Abstract
Mitochondrial dysfunction induces a strong adaptive retrograde signaling response; however, many of the downstream effectors of this response remain to be discovered. Here, we studied the shared transcriptional responses to three different mitochondrial respiratory chain inhibitors in human primary skin fibroblasts using QuantSeq 3′-RNA-sequencing. We found that genes involved in the mevalonate pathway were concurrently downregulated, irrespective of the respiratory chain complex affected. Targeted metabolomics demonstrated that impaired mitochondrial respiration at any of the three affected complexes also had functional consequences on the mevalonate pathway, reducing levels of cholesterol precursor metabolites. A deeper study of complex I inhibition showed a reduced activity of endoplasmic reticulum–bound sterol-sensing enzymes through impaired processing of the transcription factor Sterol Regulatory Element-Binding Protein 2 and accelerated degradation of the endoplasmic reticulum cholesterol-sensors squalene epoxidase and HMG-CoA reductase. These adaptations of mevalonate pathway activity affected neither total intracellular cholesterol levels nor the cellular free (nonesterified) cholesterol pool. Finally, measurement of intracellular cholesterol using the fluorescent cholesterol binding dye filipin revealed that complex I inhibition elevated cholesterol on intracellular compartments. Taken together, our study shows that mitochondrial respiratory chain dysfunction elevates intracellular free cholesterol levels and therefore attenuates the expression of mevalonate pathway enzymes, which lowers endogenous cholesterol biosynthesis, disrupting the metabolic output of the mevalonate pathway. We conclude that intracellular disturbances in cholesterol homeostasis may alter systemic cholesterol management in diseases associated with declining mitochondrial function.
Keywords: CoQ, farnesyl pyrophosphate, geranyl pyrophosphate, HMGCR, isoprenoids, mevalonate pathway, mitochondria, SQLE, SREBP2, ubiquinol
Abbreviations: AMPK, 5′ AMP-activated protein kinase; DMEM, Dulbecco's Modified Eagle Medium; ER, endoplasmic reticulum; FBS, fetal bovine serum; GO, Gene Ontology; HMGCR, 3-Hydroxy-3-Methylglutaryl-CoA reductase; HMGCS1, 3-Hydroxy-3-Methylglutaryl-CoA Synthase 1; MEF, mouse embryonic fibroblast; SCAP, SREBP cleavage activating protein; SQLE, squalene monooxygenase (squalene epoxidase); SREBP2, sterol regulatory element–binding protein 2
The functional versatility of mitochondria is reflected in the diversity of the metabolic processes they contribute to, ranging from carbohydrate to fatty acid and amino acid metabolism. Central in these processes is the generation of reducing equivalents that provide electrons to the respiratory chain. Transfer of these electrons along the respiratory chain protein complexes I to IV powers the extrusion of protons across the inner mitochondrial membrane (1). The resulting electrochemical gradient drives ATP-synthase (complex V), which is responsible for the majority of cellular ATP synthesis (2). In addition to providing ATP, mitochondria are the gatekeeper for cellular homeostasis by synthesizing substrates for cytosolic reactions, sensing cellular stresses, modulating apoptosis, controlling DNA damage, and regulating the cell cycle among others (3, 4).
In line with the integration of mitochondria in multiple aspects of cellular metabolism, disruption of mitochondrial function has widespread consequences. At the cellular level, impairment of mitochondrial function leads to a reduced capacity to produce ATP, a shift in the NAD+/NADH ratio, affects the mitochondrial membrane potential, increases reactive oxygen species formation, reduces metabolite exchange between the mitochondria and the cytosol, and perturbs Ca2+ homeostasis (5, 6). Changes in these biochemical parameters are interpreted by various cellular sensors which in turn activate downstream transcriptional programs to compensate for the changes (7). This communication from the mitochondria to the rest of the cell has been termed mitochondrial retrograde signaling (8, 9). It is the balance between the biochemical changes and adaptive retrograde signaling that determine the outcome of the primary mitochondrial defect and whether the affected tissue can cope with the primary mitochondrial defect (10, 11). Mitochondria also play important roles in cholesterol metabolism, synthesizing several metabolites such as oxysterols, steroid hormones, and bile acids (12). These sterols have the potential to act as retrograde signals, through their ability to activate nuclear receptors or directly control enzyme activities (13, 14, 15).
Cholesterol synthesis is tightly controlled by several endoplasmic reticulum (ER) membrane proteins that sense cholesterol levels (16, 17). When cholesterol levels on the ER membrane are elevated, the transcription factor sterol regulatory element–binding protein 2 (SREBP2) is retained on the ER membrane by forming a complex with the SREBP cleavage activating protein (SCAP) and insulin-induced gene (INSIG), a protein sensitive to changes in oxysterol concentrations (18, 19). Low abundance of cellular and subsequent ER cholesterol causes the SCAP–SREBP2 complex to enter COP II vesicles and be transported to the Golgi. Here SREBP2 is sequentially cleaved by site-1 and site-2 proteases to release a 60 kDa mature form. This protein enters the nucleus where it acts as a transcription factor driving expression of mevalonate pathway genes.
Cholesterol is synthesized by the mevalonate pathway, which converts acetyl-CoA to cholesterol in a series of enzymatic reactions. In addition to their transcriptional regulation by SREBP2, several enzymes of the mevalonate pathway such as 3-Hydroxy-3-Methylglutaryl-CoA reductase (HMGCR), squalene epoxidase (SQLE), and 7-dehydrocholesterol reductase (DHCR7) are posttranslationally regulated (20). These membrane proteins sense sterols through sterol sensing domains or other cholesterol sensitive sites and are rapidly degraded by the proteasome in situations of elevated ER-membrane sterols (21).
In this study, we investigated mitochondrial retrograde signaling and the associated transcriptional changes in three pharmacological models of respiratory chain dysfunction. We observed that genes of the mevalonate pathway were consistently downregulated when mitochondrial function was impaired, independent of the complex affected. Both the transcriptional and posttranslational regulation of sterol sensors such as SREBP2, HMGCR, and SQLE were affected. Consequently, several mevalonate pathway metabolite concentrations were altered. In particular, we observed a coordinated reduction of sterol intermediates downstream of squalene. We further provide evidence that impaired sterol sensing is due to elevated levels of free cholesterol on intracellular membranes. Our findings show that mitochondrial retrograde signaling affects intracellular cholesterol distribution and thereby cholesterol sensing on the ER with important consequences for the synthesis of mevalonate pathway intermediates.
Results
Pharmacological model of mitochondrial respiratory chain dysfunction in human primary fibroblasts
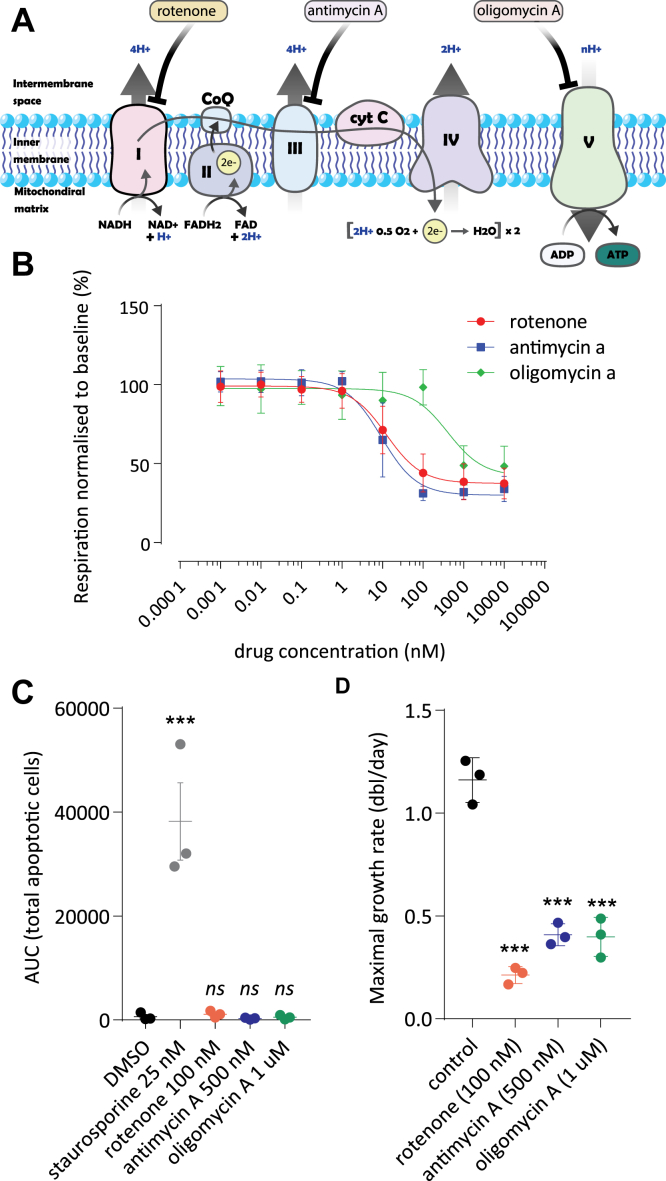
We studied the molecular mechanisms of retrograde signaling from mitochondria to the nucleus in human primary fibroblasts. To identify putative common adaptations to respiratory chain dysfunction, we treated human primary fibroblasts with three drugs that specifically inhibit different complexes of the respiratory chain: rotenone to inhibit complex I (NADH:ubiquinone oxidoreductase), antimycin A to inhibit complex III (CoQH2-cytochrome c reductase), and oligomycin A to inhibit complex V (ATP-Synthase) (Fig. 1A). Rotenone, antimycin A, and oligomycin A inhibited respiration in human fibroblasts with IC50 values of 12.39 nM, 9.33 nM, and 405.6 nM, respectively (Fig. 1B). To potently induce retrograde signaling, we aimed to severely inhibit oxygen consumption, while causing minimal effects on cellular viability. We titrated the concentration of the three respiratory chain inhibitors and measured apoptosis using Annexin V labeling. We found that 100 nM rotenone, 500 nM antimycin, and 1 μM oligomycin strongly inhibited mitochondrial respiration without significantly inducing apoptosis (Figs. 1, B and C and S1). These concentrations slow down cellular proliferation (Fig. 1D) in agreement with previously published data (22, 23, 24). These three pharmacological models were used to identify common transcriptional fingerprints in response to mitochondrial dysfunction.
Figure 1.
