Abstract
Different brain regions structurally interconnected through networks regulate behavior output. Therefore, understanding the functional organization of the brain in health and disease necessitates a foundational anatomic roadmap to its network organization. To provide this to the research community, our lab has systematically traced thousands of pathways in the mouse brain and has applied computational measures to determine the network architecture of major brain systems. Toward this effort, the brain-wide networks of the basolateral amygdalar complex (BLA) were recently generated. The data revealed uniquely connected cell types within the same BLA nucleus that were constituents of distinct neural networks. Here, we elaborate on how these connectionally unique BLA cell types fit within the larger cortico-basal ganglia and limbic networks that were previously described by our team. The significance and utility of high quality, detailed anatomic data is also discussed.
Keywords: Basolateral amygdala, connectome, neuroanatomy, brain networks, cell types, circuit tracing
COMMENT ON: Hintiryan, Houri et al. “Connectivity characterization of the mouse basolateral amygdalar complex.” Nature communications vol. 12,1 2859. 17 May. 2021, doi:10.1038/s41467-021-22915-5
Introduction
Since the first visualization of nerve cells and formulation of the Neuron Doctrine over a century ago, a comprehensive wiring diagram of the mammalian nervous system at the cell type resolution, or mesoscale, remains to be constructed. Despite the major advances in synaptic pathway tracing and imaging technologies, our understanding of the mammalian brain network organization remains fragmented and insufficient. A recent joint analysis of the mouse, non-human primate, and human primary motor cortex (MOp) by the Brain Initiative Cell Census Network (BICCN) provides a roadmap for cataloging neuronal cell types in the mammalian brain (BICCN Network, 2021) based on the combinatorial technologies of large-scale profiling of single-cell transcriptomes, spatially resolved single-cell transcriptomes, and cell-type specific connectivity.1-4 In parallel, our lab also demonstrated an anatomical approach to classifying neuron types of the anterior basolateral amygdalar nucleus (BLAa). 5 This method derives the cell type-specific wiring diagram of cell populations, from which inferences regarding their potential functional relevance can be made, thereby offering a valuable approach to cataloging neuron types.
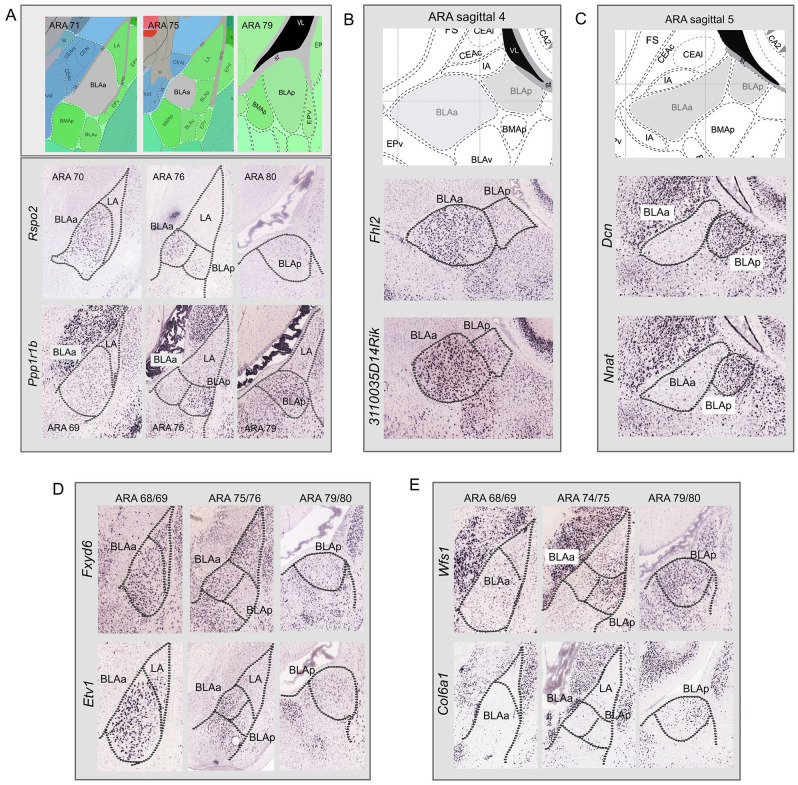
The basolateral amygdalar complex (BLA) is comprised of two major components which include the anterior (BLAa) and posterior (BLAp) BLA. The BLAa contains larger magnocellular cells, while the BLAp houses smaller parvocellular cells. 6 It is well established that these BLAa and BLAp neurons also are genetically and functionally distinct with magnocellular BLAa neurons expressing Rspo2 and parvocellular BLAp neurons expressing Ppp1r1b 7 (Figure 1A). We identified several additional genes that distinguish BLAa and BLAp neuron populations. For instance, Mef2c, Cdh11, Dkkl1, Nfib, Fhl2, Grp, Doc2a, Cyp26b1, Pde1a, Etv1, Fxyd6, Tmem178, and 3110035E14Rik all are expressed in BLAa and remain largely absent from BLAp (Figure 1B and D). Genes expressed more in BLAp than BLAa include Otof, Dcn, Wfs1, Col6a1, Gsta4, Lypd, Nnat, Tgfb2, and Marcksl1 (Figure 1C and E).
