Abstract
ASC is an essential adaptor of the inflammasome, a micrometer-size multiprotein complex that processes proinflammatory cytokines. Inflammasome formation depends on ASC self-association into large assemblies via homotypic interactions of its two death domains, PYD and CARD. ASCb, an alternative splicing isoform, activates the inflammasome to a lesser extent compared with ASC. Thus, it has been postulated that adaptor isoforms differentially regulate inflammasome function. At the amino acid level, ASC and ASCb differ only in the length of the linker connecting the two death domains. To understand inflammasome regulation at the molecular level, we investigated the self-association properties of ASC and ASCb using real-time NMR, dynamic light scattering (DLS), size-exclusion chromatography, and transmission electron microscopy (TEM). The NMR data indicate that ASC self-association is faster than that of ASCb; a kinetic model for this oligomerization results in differing values for both the reaction order and the rate constants. Furthermore, DLS analysis indicates that ASC self-associates into more compact macrostructures compared with ASCb. Finally, TEM data show that ASCb has a reduced tendency to form densely packed filaments relative to ASC. Overall, these differences can only be explained by an effect of the linker length, as the NMR results show structural equivalence of the PYD and CARD in both proteins. The effect of linker length was corroborated by molecular docking with the procaspase-1 CARD domain. Altogether, our results indicate that ASC’s faster and less polydisperse polymerization is more efficient, plausibly explaining inflammasome activation differences by ASC isoforms at the molecular level.
Keywords: inflammation, inflammasome regulation, inflammasome adaptor protein isoforms, ASC, ASCb, protein oligomerization and self-assembly, protein NMR
Abbreviations: ALR, absent in melanoma 2-like receptor; ASC, apoptosis-associated speck-like protein containing a CARD; CARD, caspase activation and recruitment domain; DAMPS, damage-associated molecular patterns; DLS, dynamic light scattering; IL-1β, interleukin-1 beta; IL-18, interleukin-18; NF-κβ, nuclear factor kappa beta; NLR, nucleotide-binding domain leucine-rich repeat containing receptor; NMR, nuclear magnetic resonance; NOD, nucleotide-binding and oligomerization domain; PAMP, pathogen-associated molecular patterns; PYD, pyrin domain; SEC, size-exclusion chromatography; TEM, transmission electron microscopy; TLR, toll-like receptor; TNF-α, tumor necrosis factor alpha
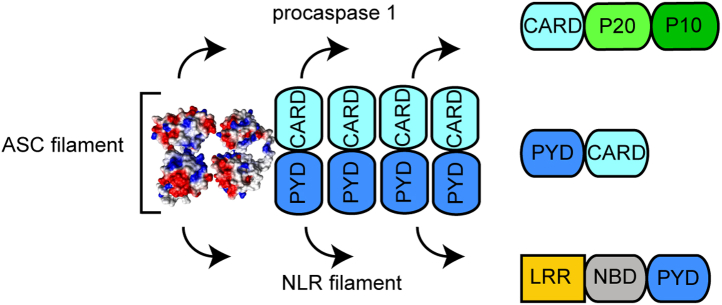
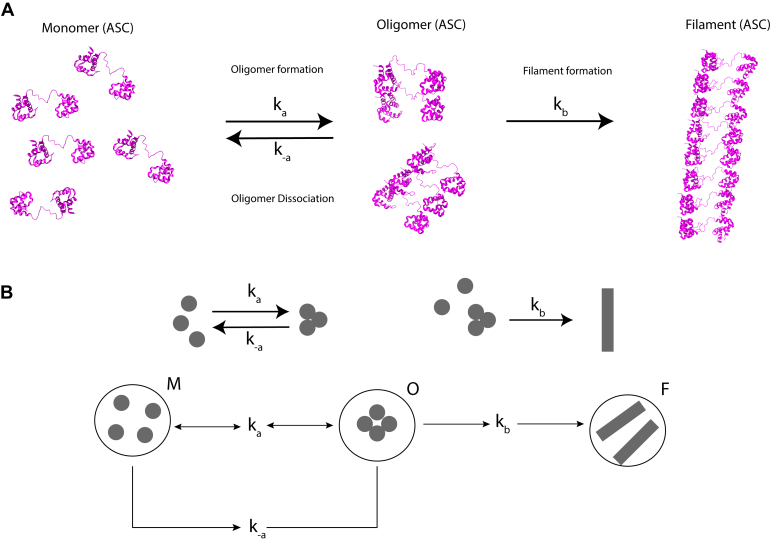
Inflammation is a vital physiological process that protects organisms from pathogens and injury. The inflammatory response is triggered by a myriad of stimuli that have been classified as pathogen- and damage-associated molecular patterns (PAMPs and DAMPs, respectively) (1, 2). The presence of these danger signals prompts the formation of a multimeric protein complex known as the inflammasome. The assembly of the inflammasome is driven by self-association and oligomerization of three types of proteins: sensor, adaptor, and effector. There are different families of inflammasome sensors that show specificity for certain PAMPS and DAMPS. Cytosolic sensors are mainly represented by the NLRs (nucleotide-binding domain leucine-rich repeat containing receptors) and ALRs (absent in melanoma 2-like receptors) families and pyrin (3, 4, 5, 6). Inflammasome sensors are modular and typically contain protein–protein interacting death domains: PYD (pyrin domain) and CARD (caspase activation and recruitment domain) (7, 8). Inflammasome sensors self-associate upon activation by danger signals and prompt the oligomerization of the inflammasome adaptor ASC (apoptosis-associated speck-like protein containing a CARD), a bimodular protein with an N-terminal PYD and a C-terminal CARD (3, 4, 9, 10). The inflammasome adaptor acts as a molecular glue by self-associating and tethering multiple copies of the sensor and the effector procaspase-1 (3, 11, 12) (Fig. 1). Inflammasome assembly leads to the activation of procaspase-1, rendering it capable of cleaving pro-IL-1β and pro-IL-18 into their active forms, which are secreted to the extracellular environment to trigger the inflammatory response. Inflammasomes are ∼0.5 μm-size perinuclear puncta formed in the cytosol of activated cells by the oligomerization and self-association of sensor, adaptor, and procaspase-1 (13).
Figure 1.
Model for ASC-dependent inflammasome assembly. ASC dimer shown as the filament minimal building block. The ASC filament shows two interacting sides, one for recruiting procaspase-1 CARD and the other for interaction with the PYD of NLRs sensor proteins (12).
The NLR member NLRP3 is one of the most well-known inflammasome sensors. NLRP3 is expressed in myeloid, muscle and endocrine cells, and neurons (14). NLRP3 remains in an autoinhibited form in the resting state and becomes activated upon stimulation, assembling into a large, micrometer-size cytosolic complex together with the adaptor ASC. NLRP3 becomes functional in a two-step process: priming and activation (15). In the priming step, Toll-like receptors (TLRs) or NODs (nucleotide binding oligomerization domain) and cytokines such as TNF-α trigger the activation of the transcription factor NF-κB promoting the expression of the inflammasome components: NLRP3, procaspase-1, and pro-IL-1β (15, 16). Subsequent posttranslation modifications (ubiquitylation, phosphorylation, and sumoylation) of NLRP3 stabilize the protein in a signal-competent, autosuppressed inactive state that transforms into an activated state upon stimulation (17). Once activated, NLRP3 assembles into the inflammasome: a mature multiprotein-complex particle also composed of ASC and procaspase-1 (the NLRP3 inflammasome), capable of activating IL-1β and IL-18.
The dysregulation of the inflammasome is linked to uncontrolled inflammation as the culprit of an increasing number of inflammatory diseases including rheumatoid arthritis, type I diabetes, asthma, psoriasis, atherosclerosis, and neurodegenerative disorders (18, 19). Therefore, gaining insight into the mechanisms involved in inflammasome regulation and assembly is critical to identify potential targets for therapeutic treatment. Protein isoforms share highly similar amino acid sequences and structures and are known to participate in regulatory roles by tuning and modulating protein function. Thus, the study of the molecular bases underpinning function modulation by protein isoforms is particularly relevant to understand regulation at the molecular level. The inflammasome adaptor ASC has four isoforms: canonical ASC, ASCb, ASCc, and ASCd, which differ in amino acid composition (20). ASC and ASCb are the most similar as both retain the two protein–protein interacting domains, PYD and CARD (Fig. 2A), and only differ in the length of the interdomain linker (23 and three amino acids for ASC and ASCb, respectively). ASCc retains the CARD and a fragment of the PYD. ASCd bears only a partial PYD and no CARD. It has been reported that only ASC and ASCb are inflammasome-activating isoforms, as both promote the release of IL-1β (21) and have been shown to colocalize with NLRP3 and procaspase-1 (20). However, ASCb activates the inflammasome to a lesser extent as determined by the amount of IL-1β released and cannot form the typical puncta characteristic of canonical inflammasomes, forming instead filamentous aggregates (20). In contrast, ASCc and ASCd do not colocalize with NLRP3 and cannot generate mature IL-1β, leading to the conclusion that these isoforms are not inflammasome activators. In fact, the release of IL-1β by ASC is diminished in the presence of ASCc, which suggests that the latter acts as an inflammasome inhibitor (20). No clear effect on inflammasome activity has been identified for ASCd, and thus, the function of this isoform is still unknown (20).
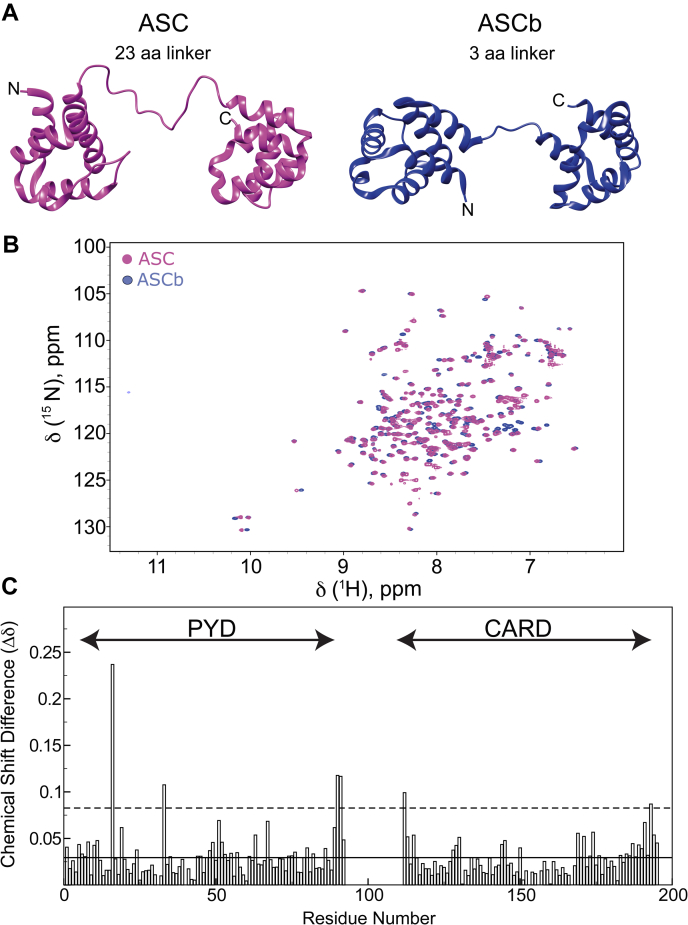
Figure 2.
