Abstract
Introduction:
Osimertinib is a third-generation EGFR tyrosine kinase inhibitor (TKI) that is approved for the use of EGFR-mutant non-small cell lung cancer (NSCLC) patients. In this study, we investigated the acquired resistance mechanisms in NSCLC patients and patient-derived preclinical models.
Methods:
Formalin-fixed paraffin-embedded tumor samples and plasma samples from 55 NSCLC patients who were treated with osimertinib were collected at baseline and at progressive disease (PD). Next-generation sequencing was performed in tumor and plasma samples using a 600-gene hybrid capture panel designed by AstraZeneca. Osimertinib-resistant cell lines and patient-derived xenografts and cells were generated and whole exome sequencing and RNA sequencing were performed. In vitro experiments were performed to functionally study the acquired mutations identified.
Results:
A total of 55 patients and a total of 149 samples (57 tumor samples and 92 plasma samples) were analyzed, and among them 36 patients had matched pre- and post-treatment samples. EGFR C797S (14%) mutation was the most frequent EGFR-dependent mechanism identified in all available progression samples, followed by EGFR G824D (6%), V726M (3%), and V843I (3%). Matched pre- and post-treatment sample analysis revealed in-depth acquired mechanisms of resistance. EGFR C797S was still most frequent (11%) among EGFR-dependent mechanism, while among EGFR-independent mechanisms, PIK3CA, ALK, BRAF, EP300, KRAS, and RAF1 mutations were detected. Among Osimertinib-resistant cell lines and patient-derived models, we noted acquired mutations which were potentially targetable such as NRAS p.Q61K, in which resistance could be overcome with combination of osimertinib and trametinib. A patient-derived xenograft established from osimertinib-resistant patient revealed KRAS p.G12D mutation which could be overcome with combination of osimertinib, trametinib, and buparlisib.
Conclusion:
In this study, we explored the genetic profiles of osimertinib-resistant NSCLC patient samples using targeted deep sequencing. In vitro and in vivo models harboring osimertinib resistance revealed potential novel treatment strategies after osimertinib failure.
Keywords: drug resistance, EGFR inhibitor, lung cancer, non-small cell lung cancer, targeted therapy
Introduction
Epidermal growth factor receptor (EGFR)-mutant lung cancers occur in higher frequencies in female East Asian and never-smoking populations. 1 The development of EGFR tyrosine kinase inhibitors (TKIs) has improved the prognosis of NSCLC patients, with higher objective response rates and increased progression-free survival (PFS) compared to former cytotoxic chemotherapy.2–4 However, previous clinical study showed that most of EGFR-mutant lung cancer patients inevitably progressed after EGFR TKI treatment. 5 Potential acquired resistance mechanisms which include EGFR T790M mutation and other secondary EGFR point mutations (50–60%) to first- and second-generation EGFR TKIs, and bypass track activations such as MET amplification, and activation of AXL,6,7 PIK3CA mutation, HER2 amplification, and epithelial to mesenchymal transition (EMT) were previously reported to be involved in acquired resistance. Histologic transformation to small cell lung cancer (SCLC) has also been reported.8,9
Third-generation EGFR TKI, osimertinib, selectively inhibits EGFR sensitizing and T790M mutations and has shown superior efficacy compared with first-generation EGFR TKI. Osimertinib demonstrated the median PFS of 18.9 months as the first-line treatment and is now recommended as a treatment option in the first-line treatment of advanced EGFR-mutant NSCLC. 10 However, acquired resistance to osimertinib is still common,10,11 and resistance mechanisms have been reported to be more heterogeneous than those of first- and second-generation EGFR TKIs. Resistance mechanisms are commonly categorized into two groups: (1) acquired tertiary EGFR resistance mutations and (2) activation of bypass receptor tyrosine kinase (RTK) signaling. EGFR C797S tertiary mutation is known as the hotspot mutation which leads to osimertinib resistance by hindering osimertinib binding.8,12,13 However, multiple previous analyses have reported that osimertinib resistance is less reliant on EGFR pathways, possibly due to more potent on-target inhibition leading to subclonal evolution. Thus, the remaining resistance mechanisms are largely due to off-target mechanisms. 14 MET amplification, HER2 amplification, and rare rearrangements in the genes such as ALK, FGFR, RET, and NTRK have also been reported. Interestingly, these alterations are potentially targetable, and there are ongoing trials to assess the efficacy of combination treatment for the patients who progressed upon osimertinib. For example, combination of c-MET inhibitors such as savolitinib with osimertinib has shown promising activity in those patients who are positive for MET after development of osimertinib resistance (NCT02143466, NCT03944772). 15 Furthermore, studies investigating novel agents and combinations to overcome acquired resistance have been conducted, such as osimertinib plus durvalumab. However, high frequency of interstitial lung disease (22%) was reported, leading to discontinuation of this combination, 16 and combination of EGFR TKI and immunotherapy remains doubtful. However, 30–40% of resistance mechanisms are still not elucidated, 17 and further studies are needed to improve outcomes in advanced EGFR-mutant NSCLC.
In this study, we investigated acquired resistance mechanisms in NSCLC patients treated with osimertinib using plasma and tumor samples collected at baseline and at the time of progression. Comprehensive genomic profiling was performed using deep targeted sequencing assays. Furthermore, acquired resistance mechanisms were investigated in patient-derived xenografts and patient-derived cells. In the end, we identified novel resistance mechanisms and validated them in vitro and in vivo.
Materials and methods
Patients
A total of 55 patients enrolled in this study were derived from the recently conducted ASTRIS study (NCT02474355), which was an open-label, single-arm, multinational, real-world treatment study that investigated the safety and efficacy of osimertinib in patients with T790M-positive advanced NSCLC, who had previously been treated with first- and second-generation EGFR TKIs. 18 All patients were positive for activating EGFR and T790M mutations and were treated with osimertinib. Tissue (formalin-fixed paraffin embedded, FFPE) and blood (plasma) samples were prospectively collected at the beginning and at the time of progression on osimertinib. Tumor response was evaluated using Response Evaluation Criteria in Solid Tumor version 1.1 (RECIST v1.1). Clinical and pathological parameters were obtained by medical chart reviews. We complied with the ethical guidelines of the 1975 Helsinki Declaration and this study was approved by the Independent Review Board of Severance Hospital, and all patients provided informed consent (IRB No. 4-2016-0001).
DNA and RNA extraction
Fresh frozen tumor DNA and RNA were extracted using High Pure PCR Template Preparation Kits, according to the manufacturer’s instruction. ctDNA was also extracted from whole blood using High Pure PCR Template Preparation Kits.
