Abstract
Introduction
The PER2 (Period circadian regulator 2) gene is related to the circadian clock, and it has been deemed as a suppressor gene in osteosarcoma and lung carcinoma. However, the part of PER2 in CRC (colorectal cancer) needs to be further determined.
Methods
First, we collected clinical samples to detect PER2 expression in CRC. Then, we used cell transfection to knock down PER2 expression in CRC cell lines and performed a series of functional experiments to elucidate the effects of PER2 on CRC cells. We next verified whether PER2 affects the epithelial-mesenchymal transformation (EMT) process in CRC by conducting quantitative real-time PCR and western blotting.
Results
In the research, we revealed that the expression of PER2 decreased in CRC clinical samples. In addition, knocking down PER2 expression caused CRC cells to acquire malignant biological features. Finally, we found that PER2 knockdown may activate the Snail/Slug axis through inhibiting p53, therefore promote the activation of the EMT pathway.
Conclusion
In conclusion, low PER2 expression reinforces migration and activates EMT in CRC, suggesting that PER2 is closely related to CRC development and could be used as a potential treatment site in the clinic.
Keywords: period 2, colorectal cancer, epithelial-mesenchymal transformation, proliferation, migration
Introduction
Colorectal cancer (CRC) is the third most common malignancy in the world, and ranks fourth in cancer-related deaths. By the end of 2030, CRC is forecasted to arise more than 2.2 million new cases and 1.1 million deaths, a total of about 60% burden for entire global.1-4 A large amount of evidence reveals that CRC is a heterogeneous disease, and its potential pathogenesis is closely related to the uncontrolled regulation of oncogenes and tumor suppressor genes in vivo.5-7 Although great efforts have been made to understand the tumorigenesis of CRC, the potential signaling pathways involved are still unclear. Exploring the underlying mechanism is of great significance for understanding the occurrence and development of CRC.
PER2 is a member of the Period gene family, which includes PER1, PER2, and PER3; members of this family are expressed in the suprachiasmatic nucleus in a circadian pattern. 3 There are at least 9 clock genes related to the function of adjustment of circadian rhythms, including Period 1 (PER1), Period 2 (PER2), Period 3 (PER3), Timeless (TIM), BMAL1, CLOCK, Cryptochrome 1 (CRY1), Cryptochrome 2 (CRY2), and Casein kinase 1e (CKe). PER2 functions by making it easier for organisms to adapt to daily changes in the environment, including temperature, pressure, and light. For example, PER2 regulates neurobiological activities, thereby controlling the osteogenic differentiation of bone marrow-derived mesenchymal stem cells.8-10 PER2 is also crucial to mammary gland development. 11 There is growing evidence that PER2 is related to the tumorigenesis and malignant biological behavior of a variety of tumors.8,10 Downregulated expression of PER2 in NSCLC is related to progression and metastasis of carcinoma, and it has an effect on the proliferation, apoptosis, and migration of osteosarcoma cells.12-14 Several studies have reported decreased expression of PER2 in various cancer cells of breast, lung, gastric, myeloid leukemia, CRC, and other cancer cells.12,13,15-18 Meantime, it is also discovered that PER2 in TME has unexpected tumorigenic effects in the liver metastasis model of colon cancer in mice. 19 So the expression of PER2 in CRC is controversial. Therefore, it is necessary to clarify the role of PER2 in CRC and the specific underlying molecular mechanism.
Epithelial-mesenchymal transition (EMT) refers to the biological process in which epithelial cells are transformed into cells with a mesenchymal phenotype through specific procedures, because it increases motility and metastatic potential, it is essential for tumorigenesis and development. 20 The hallmark of EMT is upregulated N-cadherin expression in conjunction with downregulated E-cadherin expression. 21 It was reported that the migration of CRC cells is closely related to the EMT process through the TGF-β pathway, WNT/β-Catenin pathways, and other inducers.22-24 Through EMT, epithelial cells lose cell polarity, lose their connection with basement membrane and other epithelial phenotypes, and acquire mesenchymal phenotypes like migration and invasion, anti-apoptosis, and finally lead to increased invasiveness of CRC. 25 Importantly, the key transcription factors, such as Snail and Slug, may regulate the ability of CRC cells to acquire migration and invasion, which causes the further development of cancer and poor prognosis. 24 At present, it is believed that EMT is involved in the modulation of multiple biological processes,26-29 so the mechanism of EMT process in CRC needs to be further explored. Combined with PER2 affects the proliferation and metastasis of cancer cells, we suspect PER2 can mediate EMT process in CRC.
In this study, we first used clinical samples to verify the low expression of PER2 in CRC and then stably knocked down PER2 in RKO cells, which have relatively higher expression of PER2 among all CRC cell lines. Subsequently, we performed a series of functional biological experiments, including wound-healing and clone formation assays, and confirmed that PER2 knockdown can promote the malignant biological behavior of CRC cells. At the same time, we detected the expression of EMT-related markers in PER2-knockdown cells. The results indicated that low PER2 expression was associated with the deletion of p53, which led to the activation of the Snail/Slug axis, and DOX treatment may restore the expression of p53, which in turn promoted cell apoptosis. These results indicated that PER2 is a potential target for CRC-targeted therapy.
Material and Methods
Patients and Clinical Samples
We collected 5 cases of cancer and adjacent noncancerous tissues (>2 cm) from CRC patients diagnosed at the Department of Gastrointestinal Surgery, Affiliated Zhongshan Hospital of Xiamen University, Xiamen, China. Freshly resected tissues were kept at −80°C. A part of the tissue is fixed with formalin and stored at 4°C to prepare for IHC experiment. A CRC tissue microarray (TMA) was purchased from Alenabio (BC1801, Alenabio, Xian, China), and IHC staining for PER2 was performed. The slides were scored for antibody-positive of the epithelial cells, including the percentage and the intensity of staining. Then the degree of staining was quantified by the scoring system of immunohistochemistry. All patients participating in this study provided written informed consent, and the experimental procedure was approved by the Ethics Committee of the Affiliated Zhongshan Hospital of Xiamen University, No:xmzsyyky2021197.
Cell Culture
Human colon cancer cell lines HCT116, LOVO, RKO, HT29, SW480, and SW620 and human kidney epithelial cell lines 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 IU penicillin, and 100 mg/mL streptomycin, and then placed in a 37°C humidified incubator containing 5% CO2.
Lentivirus Infection Patients and Clinical Samples
RKO and LOVO colon cancer cells in the logarithmic growth phase were plated on a 6 cm plate (5 × 106 cells/well) in advance. After the cells adhered, they were infected with lentivirus-containing PER2 shRNA (PER2 shRNA plasmid purchased from the PPL Corporation, USA). Puromycin (10 µg/mL) was used to culture for a week and we selected the positively infected cells, and then viewed them by using a fluorescence microscope When the percentage of fluorescent number under the microscope was close to 90%, the cells were collected and sub-cultured for use in experiments.