ASC and ASCb structure comparison by NMR.A, ribbon diagram of the polypeptide backbone structure of ASC (10) and ASCb (model created with the program Chimera (23) based on the PDB structure of ASC (10)). B, 2D-[1H-15N]-HSQC (NMR protein fingerprint of ASC and ASCb showing almost complete signal overlap). C, combined-average amide 1H and 15N chemical shift differences between ASC and ASCb as described in the Experimental procedures section (solid line: 1 SD; dashed line: 2 SD).
To understand the modulation of inflammasome assembly by the activating isoforms, ASC and ASCb, we have studied the self-association properties of both proteins using several biophysical and biochemical techniques. By multidimensional solution NMR, we obtained the 15N and 13C chemical shifts of ASCb’s polypeptide backbone to compare to previously published data for ASC (22), revealing that the PYD and CARD structures of both isoforms are very similar as expected due to the identical amino acid sequence. NMR signal intensity decay resulting from oligomerization was monitored by real-time NMR at the individual residue level for both proteins over a period of 65 h. Based on these studies, we determine that ASC and ASCb follow different kinetics for self-association. Moreover, NMR data indicate that the PYD and CARD of ASC behave differently during self-association, whereas no differences were observed for the two domains in ASCb. Oligomer size distribution was analyzed by dynamic light scattering in ASC and ASCb solutions as a function of time during the self-association process, revealing different tendencies in polydispersity that agree with observations from the cell studies. The effect of protein concentration, pH, and time on the oligomerization processes was studied by size-exclusion chromatography (SEC), showing that ASC has a stronger self-association capability compared with ASCb. Additionally, the characteristics of the macrostructures formed by ASCb were investigated by negative-staining transmission electron microscopy (ns-TEM) and compared with previously reported TEM data for ASC. The analysis of ns-TEM micrographs reveals that ASCb also forms filaments of similar thickness to those formed by ASC; however, ASC tends to form bundles composed of a larger number of stacked filaments. Finally, the impact of linker length on the interaction between ASC or ASCb and the CARD of procaspase-1 was studied in silico by creating structural models via molecular docking.
Results
ASC and ASCb differ only in the linker length at the structural level
The previously reported 3D NMR structure of ASC revealed that the PYD and CARD domains adopt the six-helix bundle motif common to other death domain folds. The two domains are connected by a 23 amino acid–long linker (10). We expect that the structures of the PYD and CARD of ASC and ASCb would be almost identical because the two isoforms only differ in the linker length, and the PYD and CARD of both proteins share 100% amino acid sequence identity (Fig. 2A). To compare structural information of the two isoforms derived from the NMR data, we assigned the 15N and 13C backbone chemical shifts of ASCb (Table S1) and compared this assignment with that reported for ASC (22).
In protein NMR, the [1H-15N]-HSQC spectrum correlating the amide 1H and 15N chemical shifts of the protein backbone is known as the protein fingerprint because it is unique for each protein (11). Figure 2B depicts an overlay of the [1H-15N]-HSQC spectra showing that the signals of the PYD and CARD of ASC and ASCb substantially overlap as expected. The more crowded region of the ASC spectrum around 8.5 ppm (for the 1H chemical shift) reflects the additional signals from the longer, semiflexible linker. The average difference in chemical shifts for 15N and 1H of ASC and ASCb versus the residue number is very small (Fig. 2C), indicating that both PYD and CARD in ASCb are properly folded into the six-helix bundle motif. Amino acids that are close to the linker region in the amino acid sequence or in the 3D structure are expected to show deviations in chemical shifts for both isoforms. This is the case for amino acids H90, Q91, and L112; however, T16 and R33 (ASC numbering) show significant chemical shift variations even though these amino acids are not close to the linker region. We can speculate that the origin of these changes could be related to the oligomerizing properties of ASC and ASCb. In addition, a comparison of the 13Cα and 13Cβ chemical shifts for both isoforms (Fig. S1) indicates that the secondary structure of both proteins is identical, as expected.
Different oligomerization kinetics for ASC and ASCb from real-time NMR
ASC has a strong tendency to oligomerize and polymerizes forming filaments and filament bundles at neutral pH (12). This behavior is a consequence of ASC function as the inflammasome adaptor, which involves self-association and tethering multiple copies of the inflammasome components via protein–protein interactions. It has been reported that ASC self-association is reduced at acidic pH (pH 3.8) and concentration values <200 μM; conditions used for the 3D structure determination of ASC by solution NMR techniques (10, 11). However, ASC significantly self-associates at concentrations >200 μM even under acidic conditions. As previously reported, ASC self-association can be detected by a decrease of NMR signal intensity with time (10). This observation results from the formation of large oligomeric species that are “invisible” in solution NMR due to their slow tumbling rate, hence no longer contributing to the signal arising from the monomeric species (24, 25). We have leveraged this behavior to monitor changes in signal intensity upon ASC and ASCb self-association by real-time (RT) NMR. Protein samples are in lyophilized form after purification and start to oligomerize as soon they are dissolved in the NMR buffer. A series of 2D-[1H,15N]-NMR spectra were acquired as a function of time after NMR sample preparation for 65 h. The RT-NMR kinetic experiments were performed on 15N-labeled protein samples at different concentration values (∼300–700 μM) to investigate the influence of protein concentration on the kinetics of self-association. The first 1D projection of the 2D experiment reflects the overall signal intensity of the spectra, which significantly decreases with time (Fig. 3A) for ASC and ASCb due to the formation of high-order oligomers. The slow tumbling rate of these large oligomers renders them “invisible” in NMR, resulting in signal intensity decrease upon protein self-association. However, careful analysis of the intensity decrease with time reveals that ASC oligomerizes faster than ASCb (Fig. 3B). Protein concentration is an important factor prompting polymerization for both ASC and ASCb, as self-association rates are significantly diminished at ∼300 μM versus ∼700 μM (Fig. 3B). ASC oligomerizes slightly faster than ASCb at the lowest concentration, but the difference between the two isoforms is small compared with the kinetics at ∼700 μM. It is important to note that we have not observed signal broadening nor the presence of new signals arising upon ASC or ASCb self-association (Fig. S2). There are several possible explanations for these results: 1) the oligomers formed during the kinetic experiment are already too large to be detected by NMR, 2) intermediate-size oligomers that could give rise to new signals are formed but do not live long enough to be detected.
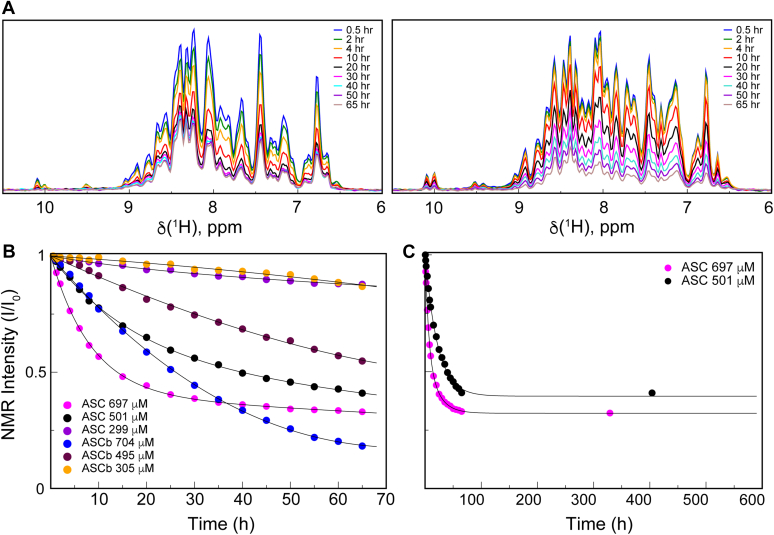
Figure 3.
Oligomerization kinetics of ASC and ASCb by RT-NMR.A, overall NMR signal decay as a function of time for the oligomerization of ASC (∼697 μM; left panel) and ASCb (∼704 μM; right panel). Time points are color-coded as indicated. B, normalized overall NMR signal intensity decay versus time for ASC and ASCb at the indicated color-coded concentrations. C, as in (B) for the same ASC samples at the concentrations indicated and time point at approximately 2 weeks after sample preparation.
The overall intensity decays were fitted to single- and double-exponential equations (Equations 1 and 2, respectively) to estimate apparent oligomerization rate constants (k1 and k2). It is important to mention that Equations 1 and 2 are logistic functions a priory not related to the underlying self-association mechanism but are particularly useful for comparing the polymerization kinetics of both proteins. Fitting of the NMR intensity decay to a double-exponential versus single-exponential improves for ASC but not for ASCb. This result suggests that ASC oligomerization involves at least two steps with different rate constants. The fitting to a double-exponential results in fast and slow rate constants for ASC: k1ASC = 0.15 ± 0.03 h−1 and k2ASC = 0.04 ± 0.01 h−1, respectively (Table 1). Thus, one kinetic phase in ASC oligomerization is 3.75 times faster than the other assuming other conditions are identical. This result could reflect the fast formation of an initial oligomer (a nucleation step) that continues growing at a slower rate forming a polymer. The overall signal decay for ASC at ∼700 μM reaches 33% of the original intensity at steady-state conditions resulting from the fraction of monomer concentration still present at the end of the kinetic experiment (65 h). Two weeks later, the intensity remains at this same value (Fig. 3C).
Table 1.
| Rate constants | Protein isoform |
|
|---|---|---|
| ASC | ASCb | |
| k1a | 0.15 ± 0.03 | 0.02 ± 0.01 |
| k2a | 0.04 ± 0.01 | 0.02 ± 0.01 |
| k1avePYD | 0.21 ± 0.04 | 0.0269 ± 0.0009 |
| k2avePYD | 0.05 ± 0.02 | 0.0269 ± 0.0009 |
| k1aveCARD | 0.12 ± 0.02 | 0.0288 ± 0.0009 |
| k2aveCARD | 0.02 ± 0.01 | 0.0288 ± 0.0009 |
| Goodness of fit | ||
| χ2 | 0.000327961 | 0.00059583 |
| r | 0.999812 | 0.999806 |
Reported values are averaged and SD obtained from fitting the logistic equations to the RT-NMR data at ∼700 and 500 μΜ for each isoform. Units for rate constants are in h−1.
