Abstract
Background: MicroRNAs (miRNAs) play an important regulatory role and serve as biomarkers in various human cancers. However, their role in the prognosis and predicting response to therapy in Indian lung cancer patients is not fully explored. Methods: We collected surgically resected tumors and paired adjacent normal lung tissues from 29 early-stage and tissue biopsies from 103 locally advanced and metastatic lung cancer patients in this prospective study. We quantified the expression levels of miR-375-3p, miR-197-3p, and miR-15a-5p using TaqMan Advanced miRNA Assays. We correlated miRNAs expression with response to therapy and survival outcomes. Results: The median age of lung cancer patients was 60 years. We found significant overexpression of miR-375-3p and miR-197-3p in the tumors compared to paired normal lung tissues. Higher expression of miR-375-3p was observed more frequently in responders compared to nonresponders. The expression of miR-375-3p and miR-197-3p was able to differentiate patients of lung adenocarcinoma from lung squamous cell carcinoma. We did not find any correlation between miRNAs expression and survival outcomes. Conclusion: Overexpression of miR-375-3p and miR-197-3p might contribute to lung carcinogenesis. The expression of miR-375-3p may assist in predicting therapeutic response. More prospective studies are warranted to evaluate the potential of miR-375-3p as a predictive biomarker of response to therapy.
Keywords: lung cancer, microRNA, miR-375-3p, miR-197-3p, miR-15a-5p, prognosis
Introduction
Lung cancer constitutes 11.6% of all cancer cases, making it the most commonly diagnosed cancer globally. 1 With 18.6% of all cancer-related deaths globally, it contributes significantly to cancer-related mortality. 1 In India, lung cancer incidence and mortality are 5.9% and 8.1%, respectively. 1 The most common histological subtype of lung cancer is nonsmall cell lung cancer (NSCLC; ∼85%) and is seen in both smokers and nonsmokers. 2 In contrast, small cell lung cancer (SCLC) is mostly smoking-related and constitutes ∼15% of all lung cancer cases. 3 Histologically, NSCLC is further categorized into lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD), and large cell carcinoma. 2 Therapeutic strategies for managing NSCLC depend on the stage, histological subtype, and the presence or absence of specific driver mutations in their tumors.4–6 The tumors of ∼50% to 60% of LUAD patients show the presence of molecular alterations in a few oncogenic driver genes such as EGFR, ALK, ROS1, BRAF, and MET.4–7 As a result, these patients become eligible to be treated by driver mutation-specific targeted therapies and show better objective response rates leading to significant improvements in their survival outcomes compared to traditional chemotherapeutic drugs.4–7 In contrast, the majority of the SCLC and LUSC patients with locally advanced and metastatic disease and LUAD patients without any targetable driver mutations are treated with chemotherapy and/or radiotherapy.4–7 However, the response rate and survival outcomes in these groups of patients are relatively inferior, which could be because, at present, there are no effective biomarkers to predict prognosis and response to therapy. 7
MicroRNAs (miRNAs) are small noncoding RNAs involved in posttranscriptional gene silencing and have emerged as important regulators of various biological processes implicated in carcinogenesis.8–11 Several studies have evaluated their utility as diagnostic, prognostic, and predictive biomarkers in various human cancers.12,13 However, there is insufficient data on the profiling of miRNA expression and their role as prognostic and predictive biomarkers in lung cancer patients from the Indian subcontinent.14–19 In our previous study, we performed small RNA sequencing to identify differentially expressed miRNAs in the serum of NSCLC patients compared to controls. 14 We observed a significant downregulation of miR-15a-5p in NSCLC patients compared to controls. 14 Further, the expression of miR-375 was downregulated in LUSC, but not in LUAD patients, compared to controls. 14 We also found differential expression of miR-197-3p in NSCLC patients using small RNA sequencing; although, we did not validate its expression in the larger sample size using qRT-PCR. 14 The main limitation of our previous study was that we did not profile the expression of miRNAs in tumor tissues. Hence, in this study, we quantified the expression levels of miR-375-3p, miR-15a-5p, and miR-197-3p in the tumors and paired normal tissues of early stage and in the tissue biopsies of locally advanced and metastatic lung cancer patients recruited prospectively.
We decided to evaluate the clinical utility of the miRNAs mentioned above in the current study based on their crucial roles in regulating the expression of an immune checkpoint protein, programmed death-ligand 1 (PD-L1), in various human cancers.20–24 Recent evidence suggests that miR-375 can negatively regulate PD-L1 expression indirectly via the JAK2/STAT3 signaling pathway in head and neck squamous cell carcinoma and gastric cancer.20,21 Fujita et al 22 revealed an essential role of the miR-197/CKS1B/STAT3 network in regulating PD-L1 expression, while ectopic overexpression of miR-197 sensitized high PD-L1 expressing drug-resistant NSCLC cells to chemotherapy. Kao et al 23 demonstrated that miR-15a could directly regulate the expression of PD-L1 in malignant pleural mesothelioma. At the same time, extracellular vesicles overexpressing miR-15a inhibited the immune evasion of colorectal cancer cells via the KDM4B/HOXC4/PD-L1 axis. 24 Since PD-L1 is highly expressed in lung tumors25,26 and we recently found differential expression of PD-L1 regulatory miRNAs in the serum of NSCLC patients, 14 we decided further to analyze their expression in the lung tumor tissues to ascertain whether these miRNAs have any clinical utility as prognostic and predictive biomarkers. To confirm the same, we correlated the expression patterns of the miRNAs mentioned above with survival outcomes, therapeutic response, and various clinicopathological parameters, including clinical stage and histology.
Materials and Methods
Recruitment of Patient and Sample Collection
We recruited 132 consecutive newly diagnosed and treatment naïve lung cancer patients prospectively between the years 2017 and 2020. The histopathological diagnosis of lung cancer was established in all the patients by hematoxylin and eosin staining. In addition, we recorded epidemiological data, including age, gender, smoking habit, histology, and clinical stage, for all the patients. The staging was done by computed tomography (CT) scans of the chest and upper abdomen. If clinically indicated, radionuclide bone scan and CT or magnetic resonance imaging of the brain were also performed for staging. The clinical staging was done according to the recommendations of the International Association for the Study of Lung Cancer Staging Committee for NSCLC. 27 Institute Ethics Committee approved this study (Ref. No. IEC-149/07.04.2017, RP-14/2017, and IEC-308/03.05.2019). We obtained informed written consent from all the patients before taking their tissue samples. Further, the identity of all the patients was kept confidential, and all the patient details were de-identified. The reporting of this study conforms to STROBE guidelines. 28
We collected surgically resected lung tumors and paired adjacent normal lung tissue samples (>5 cm away from the primary tumor) from 29 early-stage NSCLC patients undergoing surgical resection. In addition, tissue biopsies were collected from suspected lung cancer patients while undergoing fiberoptic bronchoscopy. All the specimens were collected in RNA stabilization solution (RNAlater, Thermo Fisher Scientific) and stored at −80 °C after incubation at 2 °C to 8 °C for 24 h.