Figure 1.
Gene expression in BLAa and BLAp. (A) In situ hybridization images from the Allen Brain Atlas, a genome wide gene expression atlas, 8 showing that magnocellular BLAa neurons express Rspo2, while parvocellular BLAp neurons express primarily Ppp1r1b. Additional genes highlight the genetic similarity within the BLAa and their distinction from BLAp. (B and D) Genes Fhl2, 3110035D14Rik, Fxyd6, and Etv1, among others not shown here, are expressed primarily in BLAa and not BLAp. (C and E) Genes Dcn, Nnat, Wfs1, and Col6a1, among others not shown here, are expressed primarily in BLAp and not BLAa.
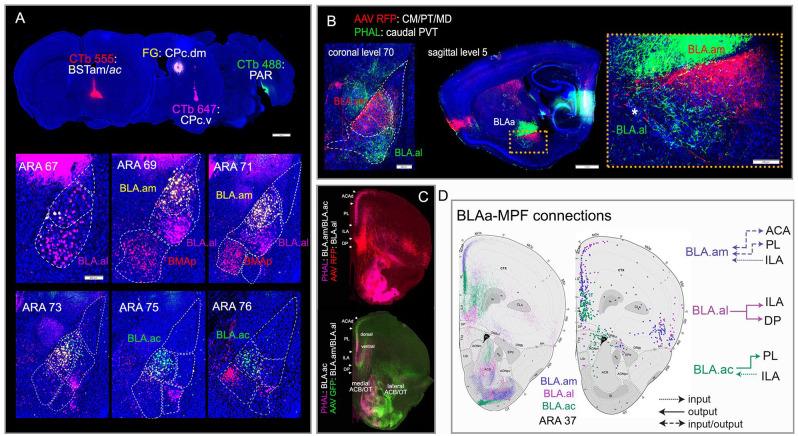
Despite the genetic similarity of BLAa neurons, studies showed that the BLAa contained different neuronal populations with distinct projection targets,9,10 which suggested that BLAa cell populations could be better cataloged based on their connectivity profile rather than on their gene expression patterns. Therefore, we set out to identify these BLAa cell types based on their whole brain connections and accordingly delineate their anatomic locations and relative boundaries. 5 To do this, we generated a comprehensive and detailed connectome of the entire mouse BLA complex, which is composed of several nuclei that include the lateral (LA), anterior and posterior basolateral (BLAa and BLAp), anterior and posterior basomedial (BMAa and BMAp), as well as the ventral (BLAv). 11 Approximately 150 neural pathways were traced to establish the brain-wide input/output organization of each BLA component. Modularity maximization analysis of annotated tracer labels was then used to generate brain-wide networks and connectivity maps for each BLA nucleus (maps available at https://mouseconnectomeproject.github.io/amygdalar/). The analysis identified three novel anatomic domains of the BLAa that housed connectionally and morphologically distinct cell types, namely the medial (BLA.am), lateral (BLA.al), and caudal (BLA.ac) (Figure 2A and B). Each domain contained unique projection-defined neuron types that displayed distinctive inputs/output connectivity patterns (Figure 2A–D). The relative boundaries of the three BLAa domains initially were demarcated by manual mapping of tracer labels. These boundaries were subsequently validated by machine learning based cytoarchitectonic parcellation of the BLAa (92% average agreement). The connectivity profile of each domain suggested that they belong to different functional networks. The BLA.am connects with brain regions involved in visual information processing, while the connections of BLA.al neurons suggest a role in gustatory/visceral information processing. The BLA.ac is the most distinct of the three cell types with exclusive connections to hippocampal ventral CA1 (CA1v), ventral subiculum (SUBv), CA3, and parasubiculum (PAR).
