Abstract
Peripheral nerve regeneration remains a significant clinical challenge due to the unsatisfactory functional recovery and public health burden. Exosomes, especially those derived from mesenchymal stem cells (MSCs), are promising as potential cell-free therapeutics and gene therapy vehicles for promoting neural regeneration. In this study, we reported the differentiation of human adipose derived MSCs (hADMSCs) towards the Schwann cell (SC) phenotype (hADMSC-SCs) and then isolated exosomes from hADMSCs with and without differentiation (i.e., dExo vs uExo). We assessed and compared the effects of uExo and dExo on antioxidative, angiogenic, anti-inflammatory, and axon growth promoting properties by using various peripheral nerve-related cells. Our results demonstrated that hADMSC-SCs secreted more neurotrophic factors and other growth factors, compared to hADMSCs without differentiation. The dExo isolated from hADMSC-SCs protected rat SCs from oxidative stress and enhanced HUVEC migration and angiogenesis. Compared to uExo, dExo also had improved performances in downregulating pro-inflammatory gene expressions and cytokine secretions and promoting axonal growth of sensory neurons differentiated from human induced pluripotent stem cells. Furthermore, microRNA (miRNA) sequencing analysis revealed that exosomes and their parent cells shared some similarities in their miRNA profiles and exosomes displayed a distinct miRNA signature. Many more miRNAs were identified in dExo than in uExo. Several upregulated miRNAs, like miRNA-132-3p and miRNA-199b-5p, were highly related to neuroprotection, anti-inflammation, and angiogenesis. The dExo can effectively modulate various peripheral nerve-related cellular functions and is promising for cell-free biological therapeutics to enhance neural regeneration.
Keywords: Anti-oxidation, Anti-inflammatory, Axon growth, Neural regeneration, microRNA
Graphical abstract
Exosomes derived from hADMSC-SCs with the SC phenotype promote peripheral nerve-related cellular functions, including anti-oxidation, angiogenesis, anti-inflammation, and sensory neuron axonal growth.
Highlights
-
•
Exosomes were isolated from hADMSCs with and without differentiation towards SC phenotype (i.e., dExo vs uExo).
-
•
hADMSC-SCs secreted more growth factors compared to hADMSCs without differentiation.
-
•
The dExo protected rat SCs from oxidative stress and enhanced endothelial cell migration and angiogenesis.
-
•
dExo promoted axonal growth of sensory neurons differentiated from hiPSCs.
-
•
miRNA sequencing analysis unveiled and compared the exosomal and cellular miRNA profiles.
1. Instruction
Peripheral nerve diseases and injuries cause substantial clinical problems and public health burden due to poor functional recovery [[1], [2], [3]]. To create new therapies that promote complete nerve regeneration, many research projects focus on strategies that could potentially aid regeneration and lead to clinical breakthroughs, such as increasing neuron survival, improving the rate of axon outgrowth, and improving the extensive growth ability of axons to overcome inhibitory signals [4,5]. However, the effective functional recovery remains insufficient and unsatisfactory, especially for severely damaged peripheral nerves [1,4,6,7]. This situation has prompted the application of stem cell-based therapies to improve the function of peripheral nerve-related cells under injury and pathological conditions.
Mesenchymal stem cells (MSCs) are multipotent stem cells and can be isolated from various tissues, such as bone marrow, the umbilical cord, and adipose tissue [8,9]. In recent years, MSCs have gradually become one of the most promising cell therapy tools for a wide range of injuries and diseases because of their self-renewal and differentiation potentials, low immunogenicity, multipotency, and mediator secretion capacity [[10], [11], [12], [13]]. At the damaged site of a peripheral nerve, the implantation of MSCs has been reported to facilitate axonal growth and remyelination as well as enhance angiogenesis and vascularization [[14], [15], [16], [17], [18]]. Notably, MSCs can be differentiated into Schwann cell (SC)-like phenotypes (MSC-SCs) by using cocktail chemical induction, electrical stimulation, or their combinations [[19], [20], [21]]. Interestingly, several studies have shown that introducing MSC-SCs promoted sciatic nerve regeneration and functional recovery, and the regeneration efficacy with MSC-SCs was superior to that with acellular or MSC counterparts [[22], [23], [24]]. Currently, the mechanism of action for how exogenous MSCs/MSC-SCs improve functional recovery is largely unknown. One possible hypothesis is that the repair of injured peripheral nerves is promoted via paracrine mechanisms by secreting various growth factors and extracellular vesicles (EVs) from the incorporated MSCs/MSC-SCs [15,[25], [26], [27]]. However, the differences between secretomes of MSCs and MSC-SCs are yet to be unveiled.
Despite the ease of isolation and availability of MSCs, their differentiation capacity into SC-like cells, and their ability to promote axonal regeneration all being appealing for tissue engineering, there exist significant challenges associated with maintaining cell viability and vitality [28,29]. In addition, after transplantation in vivo, MSCs might differentiate into unwanted and non-peripheral nerve cell-related lineages, such as tumorigenesis and calcification, in response to local stimuli or other dominant cells in the area [28]. Compared to cell-based strategies, exosomes can potentially serve as cell-free therapies, and they are more convenient to store and transport, which helps avoid many safety risks associated with cell transplantation [[30], [31], [32]]. Exosomes are nanoscale membrane EVs with diameters ranging from 30 to 200 nm [33,34]. They are produced and released by all types of cells via fusion of multivesicular bodies and the plasma membrane [35,36]. Exosomes contain proteins, nucleic acid, and lipids, which can reflect their cell of origin [[37], [38], [39]]. Exosomes also display relatively high target specificity to the receptor cells due to their surface protein profiles [40,41]. Exosomes play multiple roles in various biological processes through transferring their encapsulated cargos to recipient cells [[42], [43], [44]]. It has been reported that MSC-derived exosomes, either administrated alone or together with scaffolds (depending on the animal models), promoted the recovery of motor function and axon reconnection [25,45,46]. All these studies demonstrate the feasibility and efficacy of the application of MSC-derived exosomes for peripheral nerve regeneration. Exosomes retain some of the characteristics of their parent cells. For example, Wang et al. reported that exosomes derived from SCs, rather than fibroblasts, promoted MSC differentiation towards SC phenotypes [47]. In addition, Lopez-Verrilli et al. found that SC-derived exosomes markedly facilitated neurite growth substantially and greatly enhanced axonal regeneration in vitro and in vivo [48]. Taking the above motivations together, MSC–SC–derived exosomes have great potential for peripheral nerve regeneration due to the combining of MSC and SC characteristics, which has not been reported before. Most importantly, the methods in which the exosomes derived from MSCs with different differentiation statuses towards SCs regulate the behaviors of peripheral nerve-related cells are unknown, and the signature microRNA (miRNA) cargoes for both exosomes and their parent MSCs are unrevealed.
In this study, as shown in Scheme 1, we first differentiated human adipose derived MSCs (hADMSC) into SC-like cells and compared the secreted growth factors with undifferentiated hADMSCs. Then, the exosomes were isolated from supernatant media of undifferentiated and differentiated hADMSC (denoted as uExo and dExo). Subsequently, we explored the effects of exosomes on the modulatory functions of peripheral nerve-related cells, including rat SCs (rSCs), human macrophages, endothelial cells, and sensory neurons. Anti-oxidation and anti-inflammatory effects of exosomes were evaluated using rSCs and human monocyte derived macrophages, respectively. In addition, angiogenesis and cell migration of human umbilical vein endothelial cells (HUVECs) were compared after the addition of exosomes derived from MSCs with different differentiation statuses. We also differentiated human induced pluripotent stem cells (hiPSCs) into sensory neurons (SNs) and further generated sensory neurospheres to study the effects of different exosome origins on axonal growth. Furthermore, exosomal miRNA sequencing was conducted and analyzed to identify key miRNAs with the potential to promote the function of peripheral nerve-related cells.
Scheme 1.
Exosomes (uExo and dExo) isolated from human ADMSCs and their SC-like (hADMSC-SCs) phenotypes for regulating peripheral nerve-related cellular functions.
2. Materials and methods
2.1. hADMSCs culture and differentiation
Primary hADMSCs (Lonza, USA) before passage 6 were cultured in Dulbecco's Modified Eagle's Medium/F12 (DMEM/F12, HyClone, USA) with 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin/streptomycin (P/S, Invitrogen, USA) at 37 °C and 5% CO2. Differentiation of hADMSCs into an SC phenotype (hADMSC-SCs) could be achieved using a previously published method with some modifications [13,[49], [50], [51], [52]]. Briefly, the hADMSCs were seeded on a T-175 flask at a density of 1 × 105 cells. Differentiation was initiated in hADMSC cultures with fresh medium containing 1 mM β-mercaptoethanol (Sigma) for 24 h. After washing with sterilized PBS, the medium was replaced with DMEM/F12 with 10% FBS, 1% P/S, and 35 ng/mL all-trans retinoic acid and cells were cultured at 37 °C and 5% CO2 for 72 h. After extensively rinsing with sterilized PBS, the culture medium was replaced with fresh medium containing 5.7 μg/mL forskolin (AdipoGen Life Sicences), 10 ng/mL basic fibroblast growth factor (bFGF, PeproTech), 200 ng/mL recombinant hereglulin-β1 (PeproTech), 10 ng/mL brain derived neurotrophic factor (BDNF, PeproTech), and 5 ng/mL platelet derived growth factor AA (PDGF-AA, PeproTech). The cells were maintained in this medium for 14 days with medium changes every 3 days to establish differentiated cultures.
2.2. Immunofluorescence (IF) staining
The cultured cells (hADMSCs and hADMSC-SCs) were washed three times in PBS and fixed in 4% paraformaldehyde (PFA) at 4 °C for 4 h, respectively. Then, the samples were washed three times in PBS and permeabilized by using 0.2% Triton X-100 (Sigma) for 10 min at room temperature (RT). Subsequently, the samples were blocked in 1% BSA at 4 °C overnight after three PBS washes. For the evaluation of SC-like phenotype differentiation, the samples were then incubated at 4 °C overnight with primary SC-specific antibodies, anti-S100B (1: 200, Sigma) and anti-myelin basic protein (MBP, 1: 200, Sigma). Descriptions related to IF staining for the evaluation of exosome uptake and hiPSC differentiation can be found in the later sections. After washing three times with PBS, samples were incubated with secondary fluorescent antibodies at RT for 2 h and further incubated with DRAQ5 (1: 1000, Thermo Scientific) for 30 min at RT. Finally, the stained samples were washed one time in PBS and imaged with a confocal laser scanning microscopy (CLSM, LSM710, Carl Zeriss, Germany).
2.3. Human growth factor antibody array analysis
A human growth factor antibody array was used to evaluate and compare various cytokines secreted by the hADMSCs with and without differentiation. After 2-week culture, all of the media from the two cellular groups was collected and analyzed by utilizing a human growth factor array kit (RayBio C-Series), according to the manufacturer's protocols. This kit could detect 41 different types of human growth factors. The arrays in each group were scanned, and signal intensities were quantified by Image J software.
2.4. Exosome isolation and characterization
Exosomes were isolated from hADMSC (uExo) and hADMSC-SCs (dExo) by using ultracentrifugation. Prior to exosome collection, hADMSCs (passage 3–6) were cultured in medium with 10% exosome depleted FBS (Gibco) and 1% P/S at 37 °C and 5% CO2. The medium was collected and centrifuged at 4 °C and 300×g for 5 min, 1000×g for 20 min, and then at 10,000×g for 30 min, sequentially. Upon removing nonadherent cells and debris, the supernatant was then filtered with a 0.22 μm filter and ultracentrifuged (Sorval X+80 Ultracentrifuge, Thermo Fisher) at 4 °C and 100,000×g for 70 min. Then the exosome pellet was washed with PBS and centrifuged at 4 °C and 100,000×g for 70 min. The collected exosomes were reconstituted in PBS and then preserved at −80 °C. The sizes and concentrations of the final exosomes were examined by nanoparticle tracking analysis (NTA) using a NanoSight (NS300) measurement. The exosome morphology was measured, as previously described, by transmission electron microscopy (TEM, Hitachi H7500; Hitachi) and atomic force microscopy (AFM, Bruker) [53].
2.5. Western blot
The surface markers (CD9, CD63, TSG101, and Alix) of the isolated exosomes were correspondingly detected by Western blotting as reported previously [53,54]. The exosomes were lysed using a Mammalian Cell Lysis kit (Sigma) and quantified by a Micro BCA Protein Assay kit (Pierce). Equal amounts of the proteins were electrophoresed in a sodium dodecyl sulphate-polyacrylamide gel (10%–12%) under reducing conditions followed by transfer to polyvinylidene fluoride (PDVF) membranes. The blots were blocked with 5% non-fat dry milk in PBS. Western blots were then probed with antibodies recognizing the CD9 antibody (1:1000, Abcam), CD63 antibody (1:1000, Abcam), TSG101 (1:1000, Abcam), and calnexin (1:1000, Sigma). The secondary antibodies were alkaline phosphatase conjugated to goat anti-mouse (Santa Cruz)/rabbit lgG (1:5000, Jackson ImmunoResearch Labs)). Signals were detected by chemiluminescence and imaged on an FLA-5100 (Fujifilm) digital image scanner.
2.6. Exosome labeling and uptake in cells
Exosomes were labeled with PKH26 dye (Sigma, USA) according to the manufacturer's protocols for 5 min and then washed with PBS using ultracentrifugation for 2 h at 4 °C and 100,000×g. Pellets were resuspended in 200 μL of PBS [45,55,56]. For tracking exosome distribution in the cells, PKH26-labeled exosomes (3 × 109/mL) were added into the culture medium and incubated with the recipient cells (HUVECs, SCs and macrophages) at 37 °C overnight. After washing with sterilized PBS, the cells were fixed in 4% PFA at 4 °C for 4 h and permeabilized by using 0.2% Triton X-100 at RT for 10 min. Subsequently, HUVECs and rSCs were stained by F-actin (green) and DRAQ5 (blue). macrophages were stained by F-actin (green) and DAPI (blue). Finally, the labeled cells were imaged using a CLSM.