Real-Time Quantitative Polymerase Chain Reaction
Extract total RNA from cells by TRIzol (Lot: 183010, Ambion, USA) and reverse transcription of RNA into cDNA with a HiFi-MMLV cDNA Kit (CWBIO, Cat: CW0744M, Lot: 40420). Finally, use UltraSYBR Mixture to detect the relative expression of the target gene (Cat: CW0957H, Lot: 60409, CWBIO) with a real-time quantitative PCR system (BIO-RAD, CFX96TM Real-Time System, C1000TM Thermal Cycler). We purchased the primers from Biotech Bioengineering (Shanghai) Co., Ltd. Simultaneously, the sequences of primers are as follows:
PER2: forward: 3'-AAATCCGCTACCACCCCTTC-5'; reverse: 5'-AAGGCAGCAAAGCTGACTCTC-3'
GADPH: forward: 3'-GGAGCGAGATCCCTCCAAAAT-5'; reverse: 5'- GGCTGTTGTCATACTTCTCATGG-3'
Immunohistochemistry (IHC) and Scoring
We purchased a tissue chip (BC1801, Alenabio, Xian, China), which contains 90 matched pairs of CRC and adjacent noncancerous tissues for the next IHC analysis, and mouse monoclonal antibodies specific for human PER2 (1:50 dilution) were purchased from Santa (Cat. No. Sc-101105, Santa Cruz Biotechnology, CA, USA), after incubating overnight at 4°C, the corresponding secondary antibody was used on the next day. Scoring of IHC data was performed by 2 certified pathologists at 40 × magnification, and the scores were averaged. The intensity of staining (IS) was scored as 0: no staining; 1: mild staining; 2: moderate staining; 3: severe staining; the percentage of stained cells (0:<5%; 1:5-25%; 2:26-50%; 3:51-75%; 4:>75%).
Colony Formation Experiment
We selected RKO cells which are in the logarithmic growth phase and inoculated the prepared cell suspension into a 6-well plate (2 x 103 cells/well). Observed under the microscope, the culture was terminated when there were macroscopic clones in the petri dish after 14 days of culture. 4% paraformaldehyde was added to fix the cells and stained the colonies with GIMSA staining solution for 10 min. A digital camera was used to obtain images to count the colonies, and finally calculated the colony formation rate.
Tumorsphere Culture
Resuspending the cell pellet in serum-free DMEM/F-12 (Invitrogen, USA) supplemented with insulin (4 mg/mL, #51500-054, Gibco, USA), B-27 (1:50; catalog number: 17504-044, Invitrogen, USA), basic fibroblast growth factor (bFGF; 20 ng/mL, #AF-100-18B, PEPROTECH, USA), and EGF (20 ng/mL, #AF-100-15, PEPROTECH, USA). The cells were counted and plated in ultra-low-attachment 96-well plates (#7007, Corning Life Sciences, USA) containing 100 µl of medium. During this period, 100 µl of medium was added to the cells every 2 days. After seeding for 14 days, the sphere size and number were measured.
Cell Proliferation Experiment (CCK-8 Experiment)
RKO and LOVO cells in the logarithmic growth phase were cultured in the 96-well plate with repeated 6 replicates per group, and incubated at 37°C, 5% CO2 humidified environment for 24, 36, 48, and 72 hours. After discarding the medium, 10% of the total volume of CCK-8 (Lot. KH741, Dojindo, Japan) was added to each well. Put the culture plate back in a humid environment and continue to incubate for another 1 hour. Use a microplate reader (Thermo, USA) to measure the absorbance at 450 nm.
Cell Invasion Experiment
Counting the RKO and seeding them in 6-well plate (4 × 105 cells/ml) until the cells adhere to the wall, replace the serum-free medium and starve the cells for 12 hours, cells were digested and inoculated in 24-well Transwell plate (Lot: 24818005, 8.0 µm Polycarbonate Membrane, Corning Incorporated, USA), 200μL serum-free suspension (1 × 104 cells/well) was added to the upper chamber of the Transwell chamber, and then covering the lower chamber with 500μL culture medium containing 15% FBS. After culturing for 48 hours, the upper non-migrating inner layer cells were wiped off, and cells that migrated to the lower layer were fixed with 4% formaldehyde, then dyed with .5% crystal violet, and finally observed and counted under a microscope.
Wound-Healing Assay
Using a marker to draw a line evenly on the back of the six-well plate with a ruler, and then transfected RKO and LOVO cells were added to each well for overnight culture (2 × 106 cells/well). After the cells were overgrown the next day, a 10 μL pipette tip and a ruler were used to make a mark in the hole with a horizontal line perpendicular to the back, and cleaned the cells with preheated PBS. After adding serum-free medium, the cells were continued to culture in 37°C, 5% CO2 humidified environment, selecting 0, 24, 48, and 72 hours to take images. The ImageJ software was used to calculate the average distance between pixels on both sides of the scratch.
Flow Cytometry
The previously transfected RKO cells and control cells were collected, and the cells were washed twice with pre-cooled PBS. According to the instructions of reagents, the Annexin V-APC/7-AAD cell apoptosis detection kit (batch: BA19091, Bjbalb, Beijing, China) was used to stain the two groups of cells, and finally apoptotic cells were detected by using a KALUZA flow detector.
Western Blot Analysis
Cells were collected at 4°C and lysed in RIPA buffer (Lot: 89900, Thermo ScientificTM, USA) for 1 hour, accompanied by 1% protease inhibitor mixture and 1% phenylmethanesulfonyl fluoride (Lot: 78420, Thermo ScientificTM, USA), and collected the cell supernatant after centrifugation. The protein standard curve was developed by the BCA method, and the protein concentration of the corresponding samples was detected (Bio-Rad, Hercules, CA). Subsequently, an equivalent amount of protein (20 µg) was separated by electrophoresis on an 8% or 10% SDS gel, transferred to a PVDF membrane (Cat NO IPVH00010, Merck Millipore, Germany), and blocked with 5% skimmed milk powder. Then the membrane was incubated at 4°C overnight with primary antibodies: rabbit monoclonal antibodies against PER2 (1:1000, ab179813, Abcam, USA), GAPDH (1:1000, #21612, Signalway Antibody, USA); mouse monoclonal antibody against p53 (1:1000; #J2519; Santa Cruz; USA), β-Actin (1:1000, #3700, CST, USA); and antibodies from the Epithelial-Mesenchymal Transition Antibody Sampler Kit (1:500, #9782, CST, USA). The next day, immunoblotting was performed using the corresponding HRP-binding secondary antibody, followed by visual detection of the immune response bands using an ECL substrate liquid (Control: 102031410 Bio-Rad, USA) and an ECL system (Azure Biosystems C300, USA). ImageJ was used to detect the gray value of Western blot.
Statistical Analysis
We chose GraphPad Prism 7.00 statistical software (GraphPad Software, USA) to examine results. T test was used for comparison between groups of independent samples. Data are expressed as the means ± standard deviation (SD). Throughout the testing process, differences of P <.05 were considered statistically significant.
