Abstract
Background
The FAT atypical cadherin 1/2/3/4 (FAT1/2/3/4) has been linked to the occurrence and development of various cancers. However, the prognostic and immunological role of FAT1/2/3/4 in non-small cell lung cancer (NSCLC) has not been clarified.
Methods
The association of FAT1/2/3/4 mutations with tumor mutation burden (TMB), tumor immunity in the microenvironment, and response to ICIs in NSCLC was investigated. Whole-exome sequencing data of lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) samples from the Cancer Genome Atlas (TCGA), and an immunotherapy data set comprising mutation and survival data of 75 NSCLC patients were analyzed. Two independent pan-cancer cohorts with large samples were used to validate the prognostic value of FAT1/2/3/4 mutations in immunotherapy.
Results
A high mutation rate of FAT1/2/3/4 (57.3%, 603/1052) was observed in NSCLC patients. TMB was significantly higher in samples with mutated FAT1/2/3/4 compared to samples with wildtype FAT1/2/3/4 (P < .05). FAT2 mutation was found to be an independent prognostic biomarker in LUAD. FAT1/2/3/4 were aberrantly expressed in LUAD and LUSC, and high FAT2 expression strongly correlated with high PD-L1 levels in LUAD. Moreover, LUAD patients with FAT1 mutations showed significantly high activated dendritic cells infiltration, whereas those with FAT2/3/4 mutations had high infiltration of CD8+ T-cells, M1 macrophages, activated memory CD4+ T-cells, and helper follicular T-cells. It was also observed that FAT1/2/4 mutations were significantly associated with better enhanced objective response and durable clinical benefit, whereas FAT1/2/3 mutations correlated with longer progression-free survival in ICI-treated NSCLC cohort. FAT1/4 mutations were related to better overall survival in pan-cancer patients treated with ICIs.
Conclusions
FAT family genes are potential prognostic and immunological biomarkers and correlate with response to ICIs in NSCLC.
Keywords: FAT atypical cadherin, lung adenocarcinoma, prognosis, immune checkpoint inhibitors, tumor microenvironment
Introduction
Lung cancers are the most prevalent malignancies and the primary cause of cancer-related deaths. 1 Non-small cell lung cancer (NSCLC), which accounts for a large proportion of lung cancers, is classified into 2 broad histologic subtypes: lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). Recent studies have shown that LUAD and LUSC have different immune landscapes.2,3 Recent advancements in molecular biology have resulted in the identification of several genes involved in the pathogenesis of NSCLC. 4
In addition to chemotherapy, radiotherapy, and targeted therapy, immune checkpoint inhibitors (ICIs) therapy is also applied in the treatment of NSCLC. 5 For instance, immune checkpoint proteins like PD-1/PD-L1 and CTLA-4 are important targets for NSCLC treatment.6,7 Currently, pembrolizumab, an anti–PD-1 drug, is used as the first- or second-line treatment for patients with advanced NSCLC. 8 Nivolumab (anti–PD-1) and atezolizumab (anti–PD-L1) were approved for use as second-line treatments for NSCLC irrespective of PD-L1 expression.9,10 Durvalumab (anti–PD-L1) is recommended as maintenance treatment for patients with stage III NSCLC who are not suitable for resection. 5 According to results from a Phase III CheckMate 227 trial, the combination of nivolumab and ipilimumab was recommended (category 2A) as the first-line therapy for patients with metastatic NSCLC.11-13 However, only a limited proportion of patients with NSCLC respond well to ICIs. To enhance the benefits of immunotherapy, it is imperative to identify prognostic biomarkers that will enable individualized application of immunotherapy in NSCLC patients.
FAT atypical cadherins 1/2/3/4 (FAT1/2/3/4) are members of the cadherin superfamily of membrane proteins that contain cadherin repeat sequences.14,15 Some studies have shown that FAT1/2/3/4 expression influences the occurrence and development of various tumors.15-17 FAT1 knockdown in human cancer cell lines promoted tumor progression and cell migration.18,19 The expression of FAT1 was found to be dysregulated in invasive breast cancer and leukemia. 14 The gene signature linked to FAT1 mutation predicts poor lung cancer survival. 16 It has also been shown that silencing FAT1 in squamous cell carcinoma cells increased resistance to EGFR inhibitors, including afatinib and trametinib. 16 Moreover, FAT1 mutations have previously been linked to poor clinical outcome and resistance to CDK4/6 inhibitors in breast cancer. 20 FAT2 and FAT3 have been reported to modulate cell migration in a tissue-specific manner. 21 However, their roles in cancers are not well-known. The FAT4 codes for a single transmembrane protein consisting of 32–34 extracellular cadherin repeats. 21 FAT4 suppresses tumor development, regulate epithelial-to-mesenchymal transition (EMT), and inhibits the growth of numerous cancers, including ovarian, breast, endometrial, colorectal, and gastric cancer.17,22 However, the prognostic and immunological role of FAT family members has not been clarified in NSCLC.
In the present study, we performed a comprehensive analysis of FAT1/2/3/4 mutations on The Cancer Genome Atlas (TCGA) datasets and data from 3 independent cancer cohorts of immunotherapy.23-25 Results showed that FAT1/2/3/4 mutations are associated with high tumor mutation burden (TMB) and tumor-infiltrating immune cells (TICs) in LUAD. In addition, FAT1/2/3/4 mutations can be important predictors of response to ICIs in patients with cancers.
