Abstract
Although the laboratory information system has largely solved the problem of storing anatomic pathology reports and disseminating their contents across the healthcare system, the retrospective query of anatomic pathology reports remains an area for improvement across laboratory information system vendors. Our institution desired the ability to query our repository of anatomic pathology reports for clinical, operational, research, and educational purposes. To address this need, we developed a full-text anatomic pathology search tool using the business intelligence software, Tableau. Our search tool allows users to query the 333,685 anatomic pathology reports from our institutional clinical relational database using the business intelligence tool’s built-in regular expression functionality. Users securely access the search tool using any web browser, thereby avoiding the cost of installing or maintaining software on users’ computers. This tool is laboratory information system vendor agnostic and as many institutions already subscribe to business intelligence software, we believe this solution could be easily reproduced at other institutions and in other clinical departments.
Introduction
Electronic health records (EHRs) are a detailed source of demographic, clinical, and administrative information. The secondary use of EHR data can improve patient outcomes, enhance the quality of care, reduce cost, and advance the field of medicine.1 An unmet challenge in utilizing EHR data includes rapid searching. While efficient searching of electronic healthcare data is a problem that spans across all disciplines in medicine and all health record vendors, one particularly challenging entity is the unstructured, free-text anatomic pathology report. Not only are pathology reports difficult to access but retrieving structured information from the data within the reports remains challenging.
As early adopters of computers in medicine,2 pathology departments have electronically stored anatomic pathology reports for decades. However, many of these reports remain inaccessible. Regulations in the United States require laboratories to retain anatomic pathology reports for a minimum of 10 years,3 and many academic institutions keep reports indefinitely. Currently, no straightforward, generally accepted approaches to accessing and searching electronically stored anatomic pathology reports exist. Access is further complicated when institutions transition between laboratory information systems (LISs) and vendors. Approaches to facilitate access have included paying LIS vendors or other third parties to facilitate access to archived reports or to develop homegrown solutions. Approaches vary based upon institutions’ ability to financially invest in outsourcing the solution or leveraging in-house talent. Institutions unable to develop solutions lack the ability to query reports beyond a specific date in time. These barriers to accessing and searching archival reports limit the contribution that these reports and patients’ cases can have on the continuously evolving body of pathology knowledge.
Unfortunately, accessing individual pathology cases is only one of the barriers for the information retrieval problem. Although it is intuitive and well understood in the medical community that recording structured and discrete data into the EHR improves standardization of documentation and ease of data extraction, there remains a well-founded desire for unstructured, free-text documentation to preserve the nuance, uniqueness, and complexity of each patient and case4 and to reduce documentation burden.5 In anatomic pathology, great advances have been made in the standardization of data elements captured within anatomic pathology reports. One widely adopted initiative, the College of American Pathologists (CAP) Cancer Protocol Template, guides the minimum required elements for inclusion within reports for malignant tumors across all organ systems.6 These required elements assist with tumor staging, treatment selection, and fulfill reporting requirements to government registries. While CAP cancer templates have improved the standardization and robustness of data captured for malignant diagnoses, variability remains. The structure of an anatomic pathology report still varies across institutions (i.e., what information is or is not included in a final diagnosis, microscopic diagnosis, comment, etc.). Further, use of synonymous diagnoses varies across, and even within, institutions. Variability in documentation of diagnoses is especially pronounced when a diagnosis is not well defined, not well understood, or there is uncertainty in the diagnosis. The lack of retrieval and the variability of diagnosis documentation have made querying the anatomic pathology report a particularly daunting task.
Since the inception of capturing clinical data electronically, there has been a desire to query and retrieve these data. Solutions for querying and retrieving data from anatomic pathology reports range from manual to automated and from home-grown to vendor developed. A widely utilized manual solution entails pathologists keeping personal logs or databases of cases and manually abstracting additional information on their own or with the help of medical students, residents, or fellows. Some institutions employ data analysts capable of querying clinical data warehouses for anatomic pathology reports and clinical data on behalf of requesting pathologists. Using more automated techniques, numerous publications detail querying and extracting data from anatomic pathology reports using natural language processing (NLP), machine learning, and artificial intelligence for research questions and various initiatives.7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 Vendors have also provided an ability to query reports within their LIS with built-in NLP search functionalities.21 Additionally, different home-grown search engine tools have been developed to query an institution’s EHR data including clinical notes, radiology reports, and pathology reports.21, 22, 23, 24
Our pathology department at a large academic medical center was transitioning from the LIS by Sunquest CoPath to the Epic Beaker LIS. The faculty desired to maintain the ability for pathologists and other authorized users to perform retrospective free-text searches on anatomic pathology reports—a functionality available in CoPath (A best of breed solution), but not Epic Beaker (an EHR integrated solution). The department desired a search tool that was cost-effective, easy to develop, easy to use, easy to maintain, and highly functional. In this technical note, we detail the development and implementation of an anatomic pathology report search engine developed using a business intelligence tool. This tool is LIS agnostic, can query reports that have been generated across LIS transitions, and can be deployed without the need to maintain a completely homegrown infrastructure.
