Abstract
Objectives
This study presents the clinical assessment of the Onclarity HPV Assay (Becton Dickinson) on the novel COR high-throughput instrument (Becton Dickinson) using the international guidelines in a routine setting.
Methods
Screening samples collected in BD SurePath from women aged 30 years and older were used in this validation. Noninferiority of the Onclarity HPV Assay on the COR instrument (Onclarity-COR) was assessed with the comparator assay glycoprotein 5–positive (GP5+)/6+ enzyme immunoassay (GP-EIA) for clinical sensitivity on 122 cervical intraepithelial neoplasia 2 and greater samples. Specificity was assessed using 887 samples with twice-normal cytology. Inter- and intralaboratory reproducibility analysis was assessed using 525 samples. Finally, a time-and-motion study was performed to evaluate COR instrument performance characteristics.
Results
The Onclarity-COR was noninferior to the GP-EIA for both sensitivity (P = .0016) and specificity (P < .0001). The intralaboratory reproducibility was 98.3% (κ = 0.96), and interlaboratory agreement was 98.5 % (κ = 0.96). The daily hands-on time for the COR instrument was 58 minutes, and walk-away time was 7 hours, 2 minutes per 8-hour day shift.
Conclusions
The Onclarity-COR instrument fulfills international validation criteria on sensitivity, specificity, and laboratory reproducibility. The Onclarity assay’s extended genotyping capability, together with its high-throughput characteristics, makes the COR instrument an excellent candidate for use in human papillomavirus primary cervical cancer screening.
Keywords: HPV assays, Genotyping, Onclarity, International validation, High throughput
Key Points.
We evaluated the BD COR high-throughput platform combined with the BD Onclarity HPV assay.
Successful clinical validation was compliant with the international guidelines for human papillomavirus (HPV) assays for cervical cancer screening.
The BD COR high-throughput platform allows for unprecedented laboratory HPV test production with limited operator interaction requirements.
Introduction
Cervical cancer screening is transitioning worldwide from cytology toward primary human papillomavirus (HPV) screening. At the same time, there is a paradigm shift toward risk-based guidelines using clinical action thresholds.1 A trend in cervical cancer screening programs is a continued consolidation process toward larger, centralized laboratories with unprecedented high throughput. This shift necessitates validation of existing HPV assays and novel high-throughput HPV test instruments using well-defined clinical assay performance specifications compliant with the international criteria for HPV tests used in cervical cancer screening.2 The migration of an already-validated HPV test to a new high-throughput instrument platform can result in changed test characteristics. Validation of the new combination of assay and instrument is therefore as important as the original validation of the assay itself, especially if the new high-throughput platform entails different sample processing from the previous instrument. Operator interaction (hands-on time), workflow, and system maintenance requirements are also important determinants for the clinical performance of fully automated high-throughput HPV instruments, and these elements can differ considerably between automated systems.3,4
Commercially available high-throughput HPV instruments are only now being introduced and validated. The cobas HPV Test on the cobas 6800/8800 instruments from Roche Diagnostics,4-6 the Alinity instrument from Abbott,7,8 and the COR HPV instrument from BD Diagnostics are all branded on sample turnaround numbers surpassing their predecessors: the Roche Diagnostics cobas 4800,9 the Abbott RealTime High Risk HPV Assay,10 and the BD Viper LT system,11,12 respectively.
Besides the move toward larger instruments for existing assays, a change toward the use of HPV genotype information in management algorithms is defining cervical cancer screening. The 2019 ASCCP guidelines describe a paradigm shift to risk-based guidelines using clinical action thresholds.1,13 Here, HPV genotyping plays a role in the management of index screening samples but also, when combined over time with subsequent follow-up samples or multiple screening rounds, enables clinicians to assess HPV persistence vs new infections as an integrated element in the evaluation of risk of disease and subsequent management.
The BD Onclarity HPV Assay is an extended genotyping assay that can detect 6 individual HPV genotypes and 8 genotypes in 3 groups (individual: 16, 18, 31, 45, 51, 52; Groups: 33/58, 56/59/66, 35/39/68). The Onclarity HPV Assay has been validated according to the international guidelines for samples collected in both ThinPrep and SurePath liquid-based cytology media in several studies.11,12,14-16 It is US Food and Drug Administration (FDA) approved for clinical cervical cancer screening using the Viper LT instrument.17-19 The newly developed, fully automated high-throughput BD COR instrument runs the Onclarity HPV Assay (Onclarity-COR) with identical chemistry, aspiration, and transfer volumes as on the well-validated Viper LT instrument (Onclarity-Viper). The COR system received CE Mark approval in 2019, and a premarket approval supplement was submitted to the FDA in 2020. The COR instrument consists of 2 interconnected units: a processing unit (PX) and an analytical unit (GX). SurePath or ThinPrep vials are loaded directly into the COR system, which handles the scanning, aliquoting, preheat treatment, DNA extraction, and real-time polymerase chain reaction (RT-PCR) analysis in a fully automated workflow that performs all preanalytic and analytic assay testing in a single system.
Here, we validate the Onclarity HPV assay after migration from the Viper LT instrument to the COR high-throughput HPV instrument using SurePath screening samples from women aged 30 years and older who participated in the organized cervical cancer screening program in Denmark. The validation is performed using the samples from the fourth installment of the VALGENT validation study,20,21 where clinical sensitivity and specificity were evaluated against glycoprotein 5–positive (GP5+)/6+ enzyme immunoassay (GP-EIA), accepted as a standard comparator assay. Furthermore, the validation panel was complemented with an independently collected reproducibility panel in concordance with international validation guidelines.2 A detailed genotype concordance analysis between Onclarity-COR and Onclarity-Viper was performed as well. Finally, a time-and-motion study was conducted to assess the laboratory performance of the COR instrument in a routine 8-hour day setting.