2.7. rSC culture and proliferation assay
The rSCs were harvested from the sciatic nerves of rats according to the protocol from our previous study [21,57]. The rSCs were then seeded onto a poly-d-Lysine coated T-25 flask and cultured with DMEM medium with 10% FBS and 1% P/S in 5% CO2 at 37 °C. For rSC proliferation, the rSCs (2 × 103 cells per well) were seeded in a 96 well plate and then cultured using fresh culture medium with uExo or dExo (3 × 109/mL). The culture medium was completely refreshed with exosome containing medium (3 × 109/mL) every 2 days. After 1, 3, and 7 days, the proliferation of the rSCs was determined by a CCK8 kit assay. The culture medium was discarded, and the cells were washed with sterilized PBS. About 100 μL of culture medium containing 10 μL CCK8 solution (Abcam) were added to each well, followed by incubation for 4 h at 37 °C. The optical density (OD) of the sample was evaluated using a microplate reader (BioTek Synergy H1 model) at a wavelength of 460 nm. All experiments were performed in quintuplicate.
2.8. Intracellular reactive oxygen species (ROS) detection
The intracellular ROS were tested using a 2’,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) assay (EMD Millipore). The rSCs were seeded in 96-well culture plates with 1 × 104 cells in each well and incubated for 24 h. Subsequently, the cells were incubated with exosomes (uExo or dExo, 3 × 109/mL) overnight and then treated with H2O2 (200 μM) in the present of exosomes (3 × 109/mL). After incubation at 37 °C for 24 h, the cells were washed with PBS three times and treated with 10 μM H2DCFDA for 20 min in the dark at 37 °C. Then, the cells were washed with PBS three times. The intracellular ROS concentration was measured by detecting the dihydrodichlorofluorescein (DCF) fluorescence intensity (Ex488/Em525) through a microplate reader. The cells that were untreated by exosomes or H2O2 were used as a negative control. For imaging, cells were washed with PBS, fixed with 4% (w/v) PFA, and stained with DRAQ5. The cells were imaged using a CLSM.
2.9. HUVEC migration assay
HUVECs (Lonza) were seeded in a T-75 flask coated by collagen (RatCol®) and cultured in endothelial cell basal medium-2 (EGM-2 BulletKit, Lonza) at 37 °C and 5% CO2. The medium was replaced every 2 days. For the migration assay, HUVECs were seeded in coated 24-well plates. Upon complete confluency, wounds were made with a 200 μL sterile pipette tip. The detached cells were washed with PBS buffer and then cultured in the complete medium containing exosomes (uExo or dExo, 3 × 109/mL). The images of the scratch at different time points were taken under an inverted microscope (Leica). The migration rate was calculated and reported as the wound closure rate (%) according to the formula: wound closure rate (%) = [(wound length at 0 h – wound length at 24 h)/wound length at 0 h] × 100 [58]. At least 5 images were randomly taken for each well and then analyzed using Image J software. Each sample was performed in quintuplicate.
2.10. HUVEC tube formation assay
The tube formation assay was performed using a method similar to our previously reported one [58]. Briefly, 50 μL of cold Matrigel (Corning) were added into each well in a 96-well plate and cultured at 37 °C for 1 h. After the Matrigel coating was applied, HUVECs were seeded onto the Matrigel-coated wells with 100 μL medium at a density of 1 × 105 cells/mL and then cultured in presence of exosomes (uExo or dExo, 3 × 109/mL) for 16 h at 37 °C. Tube images (n = 5) were randomly taken under an inverted microscope. The observed total tube node branch points in each image were counted using ImageJ software.
2.11. Monocyte differentiation and anti-inflammatory activity evaluation
Monocytes isolated from peripheral blood mononuclear cells from anonymous human blood samples and were provided by the Elutriation Core Facility at the University of Nebraska Medical Center. Differentiation of monocytes into Mɸ and M1 macrophage was performed following a protocol modified from previous studies [59,60]. Briefly, 5 × 105 monocytes were seeded on sterile glass coverslips in 24-well plates and then cultured for 6 days in RPMI 1640 medium (Gibco) containing 10% FBS, 1% P/S, and 50 ng/mL of recombinant granulocyte macrophage colony-stimulating factor (GM-CSF, PeproTech). After 6 days, the cell culture medium was changed with fresh medium containing uExo or dExo (3 × 109/mL) and GM-CSF and cultured overnight. Then, Mɸ macrophages were stimulated with 100 ng/mL lipopolysaccharide (LPS) and 20 ng/mL interferon-gamma (IFN-γ) in the present of exosomes (3 × 109/mL). The experiment was divided into four groups: the negative control group (without stimulation and without exosomes), the positive control LPS group (with LPS/IFNγ stimulation), and treated groups (LPS/IFNγ + uExo, LPS/IFNγ + dExo). After overnight incubation, the culture medium was collected to assay the cytokines' levels of IL-1β, IL-6, and TNF-α with ELISA kits (bio-techne®) according to manufacturer's instructions. Real-time polymerase chain reaction (RT-PCR) was also conducted to evaluate the gene expressions of nuclear factor kappa B (NFκB), tumor necrosis factor (TNF), and nitric oxide synthase 2 (NOS2), as described in the later section. All the test results were performed in quadruplicate.
2.12. hiPSC differentiation into SNs and neurosphere formation
The differentiation of hiPSCs into SNs (hiPSC-SNs) was conducted following a modified protocol [[61], [62], [63], [64]]. Briefly, hiPSCs (WiCell®) were originally cultured in mTesR medium (STEMCELL™). Then, hiPSCs were cultured on a Matrigel coated 6-well plate at a seeding density of 3 × 105 cells/well. As shown in Fig. 6A, the neural differentiation of iPSCs (day 0–2) was initiated using KSR medium (Gibco) containing β-mercaptoethanol and two inhibitors (100 nM LDN1931189 and 10 μM SB431542) at 37 °C. Cells were fed daily, and N2 minus VA medium was added in increasing 25% increments every other day starting on day 4. N2 medium was Neurobasal medium (1X, Gibco) with 0.1% v/v P/S, 2% v/v B27 (50X, Gibco), 1% v/v GlutaMAX (100X, Gibco), and 1% v/v non-Essential Amino Acids (100X, NEAA, ScienCell). The induction of SNs was initiated with addition of the five inhibitors on days 2–6, including 100 nM LDN1931189, 10 μM SB431542, 3 μM CHIR99021 (Millipore), 10 μM SU5402, and 10 μM DAPT (Millipore). On days 6–10, the induction of SNs was initiated with addition of the three inhibitors, including 3 μM CHIR99021, 10 μM SU5402, and 10 μM DAPT. On day 10, hiPSC-SNs were dissociated with Accutase (Gibco) and seeded at a density of 2 × 105 cells/well in 150 μL of medium in a 96-well non-adherent U bottom plate (Corning) for neurosphere formation. The SN neurospheres were allowed to grow in maintain medium (neurobasal medium, 1% v/v P/S, 25 ng/mL human-b-NGF, BDNF, and glial cell line derived neurotrophic factor (GDNF)) for one week. The neurospheres were then embedded in Matrigel and transferred into a 6-well plate and cultured in neural differentiation medium for 24 h.
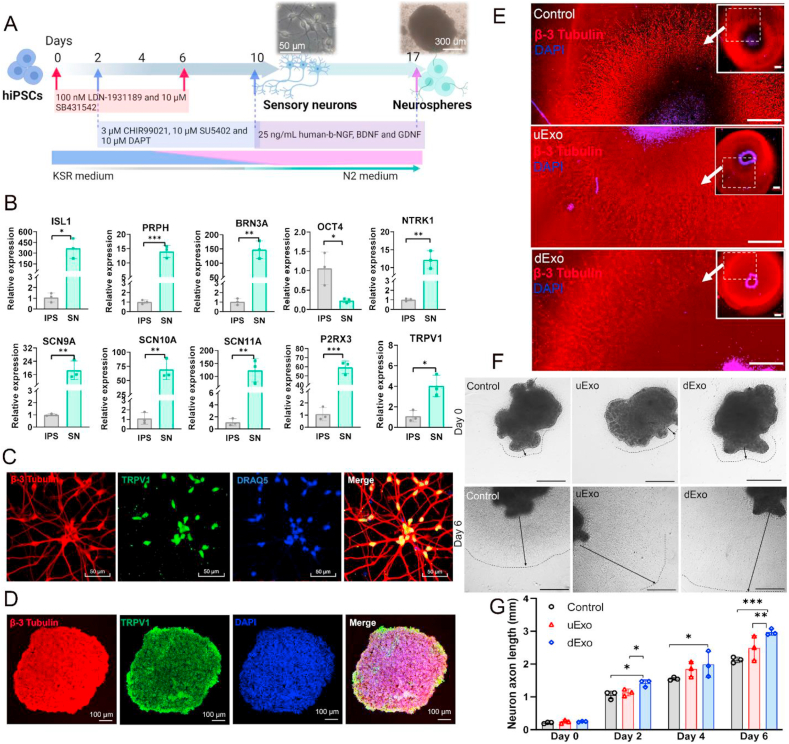
Fig. 6.
Effects of uExo and dExo on the growth of SNs derived from hiPSCs. (A) Schematic of the differentiation protocol of hiPSCs into SNs and further hiPSC-SN neurospheres. (B) Expression levels of ISL1, PRPH, BRN3A, OCT4, NTRK1, SCN9A, SCN10A, SCN11A, P2RX3, and TRPV1 for SNs were evaluated by RT-PCR. (n = 3). (C) IF staining images of hiPSC-SNs. hiPSC-SNs were positive for β-3 tubulin, TRPV1, and DRAQ5. Scale bar 50 μm. (D) IF staining images of hiPSC-SN neurospheres with β-3 tubulin, TRPV1, and DAPI. Scale bar 100 μm. (E) Representative IF images of axons from neurospheres after being encapsulated within Matrigel and treated with PBS, uExo, or dExo. Scale bar 500 μm. (F) Representative optical microscopic images of encapsulated neurospheres treated with PBS, uExo, or dExo for 6 days. Scale bar 500 μm (G) Semi-quantitative analysis of axon length of encapsulated neurospheres after treatment with uExo or dExo for 0, 2, 4, and 6 days (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001.
hiPSC-SNs and SN neurospheres were evaluated using our previously reported protocol of IF staining method [65]. The hiPSC-SNs and SN neurosphere sections were permeabilized and blocked with 0.2% Triton X-100 and 1% BSA in PBS overnight at 4 °C, followed by incubation with primary antibodies overnight at 4 °C respectively. In detail, the hiPSC-SNs and SN neurosphere sections were double stained with β-3 tubulin (1:200, Millipore), TRPV1 (1:200, Milliporem). After sequential incubation with secondary antibodies for 2 h and DRAQ5 (1:400, Thermo Scientific) for 30 min at RT, stained samples were imaged using a CLSM. For tracking exosome distribution in the hiPSC-SNs, PKH26-labeled exosomes (uExo and dExo) were added into the culture medium incubated with the recipient cells at 37 °C for 12 h. After IF staining, the labeled cells were imaged using a CLSM.
2.13. Calculation of average axon length of hiPSC-SN neurospheres
After culture in Matrigel for 24 h, the SN neurospheres were then treated with exosomes (uExo or dExo) at 3 × 109/mL and cultured for 6 days. The culture medium was completely refreshed with exosome containing medium (3 × 109/mL) every 2 days. The SN neurospheres without treatment with exosomes were employed as the control group. The images of SN neurospheres were captured using an inverted microscope on days 0, 2, 4, and 6. Three parallel samples per group were performed, and three images were captured for each sample. The average length of the longest 10 axons of each image was measured using Image J. Thirty measurements for each sample were chosen to calculate the average length of longest axon.
2.14. Quantitative RT-PCR analysis
The total RNA from the cells (macrophages or hiPSCs) was analyzed by RT-PCR. Briefly, the total RNA was isolated from cells using QIA-shredder and RNeasy mini-kits (QIAGEN). Complementary DNA (cDNA) was synthesized by following the protocol of an iScript cDNA synthesis kit (BioRad Laboratories). For the quantitative analysis of the total RNA expression, real-time PCR amplification was performed in a StepOnePlus™ Real-Time PCR System (Thermo Scientific) with the use of SYBR Green Supermix (Bio-Rad). The cDNA samples were analyzed for the genes of interest and for the housekeeping gene 18S rRNA. The relative expression of each target gene (as shown in Table S1) was calculated using the comparative Ct (2−ΔΔCt) method [58].