Figure 2.
Connectionally unique BLAa cell types and their connectivity with medial prefrontal cortex (MPF). (A) Four retrograde tracer injections placed in BSTam (CTb 555: red), dorsomedial caudal CP (FG: yellow), ventrolateral caudal CP (CTb 647: pink), and in PAR (CTb 488: green) clearly reveal uniquely connected projection neurons in the three BLAa domains: BLA.am (FG), BLA.al (CTb 647), and BLA.ac (CTb 488). BMAp neurons that target BST are also labeled (CTb 555). Note the segregation between FG (yellow) and CTb 647 (pink) labeled cells in BLA.am and BLA.al, respectively at ARA levels 69 and 71. Also note the absence of CTb 488 (green) labeled cells in BLAa at ARA levels 67-71. (B) Anterograde tracers AAV-RFP and PHAL injected in different thalamic nuclei distinctly label BLA.am and BLA.al, which is evident in coronal (left) and sagittal (right) planes. Inset shows magnified version of boxed BLAa region on sagittal section. *Denotes boundary between BLAa and IA (intercalated amygdalar nucleus). (C) Top panel: Anterograde labeling from tracer injections made primarily in BLA.am (PHAL: pink) and BLA.al (AAV RFP: red) shows their topographic projections to MPF. Bottom panel: Anterograde labeling from tracer injections made primarily in BLA.ac (PHAL: pink) and BLA.al (AAV GFP: green) shows their distinct connections with MPF, ACB, OT, and CP. Note the stronger projections from BLA.am to dorsal PL/ACA compared to projections from BLA.ac to more ventral parts of PL, which is also evident in D (left; purple and green fibers in PL). (D) Schematic summarizing how each BLAa domain shares unique input/output connections with MPF, especially layers 2/3. Left shows projections from the BLAa domains to MPF, while the right shows MPF cells that project to BLAa domains. Note (1) the reciprocal connections between the BLA.am and ACA and PL (layer 2), (2) that the BLA.al mostly projects to ILA, and (3) the unique BLA.ac connection in which it heavily projects to PL (layer 2) but receives strong input from ILA. Source: Figure adapted from Hintiryan et al. 5
Abbreviations: ac, anterior commissure; ACA, anterior cingulate cortex; ACB, nucleus accumbens; BSTam, anteromedial bed nucleus of stria terminalis; CM, central medial thalamic nucleus; CP, caudoputamen; ILA, infralimbic cortex; MD, mediodorsal thalamic nucleus; OT, olfactory tubercle; PAR, parasubiculum; PL, prelimbic cortex; PT, parataenial thalamic nucleus; PVT, paraventricular thalamic nucleus.
