovirus isolates from immunocompromised patients. J Infect Dis. 1997;175:1087–1092. doi: 10.1086/516446. [DOI] [PubMed] [Google Scholar]
- 31.Erice A, Borrell N, Li W, Miller W J, Balfour H H., Jr Ganciclovir susceptibilities and analysis of UL97 region in cytomegalovirus isolates from bone marrow recipients with CMV disease after antiviral prophylaxis. J Infect Dis. 1998;178:531–534. doi: 10.1086/517467. [DOI] [PubMed] [Google Scholar]
- 32.Field A K, Davies M E, DeWitt C, Perry H C, Liou R, Germershausen J, Karkas J D, Ashton W T, Johnston D B, Tolman R L. 9-[2-Hydroxy-1(hydroxymethyl)ethoxy]methyl guanine: a selective inhibitor of herpes group virus replication. Proc Natl Acad Sci USA. 1983;80:4139–4143. doi: 10.1073/pnas.80.13.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerna G, Baldanti F, Zavattoni M, Sarasini A, Percivalle E, Revello M G. Monitoring of ganciclovir sensitivity of multiple human cytomegalovirus strains coinfecting blood of an AIDS patient by an immediate-early antigen plaque assay. Antiviral Res. 1992;19:333–345. doi: 10.1016/0166-3542(92)90014-v. [DOI] [PubMed] [Google Scholar]
- 34.Gerna G, Sarasini A, Percivalle E, Zavattoni M, Baldanti F, Revello M G. Rapid screening for resistance to ganciclovir and foscarnet of primary isolates of human cytomegalovirus from culture-positive blood samples. J Clin Microbiol. 1995;33:738–741. doi: 10.1128/jcm.33.3.738-741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert C, Handfield J, Toma E, Lalonde R, Bergeron M G, Boivin G. Emergence and prevalence of cytomegalovirus mutations associated with ganciclovir resistance in AIDS patients. AIDS. 1998;12:125–129. doi: 10.1097/00002030-199802000-00001. [DOI] [PubMed] [Google Scholar]
- 36.Hanson M N, Preheim L C, Chou S, Talarico C L, Biron K K, Erice A. Novel mutation in the UL97 gene of a clinical cytomegalovirus strain conferring resistance to ganciclovir. Antimicrob Agents Chemother. 1995;39:1204–1205. doi: 10.1128/aac.39.5.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harada K, Eizuru Y, Isashiki Y, Ihara S, Minamishima Y. Genetic analysis of a clinical isolate of human cytomegalovirus exhibiting resistance against both ganciclovir and cidofovir. Arch Virol. 1997;142:215–225. doi: 10.1007/s007050050072. [DOI] [PubMed] [Google Scholar]
- 38.He Z, He Y, Kim Y, Chu L, Ohmstede C, Biron K K, Coen D M. The human cytomegalovirus UL97 protein is a protein kinase that autophosphorylates on serines and threonines. J Virol. 1997;71:405–411. doi: 10.1128/jvi.71.1.405-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jabs D A, Enger C, Dunn J P, Forman M. Cytomegalovirus retinitis and viral resistance: ganciclovir resistance. J Infect Dis. 1998;177:770–773. doi: 10.1086/514249. [DOI] [PubMed] [Google Scholar]
- 40.Jacobson M A, Mills J. Serious cytomegalovirus disease in the acquired immunodeficiency syndrome (AIDS) Ann Intern Med. 1988;108:585–594. doi: 10.7326/0003-4819-108-4-585. [DOI] [PubMed] [Google Scholar]
- 41.Jacobson M A. Treatment of cytomegalovirus retinitis in patients with the acquired immunodeficiency syndrome. N Engl J Med. 1997;337:105–114. doi: 10.1056/NEJM199707103370207. [DOI] [PubMed] [Google Scholar]
- 42.Kemble G, Duke G, Winter R, Spaete R. Defined large-scale alteration of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J Virol. 1996;70:2044–2048. doi: 10.1128/jvi.70.3.2044-2048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimberlin D W, Spector S A, Hill E L, Biron K K, Hay A J, Mayers D L, Whitley R J. Assays for antiviral drug resistance. Antiviral Res. 1995;26:403–413. doi: 10.1016/0166-3542(95)00028-k. [DOI] [PubMed] [Google Scholar]
- 44.Lalezari J P, Drew W L, Glutzer E, James C, Miner R C, Flaherty J, Fisher P E. (S)-1-[3-hydroxy-2-(phosphonyl-methoxy)propyl]cytosine (cidofovir): results of a phase I/II study of a novel nucleotide analogue. J Infect Dis. 1995;171:788–796. doi: 10.1093/infdis/171.4.788. [DOI] [PubMed] [Google Scholar]
