Abstract
UPLC-Q-TOF-MS was employed to analyse the non-volatile components of green teas fermented with probiotic yeast (Saccharomyces boulardii) and lactic acid bacteria (LAB, Lactiplantibacillus plantarum). The flavone glycosides in yeast-fermented and stored tea decreased significantly, together with the increases of flavone aglycones and other simple flavone glycosides. LAB-fermented tea presented different flavone glycoside profiles; in which, both C-glycosides and O-glycosides decreased and the flavone aglycones were further degraded. The profiles of flavone glycosides and aglycones in co-cultured tea differed from that in yeast- or LAB-fermented tea; less glycosides were degraded but a greater number of aglycones were produced. Two unique LAB metabolites with bioactive and antifungal properties, D-phenyllactic acid (PLA) and p-OH-PLA, were found in both L. plantarum and co-cultured teas, and the co-fermentation showed a synergic effect on the production of these two compounds that would enhance the quality and preservation of fermented teas.
Keywords: S. boulardii; L. plantarum, green tea; Fermentation; Metabolomic
Graphical abstract
Highlights
-
•
UPLC-QTOF-MS was used to analyse non-volatile components of probiotics-fermented green teas.
-
•
Changes of flavone glycosides and aglycones in monocultures and co-culture differed.
-
•
D-phenyllactic acid (PLA) and p-OH-PLA were found in both L. plantarum and co-cultured teas.
-
•
Co-fermentation showed a synergic effect on the production of PLA and p-OH-PLA.
1. Introduction
Biotransformation of tea with various microorganisms can impact the chemical components of tea, as these microorganisms would metabolize these compounds through different pathways (Ge et al., 2019; Li et al., 2017). As a consequence, numerous secondary metabolites or novel compounds can be produced after microbial fermentation (Wang et al., 2015). Lactic acid bacteria (LAB) and yeasts are the most commonly used microorganisms in food fermentation. Some species of LAB such as Limosilactobacillus fermentum, Lacticaseibacillus rhamnosus, Lactiplantibacillus plantarum and yeast Saccharomyces boulardii showed probiotic potentials in inhibiting the growth of pathogenic bacteria, neutralising enterotoxins and resisting antibiotics (Kakisu et al., 2012; Nayak, 2011).
To date, dairy-based products have been the most used carriers for probiotics, however, there are some concerns about lactose intolerance, high cholesterol levels, allergy to dairy protein for dairy-based probiotic products. Meanwhile, the demand for new flavor or taste, increasing health awareness and veganism has stimulated the trend in non-dairy matrices as potential carriers of probiotics. In addition, the low pH resulted from LAB fermentation is a challenge of the dairy-based probiotic products. The high acidity that originates from the organic acids released by metabolic activities of LAB reduces the survival of probiotic bacteria during the processing and shelf-life of fermented dairy-based products (Aspri et al., 2020). Therefore, there is a strong interest in finding new matrices and novel processing methods to enhance the survival and validity of probiotics.
There are numerous novel vegan-based probiotic products, such as probiotic vegetables, soy, and fruits. However, tea as a novel carrier for probiotics has not been explored so far. From the previous study (Chan et al., 2021), co-culturing of yeast and LAB enhanced the survival of LAB and modulated the flavor of coffee brews. We therefore conducted probiotic tea fermentation with monocultures of S. boulardii, L. plantarum, and their combination.
High-performance liquid chromatography (HPLC) coupled with ultraviolet (UV) or mass spectrometry (MS) detectors are commonly used for qualitative and quantitative analyses of tea (Tao et al., 2016; Zhang et al., 2011). However, most of these studies were conducted with analysis of a small number of targeted components, which limited the discovery of novel compounds. In recent years, ultra-performance LC-quadrupole time-of-flight MS (UPLC-Q-TOF-MS) has been receiving more attention due to its high sensitivity, high-throughput and efficiency (Jing et al., 2017). UPLC-Q-TOF-MS together with a chemometric approach enables a comprehensive analysis of chemical components in tea (Dai et al., 2015; Zhou et al., 2019). Chemometric methods have been widely used in food analysis. Principal component analysis (PCA), projection to latent structures discriminate analysis (PLS-DA), variable importance for prediction (VIP) and hierarchical cluster analysis (HCA) have been employed to extract the key metabolites from overwhelming data sets obtained from high-throughput analysis (Bevilacqua et al., 2017).
Therefore, in this study, a non-targeted UPLC–Q-TOF-MS approach was applied to investigate the variations in the metabolite profiles of fermented and stored teas obtained by fermentation with probiotic S. boulardii, L. plantarum and the combination of S. boulardii and L. plantarum. The differentiation among probiotic teas fermented with starter culture variations was explored with the aid of chemometrics. Furthermore, the changes of tea components during storage were also evaluated.
2. Materials and methods
2.1. Sample preparation
Green tea (Guizhou, China) fermentation was conducted by inoculating probiotic yeast S. boulardii CNCM I-745 (Biocodex, Inc., California, USA), L. plantarum 299V (Probi, Singapore) and their combination into tea infusions. The tea infusions were obtained by brewing tea leaves (5% w/w) in hot water (95 °C) for 10 min, then removing tea leaves. Before inoculation, the tea infusions supplemented with 0.5% glucose and 0.06% Opti-White® yeast extract (Lallemand, Edwardstown, Australia) were sterilized at 121 °C for 15 min. The sterilized tea infusions were cooled down to room temperature, then inoculated with washed yeast and LAB cells (10 mL of 0.9% (w/v) NaCl solution × 3) to achieve an initial cell count about 5–7 log CFU/mL. The inoculated tea infusions were incubated at 30 °C for 2 days, followed by storing at 25 °C for 4 weeks. Uninoculated sterilized green tea infusions were incubated under the same conditions and served as the control. All the fermentations were done in triplicate and samples were withdrawn periodically for analysis.
The samples for UPLC-Q-TOF-MS analysis were prepared as follows: 700 μL of acetonitrile were added into 700 μL of tea infusion and mixed by using a Vortex Mixer at 272×g for 10 s, then the mixture was centrifuged at 7200×g for 10 min; afterwards, it was filtered through an 0.20-μm regenerated cellulose (RC) membrane. One hundred μL of filtered tea infusion were added into 600 μL of deionised (DI) water, vortexed at 272×g for 10 s, then centrifuged at 7200×g for 10 min. The supernatant was filtered through an 0.22 μm Polytetrafluoroethylene (PTFE) filter and diluted with 0.1% aqueous formic acid solution, then subjected to UPLC-Q-TOF-MS analysis.