Guideline
This is an observational study. The reporting of this study conforms to STROBE guidelines. 30
Results
PER2 Expression is Downregulated in CRC
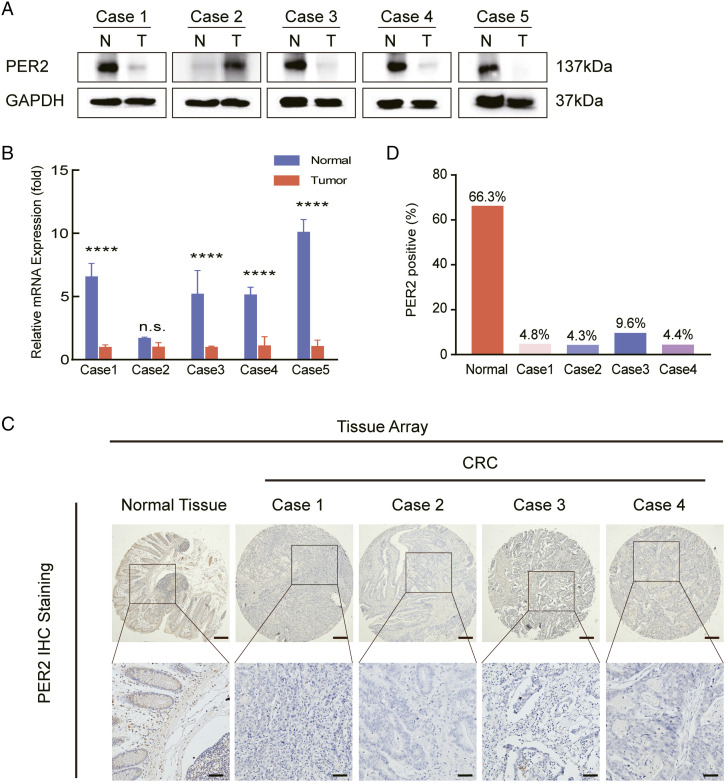
To confirm the expression of PER2 in CRC, we collected 5 clinical samples from CRC patients and found that the protein level of PER2 in CRC was extremely low compared with that in adjacent part (Figure 1A). We further confirmed the low expression of PER2 in CRC at the mRNA level, which was consistent with the western blot analysis (Figure 1B). In addition, we performed IHC staining and illustrated that the expression of PER2 in precancerous part was extremely higher than this in cancerous part (Figure 1C and 1D); detailed case information is shown in Supplementary Figure S1. These results indicated that PER2 expression is decreased in CRC patients.
Figure 1.
PER2 expression is downregulated in CRC patient samples. (A, B) The expression of PER2 was analyzed by western blot and qPCR in 5 pairs of human CRC and adjacent tissues (**** P <.0001). (C) IHC analysis of PER2 on TMAs of matched pairs of CRC tissue (n = 90) and healthy adjacent tissue (n = 90), scale bar: 200 µm (upper panel), 50 µm (lower panel). (D) Statistical results of the staining intensity of tissues subjected to IHC; ImageJ software was used for counting the staining percentage. PER2, Period 2; CRC, Colorectal cancer.
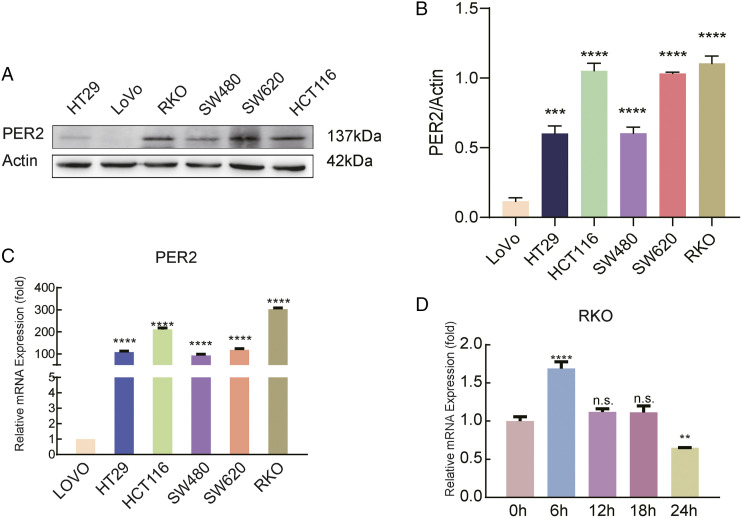
We further evaluated the expression of PER2 in the CRC cell line. Compared with other cell lines, RKO, SW620, and HCT116 cells showed higher levels of PER2 expression (Figure 2A-2C), and we chose RKO for subsequent experiments. Considering that PER2 is a protein involved in regulating circadian rhythm, we detected the protein level of PER2 at 0, 6, 12, and 24 h and found that its expression increased periodically at 6 h (Figure 2D). Therefore, in our follow-up experiments, we harvested cells at 6 h to avoid potential interference.
Figure 2.
Expression of PER2 in CRC cell lines. (A) Detect the expression of PER2 in CRC cell lines by western blot analysis; (B) statistical results of the expression of PER2 in CRC cell lines by western blot analysis; (C) PER2 expression in CRC cell lines as determined by qPCR (**** P <.0001); (D) periodic changes in PER2 expression in RKO cells as detected by qPCR (**P<.01, **** P <.0001). PER2, Period 2; CRC, Colorectal cancer.
Construction and Detection of the Period 2 Stable Knockdown Cell Line
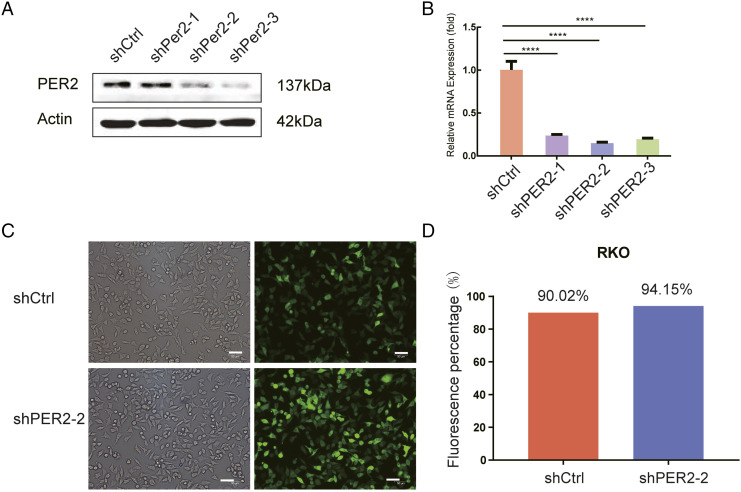
We obtained an RKO cell line with stable low expression of PER2 by constructing a plasmid with GFP. Subsequently, we used western blotting and qPCR to confirm the knockdown effect at the protein and mRNA levels, respectively (Figure 3A and 3B). The results showed that shPER2-2 and shPER2-3 worked successfully, and shPER2-2 was selected for subsequent experiments due to its superior silencing effect. Based on GFP fluorescence, we also validated the transfection effect under a fluorescence microscope (Figure 3C and 3D).