Material and Methods
Sample Collection and Histologic Follow-up
Sample collection, processing, cytology, and histology procedures have previously been described in detail.20 In short, the VALGENT4 panel was collected at the Department of Pathology, Hvidovre Hospital, Denmark (parent laboratory) in 2016 and consisted of 2 populations: 998 consecutive screening samples from routinely screened women (screening population; average age, 42.8 years [range, 30-59 years]) consisting of 947 samples negative for intraepithelial lesions or malignancy (NILM); 6 with abnormal squamous cells of undetermined significance (ASCUS); 21 with low-grade squamous intraepithelial lesions (LSILs); and 24 with high-grade SILs (HSILs), atypical glandular cells, atypical cells—cannot exclude HSIL, and adenocarcinoma in situ Table 1 . The second population was a disease-enriched cohort of 100 ASCUS, 100 LSIL, and 97 HSIL cytology samples (the enriched population; average age, 40.4 years [range, 30-59 years]). Histology was assessed from the Danish Patobank 33 months (range, 32-35 months) after baseline and revealed 122 cervical intraepithelial neoplasias (CINs) 2 or above, with the majority originating from the enriched population. In addition, 887 samples from women with consecutive cytology NILM at baseline and 12 to 24 months prior represented women without disease. Performance on the COR instrument was compared with previously obtained results on the Viper LT instrument.14
Table 1.
Characteristics of the Study Population and Human Papillomavirus Prevalence by Onclarity-COR, Onclarity-Viper, and GP-EIA
| Onclarity-COR | Onclarity-Viper | GP-EIA Assay | ||
|---|---|---|---|---|
| Total | hrHPV-Positive, No. (%) | hrHPV-Positive, No. (%) | hrHPV-Positive, No. (%) | |
| All | 1,295 | 369 (28.3) | 369 (28.4) | 396 (30.6 ) |
| Age, y | ||||
| 30-39 | 531 | 194 (36.5) | 193 (36.3) | 202 (38.0) |
| 40-49 | 519 | 124 (23.9) | 130 (25.0) | 136 (26.2) |
| 50-59 | 245 | 51 (20.8) | 51 (20.8) | 58 (23.7) |
| Cytology | ||||
| Normal | 947 | 75 (7.9) | 78 (8.2) | 105 (11.1) |
| ASCUS | 106 | 100 (94.3) | 103 (97.2) | 97 (91.5) |
| LSIL | 121 | 88 (72.7) | 88 (72.7) | 88 (72.7) |
| HSIL | 106 | 95 (89.6) | 94 (88.7) | 96 (90.6) |
| AGC/ASCH/AIS | 15 | 11 (73.3) | 11 (73.3) | 10 (66.7) |
| Histologic follow-up | ||||
| No biopsy | 946 | 108 (11.4) | 111 (11.7) | 139 (14.7) |
| NILM | 154 | 80 (51.9) | 82 (53.2) | 78 (50.6) |
| CIN1 | 73 | 66 (90.4) | 67 (91.8) | 66 (90.4) |
| CIN2 | 39 | 35 (89.7) | 34 (87.2) | 35 (89.7) |
| CIN3 | 75 | 72 (96.0) | 72 (96.0) | 70 (93.3) |
| Carcinoma | 8 | 8 (100) | 8 (100) | 8 (100) |
| ≥CIN2 | 122 | 115 (94.3) | 114 (93.4) | 113 (92.6) |
| ≥CIN3 | 83 | 80 (96.4) | 80 (96.4) | 78 (94.0) |
| 2×NILM1 | 887 | 66 (7.4) | 70 (7.9) | 95 (10.7) |
2×NILM, NILM at baseline and 12-24 months prior; AGC, atypical glandular cell; AIS, adenocarcinoma in situ; ASCH, atypical cells—cannot exclude HSIL; ASCUS, abnormal squamous cells of undetermined significance; CIN, cervical intraepithelial neoplasia; GP-EIA, glycoprotein 5–positive/6–positive enzyme immunoassay; hrHPV, high-risk human papillomavirus; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; NILM, negative for intraepithelial lesions or malignancy; Onclarity-COR, Onclarity HPV Assay on the COR instrument; Onclarity-Viper, Onclarity HPV Assay on the Viper LT instrument.
The reproducibility agreement panel contained 525 samples collected from routine samples tested with the Onclarity HPV assay on the Viper LT instrument from Danish women going for screening, with a predefined split of 32% HPV-positive and 68% HPV-negative samples, as stipulated by the international guidelines2—in total, 169 positives and 355 negatives. In total, 525 samples were tested twice for intralaboratory agreement at the parent laboratory in Copenhagen. The original SurePath vial was tested on the COR instrument twice on 2 separate runs (range, 0-6 days). For interlaboratory agreement, an aliquot of the samples, preprocessed at the parent laboratory from the original vial to instrument-compliant molecular (M) tubes, was shipped to Østfold Hospital Trust, Norway, and tested once. Only samples valid on all 3 runs were included in the analysis.
Onclarity Testing on the COR Instrument
The Onclarity assay is an RT-PCR assay with extended genotyping for 9 genotype readouts (16, 18, 31, 45, 51, 52, 33/58, 35/39/68, and 56/59/66). The Onclarity assay on the COR instrument uses the same chemistry and sample aspiration and transfer volumes as the Onclarity assay on the Viper LT instrument.11,14,22 The COR system has a flexible, modular design and consists of a centralized preanalytical PX module that performs all primary specimen conversions, including uncapping/capping, vortexing, aliquoting, heating (as needed), specimen holding (as needed), and control rehydration.
The processed specimens and controls are then sent to the analytic GX module, which is equipped with full assay automation (without user intervention). Depending on the particular needs of the laboratory, the preanalytical PX module can be connected with up to 2 independent GX modules, thus offering flexible options dependent on capacity needs. We evaluated the performance of the PX module paired with a single GX unit. Onclarity testing on the COR instrument was performed in 2019, with mean time from sample reception at the laboratory to testing of 1,185 days (range, 1176-1205 days).