In contrast, the fitting of the ASCb NMR signal intensity decay to a double-exponential does not result in any significant difference compared with a single-exponential fitting. Thus, ASCb appears to oligomerize with a single kinetic phase with an apparent rate constant kASCb = 0.02 ± 0.01 h−1. Interestingly, at the final stage of the kinetic experiment, the remaining fraction of ASCb monomeric species is 18%. The fitting of signal intensity decay for ASC and ASCb (∼700 μM) to the exponential functions includes a baseline value (A1 in Equations 1 and 2) that results in 31% and 18% of residual monomeric population for ASC and ASCb, respectively, in agreement with the experimental data. Several parameters indicative of the goodness of the fit are shown in Table 1.
| (1) |
| (2) |
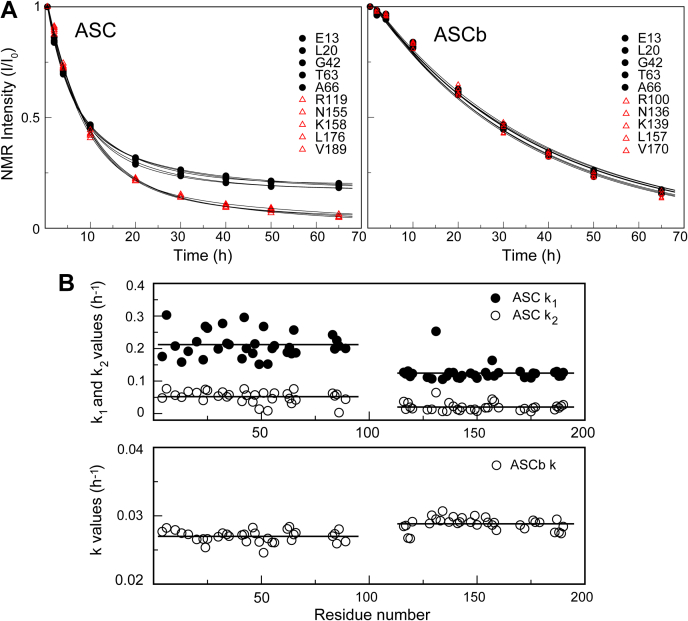
To study potentially different behaviors of the two death domains in protein polymerization, we have analyzed the signal intensity decay of the two isoforms at the amino acid level. Figure 4A shows, as examples, the fitting of several amino acids located in the PYD and CARD of ASC and ASCb to the exponential equations (Equations 1 and 2). Amino acids in the PYD and CARD of ASC show different behavior, whereas this difference is not observed for ASCb. The k1 and k2 values of individual amino acids plotted versus the residue sequence clearly show that the two death domains of ASC behave differently in the faster step (Fig. 4B). The k1 values of the PYD are larger than those of the CARD: k1avePYD = 0.21 ± 0.04 h−1 and k1aveCARD = 0.12 ± 0.02 h−1 (Table 1). However, the k2 values are similar for both domains: average value, k2avePYD = 0.05 ± 0.02 h−1 and k2aveCARD = 0.02 ± 0.01 h−1 (Table 1). This higher value for k1avePYD versus k1aveCARD suggests that PYD-PYD interactions could drive the first steps in the oligomerization reaction, and that both domains participate equally in subsequent polymerization steps. In contrast, the PYD and CARD domains of ASCb show very similar values of the oligomerization rate constant (k = 0.02 ± 0.01 h−1) (Fig. 4B and Table 1), which points to equivalent participation of both domains in the different steps of the polymerization reaction. This rate constant is slower than the slower rate constant for ASC self-association (k2), again indicating that ASCb polymerizes at a slower rate compared with ASC. The monoexponential behavior in ASCb could be explained by an initial nucleation step and subsequent polymerization with analogous rate constants. The oligomerization of ASC is a complex process as shown by our recent studies combining NMR techniques and ns-TEM, indicating that ASC dimers are the building blocks of ASC filaments and that the latter tend to form bundles comprising laterally stacked filaments. The ASC dimer involves homotypic interactions between the PYDs and CARDs of the two protomers. PYD and CARD domains have several protein–protein interacting regions that facilitate further association and polymerization.
Figure 4.
RT-NMR oligomerization kinetics of ASC and ASCb at the amino acid level.A, normalized intensity decay of ASC (left panel) and ASCb (right panel). B, rate constants of the PYD and CARD domains versus residue number for ASC (top) and ASCb (bottom).
Kinetic model for ASC and ASCb self-association
The exponential Equations 1 and 2 used for comparing the self-association behavior of ASC and ASCb report on the presence of different kinetic phases and allow the identification of similitudes and differences in the self-association process of the individual domains; however, these equations are not necessarily connected to the oligomerization kinetic mechanisms. We have attempted to propose a simple kinetic model for ASC and ASCb self-association based on previously reported models for protein oligomerization.
The oligomerization mechanisms of numerous protein systems have been studied in depth using different kinetic models of polymerization (26, 27, 28). The nucleation/polymerization model proposed by Hofrichter et al. has been frequently used and adapted to describe the oligomerization of different proteins (29, 30, 31, 32, 33). According to this model, individual kinetic steps of single-monomer additions result in the formation of an oligomer of a specific size: critical nucleus size or seed. These first steps constitute the nucleation phase of the mechanism. Once the critical nucleus is formed, the addition of new monomers to this critical-size oligomer is thermodynamically favorable, thus leading to subsequent polymerization (29). To account for the high complexity of protein self-association, this and similar models based on single-monomer addition have been complemented with other equilibria involving polymer fragmentation and association (34). In addition, other models contemplate the formation of polymers or the critical-size nucleus based on the association of smaller oligomers of potentially different size (35, 36, 37). In general, originally proposed models are typically further modified to include additional steps that allow better fitting of the resulting kinetic equations to the experimental data (34).
Recently, a model has been proposed to explain the kinetic diversity of amyloid oligomers. A Petri net (38) form is used to depict the different steps potentially involved in the oligomerization process (39). According to this model, the primary nucleation phase that results in fibril formation involves two steps: (1) the initial formation of an oligomer from the association of a certain number of monomer molecules; a process that is considered reversible, and (2) the addition of a number of monomers to this critical-size oligomer resulting in fibrils; a process that is assumed to be irreversible (39). The formed fibrils can be elongated by monomer addition to both ends of the fibril. Secondary pathways, such as fibril fragmentation and oligomer formation by clustering of monomers into preexisting fibrils, are also contemplated (39). Analytical equations derived from this model successfully describe reported experimental kinetic data on oligomer concentration and fibril mass concentration during amyloid fibril assembly. We have used a simplified version of this model, which considers the basic reactions involving the formation of fibrils (filaments) but assumes that filament elongation by single-monomer addition and secondary processes such as fragmentation are negligible. Similarly, the model is represented in Petri net form in Figure 5.
Figure 5.
Kinetic model of ASC and ASCb oligomerization.A, monomeric ASC forms oligomers at rate ka. Oligomers can dissociate back to monomers at rate k−a or further self-associate to form filaments at rate kb. B, filament formation represented in Petri net form.
The kinetic equations associated to the chemical reactions considered in Figure 5 are as follows:
| (3) |
| (4) |
| (5) |
Where M, O, and F, represent the monomer, oligomer of critical-size and filament, respectively; ka and k-a are the forward and backward reaction rate constants for the formation of the oligomer (step 1), and kb is the rate constant for the forward reaction of filament formation (irreversible step 2). The model also assumes that protein concentration does not influence the kinetic parameters of these reactions. However, it has been previously reported that the critical size of the oligomer can depend on monomer concentration (33). Analogously to other kinetic analysis of protein oligomerization, these equations become linear by assuming that the initial concentration of monomer is constant at early stages of the self-association process. Under this assumption, the solution of the differential equations results in an expression of the normalized concentration of monomer at time t ([M]t) as follows:
| (6) |
With no other assumptions and counting with the RT-NMR data reporting the decrease of monomer concentration as self-association proceeds, the fitting of the experimental data to Equation 6 allows obtaining the values of ka and k-a for the formation of the oligomer (Fig. 5) (Table 2). Both rate constants are larger for ASC than for ASCb. Specifically, ka is close to two orders of magnitude larger for ASC, whereas k-a is approximately one order of magnitude larger. This result could imply that ASC has a stronger tendency to form oligomers compared with ASCb, and that once formed, the latter has less tendency to return to monomeric form. The ka values are similar at different protein concentration (∼700 μM and ∼500 μM) as expected, whereas the k-a values show variations (Table 2). We attribute these variations to the assumption made in considering that the protein concentration will not affect oligomer critical size and thus the pertinent rate constants.
Table 2.
Kinetic parameters for the self-association of ASC and ASCb from kinetic model equations
| Conditions/Parameters | Protein isoform |
|||
|---|---|---|---|---|
| ASC | ASCb | |||
| Protein concentrationa | 697 | 501 | 704 | 495 |
| kaa | 6.05 × 10−8 | 4.74 × 10−8 | 8.69 × 10−10 | 8.60 × 10−10 |
| k-aa | 0.12 | 0.05 | 0.001 | 0.005 |
| Reaction order | 3.1 | 3.1 | 3.6 | 3.6 |
Concentration units: μM. Time units: h.
In addition, an estimate of the reaction rate order for oligomer formation can be obtained by applying the initial rate approximation. Under these conditions, the concentration of oligomer is assumed to be zero, and thus, Equation 3 is simplified to:
| (7) |
The initial velocity (V0) of the reaction was determined from the slope of the straight line formed by the first four points of the NMR signal intensity decay at the three concentration values for ASC and ASCb (Fig. 3B). Subsequently, the data at the three concentrations values were used to obtain the reaction order “m” from Equation 7 (m = 3.1 for ASC and m = 3.6 for ASCb). These results indicate that the formation of ASCb oligomers (O) has a slightly stronger dependence on the monomer concentration compared with ASC. Altogether, the results from the kinetic model help to further compare the oligomerization behavior of ASC and ASCb with more detail. However, the reaction rate constants and the order rate values are approximate as several assumptions were made.
The remaining NMR signal observed several weeks after that start of protein oligomerization (Fig. 3C) could indicate that filament formation is a reversible process. In contrast, because the second step in the proposed kinetic model is irreversible (Fig. 5A), the model seems to disagree with the NMR experimental data. However, a very small value of kb could lead to an extremely slow and barely detectable decrease of the NMR signal. In fact, the natural oligomerization of ASC and ASCb is greatly diminished under the acidic conditions of the RT-NMR experiments, which could result in the reversibility of the filament-formation step or in very small values of the rate constant of the irreversible reaction (second step in Fig. 5). Both possibilities could validly explain the observed residual NMR signal.