Characterization of pharmacological models of respiratory chain dysfunction in primary human fibroblasts.A, schematic representation of the mitochondrial respiratory chain. The target complexes of the three inhibitors rotenone (complex I), antimycin A (complex III), and oligomycin (complex V) are indicated. B, dose-dependent inhibition of oxygen consumption by respiratory chain inhibitors in primary human fibroblasts. Cells were cultured under standard conditions and prepared for Seahorse XF96 experiments (see Experimental procedures). Inhibitors were added acutely after baseline oxygen consumption measurements in the presence of 5 mM glucose. Respiratory chain inhibitors were applied at increasing concentrations as indicated for rotenone (red circles), antimycin A (blue squares), or oligomycin (green diamonds). Oxygen consumption per unit of protein was calculated for each well and expressed as percentage of basal respiration in the absence of the inhibitors. Shown is the average of three independent experiments ±standard deviation. Each experiment is the average of six measurements. C, apoptosis of human fibroblasts was followed using Annexin V Green marker in a ZOOM Live-Cell analysis system (see Experimental procedures). Apoptotic cell counts were measured after growth for 5 days in the absence or presence of respiratory chain inhibitors. Rotenone (100 nM; red circles), antimycin A (500 nM; blue circles), or oligomycin (1 μM; green circles) were used. Staurosporine (25 nM; gray circles) was included as a positive control. Results from three independent experiments ±standard deviation are shown. Significance was calculated with ANOVA followed by Tukey HSD post-hoc test. D, doubling rate of fibroblasts in the presence of respiratory chain inhibitors. Proliferation was measured during exponential growth in the absence (black circles) or the continuous presence of a concentration of respiratory chain inhibitor. Concentrations used and color code as defined in panel C. Growth rate data are expressed as doublings per day. Data from three independent experiments ±standard deviation are shown. Significance was calculated with ANOVA followed by Tukey HSD post-hoc test. ns: not significant; ∗∗∗ p < 0.001 compared to dimethyl sulfoxide (DMSO) control.
Shared transcriptional adaptations to chronic inhibition of complex I, III, or V of the respiratory chain in human fibroblasts
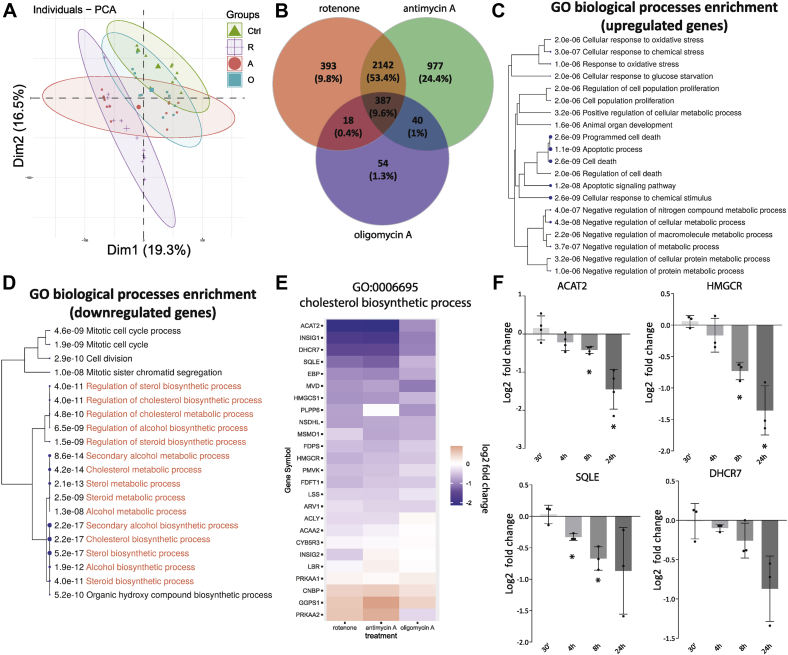
The transcriptome of the three pharmacological fibroblast models of respiratory chain dysfunction was established by QuantSeq 3′RNA-sequencing and compared to control. To capture transcriptional responses that were both rapid and long-lasting, we treated fibroblasts for 1 and 5 days, and five replicates were included for each inhibitor and timepoint. A principal component analysis shows the individual treatments cluster together, with a slight separation depending on the treatment duration (Fig. 2A). The good separation of data points between rotenone- or antimycin A-treated cells and control suggests robust mRNA expression changes at both 1 and 5 days in the presence of these two inhibitors. The oligomycin A model of mitochondrial dysfunction appears to have an intermediate phenotype. Gene expression changes after rotenone or antimycin A treatment showed a lot of similarity. Of all genes affected by rotenone, 86% were also differentially expressed after antimycin A treatment (Fig. 2B). Three hundred eighty-seven genes were shared between all three treatment groups (9.6% of total). For all three mitochondrial inhibitors, 283 genes were upregulated, and 101 genes were downregulated. Only three genes displayed varying directionality depending on the inhibitor (see Supporting Data 1). Such coherent regulation in this gene set reveals several adaptive responses to respiratory dysfunction that occur independent of the site of inhibition.
Figure 2.
Transcriptional adaptations shared in the three pharmacological models of respiratory chain dysfunction.A, PCA of QuantSeq data of human fibroblasts cultured for 1 or 5 days in the presence of respiratory chain inhibitors rotenone (100 nM; purple plus signs), antimycin A (500 nM; red circles), or oligomycin (1 μM; blue squares). Control values are from DMSO-treated cells (0.1%; green triangles). Separation along the first two dimensions is shown. Cells were plated to reach similar final confluency. B, Venn diagram illustrating the total number and overlap of the differentially expressed genes for the three treatment groups. C and D, biological processes associated with differentially expressed genes shared by all three respiratory chain inhibitors as shown in panel B (387 genes). A hierarchical clustering of the significantly enriched GO-biological process pathways is shown. For each pathway, the significance of its enrichment in the shared set of DEGs was calculated and shown in the figure. The processes most relevant for this study are highlighted in red of panel D. E, expression changes compared to DMSO (0.1%) control of genes associated with the cholesterol biosynthetic process (GO:0006695) following treatment with rotenone (left), antimycin A (middle), and oligomycin A (right). The heatmap representation reveals the consistent downregulation of most genes associated with this pathway. F, time-dependent changes in gene expression of five key mevalonate pathway genes in human fibroblasts measured by qPCR (see Experimental procedures). Cells were treated with rotenone (100 nM) for the indicated times. Data are shown as the log2-fold change of gene expression compared to control-treated human fibroblasts (0.1% DMSO). Bars show mean ± standard deviation; black circles indicate results from three independent experiments. Significance calculated as one-sample t test (H0: μ = 0). ∗ p< 0.05. DEG, differentially expressed genes; DMSO, dimethyl sulfoxide; GO, Gene Ontology.
Cholesterol biosynthesis genes are coordinately downregulated in models of mitochondrial dysfunction
To find pathways that were strongly represented in the affected genes, we performed clustered Gene Ontology (GO)-enrichment analyses on the upregulated and downregulated fractions of the co-regulated genes. The upregulated genes show a strong enrichment of GO processes linked to apoptosis, response to oxidative stress and a negative regulation of metabolism (Fig. 2C). Previous studies have investigated these stress responses in detail (25, 26, 27). Downregulated genes were strongly enriched in genes of the mevalonate pathway of cholesterol biosynthesis and other aspects of cholesterol metabolism (Fig. 2D), also noted but not further studied in earlier work (26, 28). A closer inspection of one of the central GO biological processes affected (GO: 0006695 cholesterol biosynthetic process) showed transcripts ACAT2, ACLY, CYB5R3, DHCR7, FDFT1, FDPS, HMGCR, HMGCS1, LBR, LSS, MSMO1, MVD, NSDHL, PMVK, and SQLE were significantly downregulated for each of the three respiratory chain inhibitors (Fig. 2E). These transcripts mostly encode enzymes in the mevalonate pathway (Fig. 3A). We validated the downregulation of a select number of key transcripts by qPCR after rotenone treatment. Gene expression changes were followed over a time course from 30 min up to 24 h (Fig. 2F). A significant reduction of gene expression was observed for ACAT2, HMGCS1, HMGCR, and SQLE after 8 h or less of complex I inhibition consistent with the QuantSeq data. After 24 h of complex I inhibition, expression of the selected mevalonate pathway genes was reduced by 2- to 4-fold. Rotenone may lower mevalonate pathway gene expression indirectly by slowing cell proliferation (29). We observe that confluent human fibroblasts, which arrest proliferation due to contact inhibition (30), express ACAT2 at a similar level to the proliferating cells, but HMGCR and SQLE at slightly higher levels. Rotenone lowered expression of mevalonate pathway genes in all conditions, irrespective of cellular growth rates (Fig. S2), suggesting a more direct link between mitochondrial inhibition and transcriptional regulation.
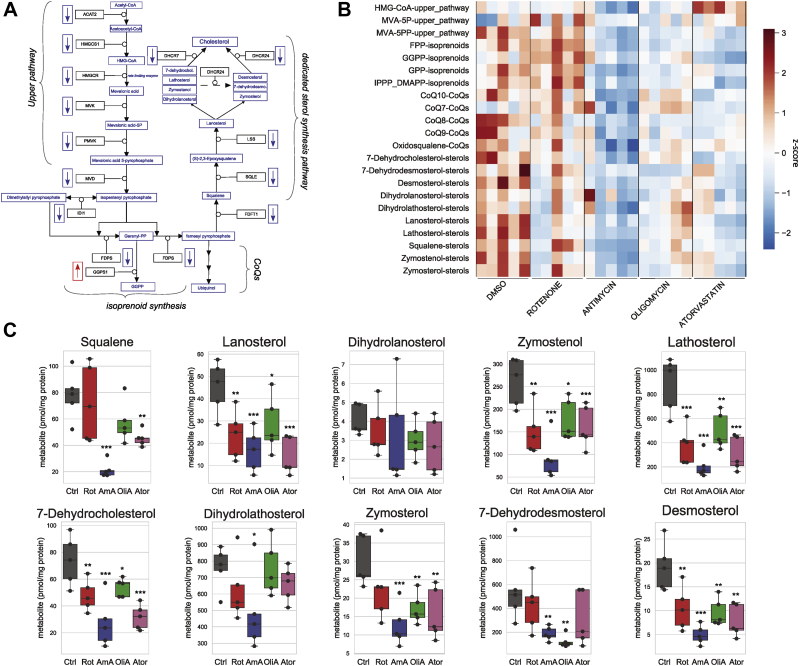
Figure 3.