Materials and Methods
TCGA NSCLC Samples
TCGA LUAD and TCGA LUSC data sets consisting of mRNA expression profiles, somatic mutation data, and patient prognosis data were downloaded from the TCGA database (https://portal.gdc.cancer.gov/). Gene expression units were normalized to log2. Strawberry-Perl-5.30.0.1 and R software (Version 3.6.3) were used to collate and normalize original data. The PFS data of TCGA LUAD and TCGA LUSC patients was obtained from cBioPortal (https://www.cbioportal.org). Patients without complete survival data and those with survival period <30 days were excluded from further analysis. Characteristics of TCGA LUAD and TCGA LUSC data sets used for survival analysis are shown in Table S1.
Available Clinical Cohorts of Cancer Patients Undergoing Immunotherapy
To explore the impact of FAT1/2/3/4 mutations on clinical benefits of ICIs, genomic and clinical data of NSCLC patients were analyzed. In addition, neoantigen data from the Hellmann cohort treated with ICIs was analyzed. The Hellmann cohort comprised 75 patients with metastatic NSCLC (59 non-squamous cases and 16 squamous cases) who were treated with anti–PD-1 (nivolumab) and anti–CTLA-4 (ipilimumab) therapy in the Phase III CheckMate 227 trial (Table S2). 23 Two independent pan-cancer cohorts, MSKCC cohort (1661 patients) 24 and MSS mixed solid tumors cohort (249 patients), 25 were used for validating the prognostic value of FAT1/2/3/4 mutations in immunotherapy. However, FAT2/3/4 mutation status were not detected in MSKCC cohort, thereby were not validated. Data of the 3 immunotherapy cohorts are derived from famous clinical studies and was downloaded from the cBioPortal. The detailed information of patients in these cohorts was described in previous published articles.23-25
Estimation of TMB
TMB refers to total somatic nonsynonymous mutation count in coding regions. For each LUAD and LUSC patient in the TCGA cohorts, TMB was calculated as previously described. 26
Estimation of TICs
The CIBERSORT method was adopted to estimate TICs from microarray data at 1000 permutations based on the LM22 signature as the reference in LUAD and LUSC samples. 27 The signature matrix constitutes 547 genes which discriminated hematopoietic cell phenotypes. Each sample was analyzed using the “Genefilter” R package, and a cutoff threshold of P < .05 was used to select samples for subsequent analysis.
Gene Set Enrichment Analysis
Next, the phenotypic and immunologic states associated with the FAT1/2/3/4 mutations were determined using GSEA analyses in samples from the TCGA LUAD dataset. GSEA 4.0 (http://www.gsea-msigdb.org/gsea/index.jsp) was employed to perform GSEA. Gene sets obtained from the MsigDB database (molecular signatures database) were subjected to enrichment analysis. A false discovery rate (FDR) of < 0.05 was used to select enriched gene sets. The normalized enrichment score (NES) is a statistical parameter used to evaluate gene set enrichment data.
Statistical Analysis
Statistical analyses and data presentation were carried out using Graphpad Prism 8.0. The mRNA and TMB data were compared using student t-test between groups if mRNA and TMB were normally distributed. Otherwise, the Mann-Whitney U test was applied. Associations between variables were analyzed with the Chi-square test. Correlation tests were determined using the Spearman’s correlation, with Spearman’s correlation coefficient of ≥0.4 and P < .01 considered significant. Kaplan–Meier, univariate and multivariate Cox regression analyses were performed to determine the relationship between mutations and survival of patients. The log-rank test was employed to calculate P values. Cox regression analysis was conducted to determine HR. All P-values were 2 tailed. P < .05 was set as the cutoff for statistical significance.
Results
FAT1/2/3/4 Mutation Landscape of NSCLC in TCGA Cohort
The prevalence of FAT family gene mutations in a TCGA dataset of NSCLC was analyzed. In the TCGA LUAD cohort (561 cases, Figure 1A), FAT3 had the highest mutation frequency (128; 22.8%), followed by FAT4 (91; 16.2%), FAT1 (58; 10.3%), and FAT2 (55; 9.8%). In the TCGA LUSC cohort (491 cases; Figure 1B), FAT3 had the highest mutation frequency (90; 18.3%), followed by FAT4 (78; 15.9%), FAT1 (72; 14.7%), and FAT2 (31; 6.3%). In the Hellmann NSCLC cohort (75 cases; Figure S1), FAT3 had the highest mutation frequency (16; 21.0%), followed by FAT2 (8; 11.0%), FAT4 (6; 8.0%), and FAT1 (5; 7.0%). In the MSKCC cohort, the mutation frequency of FAT1 is 9.9% (165/1661). In the MSS cohort, FAT4 had the highest mutation frequency (70; 28.1%), followed by FAT3 (48; 19.3%), FAT2 (29; 11.6%), and FAT1 (26; 10.4%). The type of mutations identified included missense mutations, truncating and amplification mutations, spanning over entire gene (Figure 1; Figure S1 and S2).
Figure 1.
Frequency of FAT1/2/3/4 mutations and mutation types in (A) LUAD and (B) LUSC based on the TCGA dataset.