It is also interesting to test whether potential chemical modifications in ASC and ASCb influence the kinetics of the self-association process. For this purpose, the oligomerized 15N-labeled samples were subjected to liquid chromatography (reverse phase) coupled to mass spectrometry. The mass spectrum for ASC indicates the presence of monomer and dimer with molecular weights of 23,959.9 and 47,927.6 g/mol (Fig. S3). However, the mass spectrum for ASCb only reveals a monomer with molecular weight of 22,282.8 g/mol (Fig. S3). The mass spectrometry data match the theoretical molecular weights of the monomers (23,958.7 and 22,278.9 g/mol) of 15N-labeled ASC and ASCb, respectively, thus ruling out chemical modifications during self-association. The oligomerized ASC and ASCb samples are treated with formic acid and undergo unfolding by the reverse-phase chromatography step prior to mass spectrometry, which should dissociate noncovalent oligomers into monomers. Therefore, the presence of an ASC dimer was unexpected. The mass spectrometry data indicate that ASC is capable of forming dimers stable enough to sustain the harsh unfolding conditions. This result agrees with our previous data on the oligomerization of ASC using NMR and TEM, which indicate that the ASC dimer is the building block of the ASC filament (12). Importantly, the persistence of a dimer in ASC, but not in ASCb, suggests that a potential ASCb dimer is not as stable, highlighting another difference in the self-association process of the two isoforms.
ASC has a stronger tendency to oligomerize compared with ASCb from size-exclusion chromatography analysis
Using SEC, we have attempted to study the oligomer size distribution in ASC and ASCb self-association. In particular, we have studied three factors critical in death domain oligomerization (40): time (80 min and 2 days after sample preparation), protein concentration (50 μM and 150 μM), and pH (3.8, 4.3 and 4.8). For this purpose, we have used an SEC column (Superdex 200 10/300 GL) with a molecular weight range of 10–600 kDa. The protein solutions and equilibration buffers in these experiments contained 150 mM NaCl to avoid nonspecific interactions between the proteins and the chromatographic matrix. Using NMR and analytical ultracentrifugation, we reported previously that the presence of NaCl enhances death domain oligomerization, particularly in studies involving the association of the PYD domains of ASC and of the inflammasome sensor NLRP3 (40). Thus, protein self-association significantly increases under these conditions compared with those used in the kinetic RT-NMR experiments without NaCl. Samples for chromatographic analysis are typically filtered through ≥0.2 μm; however, ASC and ASCb samples that were filtered prior to column loading did not give rise to any signal in the chromatogram under most conditions (pH > 3.8 and/or protein concentration >50 μM and/or sample preparation >2 days), thus indicating that the oligomers were trapped in the filter. Therefore, protein samples were not filtered in our SEC studies and instead were centrifuged at 5,000 rpm for 1 min to pellet down precipitated material. Precipitation and centrifugation result in lack of homogeneity in the starting conditions of the protein samples, which is reflected in variations in the absorbance values for identical protein concentration, thus rendering the interpretation of the SEC results significantly challenging.
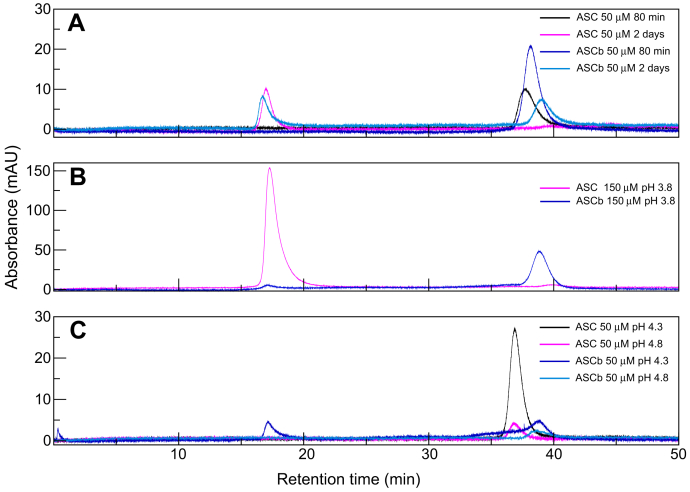
Protein samples at 50 μM concentration were injected in the SEC column 80 min after preparation, resulting in absorbance peaks (at a wavelength of 280 nm) corresponding to ASC and ASCb at an elution time of ∼38 min (Fig. 6A). According to the SEC column calibration and assuming a spherical rotor, monomeric and dimeric forms of ASC and ASCb should elute at ∼33 and ∼30 min, respectively. The discrepancy between the expected and experimental elution times is likely due to the multidomain nature of the two isoforms, thus deviating from ideal spherical conditions. The experimental elution time at ∼8 min most likely corresponds to the monomeric forms of both ASC and ASCb as the NMR data do not suggest the presence of a stable dimer (10). The protein samples were injected again 2 days after preparation and both ASC and ASCb eluted at the void SEC time (∼17 min) with similar intensity. This result indicates that the oligomers formed by ASC and ASCb are >600 kDa, which is the upper limit for the molecular weight of the SEC column (Fig. 6A). Therefore, the lower limit of the number of protomers in the ASC and ASCb oligomers is ∼30, based on the molecular weight of the monomeric proteins. ASCb, but not ASC, still shows an absorbance signal at an elution time ∼38 min in the chromatogram obtained 2 days after sample preparation, which suggests that certain population of ASCb is still monomeric under these conditions. In contrast, our RT-NMR data indicate that both ASC and ASCb are mainly monomeric at ∼300 μM in the absence of NaCl, thus corroborating our original results on the enhancing effect of NaCl in death domain self-association.
Figure 6.
ASC and ASCb oligomer size distribution by SEC.A, oligomer formation of ASC and ASCb 80 min and 2 days after sample preparation. B, effect of concentration on oligomer distribution at 150 μM. C, influence of pH on the oligomerizing capabilities of ASC and ASCb at pH 4.3 and 4.8.
The influence of protein concentration on oligomer distribution was also tested. At 150 μM protein concentration, ASC elutes mainly in the void volume (Fig. 6B) with a minor population at ∼38 min. ASCb behaves in the opposite way showing a minor peak at the void volume and a major peak at ∼38 min. This result suggests a higher tendency of ASC to oligomerize compared with ASCb. We also attempted to study the effect of pH on ASC and ASCb oligomerizing capabilities. SEC experiments on ASC and ASCb at pH values higher than 3.8 were very challenging to perform because of abundant protein precipitation; an effect originally observed for ASC (10, 40). At pH 4.3, ASC shows an absorbance signal at ∼38 min, but the signal at the void volume is no longer present (Fig. 6C). In contrast, ASCb shows two signals barely above the baseline: one appearing at ∼38 min and the other one at the void volume (Fig. 6C). At a higher pH value (pH 4.8), only one signal is still present for both isoforms appearing at ∼38 min with very low absorbance (∼2 mA units) (Fig. 6C). These results suggest that high-order oligomers formed by ASC and ASCb are more populated at higher pH values. These large oligomers mostly precipitate out of solution, explaining why they are no longer observed in the SEC chromatogram, thus resulting in single signals at ∼38 min.
The SEC data are difficult to interpret due to the dynamics of the oligomerization process and protein precipitation in the presence of NaCl and at pH > 3.8. Nonetheless, we can derive information indicative of different oligomerizing behavior for ASC and ASCb. Overall, the SEC results suggest that ASC has a higher tendency to self-associate compared with ASCb based on the stronger effect of time, pH, and protein concentration to promote oligomerization. The SEC results also indicate that oligomers of MW > 600 kDa are observed, which implies that oligomers <600 kDa are not sufficiently populated and/or have a short half-life.
ASC self-association results in less polydisperse solutions compared with ASCb
Dynamic light scattering (DLS) was used to monitor the self-association of ASC and ASCb as a function of time. The intensity of the scattered light versus particle size was obtained for ASC and ASCb solutions at different time points ranging from 30 min to 49 h after sample preparation (Fig. 7). The intensity of the observed peaks depends on the population of the different particles and is proportional to the sixth power of the particle radius. Therefore, larger particles will result in peaks with higher intensity of the scattered light compared with smaller particles at equal population. It is important to mention that a perfect spherical particle is assumed for the mathematical determination of particle size from the correlation functions derived from the dependence of photon counts versus time in DLS. ASC oligomers are not spherical but filamentous, and thus this condition is not applicable (12). Therefore, the specific dimensions obtained assuming spherical particles do not report on the real size of the oligomers. However, the data are very useful to identify particle populations of different size and their change in population during the kinetic experiment.
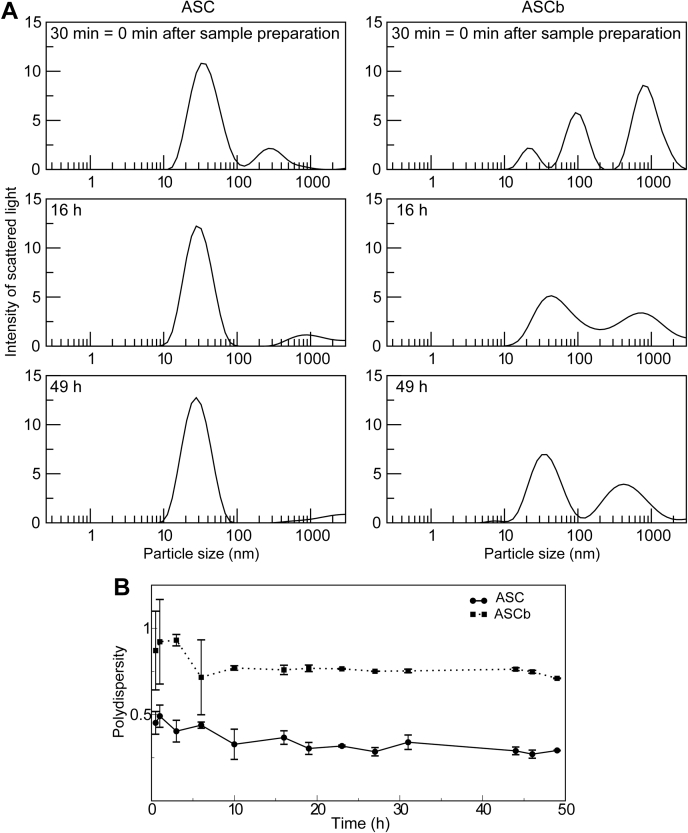
Figure 7.
Time-dependent population distribution of ASC and ASCb oligomers by dynamic light scattering.A, changes over time in population (intensity of scattered light) and apparent size of ASC and ASCb oligomers. B, changes in polydispersity values of ASC and ASCb solutions during the oligomerization process. Data in (A) and (B) are an average of three measurements and error bars in (B) represent SD. The 30 min time point can be considered as 0 min time point after sample preparation.