Inhibition of the mitochondrial respiratory chain lowers intracellular levels of sterol intermediates of cholesterol biosynthesis.A, schematic representation of the mevalonate biosynthetic pathway. Gene names coding for the enzymes catalyzing the reactions in the pathway are given in the black boxes. Mevalonate pathway metabolites are presented in blue. The direction of gene expression changes obtained by QuantSeq RNA measurements is indicated next to the enzymes for downregulated (arrows blue boxes) and upregulated (arrow red box) genes. B, heatmap representation reflecting abundance of mevalonate pathway metabolites in human fibroblasts after inhibition of the respiratory complexes or the mevalonate pathway. Cells were cultured for 48 h in DMEM with 5 mM glucose containing 5% FCS and treated for 48 h with either DMSO (0.1%), rotenone (100 nM), antimycin A (500 nM), oligomycin A (1 μM), or atorvastatin (1 μM) as indicated. Metabolites were separated from total methanol cell extract and analyzed by GC-MS (see Experimental procedures). Five independent experiments were performed, and the indicated metabolites measured. The values were normalized to total protein level in each sample and z scaled. Colors reflect relative metabolite abundance from high (red) to low (blue). C, quantification of metabolites in the dedicated sterol synthesis pathway (boxplot representation). Values from individual experiments (black circles) are given in pmol/mg protein. Ctrl (control, gray boxes), Rot (rotenone, red boxes), AmA (antimycin A, blue boxes), OliA (oligomycin A, green boxes), and Ator (atorvastatin, purple boxes). The boxes show the median and the interquartile range. Significance was calculated with ANOVA, followed by a Tukey HSD post-hoc test. ns: not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 compared to DMSO control. DMEM, Dulbecco's Modified Eagle Medium; DMSO, dimethyl sulfoxide.
Inhibition of mitochondrial respiration alters the levels of mevalonate pathway metabolites
The coordinated downregulation of mevalonate pathway genes may ultimately impact the abundance of mevalonate pathway metabolites. To investigate this, 22 mevalonate pathway metabolites were quantified in human fibroblasts treated with either of the three respiratory chain inhibitors (Fig. 3, A and B). As a positive control, the mevalonate pathway was inhibited at the level of HMGCR with atorvastatin. Consistent with the effective inhibition of HMGCR, atorvastatin increased the enzyme substrate HMG-CoA and lowered the abundance of downstream mevalonate pathway metabolites (Fig. S3).
All three respiratory chain inhibitors lowered the abundance of most metabolites in the dedicated cholesterol synthesis pathway (Fig. 3, B and C). Other parts of the pathway were differently affected by the individual inhibitors. The precursors of sterol synthesis squalene and oxidosqualene were significantly reduced after blocking respiration with antimycin A (Figs. 3C and S3). Lanosterol, the first metabolite dedicated to the synthesis of cholesterol, was significantly reduced by all three mitochondrial inhibitors (Fig. 3C). Most downstream sterol metabolites including zymosterol, zymostenol, lathosterol, and desmosterol were significantly reduced when the respiratory chain was inhibited irrespective of the complex affected (Fig. 3C). Other sterols showed a trend to be lower when human fibroblasts were inhibited with either of the three mitochondrial inhibitors (Fig. 3C). The observed reductions were comparable to the mevalonate pathway inhibiting effect of atorvastatin. This demonstrates that mitochondrial dysfunction affects the dedicated cholesterol synthesis pathway independently of the site of mitochondrial inhibition.
Metabolites in the upper part of the mevalonate pathway and the intermediates of isoprenoid synthesis and CoQ intermediates displayed variable responses to mitochondrial dysfunction depending on the respiratory chain complex affected (Fig. S3). The study of molecular mechanisms responsible for the differences observed in the three different models of mitochondrial dysfunction were out of scope for the present work. Here, we investigated the underlying mechanisms slowing cholesterol biosynthesis as suggested by the reduced levels of sterol precursors.
For further characterization of the mechanisms linking mitochondrial dysfunction to mevalonate pathway regulation, we limited ourselves to the characterization of the complex I deficiency model of respiratory chain dysfunction.
Well-known adaptive responses not primarily responsible for the downregulation of mevalonate pathway genes
Induction of the integrated stress response following mitochondrial dysfunction may be responsible for the downregulation of mevalonate pathway gene expression (31). We confirmed by qPCR that complex I inhibition induced transcription of the stress-related transcription factors ATF3, ATF4, and CHOP (Fig. S4A). Blocking eIF2α activation with ISRIB inhibited expression in both control and rotenone-treated fibroblasts. In contrast, downregulation of the mevalonate pathway genes ACAT2, HMGCR, and SQLE also observed in the presence of ISRIB (Fig. S4B). It is therefore unlikely that ISR induced by mitochondrial dysfunction is responsible for lowered transcription of mevalonate pathway enzymes following complex I inhibition.
The energy sensor 5′ AMP-activated protein kinase (AMPK) is known to lower mevalonate pathway transcripts through the phosphorylation of the upstream transcription factor SREBP2 (32). We therefore examined if mitochondrial dysfunction downregulates expression of mevalonate pathway genes through the activation of AMPK in mouse embryonic fibroblasts (MEFs) (Fig. S4C). Treatment with rotenone-activated AMPK in WT MEFs, as demonstrated by enhanced autophosphorylation of the kinase and the elevated phosphorylation of the substrate acetyl-CoA carboxylase. As expected, AMPK was completely absent in AMPK-knockout MEFs lacking functional AMPK. The ability of complex I inhibition to lower the expression of the mevalonate pathway genes ACAT2, HMGCS1, HMGCR, and SQLE was preserved in AMPK-knockout MEFs. Activation of AMPK by energy stress therefore does not explain transcriptional regulation of mevalonate genes by mitochondrial dysfunction (Fig. S4D).
Complex I inhibition elevates the intracellular free cholesterol pool
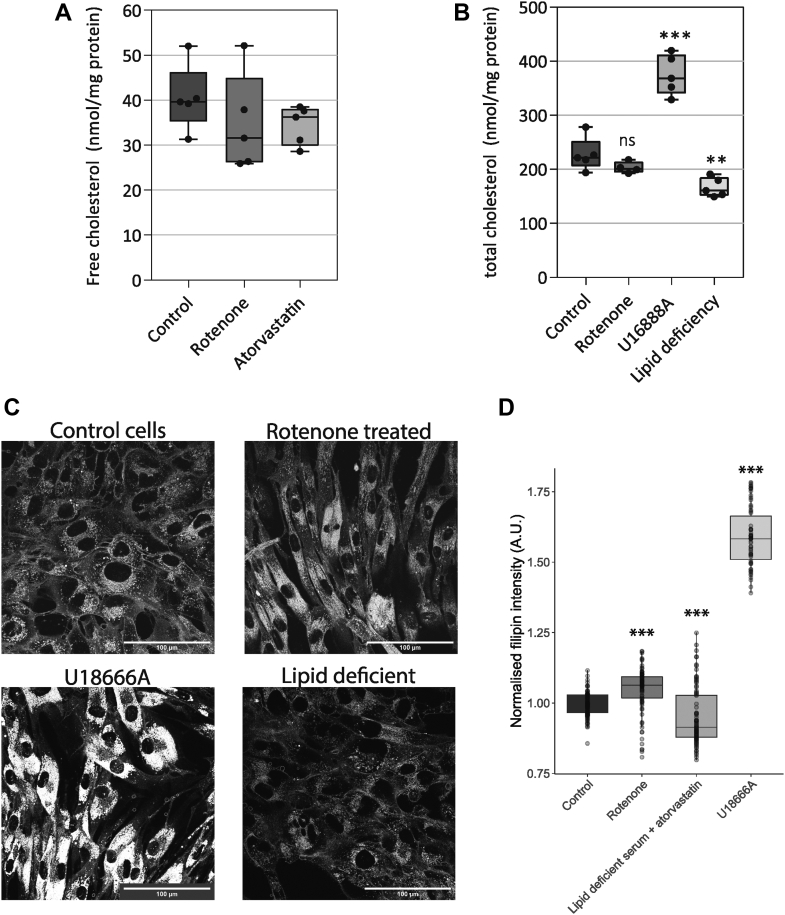
Mevalonate pathway activity is tightly controlled by changes in levels of intracellular cholesterol, which can be present in either free or esterified form. We speculated that the mevalonate pathway suppression was a consequence of elevated cholesterol caused by respiratory chain dysfunction. To assess this, we measured free and total cholesterol (free cholesterol plus cholesterol-esters) in primary fibroblasts with or without mitochondrial inhibition (Fig. 4, A and B). Free cholesterol was not elevated after 48 h of rotenone treatment (Fig. 4A). Under these conditions, cholesterol was also not reduced by atorvastatin. The results demonstrate that the levels of mevalonate pathway intermediates are much more sensitive to perturbations than the final product cholesterol. Increases in cholesterol may be buffered through the esterification and storage in lipid droplets in the cell. We therefore also measured total cholesterol. As a control, we lowered cellular cholesterol using a lipid-free medium containing atorvastatin. The cholesterol pool in cells grown in such lipid-free medium was significantly lowered. A second control was to increase cellular cholesterol with U18666a (34). Treatment with U18666a caused a marked increase of cellular cholesterol (Fig. 4B). Inhibition of complex I function did not affect total cellular cholesterol. Although our methods are sensitive to cholesterol modifying conditions, inhibition of mitochondrial function does neither affect the free nor esterified pool of cellular cholesterol.
Figure 4.
Inhibition of complex I in human fibroblasts augments intracellular free cholesterol.A, total free cellular cholesterol in primary human fibroblasts. Cells were cultured as in Figure 3. The cells were grown in control culture medium (control; black) in the presence of rotenone (100 nM; dark gray) or atorvastatin (1 μM; pale gray). The amount of free cholesterol normalized to total protein was measured by UPLC-MS. Five samples were measured for each condition; the individual data points are shown (black circles). Lines indicate minimum, maximum, median, and interquartile ranges. Significance was calculated by ANOVA with Tukey HSD posthoc test. B, total cellular cholesterol in primary human fibroblasts. Cells were cultured for 48 h under normal culture medium supplemented with either rotenone (100 nM), U18666A (500 nM), or in lipid-deficient medium (containing delipidated serum and 1 μM atorvastatin) as indicated at the bottom of the graph. Cholesterol was measured by GC-FID (see Experimental procedures). The values represent total cellular cholesterol normalized to total protein. Shown is the mean ± SD of five independent experiments; black circles show results from separate experiments. Significance was calculated by ANOVA with Tukey HSD posthoc test. C, representative images of filipin stained fibroblasts. Top left, control cells. Top right, cells treated with 100 nM rotenone. Bottom left, cells treated with U18666A. Bottom right, cells grown in lipid deficient medium. D, fluorescence quantification of intracellular-bound filipin in primary human fibroblasts as a semiquantitative read-out of intracellular free cholesterol. Cells were cultured for 48 h under normal culture conditions or lipid-free conditions and supplemented with rotenone, atorvastatin, or U18666A as described for panel C. Plasma membrane cholesterol was briefly extracted with empty cyclodextrin followed by fixation and staining with the cholesterol binding fluorescent compound filipin (see Experimental procedures). Free cholesterol was quantified by image analysis (see Experimental procedures). Boxplot showing the amount of filipin fluorescent intensity; points indicated the separate quantified images. Significance of differences versus control were calculated with ANOVA with Tukey HSD post-hoc test. ns: not significant; ∗∗ p < 0.01; ∗∗∗ p < 0.001.