2.2. UPLC-Q-TOF-MS analysis
The separation of non-volatile compounds was carried out by using Nexera X2 UPLC system (Shimadzu, Tokyo, Japan), with a Kinetex 2.6-μm Biphenyl column (2.1 mm × 100 mm,100 Å) (Phenomenex, USA) at 40 °C. The flow rate was 0.5 mL min−1 and the injection volume was 5 μL. The mobile phase A was 0.1% aqueous formic acid solution and mobile phase B was methanol/acetonitrile (v/v=30/70) with gradient elution. Mass spectrometry (MS) analysis was performed on a QTOF AB Sciex Triple TOF™ 5600 (AB SCIEX, USA), which was calibrated every five samples in a highly sensitive mode as recommended. The electrospray ionization (ESI) source was operated in negative mode, and spectra were obtained in full-scan mode. Declustering potential (DP), ion release delay (IRD) and ion release width (IRW) were set at 80 V, 67 V and 25 V, respectively. IDA MS/MS was conducted at the same collision energy (CE) of 40 V for all ions (intensity≥100 cps, ion tolerance≤10 ppm). All analyses were carried out in triplicate. Analyst 1.7.1 (Framingham, MA, USA), PeakView 2.2 and MultiQuant 3.0.2 were used for data acquisition and processing (He et al., 2020).
2.3. Data processing and statistical analysis
Raw data files acquired from the UPLC-Q-TOF-MS analysis were imported into the MS-DIAL ver. 4.38 software (RIKEN, Yokohama, Japan) to generate a peak table that included information on retention time, mass-to-charge ratio (m/z), and MS intensity of the metabolites in the sample. The signal-to-noise threshold for peak detection was set to 10. The retention time tolerance, MS 1 and MS 2 tolerances for the peak alignment were set to 0.2 min, 0.01 and 0.05 Da, respectively. PCA and partial least-squares discriminant analysis (PLS-DA) were performed using MS-DIAL after gap filling and normalization to investigate the overall tea metabolome variations caused by fermentation. Variable importance for prediction (VIP) analysis, fold change and ANOVA p-value were conducted with MS-DIAL ver. 4.38 software. Volcano plots were performed using Origin 2019 (OriginL. plantarum Corporation, Northampton, Massachusetts, USA).
2.4. Identification of compounds based on MS/MS spectra
The identification of metabolites was performed based on the accurate mass and mass spectrometric fragmentation patterns guided by the Human Metabolome Database (HMDB; http://www.hmdb.ca), MS-DIAL metabolomics MSP spectral kit (MSMS-Public-Neg-VS15 database), MassBank (https://massbank.eu), National Library of Medicine, National Center for Biotechnology Information (https://pubchem.ncbi.nlm.nih.gov/), related literatures (Dai et al., 2015; Zhou et al., 2019) and in-house databases based on commercial standards and theoretical MS2 fragments.
3. Results and discussion
3.1. Non-targeted analysis of tea samples by UPLC-Q-TOF-MS
Teas fermented and stored with S. boulardii CNCM I-745, L. plantarum 299V and a co-culture of both microorganisms were sampled and analysed for the comparison of substrate and metabolite changes in relation to the different fermentation treatments. A total of 1680 peaks were tentatively extracted and used for statistical analysis after alignment and normalization. The metabolites were identified as flavonoids, amino acids, organic acids, sugars, alkaloids, glycosides and fatty acids, etc.
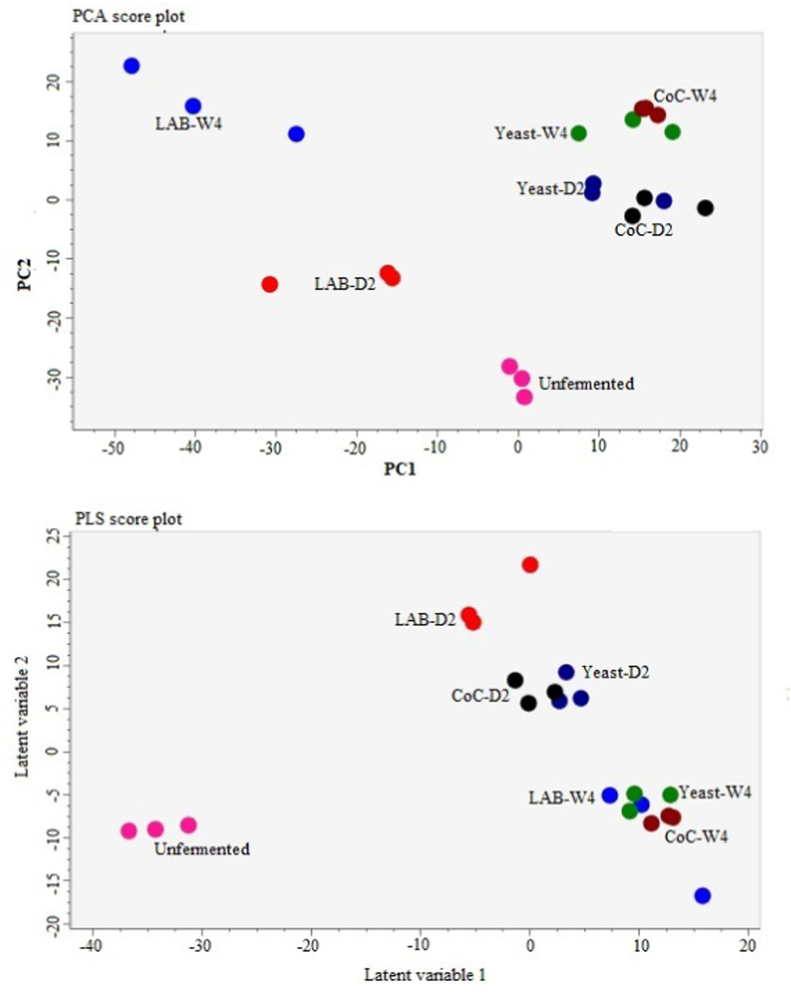
In order to evaluate the metabolic differences between fermented and stored teas with different microorganisms, unsupervised PCA method based on the UPLC-Q-TOF-MS data sets was applied to obtain an overview of the difference in the metabolite phenotypes among the tea samples. Fig. 1A shows the PCA score plot with PC1 and PC2 explaining 42.7% of the total variance (26.7% and 16.0%, respectively). It could be observed that the L. plantarum fermented tea can be completely separated from the unfermented tea, S. boulardii and co-culture fermented teas based on the metabolic profiles, which further confirmed that microbial fermentations had significant effects on the tea metabolome during fermentation. Furthermore, the clear separation of the fermented tea samples from the stored tea samples in the score plot indicated a significant effect of the storage time on the tea metabolome.
Fig. 1.
Multivariate statistical analysis of unfermented (pink), L. plantarum fermented (red), S. boulardii fermented (dark blue), coculture fermented (black), L. plantarum stored (light blue), S. boulardii stored (green), co-culture stored (brown) tea samples: (A) PCA score plot; (B) PLS-DA score plot, R2Y =57.8%, Q2 = 59.2%. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
A supervised PLS-DA analysis and variable importance for prediction (VIP) were employed to investigate the metabolites that showed the greatest difference. In total, 589 metabolites with VIP > 1 were filtered to do further analysis. As shown in the PLS-DA plot (Fig. 1B), the starter culture inoculated tea samples were separated from the unfermented sample by latent variable 1, and the stored samples were separated from the 2-day fermented samples by latent variable 2, which indicate different metabolite phenotypes for the teas fermented in different ways.