Comparator Testing
The GP-EIA was used as the standard comparator for clinical accuracy of the sensitivity detection of ≥CIN2 or ≥CIN3 and the specificity of <CIN1 using the Onclarity assay performed on the COR instrument. The high-risk HPV (hrHPV) GP-EIA assay has pooled detection of 14 hrHPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68), and the GP-EIA testing was performed as part of the VALGENT4 study described previously.20
Onclarity Testing on the Viper LT Instrument
Onclarity testing was performed on the Viper LT instrument according to the manufacturer’s recommendations and as described previously.14 Testing was performed in 2016, with mean time from sample reception at the laboratory to testing of 28 days (range, 2-70 days). The samples were stored refrigerated during this period and were subsequently stored at −20°C before COR testing.
Time-and-Motion Study
A time-and-motion study of the COR instrument workflow was performed. For 1 working week, 330 samples were loaded into the COR daily for a single-shift workflow. Time points were assessed on each of the 5 days for (1) system startup, (2) daily maintenance, (3) weekly maintenance, (4) reagent loading, and (5) sample loading. Hands-on time and time to result for each day were registered.
Two daily interactions were required with the instrument during the 8-hour shift: At startup, we loaded all the SurePath vials to be processed that day together with the required consumables and reagents (pipette tips, extraction trays, PCR plates, quality controls [QCs], diluent bottles, M tubes, and reagent troughs), and a second interaction in the afternoon was necessary to replenish pipette tips, extraction trays, M tubes, and PCR plates. The BD COR system has 6 extraction drawers (capacity = 30 patient samples and 2 QCs), 5 of which are replenished in the afternoon to allow the instrument to continue processing unattended, for a combined total of 11 × 30 = 330 specimens. Weekly maintenance was not included in the daily contact time but consisted of cleaning selected touch points on the GX and PX units with 1% bleach, which took approximately 30 minutes.
Data Analysis
A sample was considered Onclarity positive (for both instruments) if the cycle threshold value was 34.2 or less for HPV-18, 31, 45, 51, 52, 33/58, 35/39/68, and 56/59/66 and 38.3 or less for HPV-16. The GP-EIA detects all 14 HPV genotypes detected by the Onclarity assay as a pooled hrHPV positive result. Between the Onclarity assay on the Viper LT and COR instruments, the level of genotype agreement was determined by the percentage of overall agreement and κ statistics, as was the reproducibility element. The following categories were distinguished based on κ statistics: 0.00-0.20 = poor, 0.21-0.40 = fair, 0.41-0.60 = moderate, 0.61-0.80 = good, and 0.81-1.00 = excellent.23
The accuracy of the GP-EIA assay was used as a comparator for clinical validation of the Onclarity assay on the COR instrument. Noninferiority assessment of the Onclarity assay on the COR instrument compared with the GP-EIA assay was according to the international guidelines using the preset 90% and 98% benchmarks for relative sensitivity and specificity, respectively.2,24 The reproducibility element was evaluated by using the predefined setup with an 87% or greater lower confidence bound and a κ value above 0.6, as defined by the international criterion.2
Ethical Approval
Sample collection and data retrieval for the VALGENT4 study were approved by the Danish Data Inspection Agency J. No. AHH-2017-024, I-Suite: 05356.
All collected samples were cross-referenced and found eligible with the Danish register on collection, storage, and use of human biological material in health research projects (Vævsanvendelsesregistret).
Results
HPV Genotype Test Positivity and Concordance for the Onclarity HPV Assay on the COR and Viper LT Instruments
The overall HPV positivity of Onclarity-Viper and Onclarity-COR was similar, as was the concordance of the 9 individually reported genotypes Table 1 and Table 2 . Genotype detection concordance in ≥CIN2 samples showed κ values ranging from good for HPV-51 (κ = 0.76) to excellent for the other 8 genotype groups (κ = 0.90-1.00). Genotype detection concordance for 2×NILM samples showed κ values ranging from good for HPV-51 (κ = 0.80) and HPV-33/58 (κ = 0.78) to excellent for the remaining 7 genotype groups (κ = 0.85-1.00). Considering the entire cohort, the κ values were excellent for all 9 genotype groups (κ = 0.92-0.98) Table 2 .
Table 2.