Targeted sequencing analysis
Targeted next-generation sequencing (NGS)-based DNA detection was performed by 600-gene hybrid capture panel designed by AstraZeneca (AZ600) (Supplementary Table 1). A total of 149 samples were sequenced using 600-gene hybrid capture panel. BWA was used to align sequencing reads to human reference build GRCh38. Somatic single-nucleotide variant (SNV) were called using Vardict following manual guidelines. Non-synonymous somatic mutations in genes EGFR, TP53, APC, PIK3CA, ALK, KRAS, EP300, BRAF, and RAF1 were analyzed.
Cell cultures
All cells were cultured in RPMI medium supplemented with 10% FBS and 1% Antibiotic-Antimycotic solution (Thermo Fisher Scientific).The cells were incubated at 37°C and 5% CO2 in a humidified incubator. All reference compounds were purchased from Selleckchem, except osimertinib, which was provided by AstraZeneca Corporation.
Cell viability assay
Cells were seeded in triplicates in a white-bottom 96-well plate, and drugs were prepared by serial dilution. Dimethyl sulfoxide was added to control wells in the highest dilution used in the assay. The cells were treated for 72 h with the compounds following determination of ATP content as surrogate for viability by CellTiter-Glo® assay (Promega). A volume of CellTiter-Glo® reagent equal to the volume of cell culture medium was added in each well and the plate was incubated for at least 10 min and then luminescence was evaluated. Data were analyzed and plotted using PRISM.
Immunoblot analysis
EGFR, pEGFR (Y1068), MET, pMET(Y1003), AKT, pAKT (S473), ERK, pERK (T202/Y204), S6, pS6 (S240/244), and horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Cell Signaling Technology (Danvers, MA). Actin was obtained from Merck Millipore (Darmstadt, Germany). HRP-dependent luminescence was developed with SuperSignal™ West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Massachusetts, USA) and detected with LAS-4000 lumino-image analyzer system (Fujifilm, Tokyo, Japan).
Colony-forming assay
Cells were seeded at a density of 5000 cells per well on six-well plates. Cells were incubated overnight and exposed to the indicated drugs for 14 days. Medium containing drugs were replenished every 3 days. Then, cells were stained with crystal violet and counted using ImageJ software.
Establishment of resistant cell lines
Human NSCLC cell lines with acquired resistance were isolated by exposure to stepwise increasing doses of EGFR TKIs. Briefly, individual cell lines (H1975, HCC406, HCC827, PC9) were started with 10 nM osimertinib in RPMI 1640 medium containing 10% FBS. Subsequently, cells continuously exposed to increasing drug doses up to 1 µM for more than 6 months. Resistant cell lines are referred to as H1975AR, HCC4006AR, HCC827AR, PC9AR, and PC9GRAR, all resistant to osimertinib (Supplementary Figure 3). Whole exome sequencing (WES) and RNA sequencing were conducted in each of the resistant cell lines (Macrogen, Seoul, Korea).
Establishment of patient-derived xenografts and patient-derived cells
Patient-derived xenograft models were generated using 6- to 8-week-old female nude (nu/nu) and severe combined immunodeficient (NOG, NOD/Shi/scid/IL-2Rxnull) mice (OrientBio, Seoul, Korea), as described previously. 19
Patient-derived cells were established from malignant pleural effusions as previously described. 20 Briefly, samples were centrifuged at 500g for 10 min before cell pellets were suspended in phosphate-buffered saline (PBS). Then, cells were separated by density gradient centrifugation using Ficoll-PaquePLUS (GE Healthcare Bio-Sciences, Uppsala, Sweden). Mononuclear cells including tumor cells were isolated from the interphase layer, washed twice with HBSS, suspended in R10 medium, and seeded on collagen IV pre-coated culture plates at a density of approximately 1 to 2 × 106 cells per plate.
Whole exome sequencing
DNA from resistant cell lines and patient-derived models was captured using the SureSelect Human All Exon V6 (5190-8881, Agilent, Santa Clara, CA, USA). Sequencing libraries were constructed for the NovaSeq 6000 system (Illumina, San Diego, CA, USA) and sequenced using the 110-bp paired-end mode of the NovaSeq 6000 Reagent Kit (20039236, Illumina, San Diego, CA, USA). Exome sequencing reads were aligned to the hg38 reference genome using BWA-0.7.17. Putative duplications were marked by Picard (version picard-tools-2.18.2-SNAPSHOT). Sites harboring small insertions or deletions were realigned and recalibrated by employing GATK (v4.0.5.1) modules with known variant sites identified from the 1000 Genomes Project and dbSNP-151. GATK4 Mutect2 was used to call somatic mutations. The average coverage for WES was 150×.
RNA sequencing and analysis
RNA was extracted from resistant cell lines and patient-derived models using QIAGEN mini kit according to the manufacturer’s instruction. RNA quality and concentration was assessed using an Experion system (Bio-Rad). Paired reads of a length of 50 base pairs (bp) were sequenced on the HiSeq2500. Gene expression profiling from RNA-seq data was performed on ATCC resistance cell lines and eight patient-derived xenograft cells. RNA reads were aligned to the GRCh37 with STAR version 2.6.1. Read count normalization and differential gene expression analysis were performed using EdgeR. Differential expressed genes were identified using a p-value <0.05 threshold. Fusions were identified using Defuse.
Copy number variant analysis
Gene copy number status in 149 clinical samples, 5 ATCC-resistant cell lines, and 8 patient-derived xenografts were derived from DNA sequencing data using CNVkit- 21 software. Copy number segments were annotated with gene names. Copy number alterations were defined as copy number gain (copy number ⩾3) and copy number loss (Copy number 1 or 0) in this study. For the acquired copy number analysis, copy number variants (CNVs) in progression samples were assessed with respect to the matched baseline sample.
In vivo xenograft study
Female athymic BALB-c/nu mice were obtained from Orient Bio at 5–6 weeks of age. All mice were handled in accordance with the Animal Research Committee’s Guidelines at Yonsei University College of Medicine, and all facilities were approved by American Association for Accreditation of Laboratory Animal Care (AAALAC). Once the tumor volume reached approximately 150–200 mm3, mice were randomly allocated into groups of five animals to receive vehicle, osimertinib, trametinib, buparlisib, trametinib + buparlisib, and osimertinib + trametinib + buparlisib. The tumor size was measured every 2 days using calipers. The average tumor volume in each group was expressed in mm3 and calculated according to the equation for a prolate spheroid: tumor volume = 0.523 × (large diameter) × (small diameter)2.