Quantification of miRNA Expression by qRT-PCR
Total RNA enriched with miRNAs was purified from all the specimens using mirVana miRNA Isolation kit (Catalogue no. AM1560, Ambion) and stored at −80 °C until further use. The quantity and purity of total RNA were checked by Qubit RNA broad range assay (Catalogue no. Q10210, Invitrogen) using Qubit 3 fluorimeter (Invitrogen) and Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific). Total RNA was converted to cDNA using TaqMan Advanced miRNA cDNA Synthesis Kit (Catalogue no. A28007, Applied Biosystems) as per recommended protocol. The resulting cDNA was used for quantifying the expression levels of miR-15a-5p (Assay ID: 477858_mir), miR-197-3p (Assay ID: 477959_mir), and miR-375-3p (Assay ID: 478074_mir) using TaqMan Advanced miRNA Assays (Catalogue no. A25576, Applied Biosystems) and TaqMan Fast Advanced Master Mix (Catalogue no. 4444557, Applied Biosystems) on a Light Cycler 480 II instrument (Roche). The relative expression of miRNA was calculated by the 2−ΔCt method after normalizing the expression of target miRNA with the recommended endogenous control miR-423-5p assay (Assay ID: 478090_mir).
Statistical Analysis
All the analyses were performed using GraphPad v 8.0 and Stata v.11.1 (Texas, USA). Continuous variables were presented as the mean (± standard deviation) and compared using the Student's t-test. Categorical variables were presented as a frequency (percentage) and compared using the Chi-square test. We compared the relative expression levels of miRNAs in different biological samples using Mann-Whitney U-test. We used the Kaplan-Meier method for survival analysis. In addition, we performed the univariate and multivariate analyses by Cox regression models to evaluate prognostic significance. Overall survival (OS) was calculated from the date of diagnosis to death or last follow-up. Progression-free survival (PFS) was calculated from the date of diagnosis to the date of progression, relapse, or death. We also performed receiver operating characteristic (ROC) curve analysis (tumor vs paired normal tissue) for each miRNA to calculate the area under the curve (AUC), sensitivity, specificity, and likelihood ratio. The data were censored on July 31, 2021. The significance level was set at P < .05.
Results
Epidemiological and Clinical Characteristics
The demographic and clinical characteristics of 132 patients with lung cancer are summarized in Table 1. The median age of our patients was 60 years (range: 33-81 years). The majority of our patients were male (82.6%), smokers (78.8%), and with clinical-stage III-IV disease (73.2%). Histologically, the vast majority of patients were NSCLC (90.9%), with LUSC being the most common type (57.5%). We detected mutations in the EGFR (exon 19 deletion or L858R point mutation) in 11 out of 31 LUAD patients (35.5%; Table 1). Out of 132, 29 early stage-lung cancer patients underwent curative surgical resection (22%), while 48 patients (36.3%) received first-line chemotherapy or targeted therapy. The vast majority of patients (30 of 48; 62.5%) were given paclitaxel/gemcitabine + carboplatin, while 12.5% (6 of 48) of patients were given cisplatin/carboplatin + pemetrexed. The patients of SCLC (n = 7) were given cisplatin/carboplatin + etoposide, while EGFR mutation-positive LUAD patients (n = 5) received gefitinib, a tyrosine kinase inhibitor. Therapeutic responses were defined following RECIST v. 1.1 criteria and were categorized as complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). 29 Almost 57% (37 of 65) of the patients who achieved CR/PR/SD were categorized as responders, while the remaining patients who had PD were categorized as nonresponders (Table 1).
Table 1.
Baseline Characteristics of Lung Cancer Patients (n = 132).
| Clinicopathological variables | Number of patients (%) |
|---|---|
| Age (years) | |
| Median (range) | 60 (33-81) |
| Gender | |
| Male | 109 (82.6) |
| Female | 23 (17.4) |
| Histology | |
| SCLC | 12 (9.1) |
| NSCLC | 120 (90.9) |
| LUAD | 43/120 (35.8) |
| LUSC | 69/120 (57.5) |
| NSCLC-NOS | 08/120 (6.7) |
| EGFR mutations | |
| Negative | 20/31 (64.5) |
| Positive (L858R and Exon 19 deletions) | 11/31 (35.5) |
| ALK fusion | |
| Negative | 31/32 (96.9) |
| Positive | 01/32 (3.1) |
| Stage | |
| I-II | 29/108 (26.8) |
| III-IV | 79/108 (73.2) |
| Smoking status | |
| Smoker | 104 (78.8) |
| Nonsmoker | 28 (21.2) |
| Treatment received | |
| No treatment | 55/132 (41.7) |
| Surgery | 29/132 (22) |
| Chemotherapy/targeted therapy | 48/132 (36.3) |
| Therapeutic response | |
| Responders | 37/65 (56.9) |
| Nonresponders | 28/65 (43.1) |
Abbreviations: SCLC, small cell lung cancer; NSCLC, nonsmall cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; NSCLC-NOS, NSCLC not specified; EGFR, epidermal growth factor receptor, ALK, anaplastic lymphoma kinase.