Genotype Distribution and Concordance for Onclarity-COR and Onclarity-Viper, Respectively
| HPV Genotypes | COR, No. (%) | Viper LT, No. (%) | COR+/Viper LT+, No. | COR+/Viper LT−, No. | COR−/Viper LT+, No. | COR−/Viper LT−, No. | Agreement, % (95% CI) | κ |
|---|---|---|---|---|---|---|---|---|
| ≥CIN2 (n = 122) | ||||||||
| 16 | 50 (41.0) | 51 (41.8) | 50 | 0 | 1 | 71 | 99.2 (95.5-100) | 0.98 |
| 18 | 12 (9.8) | 12 (9.8) | 12 | 0 | 0 | 110 | 100 (97.0-100) | 1.00 |
| 31 | 18 (14.8) | 18 (14.8) | 18 | 0 | 0 | 104 | 100 (97.0-100) | 1.00 |
| 45 | 9 (7.4) | 10 (8.2) | 9 | 0 | 1 | 112 | 99.2 (95.5-100) | 0.94 |
| 51 | 7 (5.7) | 6 (4.9) | 5 | 2 | 1 | 114 | 97.5 (93.0-99.5) | 0.76 |
| 52 | 19 (15.6) | 18 (14.8) | 17 | 2 | 1 | 102 | 97.5 (93.0-99.5) | 0.90 |
| 33/58 | 14 (11.5) | 14 (11.5) | 14 | 0 | 0 | 108 | 100 (97.0-100) | 1.0 |
| 35/39/68 | 6 (4.9) | 7 (5.7) | 6 | 0 | 1 | 115 | 99.2 (95.5-100) | 0.92 |
| 56/59/66 | 14 (11.5) | 13 (10.7) | 13 | 1 | 0 | 108 | 99.2 (95.5-100) | 0.96 |
| 14 hrHPV | 115 (94.3) | 114 (93.4) | 114 | 1 | 0 | 7 | 99.2 (95.5-100) | 0.93 |
| 2×NILM (n = 887) | ||||||||
| 16 | 19 (2.1) | 18 (3.0) | 17 | 2 | 1 | 867 | 99.7 (99.0-99.9) | 0.92 |
| 18 | 4 (0.5) | 4 (0.5) | 4 | 0 | 0 | 883 | 100 (99.6-100) | 1.0 |
| 31 | 3 (0.3) | 4 (0.5) | 3 | 0 | 1 | 883 | 99.9 (99.4-100) | 0.86 |
| 45 | 4 (0.5) | 5 (0.6) | 4 | 0 | 1 | 882 | 99.9 (99.4-100) | 0.89 |
| 51 | 2 (0.2) | 3 (0.3) | 2 | 0 | 1 | 884 | 99.9 (99.4-100) | 0.80 |
| 52 | 12 (1.4) | 13 (1.5) | 12 | 0 | 1 | 874 | 99.9 (99.4-100) | 0.96 |
| 33/58 | 9 (1.0) | 9 (1.0) | 7 | 2 | 2 | 876 | 99.5 (98.8-99.9) | 0.78 |
| 35/39/68 | 15 (1.7) | 18 (2.0) | 14 | 1 | 4 | 868 | 99.4 (98.7-99.8) | 0.85 |
| 56/59/68 | 11 (1.2) | 11 (1.2) | 11 | 0 | 0 | 876 | 100 (99.6-100) | 1.0 |
| 14 hrHPV | 66 (7.4) | 70 (7.9) | 62 | 4 | 8 | 813 | 98.6 (97.6-99.3) | 0.90 |
| VALGENT4 panel (n = 1,295) | ||||||||
| 16 | 97 (7.5) | 97 (7.5) | 94 | 3 | 3 | 1,195 | 99.5 (99.0-99.8) | 0.97 |
| 18 | 30 (2.3) | 31 (2.4) | 30 | 0 | 1 | 1,264 | 99.9 (99.6-100) | 0.98 |
| 31 | 50 (3.9) | 51 (3.9) | 48 | 2 | 3 | 1,242 | 99.6 (99.1-99.9) | 0.95 |
| 45 | 33 (2.5) | 35 (2.7) | 33 | 0 | 2 | 1,260 | 99.8 (99.4-100) | 0.97 |
| 51 | 31 (2.4) | 30 (2.4) | 28 | 3 | 2 | 1,262 | 99.6 (99.1-99.9) | 0.92 |
| 52 | 47 (3.6) | 48 (3.7) | 45 | 2 | 3 | 1,245 | 99.6 (99.1-99.9) | 0.95 |
| 33/58 | 46 (3.6) | 46 (3.6) | 43 | 3 | 3 | 1,246 | 99.5 (99.0-99.8) | 0.93 |
| 35/39/68 | 56 (4.3) | 61 (4.7) | 54 | 2 | 7 | 1,232 | 99.3 (98.7-99.7) | 0.92 |
| 56/59/66 | 83 (6.4) | 82 (6.3) | 81 | 2 | 1 | 1,211 | 99.8 (99.3-98.7) | 0.98 |
| 14 hrHPV | 369 (28.5) | 374 (28.9) | 362 | 7 | 12 | 914 | 98.5 (97.7-99.1) | 0.96 |
2×NILM, NILM at baseline and 12-24 months prior; CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; hrHPV, high-risk human papillomavirus; Onclarity-COR, Onclarity HPV Assay on the COR instrument; Onclarity-Viper, Onclarity HPV Assay on the Viper LT instrument.
Clinical Performance of Onclarity-COR
The clinical accuracy of Onclarity-COR and the comparator assay (GP-EIA) is shown in Table 3 . The absolute sensitivity for ≥CIN2 was 94.3% for Onclarity-COR and 92.6% for GP-EIA; the relative sensitivity was 1.02 (95% confidence interval [CI], 0.99-1.04). The absolute sensitivity for ≥CIN3 was 96.4% for Onclarity-COR and 94.0% for GP-EIA; relative sensitivity was 1.03 (95% CI, 0.99-1.06). The Onclarity-COR was found to be noninferior to the comparator assay for both ≥CIN2 (P = .0016) and ≥CIN3 (P = .0005) sensitivity. The absolute specificity was 92.6% for Onclarity-COR and 89.3% for the comparator assay; relative specificity was 1.04 (95% CI, 1.02-1.05). Onclarity-COR was found to be noninferior to the comparator assay for specificity (P < .0001).
Table 3.
Clinical Accuracy of COR and GP-EIA for ≥CIN2, ≥CIN3, and <CIN1 Outcomes
| GP-EIA Results, No. | Accuracy of COR (95% CI) | Accuracy of GP-EIA (95% CI) | Relative Accuracy (COR/GP-EIA) (95% CI) | Noninferiority Test (P Value) | ||||
|---|---|---|---|---|---|---|---|---|
| Study Population | COR Results | Pos | Neg | Total | ||||
| ≥CIN2 (n = 122) | Pos | 113 | 2 | 115 | Sensitivity 94.3% (88.5-97.7) | Sensitivity 92.6% (86.5-96.6) | Sensitivity 1.02 (0.99-1.04) | .0016 |
| Neg | 0 | 7 | 7 | |||||
| Total | 113 | 9 | 122 | |||||
| ≥CIN3 (n = 83) | Pos | 78 | 2 | 80 | Sensitivity 96.4% (89.8-99.2) | Sensitivity 94.0% (86.5-98.0) | Sensitivity 1.03 (0.99-1.06) | .0005 |
| Neg | 0 | 3 | 3 | |||||
| Total | 78 | 5 | 83 | |||||
| 2×NILM3 (n = 887) | Pos | 59 | 7 | 66 | Specificity 92.6% (90.6-94.2) | Specificity 89.3% (87.1-91.2) | Specificity 1.04 (1.02-1.05) | <.0001 |
| Neg | 36 | 785 | 821 | |||||
| Total | 95 | 792 | 887 |
2×NILM, NILM at baseline and 12-24 months prior; CIN, cervical intraepithelial neoplasia; GP-EIA, glycoprotein 5–positive/6–positive enzyme immunoassay; Neg, negative; Pos, positive.