Prognostic Value of FAT1/2/3/4 Mutations in NSCLC
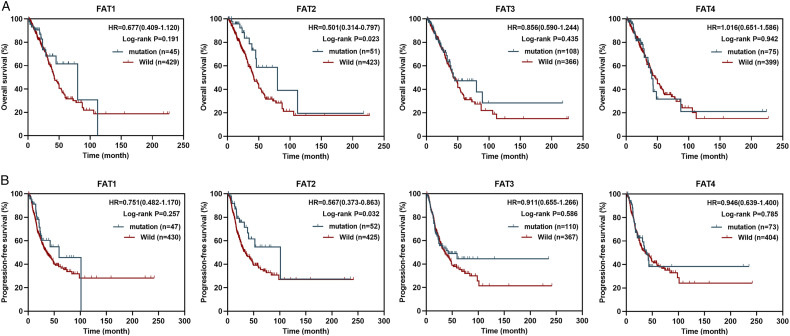
Next, we evaluated the prognostic value of FAT1/2/3/4 mutations in LUAD and LUSC patients using TCGA LUAD and TCGA LUSC data sets. In the TCGA LUAD cohort, FAT2 mutations significantly correlated with OS and PFS (Figure 2A and 2B, P < .05), and predicted good prognosis (HR < 1.0). The median OS and PFS for the FAT2 mutation type were 79.8 and 101.5 months, respectively, relative to 42.2 and 33.9 months for the wildtype FAT2 group. However, in the TCGA LUSC cohort, FAT1/2/3/4 mutations were not significantly associated with OS or PFS (Figure S3).
Figure 2.
The prognostic value of FAT1/2/3/4 mutations in TCGA LUAD cohort. Kaplan–Meier analysis was used to determine correlation between FAT1/2/3/4 mutation status and OS (A) and PFS (B) in LUAD patients.
To determine the independent prognostic value of FAT2 mutations in LUAD patients, univariate and multivariate Cox regression analyses were carried out. In these analyses, FAT2 mutation status and other clinicopathological factors such as, age, M stage gender, TNM stage, T stage, and N stage were included as covariates. It was found that even after adjusting for TNM stage and other covariates, FAT2 mutation status significantly correlated with OS and PFS (Figure 3A and 3B) in LUAD patients, indicating that FAT2 mutation status is an independent factor for the OS and PFS of LUAD.
Figure 3.
Forest plot summary of OS and PFS in the TCGA LUAD cohort. Univariate and multivariable analyses performed to determine the impact of FAT2 mutation, T stage, gender, TNM stage, age, N stage, and M stage on OS (A) and PFS (B) in LUAD patients.
Correlation of FAT Family Genes With the Predictors of Immunotherapy Efficacy
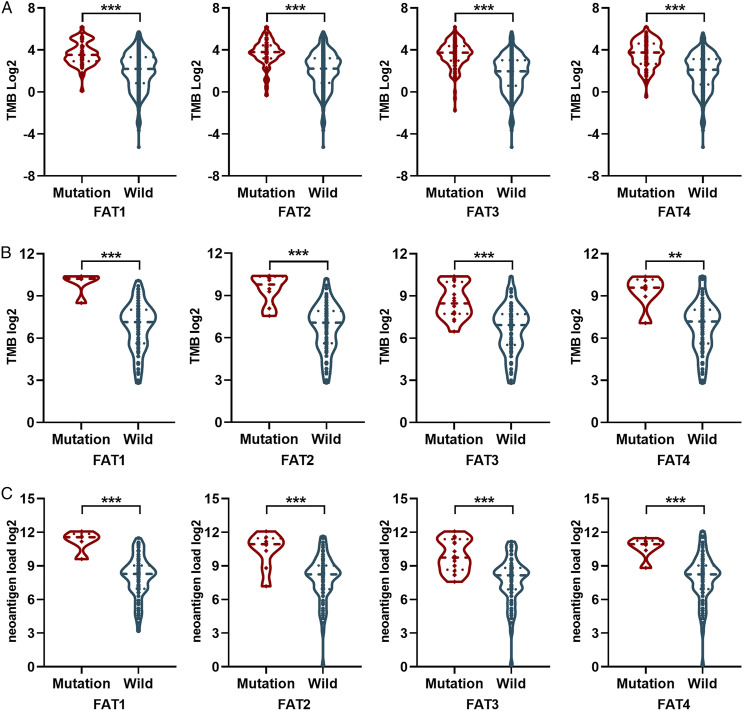
To validate the association of FAT1/2/3/4 mutations with TMB, the mutation burden of samples was compared between FAT1/2/3/4 mutations and wildtype samples. Results showed that FAT1/2/3/4 mutation was associated with high TMB in the TCGA LUAD data set (Figure 4A, P < .001). Similar results were obtained in TCGA LUAD (Figure 4B, P < .01) and Hellmann (Figure 4C, P < .01) cohorts. Additionally, a higher neoantigen load was found in patients with mutant FAT1/2/3/4 in the Hellmann data set (Figure 4D, P < .001).
Figure 4.
Correlation of TMB and neoantigen load with FAT1/2/3/4 mutation. (A) Relationship between TMB and FAT1/2/3/4 mutation status in TCGA LUAD cohort. (B) Relationship between TMB and FAT1/2/3/4 mutation status in TCGA LUSC cohort. (C) Relationship between TMB and FAT1/2/3/4 mutation status in Hellmann NSCLC cohort. (D) Comparison of neoantigen load between mutant FAT1/2/3/4 and wildtype FAT1/2/3/4 in Hellmann NSCLC cohort.