2.15. miRNA sequencing analysis
The total RNAs were isolated from hADMSC and differentiated hADMSCs using RNeasy mini kits (QIAGEN) according to the manufacturer's instructions. For exosomes (uExo and dExo), the total RNAs were extracted using a Total exosome RNA isolation kit (Thermo Scientific), following the manufacturer's protocol. The miRNA sequencing analysis was conducted by LC Sciences (Houston, TX, USA). The total RNA quality and quantity were analysis of Bioanalyzer 2100 (Agilent) with RIN number >7.0. Approximately 1 μg of total RNA were used to prepare small RNA library according to protocol of TruSeq Small RNA Sample Prep Kits (Illumina). The single-end sequencing 50 bp on an Illumina Hiseq 2500 at the LC Sciences following the vendor's recommended protocol. For bioinformatics analysis, the raw reads were subjected to an in-house program, ACGT101-miR (LC Sciences) to remove adapter dimers, junk, low complexity, common RNA families (rRNA, tRNA, snRNA, snoRNA) and repeats. Subsequently, unique sequences with length in 18–26 nucleotide were mapped to specific species precursors in miRBase 22.0 by BLAST search to identify known miRNAs and novel 3p- and 5p- derived miRNAs. Length variation at both 3′ and 5′ ends and one mismatch inside of the sequence were allowed in the alignment. The unique sequences mapping to specific species mature miRNAs in hairpin arms were identified as known miRNAs. The unique sequences mapping to the other arm of known specific species precursor hairpin opposite to the annotated mature miRNA-containing arm were considered to be novel 5p- or 3p derived miRNA candidates. To predict the genes targeted by most abundant miRNAs, two computational target prediction algorithms TargetScan and Miranda 3.3a were used to identify miRNA binding sites. The kyoto encyclopedia of genes and genomes (KEGG) pathways of these most abundant miRNAs, miRNA targets were annotated. The volcano plots were generated from log2(fold change) values of all miRNAs by a home-made R script. The heatmaps were plotted by R packages, pheatmap (v1.0.12), ggplot2 (v3.3.3) and gplots (v3.1.1), based on the values of log2(normalized_expression_value + 0.0001) in each sample to avoid any nonsense values of log2(0). The R 4.0.3 was used for both volcano and heatmap generation. The pathways for miRNA-132-3p and miRNA-199b-5p were predicted using the database miRPathDB 2.0 [66]. The networks were constructed using Cytoscape 3.8.2 using the miRNA and their interacting pathways [67].
2.16. Statistical analysis
All experiments were repeated at least thrice. Results are expressed as the mean ± standard deviation (SD). Statistical analysis was performed by GraphPad Prism using one-way analysis of variance (ANOVA). Differences were considered to be significant when *p < 0.05.
3. Results
3.1. Characterization and comparison of hADMSCs with and without differentiation into the SC-phenotype
hADMSCs were differentiated into the SC-like phenotype following our previous protocol with some modification (Fig. 1A) [21]. The morphologies and biomarkers of undifferentiated and differentiated hADMSCs were evaluated after 2-week differentiation. It was clear from optical microscopy that hADMSCs maintained a flattened fibroblast-like morphology and that hADMSC-SCs showed a spindle-shaped cell morphology (Fig. 1B), which was similar to genuine SCs. To verify the differentiation, we stained for S100B and MBP, the markers for SCs and myelinating SCs [68,69]. IF staining, shown in Fig. 1B, showed that hADMSC-SCs expressed much higher levels of S100B and MBP than that of hADMSCs, indicating successful differentiation. Moreover, to evaluate the myelination capacity of hADMSC-SCs, a co-culture of hADMSC-SCs and sensory neurons isolated from mouse dorsal root ganglion (DRG) was further employed. After a 21-day co-culture, hADMSC-SCs myelinated the axons of DRG neurons, with the expression of MBP highly localized along the axon fiber (Fig. S1A). TEM imaging also confirmed the formation of a myelin sheath (Fig. S1B). These results indicated that hADMSCs were successfully differentiated into an SC-like cell lineage and that hADMSC-SCs were able to myelinate neuronal axons.
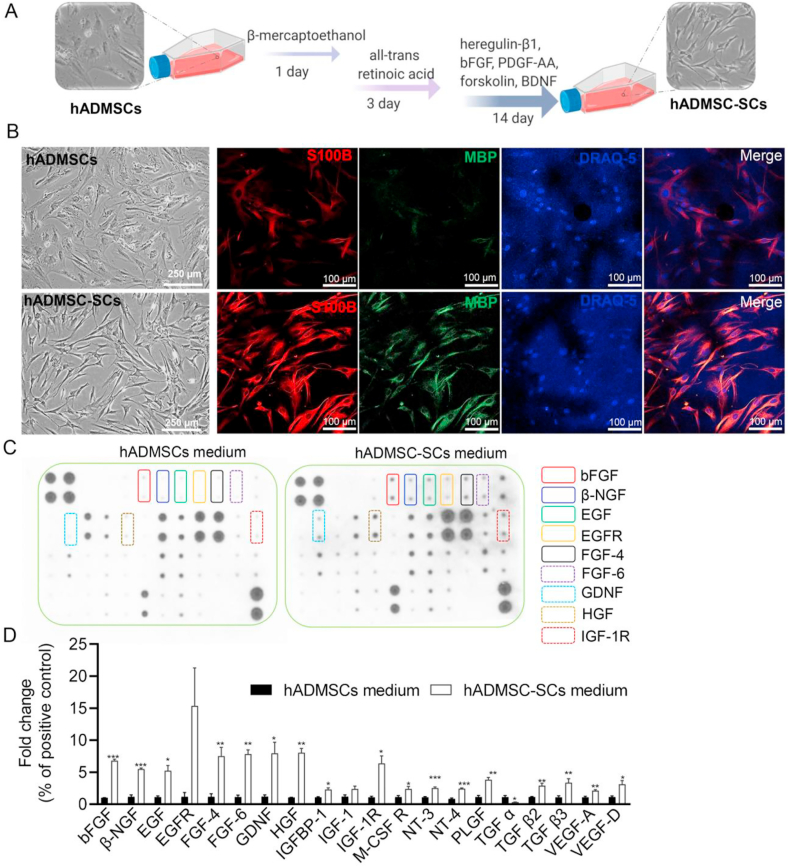
Fig. 1.
Characterization and comparison of hADMSCs with and without differentiation into SCs. (A) Schematic of the differentiation protocol of hADMSCs into SC-like cells. (B) Optical microscope and IF images of hADMSCs and hADMSC-SCs. (Optical microscope image scale bar: 250 μm, IF image scale bar: 100 μm). (C) Representative images of a human growth factor antibody array for supernatant culture media of hADMSCs and hADMSC-SCs. (D) Semi-quantitative analysis of the secreted growth factors with statistical difference. Relative expression of each targeted growth factor was presented as normalized to the positive control on the same array and relative to the hADMSC control group (n = 4, *p < 0.05, **p < 0.01, ***p < 0.001).
We also applied a human growth factor antibody array to evaluate and compare the human growth factor secretome profiling of hADMSCs with and without differentiation. As shown in Fig. 1C, the hADMSC-SCs secreted significantly more growth factors after induction, including bFGF, beta nerve growth factor (β-NGF), epidermal growth factor-receptor (EGFR), epidermal growth factor (EGF), neurotrophin (NT)-3,4, GDNF, hepatocyte growth factor (HGF), transforming growth factor (TGF) β, and vascular endothelial growth factor (VEGF) A and D. Most of these growth factors are neurotrophic factors that can support the growth and differentiation of both developing and mature neurons and protect neurons under pathological conditions [70]. Many of the growth factors mentioned above are also responsible for regulating angiogenesis and inflammation during peripheral nerve repair and regeneration [63]. Meanwhile, several other growth factors, like granulocyte colony stimulating factor (GCSF) and insulin-like growth factor 1 receptor (IGF-1R), were also highly secreted from hADMSC-SCs after induction. These factors are well-known to regulate the proliferation, differentiation, and activation of neutrophilic granulocyte lineage cells [71,72]. Interestingly, only the secretion of TGFα was decreased from hADMSC-SCs compared to hADMSCs. The reason and mechanism are still unclear. We speculate that this might be related to intrinsic cellular secretion properties [73,74].
3.2. Isolation and characterization of exosomes from hADMSC and hADMSC-SC media
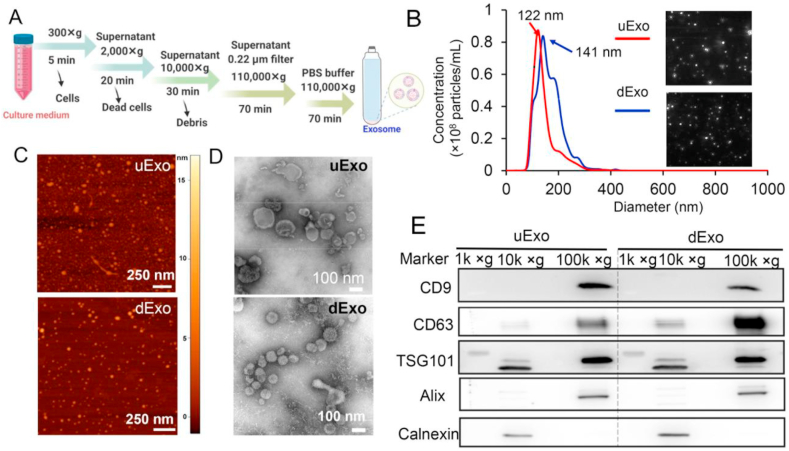
In this study, the cell culture media of both hADMSCs and hADMSC-SCs were collected, and the exosomes were further isolated by using ultracentrifugation following the protocols shown in Fig. 2A. The supernatant media went through serial ultracentrifugation, and the exosome pellets were reconstituted in PBS and stored at −80 °C before use. Then, we characterized the exosomes by using NTA, AFM, TEM, and western blotting. Herein, exosomes isolated from hADMSCs with and without differentiation are abbreviated as dExo and uExo, respectively.
Fig. 2.
Characterizations of exosomes derived from hADMSCs and hADMSC-SCs. (A) Schematic of the protocol for exosome isolation from cell culture media. (B) Size distribution of uExo and dExo was obtained from NTA test, inset showing representative exosome images captured from the NTA video frames. (C) AFM images of uExo and dExo (scale bar 250 nm). (D) TEM images of uExo and dExo (scale bar 100 nm). (E) Proteins from each centrifugal rate (1k × g : 1000 × g, 10k × g:10,000 × g, and 100k × g: 100,000 × g) were analyzed by western blot for the presence of exosome-associated proteins CD9, CD63, TSG101, and Alix.
The results shown in Fig. 2B showed that the average sizes of uExo and dExo were 144 ± 45 nm and 165 ± 48 nm, while the average concentrations of uExo and dExo were (5.62 ± 0.39) × 109/mL and (7.99 ± 0.42) × 109/mL, respectively. NTA analysis indicated that the size distribution of both uExo and dExo ranged mainly from 99 to 213 nm. The AFM images showed that the particle pellets were round-shaped vesicles in nanoscale (Fig. 1C). The TEM images showed more detailed morphologies of uExo and dExo, demonstrating that the particle pellets were cup-like vesicles with bounded membranes (Fig. 1D). Both AFM and TEM images also suggested that both uExo and dExo were nanosized. In our study, western blot analysis demonstrated that both uExo and dExo, after final ultracentrifugation at 100,000 × g, positively expressed CD9, CD63, TSG101 and Alix, which are well-known exosome biomarkers [33,34] (Fig. 2E). We also collected the supernatants during the intermediate centrifugation steps for the western blot. Compared to the final pellet under ultracentrifugation of 100,000 × g, we could not significantly detect these exosome biomarkers (except for TSG101) in the dead cells and debris under centrifugation of 1000 × g or 10,000 × g (Fig. 2E). The detected TSG101 might belong to the few EVs with large sizes that were deposited during 10,000 × g centrifugation. In contrast, Calnexin, a negative biomarker for exosomes, was undetectable in both uExo and dExo but found in the cell lysate (under centrifugal rate of 10,000 × g), verifying the purity of the exosomes. Altogether, these results implied that uExo and dExo were successfully isolated with comparable particle sizes, morphologies, purities, and biomarkers.
3.3. Effects of exosomes isolated from hADMSC with and without differentiation on the anti-oxidation and proliferation for rSCs
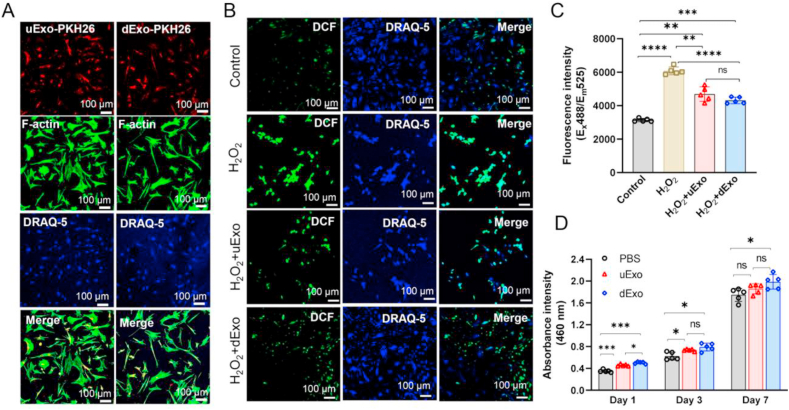
Herein, rSCs were used as a model cell type in vitro to evaluate the functional anti-oxidation and proliferation of exosomes. To determine whether exosomes could be captured by rSCs and what the distribution of exosomes was, both uExo and dExo were labeled by PKH26 and then co-cultured with rSCs. As shown in Fig. 3A, significant amounts of PKH26 labeled exosomes were found to be taken up by rSCs after a 12 h co-culture.
Fig. 3.
Exosome uptake and their effects on anti-oxidation and proliferation of rSCs. (A) Exosomes labeled with PKH26 were incubated with rSCs for 12 h, and the exosome uptake by rSCs was revealed under CLSM. Scale bar, 100 μm. (B) The mitigation of oxidative stress in rSCs was monitored via H2DCFDA staining after different treatments with H2O2, H2O2 +uExo, or H2O2 +dExo. Scale bar, 100 μm. (C) Oxidized H2DCFDA fluorescence signal intensity was quantified by a microplate reader (Ex = 488 nm, Em = 535 nm) (n = 5). (D) CCK-8 assay to detect rSC proliferation (n = 5). *p < 0.05, **p < 0.01, ***p < 0.001, ns: no significant difference.