- 45.Lalezari J P, Stagg R J, Kuppermann B D, Holland G N, Kramer F, Ives D, Youle M, Robinson M R, Drew W L, Jaffe H S. Intravenous cidofovir for peripheral cytomegalovirus retinitis in patients with AIDS. Ann Intern Med. 1997;126:257–263. doi: 10.7326/0003-4819-126-4-199702150-00001. [DOI] [PubMed] [Google Scholar]
- 46.Littler E, Stuart A D, Chee M S. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature. 1992;358:160–162. doi: 10.1038/358160a0. [DOI] [PubMed] [Google Scholar]
- 47.Lurain N S, Thompson K D, Holmes E W, Read G S. Point mutations in the DNA polymerase gene of human cytomegalovirus that result in resistance to antiviral agents. J Virol. 1992;66:7146–7152. doi: 10.1128/jvi.66.12.7146-7152.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lurain N S, Spafford L E, Thompson K D. Mutation in the UL97 open reading frame of human cytomegalovirus strains resistant to ganciclovir. J Virol. 1994;68:4427–4431. doi: 10.1128/jvi.68.7.4427-4431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lurain N S, Ammons H C, Kapell K S, Yeldandi V V, Garrity E R, O’Keefe J P. Molecular analysis of human cytomegalovirus strains from two lung transplant recipients with the same donor. Transplantation. 1996;62:497–502. doi: 10.1097/00007890-199608270-00012. [DOI] [PubMed] [Google Scholar]
- 50.McSharry J M, Lurain N S, Drusano G L, Landay A, Manischewitz J, Notka M, O’Gorman M, Shapiro H M, Weinberg A, Reichelderfer P, Crumpacker C. Flow cytometry determination of ganciclovir susceptibilities of human cytomegalovirus isolates. J Clin Microbiol. 1998;36:958–964. doi: 10.1128/jcm.36.4.958-964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller W, Flynn P, McCullough J, Balfour H H, Jr, Goldman A, Haake R, McGlave P, Ramsay N, Kersey J. Cytomegalovirus infection after bone marrow transplantation: an association with acute graft-v-host disease. Blood. 1986;67:1162–1167. [PubMed] [Google Scholar]
- 52.Pepin J M, Simon F, Dussault A, Collin G, Dazza M, Brun-Vezinet F. Rapid determination of human cytomegalovirus susceptibility to ganciclovir directly from clinical specimen primocultures. J Clin Microbiol. 1992;30:2917–2920. doi: 10.1128/jcm.30.11.2917-2920.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson P K, Balfour H H, Jr, Marker S C, Fryd D S, Howard R J, Simmons R L. Cytomegalovirus disease in renal allograft recipients: a prospective study of the clinical features, risk factors, and impact on renal transplantation. Medicine. 1980;59:283–300. [PubMed] [Google Scholar]
- 54.Reid R, Mar E C, et al. Insertion and extension of acyclic, dideoxy, and ara nucleotides by herpesviridae and human polymerases. Antimicrob Agents Chemother. 1988;25:191–194. [PubMed] [Google Scholar]
- 55.Reusser P, Cordonnier C, Einsele H, Engelhard D, Link H, Locasciulli A, Ljungman P. European survey of herpesvirus resistance to antiviral drugs in bone marrow transplant recipients. Bone Marrow Transplant. 1996;17:813–817. [PubMed] [Google Scholar]
- 56.Slavin M A, Bindra R R, Gleaves C A, Pettinger M B, Bowden R A. Ganciclovir sensitivity of cytomegalovirus at diagnosis and during treatment of cytomegalovirus pneumonia in marrow transplant recipients. Antimicrob Agents Chemother. 1993;37:1360–1363. doi: 10.1128/aac.37.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith I L, Shinkai M, Freeman W R, Spector S A. Polyradiculopathy associated with ganciclovir-resistant cytomegalovirus in an AIDS patient: phenotypic and genotypic characterization of sequential virus isolates. J Infect Dis. 1996;173:1481–1484. doi: 10.1093/infdis/173.6.1481. [DOI] [PubMed] [Google Scholar]
- 58.Smith I L, Cherrington J M, Jiles R E, Fuller M D, Freeman W R, Spector S A. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J Infect Dis. 1997;176:69–77. doi: 10.1086/514041. [DOI] [PubMed] [Google Scholar]