The relationship between FAT1/2/3/4 mutation status and mRNA expression was also investigated. For LUAD, the FAT4 mutation was correlated with low mRNA expression of FAT4 (Figure S5A, P = .007). For LUSC, the FAT1 mutation was correlated with low mRNA expression of FAT1 (Figure S5C, P = .009). Further analysis showed that FAT1/2/3 mRNA levels were significantly upregulated, while FAT4 mRNA levels were significantly downregulated in the TCGA LUAD cohort (Figure S5B). For the TCGA LUSC cohort, FAT1/2 mRNA levels were significantly upregulated while FAT3/4 mRNA levels were significantly downregulated (Figure S5D). For LUAD, the mRNA levels of immune checkpoint genes, PD-1, PD-L1, PD-L2, CTLA4, HAVCR2, and TIGIT, were markedly higher in the high FAT2 group than in the low FAT2 group (Figure 5A and 5B). Of note, spearman coefficient correlation test showed that PD-L1 had the highest r value (0.4, P < .001). Analysis of the Venn diagram of intersection between genes with significant differential expression and genes with correlation r values ≥0.4 identified only PD-L1 in LUAD (Figure 5C). A high expression of FAT2 was found to be correlated with high expression of chemokines genes; CCL2, CCL3, CCL4, CCL19, CXCL10, and CXCL11 in LUAD (Figure 5D). However, no immune checkpoint-related genes were significantly correlated with FAT1/2/3/4 in LUSC (Figure S6, r < 0.4).
Figure 5.
Correlation of immune checkpoint-related genes with FAT2 expression in the TCGA LUAD analysis. (A) Heatmap of expression correlation between 7 immune checkpoint genes and FAT1/2/3/4 expression. Correlation analysis with Spearman coefficient. (B) Violin plots showing expression of 7 immune checkpoint genes in LUAD tumor samples with low or high FAT2 expression relative to median FAT2 expression level. (C) Venn plot showing that PD-L1 was linked to FAT2 expression.
The Impact of FAT1/2/3/4 Mutations on TICs
To confirm the correlation between FAT1/2/3/4 mutations and Tumor immunity in the microenvironment (TIME), we analyzed the percentage of tumor invading immune subsets using CIBERSORT and 21 types of immune cell patterns in LUAD and LUSC (Figure S4A and 4B). For LUAD, patients with FAT1 mutations showed significantly higher infiltration of activated dendritic cells and significantly lower infiltration of memory B-cells and resting memory CD4+ T-cells relative to FAT1 wildtype patients (Figure 6A, P < 0.05). Notably, 6 kinds of TICs correlated with FAT2 mutation (Figure 6B, P < .05). Among them, CD8+ T-cells, follicular helper T-cells, M1 macrophages, and activated memory CD4+ T-cells were positively linked to FAT2 mutation, whereas monocytes and activated dendritic cells were negatively correlated with FAT2 mutation. Relative to patients with wildtype FAT3, those with FAT3 mutation had remarkably higher infiltration of CD8+ T-cells, M1 macrophages, activated memory CD4+ T-cells, and follicular helper T-cells, but significantly lower infiltration of resting CD4+ T-cells, regulatory T-cells (Tregs) and activated dendritic cells (Figure 6C, P < .05). Similarly, patients with FAT4 mutations had significantly higher infiltration of CD8+ T-cells, activated memory CD4+ T-cells, follicular helper T-cells and M1 macrophages, but significantly lower infiltration of resting memory CD4+ T-cells, monocytes, resting dendritic cells, activated dendritic cells, and resting mast cells, relative to patients with wildtype FAT1 (Figure 6D, P < .05). These findings indicate that FAT1/2/3/4 mutations could promote the infiltration of TICs in the TIME of LUAD.
Figure 6.
Correlation of TICs with FAT1/2/3/4 mutations. (A-D) Violin plot illustrating the ratio differentiation of 21 kinds of immune cells in mutant FAT1/2/3/4 and wildtype FAT1/2/3/4 in TCGA LUAD cohort. (E-H) Violin plot illustrating the ratio differentiation of 21 kinds of immune cells in mutant FAT1/2/3/4 and wildtype FAT1/2/3/4 in the TCGA LUSC cohort.
For LUSC, FAT1 mutations showed a negative correlation with infiltration level of plasma cells and CD8+ T-cells (Figure 6E, P < .05), while for FAT2 mutations, a positive correlation was found with infiltration level of follicular helper T-cells and activated NK cells (Figure 6F, P < .05). For LUSC, there was no remarkable difference in TICs between groups with mutant FAT3/4 and wildtype FAT3/4 (Figure 6G and 6H, P > .05).