Figure 3.
Construction of the PER2 stable knockdown cell line. (A) Western blot analysis of PER2 expression in RKO cells after shRNA transfection; (B) qPCR detection of PER2 mRNA expression in transfected RKO; (C) efficiency of control and shPER-2 lentiviral infection based on GFP fluorescence (approximately >90%) under an optical (left panel) or fluorescence (right panel) microscope, scale bar 50 µm. (D) The numbers of fluorescent cells were counted by ImageJ and are shown in the histogram. PER2, Period 2; RKO, human colorectal adenocarcinoma cells; qPCR: real-time quantitative polymerase chain reaction.
Period 2 Silencing Promotes Colorectal Cancer Cell Migration Through the Epithelial-Mesenchymal Transformation Pathway
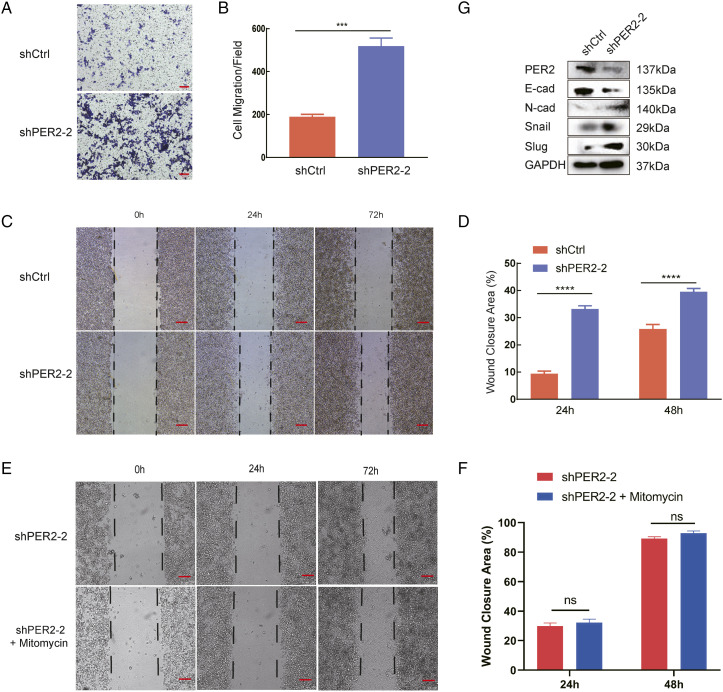
To explore the effect of PER2 on tumor migration, we used shPER2-2 stable cells. We first performed a Transwell experiment and found that compared with control cells, knocking down PER2 significantly enhanced the migration ability of RKO cells (Figure 4A and 4B). Furthermore, a wound-healing assay was performed and showed that cell migration was significantly enhanced after PER2 knockdown (Figure 4C and 4D). Since we cultured for 72 hours, we added the cell proliferation inhibitor mitomycin to eliminate the effect of proliferation on migration (Figure 4E and 4F).
Figure 4.
PER2 knockdown promotes RKO cell migration and activates the EMT process. (A) Detect the number of cell migration in the shPER2 and shControl groups by Transwell. (B) The number of migratory cells was counted by ImageJ, scale bar 100 µm (*** P <.001). (C) Determine the cell migration after PER2 knockdown after 48 and 72h by Wound-healing assay. (D) The percentage of wound area closure was analyzed by ImageJ, scale bar 200 µm, shown in the lower panel (**** P <.0001). (E) Mitomycin was added to remove the effect of proliferation on migration. (F) The percentage of wound area closure. (G) The expression of EMT-associated markers was detected by western blotting. EMT, epithelial-mesenchymal transformation.
To better understand the mechanism of enhanced migration, we then detected EMT markers in control and PER2-knockdown cells. Surprisingly, our data indicated that the expression of the epithelial marker E-cadherin was downregulated. Conversely, the expression of mesenchymal markers, including N-cadherin, Snail, and Slug, was upregulated (Figure 4G). These results indicated that the EMT pathway was activated upon PER2 knockdown.
Period 2 Reduction Promotes the Proliferation of Colorectal Cancer Cells
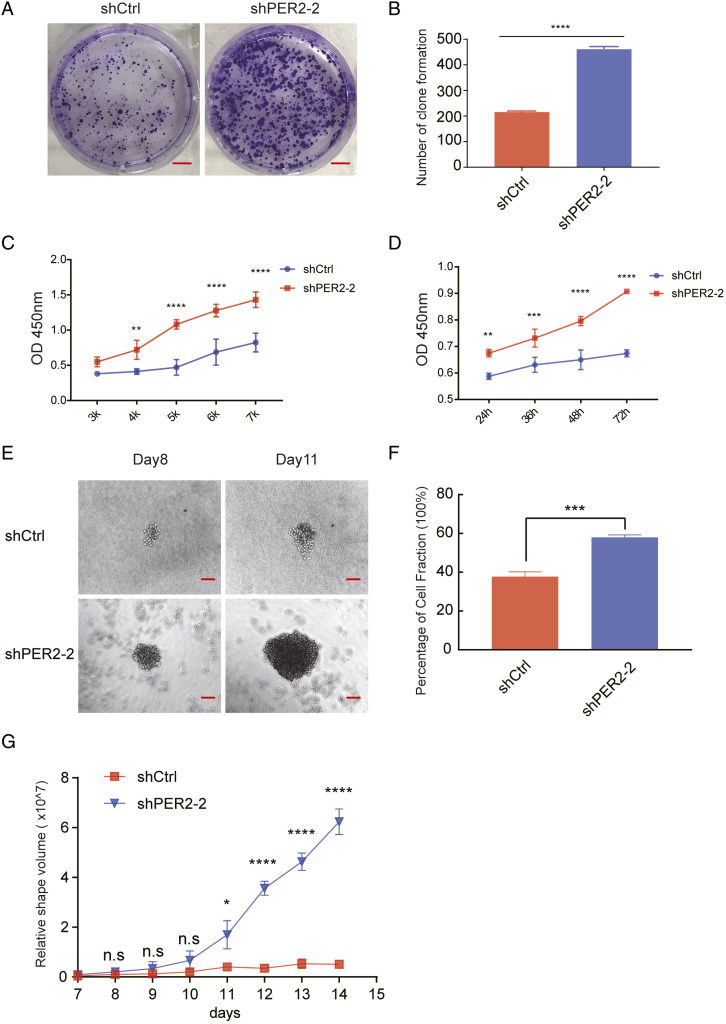
With shPER2-2 stable cells, we performed a colony formation experiment, and the results suggested that the number of clones formed in the group of shPER2 were obviously greater than shcontrol (Figure 5A and 5B). The CCK-8 experiment showed that the PER2 low expression group showed a higher OD value at 450 nm than the control group (Figure 5C and 5D), which is indicative of greater viability. We then carried out sphere formation assays, the results of which showed that the cells in the PER2-knockdown group exhibited a higher sphere-forming rate and larger spheres (Figure 5E-5G). All of our data suggested that the malignant biological performance of cells is enhanced after PER2 knockdown.