To find out whether the exosomes could protect the rSCs from H2O2-induced oxidative stress, we used H2DCFDA, a hydrogen peroxide (H2O2)-specific probe, to evaluate ROS produced by rSCs [75]. We first optimized the H2O2 dose to maximize the oxidative stress while minimizing the effects on rSC viability (Fig. S2). Based on Live/Dead and CCK-8 assay results, the concentration of H2O2 was selected to be 200 μM. To the best of our knowledge, the concentrations of exosomes ranging from 109 to 1012/mL are applied for system administration in vivo [55,[76], [77], [78]]. Thus, in order to ensure that the amount of exosomes derived from hADMSCs was sufficient, the concentration of exosomes was chosen to be 3 × 109/mL. rSCs co-incubated with H2O2 and exosomes (either uExo or dExo) showed significantly lower green fluorescence than the control group (200 μM H2O2 treatment only), suggesting that both uExo and dExo could effectively attenuate the intracellular oxidative stress (Fig. 3B). The H2O2 treatment significantly increased the oxidation of rSCs (Fig. 3C). The green fluorescence intensity of rSCs was further quantitatively confirmed to be significantly lower after the incorporation of exosomes (Fig. 3C). These results are consistent with previous reports that exosome derived from MSCs effectively mitigated oxidation [79]. However, there was no significant difference between the groups with uExo and dExo. The proliferation of rSCs was examined via a CCK-8 assay. As shown in Fig. 3D, the absorbance intensity of rSCs co-cultured with dExo was higher than the control group at days 1, 3, and 7, demonstrating that the dExo could promote rSCs proliferation. We could also observe that the average of absorbance intensity of rSCs treated with dExo was significantly higher at day 1 than that of rSCs treated with uExo, but such a trend was not detected at days 3 or 7. Meanwhile, compared to the control group, uExo only significantly increased rSC proliferation at days 1 and 3 (Fig. 3D), indicating that the effect of uExo on proliferation might be transient. These results suggest that both uExo and dExo protected rSCs from oxidative stress and dExo had more effects on improving rSC proliferation.
3.4. Effects of exosomes from hADMSCs with and without differentiation on angiogenesis of HUVECs
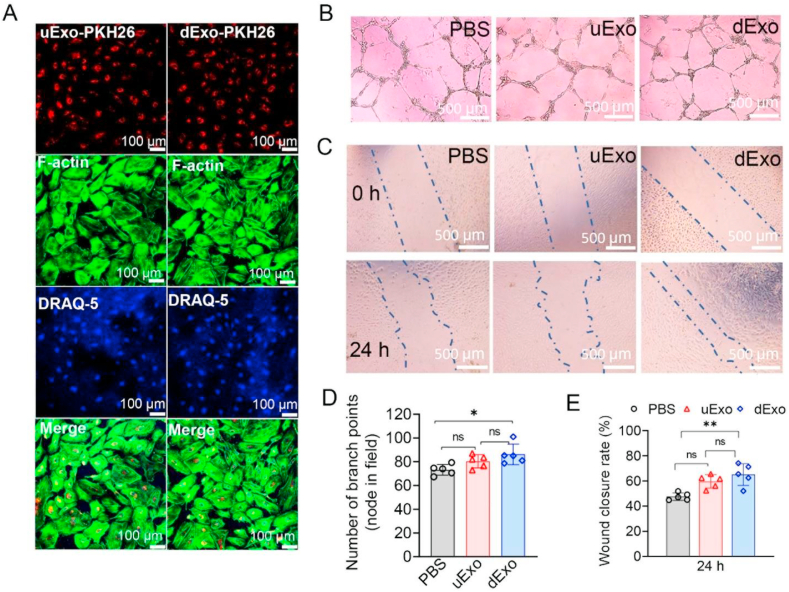
It has been reported that exosomes derived from MSCs could promote angiogenesis [80,81]. In this study, tube formation and migration assays were used to investigate the angiogenesis of HUVECs after treatment with uExo and dExo. We first examined the distribution of exosomes in the HUVECs. As shown in Fig. 4A, significant amounts of PKH26 labeled exosomes (both uExo and dExo) were captured by HUVECs compared to rSCs and other cell types, as shown previously or later in this study. This indicates that endothelial cells may be more intent to uptake and internalize exosomes in vitro.
Fig. 4.
Effects of uExo and dExo on HUVEC angiogenesis in vitro. (A) Exosomes labeled with PKH26 were incubated with HUVECs for 12 h, and the exosomes captured by HUVECs were revealed under CLSM. Scale bar, 100 μm. (B) Optical microscope images showed the tube formation with and without co-cultures with uExo or dExo. Scale bar, 500 μm. (C) Bright-field microscope images showed the migration of HUVECs after treatment with uExo or dExo. Scale bar, 500 μm. (D) Semi-quantitative measurement of the number of branch points per field in tube formation (n = 5). (E) Semi-quantitative measurement of the distance of cell migration in the wound scratch assay (n = 5). *p < 0.05, **p < 0.01, ns: no significant difference.
Subsequently, we seeded HUVECs onto Matrigel in the presence of uExo or dExo, and a vascular tubular structure was developed. As shown in Fig. 4B, the node density for HUVECs was significantly increased after incubation in the presence of uExo and dExo, compared to the normal medium alone. Moreover, the number of branches was quantified. It was found that the average number of branches for HUVECs treated by dExo (86 nodes) was higher than those incubated with PBS (73 nodes) or uExo (80 nodes) (Fig. 4D). The results indicated that dExo significantly enhanced the angiogenic activity of endothelial cells. We further evaluated how exosomes affected HUVEC migration by using a scratch wound healing assay. The scratch site was first created on the confluent monolayer of HUVECs by the mechanical removal of a portion of cells using a pipette tip. Then the HUVECs started migrating to heal the scratch site (Fig. 4C). After 24 h migration, we found that HUVECs treated with dExo showed the highest wound closure ratio, which was significantly higher than the treatment with PBS alone (Fig. 4C and E). All these data suggested that dExo released from hADMSC-SCs played positive roles in angiogenesis of HUVECs.
3.5. Anti-inflammatory effects of uExo and dExo on M1 macrophages
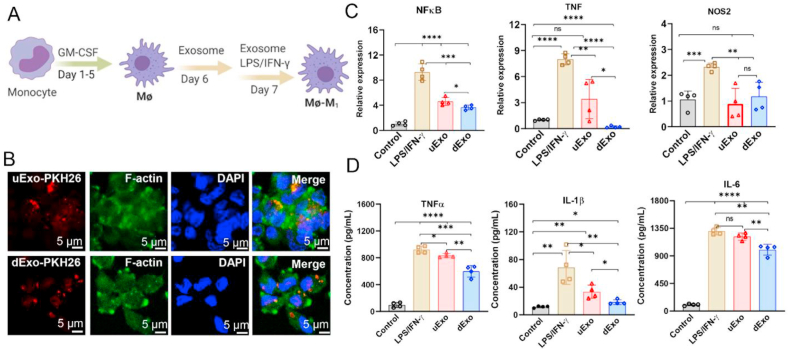
Next, we used human monocyte to differentiate into pro-inflammatory M1 macrophages, aiming at exploring how these immune cells respond to uExo and dExo. As shown in Fig. 5A, monocytes were treated with GM-CSF to facilitate their attachment and differentiation into macrophages (Mɸ) [82]. Both uExo and dExo were labeled by PKH26 and were captured by the macrophages after 12 h co-cultures (Fig. 5B).
Fig. 5.
The anti-inflammatory effects of uExo and dExo on human monocyte derived M1 macrophages. (A) Schematic of the protocol of monocyte differentiation into macrophages and M1 macrophages. (B) Exosomes labeled with PKH26 were incubated with macrophages for 12 h, and the exosome uptake by macrophages was revealed under CLSM. Scale bar, 5 μm. (C) The relative gene expressions of NFκB, TNF, and NOS2 in M1 macrophages after the indicated treatments. (n = 4). (D) Cytokine secretions of TNF-α, IL-1β, and IL-6 from the supernatants by M1 macrophages were detected by ELISA (n = 4). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns: no significant difference.
To explore the anti-inflammatory effects of uExo and dExo on M1 macrophages, the macrophages were co-cultured with exosomes and then stimulated to the M1 phenotype using LPS/IFN-γ. We quantitatively detected several gene expressions related to pro-inflammation, including nuclear factor kappa B (NFκB), tumor necrosis factor (TNF), and nitric oxide synthase 2 (NOS2), by RT-PCR (primers shown in Table S1). The relative expressions of these three genes were significantly upregulated in M1 macrophages after activation and significantly downregulated in the groups treated with exosomes (Fig. 5C). The addition of dExo statistically downregulated NFκB and TNF expressions compared to the counterparts with uExo treatment (Fig. 5C). We further measured the M1 macrophage secreted inflammatory cytokines, as shown in Fig. 5D. As expected, the secretion levels of TNF-α, interleukin (IL)-1β, and IL-6 were significantly increased after stimulation with LPS/IFN-γ. After treatment with uExo or dExo, the pro-inflammatory cytokine secretions were significantly decreased, especially for the groups with the addition of dExo (Fig. 5D). These results demonstrate that both uExo and dExo have anti-inflammatory effects by downregulating pro-inflammatory gene expressions and cytokine secretions. Moreover, dExo showed an improved outcome in modulating the anti-inflammatory activity for monocyte derived M1 macrophages.
3.6. Effects of exosomes on the growth of axons of hiPSC derived peripheral SNs
Since axon growth and reconnection are of the most importance during peripheral nerve repair and regeneration, we further investigated the effect of dExo and uExo on the growth of SNs differentiated from hiPSC. The hiPSCs were first differentiated into SN-like cells using a previously reported method with some modifications (Fig. 6A) [83], and then the hiPSC-SNs were cultured and induced into neurospheres for further encapsulation and characterization with exosomes.
After 10-day induction, the gene expressions of insulin gene enhancer protein (ISL1), peripherin (PRPH), and brain-specific homeobox/POU domain protein 3A (BRN3A), which are biomarkers of sensory and peripheral neurons, were significantly enhanced compared to hiPSCs. Meanwhile, octamer-binding transcription factor 4 (OCT4) expression, the marker of undifferentiated pluripotent stem cells, was downregulated for hiPSC-SNs. Moreover, the gene expressions of several receptors, like neurotrophic tyrosine kinase receptor type 1 (NTRK1), P2X purinoceptor 3 (P2RX3), and transient receptor potential cation channel subfamily V member 1 (TRPV1), were also upregulated after induction. These receptors are biomarkers for nociceptors that are highly related to peripheral pain responses. The expressions of sodium voltage-gated channel alpha subunit 9 (SCN9A), SCN10A, and SCN11A for hiPSC-SNs were also greatly upregulated. These genes are related to sodium channels with the function of transmitting pain signals. The expressions of all these genes indicate that some of these cells were differentiated into nociceptive neurons [83,84]. IF staining further confirmed the positive expressions of β-3 tubulin and TRPV1, which exist in sensory neurons, in the hiPSC-SNs after induction (Fig. 6C). Then hiPSC-SNs were passaged and replated in a non-adhesive plate to obtain hiPSC-SN neurospheres. As shown in Fig. 6D, SN neurospheres were successfully formed after 7-day culture, and the obtained SN neurospheres strongly expressed the markers of β-3 tubulin and TRPV1. We further encapsulated hiPSC-SN neurospheres within Matrigel. Figs. S3 and S4 showed that hiPSC-SN neurospheres grew very well within Matrigel, with extensive axon outgrowth and positive expression of BRN3A. To explore whether SNs were able to capture exosomes, we isolated mouse DRG neurons and treated them with exosomes labeled with PKH26. Different from other peripheral nerve-related cell types, exosomes were not extensively captured by DRG neuron cell bodies in vitro (Fig. S5). Instead, the exosomes were distributed around the axons of DRG neurons (Fig. S5). Other studies also showed that glial cells could absorb more MSC derived exosomes than neurons [85].
We also used PKH26 labeled exosomes to treat hiPSC-SNs. As shown in Fig. S6, PKH26 labeled exosomes (both uExo and dExo) were also captured by hiPSC-SNs. Comparing to mouse DRG neurons, slightly more exosomes were internalizated into hiPSC-SNs. We further evaluated the effects of exosomes on axon outgrowth from hiPSC-SN neurospheres. The neurospheres were encapsulated within Matrigel and then treated with PBS, uExo, or dExo. After 6-day cultures, the axon length of the neurospheres with dExo was longer than those treated with PBS or uExo (Fig. 6E). In addition, the area of axon outgrowth was larger for the neurospheres with dExo treatment (Fig. 6F). Semi-quantitative analysis showed that the neural axon length of SN spheres treated by dExo was larger than other groups on days 2, 4, and 6 (Fig. 6G). Taken together, these results indicated that dExo could promote neural axon growth.
