Abstract
Background:
Electrical impedance myography (EIM) provides insight into muscle composition and structure. We sought to evaluate its use in a mouse obesity model characterized by myofiber atrophy.
Methods:
We applied a prediction algorithm, ie, the least absolute shrinkage and selection operator (LASSO), to surface, needle array, and ex vivo EIM data from db/db and wild-type mice and assessed myofiber cross-sectional area (CSA) histologically and triglyceride (TG) content biochemically.
Results:
EIM data from all three modalities provided acceptable predictions of myofiber CSA with average root mean square error (RMSE) of 15% in CSA (ie, ±209 μm2 for a mean CSA of 1439 μm2) and TG content with RMSE of 30% in TG content (ie, ±7.3 nmol TG/mg muscle for a mean TG content of 25.4 nmol TG/mg muscle).
Conclusions:
EIM combined with a predictive algorithm provides reasonable estimates of myofiber CSA and TG content without the need for biopsy.
Keywords: electrical impedance myography, LASSO prediction algorithm, muscle triglyceride content, myofiber atrophy, myofiber size, obesity-induced sarcopenia
1 |. INTRODUCTION
Assessment of muscle health is valuable in monitoring the metabolic condition of an organism,1 and in diagnosing and managing neuromuscular diseases (NMDs). Evaluation of muscle pathology typically requires a biopsy for subsequent microscopic analysis.2,3 Improved non-invasive methods to quantify pathological changes in muscle would be a worthwhile addition to patient care and preclinical research. Dual X-ray absorptiometry,4 computed tomography,5 MRI,6 and ultrasound,7 have been used to assess muscle composition broadly; however, all have limitations, including inconvenience, high cost, limited repeatability, or insensitivity to actual cellular components.
One technique that holds promise is electrical impedance myography (EIM). In EIM, a low-intensity, high-frequency electrical current is applied to a muscle, and the consequent voltages measured from a second set of electrodes.8,9 EIM's success in providing information on compositional and histological aspects of muscle has been demonstrated in rodent models of injury,10–12 inflammation,13 aging14–16 and neuromuscular conditions,17–20 and in humans including during therapy trials in a variety of NMDs.21–26
Previously, we evaluated the prospect of using EIM to estimate myofiber size by evaluating wild-type (WT) mice ranging from postnatal day 5 to 35 using a regression model.27 We were able to predict myofiber size with an average root mean square error (RMSE) of 12% from the impedance data and the age of the animal alone. More recently, we collected EIM data from a cohort of amyotrophic lateral sclerosis (ALS) superoxide dismutase 1 (SOD1) G93A mice,28 coupled with the regression prediction technique, to obtain estimates of myofiber size with RMSE of 14%.
In addition to predicting myofiber size, our long-term goal is to determine the potential ability of EIM to estimate extra-myocellular abnormalities (eg, fibrosis) and alterations in myofiber intracellular content (eg, glycogen deposition). Here, we evaluated a group of db/db and WT mice at different ages. The db/db mice, impacted by a point mutation in the leptin receptor gene,29,30 exhibit typical obesity-associated features, including hyperglycemia, hyperinsulinemia, skeletal muscle atrophy, and fat accumulation in non-lipogenic tissues including muscle.31–35 We had two goals: (a) To determine how well EIM predicts myofiber size in this disease model, and (b) To assess the ability of EIM to estimate intramuscular triglyceride (TG) content. We performed surface, needle, and ex vivo EIM to determine how the two more direct but invasive methods (needle and ex vivo) performed compared to surface methods and whether a more extended frequency range would provide better predictions.
2 |. METHODS
2.1 |. Animals
All experimental procedures were approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center. Male WT (C57BLKS/J; Strain #000662) and db/db mice (BKS.Cg-Dock7m +/+ Leprdb/J; Strain #000642) were obtained from Jackson Labs (Bar Harbor ME), and aged to 6, 10, and 20 wk in order to evaluate the impact of increasing fat deposition and skeletal muscle atrophy, both of which occur naturally as these animals age. Mice (five db/db and five WT) were evaluated at each time point. All animals were fed standard chow ad libitum.
2.2 |. Grip strength and compound muscle action potential amplitude
Forelimb and hindlimb grip strength36 and compound muscle action potential (CMAP) amplitudes14 were each measured as previously described.36
2.3 |. EIM methods
EIM was performed with the mView impedance spectroscopy system (Myolex Inc., Boston, MA) using a frequency sweep spectroscopy technique. In total, 41 logarithmically spaced frequencies were measured from 8 to 8396 kHz. Data were collected via surface, needle, and ex vivo approaches using different arrays (depicted in Supporting Information Figure S1, which is available online).
2.3.1 |. Surface EIM
After shaving and depilating the left hindlimb, the skin was cleaned with 0.9% saline solution. A fixed rigid four-electrode impedance-measuring array was positioned over the gastrocnemius (GA) in the longitudinal direction.37 Measurements were repeated twice to ensure consistent values. The array was rotated 90°, and measurements repeated to obtain transverse values.
2.3.2 |. Needle array EIM
Measurements were made using a fixed 4 mm wide 4-electrode needle array (2 mm deep, 1 mm exposed tips) inserted along the length of the left GA. The array was assembled from a series of subdermal 27G needle electrodes (Ambu, Neuroline, Copenhagen, Denmark) with the barrel of the electrodes manually coated with standard nonconductive lacquer (e.g., standard nail polish) leaving only the tip exposed.
2.3.3 |. Ex vivo EIM
We used a Plexiglas dielectric measuring cell.38 The excised GA was first placed in the cell with the fibers oriented perpendicularly to the metal plates (for longitudinal muscle measurements), then removed and placed with the fibers parallel to the plates (for transverse muscle measurements).
2.4 |. GA muscle extraction
Mice were killed by CO2. After excision of the entire GA, its wet mass was determined using a standard analytical balance and its height with a micrometer. GA muscle was cut to approximately 5 × 5mm2 (with variable height) to fit into the dielectric cell used for ex vivo impedance measurements, described above.
2.5 |. Histology
Following ex vivo impedance measurements, GA muscles were fixed, sectioned, stained to identify myocyte cell membranes and nuclei, and the stained sections imaged and myofiber cross-sectional area (CSA) determined as previously described.15 On average, 300 myofibers (per WT muscle) and 375 myofibers (per db/db muscle) were counted per animal, for an average total number of 1487 myofibers for WT mice and 1867 myofibers for db/db mice at each timepoint.
2.6 |. Trigylceride assay
The right GA was analyzed for TG content in nmol TG/mg muscle using the Triglyceride Quantification Colorimetric Kit (Catalog # K622–100, Biovision, Inc. Milpitas CA) and a microplate reader (Fisherbrand accuSkan GO UV/Vis Microplate Spectrophotometer, Fisher Scientific) according to the manufacturer's instructions.
2.7 |. Standard statistical analyses
Statistical analyses of the physiological, histological, biochemical, and impedance data were performed using GraphPad Prism V8 (GraphPad Software, Inc. La Jolla, CA). Unless otherwise noted, all data are reported as mean ± SEM. Multiple group comparisons were performed by one-way analysis of variance (ANOVA) with Tukey's multiple comparisons test. Multifrequency EIM values were compared using the two-way ANOVA using Sidak's multiple comparison test. For correlation analyses, the Pearson correlation coefficient was calculated. Data was considered significant with P < .05.