Technical Background
Physicians at our institution provide care in more than five hospitals and a multitude of outpatient settings. The pathology department annually processes approximately 90,000 anatomic pathology cases comprised of adult surgical pathology, hematopathology, and cytopathology cases.
Our institution had used SunQuest CoPath as the LIS and pathologists, trainees, and other authorized users were accustomed to using CoPath’s built-in NLP functionality to perform retrospective text searches on anatomic pathology reports. After our laboratory’s transition to a new LIS (Epic Beaker), we desired to maintain the ability to do free-text retrospective searches.
Stakeholder Requirements
To understand the staff’s needs for a search tool better, pathologists, pathology trainees, and department leaders were surveyed about their use of a search tool and the minimum necessary and desired features (Table 1). Pathologist users delineated numerous operational, educational, and educational use cases.
Table 1.
Stakeholder input for use cases and minimum necessary features for an anatomic pathology search tool.
| Use cases and minimum necessary features |
|---|
| Operational use cases |
| To search for other/similar cases whereas signing out a difficult case to see how prior cases were handled / worded / and which IHCs were used |
| To search for other/similar cases whereas signing out a difficult case to see how prior cases were handled / worded / and which IHCs were used |
| To demonstrate the ability to find specific cases during a CAP inspection |
| Educational use cases |
| To search for cases for study sets |
| For trainees to search for cases from a particular attending to see the attending’s preferred reporting style |
| Research use case |
| To search for cases for IRB approved research projects |