DLS data for ASC 30 min after sample preparation show one major and one minor peak with an overall diameter of ∼40 nm and ∼300 nm, respectively, (Fig. 7A). The population of ASC monomers detected in the RT-NMR experiments is not observed in the DLS data likely being masked by the intense signal resulting from the large oligomeric species present in solution. The former peak slowly decreases to ∼28 nm at the end of the kinetic experiment, which could indicate that the oligomers are becoming more compact. In addition, the intensity of the peak at ∼300 nm decreases with time. Because the larger species shows very low intensity compared with the smaller species, we can conclude that the population of the ∼300 nm species is very small. At the end of the kinetic experiment, ASC shows mainly one species at ∼28 nm (Fig. 7A). The disappearance of the ∼300 nm peak is concomitant to the increase in intensity of the ∼28 nm signal. Thus, we speculate whether the ∼300 nm species has become more compact by forming filamentous rings and contributes to the intensity of the ∼28 nm species. In contrast, DLS data for ASCb 30 min after sample preparation show three main peaks at ∼20, ∼100 nm, and ∼900 nm, the latter one being more intense. As the kinetic experiment progresses, the three peaks consolidate into two major peaks with diameters of ∼30 and ∼400 nm.
In general, ASC and ASCb form species of different size and population under the same conditions during self-association. These different species tend to consolidate to a smaller number at the end of the kinetic experiment reflecting the dynamics of the polymerization process and a tendency to form oligomers of more uniform size. Importantly, the overall polydispersity values (p) for ASC and ASCb during the DLS kinetic experiments are significantly different: pASC = 0.35 ± 0.04 and pASCb = 0.79 ± 0.06 (Fig. 7B). This result is remarkable as it reflects that ASC self-associates into oligomers of very similar size, whereas ASCb oligomerizes with considerably higher polydispersity. Interestingly, it has been reported that ASC and ASCb oligomerize into different macrostructures inside the cell: ASC forms a speck of uniform size and ASCb forms clustered and disordered filaments (20). The conditions for oligomerization in the cell assays are different from our in vitro studies in terms of protein and salt concentration, pH, and other uncontrolled factors in the cell cytosol. However, cell studies agree with our DLS results indicating that ASC is capable of forming monodisperse polymers, whereas ASCb is not.
Differences in ASC and ASCb macrostructures by transmission electron microscopy
An analysis of the macrostructures formed by ASC using negative-staining Transmission Electron Microscopy (ns-TEM) has been previously reported (12). According to this study, ASC tends to form filaments with an average width of 6.4 ± 0.8 nm and length varying between 600 and 800 nm (12). The filaments form bundles of 2–7 filaments; with three- and four-filament bundles being more abundant. Bundle width is always a multiple of the width of the individual filament matching the number of filaments in the bundle, thus indicating that the bundles are formed by lateral stacking of filaments. Importantly, the TEM micrographs show that ASC filaments are formed by stacked rings with the size of an ASC dimer. In addition to forming part of the individual filaments, these rings are abundantly observed attached to preexisting filaments suggesting a possible mechanism for filament and bundle growth.
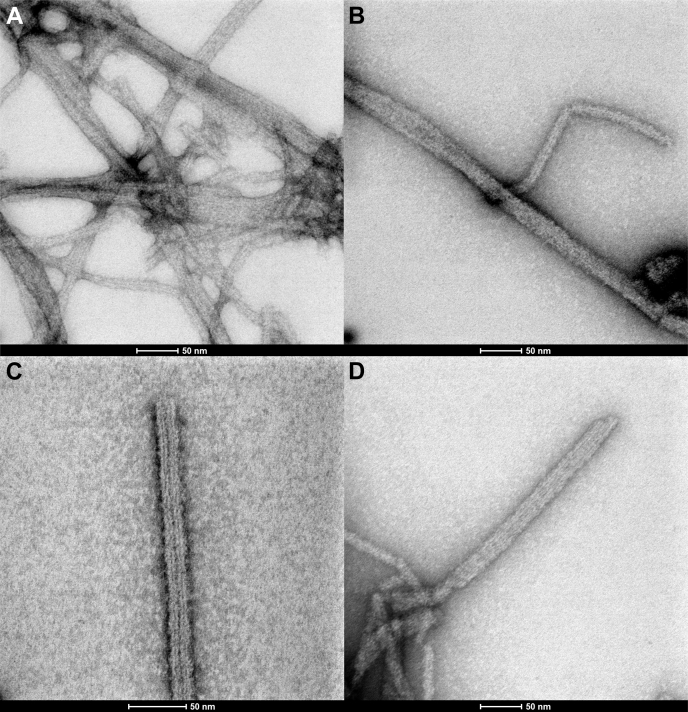
A similar ns-TEM analysis was performed for ASCb to identify potential differences in the characteristics of the macrostructures (Fig. 8). ASCb also polymerizes into filaments that tend to form bundles. However, some differences were observed: ASCb bundles are typically composed of 1-, 2-, and 3-filaments. The analysis of 72 filaments indicates the following populations: 26.3% of individual filaments, 48.7% of two-filament bundles and 25% of three-filament bundles. Thus, for ASCb, two-filament bundles are significantly more abundant. The average width of individual filaments, two- and three-filament bundles is 7.0 ± 0.2 nm, 14.1 ± 0.4 nm, and 21.2 ± 0.4 nm, respectively. These values indicate that ASCb individual filaments are of similar width compared with those observed for ASC and that bundles are also formed by lateral stacking of individual filaments. Filament length was found to be in the 200–400 nm range, thus slightly shorter than ASC filaments. Interestingly, rings attached to preexisting filaments were not observed for ASCb.
Figure 8.
Filament and filament bundles formed by ASCb.A, ns-TEM micrograph of networks of filaments of various sizes. B–D, ns-TEM images showing one-filament (B), two-filament (C), and three-filament (D) bundles of ASCb.
It is important to mention that the experimental conditions for ASC and ASCb filament formation are different from the conditions used for protein oligomerization using RT-NMR. Filament formation is enhanced at neutral pH and diminished under the acidic conditions used for NMR experiments. However, our previous TEM results indicate that ASC can form filaments under acidic conditions of similar appearance to the filaments formed at neutral pH albeit less compacted and defined (12).
Steric factors dependent on the linker length result in different interactions between ASC and ASCb with procaspase-1 CARD
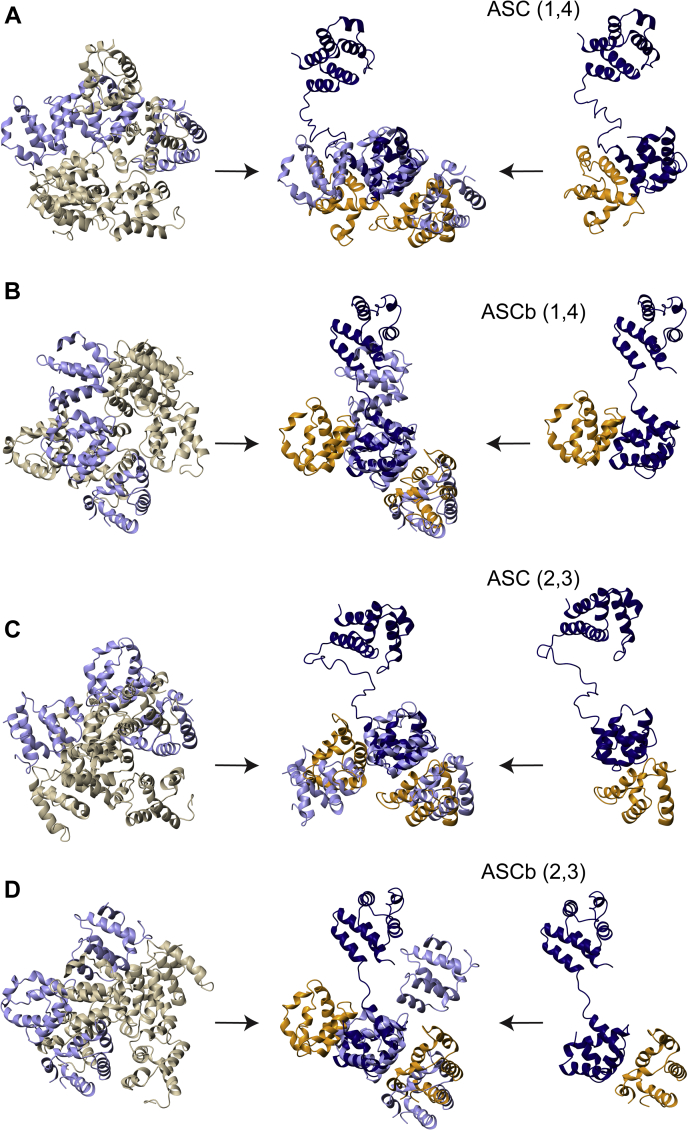
In addition to self-association, ASC and ASCb tether sensors and procaspase-1 molecules as part of their function in inflammasome assembly. Thus, both isoforms also participate in oligomerization processes involving other proteins. We investigated potential differences between ASC and ASCb in this function at the computational level. In an attempt to mimic the initial steps of inflammasome formation, we used docking and rigid body minimization protocols (41) to study the binding of ASC and ASCb to several CARD domains of procaspase-1 (procaspase-1CARD). We first generated dimers of ASC and ASCb with one procaspase-1CARD using the most prominent interactions between death domains (i.e., type Ia and type Ib interactions (42) that involve helices 1 and 4 of one domain and helices 2 and 3 of the other domain). Thus, each CARD domain in ASC or ASCb is capable of interacting with two procaspase-1CARD molecules via the surfaces composed by helices 1,4 and 2,3. The resulting dimers (ASC/procaspase-1CARD and ASCb/procaspase-1CARD) were used to generate trimers by including one additional procaspase-1CARD molecule. The structures of the trimers were analyzed and compared with the structure of a procaspase-1CARD trimer with type I interactions extracted from the cryo-EM 3D structure of the procaspase-1 CARD polymer (43).
The structures of the CARD and PYD domains of ASC and ASCb are identical, thus, conferring flexibility to the linker is critical to allow different orientations of the PYD and CARD domains that could influence the capability of interaction with procaspase-1CARD molecules. Therefore, we assigned full flexibility to the 23- and three-residue linker of ASC and ASCb, respectively, in the docking protocol used to generate all models. The structures of the dimers and trimers resulting from docking and rigid body minimization are shown in Figure 9. Briefly, molecules at the right-hand side of panels A and C represent the dimer structures of ASC with the type I interaction, ASC (helices 1, 4): procaspase-1CARD (helices 2, 3); and ASC (helices 2, 3): procaspase-1CARD (helices 1, 4), for panels A and C, respectively. Analogously, molecules at the right-hand side of panels B and D represent equivalent interactions between ASCb and procaspase-1CARD.
Figure 9.