miR-375-3p and miR-197-3p are Overexpressed in Lung Tumors
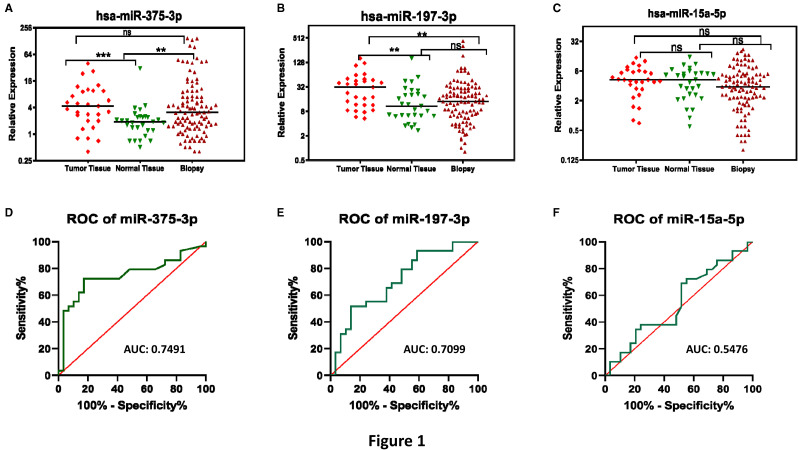
A total of 161 tissue samples, including 29 paired tumor and normal lung tissues and 103 tissue biopsies, from 132 lung cancer patients were used for quantifying the expression levels of miR-375-3p, miR-197-3p, and miR-15a-5p using TaqMan Advanced miRNA Assays (Thermo Fisher Scientific). We found a significantly higher expression of miR-375-3p (Figure 1A) and miR-197-3p (Figure 1B) in the surgically resected tumors compared to paired normal tissues (P = .0009 and P = .0056, respectively; Table 2). These findings are consistent with the data reported in The Cancer Genome Atlas (TCGA)-LUAD datasets for the expression of miR-375 (Supplemental Figure S1A) and in TCGA-LUAD and TCGA-LUSC datasets for the expression of miR-197 (Supplemental Figures S1A and S1D). Further, the expression of miR-375-3p was significantly higher in the tissue biopsies than in unpaired normal lung tissue samples (P = .007; Figure 1A and Table 2). Interestingly, the expression levels of miR-197-3p were significantly higher in surgically resected tumors of early-stage lung cancer patients than tissue biopsies of locally advanced and metastatic lung cancer patients (P = .0065; Figure 1B and Table 2). We did not find any significant differences in the expression levels of miR-15a-5p between various sample types (Figure 1C and Table 2). In contrast, the expression of miR-15a was significantly higher in the TCGA-LUAD (Supplemental Figure S1E), while significantly lower in the TCGA-LUSC tumors than controls (Supplemental Figure S1F). In addition, we checked the diagnostic performance of all 3 miRNAs (tumor vs paired normal) using ROC curve analysis (Figure 1D to F), which revealed that miR-375-3p exhibited the best AUC of 0.7491 with a sensitivity and specificity of 72.41% and 82.76%, respectively (Table 2 and Figure 1D). The miR-15a-5p showed a very low AUC (0.5476) and specificity (44.83%), indicating that it may not be a good diagnostic biomarker (Table 2 and Figure 1F).
Figure 1.
The expression of miR-375-3p and miR-197-3p is upregulated in lung tumors. The column scatter plots showing relative expression of (A) miR-375-3p, (B) miR-197-3p, and (C) miR-15a-5p in surgically resected lung tumors (n = 29), paired adjacent normal lung tissue samples (n = 29), and tissue biopsies (n = 103) of lung cancer patients. ROC curves were generated to assess the diagnostic potential of (D) miR-375-3p, (E) miR-197-3p, and (F) miR-15a-5p for lung cancer (tumor vs paired normal lung tissue). The expression of the miRNAs mentioned above was measured by TaqMan advanced miRNA assays (Thermo Fisher Scientific) and was normalized with that of miR-423-5p expression. ** P < .001; *** P < .0001; ns: not significant; AUC: area under the receiver operating characteristic curve.
Table 2.
Comparison of Relative Expression Levels of miRNAs in Different Biological Samples of Lung Cancer Patients Along With Their Diagnostic Performance Using ROC Curve Analysis.
| miR-375-3p | miR-197-3p | miR-15a-5p | |
|---|---|---|---|
| P value* (tumor vs paired normal) | .0009 | .0056 | .5392 |
| P value* (biopsy vs normal) | .007 | .5072 | .2726 |
| P value* (tumor vs biopsy) | .433 | .0065 | .0783 |
| AUC ± SE (95% CI) (tumor vs normal) | 0.7491 ± 0.0683 (0.6152-0.8830) | 0.7099 ± 0.0680 (0.5765–0.8432) | 0.5476 ± 0.0767 (0.3972-0.6979) |
| Sensitivity % (CI) | 72.41% (54.28-85.30) | 79.31% (61.61-90.15) | 72.41% (54.28-85.30) |
| Specificity % (CI) | 82.76% (65.45-92.40) | 51.72% (34.43-68.61) | 44.83% (28.41-62.45) |
| Likelihood ratio | 4.2 | 1.643 | 1.313 |
Abbreviations: ROC, receiver operating characteristic curve; AUC: area under the ROC curve; SE, standard error; CI, confidence interval.
*Mann-Whitney U-test.
We also established correlations between the expression of various miRNAs using Pearson's correlation coefficient. We found a weak positive correlation between the expression of miR-375-3p and miR-197-3p (r = .2412, P = .0021), miR-375-3p and miR-15a-5p (r = .1635, P = .0383), and a moderate positive correlation between the expression of miR-197-3p and miR-15a-5p (r = .4034, P < .001).
The Expression of miR-375-3p is Correlated With the Therapeutic Response
All lung cancer patients were categorized into high miRNA expression group and low miRNA expression group based on the median ΔCt of individual miRNA expression (Table 3). For correlating miRNA expression with therapeutic responses, lung cancer patients were categorized into 2 groups: (i) responders (those with CR/PR/SD) and (ii) nonresponders (those with PD). Interestingly, higher expression of miR-375-3p was observed more frequently in responders than nonresponders (P = .046; Table 3). On the other hand, the expression of miR-197-3p and miR-15a-5p did not correlate with response to therapy (Table 3). We also correlated the expression of miRNAs with various clinicopathological parameters. Interestingly, the expression of miR-375-3p was significantly higher, while miR-197-3p was significantly lower in LUAD patients (P < .001 and P = .009, respectively; Table 3). We also found a significant correlation of miR-197-3p expression with gender (P = .039) and smoking status (P = .011), while the expression of miR-15a-5p was significantly correlated with the stage of lung cancer patients (P = .031; Table 3).
Table 3.
Correlation of miRNA Expression With Various Clinicopathological Parameters of Lung Cancer Patients.