3.7. miRNA sequencing analysis
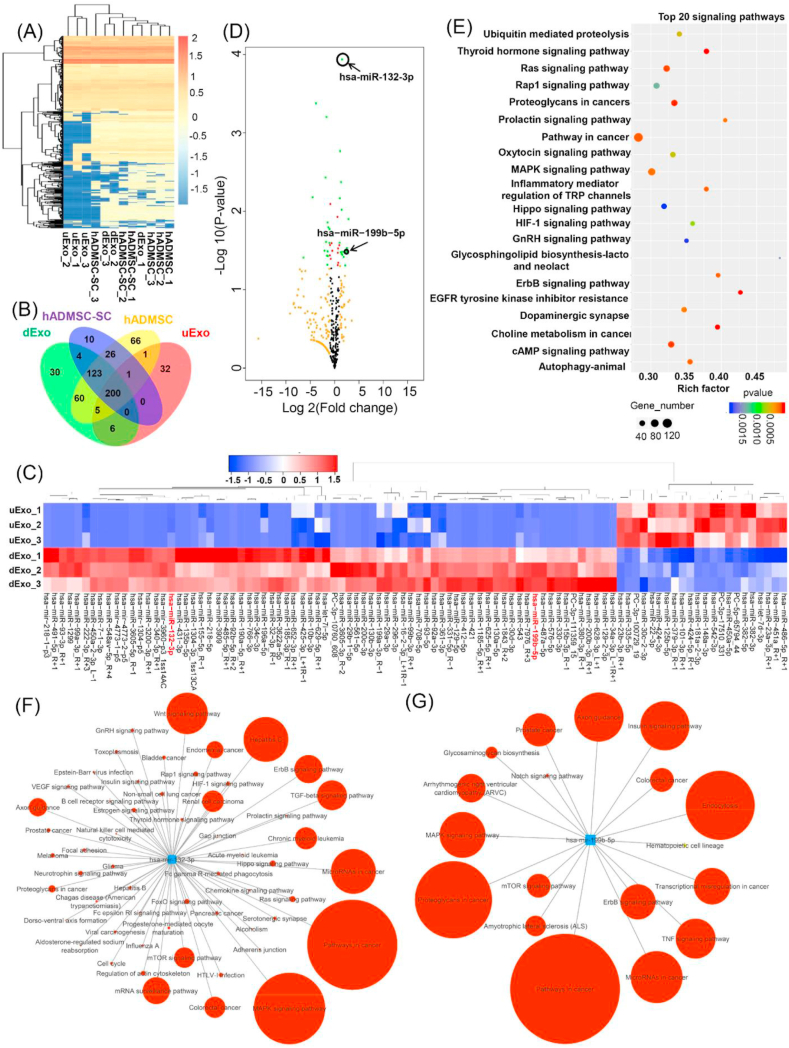
Many studies have revealed the regulatory roles of miRNAs loaded within exosomes in peripheral nerve injury [3,86] and spinal cord injury [87,88]. Therefore, we evaluated the miRNA expression profiles in hADMSCs, hADMSC-SCs, and their derived uExo and dExo via miRNA sequencing and bioinformatic analysis. To visualize overall miRNA expression differences among the four groups, an overall heatmap was generated, including all the identified miRNAs (a total of 956 miRNAs) (Fig. 7A). A number of commonalities in exosomal and intercellular secretion profiles of miRNA were observed (Fig. 7A), with uExo and hADMSC samples clustering tightly together. Interestingly, many miRNAs were either downregulated or undetectable in the uExo group compared to those in their parent cell group (i.e., hADMSC group), while the miRNA contents for dExo and hADMSC-SCs revealed high similarity. The Venn diagram showed the overlapping and nonoverlapping miRNAs in the four groups (Fig. 7B). In detail, the number of identified miRNAs for hADMSC-SCs and hADMSCs was 364 and 482, respectively. For the exosomes, the number of miRNAs for dExo and uExo was 428 and 245, respectively. Moreover, dExo contained 217 miRNAs that were not present in uExo. Interestingly, the number of miRNAs for dExo was more than that for uExo, while hADMSC-SCs contained less miRNAs than hADMSCs. Other studies also reported similar results, that the miRNAs in exosomes did not merely reflect the cellular content, and some distinct miRNAs were overrepresented in exosomes compared to the cell of origin [89]. We further focused more on the differential expression of miRNAs between uExo and dExo groups. The heatmap and volcano plot, as shown in Fig. 7C and D, displayed the differentially expressed miRNAs. The results revealed a greater number of miRNAs with a statistically significant alteration in expression in the uExo and dExo groups. KEGG pathway enrichment analysis was used to identify the significant biological pathways related to the differentially expressed miRNAs. In this study, the top 20 signaling pathways were predicted (Fig. 7E). Significantly enriched KEGG categories included the thyroid hormone signaling pathway, Ras signaling pathway, MAPK signaling pathway, Oxytocin signaling pathway, inflammatory mediator regulation of TRP channels, etc. Among various differentially expressed miRNAs, miRNA-132-3p (p = 1.16E-04, log 2 (fold change) = 1.54) and miRNA-199b-5p (p = 3.28E-02, log 2 (fold change) = 1.65) have caught our attention because both of them had high reads (>5000) in all the four groups and were significantly upregulated in the dExo group. The pathway annotations of both miRNA-132-3p and miRNA-199b-5p were predicted by miRPathDB database based on KEGG and shown in Fig. 7F and G. Each pathway is filtered based on the significant P-value < 0.05 and their expected hits, predicted hits and predicted evidence were provided in the supplementary file (Table S2 and Table S3). As shown in Fig. 7F, miRNA-132-3p is related to many targets and physiological signaling pathways. In this study, we are particularly interested in some of the targets associated with neural or axonal growth, such as the neurotrophin signaling pathway, axon guidance, VEGF signaling pathway, and TGF-β signaling pathway. miRNA-199b-5p is also associated with some special signaling pathways related to peripheral nerve repair, such as TNF signaling pathway, mTOR signaling pathway, notch signaling pathway. Both the MAPK signaling pathway and axon guidance are associated with miRNA-132-3p and miRNA-199b-5p.
Fig. 7.
miRNA sequencing analysis. (A) Heatmap and hierarchical clustering depicting the normalized Z-scaled expression of miRNA in the four groups (n = 3 for each group). (B) The Venn diagram of all differentially expressed miRNAs identified in the four groups. (C) Heatmap of significant differential expression of miRNAs in uExo and dExo groups (n = 3, <p<0.05). (D) Volcano plot showing differences in exosomal miRNAs expressed in the uExo and dExo groups. Note for spot colors: black, insignificant; red, <p≤0.05; orange, log2FoldChange ≥1 or ≤ -1; green, both red and orange criteria fulfilled. (E) The top 20 common KEGG pathways of the differentially expressed miRNAs in the uExo and dExo groups. Number of target genes enriched in KEGG pathway, p value and rich factor are shown in scatterplot. Rich factor = (number of target genes in KEGG pathway)/(total number of genes in KEGG pathway). The networks predicted by the miRPathDB database for the upregulated miRNAs (miRNA-132-3p (F) and miRNA-199b-5p (G)), showing the associated functional signaling pathways. The circle size (larger to smaller) represents the number of hits or genes in the interacting pathway.
4. Discussion
SCs are the main glial cells in the adult peripheral nervous system, and they wrap around peripheral axons of motor and sensory neurons to form the myelin sheath and are required for successful nerve regeneration [90,91]. ADMSCs have characteristics of mesenchymal tissue lineages and could differentiate into SC-like phenotypes [92]. It is widely accepted that the SC-like cells differentiated from ADMSCs have a beneficial role for the treatment of peripheral nerve injuries [19,93]. Our previous studies have reported that aligned topography, electrical stimulation, and neural differentiation media promoted hADMSC differentiation towards myelinating SCs [21,52]. In the present study, hADMSC-SCs were achieved by the differentiation of hADMSCs using previously defined induction medium plus BDNF, which has been reported to promote SC marker expressions and myelination [94,95]. We have demonstrated that the differentiation process enhanced the cellular secretory properties, showing more neurotrophic and growth factors in the hADMSC-SC secretome. In addition, the hADMSC-SCs had myelination capacity, as shown in previous studies [96,97], and formed a myelin sheath around axons after being co-cultured with rat DRG neurons. Both secretion and myelination features may explain why hADMSC-SCs had superior efficacy in treating peripheral nerve injuries.
Following the differentiation of hADMSCs, we focused on exosomes isolation from hADMSCs with and without differentiation. Some literatures have reported that MSC-derived exosomes, with MSC-sourced neuroprotective and immunomodulatory miRNAs, attenuate neuro-inflammation, promote neo-vascularization, induce neurogenesis, and reduce apoptotic loss of neural cells [55,98,99]. Therefore, exosomes, as important mediators for intercellular and intracellular communication, are ideal natural nanocarriers for (stem) cell free therapy due to their biocompatible characteristics and immune-friendly nature [[100], [101], [102]]. Thus, we hypothesize that hADMSC–SC–derived exosomes possess the functions of their parent cells, so they can regulate the peripheral nerve-related cell behaviors under physiological and pathological conditions. We thus successfully isolated exosomes from hADMSCs and hADMSC-SCs (i.e., uExo and dExo), which had nanoscale sizes and cup-like morphologies, and we systematically studied and compared the anti-ROS, anti-inflammatory, angiogenic, and axonal growth promoting properties of uExo and dExo.
Peripheral nerve injuries induce remarkable ROS production and accumulation, which in turn result in oxidative damage, neuroinflammation, and peripheral nerve injury induced neuropathic pain [103,104]. Many proteins/compounds with antioxidant activities have been effectively used to treat peripheral nerve injuries [[105], [106], [107]]. Our results demonstrated that both uExo and dExo had ROS-scavenging activities and significantly reduced the oxidative stress in rSCs. Several studies have reported that MSC-derived exosomes combat oxidative stress-induced damage through adaptive regulation of the nuclear factor erythroid 2-related factor 2 (Nrf2) defense system [59,90,91]. Both dExo and uExo significantly improved the ROS defense capacity compared to control group. The dExo also promoted SC proliferation at day 1, 3 and 7. Since neurons have relatively weak antioxidant defense mechanisms, exogenous antioxidants or antioxidants released from SCs will provide effective neuroprotection against oxidative stress [90,108].
Immediately after peripheral nerve injury, macrophages are recruited to the lesion to aid myelin debris elimination and tissue remodeling [109]. However, excessive macrophage accumulation and pro-inflammation are major obstacles for nerve repair and contribute to the initiation and maintenance of persistent nerve injury-induced pain [110]. MSC-derived exosomes, similar to their parent cells, have been reported to have anti-inflammatory effects. For example, MSC-derived exosomes attenuated brain injuries by inhibiting glial inflammation [111,112]. Our current study was consistent with previously reports that hADMSC exosomes reduced pro-inflammatory gene expressions and cytokine releases from M1 macrophages. Importantly, the differentiation process towards the SC phenotype not only did not have negative effects on anti-inflammatory activities but also enhanced the properties.
Angiogenesis and axon growth are two important key factors because the lack of blood supply and slow intrinsic growth rate of axons are the major constraints for peripheral nerve repair, especially for long-gap defects. In this study, only dExo showed significant effects on HUVECs in terms of tube formation and migration, indicating that angiogenesis was enhanced. This may be slightly inconsistent with previous reports that hADMSC-derived exosomes were shown to have the ability to promote angiogenesis of HUVECs, which could reconstruct the microvasculature network of the spinal cord injury model [80,113]. The difference may be due to the differences of donor cells and culture conditions. Similarly, we only found a significant enhancement of neurite outgrowth and axon length in the hiPSC-SN neurosphere group treated with dExo. Guo et al. reported that intranasal administration of MSC derived exosomes enhanced axonal growth and neovascularization while reducing microgliosis and astrogliosis in a rat model with a spinal cord injury [55]. Our current study indicates that dExo might have a better therapeutic efficacy for angiogenesis and axon outgrowth then that of uExo. We observed that PKH26-labeled uExo and dExo had higher internalization in HUVECs compared to other cells. We also found that the amounts of exosomes uptake by DRG neurons an hiPSC-SNs were lower compared to HUVECs and rat SCs. The detailed reason and mechanism are unclear. Haertinger et al. reported that ADMSC derived exosomes entered the SCs via endocytosis-related processes and did not bind to or fuse with the SC membrane [114]. In another study, Pulliam et al. also reported that less exosomes were captured by neural cells compared to astrocytes [115]. Ren et al. reported that exosomes could be uptake at peripheral nerve endings and transport of DRG neurons and motor neurons in the anterior horn of the spinal cord. It also showed that glia cells have more exosome uptake than neurons [116]. All these studies demonstrate that the exosome uptake efficiency depends on cell types and cell-surface receptors. More related mechanisms should be elucidated in future studies to better understand the uptake, internalization, and traffic processes of exosomes.
MSC derived exosomes retain most characteristics of their parent cells, such as the promotion of neurite outgrowth and angiogenesis, immunomodulation, and the ability to repair damaged tissue [117,118]. Exosomal miRNAs play important roles in mediating these physiologic and therapeutic effects [119].
Our miRNA sequencing results demonstrated that the exosomes and their parent hADMSCs displayed different miRNA features. hADMSC-SCs and their dExo had many similar miRNA components, whereas many miRNAs were detected in hADMSCs but not in uExo. This also demonstrates that hADMSC-SCs may be a better source for exosome isolation. Most importantly, miRNA profiling analysis also revealed that many miRNAs were significantly upregulated in dExo compared to those in uExo. This may explain why dExo had enhanced activities and functions on the tested peripheral nerve-related cells. Among various differentially expressed miRNAs, we focused on miRNA-132-3p and miRNA-199b-5p, which had high reads and significant fold changes. As previously reported, miRNA-132 is related to regulating neuronal differentiation, maturation, and function [120]. miRNA-132 overexpression in SCs significantly promoted cellular migration after sciatic nerve injury by targeting at PRKAG3 [121]. miRNA-132 can also regulate the target gene RASA1 in neuronal function to promote mouse DRG axon extension [122]. It can also regulate the vascular integrity of the brain for neurovascular communication [123]. The miRNA-132-3p target network analysis indicated that several important signaling pathways, including the neurotrophin signaling pathway, axon guidance, VEGF signaling pathway, and TGF-β signaling pathway, were activated, which may explain why axon growth and angiogenesis were promoted after the treatment with dExo. miRNA-199b-5p, on the other hand, has been reported to attenuate the inflammatory response through the GSK3β/NF-κB signaling pathways in monocytes [124]. In another study, when miRNA-199b-5p was upregulated, the inflammatory response could be suppressed by the regulation of the IκB kinase β (IKKβ)//NF-κB pathway in LPS-activated microglia [125]. In the present study, we found the upregulation of miRNA-199b-5p in dExo, and this may be reason for the decrease of pro-inflammatory gene expression and cytokine secretion. Additionally, miRNA network analysis results showed that the TNF signaling pathway, mTOR signaling pathway, and Notch signaling pathway, which are all related to neuroinflammation and oxidative stress, are regulated by miRNA-199b-5p. Inflammatory response is known to be beneficial for the recovery and regeneration of injured tissue during the acute stage. However, an overactive and chronic inflammatory response can lead to secondary damage. In this study, dExo, containing miRNA-199b-5p, might be able to regulate several signaling pathways with anti-inflammatory abilities to protect the growth and function of neuron-related cells. In another study, Lopez-Leal et al. reprogrammed rat SCs into repair SC phenotype and demonstrated that SC reprogramming upregulated exosomal miRNA-21 to promote neurite growth [126]. In current study, we also found that miRNA-21-5p expressed high levels in all the uExo, dExo, hADMSC and hADMSC-SC groups. Moreover, miRNA-21-5p was significantly upregulated (p = 4.24E-03, log 2 (fold change) = 1.53) in the hADMSC-SC group compared to hADMSC group, but no significance difference between uExo and dExo groups. It is possible that miR-21-5p might also contribute to axonal growth. In this study, we differentiated hADMSCs to SC phenotypes with more capacity for myelinating rather than repairing. This may explain why miRNA-21 expression was similar for uExo and dExo groups.