Steric hindrance in the interactions of ASCb with the CARD of procaspase-1. In all panels, Left: Different orientations of the polymeric cryo-EM structure of the CARD of procaspase-1 (procaspase-1CARD) (43). The CARD trimer formed by type I interactions is highlighted in light purple; Center: trimer structure of ASC or ASCb (navy blue) from molecular docking (41) with two molecules of procaspase-1CARD (light orange). In light purple, the CARD trimer superimposed in the same orientation shown in the left; Right: dimer structure of ASC or ASCb (navy blue) from molecular docking with one molecule of procaspase-1CARD (light orange). A and C, trimer and dimer structures starting with ASC interacting helices 1 and 4 (A), and 2 and 3 (C); B and D, trimer and dimer structures generated starting with ASCb interacting helices 1 and 4 (B), and 2 and 3 (D). The CARD trimer (purple) in the left of all panels shows different orientation to facilitate comparison to the CARD trimer superimposed to ASC(ASCb)-CARD-CARD trimers shown in the center.
ASC and ASCb are capable of binding one procaspase-1CARD molecule via type I interactions in a similar fashion (Fig. 9, right-hand side in all panels: ASC and ASCb in navy blue ribbon, procaspase-1CARD in light orange). The orientation of this procaspase-1CARD molecule is close to the expected position indicated by the cryo-EM 3D structure of the procaspase-1CARD polymer (43) (Fig. 9, left-hand side in all panels: light purple ribbon). However, when a second procaspase-1CARD molecule is included, only the trimers formed by ASC resemble the expected arrangement of the additional CARD based on the cryo-EM structure (Fig. 9, A and C). The second procaspase-1CARD molecule in the ASCb trimer is located in a position that significantly deviates from the polymeric CARD arrangement (Fig. 9, B and D). In fact, it is clear from the superimposed structures in Figure 9B (center) that the second procaspase-1CARD molecule would clash with the PYD of ASCb if located in the position of the polymeric CARD. Analogously, the ASCb trimer with type one interactions, procaspase-1CARD (helices 2, 3): ASCb (helices 1, 4): procaspase-1CARD (helices 2, 3) shown in Figure 9D, results in the position of the second procaspase-1CARD in a region where there is not interference with the PYD of ASCb, but deviates from the expected structure based on the cryo-EM trimer.
In summary, ASC and ASCb interact differently when several procaspase-1 molecules are involved in an oligomeric assembly resembling the formation of the inflammasome. The proximity between the PYD and CARD domains of ASCb hinders interactions of ASCbCARD with procaspase-1CARD. In contrast, ASC’s significantly longer linker results in lack of interference between the two domains, thus facilitating interactions with potential partners. Overall, the models generated with the docking protocols help to illustrate a tentative, simple mode of oligomerization of ASC and ASCb with procaspase-1. Preassembled ASC and ASCb could also interact with procaspase-1 during inflammasome formation, which would lead to a smaller number of interacting regions available for procaspase-1 in ASCCARD and ASCbCARD. In this situation, the hindering effect caused by PYD and CARD in ASCb might gain more relevance. Finally, based on the proximity-induced model for procaspase-1 autoactivation, a different structural arrangement of ASC versus ASCb oligomers could result in differences in the local concentration of the procaspase and thus in its autoactivation.
Discussion
It has been previously reported that ASC and its isoform ASCb are able to interact with NLRP3 and procaspase-1 (20, 21). Although both isoforms are incorporated into the inflammasome complex, ASCb leads to an overall reduction in inflammasome activity compared with ASC based on the release of smaller amounts of interleukin-1β (20). The difference in inflammasome activity between ASC and ASCb can have relevant effects on the regulation of the inflammatory response. Therefore, it is important to understand this regulatory mechanism at the molecular level. The main function of ASC as an inflammasome adaptor is to self-associate and oligomerize with the sensors and procaspase-1. Thus, we hypothesized that the modulation of function regarding inflammasome activation depends on the different self-association/interaction properties in ASC and ASCb. In this work, we have tested this hypothesis by studying the self-association behavior of ASC and ASCb using different techniques for a more thorough analysis.
The monitorization of ASC and ASCb self-association by RT-NMR allowed to identify overall kinetic phases by fitting the obtained data to single- or double-exponential equations. The results of the fitting indicate that ASC self-associates with two major kinetic phases, whereas ASCb follows a single kinetic phase or two phases with identical or very similar rate constant. Our data indicate that ASC oligomerization follows a fast kinetic phase and a slower second kinetic phase. In contrast, the single kinetic phase experienced by ASCb is slower than both kinetic phases of ASC. Furthermore, NMR allows atomic-resolution studies of the kinetics of the self-association process. We leveraged this unique quality of NMR to investigate differences in oligomerization at the atomic-residue level. Importantly, we found that the self-association capabilities of the PYD and CARD domains of ASC and ASCb are different. Specifically, the PYD and CARD of ASC show two different apparent kinetic rate constants facilitating oligomer formation. The higher k1 values of the PYD domain relative to the CARD suggest that during the initial kinetic phase, the interactions involved in the conversion from monomer to oligomer are primarily driven by PYD-PYD binding followed by CARD-CARD interactions. The second kinetic phase is dictated by the simultaneous participation of both the PYD and CARD domains as indicated by their similar k2 values. In contrast, the PYD and CARD domains of ASCb show equivalent kinetic rate constants, suggesting that both domains participate equally in the formation of the oligomeric species.
Previously reported data on ASC macrostructure characterization by ns-TEM show that ASC dimers are the building blocks of ASC polymerization into filaments (12). It is plausible that short-lived dimers are formed and rapidly assemble into oligomers of a critical nucleation size that further develop into filaments. The kinetic data obtained for ASC suggest that PYD-PYD interactions could drive dimer formation (k1avePYD > k1aveCARD), additionally stabilized by CARD-CARD binding. Contrary to the results of canonical ASC, the kinetic data for ASCb suggest that the PYD and CARD have the same oligomerization tendency. Death domain oligomerization depends on the presence of the different interacting regions in the PYD and CARD domains and on the possibility of these regions to find their interacting partners for which domain reorientation might be important. The PYD and CARD of ASC and ASCb are identical based on our structural data and the 100% amino acid sequence identity. Thus, the different behavior of the two domains in ASC is potentially related to the long linker connecting the two domains. ASC long linker will facilitate domain reorientation to a greater extent compared with ASCb. Therefore, it is reasonable to suggest that the increased oligomerization tendency observed in ASC for the PYD relative to the CARD is diminished in ASCb due to potential restrictions of domain reorientation as a result of the much shorter linker. To test the effect of the linker length, we created via molecular docking and rigid body minimization structural models of trimers composed of ASC or ASCb and two procaspase-1CARD molecules (41). The models indicate that ASCb short linker causes steric hindrance, resulting in an arrangement of the procaspase-1CARD molecules that differs from the experimental structure of the polymer (43).
The DLS kinetic study demonstrates that ASC and ASCb self-associate into different oligomer populations. Specifically, DLS data reveal the presence of a single monodisperse population present at 16 h after the start of ASC oligomerization. The low polydispersity of the protein solution suggests the formation of oligomers of similar size. In contrast, ASCb solution is more polydisperse as evidenced by the presence of two populations of oligomers 49 h after the beginning of the self-association process. These results are in agreement with previously published work on intracellular ASC and ASCb polymerization studies by fluorescence microscopy, indicating that ASC forms a filamentous speck or ring of ∼0.5 μm in diameter upon macrophage activation, whereas ASCb polymerizes forming linear filaments (13, 20).
ASC and ASCb also show differences at the macrostructural level. The ns-TEM image analysis reveals shorter filaments formed by ASCb compared with ASC and a lower tendency to assemble into bundles formed by a large number of laterally stacked filaments. This result could be related to the different polymerized macrostructures observed in the cell environment for ASC and ASCb. Furthermore, utilizing SEC we show that solution pH, oligomerization time, and protein concentration are important factors governing ASC and ASCb oligomerization. SEC studies indicate that ASC has a higher tendency to self-associate compared with ASCb. We have not observed small-size oligomers or dimers in the SEC studies but instead oligomers >600 kDa consisting of at least 30 protomers; a behavior similar to both ASC and ASCb. This result agrees with single-molecule fluorescence studies indicating that ASC massively oligomerizes into micron-size clusters of >500 proteins at above a critical concentration value in a cell-free system capable of expressing ASC at different concentrations (44). These studies also indicate that smaller oligomers are not detected (44), which matches our conclusion from RT-NMR kinetic data and SEC analysis that dimers and small oligomers have a short half-life. Nonetheless, the presence of an ASC dimer in the mass spectrum, likely resulting from the dissociation of large oligomers under the harsh unfolding conditions of the liquid chromatography–mass spectrometry experiment, points to its higher stability compared with ASCb.
Altogether, our data indicate that ASC oligomerizes faster and is capable of assembling into oligomers of more uniform size compared with ASCb. In this study, we show that these differences are only attributable to the specific structure/function of ASC and ASCb with no intervention of other proteins that could have an influence in the different polymerized macrostructures observed in the cell cytosol (20). The only difference at the amino acid level is the linker length, which seems to be optimized in ASC to enhance self-association and the formation of more compact oligomers. This different behavior intrinsic to ASC and ASCb can have important implications in the regulation of the inflammatory response. Upon macrophage activation as a result of the presence of pathogens and/or chemical signals indicative of cell dysfunction, the expression of the sensor protein, procaspase-1, and ASC is upregulated. The three proteins colocalize forming a filamentous ring-like structure, the inflammasome, consisting of self-associated and oligomerized sensor, ASC, and procaspase-1. The assembly of the inflammasome acts as platform for the activation of procaspase-1, purportedly due to an induced local increase of the caspase concentration resulting in its autoactivation (45). Therefore, ASC formation of monodisperse structures versus ASCb assembly into polydisperse filaments could be critical in procaspase activation. Our results indicate that ASC self-association is likely driven first by the PYD-PYD interactions followed by CARD-CARD binding, suggesting the possibility of domain reorientation that finally leads to the assembly of uniform oligomers. In contrast, the slower and weaker tendency of ASCb to oligomerize could delay the overall activation of the inflammasome. Furthermore, the polydisperse nature of ASCb oligomers could result in a less effective localization of increased procaspase-1 concentration, thus causing an overall reduction in inflammasome activity.