Importantly, each of these major BLAa populations displays unique and complex connections with medial prefrontal cortical areas (MPF) (Figure 2C and D) implicated in a wide range of significant roles from memory to decision making to mental health disorders.12,13 These connections are not only region specific, but also display laminar specificity. Both the BLA.am and BLA.ac project most densely to layer 2 (but also layer 5) of the prelimbic cortical area (PL) (Figure 2C and D). Topographically, BLA.am terminations are in more dorsal PL, while those from BLA.ac are to more ventral PL. In turn, layer 2 and 3 neurons of the infralimbic cortical area (ILA) provide strongest input to BLA.ac and some to BLA.am. BLA.al neurons also target MPF mainly layer 5 of ILA. This BLAa-MPF connectional specificity likely reflects functional selectivity. These connections are quite detailed, but the purpose behind generating connectivity brain maps is to provide foundational structural roadmaps to propagate hypothesis-driven functional investigations. Accordingly, for these connectivity maps to be useful, they must be thorough, reliable, and extensive so that subsequent investigations based on these maps can be performed more accurately and at a much finer resolution. Generally, we propose potential hypotheses regarding a region’s functional relevance based on what is already known in the literature about its network’s constituents. Here, we propose a gustatory/visceral role for the BLA.al, a role in visual information processing for the BLA.am, and a role in memory consolidation for the BLA.ac. However, notably these BLAa domains have vast connections that implicate them in a variety of different roles aside from those proposed here.
The BLAa and Cortico-Basal Ganglia Neural Networks
The BLA.am and BLA.al form part of a much larger cortico-basal ganglia network that has been identified in our previous work. Based on approximately 600 traced and analyzed cortical pathways, we generated the neural networks of the mouse neocortex. 14 The analysis revealed that the entire neocortex is organized into roughly a dozen subnetworks with distinct connectional and functional motifs. Among these were three somatic sensorimotor subnetworks associated with the regulation of select body regions like upper limb, lower limb/trunk, and orofacial areas. Among others, two lateral subnetworks composed of areas located along the lateral edge of the cortex, also emerged. The first consisted of the ectorhinal (ECT), perirhinal (PERI), and posterior temporal association areas (TEa), referred to as the ECT/PERI/TEa subnetwork. The second lateral subnetwork included three interconnected cortical areas located in more rostral parts of the cortex. These were the dorsal (AId), ventral (AIv), and posterior (AIp) agranular insular cortical areas, as well as the primary gustatory (GU) and visceral (VISC) areas. Next was to understand how cortical information is transposed and integrated within the basal ganglia to affect sensorimotor, cognitive, and emotional functions. Hence, a comprehensive cortico-striatal projection map was constructed: a map of cortical projections arising from the entire mouse cerebral cortex to the dorsal striatum. 15 Computational tools and graph theory applied to hundreds of traced cortico-striatal pathways revealed four major divisions of the intermediate CP (CPi), each of which was subdivided further into smaller domains based on their convergent and divergent input. Divisions and domains were also established for the rostral (CPr) and caudal (CPc) CP. To complete the cortico-basal ganglia-thalamic loop, 700 injections were made across the striatum (including the 30 CP domains), external globus pallidus (GPe), substantia nigra pars reticulata (SNr), thalamic nuclei, and cortex to generate the complete cortico-basal ganglia-thalamic wiring diagram. 16 With the cortical and striatal network data combined, a clear picture of cortico-cortical and cortico-striatal functional interactions emerges. The BLAa network can be integrated into these cortico-striatal networks, demonstrating how all these structures are connected in a functionally logical way.