2.8 |. Statistical modeling and prediction
EIM data included the resistance, reactance, and phase values at measured frequencies in both the longitudinal and transverse directions yielding a total of 246 outputs per GA muscle. A separate analysis based on a limited range of frequencies from 11 to 1027 kHz yielded a combined total of 162 outputs per GA. Prior to the formal analysis, we removed any spurious EIM data (ie, multifrequency curves exhibiting negative values over a portion of the frequency range or highly aberrant shapes). For surface EIM, only values from the left GA were included in the analysis from 6-, 10-, and 20-week-old mice, yielding a sample size of 29. For the needle array EIM, only values from the left GA from mice at 10 and 20 wk were included, yielding a sample size of 19. For the ex vivo EIM, only values from the right GA were included from mice at 6, 10, and 20 wk, yielding a sample size of 30.
We built separate models for predicting CSA and TG content for each EIM modality by implementing our previously adopted statistical approach27,28 using the least absolute shrinkage and selection operator (LASSO) to perform a penalized regression procedure, coupled with a variable tuning parameter to avoid over-fitting39 and select the most influential predictors.40 However, unlike our earlier two publications,27,28 here we introduced a second-level analysis, by incorporating a nested leave-one-out cross-validation (NLOOCV) approach41–43 using two nested loops: an inner loop to determine the ideal tuning parameter, and an outer loop to calculate the RMSE. We took this approach since, in this study, we had fewer diseased animals and used more parameters than in the earlier two studies, thus increasing the risk of “overfitting.” We chose our final model parameters (ie, individual EIM frequencies and their coefficients) based on the outer loop analysis that provided an RMSE value closest to the overall RMSE. The predictive values of the final model were converted back to the original scale using the mean and the SD to produce the raw scale RMSE40 (ie, in μm2 for CSA and in nmol/mg muscle for TG content).
3 |. RESULTS
3.1 |. Histology and physiological measurements
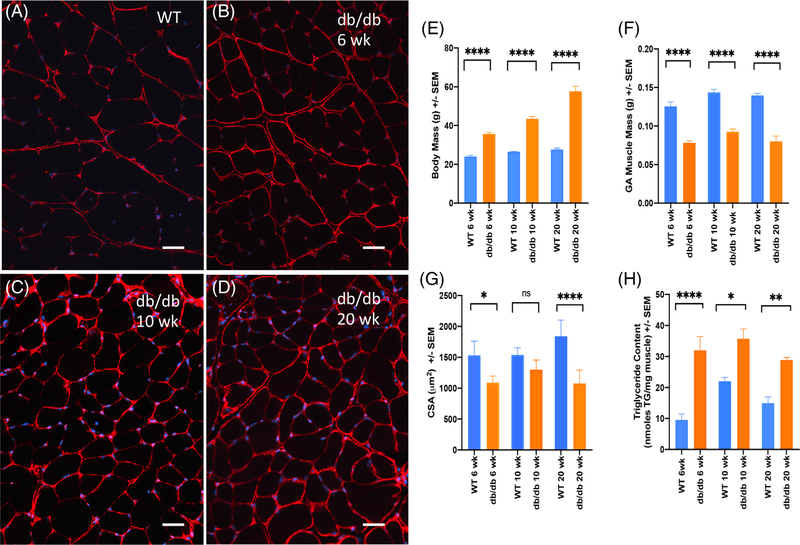
Figures 1A–D display representative histological images (each on the same scale) of GA muscle stained with anti-collagen VI antibodies (red, cell membranes) and DAPI (4'',6-diamidino-2-phenylindole; blue, nuclei) to provide a general sense of how skeletal muscle fiber characteristics change with time in this disease model as compared to WT. Figures 1E–H and Supporting Information Tables S1 and S2 provide physiological measurements and statistical analyses for both db/db and WT animals at 6, 10, and 20 wk of age. While db/db animals gained total body mass at a much faster rate than the corresponding WT animals (Figure 1E), specific measures of muscle health, including mean muscle mass (Figure 1F), forelimb and hindlimb grip strength (Supporting Information Table S1 in Supporting Information Appendix S1), and CMAP (Supporting Information Table S1 in Supporting Information Appendix S1) did not change significantly across ages. Mean myofiber CSA and TG content are shown in Figure 1G,H, respectively. Neither measure changed significantly as the db/db mice age (Table S2 in Supporting Information Appendix S1); however, there are significant differences in both CSA and TG content when db/db animals are compared to WT mice at each time point.
FIGURE 1.
Compilation of representative histological images and selected physiological parameters for db/db vs. WT mice at 6, 10, and 20 wk of age including: Muscle histology illustrating anti-collagen VI (red, cell membrane) and DAPI (blue, nuclear staining) from A, WT (C57Bl/6 at 6 wk); B, db/db at 6 wk; C, db/db 10 wk; D, db/db 20 wk. Bar = 50 μm. E, Body mass (N = 5 mice/group/time point); F, GA muscle mass (N = 10 left plus right GAs/group/time point); G, GA muscle myofiber average CSA (N = 5 left GAs/group/time point); H, GA muscle triglyceride content (N = 5 right GAs/group/time point). Mean ± sSE of the mean. Statistical significance: *P < .05; **P < .01; ***P < .001; ****P < .0001; ns not significant
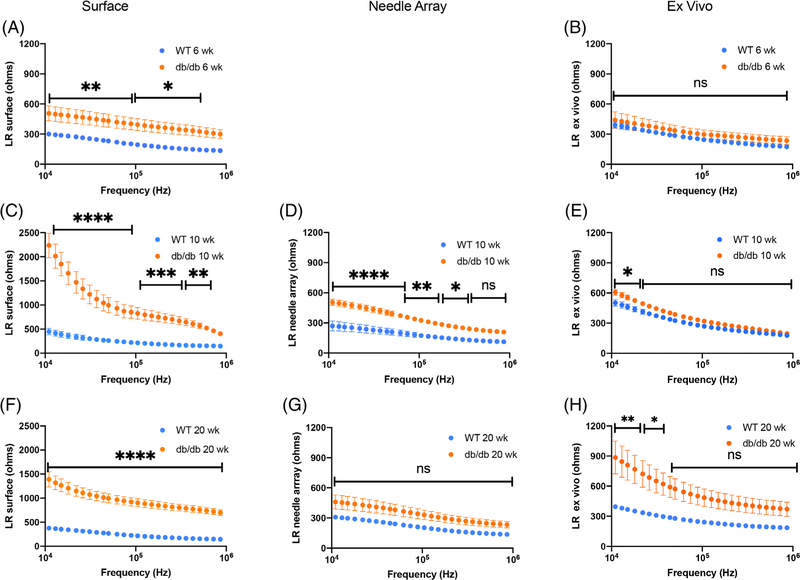
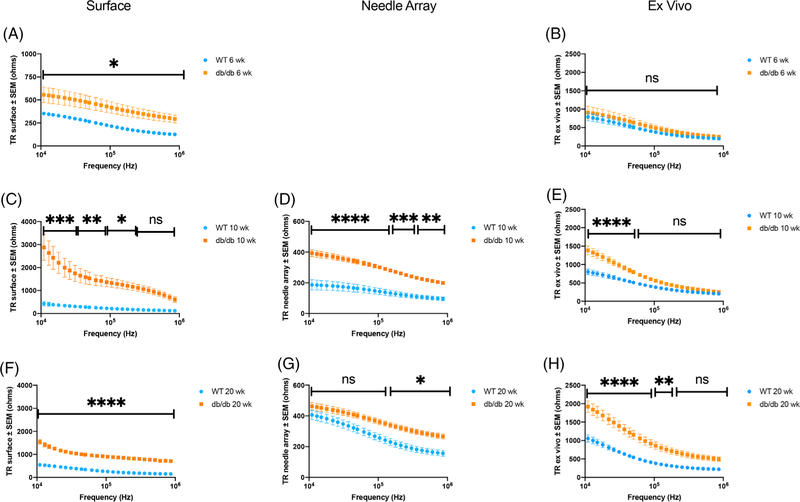
3.2 |. EIM multifrequency resistance, reactance, and phase data
Figures 2 and 3 provide graphical compilations of the surface, needle array, and ex vivo EIM multifrequency resistance data (illustrated from 10 kHz to 1 MHz) in the longitudinal and transverse directions, respectively, at the three times points examined (ie, 6, 10, and 20 wk of age). Supporting Information Figures S3 and S4 provide the analogous compilations of the longitudinal and transverse multifrequency reactance data, and Supporting Information Figures S5 and S6 show compilations of the longitudinal and transverse multifrequency phase data, at these same three time points. Although all three impedance parameters are important in our predictive algorithm, we focused on resistance since it would theoretically be most sensitive to the anticipated histological changes in this model (reduced myofiber volume and increased fat deposition). A significant increase in both longitudinal and transverse resistance over the entire frequency range in the db/db vs WT mice is apparent when impedance was determined by surface EIM at all three time points. However, when measured by needle array and ex vivo EIM, longitudinal resistance values show significant differences over a more limited range of frequencies.