Statistical analysis
All statistical analysis was performed with R version 3.4.4. PFS was defined as time from starting osimertinib until progressive disease (PD). Overall survival (OS) was defined as time from starting osimertinib until death. Kaplan–Meier method was used to estimate PFS and OS. Two-sided Fisher’s exact test was used for p value calculations.
Results
Patient characteristics
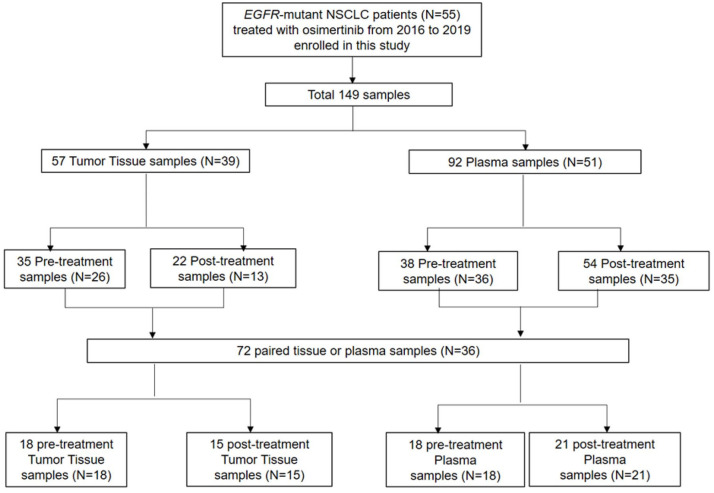
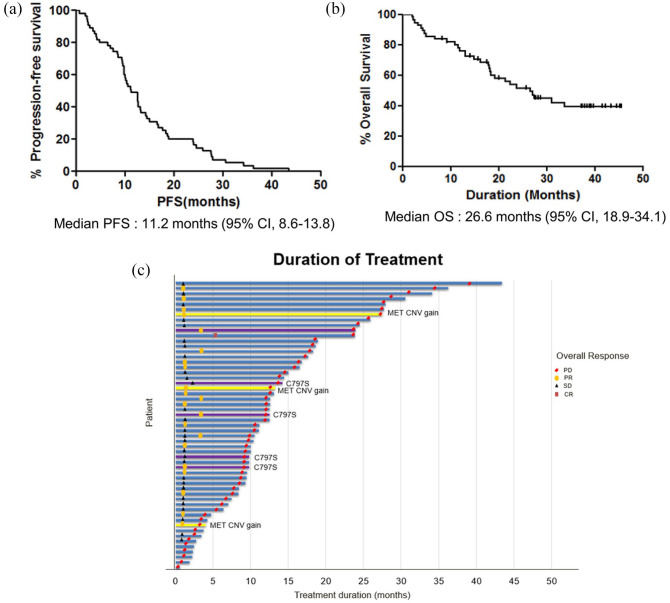
A total of 55 patients were analyzed in the final analysis. Schematic design of samples utilized in the analysis is depicted in Figure 1. Among 55 EGFR-mutant NSCLC patients treated with osimertinib from 2016 to 2019, a total of 149 samples (57 tumor, 92 plasma) were identified. For the efficacy analysis in the intention-to-treat population, all 55 patients were included. For genomic analyses, 72 samples (36 patients) which are amenable for matched pre- and post-treatment were used. Baseline patient characteristics are summarized in Supplementary Table 2. The median age was 65 years (range, 34–78), 71% were females, and 72% were never smokers. All patients had adenocarcinoma histology, and 96% had metastatic disease at diagnosis. At the time of osimertinib treatment, 62% of patients had central nervous system (CNS) disease. Patients had received 1–8 previous lines of therapy, with 1 prior therapy being the most common (44%). For the previous EGFR TKIs, 65% of patients had received gefitinib, 36% had received erlotinib, and 9% received afatinib, in the order of frequency. Two patients received osimertinib for the first-line treatment. Baseline EGFR mutation was predominantly exon 19 deletion (49%) and exon 21 L858R mutation (51%). Eight patients had exon 21 L858R mutation and L861Q compound mutations. The objective response rate of patients treated with osimertinib was 44%. For the median follow-up duration of 28 months, the median PFS was 11.2 months (95% CI, 8.6–13.8), and median OS was 26.6 months (95% CI, 18.9–34.1) (Figure 2(a) and (b)). The swimmer plot shows the duration of osimertinib treatment in all patients (Figure 2(c)). The median duration of treatment was 13.4 months, and eight patients received osimertinib beyond PD.
Figure 1.
The schematic design of study enrollment and sample disposition.
Figure 2.
(a) Kaplan–Meier curve of progression-free survival in all 55 patients. (b) Kaplan–Meier curve of overall survival in all patients. (c) Swimmer’s plot showing the duration of treatment and major acquired resistance mechanisms involving EGFR C797S mutation and MET copy number gain.