FAT1/2/3 Mutations Predict Better Response to ICIs
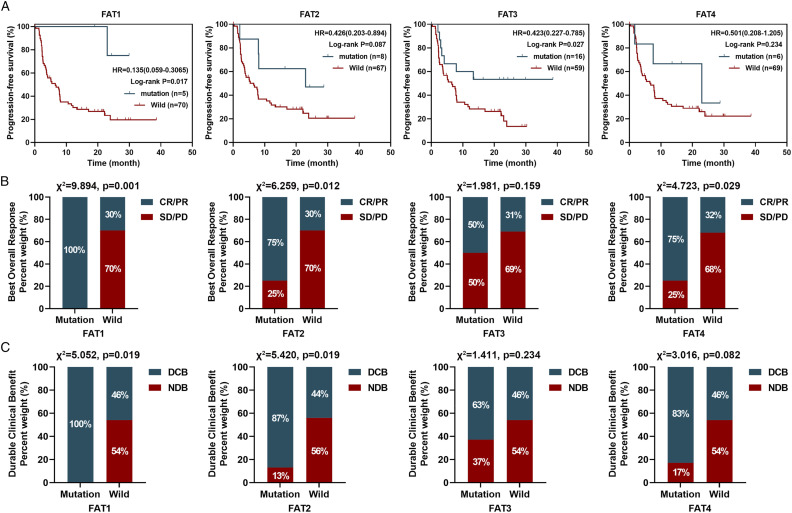
Further, the relationship between FAT1/2/3/4 mutations and efficacy of immunotherapy was explored in the Hellmann cohort consist of patients with NSCLC. Five patient who had mutant FAT1 showed better PFS compared to those with wildtype FAT1 (median, not reached (NR) vs 6.5 months; HR = 0.135; 95% CI: 0.059–0.306; P = .017, Figure 7A). The best overall response (BOR) rates of patients with a mutant FAT1 and those with wildtype FAT1 were 100.0% and 30.0%, respectively (Figure 7B, χ2 = 9.894, P = .001). The durable clinical benefit (DCB) rates of patients with mutant FAT1 and those with wildtype FAT1 were 100.0% and 46.0%, respectively (Figure 5C, χ2 = 5.502, P = .019). Relative to patients with wildtype FAT2, those with mutant FAT2 in this data set tended to have a longer PFS (median, 23.0 vs 6.5 months; HR = .426; 95% CI: 0.203–0.894; P = .087, Figure 7A), higher BOR (Figure 7B, 75.0% vs 30.0%, χ2 = 6.259, P = .012), and DCB rate (Figure7C, 87.0% vs 44.0%, χ2 = 5.420, P = .019). Moreover, mutant FAT3 was strongly linked to longer PFS relative to wildtype FAT3 (median, NR vs 6.5 months; HR = 0.423; 95% CI: 0.227–0.785; P = .027, Figure 7A) and also tended to have higher BOR rate and DCB rates (Figure 7B and 7C, P > .05). In this cohort, FAT4 mutations were significantly associated with better BOR rate (Figure 7B, χ2 = 4.723, P = .029), but not PFS (Figure 7A, P > .05) and DCB rates (Figure 7C, P > .05).
Figure 7.
Patients with FAT1/2/3/4 mutations showed better response to ICIs in Hellmann NSCLC cohort. (A) PFS Kaplan–Meier survival curves comparing the mutant FAT1/2/3/4 with wildtype FAT1/2/3/4. (B) Comparison of BOR rates between mutant FAT1/2/3/4 and wildtype FAT1/2/3/4. (C) Comparison of DCB rates between mutant FAT1/2/3/4 and wildtype FAT1/2/3/4.
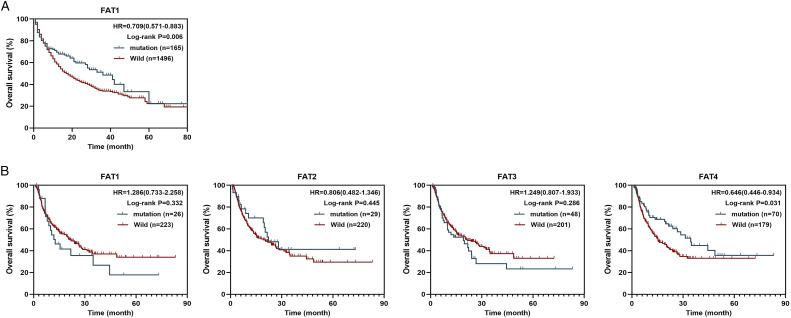
Finally, we validated the prognostic value of FAT1/2/3/4 mutations in immunotherapy using MSKCC and MSS pan-cancer cohorts. In the MSKCC cohort, patients harboring FAT1 mutation had superior OS survival (Figure 8A, HR = .709, P = .006) relative to those with wildtype FAT1. In the MSS cohort, FAT4 mutations significantly correlated with OS (Figure 8B, P = .031), and predicted good prognosis (HR = .646). However, FAT1/2/3 mutations were not significantly associated with OS in the MSS cohort (Figure 8B, P > .05). These results suggest that FAT1/2/3/4 mutations can be potentially used as biomarkers to guide ICIs treatment.
Figure 8.
The relation between FAT1/2/3/4 mutations and clinical response to ICIs in pan-cancer cohorts. (A) OS Kaplan-Meier survival curves comparing mutant FAT1 with wildtype FAT1 in MSKCC cohort. (B) OS Kaplan-Meier survival curves comparing mutant FAT1/2/3/4 with wildtype FAT1/2/3/4 in MSS cohort.
Discussion
The prevalence of FAT1/2/3/4 mutations in TCGA LUAD and LUSC cohorts was investigated in this study. The mutation frequency of 4 members of FAT family showed a similar trend in LUAD and LUSC patients. FAT3 (>18.0%) had the highest mutation frequency in both LUAD and LUSC patients, followed by FAT4 and FAT1, with FAT2 (<10%) having the lowest mutation frequency. Compared to the molecular biomarkers recommended by NCCN guidelines Version 2.2021, such as BRAF mutations, Metex14 skipping mutations, RET Rear, ALK fusions, and ROS1 fusions, we found that FAT1/2/3/4 mutations were more common in NSCLC. A high frequency of mutations in FAT family members (57.3%, 603/1052) in NSCLC means a larger pool of potential beneficiaries. A prognostic biomarker indicates patient survival independent of the treatment given because a biomarker relates to innate tumor behavior. For TCGA LUAD cohort, patients with FAT2 mutations had longer OS and PFS than those with wildtype FAT2. Moreover, FAT2 mutation was found to be an independent prognosis factor of LUAD. The results also revealed aberrant expression of FAT1/2/3/4 mRNA in LUAD and LUSC. The aforementioned results suggested that FAT family genes may be involved in the development of NSCLC.