| Minimum necessary features |
| Search functionality |
| Free-text search of anatomic pathology notes |
| Complex Search: Multiple ‘And’ or ‘Or’ |
| Implement negation |
| Search by disease (ICD-9 and ICD-10) |
| Filter by date |
| Filter by Pathologist |
| Filter by case type |
| Reasonable search time |
| Ease of use / intuitive functionality |
| Easy to review results |
| Download result data into an Excel format |
| Security |
| Multi-User access |
| Low cost / time to implement |
| Low cost / time to maintain |
| Low cost / time to train new users |
Development Approach
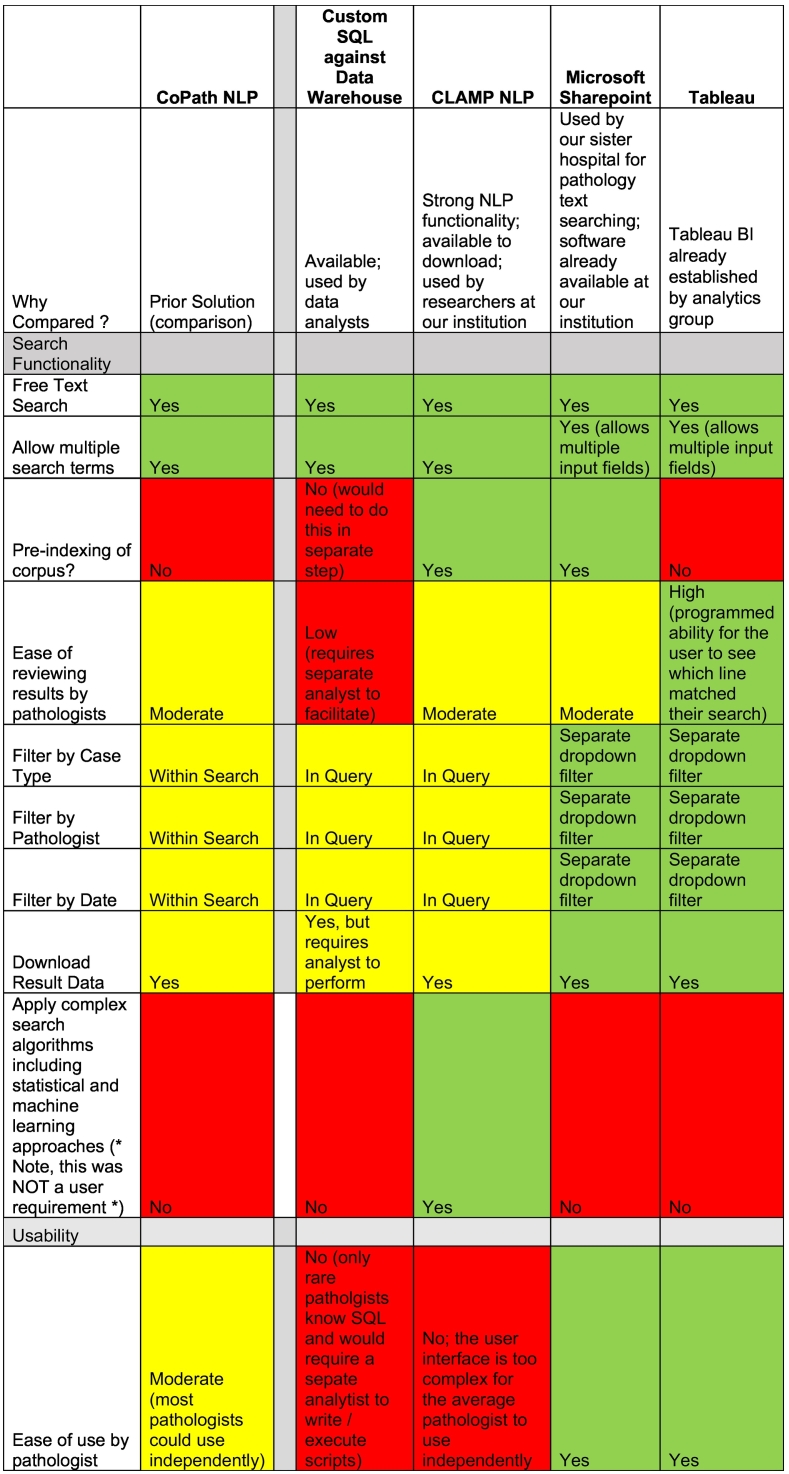
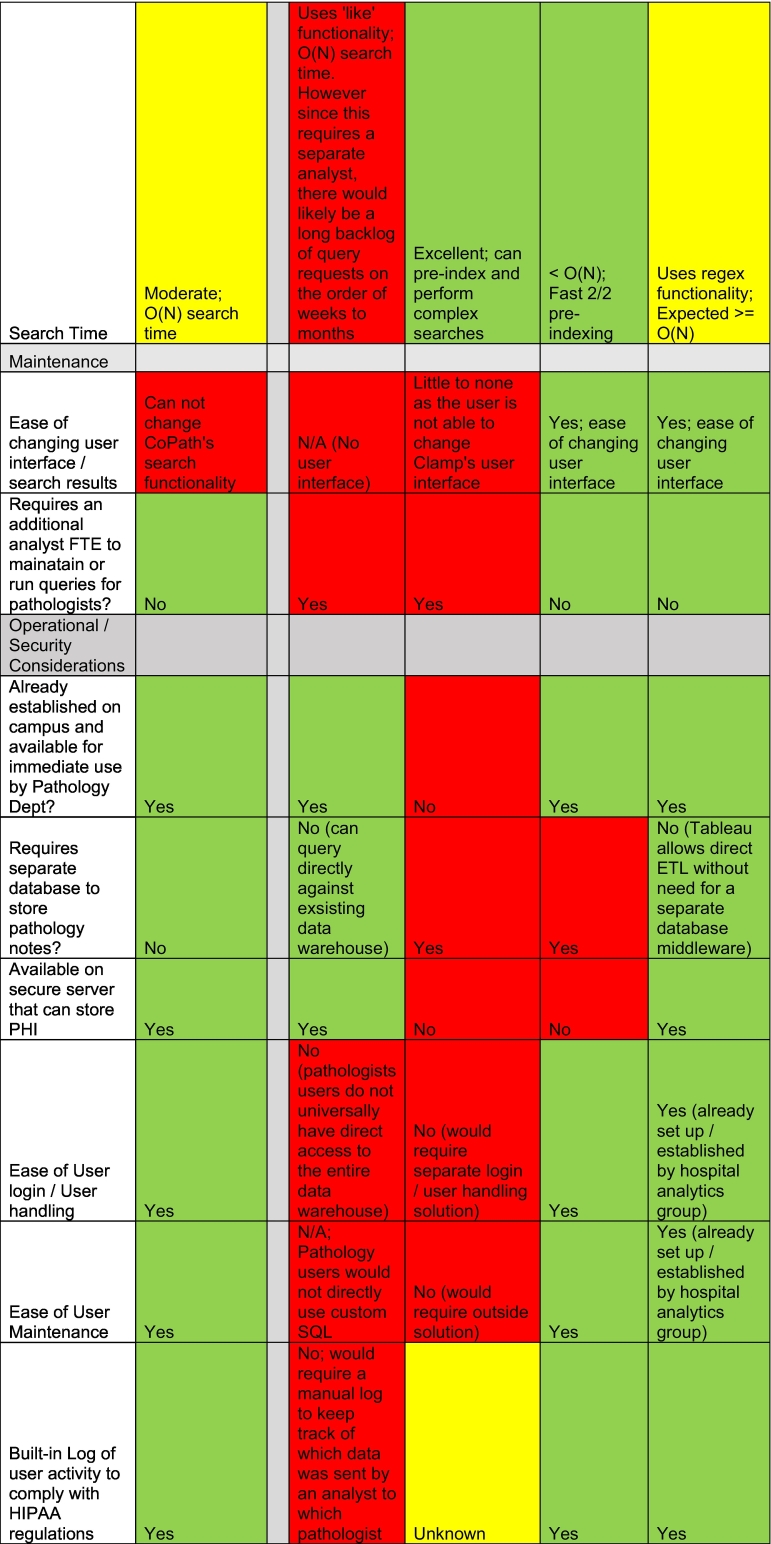
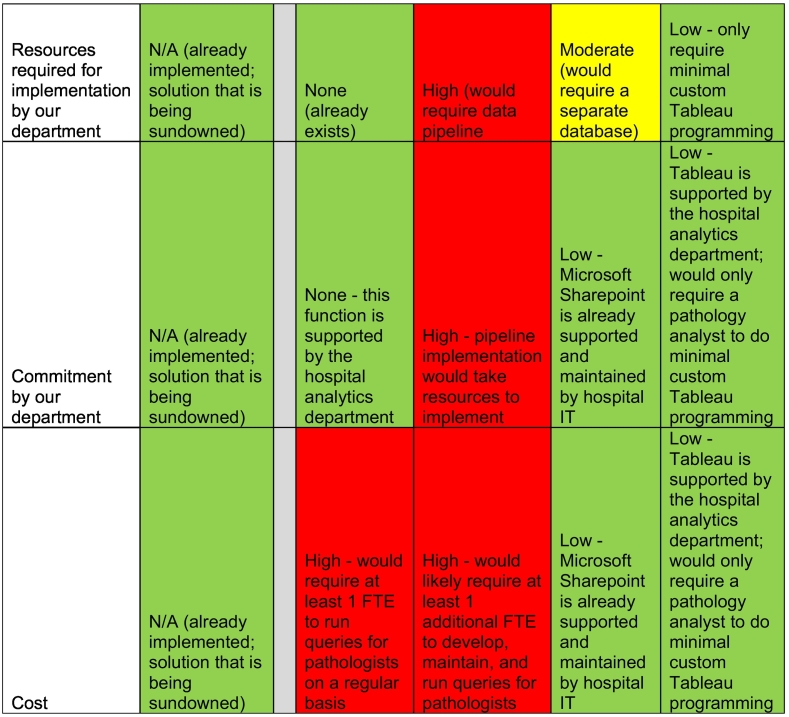
Before settling on a search platform, we carefully examined the functionality, cost, and implementation of numerous vendor tools and performed a formal comparison to guide our decision to either purchase a search tool or develop one in-house. Before implementing Tableau, our Department performed a cost–benefit analysis across potential solutions. We identified other tools, such as Clinical Language Annotation, Modeling, and Processing Toolkit (CLAMP), with exponentially more powerful NLP functionality, however we quickly realized that Tableau had sufficient search functionality to solve our user’s search needs—simple text searching—and offered significant benefits over other potential solution, including high usability, ease of user maintenance/login, security, tool maintenance, low overall cost, and low barriers to implementation (Table 2). Ultimately, we decided to develop our own search tool using the business intelligence tool Tableau. The decision was supported by the following findings:
-
•