5. Conclusion
In summary, we successfully differentiated hADMSCs towards the SC phenotype and demonstrated that the hADMSC-SCs secreted much more growth factors and neurotrophic factors compared to hADMSCs without differentiation. We further isolated uExo and dExo from hADMSCs and hADMSC-SCs, respectively. We demonstrated that all these exosomes can be captured by various peripheral nerve-related cells, like SCs, endothelial cells, macrophages, and neurons, and can be effectively internalized in HUVECs. We also showed that dExo improved the ROS scavenging capacity and promoted rSC proliferation in the early stage (day 1 and day 3). The dExo also facilitated the angiogenesis of HUVECs and significantly downregulated pro-inflammatory gene expressions and cytokine secretions of M1 macrophages. The axonal growth of hiPSC-SNs was significantly enhanced by dExo treatment. Furthermore, miRNA sequencing analysis unveiled that hADMSC-derived exosomes and their parent cells shared some similarity of their miRNA profiles, and dExo contained much more miRNA compared to uExo. Several upregulated miRNAs in dExo, like miRNA-132-3p and miRNA-199b-5p, are highly related to neuroprotection, angiogenesis, and neuroimmune modulation. Based on the combination of hADMSC and SC features, dExo is a promising cell-free biological therapeutic for promoting neural regeneration because it effectively targets various peripheral nerve-related cells. Future studies may focus more on the efficacy of dExo on neuroprotection and neural regeneration in vivo and on understanding deeper insights of specific miRNA cargo(s).
CRediT authorship contribution statement
Bo Liu: Conceptualization, Methodology, Investigation, Data curation, Visualization, Formal analysis, Writing – original draft. Yunfan Kong: Investigation of hiPSC differentiation into SNs and neurosphere formation. Wen Shi: Investigation of human growth factor antibody array analysis. Mitchell Kuss: Writing – review & editing. Ke Liao: Investigation of Western blot. Guoku Hu: Investigation of Western blot. Peng Xiao: Data curation of miRNAseq data, Writing – review & editing. Jagadesan Sankarasubramanian: Data curation of miRNAseq data, Writing – review & editing. Chittibabu Guda: Data curation of miRNAseq data, Writing – review & editing. Xinglong Wang: Manuscript review, Editing suggestions. Yuguo Lei: Manuscript review, Editing suggestions. Bin Duan: Conceptualization, Methodology, Visualization, Formal analysis, Writing – review & editing, Supervision, Project administration, Funding acquisition, All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work has been supported by Mary & Dick Holland Regenerative Medicine Program start-up grant, Mary & Dick Holland Regenerative Medicine Program pilot project grant, University of Nebraska Collaboration Initiative Grant, NIH (R21AR078439) (B.D.) and UNL and UNMC Sciences, Engineering, and Medicine Initiative funding (B.D. and G.H.)
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.11.022.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Faroni A., Mobasseri S.A., Kingham P.J., Reid A.J. Peripheral nerve regeneration: experimental strategies and future perspectives. Adv. Drug Deliv. Rev. 2015;82–83:160–167. doi: 10.1016/j.addr.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 2.Ji R.R., Xu Z.Z., Gao Y.J. Emerging targets in neuroinflammation-driven chronic pain. Nat. Rev. Drug Discov. 2014;13:533–548. doi: 10.1038/nrd4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qing L., Chen H., Tang J., Jia X. Exosomes and their microRNA cargo: new players in peripheral nerve regeneration. Neurorehabilitation Neural Repair. 2018;32:765–776. doi: 10.1177/1545968318798955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheib J., Hoke A. Advances in peripheral nerve regeneration. Nat. Rev. Neurol. 2013;9:668–676. doi: 10.1038/nrneurol.2013.227. [DOI] [PubMed] [Google Scholar]
- 5.Uz M., Das S.R., Ding S., Sakaguchi D.S., Claussen J.C., Mallapragada S.K. Advances in controlling differentiation of adult stem cells for peripheral nerve regeneration. Adv Healthc Mater. 2018;7 doi: 10.1002/adhm.201701046. [DOI] [PubMed] [Google Scholar]
- 6.Aijie C., Xuan L., Huimin L., Yanli Z., Yiyuan K., Yuqing L., Longquan S. Nanoscaffolds in promoting regeneration of the peripheral nervous system. Nanomedicine. 2018;13:1067–1085. doi: 10.2217/nnm-2017-0389. [DOI] [PubMed] [Google Scholar]
- 7.Moskow J., Ferrigno B., Mistry N., Jaiswal D., Bulsara K., Rudraiah S., Kumbar S.G. Review: bioengineering approach for the repair and regeneration of peripheral nerve. Bioact Mater. 2019;4:107–113. doi: 10.1016/j.bioactmat.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding D.C., Shyu W.C., Lin S.Z. Mesenchymal stem cells. Cell Transplant. 2011;20:5–14. doi: 10.3727/096368910X. [DOI] [PubMed] [Google Scholar]
- 9.Kariminekoo S., Movassaghpour A., Rahimzadeh A., Talebi M., Shamsasenjan K., Akbarzadeh A. Implications of mesenchymal stem cells in regenerative medicine. Artif Cells Nanomed Biotechnol. 2016;44:749–757. doi: 10.3109/21691401.2015.1129620. [DOI] [PubMed] [Google Scholar]
- 10.Uccelli A., Moretta L., Pistoia V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 11.Sun Y., Shi H., Yin S., Ji C., Zhang X., Zhang B., Wu P., Shi Y., Mao F., Yan Y., Xu W., Qian H. Human mesenchymal stem cell derived exosomes alleviate type 2 diabetes mellitus by reversing peripheral insulin resistance and relieving β-cell destruction. ACS Nano. 2018;12:7613–7628. doi: 10.1021/acsnano.7b07643. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y., Wang Y., Li Q., Liu K., Hou J., Shao C., Wang Y. Immunoregulatory mechanisms of mesenchymal stem and stromal cells in inflammatory diseases. Nat. Rev. Nephrol. 2018;14:493–507. doi: 10.1038/s41581-018-0023-5. [DOI] [PubMed] [Google Scholar]
- 13.Tohill Mel, Terenghi G. Stem-cell plasticity and therapy for injuries of the peripheralnervous system. Biotechnol. Appl. Biochem. 2004;40:17–24. doi: 10.1042/BA20030173. [DOI] [PubMed] [Google Scholar]
- 14.Ladak A., Olson J., Tredget E.E., Gordon T. Differentiation of mesenchymal stem cells to support peripheral nerve regeneration in a rat model. Exp. Neurol. 2011;228:242–252. doi: 10.1016/j.expneurol.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q., Nguyen P.D., Shi S., Burrell J.C., Cullen D.K., Le A.D. 3D bio-printed scaffold-free nerve constructs with human gingiva-derived mesenchymal stem cells promote rat facial nerve regeneration. Sci. Rep. 2018;8:6634. doi: 10.1038/s41598-018-24888-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W., Zhang L., Liu J., Zhang L., Zhang J., Tang P. Repairing sciatic nerve injury with an EPO-loaded nerve conduit and sandwiched-in strategy of transplanting mesenchymal stem cells. Biomaterials. 2017;142:90–100. doi: 10.1016/j.biomaterials.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 17.Masgutov R., Masgutova G., Mullakhmetova A., Zhuravleva M., Shulman A., Rogozhin A., Syromiatnikova V., Andreeva D., Zeinalova A., Idrisova K., Allegrucci C., Kiyasov A., Rizvanov A. Adipose-derived mesenchymal stem cells applied in fibrin glue stimulate peripheral nerve regeneration. Front. Med. 2019;6:68. doi: 10.3389/fmed.2019.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathot F., Rbia N., Bishop A.T., Hovius S.E.R., Shin A.Y. Adipose derived mesenchymal stem cells seeded onto a decellularized nerve allograft enhances angiogenesis in a rat sciatic nerve defect model. Microsurgery. 2020;40:585–592. doi: 10.1002/micr.30579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingham P.J., Kalbermatten D.F., Mahay D., Armstrong S.J., Wiberg M., Terenghi G. Adipose-derived stem cells differentiate into a Schwann cell phenotype and promote neurite outgrowth in vitro. Exp. Neurol. 2007;207:267–274. doi: 10.1016/j.expneurol.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 20.Das S.R., Uz M., Ding S., Lentner M.T., Hondred J.A., Cargill A.A., Sakaguchi D.S., Mallapragada S., Claussen J.C. Electrical differentiation of mesenchymal stem cells into Schwann-cell-like phenotypes using inkjet-printed graphene circuits. Adv Healthc Mater. 2017;6:1601087. doi: 10.1002/adhm.201601087. [DOI] [PubMed] [Google Scholar]
- 21.Wu S., Qi Y., Shi W., Kuss M., Chen S., Duan B. Electrospun conductive nanofiber yarns for accelerating mesenchymal stem cells differentiation and maturation into Schwann cell-like cells under a combination of electrical stimulation and chemical induction. Acta Biomater. 2020 doi: 10.1016/j.actbio.2020.1011.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Zhao Z., Ren Z., Zhao B., Zhang L., Chen J., Xu W., Lu S., Zhao Q., Peng J. Recellularized nerve allografts with differentiated mesenchymal stem cells promote peripheral nerve regeneration. Neurosci. Lett. 2012;514:96–101. doi: 10.1016/j.neulet.2012.02.066. [DOI] [PubMed] [Google Scholar]
- 23.Choi S.J., Park S.Y., Shin Y.H., Heo S.H., Kim K.H., Lee H.I., Kim J.K. Mesenchymal stem cells derived from Wharton's Jelly can differentiate into Schwann cell-like cells and promote peripheral nerve regeneration in acellular nerve grafts. Tissue Eng Regen Med. 2021;18:467–478. doi: 10.1007/s13770-020-00329-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W., Xiao S., Wei Z., Deng C., Nie K., Wang D. Schwann cell-like cells derived from human amniotic mesenchymal stem cells promote peripheral nerve regeneration through a microRNA-214/c-Jun Pathway. Stem Cell. Int. 2019;2019:2490761. doi: 10.1155/2019/2490761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y., Dong L., Zhou D., Li L., Zhang W., Zhen Y., Wang T., Su J., Chen D., Mao C., Wang X. Extracellular vesicles from human umbilical cord mesenchymal stem cells improve nerve regeneration after sciatic nerve transection in rats. J. Cell Mol. Med. 2019;23:2822–2835. doi: 10.1111/jcmm.14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marconi S., Castiglione G., Turano E., Bissolotti G., Angiari S., Farinazzo A., Constantin G., Bedogni G., Bedogni A., Bonetti B. Human adipose-derived mesenchymal stem cells systemically injected promote peripheral nerve regeneration in the mouse model of sciatic crush. Tissue Eng. 2012;18:1264–1272. doi: 10.1089/ten.TEA.2011.0491. [DOI] [PubMed] [Google Scholar]
- 27.Guo Z.Y., Sun X., Xu X.L., Zhao Q., Peng J., Wang Y. Human umbilical cord mesenchymal stem cells promote peripheral nerve repair via paracrine mechanisms. Neural Regen. Res. 2015;10:651–658. doi: 10.4103/1673-5374.155442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fairbairn N.G., Meppelink A.M., Ng-Glazier J., Randolph M.A., Winograd J.M. Augmenting peripheral nerve regeneration using stem cells: a review of current opinion. World J. Stem Cell. 2015;7:11–26. doi: 10.4252/wjsc.v7.i1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hundepool C.A., Nijhuis T.H., Mohseny B., Selles R.W., Hovius S.E. The effect of stem cells in bridging peripheral nerve defects: a meta-analysis. J. Neurosurg. 2014;121:195–209. doi: 10.3171/2014.4.JNS131260. [DOI] [PubMed] [Google Scholar]
- 30.Wu P., Zhang B., Shi H., Qian H., Xu W. MSC-exosome: a novel cell-free therapy for cutaneous regeneration. Cytotherapy. 2018;20:291–301. doi: 10.1016/j.jcyt.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Perets N., Hertz S., London M., Offen D. Intranasal administration of exosomes derived from mesenchymal stem cells ameliorates autistic-like behaviors of BTBR mice. Mol. Autism. 2018;9:57. doi: 10.1186/s13229-018-0240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Song B., Wei Y., Chen F., Chi Y., Fan H., Liu N., Li Z., Han Z., Ma F. Exosomes from mesenchymal stromal cells enhance imatinib-induced apoptosis in human leukemia cells via activation of caspase signaling pathway. Cytotherapy. 2018;20:181–188. doi: 10.1016/j.jcyt.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 33.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367 doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michiel Pegtel D., Gould S.J. Exosome. Annu. Rev. Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 35.El-Andaloussi S., Lee Y., Lakhal-Littleton S., Li J., Seow Y., Gardiner C., Alvarez-Erviti L., Sargent I.L., Wood M.J. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc. 2012;7:2112–2126. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 36.Li P., Kaslan M., Lee S.H., Yao J., Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colombo M., Raposo G., Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 38.Antimisiaris S.G., Mourtas S., Marazioti A. Exosomes and exosome-inspired vesicles for targeteddrug delivery. Pharmaceutics. 2018;10 doi: 10.3390/pharmaceutics10040218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., Fissell W.H., Patton J.G., Rome L.H., Burnette D.T., Coffey R.J. Reassessment of exosome composition. Cell. 2019;177:428–445. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Manrique P., Gutierrez G., Blanco-Lopez M.C. Fully artificial exosomes: towards new theranostic biomaterials. Trends Biotechnol. 2018;36:10–14. doi: 10.1016/j.tibtech.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Hong Y., Nam G.-H., Koh E., Jeon S., Kim G.B., Jeong C., Kim D.-H., Yang Y., Kim I.-S. Exosome as a vehicle for delivery of membrane protein therapeutics, PH20, for enhanced tumor penetration and antitumor efficacy. Adv. Funct. Mater. 2018;28:1703074. [Google Scholar]