Inter- and Intralaboratory Reproducibility
The intralaboratory reproducibility and interlaboratory agreement were assessed using 525 cervical cancer screening samples. The intralaboratory reproducibility of Onclarity-COR was 98.3% (95% CI, 96.8-99.2), with a κ of 0.96 Table 4 . The interlaboratory agreement was 98.5% (95% CI, 97.0-99.3), with a κ of 0.96 Table 4 .
Table 4.
Intralaboratory Reproducibility and Interlaboratory Agreement
| Assessment and Site | HPV Status | Copenhagen Laboratory Result 1, No. | Total, No. | Reproducibility/Agreement, % (95% CI) | κ | |
|---|---|---|---|---|---|---|
| hrHPV Pos | hrHPV Neg | |||||
| Intralaboratory reproducibility | ||||||
| Copenhagen laboratory result 2 | hrHPV pos | 152 | 6 | 158 | 98.3 (96.8-99.2) | 0.96 |
| hrHPV neg | 3 | 364 | 367 | |||
| Total | 155 | 370 | 525 | |||
| Interlaboratory agreement | ||||||
| Østfold laboratory result | hrHPV pos | 152 | 5 | 157 | 98.5 (97.0-99.3) | 0.96 |
| hrHPV neg | 3 | 365 | 368 | |||
| Total | 155 | 370 | 525 |
HPV, human papillomavirus; hrHPV, high-risk human papillomavirus; Neg, negative; Pos, positive.
When looking at the individual genotype concordance Table 5 , the agreement was excellent for all 9 genotype groups for both the intralaboratory reproducibility (κ = 0.89-1.00) and interlaboratory agreement (κ = 0.93-1.00).
Table 5.
Intralaboratory Reproducibility and Interlaboratory Agreement of Individual Genotyping With Onclarity-COR
| Assessment and HPV type | Concordance per Run/Laboratory, No. | Agreement, % (95% CI) | κ | |||
|---|---|---|---|---|---|---|
| Pos/Pos | Pos/Neg | Neg/Pos | Neg/Neg | |||
| Intralaboratory reproducibility (Copenhagen 1/Copenhagen 2) | ||||||
| HPV-16 | 49 | 0 | 4 | 472 | 99.2 (98.1-99.8) | 0.96 |
| HPV-18 | 8 | 1 | 0 | 516 | 99.8 (98.9-100.0) | 0.94 |
| HPV-31 | 20 | 2 | 1 | 502 | 99.4 (98.3-99.9) | 0.93 |
| HPV-45 | 7 | 0 | 0 | 518 | 100 (99.3-100.0) | 1.00 |
| HPV-51 | 8 | 0 | 0 | 517 | 100 (99.3-100.0) | 1.00 |
| HPV-52 | 11 | 0 | 1 | 513 | 99.8 (98.9-100.0) | 0.96 |
| HPV-33/58 | 20 | 2 | 1 | 502 | 99.4 (98.3-99.9) | 0.93 |
| HPV-35/39/68 | 36 | 3 | 0 | 486 | 99.4 (98.3-99.9) | 0.96 |
| HPV-56/59/66 | 27 | 3 | 3 | 492 | 98.9 (97.5-99.6) | 0.89 |
| Interlaboratory agreement (Copenhagen 1/Østfold) | ||||||
| HPV-16 | 47 | 2 | 4 | 472 | 98.9 (97.5-99.6) | 0.93 |
| HPV-18 | 8 | 1 | 0 | 516 | 99.8 (98.9-100.0) | 0.94 |
| HPV-31 | 20 | 2 | 0 | 503 | 99.6 (98.6-100.0) | 0.95 |
| HPV-45 | 7 | 0 | 0 | 518 | 100 (99.3-100.0) | 1.00 |
| HPV-51 | 8 | 0 | 0 | 517 | 100 (99.3-100.0) | 1.00 |
| HPV-52 | 11 | 0 | 1 | 513 | 99.8 (98.9-100.0) | 0.96 |
| HPV-33/58 | 21 | 1 | 0 | 503 | 99.8 (98.9-100.0) | 0.98 |
| HPV-35/39/68 | 38 | 1 | 0 | 486 | 99.8 (98.9-100.0) | 0.99 |
| HPV-56/59/66 | 28 | 2 | 1 | 494 | 99.4 (98.3-99.9) | 0.95 |
HPV, human papillomavirus; Neg, negative; Onclarity-COR, Onclarity HPV Assay on the COR instrument; Pos, positive.