Genomic landscape of osimertinib resistance
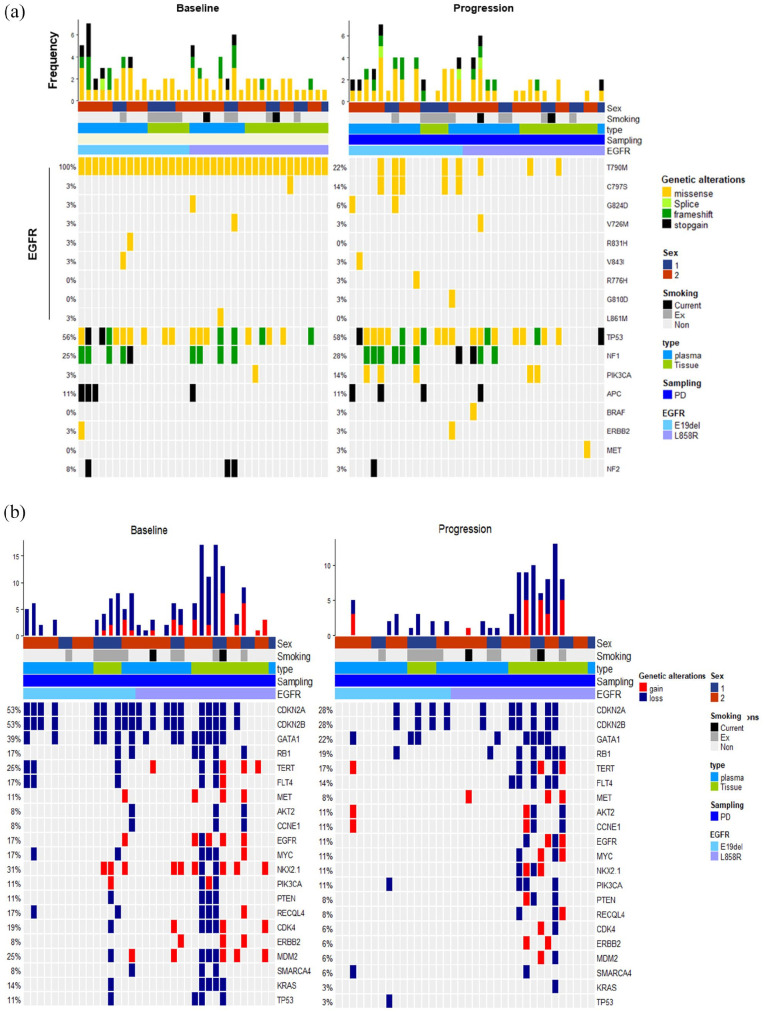
The landscape of genomic alterations identified in pre- and post-treatment samples is described in Figure 3. To characterize the potential acquired resistance mechanisms in osimertinib, we performed targeted NGS for 613 genes from FFPE tissue and plasma samples from patients. All 36 patients harbored T790M mutation at pretreatment (Figure 3(a)). At post-treatment, we identified 14% had C797S mutation all in the presence of T790M mutation, and T790M loss occurred in 78% of patients. Secondary mutations within the EGFR gene such as G824D, V726M, V843I, R775H, and G810D mutations were detected in the post-treatment samples. Besides EGFR and TP53 mutations (no difference between TP53 in before and after treatment), PIK3CA mutation was the most frequent (14%) in progression samples. TP53 gene was significantly mutated in the whole patient cohort. Other recurrent genetic missense mutations in BRAF, ERBB2, and MET were notable in three patients with acquired resistance to osimertinib. Overall, no overtly prevalent mechanism of resistance was noted. With regard to CNV, previously reported amplifications in MET, EGFR, and KRAS occurred in the post-treatment samples (Figure 3(b)). MET CNV gain was identified in 8%.
Figure 3.
Genetic landscape of baseline and progression status from enrolled 36 EGFR mutant lung cancer patients. (a) Oncoplot of somatic SNVs comparison at baseline and at the time of progression in EGFR mutant lung cancer patients. (b) Comparison of oncoplots showing copy number variations at baseline and at the time of progression.
When we analyzed the SNVs identified in the order of frequency (Supplementary Figure 1), the most commonly altered gene at progression was found to be KMT2D, a frequently aberrant epigenetic modifier in various tumors. 22 Interestingly, we identified novel acquired mutations in the post-treatment samples. Acquired GNAS mutation was identified in four (11%) patients with matched pre- and post-treatment samples, and among them two hotspot mutations (R201H, R201S) were found. HNF1A L377fs frameshift mutation was identified in two patients, and LZTR1 R362*, R340* stop gain mutations were identified in two patients, as represented in the lollipop plots in Supplementary Figure 2.
Paired-analysis of matched pre- and post-treatment samples
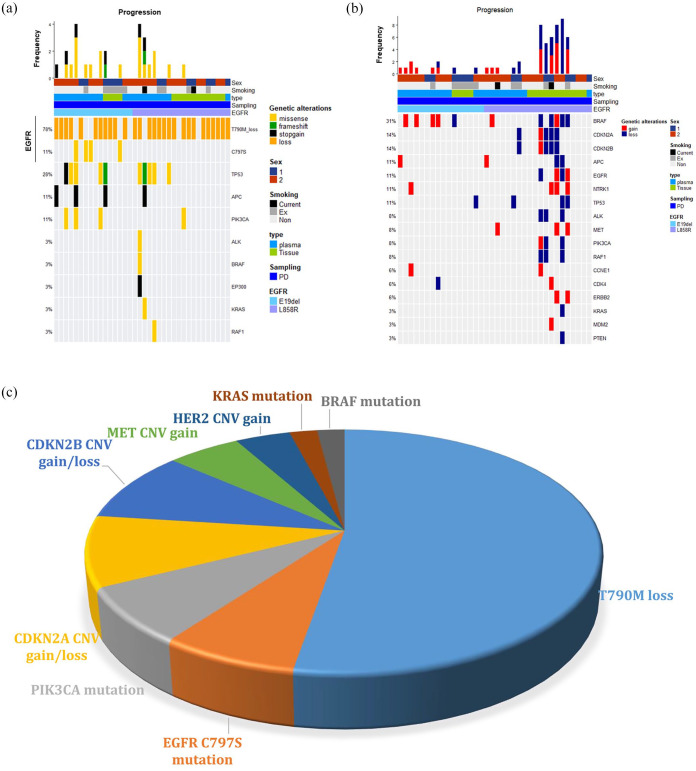
To better understand the acquired resistance mechanisms, we next selected only patients with matched pre- and post-treatment samples. There were 36 patients who had both matched samples (Supplementary Table 3). Again, the most common acquired resistance mechanism was EGFR T790M loss (78%) and EGFR C797S mutation (11%). In addition, PIK3CA mutation was also found in four patients (11%), which include E545K hotspot mutation in one patient (Figure 4(a)). MET CNV gain was identified in three patients (8%), and HER2 CNV gain was identified in two patients (6%) (Figure 4(b)). Comparison of the pre- and post-treatment samples with respect to CNV further revealed frequent alterations in the cell cycle machinery including CCNE1, CDK4, CDKN2A, and CDKN2B, with a total incidence of 5 out of 36 patients (14%) (Figure 4(b)).
Figure 4.
Acquired resistance mechanisms from pre- and post-treatment samples from 36 patients. (a) Oncoplot of acquired SNV mutations in progression samples. (b) Acquired copy number variations found at progression. (c) A pie chart depicting the frequency of acquired resistance mechanisms identified in this study.
We next compared acquired resistance mechanisms based on the baseline truncal mutation (EGFR exon 19 deletion or exon 21 L858R). EGFR C797S mutation was exclusively found in patients with exon 19 deletion (25%), whereas CNV alterations including MET were more common in patients with L858R mutations. MET amplification was identified in three patients with L858R mutation. However, four patients with L858R mutations showed no alterations among known SNVs and CNVs, which may suggest that non-genomic mechanisms of resistance could be involved. Figure 4(c) summarizes the incidences of different mechanisms of acquired resistance. The loss of T790M mutation was the most common mechanism (78%), and CDKN2A gain or loss (16%), CDKN2B gain/loss (16%), PIK3CA mutation (11%), EGFR C797S mutation (11%), MET CNV gain (8%), HER2 CNV gain (6%), KRAS mutation (3%), and BRAF mutation (3%) were identified in the order of frequency.