The free cholesterol pools on the ER membrane directly regulate of transcription of mevalonate pathway genes. In the cellular cholesterol measurements shown here (Fig. 4, A and B), potential changes of intracellular or ER cholesterol are likely masked by the large free cholesterol pool in the plasma membrane. We studied free cholesterol on intracellular compartments by confocal microscopy using the membrane cholesterol-binding fluorescent molecule filipin, following extraction of plasma membrane cholesterol (33). We find that intracellular cholesterol levels in fibroblasts grown for 48 h in lipid-deficient culture conditions were significantly lower (Fig. 4, C and D), validated by the earlier measured decrease in Figure 4B. Treatment with the compound U18666a strongly increased the filipin signal (Fig. 4, C and D) corresponding to a strong accumulation of cholesterol in endosomes (Fig. 4C). Importantly, after 48 h of rotenone treatment, the intracellular filipin staining was significantly augmented compared to the labeling intensity in control human fibroblasts, demonstrating an increase of cholesterol in the intracellular compartments (Fig. 4, C and D). Taken together, complex I inhibition elevates cholesterol on intracellular compartments without significantly affecting total cellular cholesterol.
Complex I inhibition affects multiple cholesterol sensing mechanisms in the ER
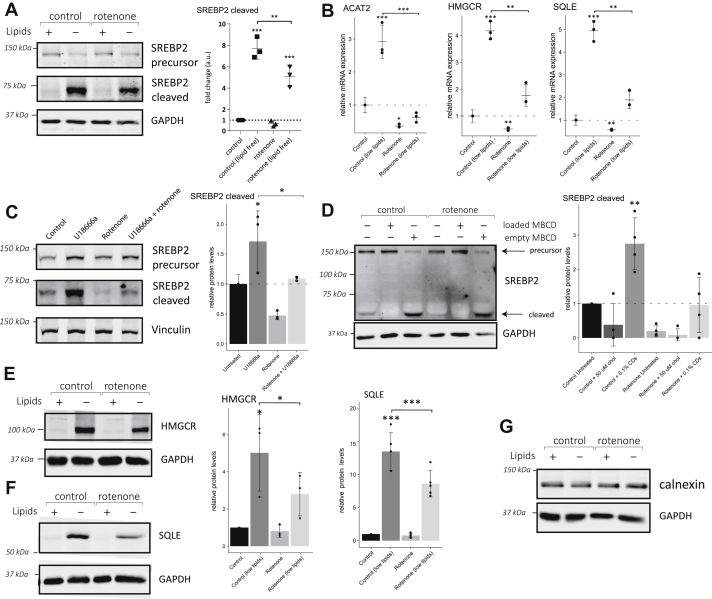
We next questioned if the observed increase in intracellular cholesterol was detected by ER sterol sensing proteins. Given the reduction of mevalonate pathway gene expression after complex I inhibition, we first investigated whether mitochondrial dysfunction inhibited sterol-dependent SREBP2 processing. Under our standard culture conditions, most SREBP2 was found in its precursor form, and only a minor fraction had undergone proteolytic processing in the Golgi (Fig. 5A). Processing of SREBP2 was induced in lipid-free growth medium containing atorvastatin. In such sterol depletion conditions, the SREBP2 precursor protein was processed, increasing the cleaved form of SREBP2 (Fig. 5A). Following inhibition of complex I, maturation of SREBP2 was reduced in both sterol conditions (Fig. 5A). This decrease was most visible in sterol-depleted conditions. In normal growth medium, the quantification of this protein band suggested lowered abundance in rotenone-treated cells, but this difference did not reach significance (Fig. 5A; quantification). The expression of mevalonate pathway genes was used as a read-out of SREBP2 transcriptional activity (Fig. 5B). As expected, when fibroblasts were cultured in cholesterol-depletion conditions, we observed a strong induction of ACAT2, SQLE, and HMGCR. Rotenone reduced the levels of SREBP2 transcriptional activity, in both normal and sterol-depleted conditions relative to the respective controls (Fig. 5B), in line with the observed decrease in protein processing. This demonstrates that complex I inhibition interferes with the ability of the cell to respond to cellular sterol levels.
Figure 5.
Complex I inhibition affects posttranslational regulation of multiple cholesterol sensors on the ER.A, expression of SREBP2 precursor and cleaved SREBP2 after maturation in human primary fibroblasts following culture in the presence of DMSO (0.1%) or rotenone (100 nM) in either normal media or lipid-deficient media for 48 h as indicated. The lipid-deficient medium contained delipidated serum and atorvastatin (1 μM). Quantification of cleaved SREBP2 from three independent experiments is shown on the right. Individual values ±standard deviation is expressed as fold change compared to control cells in standard medium. Significance was calculated by ANOVA with Tukey HSD post-hoc test. B, gene expression of ACAT2, SQLE, and HMGCR measured by qPCR in human primary fibroblasts cultured and treated as in panel A. Individual measurements (black circles) ± standard deviation from three independent experiments are presented. Significance was calculated by ANOVA with Tukey HSD post-hoc test. C, protein expression of SREBP2 precursor and cleaved form in primary human fibroblasts following retention of cholesterol in endosomes. Cells were cultured in normal conditions, supplemented with either rotenone (100 nM), U18666a (2 μM), or both. Quantification of cleaved SREBP2 from three independent experiments is shown on the right. Individual values ±standard deviation are expressed as fold change compared to control cells in standard medium. Significance was calculated by ANOVA with Tukey HSD post-hoc test. D, representative Western blot of SREBP2 protein expression in human primary fibroblasts following acute manipulation of cellular cholesterol with methyl-β-cyclodextrins (MBCD). Fibroblasts were cultured in control medium for 2 days in the absence (0.1% DMSO) or presence of rotenone (100 nM). Two hours before protein extraction, cellular cholesterol was manipulated using vehicle, empty MBCD (0.1%), or cholesterol-loaded (50 μM) MBCD. Quantification of mature form of SREBP2. Individual measurements ±standard deviation compared to control untreated cells from four independent experiments are shown (right). Significance was calculated by ANOVA with Tukey HSD post-hoc test. E–G, protein expression of HMGCR (E), SQLE (F), and calnexin (G) in primary human fibroblasts (left). Cells were cultured as described for panel A. Quantification of HMGCR and SQLE Western blot signals (right) of four independent experiments ±standard deviation normalized to loading controls are shown. GAPDH or Vinculin protein signals were used as loading control. Significance was calculated between indicated samples by ANOVA with Tukey HSD post-hoc test. ns: not significant; ∗ p < 0.05; ∗∗ p < 0.01; ∗∗∗ p < 0.001 compared to DMSO control. DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HMGCR, 3-Hydroxy-3-Methylglutaryl-CoA reductase; SQLE, squalene epoxidase; SREBP2, sterol regulatory element–binding protein 2.
A more targeted way of reducing ER cholesterol is by blocking cholesterol transport out of late endosomes with U18666a (34). In cells treated with U18666a, SREBP2 cleavage was increased, confirming a depletion of ER cholesterol (Fig. 5C). U18666a-induced SREBP2 processing was reduced in cells with complex I dysfunction. The defect in sterol sensing after mitochondrial inhibition is thus independent of enhanced retrograde transport of cholesterol from late endosomes.
Adaptation of SREBP2 processing to cholesterol levels is a rapid process. We therefore questioned whether inhibition of mitochondria affects SREBP2 processing in response to acute manipulation of intracellular cholesterol levels. To reveal this, we treated human fibroblasts with empty cyclodextrin or cholesterol-loaded cyclodextrin to extract respectively overload cells with cholesterol. After 2 h of adding exogenous cholesterol, all detectable SREBP2 was in its precursor form suggesting very slow or absent processing (Fig. 5D). Cholesterol extraction had the opposite effect: the abundance of the precursor was markedly reduced with a concurrent strong increase of processed SREBP2 compared to untreated control cells. Rotenone significantly reduced the level of mature SREBP2 in control medium, in line with earlier experiments. Augmentation of cellular cholesterol under conditions of impaired complex I was able to cause the accumulation of the SREBP2 precursor form (Fig. 5D). When complex I was inhibited, we observed less mature SREBP2 after depleting sterols with empty cyclodextrins. In conclusion, fibroblasts with complex I dysfunction show reduced mature SREBP2 and are unable to induce a normal response to low levels of cholesterol.
To test whether ER sterol sensing in general was affected, we studied HMGCR and SQLE, two additional ER-localized sterol sensors. HMGCR and SQLE are mevalonate pathway enzymes that undergo rapid sterol-dependent ubiquitination and degradation by the proteasome. Inversely, low sterol levels stabilize these enzymes, resulting in their accumulation. In human fibroblasts, both enzymes were dramatically increased in the sterol-free medium consistent with the expected stabilization (Fig. 5, E and F). When complex I function was inhibited, the abundance of these cholesterol sensing proteins was reduced compared to control conditions. In lipid containing medium, these two proteins were barely detectable at baseline and therefore a possible reduction was hard to assess. The abundance of calnexin, another ER protein unrelated to cholesterol metabolism, was unaffected by rotenone treatment; thus, the observed effects were specific to cholesterol sensing enzymes (Fig. 5G). Together, we show that several cholesterol sensors residing on the ER fail to respond fully to falling cholesterol levels. In summary, our results show that both transcriptional and posttranslational regulation of mevalonate pathway enzymes are affected in our model of respiratory dysfunction.