Time-and-Motion Study
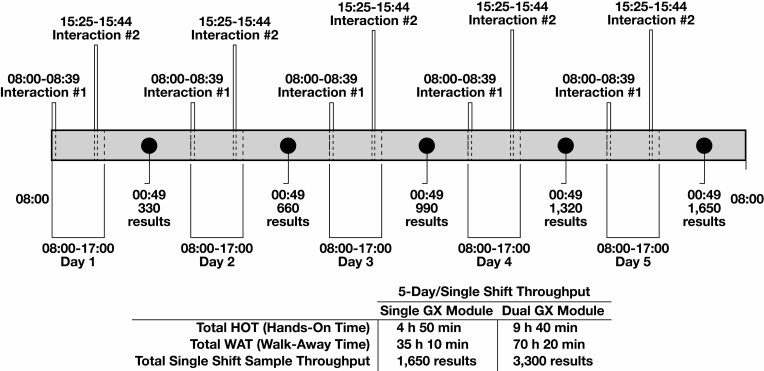
The time-and-motion study was performed to assess the sample turnaround time and overall single-shift production capacity of the COR instrument Figure 1 . The first user interaction required on average 39 minutes of hands-on time, including daily cleaning, loading of samples and reagents, and unloading samples from the day before. The average walk-away time thereafter was 7 hours, 2 minutes. The second interaction, including loading of reagents and unloading of processed samples and reagents, required on average 19 minutes of hands-on time. Time to result was 4 hours, 15 minutes for the first 30 samples loaded, whereas the successive batches of 30 test results was completed 1 hour, 13 minutes apart Figure 1 . Using this workflow, the COR system can process 330 samples in 1 8-hour shift, with up to 1,650 reported test results in 1 working week Figure 1 .
Figure 1.
Daily and weekly results output from the COR single–analytical unit (GX) and dual-GX system configurations.
Discussion
This study presents our validation of the Onclarity HPV assay on the novel high-throughput COR instrument using SurePath collected screening samples from Danish routine cervical cancer screening. Onclarity-COR clinical and analytical performance was similar to Onclarity-Viper on all parameters evaluated Table 1 and Table 2 . Onclarity-COR showed noninferior clinical sensitivity (relative sensitivity for ≥CIN2, 1.02 [95% CI, 0.99-1.04]) and noninferior and even slightly higher specificity (relative specificity, 1.04 [95% CI, 1.02-1.05]) compared with GP-EIA Table 3 . Similar results were found in the previous clinical validation of the Onclarity HPV test on the Viper LT instrument.14
Also, the intralaboratory reproducibility and interlaboratory agreement fulfilled the international validation criteria Table 4 . The intralaboratory reproducibility was 98.3% (κ = 0.96), and the interlaboratory agreement was 98.5% (κ = 0.96) using preprocessed parent laboratory aliquots from the original sample vials. In our previous study evaluating the Onclarity HPV test on the Viper LT instrument,12 the interlaboratory agreement was 96.8% (κ = 0.92), and the intralaboratory reproducibility was 97.4% (κ = 0.93). It should be noted, however, that Viper LT and COR testing was performed on 2 different reproducibility panels.
Reproducibility among the individual genotype groups showed excellent concordance for all 9 genotype groups for both intralaboratory reproducibility Table 5 (κ = 0.89-0.96) and interlaboratory agreement Table 5 (κ = 0.93-1.00).
Comparing COR performance with similar high-throughput instruments—the Roche cobas 6800/8800 and the Abbott Alinity instruments—inter- and intralaboratory analysis showed equally good results, at more than 98% reproducibility for cobas in the 2 separate studies,5,6 and the intra- and interlaboratory reproducibility of the Alinity HPV test7 was 96.7% (κ = 0.92) and 98.7% (κ = 0.97), respectively.
The time-and-motion study evaluated a workflow of 330 samples per day and a production of 1,650 test results in a regular week. In comparison, the Viper LT instrument processed 90 to 120 samples per day per instrument.22
The average daily hands-on time for a full COR workflow was 58 minutes, with 2 interactions (of 39 minutes and 19 minutes, respectively). The daily walk-away time was 7 hours, 2 minutes Figure 1 . We used a 1 GX–to–1 PX configuration, but extending the configuration to 2 GX units and 1 PX unit, the COR system can process 660 samples daily and 3,300 test results per regular 5-day week. A limitation in our analysis is the restriction to an 8-hour shift with a “cold start” each day. For high-throughput laboratories, the continuous loading function using the internal sample “hotel” storage of 480 BD SurePath liquid-based cytology vials allows “carry-over” activity between working days but was not assessed in our study. If the system is loaded to capacity with vials on day 1, additional preanalytic processing can be performed overnight, increasing throughput to approximately 500 samples on subsequent days.
A similar approach evaluating the cobas HPV assay on the cobas 6800 instrument showed comparable test turnaround figures,4 where initialization, time to first results, time to last result, and total hands-on time amounted to 24 minutes; 2 hours, 28 minutes; 7 hours, 7 minutes; and 59 minutes, respectively. In contrast to our study, the cobas 6800 study did not include preanalytical processing before testing on the cobas 6800 instrument; a recent study estimated that the combined preanalytical (cobas p480) and analytical throughput of the cobas 6800 instrument was 384 samples in an 8-hour shift. To the best of our knowledge, similar time-and-motion studies have not been published on the cobas 8800 or Alinity instruments. Hands-on time, maintenance, and cleaning operations on a high-throughput HPV instrument can greatly affect the daily workload of a laboratory.3,4,25
The 3 high-throughput HPV test instruments from Abbott, Becton Dickinson, and Roche, respectively, rely on widely different strategies with respect to reporting HPV-positive findings. HPV genotyping can play a major role in risk management of HPV-positive women, allocating women to risk tiers by HPV genotype26-28 and distinguishing new from persistent infections.29 The cobas HPV assay maintains the 2010 assay design on all cobas instruments, with individual reporting of HPV-16 and HPV-18 and reporting of the remaining 12 hrHPV genotypes as 1 group.9 The Abbott RealTime High Risk HPV Assay on the Alinity instrument is the newest design of the 3 and individually reports HPV genotypes 16, 18, and 45, whereas HPV-31/33/52/58 and HPV-35/39/51/56/59/66/68 are reported in 2 groups.10
In comparison, the Onclarity assay allows for extended genotyping on every positive sample, with individual typing of 6 genotypes (HPV-16, 18, 31, 45, 51, 52) and the remaining 8 genotypes in 3 groups (33/58, 35/39/68, 56/59/66). In the United States, Australia, many European Union countries, and other countries, partial genotyping for HPV-16 and HPV-18 is used to risk-stratify women for follow-up.28,30 The use of extended genotyping is gaining ground, however, and several studies have shown the 12 non–HPV-16/HPV-18 hr genotypes to carry distinct and markedly different risk of cervical disease.31-33 Consequently, more detailed genotyping than just HPV-16 and HPV-18 could be important in near-future triage algorithms and risk management. Of special note for the Onclarity and Alinity assays, the joint probe for HPV-33 and HPV-58 may result in overreferrals because the risk of ≥CIN2 is significantly greater for HPV-33 compared with HPV-58,26,31,34-36 and a separation of those 2 genotypes could be beneficial in improving assay specificity.