We also performed histologic evaluation in 21 patients with available post-treatment tissue (data not shown). Of 21 patients, 19 patients had maintained adenocarcinoma histology, while 1 patient showed transformation to small cell lung cancer (SCLC) and 1 patient showed poorly differentiated carcinoma. The patient who showed SCLC transformation did not harbor classical TP53, RB1 loss, 23 but rather had PIK3CA E545K hotspot mutation and CNV gain in PIK3CA, CCNE1, CDKN2A, and CDKN2B.
Serial monitoring of plasma samples
We performed serial monitoring in a patient who received osimertinib beyond progression. A 60-year-old woman with Ex19del EGFR-mutant advanced NSCLC had received six lines of previous chemotherapy regimens including EGFR TKIs (gefitinib, erlotinib) and was treated with osimertinib for 18 months. She had serial plasma collected for AZ600 panel sequencing. At baseline, she harbored EGFR del19 mutation with T790M. After 2 months of osimertinib, the patient had newly developed liver metastasis and plasma ctDNA sequencing revealed newly detected PIK3CA R38H mutation, while T790M mutation was undetectable. She received stereotactic radiotherapy for the liver lesion and continued on osimertinib. After 1 year, she had increasing primary lung lesion, but was clinically stable, and continued on osimertinib. Increased EGFR del19 mutation and EGFR C797S mutation was observed along with primary lung cancer progression at 18 months, until osimertinib was discontinued (Supplementary Figure 3). This case suggests that subclonal expansion of resistant cells may be involved in organ-specific progression.
Analysis of resistance mechanisms in in vitro and patient-derived models
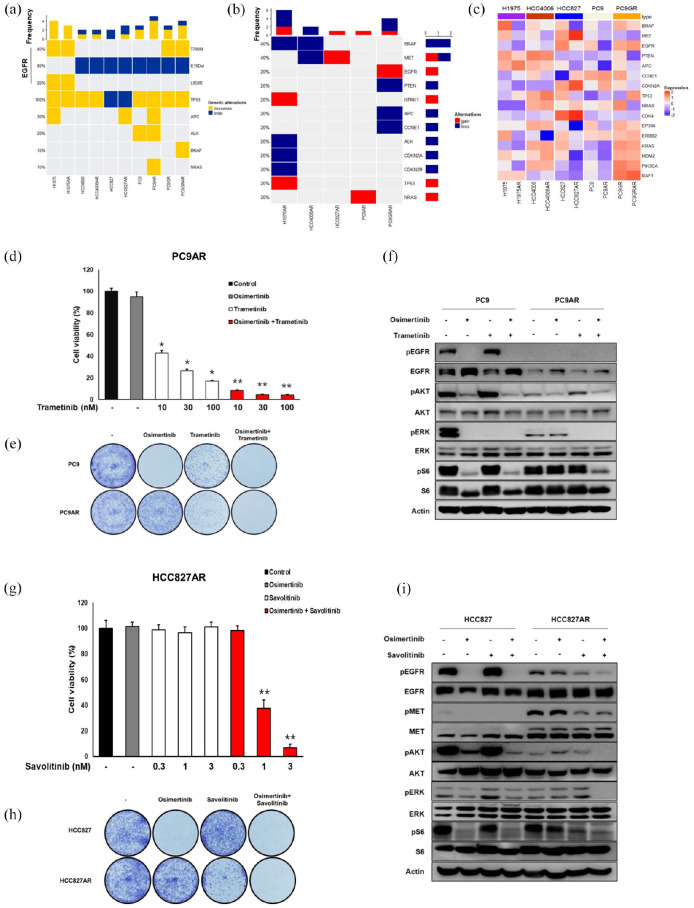
We had previously established osimertinib-resistant preclinical models using five ATCC cell lines (Supplementary Figure 4) and eight patient-derived models. We conducted WES and RNA sequencing in EGFR-mutant ATCC cell lines which have acquired resistance to osimertinib (H1975 AR, HCC4006 AR, HCC827 AR, PC9AR, PC9GRAR) and compared with their respective parental cells. Acquired resistance from WES analysis showed heterogeneous mechanisms of resistance, including recurrent TP53 and APC mutations. PC9AR cells revealed acquired NRAS mutation (p.Q61 K) with concomitant NRAS gene amplification. PC9GRAR cell line showed acquired BRAF V596C mutation (Figure 5(a)). From CNV analysis, H1975 AR and HCC4008 AR showed copy number gain in the NTRK1 gene. HCC827 AR showed CNV gain in the MET gene which is consistent with previous report, 24 and PC9GRAR had EGFR gene gain (Figure 5(b)). RNA seq analysis of ATCC cell lines revealed that MET mRNA expression showed overexpression in HCC827AR and whereas it was under-expressed in HCC4006AR, which were consistent with MET copy number variations (Figure 5(c)). NRAS mRNA overexpression was observed in PC9AR, in accordance with NRAS missense mutation and amplification, as described previously.
Figure 5.
Analysis of resistance mechanisms of osimertinib-resistant cell lines. (a) Oncoplot of somatic SNV mutations. (b) Copy number variations per samples. (c) RNA sequencing expression analysis of osimertinib-resistant cell lines. (d–f) PC9AR cells show NRAS-mediated osimertinib resistance. (d) Cell viability assay shows synergistic inhibition of resistant cell numbers after osimertinib and MEK inhibitor in PC9AR cells. *p < 0.05 and **p < 0.001. (e) Colony-forming assay confirms the efficacy of combination treatment with osimertinib and MEK inhibitor. (f) Immunoblot was performed for EGFR, AKT, ER, and S6 expression after osimertinib and MEK inhibitor treatment for 6 h. (g–i) HCC827AR cells show MET-mediated osimertinib resistance. (g) Cell viability assay shows synergistic inhibition of resistant cell numbers after osimertinib and MET inhibitor in HCC827AR cells. **p < 0.001. (h) Colony-forming assay confirms the efficacy of combination treatment with osimertinib and MET inhibitor. (i) Immunoblot was performed for MET, EGFR, AKT, ER, and S6 expression after osimertinib and MET inhibitor treatment for 6 h.