3.2. Influence of fermentation and storage on tea metabolome
3.2.1. Influence of S. boulardii fermentation
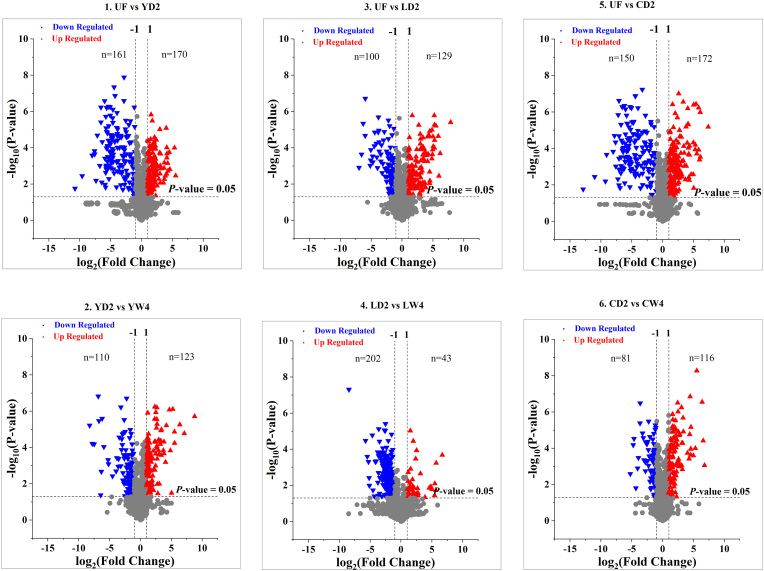
To further investigate the effects of S. boulardii on tea metabolome during fermentation and storage, a volcano-plot was employed to extract the key metabolites that contributed to the differentiation of pairwise samples (unfermented & S. boulardii fermented; 2-day fermented & 4-week storage teas). The number and degree of deviations of metabolites in each comparison group are shown in the form of volcano plots (Figs. 2–1: unfermented & 2-day fermented, Fig. 2–: 2-day fermented & 4-week storage). In total, 1680 metabolites in negative ion mode were evaluated in these two pairwise groups. The volcano plot analysis was performed with fold change > 2 and p < 0.05 using the ANOVA: 331 metabolites including organic acids, catechins, alkaloids and glycosides, etc. were significantly different in S. boulardii fermented and unfermented teas; further, 161 of 331 metabolites showed significantly lower levels in S. boulardii fermented tea and 170 metabolites exhibited higher levels. For the pair of 2-day fermented and 4-week stored teas (Fig. 2–), the number of down and up regulated metabolites was 110 and 123 respectively, which were less than that in Figs. 2–1.
Fig. 2.
Volcano-plots of unfermented, fermented and stored tea samples; (2–1)- unfermented & 2-day S. boulardii fermented; (2-2)-2-day S. boulardii fermented & 4-week S. boulardii storage; fermented and stored tea samples, (2–3)-unfermented & 2-day L. plantarum fermented; (2–4)-2-day L. plantarum fermented & 4-week L. plantarum storage; (2–5)-unfermented & 2-day co-culture fermented; (2–6)-2-day co-culture fermented & 4-week co-culture storage.
Table 1 shows the tentatively identified compounds with VIP > 1, fold change ≥ 2 and p < 0.05 in unfermented, S. boulardii fermented and stored teas. These metabolites were confirmed by comparing the RT and MS/MS fragment patterns with authentic standards and metabolomics databases. From Table 1, glucose, sucrose, fumaric acid, malic acid and most of the flavonol or flavone glycosides such as quercetin 3-O-galactosyl-rutinoside, kaempferol 3-O-galactosyl-rutinoside, kaempferol-3-O-rutinoside, kaempferol-3-O-glucoside, gallic acid hexoside and aroma glycoside (2-phenylethyl-6-O-β-D-xylopyranosyl-β-D-glucopyranoside) were reduced significantly during the two days fermentation, while the cellular metabolic intermediates including guanosine 5′-monophosphate, adenosine 3′-monophosphate, adenosine 3':5′-cyclicmonophosphate, and 2′-deoxyguanosine 5′-monophosphate were clearly generated at higher levels during fermentation and storage. The highest contents of these monophosphates appeared at the day-2 fermentation time. Meanwhile, some flavone aglycones such as eriodictyol, luteolin, naringenin, apigenin, kaempferol and flavone glycosides increased during fermentation and storage.
Table 1.
Compounds with VIP > 1, fold change ≥ 2 and p < 0.05 in unfermented, S. boulardii fermented and stored teas.