A Tableau server was already established at our institution and the cost for Tableau implementation was handled by the institution and not the pathology department.
-
•
Tableau’s “REGEX” regular expression functionality offered the ability to easily program a custom search functionality.
-
•
Tableau offered the ability to quickly modify the user interface and base functionality based on user feedback.
-
•
Tableau offered an easily deployed extract, transform, and load (ETL) functionality.
-
•
Tableau offered an easily implemented user groups / login solution.
-
•
Tableau did not require end-user software—users could access a search tool using any web browser, thereby avoiding the cost of installing or maintaining software on users’ computers.
-
•
Using Tableau as a platform for the search tool offered low-cost maintenance.
-
•
Tableau viewer user licenses were relatively affordable.
-
•
Clinical data could be stored on a secure server and accessed securely on the institution’s network or through the institution’s VPN.
Table 2.
Features of different search tool solutions already available at our institution across key variables for decision making compared to CoPath.
The data pipeline for the search tool is depicted in Fig. 1. The pipeline is easily optimized because Tableau directly interacts with our EHR relational database (Clarity) without required middleware. Data was extracted from the EHR database using a custom Structured Query Language script to capture the text from anatomic pathology reports and other structured patient data included in Table 3. Tableau allows the creation of an ETL directly into Tableau’s data model which functions similarly to a relational database. How Tableau implements their data model is proprietary to the company.
Figure 1.
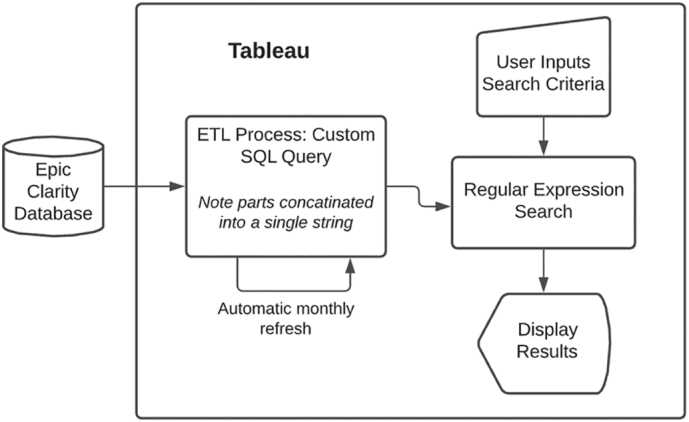
Anatomic pathology report search tool data pipeline.
Table 3.