A major strength of our study is that the Onclarity assay on the COR instrument was validated against the assay on the already-validated Viper LT instrument as well as against an internationally recognized comparator assay, the GP-EIA assay.2 In comparison, the novel cobas 6800/8800 HPV assay on the cobas 6800 instrument was validated against the cobas 4800 assay.5,6 The argument for this deviation was that the 2 internationally recognized comparator assays—GP-EIA and Hybrid Capture 2 (Qiagen)—have been more or less discontinued in clinical routine worldwide. We acknowledged the merit of this argument, and the guidelines are expected to extend the number of standard comparator tests to reflect this reality.
Another strength of our study is that the COR testing was done using the same panel of samples as previously used to validate the Onclarity assay on the Viper LT instrument. This approach makes the results directly comparable yet also confides a weakness in that the test rounds are 3 years apart. If storage had any negative impact, however, we would have expected performance differences. Such was not observed, and the Onclarity-COR concordance to the comparator assay remained high and well within validation criteria acceptance.
Conclusion
The Onclarity assay was successfully migrated from the medium-throughput BD Viper LT HPV instrument to the high-throughput BD COR instrument. The Onclarity assay on the novel COR instrument was found to be noninferior to the comparator assay for clinical sensitivity and specificity. The inter- and intralaboratory reproducibility was high. The Onclarity assay allows for extended genotyping, which, together with the COR instrument’s capacity to run a high number of samples, makes the COR instrument an excellent candidate for use in HPV primary cervical cancer screening in laboratories that use screening algorithms, including genotyping.
Acknowledgments
The authors would like to thank the laboratory staff at the Department of Pathology, Hvidovre Hospital, and Inger-Lise Håkensen, Stine Spone Marthinsen, and Tina Marie Hannestad at Østfold Hospital Trust for excellent assistance on running samples. We thank BD Diagnostics, Sparks, MD, for assistance analyzing the time and motion data.
Funding
Becton Dickinson supplied reagents for Viper LT and COR testing as well as limited cofunding. It had the opportunity to review the draft of the manuscript but had no editorial rights. M.A. was supported by the Horizon 2020 Framework Programme for Research and Innovation of the European Commission through the RISCC Network (Grant No. 847845).
Scienanso, the employer of M.A., received funding from the VALGENT as explained.21
J.B. is the principle investigator of studies funded in part by BD Diagnostics, Agena Bioscience, Genomica SAU, LifeRiver Biotech, and Qiagen. J.B. has received honoraria for lectures from BD Diagnostics, Roche Molecular Systems, Qiagen, and Genomica SAU. J.B. is an appointed member of the National Danish Cervical Screening Committee by the Danish Health Authority and a member of the Regional cervical screening steering committee of the Capital Region of Denmark.
Disclosures: D.M.E., H.P., and B.T.P. attended meetings with various HPV test manufacturers.
References
- 1. Schiffman M, Wentzensen N, Perkins RB, et al. . An introduction to the 2019 ASCCP risk-based management consensus guidelines. J Low Genit Tract Dis. 2020;24:87-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meijer CJLM, Berkhof J, Castle PE, et al. . Guidelines for human papillomavirus DNA test requirements for primary cervical cancer screening in women 30 years and older. Int J Cancer. 2009;124: 516-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ratnam S, Jang D, Gilchrist J, et al. . Workflow and maintenance characteristics of five automated laboratory instruments for the diagnosis of sexually transmitted infections. J Clin Microbiol. 2014;52:2299-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aretzweiler G, Leuchter S, Simon CO, et al. . Generating timely molecular diagnostic test results: workflow comparison of the cobas® 6800/8800 to Panther. Expert Rev Mol Diagn. 2019;19:951-957. [DOI] [PubMed] [Google Scholar]
- 5. Frayle H, Gori S, Rizzi M, et al. . HPV testing for cervical cancer screening: technical improvement of laboratory logistics and good clinical performance of the cobas 6800 in comparison to the 4800 system. BMC Womens Health. 2019;19:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saville M, Sultana F, Malloy MJ, et al. . Clinical validation of the cobas HPV test on the cobas 6800 system for the purpose of cervical screening. J Clin Microbiol 2019;57:e01239-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oštrbenk Valenčak A, Šterbenc A, Seme K, et al. . Alinity m HR HPV assay fulfills criteria for human papillomavirus test requirements in cervical cancer screening settings. J Clin Microbiol. 2019; 58:e01120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dhillon SK, Oštrbenk Valencak A, Xu L, et al. . Clinical and analytical evaluation of the Alinity m HR HPV assay within the VALGENT-3 framework. J Clin Microbiol. 2021;59:e00286-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heideman DAM, Hesselink AT, Berkhof J, et al. . Clinical validation of the cobas 4800 HPV test for cervical screening purposes. J Clin Microbiol. 2011;49:3983-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hesselink AT, Meijer CJLM, Poljak M, et al. . Clinical validation of the Abbott RealTime High Risk HPV assay according to the guidelines for human papillomavirus DNA test requirements for cervical screening. J Clin Microbiol. 2013;51:2409-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ejegod DM, Serrano I, Cuschieri KS, et al. . Clinical validation of the BD Onclarity™ HPV assay using a non-inferiority test. J Med Microbiol Diagn. 2013;S3. [Google Scholar]