- 42.Barile L., Moccetti T., Marban E., Vassalli G. Roles of exosomes in cardioprotection. Eur. Heart J. 2017;38:1372–1379. doi: 10.1093/eurheartj/ehw304. [DOI] [PubMed] [Google Scholar]
- 43.Boriachek K., Islam M.N., Moller A., Salomon C., Nguyen N.T., Hossain M.S.A., Yamauchi Y., Shiddiky M.J.A. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small. 2018;14:1702153. doi: 10.1002/smll.201702153. [DOI] [PubMed] [Google Scholar]
- 44.Farooqi A.A., Desai N.N., Qureshi M.Z., Librelotto D.R.N., Gasparri M.L., Bishayee A., Nabavi S.M., Curti V., Daglia M. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol. Adv. 2018;36:328–334. doi: 10.1016/j.biotechadv.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 45.Bucan V., Vaslaitis D., Peck C.T., Strauss S., Vogt P.M., Radtke C. Effect of exosomes from rat adipose-derived mesenchymal stem cells on neurite outgrowth and sciatic nerve regeneration after crush injury. Mol. Neurobiol. 2019;56:1812–1824. doi: 10.1007/s12035-018-1172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao F., Zhang D., Fang T., Lu C., Wang B., Ding X., Wei S., Zhang Y., Pi W., Xu H., Wang Y., Jiang B., Zhang P. Exosomes from human gingiva-derived mesenchymal stem cells combined with biodegradable chitin conduits promote rat sciatic nerve regeneration. Stem Cell. Int. 2019;2019:2546367. doi: 10.1155/2019/2546367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., Jia Y., Li J., Liu Q. Schwann cell derived exosomes induce bone marrow derived mesenchymal stem cells to express Schwann cell markers in vitro. Mol. Med. Rep. 2020;21:1640–1646. doi: 10.3892/mmr.2020.10960. [DOI] [PubMed] [Google Scholar]
- 48.Lopez-Verrilli M.A., Picou F., Court F.A. Schwann cell-derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia. 2013;61:1795–1806. doi: 10.1002/glia.22558. [DOI] [PubMed] [Google Scholar]
- 49.Dezawa M., Takahashi I., Esaki M., Takano M.H. Sawada. Sciatic nerve regeneration in rats induced by transplantation of in vitro differentiated bone-marrowstromal cells. Eur. J. Neurosci. 2001;14:1771–1776. doi: 10.1046/j.0953-816x.2001.01814.x. [DOI] [PubMed] [Google Scholar]
- 50.Caddick J., Kingham P.J., Gardiner N.J., Wiberg M., Terenghi G. Phenotypic and functional characteristics of mesenchymal stem cells differentiated along a Schwann cell lineage. Glia. 2006;54:840–849. doi: 10.1002/glia.20421. [DOI] [PubMed] [Google Scholar]
- 51.Uz M., Buyukoz M., Sharma A.D., Sakaguchi D.S., Altinkaya S.A., Mallapragada S.K. Gelatin-based 3D conduits for transdifferentiation of mesenchymal stem cells into Schwann cell-like phenotypes. Acta Biomater. 2017;53:293–306. doi: 10.1016/j.actbio.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 52.Wu S., Ni S., Jiang X., Kuss M.A., Wang H.J., Duan B. Guiding mesenchymal stem cells into myelinating Schwann cell-like phenotypes by using electrospun core-sheath nanoyarns. ACS Biomater. Sci. Eng. 2019;5:5284–5294. doi: 10.1021/acsbiomaterials.9b00748. [DOI] [PubMed] [Google Scholar]
- 53.Hu G., Liao K., Niu F., Yang L., Dallon B.W., Callen S., Tian C., Shu J., Cui J., Sun Z., Lyubchenko Y.L., Ka M., Chen X.M., Astrocyte S. Buch. EV-induced lincRNA-Cox2 regulates microglial phagocytosis: implications for morphine-mediated neurodegeneration. Mol. Ther. Nucleic Acids. 2018;13:450–463. doi: 10.1016/j.omtn.2018.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dagur R.S., Liao K., Sil S., Niu F., Sun Z., Lyubchenko Y.L., Peeples E.S., Hu G., Buch S. Neuronal-derived extracellular vesicles are enriched in the brain and serum of HIV-1 transgenic rats. J. Extracell. Vesicles. 2020;9:1703249. doi: 10.1080/20013078.2019.1703249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo S., Perets N., Betzer O., Ben-Shaul S., Sheinin A., Michaelevski I., Popovtzer R., Offen D., Levenberg S. Intranasal delivery of mesenchymal stem cell derived exosomes loaded with phosphatase and tensin homolog siRNA repairs complete spinal cord injury. ACS Nano. 2019;13:10015–10028. doi: 10.1021/acsnano.9b01892. [DOI] [PubMed] [Google Scholar]
- 56.Betzer O., Perets N., Angel A., Motiei M., Sadan T., Yadid G., Offen D., Popovtzer R. In vivo neuroimaging of exosomes using gold nanoparticles. ACS Nano. 2017;11:10883–10893. doi: 10.1021/acsnano.7b04495. [DOI] [PubMed] [Google Scholar]
- 57.Wu S., Kuss M., Qi D., Hong J., Wang H.-J., Zhang W., Chen S., Ni S., Duan B. Development of cryogel-based guidance conduit for peripheral nerve regeneration. ACS Appl. Bio Mater. 2019;2:4864–4871. doi: 10.1021/acsabm.9b00626. [DOI] [PubMed] [Google Scholar]
- 58.Zhang W., Shi W., Wu S., Kuss M., Jiang X., Untrauer J.B., Reid S.P., Duan B. 3D printed composite scaffolds with dual small molecule delivery for mandibular bone regeneration. Biofabrication. 2020;12 doi: 10.1088/1758-5090/ab906e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bessa-Goncalves M., Silva A.M., Bras J.P., Helmholz H., Luthringer-Feyerabend B.J.C., Willumeit-Romer R., Barbosa M.A., Santos S.G. Fibrinogen and magnesium combination biomaterials modulate macrophage phenotype, NF-kB signaling and crosstalk with mesenchymal stem/stromal cells. Acta Biomater. 2020;114:471–484. doi: 10.1016/j.actbio.2020.07.028. [DOI] [PubMed] [Google Scholar]
- 60.Lin H., Ji D.K., Lucherelli M.A., Reina G., Ippolito S., Samori P., Bianco A. Comparative effects of graphene and molybdenum disulfide on human macrophage toxicity. Small. 2020;16 doi: 10.1002/smll.202002194. [DOI] [PubMed] [Google Scholar]
- 61.Sikandar S., Minett M.S., Millet Q., Santana-Varela S., Lau J., Wood J.N., Zhao J. Brain-derived neurotrophic factor derived from sensory neurons plays a critical role in chronic pain. Brain : J. Neurol. 2018;141:1028–1039. doi: 10.1093/brain/awy009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muller Q., Beaudet M.J., De Serres-Berard T., Bellenfant S., Flacher V., Berthod F. Development of an innervated tissue-engineered skin with human sensory neurons and Schwann cells differentiated from iPS cells. Acta Biomater. 2018;82:93–101. doi: 10.1016/j.actbio.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 63.Jones I., Yelhekar T.D., Wiberg R., Kingham P.J., Johansson S., Wiberg M., Carlsson L. Development and validation of an in vitro model system to study peripheral sensory neuron development and injury. Sci. Rep. 2018;8:15961. doi: 10.1038/s41598-018-34280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiong C., Chua K.C., Stage T.B., Priotti J., Kim J., Altman-Merino A., Chan D., Saraf K., Canato Ferracini A., Fattahi F., Kroetz D.L. Human induced pluripotent stem cell derived sensory neurons are sensitive to the neurotoxic effects of paclitaxel. Clin. Transl. Sci. 2021;14:568–581. doi: 10.1111/cts.12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kong Y., Shi W., Zhang D., Jiang X., Kuss M., Liu B., Li Y., Duan B. Injectable, antioxidative, and neurotrophic factor-deliverable hydrogel for peripheral nerve regeneration and neuropathic pain relief. Appl. Mater. Todat. 2021;24:101090. [Google Scholar]
- 66.Kehl T., Kern F., Backes C., Fehlmann T., Stockel D., Meese E., Lenhof H.P., Keller A. miRPathDB 2.0: a novel release of the miRNA Pathway Dictionary Database. Nucleic Acids Res. 2020;48:D142–D147. doi: 10.1093/nar/gkz1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker a.T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li G., Zhao X., Zhang L., Yang J., Cui W., Yang Y., Zhang H. Anisotropic ridge/groove microstructure for regulating morphology and biological function of Schwann cells. Appl. Mater. Today. 2020;18:100468. [Google Scholar]
- 69.Xie S., Lu F., Han J., Tao K., Wang H., Simental A., Hu D., Yang H. Efficient generation of functional Schwann cells from adipose-derived stem cells in defined conditions. Cell Cycle. 2017;16:841–851. doi: 10.1080/15384101.2017.1304328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skaper S.D. Nerve growth factor: a neuroimmune crosstalk mediator for all seasons. Immunology. 2017;151:1–15. doi: 10.1111/imm.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rawat K., Syeda S., Shrivastava A. Neutrophil-derived granule cargoes: paving the way for tumor growth and progression. Cancer Metastasis Rev. 2021;40:221–244. doi: 10.1007/s10555-020-09951-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forster R., Sarginson A., Velichkova A., Hogg C., Dorning A., Horne A.W., Saunders P.T.K., Greaves E. Macrophage-derived insulin-like growth factor-1 is a key neurotrophic and nerve-sensitizing factor in pain associated with endometriosis. Faseb. J. 2019;33:11210–11222. doi: 10.1096/fj.201900797R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yasui W., Ji Z.Q., Kuniyasu H., Ayhan A., Yokozaki H., Ito H., Tahara E. Expression of transforming growth factor alpha in human tissues: immunohistochemical study and northern blot analysis. Virchows Archiv - A Pathol. Anat. Histopathol. 1992;421:513–519. doi: 10.1007/BF01606881. [DOI] [PubMed] [Google Scholar]
- 74.Togel F., Hu Z., Weiss K., Isaac J., Lange C., Westenfelder C. Administered mesenchymal stem cells protect against ischemic acute renal failure through differentiation-independent mechanisms. Am. J. Physiol. Ren. Physiol. 2005;289:F31–F42. doi: 10.1152/ajprenal.00007.2005. [DOI] [PubMed] [Google Scholar]
- 75.Wang S., Zheng H., Zhou L., Cheng F., Liu Z., Zhang H., Wang L., Zhang Q. Nanoenzyme-reinforced injectable hydrogel for healing diabetic wounds infected with multidrug resistant bacteria. Nano Lett. 2020;20:5149–5158. doi: 10.1021/acs.nanolett.0c01371. [DOI] [PubMed] [Google Scholar]
- 76.Yang Z., Shi J., Xie J., Wang Y., Sun J., Liu T., Zhao Y., Zhao X., Wang X., Ma Y., Malkoc V., Chiang C., Deng W., Chen Y., Fu Y., Kwak K.J., Fan Y., Kang C., Yin C., Rhee J., Bertani P., Otero J., Lu W., Yun K., Lee A.S., Jiang W., Teng L., Kim B.Y.S., Lee L.J. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat. Biomed. Eng. 2020;4:69–83. doi: 10.1038/s41551-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Upadhya R., Madhu L.N., Attaluri S., Gitai D.L.G., Pinson M.R., Kodali M., Shetty G., Zanirati G., Kumar S., Shuai B., Weintraub S.T., Shetty A.K. Extracellular vesicles from human iPSC-derived neural stem cells: miRNA and protein signatures, and anti-inflammatory and neurogenic properties. J. Extracell. Vesicles. 2020;9:1809064. doi: 10.1080/20013078.2020.1809064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsiapalis D., O'Driscoll L. Mesenchymal stem cell derived extracellular vesicles for tissue engineering and regenerative medicine applications. Cells. 2020;9:991. doi: 10.3390/cells9040991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li L., Zhang Y., Mu J., Chen J., Zhang C., Cao H., Gao J. Transplantation of human mesenchymal stem-cell-derived exosomes immobilized in an adhesive hydrogel for effective treatment of spinal cord injury. Nano Lett. 2020;20:4298–4305. doi: 10.1021/acs.nanolett.0c00929. [DOI] [PubMed] [Google Scholar]
- 80.Han Y., Ren J., Bai Y., Pei X., Han Y. Exosomes from hypoxia-treated human adipose-derived mesenchymal stem cells enhance angiogenesis through VEGF/VEGF-R. Int. J. Biochem. Cell Biol. 2019;109:59–68. doi: 10.1016/j.biocel.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 81.Zhang K., Zhao X., Chen X., Wei Y., Du W., Wang Y., Liu L., Zhao W., Han Z., Kong D., Zhao Q., Guo Z., Han Z., Liu N., Ma F., Li Z. Enhanced therapeutic effects of mesenchymal stem cell-derived exosomes with an injectable hydrogel for hindlimb ischemia treatment. ACS Appl. Mater. Interfaces. 2018;10:30081–30091. doi: 10.1021/acsami.8b08449. [DOI] [PubMed] [Google Scholar]