| Characteristics | miR-375-3p expression a | miR-197-3p expression a | miR-15a-5p expression a | ||||||
|---|---|---|---|---|---|---|---|---|---|
| High expression | Low expression | P value b | High expression | Low expression | P value b | High expression | Low expression | P value b | |
| Age (years) |
34 (50.7%) 32 (49.2%) |
.862 |
|||||||
| ≤ 60 | 37 (55.2%) | 30 (44.8%) | .297 | 31 (46.3%) | 36 (53.7%) | .384 | 33 (49.3%) | ||
| > 60 | 30 (46.2%) | 35 (53.8%) | 35 (53.8%) | 30 (46.2%) | 33 (50.8%) | ||||
| Gender |
56 (51.4%) 10 (43.5%) |
.491 |
|||||||
| Male | 52 (47.7%) | 57 (52.3%) | .127 | 59 (54.1%) | 50 (45.9%) | .039 | 53 (48.6%) | ||
| Female | 15 (65.2%) | 08 (34.8%) | 07 (30.4%) | 16 (69.6%) | 13 (56.5%) | ||||
|
Histology SCLC |
08 (66.7%) | 04 (33.3%) | .248 | 09 (75%) | 03 (25%) | .069 | 08 (66.7%) |
04 (33.3%) 62 (51.7%) |
.226 |
| NSCLC | 59 (49.2%) | 61 (50.8%) | 57 (47.5%) | 63 (52.5%) | 58 (48.3%) | ||||
| NSCLC |
20 (46.5%) 38 (55.1%) |
.378 |
|||||||
| LUAD | 31 (72.1%) | 12 (27.9%) | <.001 | 14 (32.6%) | 29 (67.4%) | .009 | 23 (53.5%) | ||
| LUSC | 24 (34.8%) | 45 (65.2%) | 40 (58%) | 29 (42%) | 31 (44.9%) | ||||
| Smoking status |
11 (39.3%) 55 (52.9%) |
.201 |
|||||||
| Nonsmoker | 18 (64.3%) | 10 (35.7%) | .107 | 08 (28.6%) | 20 (71.4%) | .011 | 17 (60.7%) | ||
| Smoker | 49 (47.1%) | 55 (52.9%) | 58 (55.8%) | 46 (44.2%) | 49 (47.1%) | ||||
| Stage |
09 (31%) 43 (54.4%) |
.031 |
|||||||
| I + II | 17 (58.6%) | 12 (41.8%) | .614 | 18 (62.1%) | 11 (37.9%) | .160 | 20 (69%) | ||
| III + IV | 42 (53.2%) | 37 (46.8%) | 37 (46.8%) | 42 (53.2%) | 36 (45.6%) | ||||
| EGFR |
10 (50%) 05 (45.4%) |
.809 |
|||||||
| No mutation | 15 (75%) | 05 (25%) | .505 | 03 (15%) | 17 (85%) | .408 | 10 (50%) | ||
| Mutation | 07 (63.6%) | 04 (36.4%) | 03 (27.3%) | 08 (72.7%) | 06 (54.6%) | ||||
| Therapeutic response |
13 (35.1%) 15 (53.6%) |
.137 |
|||||||
| Responders | 25 (67.6%) | 12 (32.4%) | .046 | 21 (56.8%) | 16 (43.2%) | .798 | 24 (64.9%) | ||
| Nonresponders | 12 (42.9%) | 16 (57.1%) | 15 (53.6%) | 13 (46.4%) | 13 (46.4%) | ||||
Abbreviations: SCLC, small cell lung cancer; NSCLC, nonsmall cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Patients were divided into high and low miRNA expression groups based on median ΔCt value (mean Ct of target miRNA–mean Ct of control miRNA) of miR-375-3p or miR-197-3p or miR-15a-5p.
Chi-square test.
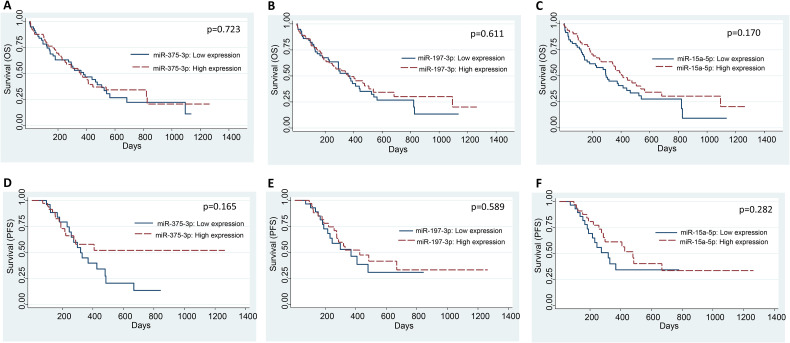
The Expression of miR-375-3p, miR-197-3p, and miR-15a-5p is not Correlated With Survival Outcomes
The median OS and PFS were 274 days (range: 1-1265 days) and 251.5 days (range: 1-1265 days), respectively. We performed the univariate and multivariate analyses by Cox regression model to find the correlation of miRNAs expression and various clinicopathological variables with OS and PFS (Tables 4 and 5). We did not find any correlation between the expression of any of the miRNAs and OS (Figure 2A to C) or PFS (Figure 2D to F) of lung cancer patients (Tables 4 and 5). The expression of miR-375 did not correlate with the OS in TCGA-LUAD and TCGA-LUSC datasets (Supplemental Figures S2A and S2B, respectively). Similarly, the expression of miR-197 and miR-15a did not correlate with the OS in the TCGA-LUAD datasets (Supplemental Figures S3A and S4A, respectively). In contrast, higher expression of miR-197 and lower expression of miR-15a significantly correlated with inferior OS in the TCGA-LUSC dataset (Supplemental Figures S3B and S4B, respectively).
Table 4.
Univariate and Multivariate Analysis of Various Factors for OS.
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR ± SE (95% CI) | P value | HR ± SE (95% CI) | P value | |
| Age (> 60 years vs ≤ 60 years) | 1.02 ± 0.24 (0.65-1.63) | .905 | - | - |
| Gender (female vs male) | 0.82 ± 0.25 (0.45-1.5) | .528 | - | - |
| Histology (NSCLC vs SCLC) | 0.49 ± 0.16 (0.26-0.95) | .033 | 0.84 ± 0.64 (0.18-3.79) | .816 |
| NSCLC histology (LUSC vs LUAD) | 1.65 ± 0.47 (0.95-2.87) | .077 | - | - |
| Smoking (smoker vs nonsmoker) | 1.57 ± 0.48 (0.86-2.86) | .139 | - | - |
| Stage (III + IV vs I + II) | 6.12 ± 2.87 (2.44-15.34) | <.001 | 4.64 ± 3.02 (1.29-16.7) | .019 |
| Therapeutic response (nonresponders vs responders) | 5.32 ± 2.24 (2.32-12.16) | <.001 | 5.02 ± 2.49 (1.9-13.28) | .001 |
| miR-375-3p expression a | 0.92 ± 0.21 (0.58-1.45) | .723 | 0.72 ± 0.34 (0.28-1.82) | .483 |
| miR-197-3p expression a | 0.89 ± 0.20 (0.56-1.4) | .611 | 0.71 ± 0.37 (0.26-1.96) | .509 |
| miR-15a-5p expression a | 0.73 ± 0.17 (0.46-1.14) | .170 | 1.84 ± 1.17 (0.53-6.39) | .339 |
Abbreviations: SCLC, small cell lung cancer; NSCLC, nonsmall cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; HR, hazard ratio; SE, standard error; CI, confidence interval.