In an attempt to further investigate the role of NRAS and BRAF mutations, we tested whether treatment with a combination of osimertinib and MEK inhibitor could overcome resistance in these settings. PC9AR cells were treated with increasing doses of osimertinib or trametinib alone or in combination (Figure 5(d)). Combination treatment of osimertinib 100 nmol/L and trametinib 10 nM/L showed synergistic inhibition of resistant cell numbers (Figure 5(d)). Colony formation assay was further performed in PC9 and PC9AR cells to test the long-term effects of osimertinib and trametinib in overcoming NRAS-driven resistance (Figure 5(e)). Combination of osimertinib (100 nM) and trametinib induced synergistic inhibition of the colony formation rates in PC9AR cells. In addition, the combination treatment showed profound reduction of p-EGFR, p-AKT, p-ERK, and p-S6 in PC9AR cells (Figure 5(f)).
We additionally investigated whether combination treatment of osimertinib and savolitinib could overcome resistance driven by MET amplification in HCC827AR. As expected, osimertinib and savolitinib significantly decreased cell viability (Figure 5(g)) and colony formation (Figure 5(h)) and also inhibited the EGFR downstream signaling molecules (Figure 5(i)).
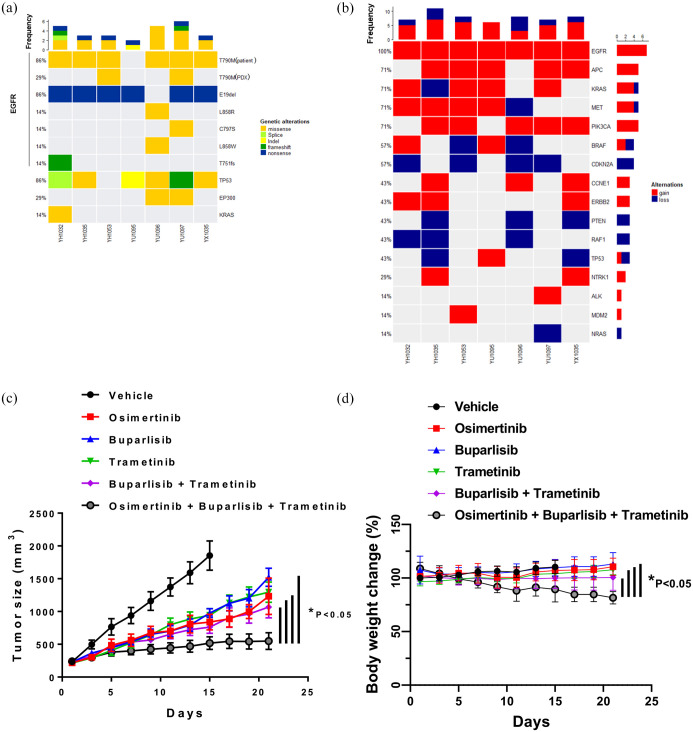
We also investigated mechanisms of acquired resistance in patient-derived xenografts (PDXs) and patient-derived cells (PDCs). A total of three PDXs and four PDCs were established previously and Supplementary Figure 5 shows the patient treatment history and Supplementary Figure 6 shows the characterization of patient-derived models analyzed. Acquired SNVs and CNVs derived from PDXs and PDCs showed both EGFR-dependent (C797S) and EGFR-independent mechanisms of resistance such as PIK3CA p.H1047R mutation, and KRAS p.G12D (Figure 6(a)). Among CNVs, amplifications in EGFR, MET, NTRK1, APC, CCNE1, PIK3CA, and ERBB2 were notable (Figure 6(b)). For a PDX harboring acquired KRAS p.G12D mutation, we investigated whether combination of osimertinib, MEK inhibitor (trametinib), and PI3K inhibitor (buparlisib) could reverse resistance. The combination of osimertinib, trametinib, and buparlisib led to significant tumor regression compared with osimertinib, buparlisib, trametinib, or buparlisib plus trametinib (Figure 6(c)). However, despite the profound antitumor activity, triple combination treatment resulted in a significant reduction in the body weight, warranting a further study to optimize the dosing of each drug.
Figure 6.
Genetic landscape of osimertinib-resistant PDXs and PDCs. (a) Oncoplot of somatic SNV mutations. (b) Copy number variations per sample. (c) The synergistic antitumor efficacy of combination of osimertinib, MEK inhibitor, and PI3K inhibitor in PDX harboring acquired KRAS G12D mutation. Mice were treated with osimertinib 25 mg/kg, trametinib 0.5 mg/kg, and buparlisib 35 mg/kg once daily for 3 weeks after the tumor volume reached 200 mm 3 . Data represent the mean ± SEM (n = 5/group). *p < 0.05. (d) The body weight change in PDX harboring acquired KRAS G12D mutation. Mice were treated with osimertinib 25 mg/kg, trametinib 0.5 mg/kg, and buparlisib 35 mg/kg once daily for 3 weeks after the tumor volume reached 200 mm 3 . Data represent the mean ± SEM (n = 5/group). *p < 0.05.
Discussion
Our study outlines the considerable inter- and intra-patient heterogeneity of resistance mechanisms developed in EGFR-mutated tumors in response to osimertinib. We identified that osimertinib resistance was associated with both EGFR-dependent and EGFR-independent genomic alterations via targeted deep sequencing of patient samples.
To comprehensively investigate the genetic landscape of osimertinib resistance, we established the seven PDCs and PDXs and five osimertinib resistance cell lines, and we observed SNV, CNV, and RNA expression patterns of osimertinib pre-treatment and post-treatment status. We identified that PC9AR cells harboring acquired NRAS mutation (p.Q61 K) with concomitant NRAS gene amplification could be sensitive to combination of osimertinib and trametinib. HCC827 AR showed CNV gain in the MET gene which is consistent with a previous report 24 and was sensitive to combination of osimertinib and savolitinib, as expected. In addition, we, for the first time, identified that acquired KRAS G12D mutation identified in PDX could be overcome by combination of osimertinib, PI3K inhibitor, and MEK inhibitor. These results suggest that patient-derived preclinical models serve as important tools in identifying resistance mechanisms and to find novel therapeutic strategy to overcome resistance.
Distant metastasis is a major challenge of treatment failure, and there is accumulating evidence suggesting that metastases are caused by the evolution of primary tumor. 25 Still, better understanding of metastatic process and evolutionary pattern in distant metastatic sites is required. Toward this goal, we conducted serial monitoring of plasma ctDNA analysis in a patient to explore genomic mutational pattern associated with treatment failure. We noted that PIK3CA R38H mutation appeared with liver progression, and EGFR C797S mutation was associated with primary lung cancer progression. However, it was a single case, and the results are limited to targeted gene panel. It would be desirable to analyze multiple metastatic lesions obtained at different stages of tumor progression over time. Recently, Shi et al. reported that MYC, YAP1, or MMP13 overexpression could increase the incidence of brain metastasis. 26 Further works on elucidating unique mutational landscape and evolutionary process in metastases might shed a light on developing precise strategy in clinical settings.