Our results were consistent with previously reported results. FAT1 has been found to be highly expressed in breast cancer, LUSC, and gastric carcinoma, and its overexpression correlates with poor patient outcomes.28,29 In mice and human squamous cell carcinoma, FAT1 mutation promoted tumor occurrence, progression, invasiveness, stemness, and metastasis due to enhanced hybrid EMT state. 16 Immunohistochemical studies demonstrated that FAT4 expression was decreased in gastric, endometrial, and colorectal cancer. 17
TIME played a critical role in the initiation and progression of tumorigenesis. To the best of our knowledge, the impact of FAT1/2/3/4 on TIME has not been sufficiently defined. This study found that FAT1/2/3/4 mutations were correlated high TMB scores in LUAD and LUSC based on TCGA cohorts. Similarly, FAT1/2/3/4 mutations correlated with high TMB and neoantigen load in Hellmann’s NSCLC cohort. It was also demonstrated that high expression of FAT2 strongly correlated with high expression of PD-L1 in LUAD. To further characterize FAT1/2/3/4 mutations in TIME, we compared differences in immune cell infiltration between FAT1/2/3/4 mutant and FAT1/2/3/4 wildtype in NSCLC. Results revealed that for LUAD, activated dendritic cells were more abundant in FAT1-mutant tumors. Notably, CD8+ T-cells, follicular helper T-cells, M1 macrophages, and activated memory CD4+ T-cells were significantly higher in LUAD samples with mutant FAT2/3/4 than in those with wildtype FAT2/3/4. Altogether, these data illustrate that FAT1/2/3/4 mutations regulate the recruitment and infiltration of immune cells in the TIME of LUAD which may influence therapeutic efficacy of ICIs.
Therefore, we investigated the relationship between FAT1/2/3/4 mutations and efficacy of immunotherapy in NSCLC. We found that NSCLC patients with FAT1/2/3 mutations were more likely to benefit from ICIs in the Hellmann NSCLC cohort. FAT4 mutations significantly corelated with improved objective response but did not affect PFS in NSCLC patients receiving ICIs. We additionally found that FAT1/4 mutations are related to better OS in pan-cancer patients treated with ICIs based 2 independent cohorts. Similar to our findings, evidence from prior studies has shown that gene mutations can be potentially used as biomarkers to guide ICIs treatment. For example, various mutations, including TP53, STK11, EGFR, KEAP1,30,31 PRKDC, 32 and ZFHX3, 33 have been reported to be important predictors of response to ICIs in NSCLC. Collectively, these discoveries indicating that patients with FAT1/2/3/4 mutations may profit from ICIs treatment.
Several parameters including TMB, PD-L1 expression, and TICs can affect the efficacy of ICIs.34-36 Hellmann et al demonstrated that TMB strongly predicts efficacy of PD-1 plus CTLA-4 blockade in patients with NSCLC. TMB is independent of other clinicopathologic features which can be used to select patients who may benefit from nivolumab alone in combination with ipilimumab.13,23 The TICs in the TIME influence efficacy of ICIs in NSCLC patients. For instance, tumor-infiltrating lymphocytes such as CD8+ T-cells and CD4+ T-cells located in the tumor and invasive margin showed the potential to predict response to ICIs in NSCLC patients.37-39 Hollern et al 40 found that follicular helper T-cells modify response to ICIs in mice models of breast cancer with high TMB. Studies have demonstrated that anti–PD-1/PD-L1 therapies directly affect inflammatory M1 macrophages. In NSCLC patients receiving anti–PD-1 therapy, high expression of PD-L1 in M1 macrophages is associated with longer OS.41,42 Thus, high TMB, PD-L1 expression and infiltrating level of TICs may explain why ICIs are more effective in patients with mutant FAT family genes.
LUAD and LUSC are distinct in disease pathology, molecular mechanisms, immunogenic features and patient outcomes.43,44 We found that, in LUSC, there was no significant correlation between patients with mutant FATs than in those with wildtype FATs in OS, PFS and immune checkpoint-related genes. Moreover, FAT1/2/3/4 mutations showed no positive correlation with infiltration level of CD8+ T-cells and CD4+ T-cells in LUSC. Therefore, these data suggest that the influence of FAT family genes in TIME between LUAD and LUSC is different.
However, this study has some limitations. First, although we found that FAT1/2/3/4 mutations affect TMB, TICs, and the outcome of patients receiving ICIs therapy, we did not confirm the extent to which FAT1/2/3/4 mutation sites contribute to the “hot” TIME and better prognosis. Second, the OS of patients who received immunotherapy was not evaluated in NSCLC. Third, the sample size of the immunotherapy cohort was small and mainly consisted of non-squamous lung cancer patients (79.0%). Therefore, further large-scale studies are needed to examine the association between FAT1/2/3/4 mutation-induced changes in the TIME and responses to ICIs therapy. Finally, in patients with mutation in FAT1/2/3/4, GSEA showed that no gene set was significantly enriched in immune response-related pathways. Thus, the molecular mechanisms through which FAT1/2/3/4 mutations affect the immune microenvironment need to be further investigated.