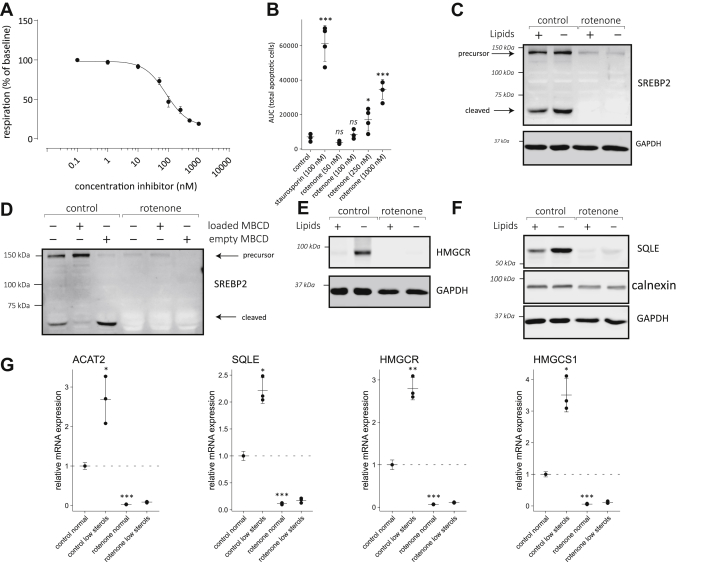
Complex I inhibition impairs the activation of ER sterol sensors in HepG2 hepatoma cell
The liver is a main tissue for the regulation of circulating cholesterol in the human body. To evaluate whether our observations in fibroblasts could be extended to this important cholesterol-regulating cell type, we studied the effect of mitochondrial dysfunction on the regulation of the mevalonate pathway in HepG2 hepatocarcinoma cells. Concentration-dependent inhibition of complex I in HepG2 cells reveals that oxygen consumption in this cell type is slightly less sensitive to rotenone. At 100 nM rotenone, the concentration used to markedly reduce respiration in human fibroblasts, basal respiration was only lowered by 50% in HepG2 cells (Fig. 6A). No significant increase of apoptosis was noted in HepG2 cells grown for 2 days in the presence of 100 nM rotenone (Fig. 6B), which was selected in further experiments as the concentration to inhibit complex I in this cell type. Higher concentrations of rotenone caused increasingly more apoptosis in HepG2 cells. Similar to our findings in fibroblasts, the transfer of HepG2 cells to a lipid-free medium promoted SREBP2 maturation (Fig. 6C). In response to complex I inhibition, HepG2 cells showed a more striking phenotype than fibroblasts. Expression of SREBP2 was strongly reduced as seen both by the reduced intensity of the precursor band and an undetectable mature form (Fig. 6C). Acute changes in cholesterol by loading or removing cholesterol using cyclodextrins lowered respectively increased the matured SREBP2 fragment (Fig. 6D), comparable to our results in fibroblasts. After complex I inhibition however, again barely any SREBP2 protein was present. The intensity changes of the SREBP2 precursor in the different conditions suggest that cholesterol sensing still occurs although at much lower expression levels. Other cholesterol sensing enzymes were similarly affected. Both HMGCR (Fig. 6E) and SQLE (Fig. 6F) were increased in sterol-deficient conditions, confirming HepG2 cells were able to sense changes in sterol levels. The expression of HMGCR and SQLE was reduced to near undetectable levels in HepG2 cells treated with rotenone (Fig. 6, E and F). The observed effect is specific to cholesterol sensing proteins, as the levels of another ER protein calnexin were unaffected (Fig. 6F). This phenotype suggests SREBP2-dependent transcription of these genes was reduced, while at the same time cholesterol-dependent protein degradation was likely activated. This strong phenotype indicates SREBP2 transcriptional activity is almost completely abolished. Indeed, after complex I inhibition, expression of all mevalonate pathway enzymes tested (ACAT2, SQLE, HMGCR, HMGCS1) were 5- to 10-fold lower than in HepG2 cells grown in control lipid containing medium (Fig. 6G). As expected, mevalonate pathway genes were induced in control cells when grown in lipid-deficient medium. In the presence of rotenone, transfer to a lipid-free medium caused little or no induction even though basal expression was very low. Taken together, in HepG2 cells, inhibition of complex I has a very severe effect on ER cholesterol sensing enzymes. SREBP2, HMGCR, and SQLE maintain their ability to sense cholesterol, but their expression is very low even under conditions where cholesterol is acutely or chronically reduced. Transcriptional regulation of mevalonate pathway genes by SREBP2 is close to abolished by rotenone in HepG2 cells.
Figure 6.
Complex I inhibition severely impairs mevalonate pathway expression in liver carcinoma cells.A, dose-dependent inhibition of oxygen consumption by complex I inhibition. The effect of increasing concentrations of rotenone on basal respiration in the presence of 5 mM glucose was studied. Data are expressed as percentage of baseline oxygen consumption as described in Figure 1B. Shown is the average ±standard deviation from five replicates. B, apoptosis in HepG2 cells caused by increasing concentrations of rotenone. Staurosporine (100 nM) was included as a positive control. Apoptosis was measured as described for Figure 1C after 2 days in culture in the presence of increasing concentrations of rotenone. Bar indicates mean four independent experiments ±standard deviation. Individual measurements shown (black circles). C and D, expression of SREBP2 precursor and cleaved form, SQLE (E), and HMGCR (F) in HepG2 cells was followed by Western blotting. HepG2 cells were treated with either dimethyl sulfoxide (0.1%) or rotenone (100 nM) in either normal media or lipid-deficient media for 2 days as described for Figure 5A. The GAPDH signal was used as a loading control. D, HepG2 cells were grown as in C. For the final 2 h, cholesterol was acutely added or removed using cholesterol (50 μM) loaded or empty cyclodextrin respectively. G, RNA expression of ACAT2, SQLE, HMGCR, and HMGCS1 in HepG2 cells grown under conditions as described for panel C. Expression changes are given as fold change compared to cells grown in normal medium without rotenone. Shown are the results from three independent experiments (black circles) ± standard deviation. Significant differences from control were calculated by t tests followed by Benjamini–Hochberg multiple testing correction. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HMGCR, 3-Hydroxy-3-Methylglutaryl-CoA reductase; SQLE, squalene epoxidase; SREBP2, sterol regulatory element–binding protein 2.
Discussion
Mitochondrial respiratory chain dysfunction induces retrograde signaling that activates transcription of nuclear genes. This retrograde signaling is intricate and not yet fully understood, as many pathways intercommunicate and are dependent on multiple environmental factors such as the nutritional state or cellular stress (8). In this study, we used QuantSeq 3′mRNA sequencing to identify transcriptional responses caused by mitochondrial dysfunction using three well-characterized respiratory chain inhibitors: rotenone, antimycin A, and oligomycin A. We were particularly interested in retrograde signaling that occurred independently of the respiratory chain complex impaired. A subset of 384 genes (10% of total) was synchronously upregulated or downregulated in all three treatments with respiratory chain inhibitors. The upregulated genes showed enrichment in cellular stress and apoptosis-related pathways such as the mitochondrial integrated stress response, consistent with earlier reports (26). Furthermore, we found a concurrent downregulation of transcripts encoding mevalonate pathway enzymes in all three treatment groups. Earlier research in cellular models of mitochondrial dysfunction revealed a similar reduction in cholesterol biosynthesis gene expression (25, 26), but the underlying molecular mechanisms have not been further investigated.
To address if the change in mevalonate pathway transcripts seen in the three models of mitochondrial dysfunction has a functional consequence on the mevalonate pathway and its output, we measured the abundance of 22 mevalonate pathway metabolites in each of the three models. Inhibition of respiration caused a marked reduction of cholesterol precursors in the dedicated cholesterol synthesis part of the mevalonate pathway, demonstrating that inhibition of respiration has functional consequences for the generation of sterols by the mevalonate pathway. Most of these metabolites were lowered to an extent similar to treatment with the HMGCR inhibitor atorvastatin and were affected similarly by all three respiratory chain inhibitors. The observed common transcriptional response is therefore mechanistically linked to the common reduction of the sterols in response to mitochondrial respiratory chain dysfunction after inhibition of either complex I, complex III, or complex V.
The metabolites in the upper mevalonate pathway, isoprenoid synthesis and ubiquinol synthesis pathway on the other hand respond differently to the three inhibitors. The complex relationship between the site of respiratory chain inhibition and the differential effects on metabolites upstream of squalene were not further addressed in the current study.
The general downregulation of mevalonate pathway genes strongly implies a perturbation of SREBP2 transcriptional activity. In this study, we observed that complex I inhibition impaired SREBP2 processing in human fibroblasts. The reduction in SREBP2 processing was even more pronounced in HepG2 cells, where also a reduction in precursor is observed. A phenotype with severely reduced SREBP2 activation also strongly reduces total SREBP2 expression as the mature SREBP2 transcription factor drives its own transcription (35). This explains why even the SREBP2 precursor levels are strongly reduced in HepG2 cells after complex I inhibition. In human fibroblasts, mature SREBP2 appears sufficiently abundant to continue driving SREBP2 transcription with close to normal expression of the SREBP2 precursor protein.
Both the fibroblasts and the HepG2 cells maintain partial sensitivity to lipid-free culture conditions by showing an increase in SREBP2 processing. They also remain responsive to rapid cyclodextrin-mediated overloading or depletion of cholesterol. This demonstrates that in our model of complex I inhibition, the SREBP2–SCAP protein complex is still able to sense changes in cholesterol levels on the ER membrane. We propose that after complex I inhibition, both primary human fibroblasts and HepG2 cells normally sense an increase in free cholesterol but are partially insensitive to lowering of intracellular cholesterol. To further investigate the observed phenotype, we asked whether other ER-residing cholesterol-sensing proteins were dysregulated as a result of respiratory chain dysfunction. The proteins HMGCR and SQLE are both regulated transcriptionally by SREBP2 (36) and posttranslationally on the ER membrane by various sterols (37, 38). They contain a membrane embedded domain which binds cholesterol in a concentration-dependent manner. Excess sterols prime these enzymes for degradation, and inversely sterol depletion stabilizes them. Consistent with this regulatory mechanism, we observed an accumulation of HMGCR and SQLE proteins when cells were cultured in medium lacking cholesterol. Accumulation of the two cholesterol sensors under low lipid conditions was strongly reduced when complex I was inhibited in human fibroblasts. In HepG2 cells, the impact of mitochondrial function on HMGCR and SQLE protein expression was again more severe. Both proteins were barely detectable even after removal of cholesterol from the medium. Taken together, we propose that inhibition of mitochondrial functions alters sterol sensing on the ER. Likely both transcriptional and posttranslational regulation of mevalonate pathway enzymes are affected by mitochondrial dysfunction (Fig. 7).
Figure 7.
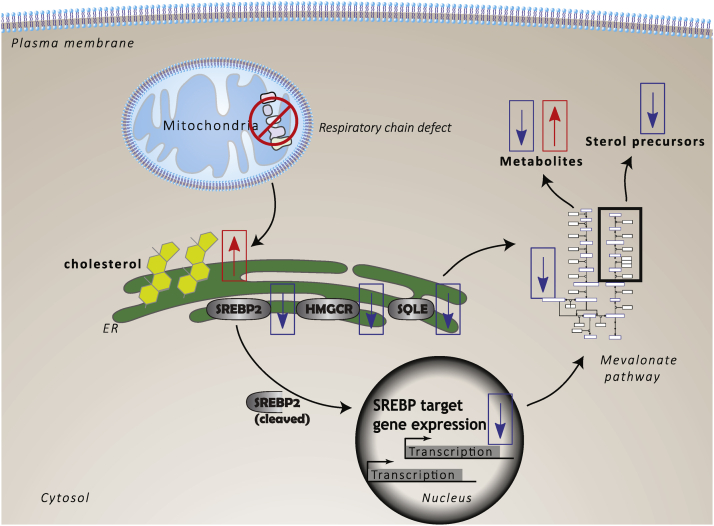
Model summarizing the alterations of cholesterol homeostasis in response to mitochondrial respiratory chain dysfunction. Mitochondrial dysfunction leads to an increase of cholesterol on the ER membrane. This increase in cholesterol downregulates the activity of SREBP2, consequently suppressing the expression of mevalonate pathway genes. The increase in cholesterol also posttranslationally regulates ER membrane–bound enzymes such as HMGCR and SQLE, reducing their abundance. This multifaceted suppression of the mevalonate pathway affects the output, lowering the abundance of sterol precursors. Other metabolites are affected in a, yet poorly understood, complex-specific manner. Indicated are the observed changes in this study. The hypothesized order of events is indicated by black arrows. The directionality of changes is indicated by boxed arrows, red indicating an upregulation or increase and blue arrows indicating a downregulation or a decrease. Proteins are indicated with gray ovals. HMGCR, 3-Hydroxy-3-Methylglutaryl-CoA reductase; SQLE, squalene epoxidase; SREBP2, sterol regulatory element–binding protein 2.