Patients were divided into high and low miRNA expression groups based on median ΔCt value (mean Ct of target miRNA–mean Ct of control miRNA) of miR-375-3p or miR-197-3p or miR-15a-5p.
Table 5.
Univariate and Multivariate Analysis of Various Factors for PFS.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variables | HR ± SE (95% CI) | P value | HR ± SE (95% CI) | P value |
| Age (>60 years vs ≤ 60 years) | 0.63 ± 0.23 (0.30-1.30) | .211 | - | - |
| Gender (female vs male) | 0.28 ± 0.17 (0.86-0.94) | .039 | 0.49 ± 0.36 (0.12-2.04) | .326 |
| Histology (NSCLC vs SCLC) | 0.25 ± 0.12 (0.09-0.65) | .004 | 4.63 ± 3.83 (0.92-23.37) | .063 |
| NSCLC histology (LUSC vs LUAD) | 1.33 ± 0.57 (0.58-3.07) | .494 | - | - |
| Smoking (smoker vs nonsmoker) | 1.89 ± 0.86 (0.77-4.61) | .162 | - | - |
| Stage (III + IV vs I + II) | 8.25 ± 3.77 (3.37-20.23) | <.001 | 17.17 ± 11.31 (4.72-62.45) | <.001 |
| Therapeutic response (nonresponders vs responders) | 13.8 ± 6.96 (5.13-37.1) | <.001 | 42.63 ± 31.41 (10.06-180.73) | <.001 |
| miR-375-3p expression a | 0.6 ± 0.22 (0.3-1.22) | .165 | 2.81 ± 1.53 (0.96-8.19) | .059 |
| miR-197-3p expression a | 0.82 ± 0.3 (0.4-1.67) | .589 | 0.65 ± 0.29 (0.27-1.56) | .332 |
| miR-15a-5p expression a | 0.68 ± 0.25 (0.33-1.38) | .282 | 3.02 ± 1.77 (0.96-9.5) | .059 |
Abbreviations: SCLC, small cell lung cancer; NSCLC, nonsmall cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; HR, hazard ratio; SE, standard error; CI, confidence interval.
Patients were divided into high and low miRNA expression groups based on median ΔCt value (mean Ct of target miRNA–mean Ct of control miRNA) of miR-375-3p or miR-197-3p or miR-15a-5p.
Figure 2.
The expression of miR-375-3p, miR-197-3p, and miR-15a-5p does not correlate with OS and PFS of lung cancer patients. Kaplan-Meier curves for correlating the expression of miR-375-3p, miR-197-3p, and miR-15a-5p with OS (A-C) and PFS (D-F) of lung cancer patients. Patients were divided into high and low miRNA expression groups based on median ΔCt value (mean Ct of target miRNA–mean Ct of control miRNA) of miR-375-3p or miR-197-3p or miR-15a-5p.
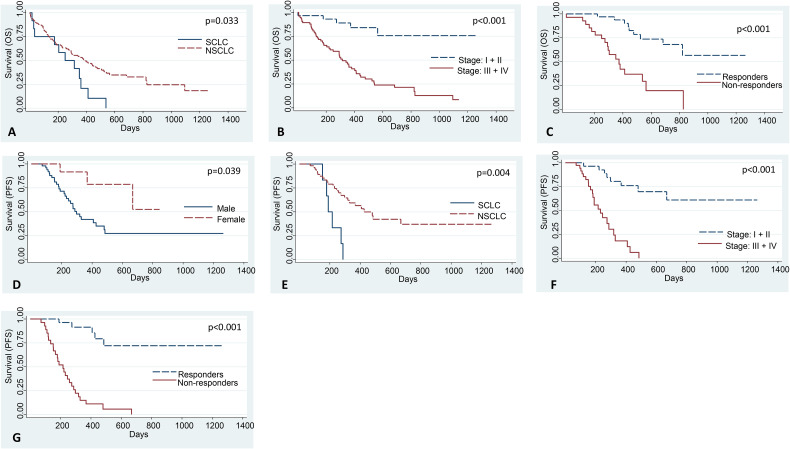
Interestingly, NSCLC patients had significantly better OS compared to SCLC patients (HR = 0.49, P = .033; Figure 3A and Table 4), while patients of stages III and IV had significantly poorer OS as compared to those with stages I and II disease (HR = 6.12, P < .001; Figure 3B). Also, nonresponders had significantly poorer OS compared to responders (HR = 5.32, P < .001; Table 4 and Figure 3C). In multivariate analysis, higher stage (HR = 4.64; P = .019) and poor response to therapy (HR = 5.02; P = 0.001) emerged as independent prognostic factors of inferior OS (Table 4). We also found a significant correlation between gender and PFS (HR = 0.28; P = .039; Figure 3D), histology and PFS (HR = 0.25; P = .004; Figure 3E), stage and PFS (HR = 8.25; P < .001; Figure 3F), and response to therapy and PFS (HR = 13.8; P < .001; Figure 3G). In multivariate analysis, higher stage (HR = 17.17; P < .001) and poor response to therapy (HR = 42.63; P < .001) emerged as independent prognostic factors of inferior PFS (Table 5).
Figure 3.
The correlation of various clinicopathological parameters with OS and PFS of lung cancer patients. Kaplan-Meier curves for correlating (A) histology, (B) stage, and (C) response to therapy with OS of lung cancer patients. Lung cancer patients of NSCLC histology, stages I and II disease, and those who responded to the therapy have significantly better OS than those with SCLC, stages III and IV disease, and nonresponders, respectively. Kaplan-Meier curves for correlating (D) gender, (E) histology, (F) stage, and (G) response to therapy with PFS of lung cancer patients. Lung cancer patients of the female gender, NSCLC histology, stages I and II disease, and those who responded to the therapy have significantly better PFS than those with male gender, SCLC, stages III and IV disease, and nonresponders, respectively.
Discussion
Identification of cancer biomarkers for prognosticating the long-term outcomes and predicting response to anti-cancer therapies is one of the biggest challenges in improving the survival outcomes of cancer patients. Towards this goal, several studies have evaluated the role of miRNAs as diagnostic, prognostic, and predictive biomarkers in various human cancers, including lung cancer.12,13 This study found significant overexpression of miR-375-3p and miR-197-3p in the tumors compared to paired normal lung tissues of early-stage lung cancer patients. Furthermore, when we correlated the miRNA expression with response to therapy, we found that higher expression of miR-375-3p was observed more frequently in responders than nonresponders, which suggests that miR-375-3p may have some potential as a biomarker to predict therapeutic responses. Unfortunately, miRNA expression did not correlate with survival outcomes meaning that miR-375-3p, miR-197-3p, and miR-15a-5p may not have any clinical relevance as prognostic biomarkers in lung cancer.