Conclusion
In conclusion, this study shows a high frequency of mutations in FAT family genes in LUAD and LUSC patients. FAT2 mutation independently predicts good prognosis of LUAD patients. FAT1/2/3/4 mutations are significantly associated with high TMB in NSCLC. Moreover, FAT2 expression positively correlates with PD-L1 mRNA levels in LUAD. The FAT1/2/3/4 mutations correlate with high TICs infiltration, including activated dendritic cells, CD8+ T-cells, CD4+ T-cells, follicular helper T-cells, and M1 macrophages in LUAD. Notably, FAT1/2/3/4 mutations predict the clinical benefit of ICIs in patients with NSCLC. Taken together, these findings suggest that FAT family genes as valuable potential prognostic and immunological biomarkers for NSCLC. Further studies should be conducted to validate these findings, and should try to establish a diagnostic model that takes into account at least some of the outcomes of both FATs mutations. This work has important implications for clinical application and offers potential predictive biomarkers to guide ICIs treatment.
Supplemental Material
Supplemental Material, sj-pdf-1-ccx-10.1177_10732748221076682 for Prognostic and Immunological Role of FAT Family Genes in Non-Small Cell Lung Cancer by Zhenxing Feng, Yan Yin, Bin Liu, Yafang Zheng, Dongsheng Shi, Hong Zhang and Jianwen Qin in Cancer Control
Supplemental Material, sj-pdf-2-ccx-10.1177_10732748221076682 for Prognostic and Immunological Role of FAT Family Genes in Non-Small Cell Lung Cancer by Zhenxing Feng, Yan Yin, Bin Liu, Yafang Zheng, Dongsheng Shi, Hong Zhang and Jianwen Qin in Cancer Control
Supplemental Material, sj-pdf-3-ccx-10.1177_10732748221076682 for Prognostic and Immunological Role of FAT Family Genes in Non-Small Cell Lung Cancer by Zhenxing Feng, Yan Yin, Bin Liu, Yafang Zheng, Dongsheng Shi, Hong Zhang and Jianwen Qin in Cancer Control
Acknowledgments
We would like to thank Hong Zhang for the financial support for article processing charge.
Appendix
Abbreviations
- BOR
Best overall response
- DCB
Durable clinical benefit
- GSEA
Gene set enrichment analysis
- ICIs
Immune checkpoint inhibitors
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- NSCLC
Non-small cell lung cancer
- PFS
Progression-free survival
- OS
Overall survival
- TCGA
The Cancer Genome Atlas
- TIME
Tumor immunity in the microenvironment
- TICs
Tumor-infiltrating immune cells
- TMB
Tumor mutational burden
Author Contributions: Zhenxing Feng and Jianwen Qin conceived the study.
Dongsheng Shi supervised data collection.
Zhenxing Feng, Yan Yin, and Bin Liu performed statistical analyses.
All authors contributed to interpretation of the results.
Zhenxing Feng and Yafang Zheng wrote the manuscript.
Zhang, Hong contributed substantially to collection, analysis and interpretation of data for the revision of our paper and revised it critically for important intellectual content.
All authors approved the final version of the manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the CAPTRA-Lung Research Funds (No. CAPTRALung2022009); Tianjin Health Science and Technology Project (Grant No. MS20015).
Ethical Approval: This study did not require an ethical board approval because it did not contain human or animal trials.
Data Availability Statement: The datasets analyzed in the study are downloaded at the cBioPortal (www.cbioportal.org/), TCGA (https://portal.gdc.cancer.gov/) and GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Supplemental Material: Supplemental material for this article is available online.
ORCID iD
Jianwen Qin https://orcid.org/0000-0002-8981-9156
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2020;70(1):7-30. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. [DOI] [PubMed] [Google Scholar]
- 3.Faruki H, Mayhew GM, Serody JS, Hayes DN, Perou CM, Lai-Goldman M. Lung adenocarcinoma and squamous cell carcinoma gene expression subtypes demonstrate significant differences in tumor immune landscape. J Thorac Oncol. 2017;12(6):943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedlaender A, Banna G, Malapelle U, Pisapia P, Addeo A. Next generation sequencing and genetic alterations in squamous cell lung carcinoma: Where are we today? Front Oncol. 2019;9:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F, Qiao M, Zhou C. The cutting-edge progress of immune-checkpoint blockade in lung cancer. Cell Mol Immunol. 2021;18(2):279-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus chemotherapy in metastatic non–small-cell lung cancer. N Engl J Med. 2018;378(22):2078-2092. [DOI] [PubMed] [Google Scholar]
- 7.Perets R, Bar J, Rasco DW, et al. Safety and efficacy of quavonlimab, a novel anti-CTLA-4 antibody (MK-1308), in combination with pembrolizumab in first-line advanced non-small-cell lung cancer. Ann Oncol. 2021;32(3):395-403. [DOI] [PubMed] [Google Scholar]
- 8.Herbst RSP, Baas PP, Kim DM, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. The Lancet (British edition). 2015;387(10027):1540-1550. [DOI] [PubMed] [Google Scholar]
- 9.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non–small-cell lung cancer. New Engl J Med. 2015;373(17):1627-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389(10066):255-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath L, Pircher A. ASCO 2020 non-small lung cancer (NSCLC) personal highlights. Magazine of European Medical Oncology. 2021;14(1):66-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellmann MD, Ciuleanu T, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. New Engl J Med. 2018;378(22):2093-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellmann MD, Paz-Ares L, Bernabe CR, et al. Nivolumab plus ipilimumab in advanced non-small-cell lung cancer. N Engl J Med. 2019;381(21):2020-2031. [DOI] [PubMed] [Google Scholar]
- 14.Xu J, Wang B, Liu Z, Lai M, Zhang M, Zheng S. miR-223-3p regulating the occurrence and development of liver cancer cells by targeting FAT1 gene. Math Biosci Eng. 2020;17(2):1534-1547. [DOI] [PubMed] [Google Scholar]