- 59.Smith I L, Taskintuna I, Rahhal F M, Powell H C, Ai E, Mueller A J, Spector S A, Freeman W E. Clinical failure of CMV retinitis with intravitreal cidofovir is associated with antiviral resistance. Arch Ophthalmol. 1998;116:178–185. doi: 10.1001/archopht.116.2.178. [DOI] [PubMed] [Google Scholar]
- 60.Smith I L, Hong C, Pilcher M L, Shapiro A M, Jiles R E, Spector S A. Program and Abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. Development of resistant cytomegalovirus genotypes during oral ganciclovir prophylaxis/preemptive therapy, abstr. H120; p. 349. [Google Scholar]
- 61.Stanat S C, Reardon J E, Erice A, Jordan M C, Drew W L, Biron K K. Ganciclovir-resistant cytomegalovirus clinical isolates: mode of resistance to ganciclovir. Antimicrob Agents Chemother. 1991;35:2191–2197. doi: 10.1128/aac.35.11.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Studies of Ocular Complications of AIDS (SOCA) in collaboration with the AIDS Clinical Trials Group. Cytomegalovirus (CMV) culture results, drug resistance, and clinical outcome in patients with AIDS and CMV retinitis treated with foscarnet or ganciclovir. J Infect Dis. 1997;176:50–58. doi: 10.1086/514039. [DOI] [PubMed] [Google Scholar]
- 63.Sullivan V, Talarico C L, Stanat S C, Davis M, Coen D M, Biron K K. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]
- 64.Sullivan V, Coen D M. Isolation of foscarnet-resistant human cytomegalovirus patterns of resistance and sensitivity to other antiviral drugs. J Infect Dis. 1991;164:781–784. doi: 10.1093/infdis/164.4.781. [DOI] [PubMed] [Google Scholar]
- 65.Sullivan V, Biron K K, Talarico C, Stanat S C, Davis M, Pozzi L M, Coen D M. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivatives. Antimicrob Agents Chemother. 1993;37:19–25. doi: 10.1128/aac.37.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Talarico C L, Stanat S C, Biron K K. Program and Abstracts of the 34th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1994. Evidence that HCMV-encoded UL97 contributes to the phosphorylation of acyclovir in HCMV-infected cells, abstr. H65; p. 206. [Google Scholar]
- 67.Tatarowicz W A, Lurain N S, Thompson K D. In situ ELISA for the evaluation of antiviral compounds effective against human cytomegalovirus. J Virol Methods. 1991;35:207–215. doi: 10.1016/0166-0934(91)90136-n. [DOI] [PubMed] [Google Scholar]
- 68.Telenti A, Smith T F. Screening with a shell vial assay for antiviral activity against cytomegalovirus. Diagn Microbiol Infect Dis. 1989;12:5–8. doi: 10.1016/0732-8893(89)90036-9. [DOI] [PubMed] [Google Scholar]
- 69.Weinberg A, Schneider S A, Bate B J, Clark J C. Program and Abstracts of the 34th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1994. Antigen reduction assay for cytomegalovirus susceptibility to antivirals, abstr. H79; p. 207. [Google Scholar]
- 70.Wolf D G, Smith I L, Lee D J, Freeman W R, Flores-Aguilar M, Spector S A. Mutations in human cytomegalovirus UL97 gene confer clinical resistance to ganciclovir and can be detected directly in patient plasma. J Clin Investig. 1995;95:257–263. doi: 10.1172/JCI117648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wolf D G, Lee D J, Spector S A. Detection of human cytomegalovirus mutations associated with ganciclovir resistance in cerebrospinal fluid of AIDS patients with central nervous system disease. Antimicrob Agents Chemother. 1995;39:2552–2554. doi: 10.1128/aac.39.11.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolf D G, Yaniv I, Honigman A, Kassis I, Schonfeld T, Ashkenazi S. Early emergence of ganciclovir-resistant human cytomegalovirus strains in children with primary combined immunodeficiency. J Infect Dis. 1998;178:535–538. doi: 10.1086/517468. [DOI] [PubMed] [Google Scholar]