| RT | M/Z | Compounds | ANOVA p-value | Fold change (Max/Min) |
|---|---|---|---|---|
| Down regulated | ||||
| 0.45 | 275.0157 | 6-Phosphogluconic acid | 0.001 | 2.72 |
| 0.46 | 165.0412 | 3-Methylxanthine | 0.001 | 2.54 |
| 0.46 | 280.1030 | 1-Methyladenosine | <0.001 | 28.00 |
| 0.46 | 179.0565 | Glucose | <0.001 | 3.48 |
| 0.47 | 341.1086 | Sucrose | <0.001 | 20.05 |
| 0.49 | 115.0041 | Fumaric acid | <0.001 | 2.63 |
| 0.49 | 133.0146 | Malic acid | <0.001 | 3.60 |
| 0.53 | 331.0678 | Gallic acid hexoside | 0.001 | 2.02 |
| 0.70 | 455.0980 | Riboflavin-5′-monophosphate | <0.001 | 38.50 |
| 0.73 | 290.0898 | N-Fructosyl pyroglutamate | <0.001 | 2.90 |
| 0.76 | 441.1104 | (−)-Catechin gallate (CG)a | 0.01 | 2.69 |
| 0.83 | 202.1114 | N6-(delta2-isopentenyl)-adenine | <0.001 | 78.67 |
| 0.84 | 292.1400 | N-Fructosyl isoleucine | <0.001 | 285.83 |
| 1.47 | 451.1260 | Aspalathin | 0.001 | 76.05 |
| 1.59 | 462.0967 | 7-Methylthioheptyl glucosinolate | 0.001 | 228.43 |
| 8.32 | 337.0910 | 4-O-p-Coumaroylquinic acid | 0.005 | 2.00 |
| 9.22 | 577.1315 | Procyanidin B1 | 0.001 | 2.08 |
| 10.18 | 635.0922 | 1,3,6-tri-O-Galloylglucose | 0.001 | 2.45 |
| 10.33 | 491.1418 | 4-Acetyl-2-hydroxyphenyl 2-O-beta-D-Fructofuranosyl-beta-D-xylopyranoside | <0.001 | 2.00 |
| 10.35 | 649.1392 | Flavonoid-7-O-glucuronides | <0.001 | 16.45 |
| 10.39 | 461.1643 | 2-Phenylethyl-6-O-β-D-xylopyranosyl- β-D-glucopyranoside | <0.001 | 2.00 |
| 10.62 | 771.2018 | Quercetin 3-O-galactosyl-rutinoside | 0.001 | 1.98 |
| 11.07 | 755.2063 | Kaempferol 3-O-galactosyl-rutinoside | 0.001 | 1.97 |
| 11.61 | 739.2065 | Kaempferol-3-galactoside-6″-rhamnoside-3'''-rhamnoside | <0.001 | 1.98 |
| 11.66 | 593.1509 | Kaempferol-3-O-rutinosidea | 0.002 | 1.96 |
| 11.79 | 447.0937 | Kaempferol-3-O-glucosidea | <0.001 | 2.25 |
| 13.70 | 306.0534 | 2-Deoxycytidine-5-monophosphoric acid (dCMP) | <0.001 | 76.24 |
| Up regulated | ||||
| 0.46 | 235.0626 | Austadiol | <0.001 | 38.54 |
| 0.48 | 362.0484 | Guanosine 5′-monophosphate (GMP) | 0.014 | 1.98 |
| 0.48 | 346.0538 | Adenosine 3′-monophosphate (AMP) | 0.001 | 1.97 |
| 0.50 | 173.0940 | Gly-Val | 0.001 | 49.87 |
| 0.69 | 328.0457 | Adenosine 3’:5′-cyclicmonophosphate | <0.001 | 7.69 |
| 0.69 | 346.0545 | 2′-Deoxyguanosine 5′-monophosphate (dGMP) | <0.001 | 19.32 |
| 0.75 | 117.0198 | Succinic acid | <0.001 | 4.10 |
| 9.34 | 416.1340 | Sorbicillactone A | 0.004 | 3.07 |
| 11.76 | 767.1836 | Unknown | 0.001 | 4.48 |
| 11.78 | 303.0503 | Haemoventosine | 0.002 | 27.37 |
| 12.49 | 389.0842 | 5,7-dihydroxy-2-(3-hydroxy-4,5-dimethoxyphenyl)-3,6-dimethoxy-4H-chromen-4-one | <0.001 | 3.92 |
| 13.00 | 287.0566 | Eriodictyola | <0.001 | 18.72 |
| 13.10 | 301.0333 | Quercetina | <0.001 | 2.04 |
| 13.15 | 285.0409 | Luteolina | 0.004 | 1.98 |
| 13.84 | 271.0619 | Naringenina | <0.001 | 5.84 |
| 13.85 | 269.0428 | Apigenina | <0.001 | 2.25 |
| 13.88 | 285.0410 | Kaempferola | 0.001 | 9.68 |
| 14.89 | 593.0840 | Biflavonoids | <0.001 | 5.82 |
| 14.96 | 425.0784 | Epiafzelechin 3-gallate | 0.001 | 11.26 |
| 15.50 | 577.2098 | Apigenin-8-C-glucoside-2′-rhamnoside | 0.001 | 9.40 |
| 15.66 | 595.0853 | Phenolic glycosides | 0.001 | 7.87 |
| 17.13 | 543.1536 | 4,5-Diferuloylquinic acid | 0.046 | 3.66 |
Note.
Confirmed with authentic standards.
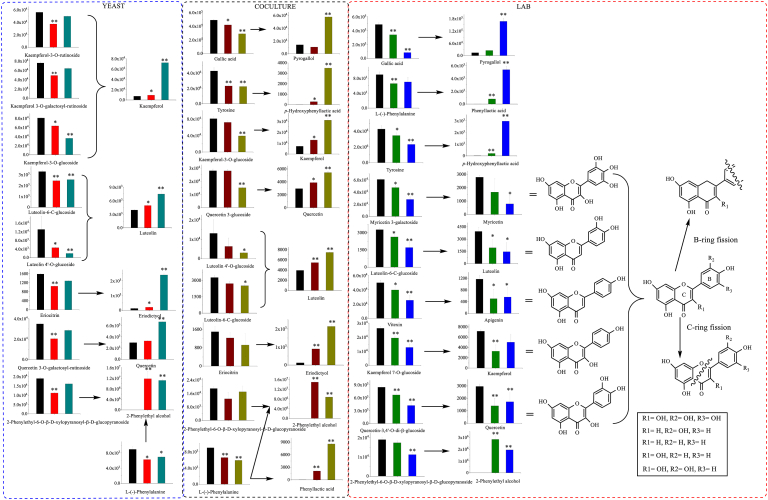
Sugars (sucrose and glucose) as the major energy sources were nearly used up in the exponential stage to support S. boulardii growth. When the available nutrients were exhausted by proliferating S. boulardii cells, they entered a stationary phase (Herman, 2002). The biochemical characteristics of the stationary phase S. boulardii cells would change since the cellular energy-consuming metabolic activity decreased to save ATP for energy-dependent stress responses (Zakrajšek et al., 2011). The volcano plot of fermented and stored teas (Fig. 2–) showed that the number of metabolites with significant changes was far less than that in the pair of unfermented and 2-day fermented tea samples. This result was in line with the decline of cell metabolism during the stationary phase (Zakrajšek et al., 2011). The decreases of flavone glycosides and the increases of flavone aglycones could be due to the S. boulardii glycosidase that broke the glycosidic bond to generate corresponding secondary metabolites (Wilkowska and Pogorzelski, 2017). Besides, several flavone glycosides were also accumulated during the long-time storage and these flavone glycosides could originate from other flavone di- and/or tri-glycosides that lost one or more glycosyl groups via glycosidase-mediated hydrolysis (Fig. 3) (Dai et al., 2015). From the UPLC-Q-TOF-MS analysis, the glycoside of 2-phenylethanol (2-phenylethyl-6-O-β-D-xylopyranosyl- β-D-glucopyranoside) was also found (Fig. 3), which further verified the results discussed in our previous study that the key flavor compound 2-phenylethanol was not only from phenylalanine metabolism but also from glycoside precursor hydrolysis (Wang et al., 2020).
Fig. 3.