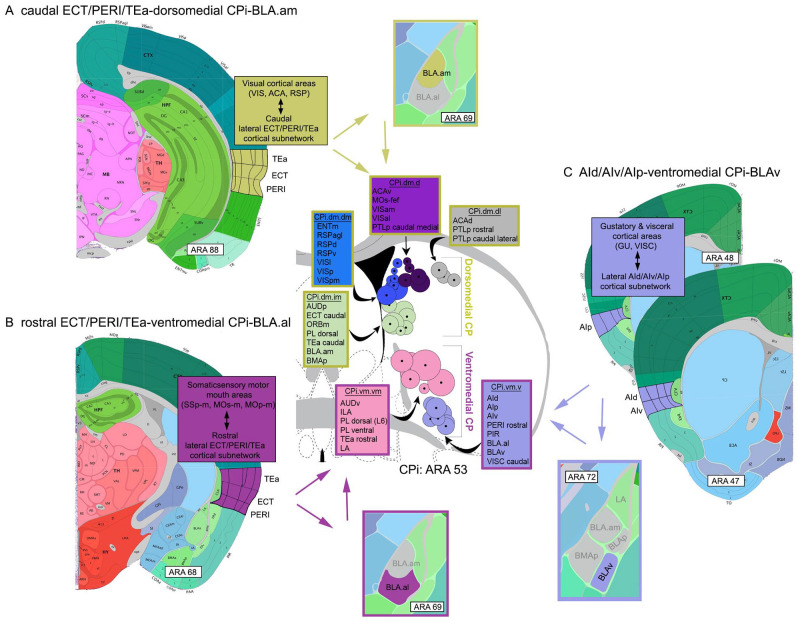
To elaborate, the caudal part of the ECT/PERI/TEa cortical subnetwork shares connections with visual cortical areas (visual [VIS], anterior cingulate [ACA], retrosplenial [RSP] areas). At the striatal level, the caudal ECT/PERI/TEa subnetwork projects to the dorsomedial division of the CPi, which also receives and integrates cortical visual information (VIS, ACA, RSP). At the level of the amygdala, the caudal ECT/PERI/TEa provides stronger input to BLA.am, which (a) connects with areas involved in visual information processing, and (b) also targets the visual dorsomedial division of the CPi (Figure 3A).
Figure 3.
Visual representations of cortico-striatal-BLA subnetworks. The intermediate CP (CPi) consists of 4 major divisions, each of which receives and integrates different cortical input. These are (1) the dorsomedial, (2) ventromedial, (3) dorsolateral, and (4) ventrolateral. Note that only the dorsomedial (yellow) and ventromedial (magenta) divisions are shown. Based on highly topographic cortico-striatal projections, each division can be subdivided into domains, smaller regions that receive convergent input from different cortical areas. Adjacent domains, particularly within a community, share integrated cortical information (for details, see Hintiryan et al 14 ). Each circle on the CPi denotes the center of terminations from a single cortical area. A group of color-coded circles represents a domain. Color-coded boxes list cortical areas that project to that domain (name of domain included, e.g., CPi.dm.dm). The stroke color of the boxes denotes the larger CPi division within which the domains interact (yellow: dorsomedial CPi; magenta: ventromedial CPi). (A) Visual representation of the caudal ECT/PERI/TEa-dorsomedial CPi-BLA.am subnetwork showing the interactions of these 3 regions potentially involved in visual information processing. At the cortical level, the caudal ECT/PERI/TEa is heavily interconnected with visual processing areas. The caudal ECT/PERI/TEa project to the dorsomedial CPi (stroke colored yellow), which also receives input from a wide variety of visual processing cortical areas. It also is the region of the CPi that BLA.am cells target. The caudal ECT/PERI/TEa also provides input to BLA.am. (B) Visual representations of the rostral ECT/PERI/TEa-ventromedial CPi-BLA.al subnetwork showing interactions among these three regions potentially involved in gustatory/visceral information processing. At the cortical level, the rostral ECT/PERI/TEa is heavily interconnected with somatic sensory motor regions that regulate the mouth. Rostral ECT/PERI/TEa project to the ventromedial CPi (stroke colored magenta), where cortical gustatory and visceral information is integrated. This is also the region in the CPi that BLA.al cells target. The rostral ECT/PERI/TEa also provides input to the BLA.al. (C) Visual representation of the AId/AIv/AIp-ventromedial CPi-BLAv subnetwork. The AId/AIv/AIp receives input from gustatory and visceral cortical areas and projects to the ventromedial CPi, which also receives input from cortical regions that process the same type of information. The BLAv also projects to the same CPi region and the AId/AIv/AIp project to the BLAv.