- 15.Katoh M. Function and cancer genomics of FAT family genes. Int J Oncol. 2012;41(6):1913-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastushenko I, Mauri F, Song Y, et al. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature. 2021;589(7842):448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malgundkar SH, Burney I, Al Moundhri M, et al. FAT4 silencing promotes epithelial-to-mesenchymal transition and invasion via regulation of YAP and β-catenin activity in ovarian cancer. BMC Cancer. 2020;20(1):374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos-de-Frutos K, Segrelles C, Lorz C. Hippo pathway and YAP signaling alterations in squamous cancer of the head and neck. J Clin Med. 2019;8(12):2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu X, Zhai Y, Kong P, et al. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer Lett. 2017;397:83-93. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Razavi P, Li Q, et al. Loss of the FAT1 Tumor Suppressor Promotes Resistance to CDK4/6 Inhibitors via the Hippo Pathway. Cancer Cell. 2018;34(6):893-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadeqzadeh E, de Bock CE, Thorne RF. Sleeping giants: Emerging roles for the fat cadherins in health and disease. Med Res Rev. 2014;34(1):190-221. [DOI] [PubMed] [Google Scholar]
- 22.Wei R, Xiao Y, Song Y, Yuan H, Luo J, Xu W. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J Exp Clin Canc Res. 2019;38(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33(5):843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samstein RM, Lee C, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao D, Margolis CA, Vokes NI, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50(9):1271-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou R, Zhang J, Zeng D, et al. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I–III colon cancer. Cancer Immunol Immunother. 2019;68(3):433-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang TT, Westcott JM, Maine EA, Kanchwala M, Xing C, Pearson GW. ΔNp63α induces the expression of FAT2 and Slug to promote tumor invasion. Oncotarget. 2016;7(19):28592-28611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Fu L, Wang H, Yan Z, Yu X, Wang Y. FAT2 is a novel independent prognostic factor for the poor prognosis of gastric carcinoma. Int J Clin Exp Patho. 2017;10(12):11603-11609. [PMC free article] [PubMed] [Google Scholar]
- 30.Papillon-Cavanagh S, Doshi P, Dobrin R, Szustakowski J, Walsh AM. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open. 2020;5(2):e706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biton J, Mansuet-Lupo A, Pécuchet N, et al. TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti–PD-1 in lung adenocarcinoma. Clin Cancer Res. 2018;24(22):5710-5723. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Li Y, Guan Y, et al. Prevalence of PRKDC mutations and association with response to immune checkpoint inhibitors in solid tumors. Mol Oncol. 2020;14(9):2096-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Zhou N, Lin A, et al. ZFHX3 mutation as a protective biomarker for immune checkpoint blockade in non-small cell lung cancer. Cancer Immunol Immunother. 2021;70(1):137-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozaki Y, Muto S, Takagi H, et al. Tumor mutation burden and immunological, genomic, and clinicopathological factors as biomarkers for checkpoint inhibitor treatment of patients with non-small-cell lung cancer. Cancer Immunol Immunother. 2020;69(1):127-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers DE, Banerji S. Biomarkers of immune checkpoint inhibitor efficacy in cancer. Curr Oncol. 2020;27(12):106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi G, Russo A, Tagliamento M, et al. Precision medicine for NSCLC in the era of immunotherapy: New biomarkers to select the most suitable treatment or the most suitable patient. Cancers. 2020;12(5):1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thommen DS, Koelzer VH, Herzig P, et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24(7):994-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spitzer MH, Carmi Y, Reticker-Flynn NE, et al. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168(3):487-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hollern DP, Xu N, Thennavan A, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 2019;179(5):1191-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Zugazagoitia J, Ahmed FS, et al. Immune cell PD-L1 colocalizes with macrophages and is associated with outcome in PD-1 pathway blockade therapy. Clin Cancer Res. 2020;26(4):970-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon SR, Maute RL, Dulken BW, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545(7655):495-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng Z, Yang F, Wang Y, et al. Significantly different immunological score in lung adenocarcinoma and squamous cell carcinoma and a proposal for a new immune staging system. OncoImmunology. 2020;9(1):1828538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zengin T, Önal-Süzek T. Comprehensive profiling of genomic and transcriptomic differences between risk groups of lung adenocarcinoma and lung squamous cell carcinoma. J Personalized Med. 2021;11(2):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-ccx-10.1177_10732748221076682 for Prognostic and Immunological Role of FAT Family Genes in Non-Small Cell Lung Cancer by Zhenxing Feng, Yan Yin, Bin Liu, Yafang Zheng, Dongsheng Shi, Hong Zhang and Jianwen Qin in Cancer Control
Supplemental Material, sj-pdf-2-ccx-10.1177_10732748221076682 for Prognostic and Immunological Role of FAT Family Genes in Non-Small Cell Lung Cancer by Zhenxing Feng, Yan Yin, Bin Liu, Yafang Zheng, Dongsheng Shi, Hong Zhang and Jianwen Qin in Cancer Control
Supplemental Material, sj-pdf-3-ccx-10.1177_10732748221076682 for Prognostic and Immunological Role of FAT Family Genes in Non-Small Cell Lung Cancer by Zhenxing Feng, Yan Yin, Bin Liu, Yafang Zheng, Dongsheng Shi, Hong Zhang and Jianwen Qin in Cancer Control