Figure 5.
PER2 knockdown promotes colorectal cancer proliferation. (A) Clone formation assay of PER2-knockdown and control cells. A total of 2 x 10^3 cells were seeded in each group and evaluated after 14 days. (B) Colony formation numbers were counted with ImageJ and are shown in the right panel (**** P <.0001). (C, D) CCK-8 was performed to detect OD 450 nm in the PER2-knockdown and control groups. The cells were seeded with varying densities (3 k, 4 k, 5 k, 6 k, and 7 k) and detected after 24 h (C) or seeded at 5 x 10^3 (5 k) cells per well and detected at 24, 36, 48, and 72 h (D) (** P <.01, *** P <.001, **** P <.0001). (E) Sphere formation experiments in the PER2-knockdown and control groups. The length and short diameter of the sphere were measured under the microscope. (F) The percentage of the cell fraction is shown. (G) The relative volume of the shapes was calculated with the formula of V = 4π/3 × R3(R = (L+W)/2) and is shown in the graph (L, length of the sphere; W, short diameter of the sphere) (*P<.05, **** P <.0001).PER2, Period 2.
Period 2 Knockdown Impairs the Chemosensitivity of Colorectal Cancer Cells
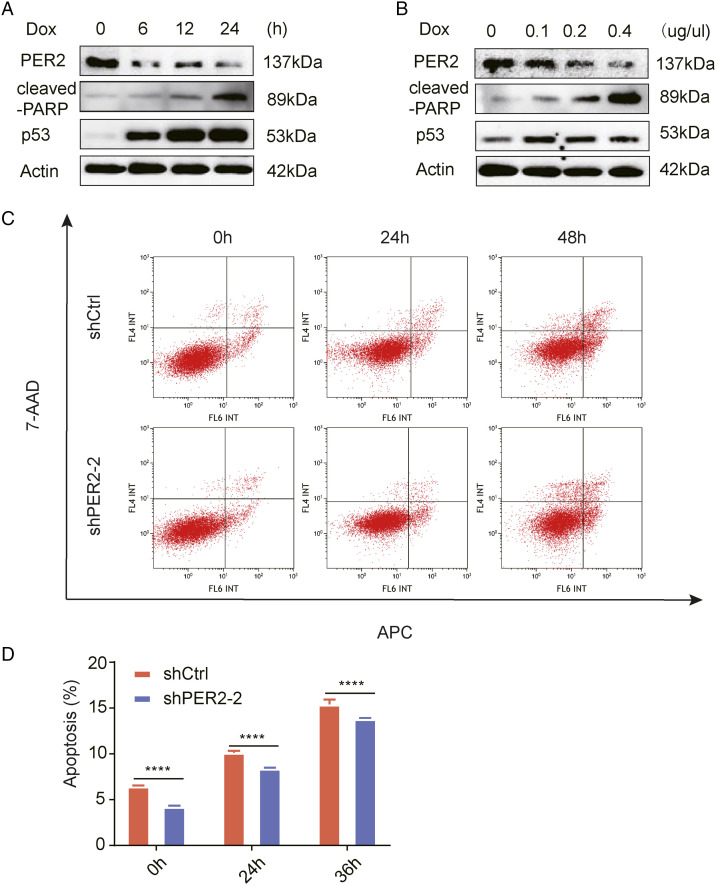
We next performed western blot analysis at different times after treatment with a concentration gradient of Dox (doxorubicin). As the drug treatment period was prolonged and the drug concentration was increased, PER2 showed a gradual decrease in expression concomitant with steady increases in p53 and cleaved-PARP expression (Figure 6A and 6B). The results showed that with increasing concentrations of Dox, PER2 expression decreased gradually, while p53 expression increased gradually in RKO cells.
Figure 6.
PER 2 reduces the sensitivity of colorectal cancer cells to doxorubicin. (A) Changes in PER2, cleaved-PARP, and p53 protein levels in RKO cells after .2 mM Dox treatment for 0, 6, 12, and 24 h. (B) Changes in PER2, cleaved-PARP, and p53 protein levels in RKO cells treated with different concentrations of Dox (0, .1 µg/µl, .2 µg/µl, and .4 µg/µl) for 24 h. (C) Representative images of flow cytometry of PER2-knockdown and control cells treated with .2 mM Dox at different times. (D) The percentage of apoptotic cells is shown (lower panel, **** P <.0001).PER2, Period 2; Dox, doxorubicin.
Finally, we analyzed the effect of PER2 on apoptosis and found that the apoptosis rate of RKO cells in shPER2 group was lower than that in control cells, showing a strong anti-apoptotic characteristic (Figure 6C and 6D). It is suggested that the survival ability of CRC cells with PER2 knockdown is stronger than that of control cells, reflecting a certain degree of drug resistance.
Discussion
Since CRC is one of the leading causes of cancer-related deaths worldwide, it brings increasing medical burden to various countries and has an obvious trend of diagnosis at a younger age, whether in developed or developing countries.6,31,32 However, due to the multistage, multistep complexity of the pathogenesis, the specific mechanism of CRC development and progression is still unclear.
The PER2 gene is located on the long arm of chromosome 2 and has 25 exons. As a cycle regulator, PER2 is considered to be related to many kinds of tumorigenesis. 33 Recently, it has been found that an imbalance in PER2 gene expression is associated with NSCLC, 12 ovarian cancer, pancreatic cancer, breast cancer, prostate cancer, thyroid cancer, and so on.16,33-35 However, it has been reported that PER2 not only has anti-cancer effects in CRC but also has carcinogenic effects,18,19 indicating that the expression of PER2 is controversial. The most important thing regarding PER2 is that the latest research shows that it is associated with the tumorigenesis and development of gastric cancer.15,36 The exact relationship between PER2 and CRC is still unknown yet. Upon detecting PER2 at a lower level in CRC tissue than in normal tissue, we constructed a cell line with stable PER2 knockdown to analyze its biological function.
The malignancy of CRC is closely related to proliferation and migration.37,38 To determine the function of PER2 in these processes, we performed CCK-8, wound healing, Transwell, and other functional experiments with stable cell lines. After knocking down PER2, malignant biological behaviors, such as tumor proliferation and migration, were enhanced, suggesting that PER2 can inhibit cancer development and confirming that PER2 suppresses proliferation and migration in CRC.