Structured patient data pulled from the clinical data warehouse that can be included in searches.
| Structured patient data |
|---|
| Patient MRN |
| Patient ID |
| Patient Name |
| Patient Gender |
| Order ID |
| Test Name |
| Authorizing Provider |
| Authorizing Provider Subspecialty |
| Order Time |
| Pathologist |
| Surgical Pathology record number |
| Full text of the pathology report |
| ICD 9 and ICD10 codes and descriptions |
Our EHR stores complete anatomic pathology report data in the form of discreet components. For example, for a single case, the “Final Diagnosis” section is stored as a separate component from the “Gross Description”. Therefore, when extracting reports from the data warehouse, all components of the pathology report must be identified, retrieved, and concatenated into one large string to recreate the original report. The recreated report captures the full text of a complete pathology report including the final, gross and intraoperative diagnoses as well as comments and addendums.
Tableau Search
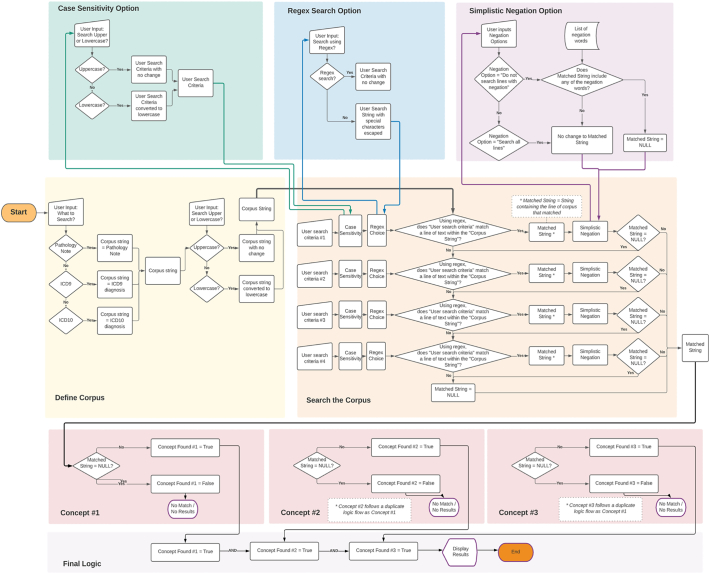
The search functionality (Fig. 2) was built entirely within Tableau using parameters for user input, nested calculated variables, and Tableau’s regular expression functionality. Search options include searching using simplistic negation, capitalization sensitive/insensitive, as well as the use of regular expression syntax. When the user performs a search, the input text parameters are taken through a series of algorithmic manipulations using Tableau’s built-in regular expression functionality to search the pathology report one “line” at a time. Each “line” is defined as a string followed by one or more line breaks.
Figure 2.
Graphical representation of parameters for user input, nested calculated variables, and Tableau’s regular expression functionality.
Programming Resources and Maintenance
The dashboard was designed in collaboration with anatomic pathology leadership and programmed by a single pathology informaticist (EA). The time required for data extraction, data validation, dashboard design, programming, and user feedback was 80 hr. An additional 40 hr were required for creation of data governance documentation and a user tutorial. Ongoing maintenance is minimal at about 1 hr a month, primarily for adding or removing users. The dashboard itself has required little to no maintenance as the ETL for data refresh, which occurs once a month, is automated.
Data Governance
To gain access to the anatomic pathology search tool, potential users must be part of the pathology department (residents, fellow, or faculty) and request access from the search tool developer. When a potential user requests access, they must sign a data use agreement. This data use agreement requires the potential user to specify how they will use the search tool. If the potential user plans to use the search tool for research purposes, they must provide an institutional review board (IRB) number for a project that has approved retrospective reviews of pathology reports and patient data and provide the IRB’s expiration date. The potential user must also agree to use the search tool in accordance with the HIPAA policies, agree to their utilization habits being randomly audited, and attest to understanding that misuse of the tool will be reported to the patient privacy team and result in potential disciplinary actions and revocation of search tool privileges. A potential user who plans to use the search tool for operations or educational purposes (Table 4) must also agree to use the search tool in accordance with the HIPAA policies, agree to utilization habits being randomly audited, and attest to understanding that misuse of the tool will be reported to the patient privacy team and result in potential disciplinary actions and revocation of search tool privileges. To comply with HIPAA policies surrounding reviewing patient-level data, Tableau offers an audit log that details when users accessed what information.
Table 4.
Data Governance use cases for the Anatomic Pathology Search Tool.
| IRB Needed | |
| Research | |
| No IRB Needed | |
Operational needs (examples)
|
|
Educational needs (examples)
|
Search Dashboard
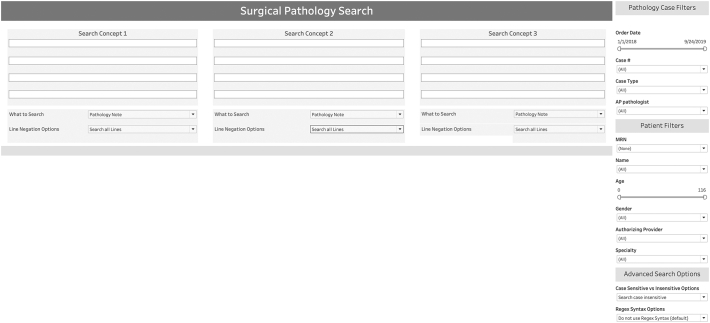
The default view for a user with access to the search tool is the search homepage (Fig. 3). The search interface allows the user to enter up to three search concepts logically combined as Boolean “AND” in the user input boxes. For each of the three concepts, the user can input up to four synonyms logically combined as Boolean “OR”.