Abbreviations: ACA, anterior cingulate cortex; AId, agranular insular cortex, dorsal part; AIp, agranular insular cortex, posterior part; AIv, agranular insular cortex, ventral part; AUD, auditory cortex; CPi, intermediate caudoputamen; d, dorsal; ECT, ectorhinal cortex; ENT, entorhinal cortex; GU, gustatory cortex; i, intermediate; l, lateral; m, medial; ORB, orbitofrontal cortex; PERI, perirhinal cortex; PIR, piriform cortex; PTLp, posterior parietal association areas; RSP, retrosplenial cortex; TEa, posterior temporal association cortex; v, ventral; VIS, visual cortex; VISC, visceral cortex.
In contrast, the rostral part of the lateral ECT/PERI/TEa cortical subnetwork heavily connects with cortical somatic sensorimotor areas associated with orofacial areas (e.g., SSp, MOp, MOs mouth regions). At the striatal level (particularly the CPi), the rostral parts of the ECT/PERI/TEa subnetwork project to the ventromedial division, which also receives and integrates input from cortical gustatory and visceral areas (AI, GU, VISC). At the amygdalar level, the rostral ECT/PERI/TEa provides input to the BLA.al, which (a) is distinctly connected with regions presumably involved in gustation/visceral processing, and (b) also projects to the ventromedial division of the CPi (Figure 3B).
Together, this suggests amygdalar interactions among the cortical and striatal networks in the form of a rostral ECT/PERI/TEa-ventromedial CPi-BLA.al subnetwork that processes gustatory/visceral information and a second caudal ECT/PERI/TEa-dorsomedial CPi-BLA.am subnetwork for visual information processing.
Interactions among the cortico-striatal-amygdalar networks also occur at the level of the BLAv. The BLAv, located ventral to the BLA.al, has a connectivity profile that is unique from all other BLA cell populations. It is also connected with regions that process gustatory and visceral information. These include the GU, AI, thalamic visceral intermediodorsal (IMD), thalamic gustatory VPMpc, and a very specific domain in the lateral entorhinal cortex (ENTl layer 5) that is also connected with gustatory and visceral areas. Cortico-striatal-amygdalar interactions through the BLAv occur via the lateral AId/AIv/AIp cortical subnetwork. The AId/AIv/AIp subnetwork provides input to the ventromedial CPi division, but also to the BLAv. The BLAv in turn projects back to the ventromedial CPi forming an AId/AIv/AIp-ventromedial CPi-BLAv subnetwork (Figure 3C).
BLAa and Hippocampal Networks
In addition to the cortex, basal ganglia, and BLA, we also assembled the connectome of the mouse hippocampus (HPF). 17 In this work, our team created the Hippocampus Gene Expression Atlas (HGEA), which refined the classic hippocampal components (DG, CA1, CA2, CA3, and subiculum) into 20 molecular domains and then assembled the brain-wide connectional profiles of each molecularly defined region. Clearly, the BLA.ac, with its connections to hippocampal regions like the ventral CA1, CA3, ventral SUB, and PAR forms part of these larger hippocampal networks. Also relevant are the connections of the BLA.ac to the MPF, specifically to the PL and ILA. These connections suggest that the BLA.ac can serve as a hub through which the MPF and HPF interact: a MPF-BLA.ac-HPF network that could modulate memory consolidation. 18 Notably, the PAR is a region rich in grid, head direction, and border cells. 19 Consequently, the BLA.ac-PAR connectivity also suggests that the BLA.ac is in a hippocampal network involved in spatial navigation and exploration.