Considering that sterol-sensing ER proteins failed to normally respond to conditions of lowered cellular sterols, we speculated that respiratory chain dysfunction may elevate the amounts of cellular cholesterol. We therefore tested whether complex I inhibition in human fibroblasts elevated the free or total cellular cholesterol pool. Free cholesterol is primarily found in the membrane lipid bilayers and is available to participate in metabolic reactions and sterol signaling (39, 40, 41). The total cellular cholesterol pool includes cholesterol esters stored in lipid droplets that do not participate in cholesterol metabolism (42). We found that neither free nor total cholesterol were significantly changed in human primary fibroblasts after inhibition of complex I.
About 90% of the free cellular cholesterol is found in the plasma membrane. We hypothesized that elevated cholesterol on intracellular membranes could be responsible for the observed effects on ER sterol sensors, and this change may be masked by the abundance of free cholesterol in the plasma membrane. To specifically study the abundance of cholesterol on intracellular compartments, we visualized the intracellular pool using the free cholesterol binding dye filipin. This method accurately detected either lowering or increasing intracellular cholesterol. Based on this semiquantitative method, we could demonstrate that blocking complex I activity elevates intracellular free cholesterol in human fibroblasts. These findings support a model where after inhibition of respiratory chain activity, cholesterol is elevated on intracellular membranes (Fig. 7). Elevated cholesterol for example on the ER may explain the observed adaptive responses slowing endogenous cholesterol biosynthesis.
These findings prompt the question about the source of the increase in intracellular free cholesterol. Enhanced uptake of exogenous cholesterol can be ruled out as removing extracellular lipids did not negate the effect of complex I inhibition on the cholesterol sensing proteins. A possible target of retrograde signaling could be lipid droplets that contain cholesterol esters. Retrograde signaling may affect mobilization of cholesterol from lipid droplets or may stimulate the transfer of cholesterol from lipid droplets to the ER at membrane contact sites.
Another possible source for the apparent increase of free membrane cholesterol is cholesterol in lipid rafts. Retrograde signaling from mitochondria may stimulate the mobilization and relocation of cholesterol to intracellular membranes. This would leave the total cholesterol pool unchanged but perceived by the cell as if cholesterol was more abundant. Alternatively, mitochondrial dysfunction may also cause a shift of cholesterol from organelles with intermediate cholesterol concentrations to membranes with low levels of cholesterol such as the ER or the mitochondria. One mechanism that may contribute to this shift is enhanced retrograde membrane trafficking of cholesterol from the endocytic recycling compartment to the ER. However, after blocking the trafficking of cholesterol from the endosomes to the ER with the NPC1 protein-inhibitor U18666a, we still observed decreased SREBP2 processing and reduced levels of cholesterol sensing proteins in our model of complex I inhibition.
Here, we have obtained novel insight into the complex signaling events that follow mitochondrial respiratory chain dysfunction. We propose that mitochondrial dysfunction perturbs intracellular cholesterol homeostasis by affecting the distribution of cholesterol into different intracellular pools. One consequence of mitochondrial inhibition at complex I is the elevation of free cholesterol on intracellular membranes. Several cholesterol sensors of the ER including SREBP, HMGCR, and SQLE partially fail to react to cholesterol lowering conditions. These results suggest cholesterol levels on the ER are increased in models of mitochondrial dysfunction. An important remaining question is what specific signal in response to mitochondrial dysfunction causes a change in intracellular cholesterol homeostasis and how cholesterol trafficking is altered by mitochondrial retrograde signaling.
Experimental procedures
Reagents and chemicals
Unless otherwise stated, all chemicals were from purchased from Merck KGaA (formerly Sigma Aldrich). Phosphate buffered saline ( pH 7.4) was purchased from Thermo Fischer (Gibco, cat# 10010023).
Cyclodextrin loading
A stock solution of 5% methyl-beta-cyclodextrins was prepared by dissolving in ultrapure water. A stock solution of 1% cholesterol was made in methanol: chloroform (2:1). To load the cyclodextrins, the 5% aqueous methyl-beta-cyclodextrin solution was heated to 80 °C to remove organic solvents. Under constant agitation, cholesterol solution was dropwise added to a final concentration of 2.5 mM cholesterol. The solution was left to agitate until transparent between each addition of cholesterol solution. To ensure full encapsulation of the sterols in the cyclodextrins, the solution was left shaking overnight.
Cell culture
Human primary fibroblasts (Cell Applications, cat# 106-05a), SV40 immortalized fibroblasts (ABM, cat# T0302), and HepG2 (ECACC, cat# 85011430) were cultured in Dulbecco's Modified Eagle Medium (DMEM, Gibco cat# 11966025), containing 4 mM glutamine in a humidified incubator with 5% CO2 at 37 °C. The DMEM was supplemented with 5 mM glucose, 10% fetal bovine serum (FBS), and 100 IU/ml penicillin and 100 IU/ml streptomycin (Gibco, cat# 10270106, 15140122). For lipid deficient growth conditions, culture media was prepared as above, but the FBS was replaced with delipidated FBS (Pan Biotech, cat# P30-3401) and was supplemented with 1 μM of atorvastatin.
Growth rate and apoptosis assays
Cells were plated at a density of 1 × 103 cells per well in a clear 96-well plate. After 24 h, 200 μl of media containing inhibitors or an equivalent volume of dimethyl sulfoxide was added to the plate. For the apoptosis assays, an additional 1:200 diluted Annexin V Green Reagent (Satorius, cat# 4642) was added to the base medium. Cellular growth and apoptosis were measured in the Incucyte ZOOM live cell analysis system. Growth metrics were calculated with the R package Growthcurver (43). Apoptosis counts were expressed as total fluorescent counts after 5 days.
Immunoblotting
Cells were routinely seeded at 1 × 106 cells in a 60 cm2 Petri dish 24 h before addition of chemical compounds. On the day of collection, cells were washed twice with ice-cold PBS on ice and lysed in 150 μl of radioimmunoprecipitation assay (Merck cat# R0278) buffer, supplemented with 10 mM sodium fluoride, 2 mM sodium orthovanadate, 0.1 μM phenylmethylsulfonyl fluoride, 1× “cOmplete Protease Inhibitor Cocktail” (Roche, Cat# 11697498001), and 1× “PhosSTOP” (Roche, Cat# 4906845001). The resulting lysate was cleared by centrifugation at 8000g for 20 min at 4 °C. The protein content of the clear supernatant was quantified by Pierce BCA Protein Assay Kit (Thermofisher, Cat# 23225). Samples were diluted to 1 μg/μl and boiled for 5 min at 95 °C in Laemmli sample buffer (Alfa Aesar, Cat# J60015). SDS-acrylamide gels were hand casted according to manufacturer’s guidelines using a Bio-Rad casting system (Bio-Rad, Cat#1658050). Routinely, 25 μg of protein sample was separated on a 10% SDS-acrylamide gel. Following separation, proteins were transferred to nitrocellulose membrane paper (Li-Cor, Cat# 926-31092) in Bjerrum Schafer-Nielsen buffer (48 mM Tris, 39 mM glycine, pH 9.2) with 10% methanol on ice. Membranes were blocked for 1 h in Odyssey blocking buffer (Li-Cor, Cat# 927-50000) and incubated with primary antibodies (see table) overnight in 5% bovine serum albumin (Sigma, Cat#A2153) in Tris-buffered saline (20 mM Tris-HCL, 500 mM NaCl, pH 7.5) with 0.05% tween 20 (TBST). Membranes were washed three times 10 min with TBST, after which the membrane was incubated with secondary antibody (ThermoFisher, Cat# A11369, A32729. 1:10,000 in 5% bovine serum albumin) for 1 h. Western blots were imaged with an Odyssey CLx imaging system (LI-COR Biosciences) and quantified with the Image Studio v5.2 software.
Real-time qPCR
Fibroblasts were seeded at 1 × 106 cells in a 60 cm2 Petri dish 24 h before addition of chemical compounds. On the day of collection, cells were washed 2× with ice-cold PBS on ice and lysed in 1 ml of TRIzol reagent (ThermoFisher, cat# 155960260). RNA was extracted from the lysate according to manufacturer’s guidelines. RNA was resuspended in 32 μl of ultrapure RNase-free water. The concentration was assessed with a NanoDrop Microvolume Spectrophotometer (Thermo Fisher Scientific). Immediately after quantification, reverse transcription was performed with the PrimeScript RT Reagent Kit (Takara Bio, cat#RR037B). Levels of mRNA were measured using SYBR Green LightCycler 1536 DNA Green Master (Roche, cat# 05573092001). Relative quantification of Ct values was calculated with the ΔΔCt method (44), normalizing the Ct values to the housekeeping gene TBP.
Gas chromatography
Fibroblasts were seeded at 200,000 cells per well in a 6-well plate. The next day, media containing the interventions were added. After 48 h, the cells were trypsinized, washed once in PBS, and the cell pellet was collected. The pellets were snap frozen at −80 °C until analysis. On the day of analysis, the cell pellet was resuspended in 600 μl of 1 M KOH in ethanol. A known amount of cholesterol was treated in the same way as calibration sample. To this solution, 600 μl of a known concentration of epicoprostanol solution in cyclohexane was added as internal standard. For complete saponification, the solution was left shaking overnight. This step ensures release of all cellular cholesterol and de-esterifies all cholesterol esters. To this solution, 600 μl of 1 M KOH in water was added and was shaken for 15 min. The resulting solution was centrifuged for 15 min at 700g. The supernatant was transferred to clean autosampler vials, and the liquid was completely dried under a nitrogen flow. To the vial, 100 μl N,O-Bis(trimethylsilyl)trifluoroacetamide and 25 μl of trimethylsilyl chloride was added, and the whole mixture was heated to 80 °C for 45 min. The vials were then cooled to room temperature and dried under a gentle nitrogen flow. Following this, 1 ml of 5% N,O-bis(trimethylsilyl)-acetamide in hexane was added, and the vials were vortexed for 2 min and 1 μl of the samples were injected into an Agilent 7890B gas chromatography system coupled to a flame ionization detector. Samples were run in high purity helium gas through a 30 m DB-5 capillary column coated with (5% phenyl)-methylpolysiloxane.