Figure 3.
Screen grab of the anatomic pathology report search tool homepage as seen by a user with a tableau viewer license.
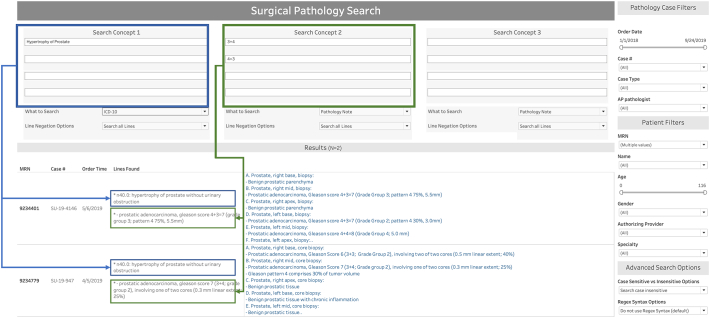
For each concept, the user can choose to search the full pathology note, ICD9, or ICD10 diagnosis. The example provided in Fig. 4 shows the user searched for all cases where the patient had an ICD-10 diagnosis of “Hypertrophy of prostate” AND for pathology cases that included the text “3+4” or “4+3”.
Figure 4.
Screen grab of the anatomic pathology report search tool illustrating a query for the search concepts ICD-10 code “Hypertrophy of the prostate” and “3 + 4” or “4 + 3” on pathology notes using fake data with results displayed. Under “Results” the “Lines Found” column allows the user to see the exact lines where the search concepts were identified.
In the simulated patient data example provided in Fig. 4, the results include two pathology cases matching the search concepts “Hypertrophy of Prostate” and “3+4” or “4+3”. When cases are identified by the search concepts, the cases are displayed under the “Results bar”, as seen in Fig. 4, and the results display the MRN, pathology case number, order time, lines found, and full pathology note text. The “lines found” section is a particularly appreciated feature among the department because it shows the user the exact line of text within the full report that matched the search concept that was provided. This allows the user to quickly assess the search results without having to look through the entire pathology report, and therefore allows the user to quickly ascertain the relevance of the returned report returned. User may click on the “Download” tab to download the entire text of the pathology report and metadata into Excel.
Results
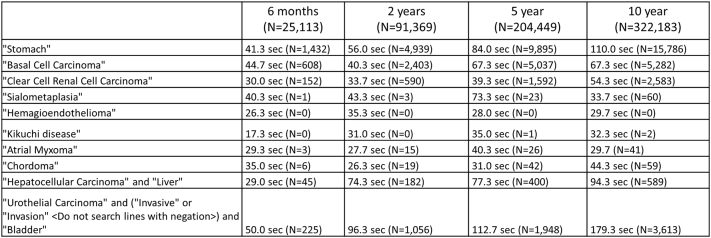
We generated 333,685 anatomic pathology reports between January 27, 2003 and September 26, 2021. Since go-live in December 2018, the search tool was made available to faculty and trainees within our department and has been widely utilized. From February 25, 2021 to September 26, 2021 (seven months), there have been 42 active users (representing approximately 40% of department members) and 627 search tool views (mean 14.9 searches per user). Figure 5 captures average search speeds for several common and uncommon diagnostic entities and simple and complex search terms across different time periods. Each term was searched three times and the average was recorded. The median search time across all terms and time periods included in Fig. 5 was 42.3 sec (IQR, 43.3 sec).
Figure 5.
Average search tool speed across a variety of search terms, including common, uncommon, and complex search terms. Each search term was searched over four time periods (previous six months, two years, five years, and ten years). The term was searched three times each and the average search time was recorded.
Discussion
Our anatomic pathology search tool was developed out of a need to maintain free-text searching of anatomic pathology reports when our institution switched to a LIS that did not offer this functionality. Search is a critical function to improve the quality of patient care and advance medical knowledge; however, this feature is not offered by all LIS vendors and, therefore, remains a deficiency for many institutions. Our solution is LIS vendor agnostic and was created using business intelligence software available at many healthcare institutions. To the best of our knowledge, this would be the first report in the peer-reviewed literature of using Tableau to search clinical notes. We were unable to find any other institutions describing similar uses in the literature. It is possible other institutions may not have considered using a BI tool, such as Tableau, for this purpose as most BI tools do not include robust NLP functionality like preindexing. Our hope is that this paper demonstrates that others may consider using their BI tools “off label” for simple text search functions.