Apart from tumor tissues derived from early-stage lung cancer patients, miR-375-3p was also significantly overexpressed in the tissue biopsies of locally advanced and metastatic lung cancer patients. Overexpression of miR-375 is also reported in the TCGA-LUAD datasets. However, there are few conflicting reports on the expression of miR-375 in various human cancers, including lung cancer. The published evidence suggests the dual role of miR-375 as an oncomiR and a tumor suppressor miRNA in carcinogenesis depending upon the cellular and biological contexts of tumors.30–32 Few studies have reported lower expression of miR-375 in the tissue and plasma of NSCLC patients.33,34 We previously reported no significant differences in the expression levels of miR-375 in the serum of NSCLC patients compared to controls. 14 Interestingly, we found a significant downregulation of miR-375 expression in the serum of LUSC patients, 14 which is consistent with the present study in which lower expression of miR-375-3p was more frequently observed in LUSC compared to LUAD tumors. Lower expression of miR-375 has also been observed in the TCGA-LUSC tumors compared to normal tissues, although the difference could not reach statistical significance. Another study reported a higher expression of miR-375 in SCLC compared with NSCLC. 35 We found a similar trend in our study; however, the analysis did not reach statistical significance, most probably due to the lower number of SCLC patients in the present study. In our study, we could not find any prognostic utility of miR-375-3p, which is in contrast to other studies which reported that lower expression of miR-375 significantly correlated with poor survival outcomes in patients with NSCLC. 34 Lower expression of miR-375 in tissue and serum or plasma is associated with poor prognosis in most human cancers. 30 In contrast, overexpression of miR-375 was associated with local recurrence in breast cancer 36 and local relapse in prostate cancer. 37 Interestingly, in our study, overexpression of miR-375-3p was observed more frequently in responders than nonresponders. To the best of our knowledge, none of the other studies has reported the potential utility of tumor miR-375 expression in predicting response to therapy. However, our observations should be interpreted with caution since patients in which response was assessed were treated with different therapeutic regimens based on their histologies and the targetable driver mutations, which can also influence the therapeutic response.
The multifaceted role of miR-197 in regulating diverse biological processes in various human cancers is well established. 38 We found overexpression of miR-197-3p in the surgically resected tumors compared to paired normal tissues of early-stage lung cancer patients as well as tissue biopsies of locally advanced and metastatic lung cancer patients. Overexpression of miR-197 is also observed in the TCGA-LUAD and TCGA-LUSC datasets. Our findings are consistent with other reports, which support that miR-197 functions as an oncomiR in lung cancer.39–42 The overexpression of miR-197 is also seen in the serum of NSCLC patients compared to controls. 43 We did not find any correlation of miR-197-3p expression with OS, PFS, and response to therapy. In contrast, higher expression of miR-197 was associated with poor prognosis in gallbladder cancer 44 and NSCLC. 45 We found a significantly lower expression of miR-197-3p in LUAD tumors compared with LUSC tumors, a finding consistent with another study in NSCLC. 45
The miR-15a/16 gene cluster, which includes miR-15a, miR-15b, miR-16-1, miR-16-2, miR-195, and miR-497, functions as a tumor suppressor in various human cancers.46,47 In our study, we did not find any significant differences in the expression levels of miR-15a-5p in the tumors and paired normal tissues of lung cancer patients. In contrast, various studies have reported downregulation of miR-15a in tumor tissues and serum/plasma of NSCLC patients.46–50 We also did not find any prognostic utility of miR-15a-5p in the present study. In contrast, many studies have reported that lower expression of miR-15a correlates with poor prognosis in many types of cancer. 51 A meta-analysis of 10 studies containing 1616 patients of 7 cancer types concluded that lower expression of miR-15a significantly predicted adverse OS. 51
The main strength of our study is that we measured miRNA expression in early stage and locally advanced and metastatic tumors of lung cancer patients. We also correlated the miRNA expression with OS, PFS, and response to therapy. Our study is the first such report from the Indian sub-continent to the best of our knowledge. Unfortunately, we could not find any prognostic relevance of miRNAs evaluated in the present study, which could be possible due to the small sample size and short follow-up time. Further, our study may be underpowered since we did not perform sample size calculations to assess the utility of studied miRNAs as prognostic and predictive biomarkers. These are the main limitations of our study.
Conclusion
In conclusion, we found significant overexpression of miR-375-3p and miR-197-3p in the tumors compared to paired normal lung tissues of lung cancer patients. The expression of miR-375-3p and miR-197-3p could differentiate patients of lung adenocarcinoma from lung squamous cell carcinoma. Furthermore, the overexpression of miR-375-3p was observed more frequently in responders than nonresponders suggesting that miR-375-3p may be used as a predictive biomarker. However, our conclusions should be interpreted with caution since patients were treated with different therapeutic regimens based on their clinical stage, histologies, and targetable driver mutations, all of which can influence the therapeutic response and survival outcomes. Hence, more prospective studies are warranted to evaluate the role of miR-375-3p and other miRNAs as biomarkers for predicting response to specific therapies, including chemotherapy and targeted therapy. It would also be clinically relevant to evaluate further their role as predictive biomarkers for immunotherapy responses since these miRNAs are known to directly or indirectly regulate the expression of PD-L1, which is an established predictive biomarker for anti-PD-1/PD-L1-based immune checkpoint inhibitors.
Supplemental Material
Supplemental material, sj-pptx-1-tct-10.1177_15330338221080981 for Analysis of miR-375-3p, miR-197-3p, and miR-15a-5p Expression and Their Clinical Relevance as Biomarkers in Lung Cancer by Sachin Kumar, Jyoutishman Saikia, Surender K. Sharawat, Prabhat S. Malik, Sunil Kumar and Anant Mohan in Technology in Cancer Research & Treatment
Acknowledgments
We acknowledge Mr Vikas Gaur (Dept. of Medical Oncology, Dr B. R. Ambedkar Institute Rotary Cancer Hospital, All India Institute of Medical Sciences, New Delhi) for assisting in preparing the column scatter plots and ROC curves.