Seahorse XF analyzer
Cellular oxygen consumption rates were measured using a Seahorse XFe96 analyzer. The following protocol was adapted from the manufacturers protocol. Cells were seeded at 70,000 cells per well in a 96-well Seahorse XF cell culture plate. Cells were left at room temperature to evenly distribute the cells according to Lundholt et al., 2003 (45). The following day, medium was exchanged for carbonate-free DMEM (Merck, cat# D5030) containing 5 mM glucose, 4 mM glutamine. Cells were left for 60 min to equilibrate in a CO2 free incubator at 37 °C, before placing in the analyzer. To measure the dose–dependent decrease of respiration, a range of concentrations of the inhibitors were injected, and the remaining respiration measured. Finally, rotenone (50 μM) and antimycin A (50 μM) were injected to measure nonmitochondrial respiration. After completion of the measurement, the supernatant was aspirated, and 25 μl of lysis buffer (1% SDS, 0.1 N NaOH) was added. The plate was briefly vortexed, and the protein content was measured with a BCA protein assay (Thermo Fischer Scientific, cat# 23225). The respiration measurements were normalized to protein content per well, and values are reported in residual respiration after compound injection, relative to baseline in each well.
3′RNAseq
The transcriptome of our studied models was measured with QuantSeq 3′ RNA sequencing. Total RNA was extracted from the treated primary fibroblasts using Agencourt RNAdvance Tissue kit (Beckman Coulter Life sciences, cat# A32646) and quantified using a Quant-iT RiboGreen RNA Assay Kit (Invitrogen, cat# R11490). RNA (250 ng) was used to generate libraries using the QuantSeq 3′ mRNA-Seq library prep kit (FWD) HT for Illumina (Lexogen). The kit uses total RNA as input, hence no prior poly(A) enrichment or rRNA depletion is needed. The libraries were amplified for 17 cycles quantified with Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, cat# P7589). Size pattern was controlled with the high sensitivity NGS Fragment Analysis kit on a Fragment Analyzer (Agilent DNF-474). Libraries were pooled at an equimolar ratio (i.e., an equal quantity of each sample library) and clustered at a concentration of 9 PM on single read sequencing flow cell (Illumina). Sequencing was performed for 65 cycles on a HiSeq 2500 (Illumina) using the Rapid SBS Kit v2 (Illumina, cat# FC-402-4021). Sequencing reads were trimmed with BBDuk (BBTools version 35.85, Bushnell B., sourceforge.net/projects/bbmap/). Mapping to the human genome (built GRCh38.p2) was performed with RNA STAR, version 2.3.0e, (46) using default parameters. Gene count was performed with HTSeq (47), version 0.5.4p3. For the statistical comparison of expression changes, we fit a negative binomial generalized linear model, estimated the dispersion across all genes and constructed a contrast matrix to assess differential expression between inhibitors, holding all other variables constant. In total, 32,041 features were identified. Of these features, 10,881 genes were maintained in the analysis after filtering for genes with at least 16 counts on a minimum of six samples. Calculation of differentially expressed genes for each treatment was identified using the R package edgeR (48). Clustering of GO biological processes was performed with the ShinyGO online tool, version 0.61 (49).
Metabolomics
Cells were plated at 3 million cells in a TPP300 peel-off flask (TPP, cat# 90303). After 24 h, 50 ml of media containing inhibitor or equivalent amounts of dimethyl sulfoxide were added to the cells. After 48 h, the cells were placed on ice, and rapidly the medium was aspirated, and the cells washed three times with ice-cold NaCl 0.9% solution. Immediately after washing, the cells were snap frozen on liquid nitrogen and placed on dry ice. Exactly, 3.5 ml ice-cold methanol was added to the flask, and all the material was collected by scratching and collected in a Falcon tube. This step was repeated once to ensure maximal recovery of cellular material. Two identical flasks were pooled in one Falcon tube and stored at −80 °C until processing. Before sample preparation, the samples were dried in a nitrogen evaporator at room temperature under a gentle nitrogen flow for 24 h.
The cell pellet of each sample was resuspended in 200 μl of 50% methanol and then transferred to a 5 ml Eppendorf tube. The samples were lysed by homogenizing with the aid of two metal balls on a MM 400 mill mixer at a frequency of 30 Hz for 1 min three times. 0.8 ml of mixed methanol/chloroform (3:1, v/v) was then added to each tube. The homogenization step was repeated three times, followed by ultrasonication in an ice-water bath for 5 min. The samples were centrifuged at 4000g for 45 min. The clear supernatants were collected, and the residues were used for protein assay using a standardized Bradford procedure.
Quantitation of HMG-CoA and isoprenyl phosphates
Two hundred fifty microliter of the supernatant of each sample was mixed with 200 μl of an internal standard solution of HMG-CoA-D3 and 100 μl of chloroform. The mixture was vortexed for 30 s, followed by centrifugal clarification for 5 min. After carful removal of the low organic phase using gel-loading tips, the upper aqueous phase was dried under a gentle nitrogen gas flow in a nitrogen evaporator. The residue was reconstituted in 200 μl of 50% methanol. A stock solution containing standard substances of HMG-CoA and isoprenyl phosphates was prepared in the internal standard solution. This standard solution was serially diluted in a ratio of 1 to 4 (v/v) to have calibration solutions in a range of 0.0001 to 10 μM. 10μL aliquots of the calibration solutions and the sample solutions were injected to run LCMRM on a Water Acquity UPLC system coupled to a Sciex QTRAP 6500 mass spectrometer operated in the negative-ion mode, using a C18 column (2.1∗150 mm, 1.8 μm) and a mobile phase composed of ammonium acetate buffer (A) and acetonitrile (B) for binary-solvent gradient elution (5% to 80% B in 12) at 0.3 ml/min and 50 °C. Concentrations of the detected compounds were calculated by interpolating the constructed linear-regression, internal-calibration curves with the peak area ratios measured from sample solutions. Note: IPPP and DMAPP cannot be separated, and their total amount in each sample was reported.
Quantitation of sterols
Two hundred microliter of the supernatant of each sample was dried under a gentle nitrogen gas flow. The residue was resuspended in 100 μl of a 13C3-cholesterol internal standard solution. Along with the sample preparation, serially diluted standard solutions of sterols were prepared in the same internal standard solution. 100 μl of each sample solution or each standard solution was mixed with 100 μl of 20 mM dansyl chloride solution and 100 μl of 25-mM dimethylaminopyridine solution. The mixture was allowed to react at 50 °C for 60 min. After reaction, the solution was dried in a speed-vac concentrator, and the residue was reconstituted in 400 μl of acetonitrile. 10 μl aliquots were injected onto a C18 UPLC column (2.1 × 50 cm, 1.7 μm) to run UPLC-MRM/MS on an Agilent 1290 UHPLC system coupled to an Agilent 6495B QQQ mass spectrometer operated in the positive-ion mode, using 0.1% formic acid in water acetonitrile/isopropanol (1:1) as the LC mobile phase for binary-solvent gradient elution at 0.35 ml/min and at 60 °C. Concentrations of individual sterols were calculated with the analyte-to-internal standard peak area ratios measured from injections of sample solutions to interpolate the constructed linear regression calibration curves of individual sterols in appropriate concentration ranges.
Quantitation of isoprenoids (dolichols, ubiquinones, squalene, oxidosqualene, and MK4)
500 μl of each supernatant was mixed with 500 μl of 50% methanol and 1000 μl of hexane. After vortex mixing and centrifugal clarification, the organic phase was collected. The extraction was repeated with 1000 μl of hexane twice. The pooled hexane extractant of each sample was dried under a gentle nitrogen gas flow. The residue was dissolved in 100 μl of isopropanol. Along with the sample preparation, a standard solution of squalene, 2,3-oxidosqualene, MK-4, CoQ 6, 8, 9, and 10, and a mixture of dolichols 13 to 21, was prepared in isopropanol and was then serially diluted in a volume ratio of 1 to 4. 20 μl of each sample solution or each standard solution was injected to run LC-MRM/MS on a Water Acquity UPLC system coupled to a Sciex QTRAP 6500 mass spectrometer with positive-ion detection, using a C8 column (2.1∗50 mm, 1.7 μm) and a mobile phase of 0.1% formic acid in water (A) and isopropanol (B) for binary solvent gradient elution (50% to 90% B in 15 min) at 0.25 ml/min and 50 °C. Molar concentrations of the detected compounds were calculated by interpolating the constructed linear-regression calibration curves from injection of calibration solutions, with the peak areas measured from sample solutions. Concentrations of dolichols were measured as the total weight amount of all detected dolichol compounds in each sample
Confocal microscopy
Experimental procedures were adapted from a published protocol (33). Cells were seeded at 100.000 in 35 mm glass-bottom microscopy dishes (MaTek, cat# P35G-1.5-14-C). After 24 h, the plating media were aspirated, and cells were washed once with PBS, and fresh media containing the interventions were added. After 48 h, a 5% cyclodextrin solution was added to a final concentration of 10 mM for 15 min to remove plasma membrane cholesterol. Cells were immediately washed three times with PBS and fixed with 4% PFA for 15 min at room temperature. After fixation, the cells were again washed three times with PBS, and a 50 μg/ml filipin solution in PBS was added. The cells were kept in the dark for 3 h under gentle agitation on a rotary shaker to ensure complete penetration of the filipin compound. After staining, the cells were again washed three times with PBS in a dark environment. Cells were kept in PBS during image acquisition. Filipin fluorescence intensity was captured on a confocal microscope (Leica, SP5) using a × 60 objective (oil; CFI Plan APO 1.4 NA). The focal plane was visually positioned to the middle of the cells in bright field. Filipin was excited with a Chameleon Ultra II multiphoton laser (Coherent) (1% transmission, 5% gain, 20% offset) at 700 nm (excitation), and the emission was collected at 380 to 535 nm. Images were acquired a scan resolution of 1024 × 1024 and a pinhole of 60 μm, at an 8 bit depth. Images were acquired at 10 different positions per coverslip. Quantification of intensity was performed using FIJI software (50). For each image, all pixels were selected above a threshold of 50. Intensity was expressed as mean intensity of the selected pixels. For each individual experiment, the intensities were normalized to the mean of the control intensities to correct for intraexperimental bias.
Antibody table
| Target protein | Supplier | Product no. |
|---|---|---|
| SREBP2 | BD Biosciences | 557,037 |
| SQLE | Cell Signaling Technology | 40659S |
| HMGCR | Thermo Fisher | MA5-31335 |
| SERCA2 | Cell Signaling Technology | 4388S |
| Vinculin | Cell Signaling Technology | 4650S |
| GAPDH | Cell Signaling Technology | 2118S |
Primers
| Gene | Fw | Rv |
|---|---|---|
| TBP | TGGTGTGCACAGGAGCCAAG | TTCACATCACAGCTCCCCAC |
| ACAT2 | TCGCCATGGGAGCAACATAG | ATTGGCTGGGGTGACTGTTC |
| HMGCS1 | TCTTGGGATGGACGGTATGC | CACGAAGCCCTCGTTCAAAA |
| HMGCR | GCCCTCAGTTCCAACTCACA | TTCAAGCTGACGTACCCCTG |
| DHCR7 | GACAACTGGATCCCACTGCT | CGATGCCCATCATGTAGTTG |
| SQLE | GTCTCCGGAAAGCAGCTATG | AAAAGCCCATCTGCAACAAC |
Data availability
All the experimental data are contained within the article and figures with the exception of the Quantseq data, which is accessible through the GEO Series accession number GSE191034.