The search tool described was designed for anatomic pathology, but may also be repurposed for other clinical specialties that have free-text reports such as radiology. Although we have not yet made formal comparisons with the performance of other search tools, anecdotally, our users reported that this tool was more user-friendly than our previous LIS’ search function. Additionally, this search tool often yields faster results than our prior LIS’ search function as many pathologists previously used an analyst to perform queries. The additional step of leveraging an analyst for search queries delays the process by days to weeks and ultimately hinders research, clinical operations, and educational activities. Regarding functionality, our users were satisfied that the Tableau search tool: (1) allows pathologists to independently use the tool, (2) performs free-text searches, (3) allows multiple search terms, (4) allows a user-friendly way of reviewing search results, (5) allows for subsequent downloading of results, (6) allows searching using multiple data filters (including date, pathologist, case #) to increase the granularity of the results.
Regular expressions and negation are two sophisticated and challenging aspects of free-text search. The average pathologist is not able to understand or write a regular expression search. Our search tool users were taught to do simple free-text searches which have been sufficient for most use cases. There have been several users who have expressed the desire to learn how to write regular expression queries for increased complexity and granularity of searches. In these situations, someone with technical expertise was required to guide them. However, most pathologist users can use our tool successfully for simple free-text searches without the need for understanding regular expressions or additional assistance.
Negation was not one of our users’ main criteria because our original tool, CoPath, did not support negation. However, we acknowledge that searches within pathology notes are frequently hindered by lack of negation due to the large number of pertinent negatives within pathology notes such as “No evidence of metastasis”. Tableau does not support negation “out of the box” and we explicitly programmed it into Tableau. The search using Tableau was programmed to search only one line at a time with a line being defined as text surrounded by line breaks. By default, the user searches without negation. If the user chooses, they can click to implement “negation”. The “negation” programmed into Tableau consisted of simply ignoring resulting lines that included the text, “negative”, “never”, “not”, “no”, “without”, “history”, “hx”, “imaging”, “clinical”, “pmh”, ”y/o”. When training users, we explicitly explain how the negation algorithm worked and explained that it would be useful to choose this option if a search was producing a high number of false positives, as is the case when searching for terms like “dysplasia” which results in extensive numbers of pathology notes with the words “No dysplasia or malignancy is identified”. Our negation function works as expected, but we have not calculated the explicit recall or precision of this coarse negation algorithm programmed into Tableau.
One area for improvement for our tool is to increase utilization within our department. At present, all users with access to the tool (40% of the department) learned about it through informal communication (e.g., word of mouth). We plan to do more systematic training and education as the next phase of our deployment. Another potential area for improvement is tin increase the search speeds. Although regular expression search is one of the most time inefficient search methodologies, our users found the search times and results to be acceptable. Using Big O notation, in comparing CoPath to Tableau, CoPath has an O(N) search time, which means that the search time rises linearly with the size of the corpus. The regular expression functionality within Tableau was expected to run at greater than or equal O(N) but surprisingly performs just under O(N) against our corpus, which is faster than CoPath’s search. How Tableau is able to accomplish less than or equal O(N) with a regular expression search is proprietary to their software. We developed a cost-effective and easy to maintain search tool for pathology reports using business intelligence software. Our department adopted this tool for operational, educational, and research use. Since many institutions already subscribe to business intelligence software, we believe this solution could be easily reproduced at other institutions and in other clinical departments.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Simone Arvisais-Anhalt: drafted and edited the manuscript, analyzed the data.
Christoph U. Lehmann: drafted and edited the manuscript.
Justin A. Bishop: assisted in the design and development of the tool, edited the manuscript.
Jyoti Balani: assisted in the design and development of the tool, edited the manuscript.
Laurie Boutte: assisted in the development of the tool, edited the manuscript.
Marjorie Morales: assisted in the development of the tool, edited the manuscript.
Jason Y. Park: drafted and edited the manuscript, analyzed the data.
Ellen Araj: designed and developed the tool, drafted and edited the manuscript, analyzed the data.
References
- 1.Meystre S.M., Lovis C., Bürkle T., Tognola G., Budrionis A., Lehmann C.U. Clinical data reuse or secondary use: current status and potential future progress. Yearb Med Inform. 2017 Aug;26(1):38–52. doi: 10.15265/IY-2017-007. Epub 2017 Sep 11. PMID: 28480475; PMCID: PMC6239225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park S., Parwani A.V., Aller R.D., et al. The history of pathology informatics: A global perspective. J Pathol Inform. 2013;4:7. doi: 10.4103/2153-3539.112689. Published 2013 May 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CLIA Regulations . 2021. Retention Requirements (42CFR§493.1105) Last accessed on Sept 15. https://www.ncleg.gov/documentsites/committees/PMC-LRC2011//December5,2012/CLIARegulationsRecordRetention(42CFR§493.1105).pdf. [Google Scholar]
- 4.Rosenbloom S.T., Denny J.C., Xu H., Lorenzi N., Stead W.W., Johnson K.B. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc. 2011;18(2):181–186. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbloom S.T., Denny J.C., Xu H., Lorenzi N., Stead W.W., Johnson K.B. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc. 2011 Mar-Apr;18(2):181–186. doi: 10.1136/jamia.2010.007237. Epub 2011 Jan 12. PMID: 21233086; PMCID: PMC3116264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CAP Cancer Protocols. https://www.cap.org/protocols-and-guidelines/cancer-reporting-tools/cancer-protocol-templates [Last accessed on 2021 Jul 17]. Available from.