FIGURE 2.
Multifrequency longitudinal EIM resistance data for db/db and WT mice including: A, C, F, surface EIM at 6, 10, and 20 wk of age, respectively; D, G, needle array EIM at 10 and 20 wk of age; and B, E, H, ex vivo EIM at 6, 10, and 20 wk of age, respectively. Mean ± SEM. Statistical significance (two-way ANOVA): *P < .05; **P < .01; ***P < .001; **** P < .0001; ns not significant
FIGURE 3.
Multifrequency transverse EIM resistance data for db/db and WT mice including: A, C, F, surface EIM at 6, 10, and 20 wk of age, respectively; D, G, needle array EIM at 10 and 20 wk of age; and B, E, H, ex vivo EIM at 6, 10, and 20 weeks of age, respectively. Mean ± SEM. Statistical significance (two-way ANOVA): *P < .05; **P < .01; ***P < .001; ****P < .0001; ns not significant
In contrast, transverse resistance values are significantly different between db/db and WT mice over an extended frequency range when analyzed by all three EIM modalities. While comparison of the longitudinal and transverse multifrequency phase values indicate few significant differences between db/db and WT mice, no matter which EIM technique was used, significant differences were observed in both longitudinal and transverse multifrequency reactance values between db/db and WT mice, particularly in the transverse direction. Files containing the multifrequency longitudinal and transverse resistance, reactance, and phase values, for the entire frequency range from 8 to 8396 kHz for all EIM modalities at all time points can be found in the Data Repository at www.rutkovelab.org.
3.3 |. Results of statistical modeling and prediction
Our first goal was to compare the ability of three different EIM modalities to predict muscle CSA. Table 1 provides a list of the frequencies identified by the LASSO penalty procedure, as well as the associated coefficient for each selected frequency, indicating its relative contribution to the prediction equation for muscle CSA. Table 1 also provides details regarding the RMSE ± confidence interval (CI) (scaled and raw) in CSA for each of these modalities based on either the full or limited multifrequency datasets. For both surface and ex vivo EIM, the predictive differences of RMSE indicate that inclusion of the higher frequencies in the analysis decreases the prediction performance to estimate CSA. RMSE increased (ie, worsened) from 14.70% to 16.90% and from 15.24% to 17.24%, for surface and ex vivo model predictions, respectively, when the full frequency range was included in the prediction model. However, for the needle array dataset, in the estimation of CSA RMSE decreased (ie, improved) from 13.59% to 12.74% when frequencies >1 MHz were included in the predictive model.
TABLE 1.
Relative contribution of individual frequencies and LASSO penalty estimates for various models, including associated RMSEs, Pearson’s correlations between observed and predicted myofiber CSA, along with 95% CI
| CSA full frequency range (8 to 8396 kHz) |
CSA limited frequency range (11 to 1027 kHz) |
|||||
|---|---|---|---|---|---|---|
| Model parameters | Surface | Needle | Ex vivo | Surface | Needle | Ex vivo |
| LP 62 kHz | ... | ... | 0.030 | ... | ... | 0.031 |
| LP 89 kHz | ... | ... | 0.112 | 0.160 | ... | 0.079 |
| LP 1027 kHz | ... | ... | ... | −0.104 | −0.143 | ... |
| LX 13 kHz | ... | −0.052 | ... | ... | ... | ... |
| LX 724 kHz | ... | −0.110 | ... | ... | ... | ... |
| LX 1027 kHz | ... | ... | ... | ... | −0.167 | ... |
| LX 8396 kHz | ... | −0.115 | −0.022 | ... | ... | ... |
| LR 150 kHz | −0.083 | ... | ... | ... | ... | ... |
| LR 510 kHz | ... | ... | −0.017 | ... | ... | ... |
| LR 724 kHz | ... | ... | ... | ... | ... | −0.061 |
| TP 74 kHz | ... | ... | ... | ... | 0.136 | ... |
| TP 89 kHz | ... | 0.267 | ... | ... | 0.248 | ... |
| TP 126 kHz | 0.369 | ... | ... | 0.445 | ... | 0.123 |
| TP 150 kHz | ... | ... | 0.067 | ... | ... | 0.148 |
| TP 178 kHz | ... | ... | 0.202 | ... | ... | ... |
| TX 1027 kHz | ... | ... | ... | ... | −0.386 | −0.478 |
| TX 1224 kHz | ... | ... | −0.015 | ... | ... | ... |
| TX 1458 kHz | ... | ... | −0.481 | ... | ... | ... |
| TX 2069 kHz | ... | −0.384 | ... | ... | ... | ... |
| TR 359 kHz | ... | ... | ... | −0.019 | ... | ... |
| TR 428 kHz | ... | ... | ... | −0.271 | ... | ... |
| TR 510 kHz | ... | ... | ... | −0.001 | ... | ... |
| TR 607 kHz | −0.197 | ... | ... | ... | ... | ... |
| TR 724 kHz | ... | ... | ... | ... | −0.123 | ... |
| RMSE, scaled | 0.696 (0.514; 0.877) | 0.542 (0.314; 0.769) | 0.753 (0.513; 0.992) | 0.635 (0.458; 0.811) | 0.576 (0.361; 0.790 | 0.666 (0.436; 0.895) |
| RMSE, original scale (μm2) | 222.191 (167.910; 276.470) | 180.396 (112.237; 248.553) | 240.601 (168.097; 313.104) | 202.802 (149.938; 255.665) | 192.583 (128.725; 256.439) | 212.502 (143.313; 281.690) |
| RMSE, % | 16.904 (12.123; 20.065) | 12.736 (7.833; 17.639) | 17.253 (12.025; 22.481) | 14.701 (10.826; 18.575) | 13.589 (8.992; 18.184) | 15.243 (10.250; 20.235) |
| Pearson correlation between observed and predicted | 0.770 (0.557; 0.888) | 0.883 (0.709; 0.956) | 0.777 (0.581; 0.889) | 0.789 (0.590; 0.898) | 0.885 (0.714; 0.957) | 0.742 (0.518; 0.871) |
Abbreviations: CSA, cross sectional area; kHz, kilohertz; LASSO, Least absolute shrinkage and selection operator; LP, longitudinal phase, LX, longitudinal reactance; LR, longitudinal resistance; RMSE, root mean square error; TP, transverse phase; TX, transverse reactance; TR, transverse resistance. Parenthetical values represent the 95% confidence intervals for each measure.