We identified novel acquired mutations which could have a role in osimertinib resistance: GNAS R201H/S and LZTR1 stopgain mutation (R362*, R340*). GNAS hotspot mutation was identified in two patients which were reported to activate ERK/MAPK pathway previously.27,28 GNAS is a proto-oncogene which encodes for the Gsα subunit of heterotrimeric G-proteins and can transduce downstream signals via the G-stimulatory pathway and increase cyclic AMP production. 29 Although therapies targeting GNAS mutations are still not available, inhibitors of MAPK pathways, such as MEK inhibitor, could be relevant to GNAS-activating mutations. Mutations at leucine zipper-like transcriptional regulator 1 (LZTR1) were seen in two patients, and loss of function in the LZTR1 gene could result in a Ras-dependent gain-of-function phenotype 30 and dysregulate RAS ubiquitination. 31 While these suggest that the LZTR1 stopgain mutations (R362*, R340*) could lead to RAS pathway activation, the putative and functional role of GNAS and LZTR1 mutations in the acquired resistance of osimertinib needs to be further validated.
It is worth noting that our study has several limitations. This is a single-center study, and the patient data was obtained from targeted sequencing; therefore, our dataset may not capture the true diversity of somatic mutational landscape. In addition, not all matched pre- and post-treatment samples were obtained from the same sample type (tissue or plasma). Although somatic mutations detected via tissue biopsy and ctDNA are highly concordant at the gene level, both sources may exhibit additional unique mutations that are missed by the other. In addition, detection of copy number alterations from plasma samples remains challenging due to low tumor content in the circulation. This may impact the observed prevalence of CNVs in this study.
In conclusion, genetic profiles of osimertinib-resistant NSCLC patient samples using targeted deep sequencing revealed that osimertinib resistance was associated with both EGFR-dependent and EGFR-independent genomic alterations. In vitro and in vivo models harboring osimertinib resistance provide insights to potential novel treatment strategies after osimertinib failure.
Supplemental Material
Supplemental material, sj-docx-1-tam-10.1177_17588359221079125 for Molecular landscape of osimertinib resistance in patients and patient-derived preclinical models by Sun Min Lim, San-Duk Yang, Sangbin Lim, Seong Gu Heo, Stetson Daniel, Aleksandra Markovets, Rafati Minoo, Kyoung-Ho Pyo, Mi Ran Yun, Min Hee Hong, Hye Ryun Kim and Byoung Chul Cho in Therapeutic Advances in Medical Oncology
Supplemental material, sj-pptx-1-tam-10.1177_17588359221079125 for Molecular landscape of osimertinib resistance in patients and patient-derived preclinical models by Sun Min Lim, San-Duk Yang, Sangbin Lim, Seong Gu Heo, Stetson Daniel, Aleksandra Markovets, Rafati Minoo, Kyoung-Ho Pyo, Mi Ran Yun, Min Hee Hong, Hye Ryun Kim and Byoung Chul Cho in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-1-tam-10.1177_17588359221079125 for Molecular landscape of osimertinib resistance in patients and patient-derived preclinical models by Sun Min Lim, San-Duk Yang, Sangbin Lim, Seong Gu Heo, Stetson Daniel, Aleksandra Markovets, Rafati Minoo, Kyoung-Ho Pyo, Mi Ran Yun, Min Hee Hong, Hye Ryun Kim and Byoung Chul Cho in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-2-tam-10.1177_17588359221079125 for Molecular landscape of osimertinib resistance in patients and patient-derived preclinical models by Sun Min Lim, San-Duk Yang, Sangbin Lim, Seong Gu Heo, Stetson Daniel, Aleksandra Markovets, Rafati Minoo, Kyoung-Ho Pyo, Mi Ran Yun, Min Hee Hong, Hye Ryun Kim and Byoung Chul Cho in Therapeutic Advances in Medical Oncology
Footnotes
Author contributions: Sun Min Lim: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration.
San-Duk Yang: Formal analysis; Investigation; Methodology; Project administration.
Sangbin Lim: Data curation; Formal analysis; Methodology; Project administration.
Seong Gu Heo: Formal analysis
Stetson Daniel: Formal analysis; Investigation; Methodology; Project administration; Supervision.
Aleksandra Markovets: Formal analysis; Investigation; Methodology; Supervision; Writing – review & editing.
Rafati Minoo: Investigation; Methodology; Supervision; Writing – review & editing.
Kyoung-Ho Pyo: Formal analysis; Investigation; Methodology; Writing – review & editing.
Mi Ran Yun: Conceptualization; Data curation; Formal analysis; Investigation; Methodology.
Min Hee Hong: Conceptualization; Data curation; Investigation; Project administration; Writing – review & editing.
Hye Ryun Kim: Project administration; Writing – review & editing.
Byoung Chul Cho: Conceptualization; Funding acquisition; Methodology; Project administration.
Conflict of interest statement: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Basic Science Research Program through the NRF funded by the Ministry of Science and ICT (2016R1A2B3016282) to B.C.C., a faculty research grant of Yonsei University College of Medicine (6-2020-0132) to S.M.L., and 7th AstraZeneca-KHIDI (Korea health industry development institute) oncology research program to S.M.L.
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Sun Min Lim, Division of Medical Oncology, Department of Internal Medicine, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Republic of Korea.
San-Duk Yang, Department of Cyber Security & AI Technology, Kyung Hee Cyber University, Seoul, Republic of Korea.
Sangbin Lim, Yonsei Cancer Research Institute, Yonsei University College of Medicine, Seoul, Republic of Korea.
Seong Gu Heo, Yonsei Cancer Research Institute, Yonsei University College of Medicine, Seoul, Republic of Korea.
Stetson Daniel, Translational Science, Oncology R&D, AstraZeneca, Boston, MA, USA.
Aleksandra Markovets, Translational Science, Oncology R&D, AstraZeneca, Boston, MA, USA.
Rafati Minoo, Translational Science, Oncology R&D, AstraZeneca, Boston, MA, USA.
Kyoung-Ho Pyo, Yonsei Cancer Research Institute, Yonsei University College of Medicine, Seoul, Republic of Korea.
Mi Ran Yun, Yonsei Cancer Research Institute, Yonsei University College of Medicine, Seoul, Republic of Korea.
Min Hee Hong, Division of Medical Oncology, Department of Internal Medicine, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Republic of Korea.
Hye Ryun Kim, Division of Medical Oncology, Department of Internal Medicine, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Republic of Korea.