Metabolic pathway changes in metabolites caused by tea fermentation. The black, red, dark cyan, wine, dark yellow, green and blue columns represent the mass intensity of metabolites in yeast, coculture, and LAB fermented tea samples, respectively; (*p < 0.05; ** < 0.01). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2.2. Influence of L. plantarum fermentation
The effects of L. plantarum fermentation and storage were evaluated in the same way as S. boulardii. A total of 1680 peaks obtained from UPLC-Q-TOF-MS were subjected to multivariate analysis to extract the key differences among unfermented, fermented and stored teas. The pairwise comparisons of two groups (unfermented & 2-day fermented teas - Fig. 2, Fig. 3; 2-day fermented & 4-week storage teas - Figs. 2–4) were performed by volcano plots to unearth the critical compounds. In total, 229 compounds were significantly different in L. plantarum fermented and unfermented teas; 129 of 229 metabolites showed significantly higher levels and 100 metabolites showed lower levels in L. plantarum fermented tea (Fig. 2, Fig. 3). For the pair of 2-day fermented and 4-week stored teas (Figs. 2–4), there were 202 metabolites that decreased and only 43 metabolites increased in 4-week stored tea. The significantly changed (fold change ≥ 2 and p < 0.05) metabolites in unfermented, L. plantarum fermented and stored teas are presented in Table 2. These metabolites were tentatively identified by comparing the MS/MS data of standards and public data base.
Table 2.
Compounds with VIP > 1, fold change ≥ 2 and p < 0.05 in unfermented, L. plantarum fermented and stored teas.
| RT | M/Z | Compound | ANOVA p-value | Fold change (Max/Min) |
|---|---|---|---|---|
| Down regulated | ||||
| 0.45 | 275.0157 | 6-Phosphogluconic acid | <0.001 | 13.06 |
| 0.46 | 165.0412 | 3-Methylxanthine | 0.001 | 4.20 |
| 0.46 | 179.0565 | Glucose | <0.001 | 3.15 |
| 0.46 | 195.0530 | Gluconic acid | 0.007 | 2.00 |
| 0.47 | 191.0565 | Citric acid | <0.001 | 6.50 |
| 0.49 | 115.0041 | Fumaric acid | 0.001 | 3.54 |
| 0.72 | 331.0672 | Gallic acid hexoside | 0.02 | 2.32 |
| 0.81 | 282.0844 | Guanosine | <0.001 | 33.46 |
| 0.84 | 292.1400 | N-Fructosyl isoleucine | <0.001 | 4.78 |
| 0.85 | 169.0150 | Gallic acida | <0.001 | 5.10 |
| 0.86 | 484.1389 | Uridine 5-triphosphate | 0.024 | 19.97 |
| 8.19 | 319.0434 | Unknown | <0.001 | 8.00 |
| 8.57 | 633.0711 | Hydrolyzable tannins | 0.001 | 71.10 |
| 9.01 | 337.0926 | 5-O-p-Coumaroylquinic acid | 0.001 | 2.59 |
| 9.22 | 577.1315 | Procyanidin B1 | 0.001 | 2.46 |
| 9.43 | 451.1244 | Aspalathin | 0.001 | 7.19 |
| 10.26 | 625.1407 | Quercetin-3,4′-O-di-beta-glucoside | 0.001 | 1.95 |
| 10.28 | 479.0825 | Myricetin 3-galactoside | 0.008 | 2.27 |
| 10.37 | 447.0920 | Luteolin-6-C-glucoside | 0.045 | 1.97 |
| 10.49 | 635.0885 | 1,3,6-tri-O-galloylglucose | 0.001 | 2.86 |
| 10.50 | 291.1266 | Curvularin | <0.001 | 388.94 |
| 10.62 | 771.2018 | Quercetin 3-O-galactosyl-rutinoside | 0.037 | 1.96 |
| 11.16 | 431.0196 | Vitexina | 0.016 | 2.26 |
| 11.19 | 463.0895 | Hirsutrin | 0.034 | 1.96 |
| 11.60 | 447.0917 | Kaempferol 7-O-glucoside | 0.020 | 2.03 |
| 11.89 | 515.0772 | 3,4-di-O-Caffeoylquinic acida | 0.036 | 33.13 |
| 14.28 | 517.0979 | Unknown | 0.001 | 126.73 |
| 15.30 | 515.1200 | 4,5-Dicaffeoylquinic acida | 0.015 | 61.77 |
| 15.37 | 467.1353 | Unknown | 0.001 | 81.39 |
| 17.10 | 393.1712 | Aloesin | <0.001 | 7.41 |
| Up regulated | ||||
| 0.50 | 173.0940 | Gly-Val | 0.001 | 40.32 |
| 0.54 | 187.0424 | 1-Hydroxy-2-naphthoic acid | <0.001 | 234.66 |
| 0.70 | 455.0980 | Riboflavin-5′-monophosphate | 0.023 | 2.15 |
| 0.96 | 125.0251 | Pyrogallol | <0.001 | 14.20 |
| 2.10 | 181.1510 | p-Hydroxy-D-phenyllactic acid | 0.008 | 97.01 |
| 7.96 | 165.0558 | D-Phenyllactic acid | <0.001 | 124.78 |
| 7.97 | 233.0434 | Anhydrobrazilic Acid | <0.001 | 122.29 |
| 9.31 | 186.1138 | 3-(2-amino-2-oxoethyl)-5-methyl-Hexanoic acid | <0.001 | 9.20 |
| 9.87 | 416.1343 | Sorbicillactone A | <0.001 | 2.31 |
| 17.05 | 337.2048 | Bavachinin A | <0.001 | 13.96 |
| 17.14 | 303.0378 | Thymidine-3′,5′-cyclic monophosphate | 0.01 | 4.55 |
Note.
Confirmed with authentic standards.
As expected, L. plantarum exerted different effects on tea metabolome from S. boulardii, and the differences could be summarized into three points. First, flavone glycosides (quercetin-3,4′-O-di-β-glucoside, quercetin 3-O-galactosyl-rutinoside, myricetin 3-galactoside, luteolin-6-C-glucoside, vitexin (apigenin-8-C-glucoside), kaempferol 7-O-glucoside) (Table 2) decreased significantly in L. plantarum fermented tea infusion; however, the individual flavone glycosides impacted by L. plantarum differed from that in S. boulardii fermentation. The significantly reduced flavone glycosides in L. plantarum fermented tea included both C-glycosides and O-glycosides, whereas only O-glycosides were affected by S. boulardii fermentation (Fig. 3). Microbes can influence the flavonoid glycosides in three ways during fermentation: cell wall absorption, flavonoid glycoside hydrolysis and formation of new metabolites. Both yeast and LAB cell walls can absorb glycosides, but the absorption is species-dependent, which could partially contribute to the variations of flavonoid glycosides in L. plantarum and S. boulardii fermented tea infusions (Devi, Aiyappaa & Waterhouse, 2019; Božič et al., 2020). Glycosidase hydrolysis plays a part in flavonoid glycosides during fermentation and the various changes of flavonoid glycosides in L. plantarum and S. boulardii cultures suggested that the enzyme active sites of glycosidases in L. plantarum and S. boulardii could be originally different or impacted by pH, since the pH of L. plantarum fermented tea was lower (pH ≈ 3.7) than S. boulardii fermented tea (pH ≈ 4.8) (Michlmayr and Kneifel, 2014). In addition, the changes of flavone aglycones exhibited opposite trends to that in S. boulardii fermented tea. For example, myricetin, luteolin, apigenin, kaempferol, and quercetin decreased during L. plantarum fermentation and storage. Following the hydrolysis of flavone glycosides, the flavone aglycones could be further degraded via B-ring and C-ring fission, thus leading to the reduction of flavone aglycones (Fig. 3); this result concurred with the research that LAB could metabolize flavone aglycones during fermentation (Guo et al., 2021).