We found that the EMT signaling pathway was activated based on changes in the expression of some key proteins. EMT, an important part of distant metastasis of tumor cells, reprograms them toward dedifferentiation and a mesenchymal phenotype. EMT and its complementary process, mesenchymal-to-epithelial transition (MET), are induced by external stimuli as the result of the interaction between tumor cells and the microenvironment at different sites and niches within the primary tumor, and it is considered to be a dynamic process.26,39 The AKT and mTOR signaling pathways are closely related to EMT, 40 but it is unclear whether these pathways are involved in the regulation of CRC. Our data indicated that hallmark of EMT, E-cadherin was downregulated and N-cadherin was upregulated. Simultaneously, two master regulators of the EMT, Snail and Slug, were also upregulated. It has been reported that Snail is essential in regulation of malignant phenotypes including stemness, cell motility, and chemotherapy resistance.41,42 Besides, Slug could also promote tumor migration and invasion, and mediate stemness, leading to malignant transformation of cells.43,44 Therefore, combined with the results of functional experiments, we suggested that the EMT pathway was activated after PER2 knockdown, and the Snail/Slug axis promotes cell growth and migration and enhances stemness, which played a leading role in the malignant phenotype of CRC.
Overexpression of PER2 is demonstrated to inhibit tumor growth in vivo 45 and to induce apoptosis in the mammary carcinoma cell and Lewis lung carcinoma cell. 46 To resolve the problem of whether PER2 affects the apoptosis of CRC cells, we treated the cell lines with the most powerful inducer of apoptosis Dox for 48 h. Dox is a longstanding chemotherapeutic agent and still represents a pillar of treatment protocols for various cancers, such as bladder, testicular, ovarian, and lung cancers,47,48 and as a DNA intercalating anthracycline antibiotic, Dox is clinically proven to be effective in treating CRC.49,50 We found that with increasing concentrations of Dox, PER2 expression decreased gradually, while p53 expression increased gradually in RKO cells. p53 has been proven to inhibit the expression of Snail and Slug and regulate EMT transformation to exert its anti-cancer effect.51,52 Conversely, Snail and Slug mediate radio-resistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in cancer cells. 53 These studies are consistent with our results. Besides, we used flow cytometry to assess apoptosis of the stable knockdown cell line, and the PER2 knockdown group showed less apoptosis than the control group at different times. This indicates that PER2 can induce apoptosis in CRC cells through p53 activation. It has been reported that p53 not only has transcriptional inhibition function but also owns transcriptional activation function.54,55 Based on this, we hypothesized that p53 may directly inhibit PER2 transcriptional activity after apoptosis, or activates genes that may inhibit PER2 expression, for example, through SIRT1/p53 axis.56,57 Combining the above findings, we speculated that after PER2 knockdown, the deletion of p53 led to the activation of the Snail/Slug axis, which made the cells presented a malignant phenotype, and DOX treatment may restore the expression of p53, which in turn promoted apoptosis.
But our experiment also has many deficiencies and limitations. Two of the biggest deficiencies are that we did not successfully constructed stable cell lines expressing PER2 and did not conducted further animal studies. We only constructed stable cell lines with PER2 knockdown, but did not successfully construct stable cell lines with overexpression of PER2. It could be a combination of things, like our technology or the molecular weight of PER2 being too high. When we attempted to explore the effect of overexpression of PER2 on the proliferation and migration of LOVO cells through transient transfection, we unfortunately found that the difference was not obvious (Supplementary Figure S2). Also, our experiment only revealed that PER2 inhibits the malignant degree of colorectal cells by inhibiting EMT progress and cell proliferation, but the specific mechanism by which PER2 activates and regulates the EMT signaling pathway through Snail/Slug axis is still unknown, and this requires further investigation.
Conclusion
In summary, we detected the role of the PER2 gene on the biological function of CRC cells and found that the ability of tumor cells to increase invasion and metastasis increased after PER2 knockdown. In addition, our data also clarify that PER2 knockdown may activate the Snail/Slug axis through inhibiting p53, therefore promote the activation of the EMT pathway, suggesting that PER2 inhibits the occurrence and progression of CRC, and these findings provide evidence that PER2 is a potential therapeutic target that may improve patient prognosis and establish a theoretical basis for clinical treatment.
Supplemental Material
Supplemental Material, sj-pdf-1-ccx-10.1177_10732748221081369 for Period 2 Suppresses the Malignant Cellular Behaviors of Colorectal Cancer Through the Epithelial-Mesenchymal Transformation Process by Yubo Xiong, Yifan Zhuang, Mengya Zhong, Wenjuan Qin, Boyi Huang, Jiabao Zhao, Zhi Gao, Jingsong Ma, Zhengxin Wu, Xuehui Hong, Zhicao Yue, and Haijie Lu in cancer control
Supplemental Material, sj-pdf-2-ccx-10.1177_10732748221081369 for period 2 suppresses the malignant cellular behaviors of colorectal cancer through the EMT process by Yubo Xiong, Yifan Zhuang, Mengya Zhong, Wenjuan Qin, Boyi Huang, Jiabao Zhao, Zhi Gao, Jingsong Ma, Zhengxin Wu, Xuehui Hong, Zhicao Yue, and Haijie Lu in cancer control
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The present study was supported by the Natural Science Foundation of Fujian Province of China (No. 2021J011335, No. 2021J011332), the Xiamen Science and Technology Plan Projects (No. 3502Z20209021), the Medical Innovation Project of Fujian Province (No. 2021CXB023), and the Health Youth Scientific Research Project of Fujian Province of China (No. 2020QNB059, No. 2020GGB055).
Ethical Approval: The experimental procedure was approved by the Ethics Committee of the Affiliated Zhongshan Hospital of Xiamen University, No:xmzsyyky2021197.
Ethical Statement: This article does not contain any studies with human or animal subjects.
Informed Consent: All patients participating in this study provided written informed consent.
Supplemental Material: Supplemental material for this article is available online.