Supporting information
This article contains supporting information
Conflict of interest
All authors were employed by Société des Produits Nestlé S.A. during the generation of this manuscript.
Acknowledgments
The authors thank Dr Jun Han and Dr David Schibli for the service of the Metabolomics Innovation Centre. A special thanks to Steve Lassueur, Magali Joffraud, Dr Philipp Rhein, Dr Margherita Springer, and Dr Robin Willows for the technical advice and discussions.
Author contributions
C. T. J. W. and J. S. D. methodology; C. T. J. W., P. D., A. W., and J. S. D. validation; C. T. J. W. and G. L. formal analysis, C. T. J. W. and S. M. investigation; C. T. J. W. and G. L. data curation, C. T. J. W., A. W., and J. S. D writing–manuscript; C. T. J. W. visualization; P. D., A. W., and J. S. D. supervision. A. W. and J. S. D. conceptualization; A. W. project administration.
Edited by Dennis Voelker
Contributor Information
Andreas Wiederkehr, Email: andreas.wiederkehr@epfl.ch.
Jaime Santo-Domingo, Email: jaime.santo-domingo@uva.es.
Supporting information
References
- 1.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 2.Zorova L.D., Popkov V.A., Plotnikov E.Y., Silachev D.N., Pevzner I.B., Jankauskas S.S., Babenko V.A., Zorov S.D., Balakireva A.v., Juhaszova M., Sollott S.J., Zorov D.B. Mitochondrial membrane potential. Anal. Biochem. 2018;552:50–59. doi: 10.1016/j.ab.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spinelli J.B., Haigis M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018;20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McBride H.M., Neuspiel M., Wasiak S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 5.Smeitink J.A., Zeviani M., Turnbull D.M., Jacobs H.T. Mitochondrial medicine: A metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 2006;3:9–13. doi: 10.1016/j.cmet.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Smeitink J.A.M., van den Heuvel L.W.P.J., Koopman W.J.H., Nijtmans L.G.J., Ugalde C., Willems P.H.G.M. Cell biological consequences of mitochondrial NADH: Ubiquinone oxidoreductase deficiency. Curr. Neurovasc. Res. 2004;1:29–40. doi: 10.2174/1567202043480224. [DOI] [PubMed] [Google Scholar]
- 7.Koopman W.J.H., Nijtmans L.G.J., Dieteren C.E.J., Roestenberg P., Valsecchi F., Smeitink J.A.M., Willems P.H.G.M. Mammalian mitochondrial complex I: Biogenesis, regulation, and reactive oxygen species generation. Antioxid. Redox Signal. 2010;12:1431–1470. doi: 10.1089/ars.2009.2743. [DOI] [PubMed] [Google Scholar]
- 8.Butow R.A., Avadhani N.G. Mitochondrial signaling: The retrograde response. Mol. Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z., Butow R.A. Mitochondrial retrograde signaling. Annu. Rev. Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 10.Suomalainen A., Battersby B.J. Mitochondrial diseases: The contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol. 2018;19:77–92. doi: 10.1038/nrm.2017.66. [DOI] [PubMed] [Google Scholar]
- 11.Quirós P.M., Mottis A., Auwerx J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 12.Martin L.A., Kennedy B.E., Karten B. Mitochondrial cholesterol: Mechanisms of import and effects on mitochondrial function. J. Bioenerg. Biomembr. 2016;48:137–151. doi: 10.1007/s10863-014-9592-6. [DOI] [PubMed] [Google Scholar]
- 13.Kulig W., Cwiklik L., Jurkiewicz P., Rog T., Vattulainen I. Cholesterol oxidation products and their biological importance. Chem. Phys. Lipids. 2016;199:144–160. doi: 10.1016/j.chemphyslip.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Brown A.J., Jessup W. Oxysterols: Sources, cellular storage and metabolism, and new insights into their roles in cholesterol homeostasis. Mol. Aspects Med. 2009;30:111–122. doi: 10.1016/j.mam.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Song B.L., Javitt N.B., DeBose-Boyd R.A. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 2005;1:179–189. doi: 10.1016/j.cmet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein J.L., DeBose-Boyd R.A., Brown M.S. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 17.Luo J., Jiang L.Y., Yang H., Song B.L. Intracellular cholesterol transport by sterol transfer proteins at membrane contact sites. Trends Biochem. Sci. 2019;44:273–292. doi: 10.1016/j.tibs.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Brown M.S., Goldstein J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 19.Sun L.P., Li L., Goldstein J.L., Brown M.S. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 20.Sharpe L.J., Brown A.J. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) J. Biol. Chem. 2013;288:18707–18715. doi: 10.1074/jbc.R113.479808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein J.L., Brown M.S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 22.García-Bartolomé A., Peñas A., Marín-Buera L., Lobo-Jarne T., Pérez-Pérez R., Morán M., Arenas J., Martín M.A., Ugalde C. Respiratory chain enzyme deficiency induces mitochondrial location of actin-binding gelsolin to modulate the oligomerization of VDAC complexes and cell survival. Hum. Mol. Genet. 2017;26:2493–2506. doi: 10.1093/hmg/ddx144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng M., Ostrovsky J., Kwon Y.J., Polyak E., Licata J., Tsukikawa M., Marty E., Thomas J., Felix C.A., Xiao R., Zhang Z., Gasser D.L., Argon Y., Falk M.J. Inhibiting cytosolic translation and autophagy improves health in mitochondrial disease. Hum. Mol. Genet. 2015;24:4829–4847. doi: 10.1093/hmg/ddv207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Distelmaier F., Valsecchi F., Liemburg-Apers D.C., Lebiedzinska M., Rodenburg R.J., Heil S., Keijer J., Fransen J., Imamura H., Danhauser K., Seibt A., Viollet B., Gellerich F.N., Smeitink J.A.M., Wieckowski M.R., et al. Mitochondrial dysfunction in primary human fibroblasts triggers an adaptive cell survival program that requires AMPK-α. Biochim. Biophys. Acta. 2015;1852:529–540. doi: 10.1016/j.bbadis.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Quirós P.M., Prado M.A., Zamboni N., D’Amico D., Williams R.W., Finley D., Gygi S.P., Auwerx J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 2017;216:2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mick E., Titov D.v., Skinner O.S., Sharma R., Jourdain A.A., Mootha V.K. Distinct mitochondrial defects trigger the integrated stress response depending on the metabolic state of the cell. Elife. 2020;9 doi: 10.7554/eLife.49178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo X., Aviles G., Liu Y., Tian R., Unger B.A., Lin Y.H.T., Wiita A.P., Xu K., Correia M.A., Kampmann M. Mitochondrial stress is relayed to the cytosol by an OMA1–DELE1–HRI pathway. Nature. 2020;579:427–432. doi: 10.1038/s41586-020-2078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kühl I., Miranda M., Atanassov I., Kuznetsova I., Hinze Y., Mourier A., Filipovska A., Larsson N.-G. Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. Elife. 2017;6 doi: 10.7554/eLife.30952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stöckl P., Hütter E., Zwerschke W., Jansen-Dürr P. Sustained inhibition of oxidative phosphorylation impairs cell proliferation and induces premature senescence in human fibroblasts. Exp. Gerontol. 2006;41:674–682. doi: 10.1016/j.exger.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Ribatti D. A revisited concept: Contact inhibition of growth. From cell biology to malignancy. Exp. Cell Res. 2017;359:17–19. doi: 10.1016/j.yexcr.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Fusakio M.E., Willy J.A., Wang Y., Mirek E.T., Baghdadi R.J.T.A., Adams C.M., Anthony T.G., Wek R.C. Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Mol. Biol. Cell. 2016;27:1536–1551. doi: 10.1091/mbc.E16-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Xu S., Mihaylova M.M., Zheng B., Hou X., Jiang B., Park O., Luo Z., Lefai E., Shyy J.Y.-J., Gao B., Wierzbicki M., Verbeuren T.J., Shaw R.J., Cohen R.A., et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilhelm L.P., Voilquin L., Kobayashi T., Tomasetto C., Alpy F. In: Intracellular Lipid Transport: Methods and Protocols. Drin G., editor. Springer New York; New York, NY: 2019. Intracellular and plasma membrane cholesterol labeling and quantification using filipin and GFP-D4; pp. 137–152. [DOI] [PubMed] [Google Scholar]
- 34.Lu F., Liang Q., Abi-Mosleh L., Das A., de Brabander J.K., Goldstein J.L., Brown M.S. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. Elife. 2015;4 doi: 10.7554/eLife.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato R., Inoue J., Kawabe Y., Kodama T., Takano T., Maeda M. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J. Biol. Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- 36.Howe V., Sharpe L.J., Prabhu A.v., Brown A.J. New insights into cellular cholesterol acquisition: Promoter analysis of human HMGCR and SQLE, two key control enzymes in cholesterol synthesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:647–657. doi: 10.1016/j.bbalip.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 37.DeBose-Boyd R.A. Feedback regulation of cholesterol synthesis: Sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 2008;18:609–621. doi: 10.1038/cr.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gill S., Stevenson J., Kristiana I., Brown A.J. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 2011;13:260–273. doi: 10.1016/j.cmet.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Simons K., Ikonen E. How cells handle cholesterol. Science. 2000;290:1721–1726. doi: 10.1126/science.290.5497.1721. [DOI] [PubMed] [Google Scholar]
- 40.Luo J., Yang H., Song B.L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020;21:225–245. doi: 10.1038/s41580-019-0190-7. [DOI] [PubMed] [Google Scholar]
- 41.Das A., Brown M.S., Anderson D.D., Goldstein J.L., Radhakrishnan A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife. 2014;3 doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walther T.C., Farese R.v. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sprouffske K., Wagner A. Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics. 2016;17:172. doi: 10.1186/s12859-016-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Lundholt B.K., Scudder K.M., Pagliaro L. A simple technique for reducing edge effect in cell-based assays. J. Biomol. Screen. 2003;8:566–570. doi: 10.1177/1087057103256465. [DOI] [PubMed] [Google Scholar]
- 46.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. Star: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S., Pyl P.T., Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xijin Ge S., Jung D., Yao R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the experimental data are contained within the article and figures with the exception of the Quantseq data, which is accessible through the GEO Series accession number GSE191034.