Second, L. plantarum degraded gallic acid and quinic acid derivatives (3,4-di-O-caffeoylquinic acid and 4,5-dicaffeoylquinic acid), which were not found in S. boulardii fermentation. L. plantarum possesses a decarboxylase that decarboxylates gallic acid to another antibacterial agent - pyrogallol (Reverón, de las Rivas, Matesanz, Muñoz, & López de Felipe, 2015), and pyrogallol was also found increasing during the fermentation and storage (Table 2). The quinic acid derivatives could be converted into caffeic acid and quinic acid by specific esterases of L. plantarum, then caffeic acid and quinic acid could be further oxidized or reduced to corresponding secondary products (Naranjo Pinta et al., 2018).
Third, two food-borne pathogen inhibitors D-phenyllactic acid (PLA) and p-hydroxy-D-phenyllactic acid (OH-PLA) increased by 125-fold and 97-fold after 4 weeks storage in L. plantarum fermented tea. PLA and p-OH-PLA were generated in L. plantarum from the degradation of phenylalanine and tyrosine (Tyr), respectively (Chesters, Walker, O'Hagan and Floss, 1996). These two compounds are known to possess effective antimicrobial activity and have the potential to be used as novel antimicrobial agents in food preservation. As such, L. plantarum fermentation may provide probiotic tea with natural and self-originated preservatives (Dao et al., 2019).
3.2.3. Influence of co-culture fermentation
Both S. boulardii and L. plantarum respectively exerted various influences on the tea metabolome as discussed above. A co-culture of both yeast and LAB was also investigated to uncover the interactive effects of the two microorganisms on tea components during fermentation and storage. Figs. 2–5 and 2-6 illustrate the significantly changed metabolites in co-cultured teas (Figure 2-5-unfermented & 2-day fermented teas, Figs. 2-6-2-day fermented & 4-week stored teas). In general, the number of metabolites which were significantly regulated was far greater in the fermentation stage (Figs. 2–5)than in the storage stage (Figs. 2–6). In total, 322 metabolites with significant changes were revealed in co-culture fermented tea; 150 of them showed lower levels and the other 172 metabolites presented higher levels after 2-day fermentation. For the storage stage (Figs. 2–6), 81 of 197 metabolites were down regulated and 116 metabolites were up regulated in the stored tea. More information on these key metabolites was extracted and is presented in Table 3. The metabolites were identified by comparing the MS/MS data with authentic standards and public data base.
Table 3.
Compounds with VIP > 1, fold change ≥ 2 and p < 0.05 in unfermented, coculture fermented and stored teas.
| RT | M/Z | Compound | ANOVA p-value | Fold change (Max/Min) |
|---|---|---|---|---|
| Down regulated | ||||
| 0.46 | 165.0412 | 3-Methylxanthine | 0.001 | 2.57 |
| 0.46 | 179.0650 | Glucose | <0.001 | 3.75 |
| 0.47 | 341.1086 | Sucrose | <0.001 | 23.48 |
| 0.49 | 115.0041 | Fumaric acid | 0.001 | 4.17 |
| 0.47 | 191.0565 | Citric acid | <0.001 | 3.31 |
| 0.49 | 133.0146 | Malic acid | <0.001 | 6.63 |
| 0.70 | 455.0980 | Riboflavin-5′-monophosphate | <0.001 | 17.29 |
| 0.73 | 290.0898 | N-Fructosyl pyroglutamate | <0.001 | 2.97 |
| 0.81 | 282.0844 | Guanosine | 0.006 | 9.20 |
| 0.83 | 202.1114 | N6-(Delta2-Isopentenyl)-adenine | <0.001 | 64.66 |
| 0.84 | 292.1400 | N-Fructosyl isoleucine | <0.001 | 146.88 |
| 9.22 | 577.1315 | Procyanidin B1 | <0.001 | 2.09 |
| 9.43 | 451.1244 | Aspalathin | <0.001 | 42.13 |
| 9.78 | 387.1655 | 12:4+3O Fatty acyl hexoside | 0.003 | 2.51 |
| 10.49 | 635.0885 | 1,3,6-tri-O-Galloylglucose | <0.001 | 9.73 |
| 11.11 | 463.0892 | Quercetin 3-glucoside | 0.05 | 1.95 |
| 11.31 | 477.0622 | Indolylmethyl glucosinolate + 1MeO | 0.004 | 2.07 |
| 11.61 | 739.2065 | Kaempferol-3-galactoside-6″-rhamnoside-3’’’-rhamnoside | 0.001 | 1.95 |
| 11.79 | 447.0937 | Kaempferol-3-O-glucosidea | <0.001 | 1.95 |
| 13.7 | 306.0534 | 2′-Deoxycytidine 5′-monophosphate (dCMP) | <0.001 | 107.51 |
| Up regulated | ||||
| 0.44 | 241.0832 | Thymidine | 0.001 | 4.01 |
| 0.48 | 346.0532 | Adenosine 3′-monophosphate | 0.001 | 2.04 |
| 0.48 | 362.0484 | Guanosine 5′-monophosphate | 0.001 | 2.29 |
| 0.50 | 173.0940 | Gly-Val | <0.001 | 58.93 |
| 0.69 | 328.0457 | Adenosine 3’:5′-cyclicmonophosphate | <0.001 | 10.72 |
| 0.69 | 346.0545 | Deoxyguanosine monophosphate (dGMP) | <0.001 | 23.46 |
| 0.75 | 117.0198 | Succinic acid | <0.001 | 4.98 |
| 2.10 | 181.0510 | p-Hydroxyphenyllactic acid | <0.001 | 112.57 |
| 7.96 | 165.0558 | Phenyllactic acid | <0.001 | 186.94 |
| 7.97 | 233.0434 | Anhydrobrazilic Acid | <0.001 | 210.08 |
| 9.31 | 186.1138 | 3-(2-Amino-2-oxoethyl)-5-methyl-hexanoic acid | <0.001 | 11.30 |
| 9.34 | 416.134 | Sorbicillactone A | 0.001 | 3.41 |
| 11.78 | 303.0503 | Haemoventosine | <0.001 | 16.11 |
| 12.49 | 389.0842 | 6-O-methylated Flavonoids | <0.001 | 2.08 |
| 13.00 | 287.0566 | Eriodictyola | <0.001 | 17.88 |
| 13.10 | 301.0333 | Quercetina | <0.001 | 2.00 |
| 13.15 | 285.0409 | Luteolina | 0.001 | 2.02 |
| 13.29 | 449.0735 | Myricetin-3-O-xyloside | 0.002 | 6.75 |
| 13.84 | 271.0619 | Naringenina | <0.001 | 5.21 |
| 13.85 | 269.0425 | Apigenina | <0.001 | 2.00 |
| 13.88 | 285.0410 | Kaempferola | <0.001 | 3.89 |
| 14.89 | 593.0840 | Biflavonoids | <0.001 | 9.47 |
| 14.96 | 425.0784 | Epiafzelechin 3-gallate | <0.001 | 14.91 |
Note.