Experimental procedures
Expression and purification of unlabeled and isotopically labelled ASC and ASCb
E. coli BL21(DE3) cells were transformed with pET15b vectors encoding for either protein, ASC or ASCb. 15N- and 15N,13C-labeled proteins were cultured in M9 minimal media, using 15NH4Cl and/or 13C-glucose as sole nitrogen and carbon sources, respectively. E. coli cells were induced using 1 mM IPTG and incubated at 37 °C for 4 h. Cells were harvested by centrifugation at 8000 rpm for 30 min at 4 °C. Cell pellet was resuspended in resuspension buffer (6 M Guanidine-HCl, 20 mM Tris, 5 mM Imidazole, 500 mM NaCl, pH 8) and was homogenized five times at 30 s intervals to lyse the cells. The cell lysate was then ultracentrifuged at 35,000 for 30 min at 4 °C. The supernatant was filtered through a 0.45 μm filter. ASC and ASCb were purified using nickel affinity HPLC. The protein was eluted using elution buffer (20 mM Tris-HCl pH 8, 500 mM NaCl, 500 mM imidazole, and 6 M guanidinium hydrochloride) and subsequently dialyzed to remove excess of imidazole, salt, and chaotropic agent. ASC and ASCb were further purified by reverse-phase HPLC with a C4 column. Eluted protein solutions were lyophilized to remove organic solvents and stored at 4 °C. Expression of unlabeled ASC and ASCb was performed in LB media, and the purification protocol was identical to the one described for the isotopically enriched proteins. Information dependent on the amino acid sequence such as molecular weight, amino acid composition, theoretical extinction coefficient, and isoelectric point was obtained from the ExPASy server.
Structure of ASC and model of ASCb
The solution structure of ASC (PDB 2KN6) (10) was used as template to create a model of the 3D structure of ASCb using the program Chimera (23). For this purpose, 20 amino acids were removed from the linker region of ASC leaving only three amino acids (QGL) corresponding to the linker region of ASCb.
Chemical shift assignment of ASCb by NMR spectroscopy
ASCb NMR samples were prepared at 200 μM in a volume of 300 μl containing 5% D2O, 20 mM glycine, and 1 mM TCEP at pH 3.8. NMR experiments were acquired at 298 K in a Bruker 800 MHz spectrometer equipped with a triple-resonance cryoprobe at UC Santa Cruz’s NIH-NMR facility. Backbone assignment of ASCb was obtained from the following experiments: 2D [1H-15N]-HSQC, 3D [1H-15N]-NOESY-HSQC, 3D HNCACB, and 3D CBCA(CO)NH (46, 47). Data were processed with NMRPipe and spectra were analyzed with Sparky (48, 49). Previously published chemical shift assignments of ASC were used to identify potential differences (22). The chemical shift differences (Δδav) between ASC and ASCb were calculated using the 1H and 15N chemical shifts according to Equation 8 (50).
| (8) |
RT-NMR experiments
ASC and ASCb samples were prepared in 5% D2O/H2O and 1 mM TCEP at pH 3.8. Oligomerization kinetics of ASC and ASCb was monitored using RT-NMR at a concentration of ∼700 μM, ∼500 μM and ∼300 μM. Exact concentration values of both ASC and ASCb were determined by absorbance at 280 nm prior to commencing the experiment. Eighteen individual 2D [1H-15N]-HSQC experiments were acquired at time points: 30 min, 1 h 10 min, 2 h, 4 h, 6 h, 8 h, 10 h, 15 h, 20 h, 25 h, 30 h, 35 h, 40 h, 45 h, 50 h, 55 h, 60 h, and 65 h after sample preparation at 298K on a Bruker Avance III 600 MHz spectrometer equipped with a z-axis gradient cryogenic probe (UC Merced NMR facility). The overall intensity of the first 1D projection of the amide region for each [1H-15N]-HSQC spectrum was used to monitor signal decay over time. 1D projections were baseline-corrected from 12.8 ppm to 4.8 ppm, and the signal intensity of each 1D projection was calculated by integration from 9.5 ppm to 6 ppm using TOPSPIN 4.1 (Bruker). Intensity values for both ASC and ASCb were normalized with respect to the first 2D experiment (with an assigned intensity of 1) and were plotted as a function of time. The decays with time of NMR signal intensity were fitted to logistic mono- and double-exponential Equations 1 and 2 (Results section) for an overall identification of potentially different kinetic phases. RT-NMR data were also fitted to the analytical Equation 6 (Results section) derived from the kinetic model (Figure 5) to obtain values of the kinetic parameters. In addition, amino-acid-specific intensity decays versus time for ASC and ASCb at ∼700 μM and ∼500 μM for the PYD and CARD domains (n = 66) were determined by obtaining the peak height value at half-height using Sparky (48). Intensity values were normalized and fitted to Equation 2. All fittings were performed with the program Qt-Grace (QTGroup, Helsinki, Finland). Goodness of fit values are shown in Table 1.
Size-exclusion chromatography
Analytical SEC was performed using a Superdex 200 Increase 10/300 GL at a flow rate of 0.5 ml/min. Prior to use, the column was calibrated with a gel filtration high molecular weight kit from GE Life Sciences. The column was equilibrated using equilibration buffer (150 mM NaCl, pH 3.8, 0.2 μm-filtered, degassed). ASC and ASCb were prepared at 50 μM and 150 μM protein concentration in 150 mM NaCl buffer, pH 3.8. Kinetic experiments of ASC and ASCb were performed at 50 μM protein concentration and loaded into the column at two different time points: 80 min and 2 days after sample preparation. The effect of protein concentration on ASC and ASCb oligomerization was tested at 150 μM (pH 3.8). Protein samples were centrifuged for 1 min at 5000 rpm and loaded into the column right after preparation. Protein absorbance values were monitored at a wavelength of 280 nm. The resulting chromatograms were plotted with QtGrace (QTGroup, Helsinki, Finland).
Dynamic light scattering
DLS experiments were performed using a Malvern Panalytical Zetasizer Pro at 25 °C, using a laser wavelength of 632.8 nm. The refractive indices of ASC and ASCb were determined to be 1.3327 using an Abbe Mark III refractometer (Reichart). The absorbance value of ASC and ASCb at 632.8 nm was 0.1089. ASC and ASCb samples were prepared at a concentration of ∼700 μM by weight in HPLC-grade water at pH 3.8. Sample concentration was accurately measured prior to each experimental run by absorbance at 280 nm for both ASC and ASCb. Samples were placed in a ZEN0040 cuvette (Malvern Panalytical) for small volumes and equilibrated for 2 min at 25 °C prior to each measurement. Measurements were taken at 30 min, 1 h, 3 h, 6 h, 10 h, 14 h, 16 h, 19 h, 23 h, 26 h, 31 h, 32 h, 44 h, 46 h, and 49 h after sample preparation. Photon counts of the scattered light versus time are fitted to correlation functions by the ZS Xplorer software, from which the size distribution and polydispersity values are obtained. Three measurements of the scattered light were taken at each time point, and the resulting data were plotted and analyzed with QtGrace (QTGroup, Helsinki, Finland).
Transmission electron microscopy
ASCb protein filaments were prepared by dissolving lyophilized ASCb (50 μM) in a buffer containing 6 M Guanidine-HCl, 20 mM Tris pH 7, and 100 mM NaCl. The buffer was filtered with 0.2 μm-pore size filter prior to dissolving ASCb. The concentration of the protein was determined by absorbance at 280 nm. The protein solution was dialyzed against 100 mM NaCl and 20 mM Tris pH 7, and the dialysis buffer was changed three times after reaching equilibrium (3 h). A volume of 4 μl of the protein solution was deposited on a carbon-coated copper grid. After 10 min, the grid was washed in three 40 μl droplets of HPLC water (10 s each) and stained in three 40 μl droplets of 2% uranyl acetate (10 s each). The copper grid was stained for 5 min and then wipe-dried to remove excess of the staining solution. Images of ASCb filaments were obtained using a Talos F200C G2 Transmission Electron Microscope equipped with a Field Emission Gun (X-FEG) at 200 kV. A Ceta 16M Camera of 4k × 4 pixels was used for image acquisition. Analysis of ASCb filaments and bundle dimensions was performed with ImageJ.
Mass spectrometry
NMR samples of 15N-labeled ASC and ASCb at ∼500 μM used for the RT-NMR experiments (several months after the start of the oligomerization process) were injected in an electrospray ionization mass spectrometer (Q-Exactive Hybrid Quadrupole-Orbitrap, Thermo) coupled to an UHPLC system (Vanquish, Thermo). The protein samples were diluted with a solution containing 95% acetonitrile, 4.9% water, 0.1% formic acid and were injected in a reverse-phase column (Acclaim 200 C18, 3 μm, Thermo) at a flow rate of 0.3 ml/min. The spectra with signal intensity versus mass/charge were analyzed with BioPharma 2.0 software (Thermo).
Molecular docking
Molecular docking was performed with the program HADDOCK (41) using full-length ASC solution structure determined by NMR (PDB code 2KN6, (10)), the ASCb model structure created with Chimera (23), and a procaspase-1 CARD protomer extracted from the 3D structure of the polymeric CARD determined by cryo-EM (43). Firstly, four sets of dimer models composed of ASC or ASCb and one procaspase-1CARD were created selecting complementary surfaces of active residues to guide the molecular docking, following the type I interactions (helices 1 and 4 of one CARD with helices 2 and 3 of the adjacent CARD and vice versa). The generated sets of dimers are dimer-set 1= ASC (helices 1, 4): procaspase-1CARD (helices 2, 3); dimer-set 2 = ASC (helices 2, 3): procaspase-1CARD (helices 1, 4); dimer-set 3 = ASCb (helices 1, 4): procaspase-1CARD (helices 2, 3); dimer-set 4 = ASCb (helices 2, 3): procaspase-1CARD (helices 1, 4).
The best dimer structure of each of the previous four sets based on the best HADDOCK score was used to create sets of trimers by the addition of another procaspase-1CARD molecule as follows: trimer-set 1 = procaspase-1CARD (helices 2, 3): ASC (helices 1, 4): procaspase-1CARD (helices 2, 3); trimer-set 2 = procaspase-1CARD (helices 1, 4): ASC (helices 2, 3): procaspase-1CARD (helices 1, 4); trimer-set 3 = procaspase-1CARD (helices 2, 3): ASCb (helices 1, 4): procaspase-1CARD (helices 2, 3); trimer-set 4 = procaspase-1CARD (helices 1, 4): ASCb (helices 2, 3): procaspase-1CARD (helices 1, 4). The best trimer structure of each of the previous four sets (based on the best HADDOCK score) is depicted in Figure 9. In all HADDOCK runs, the 23- and 3-amino acid linker of ASC and ASCb, respectively, was set fully flexible to allow for domain reorientation. The N and C termini were considered charged. The number of starting structures was 1000, out of which 200 were considered for refinement. Solvated docking was used with water as solvent for the iterations. The top clusters for the different runs were analyzed and inspected with ChimeraX (51).
Data availability
The data that support the findings of this study are contained within the article and the supporting information. All source data generated for this study are available from the corresponding author (Dr Eva de Alba; edealbabastarrechea@ucmerced.edu) upon reasonable request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to Dr Mourad Sadqi (NSF CREST Center for Cellular and Biomolecular Machines at the University of California, Merced) for mass spectrometry data. We would like to thank UC Merced Imaging Facility. We are grateful to Professor Son C. Nguyen for letting us use the dynamic light scattering equipment in his laboratory. The National Institutes of Health has funded the UC Santa Cruz NMR facility (award 1S10OD018455) used to obtain NMR data reported in this work.