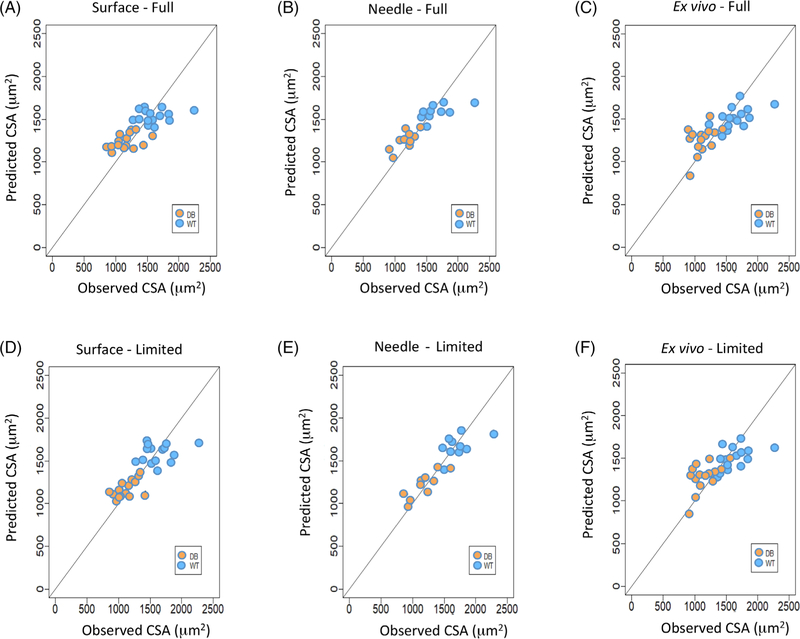
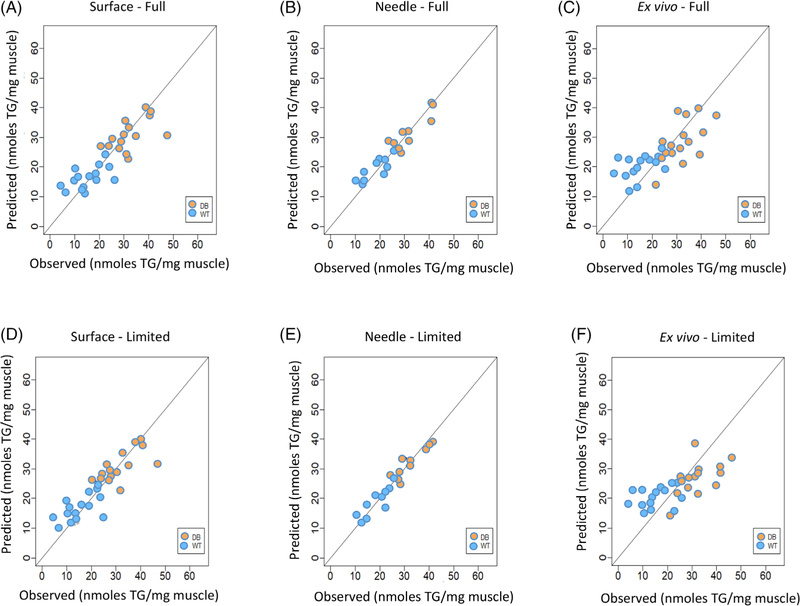
In all cases, a combination of longitudinal and transverse EIM data from only a limited number (ie, range of three to eight8) of frequencies was required to perform the prediction for mean CSA. Figure 4 illustrates plots of the observed values (abscissa) versus the predicted values (ordinate) of CSA based on the LASSO regression analysis for surface (Figure 4A), needle array (Figure 4B), and ex vivo (Figure 4C) EIM measurements for the full multifrequency EIM datasets. Figure 4D–F show the observed versus predicted CSA values for surface, needle array, and ex vivo measurements for the limited multifrequency EIM datasets. The graphical results in Figure 4 for CSA are consistent with the RMSE values presented in Table 1, in that the best agreement between the observed and predicted CSA occurred with the needle array data.
FIGURE 4.
Comparison between observed and predicted cell surface area (CSA). A, Full multifrequency values for surface. B, Full multifrequency values for needle. C, Full multifrequency values for ex vivo. D, Limited multifrequency values for surface. E, Limited multifrequency values for needle. F, Limited multifrequency values for ex vivo
Using an analogous approach, our second goal was to evaluate and compare the ability of three different EIM modalities to predict muscle TG content. Table 2 provides the selected frequency measures from the LASSO penalty procedure, indicating the relative contribution of these frequencies, as well as their performance in predicting TG content in terms of raw and scaled RMSE ± CI. Once again, based on a combination of longitudinal and transverse EIM data, only a limited number (range of 2–16) of frequencies was required to perform the prediction of TG content. For both the surface and ex vivo datasets, comparison of RMSE for the limited versus the full frequency analysis indicates that inclusion of frequencies >1 MHz actually decreases the prediction performance of the model to estimate TG content. However, for the needle array dataset, RMSE was lowered from 24.03% to 21.34% when the higher frequencies were included in the prediction model. Figures 5A–C show the observed (abscissa) and predicted (ordinate) values for TG content resulting from the LASSO regression analysis for surface, needle array, and ex vivo measurements, respectively, for the full multifrequency value sets. Figures 5D–F show the same values for surface, needle, and ex vivo measurements for the limited multifrequency datasets. The graphical results in Figure 5 are consistent with the RMSE values in Table 2, indicating that the prediction model using the needle array data yielded the best agreement between the observed and predicted TG content for the db/db and WT mice.
TABLE 2.