Confirmed with authentic standards.
Co-cultured tea shared the metabolite profiles with that of both S. boulardii and L. plantarum to some degree; meanwhile, it also showed specific metabolic characteristics. The conversion of sugars and organic acids and the production of cellular metabolic intermediates (the monophosphates) in co-cultured teas showed similar trends to that in the monocultures of S. boulardii and L. plantarum. The flavone or flavonoid glycosides were more stable in the co-culture than in the monocultures, since less glycosides decreased significantly in the co-culture. In addition, several flavone aglycones including eriodictyol, quercetin, luteolin, naringenin, apigenin and kaempferol accumulated in the co-cultured tea (Fig. 3). As discussed in Sections 3.2.1, 3.2.2, both yeast and LAB can change the flavonoid profile by cell wall absorption, glycosidase hydrolysis and generation of new metabolites. Besides, LAB can also degrade flavone aglycones by breaking B-ring and C-ring (Guo et al., 2021). In the co-culture, less flavonoid glycosides were down-regulated than in yeast and LAB monocultures, but more flavone aglycones were up-regulated than in the L. plantarum monoculture. Flavonoid glycosides could be absorbed by microbe cell walls or hydrolyzed by glycosidases to release flavone aglycones and sugar. The changes of flavonoid glycosides in the co-culture suggested that the cell wall absorption and glycosidase activities could be inhibited during co-culture fermentation. Unlike the decreases in L. plantarum monoculture, flavone aglycones accumulated in the co-culture even L. plantarum existed, suggesting that the flavones-degrading enzymes in L. plantarum could be inhibited in the co-culture.
D- Phenyllactic and p-hydroxy-D-phenyllactic acids are the products of amino acids decomposition (Phe and Tyr) of L. plantarum fermentation (Dao et al., 2019). These two aromatic acids were also found increased by 187 and 113-fold in the co-cultured tea. As discussed in L. plantarum fermentation, the increase of PLA and p-OH-PLA was 125-fold and 97-fold, respectively. Hence, the production of PLA and p-OH-PLA was enhanced significantly in the co-culture. The higher contents of PLA and p-OH-PLA in the co-culture could be due to more substrates (phenylalanine and tyrosine) provided by yeast, as S. boulardii cells contain abundant amino acids that can be released, which can support the metabolism of L. plantarum (Ponomarova et al., 2017). Most importantly, L. plantarum survived much longer in the co-culture and this would enable production of more PLA and p-OH-PLA.
Another interesting finding was the gallic acid changes in different fermentation systems. Unlike the remarkable decrease of gallic acid in L. plantarum monoculture, the gallic acid content was not significantly influenced by co-culturing. Meanwhile, the decarboxylated product of gallic acid - pyrogallol showed no significant change in the co-culture, which may suggest that the expression of decarboxylase related genes or decarboxylase activity could be inhibited in the co-culture (Reverón et al., 2015) and further research is needed to investigate the mechanism of decarboxylase expression.
In summary, this metabolomics study on tea revealed the significant impact of different cultures and their combination on the tea constituents with the discovery of numerous previously unreported bioactive compounds, especially flavonoids and amino acid derivatives. These findings would lay the foundation for developing novel fermented functional tea beverages, particularly in the emerging field of probiotics and postbiotics.
4. Conclusion
Compounds from teas fermented with different probiotic microorganisms were obtained using UPLC-Q-TOF-MS analysis, through which the differential metabolites contributing to their special metabolic profiles were identified, such as PLA and p-OH-PLA in L. plantarum associated samples, flavone glycosides and aglycones in all tea infusions. Some flavone glycosides in S. boulardii fermented tea decreased significantly and flavone aglycones and other flavone glycosides were generated during fermentation and storage. In L. plantarum fermented tea, the metabolic profile of flavone glycosides was different from that in S. boulardii fermented tea; both C-glycosides and O-glycosides were hydrolyzed and the flavone aglycones were further degraded in L. plantarum monoculture. The profiles of flavone glycosides and aglycones in co-cultured tea differed from that in S. boulardii or L. plantarum fermented tea; less glycosides were degraded but a greater number of aglycones produced in the co-culture. In general, UPLC-Q-TOF-MS analysis coupled with chemometrics presented great potential for the untargeted metabolic analysis of fermented teas.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Rui Wang: Formal analysis, Writing – original draft, conceived and designed the experiments, performed the experiments, analysed the data wrote the paper. Jingcan Sun: Formal analysis, performed the analysed the dataexperiments. Benjamin Lassabliere: conceived and designed the experiments. Bin Yu: performed the experiments.. Shao Quan Liu: Writing – original draft, conceived and designed the experiments, wrote the paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Aspri M., Papademas P., Tsaltas D. Review on non-dairy probiotics and their use in non-dairy based products. Fermentatio. 2020;6(1):30. doi: 10.3390/fermentation6010030. [DOI] [Google Scholar]
- Bevilacqua M., Bro R., Marini F., Rinnan Å., Rasmussen M.A., Skov T. Recent chemometrics advances for foodomics. Trac. Trends Anal. Chem. 2017;96:42–51. doi: 10.1016/j.trac.2017.08.011. [DOI] [Google Scholar]
- Božič J.T., Butinar L., Albreht A., Vovk I., Korte D., Vodopivec B.M. The impact of Saccharomyces and non-Saccharomyces yeasts on wine colour: a laboratory study of vinylphenolic pyranoanthocyanin formation and anthocyanin cell wall adsorption. LWT - Food Sci. Technol. (Lebensmittel-Wissenschaft -Technol.) 2020;123:109072. doi: 10.1016/j.lwt.2020.109072. 0023–6438. [DOI] [Google Scholar]