- 82.Lacey D.C., Achuthan A., Fleetwood A.J., Dinh H., Roiniotis J., Scholz G.M., Chang M.W., Beckman S.K., Cook A.D., Hamilton J.A. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J. Immunol. 2012;188:5752–5765. doi: 10.4049/jimmunol.1103426. [DOI] [PubMed] [Google Scholar]
- 83.Chambers S.M., Qi Y., Mica Y., Lee G., Zhang X.J., Niu L., Bilsland J., Cao L., Stevens E., Whiting P., Shi S.H., Studer L. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat. Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Umehara Y., Toyama S., Tominaga M., Matsuda H., Takahashi N., Kamata Y., Niyonsaba F., Ogawa H., Takamori K. Robust induction of neural crest cells to derive peripheral sensory neurons from human induced pluripotent stem cells. Sci. Rep. 2020;10:4360. doi: 10.1038/s41598-020-60036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo M., Yin Z., Chen F., Lei P. Mesenchymal stem cell-derived exosome: a promising alternative in the therapy of Alzheimer's disease. Alzheimer's Res. Ther. 2020;12:109. doi: 10.1186/s13195-020-00670-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Simeoli R., Montague K., Jones H.R., Castaldi L., Chambers D., Kelleher J.H., Vacca V., Pitcher T., Grist J., Al-Ahdal H., Wong L.F., Perretti M., Lai J., Mouritzen P., Heppenstall P., Malcangio M. Exosomal cargo including microRNA regulates sensory neuron to macrophage communication after nerve trauma. Nat. Commun. 2017;8:1778. doi: 10.1038/s41467-017-01841-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li C., Li X., Zhao B., Wang C. Exosomes derived from miR-544-modified mesenchymal stem cells promote recovery after spinal cord injury. Arch. Physiol. Biochem. 2020;126:369–375. doi: 10.1080/13813455.2019.1691601. [DOI] [PubMed] [Google Scholar]
- 88.Li D., Zhang P., Yao X., Li H., Shen H., Li X., Wu J., Lu X. Exosomes derived from miR-133b-modified mesenchymal stem cells promote recovery after spinal cord injury. Front. Neurosci. 2018;12:845. doi: 10.3389/fnins.2018.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baglio S.R., Rooijers K., Koppers-Lalic D., Verweij F.J., Perez Lanzon M., Zini N., Naaijkens B., Perut F., Niessen H.W., Baldini N., Pegtel D.M. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015;6:127. doi: 10.1186/s13287-015-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lv W., Deng B., Duan W., Li Y., Liu Y., Li Z., Xia W., Li C. Schwann cell plasticity is regulated by a weakened intrinsic antioxidant defense system in acute peripheral nerve injury. Neuroscience. 2018;382:1–13. doi: 10.1016/j.neuroscience.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 91.Balakrishnan A., Belfiore L., Chu T.H., Fleming T., Midha R., Biernaskie J., Schuurmans C. Insights into the role and potential of Schwann cells for peripheral nerve repair from studies of development and injury. Front. Mol. Neurosci. 2020;13:608442. doi: 10.3389/fnmol.2020.608442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Watanabe Y., Sasaki R., Matsumine H., Yamato M., Okano T. Undifferentiated and differentiated adipose-derived stem cells improve nerve regeneration in a rat model of facial nerve defect. J. Tissue Eng. Regen. Med. 2017;11:362–374. doi: 10.1002/term.1919. [DOI] [PubMed] [Google Scholar]
- 93.Lavorato A., Raimondo S., Boido M., Muratori L., Durante G., Cofano F., Vincitorio F., Petrone S., Titolo P., Tartara F., Vercelli A., Garbossa D. Mesenchymal stem cell treatment perspectives in peripheral nerve regeneration: systematic review. Int. J. Mol. Sci. 2021;22:572. doi: 10.3390/ijms22020572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bierlein De la Rosa M., Sharma A.D., Mallapragada S.K., Sakaguchi D.S. Transdifferentiation of brain-derived neurotrophic factor (BDNF)-secreting mesenchymal stem cells significantly enhance BDNF secretion and Schwann cell marker proteins. J. Biosci. Bioeng. 2017;124:572–582. doi: 10.1016/j.jbiosc.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 95.Lu P., Jones L.L., Tuszynski M.H. BDNF-expressing marrow stromal cells support extensive axonal growth at sites of spinal cord injury. Exp. Neurol. 2005;191:344–360. doi: 10.1016/j.expneurol.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 96.Jung N., Park S., Choi Y., Park J.W., Hong Y.B., Park H.H., Yu Y., Kwak G., Kim H.S., Ryu K.H., Kim J.K., Jo I., Choi B.O., Jung S.C. Tonsil-derived mesenchymal stem Cells differentiate into a Schwann cell phenotype and promote peripheral nerve regeneration. Int. J. Mol. Sci. 2016;17:1867. doi: 10.3390/ijms17111867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qiu X.C., Jin H., Zhang R.Y., Ding Y., Zeng X., Lai B.Q., Ling E.A., Wu J.L., Zeng Y.S. Donor mesenchymal stem cell-derived neural-like cells transdifferentiate into myelin-forming cells and promote axon regeneration in rat spinal cord transection. Stem Cell Res. Ther. 2015;6:105. doi: 10.1186/s13287-015-0100-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harrell C.R., Volarevic A., Djonov V., Volarevic V. Mesenchymal stem cell-derived exosomes as new remedy for the treatment of neurocognitive disorders. Int. J. Mol. Sci. 2021;22:1433. doi: 10.3390/ijms22031433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Riazifar M., Mohammadi M.R., Pone E.J., Yeri A., Lasser C., Segaliny A.I., McIntyre L.L., Shelke G.V., Hutchins E., Hamamoto A., Calle E.N., Crescitelli R., Liao W., Pham V., Yin Y., Jayaraman J., Lakey J.R.T., Walsh C.M., Van Keuren-Jensen K., Lotvall J., Zhao W. Stem cell-derived exosomes as nanotherapeutics for autoimmune and neurodegenerative disorders. ACS Nano. 2019;13:6670–6688. doi: 10.1021/acsnano.9b01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van den Boorn Jasper G., Martin Schlee, Coch Christoph, Hartmann G. SiRNA delivery with exosome nanoparticles. Nat. Biotechnol. 2011;29:325–326. doi: 10.1038/nbt.1830. [DOI] [PubMed] [Google Scholar]
- 101.He C., Zheng S., Luo Y., Wang B. Exosome theranostics: biology and translational medicine. Theranostics. 2018;8:237–255. doi: 10.7150/thno.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang M., Jin K., Gao L., Zhang Z., Li F., Zhou F., Zhang L. Methods and technologies for exosome isolation and characterization. Small Methods. 2018;2:1800021. [Google Scholar]
- 103.Lanza C., Raimondo S., Vergani L., Catena N., Senes F., Tos P., Geuna S. Expression of antioxidant molecules after peripheral nerve injury and regeneration. J. Neurosci. Res. 2012;90:842–848. doi: 10.1002/jnr.22778. [DOI] [PubMed] [Google Scholar]
- 104.Kim H.K., Park S.K., Zhou J.L., Taglialatela G., Chung K., Coggeshall R.E., Chung J.M. Reactive oxygen species (ROS) play an important role in a rat model of neuropathic pain. Pain. 2004;111:116–124. doi: 10.1016/j.pain.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 105.Romero F.J. Antioxidants in peripheral nerve. Free Radical Biol. Med. 1996;20:925–932. doi: 10.1016/0891-5849(95)02183-3. [DOI] [PubMed] [Google Scholar]
- 106.Qiu J., Yang X., Wang L., Zhang Q., Ma W., Huang Z., Bao Y., Zhong L., Sun H., Ding F. Isoquercitrin promotes peripheral nerve regeneration through inhibiting oxidative stress following sciatic crush injury in mice. Ann. Transl. Med. 2019;7:680. doi: 10.21037/atm.2019.11.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gonzalez-Gonzalez S., Cazevieille C., B C. Resveratrol treatment reduces neuromotor impairment and hearing loss in a mouse model of diabetic neuropathy and nerve injury. J. Diabetes Clin. Res. 2020;2:59–67. [Google Scholar]
- 108.Stavely R., Nurgali K. The emerging antioxidant paradigm of mesenchymal stem cell therapy. Stem Cells Transl. Med. 2020;9:985–1006. doi: 10.1002/sctm.19-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gaudet A.D., Popovich P.G., Ramer M.S. Wallerian degeneration: gaining perspective on inflammatory events after peripheral nerve injury. J. Neuroinflammation. 2011;8:110. doi: 10.1186/1742-2094-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu P., Peng J., Han G.H., Ding X., Wei S., Gao G., Huang K., Chang F., Wang Y. Role of macrophages in peripheral nerve injury and repair. Neural Regen. Res. 2019;14:1335–1342. doi: 10.4103/1673-5374.253510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao Y., Gan Y., Xu G., Yin G., Liu D. MSCs-derived exosomes attenuate acute brain injury and inhibit microglial inflammation by reversing CysLT2R-ERK1/2 mediated microglia M1 polarization. Neurochem. Res. 2020;45:1180–1190. doi: 10.1007/s11064-020-02998-0. [DOI] [PubMed] [Google Scholar]
- 112.Xian P., Hei Y., Wang R., Wang T., Yang J., Li J., Di Z., Liu Z., Baskys A., Liu W., Wu S., Long Q. Mesenchymal stem cell-derived exosomes as a nanotherapeutic agent for amelioration of inflammation-induced astrocyte alterations in mice. Theranostics. 2019;9:5956–5975. doi: 10.7150/thno.33872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang J.H., Yin X.M., Xu Y., Xu C.C., Lin X., Ye F.B., Cao Y., Lin F.Y. Systemic administration of exosomes released from mesenchymal stromal cells attenuates apoptosis, inflammation, and promotes angiogenesis after spinal cord injury in rats. J. Neurotrauma. 2017;34:3388–3396. doi: 10.1089/neu.2017.5063. [DOI] [PubMed] [Google Scholar]
- 114.Haertinger M., Weiss T., Mann A., Tabi A., Brandel V., Radtke C. Adipose stem cell-derived extracellular vesicles induce proliferation of Schwann cells via internalization. Cells. 2020;9:163. doi: 10.3390/cells9010163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pulliam L., Gupta A. Modulation of cellular function through immune-activated exosomes. DNA Cell Biol. 2015;34:459–463. doi: 10.1089/dna.2015.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ren R., Tan X.H., Zhao J.H., Zhang Q.P., Zhang X.F., Ma Z.J., Peng Y.N., Liu Q.B., Zhang H.Y., Li Y.Q., He R., Zhao Z.Q., Yi X.N. Bone marrow mesenchymal stem cell-derived exosome uptake and retrograde transport can occur at peripheral nerve endings. Artif. Cells Nanomed. Biotechnol. 2019;47:2918–2929. doi: 10.1080/21691401.2019.1640713. [DOI] [PubMed] [Google Scholar]
- 117.Lai R.C., Arslan F., Lee M.M., Sze N.S., Choo A., Chen T.S., Salto-Tellez M., Timmers L., Lee C.N., El Oakley R.M., Pasterkamp G., de Kleijn D.P., Lim S.K. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 118.Brennan M.Á., Layrolle P., Mooney D.J. Biomaterials functionalized with MSC secreted extracellular vesicles and soluble factors for tissue regeneration. Adv. Funct. Mater. 2020;30:1909125. doi: 10.1002/adfm.201909125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Marote A., Teixeira F.G., Mendes-Pinheiro B., Salgado A.J. MSCs-derived exosomes: cell-secreted nanovesicles with regenerative potential. Front. Pharmacol. 2016;7:231. doi: 10.3389/fphar.2016.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Qian Y., Song J., Ouyang Y., Han Q., Chen W., Zhao X., Xie Y., Chen Y., Yuan W., Fan C. Advances in roles of miR-132 in the nervous system. Front. Pharmacol. 2017;8:770. doi: 10.3389/fphar.2017.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yao C., Shi X., Zhang Z., Zhou S., Qian T., Wang Y., Ding F., Gu X., Yu B. Hypoxia-induced upregulation of miR-132 promotes Schwann cell migration after sciatic nerve injury by targeting PRKAG3. Mol. Neurobiol. 2016;53:5129–5139. doi: 10.1007/s12035-015-9449-y. [DOI] [PubMed] [Google Scholar]
- 122.Hancock M.L., Preitner N., Quan J., Flanagan J.G. MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension. J. Neurosci. 2014;34:66–78. doi: 10.1523/JNEUROSCI.3371-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu B., Zhang Y., Du X.F., Li J., Zi H.X., Bu J.W., Yan Y., Han H., Du J.L. Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell Res. 2017;27:882–897. doi: 10.1038/cr.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhu L., Xu H., Lv W., He Z., Ye P., Wang Y., Hu J. miR-199b-5p regulates immune-mediated allograft rejection after lung transplantation through the GSK3beta and NF-κB pathways. Inflammation. 2018;41:1524–1535. doi: 10.1007/s10753-018-0799-2. [DOI] [PubMed] [Google Scholar]
- 125.Gao F., Shen J., Zhao L., Hao Q., Yang Y. Curcumin alleviates lipopolysaccharide (LPS)-Activated neuroinflammation via modulation of miR-199b-5p/IκB kinase β (IKKβ)/Nuclear factor kappa B (NF-κB) pathway in microglia. Med. Sci. Mon. 2019;25:9801–9810. doi: 10.12659/MSM.918237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lopez-Leal R., Diaz-Viraque F., Catalan R.J., Saquel C., Enright A., Iraola G., Court F.A. Schwann cell reprogramming into repair cells increases miRNA-21 expression in exosomes promoting axonal growth. J. Cell Sci. 2020;133 doi: 10.1242/jcs.239004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.