Relative contribution of individual frequencies and LASSO penalty estimates for various models, including associated RMSEs, Pearson’s correlations between observed and predicted myofiber TG content, along with 95% CIs
| TG content full frequency (8 to 8396 kHz) |
TG content limited frequency (11 to 1027 kHz) |
|||||
|---|---|---|---|---|---|---|
| Model parameters | Surface | Needle | Ex vivo | Surface | Needle | Ex vivo |
| LP 8 kHz | 0.159 | ... | ... | ... | ... | ... |
| LP 15 kHz | ... | ... | ... | ... | 0.332 | ... |
| LP 26 kHz | 0.051 | ... | ... | 0.363 | ... | ... |
| LP 89 kHz | ... | −0.066 | ... | ... | −0.124 | ... |
| LP 105 kHz | ... | ... | −0.084 | ... | ... | ... |
| LP 607 kHz | ... | ... | ... | ... | 0.018 | ... |
| LP 1027 kHz | 0.665 | ... | ... | 0.92 | ... | ... |
| LP 8396 kHz | ... | 0.310 | 0.033 | ... | ... | ... |
| LX 8 kHz | 0.134 | ... | ... | ... | ... | ... |
| LX 11 kHz | ... | −0.409 | ... | ... | −0.941 | ... |
| LX 62 kHz | ... | ... | ... | ... | −0.864 | ... |
| LX 74 kHz | ... | ... | ... | −0.292 | ... | ... |
| LX 253 kHz | −0.447 | ... | ... | ... | ... | ... |
| LR 22 kHz | ... | ... | ... | −0.394 | ... | ... |
| LR 26 kHz | −0.084 | ... | ... | ... | ... | ... |
| LR 178 kHz | ... | ... | ... | ... | 0.389 | ... |
| LR 212 kHz | ... | ... | ... | ... | 0.298 | ... |
| LR 253 kHz | ... | ... | ... | ... | 0.231 | ... |
| LR 302 kHz | ... | ... | ... | ... | 0.115 | ... |
| LR 7047 kHz | ... | 0.756 | ... | ... | ... | ... |
| TP 15 kHz | ... | ... | ... | ... | 0.432 | ... |
| TP 253 kHz | −0.46 | ... | ... | −0.441 | ... | ... |
| TP 428 kHz | ... | ... | ... | ... | 0.035 | ... |
| TP 1027 kHz | ... | ... | 0.204 | ... | ... | 0.14 |
| TP 1224 kHz | ... | ... | 0.001 | ... | ... | ... |
| TP 5915 kHz | 0.147 | ... | ... | ... | ... | ... |
| TX 11 kHz | ... | ... | ... | 0.031 | −0.031 | ... |
| TX 13 kHz | ... | ... | ... | 0.009 | ... | ... |
| TX 62 kHz | ... | −0.247 | ... | ... | −0.418 | ... |
| TX 105 kHz | ... | ... | ... | ... | ... | ... |
| TX 862 kHz | ... | ... | 0.537 | ... | ... | 0.464 |
| TX 1027 kHz | ... | ... | ... | ... | 0.352 | ... |
| TX 3498 kHz | ... | 0.180 | ... | ... | ... | ... |
| TX 4168 kHz | 0.122 | ... | ... | ... | ... | ... |
| TR 13 kHz | 0.149 | ... | ... | ... | ... | ... |
| TR 302 kHz | ... | ... | ... | ... | 0.547 | ... |
| TR 428 kHz | ... | ... | ... | ... | 0.050 | ... |
| TR 1027 kHz | ... | 0.051 | ... | 0.394 | 0.014 | ... |
| TR 1224 kHz | ... | 0.136 | ... | ... | ... | ... |
| TR 8396 kHz | −0.04 | ... | ... | ... | ... | ... |
| RMSE, scaled | 0.708 (0.522; 0.892) | 0.638 (0.460; 0.813) | 0.897 (0.658; 1.134) | 0.604 (0.432; 0.774) | 0.721 (0.507; 0.934) | 0.747 (0.558; 0.935) |
| RMSE, original scale (ng TG/mg muscle) | 7.728 (5.759; 9.696) | 5.558 (3.983; 7.132) | 9.610 (7.07; 12.149) | 6.588 (4.774; 8.401) | 6.253 (4.381; 8.124) | 7.990 (6.016; 9.963) |
| RMSE, % | 32.526 (24.097; 40.954) | 21.341 (15.300; 27.382) | 40.417 (29.645; 51.187) | 27.720 (19.962; 35.477) | 24.029 (16.836; 31.222) | 33.586 (25.232; 41.939) |
| Pearson correlation between observed and predicted | 0.852 (0.704; 0.929) | 0.950 (0.870; 0.981) | 0.763 (0.567; 0.880) | 0.881 (0.757; 0.944) | 0.906 (0.762; 0.965) | 0.691 (0.434; 0.843) |
Abbreviations: kHz, kilohertz; LASSO, Least absolute shrinkage and selection operator; LP, longitudinal phase, LX, longitudinal reactance; LR, longitudinal resistance; RMSE, root mean square error; TG, triglyceride; TP, transverse phase; TX, transverse reactance; TR, transverse resistance. Parenthetical values represent the 95% confidence intervals for each measure.
FIGURE 5.
Comparison between observed and predicted muscle triglyceride content (nmol TG/mg muscle). A, Full multifrequency values for surface. B, Full multifrequency values for needle. C, Full multifrequency values for ex vivo. D, Limited multifrequency values for surface. E, Limited multifrequency values for needle. F, Limited multifrequency values for ex vivo
4 |. DISCUSSION
Rather than establishing a simple correlation between EIM values and specific histological and/or biochemical measurements or basing our analysis on a single frequency,44–46 we used the entire multifrequency range coupled with the LASSO prediction algorithm to establish the predictive power of EIM to estimate myofiber CSA and approximate muscle TG content. We also chose to collect impedance information using three different EIM modalities, ie, surface, needle array, and ex vivo. Albeit using the same measurement principle, these three approaches use different electrode designs, provide different datasets, and, based on our current analysis, generate different specific frequencies with different relative errors when used to estimate muscle CSA and TG content. Nevertheless, our results confirm the power of EIM to estimate myofiber CSA, in agreement with our two previous studies27,28, and demonstrate the potential of EIM to provide acceptable, although less accurate, information about muscle TG content in this model of obesity-induced atrophy.47
We compared three different EIM modalities using two different multifrequency datasets, one going as high as the instrument measured, 8396 kHz, and the other only up to 1027 kHz. At frequencies >1 MHz, there is distortion in the impedance values due to unavoidable problems, including inductive effects from the wires and parasitic capacitances within the hardware, which may reduce reliability and add noise to the measurements. Despite these concerns, inclusion of frequencies above 1027 kHz improved the model fit particularly for the needle array data (see Tables 1 and 2). High-frequency impedance features has been shown to differentiate slow-twitch from fast-twitch fibers.48 Therefore, inclusion of the high frequency values in this prediction strategy appears justified. Additional investigations will be required to clarify this issue. Nevertheless, the value of our approach is that researchers can use the coefficients derived in these prediction equations to approximate myofiber CSA or TG content in their samples in this disease model, provided they use the same EIM electrode array and the same frequency range used here. Importantly, this specific set of frequencies would only work for this disease model since they are pathology-specific. Other disorders would require a different set of frequencies.