ORCID iD
Xuehui Hong https://orcid.org/0000-0002-8853-7384
References
- 1.Hasakova K, Reis R, Vician M, Zeman M, Herichova I. Expression of miR-34a-5p is up-regulated in human colorectal cancer and correlates with survival and clock gene PER2 expression. Plos One. 2019;14(10):e0224396. doi: 10.1371/journal.pone.0224396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019; 16(12):713-732.doi: 10.1038/s41575-019-0189-8 [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Aranda M, Redondo M. Targeting receptor kinases in colorectal cancer. Cancers. 2019;11(4):433. doi: 10.3390/cancers11040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smyth EC, Nilsson M, Grabsch HI, van Grieken NCT, Lordick F. Gastric cancer. Lancet. 2020;396(10251):635-648. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Xu Z, Liu D. Small non-coding RNA and colorectal cancer. J Cell Mol Med. 2019;23(5):3050-3057. doi: 10.1111/jcmm.14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venugopal A, Stoffel EM. Colorectal cancer in young adults. Curr Treat Options Gastroenterol. 2019;17(1):89-98. doi: 10.1007/s11938-019-00219-4. [DOI] [PubMed] [Google Scholar]
- 7.Dai W, Zhou F, Tang D, Lin L, Zou C, Tan W, et al. Single-cell transcriptional profiling reveals the heterogenicity in colorectal cancer. Medicine. 2019;98(34):e16916. doi: 10.1097/MD.0000000000016916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Morita Y, Han B, Niemann S, Löffler B, Rudolph KL. Per2 induction limits lymphoid-biased haematopoietic stem cells and lymphopoiesis in the context of DNA damage and ageing. Nat Cell Biol. 2016;18(5):480-490. doi: 10.1038/ncb3342. [DOI] [PubMed] [Google Scholar]
- 9.Zhuo H, Wang Y, Zhao Q. The interaction between Bmal1 and per2 in mouse BMSC osteogenic differentiation. Stem Cell Int. 2018;2018:1-11. doi: 10.1155/2018/3407821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M, de la Peña JB, Cheong JH, Kim HJ. Neurobiological functions of the period circadian clock 2 gene, per2. Biomol Ther. 2018;26(4):358-367. doi: 10.4062/biomolther.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McQueen CM, Schmitt EE, Sarkar TR, Elswood J, Metz RP, Earnest D. PER2 regulation of mammary gland development. Development. 2018;145(6):dev157966. doi: 10.1242/dev.157966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang R, Cui Y, Wang Y, Xie T, Yang X, Wang Z, Circadian clock gene per2 downregulation in nonsmall cell lung cancer is associated with tumour progression and metastasis. Oncol Rep. 2018;40(5):3040-3048. doi: 10.3892/or.2018.6704. [DOI] [PubMed] [Google Scholar]
- 13.Qin T, Lu XT, Li YG, Liu Y, Yan W, Li N. Effect of period 2 on the proliferation, apoptosis and migration of osteosarcoma cells, and the corresponding mechanisms. Oncol Lett. 2018;16(2):2668-2674. doi: 10.3892/ol.2018.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng AY, Zhang Y, Mei HJ, Fang S, Ji P, Yang J. Construction of a plasmid for overexpression of human circadian gene period2 and its biological activity in osteosarcoma cells. Tumour Biol. 2015;36(5):3735-3743. doi: 10.1007/s13277-014-3013-7. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez-Rosas F, Hernandez-Oliveras A, Flores-Peredo L, Rodriguez G, Zarain-Herzberg A, Caba M. Histone deacetylase inhibitors induce the expression of tumor suppressor genes per1 and per2 in human gastric cancer cells. Oncol Lett. 2018;16(2):1981-1990. doi: 10.3892/ol.2018.8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin HH, Qraitem M, Lian Y, Taylor SR, Farkas ME. Analyses of BMAL1 and PER2 oscillations in a model of breast cancer progression reveal changes with malignancy. Integr Cancer Ther. 2019;18:1534735419836494. doi: 10.1177/1534735419836494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang MY, Yang WC, Lin PM, Hsu JF, Hsiao HH, Liu YC, et al. Altered expression of circadian clock genes in human chronic myeloid leukemia. J Biol Rhythm. 2011;26(2):136-148. doi: 10.1177/0748730410395527. [DOI] [PubMed] [Google Scholar]
- 18.Karantanos T, Theodoropoulos G, Pektasides D, Gazouli M. Clock genes: their role in colorectal cancer. World J Gastroenterol. 2014;20(8):1986-1992. doi: 10.3748/wjg.v20.i8.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaashua L, Mayer S, Lior C, Lavon H, Novoselsky A, Scherz-Shouval R. Stromal expression of the core clock gene period 2 is essential for tumor initiation and metastatic colonization. Front Cell Dev Biol. 2020;8:587697. doi: 10.3389/fcell.2020.587697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi S, Li Q, Cao Q, Diao Y, Zhang Y, Yue L, et al. EMT transcription factors are involved in the altered cell adhesion under simulated microgravity effect or overloading by regulation of E-cadherin. Int J Mol Sci. 2020;21(4):1349. doi: 10.3390/ijms21041349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loh CY, Chai JY, Tang TF, Wong WF, Sethi G, Shanmugam MK, et al. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells. 2019;8(10):1118. doi: 10.3390/cells8101118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Ashmawy NE, El-Zamarany EA, Khedr EG, Abo-Saif MA. Activation of EMT in colorectal cancer by MTDH/NF-kappaB p65 pathway. Mol Cell Biochem. 2019;457(1-2):83-91. doi: 10.1007/s11010-019-03514-x. [DOI] [PubMed] [Google Scholar]
- 23.Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18(1):91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vu T, Datta PK. Regulation of EMT in colorectal cancer: a culprit in metastasis. Cancers. 2017;9(12):171. doi: 10.3390/cancers9120171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim SH, Becker TM, Chua W, Ng WL, de Souza P, Spring KJ. Circulating tumour cells and the epithelial mesenchymal transition in colorectal cancer. J Clin Pathol. 2014;67(10):848-853. doi: 10.1136/jclinpath-2014-202499. [DOI] [PubMed] [Google Scholar]
- 26.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69-84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 27.Stefania D, Vergara D. The many-faced program of epithelial-mesenchymal transition: a system biology-based view. Front Oncol. 2017;7:274. doi: 10.3389/fonc.2017.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giudetti AM, De Domenico S, Ragusa A, Lunetti P, Gaballo A, Franck J. A specific lipid metabolic profile is associated with the epithelial mesenchymal transition program. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864(3):344-357. doi: 10.1016/j.bbalip.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21(6):341-352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007;147(8):573-577. doi: 10.7326/0003-4819-147-8-200710160-00010. [DOI] [PubMed] [Google Scholar]
- 31.Araghi M, Soerjomataram I, Jenkins M, Brierley J, Morris E, Bray F. Global trends in colorectal cancer mortality: projections to the year 2035. Int J Cancer. 2019;144(12):2992-3000. doi: 10.1002/ijc.32055. [DOI] [PubMed] [Google Scholar]