Constructing Neural Networks of the Mammalian Brain
As part of the Brain Initiative’s Mouse Connectome Project (MCP) at UCLA, our main objective is to map interconnections of all regions of the mouse brain, generating the first complete whole-brain mammalian connectome. In addition to systematically producing and collecting large-scale connectivity data, we leverage computational informatics tools to annotate and analyze the massive datasets to reveal how brain regions assemble into functional networks that potentially mediate behavioral output: the impetus for the connectome. Afterall, behavior is regulated through these networks of interconnected brain regions. Our approach to the whole-brain connectome has been to tackle, in a logical progression, one brain system at a time (e.g., cortico-basal ganglia system; classic limbic system), which has allowed us to produce the most complete and granular connectivity maps for each system. The ultimate goal however is to aggregate all the network data to ascertain the brain-wide networks of the whole brain. Here, we demonstrated how BLAa cell populations fit within the larger network motifs of the cortico-basal ganglia and hippocampus.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH/NIMH U01MH114829 (HWD), NIH/NIMH R01MH094360 (HWD), and NIH/NIMH U19MH114821 (JH/PA).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contribution: HWD and HH developed the scientific concept. HH wrote the commentary and prepared the figures, while HWD constructively guided the process.
ORCID iD: Hong-Wei Dong  https://orcid.org/0000-0001-9972-3177
https://orcid.org/0000-0001-9972-3177
References
- 1. Muñoz-Castañeda R, Zingg B, Matho KS, et al. Cellular anatomy of the mouse primary motor cortex. Nature. 2021;598:159-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. BRAIN Initiative Cell Census Network. A multimodal cell census and atlas of the mammalian primary motor cortex. Nature. 2021;598:86-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peng H, Xie P, Liu L, et al. Morphological diversity of single neurons in molecularly defined cell types. Nature. 2021;598:174-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Z, Zhou J, Tan P, et al. Epigenomic diversity of cortical projection neurons in the mouse brain. Nature. 2021;598:167-173. doi: 10.1038/s41586-021-03223-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hintiryan H, Bowman I, Johnson DL, et al. Connectivity characterization of the mouse basolateral amygdalar complex. Nat Commun. 2021;12:2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alheid GF. Extended amygdala and basal forebrain. Ann N Y Acad Sci. 2003;985:185-205. doi: 10.1111/j.1749-6632.2003.tb07082.x [DOI] [PubMed] [Google Scholar]
- 7. Kim J, Pignatelli M, Xu S, Itohara S, Tonegawa S. Antagonistic negative and positive neurons of the basolateral amygdala. Nat Neurosci. 2016;19:1636-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168-176. [DOI] [PubMed] [Google Scholar]
- 9. Beyeler A, Chang CJ, Silvestre M, et al. Organization of valence-encoding and projection-defined neurons in the basolateral amygdala. Cell Rep. 2018;22:905-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McGarry LM, Carter AG. Prefrontal cortex drives distinct projection neurons in the basolateral amygdala. Cell Rep. 2017;21:1426-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong HW. The Allen Reference Atlas: a digital color brain atlas of the C57Bl/6J male mouse. 2008. Hoboken, New Jersey: John Wiley & Sons, Inc. [Google Scholar]
- 12. Chai XJ, Whitfield-Gabrieli S, Shinn AK, et al. Abnormal medial prefrontal cortex resting-state connectivity in bipolar disorder and schizophrenia. Neuropsychopharmacology. 2011;36:2009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Euston DR, Gruber AJ, McNaughton BL. The role of medial prefrontal cortex in memory and decision making. Neuron. 2012;76:1057-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zingg B, Hintiryan H, Gou L, et al. Neural networks of the mouse neocortex. Cell. 2014;156:1096-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hintiryan H, Foster NN, Bowman I, et al. The mouse cortico-striatal projectome. Nat Neurosci. 2016;19:1100-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Foster NN, Barry J, Korobkova L, et al. The mouse cortico–basal ganglia–thalamic network. Nature. 2021;598:188-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bienkowski MS, Bowman I, Song MY, et al. Integration of gene expression and brain-wide connectivity reveals the multiscale organization of mouse hippocampal networks. Nat Neurosci. 2018;21:1628-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kitamura T, Ogawa SK, Roy DS, et al. Engrams and circuits crucial for systems consolidation of a memory. Science. 2017;356:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boccara CN, Sargolini F, Thoresen VH, et al. Grid cells in pre- and parasubiculum. Nat Neurosci. 2010;13:987-994. [DOI] [PubMed] [Google Scholar]