One somewhat unexpected aspect of the data was the relatively modest differences in the db/db mice compared to the WT as the animals aged. We had expected increasing muscle TG content and smaller CSAs in the oldest animals. In fact, the only major change that we observed was in body mass, likely representing a major deposition of extra-muscular fat.49 Thus, as would be anticipated, surface EIM shows the greatest increase in impedance values (in particular, longitudinal and transverse resistance) over time, likely reflecting increasing subcutaneous fat and not just alterations in the underlying muscle composition. As the subcutaneous fat thickness increases, the impedance data set will be increasingly enriched by fat as less and less current reaches the muscle layer. More sophisticated mathematical approaches will be needed to separate the contribution of subcutaneous fat from that of muscle in surface EIM.50,51 Alternatively, using needle array electrodes allows the operator to reach the underlying muscle and interrogate its electrical properties directly, akin to standard EMG. In fact, the idea of pursuing needle-based EIM has already been introduced.50
How do the present results compare with our earlier findings? Our initial prediction study used needle array EIM to estimate CSA as mice progressed from immaturity to adulthood.27 In that study, because of the small size of the animals, we used a narrow needle array and collected impedance data only in the longitudinal direction. In this analysis of the db/db mice, we used a different needle array with longer, more widely spaced needle electrodes designed to penetrate the layer of subcutaneous fat to reach the underlying muscle. Therefore, a direct comparison of the two data sets is not possible.
Our second prediction study focused on estimating CSA in ALS mice using surface EIM28 so, theoretically, prediction parameters from that study could be compared to the surface EIM results presented here. Compared to the animals studied here, ALS mice have a major variation in fiber size within each animal due to ongoing denervation and re-innervation. This allowed us to assess EIM's relationship to the coefficient of variation in fiber size. However, despite the fact that both ALS and db/db mice exhibit muscle atrophy, ALS mice were selected specifically because they do not display the added complication of increased fat infiltration accompanying the increased muscle atrophy characteristic of many NMDs. Critically, the db/db mouse is not a simple model of “fatty muscle” but rather, as confirmed in our histological analysis, also of obesity-induced myofiber atrophy.31,34,47 Thus, the observed differences in prediction parameters in db/db versus ALS animals do not simply represent the impact of fat deposition in and around normal muscle, but rather a combination of the effects of alterations in both CSA and TG content. Ongoing theoretical studies will be needed to separate the contributions of intramuscular fat and underlying muscle to the observed impedance.51 An alternative approach that we have undertaken separately is to use ex vivo impedance data from a number of murine disease models to calculate two intrinsic properties of muscle, ie, conductivity and relative permittivity,52 that can be properly compared.
Here we included both transverse data and longitudinal data. It is not clear why one direction would be better than another, although transverse data are more sensitive to myofiber membranes, since current crosses more myofibers. Our main goal in performing both types of measurements was to strengthen our prediction algorithms. By obtaining multidirectional data, we obtain richer information of the tissue's electrical properties from which we can build stronger prediction models.
There were several limitations to this study. First, the number of animals in each group was relatively small, especially in the needle array EIM group, since no data were obtained at 6 weeks of age, because this set of measurements had not been initially anticipated. Second, the age range of the db/db mice may have been too narrow to observe significant differences in muscle composition. Indeed this model does develop age-dependent atrophy, but at earlier times than those measured here. In these animals, obesity onset occurs are approximately 3–4 wk, hyperglycemia around 4–8 wk, and distal polyneuropathy (presumably due to hyperglycemia) at around 40 wk. Therefore, the db/db mice examined at 6 wk were already obese and had TG content significantly elevated compared to their age-matched WT counterparts. Third, we only attempted to build our model around mean values of CSA and TG rather than the entire fiber distribution, which would have been far more challenging, especially considering the relatively small number of animals studied. Fourth, the different EIM measurement techniques are not without challenges in mice. While EIM is generally straightforward to perform in humans, the animals' small size adds a number of complexities. Ensuring good electrical contact between the surface array and the skin is challenging and can lead to low-frequency artifact. Similarly, it is difficult to guarantee that the needle electrodes fully penetrate into the muscle, especially given the thickness of subcutaneous fat in the db/db model. Simple differences in wire orientation, placement, and length would reduce reliability. Finally, in ex vivo studies, it is important that the tissue be placed in the cell precisely such that true transverse and longitudinal data can be obtained. Challenges with accurate placement of such a small sample of material in a cell likely contributed to the fact that the ex vivo data did not appear stronger compared to surface or needle values.
One question that arises is what an averaged RMSE of, say, 14.7% means in practical context when estimating myofiber CSA. Simply put, it says that on average our prediction model veers from that true CSA value, by that amount. Whether that is sufficiently accurate for practical application will depend strictly on the question asked. For example, it would be possible to build a power analysis for a clinical study based on expected relative change with drug therapy based on this value and its 95% CIs. The Food and Drug Administration (FDA) views biomarkers in their specific “context of use,”53; thus, it remains conceivable that a strictly defined application could be identified that would be acceptable to the FDA. Finally, our nested predictive algorithm, used here, helps ensure that our results are sufficiently conservative so as to avoid overfitting. Indeed, had we used a non-nested model here, the RMSEs for surface measurements (limited frequency range) would have been better at 14.1% (vs 14.7% for nested) for CSA and 21.8% (vs 27.7% nested) for TG.
The major implication of this work is that the EIM modeling approach developed here can provide acceptable estimates of myofiber CSA and TG content in the db/db mouse model that would ordinarily require either relatively expensive/inconvenient approaches (eg, MRI) or invasive procedures (eg, muscle biopsy). Having such a simply obtained measure could be valuable for research purposes in which longitudinal assessment of muscle condition in obesity or other conditions (eg, various dystrophies) is needed. Studies assessing the use of this technology in humans, with comparison to other, standard approaches are a logical next step. One study already supports that basic premise.54 Finally, we plan to determine the capability of EIM paired with a statistical model to assess other compositional alterations of muscle tissue, including the presence of both extracellular and intracellular pathologies wherever relevant animal models are available, including specific muscle disease models. Our ultimate goal is to use this information to differentiate both disease type and severity.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by the National Institutes of Health grant R01 NS091159 (SBR). We acknowledge the BIDMC Morphology Core whose Histology and Epifluorescence Microscopy expertise and resources were used in this publication.