Byoung Chul Cho, Division of Medical Oncology, Department of Internal Medicine, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, 03722, Republic of KoreaYonsei Cancer Research Institute, Yonsei University College of Medicine, Seoul, Republic of Korea.
References
- 1. Sequist LV, Lynch TJ. EGFR tyrosine kinase inhibitors in lung cancer: an evolving story. Annu Rev Med 2008; 59: 429–442. [DOI] [PubMed] [Google Scholar]
- 2. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361: 947–957. [DOI] [PubMed] [Google Scholar]
- 3. Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012; 13: 239–246. [DOI] [PubMed] [Google Scholar]
- 4. Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013; 31: 3327–3334. [DOI] [PubMed] [Google Scholar]
- 5. Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013; 19: 2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drilon A, Cappuzzo F, Ou SI, et al. Targeting MET in lung cancer: will expectations finally be MET. J Thorac Oncol 2017; 12: 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012; 44: 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lim SM, Syn NL, Cho BC, et al. Acquired resistance to EGFR targeted therapy in non-small cell lung cancer: mechanisms and therapeutic strategies. Cancer Treat Rev 2018; 65: 1–10. [DOI] [PubMed] [Google Scholar]
- 9. Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011; 3: 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018; 378: 113–125. [DOI] [PubMed] [Google Scholar]
- 11. Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017; 376: 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang ZH, Lu JJ. Osimertinib resistance in non-small cell lung cancer: mechanisms and therapeutic strategies. Cancer Lett 2018; 420: 242–246. [DOI] [PubMed] [Google Scholar]
- 13. Schoenfeld AJ, Chan JM, Kubota D, et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res 2020; 26: 2654–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramalingam SS, Cheng Y, Zhou C, et al. Mechanisms of acquired resistance to first-line osimertinib: preliminary data from the phase III FLAURA study. Ann Oncol 2018; 29: IX177. [Google Scholar]
- 15. Oxnard GR, Yang JC, Yu H, et al. TATTON: a multi-arm, phase Ib trial of osimertinib combined with selumetinib, savolitinib, or durvalumab in EGFR-mutant lung cancer. Ann Oncol 2020; 31: 507–516. [DOI] [PubMed] [Google Scholar]
- 16. Liu X, Zhang X, Yang L, et al. Abstract 1320: preclinical evaluation of TQB3804, a potent EGFR C797S inhibitor. Can Res 2019; 79(Suppl. 13): S1320. [Google Scholar]
- 17. Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019; 121: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marinis F, Wu YL, De Castro G, Jr, et al. ASTRIS: a global real-world study of osimertinib in >3000 patients with EGFR T790M positive non-small-cell lung cancer. Future Oncol 2019; 15: 3003–3014. [DOI] [PubMed] [Google Scholar]
- 19. Kang HN, Kim JH, Park AY, et al. Establishment and characterization of patient-derived xenografts as paraclinical models for head and neck cancer. BMC Cancer 2020; 20: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roscilli G, De Vitis C, Ferrara FF, et al. Human lung adenocarcinoma cell cultures derived from malignant pleural effusions as model system to predict patients chemosensitivity. J Transl Med 2016; 14: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Talevich E, Shain AH, Botton T, et al. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016; 12: e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell 2013; 49: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niederst MJ, Sequist LV, Poirier JT, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 2015; 6: 6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shi P, Oh YT, Zhang G, et al. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett 2016; 380: 494–504. [DOI] [PubMed] [Google Scholar]
- 25. Turajlic S, Swanton C. Metastasis as an evolutionary process. Science 2016; 352: 169–175. [DOI] [PubMed] [Google Scholar]
- 26. Shih DJH, Nayyar N, Bihun I, et al. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat Genet 2020; 52: 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ritterhouse LL, Vivero M, Mino-Kenudson M, et al. GNAS mutations in primary mucinous and non-mucinous lung adenocarcinomas. Mod Pathol 2017; 30: 1720–1727. [DOI] [PubMed] [Google Scholar]
- 28. Wilson CH, McIntyre RE, Arends MJ, et al. The activating mutation R201C in GNAS promotes intestinal tumourigenesis in Apc(Min/+) mice through activation of Wnt and ERK1/2 MAPK pathways. Oncogene 2010; 29: 4567–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Landis CA, Masters SB, Spada A, et al. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature 1989; 340: 692–696. [DOI] [PubMed] [Google Scholar]
- 30. Bigenzahn JW, Collu GM, Kartnig F, et al. LZTR1 is a regulator of RAS ubiquitination and signaling. Science 2018; 362: 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Steklov M, Pandolfi S, Baietti MF, et al. Mutations in LZTR1 drive human disease by dysregulating RAS ubiquitination. Science 2018; 362: 1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tam-10.1177_17588359221079125 for Molecular landscape of osimertinib resistance in patients and patient-derived preclinical models by Sun Min Lim, San-Duk Yang, Sangbin Lim, Seong Gu Heo, Stetson Daniel, Aleksandra Markovets, Rafati Minoo, Kyoung-Ho Pyo, Mi Ran Yun, Min Hee Hong, Hye Ryun Kim and Byoung Chul Cho in Therapeutic Advances in Medical Oncology
Supplemental material, sj-pptx-1-tam-10.1177_17588359221079125 for Molecular landscape of osimertinib resistance in patients and patient-derived preclinical models by Sun Min Lim, San-Duk Yang, Sangbin Lim, Seong Gu Heo, Stetson Daniel, Aleksandra Markovets, Rafati Minoo, Kyoung-Ho Pyo, Mi Ran Yun, Min Hee Hong, Hye Ryun Kim and Byoung Chul Cho in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-1-tam-10.1177_17588359221079125 for Molecular landscape of osimertinib resistance in patients and patient-derived preclinical models by Sun Min Lim, San-Duk Yang, Sangbin Lim, Seong Gu Heo, Stetson Daniel, Aleksandra Markovets, Rafati Minoo, Kyoung-Ho Pyo, Mi Ran Yun, Min Hee Hong, Hye Ryun Kim and Byoung Chul Cho in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-2-tam-10.1177_17588359221079125 for Molecular landscape of osimertinib resistance in patients and patient-derived preclinical models by Sun Min Lim, San-Duk Yang, Sangbin Lim, Seong Gu Heo, Stetson Daniel, Aleksandra Markovets, Rafati Minoo, Kyoung-Ho Pyo, Mi Ran Yun, Min Hee Hong, Hye Ryun Kim and Byoung Chul Cho in Therapeutic Advances in Medical Oncology