Glossary
Abbreviations
- miRNA
microRNA
- NSCLC
nonsmall cell lung cancer
- SCLC
small cell lung cancer
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- EGFR
epidermal growth factor receptor
- ALK
anaplastic lymphoma kinase
- qRT-PCR
reverse transcription quantitative polymerase chain reaction
- PD-L1
programmed death-ligand 1
- PD-1
programmed cell death protein 1
- JAK2
Janus kinase 2
- STAT3
signal transducer and activator of transcription 3
- CKS1B
CDC28 protein kinase regulatory subunit 1B
- KDM4B
lysine-specific demethylase 4B
- HOXC4
homeobox C4
- CT
computed tomography
- RNA
ribonucleic acid
- cDNA
complementary DNA
- OS
overall survival
- PFS
progression-free survival
- ROC
receiver operating characteristic curve
- AUC
area under the ROC curve
- CR
complete response
- PR
partial response
- SD
stable disease
- PD
progressive disease
- TCGA
the cancer genome atlas
- HR
hazard ratio
- SE
standard error
- CI
confidence interval
Footnotes
Author Contributions: SK designed the study. SK and SKS performed laboratory experiments and data analysis. JS, PSM, SK, and AM provided clinical samples and epidemiological and survival data. All the authors contributed significantly to the writing of the manuscript. All the authors read and approved the final manuscript.
Ethical Disclosure: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study was approved by the ethics committee of the All India Institute of Medical Sciences, New Delhi (Ref. No. IEC-149/07.04.2017, RP-14/2017, and IEC-308/03.05.2019). Informed consent was obtained from the participants involved in the study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study is supported by an extramural research grant from the Indian Council of Medical Research (ICMR), New Delhi (Grant No. 5/13/11/2019/NCD-III) and an intramural research grant from the All India Institute of Medical Sciences, New Delhi (A-516).
Supplemental Material: Supplemental material for this article is available online.
ORCID iD: Sachin Kumar https://orcid.org/0000-0002-2594-6770
References
- 1.Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021;149(4):778‐789. doi: 10.1002/ijc.33588 [DOI] [PubMed] [Google Scholar]
- 2.Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5(3):288‐300. doi: 10.21037/tlcr.2016.06.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer. 2017;17(12):725‐737. doi: 10.1038/nrc.2017.87 [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689): 446‐454. doi: 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 5.Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer. JAMA. 2019;322(8):764. doi: 10.1001/jama.2019.11058 [DOI] [PubMed] [Google Scholar]
- 6.Singh N, Agrawal S, Jiwnani S, et al. Lung cancer in India. J Thorac Oncol. 2021;16(8):1250‐1266. doi: 10.1016/j.jtho.2021.02.004 [DOI] [PubMed] [Google Scholar]
- 7.Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol. 2019;16(6):341‐355. doi: 10.1038/s41571-019-0173-9 [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs. Cell. 2004;116(2):281‐2297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 9.Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11(12):849‐864. doi: 10.1038/nrc3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235‐246. doi: 10.1158/2159-8290.CD-15-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh DK, Bose S, Kumar S. Role of microRNA in regulating cell signaling pathways, cell cycle, and apoptosis in non-small cell lung cancer. Curr Mol Med. 2016;16(5):474‐486. doi: 10.2174/1566524016666160429120702 [DOI] [PubMed] [Google Scholar]
- 12.Iorio M V., Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143‐159. doi: 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Lin J, Kong D, et al. Current state of circulating microRNAs as cancer biomarkers. Clin Chem. 2015;61(9):1138‐1155. doi: 10.1373/clinchem.2015.241190 [DOI] [PubMed] [Google Scholar]
- 14.Kumar S, Sharawat SK, Ali A, et al. Identification of differentially expressed circulating serum microRNA for the diagnosis and prognosis of Indian non-small cell lung cancer patients. Curr Probl Cancer. 2020;44(4):100540. doi: 10.1016/j.currproblcancer.2020.100540 [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Sharawat SK, Ali A, et al. Differential expression of circulating serum miR-1249-3p, miR-3195, and miR-3692-3p in non-small cell lung cancer. Hum Cell. 2020;33(3):839 ‐ 8849. doi: 10.1007/s13577-020-00351-9 [DOI] [PubMed] [Google Scholar]
- 16.Khandelwal A, Seam RK, Gupta M, et al. Circulating micro RNA-590-5p functions as a liquid biopsy marker in non-small cell lung cancer. Cancer Sci. 2020;111(3):826‐8839. doi: 10.1111/cas.14199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khandelwal A, Sharma U, Barwal TS, et al. Circulating miR-320a acts as a tumor suppressor and prognostic factor in non-small cell lung cancer. Front Oncol. 2021;11:645475. doi: 10.3389/fonc.2021.645475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh A, Kant R, Saluja T, et al. Differential diagnosis of non-small cell lung carcinoma by circulating microRNA. J Cancer Res Ther. 2020;16(1):127. doi: 10.4103/jcrt.JCRT_872_19 [DOI] [PubMed] [Google Scholar]
- 19.Rai S, Garg P, Bhatt S, et al. The diagnostic role of microRNA 21 in patients with nonsmall cell lung cancer: an exploratory study. Lung India. 2020;37(6):501. doi: 10.4103/lungindia.lungindia_100_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Q, Zhao Y, Sun Y, Yan X, Wang P. miR-375 inhibits IFN-γ-induced programmed death 1 ligand-γ-surface expression in head and neck squamous cell carcinoma cells by blocking JAK2/STAT1 signaling. Oncol Rep. 2018. doi: 10.3892/or.2018.6177 [DOI] [PubMed]
- 21.Yan X-L, Luo Q-Y, Zhou S-N, et al. MicroRNA-375 reverses the expression of PD-L1 by inactivating the JAK2/STAT3 signaling pathways in gastric cancer. Clin Res Hepatol Gastroenterol. 2021;45(5):101574. doi: 10.1016/j.clinre.2020.10.015 [DOI] [PubMed] [Google Scholar]