Funding information
National Institutes of Health, Grant/Award Number: R01 NS091159 (SBR)
Abbreviations:
- ALS
amyotrophic lateral sclerosis
- ANOVA
analysis of variance
- CI
confidence Interval
- CMAP
compound muscle action potential
- CSA
cross-sectional area
- EIM
electrical impedance myography
- FDA
Food and Drug Administration
- GA
gastrocnemius
- kHz
kiloHertz
- LASSO
least absolute shrinkage and selection operator
- MHz
megaHertz
- MSE
mean squared error
- NLOOCV
nested leave-one-out cross-validation
- NMD
neuromuscular disease
- RMSE
root mean square error
- SOD1
superoxide dismutase 1
- TG
triglyceride
- WT
wild type
Footnotes
CONFLICTS OF INTEREST
Dr. Rutkove has equity in, and serves a consultant and scientific advisor to, Myolex, Inc., a. company that designs impedance devices for clinical and research use; he is also a member of the company's Board of Directors. The company also has an option to license patented impedance technology of which Dr. Rutkove is named as an inventor. Dr. Sanchez also serves as a consultant to Myolex, Inc., as well as Texas Instruments, Inc., Impedimed, Inc., and Gideon Health, three other companies that develop impedance technology for consumer, clinical, and research use.
5 | ETHICAL PUBLICATION STATEMENT
We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.McGregor RA, Cameron-Smith D, Poppitt SD. It is not just muscle mass: a review of muscle quality, composition and metabolism during ageing as determinants of muscle function and mobility in later life. Longev Healthspan. 2014;3(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman EP, Bronson A, Levin AA, et al. Restoring dystrophin expression in duchenne muscular dystrophy muscle: progress in exon skipping and stop codon read through. Am J Pathol. 2011;179(1):12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel HP, Syddall HE, Martin HJ, Cooper C, Stewart C, Sayer AA. The feasibility and acceptability of muscle biopsy in epidemiological studies: findings from the hertfordshire sarcopenia study (HSS). J Nutr Health Aging. 2011;15(1):10–15. 10.1007/s12603-011-0006-8. [DOI] [PubMed] [Google Scholar]
- 4.Ramírez-Campillo R, Andrade DC, Campos-Jara C, Henríquez-Olguín C, Alvarez-Lepín C, Izquierdo M. Regional fat changes induced by localized muscle endurance resistance training. J Strength Cond Res. 2013;27(8):2219–2224. 10.1519/JSC.0b013e31827e8681. [DOI] [PubMed] [Google Scholar]
- 5.Goodpaster BH, Kelley DE, Thaete FL, He J, Ross R. Skeletal muscle attenuation determined by computed tomography is associated with skeletal muscle lipid content. J Appl Physiol. 2000;89(1):104–110. 10.1152/jappl.2000.89.1.104. [DOI] [PubMed] [Google Scholar]
- 6.Finanger EL, Russman B, Forbes SC, Rooney WD, Walter GA, Vandenborne K. Use of skeletal muscle MRI in diagnosis and monitoring disease progression in Duchenne muscular dystrophy. Phys Med Rehabil Clin N Am. 2012;23:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillen S, van Alfen N, Sorenson EJ, et al. Assessing spinal muscular atrophy with quantitative ultrasound. Neurology. 2010;75(6):526–531. 10.1212/WNL.0b013e3182068eed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez B, Rutkove SB. Present uses, future applications, and technical underpinnings of electrical impedance Myography. Curr Neurol Neurosci Rep. 2017;17:86. [DOI] [PubMed] [Google Scholar]
- 9.Rutkove SB, Sanchez B. Electrical impedance methods in neuromuscular assessment: an overview. Cold Spring Harb Perspect Med. 2019; 1–12. 10.1101/cshperspect.a034405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahad M, Fogerson PM, Rosen G D, Narayanaswami P, Rutkove SB. Electrical characteristics of rat skeletal muscle in immaturity, adulthood and after sciatic nerve injury, and their relation to muscle fiber size. Physiol Meas. 2009;30(12):1415–1427. 10.1088/0967-3334/30/12/009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahad MA, Rutkove SB. Electrical properties of rat muscle after sciatic nerve injury: impact on surface impedance measurements assessed via finite element analysis. J Phys Conf Ser. 2010;224:012101. [Google Scholar]
- 12.Ahad M, Rutkove SB. Correlation between muscle electrical impedance data and standard neurophysiologic parameters after experimental neurogenic injury. Physiol Meas. 2010;31(11):1437–1448. 10.1088/0967-3334/31/11/003. [DOI] [PubMed] [Google Scholar]
- 13.Mortreux M, Semple C, Riveros D, Nagy JA, Rutkove SB. Electrical impedance myography for the detection of muscle inflammation induced by λ-carrageenan. PLoS One. 2019;14(10):e0223265. 10.1371/journal.pone.0223265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold WD, Taylor RS, Li J, Nagy JA, Sanchez B, Rutkove SB. Electrical impedance myography detects age-related muscle change in mice. PLoS One. 2017;12(10):e0185614. 10.1371/journal.pone.0185614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark-Matott J, Nagy JA, Sanchez B, Taylor R, Riveros D, Abraham NA. Altered muscle electrical tissue properties in a mouse model of premature aging. Muscle Nerve. et al. , 2019;60(6):801–810. 10.1002/mus.26714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aaron R, Esper GJ, Shiffman CA, Bradonjic K, Lee KS, Rutkove SB. Effects of age on muscle as measured by electrical impedance myography. Physiol Meas. 2006;27:953–959. 10.1088/0967-3334/27/10/002. [DOI] [PubMed] [Google Scholar]
- 17.Wang LL, Spieker AJ, Li J, Rutkove SB. Electrical impedance myography for monitoring motor neuron loss in the SOD1 G93A amyotrophic lateral sclerosis rat. Clin Neurophysiol. 2011;122:2505–2511. 10.1016/j.clinph.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Sanchez B, Pacheck A, Rutkove S. Using electrical impedance myography to predict force output in ALS: a study in the G93A SOD1 mouse. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16:111. [Google Scholar]
- 19.Li J, Geisbush TR, Rosen GD, Lachey J, Mulivor A, Rutkove SB. Electrical impedance myography for the in vivo and ex vivo assessment of muscular dystrophy (mdx) mouse muscle. Muscle Nerve. 2014;49(6): 829–835. 10.1002/mus.24086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Geisbush TR, Kumar R, Mulivor A, Rutkove SB. Electrical impedance myography for the in vivo assessment of the muscular dystrophy (mdx) mouse. Ann Neurol. 2013;74:S105. 10.1002/ana.24068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutkove SB, Caress JB, Cartwright MS, et al. Electrical impedance myography as a biomarker to assess ALS progression. Amyotroph Lateral Scler. 2012;13(5):439–445. 10.3109/17482968.2012.688837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitner ML, Kapur K, Darras BT, et al. Electrical impedance myography for reducing sample size in Duchenne muscular dystrophy trials. Ann Clin Transl Neurol. 2020;7(1):4–14. 10.1002/acn3.50958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Sung M, Rutkove SB. Electrophysiologic biomarkers for assessing disease progression and the effect of Riluzole in SOD1 G93A ALS mice. PLoS One. 2013;8:e65976. 10.1371/journal.pone.0065976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutkove S, Zaidman C, Wu J, Kapur K, Darras B. Electrical impedance myography and quantitative ultrasound as biomarkers of disease progression in duchenne muscular dystrophy: a preliminary analysis. Neurology. 2015;84. [Google Scholar]