- 32.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14(2):89-103. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morales-Santana S, Morell S, Leon J, Carazo-Gallego A, Jimenez-Lopez JC, Morell M. An overview of the polymorphisms of circadian genes associated with endocrine cancer. Front Endocrinol. 2019;10:104. doi: 10.3389/fendo.2019.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louis J, Ovrut BA. Cohomology and heterotic world-sheet anomalies. Phys Rev D: Part Fields. 1987;36(4):1119-1126. doi: 10.1103/physrevd.36.1119. [DOI] [PubMed] [Google Scholar]
- 35.Lellupitiyage Don SS, Lin HH, Furtado JJ, Qraitem M, Taylor SR, Farkas ME. Circadian oscillations persist in low malignancy breast cancer cells. Cell Cycle. 2019;18(19):2447-2453. doi: 10.1080/15384101.2019.1648957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajendran S, Benna C, Marchet A, Nitti D, Mocellin S. Germline polymorphisms of circadian genes and gastric cancer predisposition. Cancer Commun. 2020;40(5):234-238. doi: 10.1002/cac2.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tu X, Hong D, Jiang Y, Lou Z, Wang K, Jiang Y. FH535 inhibits proliferation and migration of colorectal cancer cells by regulating cyclinA2 and claudin1 gene expression. Gene. 2019;690:48-56. doi: 10.1016/j.gene.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Coronel-Hernandez J, Lopez-Urrutia E, Contreras-Romero C, Delgado-Waldo I, Figueroa-Gonzalez G, Campos-Parra AD. Cell migration and proliferation are regulated by miR-26a in colorectal cancer via the PTEN-AKT axis. Cancer Cell Int. 2019;19:80. doi: 10.1186/s12935-019-0802-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao Y, Baker D, Ten Dijke P. TGF-beta-mediated epithelial-mesenchymal transition and cancer metastasis. Int J Mol Sci. 2019;20(11):2767. doi: 10.3390/ijms20112767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karimi Roshan M, Soltani A, Soleimani A, Rezaie Kahkhaie K, Afshari AR, Soukhtanloo M. Role of AKT and mTOR signaling pathways in the induction of epithelial-mesenchymal transition (EMT) process. Biochimie. 2019;165:229-234. doi: 10.1016/j.biochi.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Wu X, Cai J, Zuo Z, Li J. Collagen facilitates the colorectal cancer stemness and metastasis through an integrin/PI3K/AKT/Snail signaling pathway. Biomed Pharmacother. 2019;114:108708. doi: 10.1016/j.biopha.2019.108708. [DOI] [PubMed] [Google Scholar]
- 42.Hojo N, Huisken AL, Wang H, Chirshev E, Kim NS, Nguyen SM. Snail knockdown reverses stemness and inhibits tumour growth in ovarian cancer. Sci Rep. 2018;8(1):8704. doi: 10.1038/s41598-018-27021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uygur B, Wu WS. SLUG promotes prostate cancer cell migration and invasion via CXCR4/CXCL12 axis. Mol Cancer. 2011;10:139. doi: 10.1186/1476-4598-10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chien MH, Lin YW, Wen YC, Yang YC, Hsiao M, Chang JL. Targeting the SPOCK1-snail/slug axis-mediated epithelial-to-mesenchymal transition by apigenin contributes to repression of prostate cancer metastasis. J Exp Clin Cancer Res. 2019;38(1):246. doi: 10.1186/s13046-019-1247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyazaki K, Wakabayashi M, Hara Y, Ishida N. Tumor growth suppressionin vivoby overexpression of the circadian component, PER2. PER2. Genes Cells. 2010;15(4):351-358. doi: 10.1111/j.1365-2443.2010.01384.x. [DOI] [PubMed] [Google Scholar]
- 46.Hua H, Wang Y, Wan C, Liu Y, Zhu B, Yang C, et al. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006;97(7):589-596. doi: 10.1111/j.1349-7006.2006.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colombo V, Lupi M, Falcetta F, Forestieri D, D’Incalci M, Ubezio P. Chemotherapeutic activity of silymarin combined with doxorubicin or paclitaxel in sensitive and multidrug-resistant colon cancer cells. Cancer Chemother Pharmacol. 2011;67(2):369-379. doi: 10.1007/s00280-010-1335-8. [DOI] [PubMed] [Google Scholar]
- 48.Yang F, Teves SS, Kemp CJ, Henikoff S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim Biophys Acta. 2014;1845(1):84-89. doi: 10.1016/j.bbcan.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee CC, Gillies ER, Fox ME, Guillaudeu SJ, Frechet JM, Dy EE. A single dose of doxorubicin-functionalized bow-tie dendrimer cures mice bearing C-26 colon carcinomas. Proc Natl Acad Sci USA. 2006;103(45):16649-16654. doi: 10.1073/pnas.0607705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li M, Tang Z, Zhang D, Sun H, Liu H, Zhang Y, et al. Doxorubicin-loaded polysaccharide nanoparticles suppress the growth of murine colorectal carcinoma and inhibit the metastasis of murine mammary carcinoma in rodent models. Biomaterials. 2015;51:161-172. doi: 10.1016/j.biomaterials.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 51.Liu X, Min S, Wu N, Liu H, Wang T, Li W, et al. miR-193a-3p inhibition of the slug activator PAK4 suppresses non-small cell lung cancer aggressiveness via the p53/Slug/L1CAM pathway. Cancer Lett. 2019;447:56-65. doi: 10.1016/j.canlet.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 52.Lee SH, Park BJ. p53 activation by blocking snail: a novel pharmacological strategy for cancer. Curr Pharm Des. 2011;17(6):610-617. doi: 10.2174/138161211795222658. [DOI] [PubMed] [Google Scholar]
- 53.Kurrey NK, Jalgaonkar SP, Joglekar AV, Ghanate AD, Chaskar PD, Doiphode RY, et al. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cell. 2009;27(9):2059-2068. doi: 10.1002/stem.154. [DOI] [PubMed] [Google Scholar]
- 54.Chao CC-K. Mechanisms of p53 degradation. Clin Chim Acta. 2015;438:139-147. doi: 10.1016/j.cca.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 55.Marcel V, Catez F, Diaz JJ. p53, a translational regulator: contribution to its tumour-suppressor activity. Oncogene. 2015;34(44):5513-5523. doi: 10.1038/onc.2015.25. [DOI] [PubMed] [Google Scholar]
- 56.Osum M, Serakinci N. Impact of circadian disruption on health; SIRT1 and telomeres. DNA Repair. 2020;96:102993. doi: 10.1016/j.dnarep.2020.102993. [DOI] [PubMed] [Google Scholar]
- 57.Park JH, Park SA, Lee YJ, Joo NR, Shin J, Oh SM. TOPK inhibition accelerates oxidative stress-induced granulosa cell apoptosis via the p53/SIRT1 axis. Int J Mol Med. 2020;46(5):1923-1937. doi: 10.3892/ijmm.2020.4712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-ccx-10.1177_10732748221081369 for Period 2 Suppresses the Malignant Cellular Behaviors of Colorectal Cancer Through the Epithelial-Mesenchymal Transformation Process by Yubo Xiong, Yifan Zhuang, Mengya Zhong, Wenjuan Qin, Boyi Huang, Jiabao Zhao, Zhi Gao, Jingsong Ma, Zhengxin Wu, Xuehui Hong, Zhicao Yue, and Haijie Lu in cancer control
Supplemental Material, sj-pdf-2-ccx-10.1177_10732748221081369 for period 2 suppresses the malignant cellular behaviors of colorectal cancer through the EMT process by Yubo Xiong, Yifan Zhuang, Mengya Zhong, Wenjuan Qin, Boyi Huang, Jiabao Zhao, Zhi Gao, Jingsong Ma, Zhengxin Wu, Xuehui Hong, Zhicao Yue, and Haijie Lu in cancer control