Author contributions
E. d. A. conceptualization; P. D-P. and E. d. A. formal analysis; E. d. A. funding acquisition; P. D-P. and E. d. A. investigation; P. D-P. and E. d. A. methodology; E. d. A. project administration; E. d. A. supervision; P. D-P. and E. d. A. validation; P. D-P. and E. d. A. visualization; P. D-P. and E. d. A. writing—original draft; P. D-P. and E. d. A. writing—review and editing.
Funding and additional information
This research was funded by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R15AI146780 to E. d. A., and by the NSF-CREST: Center for Cellular and Biomolecular Machines at the University of California, Merced (NSF-HRD-1547848) to P. D-P. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
Edited by Peter Cresswell
Supporting information
References
- 1.Kawai T., Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Man S.M., Kanneganti T.-D. Regulation of inflammasome activation. Immunol. Rev. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma M., de Alba E. Structure, activation and regulation of NLRP3 and AIM2 inflammasomes. Int. J. Mol. Sci. 2021;22:872. doi: 10.3390/ijms22020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma D., Kanneganti T.-D. The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J. Cell Biol. 2016;213:617–629. doi: 10.1083/jcb.201602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malik A., Kanneganti T.-D. Inflammasome activation and assembly at a glance. J. Cell Sci. 2017;130:3955–3963. doi: 10.1242/jcs.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroder K., Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 7.Park H.H., Lo Y.-C., Lin S.-C., Wang L., Yang J.K., Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu. Rev. Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin T., Xiao T.S. Activation and assembly of the inflammasomes through conserved protein domain families. Apoptosis. 2015;20:151–156. doi: 10.1007/s10495-014-1053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masumoto J., Taniguchi S., Ayukawa K., Sarvotham H., Kishino T., Niikawa N., Hidaka E., Katsuyama T., Higuchi T., Sagara J. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- 10.de Alba E. Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC) J. Biol. Chem. 2009;284:32932–32941. doi: 10.1074/jbc.M109.024273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz-Parga P., de Alba E. Protein interactions of the inflammasome adapter ASC by solution NMR. Methods Enzymol. 2019;625:223–252. doi: 10.1016/bs.mie.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nambayan R.J.T., Sandin S.I., Quint D.A., Satyadi D.M., de Alba E. The inflammasome adapter ASC assembles into filaments with integral participation of its two death domains, PYD and CARD. J. Biol. Chem. 2019;294:439–452. doi: 10.1074/jbc.RA118.004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Man S.M., Hopkins L.J., Nugent E., Cox S., Gluck I.M., Tourlomousis P., Wright J.A., Cicuta P., Monie T.P., Bryant C.E. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duez H., Pourcet B. Nuclear receptors in the control of the NLRP3 inflammasome pathway. Front. Endocrinol. 2021;12:630536. doi: 10.3389/fendo.2021.630536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley N., Jeltema D., Duan Y., He Y. The NLRP3 inflammasome: An overview of mechanisms of activation and regulation. Int. J. Mol. Sci. 2019;20:328. doi: 10.3390/ijms20133328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott E.I., Sutterwala F.S. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol. Rev. 2015;265:35–52. doi: 10.1111/imr.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barry R., John S.W., Liccardi G., Tenev T., Jaco I., Chen C.-H., Choi J., Kasperkiewicz P., Fernandes-Alnemri T., Alnemri E., Drag M., Chen Y., Meier P. SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat. Commun. 2018;9:3001. doi: 10.1038/s41467-018-05321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friker L.L., Scheiblich H., Hochheiser I.V., Brinkschulte R., Riedel D., Latz E., Geyer M., Heneka M.T. β-amyloid clustering around ASC fibrils boosts its toxicity in microglia. Cell Rep. 2020;30:3743–3754.e6. doi: 10.1016/j.celrep.2020.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pirzada R.H., Javaid N., Choi S. The roles of the NLRP3 inflammasome in neurodegenerative and metabolic diseases and in relevant advanced therapeutic interventions. Genes. 2020;11:131. doi: 10.3390/genes11020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryan N.B., Dorfleutner A., Kramer S.J., Yun C., Rojanasakul Y., Stehlik C. Differential splicing of the apoptosis-associated speck like protein containing a caspase recruitment domain (ASC) regulates inflammasomes. J. Inflamm. 2010;7:23. doi: 10.1186/1476-9255-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsushita K., Takeoka M., Sagara J., Itano N., Kurose Y., Nakamura A., Taniguchi S. A splice variant of ASC regulates IL-1 β release and aggregates differently from intact ASC. Mediators Inflamm. 2009;2009:1–6. doi: 10.1155/2009/287387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Alba E. 1H, 15N and 13C backbone and side chain chemical shifts of human ASC (apoptosis-associated speck-like protein containing a CARD domain) Biomol. NMR Assign. 2007;1:135–137. doi: 10.1007/s12104-007-9036-x. [DOI] [PubMed] [Google Scholar]
- 23.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Sengupta I., Bhate S.H., Das R., Udgaonkar J.B. Salt-mediated oligomerization of the mouse prion protein monitored by real-time NMR. J. Mol. Biol. 2017;429:1852–1872. doi: 10.1016/j.jmb.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Fawzi N.L., Ying J., Torchia D.A., Clore G.M. Kinetics of amyloid β monomer-to-oligomer exchange by NMR relaxation. J. Am. Chem. Soc. 2010;132:9948–9951. doi: 10.1021/ja1048253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamihira M., Naito A., Tuzi S., Nosaka A.Y., Saitô H. Conformational transitions and fibrillation mechanism of human calcitonin as studied by high-resolution solid-state 13C NMR. Protein Sci. 2000;9:867–877. doi: 10.1110/ps.9.5.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eigen M. Prionics or the kinetic basis of prion diseases. Biophys. Chem. 1996;63:A1–A18. doi: 10.1016/s0301-4622(96)02250-8. [DOI] [PubMed] [Google Scholar]
- 28.Oosawa F., Kasai M. A theory of linear and helical aggregations of macromolecules. J. Mol. Biol. 1962;4:10–21. doi: 10.1016/s0022-2836(62)80112-0. [DOI] [PubMed] [Google Scholar]
- 29.Hofrichter J., Ross P.D., Eaton W.A. Kinetics and mechanism of deoxyhemoglobin S gelation: A new approach to understanding sickle cell disease. Proc. Natl. Acad. Sci. U. S. A. 1974;71:4864–4868. doi: 10.1073/pnas.71.12.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrone F.A., Hofrichter J., Sunshine H.R., Eaton W.A. Kinetic studies on photolysis-induced gelation of sickle cell hemoglobin suggest a new mechanism. Biophys. J. 1980;32:361–380. doi: 10.1016/S0006-3495(80)84962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrone F.A., Hofrichter J., Eaton W.A. Kinetics of sickle hemoglobin polymerization: II. A double nucleation mechanism. J. Mol. Biol. 1985;183:611–631. doi: 10.1016/0022-2836(85)90175-5. [DOI] [PubMed] [Google Scholar]
- 32.Wegner A., Engel J. Kinetics of the cooperative association of actin to actin filaments. Biophys. Chem. 1975;3:215–225. doi: 10.1016/0301-4622(75)80013-5. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein R.F., Stryer L. Cooperative polymerization reactions. Analytical approximations, numerical examples, and experimental strategy. Biophys. J. 1986;50:583–599. doi: 10.1016/S0006-3495(86)83498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris A.M., Watzky M.A., Finke R.G. Protein aggregation kinetics, mechanism, and curve-fitting: A review of the literature. Biochim. Biophys. Acta. 2009;1794:375–397. doi: 10.1016/j.bbapap.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Thusius D. Mechanism of bovine liver glutamate dehydrogenase self-assembly: II. Simulation of relaxation spectra for an open linear polymerization proceeding via a sequential addition of monomer units. J. Mol. Biol. 1975;94:367–383. doi: 10.1016/0022-2836(75)90208-9. [DOI] [PubMed] [Google Scholar]
- 36.Flyvbjerg H., Jobs E., Leibler S. Kinetics of self-assembling microtubules: An “inverse problem” in biochemistry. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5975–5979. doi: 10.1073/pnas.93.12.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serio T.R., Cashikar A.G., Kowal A.S., Sawicki G.J., Moslehi J.J., Serpell L., Arnsdorf M.F., Lindquist S.L. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 38.Meisl G., Rajah L., Cohen S.A.I., Pfammatter M., Šarić A., Hellstrand E., Buell A.K., Aguzzi A., Linse S., Vendruscolo M., Dobson C.M., Knowles T.P.J. Scaling behaviour and rate-determining steps in filamentous self-assembly. Chem. Sci. 2017;8:7087–7097. doi: 10.1039/c7sc01965c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dear A.J., Michaels T.C.T., Meisl G., Klenerman D., Wu S., Perrett S., Linse S., Dobson C.M., Knowles T.P.J. Kinetic diversity of amyloid oligomers. Proc. Natl. Acad. Sci. U. S. A. 2020;117:12087–12094. doi: 10.1073/pnas.1922267117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oroz J., Barrera-Vilarmau S., Alfonso C., Rivas G., de Alba E. ASC pyrin domain self-associates and binds NLRP3 protein using equivalent binding interfaces. J. Biol. Chem. 2016;291:19487–19501. doi: 10.1074/jbc.M116.741082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A.M.J.J. The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Ferrao R., Wu H. Helical assembly in the death domain (DD) superfamily. Curr. Opin. Struct. Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu A., Li Y., Schmidt F.I., Yin Q., Chen S., Fu T.-M., Tong A.B., Ploegh H.L., Mao Y., Wu H. Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat. Struct. Mol. Biol. 2016;23:416–425. doi: 10.1038/nsmb.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gambin Y., Giles N., O'Carroll A., Polinkovsky M., Hunter D., Sierecki E. Single-molecule fluorescence reveals the oligomerization and folding steps driving the prion-like behavior of ASC. J. Mol. Biol. 2018;430:491–508. doi: 10.1016/j.jmb.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 45.Salvesen G.S., Dixit V.M. Caspase activation: The induced-proximity model. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10964–10967. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cavanagh J., Fairbrother W.J., Palmer A.G., III, Skelton N.J. Elsevier; San Diego, CA: 1995. Protein NMR Spectroscopy: Principles and Practice. [Google Scholar]
- 47.Bax A., Grzesiek S. Methodological advances in protein NMR. Acc. Chem. Res. 1993;26:131–138. [Google Scholar]
- 48.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 50.Williamson M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are contained within the article and the supporting information. All source data generated for this study are available from the corresponding author (Dr Eva de Alba; edealbabastarrechea@ucmerced.edu) upon reasonable request.