- 25.Arnold WD, McGovern VL, Sanchez B, et al. The neuromuscular impact of symptomatic SMN restoration in a mouse model of spinal muscular atrophy. Neurobiol Dis. 2016;87:116–123. 10.1016/j.nbd.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapur K, Sanchez B, Pacheck A, Darras B, Rutkove SB, Selukar R. Functional mixed-effects modeling of longitudinal Duchenne muscular dystrophy electrical impedance Myography data using state-space approach. IEEE Trans Biomed Eng. 2019;66(6):1761–1768. 10.1109/TBME.2018.2879227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapur K, Taylor RS, Qi K, et al. Predicting myofiber size with electrical impedance myography: a study in immature mice. Muscle Nerve. 2018;58(1):106–113. 10.1002/mus.26111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapur K, Nagy JA, Taylor RS, Sanchez B, Rutkove SB. Estimating Myofiber size with electrical impedance Myography: a study in amyotrophic lateral sclerosis MICE. Muscle Nerve. 2018;58(5):713–717. 10.1002/mus.26187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee GH, Proenca R, Montez JM, et al. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379(6566):632–635. 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Charlat O, Tartaglia LA, et al. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84(3):491–495. 10.1016/S0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 31.Bayley JS, Pedersen TH, Nielsen OB. Skeletal muscle dysfunction in the db/db mouse model of type 2 diabetes. Muscle Nerve. 2016;54(3): 460–468. 10.1002/mus.25064. [DOI] [PubMed] [Google Scholar]
- 32.Ostler JE, Maurya SK, Dials J, et al. Effects of insulin resistance on skeletal muscle growth and exercise capacity in type 2 diabetic mouse models. Am J Physiol Endocrinol Metab. 2014;306(6):E592–E605. 10.1152/ajpendo.00277.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campero M, Ezquer M, Ezquer F. Nerve excitability and structural changes in Myelinated axons from diabetic mice. Exp Clin Endocrinol Diabetes. 2015;123(08):485–491. 10.1055/s-0035-1559606. [DOI] [PubMed] [Google Scholar]
- 34.Campbell-Tofte J, Hansen HS, Mu H, Mølgaard P. Increased lipids in non-lipogenic tissues are indicators of the severity of type 2 diabetes in mice. Prostaglandins Leukot Essent Fatty Acids. 2007;76(1):9–18. 10.1016/j.plefa.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Mehlem A, Hagberg CE, Muhl L, Eriksson U, Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat Protoc. 2013;8(6):1149–1154. 10.1038/nprot.2013.055. [DOI] [PubMed] [Google Scholar]
- 36.Nagy JA, Kapur K, Taylor RS, Sanchez B, Rutkove SB. Electrical impedance myography as a biomarker of myostatin inhibition with ActRIIB-mFc: a study in wild-type mice. Future Sci OA. 2018;4(6): FSO308. 10.4155/fsoa-2018-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Staats WL, Spieker A, Sung M, Rutkove SB. A technique for performing electrical impedance myography in the mouse hind limb: data in normal and ALS SOD1 G93A animals. PLoS One. 2012;7:e45004. 10.1371/journal.pone.0045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Jafarpoor M, Bouxsein M, Rutkove SB. Distinguishing neuromuscular disorders based on the passive electrical material properties of muscle. Muscle Nerve. 2015;51(1):49–55. 10.1002/mus.24270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hua J, Tembe WD, Dougherty ER. Performance of feature-selection methods in the classification of high-dimension data. Pattern Recognit. 2009;42(3):409–424. 10.1016/j.patcog.2008.08.001. [DOI] [Google Scholar]
- 40.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed. Springer Science & Business Media; 2009. http://www.worldcat.org/oclc/405547558%5CnHastie. Accessed mm dd, yyyy. [Google Scholar]
- 41.Krstajic D, Buturovic LJ, Leahy DE, Thomas S. Cross-validation pitfalls when selecting and assessing regression and classification models. J Cheminform. 2014;6(1):1–15. 10.1186/1758-2946-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Bowen SR, Chaovalitwongse WA, Sandison GA, Grabowski TJ, Kinahan PE. Respiratory trace feature analysis for the prediction of respiratory-gated PET quantification. Phys Med Biol. 2014;59(4):1027–1045. 10.1088/0031-9155/59/4/1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cawley GC, Talbot NLC. On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res. 2010;11:2079–2107. [Google Scholar]
- 44.Rutkove SB, Lee KS, Shiffman CA, Aaron R. Test-retest reproducibility of 50 kHz linear-electrical impedance myography. Clin Neurophysiol. 2006;117:1244–1248. 10.1016/j.clinph.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 45.Ahad MA, Rutkove SB. Electrical impedance myography at 50 kHz in the rat: technique, reproducibility, and the effects of sciatic injury and recovery. Clin Neurophysiol. 2009;120:1534–1538. 10.1016/j.clinph.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutkove SB, Fogerson PM, Garmirian LP, Tarulli AW. Reference values for 50-kHz electrical impedance myography. Muscle Nerve. 2008;38:1128–1132. 10.1002/mus.21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalinkovich A, Livshits G. Sarcopenic obesity or obese sarcopenia: a cross talk between age-associated adipose tissue and skeletal muscle inflammation as a main mechanism of the pathogenesis. Ageing Res Rev. 2017;35:200–221. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez B, Li J, Bragos R, Rutkove SB. Differentiation of the intracellular structure of slow- versus fast-twitch muscle fibers through evaluation of the dielectric properties of tissue. Phys Med Biol. 2014;59 (10):2369–2380. 10.1088/0031-9155/59/10/2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen W, Wilson JL, Khaksari M, Cowley MA, Enriori PJ. Abdominal fat analyzed by DEXA scan reflects visceral body fat and improves the phenotype description and the assessment of metabolic risk in mice. Am J Physiol Endocrinol Metab. 2012. 10.1152/ajpendo.00078.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwon H, di Cristina JF, Rutkove SB, Sanchez B. Recording characteristics of electrical impedance-electromyography needle electrodes. Physiol Meas. 2018;39(5):055005. 10.1088/1361-6579/aabb8c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwon H, Malik WQ, Rutkove SB, Sanchez B. Separation of subcutaneous fat from muscle in surface electrical impedance Myography measurements using model component analysis. IEEE Trans Biomed Eng. 2019;66(2):354–364. 10.1109/TBME.2018.2839977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagy JA, DiDonato CJ, Rutkove SB, Sanchez B. Permittivity of ex vivo healthy and diseased murine skeletal muscle from 10kHz to 1MHz. Sci Data. 2019;6(1):1–11. 10.1038/s41597-019-0045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amur SG, Sanyal S, Chakravarty AG, et al. Building a roadmap to biomarker qualification: challenges and opportunities. Biomark Med. 2015;9:1095–1105. 10.2217/bmm.15.90. [DOI] [PubMed] [Google Scholar]
- 54.Hobson-Webb LD, Zwelling PJ, Pifer AN, et al. Point of care quantitative assessment of muscle health in older individuals: an investigation of quantitative muscle ultrasound and electrical impedance myography techniques. Geriatrics (Switzerland). 2018;3(4):92. 10.3390/geriatrics3040092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.