- Chan A.M.Z., Toh M., Liu S.Q. Growth, survival, and metabolic activities of probiotics Lactobacillus rhamnosus GG and Saccharomyces cerevisiae var. boulardii CNCM-I745 in fermented coffee brews. Int. J. Food Microbiol. 2021;350:109229. doi: 10.1016/j.ijfoodmicro.2021.109229. [DOI] [PubMed] [Google Scholar]
- Chesters N.C.J.E., Walker K., O'Hagan D., Floss H.G. The biosynthesis of tropic acid: a reevaluation of the stereochemical course of the conversion of phenyllactate to tropate in Datura stramonium. J. Am. Chem. Soc. 1996;118(4):925–926. doi: 10.1021/ja9531485. [DOI] [Google Scholar]
- Dai W., Qi D., Yang T., Lv H., Guo L., Zhang Y., Lin Z. Nontargeted analysis using ultraperformance liquid chromatography-time-of-mass spectrometry uncovers the effects of harvest season on the metabolites and taste quality of tea (Camellia sinensis L.) J. Agric. Food Chem. 2015;63(44):9869–9878. doi: 10.1021/acs.jafc.5b03967. [DOI] [PubMed] [Google Scholar]
- Dao Y., Zhang K., Lu X., Lu Z., Liu C., Liu M., Luo Y. Role of glucose and 2-oxoglutarate/malate translocator (OMT1) in the production of phenyllactic acid and p-hydroxyphenyllactic acid, two food-borne inhibitors. J. Agric. Food Chem. 2019;67(20):5820–5826. doi: 10.1021/acs.jafc.9b01444. [DOI] [PubMed] [Google Scholar]
- Ge Y., Bian X., Sun B., Zhao M., Ma Y., Tang Y., Wu J.L. Dynamic profiling of phenolic acids during pu-erh tea fermentation using derivatization liquid chromatography-mass spectrometry approach. J. Agric. Food Chem. 2019;67(16):4568–4577. doi: 10.1021/acs.jafc.9b00789. [DOI] [PubMed] [Google Scholar]
- Guo X., Guo A., Li E. Biotransformation of two citrus flavanones by lactic acid bacteria in chemical defined medium. Bioproc. Biosyst. Eng. 2021;44(2):235–246. doi: 10.1007/s00449-020-02437-y. [DOI] [PubMed] [Google Scholar]
- He G.Y., Han X., Han M., Qiu S.T., Li Y., Zhang S.l., Chen X. UPLC-QTOF-MS analysis of 13 isomers of polyphenols. Chin. J. Synth. Chem. 2020;28(10):9. [Google Scholar]
- Herman P.K. Vol. 5. Elsevier Ltd; London: 2002. pp. 602–607. (Stationary Phase in Yeast). [DOI] [PubMed] [Google Scholar]
- Jing J., Shi Y., Zhang Q., Wang J., Ruan J. Prediction of Chinese green tea ranking by metabolite profiling using ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC–Q-TOF/MS) Food Chem. 2017;221:311–316. doi: 10.1016/j.foodchem.2016.10.068. [DOI] [PubMed] [Google Scholar]
- Kakisu E., Abraham A.G., Tironi Farinati C., Ibarra C., De Antoni G.L. Lactobacillus plantarum isolated from kefir protects vero cells from cytotoxicity by type-II shiga toxin from Escherichia coli O157:H7. J. Dairy Res. 2012;80(1):64–71. doi: 10.1017/S0022029912000659. [DOI] [PubMed] [Google Scholar]
- Li Q., Huang J., Li Y., Zhang Y., Luo Y., Chen Y., Liu Z. Fungal community succession and major components change during manufacturing process of Fu brick tea. Sci. Rep. 2017;7(1):6947–6949. doi: 10.1038/s41598-017-07098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlmayr H., Kneifel W. β-Glucosidase activities of lactic acid bacteria: mechanisms, impact on fermented food and human health. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2014;352(1):1–10. doi: 10.1111/1574-6968.12348. [DOI] [PubMed] [Google Scholar]
- Naranjo Pinta M., Montoliu I., Aura A.M., Seppänen-Laakso T., Barron D., Moco S. In vitro gut metabolism of [U-13C]-quinic acid, the other hydrolysis product of chlorogenic acid. Mol. Nutr. Food Res. 2018;62(22):1800396. doi: 10.1002/mnfr.201800396. [DOI] [PubMed] [Google Scholar]
- Nayak S.K. In: Probiotics: Biology, Genetics and Health Aspects. Liong M.T., editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 2011. Biology of eukaryotic probiotics; pp. 29–55. [Google Scholar]
- Ponomarova O., Gabrielli N., Sévin D.C., Mülleder M., Zirngibl K., Bulyha K., Patil K.R. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Systems. 2017;5(4):345–357. doi: 10.1016/j.cels.2017.09.002. e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverón I., de las Rivas B., Matesanz R., Muñoz R., López de Felipe F. Molecular adaptation of Lactobacillus plantarum WCFS1 to gallic acid revealed by genome-scale transcriptomic signature and physiological analysis. Microb. Cell Factories. 2015;14(1) doi: 10.1186/s12934-015-0345-y. 160-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W., Zhou Z., Zhao B., Wei T. Simultaneous determination of eight catechins and four theaflavins in green, black and oolong tea using new HPLC-MS-MS method. J. Pharmaceut. Biomed. Anal. 2016;131:140–145. doi: 10.1016/j.jpba.2016.08.020. [DOI] [PubMed] [Google Scholar]
- Wang Q., Gong J., Chisti Y., Sirisansaneeyakul S. Fungal isolates from a Pu-erh type tea fermentation and their ability to convert tea polyphenols to theabrownins. J. Food Sci. 2015;80(4):M809–M817. doi: 10.1111/1750-3841.12831. [DOI] [PubMed] [Google Scholar]
- Wang R., Sun J., Lassabliere B., Yu B., Liu S.Q. Biotransformation of green tea (Camellia sinensis) by wine yeast Saccharomyces cerevisiae. J. Food Sci. 2020;85(2):306–315. doi: 10.1111/1750-3841.15026. [DOI] [PubMed] [Google Scholar]
- Wilkowska A., Pogorzelski E. Aroma enhancement of cherry juice and wine using exogenous glycosidases from mould, yeast and lactic acid bacteria. Food Chem. 2017;237:282–289. doi: 10.1016/j.foodchem.2017.05.120. [DOI] [PubMed] [Google Scholar]
- Zakrajšek T., Raspor P., Jamnik P. Saccharomyces cerevisiae in the stationary phase as a model organism-characterization at cellular and proteome level. J. Proteonomics. 2011;74(12):2837–2845. doi: 10.1016/j.jprot.2011.06.026. [DOI] [PubMed] [Google Scholar]
- Zhang L., Li N., Ma Z.Z., Tu P.F. Comparison of the chemical constituents of aged pu-erh tea, rpened pu-erh tea, and other teas using HPLC-DAD-ESI-MSn. J. Agric. Food Chem. 2011;59(16):8754–8760. doi: 10.1021/jf2015733. [DOI] [PubMed] [Google Scholar]
- Zhou P., Hu O., Fu H., Ouyang L., Gong X., Meng P., Wang Y. UPLC-Q-TOF/MS-based untargeted metabolomics coupled with chemometrics approach for tieguanyin tea with seasonal and year variations. Food Chem. 2019;283:73–82. doi: 10.1016/j.foodchem.2019.01.050. [DOI] [PubMed] [Google Scholar]