- 12. Ejegod D, Bottari F, Pedersen H, et al. . The BD Onclarity HPV assay on samples collected in SurePath medium meets the international guidelines for human papillomavirus test requirements for cervical screening. J Clin Microbiol. 2016;54:2267-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheung LC, Egemen D, Chen X, et al. . 2019 ASCCP risk-based management consensus guidelines: methods for risk estimation, recommended management, and validation. J Low Genit Tract Dis. 2020;24:90-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bonde JH, Pedersen H, Quint W, et al. . Clinical and analytical performance of the BD Onclarity HPV Assay with SurePath screening samples from the Danish Cervical Screening Program using the VALGENT framework. J Clin Microbiol. 2020;58:e01518-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cuschieri K, Geraets DT, Moore C, et al. . Clinical and analytical performance of the Onclarity HPV assay using the VALGENT framework. J Clin Microbiol. 2015;53:3272-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arbyn M, Snijders PJF, Meijer CJLM, et al. . Which high-risk HPV assays fulfil criteria for use in primary cervical cancer screening? Clin Microbiol Infect. 2015;21:817-826. [DOI] [PubMed] [Google Scholar]
- 17. Stoler MH, Wright TC Jr, Parvu V, et al. . The Onclarity human papillomavirus trial: design, methods, and baseline results. Gynecol Oncol. 2018;149:498-505. [DOI] [PubMed] [Google Scholar]
- 18. Stoler MH, Wright TC Jr, Parvu V, et al. . Stratified risk of high-grade cervical disease using Onclarity HPV extended genotyping in women, ≥25 years of age, with NILM cytology. Gynecol Oncol. 2019;153: 26-33. [DOI] [PubMed] [Google Scholar]
- 19. Wright TC Jr, Stoler MH, Parvu V, et al. . Detection of cervical neoplasia by human papillomavirus testing in an atypical squamous cells-undetermined significance population: results of the Becton Dickinson Onclarity trial. Am J Clin Pathol. 2019;151:53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonde J, Ejegod DM, Cuschieri K, et al. . The Valgent4 protocol: robust analytical and clinical validation of 11 HPV assays with genotyping on cervical samples collected in SurePath medium. J Clin Virol. 2018;108:64-71. [DOI] [PubMed] [Google Scholar]
- 21. Arbyn M, Depuydt C, Benoy I, et al. . VALGENT: a protocol for clinical validation of human papillomavirus assays. J Clin Virol. 2016;76(suppl 1):S14-S21. [DOI] [PubMed] [Google Scholar]
- 22. Ejegod DM, Junge J, Franzmann M, et al. . Clinical and analytical performance of the BD Onclarity™ HPV assay for detection of CIN2+ lesions on SurePath samples. Papillomavirus Res. 2016;2:31-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159-174. [PubMed] [Google Scholar]
- 24. Tang NS, Tang ML, Chan IS. On tests of equivalence via non-unity relative risk for matched-pair design. Stat Med. 2003;22:1217-1233. [DOI] [PubMed] [Google Scholar]
- 25. Loonen AJM, Huijsmans CJJ, Geurts-Giele WRR, et al. . Performance analysis of high-throughput HPV testing on three automated workflows. APMIS. 2020;128:497-505. [DOI] [PubMed] [Google Scholar]
- 26. Bonde JH, Sandri MT, Gary DS, et al. . Clinical utility of human papillomavirus genotyping in cervical cancer screening: a systematic review. J Low Genit Tract Dis. 2020;24:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Demarco M, Carter-Pokras O, Hyun N, et al. . Validation of a human papillomavirus (HPV) DNA cervical screening test that provides expanded HPV typing. J Clin Microbiol. 2018;56:e01910-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Demarco M, Egemen D, Raine-Bennett TR, et al. . A study of partial human papillomavirus genotyping in support of the 2019 ASCCP Risk-Based Management Consensus Guidelines. J Low Genit Tract Dis. 2020;24:144-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bonde J, Bottari F, Iacobone AD, et al. . Human papillomavirus same genotype persistence and risk: a systematic review. J Low Genit Tract Dis. 2021;25:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rebolj M, Brentnall AR, Mathews C, et al. ; HPV Pilot Steering Group . 16/18 genotyping in triage of persistent human papillomavirus infections with negative cytology in the English cervical screening pilot. Br J Cancer. 2019;121:455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bonde J, Bottari F, Parvu V, et al. . Bayesian analysis of baseline risk of CIN2 and ≥CIN3 by HPV genotype in a European referral cohort. Int J Cancer. 2019;145:1033-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schiffman M, Hyun N, Raine-Bennett TR, et al. . A cohort study of cervical screening using partial HPV typing and cytology triage. Int J Cancer. 2016;139:2606-2615. [DOI] [PubMed] [Google Scholar]
- 33. Schiffman M, Vaughan LM, Raine-Bennett TR, et al. . A study of HPV typing for the management of HPV-positive ASC-US cervical cytologic results. Gynecol Oncol. 2015;138:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lie AK, Tropé A, Skare GB, et al. . HPV genotype profile in a Norwegian cohort with ASC-US and LSIL cytology with three year cumulative risk of high grade cervical neoplasia. Gynecol Oncol. 2018;148:111-117. [DOI] [PubMed] [Google Scholar]
- 35. Adcock R, Cuzick J, Hunt WC, et al. ; New Mexico HPV Pap Registry Steering Committee . Role of HPV genotype, multiple infections, and viral load on the risk of high-grade cervical neoplasia. Cancer Epidemiol Biomarkers Prev. 2019;28:1816-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vintermyr OK, Andersland MS, Bjørge T, et al. . Human papillomavirus type specific risk of progression and remission during long-term follow-up of equivocal and low-grade HPV-positive cervical smears. Int J Cancer. 2018;143:851-860. [DOI] [PubMed] [Google Scholar]