- 7.Zheng S., Lu J.J., Appin C., Brat D., Wang F. Support patient search on pathology reports with interactive online learning based data extraction. J Pathol Inform. 2015;6:51. doi: 10.4103/2153-3539.166012. Published 2015 Sep 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alzu’bi A.A., Watzlaf V.J.M., Sheridan P. Electronic Health Record (EHR) abstraction. Perspect Health Inf Manag. 2021;18(Spring) 1g. Published 2021 Mar 15. [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J., Song H.J., Yoon E., et al. Automated extraction of Biomarker information from pathology reports. BMC Med Inform Decis Mak. 2018 May 21;18(1):29. doi: 10.1186/s12911-018-0609-7. PMID: 29783980; PMCID: PMC5963015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alawad M., Gao S., Qiu J.X., et al. Automatic extraction of cancer registry reportable information from free-text pathology reports using multitask convolutional neural networks. J Am Med Inform Assoc. 2020;27(1):89–98. doi: 10.1093/jamia/ocz153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen A.N., Lawley M.J., Hansen D.P., et al. Symbolic rule-based classification of lung cancer stages from free-text pathology reports. J Am Med Inform Assoc. 2010;17(4):440–445. doi: 10.1136/jamia.2010.003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giannaris P.S., Al-Taie Z., Kovalenko M., et al. Artificial intelligence-driven structurization of diagnostic information in free-text pathology reports. J Pathol Inform. 2020;11:4. doi: 10.4103/jpi.jpi_30_19. Published 2020 Feb 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Currie A.M., Fricke T., Gawne A., Johnston R., Liu J., Stein B. Automated extraction of free-text from pathology reports. AMIA Annu Symp Proc. 2006;2006:899. [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser A.P., Jordan B.J., Cohen J., Desai A., Silberman P., Meeks J.J. Automated extraction of grade, stage, and quality information from transurethral resection of bladder tumor pathology reports using natural language processing. JCO Clin Cancer Inform. 2018;2:1–8. doi: 10.1200/CCI.17.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yim W.W., Kwan S.W., Johnson G., Yetisgen M. Classification of hepatocellular carcinoma stages from free-text clinical and radiology reports. AMIA Annu Symp Proc. 2018;2017:1858–1867. Published 2018 Apr 16. [PMC free article] [PubMed] [Google Scholar]
- 16.Na H.Y., Kim J.W., Baek R.M., Hwang H., Yoo S. Transformation of pathology reports into the common data model with oncology module: use case for colon cancer. J Med Internet Res. 2020 Dec 9;22(12) doi: 10.2196/18526. PMID: 33295294; PMCID: PMC7758167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buckley J.M., Coopey S.B., Sharko J., et al. The feasibility of using natural language processing to extract clinical information from breast pathology reports. J Pathol Inform. 2012;3:23. doi: 10.4103/2153-3539.97788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson B.S., Hardy S., Pandeya N., et al. Web application for the automated extraction of diagnosis and site from pathology reports for keratinocyte cancers. JCO Clin Cancer Inform. 2020 Aug;4:711–723. doi: 10.1200/CCI.19.00152. PMID: 32755460; PMCID: PMC7469600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu J.X., Yoon H.-J., Fearn P.A., Tourassi G.D. Deep learning for automated extraction of primary sites from cancer pathology reports. IEEE J Biomed Health Inform. Jan 2018;22(1):244–251. doi: 10.1109/JBHI.2017.2700722. [DOI] [PubMed] [Google Scholar]
- 20.Burger G., Abu-Hanna A., de Keizer N., et al. Natural language processing in pathology: a scoping review. J Clin Pathol. 2016;69:949–955. doi: 10.1136/jclinpath-2016-203872. [DOI] [PubMed] [Google Scholar]
- 21.Robertson S. A novel web application for rapidly searching the diagnostic case archive. J Pathol Inform. 2020 Dec;24(11):39. doi: 10.4103/jpi.jpi_43_20. PMID: 33828897; PMCID: PMC8020840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson H.D., Weerasinghe R., Martel M., et al. Development of an electronic breast pathology database in a community health system. J Pathol Inform. 2014;5(1):26. doi: 10.4103/2153-3539.137730. Published 2014 Jul 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erinjeri J.P., Picus D., Prior F.W., Rubin D.A., Koppel P. Development of a Google-based search engine for data mining radiology reports. J Digit Imaging. 2009;22(4):348–356. doi: 10.1007/s10278-008-9110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanauer D.A., Mei Q., Law J., Khanna R., Zheng K. Supporting information retrieval from electronic health records: a report of University of Michigan’s nine-year experience in developing and using the Electronic Medical Record Search Engine (EMERSE) J Biomed Inform. 2015;55:290–300. doi: 10.1016/j.jbi.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]