- 22.Fujita Y, Yagishita S, Hagiwara K, et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther. 2015;23(4):717‐727. doi: 10.1038/mt.2015.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao SC, Cheng YY, Williams M, et al. Tumor suppressor microRNAs contribute to the regulation of PD-L1 expression in malignant pleural mesothelioma. J Thorac Oncol. 2017;12(9):1421‐1433. doi: 10.1016/j.jtho.2017.05.024 [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Yu T, Jin Y, Mai W, Zhou J, Zhao C. MicroRNA-15a carried by mesenchymal stem cell-derived extracellular vesicles inhibits the immune evasion of colorectal cancer cells by regulating the KDM4B/HOXC4/PD-L1 axis. Front Cell Dev Biol. 2021;9:629893. doi: 10.3389/fcell.2021.629893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan J, Lim KS, Mekhail T, Chang C-C. Programmed death ligand-1 (PD-L1) expression in the programmed death receptor-1 (PD-1)/PD-L1 blockade: a key player against various cancers. Arch Pathol Lab Med. 2017;141(6):851‐861. doi: 10.5858/arpa.2016-0361-RA [DOI] [PubMed] [Google Scholar]
- 26.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8(6):467‐477. doi: 10.1038/nri2326 [DOI] [PubMed] [Google Scholar]
- 27.Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The eighth edition lung cancer stage classification. Chest. 2017;151(1):193‐203. doi: 10.1016/j.chest.2016.10.010 [DOI] [PubMed] [Google Scholar]
- 28.von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007;147(8):573. doi: 10.7326/0003-4819-147-8-200710160-00010 [DOI] [PubMed] [Google Scholar]
- 29.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). European journal of cancer (Oxford, England: 1990 ). 2009;45(2):228 − 2247. doi: 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 30.Yan J-W, Lin J-S, He X-X. The emerging role of miR-375 in cancer. Int J Cancer. 2014;135(5):1011‐1018. doi: 10.1002/ijc.28563 [DOI] [PubMed] [Google Scholar]
- 31.Harrandah AM, Mora RA, Chan EKL. Emerging microRNAs in cancer diagnosis, progression, and immune surveillance. Cancer Lett. 2018;438:126‐132. doi: 10.1016/j.canlet.2018.09.019 [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Wang Q, Wen J, Wu Y, Man C. MiR-375: a novel multifunctional regulator. Life Sci. 2021;275:119323. doi: 10.1016/j.lfs.2021.119323 [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Jiang Q, Xia N, Yang H, Hu C. Decreased expression of microRNA-375 in nonsmall cell lung cancer and its clinical significance. J Int Med Res. 2012;40(5):1662‐1669. doi: 10.1177/030006051204000505 [DOI] [PubMed] [Google Scholar]
- 34.Yu H, Jiang L, Sun C, et al. Decreased circulating miR-375: a potential biomarker for patients with non-small-cell lung cancer. Gene. 2014;534(1):60‐65. http://www.ncbi.nlm.nih.gov/pubmed/24404590. [PubMed] [Google Scholar]
- 35.Lu S, Kong H, Hou Y, et al. Two plasma microRNA panels for diagnosis and subtype discrimination of lung cancer. Lung Cancer. 2018;123:44‐51. doi: 10.1016/j.lungcan.2018.06.027 [DOI] [PubMed] [Google Scholar]
- 36.Zehentmayr F, Hauser-Kronberger C, Zellinger B, et al. Hsa-miR-375 is a predictor of local control in early stage breast cancer. Clin Epigenetics. 2016;8(1):28. doi: 10.1186/s13148-016-0198-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porzycki P, Ciszkowicz E, Semik M, Tyrka M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int Urol Nephrol. 2018;50(9):1619‐11626. doi: 10.1007/s11255-018-1938-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, Chen X, Yu D, et al. miR-197: a novel biomarker for cancers. Gene. 2016;591(2):313‐319. doi: 10.1016/j.gene.2016.06.035 [DOI] [PubMed] [Google Scholar]
- 39.Du L, Schageman JJ, Subauste MC, et al. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7(8):1234‐11243. doi: 10.1158/1541-7786.MCR-08-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiori ME, Barbini C, Haas TL, et al. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ. 2014;21(5):774‐782. doi: 10.1038/cdd.2014.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang N, Tian L, Miao Z, Guo N. MicroRNA-197 induces epithelial-mesenchymal transition and invasion through the downregulation of HIPK2 in lung adenocarcinoma. J Genet. 2018;47(1):47‐53. http://www.ncbi.nlm.nih.gov/pubmed/29666324. [PubMed] [Google Scholar]
- 42.Chen Y, Yang C. Mir-197-3p-induced downregulation of lysine 63 deubiquitinase promotes cell proliferation and inhibits cell apoptosis in lung adenocarcinoma cell lines. Mol Med Rep. 2018; 17(3):3921-3927. doi: 10.3892/mmr.2017.8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sui A, Zhang X, Zhu Q. Diagnostic value of serum miR197 and miR145 in non-small cell lung cancer. Oncol Lett. 2019;17(3):3247-3252. doi: 10.3892/ol.2019.9958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng J, Tong L, Zuo H, Li J. MicroRNA-197 promotes proliferation and inhibits apoptosis of gallbladder cancer cells by targeting insulin-like growth factor-binding protein 3. Adv Clin Exp Med. 2021;30(7):661‐672. doi: 10.17219/acem/134833 [DOI] [PubMed] [Google Scholar]
- 45.Mavridis K, Gueugnon F, Petit-Courty A, et al. The oncomiR miR-197 is a novel prognostic indicator for non-small cell lung cancer patients. Br J Cancer. 2015;112(9):1527‐1535. doi: 10.1038/bjc.2015.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang E, Liu R, Chu Y. miRNA-15a/16: as tumor suppressors and more. Future Oncol. 2015;11(16):2351‐2363. doi: 10.2217/fon.15.101 [DOI] [PubMed] [Google Scholar]
- 47.Liu T, Xu Z, Ou D, Liu J, Zhang J. The miR-15a/16 gene cluster in human cancer: a systematic review. J Cell Physiol. 2019;234(5):5496‐5506. doi: 10.1002/jcp.27342 [DOI] [PubMed] [Google Scholar]
- 48.Bandi N, Zbinden S, Gugger M, et al. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69(13):5553‐5559. doi: 10.1158/0008-5472.CAN-08-4277 [DOI] [PubMed] [Google Scholar]
- 49.Yang T, Thakur A, Chen T, et al. MicroRNA-15a induces cell apoptosis and inhibits metastasis by targeting BCL2L2 in non-small cell lung cancer. Tumor Biol. 2015;36(6):4357‐4365. doi: 10.1007/s13277-015-3075-1 [DOI] [PubMed] [Google Scholar]
- 50.Tafsiri E, Darbouy M, Shadmehr MB, Zagryazhskaya A, Alizadeh J, Karimipoor M. Expression of miRNAs in non-small-cell lung carcinomas and their association with clinicopathological features. Tumor Biol. 2015;36(3):1603‐1612. doi: 10.1007/s13277-014-2755-6 [DOI] [PubMed] [Google Scholar]
- 51.Yang F, Li H, Li T, Zhao Y, Liu Z, Li X. Prognostic value of microRNA-15a in human cancers: a meta-analysis and bioinformatics. BioMed Res Int. 2019;2019:1‐12. doi: 10.1155/2019/2063823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pptx-1-tct-10.1177_15330338221080981 for Analysis of miR-375-3p, miR-197-3p, and miR-15a-5p Expression and Their Clinical Relevance as Biomarkers in Lung Cancer by Sachin Kumar, Jyoutishman Saikia, Surender K. Sharawat, Prabhat S. Malik, Sunil Kumar and Anant Mohan in Technology in Cancer Research & Treatment